Abstract
The challenge of linking intergenic mutations to target genes has limited molecular understanding of human diseases. Here we show that H3K27ac HiChIP generates high-resolution contact maps of active enhancers and target genes in rare primary human T cell subtypes and coronary artery smooth muscle cells. Differentiation of naive T cells into T helper 17 cells or regulatory T cells creates subtype-specific enhancer–promoter interactions, specifically at regions of shared DNA accessibility. These data provide a principled means of assigning molecular functions to autoimmune and cardiovascular disease risk variants, linking hundreds of noncoding variants to putative gene targets. Target genes identified with HiChIP are further supported by CRISPR interference and activation at linked enhancers, by the presence of expression quantitative trait loci, and by allele-specific enhancer loops in patient-derived primary cells. The majority of disease-associated enhancers contact genes beyond the nearest gene in the linear genome, leading to a fourfold increase in the number of potential target genes for autoimmune and cardiovascular diseases.
Gene expression programs are intimately linked to the hierarchical organization of the genome. In mammalian cells, each chromosome is organized into hundreds of megabase-sized topologically associated domains (TADs), which are conserved from early stem cells to differentiated cell types1. Within this invariant TAD scaffold, cell-type-specific enhancer–promoter interactions establish regulatory gene expression programs2. Standard methods require tens of millions of cells to obtain high-resolution interaction maps and confidently assign enhancer–promoter contacts3–5. Thus, the principles that govern enhancer–promoter conformation in disease-relevant patient samples are incompletely understood. This gap in understanding is particularly problematic for interpreting the molecular functions of inherited risk factors for common human diseases, which reside in intergenic enhancers or other noncoding DNA features in up to 90% of cases6–9. Such disease-relevant enhancers may not influence the expression of the nearest gene (often reported as the default target in the literature) and may instead act in a cell-type-specific manner on distant target genes residing up to hundreds of kilobases away2,10–14. Recently, systematic perturbations of regulatory elements in select gene loci have shown that the effects of individual regulatory elements on gene activity can be predicted from the combination of (i) enhancer activity (marked by histone H3 lysine 27 acetylation (H3K27ac) level) and (ii) enhancer–target looping5,15. Here we leverage this insight to capture the combination of these two types of information across the genome in a single assay, mapping the enhancer connectome in disease-relevant primary human cells.
RESULTS
H3K27ac HiChIP identifies functional enhancer interactions
We recently developed HiChIP, a method for sensitive and efficient analysis of protein-centric chromosome conformation16. Cohesin HiChIP in GM12878 cells identified similar numbers of loops as in situ Hi-C (~10,000) with high correlation (R = 0.83), demonstrating that HiChIP captures loops with high sensitivity and specificity. Here we evaluated the enhancer- and promoter-associated mark H3K27ac17–19 as a candidate factor to selectively interrogate enhancer–promoter interactions across the genome. We performed H3K27ac HiChIP in mouse embryonic stem (ES) cells to compare to cohesin HiChIP (Supplementary Fig. 1a and Supplementary Table 1)16. 3,552 of the 4,191 H3K27ac HiChIP loops in mouse ES cells were also identified by cohesin HiChIP. The H3K27ac-biased loops (log2 (fold change) > 1) spanned shorter distances than the cohesin-biased loops and were enriched for H3K27ac ChIP–seq peaks (78.9%; Supplementary Fig. 1b–f and Supplementary Table 2). Moreover, systematic titration of input material showed that H3K27ac HiChIP retained high signal fidelity and reproducibility when using from 25 million to 50,000 cells as input material (loop signal correlation r = 0.918; Supplementary Figs. 2 and 3). Therefore, H3K27ac HiChIP identifies high-confidence chromatin loops focused around enhancer interactions from limited cell numbers.
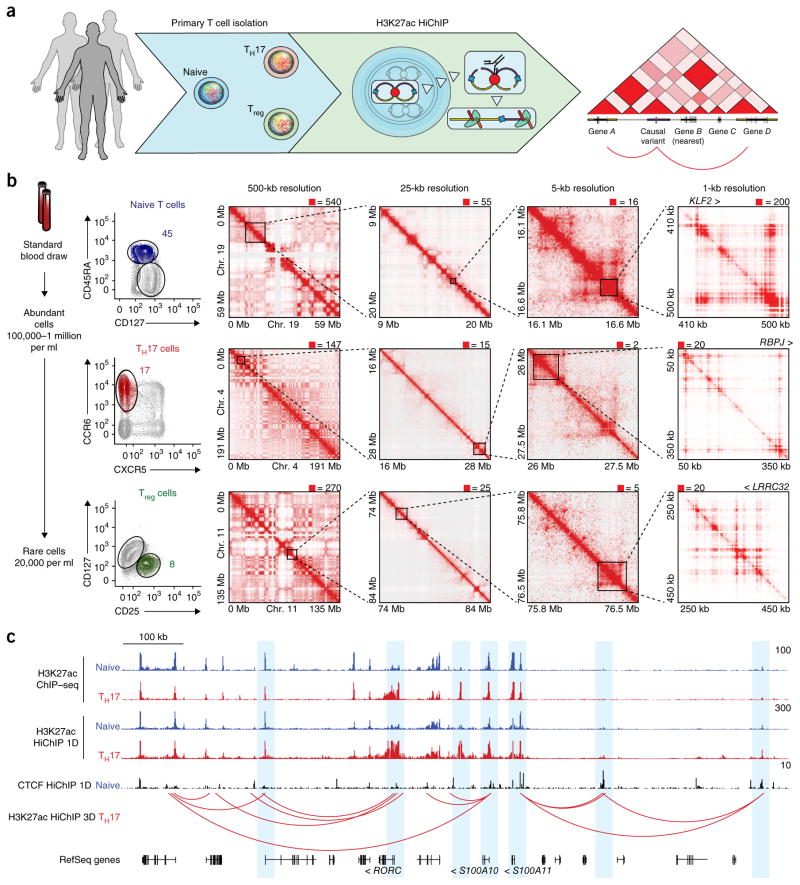
To capture (i) conformational change during T cell differentiation and (ii) cell-type-specific chromatin contacts of risk variants for autoimmune diseases in protective and pathogenic T cell types, we performed H3K27ac HiChIP on primary human naive T cells (CD4+CD45RA+CD25−CD127hi), regulatory T (Treg) cells (CD4+CD25+CD127lo), and T helper 17 (TH17) cells (CD4+CD45RA−CD25−CD127hiCCR6+CXCR5−) directly isolated from donors (Fig. 1a,b and Supplementary Fig. 4a)20,21. TH17 cells were sorted to include autoimmune disease–relevant pathogenic TH17 cells and to exclude follicular helper T cells with a distinct surface phenotype and immune function (Supplementary Fig. 4a)22–24. Peripheral blood CD4+ T cells were obtained from three healthy subjects, isolated by FACS, and subjected to H3K27ac HiChIP. The HiChIP libraries from each subset were high quality; greater than 40% of the reads represented unique paired-end tags (PETs) (Supplementary Fig. 4b–d and Supplementary Table 1). Furthermore, the libraries exhibited high 1D signal enrichment at enhancers and promoters and globally recapitulated publically available H3K27ac ChIP–seq data sets (74.7% overlap of ChIP–seq and 1D HiChIP peaks; Fig. 1c)25. Inspection of the interaction matrix at progressively higher resolution showed chromatin compartments, TADs, and focal loops, as previously reported in high-resolution Hi-C and HiChIP analyses from cell lines (Fig. 1b)4,16. Notably, H3K27ac HiChIP maps were capable of identifying focal interactions at 1-kb resolution, which is comparable to the resolution for in situ Hi-C maps generated from 100-fold more cells and sequenced to 13-fold greater depth4 (Fig. 1b).
Figure 1.
HiChIP identifies high-resolution chromosome conformation in primary human T cells. (a) Primary T cell H3K27ac HiChIP experimental outline. (b) Left, FACS strategy for naive, TH17, and Treg cells from total peripheral blood CD4+ T cells. Number represents the percentage of total CD4+ T cells within that gate. Right, Knight–Ruiz (KR) matrix–balanced interaction maps for naive, TH17, and Treg cells at 500-kb, 25-kb, and 5-kb resolution, and raw interaction maps at 1-kb resolution, centered on the KLF2, RBPJ, and LRRC32 loci. (c) HiChIP 1D and 3D signal enrichment at the RORC locus in TH17 over naive T cells.
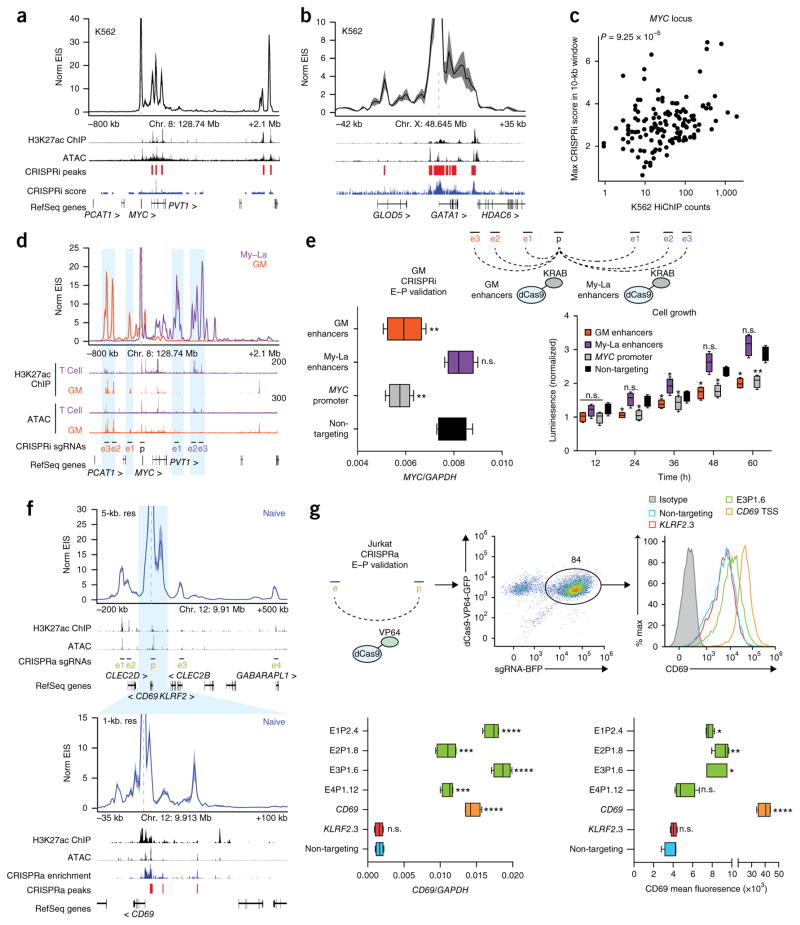
Previous saturation perturbation screens demonstrated that functional enhancers can be identified by integrating H3K27ac ChIP–seq signal with chromosome conformation contact strength (Hi-C)5. Because H3K27ac HiChIP combines these two components into one assay, we reasoned that HiChIP signal, which we term enhancer interaction signal (EIS), should identify functional regulatory elements. To validate this prediction, we first generated H3K27ac HiChIP maps in a chronic myelogenous leukemia cell line (K562) as a direct comparison to published high-resolution CRISPR interference (CRISPRi) screens5. We then examined the 3D enhancer landscape of the MYC and GATA1 loci using virtual 4C (v4C) analysis, where a specific genomic position is set as an anchor viewpoint and all interactions occurring with that anchor are visualized in 2D16. v4C analysis of the MYC promoter demonstrated that EIS in K562 cells captured all functional enhancers identified in the CRISPRi screen (Fig. 2a). Analysis of the GATA1 locus demonstrated a similar agreement between the two methods (Fig. 2b). Quantitatively, the EIS in K562 cells was significantly correlated with CRISPRi score in the same cell type, whereas the EIS in GM12878 (GM; B cell lymphoblast) cells was not correlated with the K562 CRISPRi score (Spearman’s ρ = 0.332 and 0.145; P = 9.25 × 10−5 and 0.1246; Fig. 2c).
