Abstract
Purpose of review
The purpose of this review is to discuss recent observations of epigenetic changes related to the complex pathogenesis of systemic vasculitides and their contribution to the field.
Recent findings
There have been new observations of epigenetic changes in vasculitis and their potential role in disease pathogenesis in antineutrophil cytoplasmic antibody-associated vasculitis, giant-cell arteritis, Kawasaki disease, Behçet’s disease, and IgA vasculitis. Some of this recent work has focused on the efficacy of using DNA methylation and miRNA expression as clinical biomarkers for disease activity and how DNA methylation and histone modifications interact to regulate disease-related gene expression.
Summary
DNA methylation, histone modification, and miRNA expression changes are all fruitful ground for biomarker discovery and therapeutic targets in vasculitis. Current knowledge has provided targeted and suggested effects, but in many cases, has relied upon small cohorts, cosmopolitan cell populations, and limited knowledge of functional interactions. Expanding our knowledge of how these epigenetic mechanisms interact in a disease-specific and cell-specific manner will help to better understand the pathogenesis of systemic vasculitis.
Keywords: DNA methylation, epigenetics, histone modification, microRNA, systemic vasculitis
INTRODUCTION
Systemic vasculitides are a heterogeneous group of complex inflammatory diseases of unknown cause. They are characterized by histological evidence of leukocyte infiltration, inflammation, and necrosis of the vessel wall and vascular occlusion [1]. Numerous genetic loci have been associated with increased risk of vasculitis, with human leukocyte antigen (HLA) genes encoding major histocompatibility complex (MHC) proteins being most robust and pointing toward the importance of the immune system in pathogenesis [2]. Environmental risk factors include exposure to silica dust, unknown viral or bacterial infections, drugs, farming occupations as well as complex factors like age [3–6]. The contribution of genetics alone to the pathogenesis of a systemic vasculitis varies with manifestations, but does not account for the entirety of the risk. Epigenetic mechanisms governing gene expression sit at the interface of genetic and environmental factors related to a variety of diseases [7]. Epigenetics is the study of hereditary, phenotypic traits that can alter the chromosome without changing the underlying genetic sequence [8]. DNA methylation, histone modifications, and noncoding RNA are epigenetic mechanisms that control gene expression and regulate cellular development, differentiation and activity (extensively reviewed in [9▪]).
DNA methylation is an epigenetic mechanism that consists of the addition of a methyl group to cytosines, primarily within CpG dinucleotides, catalyzed by DNA methyltransferases (DNMTs). De-novo DNA methylation is conducted primarily by DNMT3A and DNMT3B which are essential during the gestational development of mammals. Although DNMT1 is primarily responsible for maintaining established methylation patterns from cell to cell, the extent to which their functions overlap is still being explored [10]. DNMT function and targeting are regulated by complex systems including DNMT expression levels, RNA molecules, amino-acid residue modifications, genomic sequence and methylation status, histone tail signatures, transcription factor availability, and chromatin accessibility [10]. DNMT1 is also capable of recruiting histone deacetylase 1 (HDAC1) which promotes the formation of heterochromatin through the removal of acetyl groups from histone proteins, silencing gene expression and providing a link between DNA methylation and histone modification [11].
Histone modification is an epigenetic mechanism that regulates the dynamic chromatin structure and subsequently gene expression [12]. The functional opposition to HDACs, histone acetyl-transferases (HATs), promote the formation of euchromatin that is permissive to protein-DNA interactions [12]. Acetylation is only one of a multitude of histone modifications found in the genome which can include ubiquitylation, residue-specific methylation, and phosphorylation [13]. Identifying patterns of histone modifications and their relationship to gene expression has provided a way to understand how chromatin structure controls the regulation of cellular functions, and has led to the identification of ‘bivalent chromatin’ that contains both permissive (e.g., H3K4me2) and repressive (e.g., H3K27me3) histone modifications poised for expression depending on cellular requirements, which is vital during cell development [9▪].
Noncoding RNAs are transcribed but not translated into proteins, and act as an epigenetic mechanism to regulate gene expression. The most studied are microRNAs (miRNAs), around 22 nucleotides in length that regulate posttranscriptional gene silencing through translational control of mRNA molecules. miRNAs target the 3′ untranslated region of their target mRNA molecule and control their stability and protein interactions [14]. miRNA expression is controlled by other epigenetic mechanisms and itself controls these mechanisms as an ‘epigenetics-miRNA regulatory circuit’ that, when perturbed, can contribute to disease pathogenesis [15].
The focus of this review is to discuss our current knowledge of the role epigenetics in systemic vasculitis, and more specifically to highlight new developments in the field of interest to clinicians and researchers.
ANTINEUTROPHIL CYTOPLASMIC ANTIBODY-ASSOCIATED VASCULITIS
Antineutrophil cytoplasmic antibody (ANCA)-associated vasculitis (AAV) is a systemic necrotizing vasculitis of small vessels characterized by the presence of autoantigens against neutrophil cytoplasmic proteins, specifically myeloperoxidase (MPO) and proteinase 3 (PR3) [1,16]. The presence of antineutrophil cytoplasmic antibodies against MPO and PR3 is used in the classification of AAV, though not all patients have ANCA. ANCAs have been implicated in vascular damage in AAV patients. Neutrophils in AAV patients are more sensitive to activation by ANCA, as demonstrated by the production of reactive oxygen species and neutrophil extracellular traps (NETs) [17,18]. In normal neutrophils, MPO and PR3 expression primarily occurs early in cell development, contributing to the formation of intracellular granules, but AAV cells continue to express MPO and PR3 into maturity which indicates a deviation from normal gene silencing [19,20].
Ciavatta et al., Yang et al., and Jones et al. [21,22▪▪,23▪▪] have provided excellent studies of how epigenetic mechanisms control PRTN3 and MPO gene expression and their dysregulation in AAV (Fig. 1). The initial study of AAV granulocytes revealed a depletion of repressive H3K27me3 marks and an increase in mRNA expression of PRTN3 and MPO [21]. In addition, a marked demethylation of a CpG island and the promoter region of MPO in AAV were observed, although PRTN3 promoter region was constitutively demethylated in patients and controls. The authors then explored the regulatory mechanisms governing H3K27me3 and found enhancer of zeste homolog 2 (EZH2) interacted with Runt-related transcription factor 3 (RUNX3) to recruit H3K27 methyltransferase to PRTN3 and MPO. The promoter region of the RUNX3 gene was also hypermethylated in AAV granulocytes. This suggests a regulatory model whereby hypermethylation of RUNX3 and the loss of EZH2 and H3K27 methyltransferase recruitment is coupled with overexpression of H3K27me3 demethylase jumonji C domain-containing protein 3 (JMJD3) in AAV neutrophils. JMJD3 removes the H3K27me3 marks from regulatory regions of MPO and PRTN3 and increases chromatin accessibility aided by DNA demethylation allowing access to transcriptional machinery. Genomic regions containing genetic risk variants in AAV were found to be enriched for H3K27me3 marks that indicate a closed or poised state for the chromatin in Th17 cells, supporting the role of Th17 cells in AAV pathogenesis [24▪,25].
