Abstract
It is critical for bacteria to recognize surface contact and to initiate physiological changes required for surface-associated lifestyles. Ubiquitous microbial appendages called pili are involved in sensing surfaces and mediating downstream behaviors, but the mechanism by which pili mediate surface sensing remains unclear. Here we visualized Caulobacter crescentus pili undergoing dynamic cycles of extension and retraction. These cycles ceased within seconds of surface contact, which coincided with synthesis of the adhesive holdfast required for attachment. Physically blocking pili imposed resistance to pilus retraction, which was sufficient to stimulate holdfast synthesis without surface contact. Thus, resistance to pilus retraction upon surface contact is used for surface sensing.
Main Text
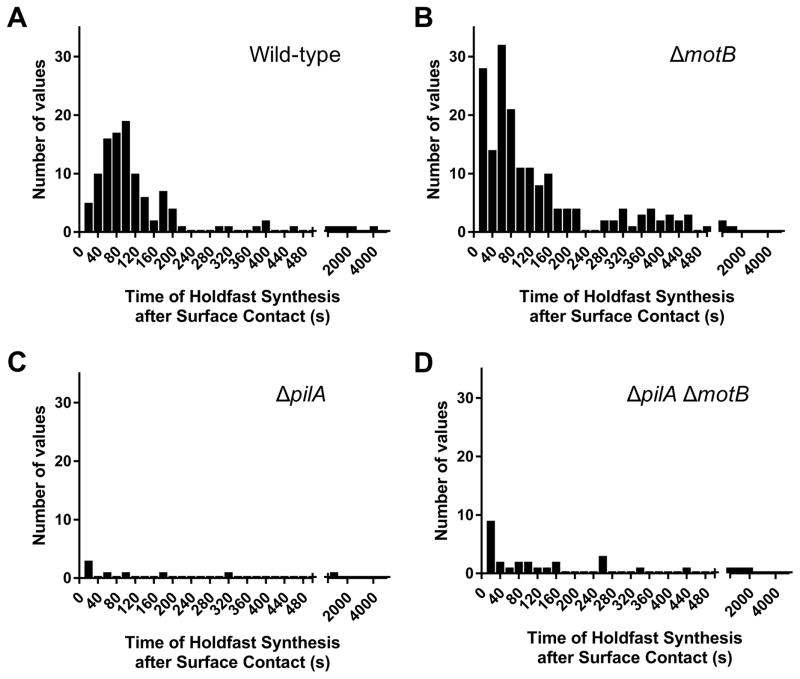
The role of pili in surface sensing by bacteria has been difficult to assess, in part due to the difficulty visualizing their dynamic activity and the time required for surface contact-induced responses. We used the rapid surface contact-stimulated synthesis of the adhesive holdfast of C. crescentus to study surface sensing. Newborn, non-reproductive swarmer cells harbor multiple tad pili and a flagellum at the same pole (Fig. S1 and S2). When swarmer cells encounter a surface, holdfast synthesis at the flagellar pole is stimulated within seconds in a tad pili-dependent process and is concurrent with an arrest of flagellum rotation (1, 2). To determine whether the flagellar motor is required for surface stimulation of holdfast synthesis, we tracked the time of holdfast synthesis by single cells after surface contact (Fig. S3A). A mutant lacking the MotB flagellar stator had wild-type levels of surface stimulation of holdfast synthesis, while a mutant lacking the pilus filament PilA subunit did not respond to surface contact (Fig. 1).
Figure 1. Tad pili are required for surface stimulation of holdfast synthesis.
(A–D) Histogram plots showing the time of holdfast synthesis for single cells after surface contact for two independent replicates of wild-type (A) n = 115, ΔmotB (B) n = 182, ΔpilA (C) n = 8, and ΔpilA ΔmotB (D) n = 33. Total number of cells tracked including cells arriving with holdfast already synthesized: wild-type n = 241, ΔmotB n = 566, ΔpilA n = 93, and ΔpilAΔmotB n = 84.