Figure 2.
Validation of regulatory elements identified by H3K27ac HiChIP with CRISPR interference and activation. (a) Interaction profile of the MYC promoter in K562 H3K27ac HiChIP at 10-kb resolution. K562 H3K27ac ChIP–seq data are from the Encyclopedia of DNA Elements (ENCODE). CRISPRi-validated regulatory regions in K562 cells are indicated5. (b) Interaction profile of the GATA1 promoter in K562 H3K27ac HiChIP at 1-kb resolution. CRISPRi-validated regulatory regions in K562 cells are indicated5. (c) Correlation of MYC K562 H3K27ac HiChIP signal with maximum CRISPRi score within the HiChIP 10-kb window. (d) Interaction profiles of the MYC promoter in GM and My-La H3K27ac HiChIP at 10-kb resolution. T cell H3K27ac ChIP–seq and ATAC–seq data are from naive T cells. (e) Top, CRISPRi validation in GM cells of GM- and My-La-biased MYC enhancers. E–P, enhancer–promoter. Bottom, MYC RNA levels by qRT–PCR and cell growth rates in CRISPRi GM cells with targeting of cell-type-biased enhancers, the MYC promoter, and a non-targeting negative control (n = 3 biological replicates, each with 2 technical replicates). (f) Interaction profile of the CD69 promoter in Jurkat H3K27ac HiChIP at 5-kb and 1-kb resolution. The 1-kb profile is focused on the window of the CRISPRa tiling screen. CRISPRa-validated regulatory regions in Jurkat cells are indicated26. (g) Top, CRISPRa validation in Jurkat cells of CD69 distal enhancers. Bottom, CD69 RNA and protein levels in CRISPRa Jurkat cells with targeting of distal enhancers, the CD69 promoter, the KLRF2 promoter as a locus negative control, and a non-targeting negative control (n = 2 biological replicates, each with 2 technical replicates). In the box plots, each box extends from the 25th to the 75th percentile with a line representing the median, and whiskers extend to the minimum and maximum values. *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001 one-way ANOVA followed by Dunnett’s multiple-comparisons test against the non-targeting control; n.s., not significant.
We found the enhancer landscapes of the MYC promoter to be highly cell type specific. v4C analysis of the MYC promoter in GM and My-La (CD4+ T cell leukemia) cells showed dramatically different regulatory interactions with the promoter as compared to K562 cells (Fig. 2d). To validate EIS specificity, we performed CRISPRi experiments in GM cells using single guide RNAs (sgRNAs) targeting enhancers identified in either GM or My-La HiChIP maps, as well as a positive-control sgRNA targeting the MYC promoter and a negative-control sgRNA targeting lambda phage sequence (Fig. 2e). As expected, we found that simultaneous CRISPRi perturbation of GM enhancers, but not My-La enhancers, impacted MYC expression and cell growth in GM cells (Fig. 2e).
Finally, we focused on the CD69 locus, where a high-resolution CRISPR activation (CRISPRa) screen identified three enhancers upstream of the transcription start site (TSS)26. These sites were also identified by H3K27ac HiChIP in naive T cells. Moreover, HiChIP identified four additional distal enhancers that were outside the region spanned by the sgRNA tiling array (Fig. 2f and Supplementary Fig. 5). To functionally validate these new enhancers, we performed CRISPRa experiments in Jurkat cells with sgRNAs targeting these enhancers, the CD69 promoter, and the KLRF2 promoter as a locus negative control, as well as a non-human-genome-targeting negative control. We observed a significant increase in CD69 RNA and protein levels in the four HiChIP enhancers as compared to the negative controls (Fig. 2g and Supplementary Fig. 5). Interestingly, two of the four new enhancers identified were within the promoter regions of distant genes. These findings are in line with previous reports that identified widespread distal gene regulatory functions of promoters across the genome27,28. Altogether, these results suggest that H3K27ac HiChIP EIS identifies functional regulatory elements and that enhancers that regulate a gene of interest can differ significantly between cell types.
Landscape of enhancer interactions in primary T cells
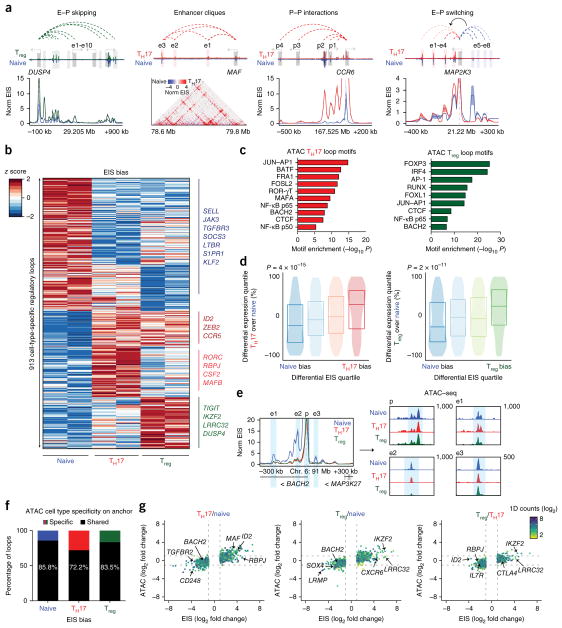
We examined global features of the enhancer connectome associated with cellular differentiation from naive T cells into either TH17 cells or Treg cells. We identified a total of 10,706 high-confidence loops in the union set of the three cell types (Supplementary Table 2). Analysis of loop read support between biological replicates demonstrated high reproducibility (Supplementary Fig. 4c), and ~91% of loop anchors were associated with either a promoter or an enhancer29, as expected, with a median distance of 130 kb (Supplementary Fig. 6a,b). Notably, high-resolution enhancer–promoter connectivity maps identified several features that could not be discerned from 1D epigenomic data (that is, H3K27ac ChIP–seq or assay for transposase-accessible chromatin using sequencing, ATAC–seq; Fig. 3a). These features included (i) ‘enhancer skipping’: enhancers that had stronger EIS with a more distal target promoter; (ii) higher-order structures such as ‘enhancer cliques’ (related to loop cliques30): multiple regulatory elements that had strong EIS with a single target promoter; (iii) promoter–promoter interactions13,31; and (iv) ‘enhancer switching’: enhancers that exhibited differential EIS with a target promoter in a cell-type-specific manner (Fig. 3a).
Figure 3.
Dynamic 3D enhancer landscapes in T cell differentiation. (a) Conformational features observed by H3K27ac HiChIP. (b) HiChIP EIS in 913 differential interactions identified in T cell subtypes. Interactions are clustered by cell type specificity. (c) Cell-type-specific motif identification from ATAC–seq peaks in biased EIS anchors. (d) EIS bias quartiles for naive to TH17 and naive to Treg differentiation, with corresponding differential RNA gene expression rankings. (e) Interaction profile of the BACH2 promoter at 5-kb resolution, demonstrating shared accessibility signal at naive-biased EIS. (f) Proportion of ATAC–seq peaks within HiChIP differential interaction anchors that are cell type specific (log2 (fold change) > 1) or shared across all three subtypes. (g) Global correlation of EIS and ATAC–seq fold change in different pairwise comparisons of T cell subsets.
We found that EIS contacts were very cell type specific. After quantile–quantile normalization of contact reads at high-confidence loops (correcting for false positives caused by 1D fragment visibility; Supplementary Note), we focused on the top and bottom 5% of EIS ranked by cell type bias for each pairwise comparison (Supplementary Figs. 6c–g and 7, and Supplementary Tables 3 and 4). Cell-type-specific enhancer loop anchors identified genes encoding canonical T cell subtype transcription factors and effector molecules (Fig. 3b and Supplementary Figs. 8 and 9). Deeper v4C analysis of shared and cell-type-specific loci pinpointed regulatory elements interacting with each gene promoter of interest as well as local conformational landscape changes (Supplementary Figs. 8 and 9). Transcription factor motifs located within cell-type-specific loop anchors were enriched for transcription factors known to drive T cell subtype differentiation and nominated new transcription factors involved in regulation (Fig. 3c). Furthermore, cell type EIS bias was associated with differential expression of genes located within corresponding EIS anchors for the same cell type (naive to TH17: Spearman’s ρ = 0.242 and P = 4 × 10−15; naive to Treg: Spearman’s ρ = 0.207 and P = 2 × 10−11); Fig. 3d).
Cell-type-specific EIS may be driven by cell-type-specific enhancer activation (based on H3K27ac ChIP–seq) or stable enhancer activation with cell-type-specific looping (Hi-C) in a gene-specific manner. We first examined H3K27ac ChIP–seq signal at differential EIS anchors and found that many biased H3K27ac HiChIP interactions also exhibited biased ChIP–seq signal, as expected. 58.5% of naive T cell–biased loops contained at least one naive T cell–biased ChIP–seq peak (log2 (fold change) > 1) located on the anchors. Similarly, 66.7% of TH17 cell–biased and 67.8% of Treg cell–biased interaction anchors were cell type specific in 1D (Supplementary Fig. 10a). Therefore, while on average ~64% of the differential EIS corresponds to changes in 1D data, ~36% is likely also driven by changes in 3D chromatin loop strength. To further assess the contribution of cell-type- specific 3D signal to EIS, we examined HiChIP 1D signal at differential EIS anchors. We found that HiChIP 1D signal correlated better with ChIP–seq signal than EIS, with a higher likelihood of differential ChIP–seq signal overlapping differential HiChIP 1D signal as compared to 3D signal, suggesting that EIS bias is in part driven by 3D changes (Supplementary Fig. 10b).
We asked whether the integration of reference cell line Hi-C data with primary T cell H3K27ac ChIP–seq data could recapitulate HiChIP EIS in primary T cells. We binned GM Hi-C loops with increasing primary T cell ChIP–seq signal at loop anchors and then determined the overlap of loops in each bin with loops derived from H3K27ac HiChIP. As expected, increased ChIP–seq signal at the Hi-C anchors led to increased overlap with the HiChIP loops. However, the overlap was lower in all T cell subtypes as compared to the same analysis performed using GM HiChIP data. These observations demonstrate that cell-type-specific 3D interactions can impact EIS independently of differences in 1D ChIP–seq signal (Supplementary Fig. 10c). Similarly, previously generated enhancer–promoter maps obtained from bulk T cells did not identify T cell subtype-specific interactions obtained using H3K27ac HiChIP. To assess the unique information obtained through cell-type-specific interaction maps, we compared promoter-capture Hi-C maps14 in bulk CD4+ T cells to H3K27ac HiChIP maps in naive T, TH17, and Treg cells. Strikingly, the most cell-type-specific loops in TH17 and Treg cells (16-fold enriched) demonstrated a low discovery rate in promoter-capture Hi-C T cells (11.83% in 415 loops and 13.83% in 373 loops, respectively; Supplementary Fig. 10d). Many of these subset-specific interactions included genomic loci encoding functionally important effector genes, such as LRRC32. The LRRC32 locus contained Treg-specific loops that were neither visualized in HiChIP maps from naive T or TH17 cells nor in bulk CD4+ promoter-capture Hi-C maps (Supplementary Fig. 10e). Because primary human TH17 and Treg cells are present in human blood at low frequencies, it would also be challenging to generate subset-specific promoter-capture Hi-C maps with published promoter-capture Hi-C protocols. In summary, EIS is derived from a combination of 1D ChIP–seq signal and 3D interaction signal and cannot be accurately predicted from 3D maps in reference cell lines or unsorted primary cell data sets.
