Abstract
Oncogenic fusion events have been identified in a broad range of tumors. Among them, RET rearrangements represent distinct and potentially druggable targets that are recurrently found in lung adenocarcinomas. Here, we provide further evidence that current anti-RET drugs may not be potent enough to induce durable responses in such tumors. We report that potent inhibitors such as AD80 or ponatinib that stably bind in the DFG-out conformation of RET may overcome these limitations and selectively kill RET-rearranged tumors. Using chemical genomics in conjunction with phosphoproteomic analyses in RET-rearranged cells we identify the CCDC6-RETI788N mutation and drug-induced MAPK pathway reactivation as possible mechanisms, by which tumors may escape the activity of RET inhibitors. Our data provide mechanistic insight into the druggability of RET kinase fusions that may be of help for the development of effective therapies targeting such tumors.
Introduction
Targeted inhibition of oncogenic driver mutations with small molecules represents the cornerstone of precision cancer medicine. RET rearrangements have been identified in a broad range of tumors including 1-2% of lung adenocarcinomas and their discovery sparked the hope for an effective treatment option in these patients (1–3). However, when compared to other oncogenic "driver" alterations such as rearranged ALK, rearranged RET seems to represent a difficult target as to date, no drug has been successfully established for the treatment of these tumors (4–6). Recent clinical data suggest that overall response rates in patients treated with currently available RET targeted drugs are rather limited and range between 18% - 53% (7–10). Improved selection of patients based on deep sequencing of individual tumors may help to increase these response rates but still progression-free survival seems to be very limited (8–11). These observations are particularly surprising from a chemical point of view since a broad spectrum of kinase inhibitors is known to bind to RET and to inhibit its kinase activity in vitro.
Based on these observations we sought to characterize rearranged RET in orthogonal cancer models to identify potent RET inhibitors with high selectivity and optimal biochemical profile to target RET-rearranged tumors.
Results
Kinase inhibitor AD80 shows extraordinary activity in RET-rearranged cancer models
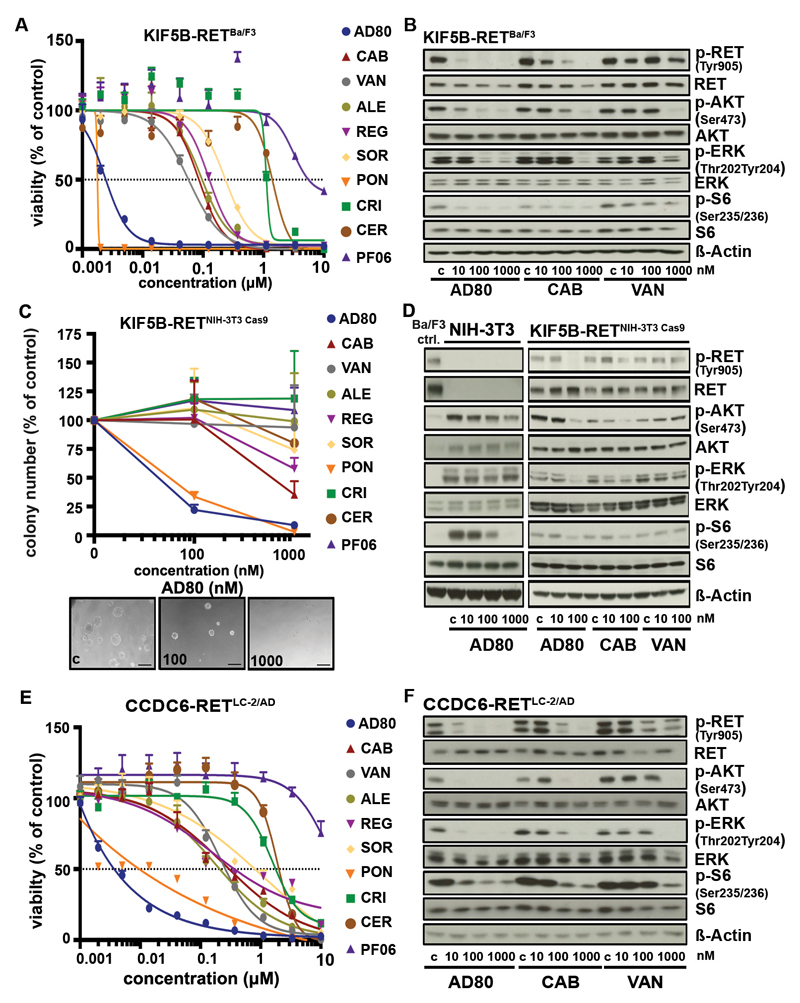
Since clinical experience with RET targeted drugs in lung cancer patients is rather disappointing we sought to test a series of clinically and preclinically available drugs with anti-RET activity in Ba/F3 cells engineered to express either KIF5B-RET or CCDC6-RET (1, 2, 12, 13). In these experiments, AD80 and ponatinib exhibited 100- to 1000-fold higher cytotoxicity compared to all other tested drugs in RET-dependent, but not IL-3 supplemented Ba/F3 cells (Fig. 1A; Fig. S1A,B). In line with these results, AD80, but not cabozantinib or vandetanib prevented phosphorylation of RET as well as of ERK, AKT and S6K at low nanomolar concentrations in KIF5B-RET expressing Ba/F3 cells (Fig. 1B, Supplementary Table 1).
Figure 1.
A) Dose-response curves (72h) as assessed for AD80, cabozantinib (CAB), vandetanib (VAN), alectinib (ALE), regorafenib (REG), sorafenib (SOR), ponatinib (PON), crizotinib (CRI), ceritinib (CER) or PF06463922 (PF06) in KIF5B-RET expressing Ba/F3 cells. B) Immunoblotting results of KIF5B-RET rearranged Ba/F3 cells after treatment are displayed (4h). C) Relative mean colony number of NIH-3T3 cells engineered with KIF5B-RET fusion via CRISPR/Cas9 was assessed in soft agar assays after 7 days under treatment. Representative pictures of colonies under AD80 treatment are depicted in the lower panel. Black bar is equal to 100µm. D) Immunoblotting of treated CRISPR/Cas9 engineered KIF5B-RET-rearranged NIH-3T3 cells with AD80, cabozantinib or vandetanib (4h). KIF5B-RET expressing Ba/F3 cells (Ba/F3 ctrl.) serve as control for RET signaling. E) Dose-response curves (72h) as assessed for different inhibitors in LC-2/AD cells are shown. F) Immunoblotting was performed in LC-2/AD cells treated with AD80, cabozantinib or vandetanib (4h).
To validate the efficacy of AD80 and ponatinib in an orthogonal model, we induced KIF5B-RET rearrangements (KIF5B exon 15; RET exon 12) in NIH-3T3 cells using CRISPR/Cas9-meditated genome editing. We confirmed their anchorage-independent growth, increased proliferation rate and their high sensitivity to AD80 and ponatinib (Fig. 1C; Fig. S1C-E) (14). Again, treatment with AD80 but not cabozantinib or vandetanib led to inhibition of phospho-RET and of downstream effectors of RET signaling at low nanomolar concentrations (Fig. 1D). Interestingly, AD80 led to dephosphorylation of S6 also in parental NIH-3T3 cells and Ba/F3myr-AKT control cells suggesting that S6 may represent an off-target at micromolar concentrations (Fig. 1D; Fig. S1F) (12).
To further substantiate our results, we next tested our panel of RET inhibitors in the CCDC6-RET rearranged lung adenocarcinoma cell line LC-2/AD (15). We observed similar activity profiles with AD80 followed by ponatinib as the most potent inhibitors compared to all other tested drugs in terms of cytotoxicity at low nanomolar concentrations (Fig. 1E) and inhibition of phospho-RET and other downstream signaling molecules (Fig. 1F).
Overall, our data suggest that in RET-rearranged cells AD80 and ponatinib are 100- to 1000-fold more effective against RET and its downstream signaling than any other clinically tested anti-RET drug.