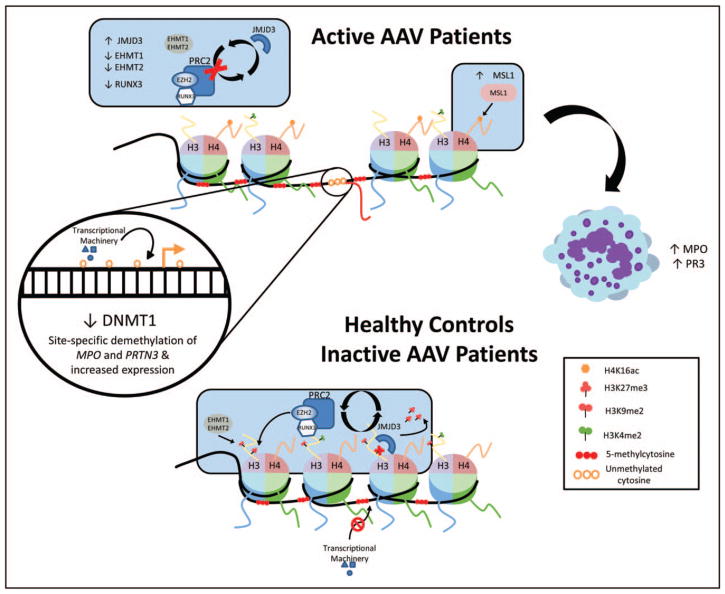
FIGURE 1.
A cartoon model of epigenetic control of MPO and PRTN3 in ANCA-associated vasculitis. Ciavatta et al. and Yang et al. suggest that histone modifications surrounding the promoter and enhancer regions of MPO and PRTN3 in AAV are in a bivalent state (presence of both repressive and permissive marks), maintaining gene silencing in mature neutrophils that is disrupted in AAV patients. In neutrophils from healthy controls and inactive patients with low MPO and PR3 expression, JMJD3 demethylates H3K27, although PRC2 remethylates it in kind to maintain a condensed silent state. EHMT1 and EHMT2 assist by maintaining H3K9me2 in the same region. Permissive H3K4me2 marks suggest an epigenetic poising and are present in both patients and controls, though the MLL2, MLL3, and MLL4 genes that regulate this mark were overexpressed in patients compared with controls. DNA methylation of the gene promoter and enhancer regions provides a second method of epigenetic control, preventing the access of transcriptional machinery, and CpG islands can be targeted by PRC2 as well for H3K27me3. In patients with active disease, some disruptive process interrupts the gene silencing and a decrease in RUNX3 expression prevents the reestablishment of H3K27me3. Decreased expression of EHMT1 and EHMT2 correlates with depletion of H3K9me2 and an increase in MSL1 expression correlates with enriched H4K16ac, a mark of gene activation. Jones et al. found that leukocytes from active AAV patients have decreased DNMT1 expression and a site-specific decrease in DNA methylation, suggesting a process that targets specific loci including MPO and PRTN3 and allows for gene expression. When AAV is inactive, methylation at these loci is returned to levels near that of healthy controls and expression is reduced. This suggests that MPO and PRTN3 DNA methylation is a disease-specific process supported by the identification of a CpG site in the PRTN3 promoter (CpG #13) that is demethylated in patients with a higher risk of relapse. AAV, ANCA-associated vasculitis; ANCA, antineutrophil cytoplasmic antibody; MPO, myeloperoxidase; proteinase 3; PR3, proteinase 3.
Yang et al. [23▪▪] investigated expression changes in genes encoding histone modification proteins and found a suite of four genes: euchromatic histone-lysine N-methyltransferase 1 and 2 (EHMT1, EHMT2) and male sex lethal 1 homolog and insulin growth factor (MSL1 and ING4) with consistent expression changes in leukocytes and granulocytes from AAV patients compared with healthy controls. EHMT1 and EHMT2 are associated with H3K9me2, a mark of gene silencing, and were found to be underexpressed in AAV leukocytes and granulocytes. MSL1 and ING4 are associated with H4K16ac, a mark of gene activation, MSL1 was found to be overexpressed in AAV leukocytes and granulocytes, although ING4 was underexpressed in leukocytes, but not significantly underexpressed in granulocytes. These expression changes were noted to be significantly different between leukocytes from active AAV patients with elevated MPO/PRNT3 expression and inactive patients with low-expression MPO/PRNT3, making them potential disease activity biomarkers. H4K16ac and H3K9me2 were, respectively, enriched and depleted at MPO and PRTN3 promoter regions in AAV granulocytes, especially in more active disease. MLL2, MLL3, and MLL4 (mixed-lineage leukemia) genes that regulate H3K4me2 were overexpressed in AAV patients compared with healthy controls. H3K4me2 is a well-recognized mark of transcriptional activation and, along with H3K27me3, is part of a bivalent chromatin signature [9▪,26]. H3K4me2 was equally enriched in both patients and controls at the MPO and PRTN3 promoter region, suggesting that they remain in a poised state at these loci even in healthy cells. Taken together, MPO and PRTN3 seem to maintain areas of bivalent chromatin that contain both permissive and repressive marks that are poised for expression with the loss of repressive marks. This occurs in AAV patients and is enhanced during active disease leading to abnormal over-production of granule proteins and neutrophil-mediated vascular damage.
Jones et al. [22▪▪] investigated DNA methylation changes in MPO and PRTN3 as potential disease biomarkers in AAV total leukocytes. They noted a decrease in DNMT1 expression but no significant reduction in global DNA methylation in AAV. However, they detected many differentially methylated genes in AAV patients, among these were MPO and PRTN3 which were both hypomethylated in patients with active disease compared with healthy controls. Methylation levels in both genes were also significantly lower in AAV patients with active compared with inactive disease. In addition, DNA methylation of both MPO and PRTN3 was negatively correlated with gene expression. By comparing AAV subsets (PR3-AAV versus MPO-AAV) using longitudinally collected samples from the same patients, they observed that MPO- and PR3-AAV patients experience a significant and near identical increase in the methylation level of PRTN3 promoter after disease remission. However, only patients with MPO-AAV, and not PR3-AAV, show evidence for a significant increase in the methylation level at MPO, suggesting that epigenetic changes at these two loci may provide a distinction between the two disease serotypes. At both PRTN3 and MOP, AAV patients who entered remission and displayed increased site-specific methylation had a significant reduction in mRNA expression of both genes, whereas those patients who experienced decreased DNA methylation upon remission displayed no change in gene expression. Perhaps the most valuable observation in this study was that demethylation of the PRTN3 promoter region was highly predictive of disease relapse in AAV patients regardless of ANCA-serotype; patients with demethylation in PRTN3 were 4.55 times more likely to relapse. This effect was narrowed down to a single CpG site in the promoter region of PRTN3. These results are very promising, but a larger study will be needed to confirm the prognostic use of this CpG site as a biomarker for relapse in AAV patients.
One drawback to this study is the use of total leukocytes in which differences in cell population proportions can mask cell-specific methylation changes from being detected. One the other hand, total leukocytes represent an easily accessible source for clinical testing and any effect that can be detected consistently in patients might have a potentially significant role in disease pathogenesis and will be valuable to develop into a disease biomarker.