Cell-type-specific EIS can occur at sites of shared chromatin accessibility. Paired chromatin accessibility profiles from ATAC–seq32 for each T cell subset showed that most cell-type-specific loop anchors had equivalent chromatin accessibility across all three cell types (Fig. 3e–g). To illustrate this finding, we examined the BACH2 promoter, which exhibited shared chromatin accessibility at enhancers but increased EIS in naive T cells (Fig. 3e). Globally, only 14.2%, 27.8%, and 16.5% of naive-, TH17-, and Treg-biased loops, respectively, contained at least one biased ATAC–seq peak (log2 (fold change) > 1) located on the anchors. Furthermore, the majority of cell-type-specific transcription factor motifs were observed in shared ATAC–seq peaks within differential interactions, highlighting the notion that these regions are functioning in T cell differentiation (Fig. 3f,g). Altogether, these results suggest that, in highly related—yet functionally distinct—cell types, a portion of transcriptional control is achieved through differential chromosome looping, rather than differential chromatin accessibility. This finding is consistent with previous studies that demonstrated that T cell subset-specific transcription factors, such as FOXP3, act predominantly at pre-accessible chromatin sites to establish subset-specific gene expression33.
Enhancer interactions link disease variants to target genes
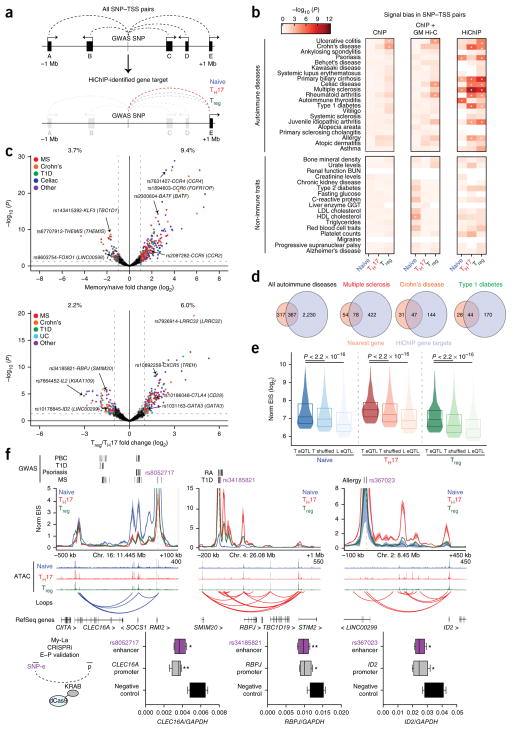
The high specificity of EIS enabled us to identify putative target genes of autoimmune disease risk loci in functionally relevant T cell subsets. To achieve this, we used a previously described list of putatively causal variants associated with 21 autoimmune diseases, known as PICS SNPs, which were fine-mapped on the basis of dense genotyping data25. We determined that PICS autoimmune disease–associated SNPs were significantly enriched in T cell loop anchors, with variants for specific autoimmune diseases showing greater than fivefold enrichment as compared to a shuffled control loop set (Supplementary Fig. 11). Next, we constructed a set of all possible connections between autoim-mune disease risk SNPs and TSSs within 1 Mb and measured the EIS for each SNP–TSS pair (Fig. 4a). We aggregated these signals to determine the overall interaction activity in each T cell subtype for each disease (Fig. 4b). We observed high interaction strength enrichments and cell type specificity for autoimmune disease–associated SNPs, but low enrichment and cell type specificity for variants associated with non-immune traits (Fig. 4b). To further visualize HiChIP bias in shared or differential enhancers, we analyzed SNP–TSS interactions grouped by their presence near H3K27ac ChIP–seq peaks (Supplementary Fig. 12a,b). We observed a large number of active SNP–TSS pairs that were present in regulatory regions that were shared by T effector cell types (Treg and TH17 cells), whereas relatively less EIS was observed for SNPs located in cell-type-specific enhancers, supporting the concept that many autoimmune disease variants impact common T cell effector/activation pathways25,34. Notably, SNPs present in enhancers shared by all three cell types could still be distinguished by HiChIP bias (Supplementary Fig. 12a,b). For example, although we could not detect cell type bias at risk loci for alopecia areata using H3K27ac ChIP–seq data (Supplementary Fig. 12a,b and ref. 3), H3K27ac HiChIP identified increased SNP–TSS activity in Treg cells at shared T cell enhancers, consistent with several studies identifying the crucial role of this cell type in disease pathogenesis35. Of note, autoimmune disease signal enrichments were not readily apparent from 1D H3K27ac ChIP–seq peaks, aggregated ChIP–seq signal within the TAD containing the SNP, or cell line H3K27ac HiChIP data sets (Fig. 4b and Supplementary Fig. 12c). Therefore, examining 3D disease variant interactions may capture cell type biases more robustly than 1D epigenomic data. Finally, to validate our findings with an orthogonal data set, we performed SNP–TSS EIS analysis on an overlapping set of autoimmune disease–associated SNPs obtained from the National Heart, Lung, and Blood Institute (NHLBI) GRASP catalog. We observed a similar pattern of enrichment for T cell subset-specific HiChIP signal in disease-associated variants (Supplementary Fig. 12d).
Figure 4.
HiChIP identifies cell type specificity and target genes of autoimmune disease–associated variants. (a) Generation of a loop set between all autoimmune disease SNPs and gene promoters within a 2-Mb region. (b) H3K27ac ChIP and HiChIP signal bias in T cell subtypes for SNP–TSS pairs. For each bin, PICS SNPs are tagged by H3K27ac only in the concordant cell type for the bias tested. SNPs are grossly divided into associations with autoimmune diseases or control, non-immune traits. Asterisks indicate FDR < 5%. (c) EIS bias of SNP–TSS loops (with nearest gene annotated) in TH17 and Treg subsets (memory T cells) versus naive T cells and TH17 versus Treg cells. Percentages represent the percentage of SNP–TSS interactions that demonstrate biased HiChIP signal in the indicated cell type (log2 (fold change) > 1). MS, multiple sclerosis. (d) Number of HiChIP gene targets versus nearest-gene predictions for all looping non-genic autoimmune disease SNPs as well as SNPs for specific diseases. (e) Global validation of HiChIP SNP gene targets. Synthetic SNP–TSS pairs were generated from each CD4+ eQTL SNP to its associated gene and compared to both a distance-matched shuffled SNP–TSS pair and a liver (L) eQTL SNP–TSS pair. P values were calculated by Kolmogorov–Smirnov test. (f) HiChIP target gene RNA levels by qRT–PCR in CRISPRi My-La cells with targeting of SNP-containing enhancers of interest, as well as positive-control sgRNAs to the HiChIP target promoters and a non-targeting negative control (n = 3 biological replicates, each with 2 technical replicates). RA, rheumatoid arthritis; PBC, primary biliary cirrhosis. *P < 0.05; **P < 0.01, two-tailed Student’s t test. In box plots, each box extends from the 25th to the 75th percentile with a line representing the median, and whiskers extend to the minimum and maximum values.
We leveraged HiChIP to identify potential gene targets of intergenic SNPs, which have classically been paired to the nearest neighboring gene. We overlapped the SNP–TSS pairs with loops to call a discrete set of target pairs. We then performed differential analysis on the SNP–TSS loops to ascertain bias for specific T cell subsets (Fig. 4c and Supplementary Table 5). Examples of biased SNP–TSS pairs included FOXO1 in naive T cells (rs9603754), BATF (rs2300604) in memory T cells, CTLA4 (rs10186048) in Treg cells, and IL2 (rs7664452) in TH17 cells (Fig. 4c and Supplementary Table 5). Next, we sought to characterize the connectivity landscape of the SNP–TSS loops. We identified an average of 1.75 gene targets per autoimmune disease–associated SNP (ranging from 0 to over 10 target genes), whereas variants for non-immune traits did not demonstrate an increase in the number of targets (0.33 genes per SNP; Supplementary Fig. 12e). For 684 autoimmune disease intergenic SNPs, we identified a total of 2,597 HiChIP target genes, representing a fourfold increase in the number of target genes for known disease-associated SNPs (Fig. 4d). Only 367 (~14%) of all targets were the nearest gene to the SNP, while approximately ~86% of SNPs skipped at least one gene to reach a predicted target TSS (Supplementary Fig. 12e). Furthermore, approximately ~45% of SNP to HiChIP target interactions had increased signal as compared to the interaction between the same SNP and the nearest gene, despite distance biases.
Target gene validation by eQTL and CRISPRi
HiChIP enhancer–target gene interactions can be validated using previously identified point mutations that alter expression at distantly located genes in T cells—that is, expression quantitative trait loci (eQTLs)36. For example, the celiac disease–associated SNP rs2058660 impacts expression of the inflammatory cytokine receptor genes IL18RAP, IL18R1, IL1RL1, and IL1RL2, which are known regulators of intestinal T cell differentiation and response37. HiChIP EIS showed contacts between rs2058660 and the promoters of each of these predicted target genes (Supplementary Fig. 13a). Similarly, the Crohn’s disease risk variant rs6890268 and the multiple sclerosis risk variant rs12946510 impact the expression of PTGER4 and IKZF3, respectively, and H3K27ac HiChIP also demonstrated clear contacts between these SNPs and their predicted promoters (Supplementary Fig. 13a). Globally, HiChIP contact signal was increased in eQTLs in T cells as compared to a distance-matched background loop set (P < 2.2 × 10−16; Fig. 4e) or to eQTLs identified in an unrelated cell type (liver; P < 2.2 × 10−16). The overlap of HiChIP and eQTL loci provides support for chromosome interactions as a physical basis for distal eQTLs10–12 and further validates the HiChIP approach to assign enhancer–target gene relationships.
We next sought to directly validate HiChIP SNP–gene target interactions using CRISPRi in My-La cells. First, we focused on three loci of interest in primary T cells and then confirmed that the SNP–TSS loops were also present in My-La cells (Fig. 4f and Supplementary Fig. 13b). We then targeted sgRNAs to these SNP-containing enhancers, as well as positive-control sgRNAs to the HiChIP target gene promoters and a non-human-genome-targeting negative control. As expected, we observed a significant reduction in RNA levels for the HiChIP target genes upon CRISPRi of the corresponding SNP-containing enhancers (Fig. 4f).