Highly specific and effective inhibition of RET fusions in DFG-out conformation
To benchmark the genotype-specific activity of AD80 and ponatinib against well described kinase inhibitors such as erlotinib, BGJ398, vandetanib, cabozantinib, regorafenib, alectinib and ceritinib in a panel of 18 cancer cell lines driven by known oncogenic lesions such as mutant EGFR or rearranged ALK, including two RET-rearranged cells (LC-2/AD and TPC-1) (Fig. S2A) (6, 13, 16). Again, we identified AD80 and ponatinib as the most effective drug and through the calculation of median on-target vs. off-target ratios also as the most specific drugs in RET-fusion positive cells (Fig. S2B; Supplementary Table 2).
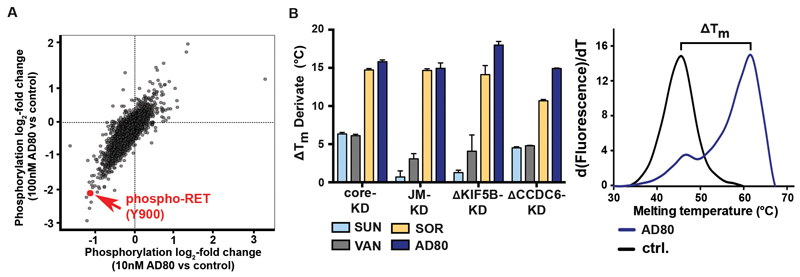
To further characterize intracellular signaling induced by a RET-inhibitor like AD80 we performed mass-spectrometry based phosphoproteomic analyses of LC-2/AD cells treated with 10nM or 100nM of AD80. In AD80 treated cells we observed a significant decrease of RET-Y900 phosphorylation with log2-fold changes of -1.07 (p=0.009; 10nM AD80) and -2.11 (p=0.0002; 100nM AD80), respectively (Fig. 2A). Among all phospho-peptides quantified under control, 10nM and 100nM conditions (n=11912), the abundance of RETY900 was among the most decreased phospho-peptides (ctrl. vs 100nM AD80; p=0.00024) and the most decreased receptor tyrosine kinases (Fig. S2C). These results highlight that in these cells RET is the primary target of AD80.
Figure 2.
A) Scatter plot of log2-fold phosphorylation change for LC-2/AD cells treated (4h) with either 10nM or 100nM AD80. Each dot represents a single phospho-site; phospho-RET (Y900) is highlighted in red. B) Difference in melting temperatures after AD80, sorafenib (SOR), vandetanib (VAN) or sunitinib (SUN) addition (ΔTm) and the respective standard errors of the mean (SEM) are shown for each construct. Thermal shift experiments were performed using independent preparations of each protein and were carried out in triplicates (left panel). Representative thermal melting curves for ΔKIF5B-KD incubated with either AD80 (1μM) or the equivalent volume of DMSO (ctrl.) are shown (right panel).
Based on these observations, we speculated that activation of RET-independent signaling pathways should largely abrogate the cytotoxic effects of AD80. To this end we supplemented LC-2/AD cells with exogenous receptor ligands and found that activity of AD80 was significantly reduced through addition of EGF, HGF and NRG1 indicating that RET is indeed the primary cellular target in RET-rearranged LC-2/AD cells (Fig. S3A).
To further characterize the high potency of AD80 and ponatinib against RET kinase fusions we expressed and purified different truncated versions of the RET core kinase and juxtamembrane-kinase domain as well as truncated forms of both CCDC6 (ΔCCDC6-KD) or KIF5B (ΔKIF5B-KD) kinase domain fusions (Fig. S3B,C) (17). We used these different RET-fusion kinase domain constructs to determine the extent to which binding of a given compound has an effect on protein thermal stability as measured by the melting temperature (Tm). The difference of melting temperature with and without drug (ΔTm) extrapolates the potency of the individual drugs against the respective constructs (17). To our surprise, we found that treatment with the type I inhibitors sunitinib or vandetanib resulted in a ΔTm of only 1-4°C whereas the type II inhibitors sorafenib, ponatinib or AD80 increased the ΔTm up to 10-18°C (Fig. 2B; Fig. S3D-H). We observed the strongest effects in ΔKIF5B-KD and ΔCCDC6-KD constructs treated with AD80 and core KD with ponatinib (Fig. 2B; Fig. S3D; Supplementary Table 3). Interestingly, this strong shift for inhibitors that stabilize the DFG-out conformation does not correlate with the differential in vitro kinase activity observed for sorafenib and other RET inhibitors (Supplementary Table 4) (6).
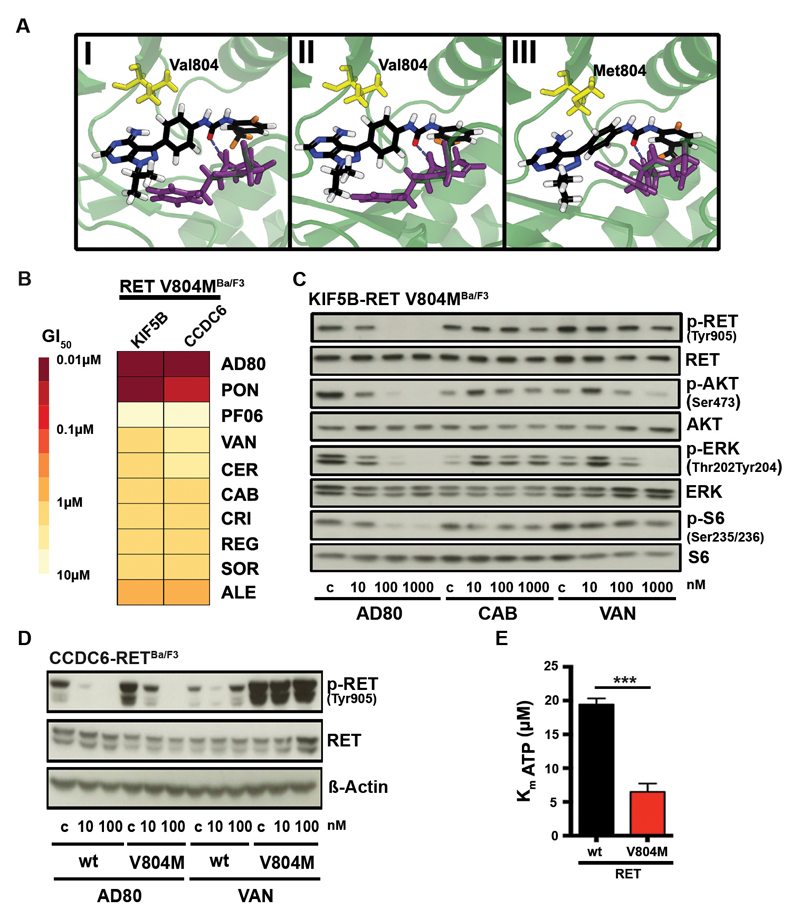
To further characterize the relevance of a DFG-out conformation for the activity of RET inhibitors we performed structural analyses. We employed homology modelling based on a VEGFR kinase (pdb code 2OH4 (18)) in the DFG-out complex, followed by extensive molecular dynamics (MD) simulation refinement, similar to a previously published methodology (19). We observed that the RMSD values remained largely stable over the time course of the MD simulation (RET-wt and RET-V804M) thus supporting our proposed model in which AD80 binds in the DFG-out conformation of the kinase (Fig. S4A). In this model AD80 forms an H-bond between the aspartate of the DFG motif that may be involved in the stabilization of the DFG-out conformation (Fig. 3A). A similar H-bond is also observed for cabozantinib, a known type II inhibitor, bound to RET-wt (Fig. S4B, see Supplementary Methods for model generation). This finding corroborates the validity of our binding mode hypothesis, though the pose is biased by construction, being based on the refined RET-wt/AD80 structure. Furthermore, we developed a binding pose model for AD57 (derivative of AD80) bound to RET-wt (see below) which, upon superimposition, displays considerable similarity with the experimentally determined structure of AD57 bound to cSrc (PDB code 3EL8) in the DFG-out form, again validating our approach (Fig. S4C, Fig. S3H). Next we performed free energy MD simulations to transform AD80 into AD57. The calculations yield a binding free energy difference of ΔΔG° = -0.21 ± 0.17 kcal mol-1 at 25°C, which compares well with the values derived from IC50 in in vitro kinase measurements. These latter concentration-based measures of binding affinity translate into an experimental estimate of the binding free energy difference of -0.41 kcal mol^-1 with IC50(AD57) of 2nM and IC50(AD80) of 4nM (see Supplementary methods) (12). Using as an alternative computational approach an integral equation approximation (see Supplementary Methods) we obtain 0.1 kcal mol1-, also in close correspondence with both MD and experimental results. Thus, these analyses further support the proposed DFG-out conformation as the preferred binding mode since such agreement between experiment and theory would not have been expected if the true and predicted binding modes were largely dissimilar.
