Abstract
Chronic pain and itch are common hypersensitivity syndromes that are affected by endogenous mediators. We applied a systems-based, translational approach to predict, discover, and characterize mediators of pain and itch that are regulated by diet and inflammation. Profiling of tissue-specific precursor abundance and biosynthetic gene expression predicted that inflamed skin would be abundant in four previously unknown 11-hydroxy-epoxy-or 11-keto-epoxy-octadecenoate linoleic acid derivatives and four previously identified 9- or 13-hydroxy-epoxy- or 9- or 13-keto-epoxy-octadecenoate linoleic acid derivatives. All of these mediators were confirmed to be abundant in rat and human skin by mass spectrometry. However, only the two 11-hydroxy-epoxy-octadecenoates sensitized rat dorsal root ganglion neurons to release more calcitonin gene–related peptide (CGRP), which is involved in pain transmission, in response to low pH (which mimics an inflammatory state) or capsaicin (which activates ion channels involved in nociception). The two 11-hydroxy-epoxy-octadecenoates share a 3-hydroxy-Z-pentenyl-E-epoxide moiety, thus suggesting that this substructure could mediate nociceptor sensitization. In rats, intradermal hind paw injection of 11-hydroxy-12,13-trans-epoxy-(9Z)-octadecenoate elicited C-fiber–mediated sensitivity to thermal pain. In a randomized trial testing adjunctive strategies to manage refractory chronic headaches, reducing the dietary intake of linoleic acid was associated with decreases in plasma 11-hydroxy-12,13-trans-epoxy-(9Z)-octadecenoate, which correlated with clinical pain reduction. Human psoriatic skin had 30-fold higher 9-keto-12,13-trans-epoxy-(10E)-octadecenoate compared to control skin, and intradermal injection of this compound induced itch-related scratching behavior in mice. Collectively, these findings define a family of endogenous mediators with potential roles in pain and itch.
INTRODUCTION
Chronic pain and itch are common sources of personal suffering, disability, and societal expense (1–3). Current treatments often provide only partial or transient relief and have substantial side effects (4–8). The discovery of previously unknown endogenous mediators and mechanisms underlying pain and itch is needed to facilitate the development of targeted, effective, and safer interventions.
As the largest sensory organ, the skin is richly innervated by cutaneous nerve endings that can sense the microenvironment (4). Linoleic acid, the most abundant polyunsaturated fatty acid in the skin (5), is an “essential fatty acid” because a small amount (about 0.5% of energy) is needed in the diet to form the outer waxy epidermal barrier that prevents transepidermal water loss (6–8). Because itch and pain are common manifestations of cutaneous inflammatory conditions (3, 9) and linoleic acid is an endogenous substrate for conversion to bioactive lipid mediators (10), linoleic acid–derived mediators could be used to modulate cutaneous itch and pain.
We have previously shown in rats that increasing dietary linoleic acid increases linoleic acid derivatives in a dose-dependent manner in many tissues including the skin (11). A small human trial has found correlations between a low–linoleic acid diet intervention and decreased headache pain (12, 13) and between reductions in circulating linoleic acid and pain relief, suggesting that linoleic acid–derived mediators might contribute to sensory signaling. However, the specific derivatives of linoleic acid that mediate or modulate sensation and the molecular pathways involved in their biosynthesis and signaling are incompletely understood.
Here, we hypothesized that previously unknown linoleic acid–derived autacoids that are abundant in the skin may play a role in the genesis of pain and itch. This hypothesis was investigated by applying a systems-based, translational approach in rats and humans to (i) predict previously unknown lipid mediators based on tissue-specific precursor abundance and transcriptomic profiling of biosynthetic genes, (ii) synthesize predicted compounds by total chemical synthesis, (iii) identify and quantitate these mediators in rat and human tissues using liquid chromatography–tandem mass spectrometry (LC-MS/MS), (iv) determine whether the abundance of these compounds could be altered by diet and by a chronic inflammatory state, and (v) examine the algesic and pruritogenic activities of these lipids using ex vivo sensory neuronal cultures and in vivo behavioral testing. This approach allowed us to identify previously unknown linoleic acid–derived lipid mediators with potential roles in inflammatory skin disorders, pruritus, and nociception.
RESULTS
Predicting mediators based on precursor abundance and biosynthetic gene expression profiles
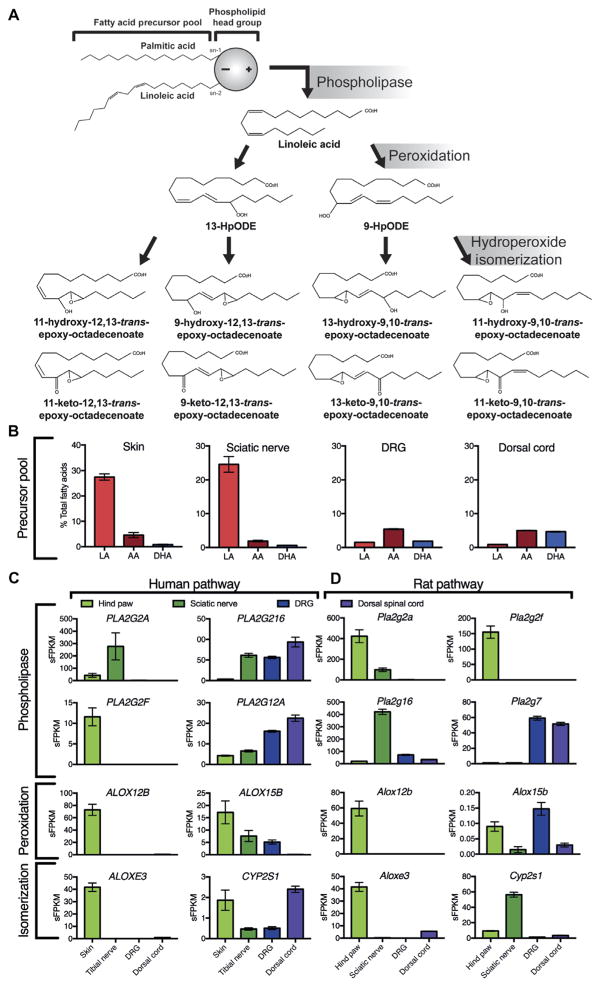
Precursor fatty acid compositions and RNA sequencing (RNA-seq) gene expression profiles of nociceptive (pain) circuit tissues were used to guide the prediction of previously unknown lipid mediators based on the biosynthetic pathway depicted in Fig. 1A. Linoleic acid was the most abundant polyunsaturated fatty acid in rat skin and sciatic nerve, accounting for 27.4 and 24.6% of total fatty acids, respectively (Fig. 1B). Linoleic acid was less abundant in sensory ganglia and in dorsal spinal cord.
Fig. 1. Proposed molecular pathways: precursor fatty acids and expression of genes coding for enzymes involved in biosynthesis of mediators in pain circuit tissues.
(A) Proposed biosynthetic pathways include lipase-mediated release of esterified linoleic acid (LA), lipoxygenase-mediated peroxidation, and hydroperoxide isomerization. (B) The abundance of precursor fatty acids in rat tissues (n = 4 rats for each tissue). (C and D) Quantitative biosynthetic gene expression in human (C) (n = 8, 8, 4, and 3 for the skin, tibial nerve, DRG, and dorsal cord, respectively) and rat (D) (n = 3, 4, 8, and 6 rats for hind paw, sciatic nerve, DRG, and dorsal cord, respectively) pain circuit tissues. Box plots and error bars in (B) to (D) indicate means and SEM. AA, arachidonic acid; DHA, docosahexaenoic acid; PLA2, phospholipase A2; Alox, lipoxygenase; Cyp, cytochrome P-450 epoxygenase; HpODE, hydroperoxyoctadecadienoate; sFPKM, standard fragments per kilobase of transcript per million mapped reads.
ALOX12B and ALOX15B genes, which encode enzymes that oxidize polyunsaturated fatty acids containing a 1,4-cis,cis-pentadiene system (14, 15), were abundantly expressed in human skin (Fig. 1C); Alox12b, but not Alox15b, was also well expressed in rat skin (Fig. 1D). ALOX15B was modestly expressed in human tibial nerve and dorsal root ganglia (DRG) but expressed to a lesser extent or absent in rat neural tissues comprising the nociceptive circuit (namely, sciatic nerve, DRG, and spinal cord dorsal horn). The ALOXE3 gene, which encodes an enzyme that isomerizes fatty acid hydroperoxides to form specific hydroxy-and keto-epoxide derivatives (16), was also abundantly expressed in rat and human skin but less abundantly expressed or absent in peripheral nerves, sensory ganglia, and dorsal cord. The CYP2S1 gene, which encodes another enzyme that isomerizes fatty acid hydroperoxides (17), was abundantly expressed in rat skin and, especially, sciatic nerve but expressed to a lesser extent or absent in human pain circuit tissues. Together, these precursor abundance and biosynthetic gene expression profiles were used to predict previously unknown lipid mediators.
