Abstract
Intestinal fungi are an important component of the microbiota, and recent studies have unveiled their potential in modulating host immune homeostasis and inflammatory disease. Nonetheless, the mechanisms governing immunity to gut mycobiota remain unknown. We identified CX3CR1+ mononuclear phagocytes (MNPs) as essential for the initiation of innate and adaptive immune responses to intestinal fungi. CX3CR1+ MNPs express antifungal receptors and activate antifungal responses in a Syk dependent manner. Genetic ablation of CX3CR1+ MNPs led to changes in the gut fungal communities and to severe colitis that was rescued by antifungal treatment. A missense mutation in the gene encoding CX3CR1 led to impaired antifungal responses in Crohn’s Disease patients. These results unravel the role of CX3CR1+ MNPs as mediators of the interactions between intestinal mycobiota and host immunity during health and disease.
Extensive studies on the intestinal bacteria have demonstrated that alterations in the microbiota have a dramatic impact on host immunity and contribute to several diseases of inflammatory origin. Fungi are present in the mammalian intestine (1–5), yet little is known about their ability to influence immune homeostasis. The recent advances in deep sequencing technologies have redefined our understanding of the fungal communities (mycobiota) colonizing the mammalian barrier surfaces(2). Intestinal fungal dysbiosis has been shown to influence colitis, alcoholic liver disease and allergic lung disease (3–6), providing evidence for its potential to influence both local and distal inflammation. Serum antibodies against Saccharomyces cerevisiae mannan (ASCA) are elevated in several inflammatory diseases including Crohn’s Disease (CD)(7–9). Systemic ASCA can develop in response to intestinal fungi (3, 7), providing a possible link between the gut mycobiota and host immunity. Despite the identification of receptors involved in the recognition and immunity to intestinal fungi (3, 10), the cell subsets that initiate and regulate mucosal immune responses to the mycobiota remain unknown.
In the intestinal lamina propria (LP) several subsets of phagocytes respond to bacterial infections or to fluctuations in the commensal bacterial communities (11–13). Among those, mononuclear phagocytes (MNPs), marked by the expression of the fractalkine receptor CX3CR1 (CX3CR1+ MNPs), and subsets of dendritic cells (DCs) marked by the differential expression of the integrins CD11b and CD103, initiate immunity and prime Th17 responses to both commensal and pathogenic bacteria in the gut (11, 12, 14). Despite their well described ability to respond to gut bacteria, their role in mucosal immunity to gut fungi is unknown.
To assess the in vivo ability of gut resident phagocytes to respond to fungi, we colonized mice with the opportunistic human commensal Candida albicans and analyzed the changes in the surface expression of the costimulatory molecules. We found that colonization with C. albicans altered the surface expression of CD40 and CD86 among CX3CR1+ MNPs but not among the other subsets (Fig. S1A, B). We thus assessed the ability of CX3CR1+ MNPs to recognize intestinal fungi. We purified CX3CR1+ MNPs from the intestinal LP, and compared their RNA-Seq expression profile to those of CD11b+CD103+ DCs (Fig. 1A, B; Fig. S2A) that have been shown to respond to lung fungal infection (14, 15). While both CD11b+CD103+ DCs and CX3CR1+ MNPs expressed genes involved in antigen presentation, CX3CR1+ MNPs showed a higher expression of genes involved in fungal recognition (Fig. 1B; Fig. S2B). Quantitative PCR and flow cytometric analysis confirmed that transcripts encoding the fungal C type lectin receptors (CLRs) dectin-1 (Clec7a), dectin-2 (Clec6a) and mincle (Clec4e) were highly present in CX3CR1+ MNPs (Fig. 1C, D; Fig. S3A,B). Further, we examined the in vivo intake of Candida by phagocytes in the murine intestine. Confocal microscopy examination revealed that Candida was efficiently recognized by intestinal phagocytes in vivo, with over 80% of all CX3CR1+ MNPs intaking Candida (Fig. 1E; Fig. S3C, D; Suppl. Movie 1). These results indicate that gut resident CX3CR1+ MNPs are equipped to efficiently recognize and respond to intestinal fungi in vivo.
Fig. 1. CX3CR1 mononuclear cells express antifungal receptors and recognize fungi in the intestine.
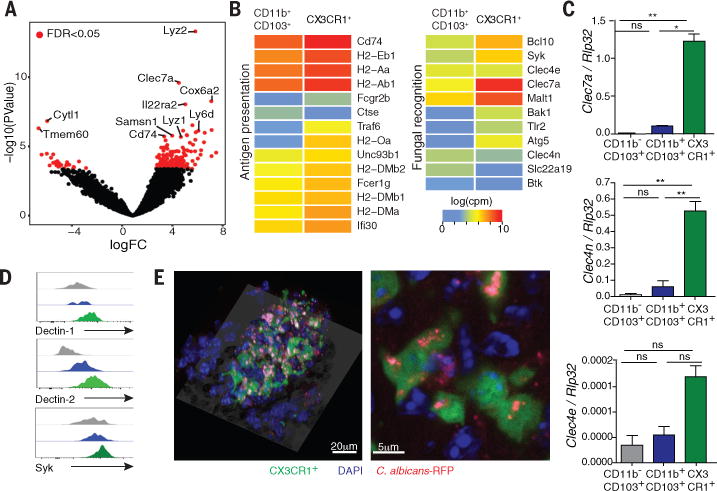
(A) RNA sequencing (RNA-seq) analysis was performed on sorted CD11b+ CD103+ dendritic cells and CX3CR1+ mononuclear phagocytes. Volcano plot of P-value versus FC comparing gene expression in the two cell subsets; red dots indicate an FDR of <0.05. (B) Logarithmic count per million (log(cpm)) normalization of genes involved in antigen presentation (left panel) or fungal recognition (right panel). (C) The expression of antifungal CLRs was confirmed by RT-qPCR. (D) Representative flow cytometry histogram of dectin-1, dectin-2 and Syk expression among CD11b− CD103+; CD11b+ CD103+ and CD11b+ CX3CR1+ cells in colons of WT mice. (E) Representative confocal imaging of the intake of C. albicans-RFP (red) by CX3CR1+ MNPs (green, CX3CR1+, DAPI+) and other cell types (blue, CX3CR1−, DAPI+) in the intestine. Bar graphs represent mean ± SEM of individual mice (N=4-7), representative of at least two independent experiments. *P<0.05, **P<0.01, one-way ANOVA.
