Abstract
Many Proteobacteria synthesize acyl-homoserine lactone (AHL) molecules for use as signals in cell density-dependent gene regulation known as quorum sensing (QS) and response. AHL detection protocols are essential to QS researchers and several techniques are available, including a 14C-AHL radiolabel assay. This assay is based on the uptake of radiolabeled methionine by living cells and conversion of the radiolabel into S-adenosylmethionine (SAM). The radiolabeled SAM is then incorporated into AHL signal by an AHL synthase enzyme. Here we describe a methodology to perform the AHL radiolabel assay, which is unbiased, relatively fast, and very sensitive compared to other AHL detection protocols.
Keywords: 14C-carboxy-methionine, quorum sensing, QS, LuxI, acyl homoserine lactone, AHL, acyl-HSL
1 Introduction
Quorum sensing overview
It is now appreciated that bacteria are able to sense one another and coordinate their group activities. Many bacteria monitor their population densities, and change their gene expression patterns as appropriate, using a process known as quorum sensing and response [1,2]. Over 100 species of Proteobacteria synthesize small diffusible chemicals, originally named ‘autoinducers’ but now called ‘quorum sensing (QS) signals’, to mediate cell density-dependent gene regulation. The QS signal accumulates in the surrounding medium during growth. Because QS signals are free to diffuse into and out of the cytoplasm into the environment [3,4], environmental and cellular concentrations are equal. At high bacterial populations, a QS signal can accumulate to high levels and once a threshold concentration is reached, the QS signal interacts with a cognate receptor protein. Once the receptor protein is bound by QS signal, it controls gene expression as activator or repressor, depending upon the particular system [1,2].
Two genes are required for such QS systems in Proteobacteria: luxR- and luxI- type genes. The luxR-encoded proteins (R proteins) are the cognate receptors for the QS signals and function as transcriptional regulators. The luxI homologs encode AHL QS signal synthases (I proteins), which use the common metabolites S-adenosylmethionine (SAM) and organic acids activated via acyl carrier protein (ACP)- or coenzyme A (CoA)-linkage [5–10] as AHL substrates (Figures 1 and 2). Most of the AHLs described have acyl side chains comprised of fatty acyl groups of varying carbon lengths (4–18) and substitutions (ACP-derived), but more recently AHL signals comprised of aromatic acid [9,11] and branched amino acid [6] side chains (CoA-derived) have been discovered (Figure 1).
Figure 1.
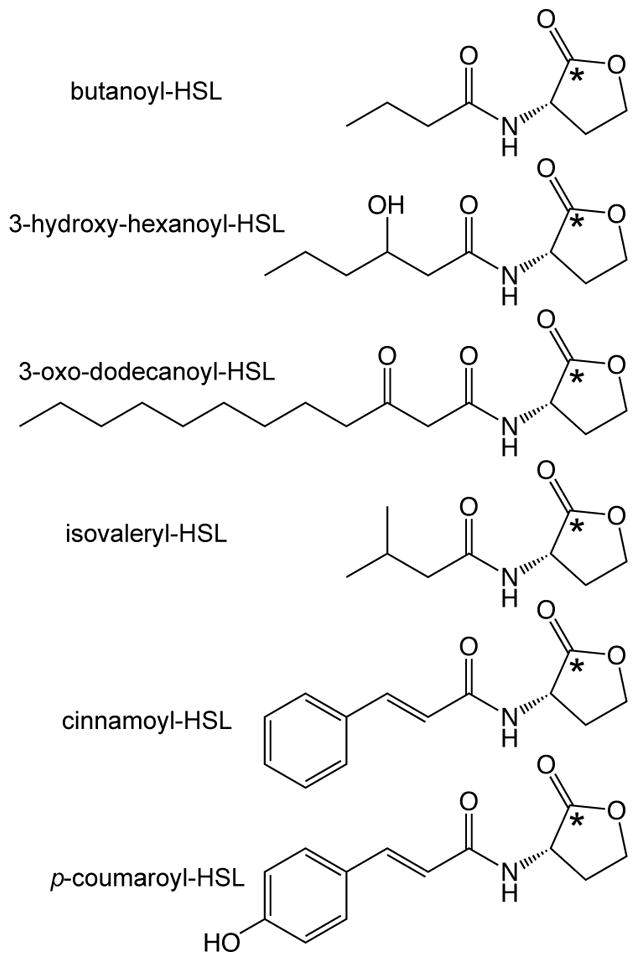
Examples of AHL QS signal structures. The top three compounds are representative of the ‘typical’ AHL molecule, which has a side chain derived from fatty acid biosynthesis (acyl-acyl carrier protein substrates). The bottom three compounds are the more recently discovered AHL signals derived from branched chain amino acid biosynthesis (isovaleryl-HSL) and aromatic acid degradation (cinnamoyl-HSL, p-coumaroyl-HSL), which utilize acyl-coenzyme A (CoA)-linked substrates [6,9]. The asterisk indicates the location of the 14C-label incorporated in the 14C-AHL during the radiolabel assay protocol described in this chapter.
