Abstract
During S phase, replicated DNA must be assembled into nucleosomes using both newly synthesized and parental histones in a process tightly coupled to DNA replication. This DNA replication-coupled process is regulated by multitude of histone chaperones as well as histone modifying enzymes. In recent years, novel insights into nucleosome assembly of new H3-H4 have been gained through studies on the classical histone chaperone CAF-1 and the identification of novel factors involved in this process. Moreover, in vitro reconstitution of chromatin replication has shed light on nucleosome assembly of parental H3–H4, a process that remains elusive. Finally, recent studies have revealed that the replication-coupled nucleosome assembly is important for the determination and maintenance of cell fate in multicellular organisms.
Keywords: DNA Replication, Nucleosome Assembly, Histone Modifications, Epigenetic Inheritance, Cell Fate Maintenance
THE NUCLEOSOME AS A CARRIER OF EPIGENETIC INFORMATION
In eukaryotic cells, chromatin structures govern a variety of cellular processes including DNA replication and gene transcription. The nucleosome, the basic repeat of chromatin, is comprised of 147bp of DNA wrapped around a histone octamer consisting of one H3–H4 tetramer and two H2A–H2B dimers. Histones are modified post-translationally. These post-translational modifications (PTMs) mark different chromatin domains and impact processes related to DNA metabolism. Recently, several studies in Caenorhabditis elegans and Schizosaccharomyces pombe have shown that some histone marks, including tri-methylation of histone H3 lysine 27 (H3K27me3) and histone H3 lysine 9 (H3K9me3), are heritable traits [1–3]. Moreover, alterations in chromatin states via mutation of histone modifying enzymes and histones in human cells play a causal role in tumorigenesis [4, 5]. Therefore, it is important to understand how chromatin states are inherited during mitotic cell divisions, a process that remains elusive.
The DNA replication-coupled nucleosome assembly, a process that links nucleosome formation with on-going DNA synthesis, is likely the first step for passage of epigenetic information. Recent studies have identified novel factors involved in nucleosome assembly and provided new insights into this process [6–10]. Moreover, mutations in factors of the replication-coupled nucleosome assembly pathways have been shown to affect cell fate maintenance and determination [11, 12]. In this review, we discuss the recent progress in understanding of mechanisms of DNA replication-coupled nucleosome assembly and the role of this process in cell identity and differentiation.
AN OVERVIEW OF DNA REPLICATION-COUPLED NUCLEOSOME ASSEMBLY
Immediately following DNA replication, nucleosomes are reassembled on the daughter DNA strands [13–15]. These two processes are fundamentally linked: the speed of the replication fork depends on the supply of new histones and an efficient nucleosome assembly [16–18]. Nucleosome reassembly, an apparently simple task, poses several challenges for the transmission of PTMs to daughter cells, and therefore requires tight regulation. First, the duplication of DNA molecules requires an additional supply of newly synthesized histones which carry PTMs distinct from those found in different chromatin domains of mature chromatin. Thus, the PTMs at distinct chromatin domains on the parental histones needs to be copied to newly synthesized histones by relevant enzymes. Second, all histone modifications are put on by “writers”, enyzmes that add the modifications and are removed by “erasers”, enzymes that remove the PTMs. The presence of erasers complicates the stable propagation of histone marks into daughter cells, which could explain why the inheritance of H3K9me3 in S. pombe can be achieved independently of DNA sequence in the absence of the H3K9me3 eraser, but requires a specific DNA sequence to recruit sufficient amount of H3K9me3 writer in the presence of the eraser [2, 3]. Finally, parental and newly synthesized H3–H4 tetramers form distinct nucleosomes onto replicating DNA in human cells, likely also in other organisms [19], and are proposed to be randomly distributed onto leading and lagging strands of DNA replication forks [20]. This random nature of the distribution of parental and new H3–H4 tetramers makes it necessary to differentiate parental and new H3–H4 tetramers in order to copy marks from parental H3–H4 to new H3–H4, and thus increases the complexity for the transmission of marks from parental histones into daughter cells. Below, we will discuss recent advances on nucleosome assembly of parental and new H3–H4 in two separate sections despite the fact that some of the factors are involved in both pathways.
REGULATION OF DE NOVO NUCLEOSOME ASSEMBLY
A. New insights on old players
Newly synthesized histones H3–H4 must travel from the cytoplasm to the nucleus for deposition on replicating DNA during S phase. Several histone chaperones likely facilitate the folding of histone proteins and prevent precocious aggregation with the DNA [6, 21] (BOX 1). In this review, we will focus on discussing downstream events after the association of the histone chaperone Asf1 (Anti-Silencing Function 1) with H3–H4 dimers.
BOX 1. BUILDING A NUCLEOSOME: THE ROLE OF HISTONE CHAPERONES.
Histone proteins have strong affinity for DNA, which under physiological conditions will lead to formation of protein-DNA aggregates instead of nucleosomes. Histone chaperones facilitate nucleosome formation by escorting histones during their transport, presenting them to histone modifying enzymes, and aid in nucleosome assembly. Apart from their functional similarity, there is no structural feature that defines a histone chaperone. In general, most histone chaperones bind either H3–H4 or H2A–H2B, and some of them, such as FACT, are chaperones for all four histones. Moreover, nucleosome formation in general is a step-wise process with the assembly of H3–H4 tetramer first followed by rapid deposition of H2A–H2B dimers. In this review, we focus on discussing nucleosome assembly of H3–H4 following DNA replication and chaperones related to these two histones. While several H2A–H2B histone chaperones have been described, it remains unclear which one functions specifically during DNA replication. For more detailed description of other histone chaperones for H2A–H2B and DNA replication independent nucleosome assembly, we direct the readers to the following recent reviews [93–95].
Asf1 is a highly conserved histone chaperone and binds H3–H4 through the H3 interface involved in the formation of H3–H4 tetramers, therefore forming a complex with a H3–H4 dimer and blocking the formation of the H3–H4 tetramer [22–24]. In budding yeast, Asf1 is essential for the acetylation of H3 lysine 56 (H3K56ac), a modification occurring predominantly on newly synthesized histone H3. It is likely that the Asf1-H3–H4 complex serves as the substrate for the H3K56 histone acetyltransferase Rtt109 [25, 26]. H3K56ac promotes the interaction of the H3–H4 dimer with the E3 ubiquitin ligase Rtt101-Mms1, which ubiquitylates the C-terminal tail of H3 including K122 [27]. This results in the destabilization of the Asf1-H3–H4 complex, facilitating the hand off of the histone dimer to downstream histone chaperones CAF-1 and Rtt106 (FIGURE 1). Moreover, in budding yeast H3K56ac also increases the binding affinity of histone chaperones CAF-1 and Rtt106 for H3 and thus promotes nucleosome assembly and genome stability [28, 29]. In humans, only a small fraction of the newly synthesized H3 contains this modification [30], and H3K56ac may not play the same critical role in nucleosome assembly as in yeast cells, although ubiquitylation of H3K122, catalyzed by Cul4/DDB1 in human cells, likely also negatively regulates the Asf1-H3 interaction [27] for hand off of H3–H4 from Asf1 to downstream chaperones including CAF-1 based on in vivo studies. However, it remains to be determined how H3K56ac and H3K122ub impact the association of histones with histone chaperones in vitro.