Both CX3CR1+ MNPs and CD11b+CD103+ DCs have been shown to play a role in the regulation of adaptive immunity to commensal and pathogenic bacteria (12, 14, 16, 17) . CX3CR1+ MNPs are involved in the induction of Th17 responses to intestinal bacteria (18) and are essential for the killing of Candida in the kidneys during systemic infection (19). Conversely, several studies have suggested a central role for IRF4-dependent CD11b+CD103+ DCs in intestinal Th17 cell differentiation, as well as Th17-induced bacterial and fungal clearance in the lung (12, 14). Further, conventional migratory CD11b− CD103+ DCs have been shown to be a cellular entry point to opportunistic pathogens and are absent in Batf3−/− mice (20) (Fig S10A, B). To directly assess the role of different phagocytic subsets in the induction of anti-fungal immune responses, we crossed flox inducible Irf4fl/fl allele mice or flox inducible Cx3cr1DTR mice (11), with transgenic Cd11c-Cre mice. The first strategy allowed the specific ablation of Irf4 in DCs leading to the loss of intestinal CD11b+CD103+ DCs (referred as ΔIrf4 mice), but intact CX3CR1+ MNPs (14) (Fig. S4A, B). The second strategy allowed for the selective depletion of intestinal CX3CR1+ MNPs upon administration of diphtheria toxin (DT, mice referred as ΔCX3CR1), without affecting CD11b+ CD103+ DCs (11) (Fig. S4C–E).
Th17 cells are crucial for the control of fungi at other gastrointestinal sites such as the oral cavity, while Treg cells suppress fungal infection-related host damage (21, 22). Upon colonization with C. albicans we observed a strong Th17 response in the colon and mesenteric lymph nodes (mLNs) that was consistent with other studies (Fig. 2A, B; Fig. S5A) (23) while the frequency of Foxp3+ Treg cells was not affected (Fig. S5B). We next determined whether specific phagocytic subsets are involved in Th17 responses to intestinal fungi. Candida colonization induced a consistent increase in Th17 cell frequencies that were not affected by depletion of CD11b+CD103+ DCs (Fig. 2A–B; Fig. S5C, S6A) or lack of CD11b−CD103+ DCs (Fig. S10 C–J). In contrast, a dramatic decrease in Th17 cells was observed upon depletion of CX3CR1+ MNPs in the colon and mLN (Fig. 2C–D; Fig S6B) but not in the small intestine (Fig. S7A, B). The observed Th17 induction was independent from segmented filamentous bacteria (SFB) that were absent in our colony (Fig. S8A–C). Foxp3+ Treg cells were not affected by the absence of either phagocytic subset (Fig. S5D; Fig. S6C; Fig. S7C; Fig. S10 F, J).
Fig. 2. CX3CR1+ MNPs control gut antifungal immunity.