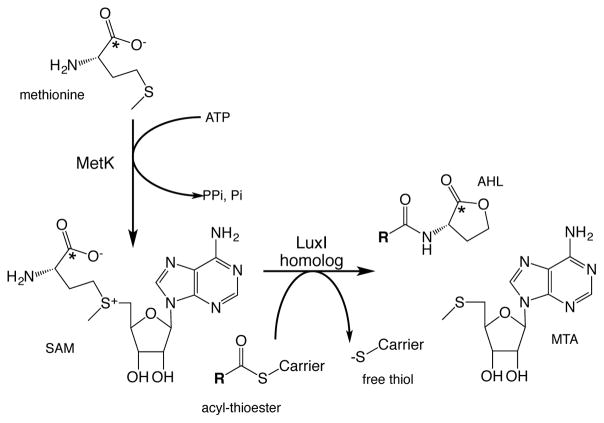
Figure 2.
Pathway illustrating flow of radiolabel into AHL from methionine, via S-adenosylmethionine (SAM). The radiolabeled carbon is indicated by asterisk. Bacteria convert some labeled methionine to SAM via the enzymatic action of MetK, a SAM synthetase. A portion of the 14C-SAM is used by a LuxI-type AHL synthase as substrate for AHL synthesis. The acyl-thioester carrier is either acyl carrier protein or coenzyme A, depending upon the LuxI enzyme. MTA is the co-product 5′-methylthioadenosine.
Techniques to detect and identify AHL compounds
A variety of protocols have been developed to detect and quantify AHL signals: bioassays [12], enzyme-linked immunosorbant assay (ELISA) [13], mass spectrometry [14], and the radiolabel assay [15,16] described here. Each assay has strengths and weaknesses, and in practice all of these techniques are used in some combination by the field. Detailed descriptions of AHL detection by bioassay (Chapter 1), mass spectrometry (Chapter 4), and ELISA (Chapter 5) are provided elsewhere in this volume, but we briefly discuss these techniques here in order to better understand the relative benefits (and limitations) of the radiolabel assay.
Most often, AHLs are detected using a series of bioassay reporter strains. These strains contain a LuxR-type regulator and a gene promoter fusion that requires R protein bound to an AHL signal for expression (for examples see Chapter 1 and references [6,9,12,17]). Bioassays can quantitate the total AHL levels present in a sample if a standard curve is generated using known amounts of purified AHL compound. The simple-to-perform and inexpensive bioassay reporters are important tools for the QS field, but they do have limitations. Each reporter responds only to a subset of known AHLs, therefore multiple reporters must be employed to screen for an undefined AHL signal. Also when the non-fatty acyl-linked AHLs were first identified, the p-coumaroyl- and isovaleryl-linked AHLs (Figure 1), they were not detected by the existing repertoire of bioassay reporters [6,9]. There are also examples of non-HSL compounds activating reporter strains [18,19], which could confound results. Additionally, bioassays are sensitive to inhibition by other compounds sometimes present in sample extracts (e.g. we had difficulties detecting AHLs in cystic fibrosis patient sputum extracts using bioassays, but not using the radiolabel assay described here [15]).
Mass spectrometry (MS) analyses are essential to QS research (see Chapter 4), especially with regards to structural elucidation of undefined AHL compounds, and can be quantitative when an internal deuterated AHL standard is included [14]. However, not all laboratories have ready access to a mass spectrometer and many of the MS techniques rely on the presence of a fragment ion at (m/z+H) 102 (corresponding to aminobutyrolactone), which is observed in most, but not all [9], AHLs. Recently ELISA techniques (see Chapter 5) have been used to detect acyl-HSLs, but require custom monoclonal antibodies and cannot differentiate between closed HSL ring compounds (acyl-HSL, active QS signal) and lactonized ones (acyl-homoserine, inactive QS signal) [13].
The 14C-radiolabel assay detailed here is similar to that first described by Eberhard et al [20]. AHL-synthesizing bacteria are fed radiolabeled L-[1-14C]-methionine (14C-met), which is transported into the cell and a portion is converted into SAM via enzymes like the Escherichia coli MetK SAM synthetase. The 14C-SAM is used by LuxI-type synthases as the substrate for the conserved HSL ring in the AHL QS signal (diagrammed in Figure 2). 14C-AHLs are solvent extracted from cell-free supernatants, separated by HPLC and radioactivity in the HPLC fractions is measured by liquid scintillation counting. Because all LuxI-synthesized QS signals utilize SAM as the HSL ring substrate, all HSL-type signals (including novel AHL structures) are detected by the radiolabel technique so long as the bacterium tested is capable of assimilating exogenous 14C-met. The presence of a single radiolabeled carbon in the HSL ring, regardless of side chain moiety, allows for easy comparison of relative HSL compounds in a single sample (Figure 3 illustrates the detection of both major and minor AHL compounds present in Pseudomonas cultures). It is possible that some bacteria could incorporate 14C-met into other non-AHL, extracellular, solvent-extractable compounds; however, we have not found this to be the case for the dozens of Proteobacteria species we have tested using the radiolabel assay.
