Abstract
The control of lymphoid homeostasis is the result of a very fine balance between lymphocyte production, proliferation, and apoptosis. Here, we focused on the role of T cells in the maintenance/survival of the mature naïve peripheral B cell population. We show that naïve B and T cells interact via the signaling lymphocyte activation molecule (SLAM) family receptor, SLAMF6. This interaction induces cell type specific signals in both cell types, mediated by the SLAM-associated protein (SAP) family of adaptors. This signaling results in an upregulation of the expression of the cytokine MIF in the T cells and augmented expression of its receptor CD74 on the B-cell counterparts, consequently enhancing B-cell survival. Furthermore, in XLP patients, SAP deficiency reduces CD74 expression resulting in the perturbation of B cell maintenance from the naïve stage. Thus, naïve T cells regulate B cell survival in a SLAMF6 and SAP dependent manner.
Introduction
The survival of peripheral naïve mature B cells is dependent on three key cascades: (1) B cell receptor (BCR) tonic signals (e.g., Igα and Syk) (1, 2), (2) the B cell activating factor receptor (BAFFR), which binds the B cell activating factor, belonging to the TNF family (BAFF; also known as BLyS/TALL-1/THANK/zTNF4) (3), and (3) CD74 (invariant chain, Ii) expressed on B cells, and its cognate ligand, macrophage migration inhibitory factor (MIF), which is secreted by almost cell types. These pathways have complementary roles in B cell survival (4, 5).
CD74 is a type II integral membrane protein that acts as a chaperone for MHC class II protein expression (6). A small proportion of CD74 is modified by the addition of chondroitin sulfate (CD74-CS), and this form of CD74 is expressed on the surface of antigen presenting cells (including monocytes and B cells) and epithelial cells (7). It was previously shown that macrophage migration inhibitory factor (MIF) binds to the CD74 extracellular domain, a process that results in the initiation of a signaling pathway in these cells (8).
CD74 stimulation by MIF induces a signaling cascade leading to NF- κB activation, and transcription of genes that regulate the entry of the stimulated B cells into the S phase, an increase in DNA synthesis, cell division, and augmented expression of anti-apoptotic proteins (5, 9, 10). The CD74 receptor induces a similar survival cascade in oncogenically transformed cells derived from chronic lymphocytic leukemia (CLL) patients (11). To define the molecules whose expression is modulated by CD74 to regulate CLL cell survival, we previously screened for CD74 target genes. One molecule, whose expression was strongly upregulated by CD74 activation, is SLAMF5 (CD84), a member of the Signaling lymphocytic activation molecule (SLAM) immunoglobulin superfamily (12).
The SLAM family of receptors includes homophilic and heterophilic receptors that modulate the behavior of immune cells (13-15). These receptors share a common ectodomain organization: a membrane-proximal immunoglobulin (Ig)-like constant domain, and a membrane-distal Ig variable domain that is responsible for ligand recognition. SLAM receptors interact with SLAM-associated protein (SAP)-related molecules, a group of SRC homology 2 (SH2) domain adaptors. The SAP family is comprised of three members: SAP, Ewing’s sarcoma-associated transcript-2 (EAT2), and in rodents, EAT2-related transducer (ERT) (16, 17). SAP controls signal transduction pathways downstream of the SLAM family receptors, and is a key regulator of normal immune function in T, natural killer (NK), and NKT cells (15, 18). However, B cells do not express SAP (19), and EAT2 was suggested to serve as its functional homologue in these cells (20, 21).
The SLAM receptors and their adaptor molecules were shown to be required for germinal center development and humoral memory (22-24). However, their role in naïve B cell maintenance has not been assessed in detail. Lymphocyte populations derived from SAP-deficient mice are grossly normal, although occasional mutant animals exhibit a higher percentage of T and NK cells, and a lower percentage of B cells in the spleen (25).
In the current study, we hypothesized that the SLAM family might be involved in the regulation of naïve B cell survival in the cross-talk between naïve B and naïve T cells in an antigen independent environment. Our findings demonstrate that interaction of B cells with T cells in a SLAMF6/SAP mediated manner upregulates CD74 cell surface expression on B cells, inducing their survival in vitro and in vivo. This study highlights a crucial role of SAP expression on T cells in regulating B cell survival and provides new insights into the requirements for cell collaboration during homeostasis.