Fine-mapping of disease-associated DNA variants
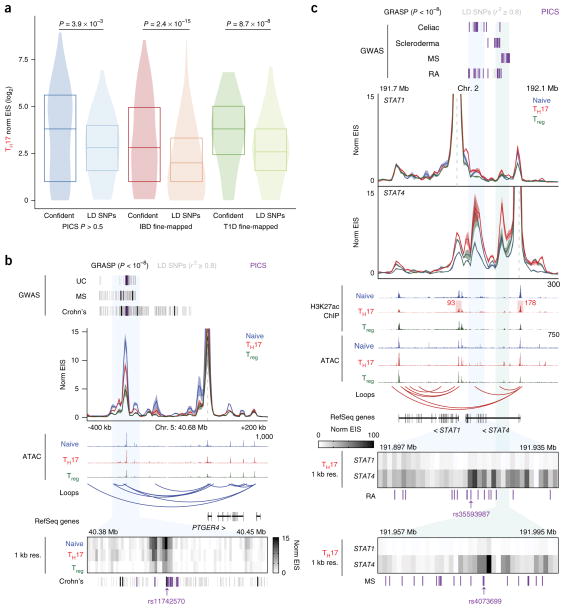
As SNP–TSS HiChIP signal is capable of identifying the target genes of candidate SNPs, we asked whether TSS–SNP HiChIP signals could also be used to nominate functional causal variants within haplotype blocks in a reciprocal manner. We first performed a proof-of-principle analysis using fine-mapped SNPs associated with inflammatory bowel disease (IBD)38 or type 1 diabetes (T1D)39 as well as high-confidence PICS SNPs and examined EIS from putatively causal SNPs to all gene promoters within 300 kb. EIS from putatively causal SNPs to gene promoters was significantly higher than EIS from a distance-matched set of SNPs within the same linkage disequilibrium (LD; r2 ≥ 0.8) block to gene promoters (P = 2.4 × 10−15, 8.7 × 10−8, and 3.9 × 10−3 for IBD fine-mapped SNPs, T1D fine-mapped SNPs, and high-confidence PICS SNPs, respectively; Fig. 5a and Supplementary Fig. 14a). Next, we assessed the fine-mapping ability of HiChIP EIS at individual loci of interest. We focused on IBD- and multiple sclerosis–associated SNPs neighboring the PTGER4 and SATB1 loci and performed v4C analysis anchored at the gene promoters. We calculated EIS signal at 1-kb resolution and identified specific regions within the LD blocks that contained the highest EIS to the target promoters, positioning the likely causal SNPs within these regions (Fig. 5b and Supplementary Fig. 14b). For example, at the PTGER4 locus (Fig. 5b), the ~160-kb genomic interval spanned by LD SNPs in association with Crohn’s disease was refined to two bins of 3 kb and 4 kb, which both contained PICS SNPs.
Figure 5.
Fine-mapping of GWAS-identified variants using H3K27ac HiChIP. (a) Global validation of HiChIP signal at putatively causal SNPs (confident) versus corresponding SNPs in LD (LD SNPs; r2 ≥ 0.8) for TH17 cells. SNP–TSS pairs were generated from published fine-mapping data sets, in comparison to a distance-matched SNP–TSS pair set in the same LD block. P values were calculated by Kolmogorov–Smirnov test. (b) Interaction profile of the PTGER4 promoter and a 1-kb-resolution visualization of the SNP-containing enhancer of interest. SNPs in LD (r2 ≥ 0.8) correspond to GRASP SNPs (genome-wide significance P < 1 × 10−8). The highlighted SNP was identified in both the high-confidence PICS and GRASP data sets. UC, ulcerative colitis. (c) Interaction profiles of the STAT1 and STAT4 promoters, with 1-kb-resolution visualizations of the SNP-containing enhancers of interest. 1D signal contributions at the STAT1 and STAT4 promoters are highlighted. Highlighted SNPs are PICS SNPs closest to focal EIS for STAT4.
We asked whether complex disease–associated loci containing more than one gene could be fine-mapped using HiChIP. We focused on two disease-associated enhancers in between the STAT1 and STAT4 gene promoters (Fig. 5c). These two genes encode transcription factors with distinct roles in immune regulation. Signal transducer and activator of transcription 1 (STAT1) is critical for type I interferon (IFN) and IFN-γ signaling, whereas STAT4 induces TH1 differentiation and IFN-γ expression40. We investigated bias of these enhancers to STAT1 and STAT4 and found that, despite comparable linear distances and 1D signals at the promoters, the enhancers were biased to interact with STAT4. Next, we fine-mapped the disease-associated SNPs within this locus using 1-kb-resolution EIS from the STAT4 promoter and narrowed down candidate functional variants within the two enhancers (Fig. 5c). In summary, HiChIP EIS can nominate functional causal variants within haplotype blocks, and two-way analysis of target gene identification from an enhancer of interest and high-resolution interaction maps of that enhancer with its target gene can be used to fine-map disease-associated loci containing several candidate genes.
Allelic target gene bias of cardiovascular disease variants
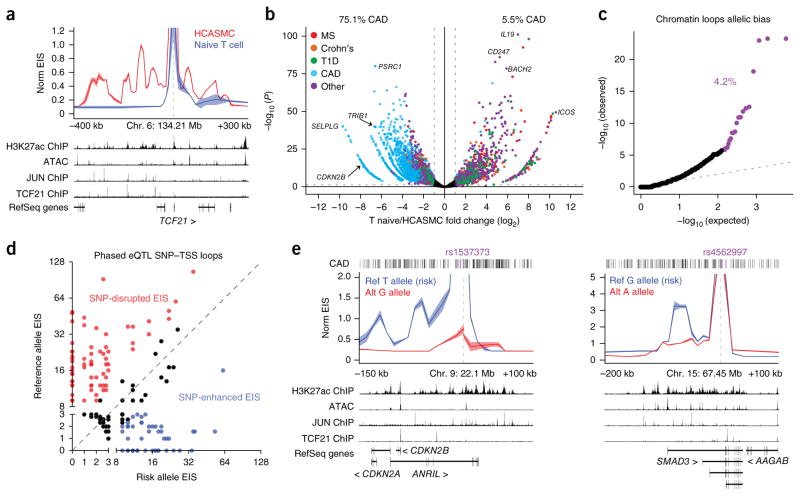
Finally, we asked whether this approach could be applied broadly to other categories of human disease and whether we could directly test SNP–TSS associations using allele-specific HiChIP. We generated high-resolution enhancer–promoter maps from primary human coronary artery smooth muscle cells (HCASMCs), which can be used to inform on variants linked to cardiovascular diseases41. First, to validate cell type specificity, we examined the gene promoter for TCF21, a transcription factor required for the differentiation of HCASMCs42, and observed enrichment of HCASMC EIS relative to naive T cells (Fig. 6a). We next examined the 9p21.3 locus, which harbors risk associations with several cardiovascular disorders43–45. We found that the promoters of all three genes in the locus interacted with one another and with CAD-variant-containing enhancers located approximately 100 kb upstream of the CDKN2B promoter (Supplementary Fig. 15). We then generated SNP–TSS target lists using CAD-associated SNPs identified in the CARDIoGRAMplusC4D study46. We again performed differential analysis on the SNP–TSS loops to ascertain bias for HCASMCs versus naive T cells (Fig. 6b). Overall, 75.1% of HCASMC-biased SNP–TSS pairs involved CAD-associated SNPs, whereas only 5.5% of naive T cell–biased SNP–TSS pairs were CAD SNP–TSS loops. Next, we examined the connectivity of the HCASMC SNP–TSS contacts and identified 1,062 gene targets, of which only 120 (~11%) mapped to the nearest gene. Furthermore, approximately 89% skipped at least one gene to reach a predicted target TSS, and 64% of SNPs were mapped to more than a single gene target.
Figure 6.
HiChIP identifies allelic bias to target genes for cardiovascular disease risk variants. (a) Interaction profile of the TCF21 gene promoter for H3K27ac HiChIP of HCASMCs and naive T cells. (b) EIS bias between HCASMCs and naive T cells in a union set of CARDIoGRAMplusC4D CAD and PICS autoimmune disease SNP–TSS loops. Percentages represent the percent of SNP–TSS interactions that demonstrate biased HiChIP signal in the indicated cell type (log2 (fold change) > 1). (c) Quantile–quantile plot of allelic EIS imbalance in high-confidence loops. Allelic mapping biased loops were identified through simulation and removed before EIS analysis. (d) EIS bias between CAD risk variants and their alternative alleles to eQTL-associated target genes. (e) Allele-specific HiChIP interaction profiles at the 9p21.3 and SMAD3 loci at 10-kb resolution to examine the functional consequence of a risk variant as compared to its alternative allele. ATAC–seq and ChIP–seq data sets shown are from ref. 41.
We took advantage of genome phasing information in HCASMCs to measure enhancer–promoter interactions at allele-specific CAD-associated SNPs, allowing us to examine the functional consequence of a risk variant as compared to its alternative allele in the same nucleus. First, 4.2% of high-confidence loops in HCASMCs with no observed mapping bias in the anchors exhibited significant allelic bias (FDR < 0.05; Fig. 6c), consistent with the frequency of allelic imbalance of RNA expression and prior evidence of allele-specific regulation of specific enhancer–promoter interactions47,48. We leveraged this global enhancer–promoter allelic bias to examine the effect of a risk-associated variant as compared to its control alternative allele for a set of CAD-associated SNP–target gene pairs (Fig. 6d)49. We found that many risk alleles disrupted enhancer–target gene interactions, but a subset of pathogenic SNPs increased enhancer–target gene interaction. At CAD risk variant rs1537373 in the 9p21.3 locus, the risk allele (T) showed increased EIS to the CDKN2A promoter as well as an additional enhancer within the long noncoding RNA (lncRNA) ANRIL gene relative to the reference allele (G) (Fig. 6e). We further observed increased EIS of the CAD risk variant rs4562997 to an additional SMAD3 enhancer 10 kb downstream of the TSS (Fig. 6e). The ability to resolve enhancer connectomes of the risk and reference alleles in the same nucleus demonstrates that the mutated base in the risk allele suffices to alter enhancer looping in cis in disease-relevant primary cells.
DISCUSSION
Here we developed an approach to define the high-resolution landscape of enhancer–promoter regulation in primary human cells. We find that enhancer–promoter contacts are highly dynamic in related cell types and often involve genomic elements with shared accessibility. Accordingly, many complex features of the 3D enhancer connectome cannot simply be predicted from 1D data, demonstrating that mapping conformation in primary cells can identify new regulatory connections underlying gene function in human disease. We take advantage of this principle to chart the connectivity of autoimmune and cardiovascular disease genome-wide association study (GWAS)-identified SNPs and link SNPs to hundreds of potential target genes. Although non-genic SNPs have previously been paired with their closest neighboring gene, we find that the majority of these variants can engage in long-distance interactions, including skipping several promoters to predicted target genes, connecting to multiple genes, or acting in concert with enhancer cliques to contact a single gene. Further use of this approach will help to clarify hidden mechanisms of human disease that are driven by genetic perturbations in non-protein-coding DNA elements, which can now be linked to their cognate gene targets in primary cells.
ONLINE METHODS
Human subjects
This study was approved by the Stanford University Administrative Panels on Human Subjects in Medical Research, and written informed consent was obtained from all participants.