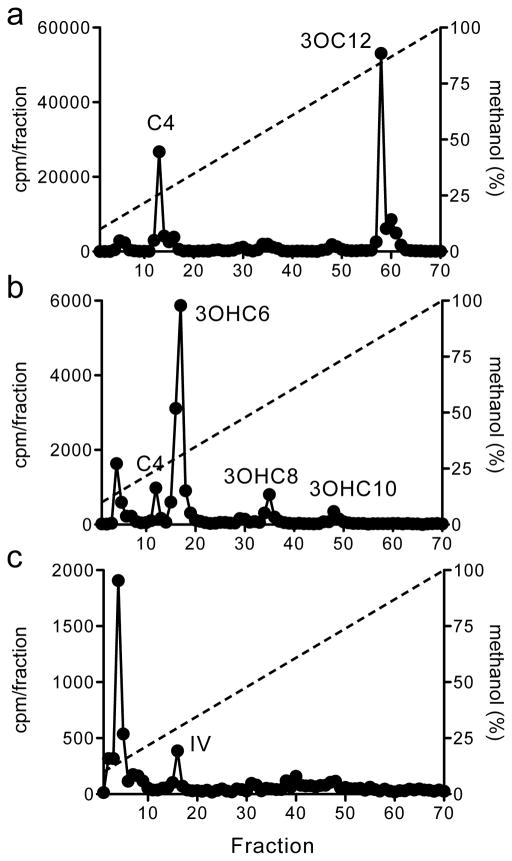
Figure 3.
HPLC profiles of 14C-AHLs synthesized by (A) Pseudomonas aeruginosa PAO1 [25] (labeled for 30 min), (B) Pseudomonas chloroaphis GM17 [26] (labeled for 2 hours), and (C) Bradyrhizobium japonicum USDA110 [27] (labeled for 1 day). In all graphs the x-axis indicates the fraction numbers that were collected over a 10–100% methanol-in-water gradient. The left y-axis denotes the counts per minute (CPM) of radiolabel in each fraction (black circles) and the right y-axis indicates the methanol concentration of the HPLC run (dashed line). The synthetic AHL compound that co-elutes with the observed radiolabel peak is abbreviated as follows: butanoyl-HSL (C4), 3-oxo-dodecanoyl-HSL (3OC12), 3-hydroxy-hexanoyl-HSL (3OHC6), 3-hydroxy-octanoyl-HSL (3OHC8), 3-hydroxy-decanoyl-HSL (3OHC10), and isovaleryl-HSL (IV). Radioactivity that eluted in the column void volume (fraction 4) is presumed to be unincorporated methionine.
Although some labs prefer to avoid using radioactivity due to licensing and disposal cost issues, the radiolabel protocol for monitoring AHL production is a powerful technique that shows no bias with regards to AHL side chain and is faster and more sensitive than many traditional bioassays. Variations of this protocol have been applied in many ways: to detect novel AHL signals [6,11]; to detect AHL production in situ for complex environments such as cystic fibrosis patient sputum and biofilm reactors [15,16]; to generate significant amounts of radiolabeled AHL compound [21]; to test for positive autoregulation of AHL synthase genes [20,22]; to study AHL synthesis patterns during bacterial growth [23]; to identify preferred side chain substrates [11], and to assess inhibitors of AHL synthesis in whole bacterial cells [24]. Here we provide detailed protocols for AHL detection during bacterial growth and identification of preferred side chain substrates using the radiolabel assay.
