Summary
We analyzed the in silico purified DNA methylation signatures of 82 mantle cell lymphomas (MCL) in comparison with cell subpopulations spanning the entire B cell lineage. We identified two MCL subgroups, respectively carrying epigenetic imprints of germinal center-inexperienced and germinal center-experienced B cells, and we found that DNA methylation profiles during lymphomagenesis are largely influenced by the methylation dynamics in normal B cells. An integrative epigenomic approach revealed 10,504 differentially methylated regions in regulatory elements marked by H3K27ac in MCL primary cases, including a distant enhancer showing de novo looping to the MCL oncogene SOX11. Finally, we observed that the magnitude of DNA methylation changes per case is highly variable and serves as an independent prognostic factor for MCL outcome.
Introduction
The existence of alterations in the DNA methylome of cancer cells is known since the early 1980s (Feinberg and Vogelstein, 1983; Gama-Sosa et al., 1983). In spite of the widely reported role of DNA methylation in cancer (Baylin and Jones, 2011; Esteller, 2008), the analyses of whole DNA methylomes are now questioning the accepted view of DNA methylation as a major player in gene deregulation. These analyses are revealing that the roles of this epigenetic mark are more variable and context-dependent than previously appreciated (Jones, 2012; Kulis et al., 2013) and that a large fraction of the DNA methylation changes in cancer do not seem to have any apparent functional effect (Agirre et al., 2015; Keshet et al., 2006; Ziller et al., 2013). So far, cancer epigenomics studies have detected tumor-specific changes by comparing tumor cells versus their normal counterparts, or by comparing longitudinal tumor samples. However, it is becoming increasingly evident that regions with dynamic DNA methylation levels in normal cells seem to be prone to be altered upon neoplastic transformation (Feinberg, 2014; Hansen et al., 2011; Kulis et al., 2015). Thus, a detailed analysis of a specific tumor type in the context of the entire differentiation program of its normal cellular counterpart shall reveal new insights into the role of DNA methylation in tumorigenesis.
Mantle cell lymphoma (MCL) is a B cell lymphoma that shows a broad spectrum of clinical behaviors and biological features (Jares et al., 2012). In spite of the heterogeneity, the unifying factor in MCL is the t(11;14)(q13;q32) translocation leading to the cyclin D1 gene (CCND1) deregulation, which is considered to be a primary driver event in this disease (Jares et al., 2007). Most MCLs have an aggressive clinical behavior with poor survival rates. However, some cases classified as leukemic non-nodal MCLs show a rather indolent clinical course even in the long-term absence of chemoterapy (Royo et al., 2012). Aggressive cases are highly proliferative and seem to be associated with lack or low levels of somatic mutations in the IGHV locus and de novo expression of SOX11, which is not expressed in normal B cells. In contrast, leukemic non-nodal MCLs show a very low proliferation index, have high levels of somatic mutations in the IGHV locus and lack SOX11 expression. However, some of these SOX11-negative MCLs can acquire oncogenic mutations and progress towards a fatal clinical outcome (Jares et al., 2012).
The DNA methylome of MCL remains largely unknown, as it has only been analyzed in promoter regions (Enjuanes et al., 2013; Halldorsdottir et al., 2012; Leshchenko et al., 2010; Rahmatpanah et al., 2006). To obtain deeper insights into MCL epigenetics, we have here we applied an analytic strategy to deconstruct the DNA methylome of MCL in the light of the complete normal B cell differentiation program (Kulis et al., 2015).
Results
Deconvolution and in silico purification of MCL DNA methylation signatures
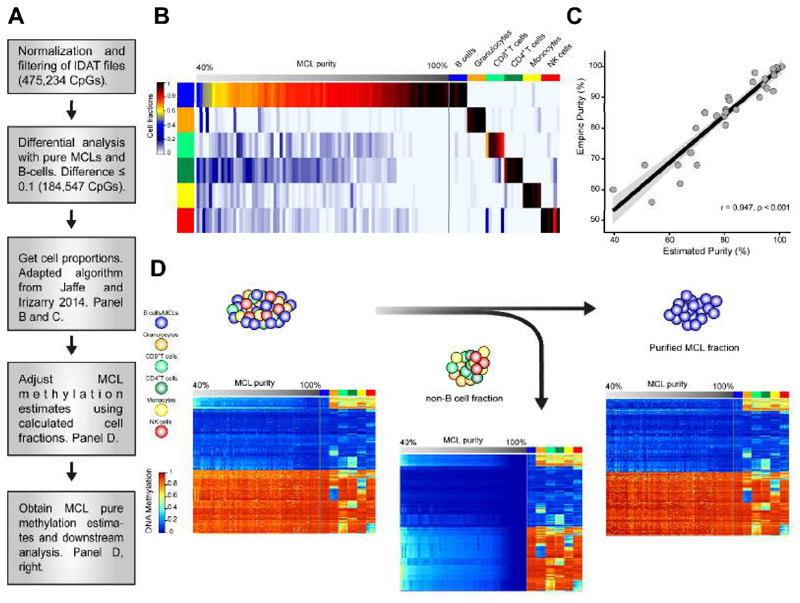
We generated genome-wide DNA methylation profiles of 82 MCL samples using the HumanMethylation450 BeadChip (Illumina Inc.) (Bibikova et al., 2011). Biological and clinical information of the analyzed cases is shown in Table S1. As normal controls, we used 67 samples from 10 different cell subpopulations spanning the entire B cell lineage (Kulis et al., 2015). We considered two potential confounding variables that may affect our epigenomic analyses, i.e. the biological origin of the samples (lymph node vs. peripheral blood) and the tumor cell content. We did not identify any consistent differential methylation pattern between lymph node and peripheral blood samples (data not shown). However, in spite of the generally high tumor cell content of the selected MCL samples (median 89%, range = 56 to 100%, Table S1), purity affected the DNA methylation analyses (Figure S1). Therefore, we developed a strategy to deconvolute the DNA methylation signal of mixed subpopulations and to isolate in silico the DNA methylation levels of the tumor cells (Figure 1A). To that end we adapted a published algorithm (Houseman et al., 2012; Jaffe and Irizarry, 2014) to estimate the fractions of 6 different hematopoietic cell types (Reinius et al., 2012) in our tumor samples (Figure 1B). The normal B cell fraction in MCL samples is estimated to be very low (0-0.3%) (Saba et al., 2016), therefore the total B cell fraction was taken as a measure for the tumor fraction. Using the adapted algorithm, we calculated the proportion of each cell type in our samples. We validated the approach by comparing the in silico estimated tumor B cell fraction with the sample purity measured by flow cytometry in 32 MCL samples (Pearson r = 0.947, Figure 1C). Finally, we used the DNA methylation estimates of the normal non-B cell subtypes together with their respective proportions to extract the DNA methylation signature derived from the tumor B cells in each MCL sample (Figure 1D). These pure DNA methylation estimates of the tumor fraction were used for all downstream analyses.
Figure 1. Deconvolution of DNA methylation data and in silico purification of MCL methylation estimates.
(A) Work flow of the deconvolution process in MCL samples. (B) Estimation of the proportion of hematopoietic cell subpopulations in MCL samples and in sorted B cells, CD8+ T cells, CD4+ T cells, NK cells, monocytes and granulocytes. Sorted cell subpopulations (right part of the heatmap) are correctly predicted and MCLs show a gradient from lower to higher proportion of B cells (left part of the heatmap) (C) The proportion of B cells in MCL samples as detected by flow cytometry and by the in silico prediction are highly correlated. (D) Heatmaps of the CpGs representative of each cell type (n=580) showing the initial methylation estimates the MCL samples (left), the extraction of the DNA methylation signature from contaminating non-B cells (middle) and the final in silico purification of the DNA methylation estimates from MCL cells (right). See also Figure S1 and Table S1.