Materials and methods
Mice
C57BL/6, CD74 deficient (26), MIF deficient (27), SLAMF5 deficient (24), SH2D1A (SAP) deficient (25) SLAMF6 deficient (28), and SLAMF1 deficient mice (29) used in this study. All animal procedures were approved by the Animal Research Committee at the Weizmann Institute of Science. The animals in each experiment were aged and sex matched.
Cells
Human peripheral blood (PB) lymphocytes
XLP patient samples were provided in compliance with the Institutional Review Board of the Hadassah Hospital. Healthy control samples were provided by the “Magen David Adom in Israel” blood bank.
Cells were separated as previously described (11). B or T cell lymphocytes were enriched by negative and positive selection using the anti-CD3 Milteny MACS beads according to the manufacturer’s protocol.
Mouse splenic lymphocytes
B lymphocytes were enriched by positive selection using the anti-B220 beads (BD Bioscience), as previously described (9). The CD4 T cell population was enriched from the B220 negative fraction by positive selection using the CD4 (L3T4) beads (Milteny) according to the manufacturer’s protocol. The CD8 enriched population was obtained as the negative fraction of the CD4 enrichment.
Flow cytometry
FACS analysis was performed using FACS Canto (BD Biosciences). Antibodies from the following sources were used: anti-CD74, Ly108, CD40L (eBioscience), CD19, B220, CD4, CD3, CD21, CD24, CD23, SLAMF5, SLAMF1 (Biolegend), anti-MIF (Abcam) and anti-EAT- 2 (Proteintech). For assessment of apoptosis, Annexin V and 7-AAD staining was performed (BD Pharmingen). Flow cytometry data analysis was performed using FloJo software (Treestar).
Cell Conjugation assay
Naïve WT B cells (5 × 106) were cultured with WT or SAP−/− T cells (5 × 106) and incubated for 24 hrs in 24-well plates. Conjugates were enumerated by flow cytometry after the cell mixture was stained for CD3/CD4/CD8 and B220 at 4°C.
Quantification of B and T cell conjugates by Imaging Flow Cytometry
Cells were examined by Imaging Flow Cytometry using the ImageStreamX mark II (Amnis-part of EMD Millipore, Seattle, WA). At least 105 cells were collected from each sample. Images were analyzed using IDEAS 6.0 software (Amnis). Cells were gated for the cells in focus using the Gradient RMS feature (30)
B220+ B/CD3+ T cell conjugates were gated according to their area and aspect ratio to include doublets of relevant size. To eliminate doublets that did not consist of viable cells, or cells that were too far apart, only cells with the correct distance (3-7 μm) were gated. All the gates were visually inspected to verify the classifications.
SAP and SLAMF6 downregulation by siRNA
Human cells
Purified T cells and B cells (5×106) were subjected to electroporation as previously described (12). The cells were treated with the following siRNAs: SAP, NTB- A, CD74 (ON-TARGET plus SMART pool) and negative control siRNA (ON-TARGET plus non-targeting pool) (Dharmacon).
Mouse B cells
B cells (5×106) were subjected to electroporation as previously described (12).The cells were treated with the following siRNAs: EAT2 (ON-TARGET plus SMART pool) and negative control siRNA (ON-TARGET plus non-targeting pool) (Dharmacon).
Real-time reverse transcription-PCR analysis
RNA extraction and qRT-PCR were performed as previously described (31). Primers are summarized in Table1.
Table 1.
Primer sets for qRT-PCR
| Gene | Direction | Primer Sequence |
|---|---|---|
| Bcl-2 | Forward | GCCAACTGAGCAGAGTCTC |
| Reverse | GGACAACATCGCCCTGTG | |
| CD74 | Forward | GGAGTACCCGCAGCTGAAGGGG |
| Reverse | GAAGATAGGTCTTCCATGTCCAGTG | |
| SAP | Forward | TCATGGGGCTTTCATTTCAGGCAGACATCAGG |
| Reverse | GACGCAGTGGCTGTGTAT | |
| NTB-A | Forward | TCCTTCTCTGTCTCTGCCCA |
| Reverse | GCCCTGTGTTCGCTGAGTAG |
Stimulation murine B cells with CD74
Activation of CD74 was performed as previously described (9).