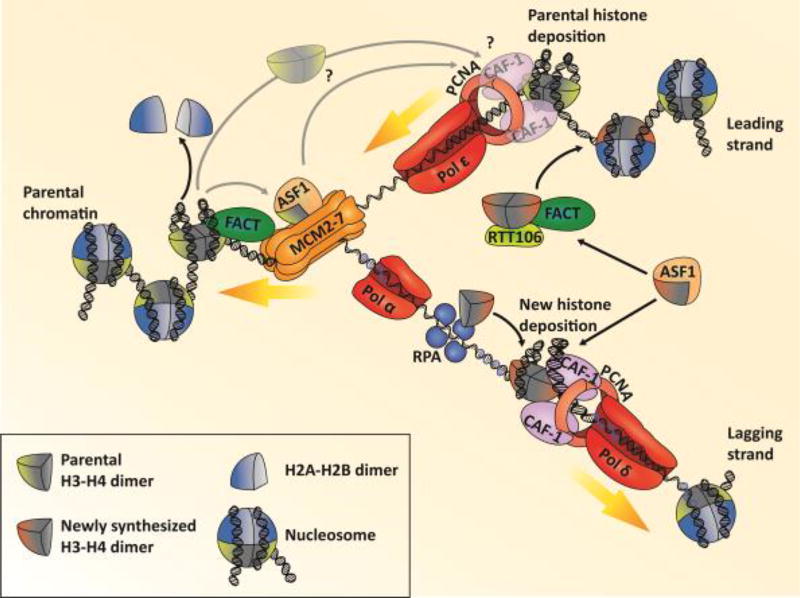
Figure 1. Nucleosome assembly at DNA replication forks.
Ahead of the replication fork, the MCM helicase unwinds the parental double-stranded DNA. The histone chaperone FACT, which interacts with MCM, presumably helps the advancement of the replication fork by disassembling parental nucleosomes. Parental H3–H4 tetramers could then be transferred to Mcm2 and FACT. However, how parental H3–H4 tetramers are shuttled to replicating DNA behind DNA replication forks and how they are deposited is currently unknown. The leading strand is synthesized in the same direction as the advancing replication fork by the DNA polymerase ε, which is in close contact with the CMG helicase. Each of the Okazaki fragments at the lagging strand requires an RNA primer first synthesized by DNA polymerase α. The remainder of the Okazaki fragment is synthesized by DNA polymerase δ, accompanied by PCNA. Newly synthesized H3–H4 dimers are escorted by Asf1 and transferred to downstream histone chaperones including CAF-1, which interacts with PCNA and DNA to deposit H3–H4 tetramers, the first step in nucleosome assembly of new H3–H4. Additionally, in yeast cells Asf1 transfers newly synthesized H3–H4 dimers to Rtt106, which deposits them cooperatively with FACT. RPA binds to ssDNA predominantly at the lagging strand template and regulates nucleosome assembly at the adjoining dsDNA by interacting with H3–H4 and several histone chaperones.
The conserved histone chaperone CAF-1 (Chromatin Assembly Factor 1), comprised of subunits Cac1, Cac2, and Cac3 in yeast (p150, p60, and p48 in humans), is recruited to replication forks through its interaction with the sliding-clamp PCNA [31–34] (FIGURE 1). Recent studies have gained insight into the role of CAF-1’s ability to bind DNA for its stabilization at DNA replication forks and H3–H4 tetramer formation [32, 35, 36]. The Cac1 subunit is responsible for the association with H3–H4 tetramers and acts as the scaffold for the complex in vitro [37, 38]. This largest subunit of CAF-1 in both yeast and human cells contains a Winged-Helix Domain (WHD), which has been shown to be required for budding yeast CAF-1 to bind DNA in a sequence independent manner [32]. Mutations at the WHD in combination with mutations at the Cac1 PCNA binding motif result in reduced chromatin association of CAF-1 with replication forks during S phase [32]. Therefore, the ability of CAF-1 to bind PCNA as well as DNA is important to stabilize CAF-1 at DNA replication forks for nucleosome assembly. Through a series of in vitro studies, several groups uncovered molecular insight into the functions of the DNA binding ability of CAF-1 in this process. One CAF-1 heterotrimer binds a H3–H4 dimer, which will induce a conformational change and unmask the WHD of Cac1 triggering the DNA binding activity of CAF-1 [35–37]. The subsequent association with DNA promotes the formation of a complex containing two CAF-1 and two H3–H4 dimers. Therefore, it is likely that CAF-1 dimerization facilitates the formation of H3–H4 tetramers for nucleosome formation, consistent with a previous finding that dimerization of human CAF-1 complex is important for the function of CAF-1 in nucleosome assembly [39]. Interestingly, mutant histone proteins with impaired ability to form tetramers fail to release from CAF-1 in vitro [36] and the WHD domain also interacts with the H3 N tail based on cross-linking studies. Taken together, these results indicate that the deposition of H3–H4 tetramers by CAF-1 requires the cooperation of DNA binding activity and self-dimerization of CAF-1 as well as H3–H4 tetramerization. This coordination may provide a mechanism to ensure that parental H3–H4 and newly synthesized H3–H4 tetramers form distinct nucleosomes. It has been shown previously that histone chaperone Rtt106 in budding yeast also contains a DNA binding motif and mutations at this motif impact Rtt106’s functions [40]. Moreover, Rtt106 also forms a dimer [29]. Therefore, it would be interesting to determine whether Rtt106 utilizes similar coordinated mechanisms for nucleosome assembly and deposition. Further, future complementary investigations and structural studies on the complex containing CAF-1, histones and possibly DNA will provide additional insights into the molecular mechanism of CAF-1 mediated nucleosome assembly and subsequent release of histone chaperones from H3–H4 for the assembly of tetrasomes.
B. New players in the old puzzle
In the last several years, we have also witnessed discoveries highlighting the roles of new players in the de novo nucleosome assembly. For instance, it has been shown that FACT is involved in nucleosome assembly of new H3–H4 in budding yeast [8]. The histone chaperone complex FACT (FAcilitates Chromatin Transactions) consists of Spt16 and SSRP1 (Pob3 in yeast) and binds to all four canonical histones [41–43]. FACT is involved in nucleosome reorganization in part through its interactions with H2A–H2B and is best known for its role in both gene transcription and DNA replication [43–45]. Using the spt16-m mutant allele, a partial separation of function mutant that is defective in DNA replication, but not in transcription per se, Yang et al. found that FACT functions along with CAF-1 and Rtt106 in nucleosome assembly. Biochemically, new histones H3–H4 are handed-off from Rtt106 to FACT, a transfer promoted by H3K56ac. Genetically, the spt16-m mutant allele exhibits synthetic defects with mutation lacking CAF-1, Rtt106. Therefore, it is likely that CAF-1, Rtt106 and FACT function redundantly as well as in cooperation for the deposition of new H3–H4 histones on the nascent DNA in budding yeast [8] (FIGURE 1).