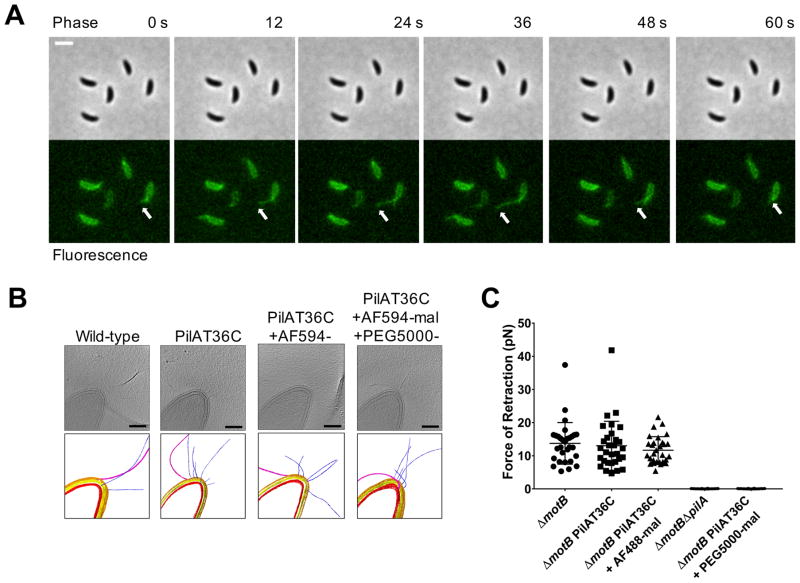
For systems where pili are required for surface-stimulated phenotypes, it has been proposed that the tension imparted on retracting pili upon their attachment to a surface is used for surface sensing (1, 3–5). Because tad pili were not known to retract, and they lack a homologue of known retraction ATPases, we sought to label them fluorescently to study their behavior. Inspired by a technique for labeling flagellar filaments (6), we replaced a native residue within the major pilin subunit with a cysteine for subsequent labeling with thiol-reactive maleimide dyes (AF488- or AF594-mal) (Fig. S3B and Fig. S4, C and E). The C. crescentus PilAT36C strain had wild-type levels of attachment as well as sensitivity to the pilus-dependent bacteriophage ɸCbK (Fig. S5, A and B). Cryo-electron tomography (cryo-ET) showed that neither the introduction of the cysteine residue nor the addition of maleimide dye affected the structure of the pilus fiber or the cell envelope (Fig. 2B and Movie S1). Fluorescently labeled tad pili were highly dynamic and capable of both extension and retraction (Fig. 2A and Movie S2).
Figure 2. Tad type IVc pili undergo dynamic cycles of extension and retraction.
(A) Time-lapse of labeled, synchronized PilAT36C swarmer cells extending and retracting pili after labeling with AF488-mal dye. White arrow follows the most prominent extension and retraction event for a single cell, though all cells shown extend and retract pili. Scale bar is 2 μm. (B) Slices from tomograms and corresponding 3D segmentations of wild-type, PilAT36C, PilAT36 labeled with AF594-mal, and PilAT36C blocked with PEG5000-mal and labeled with AF594-mal. In 3D segmentation volumes, flagella are pink, pili are blue, S-layer is gold, outer membrane is yellow, and inner membrane is red. Scale bars are 200 nm. (C) Micropillars assay force measurements of retraction of tad type IVc pili in flagellar motor mutant ΔmotB strains. Flagellar motor mutants exhibiting paralyzed flagella were used to ensure all measurements obtained were dependent solely on pilus activity. Error bars show mean ± SD from 30 cells for each dataset.