Tissue-specific distributions of hydroxy-epoxy- and keto-epoxy-octadecenoates
On the basis of the high concentrations of linoleic acid and the moderate-to-high expression of genes encoding the biosynthetic enzymes noted above (Fig. 1), we predicted thattwopreviouslyunknown11-hydroxy-trans-epoxy-octadecenoates [11-hydroxy-12,13-trans-epoxy-(9Z)-octadecenoate (11H-12,13E-LA) and 11-hydroxy-9,10-trans-epoxy-(12Z)-octadecenoate (11H-9,10E-LA)], two previously unknown 11-keto-trans-epoxy-octadecenoates [11-keto-12,13-trans-epoxy-(9Z)-octadecenoate (11K-12,13E-LA) and 11-keto-9,10-trans-epoxy-(12Z)-octadecenoate(11K-9,10E-LA)], and four previously identified 9- or 13-hydroxy- or 9- or 13-keto-trans-epoxy-octadecenoates [9-hydroxy-12,13-trans-epoxy-(10E)-octadecenoate (9H-12,13E-LA), 13-hydroxy-9,10-trans-epoxy-(11E)-octadecenoate (13H-9,10E-LA), 9-keto-12,13-trans-epoxy-10E-octadecenoate (9K-12,13E-LA), and 13-keto-9,10-trans-epoxy-11E-octadecenoate (13K-9,10E-LA)] would be abundant in human and rat skin.
After total chemical synthesis of these eight linoleic acid derivatives for use as authentic standards (figs. S1 and S2, A to H), we used LC-MS/MS to quantify these mediators in rat and human tissues. Five of the eight mediators were present in rat skin but not in rat dorsal horn (table S1), indicating tissue specificity in accordance with our predictions (Fig. 1). All eight mediators were detected in human skin (table S2); seven of these eight mediators were confirmed by matching the ion spectra of the authentic standards and human skin extracts at characteristic retention times (fig. S2).
Increased abundance of free mediators in inflamed psoriatic human skin
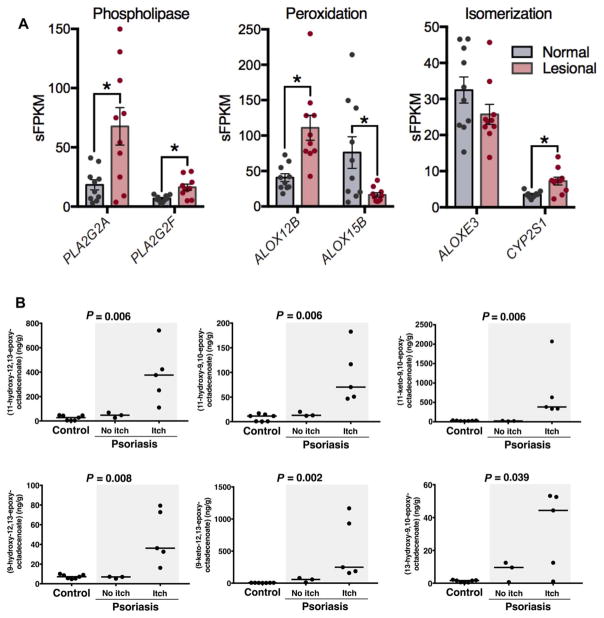
Psoriatic lesions exhibited higher expression of genes coding for lipase-mediated release(PLA2G2AandPLA2G2F), enzymatic peroxidation (ALOX12B), and hydro-peroxide isomerization (CYP2S1), compared to nonlesional psoriatic skin (Fig. 2A). Thus, increases in both local biosynthesis and release of esterified, preformed lipids could potentially contribute to the higher concentrations of hydroxy-epoxy- and keto-epoxy-octadecenoates observed in psoriatic lesions.
Fig. 2. Free hydroxy-epoxy- and keto-epoxy-octadecenoates are increased in human psoriatic skin lesions in participants reporting itch.
(A) Expression of genes coding the phospholipases, ALOX12B and CYP2S1 enzymes in human psoriatic skin lesions compared to nonlesional psoriatic skin (n = 10 participants per group). (B) Concentrations of free hydroxy-epoxy- and keto-epoxy-octadecenoates in psoriatic lesions compared to control human skin. Statistical analysis was performed using the Kruskal-Wallis test [n = 7, 3, and 5 participant specimens for control skin, psoriasis lesion (no itch), and psoriasis lesion (itch), respectively].
In human psoriatic skin lesions and nonpsoriatic control skin, we measured these mediators in both the nonesterified (free) and total (sum of free plus esterified) lipid fractions. There were no significant differences between psoriatic skin lesions and control skin in the total lipid fraction (table S2). However, six of the mediators (11H-12,13E-LA, 11H-9,10E-LA, 11K-9,10E-LA, 9H-12,13E-LA, 9K-12,13E-LA, and 13H-9,10E-LA) were markedly increased as free acids (the bio-active pool) in psoriatic lesions compared to control skin. The concentrations of free 11H-12,13E-LA and 9K-12,13E-LA were >6-fold and >30-fold higher in psoriatic lesions compared to control skin, respectively. The highest concentrations occurred in the lesions of psoriasis patients who reported itch (Fig. 2B).
The percentage of each mediator that was present as a bioavailable free acid differed according to the mediator. In control human skin, the percentage as free acid ranged from 0.05% for 13H-9,10E-LA to 44.4% for 11H-12,13E-LA. In psoriatic skin lesions, the percentage as free acid was significantly higher than control skin for 11H-12,13E-LA, 11K-9,10E-LA, 9K-12,13E-LA, and 13H-9,10E-LA (table S3). These findings support the hypothesis that there is increased enzymatic synthesis and/or release of these free acids from esterified lipids in chronic epidermal inflammation.
To determine whether measurements obtained from the circulating blood could provide surrogate markers of skin inflammation, we next quantified these mediators in serum from psoriatic patients and nonpsoriatic controls. Unlike the skin, the serum concentrations of these eight mediators did not differ by disease status (table S4).
Stimulation of rat sensory neurons by linoleic acid derivatives
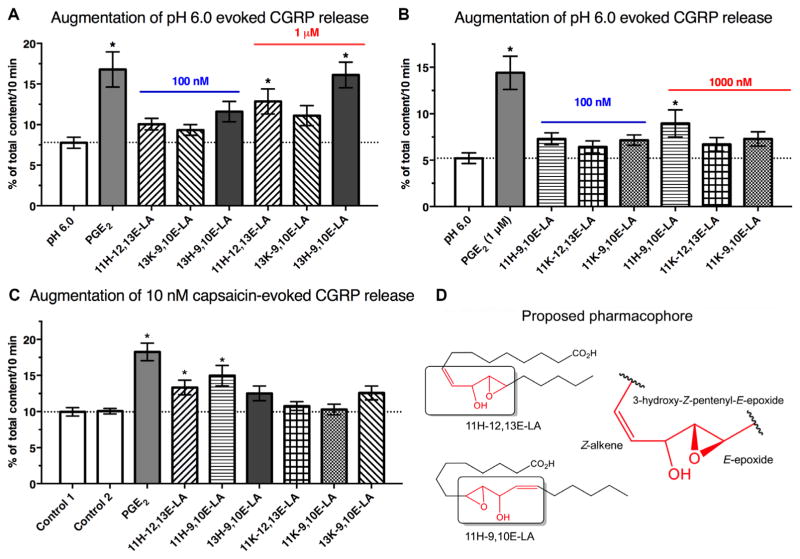
The release of calcitonin gene–related peptide (CGRP) by sensory neurons is implicated in the pathophysiology of numerous pain disorders (18, 19). The classic nociceptive mediator prostaglandin E2 (PGE2) does not significantly alter CGRP release at 1 μM concentrations at neutral pH (20, 21); however, PGE2 augments CGRP release evoked by low pH (which mimics an inflammatory state) or capsaicin (which activates ion channels involved in nociception) (20, 21), indicating DRG sensitization. To determine whether these linoleic acid derivatives sensitize DRG neurons, we tested each mediator in adult rat DRGs using a CGRP release assay, with PGE2 serving as a positive control. At neutral pH, neither PGE2 nor any of the other tested compounds directly stimulated CGRP release (fig. S3, A and B). However,11H-12,13E-LAand11H-9,10E-LAsignificantly increased both low pH–evoked and capsaicin-evoked CGRP release. 13H-9,10E-LA significantly increased CGRP release at low pH but not in response to capsaicin. Neither 9H-12,13E-LA nor any of the tested keto-epoxy-octadecenoates augmented low pH–evoked or capsaicin-evoked CGRP release (Fig. 3, A to C). These observations indicate that octadecenoate-induced sensitization was regioselective, with the most robust effects observed for compounds containing both a hydroxyl group at carbon 11 and an adjacent epoxide group. These two compounds share a 3-hydroxy-Z-pentenyl-E-epoxide moiety, identifying this substructure as a possible pharmacophore that mediates nociceptor sensitization (Fig. 3D).
Fig. 3. Regioselective augmentation of CGRP release from adult rat DRG neurons.
