Abstract
Neurons in the olfactory epithelium (OE) each express a single dominant olfactory receptor (OR) allele from among roughly 1,000 different OR genes. While monogenic and monoallelic OR expression has been appreciated for over two decades, regulators of this process are still being described; most recently, epigenetic modifiers have been of high interest as silent OR genes are decorated with transcriptionally repressive trimethylated histone 3 lysine 9 (H3K9me3) whereas active OR genes are decorated with transcriptionally activating trimethylated histone 3 lysine 4 (H3K4me3). The lysine specific demethylase 1 (LSD1) demethylates at both of these lysine residues and has been shown to disrupt neuronal maturation and OR expression in the developing embryonic OE. Despite the growing literature on LSD1 expression in the OE, a complete characterization of the timing of LSD1 expression relative to neuronal maturation and of the function of LSD1 in the adult OE have yet to be reported. To fill this gap, the present study determined that LSD1 (1) is expressed in early dividing cells before OR expression and neuronal maturation and decreases at the time of OR stabilization; (2) colocalizes with the repressor CoREST (also known as RCOR1) and histone deacetylase 2 in these early dividing cells; and (3) is required for neuronal maturation during a distinct time window between activating reserve stem cells (horizontal basal cells) and Neurogenin1 (+) immediate neuronal precursors. Thus, this study clarifies the role of LSD1 in olfactory neuronal maturation.
Keywords: LSD1, CoREST, Epigenetics, Olfactory Receptors, Neuronal Maturation, RRID: AB_10979409, RRID: AB_307210, RRID: AB_2134831, RRID: AB_2301051, RRID: AB_1310252, RRID: AB_300798, RRID: AB_2118547, RRID: AB_11145494, RRID: AB_2158008, RRID: AB_2636804, RRID: AB_2209751, RRID: AB_10063408
1 INTRODUCTION
Olfactory receptors (ORs) make up the largest gene family in mammals, subserving the detection of and discrimination among thousands of different odorants and trillions of odorant mixtures (Buck & Axel, 1991; Lander et al., 2001; Zhang & Firestein, 2002; Bushdid, Magnasco, Vosshall, & Keller, 2014). Following the initial discovery of the ~1,000 gene OR family (Buck & Axel, 1991), it was proposed that each olfactory sensory neuron (OSN) expresses only one OR in both a monoallelic and monogenic fashion. Evidence for monogenic OR expression has largely relied on double OR in situ hybridizations and amplification of ORs via single cell RT-PCR (Buck & Axel, 1991; Ressler, Sullivan, & Buck, 1993; Vassar, Ngai, & Axel, 1993). Most recently, single-cell RNA-seq experiments revealed that multiple ORs can exist in mature OSNs, although a single dominant OR allele prevails over lowly transcribed accessory ORs (Hanchate et al., 2015; Tan, Li, & Xie, 2015; Scholz et al., 2016). The question remains as to how a single OR allele is selected for dominant expression out of over 1,000 OR genes.
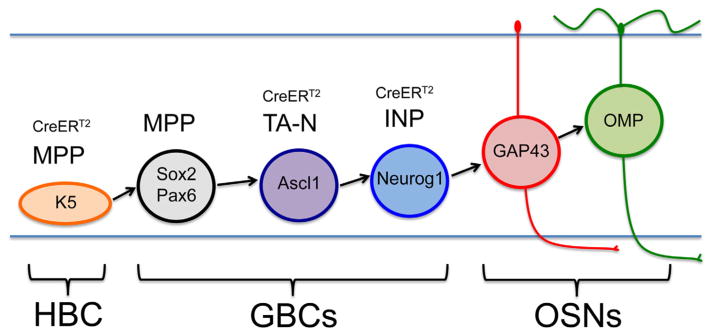
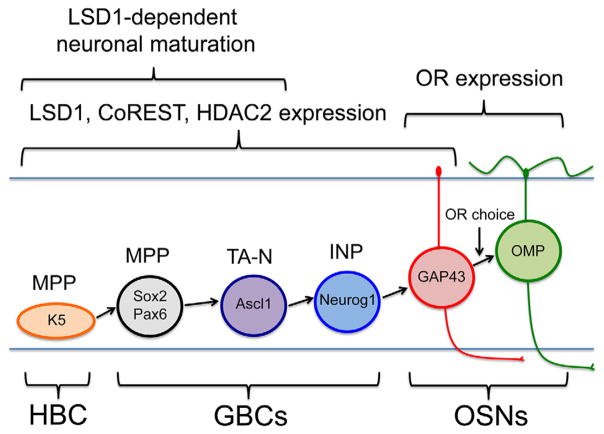
In considering the process of OR gene selection, it is essential to understand the timing of OR gene choice during OSN maturation. In the adult olfactory epithelium (OE), there are two stem cell populations that can give rise to mature OSNs: the reserve horizontal basal cells (HBCs) dormant in the uninjured OE and activated only by severe injury (Leung, Coulombe, & Reed, 2007), and the active stem cell population, the globose basal cells (GBCs) (Huard, Youngentob, Goldstein, Luskin, & Schwob, 1998; Chen, Fang, & Schwob, 2004; Schnittke et al., 2015; Schwob et al., 2017). GBCs themselves can be subdivided into distinct stages progressing from multipotency to the sole production of neurons as follows: Sox2 (+)/Pax6 (+) multipotent progenitors (GBCMPPs), to Ascl1 (+) transit-amplifying progenitors that make neurons in the uninjured OE (GBCTA-Ns), to the final Neurogenin 1 (+) (Neurog1) immediate neuronal precursors (GBCINPs), which are thought to give rise to OSNs directly (Gordon, Mumm, & Davis, 1995; Cau, Gradwohl, Fode, & Guillemot, 1997; Cau, Casarosa, & Guillemot, 2002; Manglapus, Youngentob, & Schwob, 2004; Guo et al., 2010; Packard, Giel-Moloney, Leiter, & Schwob, 2011a). After terminal mitosis, immature OSNs express the growth-associated protein-43 (GAP-43) (Verhaagen et al., 1989), while full functional maturation apparently coincides with the expression of the olfactory marker protein (OMP) (Fig. 1) (Farbman & Margolis, 1980; Schwob, Szumowski, Stasky, Mieleszko Szumowski, & Stasky, 1992). Placing OR expression within the process of neuronal maturation, OR mRNA is first detectable in immature GAP-43 (+) OSNs by in situ hybridization, but quantitative analysis suggests that OR detection lags the initiation of GAP-43 expression (Iwema & Schwob, 2003).
FIGURE 1.
Stem and progenitor progression in the neuronal lineage. Horizontal basal cells (HBCs) are K5 (+) multipotent progenitors (MPPs) able to give rise to all cell types of the OE following activation. The GBC population can be subdivided into stages of progenitor capacity (Schwob et al., 2017): GBCs that function as MPP express Sox2 and Pax6; GBCs that function as transit-amplifying neuronally committed cells (TA-N) express Ascl1; GBCs that function as immediate neuronal precursors (INP) express Neurog1. Immature OSNs express GAP-43 and mature OSNs express OMP. This study used CreERT2 drivers to conditionally delete Lsd1 in K5 (+) HBCsMPP., Ascl1 (+) GBCsTA-N and Neurog1 (+) GBCsINP
With regard to OR expression during neuronal differentiation, Greer and colleagues determined that ORs first appear at 4 days post cell division, lagging 3 days behind GAP43 expression (Rodriguez-Gil et al., 2015). These data suggest that the latest possible time for OR choice is at the immature OSN stage, concomitant with OR mRNA expression. However, OR choice may precede OR mRNA expression, and could occur earlier within the GBC progression. Other data suggest that choice is not fixed at that stage. For example, OR switching apparently occurs in immature OSNs; moreover, mRNA from multiple ORs is present in immature OSNs by single-cell RNA-seq (Lewcock & Reed, 2004; Shykind et al., 2004; Hanchate et al., 2015; Saraiva et al., 2015; Tan et al., 2015; Scholz et al., 2016). In fact, single-unit recordings have shown that immature OSNs are more broadly responsive to odorants before becoming increasingly selective during the transition from immature to mature OMP (+) OSNs (Gesteland, Yancey, & Farbman, 1982). In sum, the choice of OR, if made and implemented at the stage when neurons are still maturing, is impermanent or still in progress at that time.
The mechanisms underlying singular OR gene choice are likely to be multiple but will almost certainly involve both short and long range chromosomal interactions (including promoter sequences and enhancer regions) (Vassalli, Rothman, Feinstein, Zapotocky, & Mombaerts, 2002; Serizawa et al., 2003; Rothman, Feinstein, Hirota, & Mombaerts, 2005; Lomvardas et al., 2006) as well as negative feedback after the choice is made (Serizawa et al., 2003; Lewcock & Reed, 2004; Dalton, Lyons, & Lomvardas, 2013). However, none of these mechanisms fully accounts for the initial selection of a single OR allele. More recently, epigenetic silencing of non-expressed ORs has been described and implicated in singular OR gene choice (Magklara et al., 2011). Specifically, OR gene clusters are decorated with heterochromatic markers H3K9me3 and H3K20me3, and these transcriptional-silencing chromatin modifications are located on ORs in OSNs, sustentacular cells (Sus cells; supporting cells of the OE), and in Neurog1 (+) GBCs and immature neurons harvested from Neurog1-eGFP mice, but not in the reserve stem cell population, the HBCs (Magklara et al., 2011). The fact that ORs sit within heterochromatin in non-neuronal Sus cells suggests that epigenetic OR silencing occurs before neuronal commitment, and therefore likely before OR gene choice. Importantly, actively expressed ORs lack H3K9me3 and are instead marked with the transcriptionally activating chromatin modification, H3K4me3 (Magklara et al., 2011). Thus, the model has emerged where all ORs are initially silenced until only one OR allele is de-repressed and concomitantly activated (Magklara et al., 2011). To flesh out this model fully, the key demethylases and methytransferases need to be identified that determine the chromatin state at OR alleles.
The lysine specific demethylase 1 (LSD1) has been nominated as a candidate epigenetic regulator of OR genes (Lyons et al., 2013) as it demethylates at both H3K4me1/2 (functioning to repress) and H3K9me1/2 (having an activating effect) (Shi et al., 2004; Metzger et al., 2005; Garcia-Bassets et al., 2007; Laurent et al., 2015). LSD1 activity on H3K4 vs. H3K9 is dictated by the binding partners with which it complexes: LSD1 can demethylate at H3K4 when associated with the REST corepressor 1 (CoREST1) (Lee, Wynder, Cooch, & Shiekhattar, 2005) and can demethylate at H3K9 in association with the androgen receptor, estrogen receptor, or supervillin (Metzger et al., 2005; Garcia-Bassets et al., 2007; Laurent et al., 2015). It was recently reported that CoREST colocalizes with LSD1 in both the uninjured OE (Krolewski, Packard, & Schwob, 2013; Kilnic, Savarino, Coleman, Schwob & Lane, 2016) and in nuclear compartments in an olfactory placode-derived cell line (Kilinc et al., 2016), suggesting that the LSD1/CoREST complex could act at H3K4 to repress transcription.
LSD1 is demonstrable in the OE by immunohistochemistry, RNA-seq, and microarray analysis and appears to be most abundant in GBCs (Krolewski et al., 2013; Lyons et al., 2013; Kilinc et al., 2016). Therefore, LSD1 is likely present before, and potentially during, OR gene choice. To test for a role for LSD1 in OR gene choice, Lomvardas and colleagues knocked-out Lsd1 in mice under the control of three cell-specific drivers: Foxg1-Cre, MOR28-Cre, and OMP-Cre (Lyons et al., 2013). Lsd1 knockout in MOR28 (+) and OMP (+) mature OSNs using the latter two drivers had no effect on OR gene choice, while knockout using the Foxg1-Cre driver, which is broadly activated in the embryonic OE, aborted OSN differentiation at the immature OSN stage and blocked OR expression. Ostensibly full maturation of a few OSNs could be rescued by transgene-driven overexpression of ORs, suggesting that the halt in neuronal maturation was secondary to the inability to express ORs (Lyons et al., 2013). However, Foxg1-driven deletion of Lsd1 results in perinatal lethality, limiting these experiments to embryonic development. Consequently, we sought to further characterize a role for LSD1 in the adult OE by knocking out LSD1 using tamoxifen-inducible cell-specific drivers utilizing Cre recombinase fused to a mutated estrogen receptor fragment (CreERT2).
To enhance understanding of LSD1 function in the OE we focused on three main areas: (1) The timing of LSD1 expression relative to OSN maturation and OR expression; (2) The presence of LSD1 binding partners in LSD1 (+) cells in the mouse OE; and (3) The effect of inducible Lsd1 knockout in three basal cell populations: K5 (+) HBCMPPs, Ascl1 (+) GBCTA-Ns, and Neurog1 (+) GBCINPs. These experiments demonstrate that LSD1 is expressed in early dividing cells prior to OR expression and neuronal maturation, that LSD1 co-localizes with CoREST and histone deacetylase 2 (HDAC2) in these early dividing cells, and that LSD1 is required for neuronal maturation during the time window between activating HBCs and Neurog1 (+) immediate neuronal precursors.