Cells synthesized an average of two pili per minute at an average length of 1.08 μm (Fig. S6, A and B). To measure the strength of pilus retraction, we used an elastic micropillars assay (7). In this assay, pili from the same cell bind adjacent micropillars, and their subsequent retraction causes micropillar bending, which allows calculation of a retraction rate and force (Fig. S7A, Movie S3, Movie S4, Movie S5, Movie S6, and Movie S7). C. crescentus pilus retraction generated an average force of 14 pN and labeling did not affect retraction force (Fig. 2C). Extension and retraction rates of labeled pili under agar pads averaged 0.14 and 0.16 μm/sec respectively, which correlate with retraction rates measured by the micropillars assay (Fig. S7, B and C). Thus, tad pili retract, and labeled tad pili extend and retract normally. Tad pili, which we refer to as type IVc (Fig. S1, A and B), are more similar to the type IV secretion system and the archaellum than to other type IV pili. Additionally, we labeled the Vibrio cholerae type IVa MSHA (mannose-sensitive hemagglutanin) pili and the V. cholerae type IVb TCP (toxin co-regulated pilus) pili (Fig. S4, A, B, D and Fig. S8, A and B), demonstrating that this pilus labeling method is broadly applicable to diverse pilus systems (8).
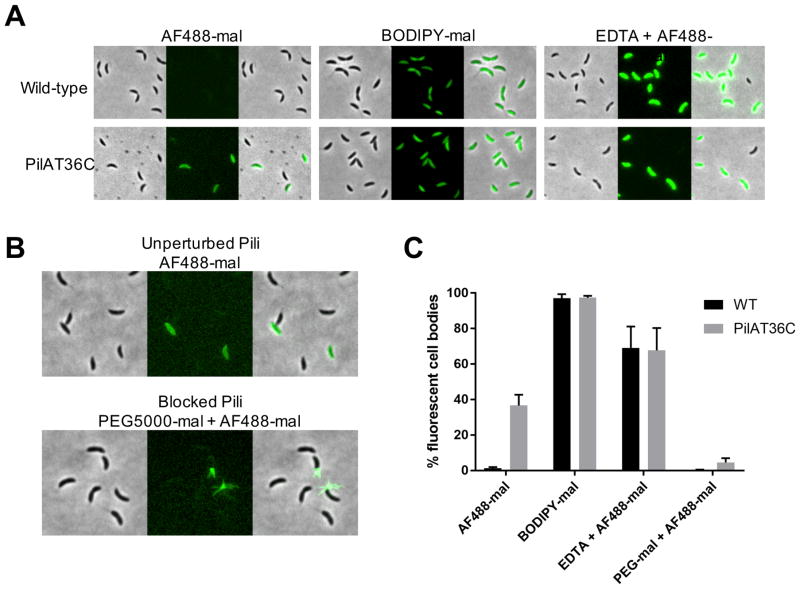
Mutants lacking pilus retraction machinery exhibit changes in pilin distribution (9). However, direct demonstration of pilin subunit recycling has proven difficult due to limitations in techniques for studying their real-time dynamics. Upon labeling with AF488-mal, we observed that piliated swarmer cells exhibited fluorescent cell bodies, suggesting that externally labeled pilins might be internalized and recycled during pilus retraction. Using the principle of size-based exclusion by the outer membrane (OM), we labeled both wild-type and the PilAT36C strain using either the large, OM-excluded, AF488-mal (720.66 Da) dye or a small, OM-permeable, BODIPY-mal (414.22 Da) dye to quantify fluorescent cell bodies in each population (Fig. S3C and Fig. 3, A and B). The OM-permeable BODIPY-mal labeled all cell types, including stalked and predivisional cells lacking pili, from either wild-type or PilAT36C strains. In contrast, the OM-excluded AF488-mal did not label wild-type cells or cells lacking pili within the PilAT36C population unless their outer membrane was compromised (Fig. 3, A and C). Thus, we hypothesized that cell body staining with AF488-mal was due to pilus retraction into the periplasm. To test this hypothesis, we investigated the impact of adding a large polyethylene glycol maleimide conjugate of ~5,000 Da (PEG5000-mal). Upon co-labeling pili with PEG5000-mal and AF488-mal, labeled pili no longer exhibited dynamic activity and could not cause micropillar bending (Fig. 3B and 2C). We also observed a large reduction in the number of swarmer cells with fluorescent cell bodies and a concomitant increase in the number of cells with multiple fluorescent pili (Fig. 3, B and C). Cryo-ET of cells labeled with PEG5000-mal showed no effect on pilus structure or the cell envelope (Fig. 2B). Thus, AF488-mal is excluded by the OM, externally labeled pilins are retracted into an intracellular pool of subunits, and pilus retraction can be physically obstructed. Because newly extended pili are still labeled after removal of excess dye, the results also indicate that pilins are recycled and reused.