GIANT-CELL ARTERITIS
Giant-cell arteritis (GCA) is an inflammatory disease of the large and medium arteries, occurring almost exclusively in people over 50 years of age, and characterized by granulomatous involvement which can lead to ischemia and necrosis or vision loss [27]. The arterial microenvironment at the site of inflammation in GCA is considerably complex with vessel-residing dendritic cells (DCs) acting as pathogen detectors and guiding T cell stimulation and the local inflammatory response, and Th1 and Th17 cells providing proinflammatory signals [28,29]. An exploration of DNA methylation changes occurring within the temporal artery of GCA patients revealed a strong T cell-specific signature consisting of hypomethylated loci in genes involved in TCR/CD28 signaling and calcineurin (Ca)/NFAT-regulatory pathways [30▪]. NFAT is a transcription factor regulating cytokine expression in T cells, including IFNG and TNF, and CD40LG expression, which were also demethylated in affected arterial tissue from GCA patients. NFAT1 was also found to be localized to the nucleus of cells (suggesting dephosphorylation and activation) in the vessel wall of GCA biopsies by immunohistochemistry. Hypomethylation of cytokine genes for Th1 (IFNG) and Th17 (IL6, IL21) cells, macrophages (TNF), and DCs (CCR7, CCL18) supported their presence in immune infiltration of the vessel wall. The Ca/NFAT pathway presents an intriguing therapeutic target. Dipyridamole, a highly specific calcineurin inhibitor, is suitable for targeting NFAT-regulated expression and has been shown to inhibit the production of interferon-gamma (IFNγ), IL-17, and IL-6 in T cells from MRL/lpr lupus mouse model [31]. Many proinflammatory genes regulated by NFAT were hypomethylated in GCA-affected arteries, and there is evidence that NFAT can interact with HDAC proteins to control histone modifications in specific contexts [32].
Croci et al. [33▪] identified miRNAs overex-pressed in GCA tissue by comparing active, non-active, and normal artery tissue. Of these miRNAs, miR-146a, miR-155, and miR-21 were overexpressed in inflamed temporal artery tissue compared with noninflamed and normal tissue. These miRNAs play a role in the regulation of the inflammatory response in T cells, macrophages, and DCs. They are also overexpressed in abdominal aortic aneurysms and atherosclerotic plaques and might play a role in vascular remodeling [34,35]. Although none of the known protein targets of these miRNAs were differentially expressed, miR-21 expression was found to be localized to cells in the medial-intimal layer of the artery in this study.
Age itself is a considerable risk factor for GCA and is likely due in part to changes in the immune function throughout the lifespan, which is known as immunosenescence [36,37]. Age-related DNA methylation changes in CD4+ T cells suggest a pro-inflammatory epigenetic architecture with age [38]. The miRNAs highlighted by Croci et al. [33▪,39] have also been implicated in immunesenescence and their increased expression in GCA tissue perhaps reflects an accelerated biological age that will need to be explored further.
KAWASAKI DISEASE
Kawasaki disease (KD) is a medium-vessel vasculitis that primarily occurs in children between ages 8 months and 5 years. It is characterized by inflammation of the coronary arteries, and is the leading cause of acquired heart disease in children from developed regions [40].
DNA methylation studies of KD have revealed a relationship between FCGR2A methylation and response to intravenous immunoglobulin (IVIG) treatment. FCGR2A encodes the low-affinity immunoglobulin gamma Fc region receptor II-a protein that is expressed on the surface of macrophages, neutrophils, monocytes, and DCs, and acts to increase phagocytosis and inflammatory mediator production and contains a genetic risk variant for KD [41]. CpG sites within the promotor region of FCGR2A were hypomethylated in whole blood cells from KD patients compared with controls, and especially in patients resistant to IVIG treatment [42]. Another small-scale study found genome-wide, site-specific hypomethylation changes enriched in genes associated with the inflammatory immune response including FCGR2A [43]. This study demonstrated significant changes in DNA methylation patterns following IVIG treatment in KD, including reversal of the disease-associated hypomethylation in FCGR2A [43].
Toll-like receptors are a group of proteins that recognize molecular patterns, both exogenous and endogenous, and can interact with FCGR2A to induce a proinflammatory response [44,45]. A suite of TLR genes encoding TLR1, TLR2, TLR4, TLR6, TLR8, and TLR9 were found to be hypomethylated in KD patients compared with healthy and febrile controls [46]. The methylation levels of these genes were recovered within 3-week post-IVIG therapy, and mRNA expression levels maintained a negative correlation with DNA methylation.
Regulatory T cells (Tregs) play an important role in suppressing the proinflammatory activity and cytokine expression of Th17 cells through physical interactions or by releasing cytokines like IL-10 and TGF-beta. This regulatory balance seems to be skewed toward proinflammatory Th17 cells in acute KD patients where there is a reduction in FoxP3 expression, a critical transcription factor in Treg activity [47]. miR-31 expression was increased in Tregs from acute KD patients and suppresses FoxP3 expression, although miR-155, which promotes FoxP3 expression, was found to be decreased in Tregs from patients [48]. IVIG treatment partially recovered the abnormal expression of miR-31 and miR-155. Furthermore, miR-145, which might be involved in modulating TGF-beta signaling, was increased in whole blood and detected in plasma exosomes isolated from acute KD patients [49]. Exosomes are extracellular vesicles released from cells that can be taken in by other cells and are capable of transporting miRNAs as a theorized form of cell-to-cell communication [50,51]. Other proinflammatory microRNAs that also potentially target TGF-beta signaling and are involved in KD are miR-200c and miR-371-5p. miR-200c promotes endothelial cell apoptosis, inducing vascular smooth muscle cell inflammatory response and modulating TLR4 response [52]. Both miR-200c and -371-5p were shown to effectively distinguish between KD and healthy controls as well as IVIG-responsive and nonresponsive patients [53]. More recent research has identified new miRNAs with disease activity that potentially target vessel endothelial cell functions in KD patients [54–58]. Jia et al. [55] performed a biomarker discovery screen on serum samples to detect exosome miRNAs in KD patients. After normalizing to internal control miRNA expression and recruiting an independent validation cohort, they identified two miRNA pairs (miR-1246/miR-4436b-5p and miR-197-3p/miR-671-5p) that when combined differentiated KD patients from healthy and febrile disease controls. These miRNA discovery studies are promising but generally relied upon small phenotypically varied cohorts of patient and controls. More work is required to validate these findings in KD, and to understand their cell-specific function and evaluate their efficacy as biomarkers.
BEHÇET’S DISEASE
Behçet’s disease (BD) is a systemic, variable-vessel vasculitis of unknown cause characterized by recurrent acute inflammatory episodes with oral and genital ulcers, eye involvement, and skin involvement [59,60]. Although its pathogenesis is still currently under investigation, evidence points toward a combination of genetic and environmental triggers as contributing factors to the development of BD [59]. Genetic susceptibility for BD shows a very strong association with the HLA-B/MICA region, though non-MHC risk factors have been identified as well that support the involvement of Th1 and Th17 cells (IL10, IL12A, STAT4, and IL23R-IL12RB2 locus) in pathogenesis [2,61–63]. This is supported by research showing that Th17, Th1, and Treg cell populations and cytokine production change with the disease state and can be found at inflammatory sites of BD patients [64–66].