2 MATERIALS AND METHODS
2.1 Animals
Wild-type F1 mice bred in-house from C57/BJ6 and 129S1/Sv1MJ stocks or ordered from Jackson Laboratories (stock #101043) were used for LSD1 and CoREST co-labeling immunohistochemistry, Western blotting and immunoprecipitation, and for generation of control methyl bromide (MeBr)-lesioned tissue. Neurog1-eGFP BAC transgenic mice generated by the GENSAT project (Gong et al., 2003) were harvested for Neurog1 staining when uninjured or following olfactory bulbectomy. All EdU pulse-chase experiments were performed on bigenic M72-IRES-tauGFP/P2-IRES-taulacZ mice bred to congenicity with C57/Bl6 mice, and originally gifted by Dr. Peter Mombaerts (Mombaerts et al., 1996; Feinstein & Mombaerts, 2004). Floxed Lsd1 mice were generously provided by Dr. Michael Rosenfeld (Wang et al., 2007) and bred in house to the Cre reporter strain R26Rfl(stop)TdTomato from Jackson Laboratories (stock #007909); these bigenic mice were bred to three different cell-specific CreERT2 drivers: K5-CreERT2 generously provided by Dr. Pierre Chambon and Dr. Randall Reed (Indra et al., 1999), Ascl1-CreERT2 from Jackson Laboratories (stock #012882), and Neu-rog1-CreERT2 generously provided by Dr. Lisa Goodrich (Koundakjian, Appler, & Goodrich, 2007). All mice were maintained on ad libitum rodent chow and water and were housed in a heat- and humidity-controlled, AALAC-accredited vivarium operating under a 12:12-hr light:dark cycle. All protocols for the use of vertebrate animals were approved by the Committee for the Humane Use of Animals at Tufts University School of Medicine.
2.2 MeBr and methimazole lesion
Eight-week old male mice were exposed to MeBr gas (Matheson Gas Products, East Rutherford, NJ) for 8 hr at a concentration of 170 ppm in purified air at a rate of 10 L/min as previously described (Schwob, Youngentob, & Mezza, 1995). Unilateral exposures were performed by plugging the left naris with a 5-mm piece of PE10 tubing and the lumen of the tube was obstructed with a knotted 5.0 suture thread (Schwob, Youngentob, Ring, Iwema, & Mezza, 1999). Following lesion, mice were kept on the same feeding and housing schedule and were sacrificed at 2 days or 3 weeks after exposure.
Methimazole lesion (Bergman, Östergren, Gustafson, & Brittebo, 2002) was performed on male and female mice with a single intraperitoneal injection of 50 mg/kg methimazole in saline at a concentration of 10 mg/ml. Following injection, mice were kept on the same feeding and housing schedule and were perfused 2-weeks post lesion.
2.3 Olfactory bulbectomy
Male and female mice were anesthetized with a double cocktail of ketamine (6.25 mg/kg) and xylazine (12.5 mg/kg). Once animals reached a surgical anesthetic plane, they were immobilized in a stereo-tactic head mount resting on a 37°C heating pad. A single incision was made on shaved, disinfected skin to expose the overlying frontal bone and a bone drill was used to expose the right olfactory bulb; the bulb was removed by aspiration and oxycel was placed into the empty cavity to achieve hemostasis. Lastly, the overlying skin was sutured, mice were given 1 ml of sterile saline subcutaneously and placed on a warm pad for recovery. Tissue was harvested 3 weeks after the surgery.
2.4 EdU and tamoxifen administration
To label dividing cells, EdU (ThermoFisher catalog # A10044) dissolved in sterile PBS was injected subcutaneously in 10-week old mice at 50 mg/kg body weight. Animals were sacrificed at 1, 2, 3, 5, 7, and 9 days post-EdU injection, and EdU was detected in 10 μm coronal sections via Click-iT® chemistry according to the manufacturer’s instructions (ThermoFisher catalog # C10339).
Tamoxifen purchased from Sigma (#T5648) was dissolved in sterile USP-grade corn oil at 30 mg/ml by gentle inversion at 60°C for 10 min; the dissolved tamoxifen was injected intraperitoneally at 150 mg/kg into 6-week-old mice once a day for two successive days. In lineage tracing experiments utilizing Ascl1 and Neurog1 CreERT2 drivers, mice were injected with methimazole the day after the second dose of tamoxifen. Previous work from our lab demonstrates that further delay between recombination and lesion means that the newly labeled progenitors will progress to terminal neuronal differentiation (Lin, Hewitt, Peterson & Schwob, 2015). Thus, methimazole lesion immediately following tamoxifen is necessary to modify the labeled Ascl1 (+) or Neurog1 (+) progenitors of interest.
2.5 Tissue processing
Mice were anaesthetized by intraperitoneal injection of a lethal dose of a triple cocktail of ketamine (37.5 mg/kg), xylazine (7.5 mg/kg), and acepromazine (1.25 mg/kg). Anaesthetized animals were transcardially flushed with PBS and perfused with phosphate-buffered 4% PFA, pH 7.2 for LSD1 and CoREST costains or with phosphate-buffered 1% PLP (paraformaldehyde-lysine-periodate) fixative (McLean & Nakane, 1974) for all other antibody markers. Following perfusion, the cranium and bones overlying the nose were removed; the remaining nasal tissue was post-fixed under vacuum for 1 hr, rinsed in PBS and decalcified in saturated EDTA overnight. Tissue was cryoprotected in 30% sucrose overnight, immersed in OCT® compound (Miles Inc., Elkhart, IN) and snap frozen in liquid nitrogen. 10 μm coronal sections were generated on a Leica cryostat and mounted on Ultraclear Plus charged slides (Denville Scientific) and stored at −20°C until needed.
2.6 Antibody specificity
Primary antibodies used in this study are shown in Table 1. Information on the antibodies is derived from the manufacturers’ description, the literature, and our own data.
TABLE 1.
Antibodies used in this study
| Primary Antibody | Immunogen | Source, species, clonality, catalog#, RRID | Dilution, staining protocol |
|---|---|---|---|
| beta-actin | Beta-actin N-terminal peptide | ThermoFisher, mouse monoclonal, cat # MA5–15739 (clone BA3R), RRID: AB_10979409 | 1:1,000 for Western blot |
| beta-gal | Bacterial beta-galactosidase | Abcam, chicken polyclonal, cat # ab9361, RRID: AB_307210 | 1:750 directly conjugated (5 min trypsin digest) |
| CK14 | Recombinant human KRT14 | Proteintech, rabbit polyclonal, cat # 10143–1-AP, RRID: AB_2134831 | 1:1,000 directly conjugated (5 min 3% H2O2 in methanol + 10 min steam) |
| CoREST | Recombinant fusion protein to a region of human Co-REST | NeuroMab, Mouse monoclonal, cat#75–039 (clone K72/8), RRID: AB_2301051 | 1:500 TSA (5 min 3% H2O2 in methanol +10 min steam) |
| GAP43 | Synthetic peptide corresponding to residues on the C-terminus of human GAP43 | Abcam, rabbit monoclonal, cat #ab75810, RRID: AB_1310252 | 1:1,000 TSA (5 min 3% H2O2 in methanol) |
| GFP | Recombinant full length protein | Abcam, chicken polyclonal, cat #ab13970, RRID: AB_300798 | 1:250 directly conjugated (tertiary for M72-GFP only) |
| HDAC2 | Synthetic peptide corresponding to amino acids 417–488 of human HDAC2 | Abcam, mouse monoclonal, cat #ab12169, RRID: AB_2118547 | 1:100 directly conjugated (5 min 3% H2O2 in methanol + 10 min steam) |
| LSD1 | Synthetic peptide corresponding to Human KDM1/LSD1 aa 50–150 | Abcam, rabbit monoclonal, cat #ab129195, RRID: AB_11145494 | 1:20,000 TSA (5 min 3% H2O2 in methanol +10 min steam) |
| OMP | Synthetic peptide mapping within an internal region of human OMP | Santa Cruz Biotechnology Inc, goat polyclonal, cat# sc-49070, RRID: AB_2158008 | 1:200 directly conjugated in steamed tissue, and 1:200 with TSA in non-steamed tissue |
| MOR28 | MOR28 extracellular epitope (amino acids 167–182) | Gilad Barnea PhD Brown University, rabbit polyclonal, RRID: AB_2636804 | 1:5000 tertiary (5 min 3% H202 in methanol +10 min steam) |
| TdT | Recombinant fusion protein to full length aa sequence from the mush-room polyp coral Discosoma | Rockland Antibodies, rabbit polyclonal, cat #600–401-379, RRID: AB_2209751 | 1:200 directly conjugated (5 min 3% H2O2 in methanol + 10 min steam) |
| Tuj1 | Rat brain microtubules | BioLegend, mouse monoclonal, cat # 801202 (clone TUJ1), RRID: AB_10063408 | 1:200 directly conjugated (5 min 3% H2O2 in methanol + 10 min steam) |
Anti-β-actin from ThermoFisher was used for a loading control in Western blots. This antibody recognizes a single 42 kD band and has been used extensively in the literature (see ThermoFisher datasheet, cat #MA5–15379).
An anti-β-galactosidase antibody was used to detect lacZ-tagged P2 OSNs; the observed P2 expression patterns match previous descriptions closely (Mombaerts et al., 1996).
The rabbit anti-cytokeratin 14 (CK14) from ProteinTech robustly stains the HBCs and perfectly matches previous staining patterns reported for CK14 (+) cells in the OE, which expression was confirmed by 2-D Western blot (Holbrook, Szumowski, & Schwob, 1995).
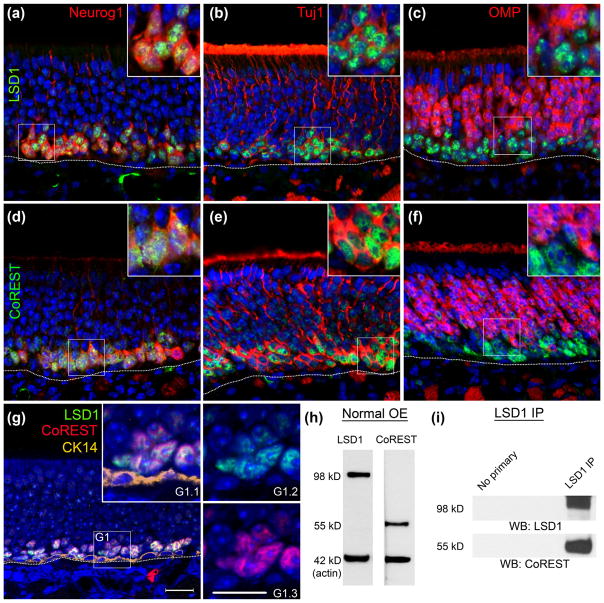
Mouse anti-CoREST from NeuroMab recognizes a single band at 55 kD by western blot of the OE (Fig. 4h) and has been verified in CoREST transfected Cos-1 cells (see NeuroMab datasheet, clone K72/8). Staining patterns using this antibody match previously reported CoREST staining patterns in the OE (Krolewski et al., 2013).
FIGURE 4.
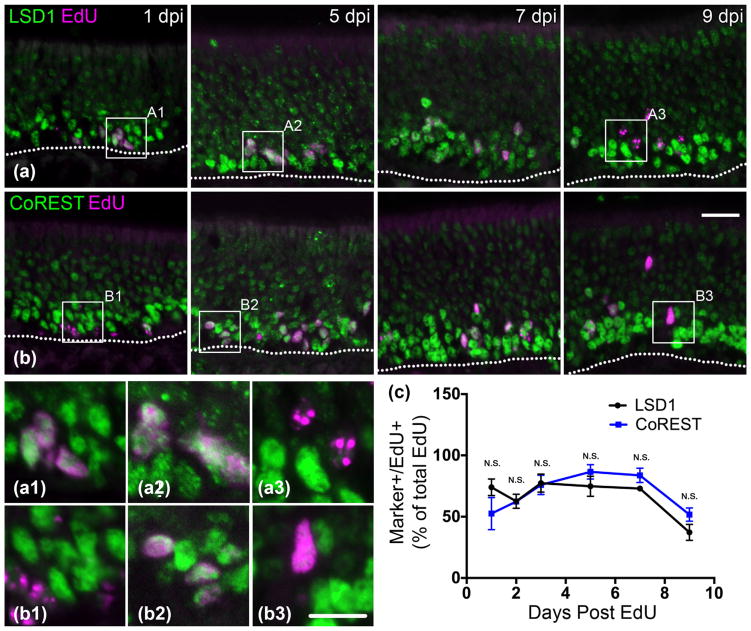
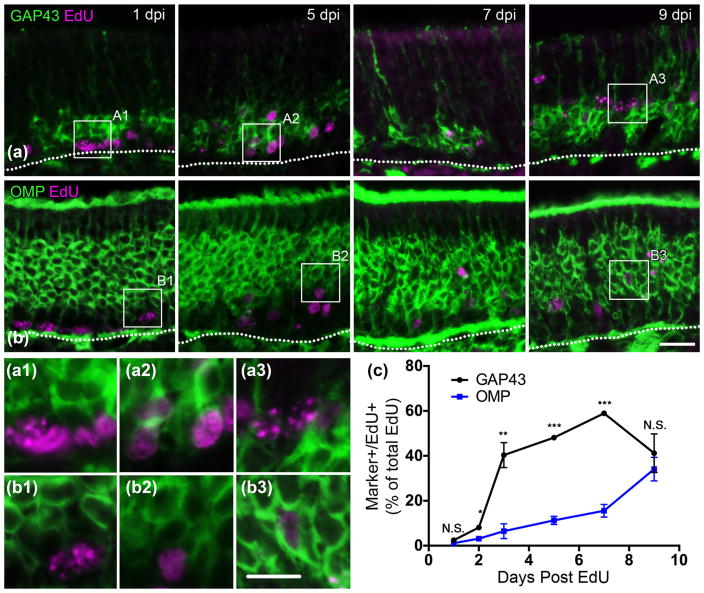
LSD1 and CoREST are prevalent in early dividing cells and begin to decrease starting at 7 days post basal cell division. (a) LSD1 and EdU co-staining at 1, 5, 7, and 9 days post-EdU injection. Co-immunoabeled cells are abundant until 9 days post injection (DPI). (b) CoREST and EdU costaining. Co-immunoabeled cells are abundant until 9 DPI. (c) Quantification of marker (+)/Edu (+) cells out of total counted EdU (+) cells; the mean and SEM are plotted (n = 3). Dotted white line indicates the basal lamina, scale bar in (b) represents 20 μm and applies to (a) as well, while scale bar in (b3) represents 10 μm and applies to all other insets as well
A rabbit monoclonal GAP43 antibody (Abcam) used to identify immature OSNs has been verified for staining in human brain tissue, mouse ear tissue, and in the rat dorsal root ganglion (Abcam datasheet, ab75810). The staining pattern observed for this antibody in the OE matches that of previously reported GAP43 antibodies (Verhaagen et al., 1989; Meiri, Bickerstaff, & Schwob, 1991).