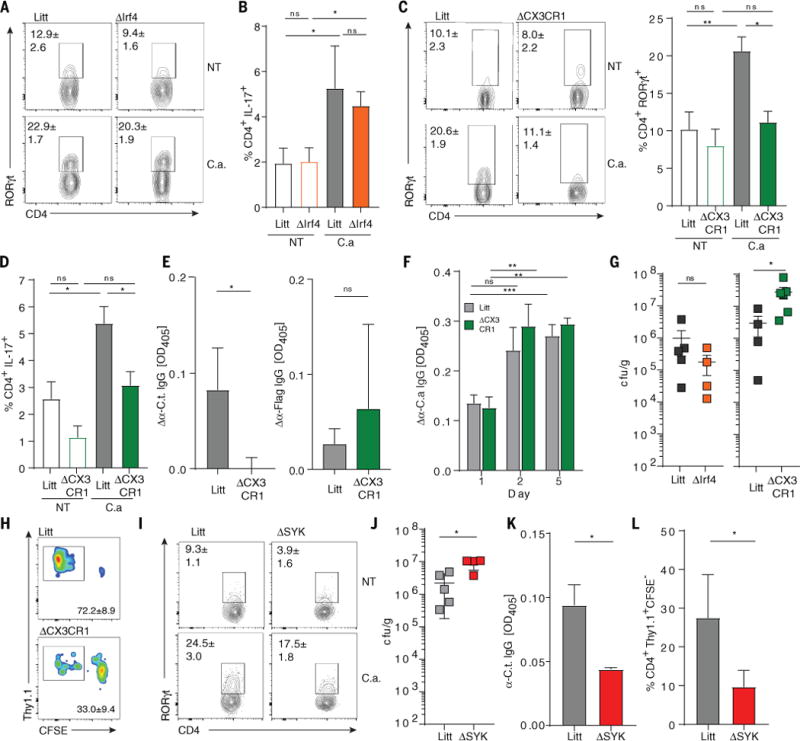
Colonic lamina propria cells were collected from ΔIrf4 mice (orange bars) or littermates (lit, grey bars) fed (C.a) or not (NT) with 5.107 C. albicans at day 10. (A and B) Expression of RORγt and IL-17 by CD4+ T cells (pooled from two independent experiments). Cd11c-Cre+/− CX3CR1DTR mice (ΔCX3CR1, green bars) or Cd11c-Cre−/− CX3CR1DTR littermates (lit, grey bars) were treated with diphtheria toxin (DT), and fed with C. albicans. (C and D) RORγt and IL-17 expression by the CD4+ T cells in the colon (pooled from three independent experiments). (E) IgG against the commensal C. tropicalis and flagellin. (F) Systemic IgG responses following i.p injection with C. albicans at day 1, 2 and 5 (pooled from two independent experiments). (G) C. albicans in the feces of control, ΔIrf4 and ΔCX3CR1 mice at day 10 (dots represent individual mice). ΔCX3CR1 mice and littermates were transferred with purified CD4+ Thy1.1+ OT-II cells, fed C. albicans-OVA and sacrificed after 10 days. (H) Representative plots of CD4+ Thy1.1+ OT-II cells proliferation in the colon (pooled from three independent experiments). Cx3cr1-Cre-ERT+/− Sykfl/fl mice (ΔSyk) or littermates (Litt) treated with tamoxifen and fed with C. albicans. (I) RORγt expression by CD4+ T cells in the colon (pooled from two independent experiments). (J) C. albicans in the feces at day 10 (dots represent individual mice). (K) IgG responses against C. tropicalis at day 10 (n=5 per group). (L) Quantification of proliferating CFSE− CD4+ Thy1.1+ OT-II cells in the colon (pooled from two independent experiments). Statistical analysis: Data are presented as mean +/− SEM; *P<0.05, **P<0.01, ***P<0.001 (Mann-Whitney Test (E, G, J, K, L), one-way ANOVA (B, C, D, F)).
Besides Th17 responses, the development of systemic ASCA during intestinal inflammation is another hallmark of adaptive immunity activation in response to intestinal fungi (3, 7). In addition to S. cerevisiae, Candida can act as an immunogen for ASCA production (7). Thus we assessed whether lack of CX3CR1+ MNPs also alters the production of systemic IgG antibodies against the commensal fungus C. tropicalis (3) commonly found in our colony during steady state (Fig. S8D). We found that CX3CR1+ MNPs depletion reduced IgG-antibody responses against C. tropicalis without affecting the response against the commensal bacterial antigen flagellin (Fig. 2E). To assess whether the induction of antifungal IgG is dependent on recognition of luminal fungal antigens by CX3CR1+ MNPs, we compared the induction of systemic IgG following intestinal colonization or systemic infection with C. albicans. While both approaches led to the generation of systemic anti-C. albicans IgG in WT mice, depletion of CX3CR1+ MNPs led to a significant decrease of antibody production against intestinal C. albicans (Fig. S9) without affecting the antibody production upon systemic infection (Fig. 2F). This suggests that the defect in antifungal antibody production in ΔCX3CR1 mice is specific to the gut. In contrast, neither the depletion CD11b+CD103+ DCs nor the lack of CD11b−CD103+ DCs influenced systemic anti-Candida antibody production (Fig. S9; Fig. S10K). Consistent with the decreased antifungal responses, C. albicans burdens are increased in ΔCX3CR1 mice, but not in ΔIrf4 and Batf3−/− mice (Fig. 2G; Fig. S10L).
To further assess the role of CX3CR1+ MNPs in the induction of responses to antigens delivered by intestinal fungi, we fed ΔCX3CR1 mice and control littermates a recombinant C. albicans strain expressing a model MHC-II-restricted OT-II peptide (C.a-OVA, Fig. S11) and adoptively transferred mice with CFSE-labelled CD4+ Thy1.1+ OT-II T-cells (Fig. S12A, B). CX3CR1+ MNPs depletion lead to decreased antigen specific proliferation of Th17 cells in response to C. albicans in both the colon and the mLN (Fig. 2H; Fig. S12C–E). CX3CR1+ MNPs might either directly transport fungal antigens to the mLNs for T cell priming or pass the antigens to other migratory phagocytes, although both scenarios are possible (11, 24). Nevertheless, those data demonstrate that CX3CR1+ MNPs play an important role in the induction of Th17 and antibody responses to intestinal fungi, while CD11b+ CD103+ DCs and CD11b− CD103+ DCs are not crucial.
Spleen tyrosine kinase (Syk) is crucial for the initiation of the signaling cascade downstream of several CLRs (Fig. S2) and is highly expressed in CX3CR1+ MNPs (Fig. 1B, D; Fig S3B). We thus generated ΔSyk CX3CR1 mice (Sykfl/fl x Cx3cr1-cre/ERT2+/−) to selectively delete Syk in CX3CR1+ cells (Fig S13). Consistently, we observed fungal outgrowth but impaired Th17 and antifungal antibody responses to intestinal Candida colonization (Fig 2 I–L; Fig S12F; Fig S14), suggesting that intact CLR signaling in CX3CR1+ cells is required to control fungal colonization and to induce effective adaptive immunity to fungi in the gut.