DNA methylation of CD4+ T cells and monocytes extracted from the peripheral blood of BD patients are hypomethylated at genes associated with cytoskeletal remodeling processes such as actin and microtubule processing and cell adhesion [67]. Interestingly, some of the methylation deficiencies observed were returned to near those of healthy control levels at specific genes after treatment and disease remission. This recovery was more pronounced in monocytes than in T cells, but genes involved in microtubule formation and organization (KIFA2 and TPPP) were affected in both cell types making them intriguing targets for clinical biomarkers and therapeutics.
Research into changes in miRNA expression in BD has revealed a variety of potential targets and biomarkers. Regulation of Th17 cell activity has shown up as a theme in miRNA research in BD. miR-23b was underexpressed in CD4+ T cells from active BD patients [68]. When transfected into CD4+ T cells in vitro, miR-23b reduced the expression of Notch pathway genes and production of IFNγ and IL-17 [68]. As an example of genetic–epigenetic interaction, genetic variants also play a role in miRNA functions including expression and protein targeting [69]. Two such variants have been identified in BD patients: rs2910164 (MIR146A; miR-146a) and rs11614913 (MIR196A2; miR-196a2) [70,71]. Carriers of the rs2910164 CC genotype displayed a reduction in mature miR-146a transcripts and IL-17, TNF-alpha, and IL-1 beta at the protein level in PBMCs as compared with the GG genotype which was more frequent in BD patients [70]. Carriers of the rs11614913 TT allele had reduced expression of miR-196a2 in PBMCs and the T allele was significantly more frequent among BD patients compared with healthy and disease (Vogt–Koyanagi–Harada syndrome and acute anterior uveitis associated with ankylosing spondylitis) controls and more frequent among BD patients with arthritis compared with other subgroups [71]. Reduced expression of miR-196a2 coincided with a reduction in the target protein Bach1 and an increase in proinflammatory IL-1 beta and MCP-1 cytokines [71].
IGA VASCULITIS (HENOCH–SCHÖNLEIN PURPURA)
IgA vasculitis (IgAV) primarily targets small vessels and is characterized by the deposition of IgA immune complexes in the vessel wall, and disease onset is often associated with an infection of the upper airway or gastrointestinal tract [60]. IgAV is considered a geographically and ethnically ubiquitous disease predominantly of infants and children between 3 and 12 years of age [72]. Ascertaining genetic risk is difficult because of case studies of insufficient size, but a meta-analysis confirmed the risk associated with HLA-DRB1*01 and HLA-DRB1*07 variants [2].
Luo et al. [73,74] observed a genome-wide increase in H3 acetylation and H3K4 methylation, which are both marks of open and transcriptionally accessible chromatin, in PBMCs isolated from IgAV patients. These marks were positively correlated with disease activity and were significantly enriched in IgAV patients with renal involvement compared with IgAV patients without renal involvement and healthy controls [73]. Coinciding with this was an increase in HATs and histone methyltransferases in IgAV patients and a decrease in the opposing histone deacetylases and histone demethylases, indicating a shift in the transcriptional profile to support an abnormal transcriptionally active state [73]. IgAV patients with renal involvement had a marked increase in IL-4, IL-6, and IL-13 at the mRNA and protein levels [73]. The authors found an enrichment of H3 acetylation and H3K4me3 at promoter and enhancer regions of IL4, a Th2 cytokine, in CD4+ T cells from IgAV patients compared with controls and an increased expression of TIM-1, a suggested regulator of the Th2 response [73]. By comparison, IFNG, encoding IFNγ cytokine of Th1 cells, displayed no enrichment for these histone marks or elevated expression [73]. An IgAV-specific global increase in open chromatin marks coupled with the open chromatin state and overexpression of Th2-related genes points toward Th2 cells as being potential effectors in the pathogenesis of IgAV. Future studies would benefit from next-generation sequencing to gain a more holistic view in which these chromatin changes are occurring in both circulating immune cells as well as those residing in the kidney, which may have different disease-specific epigenetic profiles.
CONCLUSION
Epigenetic mechanisms provide a means to understand the pathogenesis of vasculitis, improve diagnosis, monitor disease progression, and the potential identification of novel therapeutic targets (Table 1). The highly interconnected nature of these mechanisms places the emphasis of future epigenetics research in systemic vasculitis on integrating data from disease-specific and cell-specific DNA methylation states, histone modifications, and miRNA activity.
Table 1.
Current knowledge of disease-specific epigenetic mechanisms in vasculitis
| Epigenetic mechanism | Region (s) of interest | Disease-specific relationship | Function | Source | Reference |
|---|---|---|---|---|---|
| ANCA-associated vasculitis (AAV) | |||||
| Histone modification (H3K27me3) | PRTN3, MPO | Depleted H3K27me3 in AAV and corresponding increased mRNA expression; a mark of gene silencing | Neutrophil granule protein | Granulocytes | [21] |
| Histone modification (H3K9me2) | PRTN3, MPO | Depleted H3K9me2 at gene promoter in active AAV patients with high mRNA expression compared with healthy controls and inactive patients; regulated by EHMT1 and EHMT2 and a mark of gene silencing | Neutrophil granule protein | Neutrophils | [23▪▪] |
| Histone modification (H4K16ac) | PRTN3, MPO | Enriched H4K16ac at gene promoter in active AAV patients with high mRNA expression compared with healthy controls and inactive patients; regulated by ING4 and MSL1 and a mark of gene activation | Neutrophil granule protein | Neutrophils | [23▪▪] |
| Histone modification (H3K4me2) | PRTN3, MPO | H3K4me2 at promoter regions of both PRTN3 and MPO in patients and controls; a mark of gene activation and suggests bivalent chromatin state | Neutrophil granule protein | Neutrophils | [23▪▪] |
| DNA methylation (promoter/CpG island) | MPO | Hypomethylation of promoter/CpG island of MPO in AAV patients | Neutrophil granule protein | Granulocytes | [21] |
| DNA methylation (promoter) | RUNX3 | Hypermethylation of promoter region and reduced RUNX3 mRNA expression in AAV patients | Recruits PRC2 that regulates H3K27 methylation of MPO and PRTN3 | Granulocytes | [21] |
| DNA methylation (CGI/Exon 5–6) | MPO | Hypomethylation of CpG island/exon 5–6 of MPO gene of AAV patients; typically increased with disease remission and corresponded with decreased MPO mRNA | Neutrophil granule protein | Total leukocytes | [22▪▪] |
| DNA methylation (promoter: CpG #13) | PRTN3 | Hypomethylation of promoter of PRTN3 gene in AAV patients; typically increased with disease remission and corresponded with decreased PRTN3 mRNA; hypomethylation of CpG #13 is associated with increased risk of disease relapse | Neutrophil granule protein | Total leukocytes | [22▪▪] |
| Giant-cell arteritis (GCA) | |||||
| DNA methylation (various loci) | PPP3CC, NFATC1, NFATC2 | Hypomethylated in GCA temporal artery; nuclear localization of NFAT1 protein in GCA-positive biopsies | Calcineurin/NFAT-signaling pathway | Temporal artery biopsy | [30▪] |