Blocking SLAMF6 on human B cells
Human B cells (1×107) were incubated in the presence of 20 μg/ml monoclonal anti- NTB-A Ab (clone NT-7) or mouse IgG1 κ isotype control Abs (BioLegend), as previously described (32).
Co-culture
Co-culture of murine primary B and T cells was performed in 24 well plates with 10% FCS w/v Iscov medium. Transwell experiments were performed in 24 well plates containing an insert with a 5.0 polycarbonate membrane. B cells were seeded in the well, and T cells in the insert.
Immunoprecipitation and western blot analysis
Immunoprecipitation of endogenously expressed proteins and their detection were performed on fresh mouse lymphocytes as previously described (12). For IP and Blotting of EAT2, anti-EAT-2 Ab (N-14; Santa cruz) was used. pTYR was detected with the anti-pTyr (FL-293; Santa Cruz) antibody. IP of Ly108 was performed with theanti- Ly108 (13G3-19D; eBioscience) antibody.
RAG adoptive transfer
A mixture of B and T cells at a 1:1 ratio (total of 1 × 107 cells) in 300 μl PBS per mouse was injected i.v. to the tail vein of RAG1−/− mice.
CD4 depletion
WT mice were injected with 150 μg/ml anti-CD4 mAb GK1.5 (BioXcell BE0003-1) diluted in sterile PBS to a final volume of 200 μl, i.v to the tail vein.
Statistics
Data analysis was performed as previously described (33), using Graphpad Prism (Version 6.0f, GraphPad Software Inc., La Jolla, CA, USA). Data are reported as mean +/− SEM. To determine significance of differences, we used either the unpaired Student’s t- test or one-way ANOVA test for mouse group experiments, and ratio paired Student’s t- test for treatments of human samples.
Results
Direct contact between B and T cells is essential for B cell survival
To determine the role of SLAMs and their adaptor molecule, SAP, which expressed in T cells, in the regulation of B cell maintenance, a co-culture experiment was first performed. In this experiment, wild type (wt) B220+ cells, which are mostly naïve mature B cells, were cultured alone or in the presence of wt or SAP-deficient naïve T cells. As shown in Fig. 1A, the presence of wt T cells significantly increased the survival of naïve B cells compared to B cells cultured alone by about three-fold, while incubation with SAP deficient T cells supported less B cell survival. No significant change was observed in wt and SAP−/− T cell survival rates in this co-culture setting (Supplementary Fig. 1A). These results suggest a SAP dependent beneficial effect of naïve T cells on naïve B cell survival.
Figure 1. Direct B/T contact regulates naïve B cell survival.
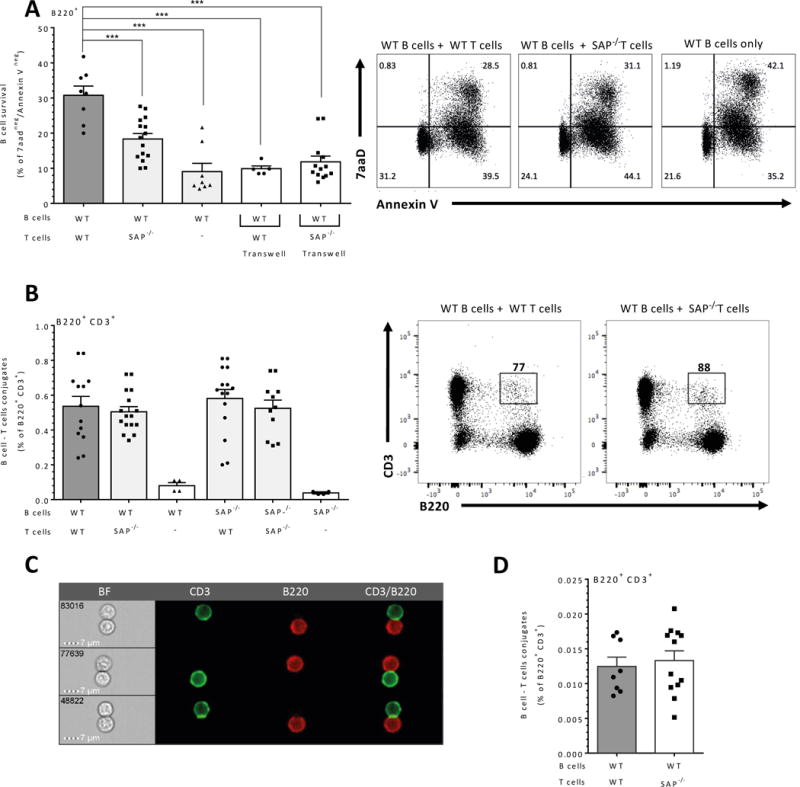
(A) Purified naïve wt B cells were cultured alone or at 1:1 ratio with 5×106 wt or SAP−/− T cells in regular or Transwell apparatus wells (white bars). After 24 hrs, B cells were analyzed by flow cytometry for B cell survival by Annexin V/7AAD staining. Results are shown as the percentage of live B cells (double negative Annexin V/7AAD); right panels exhibit representative dot plots of Annexin V/7AAD staining. N=7 (B) Purified naïve wt or SAP−/− derived B cells were cultured alone or at a 1:1 ratio with 5×106 wt or SAP−/− T cells. After 24 hrs, the cells were analyzed for T/B cell conjugates by flow cytometry of double positive staining for B220+ and CD3+. Results are shown as the percentage of conjugates. Right