Genome-wide DNA methylation analysis reveals two major MCL subgroups with distinct clinico-biological features
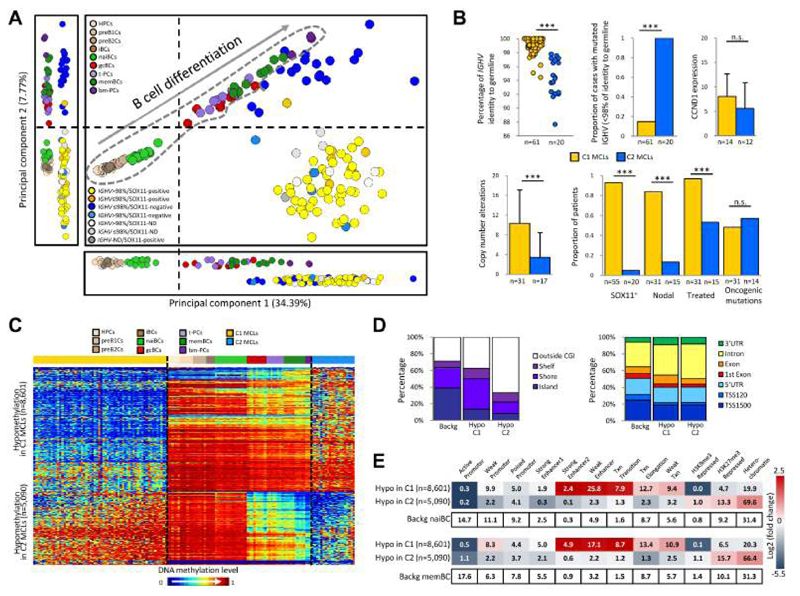
We performed an unsupervised principal component analysis (PCA) of DNA methylation data from normal B cell subpopulations and MCL samples (Figure 2A). The two first components ordered normal B cells according to their maturation stage, mainly separating germinal center-inexperienced B cells (uncommitted precursors, pre-B cells and naive B cells) from germinal center-experienced B cells (germinal center B cells, memory B cells and plasma cells). Principal component 1 showed that all MCLs are globally more similar to germinal center-experienced B cells (i.e. antigen experienced). In contrast, principal component 2 split MCLs into two subgroups: cluster 1 (C1) (n=62) and cluster 2 (C2) (n=20), which respectively showed a DNA methylation pattern more similar to germinal center-inexperienced B cells and germinal center-experienced B cells. These subgroups showed significant clinico-biological differences (p < 0.001) in e.g. IGHV mutation levels, SOX11 expression, number of copy number alterations, nodal presentation, and requirement of treatment at diagnosis (Figure 2B). Furthermore, C1 cases showed a significantly worse overall survival than C2 (p = 0.026) (Figure S2A).
Figure 2. Identification of two MCL subgroups based on DNA methylation profiling.
(A) Unsupervised PCA of 82 MCLs and 67 normal B cell subpopulations using the adjusted methylation values of all CpGs analyzed with the 450K array. The two main principal components are shown together in a 2D plot and separately. Vertical and horizontal dotted lines point to the cut-off value separating germinal center-inexperienced and -experienced B cells. Normal B cells are surrounded by a dotted grey line. (B) Comparison of biological and clinical features between the two epigenetic subgroups (i.e. C1 and C2). The presence of oncogenic mutations is defined as having a mutation in at least one of the following genes: BIRC3, MEF2B, NOTCH2, TLR2, TP53 and WHSC1. Data show mean ± SD. ***p < 0.001 (Fisher's exact test or t-test for independent samples). (C) Heatmap of the CpGs differentially methylated in C1 as compared to C2. (D) Location of the hypo- and hypermethylated CpGs between C1 and C2 MCLs in the context of CpG islands (CGI) and gene-related regions. (E) Chromatin states of naive (upper panel) and memory (lower panel) B cells of the differentially methylated CpGs between C1 and C2 MCLs. The numbers inside each cell point to the percentage of CpGs belonging to a particular chromatin state. The differentially methylated CpGs annotated in panels D and E are the same as those shown in panel C. HPCs, hematopoietic progenitor cells; preB1Cs, pre-BI cells; preB2Cs, pre-BII cells; iBCs, immature B cells; naiBCs, naive B cells from peripheral blood; gcBCs, germinal center B cells; t-PCs, plasma cells from tonsil; memBCs, memory B cells from peripheral blood; bm-PCs, plasma cells from bone marrow; C1 MCLs, germinal center-inexperienced MCLs; C2 MCLs, germinal center-experienced MCLs; Backg, background; Hypo, hypomethylation; Hyper, hypermethylation; TSS, transcriptional start site, UTR, untranslated region. See also Figure S2 and Table S2.
Next, we compared C1 and C2 MCLs, and identified 13,691 differentially methylated CpGs (Figure 2C). Most CpGs hypomethylated in C2 MCLs linked C1 cases to germinal center-inexperienced cells and C2 cases to germinal-center experienced B cells (Figures 2C), further supporting the concept shown in the second component of the PCA analysis (Figure 2A). In contrast, hypomethylation in C1 was predominantly a de novo event targeting regions that are highly methylated both in C2 MCLs and normal B cells (Figure 2C). These regions frequently targeted CpG island shores and gene bodies (Figure 2D). Furthermore, we performed ChIP-seq with 6 histone marks and generated chromatin states from sorted naive and memory B cells from healthy donors, and observed that hypomethylated regions in C1 MCLs were enriched for enhancers and transcribed regions (Figure 2E). Naive and memory B cells have been previously suggested as potential cells of origin of respectively IGHV unmutated and mutated MCLs (Navarro et al., 2012). Therefore, in this study we used them as normal counterparts of C1 MCLs and C2 MCLs, respectively. Interestingly, the genes affected by hypomethylation in C1 MCLs were significantly enriched (adjusted p < 0.05) in several pathways (Table S2), such as NOTCH signaling (Figure S2B), which has been previously linked to MCL pathogenesis of the IGHV unmutated/SOX11-positive subgroup (i.e. C1) (Bea et al., 2013; Kridel et al., 2012).
Comparing MCL groups with their normal cell counterparts reveals a major epigenetic link with normal B cell differentiation
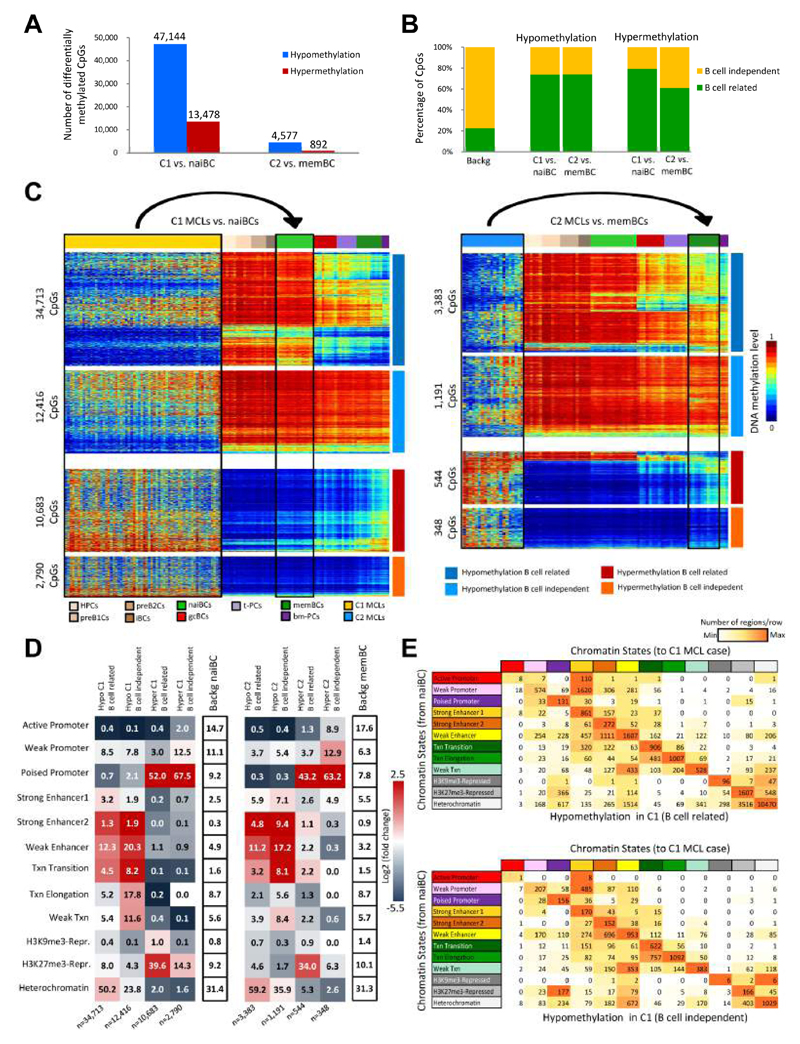
Next, we sought to detect epigenetic differences in the MCL subgroups as compared with their respective putative normal counterparts. We observed 60,622 differentially methylated CpGs in C1 MCLs (78% hypomethylated) in comparison with naive B cells and 5,469 CpGs in C2 MCLs (84% hypomethylated) in comparison with memory B cells (Figure 3A). Interestingly, we found that 61 to 79% of these CpGs overlapped with those previously described to show variable DNA methylation levels during normal B cell differentiation (Kulis et al., 2015) (Figures 3B and 3C). This finding suggests that only a fraction of the DNA methylation changes in MCLs as compared with their normal counterparts is unrelated to normal B cell differentiation and thus, strictly tumor-specific. Those CpGs dynamically methylated both in MCL and B cell differentiation (from now on called B cell-related CpGs) and those exclusively changing in MCL (from now on called B cell-independent CpGs) were in part enriched in different chromatin states defined in naive and memory B cells (Figure 3D). Overall, hypomethylation in MCL in both the B cell-related and independent fractions was enriched for enhancer elements. On the contrary, B cell-related hypermethylated CpGs in MCL were located both in H3K27me3-repressed and poised promoters, whereas those in the B cell-independent fraction were mostly associated with poised promoters (Figure 3D). To identify chromatin state transitions in relationship with DNA methylation changes, we generated ChIP-seq profiles and chromatin states from two MCL cases representative for C1 and C2, and compared them to those from naive and memory B cells, respectively (Figures 3E, S3A and S3B). Overall, 56% of the regions did not seem to change their chromatin state in MCL upon a DNA methylation alteration. However, we observed that repressed regions losing DNA methylation tend to change towards chromatin states related to activating histone modifications (especially H3K4me3 in poised promoters and H3K4me1 in weak enhancers) (Figure 3E), being this phenomenon more prominent in the B cell independent than in the B cell related fraction in C1 (52% vs. 18%, p < 0.001). In the case of hypermethylated regions in C1 MCLs, we observed that active and weak promoters in naive B cells turn into poised promoters in MCL and that poised promoters turn into H3K27me3-repressed regions (Figure S3A).