| DNA methylation (cg09993145) | RUNX3 | Hypomethylated in GCA temporal artery | Combines with T-bet for expression of IFNγ in Th1 cells | Temporal artery biopsy | [30▪,75] |
| DNA methylation (various loci) | CD40LG, IL21, IL21R | Hypomethylated in GCA temporal artery; protein present in GCA-positive biopsies | NFAT-regulated genes; involved in activation and maturation of Th cell lineages (CD40LG) and production of IL-17 in Th17 cells (IL21, IL21R) | Temporal artery biopsy | [30▪] |
| DNA methylation (various loci) | CCR7, CCL18 | Hypomethylated in GCA temporal artery | Present on mature dendritic cells (CCR7) and attract naive T cells (CCL18) | Temporal artery biopsy | [30▪] |
| DNA methylation (various loci) | CD3E, CD3G, CD3D, CD3Z, CD28, ZAP70 | Hypomethylated in GCA temporal artery | TCR/CD28-regulated T cell activation | Temporal artery biopsy | [30▪] |
| DNA methylation (various loci) | TNF, IL2, IL1B, IL18, IFNG, LTA, LTB | Hypomethylated in GCA temporal artery | Proinflammatory proteins and key Th1 cell cytokine (IFNG) | Temporal artery biopsy | [30▪] |
| Noncoding RNA (microRNA) | MIR146A, MIR155, MIR21 | MicroRNAs overexpressed in inflamed GCA temporal artery | Regulation of inflammatory and vascular remodeling networks; miR-21 is localized to medial/intimal layers | Temporal artery biopsy | [33▪] |
| Kawasaki disease (KD) | |||||
| DNA methylation (promoter: cg24422489) | FCGR2A | Hypomethylated in KD patients, more so in IVIG-resistant cases; methylation increased after IVIG treatment and mRNA expression was reduced | Ig Fc receptor that regulates an array of immune responses and is a potent proinflammatory gene; genetic risk locus for KD | Whole blood | [42,43] |
| DNA methylation (various loci) | TLR1, TLR2, TLR4, TLR6, TLR8, TLR9 | Hypomethylated in KD patients; methylation is restored with IVIG treatment and mRNA expression is reduced | Recognition of bacterial products and inflammatory signals | Total leukocytes | [46] |
| Noncoding RNA (microRNA) | MIR145 | Increased miR-145 in whole blood of acute KD patients; found in extracellular vesicles from patient plasma | Differentiation of neutrophils and vascular smooth muscle cells and targets regulatory genes in TGF beta signaling pathway | Whole blood & plasma | [49] |
| Noncoding RNA (microRNA) | MIR125A | Increased miR-125a-5p circulating in acute and convalescent KD patient plasma | Inhibition of MKK7 expression which promotes caspase-3 expression and apoptosis in endothelial cells | Plasma | [54] |
| Noncoding RNA (microRNA) | MIR1246/MIR4436B1 & MIR197/MIR671 | MiRNA pairs that, when combined, can differentiate KD patients from controls and non-KD febrile cases | miR-197 is predictive of death in symptomatic coronary artery disease and miR-1246 is a biomarker for diastolic dysfunction | Serum exosomes | [55] |
| Noncoding RNA (microRNA) | MIR155, MIR31 | Decreased miR-155 and increased miR-31 expression in CD4 + CD25 + Treg cells of acute KD patients; IVIG partially reverses expression difference | miRNAs expression correlated with FoxP3 mRNA in CD4 + CD25 + Treg cells suggesting common regulatory factors | CD4 + CD25 + Treg cells | [48] |
| Noncoding RNA (microRNA) | MIR223 | Increased miR-223 in KD patient serum especially those with coronary artery lesions; identified as part of KD diagnostic miRNA panel in total leukocytes | Released by bone-marrow derived blood cells into serum; promotes apoptosis in endothelial cells by targeting IGF1R and suppresses cell proliferation | Serum/total leukocytes | [56▪,57] |
| Noncoding RNA (microRNA) | MIR200C, MIR371 | Circulating miR-200c and miR-371-5p are increased in KD patients; reduced after IVIG therapy | Involved in proinflammatory response | Serum | [52,53] |
| Noncoding RNA (microRNA) | MIR93 | Increased miR-93 in patients responding to IVIG; inverse correlation with VEGFA mRNA expression | Related to expression of VEGF-A | PBMCs | [58] |
| Behçet’s disease (BD) | |||||
| DNA methylation | RAC1, ARHGAP24, FSCN2, BAIAP2L1, FILIP1 SSH, ANK1, MYH15, MYO1C, MYO1D, MPRIP | Methylation changes varies with cell type, but is distinct in BD patients; methylation at some sites restored after therapy | Actin processing and cytoskeletal remodeling | CD4 + T cells/monocytes | [67] |
| DNA methylation | TBCD, KIF1B, DNAH3, TUBB8, RGS14, TUBA3C, TUBA3D, TPPP, KIF2A | Methylation changes varies with cell type, but is distinct in BD patients; methylation at some sites restored after therapy | Microtubule processing | CD4 + T cells/monocytes | [67] |
| Noncoding RNA (microRNA) | MIR155 | Conflicting reports of expression changes in BD patients | Overexpression inhibits production of IL-6 and IL-1 beta and promotes IL-10 in DCs which reduced CD4 + T cell expression of IL-17; targets Ets-1 which inhibits IL-17 production | PBMCs/DCs/CD4 + T cells | [76,77] |
| Noncoding RNA (microRNA) | MIR146A | Decreased miR-146a expression associated with rs2910164 (C >G) C allele which is less frequent in BD patients | Regulates proinflammatory cytokine production (IL-17, TNF-alpha, IL-1 beta) | PBMCs | [70] |
| Noncoding RNA (microRNA) | MIR196A2 | Decreased miR-196a2 expression associated with rs11614913 (C >T) T allele which is more frequent in BD patients (specifically arthritis subgroup) | Presence of variant associated with decreased miR-196a expression and an increase in target gene BACH1; increased production of proinflammatory cytokines IL-1 beta and MCP-1 | PBMCs | [71] |
| Noncoding RNA (microRNA) | MIR638, MIR448, MIR3591 | Decreased miR-638 and miR-4488 expression in stable BD patients; Increased miR-3591-3p expression in active BD patients | Regulation of IL-6 production; differential expression associated with cancers, lupus and viral infection | CD11b + PBMCs | [78] |
| Noncoding RNA (microRNA) | MIR23B | Decreased miR-23b expression in CD4 + T cells of active BD patients | Regulation of Notch pathway and decreased expression of IL-17 and IFNγ in CD4 + T cells | CD4 + T cells | [68] |
| Noncoding RNA (microRNA) | MIR139, MIR720 | Increased miR-139-3p and miR-720 expression in BD patients regardless of disease activity | Target Toll-like receptor and T cell receptor signaling pathways | PBMCs | [79] |
| IgA vasculitis (IgAV; Henoch–Schönlein purpura) | |||||
| Histone modification (H3 acetylation) | IL4 | Increased H3 acetylation at IL4 promoter and enhancer regions of CD4 + T cells from IgAV patients | Th2 cell cytokine | CD4 + T cells | [73] |
| Histone modification (H3K4me3) | IL4 | Increased H3K4me3 at IL4 promoter and enhancer regions of CD4 + T cells from IgAV patients | Th2 cell cytokine | CD4 + T cells | [73] |
| Histone modification (H3K4me) | Genome-wide | Globally increased H3K4me in PBMCs of IgAV patients with kidney disease; positive correlation with disease activity | Suggests that abnormal levels of H3K4me and maintenance proteins contributes to kidney disease in IgAV patients | PBMCs | [73] |
| Histone modification (H3 acetylation) | Genome-wide | Globally increased H3 acetylation in PBMCs of IgAV patients with kidney disease; positive correlation with disease activity | Suggests that abnormal levels of H3 acetylation and maintenance proteins contributes to kidney disease in IgAV patients | PBMCs | [73] |
ANCA, antineutrophil cytoplasmic antibody.