Cell culture and primary T cell isolation
Mouse ES cells (v6.5, Novus Biologicals, NBP1-41162) were cultured in Knockout DMEM (Gibco) supplemented with 15% FBS and leukemia inhibitory factor (LIF; Millipore) to 80% confluence. GM12878 (Coriell), Jurkat, and My-La (CD4+) cells (ATCC) were grown in RPMI 1640 (Gibco) supplemented with 15% FBS to a concentration of 500,000 to 1 million cells/ml. Normal donor human peripheral blood cells were obtained fresh from AllCells. CD4+ T cells were enriched from peripheral blood using RosetteSep Human CD4+ T Cell Enrichment Cocktail (StemCell Technology). For CD4+ T helper cell subtypes, naive T cells were sorted as CD4+CD25−CD45RA+ cells, TH17 cells were sorted as CD4+CD25-CD45RA−CCR6+CXCR5− cells, and Treg cells were sorted as CD4+CD25+CD127lo cells. Antibodies used for FACS included the following: PerCP/Cy5.5 anti-CD45RA (BioLegend, 304122), Brilliant Violet 510 anti-CD127 (BioLegend, 351331), APC/Cy7 anti-CD4 (BioLegend, 344616), PE anti-CCR6 (BioLegend, 353410), FITC anti-CD25 (BioLegend, 302603), Brilliant Violet 421 anti-CXCR3 (BioLegend, 353715), and BB515 anti-CXCR5 (BD Biosciences, 564625). For HiChIP experiments, 500,000 to 1 million cells were sorted into RPMI medium supplemented with 10% FCS. For ATAC–seq experiments, 55,000 cells were sorted into RPMI medium supplemented with 10% FCS. Post-sort purities of >95% were confirmed by flow cytometry for each sample.
A primary HCASMC line derived from a normal human donor heart was purchased from Cell Applications (350-05A) and cultured in smooth muscle growth medium (Lonza, CC-3182) supplemented with hEGF, insulin, hFGF-b, and 5% FBS. Cells were grown according to Lonza’s instructions.
Cell fixation
Detached cell lines or sorted CD4+ T cells were pelleted and resus-pended in fresh 1% formaldehyde (Thermo Fisher) at a volume of 1 ml of formaldehyde per 1 million cells. Cells were incubated at room temperature for 10 min with rotation. Glycine was added at a final concentration of 125 mM to quench the formaldehyde, and cells were incubated at room temperature for 5 min with rotation. Finally, cells were pelleted and washed with PBS, pelleted again, and stored at −80 °C or immediately taken into the HiChIP protocol.
HiChIP
The HiChIP protocol was performed as previously described, using antibody to H3K27ac (Abcam, ab4729) or CTCF (Abcam, ab70303)16 with the following modifications. For primary T cells, we performed HiChIP on as many cells as we could obtain from a blood donation—approximately 500,000 to 1 million cells per T cell subtype per replicate. We performed 2 min of sonication, did not carry out Protein A bead preclearing, used 4 μg of antibody to H3K27ac (Abcam, ab4729), and captured the chromatin–antibody complex with 34 μl of Protein A beads (Thermo Fisher). Qubit quantification following ChIP ranged from 5–25 ng depending on the cell type and amount of starting material. The amount of Tn5 used and number of PCR cycles performed were based on the post-ChIP Qubit amounts, as previously described16.
Twenty-five million cell line libraries were generated as previously described16. For mouse ES cell samples with low cell numbers, we performed 2 min of sonication and did not carry out Protein A bead preclearing. Either 4 μg or 2 μg of antibody to H3K27ac (Abcam, ab4729) was used for ChIP in 500,000 or 100,000/50,000 cells, respectively, and the chromatin–antibody complex was captured with 34 (500,000 cells) or 20 (100,000/50,000 cells) μl of Protein A beads. Post-ChIP Qubit quantification for the 25 million cell samples was approximately 1.5 μg. For lower cell numbers, quantification was 30, 10, and 5 ng for 500,000, 100,000, and 50,000 cells, respectively. The amount of Tn5 used and the number of PCR cycles performed were based on the post-ChIP Qubit amounts, as previously described.
HiChIP samples were size selected by PAGE purification (300–700 bp) for effective paired-end tag mapping and were therefore removed of all primer contamination that would contribute to recently reported ‘index switching’ on the Illumina HiSeq 4000 sequencer50. All libraries were sequenced on the Illumina HiSeq 4000 instrument to an average depth of 500–600 million total reads.
HiChIP data processing
HiChIP paired-end reads were aligned to the hg19 or mm9 genome using the HiC-Pro pipeline51. Default settings were used to remove duplicate reads, assign reads to MboI restriction fragments, filter for valid interactions, and generate binned interaction matrices. HiC-Pro filtered reads were then processed into a .hic file using the hicpro2juicebox function. The Juicer pipeline HiCCUPS tool was used to identify high-confidence loops4 using the same parameters as for the GM12878 in situ Hi-C map: hiccups -m 500 -r 5000,10000 -f 0.1,0.1 -p 4,2 -i 7,5 -d 20000,20000 .hic_input HiCCUPS_output. For T cell Juicer loops, performing the default Juicer calls resulted in a high rate of false positives upon visual inspection of the interaction matrix. We therefore called loops with the same HiCCUPS parameters in two biological replicates for each T cell subtype and then filtered loops for those that were reproducibly called in both replicates. In addition, we removed all loops greater than 1 Mb in length.
1D signal enrichment and peak calling were generated from the HiC-Pro filtered contacts file. Intrachromosomal contacts were filtered, and both anchors were extended by 75 bp. The combined bed file containing both anchors was then used to generate bigwigs for visualization in the WashU Epigenome Browser or call peaks using MACS2.
Allele-specific HiChIP data processing was achieved using HiC-Pro’s allele-specific analysis features51. First, HCASMC phasing data41 were used to mask the hg19 genome and make indexes. HiC-Pro settings were similar to those described above, with the exception that reads were aligned to the masked genome and then assigned to a specific allele on the basis of phasing data.
Interaction matrices and virtual 4C visualization
HiChIP interaction maps were generated with Juicebox using KR matrix balancing and visualized using Juicebox software at 500-kb, 25-kb, 10-kb, and 5-kb resolution as indicated in each analysis4. For 1-kb profiles, raw matrix counts were visualized in Java TreeView.
v4C plots were generated from dumped matrices generated with Juicebox. The Juicebox tools dump command was used to extract the chromosome of interest from the .hic file. The interaction profile of a specific 5-kb or 10-kb bin containing the anchor was then plotted in R. Replicate reproducibility was visualized with the mean profile shown as a line and the shading surrounding the mean representing the s.d. between replicates. For the HCASMC data, we observed low read coverage for allele-specific v4Cs at loci of interest. This is due to the density of SNPs for this genotype and a low number of reads containing a phased SNP. We thus could not observe interaction profiles when visualizing separate replicates with s.d. We therefore used pseudoreplicates for the HCASMC v4C visualizations52.
High-confidence Juicer loop calls were loaded into the WashU Epigenome Browser along with corresponding ATAC–seq profiles and publically available H3K27ac ChIP–seq data from the Roadmap Epigenomics Project. Browser shots from WashU track sessions were then included in v4C and interaction map anecdotes.
Differential analysis of HiCCUPS loop calls
Juicer loop calls from the three T cell subtypes were initially combined into a union set of T cell loops. Loop signal was then obtained for the biological replicates of each T cell subtype. Vanilla coverage square root (VCsqrt) normalized signal for the interaction matrix of each biological replicate using the Juicebox tools dump command. Normalized signal was then assigned to the union loop set in each replicate.
VCsqrt signal per sample was quantile–quantile normalized under the assumption that overall signal was identically distributed across all samples. Following normalization, samples for naive T, TH17 and Treg cells had Pearson correlations of 0.938, 0.942, and 0.934, respectively. Principal-component analysis (PCA) was performed using the prcomp function in R, which demonstrated that the first principal component, which exhibited nearly identical loadings across the six samples, explained 93% of the variance across the six samples. This was taken to represent the shared signal across cell types. Principal components 2–4 explained 2.2%, 2.0%, and 1.4%, respectively.
To study cell-type-specific looping, the residual signal per loop was taken after projecting the loop onto the unit vector along the diagonal (equal signal per cell type). Cell-type-specific and differential looping analysis were performed with the top and bottom 5% of the distributions of either residual signal or differences between cell type residual signals. Hierarchical clustering was performed using the union of all differential loops in these extremes and using 1 minus the Pearson correlation as the distance metric. Quantile– quantile plots were generated by permuting residuals from the same cell type or individual and summing them and using this distribution to calculate P values for the observed sums.
In parallel, differential loops were called using edgeR for both the mouse ES and T cell data sets. Again, biological replicate loop signal was obtained across a union set of Juicer loops. We then used edgeR to identify loops with significant changes in signal among pairwise comparisons (FDR < 0.1, log2 (fold change) > 1). Notably, inspection of differential loops identified from the two methods showed high concordance.
Gene density was calculated from Ensembl gene annotations. GC content was calculated per 10-kb bin using the BEDTools nuc function and aggregated as needed. Notably, Spearman correlation between the gene density of an entire chromosome and the number of differential loop anchors (ρ = 0.914) was much higher than the correlations between the variance in cell type signal per loop anchor and number of genes per 10-kb window (ρ = 0.322) and between differential loop anchors and gene density per 100-kb section (ρ = 0.083). Correlations between GC content and number of differential loops were similar at both the chromosome (ρ = 0.729) and 100-kb (ρ = 0.148) levels, but, while local GC content is likely to confound relative abundance, it is unclear how chromosome-wide GC content could have the same effect.
For mouse ES cell analysis of H3K27ac- and cohesin-mediated HiChIP, we performed edgeR to obtain the biased loops for each factor. To determine the functional bias of the top loops, overlap was determined between edgeR differential loop anchors and relevant ChIP–seq peaks. SMC1A ChIP–seq peaks were obtained from a published data set53. CTCF, RNA polymerase II, and H3K27ac ChIP–seq peaks were obtained from the mouse ENCODE repository54.
RNA expression analysis
Previously generated RNA-seq data55 from naive T, TH17 and Treg cells were downloaded as fastq files from ArrayExpress. Illumina adaptors were trimmed using CutAdapt, and Ensembl cDNA transcripts were quantified using kallisto. Sleuth was used to identify transcripts that were differentially expressed across cell types with FDR controlled at 5%. The mean TPM was calculated per cell type, and TSS differential looping quantiles at genes with nonzero expression were correlated with differential expression quantiles of the same genes. Only 10-kb segments of the genome that contained a single annotated gene were considered to avoid errors in attribution of looping signal per 10-kb bin. For genes with multiple annotated TSSs, the 10-kb bin corresponding to the median TSS was used. Significance was assessed by the cor.test function in R.
Distance-matched eQTL SNP–TSS comparisons
We obtained three groups of eQTL SNP–TSS pairs within 1 Mb for HiChIP EIS comparisons. The treatment group contained CD4+ T cell eQTL–TSS targets. We had two distance-matched groups as controls. The first control group contained CD4+ T cell eQTL SNP–random TSS pairs such that the distance between the eQTL SNP and random TSS differed by at most 5 kb with the treatment group. The second control group contained liver eQTL SNP–TSS targets that were also distance matched with the treatment group. The random eQTL SNP–TSS pairs were generated by individual chromosome, such that the numbers of control pairs and treatment pairs were the same for every chromosome. In total, there were 158,482 distance-matched eQTL–TSS pairs. We compared the 5-kb-resolution EIS values among the three eQTL SNP–TSS groups for all three T cell subtypes. Results showed that, in all cases, the EIS values between CD4+ T cell eQTL–TSS targets were significantly higher than the two control groups (P < 1 × 10−16, Kolmogorov–Smirnov test).