Recently, the single-stranded DNA (ssDNA)-binding protein replication factor A (RPA) has been implicated in both DNA replication-coupled and replication-independent nucleosome assembly of new H3–H4. RPA consists of three subunits, Rfa1, Rfa2, and Rfa3 in yeast (RPA1, RPA2, and RPA3 in humans) and coats ssDNA generated during replication or repair. RPA is essential for these processes [46]. It has been reported recently that budding yeast RPA binds H3–H4, as well as three histone chaperones (CAF-1, FACT, and Rtt106) known to be involved in nucleosome assembly of new H3–H4. An rfa1 mutant defective in H3–H4 association exhibits impaired nucleosome assembly of new H3–H4. In vitro, RPA binds to ssDNA and promotes nucleosome assembly of H3–H4 on the adjacent double-stranded DNA [9]. Based on these results, the authors propose that RPA serves as a binding platform for several histone chaperones and H3–H4 molecules for their assembly on the adjoining nascent DNA. Human RPA was also found to co-purify with histone H3.1, which is assembled into nucleosomes in a replication-coupled process [47]. Therefore, it is tempting to speculate that human RPA likely also facilitates DNA replication-coupled nucleosome assembly. In addition, human RPA was also found to interact with histone chaperone HIRA in the deposition of H3.3 at gene regulatory elements [47]. Gene regulatory elements are enriched with R-loops [48] and the ability of RPA to bind ssDNA is important for deposition of H3.3. Therefore, it is proposed that RPA binds ssDNA in R-loops and likely promotes HIRA-mediated nucleosome assembly of H3.3 at gene regulatory elements. Thus, the RPA complex has a role in both DNA replication-coupled and replication-independent nucleosome assembly. Previously, H3.3 was proposed to have a gap filling role during DNA replication-coupled nucleosome assembly [49]. It would be interesting to determine whether the RPA-HIRA-H3.3 deposition axis is involved in this process. However, during S phase, RPA binds ssDNA at all DNA replication forks and nucleosome assembly of H3.1 dominates over H3.3. Therefore, if the RPA-HIRA-H3.3 deposition pathway participates in gap filling during S phase, this pathway must be restricted to facilitate CAF-1 mediated nucleosome assembly of H3.1.
The TONSL-MMS22L complex, first described for its role in homologous recombination [50–52], is a newly discovered histone chaperone for H3.1, at least in vitro. Using a quantitative mass spectrometry approach, the TONSL-MMS22L complex was found to co-purify with H3.1 as well as histone chaperone Asf1b and three subunits of replicative helicase MCM [6, 7]. It is proposed that TONSL-MMS22L likely interact with the inactive form of the MCM helicase based on the observation that Cdc45 and GINS, two key components of the active replicative helicase complex CMG (Cdc45, MCM and GINS) were not found to interact with TONSL-MMS22L [6]. In vitro, the TONSL-MMS22L complex has histone chaperone activity and the ankyrin repeat domain (ARD) at the N-terminus of TONSL mediates its interaction with histone H3–H4 [6, 7]. The structural analysis of the ARD domain in complex with the Mcm2 histone binding domain (HBD) (see below) reveals that ARD recognizes unmethylated H4K20, which characterizes newly synthesized H4 [7]. The ability of TONSL to bind H4K20me0 is important for TONSL-MMS22L to bind replication forks. Therefore, it is proposed that in addition to its role as a histone chaperone for H3.1, the TONSLMMS22L also functions in labelling nascent chromatin containing H4K20me0. These studies raise several questions. First, how does the TONSL-MMS22L complex recognize H3.1 over H3.3? Second, does the TONSL-MMS22L complex have a role in nucleosome assembly of H3.1 during normal DNA replication? Third, are the roles of the TONSL-MMS22L complex in nucleosome assembly linked to its roles in homologous recombination at stalled forks?
NUCLEOSOME ASSEMBLY OF PARENTAL H3–H4
Compared to newly synthesized H3–H4, reassembly of parental H3–H4 tetramers into nucleosomes remains poorly understood. Like new H3–H4 tetramers, it is highly likely that parental histones are escorted by histone chaperones, some of which are proteins directly involved in DNA replication, once nucleosomes are disrupted ahead of the replisome. Without an active mechanism for transferring parental H3–H4 ahead of DNA replication forks to the adjacent replicating DNA behind the fork, parental histones would diffuse away, with the consequent loss of locus-specific epigenetic information [53]. The challenge remains to identify proteins involved in nucleosome assembly of parental H3–H4 and elucidate their functions. Recent studies have identified several candidates with a potential role in this process.
Early in vitro studies showed that human Mcm2, a subunit of the MCM helicase complex (BOX 2), contains a histone binding domain (HBD) that binds histones H3–H4 [54–56]. In budding yeast cells, this domain is not required for helicase activity and mutations at the HBD of Mcm2, while having little effect on DNA replication, lead to defects in transcriptional silencing, supporting a role of this domain in chromatin function [57]. If Mcm2 is involved in deposition of parental H3–H4, it is likely that Mcm2 functions with other proteins to do so as yeast cells expressing the Mcm2 histone binding defect mutation are viable.
BOX 2. DNA REPLICATION.
During S phase of the cell cycle, the whole genome is duplicated from origins of replication, DNA sequences with the capacity to initiate DNA replication. In G1 phase, these origins of replication are first bound by the origin recognition complex (ORC, consisting of ORC1–6), followed by two mini-chromosome maintenance (MCM) helicase complexes with the help of CDC6 and CDT1 to form the pre-replication complex. This complex is converted to the pre-initiation complex by the action of kinases DDK and CDKs at the G1 to S phase transition. These kinases phosphorylate numerous replication factors for their activation (MCM complex) or for their recruitment to origins (MCM10, CDC45, GINS, and DNA polymerase ε among others). Two active CMG helicase complexes, comprised of CDC45, MCM complex, and GINS, start their DNA-unwinding activity in opposite directions from the replication origin. At the same time, activation of CMG helps recruit the rest of the replisome components, namely the replication factor C (RFC) complex, which loads the sliding clamp proliferating cell nuclear antigen (PCNA), the ssDNA-binding protein RPA, and DNA polymerases α and δ. Each advancing replication fork unwinds the double-stranded DNA into the leading strand whose continuous replication follows the direction of the fork and the lagging strand, which is discontinuously replicated in the opposite direction of the fork. Replication on both strands starts with the synthesis of an RNA primer by the Pol α primase complex, which also elongates the first few nucleotides. Leading strand DNA replication is mediated by the DNA polymerase ε bound to its processivity factor PCNA as well as the CMG complex. In contrast, the lagging strand is synthesized in short fragments called Okazaki fragments, each requiring the action of both DNA polymerase α and δ for elongation and PCNA [96] (FIGURE 1).