KEY POINTS.
Epigenetic characterization in vasculitis can provide insights into disease pathogenesis and identify novel potential therapeutic targets.
The autoantigen gene loci in ANCA-associated vasculitis PRTN3 and MPO undergo epigenetic changes that correlate with disease activity and with PR3 and MPO expression.
Epigenetic changes and microRNA expression profiles can be developed into disease biomarkers in vasculitis, but larger cohorts and validation studies are needed.
Cell-type specific epigenetic signatures can be used to map causal variants in genetic risk loci and identify genetic-epigenetic interactions in vasculitis.
Acknowledgments
Financial support and sponsorship
This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) of the National Institutes of Health (grant number R01AR070148).
Footnotes
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
- 1.Scott DG, Watts RA. Epidemiology and clinical features of systemic vasculitis. Clin Exp Nephrol. 2013;17:607–610. doi: 10.1007/s10157-013-0830-8. [DOI] [PubMed] [Google Scholar]
- 2.Carmona FD, Martin J, Gonzalez-Gay MA. Genetics of vasculitis. Curr Opin Rheumatol. 2015;27:10–17. doi: 10.1097/BOR.0000000000000124. [DOI] [PubMed] [Google Scholar]
- 3.Chen M, Kallenberg CG. The environment, geoepidemiology and ANCA-associated vasculitides. Autoimmun Rev. 2010;9:A293–298. doi: 10.1016/j.autrev.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Lane SE, Watts RA, Bentham G, et al. Are environmental factors important in primary systemic vasculitis? A case-control study. Arthritis Rheum. 2003;48:814–823. doi: 10.1002/art.10830. [DOI] [PubMed] [Google Scholar]
- 5.Nordborg E, Nordborg C. Giant cell arteritis: epidemiological clues to its pathogenesis and an update on its treatment. Rheumatology (Oxford) 2003;42:413–421. doi: 10.1093/rheumatology/keg116. [DOI] [PubMed] [Google Scholar]
- 6.Mumcu G, Inanc N, Yavuz S, Direskeneli H. The role of infectious agents in the pathogenesis, clinical manifestations and treatment strategies in Behcet’s disease. Clin Exp Rheumatol. 2007;25:S27–33. [PubMed] [Google Scholar]
- 7.Liu L, Li Y, Tollefsbol TO. Gene-environment interactions and epigenetic basis of human diseases. Curr Issues Mol Biol. 2008;10:25–36. [PMC free article] [PubMed] [Google Scholar]
- 8.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9▪.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. Summary of the fundamental mechanisms of cellular epigenetics and their history highlighting current concepts of how they function and contribute to regulatory aberration in disease. [DOI] [PubMed] [Google Scholar]
- 10.Jeltsch A, Jurkowska RZ. New concepts in DNA methylation. Trends Biochem Sci. 2014;39:310–318. doi: 10.1016/j.tibs.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Fuks F, Burgers WA, Brehm A, et al. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 12.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Esteller M. Noncoding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 15.Sato F, Tsuchiya S, Meltzer SJ, Shimizu K. MicroRNAs and epigenetics. FEBS J. 2011;278:1598–1609. doi: 10.1111/j.1742-4658.2011.08089.x. [DOI] [PubMed] [Google Scholar]
- 16.Hilhorst M, van Paassen P, Tervaert JW, Limburg Renal R. Proteinase 3-ANCA vasculitis versus myeloperoxidase-ANCA vasculitis. J Am Soc Nephrol. 2015;26:2314–2327. doi: 10.1681/ASN.2014090903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohlsson SM, Ohlsson S, Soderberg D, et al. Neutrophils from vasculitis patients exhibit an increased propensity for activation by antineutrophil cytoplasmic antibodies. Clin Exp Immunol. 2014;176:363–372. doi: 10.1111/cei.12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao H, Heeringa P, Hu P, et al. Antineutrophil cytoplasmic autoantibodies specific for myeloperoxidase cause glomerulonephritis and vasculitis in mice. J Clin Invest. 2002;110:955–963. doi: 10.1172/JCI15918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cowland JB, Borregaard N. The individual regulation of granule protein mRNA levels during neutrophil maturation explains the heterogeneity of neutrophil granules. J Leukoc Biol. 1999;66:989–995. doi: 10.1002/jlb.66.6.989. [DOI] [PubMed] [Google Scholar]
- 20.Yang JJ, Pendergraft WF, Alcorta DA, et al. Circumvention of normal constraints on granule protein gene expression in peripheral blood neutrophils and monocytes of patients with antineutrophil cytoplasmic autoantibody-associated glomerulonephritis. J Am Soc Nephrol. 2004;15:2103–2114. doi: 10.1097/01.ASN.0000135058.46193.72. [DOI] [PubMed] [Google Scholar]
- 21.Ciavatta DJ, Yang J, Preston GA, et al. Epigenetic basis for aberrant upregulation of autoantigen genes in humans with ANCA vasculitis. J Clin Invest. 2010;120:3209–3219. doi: 10.1172/JCI40034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22▪▪.Jones BE, Yang J, Muthigi A, et al. Gene-specific DNA methylation changes predict remission in patients with ANCA-associated vasculitis. J Am Soc Nephrol. 2017;28:1175–1187. doi: 10.1681/ASN.2016050548. This study found that reduced DNMT1 expression in AAV leukocytes did not lead to global demethylation, but gene-specific hypomethylation including MPO and PRTN3. Patients who showed increased methylation with remission had an associated reduction in MPO and PR3 expression, and hypomethylation of PRTN3 promoter region was highly predictive of disease relapse. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23▪▪.Yang J, Ge H, Poulton CJ, et al. Histone modification signature at myeloperoxidase and proteinase 3 in patients with antineutrophil cytoplasmic autoanti-body-associated vasculitis. Clin Epigenetics. 2016;8:85. doi: 10.1186/s13148-016-0251-0. This study used AAV-relevant expression changes in genes associated with specific histone modifications in AAV neutrophils to identify disease-specific epigenetic changes. The study identified correlations between H4K16ac (permissive) and H3K9me2 (repressive) marks with expression of MPO and PRTN3 and AAV disease activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24▪.Sawalha AH, Dozmorov MG. Epigenomic functional characterization of genetic susceptibility variants in systemic vasculitis. J Autoimmun. 2016;67:76–81. doi: 10.1016/j.jaut.2015.10.002. This study used enrichment patterns of poised and primed enhancer epigenetic marks and DNase I hypersensitivity data to identify specific immune cell types likely affected by the genetic risk within identified susceptibility loci in vasculitis. [DOI] [PubMed] [Google Scholar]
- 25.Nogueira E, Hamour S, Sawant D, et al. Serum IL-17 and IL-23 levels and autoantigen-specific Th17 cells are elevated in patients with ANCA-associated vasculitis. Nephrol Dial Transplant. 2010;25:2209–2217. doi: 10.1093/ndt/gfp783. [DOI] [PubMed] [Google Scholar]