panels exhibit representative dot plots of B220/CD3 staining. N=4. (C–D) WT B cells were cultured together with WT or SAP−/− T cells. B/T cell conjugates were analyzed by imaging flow cytometry (Image Stream, n=6). (C) Representative images of wt B cells co-cultured with wt T cells. (D) Percentage of B/T doublets of total B and T cell populations. Results are shown as the percentage of T/B cell conjugates. N=4. Each dot represents a biological repeat, and bars show SEM. ns p≥ 0.05, * p<0.05, **p<0.01, ***p<0.001, ****p<0.0001
To examine whether cell-cell contact or soluble factors are responsible for the T cell dependent regulation of B cell survival, similar cultures were established, but direct B/T cell interactions were prevented by a Transwell membrane. As shown in Fig. 1A (white bars), B and T cell contacts were required for the T cell control of B cell live population. Abrogation of cell contacts by the Transwell membrane reduced cell survival to levels observed in B cells co-cultured with SAP-deficient T cells.
It was previously shown that SAP−/− T cells show impaired ability to generate stable interactions with B cells following immunization (22). To determine whether SAP deficiency affects naïve B/T conjugates, wt or SAP−/− derived B cells were cultured with wt or SAP−/− T cells (25). After 24 hrs, the cells were analyzed for double positive staining of CD3+ and B220+, as was previously described (22). Surprisingly, as seen in Fig. 1B, SAP−/− T cells formed similar numbers of T/B conjugates as wt T cells. We further validated our results and the existence of naïve T cell/B cell conjugates by looking for naïve T cell-B cell doublets using image-based quantification. B cells were co-cultured with wt T cells for 24 hrs, and the co-cultures stained for CD3 and B220. As shown in Fig. 1 C-D, T cell (CD3+)/B cell (B220+) doublets were detected in this setting, suggesting direct contact between the cells. To further resolve the role of SAP in this interaction, and to determine whether formation of doublets is inhibited in the absence of SAP in T cells, B cells were co-cultured with SAP deficient T cells and the number of doublets was analyzed. As shown in Fig. 1D, SAP deficiency did not affect the percentage of T cell-B cell doublets, suggesting that in the absence of SAP, T cells can still interact with the B cells but their downstream cascade is altered. Since SAP deficiency in B cells did not cause any intrinsic defect on their cell survival (Supplementary Fig. 1B), these results suggest that T cell support B maintenance in a SAP dependent manner.
A SAP dependent B and T cell interaction regulates CD74 surface expression on B cells
We next followed the SAP-dependent molecular mechanism regulating splenic B cell survival. Our hypothesis was that since CD74 is a survival receptor expressed on B cells, its expression or function might be modulated by a SAP dependent T/B interaction. Therefore, CD74 cell surface expression was analyzed on B cell splenic populations derived from wt and SAP deficient mice. A significant reduction in surface CD74 expression was detected on total B cells, mature (B220+, CD21intCD24low), and transitional B cell (Transitional 1; B220+, CD21lowCD24high, Transitional 2: CD21highCD24highCD23high) populations derived from SAP-deficient mice (Fig. 2A).