2 Materials
2.1 14C-labeling of AHLs during bacterial growth
L-[1-14C]-methionine in sterile water (50–60 mCi/mmol, see Notes 1 and 2)
Bacterium of interest (see Note 3)
Methionine-free medium appropriate for growing your bacterium of interest (see Notes 4 and 5)
(Optional, for aromatic substrate specificity assay) Mixture of aromatic compounds at a stock concentration of 0.1 M (see Note 6) or other naturally occurring mixtures (see Note 7)
2.2 Extraction and separation 14C-AHLs
Acidified ethyl acetate (0.1 ml glacial acetic acid per 1 liter ethyl acetate)
HPLC-grade methanol and water solvents
1.5-ml plastic snapcap tubes
15-ml plastic centrifuge tubes (ethyl acetate-resistant)
3-ml polyethylene transfer pipettes (ethyl acetate-resistant)
N-evap nitrogen evaporator; alternatively incubate your extract-containing tubes in a warm water bath without cap in a fume hood until solvent has evaporated
High pressure liquid chromatography (HPLC) system (200 μl loading loop) using a C18-reverse phase HPLC column
Fraction collector outfitted with solvent-resistant, 7-ml polyethylene scintillation vials
AHL standards or defined AHL-positive bacteria (see Note 8)
2.3 Detection of 14C-AHLs
Scintillation counting cocktail (see Note 9)
Liquid scintillation counter
(Alternative) Some laboratories have an in-line scintillation detector, which can be used in lieu of fraction collection and liquid scintillation counting
3 Methods
Protocol for AHL detection during bacterial growth
This is a general protocol for 14C-radiolabeling of AHL signals during bacteria growth, but it should be modified according to the needs of the particular strain as noted below. To illustrate the range of labeling times and 14C-HSL production among strains, we performed the radiolabel assay using three different bacteria (Figure 3, see Note 4): Pseudomonas aeruginosa PAO1 [25], Pseudomonas chloroaphis GM17 [26], and Bradyrhizobium japonicum USDA110 [27].
Grow your bacterium of interest in a small volume (usually 5-ml) of methionine-free medium with the appropriate growth conditions (e.g. temperature, aerobic with shaking, anaerobic, photosynthetic, etc.) until the culture reaches desired density, usually late logarithmic phase or early stationary phase (see Note 10).
-
Radiolabel cells by adding 5 μCi (~90 nmol, see Note 1) of [1-14C]-methionine (14C-met) to the culture for an appropriate labeling time. The radiolabeling duration can vary greatly, from minutes to days, depending on the organism and growth conditions. For the experiments in Figure 3 shorter labeling times used for the fast-growing bacteria P. aeruginosa (30 minutes) and P. chloroaphis (2 hours), while a longer labeling time was required for the slow-growing B. japonicum (1 day).
You can test whether the labeling incubation time is sufficient by monitoring the amount of 14C-met incorporated into the cell pellet: i) sample a small amount (50 μl) of culture and aliquot to a 1.5-ml snapcap tube, ii) determine the amount of radioactivity in culture by pipetting a 5 μl sample into a scintillation vial containing 4-ml of scintillation cocktail fluid and then count 14C-radioactivity using a liquid scintillation detector, iii) take the remaining 45 μl of culture and pellet the cells by centrifugation, aliquot 5 μl of the cell-free supernatant to a scintillation vial containing 4-ml of scintillation cocktail fluid and count 14C-radioactivity using a liquid scintillation detector, iv) compare 14C-counts from the cell-free supernatant vs. the total culture to estimate the amount of radioactivity incorporated into cell material. If ~10% (or more) of the 14C-counts are associated with the cell pellet, this is usually sufficient for a successful radiolabel assay. However, longer incubation times typically yield increased 14C-AHL-product. This step also confirms that your organism is capable of 14C-methionine transport (see Note 11).
After labeling for a sufficient time (determined as described in the previous step), you are ready to extract the 14C-AHLs from the bacterial culture. Pellet cells by centrifugation and transfer the cell-free supernatant to a solvent-resistant tube using a 3-ml polypropylene transfer pipette. Extract the supernatant twice with equal volumes of acidified ethyl acetate (EtAc), collect and combine both organic phases.
EtAc solvent is evaporated and AHLs dried under a gentle stream of N2 gas with warming (using N-evap nitrogen evaporator or similar). Resuspend the dried sample in 100 μl of 50% methanol-in-water.
Separate the 100-μl sample by HPLC on a C18-reverse phase column by using a 10–100% (vol/vol) methanol in water gradient (1 ml/min flow, 70-min profile). Collect seventy 1-ml (1 minute) fractions in in scintillation vials using an automated fraction collector.
Add 4-ml of scintillation cocktail to each fraction and count with a scintillation detector. AHL assignment is determined by co-elution with known AHL standards. As illustrated in Figures 3 and 4, the amount 14C-AHL produced can vary by ~10,000-fold depending upon the bacterium, labeling time, and experimental setup (see Note 12).
Figure 4.