(A to C) Ex vivo CGRP release measured from adult rat DRG neuronal cultures in response to low pH (A and B) or capsaicin (C) [n = 9 wells (A) or 12 wells (B and C), each from three separate harvests]. (D) The shared 3-hydroxy-Z-pentenyl-E-epoxide moiety that is unique to these two lipids is the proposed pharmacophore mediating the effects of 11H-12,13E-LA and 11H-9,10E-LA. *P < 0.05 using analysis of variance (ANOVA) with Tukey’s post hoc test.
Pain- and itch-related behaviors in rodents after intradermal injection of mediators
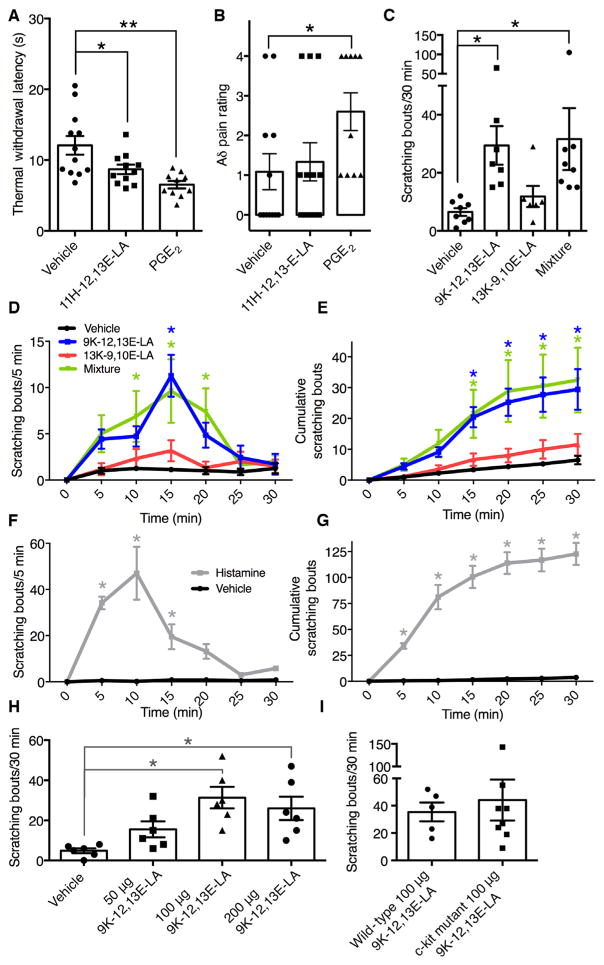
We next tested behavioral pain responses after intradermal injections of 11H-12,13E-LA because it was abundant as a free acid in inflamed human skin and augmented capsaicin and pH-stimulated CGRP release in rat sensory neurons. For these experiments, we compared the effects of the linoleic acid derivatives to vehicle and to the classic inflammatory mediator PGE2, which served as a positive control. After injection, C-fiber withdrawal latencies were decreased by 28 and 46% for 11H-12,13E-LA and PGE2, respectively, indicating nociceptive hyper-sensitivity (Fig. 4A). Intradermal injection of PGE2, but not 11H-12,13E-LA, also significantly enhanced the proportion of withdrawal responses after stimulation with a laser tuned to selectively excite Aδ sensory neuron fibers by generating a high rate of heating (Fig. 4B).
Fig. 4. Pain- and itch-related rodent behavior responses after intradermal injections.
(A and B) C-fiber withdrawal latency responses (A) and Aδ fiber stimulation responses after injection of mediators or vehicle control (B) (30 μg per injection; n = 12, 11, and 10 rats for vehicle, 11H-12,13E-LA, and PGE2, respectively). (C) Scratching bouts after injection of mediators (100 μg per mediator; n = 8, 7, 6, and 8 mice for vehicle, 9K-12,13E-LA, 13K-9,10E-LA, and the mixture, respectively). (D and E) Time course of scratching responses (D) and cumulative scratching bouts (E) evoked by the various mediators. (F and G) Time course (F) and scratching responses and cumulative scratching bouts (G) evoked by histamine (50 μg; n = 6 mice each for histamine and control). (H) Dose response for scratching bouts after injection of 9K-12,13E-LA (n = 6 mice for each dose). (I) Cumulative scratching bouts after injection of 9K-12,13E-LA in wild-type compared to c-Kit mutant mice (n = 5 and 8 for wild-type and c-Kit mutant mice, respectively). C-fiber withdrawal responses were compared using one-way ANOVA followed by Dunnett’s multiple comparisons test. Aδ pain ratings and itch responses were analyzed by Kruskal-Wallis test followed by Dunn’s multiple comparisons test. Time course itch-related scratching data (D to G) were analyzed by two-way repeated-measures ANOVA followed by Dunnett’s multiple comparison test to compare each treatment at each time point. *P < 0.05, **P < 0.01.
Next, to examine the effects of these eight mediators on itch, we used a mouse model quantifying itch-related scratching bouts over the first 30 min after intradermal injection into the nape of the neck. We observed that 9K-12,13E-LA, but not 13K-9,10E-LA, induced itch-related scratching behavior (Fig. 4C). Injection of a combination of 9K-12,13E-LA and 13K-9,10E-LA also significantly increased scratching behavior compared to vehicle but to the same degree as observed with 9K-12,13E-LA alone. Scratching responses evoked by 9K-12,13E-LA had a slower onset and more gradual tapering than those observed for histamine (Fig. 4, D to G), with the maximal responses observed at the 100-μg dose (Fig. 4H). Scratching responses evoked by 9K-12,13E-LA were comparable in wild-type and mast cell knockout (c-Kit mutant) mice (Fig. 4I). Together with our results showing that 9K-12,13E-LA was exclusively increased in psoriatic lesional skin of those with itch (Fig. 2B), these behavioral findings suggest that 9K-12,13E-LA may represent a novel itch mediator.
Regulation of mediators by dietary linoleic acid and correlation with clinical pain reduction
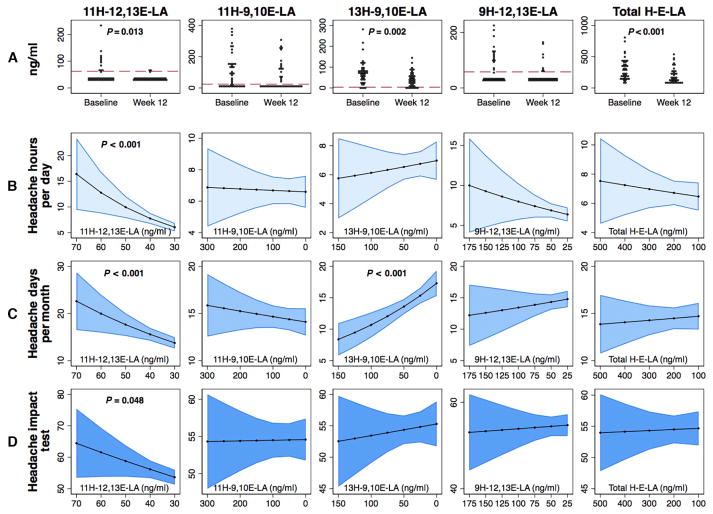
Next, to determine whether these mediators can be decreased by lowering the amount of their precursor linoleic acid in the diet, we used plasma samples from a completed randomized clinical trial testing a 12-week linoleic acid–lowering diet in patients with severe chronic daily headache (CDH) (12). We observed that five of these eight mediators were present in plasma (Fig. 5A). Two mediators—11H-12,13E-LA and 13H-9,10E-LA—were significantly decreased by the linoleic acid–lowering intervention; the sum of the four hydroxy-epoxide-octadecenoates was reduced by 41% (Fig. 5A).
Fig. 5. Association between diet-induced changes in plasma linoleic acid derivatives and pain-related end points in the CDH trial.
(A) Changes in plasma concentrations of individual and total hydroxy-epoxy-octadecenoates (H-E-LA) after decreased dietary intake of linoleic acid for 12 weeks in patients with CDH (n = 44 participants). Red dashed lines indicate the limit of quantitation; P values based on Wilcoxon matched-pairs signed-rank test. (B to D) Associations between diet-induced reductions in plasma hydroxy-epoxy-octadecenoate concentrations and headache hours per day (B) (n = 40 participants), headache days per month (C) (n = 44 participants), and headache impact (D) (n = 44 participants). Graphs include the headache outcomes (y axes) compared to the mediator concentrations at week 12 (x axis) based on a Poisson regression model controlling for each outcome and mediator concentration at baseline. 95% confidence intervals are shown in blue.
Moreover, we observed that diet-induced reductions in one of these mediators (11H-12,13E-LA), but not the others, were closely correlated with decreases in headache hours per day (Fig. 5B) and headache days per month (Fig. 5C). Each SD decrease in 11H-12,13E-LA was associated with 25 and 11% decreases in headache hours per day and headache days per month, respectively (Fig. 5D). Reduction in 11H-12,13E-LA also tended to correlate with improvements in overall headache impact (Fig. 5D) and physical function but was not related to psychological distress (table S5).