Figure 2. SAP dependent B and T cell interactions regulate CD74 expression on the B cell surface.
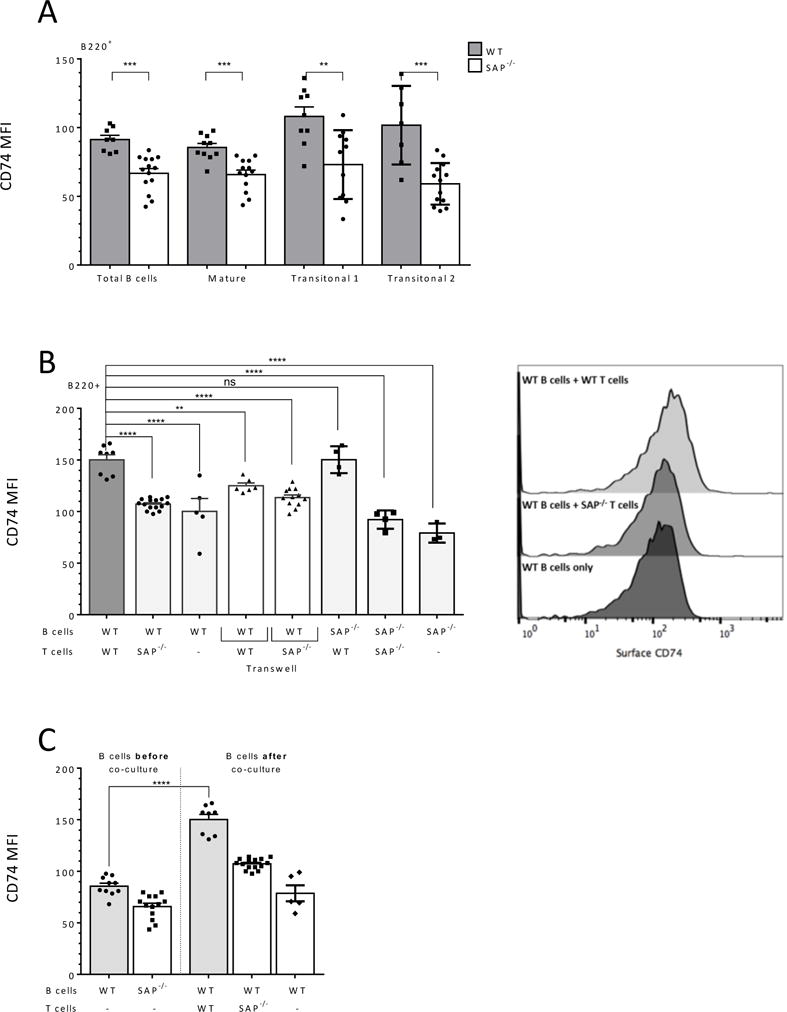
(A) Wt and SAP−/− splenocytes were analyzed by flow cytometry for surface CD74 expression on B cell populations. Total B cells (B220+); mature population (B220+, CD21intCD24low), Transitional 1 (B220+, CD21lowCD24high) and Transitional 2 (B220+, CD21highCD24+CD23+); graph show CD74 MFI levels. N=4. (B) Naïve wt and SAP−/− derived B cells and wt or SAP−/− T splenocytes were isolated. B cells were cultured alone or at a 1:1 ratio with 5×106 wt or SAP−/− T cells in regular or Transwell apparatus wells. After 24 hrs, cells were analyzed by flow cytometry for CD74 surface expression on B cells. Graph shows CD74 MFI levels. Right panels exhibit representative histograms of CD74 staining on B220+ cells. N=4. (C) Purified wt or SAP−/− were cultured for 24 hours alone or at 1:1 ratio with 5×106 wt or SAP−/− T cells were analyzed for CD74 surface expression before (“B cells before co-culture”) or after (“B cells after co-culture”) co-culture. CD74 MFI levels are shown in the graph. In all the graphs, each dot represents a biological repeat. N=3. Bars showing SEM. ns p≥ 0.05, * p<0.05, **p<0.01, ***p<0.001, ****p<0.0001
Next, wt or SAP−/− derived B cells were cultured alone or with naïve wt or SAP deficient T cells for 24 hrs and expression of surface CD74 was analyzed. B cells (wt or SAP−/−) cultured alone or with SAP-deficient T cells exhibited significantly lower cell surface expression levels of surface CD74 compared to B cells co-cultured with wt T cells (Fig. 2B). Furthermore, lower levels of CD74 were observed in the Transwell setting, in which direct interaction between wt B and T cells was inhibited (Fig. 2B white bars).