Figure 3. DNA methylation of MCL subgroups versus their respective normal B cell counterpart.
(A) Number of differentially methylated CpGs between C1 and naiBCs, and between C2 and memBCs. (B) Percentage of B cell-related and B cell-independent CpGs differentially methylated in each comparison. (C) Heatmaps of differentially methylated CpGs in C1 MCLs as compared to naiBCs (left) and in C2 MCLs as compared to memBCs (right) in the context of normal B cell differentiation. (D) Chromatin states in naiBCs and memBCs of the differentially methylated CpGs between C1 and naiBCs (left panel), and between C2 and memBCs (right panel), respectively. The numbers inside each cell point to the percentage of CpGs belonging to a particular chromatin state. (E) Transition of the chromatin states from naiBCs to a C1 MCL case in the B cell-related and B cell-independent hypomethylated CpGs. The numbers inside each cell point to the total number of CpGs in each transition. See also Figure S3.
Individual epigenetic heterogeneity in MCL
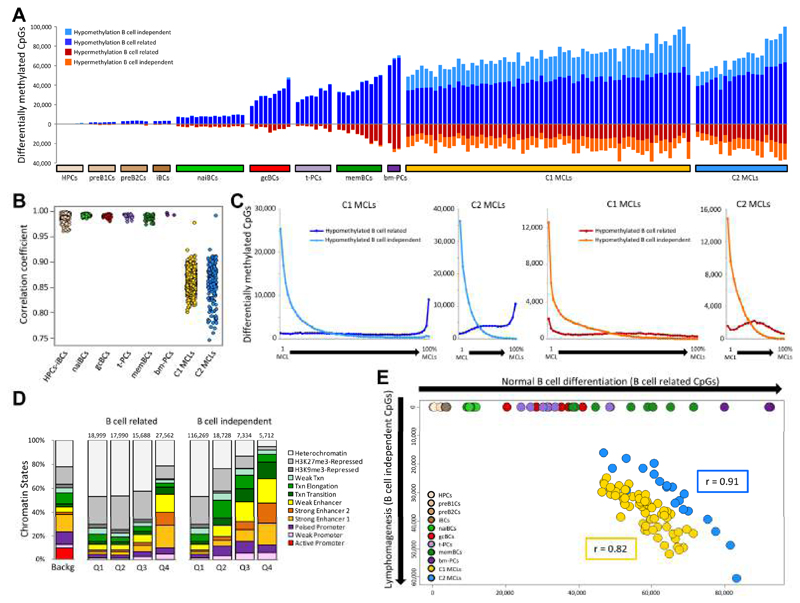
The data presented in Figures 2 and 3 suggest that both MCL groups are epigenetically heterogeneous. Based on these observations, we have applied a second analytic strategy to tackle individual epigenetic variation of MCL cases in the context of the entire B cell maturation program. We compared the DNA methylome of each individual MCL case with the hematopoietic progenitor cells (HPCs) (using as cut-off an absolute difference of methylation values of at least 0.25). This seemingly unorthodox approach has the advantage that it uses a fixed reference point for B cell neoplasms with different normal counterparts, and allows us not only to precisely dissect but also to compare the DNA methylation modulation of each individual MCL sample from the moment of B cell commitment up to and beyond its cell of origin. We observed that the total number of changes per case is highly variable both in C1 and C2 MCLs (ranging from 62,888 to 143,925 CpGs) (Figure 4A). Furthermore, we identified that the DNA methylation levels of the MCLs correlate less among each other than within normal B cells, showing that the inter-sample heterogeneity is much higher in MCLs than in normal B cells (Figure 4B). Additionally, we saw that 318,659 unique CpGs (98% of the 106,552 B cell-related and 53% of the 368,442 B cell-independent CpGs measured by the 450k array) showed a DNA methylation change as compared to HPCs in at least one MCL case, suggesting that a large fraction of the human methylome can be modulated in normal and neoplastic B cells.
Figure 4. Association between B cell-related and B cell-independent DNA methylation changes in MCL.
(A) Number of differentially methylated CpGs for each individual normal B cell subpopulation and MCL as compared to HPCs. (B) Correlation coefficient among samples of the different groups. (C) Number of B cell-related and B cell-independent differentially methylated CpGs based on their level of recurrence in C1 (1st and 3rd panel) and C2 (2nd and 4th panel). (D) Chromatin states, defined in a MCL primary case representative of C1 cases, of the hypomethylated CpGs between C1 and naiBCs divided into quartiles based on their level of recurrence. Q1, recurrent in 0-25% of patients; Q2, recurrent in 25-50% of patients; Q3, recurrent in 50-75% of patients; Q4, recurrent in 75-100% of patients. (E) Scatter plot showing the number of B cell-related (x-axis) and B cell-independent (y-axis) CpGs differentially methylated in individual MCLs and normal B cells as compared to HPCs. See also Figure S4.
Next, to identify regions that may play a role in MCL development, we sought to identify B cell-independent CpGs with recurrent differential methylation in C1 and C2 MCLs. We detected that the majority of the differentially methylated sites between MCL and HPC were present in one or few MCLs, and that highly recurrent changes were rare events (Figure 4C). Furthermore, the relative proportion of differentially methylated regions marked by particular chromatin states (as defined in primary MCL cases), such as heterochromatin and enhancers, was related to the level of recurrence of the DNA methylation changes (Figures 4D, S4A and S4B). These findings suggest that most B cell-independent changes in individual MCLs seem to target non-functional regions (i.e. heterochromatin) while commonly altered CpGs, although rare, target regulatory elements (i.e. enhancers).
An additional interesting aspect of this analysis of individual variation was that the number of B cell-related and B cell-independent differentially methylated CpGs per MCL case were linearly related (Pearson r = 0.82 and 0.91 for C1 and C2 MCLs, respectively; p < 0.001) (Figure 4E). This association suggests that the mechanisms underlying differentially methylation in B cell-related and B cell-independent CpGs are shared, even though different cases show different degrees of epigenetic changes. Additionally, in C1 MCLs, we detected 6,245 CpGs with an inverse correlation between their DNA methylation levels and the percentage of IGHV somatic hypermutation (SHM) (Figures S4C to S4F), a phenomenon not observed in C2 cases. The fact that some C1 MCLs concurrently show some degree of SHM and DNA demethylation suggests that C1 MCLs may be derived from germinal center-inexperienced B cells at different maturation stages, ranging from those lacking SHM to those showing low but variable degrees of SHM, which correlate with epigenetic changes (Kolar et al., 2007; Sims et al., 2005).