In human cells, Asf1 was found to interact with MCM helicase and this interaction is bridged through histones H3–H4 [58]. Moreover, the H3–H4 molecules that co-purified with MCM helicase contained modifications found on parental histones. Thus, it was proposed that Asf1 and the MCM helicase function in parental histone deposition. Recent structural studies revealed that Mcm2 can form two distinct complexes with histones H3–H4: two Mcm2 proteins bound to a H3–H4 tetramer or one Mcm2 associated with a H3–H4 dimer with Asf1 [59, 60]. When bound to a tetramer, Mcm2 blocks both the DNA- and H2A–H2B-docking sites of H3–H4 and therefore prevents their spurious assembly into nucleosomes, consistent with an early observation that parental H3–H4 remains as tetramers after DNA replication [61]. The authors and others propose a model where Mcm2 captures the evicted H3–H4 tetramer but is then joined by Asf1, which binds the H3 interface involved in the formation of H3–H4 tetramers [23, 24, 62]. In this model, parental H3–H4 tetramers would transiently split into dimers. As stated above, parental H3–H4 tetramers are deposited as a unit during the S phase of the cell cycle. Therefore, it remains to be seen whether splitting these parental tetramers is necessary for their reassembly following DNA replication.
Recently, using chromatin as a template, two groups reconstituted chromatin replication using purified proteins in budding yeast [10, 63]. In this in vitro system, nucleosomes are assembled efficiently following DNA replication. Moreover, newly synthesized H3–H4 was not added in the reaction. Therefore, this system likely contains the minimal components that are essential for the transfer of parental H3–H4 tetramers onto replicating DNA immediately following DNA replication and suggests that nucleosome assembly of parental H3–H4 is separable from nucleosome assembly of new H3–H4. Using this system, it has been shown that FACT is an essential for DNA synthesis, whereas two histone acetyltransferases, Gcn5 and Esa1, also stimulate DNA synthesis [10]. Previously, FACT was found to co-purify with the MCM helicase and is thought to deposit parental histones H3–H4 [57, 64, 65]. In the reconstituted in vitro transcription on chromatin template, FACT is proposed to disrupt nucleosomes to facilitate gene transcription [66, 67]. Therefore, it remains to be determined whether the essential function of FACT in the in vitro DNA replication system is related to FACT’s role in nucleosome reorganization, deposition of parental H3–H4 and/or both (FIGURE 1).
A recent study indicates that different chromatin remodelling complexes utilize distinct histone modifications to determine substrate preference [68]. Moreover, chromatin remodelling complexes have distinct roles in DNA replication in vitro [69]. Thus, it is possible that Gcn5 and Esa1 modify parental histones to guide chromatin remodelling during DNA replication. Gcn5 has been shown previously to acetylate lysine residues at the H3 N terminus and functions in DNA replication-coupled nucleosome assembly of newly synthesized H3–H4 in budding yeast [70]. Therefore, it is likely that Gcn5, like FACT, functions in both nucleosome assembly of parental and newly synthesized H3–H4. In summary, several important factors implicated in nucleosome assembly of parental H3–H4 tetramers have been identified. Future studies are needed to address whether they are indeed involved in transfer of parental H3–H4, and if they are, by which mechanism, using a defined method in vivo and/or site-specific mutations. Further, it would be of great interest to determine whether any other replisome components are needed for nucleosome assembly of parental H3–H4 and to what extent nucleosome assembly of parental H3–H4 coordinates with de novo nucleosome assembly of newly synthesized H3–H4.
CHROMATIN MATURATION FOLLOWING DNA REPLICATION: KEEPING NUCLEOSOMES IN THEIR PLACE
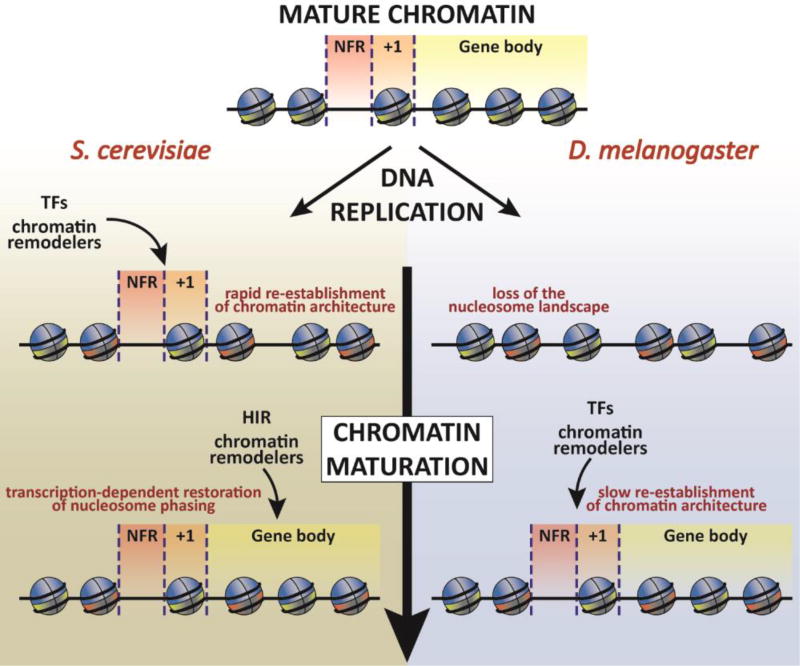
Following nucleosome assembly, initially randomly positioned nucleosomes are then moved to the original DNA positions in a process that is broadly called chromatin maturation. In both yeast and human cells, the time it takes to re-establish nucleosome positions after the passing of the replication fork varies depending on the genomic region. The characteristic nucleosome architecture observed at promoters (the nucleosome-free region (NFR) immediately upstream of the transcription-start site) is reproduced minutes after replication in budding yeast cells [71–73] (FIGURE 2). Two models are proposed for the rapid restoration of NFRs in budding yeast. First, transcription factors and chromatin remodelling complexes rapidly bind to the nascent DNA and act as a reference point for nucleosome deposition [73]. Alternatively, chromatin remodelers cooperate with histone chaperones for the deposition of nucleosomes at the right positions [71]. In contrast to budding yeast, nucleosome positions observed in Drosophila melanogaster late stage embryonic cells follow a different dynamics. After DNA replication, regulatory elements such as promoters and enhancers lose their characteristic NFR features, and are not restored until an hour after the passing of the replication fork [74]. Restoration of the NFRs correlates with binding of transcription factors and chromatin remodelers which probably guide the newly deposited nucleosomes back to their original positions (FIGURE 2). The authors propose that this slower maturation forces transcription factors to compete with nucleosomes for DNA binding [75]. It is also possible that this provides a window of opportunity for alterations in chromatin state and gene transcription. However, in cells from early embryos, the reestablishment of chromatin accessibility patterns was observed minutes after DNA replication [76]. This suggests that chromatin maturation after the passage of the replication fork may depend on the developmental stage in D. melanogaster. In contrast to the NFR regions, the nucleosome landscape at gene bodies displays a slower and variable rate of maturation in budding yeast. Highly transcribed genes show faster maturation after replication pointing towards an active role for transcription in the re-establishment of the nucleosome landscape [72]. Moreover, the HIR complex, involved in replication-independent nucleosome assembly [77], is also required for the faster chromatin maturation observed at active genes (FIGURE 2). Notably, some genes display a distinct rate of maturation between each one of the two copies: when the template for transcription is the newly synthesized DNA strand, either at the leading or lagging strands, the maturation is slower. Since this coincides with the direction of transcription, with the slower maturating copy being transcribed in the opposite direction of the advancing replication fork, the authors propose that a mechanism would suppress transcription from this gene copy to avoid collisions between the replication and transcription machineries [72]. In addition to distinct dynamics in the maturation of nucleosomes at different chromatin regions, different histone modifications appear to be restored at different rates after DNA replication. Through analysis of various histone modifications at newly synthesized and mature chromatin following DNA replication by mass spectrometry, it has been found that the total levels of most PTMs are restored before cells enter the next S phase [78]. Intriguingly, two distinct modes of restoration of histone marks on new H3 have been observed: some histone marks are copied efficiently from old histones to newly synthesize ones, whereas other marks need several generations to allow modifications on newly synthesized H3 to reach the same levels as parental H3. For instance, while monomethylation marks are acquired by new histones before the next cell cycle, trimethylation marks such as H3K9me3 and H3K27me3 take much longer to be restored and require continuous modification of both new and old histones. Together, these studies highlight the dynamic and context-dependent nature for the restoration of chromatin states following DNA replication.