Chicken anti-GFP was used to detect M72-GFP OSNs and Neurog1-eGFP GBCs. This antibody has been used extensively for immunohistochemistry in the brain, olfactory bulb, and olfactory epithelium (Abcam datasheet, ab13970). M72 expression patterns using this antibody match those seen in our hands with other GFP antibodies as well as previously reported M72 expression patterns (Feinstein & Mombaerts, 2004). Neurog1-eGFP staining patterns match that of the native GFP visualized prior to steam pretreatment, which destroys GFP fluorescence.
The mouse HDAC2 antibody recognizes a single 58 kD band in cell lysates from several cell lines including H3k293T, HeLa, and Neuro-2a and stains human glioblastoma cell lines by immunofluorescence (Abcam datasheet, ab12169).
The rabbit polyclonal LSD1 antibody recognizes a single 98 kD band in the OE by Western blot (see Fig. 4h). This antibody encompasses previously reported LSD1 staining patterns in the OE (Krolewski et al., 2013), but is more sensitive than the antibody used in the prior publication. The currently used antibody was also utilized in other recent olfactory publications (Kilinc et al., 2016). Cells that have undergone conditional LSD1 knock-out do not stain with this antibody (see Figs. 6–10).
FIGURE 6.
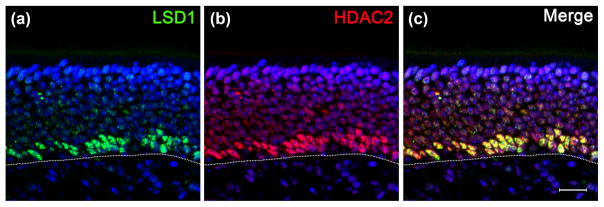
(a–c) LSD1 and HDAC2 are co-expressed in the basal layers of the adult OE. Dotted white line indicates the basal lamina, scale bar in (c) represents 20 μm
FIGURE 10.
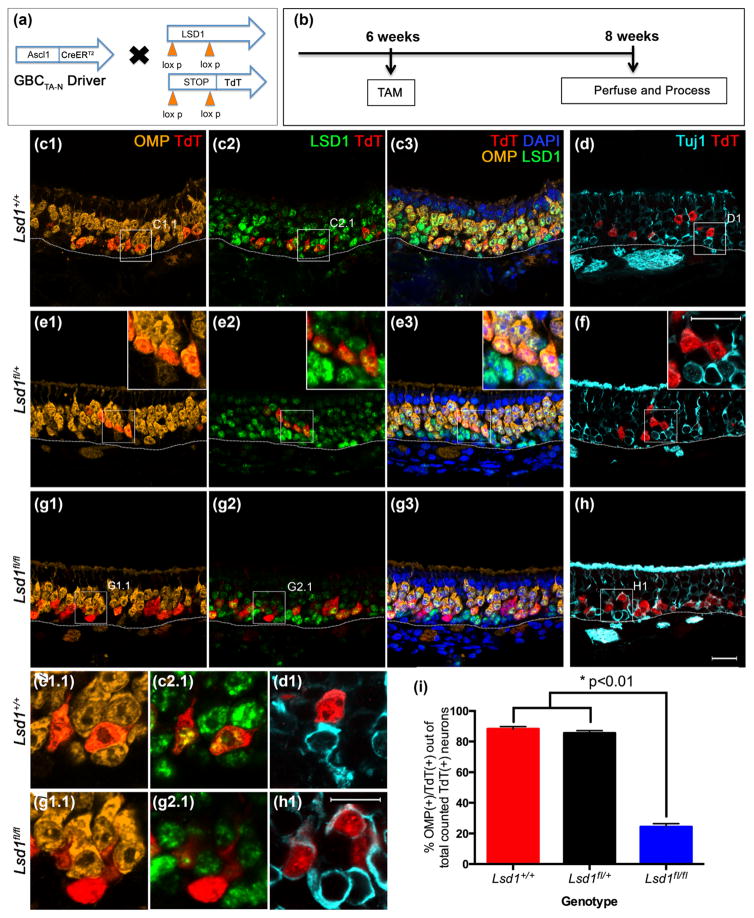
Neuronal maturation is halted in maturing transit-amplifying Ascl1 (+) GBCs when Lsd1 is excised. (a, b) Transgenic mice and experimental design. (c–f) Ascl1(+) GBCsTA-N from Lsd1+/+ and Lsd1fl/+ mice give rise to neurons that mature and express OMP by 2 weeks post-tamoxifen administration. (g, h) Ascl1 (+) GBCs TA-Ns from Lsd1fl/fl mice are unable to mature into OMP (+) OSNs and instead halt their differentiation as Tuj1 (+) immature OSNs. D, F, H are taken from sections adjacent to (c), (e), and (g), respectively. (i) Quantitation of the percent of Ascl1 (+) GBCsTA-N progeny that are OMP (+) cells out of the total number of counted lineage traced neurons (both OMP (+) and Tuj1 (+) OSNs). Mean percent and SEM are plotted for each genotype (n = 3 per genotype). Dotted white line indicates the basal lamina, scale bar in (h) represents 20 μm and applies to (c–g) as well while scale bar in inset (f) represents 20 μm and applies to insets in (e) as well. Scale bar in (h1) represents 20 μm and applies to (c1.1–h1)
Goat anti-OMP from Santa Cruz Biotechnology was used to identify mature OSNs (Farbman & Margolis, 1980). OMP (+) cells marked by this antibody are co-extensive with GFP (+) cells in OMP-GFP transgenic mice in our hands as previously reported (Krolewski et al., 2013).
The rabbit MOR28 antibody was generously provided by Gilad Barnea, Brown University (Barnea et al., 2004). This antibody robustly stains MOR28 (+) cells and matches MOR28 patterns shown by in situ hybridization both in our hands and others (Barnea et al., 2004).
Rabbit polyclonal anti-TdTomato (TdT) was used for lineage tracing experiments to detect TdT (+) cells after steam pretreatment; cells that stain for TdT by immunohistochemistry are the same as those identifiable by native fluorescence of the TdT reporter prior to steaming.
The mouse monoclonal antibody Tuj1 is highly selective for neuron-specific Class III β-tubulin and does not label other classes of β-tubulin. This antibody has been rigorously shown to stain immature OSNs (Schwob et al., 1995; Roskams, Cai, & Ronnett, 1998). The BioL-egend antibody used in this study is the same Covance Tuj1 antibody used by our lab previously (Krolewski et al., 2013; Jang, Chen, Flis, Harris, & Schwob, 2014).
The following secondary antibodies were used: AlexaFlour 488-donkey anti-chicken IgG, Cy3-donkey anti-chicken IgG, Cy3-donkey anti-goat IgG, Cy3-donkey anti-mouse IgG, Cy3-donkey anti-rabbit IgG, AlexaFlour 647-donkey anti-goat IgG, AlexaFlour 647-donkey anti-mouse IgG, AlexaFlour 647-donkey anti-rabbit IgG, Biotin-donkey anti-chicken IgG, Biotin-donkey anti-goat IgG, Biotin-donkey anti-mouse IgG, and Biotin-donkey anti-rabbit IgG, HRP-donkey anti-mouse IgG, HRP-donkey anti-rabbit IgG; all were used at 1:100 for immunohistochemistry or 1:1,000 for Western blotting. Additionally, horseradish peroxidase conjugated streptavidin (HRP-SA) was used at 1:400 and Cy3 conjugated streptavidin was used at 1:100. All secondary and tertiary antibodies were obtained from Jackson ImmunoReasearch where they were tested for minimal cross-reactivity.
2.7 Immunohistochemistry
Prior to immunostaining, rubber cement was applied to encircle each section and dried onto the slide for 15 min on a hot plate; then slides were rinsed in PBS to remove OCT. Sections were subjected to antibody-specific pretreatments (Table 1, where marked) for antigen retrieval including enzymatic digestion, hydrogen peroxide incubation (5 min in 3% H2O2 in MeOH), and “steaming” in 0.01M, pH 6 citric acid buffer heated in a commercial food-steamer. Non-specific-binding was blocked by incubating with 10% donkey serum/5% non-fat dry milk/4% BSA/0.1% TritonX-100 in PBS. Primary antibodies were applied overnight at 4°C. Subsequent visualization may entail one of an methods as indicated in Table 1 and including: (1) staining with directly conjugated secondary antibodies; (2) use of biotinylated secondary antibodies and fluorescently conjugated streptavidin; (3) enhancement using Tyramide signal amplification (TSA), all as described (Guo et al., 2010; Packard et al., 2011a; Schnittke et al., 2015).
After labeling, slides were counter-stained in DAPI for 10 min, mounted in N-propyl gallate anti-fadent media, and cover-slipped. Slide edges were sealed with a thin layer of commercial-grade nail polish and stored at −20°C until imaging.
2.8 Imaging and quantification
Images of EdU-marked cells were obtained with a Spot RT2 color digital camera attached to a Nikon 800E epiflourescent microscope. The remaining images were acquired on a Zeiss 510 confocal microscope in multi-track mode. Confocal images were first processed to adjust color palette, balance, and contrast using the Fiji package of ImageJ 1.50i software applied to the entire image before cropping and figure assembly in Adobe Photoshop CS5.1. Images obtained using the Nikon photomicroscope were assembled, cropped, and adjusted for brightness and contrast applied to the entire image in Photoshop directly.
Labeled cells were counted directly using the Nikon 800E epifluorescent microscope with a dual red/green fluorescent filter to allow simultaneous counting of single- and double-labeled cells. All cells included in counts contained nuclear profiles demonstrated via DAPI staining. For P2/EdU co-labeling experiments, eight evenly spaced sections through the anterior to posterior epithelium were counted; for the rest of the EdU co-staining experiments, four evenly spaced sections from levels 3 to 6 were analyzed. All cell counts were made in the P2-expressing regions of the OE excluding counts of OR28 (+) and M72 (+) cells which were taken in OR28- and M72-expressing regions, respectively. All co-immunolabeled cells were normalized to the total number of EdU (+) cells counted per region and in the case of the ORs, we normalized to both EdU (+) and OR (+) cells counted per region. For graphical representation, we summed the total cells counted within each animal and averaged count from three animals, shown as a line graph with a standard deviation based on biological replicates at each time-point.
For assaying neuronal maturation following LSD1 knockout, we again counted four evenly spaced sections of the OE from a level just rostral to the tip of endoturbinate III anteriorly to a level just caudal to the fusion of endoturbinate II with the cribriform plate posteriorly. Due to a reduced efficiency of recombination, TdT-positivity did not always signify LSD1 knockout in these animals. In other words, some TdT (+) cells in LSD1fl/fl animals were co-stained with the anti-LSD1 antibody. The extent of incomplete recombination varied from region to region across the epithelium. Consistently, the dorsolateral OE was characterized by efficient knockout of LSD1. As a consequence, we selected this area for purposes of quantification. Counts of OMP (+)/TdT (+) cells were normalized to the total number of counted OMP (+)/TdT (+) and Tuj1 (+)/TdT (+) cells (total counted neurons) for all animals. For graphical representation, we summed counts within animals and plotted the average and SEM of the three animals. Prism software was used to perform one-way ANOVAs; if significant, a post-hoc Tukey’s multiple comparison was performed to determine p-values between groups. For pairwise tests in Figures 2–4, a Shapiro-Wilk Normality test was first performed to determine whether a t-test or Mann-Whitney rank sum test was appropriate.
FIGURE 2.
Accelerated transition from immature to mature OSNs begins at 7 days post basal cell division. (a) GAP-43 and EdU co-staining at 1, 5, 7, and 9 days post-EdU injection (DPI). Co-immunolabeled cells are rare at 1 DPI, abundant at 3, 5, and 7 DPI, and decrease by 9 DPI. (b) OMP and EdU co-staining. Co-immunolabeled cells are most abundant at 9 DPI. (c) Quantification of marker (+)/EdU (+) cells out of the total number of counted EdU (+) cells; the mean and SEM are plotted (n = 3). Dotted white line indicates the basal lamina, scale bar in B represents 20 μm and applies to A as well, while scale bar in inset B3 represents 10 μm and applies to all insets. * p <.05, ** p <.01, *** p <.001
2.9 Western Blotting and immunoprecipitation
Flash-frozen OE was suspended in lysis buffer (1M Tris pH 7.5, 0.5M EDTA, 1Xprotease and phosphatase inhibitors (ThermoFisher cat #79440), 20% Triton X-100, 0.5M NA3VO4, 2M NaCl) at 60 μl/mg of tissue, and sonicated until the tissue was homogenized. Membranes were removed by spinning at 14,000 rpm and a bicinchoninic acid assay (BCA) protein assay was used to determine protein concentrations; stocks of 2 mg/ml were made up in 1X SDS sample buffer (ThermoFisher cat # B0008) and 1X reducing agent (ThermoFisher cat #B0009) and frozen at −80°C until used for Western blotting. Samples were denatured at 95°C for 5 min and loaded at 40 μg/lane. Electrophoretically separated proteins were transferred to PVDF transfer using the Bolt Mini Blot Module (ThermoFisher cat #B1000). The membranes were blocked in 1X TBST (20 mM Tris, 0.15 NaCl, 0.1% Tween-20) with 5% Carnation Instant Non-fat Dry Milk for 1 hr at room temperature. Primary antibodies were incubated overnight at 4°C at the following concentrations: CoREST, 1:500; LSD1, 1:20,000; β-actin 1:1,000. The following day, membranes were washed in 1X TBST/5% milk 6 times for 5 min and secondary HRP conjugated antibodies (Jackson ImmunoResearch) were incubated at 1:1,000 for 1 hr at room temperature. After two 30-min washes, the membrane was incubated in ECL substrate (ThermoFisher cat #34080) and developed.