- 26.Voigt P, LeRoy G, Drury WJ, 3rd, et al. Asymmetrically modified nucleosomes. Cell. 2012;151:181–193. doi: 10.1016/j.cell.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Smit E, Palmer AJ, Hewitt AW. Projected worldwide disease burden from giant cell arteritis by 2050. J Rheumatol. 2015;42:119–125. doi: 10.3899/jrheum.140318. [DOI] [PubMed] [Google Scholar]
- 28.Weyand CM, Ma-Krupa W, Pryshchep O, et al. Vascular dendritic cells in giant cell arteritis. Ann N Y Acad Sci. 2005;1062:195–208. doi: 10.1196/annals.1358.023. [DOI] [PubMed] [Google Scholar]
- 29.Terrier B, Geri G, Chaara W, et al. Interleukin-21 modulates Th1 and Th17 responses in giant cell arteritis. Arthritis Rheum. 2012;64:2001–2011. doi: 10.1002/art.34327. [DOI] [PubMed] [Google Scholar]
- 30▪.Coit P, De Lott LB, Nan B, et al. DNA methylation analysis of the temporal artery microenvironment in giant cell arteritis. Ann Rheum Dis. 2016;75:1196–1202. doi: 10.1136/annrheumdis-2014-207116. Identified DNA hypomethylation of cell-specific cytokine genes in the temporal artery of GCA patients that suggested the presence of DCs, Th1 and Th17 cells and macrophages. The hypomethylated calcineurin/NFAT1 signaling pathway is a promising target for current therapeutic compounds like dipyridamole in GCA. [DOI] [PubMed] [Google Scholar]
- 31.Kyttaris VC, Zhang Z, Kampagianni O, Tsokos GC. Calcium signaling in systemic lupus erythematosus T cells: a treatment target. Arthritis Rheum. 2011;63:2058–2066. doi: 10.1002/art.30353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choo MK, Yeo H, Zayzafoon M. NFATc1 mediates HDAC-dependent transcriptional repression of osteocalcin expression during osteoblast differentiation. Bone. 2009;45:579–589. doi: 10.1016/j.bone.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33▪.Croci S, Zerbini A, Boiardi L, et al. MicroRNA markers of inflammation and remodelling in temporal arteries from patients with giant cell arteritis. Ann Rheum Dis. 2016;75:1527–1533. doi: 10.1136/annrheumdis-2015-207846. Identified the tissue-specific overexpression of miRNAs that have been previously associated with inflammation and age-related immunosenescence, which may be related to GCA’s almost exclusive presence in patients over 50 years of age. [DOI] [PubMed] [Google Scholar]
- 34.Kin K, Miyagawa S, Fukushima S, et al. Tissue- and plasma-specific MicroRNA signatures for atherosclerotic abdominal aortic aneurysm. J Am Heart Assoc. 2012;1:e000745. doi: 10.1161/JAHA.112.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raitoharju E, Lyytikainen LP, Levula M, et al. miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis. 2011;219:211–217. doi: 10.1016/j.atherosclerosis.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 36.Mohan SV, Liao YJ, Kim JW, et al. Giant cell arteritis: immune and vascular aging as disease risk factors. Arthritis Res Ther. 2011;13:231. doi: 10.1186/ar3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yung RL, Julius A. Epigenetics aging, and autoimmunity. Autoimmunity. 2008;41:329–335. doi: 10.1080/08916930802024889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dozmorov MG, Coit P, Maksimowicz-McKinnon K, Sawalha AH. Age-associated DNA methylation changes in naive CD4+ T cells suggest an evolving autoimmune epigenotype in aging T cells. Epigenomics. 2017;9:429–445. doi: 10.2217/epi-2016-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olivieri F, Rippo MR, Monsurro V, et al. MicroRNAs linking inflammaging, cellular senescence and cancer. Ageing Res Rev. 2013;12:1056–1068. doi: 10.1016/j.arr.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 40.Harnden A, Takahashi M, Burgner D. Kawasaki disease. BMJ. 2009;338:b1514. doi: 10.1136/bmj.b1514. [DOI] [PubMed] [Google Scholar]
- 41.Duan J, Lou J, Zhang Q, et al. A genetic variant rs1801274 in FCGR2A as a potential risk marker for Kawasaki disease: a case-control study and meta-analysis. PLoS One. 2014;9:e103329. doi: 10.1371/journal.pone.0103329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuo HC, Chang JC, Kuo HC, et al. Identification of an association between genomic hypomethylation of FCGR2A and susceptibility to Kawasaki disease and intravenous immunoglobulin resistance by DNA methylation array. Arthritis Rheumatol. 2015;67:828–836. doi: 10.1002/art.38976. [DOI] [PubMed] [Google Scholar]
- 43.Li SC, Chan WC, Huang YH, et al. Major methylation alterations on the CpG markers of inflammatory immune associated genes after IVIG treatment in Kawasaki disease. BMC Med Genomics. 2016;9(Suppl 1):37. doi: 10.1186/s12920-016-0197-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ellerman JE, Brown CK, de Vera M, et al. Masquerader: high mobility group box-1 and cancer. Clin Cancer Res. 2007;13:2836–2848. doi: 10.1158/1078-0432.CCR-06-1953. [DOI] [PubMed] [Google Scholar]
- 45.Vogelpoel LT, Hansen IS, Visser MW, et al. FcgammaRIIa cross-talk with TLRs, IL-1R, and IFNgammaR selectively modulates cytokine production in human myeloid cells. Immunobiology. 2015;220:193–199. doi: 10.1016/j.imbio.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 46.Huang YH, Li SC, Huang LH, et al. Identifying genetic hypomethylation and upregulation of toll-like receptors in Kawasaki disease. Oncotarget. 2017;8:11249–11258. doi: 10.18632/oncotarget.14497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jia S, Li C, Wang G, et al. The T helper type 17/regulatory T cell imbalance in patients with acute Kawasaki disease. Clin Exp Immunol. 2010;162:131–137. doi: 10.1111/j.1365-2249.2010.04236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ni FF, Li CR, Li Q, et al. Regulatory T cell microRNA expression changes in children with acute Kawasaki disease. Clin Exp Immunol. 2014;178:384–393. doi: 10.1111/cei.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimizu C, Kim J, Stepanowsky P, et al. Differential expression of miR-145 in children with Kawasaki disease. PLoS One. 2013;8:e58159. doi: 10.1371/journal.pone.0058159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turchinovich A, Samatov TR, Tonevitsky AG, Burwinkel B. Circulating miR-NAs: cell-cell communication function? Front Genet. 2013;4:119. doi: 10.3389/fgene.2013.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edgar JR. Q&A: What are exosomes, exactly? BMC Biol. 2016;14:46. doi: 10.1186/s12915-016-0268-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yun KW, Lee JY, Yun SW, et al. Elevated serum level of microRNA (miRNA)-200c and miRNA-371-5p in children with Kawasaki disease. Pediatr Cardiol. 2014;35:745–752. doi: 10.1007/s00246-013-0846-6. [DOI] [PubMed] [Google Scholar]