Having determined that CX3CR1+ MNPs recognize intestinal fungi and control gut antifungal immunity, we next explored whether these cells play a role in shaping the composition of gut fungal communities. We thus characterized the microbiota upon CX3CR1+ MNPs depletion using high-throughput ITS1 and 16S sequencing of fungal and bacterial rDNA. Despite their role in the control of commensal and pathogenic bacteria (11, 16, 25), ablation of CX3CR1+ MNPs did not affect beta and alpha diversity of the intestinal bacterial community (Fig. S15A–E; Fig S16A). In contrast, ablation of CX3CR1+ MNPs, but not CD11b+ CD103+ DCs, altered the fungal community composition of these mice as compared to control littermates (Fig. 3A; Fig. S15B; Fig. S16B–D; Fig. S17; Fig. S18). CX3CR1+ MNPs depletion also led to an increase in fungal alpha diversity, that was mainly driven by an increased abundance and diversity among the Ascomycota phylum (Fig. S15B, E; Fig. S16C–D). This suggest that CX3CR1+ MNPs play a role in immune surveillance and maintenance of a balanced gut fungal community.
Fig. 3. Depletion of CX3CR1+ MNP affects gut mycobiota and results in exacerbated intestinal disease.
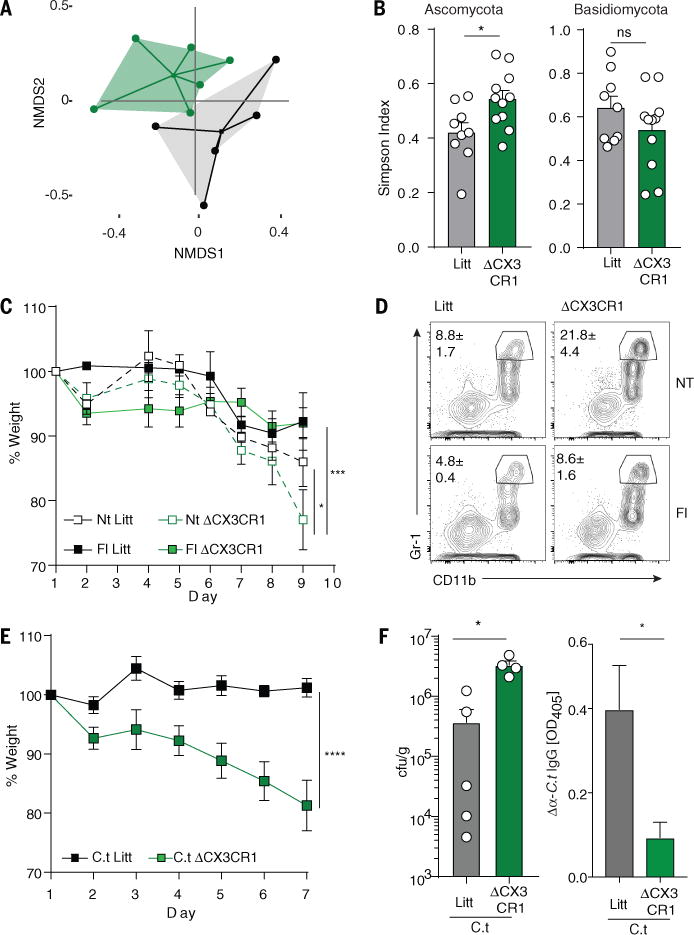
Feces from ΔCX3CR1 or WT littermates (Litt) mice were collected 7 days after administration of the first DT dose. (A) NMDS plot of distance ordination based on Bray-Curtis dissimilarities in the colon for fungal OTUs (WT n=5; ΔCX3CR1 n=6). (B) Alpha diversity (Simpson diversity index) among the Ascomycota (left) and Basidiomycota (right) phyla; pooled from two independent experiments, mean +/− SEM, each circle denotes one mouse. (C) Weight change during DSS colitis in ΔCX3CR1 or control littermates following short-term treatment with fluconazole (Fl) or no treatment (NT) (pooled from two independent experiments). (D) Representative plots of neutrophil infiltration (CD11b+ Gr-1high) in the colon following DSS administration. (E) Weight change during DSS colitis in ΔCX3CR1 or control littermates fed with C. tropicalis (C.t) (mean +/− SEM, lit n=5; ΔCX3CR1 n=5). (F) C. tropicalis cfu/g in the feces of ΔCX3CR1 or control littermates at day 7 (dots represent individual mice, mean +/− SEM). (K) Systemic IgG responses against C. tropicalis were assessed by ELISA. Statistical analysis: *P<0.05, **P<0.01, ***P<0.001 (Mann-Whitney Test (B, F), two-way ANOVA (C, E)).