For immunoprecipitation, samples were lysed by sonication in RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS< 50 mM Tris, pH 8.0). Following BCA protein quantification, the protein was diluted to 1 mg/300 μl and incubated with anti-LSD1 primary antibody at 1:100 overnight at 4°C with rotation. The NAb Protein G spin kit was used for immunoprecipitation as described by the manufacturer (ThermoFisher cat#89949). During Western blotting, primary antibodies were used at 1:5,000 and 1:500 for LSD1 and CoR-EST, respectively; secondary HRP conjugates were used at 1:2,000. Gel electrophoresis, transfer, and development were performed as described above.
3 RESULTS
3.1 OR expression during OSN maturation
To analyze the relative timing of OR and LSD1 expression, we first characterized OSN maturation using EdU pulse-chase experiments, comparing EdU staining with that for GAP-43 to mark immature OSNs, and with that for OMP to mark mature OSNs, respectively (Fig. 2). GBCs are the predominant cycling cell within the OE, representing over 90% of the cells that incorporate thymidine analogues (including EdU) during S phase (Huard & Schwob, 1995; Salic & Mitchison, 2008). In the normal unlesioned OE, GBCs give rise exclusively to neurons, and incorporated thymidine analogue label can be chased from the S-phase progenitors into the neuronal population (Schwartz Levey, Chi-karaishi, & Kauer, 1991; Caggiano, Kauer, & Hunter, 1994; Schwob, Huard, Luskin, & Youngentob, 1994; Huard & Schwob, 1995). Because some among the GBC population may divide more than once (Huard & Schwob, 1995), the time at which the leading edge of the double-labeled cells appears represents the shortest interval between last mitosis and that stage in differentiation. All of the numbers listed below report the percent of double-labeled cells among the group of target cells that were counted.
For pulse-chase experiments, we injected EdU into 8-week old mice and waited 1, 2, 3, 5, 7, and 9 days post injection (DPI) to collect tissue for immunohistochemistry. GAP-43 (+)/EdU (+) cells were found in low numbers as early as 1 DPI, when an average of 2.5% of EdU (+) were GAP-43 (+)/EdU (+) (Fig. 2c). At 2 DPI, this percent increased to 8%, and a drastic jump occurred at 3 DPI with 40% of EdU (+) cells being co-immunolabeled for GAP-43 and EdU (Fig. 2c). GAP-43 (+)/EdU (+) cells peaked at 7 DPI to ~60% before declining by 9 DPI back to 40%, likely reflecting the timeframe when GAP-43 (+) OSNs transition to OMP (+) OSNs (Fig. 2a, c). By comparison, OMP (+)/EdU (+) cells were rare for the first few DPI, and then gradually increased to 11% at 5 DPI and 34% at 9 DPI, again demonstrating that the emergence of OMP (+) OSNs is beginning to accelerate at the end of that period (Fig. 2b, c). For OMP and GAP43 EdU incorporation experiments, an average of 432 EdU (+) cells were counted for the group of animals at 1 DPI, 402 at 2 DPI, 348 at 3 DPI, 368 at 5 DPI, 411 at 7 DPI, and 359 at 9 DPI. These counts represent the average number of EdU (+) cells across four sections evenly spaced along the axis from the anterior to the posterior OE in the three animals per group (see Materials and Methods).
In like fashion, the timing of OR expression following basal cell division was determined by analyzing numbers of OR (+)/EdU (+) cells following EdU incorporation (Fig. 3). In these experiments, we assayed three different ORs: (1) the P2 receptor, a Class II OR located on chromosome 7 and expressed in a ventral to ventrolateral swath of OE (Mombaerts et al., 1996) (Fig. 3a); (2) MOR28, a Class II OR located on Chromosome 14 and concentrated in the dorsolateral OE (Serizawa et al., 2003) (Fig. 3b); and (3) M72, a Class II OR located on Chromosome 9 and expressed in the dorsomedial OE (Feinstein & Mombaerts, 2004) (Fig. 3c). In these experiments, we compared the number of OR (+)/Edu (+) cells to both OR (+) and EdU (+) cells, respectively.
FIGURE 3.
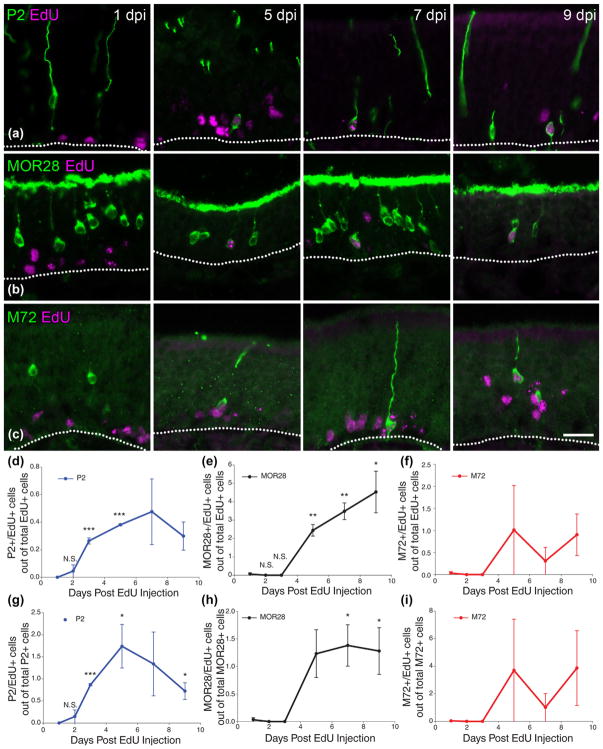
ORs become prominent at 5 days post basal cell division. (a) P2 and EdU co-staining at 1, 5, 7, and 9 days post EdU injection. Co-immunolabeled cells can be found at 5 day post injection (DPI). (b) MOR28 and EdU co-staining. (c) M72 and EdU co-staining. (d–f) Quantification of OR (+)/EdU (+) cells out of total counted EdU (+) cells. P2 co-immunolabeled cells increase prior to MOR28 co-immunolabeled cells and begin to decrease starting at 7 DPI whereas MOR28 co-immunolabeled cells trend toward increases (or remain constant) from 5 DPI to 9 DPI. M72 co-immunolabeled cells appear similarly to MOR28 co-immunolabeled cells, but are highly variable afterward. (g–i) Quantification of OR (+)/EdU (+) cells out of total counted OR (+) cells. (d–i) the mean and SEM are plotted (n = 3). Dotted white line marks the basal lamina, scale bar in (c) represents 20 μm and applies to (a) and (b) as well
P2 (+)/Edu (+) cells were found as early as 2 DPI at an average of 0.04% of EdU (+) cells and an average of 0.15% of P2 (+) cells (Fig. 3d, g). P2 (+)/EdU (+) cells peaked at 5–7 DPI to 0.48% of EdU (+) cells and to 1.74% of P2 (+) cells (Fig. 3d, g). At 9 DPI, the percentage of P2 (+)/EdU (+) cells decreased, potentially reflecting either cell death or a switch in OR expression around the time of full OSN maturation (Fig. 3d, g). For P2/EdU incorporation experiments, an average of 895 EdU (+) cells were counted in the group of animals at 1 DPI, 834 at 2 DPI, 768 at 3 DPI, 788 at 5 DPI, 779 at 7 DPI, and 667 at 9 DPI. An average of 247 P2 (+) cells were counted per animal. These counts represent the average number of EdU (+) cells across eight evenly spaced sections from the anterior to the posterior OE in the three animals per group (see Materials and Methods).
The first MOR28 (+)/EdU (+) cell were found as early as 1 DPI at 0.06% of EdU (+) cells and 0.03% of MOR28 (+) cells. Following this rare event, additional MOR28 (+)/EdU (+) cells were not apparent until 5 DPI when they constituted 2.44% of EdU (+) cells and 1.23% of MOR28 (+) cells (Fig. 3e, h). After this time-point, the number of MOR28 (+)/EdU (+) cells increased as a percentage of EdU (+) cells but remained constant when measured by comparison to MOR28 (+) cells (Fig. 3e, h). Thus, it appears that MOR28 (+) cells appear in a burst around 5 days post-mitosis and remain constant as OSNs shift from immature to mature OSNs. For the MOR28/EdU incorporation experiments, an average of 593 EdU (+) cells were counted in the group of animals at 1 DPI, 528 at 2 DPI, 353 at 3 DPI, 425 at 5 DPI, 561 at 7 DPI, and 324 at 9 DPI. An average of 1104 MOR28 (+) cells were counted per animal. These counts represent the average number of EdU (+) cells across eight evenly spaced sections from the anterior to the posterior OE in the three animals per group (see Materials and Methods). M72 (+)/EdU (+) cells were found as early as 5 DPI when they constituted 1% of EdU (+) cells and 4% of M72 (+) cells (Fig. 3f, i). The number of M72 (+)/EdU (+) cells remained consistent from 5 to 9 DPI again suggesting that this OR reaches detectable levels of expression by 5 days after terminal mitosis. Of the three ORs assessed, M72 was the least expressed, with an average of ~10 OR counted per section (unilaterally) making our analysis quite variable, but nonetheless consistent with an onset of OR expression at 5 DPI (Fig. 3f, i).
3.2 LSD1 expression during OSN maturation
A hypothesized role for LSD1 in epigenetic regulation of OR gene transcription requires that its expression coincides with or precedes receptor expression. LSD1 immunostaining patterns have been published with several different anti-LSD1 antibodies (Krolewski et al., 2013; Lyons et al., 2013; Kilinc et al., 2016). The described staining patterns are overlapping, though not completely concordant. Previous work from this lab suggested that LSD1 was limited to GBCs and immature OSNs, and that LSD1 was not detectable in mature OSNs. We reassessed the distribution of LSD1 tissue staining using the rabbit polyclonal antibody that was utilized for the immunoprecipitation experiments described below (Abcam ab#129195). The monospecificity of this new Abcam antibody for LSD1 was validated by the lack of staining on LSD1 knock-out tissue, as well as its labeling of a single band on Western blots (see below). In contrast with our previous report, the Abcam LSD1 antibody labels mature OSNs as well, albeit more weakly than GBCs and more immature neurons (Fig. 4). Taking advantage of the specificity and enhanced sensitivity, we re-examined when and where LSD1 is expressed during OSN maturation using EdU pulse-chase experiments and whether LSD1 associates with its potential binding partner, CoREST, in the OE by co-immunostaining and co-immunoprecipitation.
With respect to the timing of LSD1 expression, EdU pulse-chase experiments demonstrate that LSD1 (+)/EdU (+) cells are abundant at 1 DPI encompassing 74% of the EdU (+) cells (Fig. 4a, c). With respect to subsequent stages, LSD1 (+)/EdU (+) cells remain high until 9 DPI at the time of OSN maturation to OMP (+) neurons (Figs. 2c, 4a, c). For LSD1 and CoREST EdU-incorporation experiments, an average of 287 EdU (+) cells were counted in the group of animals at 1 DPI, 310 at 2 DPI, 254 at 3 DPI, 230 at 5 DPI, 224 at 7 DPI, and 219 at 9 DPI. These counts represent the average number of EdU (+) cells across four evenly spaced sections from the anterior to the posterior OE in the three animals per group (see Materials and Methods). CoREST expression as a function of time after injection of EdU mimics that of LSD1 suggesting that the two proteins are likely expressed in the same cell populations (Fig. 4b, c). The surmised LSD1/CoREST co-localization was directly assessed and validated (Fig. 5).
FIGURE 5.
LSD1 and CoREST are expressed in the same population of cells and complex together in the mouse OE. (a–c) Anti-LSD1 co-labels with GFP expressed from a Neurog1-eGFP BAC transgene in GBCs and immature OSNs, Tuj1, and OMP. Highest LSD1 expression is found in Neurog1-GFP (+) and Tuj1 (+) cells but not in OMP (+) OSNs. (d, e) In the same tissue CoREST co-stains with Neurog1-GFP, Tuj1, and OMP. (g) LSD1 and CoREST are expressed in overlapping cells and are not found in CK14 (+) HBCs. (h) Western blots stained with anti-LSD1 and anti-CoREST demonstrate abundant expression in the mouse OE with molecular weights of 98 and 55 kD, respectively. (i) Blots from anti-LSD1 immunoprecipitations are positive for CoREST, demonstrating that the two proteins are complex together in the OE. Dotted white line indicates the basal lamina, scale bar in (g) represents 20 μm and applies to all non-inset panels, while scale bar in (g1.3) represents 10 μm and applies to all insets
Both proteins are co-expressed in the vast majority of LSD1-labeled basal cells in the normal OE, which are situated immediately superficial to the CK14 (+)/LSD1 (−)/CoREST (−) HBCs (Fig. 5G). For example, LSD1 and CoREST co-label Neurog1-eGFP (+) GBCINPs and Tuj1 (+) immature OSNs (Fig. 5a, b, d, e). By immunohistochemistry, LSD1 and CoREST are also co-expressed at low levels in OMP (+) mature OSNs (Fig. 5c, f). While the co-expression in mature OSNs was not previously reported (Krolewski et al., 2013), our novel observation here, no doubt, reflects the enhanced sensitivity of the new LSD1 antibody.
The immunostaining results are supported by the demonstration of both proteins in Western blots of normal OE tissue (Fig. 5h). By comparison with our data on the timing of OR expression, LSD1 and CoREST are detectable prior to OR expression (as defined by either co-expression of a marker protein or immunostaining for the OR) and full neuronal maturity, i.e., OMP expression (cf. Figs. 2–4). Finally, we demonstrated that LSD1 and CoREST associate directly with one another in experiments that utilize immunoprecipation for LSD1 followed by Western blotting for CoREST (Fig. 5h, i).
To characterize further the LSD1/CoREST complex that is present in GBCINPs, we immunostained for HDAC2, another member of this epigenetic silencing complex, and found that its expression is also almost completely co-extensive with that of LSD1 (Fig. 6) (Lee et al., 2005).