Deep characterization of the MCL methylome by whole-genome bisulfite sequencing
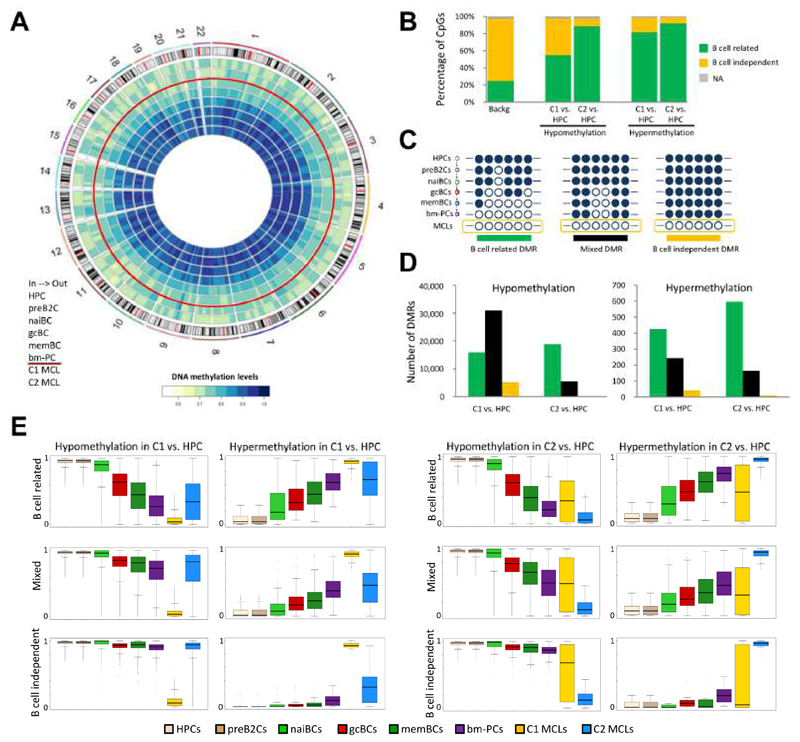
We sequenced the entire DNA methylome of two highly pure (95 and 99% tumor cells) representative MCLs previously analyzed by 450k microarrays (one from MCL C1 and one from MCL C2, Figures S5A to S5C) at a single base pair resolution (48x mean coverage, Table S3), and we analyzed them in the context of the DNA methylome of the B cell lineage (Kulis et al., 2015) (Figures 5A, S5D and S5E). The methylation estimates obtained by the two methods were highly comparable (Pearson r = 0.97 for both cases, Figure S5C). We compared each MCL vs. HPCs as fixed reference and we defined both differentially methylated CpGs (DMCs) and differentially methylated regions (DMRs). Determining DMRs increased the detection of regulatory regions as compared to detecting DMCs, and therefore we continued our analyses using the DMR strategy (Figures S5F to S5I). Subsequently, we split the CpGs within DMRs into B cell-related and B cell-independent CpGs and we observed that 55-92% overlapped with those modulated during normal B cell differentiation (Figure 5B). Intriguingly, most DMRs in MCL either contained only B cell-related or a mixture of both B cell-related and B cell-independent CpGs, and few were exclusively B cell independent (Figures 5C to 5E). More specifically, in the C1 MCL case, only 9.3% of the hypomethylated and 5.6% of the hypermethylated DMRs were B cell-independent, and these numbers respectively dropped to 1% and 1.2% in the C2 MCL case (Figure 5D). This analysis suggests that those regions prone to acquire differential methylation during normal B cell differentiation seem to be predisposed to be further altered in the context of malignant transformation, and that regions with pure tumor-specific DMRs seem to be a rare phenomenon.
Figure 5. Analysis of the MCL methylome by WGBS.
(A) Circular representation of the DNA methylation levels for HPC, preB2C, naiBC, gcBC, memBC and bm-PC, as well as two MCLs representative for C1 and C2, respectively. CpG methylation levels are averaged over 10 Mb genomic windows. (B) Percentage of B cell-related and B cell-independent CpGs differentially methylated in C1 MCL and C2 MCL versus HPC. (C) Graphical representation of the different DMR types: DMRs with only B cell-related CpGs are defined as B cell-related DMRs (left), DMRs containing both B cell-related and B cell-independent CpGs are defined as mixed DMRs (middle), and DMRs with only B cell-independent CpGs are defined as B cell-independent DMRs (right). (D) Number of B cell-related, mixed and B cell-independent DMRs between C1 versus HPC and between C2 versus HPC. (E) Distribution of DNA methylation levels for the different DMRs types defined between C1 MCL and HPC and between C2 MCL and HPC. Box plots show upper and lower quartiles and the median, and whiskers represent minimum and maximum, with outer points indicating outliers. See also Figure S5 and Table S3.
Identification of potential epigenetic drivers in MCL and detection of distant SOX11 enhancers
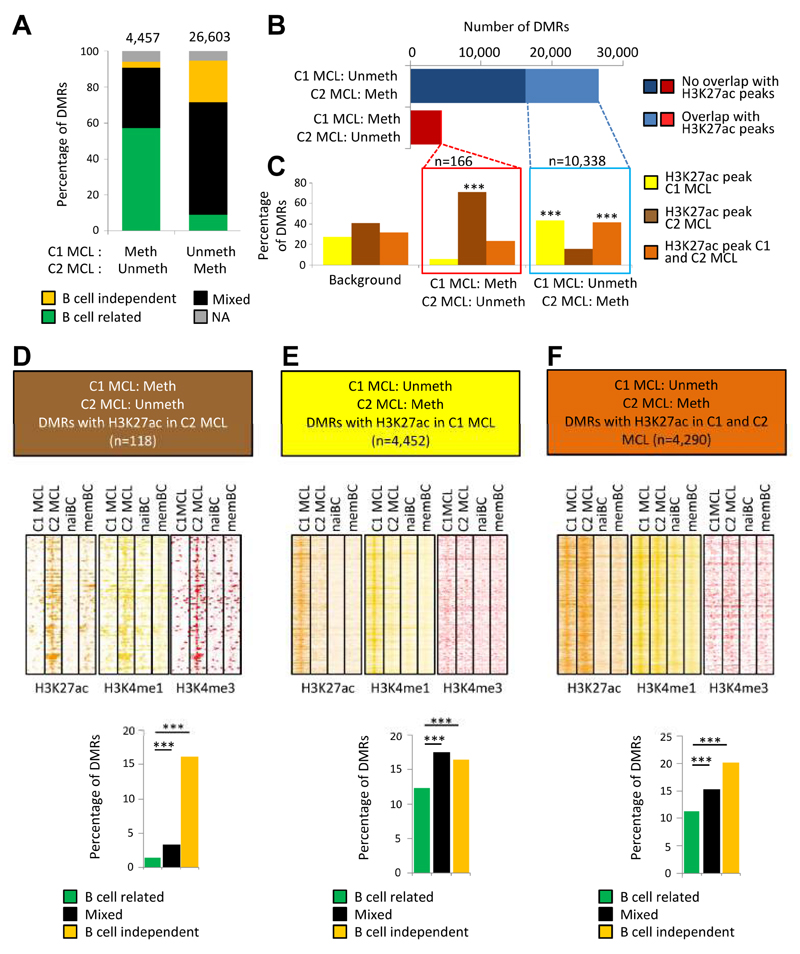
Next, we aimed to study whether DMRs between C1 and C2 MCLs can lead to the detection of potential functional regulatory regions that are differentially active in these two groups. By comparing them, we detected 26,603 DMRs hypomethylated in C1 and 4,457 DMRs hypomethylated in C2. Approximately 60% of these DMRs contained a mixed pattern of B cell-related and B cell-independent CpGs (Figure 6A). Subsequently, we generated ChIP-seq profiles of the same MCL cases studied by WGBS and overlapped the detected DMRs with the genomic regions simultaneously containing H3K27ac, which marks active regulatory elements (Heintzman et al., 2009). We observed that hypomethylated DMRs in the C1 MCL case had a substantial overlap (39%) with H3K27ac peaks, which were predominantly present either in the MCL C1 case only or in both MCL cases (Figures 6B and 6C, and Table S4).
Figure 6. Integrative analysis of differentially methylated regions and histone modifications.
(A) Distribution of differentially methylated regions (DMRs) defined by WGBS between the MCL cases representative of C1 (SOX11-positive) and C2 (SOX11-negative) into three different DMR types (B cell-dependent, B cell-independent or mixed DMRs; NA = non-assigned). (B) Number of DMRs between the C1 and C2 MCL cases and their overlap with H3K27ac peaks in these MCL cases. (C) Distribution of the DMRs showing an overlap with H3K27ac peaks in the C1 MCL case only, the C2 MCL case only or in both cases. The background represents all H3K27ac peaks in the C1 and C2 MCL case, and shows which percentage is unique for these cases (yellow and darkbrown) and which percentage overlaps (lightbrown). ***p < 0.001 (Fisher's test). (D-F) Heatmaps showing the read density of H3K27ac, H3K4me1 and H3K4me3 chIP-seq in the C1 MCL case, C2 MCL case, naive B cells (NBC) and memory B cells (MBC) at selected DMRs (±10 Kb). Only the DMRs showing significant differences versus the background in panel C were used for these heatmaps, i.e. unmethylated regions in the C2 case that overlap with H3K27ac peaks in the C2 case only (D), unmethylated regions in the C1 case that overlap with H3K27ac peaks in the C1 case only (E), or with H3K27ac peaks in both the C1 and C2 case (F). In the lower part of these panels, the percentage of these respective DMRs within the B cell-related, mixed and B cell-independent DMRs is represented (***p < 0.001). See also Figure S6, and Tables S4 and S5.