Figure 3. C. crescentus tad pilus retraction internalizes labeled pilins into a recyclable pool of subunits.
(A) Representative images of wild-type or PilAT36C cells labeled with AF488-mal, BODIPY-mal, or AF488-mal after OM permeabilization with 20 mM EDTA. (B) Representative images of PilAT36C cells labeled with AF488-mal ± PEG5000-mal. (C) Quantification of fluorescent cell bodies in populations of cells from images shown in (A and B). A minimum of 398 cells from each of 3 independent biological replicates was quantified. Error bars show mean ± SD.
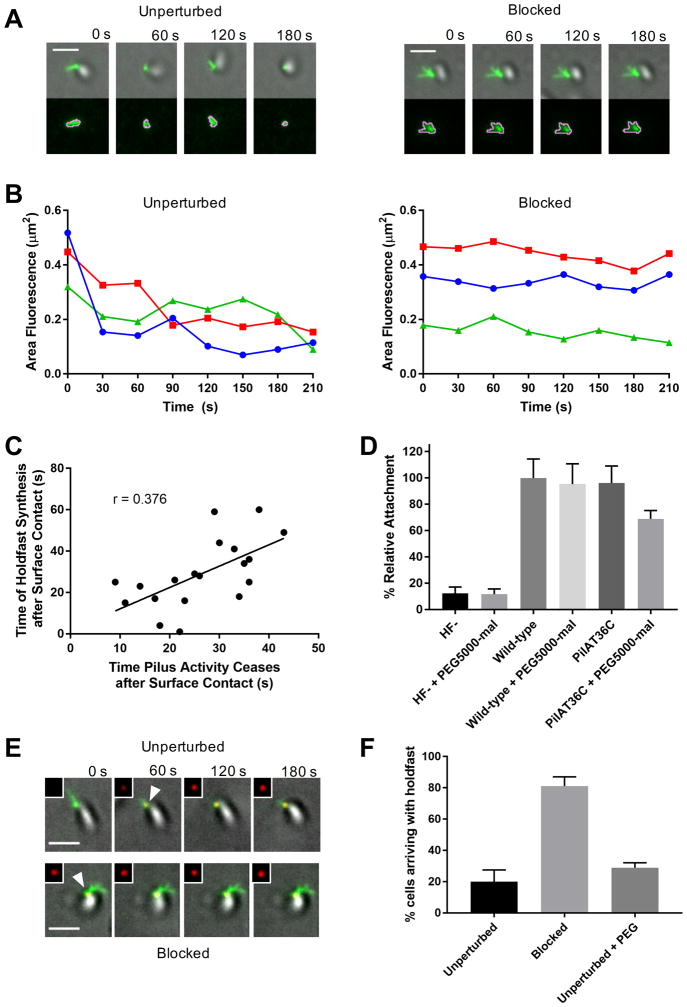
The inability to observe pilus behavior directly upon cell contact with a surface has complicated analysis of the role of pilus dynamics in surface sensing. This has required the use of pilus synthesis and/or retraction mutants to infer their function by observing the behavior of a non-functional or genetically modified system. To bypass these problems and determine the role of pilus retraction in surface sensing, we combined our abilities to observe and physically perturb fully functional pili. We first examined the dynamics of pilus extension and retraction upon surface contact. Cells were added to microfluidic well devices and those that were detected at the glass-liquid interface by differential interference contrast (DIC) microscopy were analyzed for pilus activity. We defined cessation of pilus activity as the first frame after which fluorescence area neither increased nor declined. Cells with unperturbed pili exhibited dynamic pilus activity, whereas cells blocked for pilus retraction exhibited no dynamic activity, as indicated by tracking fluorescence area of pili associated with single cells over time (Fig. 4, A and B and Movie S8). In unperturbed cells, upon surface contact, the cessation of dynamic pilus extension and retraction was positively correlated with the stimulation of holdfast synthesis (Fig. 4C). While holdfast synthesis did not always occur immediately upon cessation of pilus activity, the correlation between these two events implicate perturbation of pilus dynamics, and not necessarily cessation of activity, in the surface-sensing process.