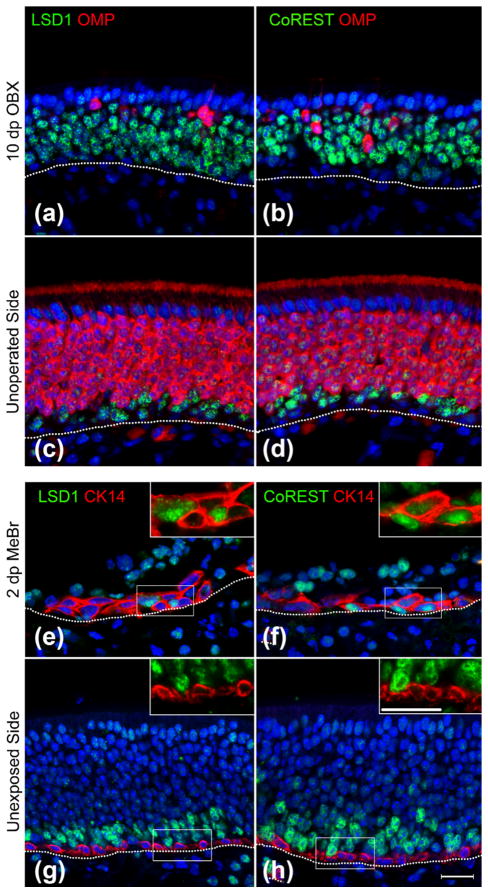
Previous descriptions suggested that LSD1 expression patterns change following OE lesion (Krolewski et al., 2013). Therefore, we assessed LSD1 and CoREST patterns in two injury models: olfactory bulbectomy (OBX), a lesion model that results in a loss of mature OSNs and an expansion of progenitors, and methylbromide (MeBr)-lesion, a lesion model that activates the HBC reserve stem cell population (Fig. 7). As expected, LSD1 (+) and CoREST (+) cells increased post OBX (Fig. 7a–d), in line with previous publications demonstrating increases in Neurog1-eGFP (+) GBCs and immature OSNs following bulbectomy (Verhaagen et al., 1990; Schwob et al., 1992; Iwema & Schwob, 2003; Krolewski et al., 2013). Both LSD1 and CoREST appear in CK14 (+) HBCs by 2 days after MeBr lesion (Fig. 7e–h), a finding previously reported by our lab using a different LSD1 antibody (Krolewski et al., 2013). Thus, activated HBCs that are transitioning toward GBCs represent the earliest cell type co-expressing LSD1 and CoREST in the adult mouse OE.
FIGURE 7.
LSD1 and CoREST populations expand following bulbectomy and appear in activating HBCs at 2 days post MeBr lesion. (a–d) LSD1 and CoREST populations expand on the ablated side at 10 days post-OBX (a, b) as compared to the contralateral control side (c, d). (e–h) LSD1 and CoREST appear in CK14 (+) HBCs at 2 days post-unilateral MeBr lesion (e, f) whereas on the contralateral, unlesionsed side, LSD1, and CoREST are not found in CK14 (+) HBCs (g, h). Dotted white line indicates the basal lamina, scale bar in (h) represents 20 μm and applies to all non-inset panels, while scale bar in inset for (h) represents 20 μm and applies to all inset panels
3.3 LSD1 knockout in HBCs in anticipation of activation and in Ascl1-CreERT2 GBCs aborts OSN maturation while knockout driven by Neurog1-CreERT2 does not
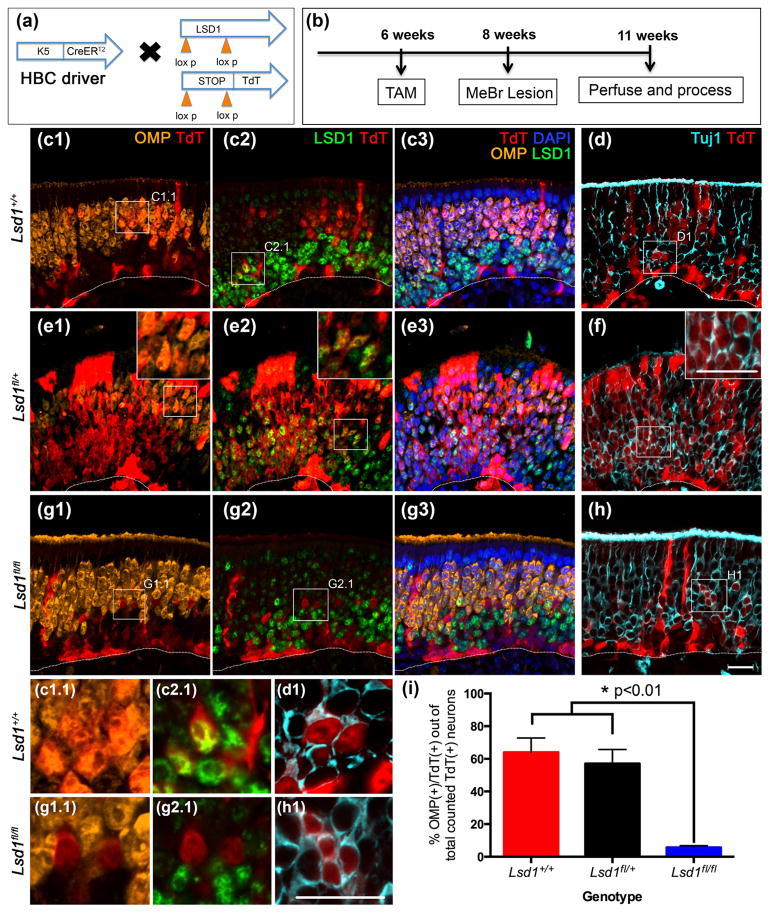
Previous studies of LSD1 function in the OE used a Foxg1-Cre driver to knock out Lsd1 as early as E10.5 in the olfactory placode (Duggan & DeMaria, 2008; Lyons et al., 2013). Because elimination of Lsd1 precedes all but the very earliest stage in GBC and neuronal differentiation, one cannot draw any conclusion regarding the specific cell types in which the gene is functioning and how Foxg1 expression relates to the timing of OR expression and OR choice. Here, we take advantage of existing CreERT2 drivers in order to excise Lsd1 at distinct, defined stages in the emergence of OSNs from the stem and progenitor populations. Specifically, we used a K5-CreERT2 driver to excise Lsd1 in the population of reserve stem cells, i.e., the HBCMPPs, prior to their activation by injury. Additionally, we used an Ascl1-CreERT2 driver to accomplish excision and protein depletion at or after the GBCTA-N stage and a Neurog1-CreERT2 to do the same at or after the GBCINP stage.
3.3.1 K5-CreERT2 driver
For the first set of experiments, mice with the following genotype K5-CreERT2; R26Rfl(stop)TdTomato (TdT);Lsd1fl/fl, were given tamoxifen at 6 weeks of age to excise Exon 6 of Lsd1 and induce heritable expression of the TdT reporter (Fig. 8a) (Wang et al., 2007). Two weeks later, the epithelium was lesioned by exposure to MeBr in order to activate dormant HBCs to multipotency and differentiation into GBCs. The tissue was then harvested 3 weeks post-injury, which allows adequate time for large numbers of HBC-derived TdT (+) progeny to mature into OMP (+) OSNs (Fig. 8b, c) (Schwob et al., 1995). Close inspection of the tissue indicates that some TdT (+) cells retain detectable levels of LSD1 protein in mice that are homozygous for the conditional knockout allele, indicating variability in the efficiency of Cre recombination at the Lsd1 and ROSA26-fl(stop)TdT loci. Importantly, staining for LSD1 in the tamoxifen-treated, homozygous conditional knockout mice demonstrated also that the efficiency of recombination varied across the plane of the epithelium. Lateral regions of the epithelium consistently exhibit a greater overall degree of recombination, as shown by a higher percentage of TdT (+) HBCs, and fewer TdT (+) cells retained LSD1 immunoreactivity there. Thus, in order to minimize the impact of incomplete recombination on the findings, we limited the quantitative analysis of the extent of neuronal maturation to regions in which Lsd1 knock out in TdT (+) cells was more complete. In these areas, we observed a 93% efficiency in homozygous deletion of Lsd1 using the K5 driver, based on the absence of LSD1 from that percentage of TdT (+) basal cells and neurons (both mature and immature). In areas such as these, TdT (+)/OMP (+) and TdT (+)/Tuj1 (+) neurons were counted in matching locations on the adjacent sections (simultaneous screening of TdT/LSD/OMP/Tuj1 on the same section was not possible due to antibody incompatibility). While the variable efficiency of Cre-mediated recombination complicated cell counting, the intermingling of TdT (+)/LSD1 (+) and TdT (+)/LSD1 (−) cells in the same section presented a within-animal control.
FIGURE 8.
Neuronal maturation is halted in activated HBCs lacking LSD1. (a, b) Transgenic mice and experimental design. (c–f) Activated HBCMPPs from Lsd1+/+ and Lsd1fl/+ mice mature into OMP (+) OSNs by 3 weeks post-MeBr lesion. (d, f) are taken from sections adjacent to (c), (e), respectively. (g–h) Activated HBCMPPs from Lsd1fl/fl mice are unable to mature into OMP (+) OSNs and instead halt in maturation as Tuj1 (+) immature OSNs. H is taken from a section adjacent to (g). (i) Quantitation of the percent of activated HBCs that are OMP (+) cells out of the total number of counted lineage-traced neurons (OMP (+) and Tuj1 (+) OSNs inclusive). Mean percent and SEM are plotted for each genotype (n = 3 per genotype). Dotted white line indicates the basal lamina, scale bar in (h) equals 20 μm and applies to (c–g) as well while scale bar in inset (f) represents 20 μm and applies to insets (e1), (e2), and (f). Scale bar in (h1) represents 20 μm and applies to insets (c1.1–h1)
For animals that are either wild-type (Lsd1+/+) or heterozygous (Lsd1fl/+) at the Lsd1 locus, roughly 60% of the total number of counted TdT (+) neurons (OMP (+) and Tuj1 (+) cells inclusive) on average were OMP (+) at 3 weeks after MeBr lesion (Fig. 8c–f, i). In contrast, in the homozygous conditional knockout animals (Lsd1fl/fl), only about of 6% of counted TdT (+) neurons were OMP (+) (Fig. 8g–i). An average of ~1,400 TdT (+) cells were counted per animal (n = 3) across four sections that were evenly spaced from the anterior to the posterior OE (see Materials and Methods).
Despite taking the precaution of examining areas of the epithelium where recombination was maximally efficient, it remains possible that the counts include a few TdT (+) cells that retain one or more copies of the intact Lsd1 allele and potentially retain the ability to mature into OMP (+) OSNs. Thus, as noted above, the lower rate of recombination at the Lsd1 gene locus (as compared to the ROSA26-TdT allele) leads to the interpretation that the small numbers of TdT (+) cells that reach full maturation and OMP expression in the Lsd1fl/fl mice most likely reflect the failure to delete one or both alleles at the Lsd1 locus, i.e., the small minority of the TdT (+) cells that achieve OMP expression in the homozygous mice are not likely to be null for Lsd1.
Our results demonstrate that LSD1 is required for the progression from activated HBC to fully mature, OMP (+) OSNs. Our findings phenocopy the results obtained with Foxg1-driven excision of LSD1 in early development (Lyons et al., 2013). Within the Lsd1 animals, we did not observe a deficiency in Sus cell production, suggesting that LSD1 is specifically required for neuronal maturation within the OE.
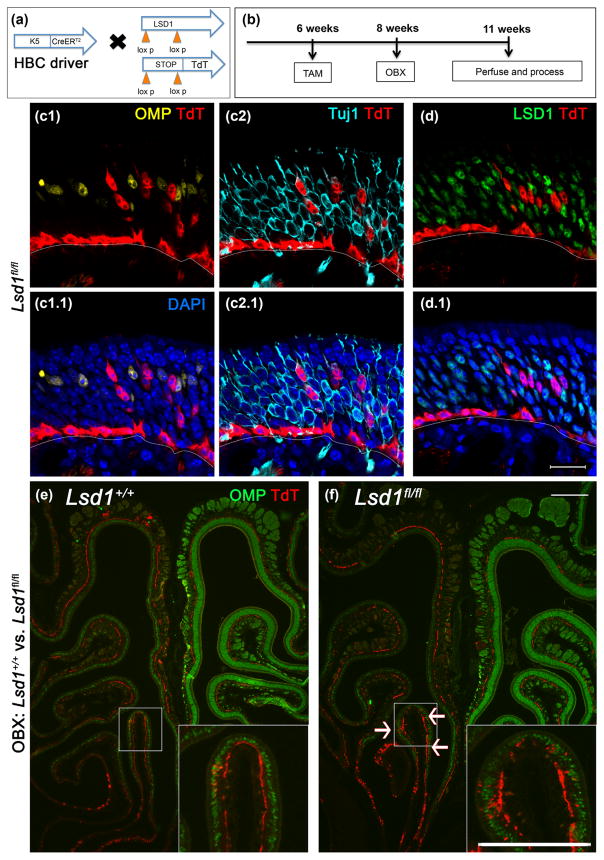
To rule out any contribution that MeBr lesion might make specifically to the failure of neuronal maturation, we took advantage of the very low, but noticeable activation of HBCs following ablation of the olfactory bulb. Thus, we bulbectomized K5-CreERT2; R26Rfl(stop)TdTomato (TdT); Lsd1fl/fl mice (Fig. 9). Once again, in the Lsd1fl/fl animals, neurons failed to mature to express OMP while adjacent cells were able to express OMP (Fig. 9c–f).
FIGURE 9.