We then analyzed the chromatin architecture of the DMRs within H3K27ac peaks in further detail by taking into account H3K4me1, mostly marking enhancers, and H3K4me3, marking promoters (Figures 6D to 6F). The hypomethylated DMRs in the C2 MCL case that are located within H3K27ac peaks (n=118, 2.6%) in the corresponding MCL case but not in the C1 MCL case, normal naive or memory B cells, showed simultaneous presence of H3K4me1 and H3K4me3 (Figure 6D), suggesting that these regions represent de novo active promoters. The hypomethylated DMRs (n=4,452, 16.7%) in the C1 MCL case within H3K27ac peaks only in the corresponding MCL case and not in MCL C2, normal naive or memory B cells, showed enrichment for H3K4me1 (Figure 6E), pointing towards de novo activation of enhancers at these regions. Furthermore, the DMRs within H3K27ac peaks appeared to be significantly enriched (p < 0.001) in mixed and B cell-independent DMRs (Figures 6D to 6F). Similar results were obtained analyzing the overlap between DMRs and super enhancers (Figure S6 and Table S5). Overall, these results show that DMRs between C1 and C2 MCLs may point towards differential active enhancers and promoters in these samples, especially when they contain CpGs that change only in MCL (i.e. mixed/B cell independent DMRs).
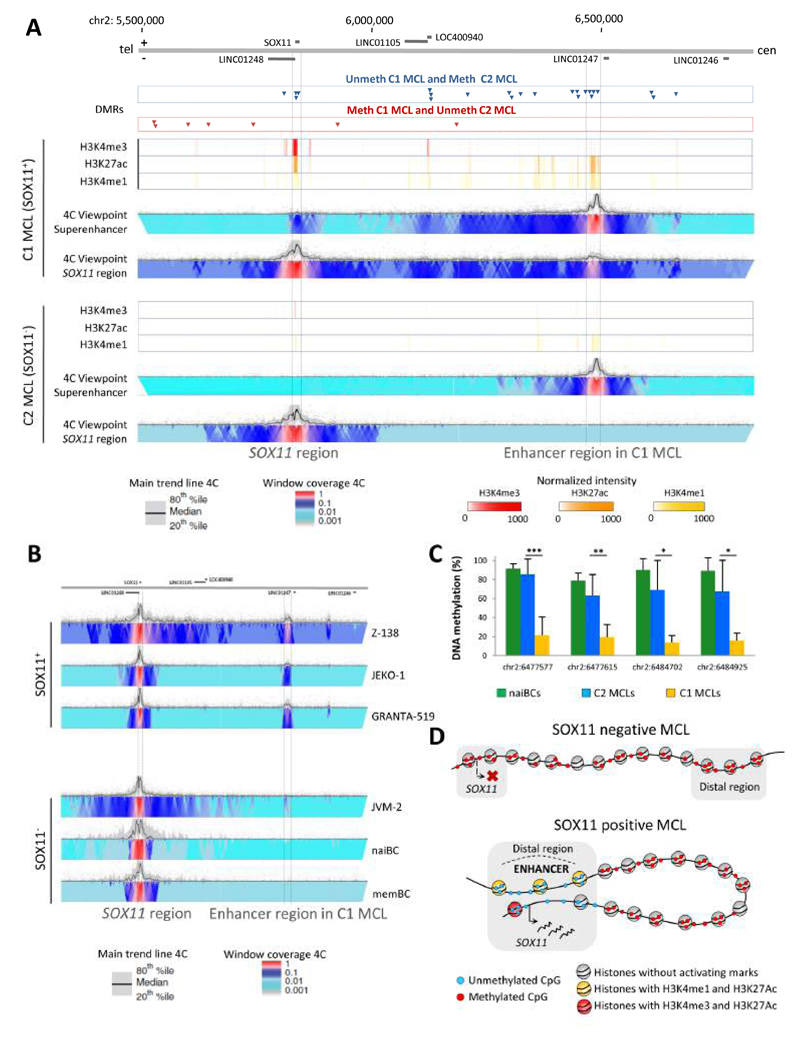
One striking example is a cluster of mixed DMRs hypomethylated in the C1 MCL case overlapping with an enhancer region located 624-653 Kb downstream of SOX11 only in the SOX11-expressing MCL C1 case (Figure 7A). A set of 4C-seq analyses (Simonis et al., 2007; van de Werken et al., 2012) showed that this region presents high contact frequencies with the SOX11 gene in 3-dimensional (3D) space in the representative C1 primary MCL case and 3 SOX11-positive MCL cell lines but not in the C2 MCL case, in the SOX11-negative MCL cell line JVM-2, or in normal naive and memory B cells (Figures 7A and 7B). To investigate whether the association between DNA hypomethylation of this distant enhancer and the expression of SOX11 is a recurrent phenomenon in MCL primary cases, we analyzed the DNA methylation status of this region by bisulfite pyrosequencing in additional primary SOX11-positive (n=12) and SOX11-negative MCL cases (n=10). In this way, we confirmed that the identified regulatory region is de novo demethylated in SOX11-positive (average methylation level 14-21%) as compared to SOX11-negative cases (average methylation level 63-85%, p < 0.01) or naive B cells (average methylation level 79-91%) (Figure 7C and Table S6). However, whether this demethylation is a cause or a consequence of the enhancer activation and SOX11 expression remains to be elucidated. Altogether, these data suggest a model in which aberrant SOX11 expression in MCL is associated with a de novo activation of a distant enhancer element that interacts with the SOX11 locus in 3D space (Figure 7D).
Figure 7. Analysis of the epigenetic and 3D structure of the SOX11 locus.
(A) Differentially methylated regions (DMRs), ChIP-seq levels and 4C-seq signals around the SOX11 locus. The represented region covers chr2:5,492,778-6,834,378 (hg19). Unmethylated DMRs in respectively the C1 (SOX11-positive) and C2 (SOX11-negative) MCL cases are represented in the upper part of the panel by the blue and red arrows. In the lower 2 panels, normalized chIP-seq intensities for H3K4me3, H3K4me1 and H3K27ac are depicted for the C1 and C2 MCL case. Furthermore, normalized 4C-seq intensities are indicated using the enhancer in MCL C1 (chr2:6,465,559-6,496,708, hg19) or the SOX11 region as viewpoint. tel, telomere; cen, centromere. (B) Normalized 4C-seq intensities taking the SOX11 region as viewpoint in 3 SOX11-positive MCL cell lines (Z-138, JEKO-1, GRANTA-519), one SOX11-negative MCL cell line (JVM-2) and in normal naive and memory B cells (naiBCs and memBCs). (C) Mean methylation levels of 4 CpGs within the SOX11-positive MCL enhancer region in naive B cells (green, n=4), SOX11-negative (blue, n=10) and SOX11-positive (orange, n=12) MCLs as analyzed by bisulfite pyrosequencing. Data show mean ± SD. *p < 0.01, **p < 0.001, ***p < 0.0001 (Wilcoxon test for independent samples). (D) Model of the SOX11 locus in SOX11-negative MCL (upper) and SOX11-positive MCL (lower). See also Table S6.