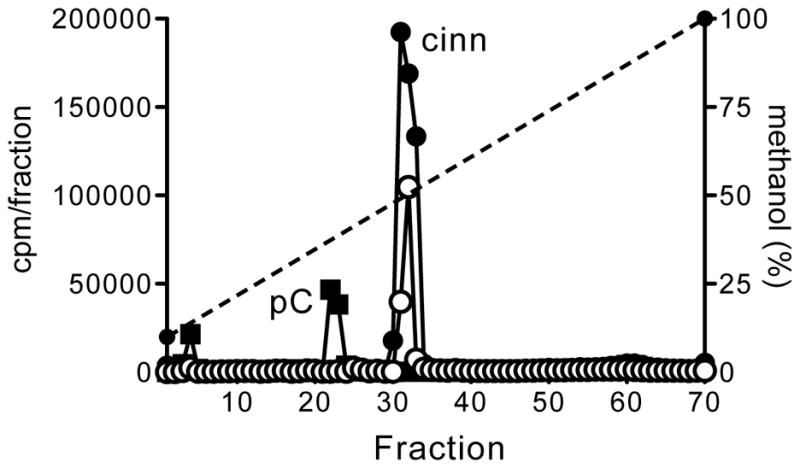
HPLC profiles of 14C-AHLs synthesized by AHL synthases when grown in presence of potential substrates added exogenously. Cells expressing the LuxI-type AHL synthases from either Rhodopseudomonas palustris CGA009 [9] (RpaI, squares) or Bradyrhizobium ORS278 [11] (BraI, circles) were grown in the presence of a mixture of 13 aromatic acids (black-filled shapes) or cinnamic acid only (white-filled shapes) (see Note 13). The x-axis indicates the fraction numbers that were collected over a 10–100% methanol-in-water gradient. The left y-axis denotes the counts per minute (CPM) of radiolabel in each fraction (circles or squares) and the right y-axis indicates the methanol concentration of the HPLC run (dashed line). The synthetic AHL compound that co-elutes with the observed radiolabeled peak is abbreviated as follows: p-coumaroyl-HSL (pC) and cinnamoyl-HSL (cinn). Radioactivity that eluted in the column void volume (fraction 4) is presumed to be unincorporated methionine.
Protocol for aromatic substrate specificity assay
Some bacteria, such as Rhodopseudomonas [9] and Bradyrhizobium [11] species, utilize exogenous aromatic substrates as the AHL side chain (Figure 1). To screen for the preferred aromatic substrate of a particular LuxI homolog, the radiolabel assay can be performed using cells grown in the presence of aromatic acid mixtures. This allows the AHL synthase enzyme to select its preferred side-chain substrate [11]. To illustrate this technique here (Figure 4), we used a heterologous host (see Note 13) to express the AHL synthase from either R. palustris CGA009 (RpaI) or Bradyrhizobium sp. ORS278 (BraI).
Grow the bacterium in a small volume (usually 5-ml) of methionine-free, minimal medium with the appropriate growth conditions (e.g. temperature, aerobic with shaking, anaerobic, photosynthetic, etc.) until the appropriate culture density is reached (mid-logarithmic phase, A660 of 0.5 for the example shown in Figure 4).
Dilute the culture (~1:5) in fresh medium. For each experimental comparison condition (e.g. no addition, aromatic acid mixture addition, etc.) use 5-ml of freshly diluted culture in a 15-ml plastic tube and then add the potential substrates to be tested to the culture (see Note 7). For the experiments represented in Figure 4, we added either a mixture of 13 aromatic acids (see Note 6) or cinnamate alone (0.1 mM final concentrations).
-
Radiolabel cells by adding 5 μCi (~90 nmol, see Note 1) of [1-14C]-methionine (14C-met) to the culture for an appropriate labeling time. As described above, the labeling duration can vary greatly depending on the bacterium and growth conditions. For the experiments in Figure 4 we radiolabeled cells for 20 hours.
You can test whether the labeling incubation time is sufficient by monitoring the amount of 14C-met incorporated into the cell pellet: i) sample a small amount (50 μl) of culture and aliquot to a 1.5-ml snapcap tube, ii) determine the amount of radioactivity in culture by pipetting a 5 μl sample into a scintillation vial containing 4-ml of scintillation cocktail fluid and then count 14C-radioactivity using a liquid scintillation detector, iii) take the remaining 45 μl of culture and pellet the cells by centrifugation, aliquot 5 μl of the cell-free supernatant to a scintillation vial containing 4-ml of scintillation cocktail fluid and count 14C-radioactivity using a liquid scintillation detector, iv) compare 14C-counts from the cell-free supernatant vs. the total culture to estimate the amount of radioactivity incorporated into cell material. If ~10% (or more) of the 14C-counts are associated with the cell pellet, this is usually sufficient for a successful radiolabel assay. However, longer incubation times typically yield increased 14C-AHL-product. This step also confirms that your organism is capable of 14C-methionine transport (see Note 11).
After labeling for a sufficient time (determined as described in the previous step), you are ready to extract the 14C-AHLs from the bacterial culture. Pellet cells by centrifugation and transfer the cell-free supernatant to a solvent-resistant tube using a 3-ml polypropylene transfer pipette. Extract the supernatant twice with equal volumes of acidified ethyl acetate (EtAc), collect and combine both organic phases.
EtAc solvent is evaporated and AHLs dried under a gentle stream of N2 gas with warming (using N-evap nitrogen evaporator or similar). Resuspend the dried sample in 100 μl of 50% methanol-in-water.