Distance-matched fine-mapped SNP–TSS comparisons
We obtained a list of putatively causal SNPs from the PICS SNP list25 (PICS probability > 0.5), as well as fine-mapped SNPs associated with IBD38 or T1D39. Next, we obtained all SNPs in LD with each putatively causal SNP using European LD blocks determined by all SNPs with r2 ≥ 0.8 with the SNP being considered. For the fine-mapped (T1D/IBD) sets, using SNPs in LD with highly significant GWAS SNPs might mean that there are several SNPs of equal or greater significance in the control set, but we still expect enrichment relative to the LD block.
We collected all the synthetic pairs between the putatively causal immune disease–related SNPs (IBD, T1D, and PICS) and nearby genes within 300 kb. To perform the distance-matched EIS comparisons, for each fine-mapped SNP category, we selected the SNP–TSS control pairs that satisfied two constraints: (i) the selected control SNP was positioned at least 5 kb away from the fine-mapped SNP in the same LD block and (ii) the distance of the SNP–TSS control pair differed with the fine-mapped SNP and the target gene by at most 5 kb.
SNP–TSS loop analyses
We obtained 7,747 PICS SNPs that are associated with autoimmune disease or non-immune traits25. 4,331 (55.9%) were associated with autoimmune disease, and 3,416 (44.1%) were associated with non-immune traits. In addition, we obtained a set of SNPs associated with six overlapping autoimmune diseases using the GRASP catalog (genome-wide significance P < 1 × 10−8).
We constructed a synthetic loop set for immune and non-immune SNPs and any TSS within 1 Mb of each SNP. We then assigned VCsqrt signal in each biological replicate of the three T cell subtypes to the synthetic loop set, as described above.
VCsqrt signal per sample was quantile–quantile normalized as above. In this analysis, we did not restrict to HiCCUPS-identified loops but instead examined all possible interactions between a SNP and a TSS within 1 Mb. Many of these interactions do not exist and therefore had little or no matrix-balanced signal supporting them. While we removed all SNP–TSS pairs below an average of 1 normalized read per sample from subsequent analyses, in general, these false interactions contributed little to the overall differential signal for a trait.
H3K27ac data were downloaded from the WashU Roadmap repository. PICS SNPs were taken from Farh et al.25. Rather than requiring strict membership within H3K27ac peaks, PICS SNPs were labeled as active if they were within 8 kb of a peak, increasing the number of nominally functional SNPs from ~700 to ~3,200 per cell type, out of 7,735 total candidate SNPs.
Differential looping across cell types was assessed by one-sided t test per trait and activity partition if there were at least eight PICS SNP–TSS pairs in the partition. TH17 bias was defined as TH17 total loop signal minus naive T cell total loop signal; Treg bias was defined as Treg total loop signal minus naive T cell total loop signal; and naive T cell bias was defined as naive T cell total loop signal minus one-half times the TH17 and Treg total loop signals. For Supplementary Figure 12a,b,d, naive T cell bias was assessed only using SNPs that were active in naive T cells, TH17 bias from SNPs active in TH17 and not naive T cells, and Treg bias from SNPs active in Treg and not naive T cells. P values were corrected for multiple-hypothesis testing by the Holm method using the p.adjust function in R. Bias assessed from SNPs with the opposing cell type specificities (for example, naive T cell bias using SNPs active in TH17 and Treg cells but not naive T cells) yielded no significant hits after correction.
ATAC–seq
Cells were isolated and subjected to ATAC–seq as previously described16. Briefly, 55,000 cells were pelleted, resuspended in 50 μl of lysis buffer (10 mM Tris-HCl, pH 7.4, 3 mM MgCl2, 10 mM NaCl, 0.1% NP-40 (Igepal CA-630)), and immediately centrifuged at 500 r.c.f. for 10 min at 4 °C. The nuclei pellets were resuspended in 50 μl of transposition buffer (25 μl of 2× TD buffer, 22.5 μl of distilled water, 2.5 μl of Illumina Tn5 transposase) and incubated at 37 °C for 30 min. Transposed DNA was purified with the MinElute PCR Purification kit (Qiagen), and eluted in 10 μl of EB buffer.
ATAC–seq data processing
Adaptor sequence trimming using SeqPurge and mapping to hg19 using Bowtie2 were performed. The reads were then filtered for mitochondrial reads, low-quality reads, and PCR duplicates. The filtered reads for each sample were merged, and peak calling was performed by MACS2. The reads in peaks for each individual sample were quantified using BEDTools intersect with the MACS2 narrow peaks. Peak counts were then combined into an N × M matrix where N represents called peaks, M represents the samples, and each value Di,j represents the peak intensity for respective peak i in sample j. This matrix was then normalized using the ‘CQN’ package in R to minimize bias in GC content and length.
CRISPRi validation of HiChIP targets
For virus production, 5 × 106 HEK293T cells were plated per 10-cm plate. The following day, plasmid encoding lentivirus was cotransfected with pMD2.G and psPAX2 into the cells using Lipofectamine 3000 (Thermo Fisher, L3000) according to the manufacturer’s instructions. Supernatant containing viral particles was collected 48 h after transfection and filtered. For lentivirus encoding individual sgRNAs, virus was concentrated tenfold using Lenti-X concentrator (Clontech, 631232) and stored at −80 °C.
To generate a My-La cell line expressing CRISPRi, 2 × 106 My-La cells were plated per T75 flask. A dCas9-BFP-KRAB-2A-Blast construct was generated by inserting a 2A-Blast cassette into dCas9-BFP-KRAB (Addgene, 46911). 24 h after plating, lentivirus harboring the dCas9-KRAB construct was added with polybrene (4 μg/ml). The medium was changed 24 h after infection and then again 48 h after infection with blasticidin (Thermo Fisher, A1113903) at a 4 μg/ml concentration. Blasticidin-resistant cells were selected for 8 d with the medium changed every other day.
Three different U6 sequences were used for transcription of three different sgRNAs targeting the candidate enhancers, as previously described56. For the MYC locus CRISPRi experiments, each enhancer was targeted by one guide, and all MYC GM or My-La enhancers together were therefore targeted in one experiment. For the PICS SNP CRISPRi experiments, three guides were targeted to a single SNP-containing enhancer. One of three sgRNAs was cloned into a lentiviral vector with a human (pMJ117, Addgene, 85997), mouse (pMJ179, Addgene, 85996), or bovine (pMJ114, Addgene, 85995) U6 promoter. These U6-sgRNA constructs were then combined into a lentivirus with a Puromycin-2A-mCherry vector, which was modified from Addgene 46914. My-La-CRISPRi cells were infected with lentivirus harboring three sgRNAs and selected by puromycin (Thermo Fisher, A11138) at a final concentration of 1 μg/ml. Previously reported sgRNAs targeting VPS54 or SEC24C were used for validating CRISPRi functionality in the My-La cell line57.
For readout of CRISPRi validation, we performed qRT–PCR and cell growth assays on three biological and two technical replicates. For qRT–PCR, RNA was extracted with TRIzol (Thermo Fisher, 15596026) and purified using the Zymo RNA Clean and Concentrator kit (Zymo Research, R1016). qRT–PCR was performed with Brilliant qRT–PCR Mastermix (Agilent, 600825). Ct values were measured using a LightCycler 480 instrument (Roche), and the relative expression level was calculated by the Δ ΔΔCt method in comparison to a GAPDH control. Primer sequences are listed in Supplementary Table 6. For cell growth, we used the CellTiter-Glo kit (Promega, G7572) according to the manufacturer’s instructions. Statistics for both RNA and cell growth changes were calculated using a Student’s t test against the non-targeting control.
CRISPRa validation of HiChIP targets
Jurkat cells were transduced with a lentiviral dCas9-VP64-2A-GFP expression vector (Addgene, 61422). Single GFP+ cells were sorted by FACS into the wells of a 96-well plate, and a clone with bright, uniform GFP expression were selected for use in future experiments.
sgRNAs were cloned in arrayed format for CD69 HiChIP peaks falling outside the range of the tiling CRISPRa screen26. sgRNAs were chosen on the basis of high predicted on-target activity58 and low predicted off-target activity59. sgRNAs were cloned into the lentiviral expression vector pCRISPRia-v2 (Addgene, 84832) as described in Horlbeck et al.60. Lentivirus was produced by transfecting HEK293T cells with standard packaging vectors using TransIT-LTI Transfection Reagent (Mirus, MIR 2306). The medium was changed 24 h after transfection. Viral supernatant was harvested at 48 and 72 h following transfection and immediately used for infection of Jurkat-dCas9-VP64 cells.
Jurkat-dCas9-VP64 cells were infected with lentiviral sgRNAs by resus-pending cells in a 1:1 mix of fresh medium and lentiviral supernatant at a final concentration of 0.25 × 106 cells/ml with 5 μg/ml polybrene. Cells were spinfected for 1 h at 1,000 r.c.f. at 32 °C. The next day, half of the medium was removed and replaced with fresh lentiviral supernatant and the spinfection was repeated. The next day, the cells were resuspended in fresh medium with 1.5 μg/ml puromycin and cultured for 2 d to remove uninfected cells. For readout of CRISPRa validation, we performed qRT–PCR and FACS on two biological and two technical replicates. RNA extraction and qRT–PCR were performed as described above. Expression of CD69 on infected cells (GFP+BFP+) was analyzed by flow cytometry with an Attune NxT flow cytometer (Life Technologies). Statistics for both RNA- and protein-level changes were calculated with one-way ANOVA followed by a Dunnett’s multiple- comparisons test against the non-targeting control.
Additional methods are provided in the Supplementary Note.
Data availability
Raw and processed data are available at the Gene Expression Omnibus (GEO) under accession GSE101498. T cell ATAC–seq and HiChIP data sets can be visualized in the WashU Epigenome Browser with the following link: 98051079. A Life Sciences Reporting Summary is available.
Supplementary Material
Supplementary Figure 1. H3K27ac HiChIP enriches enhancer–promoter-associated chromatin contacts.
(a) Schematic of chromatin contacts captured in H3K27ac HiChIP. (b) Loop call overlap for cohesin HiChIP and H3K27ac HiChIP in mES cells. (c) Contact distance distribution for loops that are biased in cohesin versus H3K27ac HiChIP. (d) Proportion of cohesin- and H3K27ac-biased HiChIP loops that have cohesin, CTCF, H3K27ac, and RNA polymerase II binding in at least one loop anchor. (e) Virtual 4C interaction profile of an H3K27ac-biased loop focused at the Malat1 promoter. (f) Virtual 4C interaction profile of a cohesin-biased loop with low transcriptional activity associated.
Supplementary Figure 2. H3K27ac HiChIP achieves high chromatin loop signal over background at low cell inputs.
(a) KR balanced interaction matrices focused around the Etv5 locus in mES cells with decreasing cellular starting material. (b) Read support reproducibility of loops in H3K27ac HiChIP libraries from 25 million cells as compared to HiChIP in lower cell input libraries. (c) Aggregate peak analysis of loops in mES H3K27ac HiChIP libraries.
Supplementary Figure 3. H3K27ac HiChIP generates reproducible chromatin loop signals at low cell inputs.