Rare LSD1-negative HBCs activated by OBX do not mature into OMP (+) OSNs. (a, b) Transgenic mice and experimental design. (c, d) Rare HBCsMPP activated by OBX and lacking LSD1 do not mature into OMP (+) OSNs. (d) is the taken from the section adjacent to (c). (e) Wild-type tissue (Lsd1+/+) from a TdT reporter mouse showing no activation upon bulbectomy. (f) Homozygous Lsd1 conditional knockout tissue (Lsd1fl/fl) is marked by rare occurrences of HBCsMPP activation following olfactory bulbectomy (white arrows). Scale bar in (d.1) equals 20 μm and applied to (c) as well; scale bar in (f) represents 200 μm and applies to (e) as well. Scale bar in inset (f) represents 200 μm and applies to inset in (e) as well
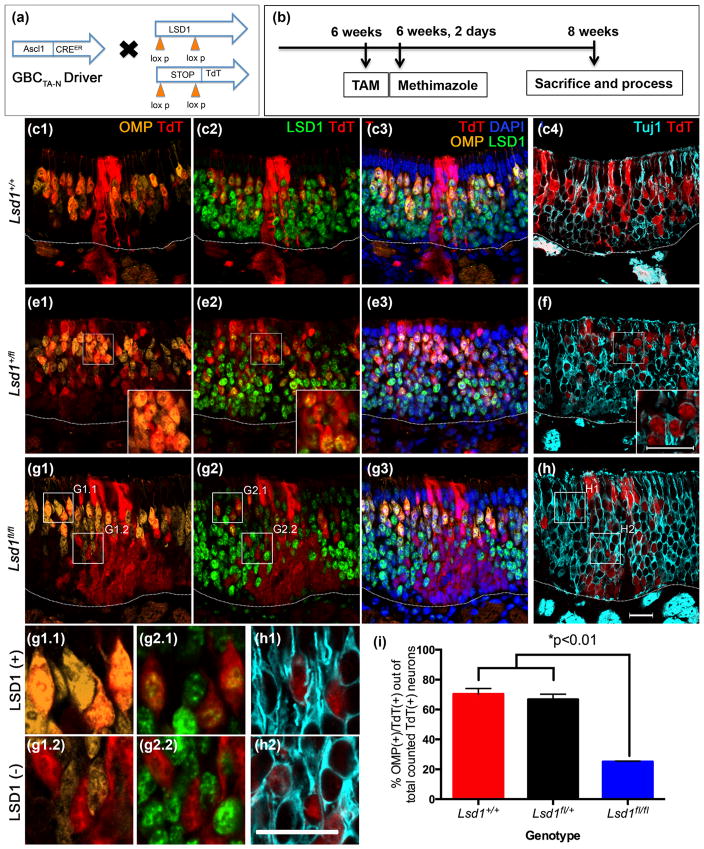
3.3.2 Ascl1-CreERT2 driver
For the second set of experiments, mice with the following genotype, Ascl1-CreERT2; R26Rfl(stop)TdTomato (TdT); Lsd1fl/fl, were also given tamoxifen at 6 weeks of age to excise Exon 6 of Lsd1 and induce heritable expression of the TdT reporter, in order to interrogate LSD1 function at and after the Ascl1 (+) GBCTA-N stage (Figs. 10–11). In these experiments, we assessed neuronal maturation both in the unlesioned OE (Fig. 10) and following direct epithelial injury by injection of methimazole (Bergman et al., 2002) (Fig. 11); methimazole was used because the mixed strain background of the transgenic mice renders them resistant to the olfactotoxic effects of MeBr. Notably, tamoxifen administration to Ascl1-CreERT2; R26Rfl(stop)TdTomato (TdT) mice labels GBCTA-N cells that are actively advancing within the neurogenic progression; for this reason, we injured the OE immediately following tamoxifen administration in order to assess the consequences of epithelial damage on the behavior of the targeted GBCsTA-N while they remained progenitors. The lesioned mice were euthanized 2 weeks post-methimazole injury by which time OMP (+) mature neurons has recovered; the time of tissue harvest differs between methimazole and MeBr because the recovery following MeBr lesion is relatively delayed. As was true of the K5-driver, we observed incomplete recombination at the Lsd1 locus in the Ascl-CreERT2 mice despite robust recombination at the ROSA26 locus, such that some of the TdT (+) basal cells and neurons retained detectable LSD1 in the mice bearing the homozygous knockout allele; however, 93% of the marked neurons did lack detectable LSD1 and were often found in contiguous swathes of the epithelium, particularly in methimazole-lesioned animals (Figs. 10g2, 11g2). Thus, for purposes of determining the likelihood of neuronal maturation following Lsd1 knock out, we again screened for TdT (+)/LSD1 (−) regions and counted adjacent sections for TdT, OMP, and Tuj1. As for the K5 driver, the percentage of TdT (+)/OMP (+) neurons (as proportion of total OSN counts) was markedly reduced in the Lsd1 homozygous conditional knock-out animals under both unlesioned and lesioned conditions as compared to mice that were wild-type or heterozygous for the floxed Lsd1 allele (Figs. 10i, 11i). In animals that were either wild-type or heterozygous at the Lsd1 locus, an average of ~85% (under non-lesioned conditions) and ~70% (under lesioned conditions) of the total number of counted TdT (+) OSNs were OMP (+) at 2 weeks post-tamoxifen/lesion (Figs. 10i, 11i). In contrast, in the homozygous conditional knockout animals (Lsd1fl/fl), only ~25% of total counted TdT (+) OSNs were OMP (+) under both unlesioned and lesioned conditions (Figs. 10i, 11i). Roughly 830 TdT (+) neurons were counted per uninjured animal and 930 TdT (+) cells were counted per lesioned mouse across four sections evenly spaced along the AP axis of the nose (n = 3) (see Materials and Methods). As do the results following recombination in HBCs, these data demonstrate that cells that lack LSD1 expression during the GBC stage fail to mature and do not express OMP, whereas cells that retain LSD1 immunoreactivity do (Fig. 11g).
FIGURE 11.
Neuronal maturation of progeny from transit-amplifying Ascl1 (+) GBCs is also blocked following olfactory lesion when Lsd1 is excised. (a, b) Transgenic mice and experimental design. (c–f) In Lsd1+/+ and Lsd1fl/+ mice, neuronal progeny from Ascl1 (+) GBCsTA-N mature into OMP (+) OSNs by 2 weeks post-methimazole lesion. (g–h) In contrast, in Lsd1fl/fl mice, neuronal progeny from Ascl1 (+) GBCsTA-N do not mature into OMP (+) OSNs for the most part and instead are Tuj1 (+) immature OSNs. The small percentage of OMP (+) OSNs that emerge remain LSD1 (+) (g1.1, 2.1) in contrast to those that lack detectable LSD1 (g1.2, 2,2); antibody incompatibility prevents us from determining the status of LSD1 staining in the neurons that lack Tuj1 labeling (h1). (i) Quantitation of the percent of Ascl1 (+) GBCsTA-N progeny that are OMP (+) cells out of the total number of counted TdT (+) neurons (both OMP (+) and Tuj1 (+) OSNs). Mean percent and SEM are plotted for each genotype (n = 3 per genotype). Dotted white line indicates the basal lamina, scale bar in (h) represents 20 μm and applies to (c–g) as well while scale bar in inset (f) represents 20 μm and applies to insets in (e) as well. Scale bar in (h1) represents 20 μm and applies to (c1.1–h1)
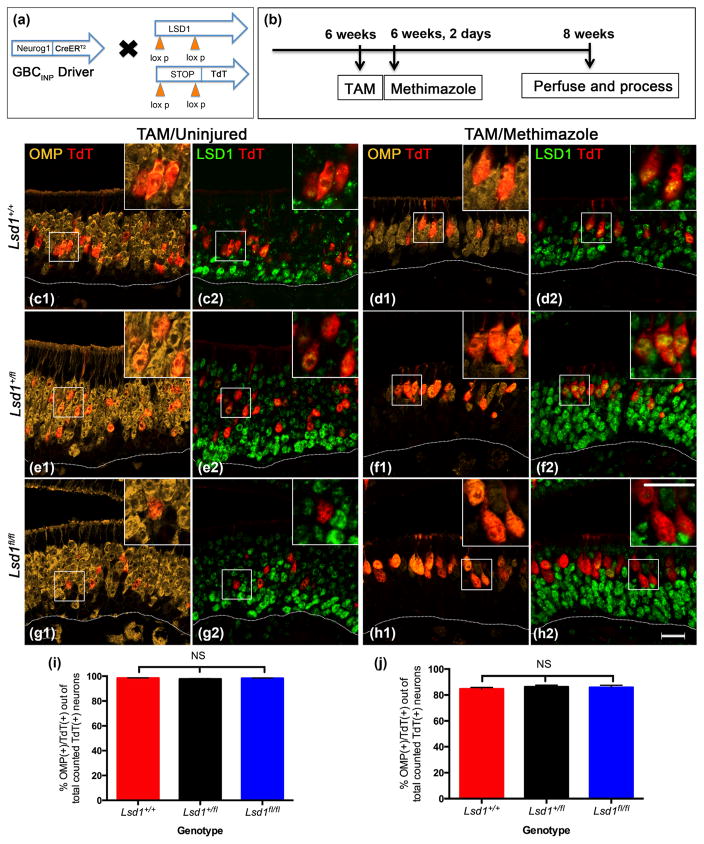
3.3.2 Neurog1-CreERT2 driver
For the third set of experiments, mice with the following genotype Neurog1-CreERT2; R26Rfl(stop)TdTomato (TdT); Lsd1fl/fl, were given tamoxifen at 6 weeks of age to interrogate LSD1 function at and after the Neurog1 (+) GBCINP stage (Fig. 12). In like fashion to the Ascl1-CreERT2; R26Rfl(stop)TdTomato (TdT) experiments, we lesioned the OE by injection of methimazole immediately following tamoxifen administration to assess the consequences of epithelial damage on the behavior of the maturing GBCsINP while they were progenitors. Once again, we observed differential Cre recombination at Lsd1 and the ROSA26-TdT reporter loci whereby the LSD1 was eliminated with an efficiency of 85% in the homozygous conditional knockout mice, for which we compensated as described above.
FIGURE 12.
Neurog1-GBCs lacking LSD1 are capable of maturing into OMP (+) OSNs under both basal and lesioned conditions. (a, b) Transgenic mice and experimental design. (c, e, g) Neurog1 (+) GBCsINP from from Lsd1 +/+, Lsd1fl/+, and Lsd1fl/fl mice, respectively, give rise to progeny that mature into OMP (+) OSNs by 2 weeks post-tamoxifen administration in the uninjured OE. These TdT (+) mature OSNs lack LSD1 (g1, g2, and insets). (d, f, h) Neurog1-GBCs from Lsd1+/+, Lsd1fl/+, and Lsd1fl/fl mice give rise to progeny that mostly mature into OMP (+) OSNs by 2 weeks post-methimazole lesion. These TdT (+) mature OSNs also lack LSD1 (h1, h2, and insets). (i, j) Quantitation of the percent of Neurog1-GBC INP progeny that are OMP (+) cells out of the total number of counted TdT (+) neurons (OMP (+) and Tuj1 (+) OSNs inclusive) for wild-type (Lsd1+/+), heterozygous conditional knockout (Lsd1fl/+), and homozygous conditional knockout (Lsd1fl/fl) mice in both tamoxifen-only and tamoxifen plus methimazole conditions. Mean percent and SEM are plotted for each genotype (n = 3 per genotype). Dotted white line indicates the basal lamina, scale bar in (h2) represents 20 μm and applies to (c–g) as well while scale bar in inset (h2) represents 20 μm and applies to all insets (c1–h1) as well
Lsd1 knockout in Neurog1 (+) GBCsINP under both basal and lesioned conditions did not interfere with the maturation of the OSNs that were progeny of the targeted GBCs (Fig. 12). Unlike either dormant HBCs or in Ascl1 (+) GBCsTA-N, Neurog1 (+) GBCsINP following Lsd1 excision were capable of maturing into OMP (+) OSNs during the same timeframe to the same extent as in both Lsd1+/+ and Lsd1fl/+ control animals (Fig 12c–j). Roughly 930 TdT (+) cells were counted per uninjured animal and 890 TdT (+) cells were counted per lesioned animal across four sections evenly spaced along the AP axis of the nose (n = 3) (see Materials and Methods). In mice that are homozygous for the floxed Lsd1 allele, nearly 100% of TdT (+) neurons in the unlesioned OE and greater than 80% of TdT (+) neurons in the methimazole-lesioned/recovered OE express OMP (Fig. 12g, h).
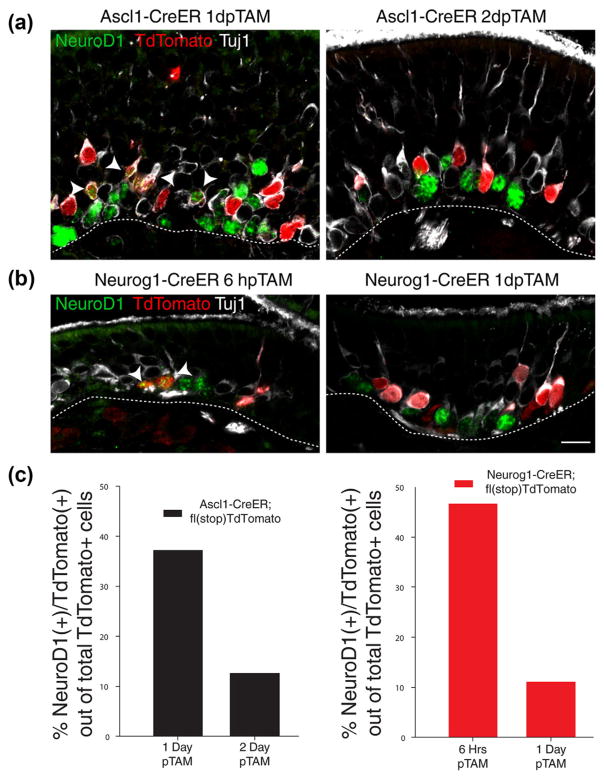
We examined the kinetics of Cre recombination in both the Ascl1-CreERT2 and Neurog1-CreERT2 driver strains by monitoring TdT labeling at short survivals post-tamoxifen. The appearance of TdT serves as a surrogate for the assessing the timing of Lsd1 exon 6 gene excision relative to the progression of Ascl1 (+) and Neurog1 (+) GBC populations toward terminal neuronal differentiation (Fig. 13). With the Ascl1-CreERT2 driver, we found that at 1 day post-tamoxifen administration, 37% of the TdT (+) cells remained within the GBC population and expressed NeuroD1 protein (37%), dropping to 12% by 2 days post-tamoxifen administration (Fig. 13a, c). As expected, the Neurog1-CreERT2 driver progressed more rapidly to neuronal differentiation, with 47% being NeuroD1 (+) at 6 hr after injection, and dropping to 11% by 1 day post-tamoxifen administration (Fig. 13b, c). Thus, a
FIGURE 13.