Figure 3.
A) Optimized structures after extensive MD refinement followed by ALPB optimization. (I) RET-wt/AD80 after 102 ns, (II) RET-wt/AD57 after 202 ns (92 ns from RET-wt/AD80 simulation followed by 110 ns TI-MD), (III) RET-V804M/AD80 after 107 ns (side view). The DFG motif is shown in violet. Distances from central phenyl’s center: 4.77 Å to Val804-C(wt), 3.90 Å to Ile788-C(wt) and 4.29 Å to Met804-S(V804M). Dashed lines indicate the marked H-bond between the bound ligands and aspartate of the DFG motif. B) Heatmap of mean GI50-values (from n≥3) of Ba/F3 cells expressing CCDC6-RETV804M or KIF5B-RETV804M after 72h of treatment as assessed for various inhibitors is shown. C) Immunoblotting of AD80, cabozantinib or vandetanib treated (4h) KIF5B-RETV804M Ba/F3 cells is displayed. D) Immunoblotting of Ba/F3 cells expressing CCDC6-RET-RETwt or CCDC6-RETV804M under AD80 or vandetanib treatment (4h). E) Calculated Km values of ATP binding to RETwt or RETV804M-mutant from three independent experiments are displayed. ***, p<0.001.
Overall, our cellular screening, phosphoproteomic, biochemical and structural data indicate that potent type II inhibitors like AD80 or ponatinib have an optimal RET specific profile that distinguishes them from currently available anti-RET drugs.
Differential activity of RET inhibitors against RET kinase gatekeeper mutation
Secondary resistance mutations frequently target a conserved residue, termed gatekeeper that controls access to a hydrophobic subpocket of the kinase domain (20). To test the impact of the gatekeeper resistance mutations on RET inhibitors we established Ba/F3 cells expressing KIF5B-RETV804M or CCDC6-RETV804M and tested them against a panel of different drugs. As expected only ponatinib but also AD80 showed high activity in these gatekeeper mutant cells (Fig. 3B) (21). Similar activity was observed when testing the AD80 derivatives AD57 and AD81 for their inhibitory potential on Ba/F3 cells expressing wild type and V804M mutated KIF5B-RET or CCDC6-RET (Fig. S5A). This effect was also evident in the ability of AD80 to inhibit phosphorylation of RET as well as of ERK, AKT and S6K in these cells (Fig. 3C, Supplementary Table S1). Next, we used computational homology modeling coupled with MD refinement of AD80 in RETwt in comparison with RETV804M-mutant kinases. In line with our in vitro results, this analysis revealed high structural similarity and similar binding free energy estimates for both variants (-2.5 kcal mol-1 for transforming RETwt to RETV804M bound to AD80 from the integral equation model) (Fig. 3A, Supplementary Methods).
In parallel, we noticed that independent of the individual treatment, RET phosphorylation levels tended to be higher in gatekeeper mutant cells when compared to wild type RET (Fig. 3D). To further characterize these differences we performed in vitro kinase assays and found that the introduction of the RETV804M mutation significantly (p<0.001) increases the affinity of the recombinant receptor for ATP when compared to the recombinant wild type receptor (Fig. 3E).
Thus, similar to gatekeeper-induced effects on ATP affinity observed for EGFRT790M mutations, our data suggests that these effects may be of relevance for the activity of RET inhibitors in KIF5B-RETV804M and CCDC6-RETV804M cells (22).
Saturated mutagenesis screening identifies novel CCDC6-RETI788N drug resistance mutation
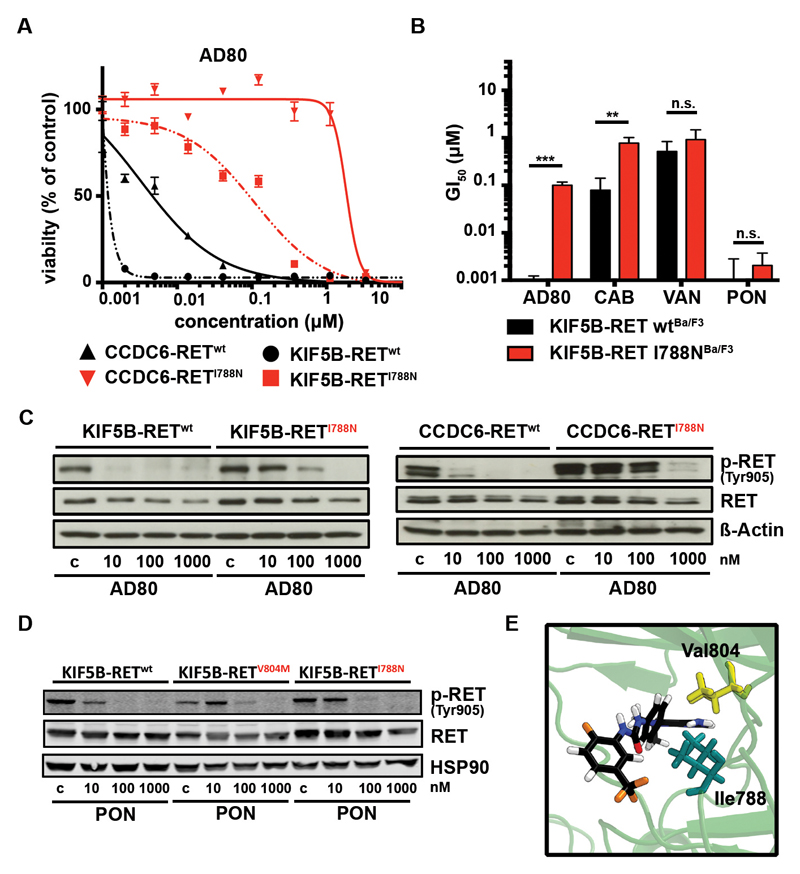
To identify novel RET kinase mutations that may be associated with resistance against targeted therapy we performed accelerated mutagenesis of RET-fusion plasmids (23, 24). We identified the CCDC6-RETI788N mutation by selection of an AD80 resistant cell population (Supplementary Table 5). To validate this finding we engineered Ba/F3 cells expressing KIF5B-RETI788N or CCDC6-RETI788N and, in fact, observed a robust shift in cytotoxicity in response to AD80 treatment (Fig. 4A), as well as the other RET inhibitors cabozantinib, vandetanib but not ponatinib (Fig. 4B,C; Fig. S5B). Immunoblotting confirmed that the introduction of the KIF5B-RETI788N mutation had a minor effect on the efficacy of ponatinib but a major impact on AD80 as measured by phospho-RET levels (Fig. 4D). Computational binding mode analysis (Fig. 3C and 4E) suggests that both positions 804 and 788 are adjacent to the location of AD80’s central phenyl ring; characteristic distances between phenyl center of mass and nearest adjacent protein non-hydrogen sites are 4.77 Å to Val804-C(wt), 3.90 Å to Ile788-C(wt), 4.29 Å to Met804-S(V804M), and 4.61 Å to Ile788-C(V804M). However, since V804M and I788N mutants respond differently to AD80, a clear conclusion about the molecular origin is not possible based on structural analysis alone, requiring further investigations.
Figure 4.