DISCUSSION
Here, we applied an interdisciplinary, translational approach in rodents and humans to discover and characterize a new family of endogenous lipid mediators of pain and itch. On the basis of our own findings and previous reports indicating that the skin is enriched in both linoleic acid and enzymes capable of peroxidation and hydro-peroxide isomerization, we predicted that the skin would have high concentrations of four previously unknown endogenous 11-hydroxy-trans-epoxy- or 11-keto-trans-epoxy-octadecenoates and four previously identified 9- or 13-hydroxy- or 9- or 13-keto-trans-epoxy-octadecenoates. As predicted, we measured substantial concentrations of 11H-12,13E-LA, 11H-9,10E-LA, 11K-12,13E-LA, and 11K-9,10E-LA in human skin. Notably, 11H-12,13E-LA was increased in inflamed psoriatic human skin, sensitized primary afferent DRG neurons in ex vivo CGRP release assays, and induced C-fiber–mediated pain-related hypersensitivity in rats. Moreover, plasma 11H-12,13E-LA correlated with headache frequency and impact in humans and was reduced by lowering the amount of its precursor linoleic acid in diet. In aggregate, these findings suggest that 11H-12,13E-LA could potentially be a mediator of pain modulated by diet and inflammation. 11H-9,10E-LA, which shares a 3-hydroxy-Z-pentenyl-E-epoxide moiety with 11H-12,13E-LA, was also increased in inflamed human skin and sensitized rat sensory neurons, suggesting that it might also contribute to inflammation-related sensitization. Our findings also confirm the presence of previously identified hydroxy-epoxy- and keto-epoxy-octadecenoates in human skin and provide insights into their potential biological actions (5, 8, 14, 15, 22–25). Consistent with previous findings, we observed relatively high concentrations of 13H-9,10E-LA in rodent and human skin (23, 24). In humans, 13H-9,10E-LA was found almost exclusively in the esterified lipid pool (table S3), but concentrations of free 13H-9,10E-LAwereninefold higher in psoriatic lesions as compared to control skin. Together with our finding that 13H-9,10E-LA augmented sensory neuron CGRP release in a low-pH environment, higher concentrations of this free acid in psoriatic skin suggest that it could potentially contribute to the hypersensitivity that accompanies cutaneous inflammation.
Another key finding reported here is the identification of 9K-12,13E-LA as an endogenous pruritogen that was increased in psoriatic lesions from patients who reported chronic itch but was not increased in lesions without itch. 9K-12,13E-LA has previously been detected in human plasma (26). In control human skin, >99% of 9K-12,13E-LA was in the esterified lipid fraction. The 30-fold higher concentration of 9K-12,13E-LA in the free fatty acid pool of psoriatic lesions compared to control skin suggests that 9K-12,13E-LA could potentially act as a signaling molecule in cutaneous inflammation. Consistent with this notion, we observed that injection of free 9K-12,13E-LA into mouse dermis caused mast cell–independent, itch-related scratching behavior. Because 9K-12,13E-LA is enriched in esterified skin lipids, preformed 9K-12,13E-LA can be released by lipases to directly stimulate pruritus, obviating the need for de novo biosynthesis. In this regard, we detected high expression of PLAG2A and PLAG2F in rat skin and especially inflamed human skin that could serve the relevant lipase function. Future studies are needed to delineate the specific molecular mechanisms and receptors mediating the pruritogenic properties of 9K-12,13E-LA.
We previously demonstrated in rats (11) that increasing dietary linoleic acid markedly increased the abundance of linoleic acid and its oxidized derivatives [for example, hydroxyoctadecadienoates (HODEs)] in tissues associated with idiopathic pain syndromes, including the skin. Moreover, in a trial involving 67 patients with chronic headaches, a linoleic acid–lowering dietary intervention decreased headache pain (12), and decreases in circulating linoleic acid were associated with clinical pain reduction (27), suggesting that linoleic acid or its derivatives could potentially contribute to pain in humans. Here, our finding that diet-induced reduction in circulating 11H-12,13E-LA was closely correlated with clinical pain reduction raises the possibility that high linoleic acid intakes could contribute to a biochemical susceptibility to develop chronic pain, in part by increasing the abundance of hydroxy-epoxy- and keto-epoxy-octadecenoates. Future trials are needed to determine whether lowering dietary linoleic acid can reduce these compounds in other tissues more directly implicated in nociception and to confirm whether such changes decrease chronic pain.
The current report introduces new mediators to the growing field of lipid mediators of pain and itch (10, 12, 28–33). Most of the work in this field has focused on mediators derived from arachidonic acid (34–36). Because linoleic acid is more abundant than arachidonic acid and other polyunsaturated fatty acids in the skin and epithelial tissues (11) and is also a substrate for enzymatic conversion to oxidized mediators (14, 15), linoleic acid–derived mediators may be uniquely positioned to regulate nociceptive and pruriceptive responses in these tissues. Patwardhan et al. (28, 30) have previously implicated HODEs and other well-known oxidized linoleic acid derivatives in peripheral and central nervous system nociceptive responses. In vivo cutaneous inflammatory responses are characterized by low pH and concurrent increases in numerous lipid and nonlipid mediators, which, together, are implicated in inflammation-associated hypersensitivity (4, 37). Under these conditions, HODEs could potentially be converted by cytochrome P-450 epoxygenases or lipoxygenases to form 9H-12,13E-LA, 13H-9,10E-LA, and other bioactive linoleic acid–derived mediators. Future studies are needed to determine which mediators or combinations are most responsible for the observed nociceptive responses.
This study has several important limitations. Because each of these eight linoleic acid derivatives was synthesized as a racemic mixture, future studies are needed to determine the tissue distributions and actions of individual stereoisomers. These compounds could potentially be converted in vivo to trihydroxyoctadecenoates (24) and dihydroxy-keto-octadecenoates; future studies are needed to determine whether some of the observed effects are due to these mediators. It is also important to point out that although higher concentrations of these compounds were observed in chronic psoriatic lesions, we used acute rodent behavioral testing models to assess their effects. Additional studies using infusions or repeated injections may be informative to assess the effects of chronic exposure to these mediators. Finally, future studies are needed to determine the specific cell types, receptors, and signaling pathways responsible for the actions of these mediators and to determine whether the effects differ by species or sex. These findings define a new family of endogenous lipid mediators regulated by diet and chronic inflammation and provide proof of concept for this systems-based, translational approach for discovering new lipid mediators of itch and pain.
MATERIALS AND METHODS
Sample preparation and data acquisition
Clinical sample preparation, rodent behavioral testing, ex vivo CGRP release assays, and all laboratory analyses were performed by investigators who were blinded to clinical data and treatment groups.
Data analysis
Normally distributed data were expressed as means ± SE and compared using Student’s t test (two groups) or one-way ANOVA (multiple groups), with corrections for multiple comparisons as described in figure and table legends. Data that were not normally distributed were expressed as median and interquartile ranges and compared using Wilcoxon rank-sum test (two groups) and Kruskal-Wallis test (multiple groups), with corrections for multiple comparisons as described in figure and table legends. P < 0.05 when adjusted for multiple comparisons was considered significant.
Rat tissue collection
The rat tissues analyzed in this study were obtained under protocols approved by the Institutional Animal Care and Use Committees of the National Institute of Dental and Craniofacial Research and the Clinical Center, National Institutes of Health (NIH). Male Sprague-Dawley rats were housed in pairs and given access to Rodent NIH-31M modified formula chow (Zeigler) and water ad libitum. To obtain hind paw, sciatic nerve, DRG, trigeminal ganglia, and dorsal horn tissue, rats were anesthetized with isoflurane and decapitated, and tissues were immediately dissected. Sections of the plantar surface of the hind paw were collected using a scalpel. Sciatic nerves were dissected starting from just distal to the sciatic notch and extending to just above the sciatic trifurcation. L4 and L5 DRGs were removed after laminectomy. The spinal cord was ejected from the vertebral column by hydraulic force using a syringe and Hanks’ balanced salt solution, and the left and right dorsal quadrants were isolated. Tissues were frozen immediately on dry ice and stored at −80°C until processed. Rat DRG and sciatic nerve RNA-seq data are available under project PRJNA313202 in the Sequence Read Archive (SRA) database (38).
Precursor fatty acid analysis of rat pain circuit tissues
Tissue fatty acids were analyzed as previously described (11). Briefly, samples were thawed, weighed, and homogenized in butylated hydroxytoluene (BHT)/methanol for fatty acid extraction according to the method of Folch et al. (39). BHT was added in methanol to reduce lipid oxidation during the procedures. The internal standard methyl tricosanoate (23:0) was added to each sample. This was followed by methylation with 14% BF3/methanol. The hexane extracts were concentrated to a small volume with a stream of nitrogen and transferred to microvials for gas chromatography (GC) analysis. Fatty acid methyl esters were analyzed with an HP-7890A GC equipped with a flame ionization detector (Hewlett-Packard) and a fused silica capillary column [DB-FFAP; 15 m × 0.100 mm (internal diameter) × 0.10 μm (film thickness), J&W Scientific]. The detector and injector temperatures were set to 250°C. The oven temperature program began at 150°C for 0.25 min and increased to 200°C at a rate of 10°C/min, then to 225°C at a rate of 3.5°C/min for 0.5 min, and finally increased to 245°C at a rate of 40°C/min, with a final hold for 15 min. Hydrogen was used as carrier gas at a linear velocity of 50 cm/s. A custom-mixed, 30-component, quantitative methyl ester standard containing 10 to 24 carbons and 0 to 6 double bonds was used to assign retention times and to ensure accurate quantification (Nu-Chek Prep 462). Fatty acid data were expressed as percentage of total peak area, which corresponded to weight % to within 5%, as demonstrated by quantitative standard mixtures. Internal standards were used to calculate tissue fatty acid concentrations. Fatty acid data were expressed as percentage of total fatty acids (%FA).