The lower cell surface expression of CD74 seen in the absence of SAP expressing T cells might result from an upregulation of its levels on B cells in the presence of T cells or by downregulation of its levels due to the lack of a proper signal from the SAP−/− T cells. To address this point, the levels of cell surface CD74 on freshly isolated splenic B cells were compared to its levels detected on B cells co-cultured for 24 hrs with T cells. As seen in Fig. 2C, following co-culture with wt T cells, elevated levels of cell surface CD74 were detected on the B cells, while incubation with SAP−/− T cells resulted in a milder elevation. These results suggest a mechanism whereby T cells upregulate CD74 cell surface levels on B cells in an SAP dependent manner.
CD74 regulation by SAP signaling is mediated by SLAMF6
SAP transmits signals induced by the SLAM family of receptors (16). To determine which SLAM member regulates the T and B cell contact that results in survival of B cells, the effect of three SLAM members, SLAMF6 (Ly108 or NTB-A in human), SLAMF5 (CD84), and SLAMF1 (CD150) were analyzed. These receptors were shown to work in synergy in the regulation of antibody responses, and were identified as mediators of the B/T cell contact in germinal centers (24, 34). We first analyzed the expression of these receptors on splenic B cells. As shown in Fig. 3A, high levels of SLAMF6 were detected on the surface of B cells while, SLAMF1 and SLAMF5 levels were significantly lower. In addition, SLAMF6 expression levels on B cells derived from SAP deficient mice were slightly reduced (Supplementary Fig. 1C) while almost no differences were detected in SLAMF5 (Supplementary Fig. 1D) and SLAMF1 (Supplementary Fig. 1E) cell surface levels. These results suggest that SLAMF6 may have a more important role in support of B cells by naïve T cells.
Figure 3. SAP dependent B and T cell interaction regulates SLAMF6 expression and function in B cells.
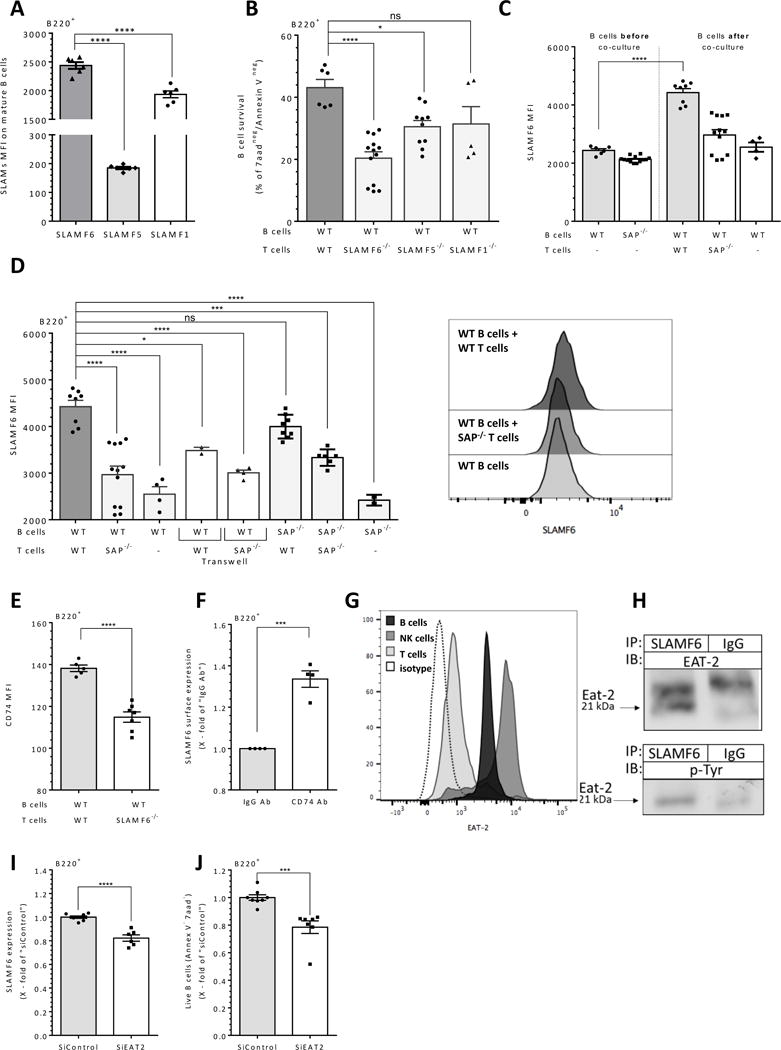
(A) Fresh wt mature naïve B cells were analyzed by flow cytometry for SLAMF6, SLAMF5 and SLAMF1 surface expression. Results are shown as the levels of SLAM receptor MFI. N=3. (B) Wt B cells were cultured at a 1:1 ratio with 5×106 wt/SLAMF6−/−/SLAMF5−/−/or SLAMF1−/− T cells. After 24 hrs, B cells were analyzed by flow cytometry for B cell survival by Annexin V/7AAD staining. Results are shown as the percentage of live B cells (double negative for Annexin V/7AAD). N=6. _(C) Fresh wt or SAP−/− (“B cells before co-culture”) and purified naïve wt B cells were