A) Dose-response curves as assessed for AD80 against Ba/F3 cells expressing KIF5B-RETwt (black) or KIF5B-RETI788N (red) and CCDC6-RETwt (black dashed) or CCDC6-RETI788N (red dashed). B) Column chart of mean GI50-values + SD (from n=3) of KIF5B-RETwt or KIF5B-RETI788N Ba/F3 cells treated (72h) with AD80, cabozantinib (CAB), vandetanib (VAN) or ponatinib (PON). p-values are given as ”*”. C) Immunoblotting of KIF5B-RETwt (left panel) or KIF5B-RETI788N (right panel) and CCDC6-RETwt or CCDC6-RETI788N (lower panel) Ba/F3 treated (4h) with AD80 are displayed (4h). D) Immunoblotting of KIF5B-RETwt, KIF5B-RETV804M or KIF5B-RETI788N Ba/F3 cells treated (4h) with ponatinib are shown. HSP90 is used as loading control. E) Optimized structure after extensive MD refinement followed by ALPB optimization. RET-wt/AD80 after 102 ns (side view). Distance from central phenyl’s center: 4.61 Å to Ile788-C(V804M).
Thus, our data uncover a novel resistance mutation RETI788N that may arise in RET-rearranged tumors under RET inhibitor treatment and that retains sensitivity against ponatinib.
Feedback-induced activation of MAPK signaling modulates activity of RET inhibitors
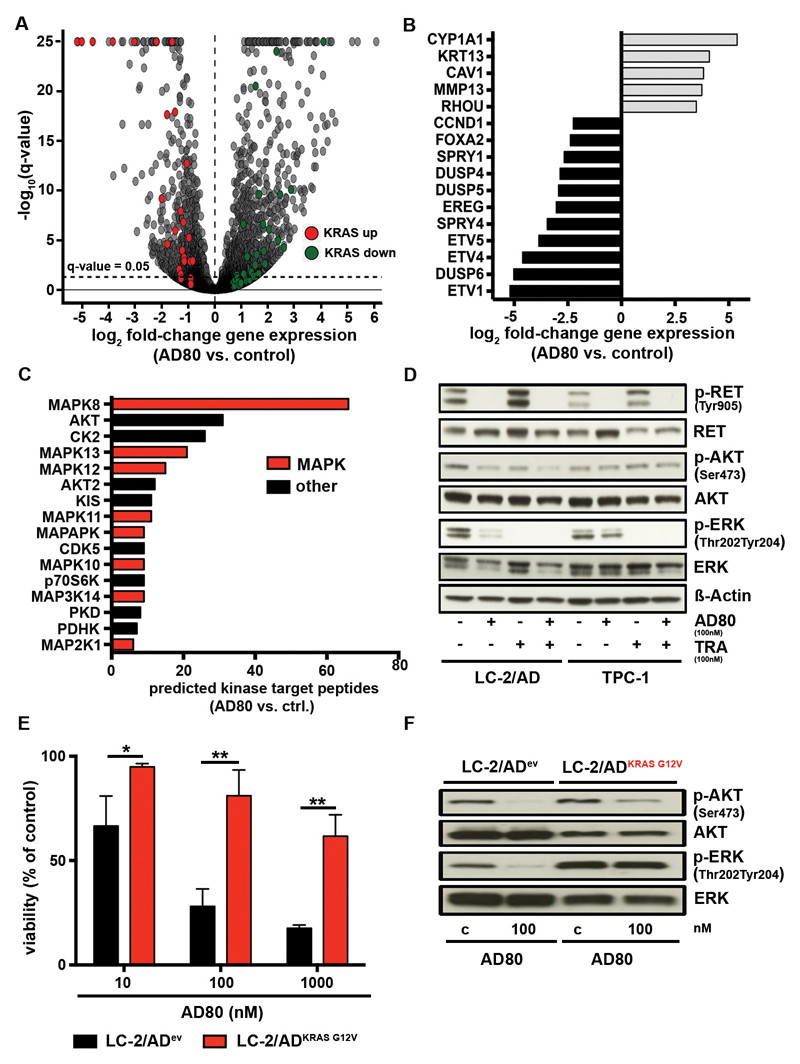
Beyond the acquisition of secondary mutations drug treatment of cancer cells may also lead to the release of feedback loops that override the activity of targeted cancer treatment (25, 26). To systematically characterize these effects we analyzed altered gene expression by RNA-sequencing of LC-2/AD cells under AD80 treatment and performed gene set enrichment analysis (GSEA) (27). Our analyses revealed that treatment with AD80 lead to upregulation of genes that are typically repressed by active KRAS (KRAS down; adj. p<0.0001). On the contrary, genes that are known to be activated by KRAS were downregulated (KRAS up; adj. p=0.003) (Fig. 5A). Accordingly, the list of significantly downregulated genes contained DUSP6 (adj. p<1x10-250), SPRY4 (adj. p=5.75x10-89), DUSP5 (2,52x10-38) and other genes that are known to buffer MAPK pathway (Fig. 5B). This transcriptional deregulation of MAPK signaling was accompanied by residual phospho-ERK staining in immunoblotting analyses of RET-rearranged LC-2/AD cells after 24h of inhibitor treatment (Fig. S5C). Using a Group-based Prediction System (GPS 2.12) to identify kinase specific phospho-sites that are perturbed in AD80 treated LC-2/AD cells assessed in our mass-spectrometry based analysis we identified a striking enrichment of phospho-sites known from different families of non-canonical MAPK kinases, such as MAPK8 (66 phospho-sites) MAPK13 (21 phospho-sites) or MAPK12 (15 phospho-sites) (Fig. 5C).
Figure 5.
A) RNA-Seq results of LC-2/AD cells treated (48h) with 100nM AD80 are shown. Genes contained within the core enrichments following GSEA against the hallmark gene sets with genes upregulated (KRAS up) or downregulated (KRAS down) by active KRAS are highlighted by red and green, respectively. The dashed line represents FDR-adjusted q-value= 0.05. B) Relevant genes from the top-50 genes with strongest and significant change in RNAseq after AD80 treatment (100nM/48h) are shown. C) The predicted number of down regulated phosphorylation sites for each kinase is shown. All kinases with ≥6 down regulated phosphorylation sites are shown in hierarchical order. Kinases associated with MAPK pathway signaling are highlighted in red. D) In immunoblotting assays RET signaling was monitored in LC-2/AD and TPC-1 cells, treated (48h) with either AD80 (0.1µM), trametinib (TRA) (0.1µM) or a combination of both inhibitors (combo). E) LC-2/ADev or LC-2/ADKRAS G12V cells were treated (72h) with AD80. Results are shown as mean + SD (n=3). p-values given as ”*” are displayed. F) Immunoblotting of LC-2/ADev or LC-2/ADKRAS G12V cells under AD80 treatment (100nM/4h) is shown.
We next tested the relevance of RAS-MAPK pathway reactivation in RET-rearranged cells and treated with either AD80 alone or a combination of AD80 and the MEK inhibitor trametinib. In TPC-1 cells with limited vulnerability to RET inhibition we observed a pronounced phospho-ERK signal in cells after inhibition with AD80 when compared to LC-2/AD cells (Fig. S5D). The combination of AD80 and trametinib fully abrogated MAPK signaling and depleted outgrowth of resistant cells in clonogenic assays and enhanced reduction of viability (Fig. 5D and Fig. S5E,F).
To formally test the relevance of MAPK pathway activation in the context of resistance to RET targeted therapies in RET-rearranged cells, we stably transduced LC-2/AD cells with lentiviral KRASG12V. Indeed, introduction of the oncogenic KRAS allele into LC-2/AD cells largely eliminated activity of AD80 as measured in viability assays and by staining of phospho-ERK (Fig. 5E,F).
Overall, our data suggest that drug-induced transcriptional and post-translational reactivation of RAS-MAPK signaling may modulate the activity of RET targeted inhibitors in RET-rearranged cells.