Link among epigenetic burden, genetic changes and clinical outcome of MCL patients
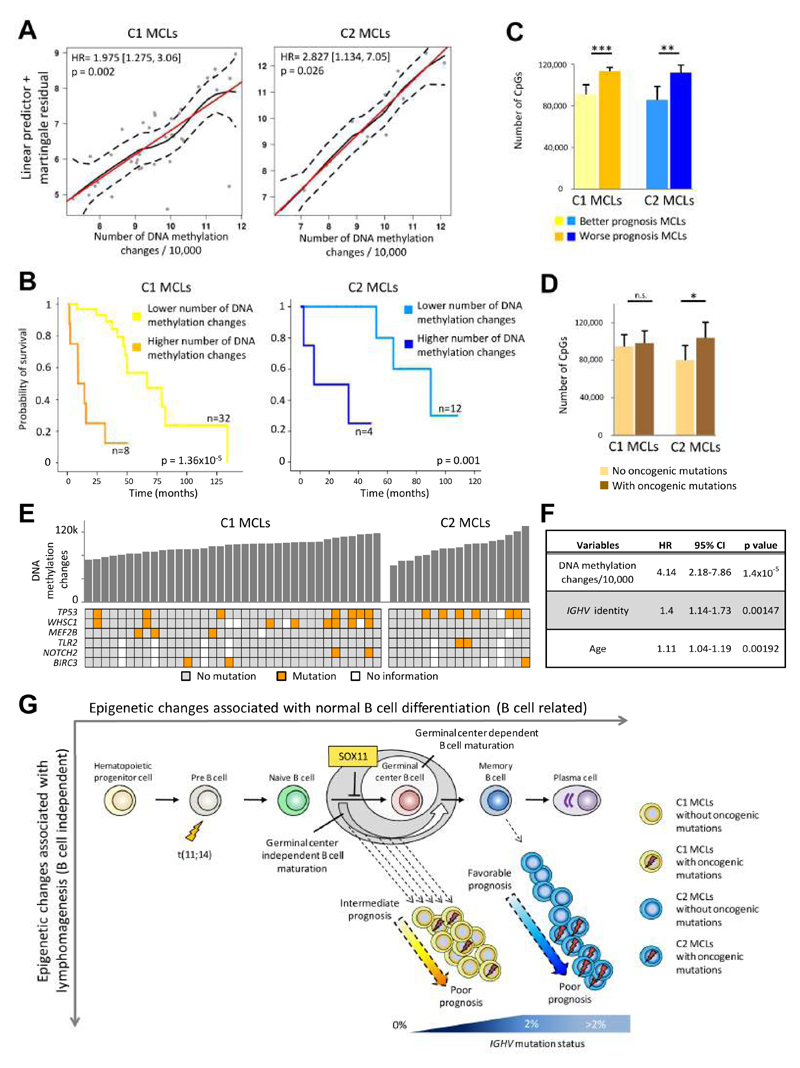
In addition to the significant survival difference between C1 and C2 MCLs (Figure S2A), we postulated that the epigenetic burden (i.e. number of differentially methylated sites regardless of their relationship to normal B cells) may also be associated with clinical behavior. Indeed, in both MCL subgroups we found that the number of DNA methylation changes as compared to HPCs showed a significant linear association with the clinical outcome, approximately doubling the risk of death with each 10,000 methylation changes (Figures 8A and S7A). Beyond this quantitative association, we also calculated the threshold of DNA methylation changes that maximizes the difference in clinical outcome between two subsets of patients (Figures 8B and 8C as well Figures S7B to S7D). Furthermore, we compared DNA methylation changes with the presence of mutations using a set of six recurrent driver genes in MCL (Bea et al., 2013). We observed that cases with gene mutations in C2 MCLs, but not C1 MCLs, displayed a significantly higher number of CpG methylation changes (Figures 8D and 8E). To determine whether these observations can be linked to cell proliferation, we calculated the proliferation signature in 25 of our MCL cases (Navarro et al., 2012). As expected, MCL C1 cases are in general more proliferative than C2 cases (Figure S7E), but the proliferation signature was positively correlated with the number of epigenetic changes only in C2 MCLs (Figure S7F). Finally, we performed a multivariate Cox regression model with 6 variables related to MCL prognosis (supplementary methods) and identified that the number of DNA methylation changes was the strongest independent prognostic factor in our MCL series (p = 1.4x10-5) followed by IGHV identity levels (p = 0.0015) and age (p = 0.0019) (Figure 8F). Altogether, these data suggest that patients with more epigenetic changes have a worse clinical outcome and that in C2 MCLs, this correlates with the acquisition of genetic changes and increased cell proliferation.
Figure 8. Link between the number of DNA methylation changes and prognosis.
(A) Relationship between the number of epigenetic changes and overall survival through a linear predictor. Red line: perfect linear relationship; black line: observed regression line; dash line: 95% confidence interval of observed regression. (B) Kaplan-Meier plots of MCLs with lower vs. higher number of differentially methylated CpGs compared to HPCs in C1 and C2 MCLs. (C) Number of differentially methylated CpGs between the subgroups with different prognosis defined in panel B. Data show mean ± SD. ***p < 0.001 (t-test for independent samples). (D) Association between the number of differentially methylated CpGs and the presence of oncogenic mutations (in BIRC3, MEF2B, NOTCH2, TLR2, TP53 and WHSC1 genes) for both C1 and C2 MCLs. Data show mean ± SD. *p < 0.05; n.s. not significant (t-test for independent samples). For cases without or with mutations, the sample sizes are, respectively: C1 (n=16 and n=15) and C2 (n=6 and n=8). (E) Representation of epigenetic changes and the presence of oncogenic mutations in both C1 and C2 subgroups. (F) Results of the multivariate Cox regression model. (G) Proposed epi(genetic) model of MCL pathogenesis. See also Figure S7.
Discussion
Recent reports using unbiased genome-wide approaches are reshaping our perception of the role of DNA methylation in cancer (Agirre et al., 2015; Berman et al., 2012; Hansen et al., 2011; Jones, 2012; Kulis et al., 2012; Ziller et al., 2013). Here, we have analyzed the DNA methylome of MCL, a heterogeneous B cell neoplasm, and decoded its clinico-biological impact in the context of the DNA methylome of the entire B cell lineage. This analytic strategy has allowed us to obtain insights into not only the pathogenesis and clinical behavior of MCL but also the general role of DNA methylation and its significance in cancer. An important aspect of our study was the initial deconvolution of the methylation estimates and in silico extraction of the methylation levels of tumor cells. Thus, the results obtained were not influenced by the composition of non-tumoral cells within the MCL samples. We believe that this strategy can be highly valuable for other epigenetic studies in which purified tumor cells cannot be obtained.
Our results indicate a major link between the dynamic DNA methylome during B cell maturation and MCL tumorigenesis from various perspectives. From the biological point of view, our findings put together two previous observations in MCL. First, most MCLs are derived from antigen experienced B lymphocytes (Hadzidimitriou et al., 2011; Xochelli et al., 2015), which is reflected by the fact that all MCLs in our study have a DNA methylation profile more similar to antigen-experienced cells. Second, MCLs with unmutated and mutated IGHV may actually reflect a different cellular origin (Navarro et al., 2012). In the case of unmutated MCLs (C1), they retain an imprint of B cells preceding the germinal center, and its cellular origin may range from naive B cells lacking somatic hypermutation to pro-germinal center B cells with modest somatic hypermutation (Kolar et al., 2007). In contrast, mutated MCLs (C2) clearly show an imprint of B cells that have experienced the germinal center reaction. This phenomenon has also been observed in chronic lymphocytic leukemia (CLL), in which three distinct clinico-biological entities can be defined based on DNA methylation patterns of B cell subpopulations at different maturation stages (Bhoi et al., 2016; Kulis et al., 2012; Oakes et al., 2016; Queiros et al., 2015). Overall, we propose an (epi)genetic model of MCL pathogenesis (Figure 8G) in which C1 MCL cases derive from a range of germinal center-inexperienced B cells that carry the t(11;14) translocation and show absence or low levels of IGHV somatic hypermutation (Navarro et al., 2012). Early during transformation, these cells acquire genetic and epigenetic changes and show expression of SOX11, which prevents these cells to enter the germinal center (Palomero et al., 2015). C2 MCLs also carry the t(11;14) translocation but, in contrast to cases from C1, they lack SOX11 expression and show high levels of IGHV somatic hypermutation. This fits with the finding that they seem to be derived from germinal center-experienced B cells, most likely memory B cells (Navarro et al., 2012). C2 MCLs with an indolent clinical course lack oncogenic mutations and acquire few epigenetic changes whereas C2 MCLs with a more aggressive clinical behavior acquire mutations and present extensive DNA methylation changes. The accumulation of DNA methylation changes may suggest the presence of an epigenetic drift derived from enhanced proliferation induced by oncogenic mutations. However, this finding may also point to a co-evolution of genetic and epigenetic aberrations, as previously reported in CLL (Oakes et al., 2014).
Like within the genetics field, a major question in cancer epigenomics is how to detect potential drivers within a widespread alteration of the DNA methylation landscape. Extrapolating from recent cancer genomic studies, in which the number of potential driver mutations is low as compared to the entire mutational burden (Puente et al., 2015; Schuster-Bockler and Lehner, 2012), the proportion of epigenetic drivers may also be low. This is also supported by our data comparing chromatin states between normal B cells and primary cases, in which overall 56% of the regions that undergo DNA methylation changes maintain a stable chromatin environment, and therefore, the function of these regions is most likely not altered. Furthermore, we showed that the majority of CpGs with methylation changes in MCL are affected in only one or few cases. Most likely, this low frequency of recurrent patterns highlights the epigenetic heterogeneity of cancer and reflects that DNA methylation changes globally follow a stochastic model, as previously observed (Landan et al., 2012; Landau et al., 2014; Shipony et al., 2014).
In spite of the above mentioned heterogeneity, an integrative approach combining the DNA methylome and histone modification patterns in primary MCL cases allowed us to identify DMRs with potential functional impact. We propose that epigenetic drivers should be searched in recurrent DMRs containing at least some B cell-independent CpGs and showing a concurrent change in the chromatin activation state. This approach is exemplified by our findings related to the SOX11 oncogene. With the exception of activating histone marks in its promoter region (Vegliante et al., 2011), no other epigenetic or genetic alterations have been described to account for its de novo upregulation in MCL. Here, we have identified a connection between SOX11 expression and a cluster of hypomethylated DMRs located 650 Kb downstream of SOX11, a phenomenon that has been observed previously for other cancer-related genes (Aran and Hellman, 2013; Aran et al., 2013). This region showed the canonical elements of an enhancer element such as the presence of nucleosomes containing H3K4me1 and H3K27ac. Furthermore, this region showed high interaction frequencies with the SOX11 promoter at the 3D level exclusively in SOX11-expressing MCLs, strongly suggesting that it represents an important SOX11 regulatory region in MCL.