Collection of tissue and RNA purification for RNA-seq analyses
Four human L3 DRGs (three females and one male) were purchased from AnaBios from four different normal organ donors. Detailed information about these donors has been previously described (38, 40). Three female human medullary dorsal horn samples were collected at the level of the pyramidal decussation, and gray matter of the dorsal horn was isolated from fresh tissue by dissection as part of the collection procedure from the National Institute of Mental Health Human Brain Collection Core as described in (40). Rat and human samples were homogenized in QIAzol reagent (Qiagen Inc.) using a FastPrep-24 homogenizer (MP Biomedicals) or using a Polytron homogenizer (IKA) and purified using the RNeasy Mini Kit (Qiagen Inc.) with deoxyribonuclease digestion. RNA integrity number (RIN) was assessed after gel electrophoresis using an Agilent Bioanalyzer (Agilent Technologies). For rat tissues, samples with a RIN above 8.5 were sequenced. For human DRGs, samples with a RIN above 7 were sequenced. For other human samples, the highest possible RIN was obtained. The lowest sample included in this study was 5.5.
Alignment and quantification of RNA-seq count data
Rat RNA-seq data were aligned by STAR (version 2.4.2a) (41) using the rn6 genome build (Ensembl). Bam files resulting from this analysis were quantified using QoRTs (version 0.3.18) (42) and converted to raw read counts and FPKM. Data from human skin of the lower leg (one female and seven males) and tibial nerve (six males and two females) were accessed by selecting eight samples of high quality (based on RIN) from the Genotype-Tissue Expression repository. RPKM values were directly mined from data files available through the consortium (43). Psoriatic skin samples were accessed from the SRA database (PRJNA236547) (44). SRA and other human data were aligned and quantified using the MAGIC pipeline (45) and a genome target built in March 2016 based on RefSeq and AceView annotations (46). Genomic target files for MAGIC alignment are available upon request. Quantification and normalization of gene counts were performed by MAGIC and are reported in sFPKM.
Total chemical synthesis of hydroxy-epoxy- and keto-epoxy-octadecenoates
Total synthesis of each compound was performed by Cayman Chemical. Synthesized compounds were purified with flash chromatography and/or normal-phase high-performance liquid chromatography. Nuclear magnetic resonance (NMR) analysis indicated chemical shifts and coupling constants consistent with each chemical structure. Hydroxy-epoxy- or keto-epoxy-octadecenoates were analyzed by proton NMR in deuterated chloroform as their free acids or methyl esters as indicated (fig. S1).
Identification and quantitation of hydroxy- and keto-epoxide-octadecenoates with LC-MS/MS
Authentic standards prepared by total synthesis were used to identify and quantitate these eight compounds in human and rat tissues using UPLC-MS/MS. Briefly, solid-phase extraction (SPE) of oxylipins from biological matrices was performed using Strata X cartridges (33u, 200 mg/6 ml; Phenomenex). The cartridges were conditioned with 6 ml of methanol, followed by 6 ml of water before samples were extracted. Samples were washed with 6 ml of 10% methanol. The oxylipins were eluted with 6 ml of methanol into a glass tube containing 10 μl of 30% glycerol in methanol. The eluate was evaporated to dryness under a stream of nitrogen and reconstituted with 40 μl of methanol, and an aliquot (10 μl) was injected into the LC-MS/MS system. A UPLC (Shimadzu Scientific Instruments) coupled with a QTRAP 5500 (AB Sciex) was used for qualitative and quantitative analysis. Briefly, separation was performed on a ZORBAX RRHD Eclipse Plus C18 column (100 mm × 4 mm; 1.8 μm) (Agilent Corporation) consisting of (A) 12 mM ammonium acetate solution and acetic acid [100:0.02 (v/v)] and (B) 12 mM ammonium acetate and was composed of acetonitrile/water/acetic acid [90:10:0.02 (v/v/v)]. The flow rate was 0.5 ml/min. The column oven temperature was set at 30°C. The elution gradient conditions were as follows: 25 to 40% B from 0 to 2.0 min, 40 to 46% B from 2 to 8 min, 46 to 57% B from 8 to 9 min, 57 to 66% B from 9 to 20 min, 66 to 76% B from 20 to 22 min, 76 to 100% B from 22 to 27 min, held at 100% B from 27 to 33 min, and 100 to 25% B from 33.1 to 35 min. The mass spectrometer was operated in electrospray negative ionization using scheduled multiple reaction monitoring (sMRM) acquiring MRM data for each analyte with a retention time window of 90 s. The source parameters were set as follows: ion spray voltage, −4500 V; nebulizer gas (GS1), 65 psi; turbo gas (GS2), 70 psi; and turbo ion spray source temperature (TEM), 500°C. The analytes were quantified using MRM. MRM transitions, retention time, declustering potential, collision energy, and collision cell exit potential of each target analyte are listed in table S6. For hydroxy-epoxy-octadecenoates and keto-epoxy-octadecenoates with two or three isomeric peaks in the synthesized standards (fig. S2), quantitation was performed by sum peak area ratios of its related peak area component/peak area internal standard generated from Analyst 1.6.2 and plotting the best fit of total peak area ratios of analyte/peak area internal standard versus concentration in Microsoft Excel and was fitted to the equation y = ax + b. The MS/MS spectra were obtained by using enhanced product ion scan mode at a scan speed of 1000 Da/s. Collision-induced dissociation was performed using a collision energy of 35 V with a collision energy spread of 10. Data processing was performed using analyst software (version 1.6.2, AB Sciex). The identification of seven of the eight predicted compounds was confirmed by matching of the MS/MS spectra and retention times of endogenous linoleic acid derivatives from psoriatic skin samples with synthetic material using total ion mode (fig. S2).
Human studies with sample collection
Skin biopsies and serum collected from eight consecutive psoriasis participants and seven nonpsoriatic controls were included in the study (age range, 26 to 82 years) enrolled in an ongoing NIH observational study of psoriasis and cardiometabolic diseases (NCT01778569). The demographic and clinical characteristics of the study groups are shown in table S7. Study procedures were approved by the National Heart, Lung, and Blood Institute Institutional Review Board. All participants submitted written informed consent before enrollment. Briefly, a diagnosis of psoriasis was confirmed and quantified by a dermatologist using the Psoriasis Area Severity Index. The presence or absence of substantial itch was documented using a self-reported questionnaire. Corresponding controls were consecutively recruited to undergo the same testing as the psoriasis participants. All participants were free of any systemic antipsoriatic treatments or topical therapy within 2 weeks before biopsy. At baseline, 4-mm punch biopsies were obtained under local anesthesia from psoriatic plaque and unaffected skin. Biopsy sites were selected on the basis of active plaques and varied between subjects. However, biopsies of unaffected and control skin were predominantly from the buttocks. Whole blood from the same participants was collected in serum separator tubes, centrifuged, and immediately stored at −20°C until analysis.
The CDH trial was a randomized, 12-week trial designed to test the clinical and biochemical effects of diets low in linoleic acid (L6 intervention) with or without a concurrent increase in n–3 fatty acids (H3-L6 intervention) in a population with CDH. The trial was conducted at the University of North Carolina at Chapel Hill (UNC) from April 2009 to November 2011. Trial procedures were approved by the UNC Institutional Review Board, and the trial protocol, dietary compositions, and primary clinical and some biochemical findings were previously described (12, 47, 48). Briefly, adults meeting the CDH criteria of headaches >4 hours per day and >15 days per month for at least 3 months and a headache history of >2 years were recruited to participate. The demographic and clinical characteristics of the study groups are shown in table S8. Participants continued their usual physician care and were advised to continue taking medications as needed throughout the study. During the 4-week pre-intervention phase, participants continued their habitual diets and recorded headache characteristics in a daily headache diary. On completion of the run-in phase, participants were randomized to one of the two study diets, which lasted for 12 weeks. Linoleic acid was reduced in the study diets by restricting consumption of vegetable oils and other rich sources of linoleic acid and replacing them with vegetable oils and foods rich in monounsaturated and saturated fats. Plasma was collected at baseline and at the conclusion of the 12-week diet phase. We previously reported that the H3-L6 intervention produced marked reductions in headache frequency and severity and enhanced quality of life and function while reducing the use of acute pain medications (12, 13). Diet-induced changes in one or more families of n–6– or n–3–derived lipid autacoids likely contributed to these clinical benefits; however, the specific mechanisms responsible for these effects are unknown. Here, we use pre- and post-intervention plasma samples from this study to investigate (i) whether the study diets altered plasma concentrations of hydroxy- and keto-epoxide derivatives of linoleic acid using the Wilcoxon matched-pairs signed-rank test and (ii) whether changes in mediator concentrations correlated with clinical pain reduction using regression models adjusted for the baseline values of each outcome and mediator.