AD80 potently shrinks RET-rearranged tumors in patient-derived xenografts
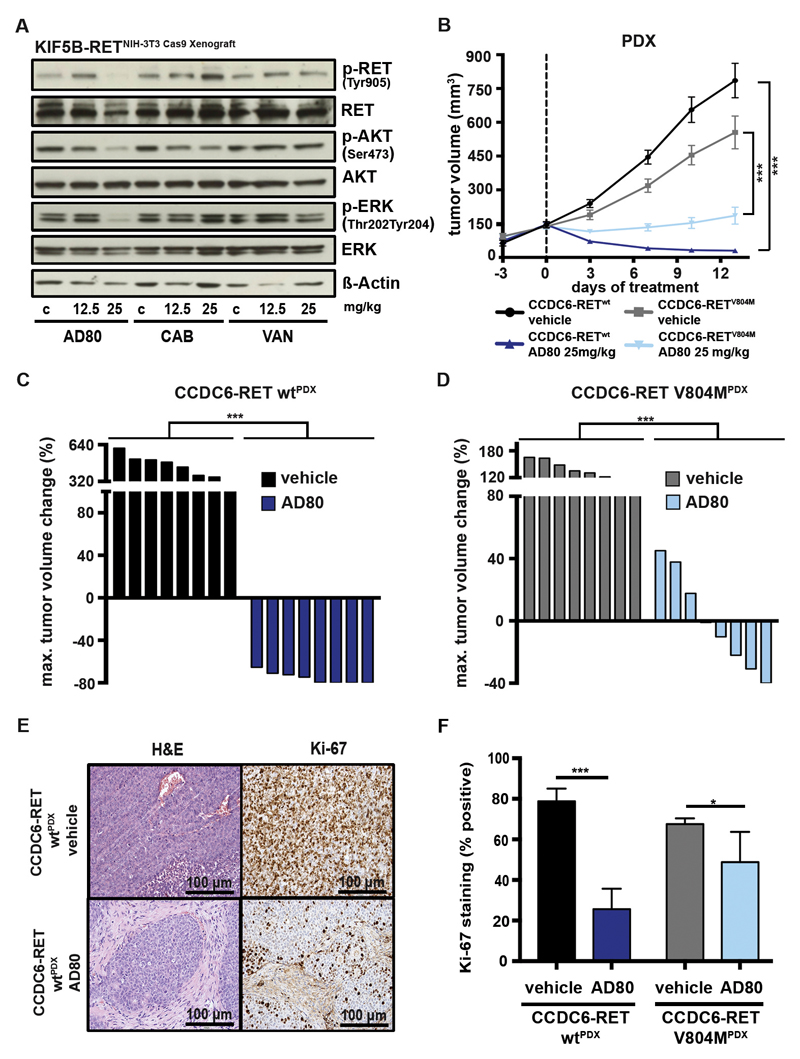
To compare the in vivo efficacy of AD80 head-to-head with other RET inhibitors we engrafted NIH-3T3 cells driven by CRISPR/Cas9-induced KIF5B-RET rearrangements into NSG mice. After development of tumors, mice were treated with either vehicle or 12.5 to 25mg/kg of AD80, cabozantinib or vandetanib and tumors were explanted 4 hours later (28, 29). We observed a pronounced reduction of phosphorylation of RET as well as AKT and ERK in tumors treated with 25mg/kg AD80, but not in tumors treated with cabozantinib or vandetanib (Fig. 6A). Encouraged by these results, we next treated a cohort (n=16) of patient-derived xenograft (PDX) mice engrafted with tumor tissue from a CCDC6-RET rearranged colorectal cancer (CRC) patient either with vehicle or with 25mg/kg of AD80. Treatment with AD80 induced significant (p<0.001) tumor shrinkage in CCDC6-RET PDXwt (Fig. 6B,C; Fig. S6A) (30). In line with our in vitro data of cells harboring RET gatekeeper mutations, tumor shrinkage (p<0.01) was robust but less pronounced when we treated PDX mice (n=16) engrafted with CRC tissue that had developed a CCDC6-RETV804M gatekeeper mutation under ponatinib treatment (Fig. 6B,D; Fig. S6B) (31). Consequently, we observed a robust reduction of cellular proliferation (CCDC6-RET wt: p<0.001; V804M: p<0.05) as measured by KI-67 staining in CCDC6-RETwt and CCDC6-RETV804M tumors (Fig. 6E,F). Of importance, AD80 treatment did not lead to body weight loss in both PDX models over the course of the study (Fig. S6C,D). Taken together our data indicate that AD80 represents a highly potent RET inhibitor with a favorable pharmacokinetic profile in clinically relevant RET fusion driven tumor models.
Figure 6.
A) Immunoblotting of tumor tissue from CRISPR/Cas9 induced NIH-3T3KIF5B-RET Xenografts was performed. Mice were treated (4h) with vehicle control, 12.5 or 25 mg/kg AD80, CAB or VAN and sacrificed. B) Median tumor volume was assessed using consecutive measurements of patient-derived xenograft (PDX) tumors driven by CCDC6-RETwt or CCDC6-RETV804M rearrangements under treatment with either 25 mg/kg AD80 (14d) or vehicle-control (14d). Treatment starts at day 0. C) Waterfall plot for each CCDC6-RETwt fusion positive PDX depicting best response (14d) under AD80 or vehicle-control treatment is displayed. ***, p<0.001. D) Waterfall plot for each CCDC6-RETV804M positive PDX depicting best response (7d) under AD80 or vehicle-control treatment is displayed. ***, p<0.001. E) Representative IHC stainings for H&E and Ki-67 of AD80 or vehicle control treated CCDC6-RETwt derived PDX. Scale bar represents 100 µm. F) Ki-67 IHC staining and the plotted values are shown. ***, p<0.001.
Discussion
Our chemical-genomic and chemical-proteomic analyses revealed three interesting findings with major implications for the development of effective therapies against RET-rearranged tumors: i) RET-rearranged tumors show exquisite vulnerability to a subset of type II inhibitors that target the DFG-out conformation of RET kinase, ii) compound specificity and compound activity can be only faithfully determined in orthogonal in vitro and in vivo models of rearranged RET and iii) resistance mechanisms against targeted inhibition of RET may involve RETI788N mutations and the reactivation of MAPK signaling.
The repurposing of crizotinib for the targeted treatment of ALK-rearranged tumors enabled a fast-track introduction of precision cancer medicine for this group of cancer patients and raised hopes that this approach may be a blueprint for the targeted treatment of other driver oncogenes, such as RET (32). Although initial clinical response rates were promising in selected patients, a median progression-free survival of less than 6 months and response rates of only about 18% in retrospective studies indicated that RET may be a difficult drug target after all (7–9, 33). These data are in line with our own retrospective analysis where out of four patients with RET-rearranged tumors we observed only one PR in a patient receiving vandetanib (P2) as first-line treatment (Fig. S7, Supplementary Table 6A,B). Interestingly, sequencing of rebiopsy samples did not reveal candidate drug resistance mutations, suggesting that the target had been insufficiently inhibited (Supplementary Methods; Supplementary Table 6C).
Our systematic characterization of anti-RET drugs revealed unique activity and specificity profiles for the type II kinase inhibitors AD80 and ponatinib across orthogonal in vitro and in vivo models across different lineages of RET-rearranged cancer. This finding is noteworthy, since the biochemical profiling of these compounds and structurally related compounds would have suggested a broad spectrum of kinase targets (12, 34, 35). Our data also suggest that a tight RET inhibitor binding in the DFG-out conformation of RET as measured by thermal shift assays only partially tracks with potent in vitro kinase activity and that an optimal combination of both drug target interactions may open up a unique opportunity for an effective inhibition of RET signaling. It remains to be seen how much drug residence time or structural kinetics that capture additional features of drug target interactions may contribute to the overall activity of type II inhibitors like sorafenib and other RET inhibitors (19, 36).
Of note, we identify a novel CCDC6-RETI788N resistance mutation that renders a number of tested RET inhibitors ineffective while retaining vulnerability against ponatinib. These findings resemble the experience with ALK inhibitors in ALK-rearranged tumors where the availability of potent inhibitors allows a mutant specific selection of inhibitors to overcome drug resistance (37). In addition, our results suggest that the reactivation of intracellular networks including MAPK signaling may contribute to drug tolerance and over time may modulate the efficacy of RET kinase inhibitors in RET-rearranged tumors. Given the evident clinical need for effective targeted drugs against RET our results provide a strong rationale for optimization of current therapeutic strategies and development of RET inhibitors for an effective treatment of RET-rearranged cancers.