Kinetics of Ascl1-CreERT2 and Neurog1-CreERT2 activity. Both Ascl1-CreERT2 and Neurog1-CreERT2 mice retain some NeuroD1 (+)/TdT (+) GBCs at 1-day post-tamoxifen (TAM) administration. (a, b) Immunohistochemistry of TdTomato, NeuroD1, and Tuj1 in Ascl1-CreERT2 and Neurog1-CreERT2 driver mice, respectively, at various time points after last tamoxifen administration. (c) Quantification of TdTomato (+) and NeuroD1 (+) double positive GBCs shown above. Counts are from three animals, pooled together to control for tamoxifen labeling efficiency, obtained evenly from across the anterior–posterior axis. Arrowheads indicate triple-labeled cells; at this level of staining sensitivity, Tuj1 will label dividing, NeuroD1 (+) GBCs (Packard et al., 2011a). Scale bar in (b) represents 10 μm and applies to (a) as well substantial proportion of the cells that have undergone recombination using the Neurog1-driver remain as GBCs for more than a day after.
4 DISCUSSION
The results presented here demonstrate the timing of critical events in the maturation of OSNs in the adult OE subsequent to the incorporation of EdU by proliferating GBC progenitors, including the initiation of neuronal differentiation as marked by the expression of GAP-43, neuronal maturation as marked by OMP, and the emergence of immunodetectable or marker-associated OR expression (Fig. 14). For each stage, there tends to be a gradual increase and then a relatively abrupt inflection of the labeling index as a function of time. Thus, although a few differentiating neurons are seen 1 day after the EdU pulse, their numbers sharply increase between 2 and 3 days after injection. Likewise, mature OSNs are apparent, though sparse, by 3 days followed by the major accumulation between 7 and 9 days of pulse-chase. As expected from previous work from this lab and others (Iwema & Schwob, 2003; Rodriguez-Gil et al., 2015), OR (+) neurons become evident between those two time-points, offset by about 2 days relative to the curve mapping GAP-43 (+) neurons as a function of time. These data present more granularity than available from previous studies of the adult OE (Iwema & Schwob, 2003), and their concordance with data obtained in the context of early development (Rodriguez-Gil et al., 2015) suggests that neuronal differentiation progresses similarly in both settings. Notably, we report earlier expression of the P2 receptor as compared to previous reports of the timing of P2 expression relative to a thymidine pulse (Rodriguez-Gil et al., 2015); this discrepancy may be due to the fact that the experiments were performed at different developmental time points—10-week old adult mice in our hands versus 7-day old mice (Rodriguez-Gil et al., 2015). Also important is the opportunity to compare the timing of these events to the expression of LSD1, given the suggestion that it acts as a key regulator of OR expression and singularity (Lyons et al., 2013).
FIGURE 14.
LSD1 in neuronal maturation: results summary. LSD1 is expressed in early dividing cells before OR expression and neuronal maturation and decreases at the time of OR stabilization. LSD1 co-localizes with CoREST and HDAC2 and is required for neuronal maturation during a distinct time window between activating reserve stem cells (HBCs) and Neurog1 (+) GBC immediate neuronal precursors (INPs)
Within the context of neuronal maturation and OR expression, both LSD1 and one of the proteins with which it complexes, CoREST, are highly expressed in dividing GBCs, reach peak expression 5 days post-EdU pulse and then begin to decline by 7 days, which is around the time immature, GAP-43 (+) OSNs transition to mature, OMP (+) OSNs (Fig. 14). Thus, both proteins anticipate terminal mitosis and the initiation of OR expression, and decrease at full neuronal maturation. Thus, on temporal grounds, LSD1 could play a role prior to, at the time of, or during a short time-window following OR gene expression. However, LSD1 may not play a role achieving or maintaining OR singularity, since its expression declines precipitously when neurons express OMP, preceding the expression of a single OR (Shykind et al., 2004; Hanchate et al., 2015; Tan et al., 2015).
That LSD1 and CoREST anticipate OR expression by several days is consistent with a hypothesized role in suppressing OR transcription prior to OR gene choice. For example, the association of LSD1 and CoREST demonstrated by co-immunoprecipitation and, putatively, of HDAC2, is most consistent with a suppressive effect given the established role for the complex in other settings (Lee et al., 2005; Shi et al., 2005). However, the initial multiplicity of OR expression in OMP (+) OSNs, at which point LSD1 levels have already decreased, suggest additional players are involved in establishing the final singularity. Nonetheless, the appearance of LSD1 and CoREST in HBCs that are undergoing activation in response to epithelial injury is similarly suggestive that some type of early suppression is underway as the HBCs transition into GBCs. Consistent with that notion is the absence of H3K9me3 at OR loci in HBCs, which contrasts with their deposition in GBCsINP and immature OSNs (Magklara et al., 2011). Unfortunately, the methylation status of OR-associated H3K4s has not been defined in HBCs.
It should be noted, however, that LSD1 is not exclusively associated with gene repression. Indeed, LSD1/CoREST complexes that replace HDAC2 with C-terminal binding protein (CtBP) have been found to activate transcription at select genes (Ray, Li, Metzger, Schule, & Leiter, 2014). Thus, a second role for LSD1 in activating the OR locus eventually selected for singular expression has also been posited (Lyons et al., 2013). Furthermore, LSD1 compartmentalizes in early post-mitotic cells (immature OSNs) and co-localizes with 1–3 OR loci during this timeframe, suggesting that LSD1 may in fact modify specific OR alleles at the time of OR gene choice (Kilinc et al., 2016). Given the duality of LSD1 function at both H3K4 and H3K9, it is very possible that LSD1 plays a key role in both repression and activation of OR genes at different time-points during OSN maturation. This idea was previously posited based on observations that LSD1 overexpression in mature OSNs decreased OR expression (Lyons et al., 2013); however, the concept of LSD1 repression of OR genes prior to OR gene choice has received little attention.
In light of the complexities of LSD1 and CoREST expression in the OE and in other tissues, we knocked out Lsd1 in adult epithelium using three different progenitor cell drivers: K5, which is expressed in HBCs, the population of dormant reserve stem cells (Leung et al., 2007; Fletcher et al., 2011; Packard, Schnittke, Romano, Sinha, & Schwob, 2011b; Schnittke et al., 2015; Schwob et al., 2017); Ascl1, which is expressed in GBCs that are usually described as transit amplifying progenitors (Cau et al., 1997; Manglapus et al., 2004; Schwob et al., 2017); Neurog1, which is expressed in GBCs that are usually described as progenitors that are immediate precursors of post-mitotic, differentiating, immature OSNs (Cau et al., 2002; Manglapus et al., 2004; Schwob et al., 2017). We found that Lsd1 deletion in HBCs and in GBCs driven by Ascl1 aborts OSN maturation at the immature neuronal stage, which complements our observation that LSD1 and CoREST expression remain high through the period of neuronal immaturity (Fig. 14). Thus, LSD1 is playing an important role in gene regulation during neurogenesis in the adult OE, as in the embryo (Lyons et al., 2013).
Lsd1 deletion driven by Neurog1, on the other hand, did not disrupt OSN maturation, as cells with Lsd1 deletion at or after this stage make the transition into OMP (+) OSNs (Fig. 14). It is perhaps surprising that Lsd1 excision in the immediate neuronal precursors does not share the maturational phenotype given the abundant expression of LSD1 in this cell type as well as immature neurons (Krolewski et al., 2013; Lyons et al., 2013; Kilinc et al., 2016). However, as previously discussed, LSD1 may be playing multiple roles in gene regulation at different maturational stages, with later roles not as critical for neuronal differentiation. The identification of the stage of neurogenic differentiation at which loss of LSD1 no longer blocks full neuronal maturation needs to take into account the half-life of the protein following successful gene deletion. In more specific terms, has neuronal maturation become independent of LSD1 at the GBCINP stage, or is it required until the neuron emerges as post-mitotic? Although a definitive answer cannot be provided at this point, some previously published and currently presented data suggest that LSD1 independency for OSN maturation emerges among cell that are still GBCs. First, the half-life of LSD1 in cells is 6–8 hr (Cao et al., 2016), suggesting that by 24 hr after excision, the abundance of the protein is likely reduced below the concentration required for efficacy. Second, some GBCs remain TdT (+) beyond 1 day after tamoxifen administration to Neu-rog1-CreERT2 mice, yet 100% of the neurons mature in the uninjured OE (with extremely low variability). Furthermore, that only 85% mature in the context of the methimazole-lesioned OE fits with data indicating that a proportion of Neurog1 (+) GBCs assume multipotency in that setting (Lin et al., 2015; Schwob et al., 2017) and is consistent with locating the blockade to differentiation upstream of the immediate neuronal precursor stage. Third, following recombination in the Ascl1-CreERT2 mice, some GBCs have progressed to the immediate neuronal precursor stage in the unlesioned OE 1 day after tamoxifen treatment; that extent of neurogenic progression parallels the finding that a quarter of the TdT (+) neurons generated in these mice do mature fully. Taken together, our data demonstrate a critical time window for LSD1-dependent neuronal maturation that begins as HBCs activate and terminates prior to the high-level expression of LSD1 in immature OSNs (Fig. 14). The results also suggest, but do not fully prove that the transition to LSD1 independency for purposes of OSN maturation occurs at the Neurog1 (+) GBCINP stage.
It seems unlikely that the disruption in neuronal maturation caused by Lsd1 deletion at an early stage in the progenitor cell hierarchy is due exclusively to abnormalities of OR gene expression; that hypothesis is not especially consistent with either the substantial interval between recombination in the progenitors and OR expression by immature OSNs or the lack of effect of LSD1 depletion when ORs are first detected in immature neurons (Iwema & Schwob, 2003; Shykind et al., 2004). At the very least, given that the amelioration of the phenotype anticipates OR expression, these data suggest that LSD1 is playing a role in repressing OR expression. For example, if LSD1 acts to remove methyl groups at H3K4me1/2 in upstream neuronal progenitors, the halt in maturation may be due to the overexpression of multiple OR genes. This multiplicity, in turn, might have consequences for axon targeting to and on the olfactory bulb, thereby disrupting the transition to OMP expression and/or depriving the mutant OSNs of the trophic support needed for long-term survival. Further investigation into OR multiplicity under the background of Lsd1 knockout and into the status of H3K4 epigenetic marks in HBCs and upstream progenitors in the presence and absence of LSD1 will be needed to clarify the nature of the defect.
Furthermore, the effect of Lsd1 knockout on neuronal maturation may be completely independent of an OR phenotype. In fact, LSD1 has been shown to be required for terminal differentiation (from precursor cells) in several cell types including pituitary cells, adipocytes, photoreceptor cells, gastrointestinal endocrine cells, hematopoietic cells, and neurons (Saleque, Kim, Rooke, & Orkin, 2007; Wang et al., 2007; Musri et al., 2010; Sun et al., 2010; Ray et al., 2014; Laurent et al., 2015; Popova, Pinzon-Guzman, Salzberg, Zhang, & Barnstable, 2015). Thus, future studies are necessary to fill in the gaps between LSD1 removal and the halt in OSN maturation.
In summary, the current data demonstrate that LSD1 and its CoR-EST complex components are abundantly expressed before OR expression and OSN maturation, and that LSD1 is required for terminal differentiation during a distinct developmental time-frame extending from the point when upstream GBCs emerge from activated HBCs to the point at which GBCs reach the stage of functioning as immediate neuronal precursors. Future studies will reveal whether this maturational phenotype in the adult OE is dependent on OR gene regulation.
Acknowledgments
Funding information
R01 DC002167 (JES), F31 DC014398 (JHC), F31 DC014637 (BL).
The authors thank Po Kwok-Tse for her excellent technical contributions to the work.