Fungi are present at highest densities in the colon (3) where fungal colonization induced a strong Th17 response (Fig. S5A). Thus, we next tested whether ΔCX3CR1 mice are more susceptible to DSS-induced colitis. Consistent with their inability to mount an efficient antifungal response, we found that ΔCX3CR1 mice were more susceptible to DSS colitis as compared to their littermate controls (Fig. 3C, D; Fig S19A, B). Fluconazole targets most Candida species and other dimorphic fungi, and ameliorates colitis in mice with defects in antifungal immunity when used during the onset of intestinal disease (2, 3, 10). Since CX3CR1+ MNPs depletion had a strong effect on Ascomycota, we treated ΔCX3CR1 mice with fluconazole. Notably, fluconazole treatment significantly ameliorated intestinal disease in ΔCX3CR1 mice (Fig. 3C, D; Fig S19A, B). Transfer of feces from ΔCX3CR1 mice did not affect the outcome of colitis in ex-germ free mice (Fig S19C–H), indicating that ΔCX3CR1 microbiota is not per-se pathogenic. Further supplementation with C. tropicalis, previously shown to affect intestinal conditions in dectin-1 and dectin-3 deficient mice (3, 10, 26), or with C. albicans, led to severe wasting disease, colon shortening and fungal overgrowth in the intestines of ΔCX3CR1 mice without worsening the disease in littermate controls (Fig. 3E; Fig. S20A, B). Despite the increased disease susceptibility and augmented Candida burden, ΔCX3CR1 mice failed to mount a systemic antifungal IgG antibody response (Fig. 3F; Fig. S20C, D), consistent with the defective antifungal immunity that we observed during the steady-state. These results indicate that CX3CR1+ MNPs play a crucial role in controlling fungal microbiota during intestinal disease.
Having established a role for CX3CR1+ MNPs in the control of gut fungi during intestinal disease, and finding that CX3CR1+ MNPs have the potential to intake other species of mouse and human commensal fungi (Fig. 4A), we explored whether genetic variations of the human CX3CR1 gene affect immunity to fungi in inflammatory bowel disease (IBD). Defects in CX3CR1 have been shown to increase susceptibility of mice and humans to systemic candidiasis (19), but not to vulvovaginal and oral candidiasis (27). In contrast, the role of CX3CR1 in the initiation of anti-fungal responses in the gut is unknown. We focused on polymorphisms in the coding region of CX3CR1 gene (Fig. S21) that have been previously associated with human inflammatory diseases such as arteriosclerosis and coronary artery disease (28, 29). While none of these polymorphisms were associated with a predisposition to IBD (Table S1), we identified a striking association of CX3CR1 T280M (rs3732378) polymorphism specifically with IgG ASCA positivity in a cohort of 503 CD patients (Fig. 4B, OR=0.59, logistic regression p=3.73E−03). In contrast, antibodies against bacterial and host antigens previously shown to increase in IBD (7) were not affected by this mutation (Fig. 4B, Table S2). Since ASCA antibodies are directed against both S. cerevisiae and C. albicans (7), we next assessed whether antifungal antibody responses to common human commensals are also affected by CX3CR1 T280M. Compared to non-affected individuals, patients homozygous for CX3CR1 T280M were severely impaired in their ability to generate systemic IgG recognizing a variety of gut relevant fungi belonging to the phylum Ascomycota, while producing normal antibody responses against the bacterial antigen flagellin (Fig. 4C, D; Fig. S22) consistent with the hypothesis that this mutation in CX3CR1 leads to impaired responses to fungi in the gut.
Fig. 4. Polymorphisms in the coding region of the CX3CR1 gene is associated with decreased anti-fungal IgG responses in CD patients.
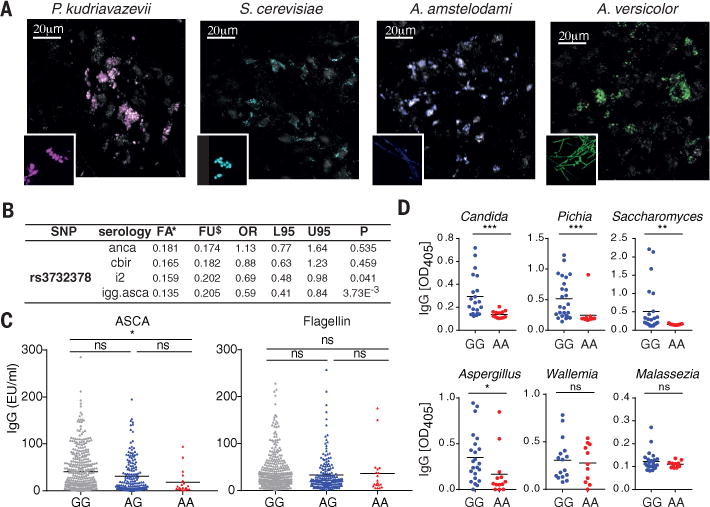
(A) Representative pictures of the intake of fungal species (colored) by CX3CR1+ MNPs (grey) in the colon. (B) Association between the missense mutation rs3732378 and the systemic serologic markers anti-neutrophil cytoplasmic antibodies (anca), flagellin (cbir), Pseudomonas fluorescens – associated sequence I-2 (i2), and anti S. cerevisiae IgG antibodies (igg.asca) among 503 CD patients. FA, frequency affected; FU, frequency unaffected; L95 and U95, lower and upper 95th confidence interval. (C) IgG ASCA and anti-flagellin (cbir) IgG responses were assessed in the sera from rs3732378 homozygous (AA), heterozygous (AG) and control (GG) CD patients by ELISA (dots represent individual patients, bar represent mean). (D) IgG responses against different commensal fungi (Candida albicans, Pichia kudrazevii, Saccharomyces cerevisiae, Aspergillus amstellodamii, Wallemia sebi, Malassezia restricta were assessed (dots represent individual patients, bar represent mean). Statistical analysis: *P<0.05, **P<0.01, ***P<0.001 (Mann-Whitney test (D), one-way ANOVA (C)).