From the clinical perspective, our results suggest that the magnitude of DNA methylation changes is the most relevant independent prognostic factor in our MCL series. However, a more clinically-oriented study with a better characterized and homogeneously treated series is required to validate our findings. The extensive epigenetic changes observed in MCL suggest that patients may benefit from the administration of epigenetic drugs (Fiskus et al., 2012). However, the epigenetic heterogeneity observed in MCL may influence efficacy and it should be taken into account as potential means to stratify patients.
In conclusion, the analytic strategy presented in this study highlights the significance of taking into account the dynamics of the DNA methylome during normal differentiation to better understand the cancer epigenome and its clinical implications. Furthermore, our study underlines the importance of performing an integrative whole-genome analysis, as proposed by international consortia (Adams et al., 2012; Bernstein et al., 2010), combining DNA methylation, histone modifications, 3D looping and gene expression to detect distant regulatory elements associated with cancer.
Experimental Procedures
The following experimental procedures represent a succinct summary of the extensive materials and methods applied in the present study. Please be referred to the Supplemental Experimental Procedures for further details.
Samples studied
A total of 82 MCL samples, 4 MCL cell lines (Z-138, JVM-2, JEKO-1 and Granta-519) and 67 samples from B cell subpopulations at different maturation stages (Kulis et al., 2015) were used in the present study. Clinical and biological features of the MCL patients are shown in Table S1. Patients gave their written informed consent, and the study was approved by the clinical research ethics committee of the Hospital Clinic of Barcelona (number 2009/5069) and the internal review board of the University of Kiel (number 447/10).
Deconvolution of DNA methylation values
We estimated the proportion of B cells, CD8+ T cells, CD4+ T cells, NK cells, Monocytes and Granulocytes in the MCL samples (algorithm adapted from (Houseman et al., 2012; Jaffe and Irizarry, 2014)), and purified the methylation values of the B cell (i.e. tumor) fraction by subtracting the methylation estimates of the non-B cell fractions.
Epigenomic analyses
We applied a range of different epigenomic methods, including the HumanMethylation BeadChip (Illumina) in 82 primary MCLs and 67 normal B cell subpopulations (Kulis et al., 2015); WGBS in 2 MCLs; bisulfite pyrosequencing in 22 MCL cases and naive B cells; ChIP-seq with 6 different histone modifications in the 2 MCLs analyzed by WGBS as well as naive and memory B cells, and finally 4C-seq in the 2 MCLs analyzed by WGBS, 4 MCL cell lines as well as naive and memory B cells.
Statistical analysis
The relationships between MCL subgroups and clinical and biological variables of patients was evaluated using the Fisher’s exact test or t-tests, and statistical significance was defined as p < 0.05 (corrected for multiple testing if necessary). Univariate and multivariate survival analyses were used to measure the impact of DNA methylation changes in the clinical behavior of MCL patients. All analyses were performed with the IBM-SPSS Statistics version 20 or various packages within the R software.
Supplementary Material
Significance.
Recent studies on the DNA methylome of cancer cells are reshaping our perception on the pathogenic role of this epigenetic mark, including reports that suggest a major link between the dynamic DNA methylation landscape of normal cell differentiation and neoplastic transformation. Here, we performed a detailed epigenomic analysis of mantle cell lymphoma (MCL), a heterogeneous B cell tumor, in the context of the DNA methylome of the entire normal B cell maturation program. Our results provide insights into the cellular origin, pathogenetic mechanisms and clinical behavior of MCL, and we highlight that integrative analyses of the DNA methylome, histone modifications and three-dimensional interactions in cancer cells can identify potential epigenetic drivers at distant regulatory elements of key oncogenes.
Acknowledgements
This work was funded by the European Union's Seventh Framework Program through the Blueprint Consortium (grant agreement 282510), the European Hematology Association (Non-Clinical Advanced Research Fellowships to J.I.M.-S.), the Worldwide Cancer Research (grant number 16-1285, to J.I.M.-S.), Spanish Ministerio de Economía y Competitividad (MINECO), grant no. SAF2015-64885-R (to E.C.) and Generalitat de Catalunya Suport Grups de Recerca AGAUR 2014-SGR-795 (to E.C.). Methylation microarrays were outsourced to the Spanish Centro Nacional de Genotipado (CEGEN-ISCIII). We are indebted to the Genomics core facility of the Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS) for technical help. This work was partially developed at the Centro Esther Koplowitz (CEK; Barcelona, Spain). J.I.M.-S. is a Ramón y Cajal researcher of MINECO, E.C. is an Academia Researcher of the “Institució Catalana de Recerca i Estudis Avançats” (ICREA) of the Generalitat de Catalunya, A.C.Q. is supported by a Portuguese Fundação para a Ciência e a Tecnologia (FCT) fellowship, R.B. by fellowships from the Netherlands Organisation for Scientific Research (NWO) and the EU (Marie Curie), and R.V.-B by a pre-doctoral fellowship of the MINECO. Paul Flicek is a member of the Scientific Advisory Board for Omicia, Inc.
Footnotes
Accession Numbers
Whole-genome bisulfite sequencing, ChIP-seq and microarray data have been deposited in the European Genome-phenome Archive (EGA) under accession numbers EGAS00001001638, EGAD00001002655 and EGAS00001001637, respectively.
Author Contributions
A.C.Q., R.B., M.D.-F and M.K. analyzed DNA methylation arrays and integrated the data. R.V.-B., R.B., N.V.-D., A.C.Q. and H.J.G.v.d.W performed and/or analyzed 4C-seq experiments. A.C.Q., R.B., A.M., E.R., M.D.-F and S.H.. processed and analyzed WGBS data. N.R., A.B. and J.M. performed ChIP-seq experiments. S.B., A.N., P.J., A.E., M.J.C., I.V., I.S., W.H.W., W.K., C.P., R.S. and E.C. provided study materials and biological information. A.C.Q., R.B., G.Ca and G.Cl functionally characterized differentially methylated regions and performed statistical analysis. E.G., A.L.-G. W.K., C.P., and E.C. reviewed the pathologic and clinical data and confirmed diagnoses. A.D., P.F., and R.R. were in charge of data management. S.H. and I.G.G. coordinated sequencing efforts and performed primary data analysis. M.K., H.G.S., R.S. and E.C. participated in the study design and data interpretation together with A.C.Q, R.B., R.V.-B., and J.I.M.-S. E.C. and J.I.M.-S. conceived the study and led the experiments. A.C.Q, R.B., R.V.-B., and J.I.M.-S wrote the manuscript.