Preparation of solid tissues for LC-MS/MS analysis
Solid tissues (human skin, rat hind paw, or rat dorsal horn) were transferred into FastPrep Lysing Matrix tubes on ice (Lysing Matrix A for the skin and hind paw and Lysing Matrix D for the dorsal horn; MP Biomedicals), and at least eight times greater volume of ice-cold methanol with 0.02% (v/v) BHT and 0.02% (v/v) EDTA was immediately added to each tube. A known amount of internal standards was added to each sample, and samples were homogenized using a FastPrep-24 homogenizer (MP Biomedicals). Tissue homogenates were transferred to −80°C for 1 hour to precipitate proteins. Homogenates were centrifuged at 17,000g in 4°C for 10 min, and the supernatant was transferred to a new test tube. Half the supernatant was stored in −80°C until SPE purification and LC-MS/MS analysis (see above). To allow for the analysis of total lipid pools, we saponified the other half of the supernatant with 2.6% sodium carbonate (by weight) at 60°C for 30 min under gentle shaking. The solution was then neutralized (pH 5 to 7) using acetic acid and stored in −80°C overnight. Immediately before purification by SPE and LC-MS/MS analysis, lipid extracts (free and saponified total) were added to a ninefold greater volume of ice-cold water.
Preparation of plasma and serum for LC-MS/MS analysis
Two hundred microliters of plasma or serum was transferred to 500 μl of ice-cold methanol with 0.02% BHT and 0.02% EDTA and transferred to −80°C to precipitate proteins (as described above). A known amount of internal standards was then added, samples were centrifuged, and the supernatant was collected as described above. The supernatant was then added to a ninefold greater volume of ice-cold water, purified with SPE, and analyzed by LC-MS/MS, as described above.
Ex vivo sensory neuron sensitization assays (CGRP release assays)
For release experiments, the work was approved by the Animal Care and Use Committee at the Indiana University School of Medicine, Indianapolis, IN. Adult rat sensory neuronal cultures were prepared as previously described (49, 50). Cells were maintained for 10 to 12 days in F-12 medium (Invitrogen) supplemented with 10% horse serum, 2 mM glutamine, normocin (100 μg/ml), penicillin (50 μg/ml), streptomycin (50 μg/ml), 50 μM 5-fluoro-2′-deoxyuridine (Invitrogen), 150 μM uridine, and nerve growth factor (30 ng/ml) (Harlan Bioproducts for Science Inc.) in 3% CO2 at 37°C. On the day of the release experiments, cultures were washed with Hepes buffer [25 mM Hepes, 135 mM NaCl, 3.5 mM KCl, 2.5 mM CaCl2, 1 mM MgCl2, 3.3 mM D-glucose, and 0.1% bovine serum albumin (pH 7.4)] and maintained at 37°C. Cultures were then incubated with 0.4 ml of the same buffer in the absence or presence of drugs. Basal release was determined by exposing the cells to Hepes buffer alone for 10 min and then to buffer in the presence of mediators for 10 min to ascertain whether the compounds stimulated release. Cultures were then exposed to buffer containing 10 nM capsaicin or buffer with the pH adjusted to 6.0 in the absence or presence of mediators. Cells were then re-exposed to Hepes buffer without drugs for a 10-min incubation to reestablish basal release. After each incubation, the buffer was removed to measure the amount of CGRP using radio-immunoassay as previously described (51). At the end of each release experiment, cells were hypotonically lysed by exposing the cultures to 0.1 M HCl for 10 min and an aliquot was taken to measure total CGRP content in the cultures using radioimmunoassay. Release data are presented in femtomoles per well of cell per 10 min from three independent experiments from separate harvests. Statistical analysis was performed using ANOVA with Tukey’s post hoc test.
Rodent behavioral assays
To analyze pruriceptive (itch) behavior, we intradermally injected hydroxy-and keto-epoxide derivatives of linoleic acid (100 μg) or histamine (50 μg) into the nape of the neck of the mice (C57BL/6J and c-Kit mast cell–deficient mice, both from the Jackson Laboratory). Linoleic acid derivatives (9K-12,13E-LA or 13K-9,10E-LA) were injected independently and in combination [9K-12,13E-LA plus 13K-9,10E-LA (100 μg of each)] except in the c-Kit study in which 9K-12,13E-LA was injected alone. Pruriceptive behavior was quantified as the number of scratching bouts assessed over 30 min, as previously described (52). For the dose-response study, we injected 0, 50, 100, or 200 μg of 9K-12,13E-LA in 20 μl of vehicle and counted scratching bouts for 30 min.
To analyze nociceptive (pain) behavior, we intradermally injected 11H-12,13E-LA (30 μg) into the hind paw of male Sprague-Dawley rats. Baseline measurements were taken for all tests before injection. Aδ fiber– and C-fiber–mediated hind paw withdrawal responses were measured as previously described (53) by the delivery of thermal stimuli using a 980-nm infrared diode laser with a visible guide beam (LASS-10 M, Lasmed). Aδ fibers were selectively activated using a 100-ms 1.6-mm heat pulse that generates a high rate of heating (54). This stimulation paradigm results in a fast reflex response that was scored for intensity using a previously validated scale (55). Briefly, 1 is a simple withdrawal, 2 is a repeated tapping of the paw, 3 is orienting toward the paw, and 4 is a paw lick. C-fiber–mediated responses were measured after the delivery of a 5-mm-diameter slow temperature ramp by using a lower-intensity heating beam (53, 55).
Supplementary Material
Fig. S1. Proton NMR data for authentic standards.
Fig. S2. Identification of hydroxy-epoxy-octadecenoate and keto-epoxy-octadecenoates.
Fig. S3. Basal and low pH–evoked CGRP release in response to lipid mediators.
Table S1. Rat tissue concentrations of hydroxy-epoxy- and keto-epoxy-octadecenoates.
Table S2. Concentrations of hydroxy-epoxy- and keto-epoxy-octadecenoates in control skin and psoriatic skin.
Table S3. Percentage of total hydroxy-epoxy- and keto-epoxy-octadecenoates as free acids in control skin and psoriatic skin.
Table S4. Serum concentrations of free hydroxy-epoxy- and keto-epoxy-octadecenoates in control and psoriatic patients.
Table S5. Association between diet-induced changes in plasma linoleic acid derivatives and pain-related end points in the CDH trial.
Table S6. Mass spectrometry parameters established for the sMRM mode.
Table S7. Demographic and clinical characteristics of psoriatic patients and controls.
Table S8. Baseline demographic and clinical characteristics of patients with CDH.
Acknowledgments
We thank the patients who participated in the studies. We also thank C. Guo for her technical support in completing neuronal culture studies, J. DeBrecht for technical support with itch behavioral assays, J. Loewke for completing gas chromatography fatty acid analyses, and the UNC CDH trial investigators including J. Douglas Mann, K. Faurot, B. MacIntosh, and C. Lynch for contributions to the study that provided plasma samples and C. Serhan for expertise in lipid mediator profiling.
Funding: This work was supported in part by the Intramural Programs of the National Institute on Aging (NIA), the National Institute on Alcohol Abuse and Alcoholism, the National Heart, Lung, and Blood Institute, and the NIH Clinical Center Department of Perioperative Medicine; by a North Carolina State College of Veterinary Medicine startup fund (S.K.M); and by the NIH (NS069915 to M.R.V). The ex vivo studies were conducted in part in a facility constructed with the support from the Research Facilities Improvement Program grant number C06 RR015481-01 from the National Center for Research Resources, NIH. A.D. was supported by the Center for Neuroscience and Regenerative Medicine (grant to C.E.R.).
Footnotes
Author contributions: C.E.R. designed and directed the project, predicted the previously unknown mediators, helped with experiments, and wrote the first draft of the manuscript. Z.-X.Y. developed the LC-MS/MS methods for quantitation of lipid mediators, metabolite identification, and contributed to manuscript writing and revision. A.F.D. developed the methods for quantitation of total (free plus esterified) mediators in the skin and dorsal cord, contributed to experiments and data analysis, and helped write and revise the manuscript. G.S.K. performed the total chemical synthesis and purification and helped with manuscript preparation and revision. M.R.S. performed transcriptomic analyses. M.R.S., J.J.W., and S.K.M. conducted blinded behavior experiments and contributed to manuscript preparation and revision. D.M.L. collected and processed rodent tissue samples and revised the manuscript. N.N.M., A.V.S., and A.D. collected skin psoriasis samples and psoriatic clinical data and revised the manuscript. M.S.H., J.M.D., and D.Z. contributed to analysis and graphical depiction and helped revise the manuscript. S.M.-H. revised and referenced the manuscript. J.R.G. made the molecular pathway illustration and revised the manuscript. M.R.V. completed the blinded ex vivo DRG assays and contributed to manuscript preparation and revision. M.J.I. and A.J.M. designed and supervised the experiments, participated in tissue collection, and contributed to the manuscript preparation.