In summary, we identified a specific subset of gut resident phagocytes, namely the CX3CR1+ MNPs, which are able to recognize and respond to the gut mycobiota in a Syk-dependent manner. CX3CR1+ MNPs can influence adaptive immune responses to gut fungi and control the mycobiota during experimental colitis. In humans, we found that a CX3CR1 polymorphism is strongly associated with a decrease of antifungal antibody responses in CD patients. CX3CR1 T280M is a common polymorphism (23.3% heterozygous and 4.4% homozygous (30)) and has been previously associated with extra-intestinal inflammatory diseases (28, 29). Conceivably, gut mycobiota and CX3CR1-dependent immune responses might further contribute to extra-intestinal manifestations of inflammatory diseases. In support to this notion, antifungal antibodies are increased in patients with alcoholic liver disease, spondyloarthritis, Graves’ disease and systemic lupus erythematosus (6–9). Altogether, our findings provide evidence for the influence of gut mycobiota on both local and systemic immunity which is mediated by the recognition of intestinal fungi by CX3CR1+ MNPs.
Supplementary Material
Acknowledgments
This work was funded by the US National Institutes of Health (grants DK113136, DK098310 and AI123819 to I.D.I); (P01 DK046763, U01 DK062413 to D.P.B.M.), Infect-ERA FUNCOMPATH (PCIN-2014-052 to J.P), MINECO (BIO2015-6477-P to J.P.), Swiss National Science Foundation (fellowship P2ZHP3_164850 to I.L), Kenneth Rainin Foundation (Innovator and Breakthrough awards to I.D.I), and support from the Jill Roberts Institute for Research in IBD. The data presented in this study are tabulated in the main text and supplementary materials. Microbiota sequencing data are deposited in NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra, run number SRP124782, SRP124783, SRP124736, SRP124742). RNAseq data are deposited in GEO (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE106594.
Footnotes
Supplementary Materials for this manuscript include
Materials and Methods
Captions for Movie S1
Supplementary References 31 to 52 Movie S1
References and Notes
- 1.Scupham AJ, Presley LL, Wei B, Bent E, Griffith N, McPherson M, Zhu F, Oluwadara O, Rao N, Braun J, Borneman J. Abundant and diverse fungal microbiota in the murine intestine. Appl Environ Microbiol. 2006;72:793–801. doi: 10.1128/AEM.72.1.793-801.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iliev ID, Leonardi I. Fungal dysbiosis: immunity and interactions at mucosal barriers. Nat Rev Immunol. 2017;17:635–646. doi: 10.1038/nri.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, Rotter JI, Wang HL, McGovern DP, Brown GD, Underhill DM. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, Cosnes J, Seksik P, Langella P, Skurnik D, Richard ML, Beaugerie L. Fungal microbiota dysbiosis in IBD. Gut. 2017;66:1039–1048. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wheeler ML, Limon JJ, Bar AS, Leal CA, Gargus M, Tang J, Brown J, Funari VA, Wang HL, Crother TR, Arditi M, Underhill DM, Iliev ID. Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host Microbe. 2016;19:865–873. doi: 10.1016/j.chom.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang AM, Inamine T, Hochrath K, Chen P, Wang L, Llorente C, Bluemel S, Hartmann P, Xu J, Koyama Y, Kisseleva T, Torralba MG, Moncera K, Beeri K, Chen CS, Freese K, Hellerbrand C, Lee SM, Hoffman HM, Mehal WZ, Garcia-Tsao G, Mutlu EA, Keshavarzian A, Brown GD, Ho SB, Bataller R, Starkel P, Fouts DE, Schnabl B. Intestinal fungi contribute to development of alcoholic liver disease. J Clin Invest. 2017;127:2829–2841. doi: 10.1172/JCI90562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Standaert-Vitse A, Jouault T, Vandewalle P, Mille C, Seddik M, Sendid B, Mallet JM, Colombel JF, Poulain D. Candida albicans is an immunogen for anti-Saccharomyces cerevisiae antibody markers of Crohn’s disease. Gastroenterology. 2006;130:1764–1775. doi: 10.1053/j.gastro.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 8.Mankai A, Thabet Y, Manoubi W, Achour A, Sakly W, Ghedira I. Anti-Saccharomyces cerevisiae antibodies are elevated in Graves’ disease but not in Hashimoto’s thyroiditis. Endocr Res. 2013;38:98–104. doi: 10.3109/07435800.2012.723293. [DOI] [PubMed] [Google Scholar]
- 9.Riente L, Chimenti D, Pratesi F, Delle Sedie A, Tommasi S, Tommasi C, Bombardieri S, Migliorini P. Antibodies to tissue transglutaminase and Saccharomyces cerevisiae in ankylosing spondylitis and psoriatic arthritis. J Rheumatol. 2004;31:920–924. [PubMed] [Google Scholar]