- 53.Zhang W, Wang Y, Zeng Y, et al. Serum miR-200c and miR-371-5p as the useful diagnostic biomarkers and therapeutic targets in Kawasaki disease. Biomed Res Int. 2017;2017:8257862. doi: 10.1155/2017/8257862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Z, Jiang J, Tian L, et al. A plasma mir-125a-5p as a novel biomarker for Kawasaki disease and induces apoptosis in HUVECs. PLoS One. 2017;12:e0175407. doi: 10.1371/journal.pone.0175407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jia HL, Liu CW, Zhang L, et al. Sets of serum exosomal microRNAs as candidate diagnostic biomarkers for Kawasaki disease. Sci Rep. 2017;7:44706. doi: 10.1038/srep44706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56▪.Chu M, Wu R, Qin S, et al. Bone marrow-derived MicroRNA-223 works as an endocrine geneticsignalin vascular endothelial cells and participates in vascular injury from Kawasaki disease. J Am Heart Assoc. 2017;6 doi: 10.1161/JAHA.116.004878. pii: e004878. Used in vitro experiments to provide a mechanism for how the overexpression of miR-223 in the serum of KD patients can influence endothelial cells. Endothelial cells are able to take-in extracellular serum vesicles from KD patients that contain miR-223 produced by bone marrow cells and promote cell apoptosis by targeting IGF1R which can be blocked by miR-223 inhibitor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuo HC, Hsieh KS, Ming-Huey Guo M, et al. Next-generation sequencing identifies micro-RNA-based biomarker panel for Kawasaki disease. J Allergy Clin Immunol. 2016;138:1227–1230. doi: 10.1016/j.jaci.2016.04.050. [DOI] [PubMed] [Google Scholar]
- 58.Saito K, Nakaoka H, Takasaki I, et al. MicroRNA-93 may control vascular endothelial growth factor A in circulating peripheral blood mononuclear cells in acute Kawasaki disease. Pediatr Res. 2016;80:425–432. doi: 10.1038/pr.2016.93. [DOI] [PubMed] [Google Scholar]
- 59.Zeidan MJ, Saadoun D, Garrido M, et al. Behcet’s disease physiopathology: a contemporary review. Auto Immun Highlights. 2016;7:4. doi: 10.1007/s13317-016-0074-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jennette JC, Falk RJ, Bacon PA, et al. 2012 Revised international chapel hill consensus conference nomenclature of vasculitides. Arthritis & Rheumatism. 2013;65:1–11. doi: 10.1002/art.37715. [DOI] [PubMed] [Google Scholar]
- 61.Gul A. Pathogenesis of Behcet’s disease: autoinflammatory features and beyond. Semin Immunopathol. 2015;37:413–418. doi: 10.1007/s00281-015-0502-8. [DOI] [PubMed] [Google Scholar]
- 62.Takeuchi M, Kastner DL, Remmers EF. The immunogenetics of Behcet’s disease: a comprehensive review. J Autoimmun. 2015;64:137–148. doi: 10.1016/j.jaut.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hughes T, Coit P, Adler A, et al. Identification of multiple independent susceptibility loci in the HLA region in Behcet’s disease. Nat Genet. 2013;45:319–324. doi: 10.1038/ng.2551. [DOI] [PubMed] [Google Scholar]
- 64.Geri G, Terrier B, Rosenzwajg M, et al. Critical role of IL-21 in modulating TH17 and regulatory T cells in Behcet disease. J Allergy Clin Immunol. 2011;128:655–664. doi: 10.1016/j.jaci.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 65.Kim J, Park JA, Lee EY, et al. Imbalance of Th17 to Th1 cells in Behcet’s disease. Clin Exp Rheumatol. 2010;28:S16–19. [PubMed] [Google Scholar]
- 66.Deniz R, Tulunay-Virlan A, Ture Ozdemir F, et al. Th17-inducing conditions lead to in vitro activation of both Th17 and Th1 responses in Behcet’s disease. Immunol Invest. 2017;46:518–525. doi: 10.1080/08820139.2017.1306865. [DOI] [PubMed] [Google Scholar]
- 67.Hughes T, Ture-Ozdemir F, Alibaz-Oner F, et al. Epigenome-wide scan identifies a treatment-responsive pattern of altered DNA methylation among cytoskeletal remodeling genes in monocytes and CD4+ T cells from patients with Behcet’s disease. Arthritis Rheumatol. 2014;66:1648–1658. doi: 10.1002/art.38409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qi J, Yang Y, Hou S, et al. Increased Notch pathway activation in Behcet’s disease. Rheumatology (Oxford) 2014;53:810–820. doi: 10.1093/rheumatology/ket438. [DOI] [PubMed] [Google Scholar]
- 69.Cammaerts S, Strazisar M, De Rijk P, Del Favero J. Genetic variants in microRNA genes: impact on microRNA expression, function, and disease. Front Genet. 2015;6:186. doi: 10.3389/fgene.2015.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou Q, Hou S, Liang L, et al. MicroRNA-146a and Ets-1 gene polymorphisms in ocular Behcet’s disease and Vogt-Koyanagi-Harada syndrome. Ann Rheum Dis. 2014;73:170–176. doi: 10.1136/annrheumdis-2012-201627. [DOI] [PubMed] [Google Scholar]
- 71.Qi J, Hou SP, Zhang Q, et al. A functional variant of premiRNA-196a2 confers risk for Behcet’s disease but not for Vogt-Koyanagi-Harada syndrome or AAU in ankylosing spondylitis. Human Genetics. 2013;132:1395–1404. doi: 10.1007/s00439-013-1346-8. [DOI] [PubMed] [Google Scholar]
- 72.Piram M, Mahr A. Epidemiology of immunoglobulin A vasculitis (Henoch-Schonlein): current state of knowledge. Curr Opin Rheumatol. 2013;25:171–178. doi: 10.1097/BOR.0b013e32835d8e2a. [DOI] [PubMed] [Google Scholar]
- 73.Luo S, Liang G, Zhang P, et al. Aberrant histone modifications in peripheral blood mononuclear cells from patients with Henoch-Schonlein purpura. Clin Immunol. 2013;146:165–175. doi: 10.1016/j.clim.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 74.Yan C, Boyd DD. Histone H3 acetylation and H3 K4 methylation define distinct chromatin regions permissive for transgene expression. Mol Cell Biol. 2006;26:6357–6371. doi: 10.1128/MCB.00311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Djuretic IM, Levanon D, Negreanu V, et al. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat Immunol. 2007;8:145–153. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- 76.Zhou Q, Xiao X, Wang C, et al. Decreased microRNA-155 expression in ocular Behcet’s disease but not in Vogt Koyanagi Harada syndrome. Invest Ophthalmol Vis Sci. 2012;53:5665–5674. doi: 10.1167/iovs.12-9832. [DOI] [PubMed] [Google Scholar]
- 77.Na SY, Park MJ, Park S, Lee ES. MicroRNA-155 regulates the Th17 immune response by targeting Ets-1 in Behcet’s disease. Clin Exp Rheumatol. 2016;34:S56–S63. [PubMed] [Google Scholar]
- 78.Woo MY, Yun SJ, Cho O, et al. MicroRNAs differentially expressed in Behcet disease are involved in interleukin-6 production. J Inflamm (Lond) 2016;13:22. doi: 10.1186/s12950-016-0130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Erre GL, Piga M, Carru C, et al. Global microRNA profiling of peripheral blood mononuclear cells in patients with Behcet’s disease. Clin Exp Rheumatol. 2015;33:S72–79. [PubMed] [Google Scholar]