Figure 2. Chromatin maturation after DNA replication.
Mature chromatin has a characteristic nucleosome pattern at gene regulatory elements such as the nucleosome-free region (NFR) upstream of the transcription start site (TSS) and the strongly positioned nucleosome (+1) immediately downstream of the TSS. The coding sequence of active genes also displays tight nucleosome spacing. In Saccharomyces cerevisiae, the NFR and the +1 nucleosome are rapidly restored soon after DNA replication by the action of transcription factors (TFs) and chromatin remodelers (left panel). The nucleosome phasing observed at gene bodies is restored later and is dependent on the transcription rate of a given gene and the activity of the HIR complex and chromatin remodelers. In contrast, the NFRs and the +1 nucleosomes are lost during the replication-coupled nucleosome assembly in Drosophila melanogaster (right panel). The nucleosome landscape at active regions is restored, more than an hour after DNA replication by TFs and chromatin remodelers. This may create a window of opportunity for alterations in chromatin state and cell identity.
ROLE OF DNA REPLICATION-COUPLED NUCLEOSOME ASSEMBLY IN CELL FATE DETERMINATION AND MAINTENANCE
In simpler eukaryotic systems including budding yeast and Drosophila, mutations in factors that are required for replication-coupled nucleosome assembly, including CAF-1 and PCNA, reduce transcriptional silencing [79]. It was proposed that these factors are important for inheritance of the silent chromatin state. In multicellular organisms, the differentiated state of a given cell type is sustained by maintaining a specific transcriptional program. Chromatin structures and histone PTMs play an important role in this process [80]. Consequently, faithful transmission of the epigenetic information encoded in chromatin in each given cell type helps maintain the patterns of gene expression through cell divisions. Recently, the impacts of nucleosome assembly defects on cell identity and differentiation have come to light through studies in multiple systems.
In a screen for proteins that when depleted increase the reprograming efficiency of mouse fibroblast cells into induced pluripotent stem cells by transcription factors including Oct4 [81], the most prominent hits were in the CAF-1 complex subunits [11]. The authors found that a reduction of the levels of CAF-1 complex accelerated the formation of induced pluripotent stem cells from various differentiated cell types. The absence of CAF-1 increases chromatin accessibility at enhancer elements that were otherwise unavailable to transcription factors, promoting the reprograming of these cells [11]. In another independent study, CAF-1 has also been found to suppress the emergence of totipotent cells that resemble 2-cell-stage (2C) blastomere [12]. In mice, only the zygote and the 2-cell-stage blastomeres are considered totipotent (able to generate extraembryonic lineages), a characteristic lost in pluripotent stem cells, which display less cellular plasticity. However, impairment of CAF-1-mediated nucleosome assembly activity (by depletion or by expression of mutant versions defective in chromatin assembly) causes some pluripotent stem cells to revert to the 2C like state. Depletion of this histone chaperone leads to changes in chromatin accessibility that induce the expression of totipotency markers [12]. The increase in chromatin accessibility in cells depleted of CAF-1 likely presents an opportunity for transcription factors and chromatin regulators to reset chromatin states. These studies suggest that CAF-1 could act as a gatekeeper of cell identity by maintaining chromatin patterns through cell division, thus maintaining the transcriptional program of a specific cell type. Early studies indicate that the CAF-1 complex interacts with the heterochromatin protein HP1 during DNA replication, suggesting a mechanism by which this histone chaperone regulates the propagation of the heterochromatin mark H3K9me3 [82–85]. In a separate study, CAF-1 has been reported to be important for the transmission of the repressive mark H3K27me3 and the silencing of genes responsible for the activation of stem cells in plants [86]. CAF-1 interacts with PRC2, the H3K27me3 methyltransferase, and it is proposed that CAF-1 recruits PRC2 to DNA replication forks for copying the H3K27me3 mark from parental H3 to newly synthesized histone H3. This replication-coupled restoration of H3K27me3 in differentiating plant cells contrasts with the slow propagation of this silencing mark observed in adult human cells ([78], see previous section). Whether CAF-1 also plays an active role in the transmission of H3K27me3 by recruiting histone methyltransferases in mammalian cells during development will need to be determined in future experiments. Remarkably, the histone chaperone Asf1 is also involved in cell fate maintenance and determination. Asf1a was identified as a reprogramming factor in oocytes and is necessary to reprogram human adult dermal fibroblasts into induced pluripotent stem cells [87, 88]. How Asf1a functions in this process remains to be determined.