References
- Barnea G, O’Donnell S, Mancia F, Sun X, Nemes A, Mendelsohn M, Axel R. Odorant receptors on axon termini in the brain. Science. 2004;304:1468. doi: 10.1126/science.1096146. [DOI] [PubMed] [Google Scholar]
- Bergman U, Östergren A, Gustafson AL, Brittebo E. Differential effects of olfactory toxicants on olfactory regeneration. Archives of Toxicology. 2002;76:104–112. doi: 10.1007/s00204-002-0321-2. [DOI] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Bushdid C, Magnasco MO, Vosshall LB, Keller A. Humans can discriminate more than one trillion olfactory stimuli. HHS Public Access. 2014;343:1370–1372. doi: 10.1126/science.1249168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caggiano M, Kauer JS, Hunter DD. Globose basal cells are neuronal progenitors in the olfactory epithelium: A lineage analysis using a replication-incompetent retrovirus. Neuron. 1994;13:339–352. doi: 10.1016/0896-6273(94)90351-4. [DOI] [PubMed] [Google Scholar]
- Cao C, Vasilatos SN, Bhargava R, Fine JL, Oesterreich S, Davidson NE, Huang Y. Functional interaction of histone deacetylase 5 (HDAC5) and lysine-specific demethylase 1 (LSD1) promotes breast cancer progression. Oncogene. 2016;5:1–13. doi: 10.1038/onc.2016.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cau E, Casarosa S, Guillemot F. Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development. 2002;129:1871–1880. doi: 10.1242/dev.129.8.1871. [DOI] [PubMed] [Google Scholar]
- Cau E, Gradwohl G, Fode C, Guillemot F. Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development. 1997;124:1611–1621. doi: 10.1242/dev.124.8.1611. [DOI] [PubMed] [Google Scholar]
- Chen X, Fang H, Schwob JE. Multipotency of purified, transplanted globose basal cells in olfactory epithelium. Journal of Comparative Neurology. 2004;469:457–474. doi: 10.1002/cne.11031. [DOI] [PubMed] [Google Scholar]
- Dalton RP, Lyons DB, Lomvardas S. Co-opting the unfolded protein response to elicit olfactory receptor feedback. Cell. 2013;155:321–332. doi: 10.1016/j.cell.2013.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan C, DeMaria S. Foxg1 is required for development of the vertebrate olfactory system. Journal of Neuroscience. 2008;28:5229–5239. doi: 10.1523/JNEUROSCI.1134-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farbman AI, Margolis FL. Olfactory marker protein during ontogeny: Immunohistochemical localization. Developmental Biology. 1980;74:205–215. doi: 10.1016/0012-1606(80)90062-7. [DOI] [PubMed] [Google Scholar]
- Feinstein P, Mombaerts P. A contextual model for axonal sorting into glomeruli in the mouse olfactory system. Cell. 2004;117:817–831. doi: 10.1016/j.cell.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Fletcher RB, Prasol MS, Estrada J, Baudhuin A, Vranizan K, Choi YG, Ngai J. P63 regulates olfactory stem cell self-renewal and differentiation. Neuron. 2011;72:748–759. doi: 10.1016/j.neuron.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, … Rosenfeld MG. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland RC, Yancey RA, Farbman AI. Development of olfactory receptor neuron selectivity in the rat fetus. Neuroscience. 1982;7:3127–3136. doi: 10.1016/0306-4522(82)90235-4. [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, … Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Gordon M, Mumm J, Davis R. Dynamics of MASH1 expression in vitro and in vivo suggest a non-stem cell site of MASH1 action in the olfactory receptor neuron lineage. Molecular and Cellular Neurosciences. 1995;6:363–379. doi: 10.1006/mcne.1995.1028. [DOI] [PubMed] [Google Scholar]
- Guo Z, Packard A, Krolewski RC, Harris MT, Manglapus GL, Schwob JE. Expression of pax6 and sox2 in adult olfactory epithelium. Journal of Comparative Neurology. 2010;518:4395–4418. doi: 10.1002/cne.22463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanchate NK, Kondoh K, Lu Z, Buck LB, Kuang D, Ye X, … Buck LB. Single-cell transcriptomics reveals receptor transformations during olfactory neurogenesis. Science (80-) 2015;350:1251–1255. doi: 10.1126/science.aad2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook EHE, Szumowski KEM, Schwob JE. An immunochemical, ultrastructural, and developmental characterization of the horizontal basal cells of rat olfactory epithelium. Journal of Comparative Neurology. 1995;146:129–146. doi: 10.1002/cne.903630111. [DOI] [PubMed] [Google Scholar]
- Huard JM, Schwob JE. Cell cycle of globose basal cells in rat olfactory epithelium. Developmental Dynamics. 1995;203:17–26. doi: 10.1002/aja.1002030103. [DOI] [PubMed] [Google Scholar]
- Huard JM, Youngentob SL, Goldstein BJ, Luskin MB, Schwob JE. Adult olfactory epithelium contains multipotent progenitors that give rise to neurons and non-neural cells. Journal of Comparative Neurology. 1998;400:469–486. [PubMed] [Google Scholar]
- Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: Comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Research. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwema CL, Schwob JE. Odorant receptor expression as a function of neuronal maturity in the adult rodent olfactory system. Journal of Comparative Neurology. 2003;459:209–222. doi: 10.1002/cne.10583. [DOI] [PubMed] [Google Scholar]
- Jang W, Chen X, Flis D, Harris M, Schwob JE. Label-retaining, quiescent globose basal cells are found in the olfactory epithelium. Journal of Comparative Neurology. 2014;522:731–749. doi: 10.1002/cne.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilinc S, Savarino A, Coleman JH, Schwob JE, Lane RP. Lysine-specific demethylase-1 (LSD1) is compartmentalized at nuclear chromocenters in early post-mitotic cells of the olfactory sensory neuronal lineage. Molecular and Cellular Neuroscience. 2016;74:58–70. doi: 10.1016/j.mcn.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koundakjian EJ, Appler JL, Goodrich LV. Auditory neurons make stereotyped wiring decisions before maturation of their targets. Journal of Neuroscience. 2007;27:14078–14088. doi: 10.1523/JNEUROSCI.3765-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krolewski RC, Packard A, Schwob JE. Global expression profiling of globose basal cells and neurogenic progression within the olfactory epithelium. Journal of Comparative Neurology. 2013;521:833–859. doi: 10.1002/cne.23204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, … Szustakowki J. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Laurent B, Ruitu L, Murn J, Hempel K, Ferrao R, Xiang Y, … Shi Y. A specific LSD1/KDM1A isoform regulates neuronal differentiation through H3K9 demethylation. Molecular Cell. 2015;57:957–970. doi: 10.1016/j.molcel.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- Leung CT, Coulombe PA, Reed RR. Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nature Neuroscience. 2007;10:720–726. doi: 10.1038/nn1882. [DOI] [PubMed] [Google Scholar]
- Lewcock JW, Reed RR. A feedback mechanism regulates monoallelic odorant receptor expression. Pnas. 2004;101:1069–1074. doi: 10.1073/pnas.0307986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B, Hewitt J, Peterson J, Schwob JE. Program no: 291.11, 2015 Neuroscience Meeting Planner. Washington, DC: Society for Neuroscience. Online; 2015. Injury can induce neuronally committed Neurog1+ progenitors to become multi-potent. [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Lyons DB, Allen WE, Goh T, Tsai L, Barnea G, Lomvardas S. An epigenetic trap stabilizes singular olfactory receptor expression. Cell. 2013;154:325–336. doi: 10.1016/j.cell.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, … Lomvardas S. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145:555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manglapus GL, Youngentob SL, Schwob JE. Expression patterns of basic helix-loop-helix transcription factors define subsets of olfactory progenitor cells. Journal of Comparative Neurology. 2004;479:216–233. doi: 10.1002/cne.20316. [DOI] [PubMed] [Google Scholar]
- McLean IW, Nakane PK. Periodate lysine paraformadehyde fixation. A new fixation for immunoelectron microscopy. Journal of Histochemistry & Cytochemistry. 1974;22:1077–1083. doi: 10.1177/22.12.1077. [DOI] [PubMed] [Google Scholar]
- Meiri KF, Bickerstaff LE, Schwob JE. Monoclonal antibodies show that kinase c phosphorylation of GAP-43 during axogenesis is both spatially and temporally restricted in vivo. Journal of Cell Biology. 1991;112:991–1005. doi: 10.1083/jcb.112.5.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AHFM, … Schüle R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, … Axel R. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- Musri MM, Carmona MC, Hanzu FA, Kaliman P, Gomis R, Párrizas M. Histone demethylase LSD1 regulates adipogenesis. Journal of Biological Chemistry. 2010;285:30034–30041. doi: 10.1074/jbc.M110.151209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packard A, Giel-Moloney M, Leiter A, Schwob JE. Progenitor cell capacity of NeuroD1-expressing globose basal cells in the mouse olfactory epithelium. Journal of Comparative Neurology. 2011a;519:3580–3596. doi: 10.1002/cne.22726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packard A, Schnittke N, Romano RA, Sinha S, Schwob JE. DeltaNp63 regulates stem cell dynamics in the mammalian olfactory epithelium. Journal of Neuroscience. 2011b;31:8748–8759. doi: 10.1523/JNEUROSCI.0681-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova EY, Pinzon-Guzman C, Salzberg AC, Zhang SS, Barnstable CJ. LSD1-mediated demethylation of H3K4me2 is required for the transition from late progenitor to differentiated mouse rod photoreceptor. Molecular Neurobiology. 2015;53:4563–4581. doi: 10.1007/s12035-015-9395-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray SK, Li HJ, Metzger E, Schule R, Leiter AB. CtBP and associated LSD1 are required for transcriptional activation by NeuroD1 in gastrointestinal endocrine cells. Molecular and Cellular Biology. 2014;34:2308–2317. doi: 10.1128/MCB.01600-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Gil DJ, Bartel DL, Jaspers AW, Mobley AS, Imamura F, Greer CA. Odorant receptors regulate the final glomerular coalescence of olfactory sensory neuron axons. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:5821–5826. doi: 10.1073/pnas.1417955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskams AJ, Cai X, Ronnett GV. Expression of neuron-specific beta-III tubulin during olfactory neurogenesis in the embryonic and adult rat. Neuroscience. 1998;83:191–200. doi: 10.1016/s0306-4522(97)00344-8. [DOI] [PubMed] [Google Scholar]
- Rothman A, Feinstein P, Hirota J, Mombaerts P. The promoter of the mouse odorant receptor gene M71. Molecular and Cellular Neuroscience. 2005;28:535–546. doi: 10.1016/j.mcn.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Saleque S, Kim J, Rooke HM, Orkin SH. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Molecular Cell. 2007;27:562–572. doi: 10.1016/j.molcel.2007.06.039. [DOI] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva L, Ibarra-Soria X, Khan M, Omura M, Scialdone A, Mombaerts P, … Logan D. Hierarchical deconstruction of mouse olfactory sensory neurons: From whole mucosa to single-cell RNA-seq. Scientific Reports. 2015;5:18178. doi: 10.1038/srep18178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittke N, Herrick DB, Lin B, Peterson J, Coleman JH, Packard AI, … Schwob JE. Transcription factor p63 controls the reserve status but not the stemness of horizontal basal cells in the olfactory epithelium. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E5068–E5077. doi: 10.1073/pnas.1512272112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz P, Kalbe B, Jansen F, Altmueller J, Becker C, Mohrhardt J, … Osterloh S. Transcriptome analysis of murine olfactory sensory neurons during development using single cell RNA-Seq. Chemical Senses. 2016;41:313–323. doi: 10.1093/chemse/bjw003. [DOI] [PubMed] [Google Scholar]
- Schwartz Levey M, Chikaraishi DM, Kauer JS. Characterization of potential precursor populations in the mouse olfactory epithelium using immunocytochemistry and autoradiography. Journal of Neuroscience. 1991;11:3556–3564. doi: 10.1523/JNEUROSCI.11-11-03556.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob JE, Huard JM, Luskin MB, Youngentob SL. Retroviral lineage studies of the rat olfactory epithelium. Chemical Senses. 1994;19:671–682. doi: 10.1093/chemse/19.6.671. [DOI] [PubMed] [Google Scholar]
- Schwob JE, Jang W, Holbrook EH, Lin B, Herrick DB, Peterson JN, Hewitt Coleman J. Stem and progenitor cells of the mammalian olfactory epithelium: Taking poietic license. Journal of Comparative Neurology. 2017;525:1034–1054. doi: 10.1002/cne.24105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob JE, Szumowski KE, Stasky AA, Mieleszko Szumowski KE, Stasky AA. Olfactory sensory neurons are trophically dependent on the olfactory bulb for their prolonged survival. Journal of Neuroscience. 1992;12:3896–3919. doi: 10.1523/JNEUROSCI.12-10-03896.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob JE, Youngentob SL, Mezza RC. Reconstitution of the rat olfactory epithelium after methyl bromide-induced lesion. Journal of Comparative Neurology. 1995;359:15–37. doi: 10.1002/cne.903590103. [DOI] [PubMed] [Google Scholar]
- Schwob JE, Youngentob SL, Ring G, Iwema CL, Mezza RC. Reinnervation of the rat olfactory bulb after methyl bromide-induced lesion: Timing and extent of reinnervation. Journal of Comparative Neurology. 1999;412:439–457. doi: 10.1002/(sici)1096-9861(19990927)412:3<439::aid-cne5>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science (80-) 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Shi YJJY, Matson C, Lan F, Iwase S, Baba T, Shi YJJY. Regulation of LSD1 histone demethylase activity by its associated factors. Molecular Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shykind BM, Rohani SC, O’Donnell S, Nemes A, Mendelsohn M, Sun Y, … Barnea G. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117:801–815. doi: 10.1016/j.cell.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Sun G, Alzayady K, Stewart R, Ye P, Yang S, Li W, Shi Y. Histone demethylase LSD1 regulates neural stem cell proliferation. Molecular and Cellular Biology. 2010;30:1997–2005. doi: 10.1128/MCB.01116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Li Q, Xie XS. Olfactory sensory neurons transiently express multiple olfactory receptors during development. Molecular Systems Biology. 2015;11:844. doi: 10.15252/msb.20156639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassalli A, Rothman A, Feinstein P, Zapotocky M, Mombaerts P. Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb. Neuron. 2002;35:681–696. doi: 10.1016/s0896-6273(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Vassar R, Ngai J, Axel R. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell. 1993;74:309–318. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- Verhaagen J, Oestreicher AB, Gispen WH, Margolis FL. The expression of the growth associated protein B50/GAP43 in the olfactory system of neonatal and adult rats. Journal of Neuroscience. 1989;9:683–691. doi: 10.1523/JNEUROSCI.09-02-00683.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaagen J, Oestreicher AB, Grillo M, Khew-Goodall YS, Gispen WH, Margolis FL. Neuroplasticity in the olfactory system: Differential effects of central and peripheral lesions of the primary olfactory pathway on the expression of B-50/GAP43 and the olfactory marker protein. Journal of Neuroscience Research. 1990;26:31–44. doi: 10.1002/jnr.490260105. [DOI] [PubMed] [Google Scholar]
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, … Rosenfeld MG. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nature Neuroscience. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]