Materials and Methods
CRISPR/Cas9
CRISPR technology was used via a pLenti vector containing Cas9-IRES-blasticidine and two U6 promoters for expression of individual sgRNAs (sgRNA1 (intron 15 murine KIF5B): GGCACCAAACACTTCACCCC; sgRNA2 (intron 11 murine RET): GGGTGTAGCGAAGTGTGCAT) (14). 24 hours after transfection the media was changed to media supplemented with blasticidine (10µg/ml) (Life Technologies) for 4 days.
Immunoblot analyses
Immunoblot analyses were performed as previously described (38). The individual antibodies are specified in the Supplementary Data. Detections of proteins was performed via horseradish peroxidase or via near-infrared fluorescent antibodies using a LI-COR Odyssey® CLx imaging system.
Phosphoproteomic analyses
LC-2/AD cells were treated with 0, 10 or 100 nM AD80, lysed, proteolytically digested with Trypsin and labeled with an Isobaric Mass Tag (TMT10plex, Thermo Fisher Scientific). Peptides for global proteome analysis were fractionated by high-pH reversed phase chromatography. Phosphopeptides were enriched via TiO2 beads and fractionated using hydrophilic interaction chromatography (39). Fractions were analyzed by nano LC-MS/MS on a Q-Exactive HF mass spectrometer (Thermo Scientific) and data were analyzed using the Proteome Discoverer 1.4 software (Thermo Scientific).
Protein Thermal Shift assay
Different variants of RET kinase domain were designed and ordered from Geneart (Life Technologies). RET variants were expressed in SF21 cells and harvested 72h post transfection. Subsequently proteins were purified and phosphorylated. For determining the protein thermal shift protein variants were incubated with DMSO or 1µM compound. Sypro-Orange dye (Life Technologies) was added to each drug treatment and thermal shift was measured in a 7500 Fast RT PCR machine (Applied Biosystems) in a temperature range of 25 – 90°C. Subsequent analysis was performed using Protein Thermal Shift Software v1.2 (Applied Biosystems).
Computational binding mode modeling
Briefly, the VEGFR was taken as template for modeling and filling of sequence gaps, representing the relevant part of the wt RET protein. All ligand-bound models were created by superpositioning followed by extensive MD simulations and energy minimization to relax the structures (RETwt/AD80, RETV804M/AD80, RETwt/Cabozantinib). For comparison with experimentally determined IC50 ratios, the binding free energy difference between RETwt/AD80 and RETwt/AD57 was further estimated by MD simulations and integral equation calculations (40), the latter approach was also used for approximate determination of the impact of the V804M mutation on the binding affinity of AD80. A detailed description can be found in the supplementary methods section.
ATP binding constant determination
ATP Km determination for RET wt and V804M mutant was performed using the HTRF KinEASE-TK assay (Cisbio) according to manufacturer’s instructions. To determine ATP Km, wt and V804M mutant were incubated with different ATP concentrations (300µM - 1.7nM) for 20min (RETwt) and 15min (RETV804M). Phosphorylation of the substrate peptide was determined by FRET between europium cryptate and XL665. ATP Km (app) was calculated using a Michaelis-Menten plot.
PDX
Tumor fragments from stock mice inoculated with CCDC6-RET fusion positive patient-derived tumor tissues were harvested and used for inoculation into BALB/c nude mice. Mice were randomly allocated into vehicle and AD80 (25mg/kg) treated groups when the average tumor volume reached 100-200mm3. Tumor volume was measured twice weekly in two dimensions using a caliper, and the volume is expressed in mm3 (TV = 0.5 a × b2, a/b represent long and short diameter).
Immunohistochemistry
Immunohistochemistry was performed on Leica Bond automated staining systems suing Ki-67 and Mib-1 (Dako) antibodies according to the manufacturer's instructions. Ki-67 labeling index was determined by manually counting 100 tumor cells in the area of the highest proliferation.
Supplementary Material
One Sentence Summary.
Our work provides mechanistic insights into the activity profiles of RET inhibitors that bind in the DFG-out conformation of RET and may be of importance for the effective treatment of RET-rearranged cancers.
Acknowledgements
We thank Dr. Thomas Zillinger from the University Hospital Bonn for sharing the Cas9 expression and the backbone of the pLenti-IRES-blasticidine vector system, the members of the Sos lab and Thomas lab for technical support, Alexandra Florin and Ursula Rommerscheidt-Fuß for supporting us with IHC staining and Patrick Kibies and Lukas Eberlein as well as Ludger Goeminne and Lieven Clement for supporting the computational modeling. We thank AstraZeneca (Zug, Switzerland) for supporting vandetanib off label use, SOBI (Lyngby, Denmark) for providing cabozantinib compassionate use, and Franziska Aebersold and Astrid Hirschmann (Luzern, Switzerland) for diagnostic work.
Funding: This work was supported by the German federal state North Rhine Westphalia (NRW) and by the European Union (European Regional Development Fund: Investing In Your Future) as part of the PerMed.NRW initiative (grant 005-1111-0025 to R.K.T., J.W. and R.B) as well as the EFRE initiative (grant LS-1-1-030 to R.B., J.W., R.K.T. and M.L.S) and by the German Ministry of Science and Education (BMBF) as part of the e:Med program (grant no. 01ZX1303 to M.P. and 01ZX1603, R.K.T., J.W., R.B and grant no. 01ZX1406 to M.P. and M.L.S), by the Deutsche Forschungsgemeinschaft (DFG; through TH1386/3-1 to R.K.T and M.L.S. and KA1381/5-1 to S.M.K) and by the German Consortium for Translational Cancer Research (DKTK) Joint Funding program. V.T. is the recipient of a joint ERS/EMBO Long-Term Research fellowship n° LTRF 2014-2951. N.Q.M. acknowledges that this work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001115), the UK Medical Research Council (FC001115) and the Wellcome Trust (FC001115); by the NCI/NIH (grant reference 5R01CA197178); by the Association for Multiple Endocrine Neoplasia Disorders MTC Research Fund. Financial support by the Ministerium für Innovation, Wissenschaft und Forschung des Landes Nordrhein-Westfalen, the Senatsverwaltung für Wirtschaft, Technologie und Forschung des Landes Berlin (O.P., R.P.Z.) and the Bundesministerium für Bildung und Forschung (O.P, R.P.Z.) is gratefully acknowledged.
Footnotes
Authors’ Contribution: D.P., M.R., J.B., M.D., C.L. and D.S. performed the cloning, experiments and cell culture experiments. V.T., A.H.S. and R.B. analyzed IHC and FISH images. Y.S. was responsible for PDX establishment and measurements. J. S., F.M., Y.A. and S.M.K performed computational modeling. O.P. and R.P.Z. performed quantitative phosphoproteomics and data analysis. M.K., M.B., A.R., J.S., J.E., M.A. and K.G. performed in vitro kinase experiments and analyses. R.C., P.P.K, and N.Q.M. purified recombinant RET fusion proteins and carried out the thermal shift analyses. J.D., G.P. and O.G. contributed the clinical patient data. F.L. and J.M.H. were responsible for NGS of RET. J.B. and M.P. analyzed RNAseq data. K.M.S. provided compounds. D.P., M.R., J.B., M.D., F.L., J.W., N.Q.M., K.M.S., R.K.T and M.L.S. interpreted the data and performed statistical analyses. D.P., M.R., S.M.K., R.K.T., O.G. and M.L.S. wrote the manuscript.
Competing interest: R.K.T. is a founder and consultant of NEO New Oncology GmbH, received commercial research grants from AstraZeneca, EOS and Merck KgaA and honoraria from AstraZeneca, Bayer, NEO New Oncology AG, Boehringer Ingelheim, Clovis Oncology, Daiichi-Sankyo, Eli Lilly, Johnson & Johnson, Merck KgaA, MSD, Puma, Roche and Sanofi. F.L. and J.M.H. are employee of NEO New Oncology GmbH. M.L.S received commercial research grants from Novartis. K.M.S and M.L.S. are both patent holders for the compound AD80.
Data and materials availability: RNA-seq data was deposited at the European Genome-phenome Archive (https://www.ebi.ac.uk/ega/, accession number EGAS00001002335). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD006006.