Under certain conditions, stem cells undergo asymmetric cell division, producing one stem cell and one differentiated cell [89]. Factors involved in DNA replication-coupled nucleosome assembly also play a role in asymmetric cell divisions. For instance, in the worm Caenorhabditis elegans, mutations that hinder replication-coupled nucleosome assembly, including mutations at the tetramerization interface of H3 or depletion of CAF-1, abolish asymmetric cell division necessary for the generation of the neural bilateral asymmetry [90]. The authors propose that this asymmetry may arise from the differential nucleosome deposition between the leading and lagging strands at DNA replication forks. PCNA is enriched at the lagging strand (FIGURE 1), which could favour CAF-1-mediated nucleosome deposition, generating sister chromatids with different nucleosome densities that would affect the expression of genes required for neuronal fate on daughter cells. Asymmetric histone segregation has also been observed in the germline stem cells of Drosophila melanogaster, which produce one stem cell enriched with parental histone H3.1 and one differentiated gonialblast enriched in newly synthesized histone H3.1 [91]. In contrast, H3.3, which is assembled into nucleosomes in a replication-independent reaction, does not show this asymmetric segregation pattern. These results indicate that the replication-coupled nucleosome assembly pathway likely plays a role in this drastic asymmetric histone distribution between a stem cell and a differentiated cell.
CONCLUDING REMARKS
The discovery of CAF-1, the classical histone chaperone involved in DNA replication-coupled nucleosome assembly [92], has fuelled progress in our understanding of this important cellular process in the last 25 years. In the past years, many proteins with diverse functions have joined the long list of factors regulating replication-coupled nucleosome assembly. It has become evident that DNA replication and nucleosome assembly are so closely interconnected that some proteins involved in DNA synthesis, such as Mcm2 and RPA, also have a direct and separate function in nucleosome assembly. Structural studies of histone chaperone complexes have revealed new binding modes and interactions between these proteins and their cargo, shedding light into the molecular mechanisms of histone shuttling and deposition. Furthermore, in vitro studies using reconstituted chromatin replication have proven to be an invaluable tool to investigate nucleosome assembly. In contrast with the complex network of histone chaperones that regulate the nucleosome assembly of newly synthesized histones, the exact mechanism governing the transfer of parental histones, the ones carrying the epigenetic information, remains to be unravelled (see Outstanding Questions). One major challenge to answer this and other fundamental questions related to DNA replication-coupled nucleosome assembly is in part due to technical challenges to monitor the process. With the development of new technologies including Cryo-EM technology suitable for the determination of the structures of large complexes, and the eSPAN method that can detect proteins at leading and lagging strands of DNA replication forks genome-wide, as well as potential single molecule technology, we expect to gain new mechanistic insight into the nucleosome assembly process. Finally, with the development of genome editing technologies such as CRISPR/Cas9 that edit genes with precision, we also expect to gain additional insights into the roles of factors in replication-coupled nucleosome assembly in cell differentiation and development in multicellular organisms.
OUTSTANDING QUESTIONS BOX.
-
-
When does the tetramerization of H3–H4 dimers occur when bound to different histone chaperones?
-
-
How is a histone chaperone released from histones for nucleosome formation?
-
-
Which factors are responsible for parental H3–H4 deposition onto replicating DNA?
-
-
In view of the fact that FACT is involved in nucleosome disassembly and reassembly, could this histone chaperone escort parental histones during the process of histone recycling?
-
-
Is nucleosome assembly regulated distinctly at leading and lagging strands of DNA replication forks?
-
-
What are the exact roles of transcription factors and chromatin remodelers in replication-coupled nucleosome assembly?
-
-
Does a transient loss of the nucleosome landscape have any biological meaning in the maintenance of cell identity during division?
-
-
How, and to what extent, does DNA replication-coupled nucleosome assembly regulate cell fate and differentiation? Do factors responsible for distinct steps of nucleosome assembly have different roles in cell identity maintenance and differentiation? What are the implications of deregulation of these mechanisms in diseases such as cancer?
-
-
Is the nucleosome assembly machinery involved in the asymmetrical cell divisions that give rise to two distinct daughter cells? How common is this mode of histone segregation in multicellular organisms?
-
-
Are the different histone marks transmitted with distinctive fidelity? How are they re-established after mitosis?
HIGHLIGHTS.
Replication-coupled nucleosome assembly is the process by which newly synthesized DNA is assembled into nucleosomes immediately following DNA replication using both parental and newly synthesized histones.
In vitro reconstituted DNA replication system using chromatin template identifies minimal factors involved in parental histone transfer.
New roles for well-known proteins including the ssDNA-binding complex RPA in both DNA replication-coupled and DNA replication-independent nucleosome assembly.
Transcription factors and chromatin remodelers cooperate to re-establish chromatin architecture at gene regulatory elements and gene bodies after the passing of the replication fork.
The histone chaperone CAF-1 is involved in the maintenance of cell fate and differentiation.
Acknowledgments
We thank Dr. Jessica Tyler for helpful discussion and suggestions. This work is supported by a grant from NIH R35 GM118015. We apologize to those whose work cannot be cited because of word limitation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gaydos LJ, et al. Gene repression. H3K27me and PRC2 transmit a memory of repression across generations and during development. Science. 2014;345(6203):1515–8. doi: 10.1126/science.1255023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ragunathan K, et al. Epigenetics. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science. 2015;348(6230):1258699. doi: 10.1126/science.1258699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Moazed D. DNA sequence-dependent epigenetic inheritance of gene silencing and histone H3K9 methylation. Science. 2017;356(6333):88–91. doi: 10.1126/science.aaj2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Audia JE, Campbell RM. Histone Modifications and Cancer. Cold Spring Harb Perspect Biol. 2016;8(4):a019521. doi: 10.1101/cshperspect.a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones C, Baker SJ. Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nat Rev Cancer. 2014;14(10) doi: 10.1038/nrc3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campos EI, et al. Analysis of the Histone H3.1 Interactome: A Suitable Chaperone for the Right Event. Mol Cell. 2015;60(4):697–709. doi: 10.1016/j.molcel.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saredi G, et al. H4K20me0 marks post-replicative chromatin and recruits the TONSL-MMS22L DNA repair complex. Nature. 2016;534(7609):714–718. doi: 10.1038/nature18312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, et al. The Histone Chaperone FACT Contributes to DNA Replication-Coupled Nucleosome Assembly. Cell Rep. 2016;14(5):1128–1141. doi: 10.1016/j.celrep.2015.12.096. [DOI] [PubMed] [Google Scholar]
- 9.Liu S, et al. RPA binds histone H3-H4 and functions in DNA replication-coupled nucleosome assembly. Science. 2017;355(6323):415–420. doi: 10.1126/science.aah4712. [DOI] [PubMed] [Google Scholar]