Separate the 100-μl sample by HPLC on a C18-reverse phase column by using a 10–100% (vol/vol) methanol in water gradient (1 ml/min flow, 70-min profile). Collect seventy 1-ml (1 minute) fractions in in scintillation vials using an automated fraction collector.
Add 4-ml of scintillation cocktail to each fraction and count with a scintillation detector. Examples of 14C-radiolabel profiles of cells grown in the presence of exogenous aromatic substrates are shown in Figure 4. AHL assignment is determined by co-elution with known AHL standards.
Acknowledgments
This work was sponsored by the following funding sources: i) the Agricultural and Food Research Initiative Competitive Grants Program 2010-65108-20536 from the US Department of Agriculture, ii) the US Army Research Office Grant W911NF0910350, iii) the US Public Health Service (USPHS) Grant GM59026, and iv) the Genomic Science Program, U.S. Department of Energy, Office of Science, Biological and Environmental Research, as part of the Plant Microbe Interfaces Scientific Focus Area (http://pmi.ornl.gov). Oak Ridge National Laboratory is managed by UT-Battelle LLC, for the U.S. Department of Energy under contract DE-AC05-00OR22725.
Footnotes
Previous experiments using similar protocols have used [2-14C]-methionine [20] or uniformly labeled 3H-methionine [22], but these compounds are no longer commercially available. To our knowledge, the only current supplier of L-[1-14C]-methionine is American Radiolabel Chemicals, St. Louis, MO (ARC-0271A, 50–60 mCi/mmol, 0.1 mCi/ml).
Confirm the purity of the 14C-methionine when you first receive a new lot by separating a small amount (1 μl) over the HPLC column; all radioactive counts should pass through the column in the void volume.
LuxI-type AHL synthases have only been identified in members of the Proteobacteria. As the number of bacterial genomes sequenced increase, so does the number of potentially interesting AHL systems. We often identify potentially interesting strains for AHL screening by searching their sequenced genomes for the AHL synthase motif pfam00765 [28].
The growth medium is specific to the bacterium you are testing and, if possible, should be methionine-free. For the experiments used in this chapter (Figures 3 and 4) we used the following media: Jensen’s medium with 0.3% glycerol [29] (Pseudomonas aeruginosa PAO1), M9 minimal medium [30] with 10 mM succinate (Pseudomonas chloroaphis GM17), AG medium [31] (Bradyrhizobium japonicum USDA110), and PM medium with 10 mM succinate [32] (Rhodopseudomonas palustris CGA009).
In some cases, we have also grown bacteria in rich (methionine-replete) medium, pelleted cells by centrifugation, and resuspended the cell pellet in phosphate buffered saline with a carbon and energy source (e.g. 10 mM glucose) for the 14C-labeling reaction [21].
We made filter-sterilized stock solutions of 13 individual aromatic salts, at a concentration of 0.1 M and pH 7, including: benzoate, p-coumarate, m-coumarate, o-coumarate, cinnamate, caffeate, ferulate, methoxycinnamate, sinapate, p-hydroxybenzoate, vanillate, phenylalanine, and tryptophan. Compounds were diluted 1000-fold in culture medium to a final concentration of 0.1 mM for substrate labeling experiments.
Although here we use defined aromatic acid mixtures, one could imagine using naturally occurring mixtures of potential substrates such as extracts of plant tissue or sediments.
Many synthetic AHL standards can be purchased from variety of vendors including Sigma-Aldrich (St. Louis, MO), Caymen Chemical Co. (Ann Arbor, MI), or the University of Nottingham (Nottingham, UK, https://www.nottingham.ac.uk/quorum/compounds.htm). Alternatively, one could extract AHLs from bacteria with well-defined AHL QS systems, such as P. aeruginosa PAO1 (Figure 3A).
Over the years we have tested a variety of scintillation counting cocktails, including those marketed as non-hazardous or biodegradable fluids, but we obtained the best results using a xylene-based scintillation fluid
Keep in mind that the AHL synthase must be expressed under the laboratory growth conditions employed for the 14C-radiolabel (and other AHL detection methods) to work. Some AHL-type systems have additional regulatory controls, such as the presence of plant metabolites [9,33] or host redox conditions [34], that must be satisfied before AHL signals will accumulate. If the genome sequence is available for the bacterium of interest, cloning the LuxI-homolog gene into a heterologous host under a constitutive or inducible promoter can circumvent some of these issues.
There are some bacteria that cannot transport exogenous methionine, for example the chemolithoautotroph Acidithiobacillus ferrooxidans. Although this organism synthesizes an AHL molecule, we were unable to utilize the radiolabel protocol to detect AHL in this organism because the 14C-met is not assimilated into the AHL signal.
The amount of 14C-AHL produced is influenced by several things including rates of bacterial growth and methionine incorporation, presence of AHL-degrading enzymes [35], and AHL synthesis and accumulation rates. For example, P. aeruginosa PAO1 typically produces low μM levels of its AHL signals [36], while B. japonicum USDA110 maximally produces isovaleryl-HSL (IV-HSL) in the low nM range [6].