References
- 1.Lipson D, Capelletti M, Yelensky R, Otto G, Parker A, Jarosz M, Curran JA, Balasubramanian S, Bloom T, Brennan KW, Donahue A, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, Hatano S, Asaka R, Hamanaka W, Ninomiya H, Uehara H, Lim Choi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18:378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 3.Kohno T, Ichikawa H, Totoki Y, Yasuda K, Hiramoto M, Nammo T, Sakamoto H, Tsuta K, Furuta K, Shimada Y, Iwakawa R, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;18:375–377. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kodama T, Tsukaguchi T, Satoh Y, Yoshida M, Watanabe Y, Kondoh O, Sakamoto H. Alectinib shows potent antitumor activity against RET-rearranged non-small cell lung cancer. Molecular Cancer Therapeutics. 2014;13:2910–2918. doi: 10.1158/1535-7163.MCT-14-0274. [DOI] [PubMed] [Google Scholar]
- 5.Kurzrock R, Sherman SI, Ball DW, Forastiere AA, Cohen RB, Mehra R, Pfister DG, Cohen EEW, Janisch L, Nauling F, Hong DS, et al. Activity of XL184 (Cabozantinib), an oral tyrosine kinase inhibitor, in patients with medullary thyroid cancer. J Clin Oncol. 2011;29:2660–2666. doi: 10.1200/JCO.2010.32.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borrello MG, Ardini E, Locati LD, Greco A, Licitra L, Pierotti MA. RET inhibition: implications in cancer therapy. Expert Opinion on Therapeutic Targets. 2013;17:403–419. doi: 10.1517/14728222.2013.758715. [DOI] [PubMed] [Google Scholar]
- 7.Gautschi O, Wolf J, Milia J, Filleron T, Carbone DP, Camidge DR, Shih J-Y, Awad MM, Cabillic F, Peled N, van den Heuvel M, et al. Targeting RET in patients with RET-rearranged lung cancers: Results from a global registry. 2016 doi: 10.1200/JCO.2016.70.9352. meetinglibrary.asco.org. [DOI] [PMC free article] [PubMed]
- 8.Drilon A, Wang L, Hasanovic A, Suehara Y, Lipson D, Stephens P, Ross J, Miller V, Ginsberg M, Zakowski MF, Kris MG, et al. Response to Cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discovery. 2013;3:630–635. doi: 10.1158/2159-8290.CD-13-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoh K, Seto T, Satouchi M, Nishio M, Yamamoto N, Murakami H, Nogami N, Matsumoto S, Kohno T, Tsuta K, Tsuchihara K, et al. Vandetanib in patients with previously treated RET-rearranged advanced non-small-cell lung cancer (LURET): an open-label, multicentre phase 2 trial. The Lancet Respiratory Medicine. 2017;5:42–50. doi: 10.1016/S2213-2600(16)30322-8. [DOI] [PubMed] [Google Scholar]
- 10.Falchook GS, Ordóñez NG, Bastida CC, Stephens PJ, Miller VA, Gaido L, Jackson T, Karp DD. Effect of the RET Inhibitor Vandetanib in a Patient With RET Fusion–Positive Metastatic Non–Small-Cell Lung Cancer. Journal of Clinical Oncology. 2016 doi: 10.1200/JCO.2013.50.5016. page:string:Article/Chapter. [DOI] [PubMed] [Google Scholar]
- 11.Cascone T, Hess KR, Piha-Paul S, Hong DS, Subblah IM, Bhatt T, Lui A, Fu S, Naing A, Janku F, Karp DD, et al. Safety, toxicity and activity of multi-kinase inhibitor vandetanib in combination with everolimus in advanced solid tumors. 2016 doi: 10.1016/j.esmoop.2021.100079. meetinglibrary.asco.org. [DOI] [PMC free article] [PubMed]
- 12.Dar AC, Das TK, Shokat KM, Cagan RL. Chemical genetic discovery of targets and anti-targets for cancer polypharmacology. Nature. 2012;486:80–84. doi: 10.1038/nature11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song M. Progress in Discovery of KIF5B-RET Kinase Inhibitors for the Treatment of Non-Small-Cell Lung Cancer. J Med Chem. 2015;58:3672–3681. doi: 10.1021/jm501464c. [DOI] [PubMed] [Google Scholar]
- 14.Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nature Communications. 2014;5:3728. doi: 10.1038/ncomms4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki M, Makinoshima H, Matsumoto S, Suzuki A, Mimaki S, Matsushima K, Yoh K, Goto K, Suzuki Y, Ishii G, Ochiai A, et al. Identification of a lung adenocarcinoma cell line with CCDC6-RET fusion gene and the effect of RET inhibitors in vitro and in vivo. Cancer Science. 2013;104:896–903. doi: 10.1111/cas.12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sos ML, Michel K, Zander T, Weiss J, Frommolt P, Peifer M, Li D, Ullrich R, Koker M, Fischer F, Shimamura T, et al. Predicting drug susceptibility of non–small cell lung cancers based on genetic lesions. J Clin Invest. 2009;119:1727–1740. doi: 10.1172/JCI37127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knowles PP, Murray-Rust J, Kjær S, Scott RP, Hanrahan S, Santoro M, Ibáñez CF, McDonald NQ. Structure and chemical inhibition of the RET tyrosine kinase domain. J Biol Chem. 2006;281:33577–33587. doi: 10.1074/jbc.M605604200. [DOI] [PubMed] [Google Scholar]
- 18.Hasegawa Masaichi, Nishigaki Naohiko, Washio Yoshiaki, Kano Kazuya, Harris Philip A, Sato Hideyuki, Mori Ichiro, West Rob I, Shibahara Megumi, Toyoda Hiroko, Wang Liping, et al. Discovery of Novel Benzimidazoles as Potent Inhibitors of TIE-2 and VEGFR-2 Tyrosine Kinase Receptors. J Med Chem. 2007;50:4453–4470. doi: 10.1021/jm0611051. [DOI] [PubMed] [Google Scholar]
- 19.Frett B, Carlomagno F, Moccia ML, Brescia A, Federico G, De Falco V, Admire B, Chen Z, Qi W, Santoro M, Li HY. Fragment-Based Discovery of a Dual pan-RET/VEGFR2 Kinase Inhibitor Optimized for Single-Agent Polypharmacology. Angewandte Chemie International Edition. 2015;54:8717–8721. doi: 10.1002/anie.201501104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlomagno F, Guida T, Anaganti S, Vecchio G, Fusco A, Ryan AJ, Billaud M, Santoro M. Disease associated mutations at valine 804 in the RET receptor tyrosine kinase confer resistance to selective kinase inhibitors. Oncogene. 2004;23:6056–6063. doi: 10.1038/sj.onc.1207810. [DOI] [PubMed] [Google Scholar]
- 21.Mologni L, Redaelli S, Morandi A, Plaza-Menacho I, Gambacorti-Passerini C. Ponatinib is a potent inhibitor of wild-type and drug-resistant gatekeeper mutant RET kinase. Molecular and Cellular Endocrinology. 2013;377:1–6. doi: 10.1016/j.mce.2013.06.025. [DOI] [PubMed] [Google Scholar]
- 22.Yun C-H, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong K-K, Meyerson M, Eck MJ. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci USA. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azam M, Latek RR, Daley GQ. Mechanisms of Autoinhibition and STI-571/Imatinib Resistance Revealed by Mutagenesis of BCR-ABL. Cell. 2003;112:831–843. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 24.Heuckmann JM, Holzel M, Sos ML, Heynck S, Balke-Want H, Koker M, Peifer M, Weiss J, Lovly CM, Grutter C, Rauh D, et al. ALK Mutations Conferring Differential Resistance to Structurally Diverse ALK Inhibitors. Clinical Cancer Research. 2011;17:7394–7401. doi: 10.1158/1078-0432.CCR-11-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sos ML, Levin RS, Gordan JD, Oses-Prieto JA, Webber JT, Salt M, Hann B, Burlingame AL, McCormick F, Bandyopadhyay S, Shokat KM. Oncogene Mimicry as a Mechanism of Primary Resistance to BRAF Inhibitors. Cell Rep. 2014;8:1037–1048. doi: 10.1016/j.celrep.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandarlapaty S. Negative Feedback and Adaptive Resistance to the Targeted Therapy of Cancer. Cancer Discovery. 2012;2:311–319. doi: 10.1158/2159-8290.CD-12-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Boer R, Humblet Y, Wolf J, Nogová L, Ruffert K, Milenkova T, Smith R, Godwood A, Vansteenkiste J. An open-label study of vandetanib with pemetrexed in patients with previously treated non-small-cell lung cancer. Ann Oncol. 2009;20:486–491. doi: 10.1093/annonc/mdn674. [DOI] [PubMed] [Google Scholar]