Figure 4. Resistance to C. crescentus tad pilus retraction triggers surface stimulation of holdfast synthesis.
(A) Representative TIRF images of unperturbed cell labeled with AF488-mal exhibiting dynamic pilus activity and blocked cell labeled with both AF488-mal and PEG5000-mal exhibiting no dynamic pilus activity with overlays of gray cell body and green fluorescent pili. The bottom image panel shows green fluorescent pili surrounded by pink MicrobeJ overlay used to measure changes in fluorescence area of pili (μm2) over time shown in graphs (B). (B) Graphs showing changes in fluorescence area occupied by pili over time by cells shown in (A). (C) Plot showing correlation between the time of holdfast synthesis after surface contact and the time of cessation of dynamic pilus activity after surface contact for 19 cells. (D) Relative attachment assay showing binding efficiency of holdfast minus (HF-) and PilAT36C strains compared to wild-type after 30 min binding ± PEG5000-mal. Data are representative of binding from 3 independent cultures normalized to wild-type binding levels. Error bars show mean ± SD. PilAT36C + PEG-mal is significantly different from all other treatments based on unpaired, two-tailed T-test (P < 0.03). (E) Representative TIRF microscopy images of cells unperturbed or blocked for pilus retraction upon surface contact in the presence of AF594-WGA where time = 0 s is time of surface contact. The cell body is gray, labeled pili are green, and HF are red, and shown in upper right inset of each image. White arrowheads represent first appearance of holdfast for cell depicted. Scale bars are 2 μm. (F) Quantification of cells after labeling with AF488-mal (unperturbed); AF488-mal + PEG5000-mal (blocked); or AF488-mal + PEG5000 (unperturbed + PEG). A minimum of 30 cells from each of 3 independent biological replicates was quantified. Error bars show mean ± SD. Scale bars are 2 μm.
The above observations are consistent with the hypothesis that resistance on retracting surface-bound pili provides a surface-sensing signal to trigger holdfast synthesis. To test this hypothesis, we blocked pilus retraction by treatment with PEG5000-mal and measured surface attachment (Fig. 4D). Cultures in which pilus retraction was blocked experienced a 27% reduction in adhesion compared to unperturbed cells, suggesting that dynamic pilus activity is important for mediating attachment. Next, we used TIRF (total internal reflection fluorescence) microscopy to track holdfast synthesis upon surface contact in piliated swarmer cells with either unperturbed or blocked pili (Fig. 4E and Fig. S9, A and B and Movie S9, Movie S10). While only 20% of unperturbed cells arrived on the surface with a holdfast synthesized prior to surface contact, 81% of cells blocked for pilus retraction had synthesized a holdfast prior to reaching the surface, indicating that blocking pilus retraction in planktonic cells was sufficient to stimulate holdfast synthesis in the absence of surface contact (Fig. 4F). The addition of PEG5000-mal had no effect on cell swimming, indicating that blocking pili retraction does not impede flagellum rotation (Fig. S10A). While a ΔmotB mutant was deficient in surface colonization because it was unable to reach the surface efficiently (Fig. S10B), it was stimulated for holdfast synthesis prior to surface contact after pilus retraction was blocked (Fig. S10, C and D). Thus, blocking pili retraction of ΔmotB phenocopied blocking pili retraction in wild-type, demonstrating that rotating flagella were not required for this process. We note that the ΔmotB mutation increased the number of piliated cells with a holdfast in the medium (Fig. S10D), suggesting a regulatory interaction between the flagellum and holdfast synthesis that may reflect a role for flagellum rotation in a parallel surface sensing pathway. These data support a model in which resistance on retracting, surface-bound pili generates a surface-sensing signal, expanding previous work in which mutants deficient in pilus retraction and extension could not sense surface contact (4, 5).