For the experiments represented in Figure 4, we used an rpaI-mutant of R. palustris [9] as the heterologous host because this bacterium can metabolize a wide variety of aromatic compounds, no longer produces any endogenous AHL signal, and can constitutively express LuxI homologs using the vector pBBR1MCS-5 [37]. Cells were grown photoheterotrophically in PM-succinate plus 100 μg/ml of gentamycin [9].
References
- 1.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication. Annu Rev Cell Dev Biol. 2005;21:19–46. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 2.Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GP. Quorum-sensing in gram-negative bacteria. FEMS Microbial Review. 2001;25:365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x. [DOI] [PubMed] [Google Scholar]
- 3.Kaplan HB, Greenberg EP. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. Journal of Bacteriology. 1985;163:1210–1214. doi: 10.1128/jb.163.3.1210-1214.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearson JP, VanDeldon C, Iglewski BH. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. Journal of Bacteriology. 1999;181:1203–1210. doi: 10.1128/jb.181.4.1203-1210.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanzelka BL, Greenberg EP. Quorum sensing in Vibrio fischeri: evidence that S-adenosylmethionine is the amino acid substrate for autoinducer synthesis. J Bacteriol. 1996;178:5291–5294. doi: 10.1128/jb.178.17.5291-5294.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindemann A, Pessi G, Schaefer AL, Mattmann ME, Christensen QH, Kessler A, Hennecke H, Blackwell HE, Greenberg EP, Harwood CS. Isovaleryl-homoserine lactone, an unusual branched-chain quorum-sensing signal from the soybean symbiont Bradyrhizobium japonicum. Proc Natl Acad Sci USA. 2011;108:16765–16770. doi: 10.1073/pnas.1114125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.More MI, Finger LD, Stryker JL, Fuqua C, Eberhard A, Winans SC. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science. 1996;272:1655–1658. doi: 10.1126/science.272.5268.1655. [DOI] [PubMed] [Google Scholar]
- 8.Parsek MR, Val DL, Hanzelka BL, Cronan JEJ, Greenberg EP. Acyl homoserine-lactone quorum-sensing signal generation. Proc Natl Acad Sci USA. 1999;96:4360–4365. doi: 10.1073/pnas.96.8.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaefer AL, Greenberg EP, Oliver CM, Oda Y, Huang JJ, Bittan-Banin G, Peres CM, Schmidt S, Juhaszova K, Sufrin JR, Harwood CS. A new class of homoserine lactone quorum-sensing signals. Nature. 2008;454(7204):595–599. doi: 10.1038/nature07088. nature07088 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Schaefer AL, Val DL, Hanzelka BL, Cronan JEJ, Greenberg EP. Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc Natl Acad Sci USA. 1996;93:9505–9509. doi: 10.1073/pnas.93.18.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahlgren NA, Harwood CS, Schaefer AL, Giraud E, Greenberg EP. Aryl-homoserine lactone quorum sensing in stem-nodulating photosynthetic bradyrhizobia. Proc Natl Acad Sci USA. 2011;108:7183–7188. doi: 10.1073/pnas.1103821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaefer AL, Hanzelka BL, Parsek MR, Greenberg EP. Detection, purification, and structural elucidation of the acylhomoserine lactone inducer of Vibrio fischeri luminescence and other related molecules. Meth Enzymology. 2000;304:288–301. doi: 10.1016/s0076-6879(00)05495-1. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Kremmer E, Gouzy MF, Clausen E, Starke M, Wullner K, Pfister G, Hartmann A, Kramer PM. Development and characterization of rat monoclonal antibodies for N-acylated homoserine lactones. Anal Bioanal Chem. 2010;398:2655–2667. doi: 10.1007/s00216-010-4017-9. [DOI] [PubMed] [Google Scholar]
- 14.Gould TA, Herman J, Krank J, Murphy RC, Churchill ME. Specificity of acyl-homoserine lactone synthases examined by mass spectrometry. J Bacteriol. 2006;188:773–783. doi: 10.1128/JB.188.2.773-783.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Quorum-sensing signals indicate the cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;12:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 16.Schaefer AL, Greenberg EP, Parsek MR. Acylated homoserine lactone detection in Pseudomonas aeruginosa biofilms by radiolabel assay. Meth Enzymology. 2001;336:41–47. doi: 10.1016/s0076-6879(01)36576-x. [DOI] [PubMed] [Google Scholar]