- 29.Bentzien F, Zuzow M, Heald N, Gibson A, Shi Y, Goon L, Yu P, Engst S, Zhang W, Huang D, Zhao L, et al. In vitro and in vivo activity of cabozantinib (XL184), an inhibitor of RET, MET, and VEGFR2, in a model of medullary thyroid cancer. Thyroid. 2013;23:1569–1577. doi: 10.1089/thy.2013.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gozgit JM, Chen T-H, Clackson T, Rivera VM. Abstract 2726: RET fusions identified in colorectal cancer PDX models are sensitive to the potent RET inhibitor ponatinib. Cancer Research. 2014;74:2726–2726. [Google Scholar]
- 31.Yang M, Cai J, Guo S, Wery J-P, Li HQ. Abstract 3581: Rapid conversion to resistance, of a colon PDX with ret-fusion, by ponatinib treatment could potentially be attributed to the introduction of the gate keeper mutation V804M. Cancer Research. 2015;75:3581–3581. [Google Scholar]
- 32.Solomon BJ, Mok T, Kim D-W, Wu Y-L, Nakagawa K, Mekhail T, Felip E, Cappuzzo F, Paolini J, Usari T, Iyer S, et al. First-Line Crizotinib versus Chemotherapy in ALK-Positive Lung Cancer. N Engl J Med. 2014;371:2167–2177. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 33.Gautschi O, Zander T, Keller FA, Strobel K, Hirschmann A, Aebi S, Diebold J. A Patient with Lung Adenocarcinoma and RET Fusion Treated with Vandetanib. Journal of Thoracic Oncology. 2013;8:e43–e44. doi: 10.1097/JTO.0b013e31828a4d07. [DOI] [PubMed] [Google Scholar]
- 34.Dar AC, Lopez MS, Shokat KM. Small Molecule Recognition of c-Src via the Imatinib-Binding Conformation. Chemistry & Biology. 2008;15:1015–1022. doi: 10.1016/j.chembiol.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, Wang F, Adrian LT, Zhou T, Huang W-S, Xu Q, Metcalf CA, III, et al. AP24534, a Pan-BCR-ABL Inhibitor for Chronic Myeloid Leukemia, Potently Inhibits the T315I Mutant and Overcomes Mutation-Based Resistance. Cancer Cell. 2009;16:401–412. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carlomagno F, Anaganti S, Guida T, Salvatore G, Troncone G, Wilhelm SM, Santoro M. BAY 43-9006 Inhibition of Oncogenic RET Mutants. J Natl Cancer Inst. 2006;98:326–334. doi: 10.1093/jnci/djj069. [DOI] [PubMed] [Google Scholar]
- 37.Gainor JF, Dardaei L, Yoda S, Friboulet L, Leshchiner I, Katayama R, Dagogo-Jack I, Gadgeel S, Schultz K, Singh M, Chin E, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discovery. 2016;6:1118–1133. doi: 10.1158/2159-8290.CD-16-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandez-Cuesta L, Plenker D, Osada H, Sun R, Menon R, Leenders F, Ortiz-Cuaran S, Peifer M, Bos M, Daßler J, Malchers F, et al. CD74-NRG1 Fusions in Lung Adenocarcinoma. Cancer Discovery. 2014;4:415–422. doi: 10.1158/2159-8290.CD-13-0633. [DOI] [PubMed] [Google Scholar]
- 39.Dickhut C, Radau S, Zahedi RP. Fast, efficient, and quality-controlled phosphopeptide enrichment from minute sample amounts using titanium dioxide. Methods Mol Biol. 2014;1156:417–430. doi: 10.1007/978-1-4939-0685-7_28. [DOI] [PubMed] [Google Scholar]
- 40.Heil J, Kast SM. 3D RISM theory with fast reciprocal-space electrostatics. The Journal of Chemical Physics. 2015;142:114107. doi: 10.1063/1.4914321. [DOI] [PubMed] [Google Scholar]
- 41.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Meth. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 42.Kollipara L, Zahedi RP, Zahedi R, Sickmann A, editors. Protein carbamylation: In vivo modification or in vitro artefact? PROTEOMICS. 2013;13:941–944. doi: 10.1002/pmic.201200452. [DOI] [PubMed] [Google Scholar]
- 43.Burkhart JM, Schumbrutzki C, Wortelkamp S, Sickmann A, Zahedi RP. Systematic and quantitative comparison of digest efficiency and specificity reveals the impact of trypsin quality on MS-based proteomics. Journal of Proteomics. 2012;75:1454–1462. doi: 10.1016/j.jprot.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 44.Engholm-Keller K, Birck P, Størling J, Pociot F, Mandrup-Poulsen T, Larsen MR. TiSH — a robust and sensitive global phosphoproteomics strategy employing a combination of TiO2, SIMAC, and HILIC. Journal of Proteomics. 2012;75:5749–5761. doi: 10.1016/j.jprot.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 45.Taus T, Köcher T, Pichler P, Paschke C, Schmidt A, Henrich C, Mechtler K. Universal and Confident Phosphorylation Site Localization Using phosphoRS. J Proteome Res. 2011;10:5354–5362. doi: 10.1021/pr200611n. [DOI] [PubMed] [Google Scholar]
- 46.Spivak M, Weston J, Bottou L, Käll L, Noble WS. Improvements to the Percolator Algorithm for Peptide Identification from Shotgun Proteomics Data Sets. J Proteome Res. 2009;8:3737–3745. doi: 10.1021/pr801109k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xue Y, Ren J, Gao X, Jin C, Wen L, Yao X. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics. 2008;7:1598–1608. doi: 10.1074/mcp.M700574-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.modeller. salilab.org (available at https://salilab.org/modeller/).
- 49.Sigalov G, Fenley A, Onufriev A. Analytical electrostatics for biomolecules: Beyond the generalized Born approximation. The Journal of Chemical Physics. 2006;124:124902. doi: 10.1063/1.2177251. [DOI] [PubMed] [Google Scholar]
- 50.Amber. ambermd.org, doi: 10.1002/jcc.23031/abstract.
- 51.Engel J, Richters A, Getlik M, Tomassi S, Keul M, Termathe M, Lategahn J, Becker C, Mayer-Wrangowski S, Grütter C, Uhlenbrock N, et al. Targeting Drug Resistance in EGFR with Covalent Inhibitors: A Structure-Based Design Approach. J Med Chem. 2015;58:6844–6863. doi: 10.1021/acs.jmedchem.5b01082. [DOI] [PubMed] [Google Scholar]
- 52.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics. 1983;79:926–935. [Google Scholar]
- 53.NAMD. ks.uiuc.edu, doi: 10.1002/jcc.23422/abstract.
- 54.salilab.org (; https://salilab.org).
- 55.Beglov D, Roux B. An Integral Equation To Describe the Solvation of Polar Molecules in Liquid Water. J Phys Chem B. 1997;101:7821–7826. [Google Scholar]
- 56.Kovalenko A, Hirata F. Three-dimensional density profiles of water in contact with a solute of arbitrary shape: a RISM approach. Chemical Physics Letters. 1998;290:237–244. [Google Scholar]
- 57.Mrugalla F, Kast SM. Designing molecular complexes using free-energy derivatives from liquid-state integral equation theory. J Phys: Condens Matter. 2016;28:344004. doi: 10.1088/0953-8984/28/34/344004. [DOI] [PubMed] [Google Scholar]
- 58.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ, Prlic A, editors. Software for Computing and Annotating Genomic Ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:31. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehár J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, et al. PGC-1|[alpha]|-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 63.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, Faraoni R, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 64.Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, Zarrinkar PP. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.