(a) Comparison of KR balanced interaction maps in H3K27ac HiChIP biological replicates. Each replicate corresponds to one side of the interaction map, separated by the diagonal. (b) Read support reproducibility of loops between H3K27ac HiChIP biological replicates. (c) HiCCUPS loop call overlap between H3K27ac HiChIP libraries from 25 million and 50,000 mES cells. (d) Preseq library complexity estimation of H3K27ac HiChIP libraries from 25 million and 50,000 mES cells.
Supplementary Figure 4. H3K27ac HiChIP biological replicates from primary sorted T cells are highly reproducible.
(a) cells starting from human peripheral blood. Post-sort validation was used to ensure purity of FACS strategy for naive, TH17, and Treg T cell subtypes. Number represents the percentage of cells within that gate. (b) KR balanced interaction map of T cell subtype biological replicates. Each replicate corresponds to one side of the interaction map, separated by the diagonal. (c) Read support reproducibility of loops between H3K27ac HiChIP biological replicates in naive, TH17, and Treg cells. (d) Aggregate peak analysis of loops in naive, TH17, and Treg H3K27ac HiChIP libraries.
Supplementary Figure 5. Validation of HiChIP-identified CD69 distal enhancers with CRISPR activation.
CRISPRa validation in Jurkat cells of CD69 distal enhancers. CD69 protein levels are shown for individual sgRNAs tiling H3K27ac HiChIP-identified distal CD69 enhancers relative to the KLRF2 promoter as a locus negative control and a non-targeting negative control.
Supplementary Figure 6. Global enhancer connectome characterization in T cell differentiation.
(a) ChromHMM classification of union T cell loop anchors. (b) Contact distance distribution of union T cell loops. (c) Left, agreement in signal observed per loop between samples. The quantile–quantile plot shows modest enrichment above random pairings. Right, PCA on residual signal clusters samples first by naive versus memory cell types (PC1) and then by donor identity (PC2, PC3). (d) Overlap of differential interactions between naive, TH17, and Treg subtypes. Biased interactions were obtained by performing pairwise comparisons between T cell types and analyzing the top 5% enriched and top 5% depleted EIS in each T cell subtype. (e) ChromHMM annotation of total loops, differential loops, and shared loops in all three T cell subtypes. O corresponds to other loop anchors not classified as enhancer or promoter. (f) Number of connections for different classes of loop elements. (g) Quantification of promoters skipped before an enhancer reaches its loop target.
Supplementary Figure 7. Positioning of differential HiChIP contacts in gene-dense chromosomes.
(a) Distribution of T cell subtype differential HiChIP contacts across different chromosomes in comparison to the distribution of all loops.
(b) Correlation of differential loop density with gene density and GC content.
Supplementary Figure 8. Characterization of conformational landscapes surrounding key T cell regulatory factors.
(a–c) Virtual 4C interaction profiles anchored at the promoters of canonical naive (a), TH17 (b), and Treg (c) regulatory factors.
Supplementary Figure 9. Chromosome conformation dynamics of canonical T cell regulatory factors.
(a–c) Delta interaction maps focused around known naive (a), TH17 (b), and Treg (c) regulatory factors.
Supplementary Figure 10. Contribution of H3K27ac ChIP and chromosome conformation to HiChIP EIS.
(a) Left, proportion of H3K27ac ChIP peaks within EIS differential loop anchors that are cell type specific (log2 (fold change) > 1) or shared across all three subtypes. Right, global correlation of EIS and H3K27ac ChIP fold change in different T cell subset pairwise comparisons. (b) Same as a, but using HiChIP 1D differential signal at EIS biased loop anchors. (c) Overlap of H3K27ac HiChIP and bins of Hi-C loops with increasing T cell subset and GM H3K27ac ChIP–seq signal. (d) Overlap of CD4+ capture Hi-C14 with total and differential T cell subset HiChIP loops. Naïve 4X and 16X corresponds to EIS fold-enrichment over TH17 and Treg, TH17 and Treg 4X and 16X corresponds to EIS fold-enrichment over Naïve. (e) Treg-specific loops at the LRRC32 promoter not observed in other H3K27ac HiChIP T cell subsets nor in CD4+ capture Hi-C data14.
Supplementary Figure 11. Enrichment of autoimmune disease–associated SNPs in T cell HiChIP loop anchors.
(a) Enrichment of specific PICS autoimmune disease and non-immune SNPs in anchors of loops called by Juicer and Fit-Hi-C in comparison to a background shuffled loop set. (b) Enrichment of all PICS autoimmune disease and non-immune SNPs in T cell subset–biased loop anchors and all anchors in comparison to a background shuffled loop set.
Supplementary Figure 12. T cell subtype HiChIP specificity of autoimmune disease–associated SNPs.
(a) H3K27ac HiChIP signal bias in T cell subsets for PICS SNP–TSS pairs grouped by each SNP’s presence in cell-type-specific or shared H3K27ac ChIP peaks up to 8 kb away. (b) H3K27ac HiChIP signal bias in T cell subsets for PICS SNP–TSS pairs grouped by each SNP’s presence in cell-type-specific or shared H3K27ac ChIP peaks up to 2.5 kb away. (c) H3K27ac HiChIP signal bias in GM, K562, and My-La cell lines for PICS SNP–TSS pairs grouped by each SNP’s presence in T cell subset-specific or shared H3K27ac ChIP peaks up to 8 kb away. (d) H3K27ac HiChIP signal bias in T cell subsets for GRASP SNP–TSS pairs grouped by each SNP’s presence in cell-type-specific or shared H3K27ac ChIP peaks up to 2.5 kb away. (e) Left, average number of HiChIP gene targets for non-genic autoimmune disease and non-immune SNPs. Middle, quantification of SNP HiChIP gene targets in autoimmune disease. Right, quantification of promoters skipped before a SNP reaches its gene target.
Supplementary Figure 13. Validation of HiChIP signal at SNP–eQTL contacts.
(a) Validation of HiChIP signal at SNP–TSS pairs using interaction profiles of eQTL SNPs to ensure they contact their associated target gene promoter. (b) Interaction profiles of CRISPRi-validated loci in My-La cells.
Supplementary Figure 14. H3K27ac HiChIP fine-mapping of GWAS variants in haplotype blocks.
(a) Global validation of HiChIP signal at putatively causal SNPs versus corresponding SNPs in LD (r2 ≥ 0.8) for naive and Treg cell subtypes. SNP–TSS pairs were generated from published fine-mapped data sets, in comparison to a distance-matched SNP–TSS pair set in the same LD block. (b) Interaction profile of the SATB1 promoter, and a 1-kb resolution visualization of the SNP-containing enhancer of interest. Differences in EIS bias between 5-kb and 1-kb resolution reflect high signal in a single 1 kb bin in TH17 cells around the SATB1 TSS and specific SNPs within the enhancer. LD SNPs (r2 ≥ 0.8) correspond to GRASP SNPs (genome-wide significance P < 10−8). The highlighted SNP is a PICS closest to focal EIS to SATB1.
Supplementary Figure 15. Chromatin interaction landscape of the 9p21.3 cardiovascular disease risk locus.
HCASMC v4C interaction profiles focused around the promoters of CDKN2A, CDKN2B, and ANRIL within the 9p21.3 locus.
Acknowledgments
We thank members of the Chang and Greenleaf laboratories for helpful discussions and J. Tumey for artwork. We thank J. Engreitz, M. Pjanic, and C. Miller for assistance interpreting their published data sets. We thank X. Ji and J. Coller at the Stanford Functional Genomics Facility. We thank Agilent Technologies for generating oligonucleotide pools for cloning of the CRISPRa gRNAs. We thank the UC Berkeley High-Throughput Screening Facility and Flow Cytometry Facility. This work was supported by US National Institutes of Health (NIH) grants P50HG007735 (H.Y.C. and W.J.G.) U19AI057266 (W.J.G.), and 1UM1HG009436 (W.J.G.), the Human Frontier Science Program (W.J.G.), the Rita Allen Foundation (W.J.G.), and the Scleroderma Research Foundation (H.Y.C.). M.R.M. and E.A.B. acknowledge support from the National Science Foundation Graduate Research Fellowship. A.T.S. is a Cancer Research Institute Irvington Fellow supported by the Cancer Research Institute. B.G.G. was supported by an IGI-AstraZeneca Postdoctoral Fellowship. M.R.C. is supported by a grant from the Leukemia & Lymphoma Society Career Development Program. J.E.C. was supported by the Li Ka Shing Foundation and the Heritage Medical Research Institute. W.J.G. and A.M. are Chan Zuckerberg Biohub investigators. Sequencing was performed by the Stanford Functional Genomics Facility (NIH S10OD018220).
Footnotes
AUTHOR CONTRIBUTIONS
M.R.M., A.T.S., W.J.G., and H.Y.C. conceived the project. M.R.M., A.T.S., J.T., and R.L. performed all genomics assays with help from T.N., M.R.C., N.S., and R.A.F. A.T.S. performed all sorting for experiments. B.G.G., S.W.C., M.R.M., M.L.N., K.R.K., and D.R.S. performed all CRISPR validation experiments. E.A.B., C.D., M.R.M., and J.X. analyzed HiChIP data. J.G., A.T.S., and Y.W. analyzed ATAC–seq data. A.J.R. and P.G.G. analyzed GWAS SNPs in HiChIP data. A.K., P.A.K., A.M., J.E.C., T.Q., W.J.G., and H.Y.C. guided experiments and data analysis. M.R.M., A.T.S., E.A.B., C.D., W.J.G., and H.Y.C. wrote the manuscript with input from all authors.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Arensbergen J, van Steensel B, Bussemaker HJ. In search of the determinants of enhancer–promoter interaction specificity. Trends Cell Biol. 2014;24:695–702. doi: 10.1016/j.tcb.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rao SSP, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fulco CP, et al. Systematic mapping of functional enhancer–promoter connections with CRISPR interference. Science. 2016;354:769–773. doi: 10.1126/science.aag2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar V, et al. Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS Genet. 2013;9:e1003201. doi: 10.1371/journal.pgen.1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards SL, Beesley J, French JD, Dunning AM. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet. 2013;93:779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP–trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smemo S, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Claussnitzer M, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grubert F, et al. Genetic control of chromatin states in humans involves local and distal chromosomal interactions. Cell. 2015;162:1051–1065. doi: 10.1016/j.cell.2015.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mifsud B, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 14.Javierre BM, et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell. 2016;167:1369–1384. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanjana NE, et al. High-resolution interrogation of functional elements in the noncoding genome. Science. 2016;353:1545–1549. doi: 10.1126/science.aaf7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mumbach MR, et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods. 2016;13:919–922. doi: 10.1038/nmeth.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 18.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferraro A, et al. Interindividual variation in human T regulatory cells. Proc Natl Acad Sci USA. 2014;111:E1111–E1120. doi: 10.1073/pnas.1401343111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arvey A, et al. Genetic and epigenetic variation in the lineage specification of regulatory T cells. eLife. 2015;4:e07571. doi: 10.7554/eLife.07571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bettelli E, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 23.Acosta-Rodriguez EV, et al. Surface phenotype and antigenic specificity of human interleukin 17–producing T helper memory cells. Nat Immunol. 2007;8:639–646. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- 24.Morita R, et al. Human blood CXCR5+CD4+ T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34:108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farh KKH, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simeonov DR, et al. Discovery of stimulation-responsive immune enhancers with CRISPR activation. Nature. 2017;549:111–115. doi: 10.1038/nature23875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engreitz JM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dao LTM, et al. Genome-wide characterization of mammalian promoters with distal enhancer functions. Nat Genet. 2017;49:1073–1081. doi: 10.1038/ng.3884. [DOI] [PubMed] [Google Scholar]
- 29.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanborn AL, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci USA. 2015;112:E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li G, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samstein RM, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151:153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye CJ, et al. Intersection of population variation and autoimmunity genetics in human T cell activation. Science. 2014;345:1254665. doi: 10.1126/science.1254665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petukhova L, et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature. 2010;466:113–117. doi: 10.1038/nature09114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raj T, et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344:519–523. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schiering C, et al. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature. 2014;513:564–568. doi: 10.1038/nature13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang H, et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature. 2017;547:173–178. doi: 10.1038/nature22969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Onengut-Gumuscu S, et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. 2015;47:381–386. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Shea JJ, Lahesmaa R, Vahedi G, Laurence A, Kanno Y. Genomic views of STAT function in CD4+ T helper cell differentiation. Nat Rev Immunol. 2011;11:239–250. doi: 10.1038/nri2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller CL, et al. Integrative functional genomics identifies regulatory mechanisms at coronary artery disease loci. Nat Commun. 2016;7:12092. doi: 10.1038/ncomms12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nurnberg ST, et al. Coronary artery disease associated transcription factor TCF21 regulates smooth muscle precursor cells that contribute to the fibrous cap. PLoS Genet. 2015;11:e1005155. doi: 10.1371/journal.pgen.1005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McPherson R, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Helgadottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 45.Clarke R, et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N Engl J Med. 2009;361:2518–2528. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 46.CARDIoGRAMplusC4D Consortium. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leung D, et al. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature. 2015;518:350–354. doi: 10.1038/nature14217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dixon JR, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Franzén O, et al. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science. 2016;353:827–830. doi: 10.1126/science.aad6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sinha R, et al. Index switching causes “spreading-of-signal” among multiplexed samples in Illumina HiSeq 4000 DNA sequencing. Preprint at. bioRxiv. 2017 http://dx.doi.org/10.1101/125724.