cultured for 24 hours alone at 1:1 ratio with 5×106 wt or SAP−/− T cells (“B cells after co-culture”), were analyzed for SLAMF6 expression. SLAMF6 MFI levels are shown in the graph. N=3. (D) Naïve wt or SAP−/− derived B cells and wt or SAP−/− T splenocytes were cultured alone or at a 1:1 ratio with 5×106 wt or SAP−/− T cells in regular or Transwell apparatus (white bars) wells. After 24 hrs, cells were analyzed by flow cytometry for SLAMF6 expression on B cells (B220+ gate). Results are shown as SLAMF6 MFI; right panels display representative histograms of SLAMF6 staining on B220+ cells. N=4. (E) Naïve wt B splenocytes were cultured alone or at a 1:1 ratio with 5×106 wt or SLAMF6−/− T cells. After 24 hrs, cells were analyzed by flow cytometry for CD74 expression, shown by CD74 MFI. N=3 (F) Naïve mature wt B cells were activated with anti-CD74 or IgG control antibodies for 18 hrs. Cells were then analyzed for SLAMF6 expression on the B220+ gate. N=3 (G) Fresh B cells, NK cells and T cells were analyzed for EAT-2 expression by intra-cellular staining and flow cytometry analysis. Results are shown as representative histograms of EAT-2 expression on the various populations. (H) Purified wt B splenocytes were lysed, and SLAMF6 was immunoprecipitated. Proteins were separated by 12% SDS-PAGE and analyzed for EAT-2 expression (Top panel N=3), and for p-Tyr (G-bottom, N=2). (I-J) siRNA control or EAT2 treated naive B cells were were co-cultured with untreated naive T cells. After 48 hrs, the cells were stained for SLAMF6 (I) and annexin V and 7aad (J). N=2. In all graphs, each dot represents a biological repeat. N represents the number of experiments._Bars showing SEM. ns p≥ 0.05, * p<0.05, **p<0.01, ***p<0.001, ****p<0.0001
We then analyzed the role of these receptors expressed on T cells in regulation of naïve B cell survival. Naïve B cells were co-cultured with naïve WT or SLAMF6/SLAMF5/SLAMF1 (24, 28, 29) deficient T cells for 24 hrs, and B cell survival was then analyzed. While deficiency of each of these receptors interfered with the support induced by T cells, SLAMF6 deficiency caused the most significant reduction in B cell survival (Fig. 3B). In addition, the abrogation of T cell support to B cells, following disruption of interaction was independent of whether the B cells or the T cells were SLAMF6 deficient (Supplementary Fig. 1F), SLAMF5 (Supplementary Fig. 1G) or SLAMF1 (Supplementary Fig. 1H) deficient. Based on these results, we decided to further focus on the role of SLAMF6 in the T cell induced survival support.