How is the surface sensing signal transduced? Holdfast synthesis was stimulated normally in the absence of RNA or protein synthesis (Fig. S11), suggesting that it could be mediated by a second messenger molecule such as cyclic-di-GMP (cdG). Chemical signaling would be compatible with the rapidity of the response. Deletion of the diguanylate cyclase PleD reduces cdG concentration by approximately a third (10), but a ΔpleD mutant still synthesizes pili and holdfasts (10–13). However, the ΔpleD mutant was deficient in synthesizing holdfast in response to surface contact (Fig. S12A). The pleD mutant was stimulated for holdfast synthesis upon blocking pilus retraction, albeit not to the same extent as wild-type cells (Fig. S12B). These data implicate cdG levels in the surface stimulation of holdfast synthesis.
In conclusion, we have developed a broadly applicable pilus labeling method enabling real-time observation of pilus dynamics and its targeted physical obstruction. Tad pili can retract despite the absence of an orthologous retraction ATPase; pilins are internalized upon retraction into a pilin pool that is recycled and reused; resistance to pilus retraction upon surface binding functions in surfacing sensing; and cdG signaling may be involved in surface sensing. Although a rotating flagellum was not required for surface stimulation of holdfast synthesis in the absence of flow, obstruction of flagellum rotation may be involved in surface sensing through an unknown mechanism (14, 15). Whether these two mechanosensors use the same signaling pathways and whether they are used to sense surfaces in different conditions remains unclear.
Supplementary Material
Acknowledgments
We thank members of the Brun laboratory, C. Fuqua, and D. Kearns for critical comments on the manuscript. We thank members of the Dalia laboratory C. Hayes and T. Dalia for strain construction. This work was supported by grants R01GM102841, R01GM51986, and R35GM122556 from the National Institutes of Health to YVB, by grant AI118863 from the National Institutes of Health to ABD, by National Science Foundation fellowship 1342962 to CKE, and by grant AI116566 from the National Institutes of Health to NB. This work was supported in part by grants R01GM104540 and R01GM104540-03S1 from the National Institutes of Health, NSF grant 0923395, and grants from Emory University, Children’s Healthcare of Atlanta, the Georgia Research Alliance, the Center for AIDS Research at Emory University (P30 AI050409), James B. Pendleton Charitable Trust to E.R.W. We thank the Robert P. Apkarian Integrated Electron Microscopy Core of Emory University for microscopy services and support. An early version of this work can be found on bioRxiv at https://doi.org/10.1101/157727. Data are available in the manuscript and supplementary materials. The authors declare no competing interests.