- 51.Servant N, et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gjoneska E, et al. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature. 2015;518:365–369. doi: 10.1038/nature14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yue F, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonnal RJP, et al. De novo transcriptome profiling of highly purified human lymphocytes primary cells. Sci Data. 2015;2:150051. doi: 10.1038/sdata.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adamson B, et al. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell. 2016;167:1867–1882. doi: 10.1016/j.cell.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gilbert LA, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR–Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Horlbeck MA, et al. Nucleosomes impede Cas9 access to DNA in vivo and in vitro. eLife. 2016;5:e12677. doi: 10.7554/eLife.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. H3K27ac HiChIP enriches enhancer–promoter-associated chromatin contacts.
(a) Schematic of chromatin contacts captured in H3K27ac HiChIP. (b) Loop call overlap for cohesin HiChIP and H3K27ac HiChIP in mES cells. (c) Contact distance distribution for loops that are biased in cohesin versus H3K27ac HiChIP. (d) Proportion of cohesin- and H3K27ac-biased HiChIP loops that have cohesin, CTCF, H3K27ac, and RNA polymerase II binding in at least one loop anchor. (e) Virtual 4C interaction profile of an H3K27ac-biased loop focused at the Malat1 promoter. (f) Virtual 4C interaction profile of a cohesin-biased loop with low transcriptional activity associated.
Supplementary Figure 2. H3K27ac HiChIP achieves high chromatin loop signal over background at low cell inputs.
(a) KR balanced interaction matrices focused around the Etv5 locus in mES cells with decreasing cellular starting material. (b) Read support reproducibility of loops in H3K27ac HiChIP libraries from 25 million cells as compared to HiChIP in lower cell input libraries. (c) Aggregate peak analysis of loops in mES H3K27ac HiChIP libraries.
Supplementary Figure 3. H3K27ac HiChIP generates reproducible chromatin loop signals at low cell inputs.
(a) Comparison of KR balanced interaction maps in H3K27ac HiChIP biological replicates. Each replicate corresponds to one side of the interaction map, separated by the diagonal. (b) Read support reproducibility of loops between H3K27ac HiChIP biological replicates. (c) HiCCUPS loop call overlap between H3K27ac HiChIP libraries from 25 million and 50,000 mES cells. (d) Preseq library complexity estimation of H3K27ac HiChIP libraries from 25 million and 50,000 mES cells.
Supplementary Figure 4. H3K27ac HiChIP biological replicates from primary sorted T cells are highly reproducible.
(a) cells starting from human peripheral blood. Post-sort validation was used to ensure purity of FACS strategy for naive, TH17, and Treg T cell subtypes. Number represents the percentage of cells within that gate. (b) KR balanced interaction map of T cell subtype biological replicates. Each replicate corresponds to one side of the interaction map, separated by the diagonal. (c) Read support reproducibility of loops between H3K27ac HiChIP biological replicates in naive, TH17, and Treg cells. (d) Aggregate peak analysis of loops in naive, TH17, and Treg H3K27ac HiChIP libraries.
Supplementary Figure 5. Validation of HiChIP-identified CD69 distal enhancers with CRISPR activation.
CRISPRa validation in Jurkat cells of CD69 distal enhancers. CD69 protein levels are shown for individual sgRNAs tiling H3K27ac HiChIP-identified distal CD69 enhancers relative to the KLRF2 promoter as a locus negative control and a non-targeting negative control.
Supplementary Figure 6. Global enhancer connectome characterization in T cell differentiation.
(a) ChromHMM classification of union T cell loop anchors. (b) Contact distance distribution of union T cell loops. (c) Left, agreement in signal observed per loop between samples. The quantile–quantile plot shows modest enrichment above random pairings. Right, PCA on residual signal clusters samples first by naive versus memory cell types (PC1) and then by donor identity (PC2, PC3). (d) Overlap of differential interactions between naive, TH17, and Treg subtypes. Biased interactions were obtained by performing pairwise comparisons between T cell types and analyzing the top 5% enriched and top 5% depleted EIS in each T cell subtype. (e) ChromHMM annotation of total loops, differential loops, and shared loops in all three T cell subtypes. O corresponds to other loop anchors not classified as enhancer or promoter. (f) Number of connections for different classes of loop elements. (g) Quantification of promoters skipped before an enhancer reaches its loop target.
Supplementary Figure 7. Positioning of differential HiChIP contacts in gene-dense chromosomes.
(a) Distribution of T cell subtype differential HiChIP contacts across different chromosomes in comparison to the distribution of all loops.
(b) Correlation of differential loop density with gene density and GC content.
Supplementary Figure 8. Characterization of conformational landscapes surrounding key T cell regulatory factors.
(a–c) Virtual 4C interaction profiles anchored at the promoters of canonical naive (a), TH17 (b), and Treg (c) regulatory factors.
Supplementary Figure 9. Chromosome conformation dynamics of canonical T cell regulatory factors.
(a–c) Delta interaction maps focused around known naive (a), TH17 (b), and Treg (c) regulatory factors.
Supplementary Figure 10. Contribution of H3K27ac ChIP and chromosome conformation to HiChIP EIS.
(a) Left, proportion of H3K27ac ChIP peaks within EIS differential loop anchors that are cell type specific (log2 (fold change) > 1) or shared across all three subtypes. Right, global correlation of EIS and H3K27ac ChIP fold change in different T cell subset pairwise comparisons. (b) Same as a, but using HiChIP 1D differential signal at EIS biased loop anchors. (c) Overlap of H3K27ac HiChIP and bins of Hi-C loops with increasing T cell subset and GM H3K27ac ChIP–seq signal. (d) Overlap of CD4+ capture Hi-C14 with total and differential T cell subset HiChIP loops. Naïve 4X and 16X corresponds to EIS fold-enrichment over TH17 and Treg, TH17 and Treg 4X and 16X corresponds to EIS fold-enrichment over Naïve. (e) Treg-specific loops at the LRRC32 promoter not observed in other H3K27ac HiChIP T cell subsets nor in CD4+ capture Hi-C data14.
Supplementary Figure 11. Enrichment of autoimmune disease–associated SNPs in T cell HiChIP loop anchors.
(a) Enrichment of specific PICS autoimmune disease and non-immune SNPs in anchors of loops called by Juicer and Fit-Hi-C in comparison to a background shuffled loop set. (b) Enrichment of all PICS autoimmune disease and non-immune SNPs in T cell subset–biased loop anchors and all anchors in comparison to a background shuffled loop set.
Supplementary Figure 12. T cell subtype HiChIP specificity of autoimmune disease–associated SNPs.
(a) H3K27ac HiChIP signal bias in T cell subsets for PICS SNP–TSS pairs grouped by each SNP’s presence in cell-type-specific or shared H3K27ac ChIP peaks up to 8 kb away. (b) H3K27ac HiChIP signal bias in T cell subsets for PICS SNP–TSS pairs grouped by each SNP’s presence in cell-type-specific or shared H3K27ac ChIP peaks up to 2.5 kb away. (c) H3K27ac HiChIP signal bias in GM, K562, and My-La cell lines for PICS SNP–TSS pairs grouped by each SNP’s presence in T cell subset-specific or shared H3K27ac ChIP peaks up to 8 kb away. (d) H3K27ac HiChIP signal bias in T cell subsets for GRASP SNP–TSS pairs grouped by each SNP’s presence in cell-type-specific or shared H3K27ac ChIP peaks up to 2.5 kb away. (e) Left, average number of HiChIP gene targets for non-genic autoimmune disease and non-immune SNPs. Middle, quantification of SNP HiChIP gene targets in autoimmune disease. Right, quantification of promoters skipped before a SNP reaches its gene target.
Supplementary Figure 13. Validation of HiChIP signal at SNP–eQTL contacts.
(a) Validation of HiChIP signal at SNP–TSS pairs using interaction profiles of eQTL SNPs to ensure they contact their associated target gene promoter. (b) Interaction profiles of CRISPRi-validated loci in My-La cells.
Supplementary Figure 14. H3K27ac HiChIP fine-mapping of GWAS variants in haplotype blocks.
(a) Global validation of HiChIP signal at putatively causal SNPs versus corresponding SNPs in LD (r2 ≥ 0.8) for naive and Treg cell subtypes. SNP–TSS pairs were generated from published fine-mapped data sets, in comparison to a distance-matched SNP–TSS pair set in the same LD block. (b) Interaction profile of the SATB1 promoter, and a 1-kb resolution visualization of the SNP-containing enhancer of interest. Differences in EIS bias between 5-kb and 1-kb resolution reflect high signal in a single 1 kb bin in TH17 cells around the SATB1 TSS and specific SNPs within the enhancer. LD SNPs (r2 ≥ 0.8) correspond to GRASP SNPs (genome-wide significance P < 10−8). The highlighted SNP is a PICS closest to focal EIS to SATB1.
Supplementary Figure 15. Chromatin interaction landscape of the 9p21.3 cardiovascular disease risk locus.
HCASMC v4C interaction profiles focused around the promoters of CDKN2A, CDKN2B, and ANRIL within the 9p21.3 locus.
Data Availability Statement
Raw and processed data are available at the Gene Expression Omnibus (GEO) under accession GSE101498. T cell ATAC–seq and HiChIP data sets can be visualized in the WashU Epigenome Browser with the following link: 98051079. A Life Sciences Reporting Summary is available.