- 10.Kurat CF, et al. Chromatin Controls DNA Replication Origin Selection, Lagging-Strand Synthesis, and Replication Fork Rates. Mol Cell. 2017;65(1):117–130. doi: 10.1016/j.molcel.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheloufi S, et al. The histone chaperone CAF-1 safeguards somatic cell identity. Nature. 2015;528(7581):218–24. doi: 10.1038/nature15749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishiuchi T, et al. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat Struct Mol Biol. 2015;22(9):662–71. doi: 10.1038/nsmb.3066. [DOI] [PubMed] [Google Scholar]
- 13.McKnight SL, Miller OL., Jr Electron microscopic analysis of chromatin replication in the cellular blastoderm Drosophila melanogaster embryo. Cell. 1977;12(3):795–804. doi: 10.1016/0092-8674(77)90278-1. [DOI] [PubMed] [Google Scholar]
- 14.Sogo JM, et al. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J Mol Biol. 1986;189(1):189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
- 15.Kaufman PD, et al. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81(7):1105–14. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- 16.Klapholz B, et al. CAF-1 is required for efficient replication of euchromatic DNA in Drosophila larval endocycling cells. Chromosoma. 2009;118(2):235–48. doi: 10.1007/s00412-008-0192-2. [DOI] [PubMed] [Google Scholar]
- 17.Smith DJ, Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483(7390):434–8. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mejlvang J, et al. New histone supply regulates replication fork speed and PCNA unloading. J Cell Biol. 2014;204(1):29–43. doi: 10.1083/jcb.201305017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu M, et al. Partitioning of histone H3–H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328(5974):94–8. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- 20.Almouzni G, Cedar H. Maintenance of Epigenetic Information. Cold Spring Harb Perspect Biol. 2016;8(5) doi: 10.1101/cshperspect.a019372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campos EI, et al. The program for processing newly synthesized histones H3.1 and H4. Nat Struct Mol Biol. 2010;17(11):1343–51. doi: 10.1038/nsmb.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mousson F, et al. Structural basis for the interaction of Asf1 with histone H3 and its functional implications. Proc Natl Acad Sci U S A. 2005;102(17):5975–80. doi: 10.1073/pnas.0500149102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.English CM, et al. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127(3):495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Natsume R, et al. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446(7133):338–41. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- 25.Han J, et al. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315(5812):653–5. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- 26.Masumoto H, et al. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436(7048):294–8. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 27.Han J, et al. A Cul4 E3 ubiquitin ligase regulates histone hand-off during nucleosome assembly. Cell. 2013;155(4):817–29. doi: 10.1016/j.cell.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Q, et al. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134(2):244–55. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su D, et al. Structural basis for recognition of H3K56-acetylated histone H3–H4 by the chaperone Rtt106. Nature. 2012;483(7387):104–7. doi: 10.1038/nature10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jasencakova Z, et al. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol Cell. 2010;37(5):736–43. doi: 10.1016/j.molcel.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 31.Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96(4):575–85. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 32.Zhang K, et al. A DNA binding winged helix domain in CAF-1 functions with PCNA to stabilize CAF-1 at replication forks. Nucleic Acids Res. 2016;44(11):5083–94. doi: 10.1093/nar/gkw106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, et al. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature. 2000;408(6809):221–5. doi: 10.1038/35041601. [DOI] [PubMed] [Google Scholar]
- 34.Moggs JG, et al. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol Cell Biol. 2000;20(4):1206–18. doi: 10.1128/mcb.20.4.1206-1218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mattiroli F, et al. DNA-mediated association of two histone-bound complexes of yeast Chromatin Assembly Factor-1 (CAF-1) drives tetrasome assembly in the wake of DNA replication. Elife. 2017;6 doi: 10.7554/eLife.22799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sauer PV, et al. Insights into the molecular architecture and histone H3–H4 deposition mechanism of yeast Chromatin assembly factor 1. Elife. 2017;6 doi: 10.7554/eLife.23474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu WH, et al. The Cac1 subunit of histone chaperone CAF-1 organizes CAF-1-H3/H4 architecture and tetramerizes histones. Elife. 2016;5 doi: 10.7554/eLife.18023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim D, et al. Molecular Architecture of Yeast Chromatin Assembly Factor 1. Sci Rep. 2016;6:26702. doi: 10.1038/srep26702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quivy JP, et al. Dimerization of the largest subunit of chromatin assembly factor 1: importance in vitro and during Xenopus early development. EMBO J. 2001;20(8):2015–27. doi: 10.1093/emboj/20.8.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, et al. Structural analysis of Rtt106p reveals a DNA binding role required for heterochromatin silencing. J Biol Chem. 2010;285(6):4251–62. doi: 10.1074/jbc.M109.055996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCullough L, et al. Insight into the mechanism of nucleosome reorganization from histone mutants that suppress defects in the FACT histone chaperone. Genetics. 2011;188(4):835–46. doi: 10.1534/genetics.111.128769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hondele M, et al. Structural basis of histone H2A–H2B recognition by the essential chaperone FACT. Nature. 2013;499(7456):111–4. doi: 10.1038/nature12242. [DOI] [PubMed] [Google Scholar]
- 43.Tsunaka Y, et al. Integrated molecular mechanism directing nucleosome reorganization by human FACT. Genes Dev. 2016;30(6):673–86. doi: 10.1101/gad.274183.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Belotserkovskaya R, et al. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301(5636):1090–3. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 45.Kemble DJ, et al. FACT Disrupts Nucleosome Structure by Binding H2A–H2B with Conserved Peptide Motifs. Mol Cell. 2015;60(2):294–306. doi: 10.1016/j.molcel.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marechal A, Zou L. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015;25(1):9–23. doi: 10.1038/cr.2014.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang H, et al. RPA Interacts with HIRA and Regulates H3.3 Deposition at Gene Regulatory Elements in Mammalian Cells. Mol Cell. 2017;65(2):272–284. doi: 10.1016/j.molcel.2016.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ginno PA, et al. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45(6):814–25. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ray-Gallet D, et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol Cell. 2011;44(6):928–41. doi: 10.1016/j.molcel.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 50.Duro E, et al. Identification of the MMS22L-TONSL complex that promotes homologous recombination. Mol Cell. 2010;40(4):632–44. doi: 10.1016/j.molcel.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 51.O'Donnell L, et al. The MMS22L-TONSL complex mediates recovery from replication stress and homologous recombination. Mol Cell. 2010;40(4):619–31. doi: 10.1016/j.molcel.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Connell BC, et al. A genome-wide camptothecin sensitivity screen identifies a mammalian MMS22L-NFKBIL2 complex required for genomic stability. Mol Cell. 2010;40(4):645–57. doi: 10.1016/j.molcel.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Radman-Livaja M, et al. Patterns and mechanisms of ancestral histone protein inheritance in budding yeast. PLoS Biol. 2011;9(6):e1001075. doi: 10.1371/journal.pbio.1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ishimi Y, et al. Binding of human minichromosome maintenance proteins with histone H3. J Biol Chem. 1996;271(39):24115–22. doi: 10.1074/jbc.271.39.24115. [DOI] [PubMed] [Google Scholar]