Competing interests: The NIA (NIH) has claimed intellectual property related to this discovery (application no. 62/529,846), with C.E.R. and G.S.K. named as inventors. All other authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.IOM Report. Relieving Pain in America: A Blueprint for Transforming Prevention, Care, Education, and Research. Institute of Medicine of the National Academies; 2011. www.ncbi.nlm.nih.gov/books/NBK914971. [PubMed] [Google Scholar]
- 2.Napolitano M, Megna M, Patruno C, Gisondi P, Ayala F, Balato N. Adult atopic dermatitis: A review. G Ital Dermatol Venereol. 2016;151:403–411. [PubMed] [Google Scholar]
- 3.Leader B, Carr CW, Chen SC. Pruritus epidemiology and quality of life. Handb Exp Pharmacol. 2015;226:15–38. doi: 10.1007/978-3-662-44605-8_2. [DOI] [PubMed] [Google Scholar]
- 4.Han SK, Simon MI. Intracellular signaling and the origins of the sensations of itch and pain. Sci Signal. 2011;4:pe38. doi: 10.1126/scisignal.2002353. [DOI] [PubMed] [Google Scholar]
- 5.Bowser PA, Nugteren DH, White RJ, Houtsmuller UM, Prottey C. Identification, isolation and characterization of epidermal lipids containing linoleic acid. Biochim Biophys Acta. 1985;834:419. doi: 10.1016/0005-2760(85)90016-5. [DOI] [PubMed] [Google Scholar]
- 6.Choque B, Catheline D, Delplanque B, Guesnet P, Legrand P. Dietary linoleic acid requirements in the presence of α-linolenic acid are lower than the historical 2 % of energy intake value, study in rats. Br J Nutr. 2015;113:1056–1068. doi: 10.1017/S0007114515000094. [DOI] [PubMed] [Google Scholar]
- 7.Guesnet P, Lallemand SM, Alessandri JM, Jouin M, Cunnane SC. α-Linolenate reduces the dietary requirement for linoleate in the growing rat. Prostaglandins Leukot Essent Fatty Acids. 2011;85:353–360. doi: 10.1016/j.plefa.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Hansen AE, Haggard ME, Boelsche AN, Adam DJ, Wiese HF. Essential fatty acids in infant nutrition. III. Clinical manifestations of linoleic acid deficiency. J Nutr. 1958;66:565–576. doi: 10.1093/jn/66.4.565. [DOI] [PubMed] [Google Scholar]
- 9.Brooks JP, Malic CC, Judkins KC. Scratching the surface—Managing the itch associated with burns: A review of current knowledge. Burns. 2008;34:751. doi: 10.1016/j.burns.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 10.Shapiro H, Singer P, Ariel A. Beyond the classic eicosanoids: Peripherally-acting oxygenated metabolites of polyunsaturated fatty acids mediate pain associated with tissue injury and inflammation. Prostaglandins Leukot Essent Fatty Acids. 2016;111:45–61. doi: 10.1016/j.plefa.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Ramsden CE, Ringel A, Majchrzak-Hong SF, Yang J, Blanchard H, Zamora D, Loewke JD, Rapoport SI, Hibbeln JR, Davis JM, Hammock BD, Taha AY. Dietary linoleic acid-induced alterations in pro- and anti-nociceptive lipid autacoids: Implications for idiopathic pain syndromes? Mol Pain. 2016;12:1744806916636386. doi: 10.1177/1744806916636386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramsden CE, Faurot KR, Zamora D, Suchindran CM, MacIntosh BA, Gaylord S, Ringel A, Hibbeln JR, Feldstein AE, Mori TA, Barden A, Lynch C, Coble R, Mas E, Palsson O, Barrow DA, Mann DJ. Targeted alteration of dietary n-3 and n-6 fatty acids for the treatment of chronic headaches: A randomized trial. Pain. 2013;154:2441–2451. doi: 10.1016/j.pain.2013.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramsden CE, Faurot KR, Zamora D, Palsson OS, MacIntosh BA, Gaylord S, Taha AY, Rapoport SI, Hibbeln JR, Davis JM, Mann JD. Targeted alterations in dietary n-3 and n-6 fatty acids improve life functioning and reduce psychological distress among patients with chronic headache: A secondary analysis of a randomized trial. Pain. 2015;156:587–596. doi: 10.1097/01.j.pain.0000460348.84965.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boeglin WE, Kim RB, Brash AR. A 12R-lipoxygenase in human skin: Mechanistic evidence, molecular cloning, and expression. Proc Natl Acad Sci USA. 1998;95:6744–6749. doi: 10.1073/pnas.95.12.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brash AR, Boeglin WE, Chang MS. Discovery of a second 15S-lipoxygenase in humans. Proc Natl Acad Sci USA. 1997;94:6148–6152. doi: 10.1073/pnas.94.12.6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Z, Schneider C, Boeglin WE, Marnett LJ, Brash AR. The lipoxygenase gene ALOXE3 implicated in skin differentiation encodes a hydroperoxide isomerase. Proc Natl Acad Sci USA. 2003;100:9162–9167. doi: 10.1073/pnas.1633612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bui P, Imaizumi S, Beedanagari SR, Reddy ST, Hankinson O. Human CYP2S1 metabolizes cyclooxygenase- and lipoxygenase-derived eicosanoids. Drug Metab Dispos. 2011;39:180–190. doi: 10.1124/dmd.110.035121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schou WS, Ashina S, Amin FM, Goadsby PJ, Ashina M. Calcitonin gene-related peptide and pain: A systematic review. J Headache Pain. 2017;18:34. doi: 10.1186/s10194-017-0741-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vasko MR. Prostaglandin-induced neuropeptide release from spinal cord. Prog Brain Res. 1995;104:367–380. doi: 10.1016/s0079-6123(08)61801-4. [DOI] [PubMed] [Google Scholar]
- 20.Southall MD, Vasko MR. Prostaglandin E2-mediated sensitization of rat sensory neurons is not altered by nerve growth factor. Neurosci Lett. 2000;287:33–36. doi: 10.1016/s0304-3940(00)01158-7. [DOI] [PubMed] [Google Scholar]
- 21.Vasko MR, Campbell WB, Waite KJ. Prostaglandin E2 enhances bradykinin-stimulated release of neuropeptides from rat sensory neurons in culture. J Neurosci. 1994;14:4987–4997. doi: 10.1523/JNEUROSCI.14-08-04987.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krieg P, Marks F, Fürstenberger G. A gene cluster encoding human epidermis-type lipoxygenases at chromosome 17p13.1: Cloning, physical mapping, and expression. Genomics. 2001;73:323–330. doi: 10.1006/geno.2001.6519. [DOI] [PubMed] [Google Scholar]
- 23.Zheng Y, Yin H, Boeglin WE, Elias PM, Crumrine D, Beier DR, Brash AR. Lipoxygenases mediate the effect of essential fatty acid in skin barrier formation: A proposed role in releasing omega-hydroxyceramide for construction of the corneocyte lipid envelope. J Biol Chem. 2011;286:24046–24056. doi: 10.1074/jbc.M111.251496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiba T, Thomas CP, Calcutt MW, Boeglin WE, O’Donnell VB, Brash AR. The precise structures and stereochemistry of trihydroxy-linoleates esterified in human and porcine epidermis and their significance in skin barrier function: Implication of an epoxide hydrolase in the transformations of linoleate. J Biol Chem. 2016;291:14540–14554. doi: 10.1074/jbc.M115.711267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nugteren DH, Christ-Hazelhof E, van der Beek A, Houtsmuller UM. Metabolism of linoleic acid and other essential fatty acids in the epidermis of the rat. Biochim Biophys Acta. 1985;834:429–436. doi: 10.1016/0005-2760(85)90017-7. [DOI] [PubMed] [Google Scholar]
- 26.Goodfriend TL, Ball DL, Egan BM, Campbell WB, Nithipatikom K. Epoxy-keto derivative of linoleic acid stimulates aldosterone secretion. Hypertension. 2004;43:358–363. doi: 10.1161/01.HYP.0000113294.06704.64. [DOI] [PubMed] [Google Scholar]