- 10.Wang T, Pan D, Zhou Z, You Y, Jiang C, Zhao X, Lin X. Dectin-3 Deficiency Promotes Colitis Development due to Impaired Antifungal Innate Immune Responses in the Gut. PLoS Pathog. 2016;12:e1005662. doi: 10.1371/journal.ppat.1005662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diehl GE, Longman RS, Zhang JX, Breart B, Galan C, Cuesta A, Schwab SR, Littman DR. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 2013;494:116–120. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Persson EK, Uronen-Hansson H, Semmrich M, Rivollier A, Hagerbrand K, Marsal J, Gudjonsson S, Hakansson U, Reizis B, Kotarsky K, Agace WW. IRF4 transcription-factor-dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity. 2013;38:958–969. doi: 10.1016/j.immuni.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 13.Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl JP, Ricciardi-Castagnoli P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 14.Schlitzer A, McGovern N, Teo P, Zelante T, Atarashi K, Low D, Ho AW, See P, Shin A, Wasan PS, Hoeffel G, Malleret B, Heiseke A, Chew S, Jardine L, Purvis HA, Hilkens CM, Tam J, Poidinger M, Stanley ER, Krug AB, Renia L, Sivasankar B, Ng LG, Collin M, Ricciardi-Castagnoli P, Honda K, Haniffa M, Ginhoux F. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. 2013;38:970–983. doi: 10.1016/j.immuni.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kinnebrew MA, Buffie CG, Diehl GE, Zenewicz LA, Leiner I, Hohl TM, Flavell RA, Littman DR, Pamer EG. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 2012;36:276–287. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, Vyas JM, Boes M, Ploegh HL, Fox JG, Littman DR, Reinecker HC. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 17.Aychek T, Mildner A, Yona S, Kim KW, Lampl N, Reich-Zeliger S, Boon L, Yogev N, Waisman A, Cua DJ, Jung S. IL-23-mediated mononuclear phagocyte crosstalk protects mice from Citrobacter rodentium-induced colon immunopathology. Nature communications. 2015;6:6525. doi: 10.1038/ncomms7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panea C, Farkas AM, Goto Y, Abdollahi-Roodsaz S, Lee C, Koscso B, Gowda K, Hohl TM, Bogunovic M, Ivanov II. Intestinal Monocyte-Derived Macrophages Control Commensal-Specific Th17 Responses. Cell Rep. 2015;12:1314–1324. doi: 10.1016/j.celrep.2015.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lionakis MS, Swamydas M, Fischer BG, Plantinga TS, Johnson MD, Jaeger M, Green NM, Masedunskas A, Weigert R, Mikelis C, Wan W, Lee CC, Lim JK, Rivollier A, Yang JC, Laird GM, Wheeler RT, Alexander BD, Perfect JR, Gao JL, Kullberg BJ, Netea MG, Murphy PM. CX3CR1-dependent renal macrophage survival promotes Candida control and host survival. J Clin Invest. 2013;123:5035–5051. doi: 10.1172/JCI71307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edelson BT, Bradstreet TR, Hildner K, Carrero JA, Frederick KE, Kc W, Belizaire R, Aoshi T, Schreiber RD, Miller MJ, Murphy TL, Unanue ER, Murphy KM. CD8alpha(+) dendritic cells are an obligate cellular entry point for productive infection by Listeria monocytogenes. Immunity. 2011;35:236–248. doi: 10.1016/j.immuni.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conti HR, Shen F, Nayyar N, Stocum E, Sun JN, Lindemann MJ, Ho AW, Hai JH, Yu JJ, Jung JW, Filler SG, Masso-Welch P, Edgerton M, Gaffen SL. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med. 2009;206:299–311. doi: 10.1084/jem.20081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trautwein-Weidner K, Gladiator A, Kirchner FR, Becattini S, Rulicke T, Sallusto F, LeibundGut-Landmann S. Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis. PLoS Pathog. 2015;11:e1005164. doi: 10.1371/journal.ppat.1005164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, Ishikawa E, Shima T, Hara T, Kado S, Jinnohara T, Ohno H, Kondo T, Toyooka K, Watanabe E, Yokoyama S, Tokoro S, Mori H, Noguchi Y, Morita H, Ivanov II, Sugiyama T, Nunez G, Camp JG, Hattori M, Umesaki Y, Honda K. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 2015;163:367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazzini E, Massimiliano L, Penna G, Rescigno M. Oral tolerance can be established via gap junction transfer of fed antigens from CX3CR1(+) macrophages to CD103(+) dendritic cells. Immunity. 2014;40:248–261. doi: 10.1016/j.immuni.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 25.Medina-Contreras O, Geem D, Laur O, Williams IR, Lira SA, Nusrat A, Parkos CA, Denning TL. CX3CR1 regulates intestinal macrophage homeostasis, bacterial translocation, and colitogenic Th17 responses in mice. J Clin Invest. 2011;121:4787–4795. doi: 10.1172/JCI59150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang C, Kamiya T, Liu Y, Kadoki M, Kakuta S, Oshima K, Hattori M, Takeshita K, Kanai T, Saijo S, Ohno N, Iwakura Y. Inhibition of Dectin-1 Signaling Ameliorates Colitis by Inducing Lactobacillus-Mediated Regulatory T Cell Expansion in the Intestine. Cell Host Microbe. 2015;18:183–197. doi: 10.1016/j.chom.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Break TJ, Jaeger M, Solis NV, Filler SG, Rodriguez CA, Lim JK, Lee CC, Sobel JD, Netea MG, Lionakis MS. CX3CR1 is dispensable for control of mucosal Candida albicans infections in mice and humans. Infect Immun. 2015;83:958–965. doi: 10.1128/IAI.02604-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moatti D, Faure S, Fumeron F, Amara Mel W, Seknadji P, McDermott DH, Debre P, Aumont MC, Murphy PM, de Prost D, Combadiere C. Polymorphism in the fractalkine receptor CX3CR1 as a genetic risk factor for coronary artery disease. Blood. 2001;97:1925–1928. doi: 10.1182/blood.v97.7.1925. [DOI] [PubMed] [Google Scholar]