- 55.Ishimi Y, et al. Biochemical function of mouse minichromosome maintenance 2 protein. J Biol Chem. 1998;273(14):8369–75. doi: 10.1074/jbc.273.14.8369. [DOI] [PubMed] [Google Scholar]
- 56.Ishimi Y, et al. Biochemical activities associated with mouse Mcm2 protein. J Biol Chem. 2001;276(46):42744–52. doi: 10.1074/jbc.M106861200. [DOI] [PubMed] [Google Scholar]
- 57.Foltman M, et al. Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Rep. 2013;3(3):892–904. doi: 10.1016/j.celrep.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 58.Groth A, et al. Regulation of replication fork progression through histone supply and demand. Science. 2007;318(5858):1928–31. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- 59.Richet N, et al. Structural insight into how the human helicase subunit MCM2 may act as a histone chaperone together with ASF1 at the replication fork. Nucleic Acids Res. 2015;43(3):1905–17. doi: 10.1093/nar/gkv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang H, et al. A unique binding mode enables MCM2 to chaperone histones H3-H4 at replication forks. Nat Struct Mol Biol. 2015;22(8):618–26. doi: 10.1038/nsmb.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jackson V. Deposition of newly synthesized histones: hybrid nucleosomes are not tandemly arranged on daughter DNA strands. Biochemistry. 2002;27(6):2109–2120. doi: 10.1021/bi00406a044. [DOI] [PubMed] [Google Scholar]
- 62.Clement C, Almouzni G. MCM2 binding to histones H3-H4 and ASF1 supports a tetramer-to-dimer model for histone inheritance at the replication fork. Nat Struct Mol Biol. 2015;22(8):587–9. doi: 10.1038/nsmb.3067. [DOI] [PubMed] [Google Scholar]
- 63.Devbhandari S, et al. Chromatin Constrains the Initiation and Elongation of DNA Replication. Mol Cell. 2017;65(1):131–141. doi: 10.1016/j.molcel.2016.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan BC, et al. Functional cooperation between FACT and MCM helicase facilitates initiation of chromatin DNA replication. EMBO J. 2006;25(17):3975–85. doi: 10.1038/sj.emboj.7601271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gambus A, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8(4):358–66. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 66.LeRoy G, et al. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282(5395):1900–4. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- 67.Pavri R, et al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125(4):703–17. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 68.Dann GP, et al. ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference. Nature. 2017;548(7669):607–611. doi: 10.1038/nature23671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Azmi IF, et al. Nucleosomes influence multiple steps during replication initiation. Elife. 2017;6 doi: 10.7554/eLife.22512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burgess RJ, et al. A role for Gcn5 in replication-coupled nucleosome assembly. Mol Cell. 2010;37(4):469–80. doi: 10.1016/j.molcel.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yadav T, Whitehouse I. Replication-Coupled Nucleosome Assembly and Positioning by ATP-Dependent Chromatin-Remodeling Enzymes. Cell Rep. 2016;15:715–723. doi: 10.1016/j.celrep.2016.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vasseur P, et al. Dynamics of Nucleosome Positioning Maturation following Genomic Replication. Cell Rep. 2016;16(10):2651–2665. doi: 10.1016/j.celrep.2016.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fennessy RT, Owen-Hughes T. Establishment of a promoter-based chromatin architecture on recently replicated DNA can accommodate variable internucleosome spacing. Nucleic Acids Res. 2016;44(15):7189–203. doi: 10.1093/nar/gkw331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ramachandran S, Henikoff S. Transcriptional Regulators Compete with Nucleosomes Post-replication. Cell. 2016;165(3):580–92. doi: 10.1016/j.cell.2016.02.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ramachandran S, et al. Capitalizing on disaster: Establishing chromatin specificity behind the replication fork. Bioessays. 2017;39(4) doi: 10.1002/bies.201600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blythe SA, Wieschaus EF. Establishment and maintenance of heritable chromatin structure during early Drosophila embryogenesis. Elife. 2016;5 doi: 10.7554/eLife.20148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Green EM, et al. Replication-independent histone deposition by the HIR complex and Asf1. Curr Biol. 2005;15(22):2044–9. doi: 10.1016/j.cub.2005.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alabert C, et al. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 2015;29(6):585–90. doi: 10.1101/gad.256354.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Q, Zhang Z. Linking DNA replication to heterochromatin silencing and epigenetic inheritance. Acta Biochim Biophys Sin (Shanghai) 2012;44(1):3–13. doi: 10.1093/abbs/gmr107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen T, Dent SY. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet. 2014;15(2):93–106. doi: 10.1038/nrg3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 82.Murzina N, et al. Heterochromatin Dynamics in Mouse Cells. Molecular Cell. 1999;4(4):529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- 83.Quivy JP, et al. A CAF-1 dependent pool of HP1 during heterochromatin duplication. EMBO J. 2004;23(17):3516–26. doi: 10.1038/sj.emboj.7600362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Loyola A, et al. The HP1alpha-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 2009;10(7):769–75. doi: 10.1038/embor.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roelens B, et al. Maintenance of Heterochromatin by the Large Subunit of the CAF-1 Replication-Coupled Histone Chaperone Requires Its Interaction with HP1a Through a Conserved Motif. Genetics. 2017;205(1):125–137. doi: 10.1534/genetics.116.190785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jiang D, Berger F. DNA replication-coupled histone modification maintains Polycomb gene silencing in plants. Science. 2017;357(6356):1146–1149. doi: 10.1126/science.aan4965. [DOI] [PubMed] [Google Scholar]
- 87.Awe JP, Byrne JA. Identifying candidate oocyte reprogramming factors using cross-species global transcriptional analysis. Cell Reprogram. 2013;15(2):126–33. doi: 10.1089/cell.2012.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gonzalez-Munoz E, et al. Cell reprogramming. Histone chaperone ASF1A is required for maintenance of pluripotency and cellular reprogramming. Science. 2014;345(6198):822–5. doi: 10.1126/science.1254745. [DOI] [PubMed] [Google Scholar]
- 89.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132(4):583–97. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 90.Nakano S, et al. Replication-coupled chromatin assembly generates a neuronal bilateral asymmetry in C. elegans. Cell. 2011;147(7):1525–36. doi: 10.1016/j.cell.2011.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tran V, et al. Asymmetric division of Drosophila male germline stem cell shows asymmetric histone distribution. Science. 2012;338(6107):679–82. doi: 10.1126/science.1226028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58(1):15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 93.Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol. 2013;20(1):14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hammond CM, et al. Histone chaperone networks shaping chromatin function. Nat Rev Mol Cell Biol. 2017;18(3):141–158. doi: 10.1038/nrm.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Talbert PB, Henikoff S. Histone variants on the move: substrates for chromatin dynamics. Nat Rev Mol Cell Biol. 2017;18(2):115–126. doi: 10.1038/nrm.2016.148. [DOI] [PubMed] [Google Scholar]
- 96.Burgers PMJ, Kunkel TA. Eukaryotic DNA Replication Fork. Annu Rev Biochem. 2017;86:417–438. doi: 10.1146/annurev-biochem-061516-044709. [DOI] [PMC free article] [PubMed] [Google Scholar]