- 17.Zhu J, Chai Y, Zhong Z, Li S, Winans SC. Agrobacterium bioassay strain for ultrasensitive detection of N-acylhomoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium huakuii. Appl Environ Microbiol. 2003;69:6949–6953. doi: 10.1128/AEM.69.11.6949-6953.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muh U, Hare BJ, Duerkop BA, Schuster M, Hanzelka BL, Heim R, Olson ER, Greenberg EP. A structurally unrelated mimic of a Pseudomonas aeruginosa acyl-homoserine lactone quorum-sensing signal. Proc Natl Acad Sci USA. 2006;103:16948–16952. doi: 10.1073/pnas.0608348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajamani S, Bauer WD, Robinson JB, Farrow JM, Pesci EC, Teplitski M, Gao M, Sayre RT, Phillips DA. The vitamin riboflavin and its derivative lumichrome activate the LasR bacterial quorum sensing receptor. Mol Plant Microbe Interact. 2013;21:1184–1192. doi: 10.1094/MPMI-21-9-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eberhard A, Longin T, Widrig CA, Stranick SJ. Synthesis of the lux gene autoinducer in Vibrio fischeri is positively autoregulated. Archives of Microbiology. 1991;155:294–297. [Google Scholar]
- 21.Leadbetter JR, Greenberg EP. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J Bacteriol. 2000;182:6921–6926. doi: 10.1128/jb.182.24.6921-6926.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gray KM, Greenberg EP. Physical and functional maps of the luminescence gene cluster in an autoinducer-deficient Vibrio fischeri strain isolated from a squid light organ. J Bacteriol. 1992;174:4384–4390. doi: 10.1128/jb.174.13.4384-4390.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blosser-Middleton RS, Gray KM. Multiple N-acyl homoserine lactone signals of Rhizobium leguminosarum are synthesized in a distinct temporal pattern. J Bacteriol. 2001;183:6771–6777. doi: 10.1128/JB.183.23.6771-6777.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christensen QH, Grove TL, Booker SJ, Greenberg EP. A high-throughput screen for quorum-sensing inhibitors that target acyl-homoserine lactone synthases. Proc Natl Acad Sci USA. 2013;110:13815–13820. doi: 10.1073/pnas.1313098110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stover CK, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:960–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 26.Brown SD, Utturkar SM, Klingeman DM, Johnson CM, Martin SL, Land ML, Lu TS, Schadt CW, Doktycz MJ, Pelletier DA. Twenty-one Pseudomonas genomes and ninteen genomes from diverse bacteria isolated from the rhizophere and endosphere of Populus deltoides. Journal of Bacteriology. 2012;194:5991–5993. doi: 10.1128/JB.01243-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taneko T, et al. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Research. 2002;9:189–197. doi: 10.1093/dnares/9.6.189. [DOI] [PubMed] [Google Scholar]
- 28.Schaefer AL, Lappala CR, Morlen RP, Pelletier DA, Lu TY, Lankford PK, Harwood CS, Greenberg EP. LuxR- and LuxI-type quorum-sensing circuits are prevalent in members of the Populus deltoides microbiome. Appl Environ Microbiol. 2013;79:5745–5752. doi: 10.1128/AEM.01417-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jensen JE, Fecyca T, Campell JN. Nutritional factors controlling exocellular protease production by Pseudomonas aeruginosa. J Bacteriol. 1980;144:844–847. doi: 10.1128/jb.144.2.844-847.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller JH. Experiments in Molecular Genetics. Vol. 433. Cold Spring Harbor, NY: 1972. [Google Scholar]
- 31.Sadowsky MJ, Tully RE, Cregan PB, Keyser HH. Genetic diversity in Bradyrhizobium japonicum serogroup 123 and its relation to genetype-specific nodulation of soybean. Appl Environ Microbiol. 1987;53:2624–2630. doi: 10.1128/aem.53.11.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim M-K, Harwood CS. Regulation of benzoate-CoA ligase in Rhodopseudomonas palustris. FEMS Microbial Lett. 1991;83:199–204. [Google Scholar]
- 33.Fuqua WC, Winans SC. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol. 1994;176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bose JL, Kim U, Barkowski W, Gunsalus RP, Overley AM, Stabb EV. Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol Microbiol. 2007;65:538–553. doi: 10.1111/j.1365-2958.2007.05809.x. [DOI] [PubMed] [Google Scholar]
- 35.Huang JJ, Peterson A, Whiteley M, Leadbetter JR. Identification of QuiP, the product of gene PA1032, as the second acyl-homoserine latone acylase of Pseudomonas aeruginosa PAO1. Appl Environ Microbiol. 2006;72:1190–1197. doi: 10.1128/AEM.72.2.1190-1197.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chugani S, Kim BS, Phattarasukol S, Brittnacher MJ, Choi SH, Harwood CS, Greenberg EP. Strain-dependent diveresity in the Pseudomonas aeruginosa quorum-sensing regulon. Proc Natl Acad Sci USA. 2012;109:E2823–E2831. doi: 10.1073/pnas.1214128109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RMI, Peterson KM. Four new derivatives of the broad-host-range clining vector pBBR1MCS, carrying different antibiotic resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]