- 29.McDermott DH, Halcox JP, Schenke WH, Waclawiw MA, Merrell MN, Epstein N, Quyyumi AA, Murphy PM. Association between polymorphism in the chemokine receptor CX3CR1 and coronary vascular endothelial dysfunction and atherosclerosis. Circ Res. 2001;89:401–407. doi: 10.1161/hh1701.095642. [DOI] [PubMed] [Google Scholar]
- 30.Brand S, Hofbauer K, Dambacher J, Schnitzler F, Staudinger T, Pfennig S, Seiderer J, Tillack C, Konrad A, Goke B, Ochsenkuhn T, Lohse P. Increased expression of the chemokine fractalkine in Crohn’s disease and association of the fractalkine receptor T280M polymorphism with a fibrostenosing disease Phenotype. Am J Gastroenterol. 2006;101:99–106. doi: 10.1111/j.1572-0241.2005.00361.x. [DOI] [PubMed] [Google Scholar]
- 31.Whitfield J, Littlewood T, Soucek L. Tamoxifen administration to mice. Cold Spring Harb Protoc. 2015;2015:269–271. doi: 10.1101/pdb.prot077966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prieto AD, Román E, Correia I, Pla J. The HOG pathway is critical for the colonization of the mouse gastrointestinal tract by Candida albicans. PLoS ONE. 2014;9:e87128. doi: 10.1371/journal.pone.0087128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morschhäuser J, Michel S, Hacker J. Expression of a chromosomally integrated, single-copy GFP gene in Candida albicans and its use as a reporter of gene regulation. Molecular and General Genetics. 1998;257:412–420. doi: 10.1007/s004380050665. [DOI] [PubMed] [Google Scholar]
- 34.Park YN, Morschhauser J. Tetracycline-inducible gene expression and gene deletion in. Candida albicans Eukaryotic Cell. 2005;4:1328–1342. doi: 10.1128/EC.4.8.1328-1342.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niess JH, Adler G. Enteric flora expands gut lamina propria CX3CR1+ dendritic cells supporting inflammatory immune responses under normal and inflammatory conditions. J Immunol. 2010;184:2026–2037. doi: 10.4049/jimmunol.0901936. [DOI] [PubMed] [Google Scholar]
- 37.Li D, Achkar JP, Haritunians T, Jacobs JP, Hui KY, D’Amato M, Brand S, Radford-Smith G, Halfvarson J, Niess JH, Kugathasan S, Buning C, Schumm LP, Klei L, Ananthakrishnan A, Aumais G, Baidoo L, Dubinsky M, Fiocchi C, Glas J, Milgrom R, Proctor DD, Regueiro M, Simms LA, Stempak JM, Targan SR, Torkvist L, Sharma Y, Devlin B, Borneman J, Hakonarson H, Xavier RJ, Daly M, Brant SR, Rioux JD, Silverberg MS, Cho JH, Braun J, McGovern DP, Duerr RH. A Pleiotropic Missense Variant in SLC39A8 Is Associated With Crohn’s Disease and Human Gut Microbiome Composition. Gastroenterology. 2016;151:724–732. doi: 10.1053/j.gastro.2016.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landers CJ, Cohavy O, Misra R, Yang H, Lin YC, Braun J, Targan SR. Selected loss of tolerance evidenced by Crohn’s disease-associated immune responses to auto- and microbial antigens. Gastroenterology. 2002;123:689–699. doi: 10.1053/gast.2002.35379. [DOI] [PubMed] [Google Scholar]
- 39.Matteoli G, Mazzini E, Iliev ID, Mileti E, Fallarino F, Puccetti P, Chieppa M, Rescigno M. Gut CD103+ dendritic cells express indoleamine 2,3-dioxygenase which influences T regulatory/T effector cell balance and oral tolerance induction. Gut. 2010;59:595–604. doi: 10.1136/gut.2009.185108. [DOI] [PubMed] [Google Scholar]
- 40.Tang J, Iliev ID, Brown J, Underhill DM, Funari VA. Mycobiome: Approaches to analysis of intestinal fungi. J Immunol Methods. 2015;421:112–121. doi: 10.1016/j.jim.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 42.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 43.Shannon CE. The mathematical theory of communication. 1963. MD Comput. 1997;14:306–317. [PubMed] [Google Scholar]
- 44.Simpson EH. Measurement of Diversity. Nature. 1949;163:688–688. [Google Scholar]
- 45.Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, Fagarasan S. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci U S A. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loeffler J, Henke N, Hebart H, Schmidt D, Hagmeyer L, Schumacher U, Einsele H. Quantification of fungal DNA by using fluorescence resonance energy transfer and the light cycler system. J Clin Microbiol. 2000;38:586–590. doi: 10.1128/jcm.38.2.586-590.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schabereiter-Gurtner C, Selitsch B, Rotter ML, Hirschl AM, Willinger B. Development of novel real-time PCR assays for detection and differentiation of eleven medically important Aspergillus and Candida species in clinical specimens. J Clin Microbiol. 2007;45:906–914. doi: 10.1128/JCM.01344-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nikolcheva LG, Bourque T, Barlocher F. Fungal diversity during initial stages of leaf decomposition in a stream. Mycol Res. 2005;109:246–253. doi: 10.1017/s0953756204001698. [DOI] [PubMed] [Google Scholar]
- 50.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