- 27.Ramsden CE, Zamora D, Makriyannis A, Wood JT, Mann JD, Faurot KR, MacIntosh BA, Majchrzak-Hong SF, Gross JR, Courville AB, Davis JM, Hibbeln JR. Diet-induced changes in n-3- and n-6-derived endocannabinoids and reductions in headache pain and psychological distress. J Pain. 2015;16:707–716. doi: 10.1016/j.jpain.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patwardhan AM, Akopian AN, Ruparel NB, Diogenes A, Weintraub ST, Uhlson C, Murphy RC, Hargreaves KM. Heat generates oxidized linoleic acid metabolites that activate TRPV1 and produce pain in rodents. J Clin Invest. 2010;120:1617–1626. doi: 10.1172/JCI41678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji RR, Xu ZZ, Strichartz G, Serhan CN. Emerging roles of resolvins in the resolution of inflammation and pain. Trends Neurosci. 2011;34:599–609. doi: 10.1016/j.tins.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patwardhan AM, Scotland PE, Akopian AN, Hargreaves KM. Activation of TRPV1 in the spinal cord by oxidized linoleic acid metabolites contributes to inflammatory hyperalgesia. Proc Natl Acad Sci USA. 2009;106:18820–18824. doi: 10.1073/pnas.0905415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morisseau C, Inceoglu B, Schmelzer K, Tsai HJ, Jinks SL, Hegedus CM, Hammock BD. Naturally occurring monoepoxides of eicosapentaenoic acid and docosahexaenoic acid are bioactive antihyperalgesic lipids. J Lipid Res. 2010;51:3481–3490. doi: 10.1194/jlr.M006007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Z-Z, Liu X-J, Berta T, Park C-K, Lü N, Serhan CN, Ji R-R. Neuroprotectin/protectin D1 protects against neuropathic pain in mice after nerve trauma. Ann Neurol. 2013;74:490–495. doi: 10.1002/ana.23928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dalli J, Zhu M, Vlasenko NA, Deng B, Haeggström JZ, Petasis NA, Serhan CN. The novel 13S,14S-epoxy-maresin is converted by human macrophages to maresin 1 (MaR1), inhibits leukotriene A4 hydrolase (LTA4H), and shifts macrophage phenotype. FASEB J. 2013;27:2573–2583. doi: 10.1096/fj.13-227728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davies P, Bailey PJ, Goldenberg MM, Ford-Hutchinson AW. The role of arachidonic acid oxygenation products in pain and inflammation. Annu Rev Immunol. 1984;2:335–357. doi: 10.1146/annurev.iy.02.040184.002003. [DOI] [PubMed] [Google Scholar]
- 35.Antonova M, Wienecke T, Olesen J, Ashina M. Prostaglandin E2 induces immediate migraine-like attack in migraine patients without aura. Cephalalgia. 2012;32:822–833. doi: 10.1177/0333102412451360. [DOI] [PubMed] [Google Scholar]
- 36.Chen L, Yang G, Grosser T. Prostanoids and inflammatory pain. Prostaglandins Other Lipid Mediat. 2013;104–105:58–66. doi: 10.1016/j.prostaglandins.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 37.Sun WH, Chen CC. Roles of proton-sensing receptors in the transition from acute to chronic pain. J Dent Res. 2016;95:135–142. doi: 10.1177/0022034515618382. [DOI] [PubMed] [Google Scholar]
- 38.Sapio MR, Goswami SC, Gross JR, Mannes AJ, Iadarola MJ. Transcriptomic analyses of genes and tissues in inherited sensory neuropathies. Exp Neurol. 2016;283:375–395. doi: 10.1016/j.expneurol.2016.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 40.Goswami SC, Thierry-Mieg D, Thierry-Mieg J, Mishra S, Hoon MA, Mannes AJ, Iadarola MJ. Itch-associated peptides: RNA-Seq and bioinformatic analysis of natriuretic precursor peptide B and gastrin releasing peptide in dorsal root and trigeminal ganglia, and the spinal cord. Mol Pain. 2014;10:44. doi: 10.1186/1744-8069-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartley SW, Mullikin JC. QoRTs: A comprehensive toolset for quality control and data processing of RNA-Seq experiments. BMC Bioinformatics. 2015;16:224. doi: 10.1186/s12859-015-0670-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swindell WR, Remmer HA, Sarkar MK, Xing X, Barnes DH, Wolterink L, Voorhees JJ, Nair RP, Johnston A, Elder JT, Gudjonsson JE. Proteogenomic analysis of psoriasis reveals discordant and concordant changes in mRNA and protein abundance. Genome Med. 2015;7:86. doi: 10.1186/s13073-015-0208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang W, Yu Y, Hertwig F, Thierry-Mieg J, Zhang W, Thierry-Mieg D, Wang J, Furlanello C, Devanarayan V, Cheng J, Deng Y, Hero B, Hong H, Jia M, Li L, Lin SM, Nikolsky Y, Oberthuer A, Qing T, Su Z, Volland R, Wang C, Wang MD, Ai J, Albanese D, Asgharzadeh S, Avigad S, Bao W, Bessarabova M, Brilliant MH, Brors B, Chierici M, Chu TM, Zhang J, Grundy RG, He MM, Hebbring S, Kaufman HL, Lababidi S, Lancashire LJ, Li Y, Lu XX, Luo H, Ma X, Ning B, Noguera R, Peifer M, Phan JH, Roels F, Rosswog C, Shao S, Shen J, Theissen J, Tonini GP, Vandesompele J, Wu PY, Xiao W, Xu J, Xu W, Xuan J, Yang Y, Ye Z, Dong Z, Zhang KK, Yin Y, Zhao C, Zheng Y, Wolfinger RD, Shi T, Malkas LH, Berthold F, Wang J, Tong W, Shi L, Peng Z, Fischer M. Comparison of RNA-seq and microarray-based models for clinical endpoint prediction. Genome Biol. 2015;16:133. doi: 10.1186/s13059-015-0694-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thierry-Mieg D, Thierry-Mieg J. AceView: A comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(suppl 1):S12. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramsden CE, Mann JD, Faurot KR, Lynch C, Imam ST, MacIntosh BA, Hibbeln JR, Loewke J, Smith S, Coble R, Suchindran C, Gaylord SA. Low omega-6 vs. low omega-6 plus high omega-3 dietary intervention for chronic daily headache: Protocol for a randomized clinical trial. Trials. 2011;12:97. doi: 10.1186/1745-6215-12-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacIntosh BA, Ramsden CE, Faurot KR, Zamora D, Mangan M, Hibbeln JR, Mann JD. Low-n-6 and low-n-6 plus high-n-3 diets for use in clinical research. Br J Nutr. 2013;110:559–568. doi: 10.1017/S0007114512005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burkey TH, Hingtgen CM, Vasko MR. Isolation and culture of sensory neurons from the dorsal-root ganglia of embryonic or adult rats. Methods Mol Med. 2004;99:189–202. doi: 10.1385/1-59259-770-X:189. [DOI] [PubMed] [Google Scholar]
- 50.Kelley MR, Jiang Y, Guo C, Reed A, Meng H, Vasko MR. Role of the DNA base excision repair protein, APE1 in cisplatin, oxaliplatin, or carboplatin induced sensory neuropathy. PLOS ONE. 2014;9:e106485. doi: 10.1371/journal.pone.0106485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen JJ, Barber LA, Dymshitz J, Vasko MR. Peptidase inhibitors improve recovery of substance P and calcitonin gene-related peptide release from rat spinal cord slices. Peptides. 1996;17:31–37. doi: 10.1016/0196-9781(95)02091-8. [DOI] [PubMed] [Google Scholar]
- 52.Mishra SK, Hoon MA. The cells and circuitry for itch responses in mice. Science. 2013;340:968–971. doi: 10.1126/science.1233765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitchell K, Lebovitz EE, Keller JM, Mannes AJ, Nemenov MI, Iadarola MJ. Nociception and inflammatory hyperalgesia evaluated in rodents using infrared laser stimulation after Trpv1 gene knockout or resiniferatoxin lesion. Pain. 2014;155:733–745. doi: 10.1016/j.pain.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blivis D, Haspel G, Mannes PZ, O’Donovan MJ, Iadarola MJ. Identification of a novel spinal nociceptive-motor gate control for Aδ pain stimuli in rats. eLife. 2017;6:e23584. doi: 10.7554/eLife.23584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mitchell K, Bates BD, Keller JM, Lopez M, Scholl L, Navarro J, Madian N, Haspel G, Nemenov MI, Iadarola MJ. Ablation of rat TRPV1-expressing Aδ/C-fibers with resiniferatoxin: Analysis of withdrawal behaviors, recovery of function and molecular correlates. Mol Pain. 2010;6:94. doi: 10.1186/1744-8069-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Proton NMR data for authentic standards.
Fig. S2. Identification of hydroxy-epoxy-octadecenoate and keto-epoxy-octadecenoates.
Fig. S3. Basal and low pH–evoked CGRP release in response to lipid mediators.
Table S1. Rat tissue concentrations of hydroxy-epoxy- and keto-epoxy-octadecenoates.
Table S2. Concentrations of hydroxy-epoxy- and keto-epoxy-octadecenoates in control skin and psoriatic skin.
Table S3. Percentage of total hydroxy-epoxy- and keto-epoxy-octadecenoates as free acids in control skin and psoriatic skin.
Table S4. Serum concentrations of free hydroxy-epoxy- and keto-epoxy-octadecenoates in control and psoriatic patients.
Table S5. Association between diet-induced changes in plasma linoleic acid derivatives and pain-related end points in the CDH trial.
Table S6. Mass spectrometry parameters established for the sMRM mode.
Table S7. Demographic and clinical characteristics of psoriatic patients and controls.
Table S8. Baseline demographic and clinical characteristics of patients with CDH.