References
- Adams D, Altucci L, Antonarakis SE, Ballesteros J, Beck S, Bird A, Bock C, Boehm B, Campo E, Caricasole A, et al. BLUEPRINT to decode the epigenetic signature written in blood. Nat Biotechnol. 2012;30:224–226. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]
- Agirre X, Castellano G, Pascual M, Heath S, Kulis M, Segura V, Bergmann A, Esteve A, Merkel A, Raineri E, et al. Whole-epigenome analysis in multiple myeloma reveals DNA hypermethylation of B cell-specific enhancers. Genome Res. 2015;25:478–487. doi: 10.1101/gr.180240.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D, Hellman A. DNA methylation of transcriptional enhancers and cancer predisposition. Cell. 2013;154:11–13. doi: 10.1016/j.cell.2013.06.018. [DOI] [PubMed] [Google Scholar]
- Aran D, Sabato S, Hellman A. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol. 2013;14:R21. doi: 10.1186/gb-2013-14-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bea S, Valdes-Mas R, Navarro A, Salaverria I, Martin-Garcia D, Jares P, Gine E, Pinyol M, Royo C, Nadeu F, et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:18250–18255. doi: 10.1073/pnas.1314608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2012;44:40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoi S, Ljungstrom V, Baliakas P, Mattsson M, Smedby KE, Juliusson G, Rosenquist R, Mansouri L. Prognostic impact of epigenetic classification in chronic lymphocytic leukemia: The case of subset #2. Epigenetics. 2016;11:449–455. doi: 10.1080/15592294.2016.1178432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Enjuanes A, Albero R, Clot G, Navarro A, Bea S, Pinyol M, Martin-Subero JI, Klapper W, Staudt LM, Jaffe ES, et al. Genome-wide methylation analyses identify a subset of mantle cell lymphoma with a high number of methylated CpGs and aggressive clinicopathological features. Int J Cancer. 2013;133:2852–2863. doi: 10.1002/ijc.28321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Feinberg AP. Epigenetic stochasticity, nuclear structure and cancer: the implications for medicine. J Intern Med. 2014;276:5–11. doi: 10.1111/joim.12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- Fiskus W, Rao R, Balusu R, Ganguly S, Tao J, Sotomayor E, Mudunuru U, Smith JE, Hembruff SL, Atadja P, et al. Superior efficacy of a combined epigenetic therapy against human mantle cell lymphoma cells. Clin Cancer Res. 2012;18:6227–6238. doi: 10.1158/1078-0432.CCR-12-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Sosa MA, Slagel VA, Trewyn RW, Oxenhandler R, Kuo KC, Gehrke CW, Ehrlich M. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983;11:6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadzidimitriou A, Agathangelidis A, Darzentas N, Murray F, Delfau-Larue MH, Pedersen LB, Lopez AN, Dagklis A, Rombout P, Beldjord K, et al. Is there a role for antigen selection in mantle cell lymphoma? Immunogenetic support from a series of 807 cases. Blood. 2011;118:3088–3095. doi: 10.1182/blood-2011-03-343434. [DOI] [PubMed] [Google Scholar]
- Halldorsdottir AM, Kanduri M, Marincevic M, Mansouri L, Isaksson A, Goransson H, Axelsson T, Agarwal P, Jernberg-Wiklund H, Stamatopoulos K, et al. Mantle cell lymphoma displays a homogenous methylation profile: a comparative analysis with chronic lymphocytic leukemia. Am J Hematol. 2012;87:361–367. doi: 10.1002/ajh.23115. [DOI] [PubMed] [Google Scholar]
- Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee L, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014;15:R31. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jares P, Colomer D, Campo E. Genetic and molecular pathogenesis of mantle cell lymphoma: perspectives for new targeted therapeutics. Nat Rev Cancer. 2007;7:750–762. doi: 10.1038/nrc2230. [DOI] [PubMed] [Google Scholar]
- Jares P, Colomer D, Campo E. Molecular pathogenesis of mantle cell lymphoma. J Clin Invest. 2012;122:3416–3423. doi: 10.1172/JCI61272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, Pikarski E, Young RA, Niveleau A, Cedar H, Simon I. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- Kolar GR, Mehta D, Pelayo R, Capra JD. A novel human B cell subpopulation representing the initial germinal center population to express AID. Blood. 2007;109:2545–2552. doi: 10.1182/blood-2006-07-037150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kridel R, Meissner B, Rogic S, Boyle M, Telenius A, Woolcock B, Gunawardana J, Jenkins C, Cochrane C, Ben-Neriah S, et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood. 2012;119:1963–1971. doi: 10.1182/blood-2011-11-391474. [DOI] [PubMed] [Google Scholar]
- Kulis M, Heath S, Bibikova M, Queiros AC, Navarro A, Clot G, Martinez-Trillos A, Castellano G, Brun-Heath I, Pinyol M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44:1236–1242. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- Kulis M, Merkel A, Heath S, Queiros AC, Schuyler RP, Castellano G, Beekman R, Raineri E, Esteve A, Clot G, et al. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat Genet. 2015;47:746–756. doi: 10.1038/ng.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulis M, Queiros AC, Beekman R, Martin-Subero JI. Intragenic DNA methylation in transcriptional regulation, normal differentiation and cancer. Biochim Biophys Acta. 2013;1829:1161–1174. doi: 10.1016/j.bbagrm.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Landan G, Cohen NM, Mukamel Z, Bar A, Molchadsky A, Brosh R, Horn-Saban S, Zalcenstein DA, Goldfinger N, Zundelevich A, et al. Epigenetic polymorphism and the stochastic formation of differentially methylated regions in normal and cancerous tissues. Nat Genet. 2012;44:1207–1214. doi: 10.1038/ng.2442. [DOI] [PubMed] [Google Scholar]
- Landau DA, Clement K, Ziller MJ, Boyle P, Fan J, Gu H, Stevenson K, Sougnez C, Wang L, Li S, et al. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell. 2014;26:813–825. doi: 10.1016/j.ccell.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshchenko VV, Kuo PY, Shaknovich R, Yang DT, Gellen T, Petrich A, Yu Y, Remache Y, Weniger MA, Rafiq S, et al. Genomewide DNA methylation analysis reveals novel targets for drug development in mantle cell lymphoma. Blood. 2010;116:1025–1034. doi: 10.1182/blood-2009-12-257485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Clot G, Royo C, Jares P, Hadzidimitriou A, Agathangelidis A, Bikos V, Darzentas N, Papadaki T, Salaverria I, et al. Molecular subsets of mantle cell lymphoma defined by the IGHV mutational status and SOX11 expression have distinct biologic and clinical features. Cancer Res. 2012;72:5307–5316. doi: 10.1158/0008-5472.CAN-12-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes CC, Claus R, Gu L, Assenov Y, Hullein J, Zucknick M, Bieg M, Brocks D, Bogatyrova O, Schmidt CR, et al. Evolution of DNA methylation is linked to genetic aberrations in chronic lymphocytic leukemia. Cancer Discov. 2014;4:348–361. doi: 10.1158/2159-8290.CD-13-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes CC, Seifert M, Assenov Y, Gu L, Przekopowitz M, Ruppert AS, Wang Q, Imbusch CD, Serva A, Koser SD, et al. DNA methylation dynamics during B cell maturation underlie a continuum of disease phenotypes in chronic lymphocytic leukemia. Nat Genet. 2016;48:253–264. doi: 10.1038/ng.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero J, Vegliante MC, Eguileor A, Rodriguez ML, Balsas P, Martinez D, Campo E, Amador V. SOX11 defines two different subtypes of mantle cell lymphoma through transcriptional regulation of BCL6. Leukemia. 2015 doi: 10.1038/leu.2015.355. [DOI] [PubMed] [Google Scholar]
- Puente XS, Bea S, Valdes-Mas R, Villamor N, Gutierrez-Abril J, Martin-Subero JI, Munar M, Rubio-Perez C, Jares P, Aymerich M, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526:519–524. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- Queiros AC, Villamor N, Clot G, Martinez-Trillos A, Kulis M, Navarro A, Penas EM, Jayne S, Majid A, Richter J, et al. A B-cell epigenetic signature defines three biologic subgroups of chronic lymphocytic leukemia with clinical impact. Leukemia. 2015;29:598–605. doi: 10.1038/leu.2014.252. [DOI] [PubMed] [Google Scholar]
- Rahmatpanah FB, Carstens S, Guo J, Sjahputera O, Taylor KH, Duff D, Shi H, Davis JW, Hooshmand SI, Chitma-Matsiga R, Caldwell CW. Differential DNA methylation patterns of small B-cell lymphoma subclasses with different clinical behavior. Leukemia. 2006;20:1855–1862. doi: 10.1038/sj.leu.2404345. [DOI] [PubMed] [Google Scholar]
- Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlen SE, Greco D, Soderhall C, Scheynius A, Kere J. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One. 2012;7:e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo C, Navarro A, Clot G, Salaverria I, Gine E, Jares P, Colomer D, Wiestner A, Wilson WH, Vegliante MC, et al. Non-nodal type of mantle cell lymphoma is a specific biological and clinical subgroup of the disease. Leukemia. 2012;26:1895–1898. doi: 10.1038/leu.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba NS, Liu D, Herman SE, Underbayev C, Tian X, Behrend D, Weniger MA, Skarzynski M, Gyamfi J, Fontan L, et al. Pathogenic role of B-cell receptor signaling and canonical NF-kappaB activation in mantle cell lymphoma. Blood. 2016 doi: 10.1182/blood-2015-11-681460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- Shipony Z, Mukamel Z, Cohen NM, Landan G, Chomsky E, Zeliger SR, Fried YC, Ainbinder E, Friedman N, Tanay A. Dynamic and static maintenance of epigenetic memory in pluripotent and somatic cells. Nature. 2014;513:115–119. doi: 10.1038/nature13458. [DOI] [PubMed] [Google Scholar]
- Simonis M, Kooren J, de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nat Methods. 2007;4:895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- Sims GP, Ettinger R, Shirota Y, Yarboro CH, Illei GG, Lipsky PE. Identification and characterization of circulating human transitional B cells. Blood. 2005;105:4390–4398. doi: 10.1182/blood-2004-11-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Werken HJ, Landan G, Holwerda SJ, Hoichman M, Klous P, Chachik R, Splinter E, Valdes-Quezada C, Oz Y, Bouwman BA, et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat Methods. 2012;9:969–972. doi: 10.1038/nmeth.2173. [DOI] [PubMed] [Google Scholar]
- Vegliante MC, Royo C, Palomero J, Salaverria I, Balint B, Martin-Guerrero I, Agirre X, Lujambio A, Richter J, Xargay-Torrent S, et al. Epigenetic activation of SOX11 in lymphoid neoplasms by histone modifications. PLoS One. 2011;6:e21382. doi: 10.1371/journal.pone.0021382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xochelli A, Sutton LA, Agathangelidis A, Stalika E, Karypidou M, Marantidou F, Lopez AN, Papadopoulos G, Supikova J, Groenen P, et al. Molecular evidence for antigen drive in the natural history of mantle cell lymphoma. Am J Pathol. 2015;185:1740–1748. doi: 10.1016/j.ajpath.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.