Footnotes
References and Notes
- 1.Hoffman MD, et al. Timescales and Frequencies of Reversible and Irreversible Adhesion Events of Single Bacterial Cells. Anal Chem. 2015 doi: 10.1021/acs.analchem.5b02087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li G, et al. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol Microbiol. 2012;83:41–51. doi: 10.1111/j.1365-2958.2011.07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Utada AS, et al. Vibrio cholerae use pili and flagella synergistically to effect motility switching and conditional surface attachment. Nat Commun. 2014;5:4913. doi: 10.1038/ncomms5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siryaporn A, Kuchma SL, O’Toole GA, Gitai Z. Surface attachment induces Pseudomonas aeruginosa virulence. Proc Natl Acad Sci U S A. 2014;111:16860–5. doi: 10.1073/pnas.1415712111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Persat A, Inclan YF, Engel JN, Stone HA, Gitai Z. Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc Natl Acad Sci. 2015;112:7563–7568. doi: 10.1073/pnas.1502025112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science. 2008;320:1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- 7.Biais N, Higashi D, So M, Ladoux B. Techniques to measure pilus retraction forces. Methods Mol Biol. 2012;799:197–216. doi: 10.1007/978-1-61779-346-2_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burrows LL. Twitching Motility: Type IV Pili in Action. Annu Rev Microbiol. 2012;66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- 9.Morand PC, et al. Type IV pilus retraction in pathogenic Neisseria is regulated by the PilC proteins. EMBO J. 2004;23:2009–17. doi: 10.1038/sj.emboj.7600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abel S, et al. Bi-modal Distribution of the Second Messenger c-di-GMP Controls Cell Fate and Asymmetry during the Caulobacter Cell Cycle. PLoS Genet. 2013;9:e1003744. doi: 10.1371/journal.pgen.1003744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aldridge P, Jenal U. Cell cycle-dependent degradation of a flagellar motor component requires a novel-type response regulator. Mol Microbiol. 1999;32:379–391. doi: 10.1046/j.1365-2958.1999.01358.x. [DOI] [PubMed] [Google Scholar]
- 12.Hecht GB, Newton A. Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J Bacteriol. 1995;177:6223–9. doi: 10.1128/jb.177.21.6223-6229.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levi A, Jenal U. Holdfast formation in motile swarmer cells optimizes surface attachment during Caulobacter crescentus development. J Bacteriol. 2006;188:5315–8. doi: 10.1128/JB.01725-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarter L, Hilmen M, Silverman M. Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell. 1988;54:345–51. doi: 10.1016/0092-8674(88)90197-3. [DOI] [PubMed] [Google Scholar]
- 15.Belas R. Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol. 2014 doi: 10.1016/j.tim.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Poindexter JS. Biological properties and classification of the Caulobacter group. Bacteriol Rev. 1964;28:231–95. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonin M, Quardokus EM, O’Donnol D, Maddock J, Brun YV. Regulation of stalk elongation by phosphate in Caulobacter crescentus. J Bacteriol. 2000;182:337–47. doi: 10.1128/jb.182.2.337-347.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ely B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- 19.Petersen B, et al. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct Biol. 2009;9:51. doi: 10.1186/1472-6807-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 21.Dalia AB, McDonough E, Camilli A. Multiplex genome editing by natural transformation. Proc Natl Acad Sci U S A. 2014;111:8937–42. doi: 10.1073/pnas.1406478111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–82. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 25.Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol. 1994;39:306–14. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- 26.Jiang C, Brown PJB, Ducret A, Brun YV. Sequential evolution of bacterial morphology by co-option of a developmental regulator. Nature. 2014;506:489–493. doi: 10.1038/nature12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ducret A, et al. Microbe J, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol. 2016;1:16077. doi: 10.1038/nmicrobiol.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Kremer JR, Mastronarde DN, McIntosh JR. Computer Visualization of Three-Dimensional Image Data Using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 30.Mastronarde DN. Dual-Axis Tomography: An Approach with Alignment Methods That Preserve Resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- 31.Bodenmiller D, Toh E, Brun YV. Development of Surface Adhesion in Caulobacter crescentus. J Bacteriol. 2004;186:1438–1447. doi: 10.1128/JB.186.5.1438-1447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skerker JM, Shapiro L. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 2000;19:3223–3234. doi: 10.1093/emboj/19.13.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marks ME, et al. The genetic basis of laboratory adaptation in Caulobacter crescentus. J Bacteriol. 2010;192:3678–88. doi: 10.1128/JB.00255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dalia AB, Lazinski DW, Camilli A. Identification of a membrane-bound transcriptional regulator that links chitin and natural competence in Vibrio cholerae. MBio. 2014;5:e01028–13. doi: 10.1128/mBio.01028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.West L, Yang D, Stephens C. Use of the Caulobacter crescentus genome sequence to develop a method for systematic genetic mapping. J Bacteriol. 2002;184:2155–66. doi: 10.1128/JB.184.8.2155-2166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.