Next, to determine the role of SLAMF6 expression in the T cell dependent B cell survival, murine splenic wt or SAP−/− derived B cells were cultured alone or with naïve wt or SAP deficient T cells for 24 hrs, and expression of SLAMF6 was analyzed. SLAMF6 expression was significantly upregulated on B cells cultured with T cells, compared to its levels on freshly isolated B cells (Fig. 3C). This elevation did not occur in the presence of SAP−/− T cells (Fig. 3 C-D). Moreover, no change in SLAMF6 expression levels on T cells was detected under these conditions (Supplementary Fig. 1I). These results suggest a T cell dependent elevation of SLAMF6 expression on B cells mediated through SAP. When B/T cell interactions were prevented by a Transwell membrane, CD74 cell surface levels on B cells remained at the low levels observed in B cells incubated with SAP-deficient T cells (Fig. 3D, white bars).
We further investigated the role of SLAMF6 in the T-cell dependent regulation of CD74 expression and B cell survival. First, CD74 expression levels were analyzed on the SLAMF6−/− cells. Similar surface levels of CD74 were detected on freshly isolated wt and SLAMF6−/− B cell populations (Supplementary Fig. 1J). Next, splenic wt or SLAMF6−/− B cells were co-cultured with splenic wt or SLAMF6−/− T cells and levels of CD74 and B cell survival were analyzed (Fig. 3E, and Supplementary Fig. 1K). Abolishment of the SLAMF6-mediated interaction between T and B cells led to a significant decrease in CD74 surface expression on B cells, accompanied by a downregulation in B cell survival (Supplementary Fig. 1F). SLAMF5 deficiency also resulted in a reduction in CD74 upregulation (Supplementary Fig. 1L), while SLAMF1 deficiency showed no effect at all (Supplementary Fig. 1M).
CD74 activation induces B cell survival ((9, 10) and Supplementary Fig. 2A), and upregulates SLAMF6 expression on B cells (Fig. 3F). SAP−/− mice exhibit lower levels of CD74 on B cells (Fig. 2A) and a lower response to CD74 Ab stimulation (data not shown). These results suggest that the SLAMF6 B/T interaction elevates CD74 expression on B cells, which results in B cell survival. In addition, CD74 induces SLAMF6 levels, which can further induce/enhance the process.
EAT-2 is recruited to SLAMF6
B cells do not express SAP, and instead express the mRNA of the adaptor molecule, EAT-2 (16, 35). To characterize SLAMF6 induced cascade in B cells, we wished to determine whether EAT-2 plays a role in the B cell side of this cascade. To this end, we first established the expression of EAT-2 protein in splenic B cells. Using a flow cytometry analysis, EAT-2 levels in B cells were compared to its levels in NK cells, which are known to express EAT-2, and to T cells which express low levels of the adaptor (15, 18). As shown in Fig. 3G, B cells express the EAT-2 protein. Next, to determine whether EAT-2 is involved in the interaction via SLAMF6, a co-immunoprecipitation (Co-IP) experiment was performed, in which SLAMF6 was pulled down by anti-SLAMF6 Ab from B cell lysates and EAT2 levels were analyzed in the immunoprecipitated proteins. As seen in Fig. 3H (top panel), SLAMF6 recruited EAT-2 in B cells, suggesting that EAT-2 transmits the SLAMF6 signal in B cells. Following the recruitment of EAT-2 to the receptor, it is phosphorylated on tyrosine residues, which activates it to transmit downstream signaling (14). We next wished to identify the presence of phosphorylated EAT-2 in naïve B cells. EAT-2 was pulled down from naïve B cell lysate, and the immunoprecipitated proteins were analyzed for pTyr. As demonstrated in Supplementary Fig. 2B, phosphorylated EAT-2 was detected in the lysates of naïve B cells. To directly demonstrate recruitment of phosphorylated EAT-2 to SLAMF6, SLAMF6 was pulled down and the immunoprecipitated proteins were analyzed. Phosphorylated EAT-2 was pulled down with SLAMF6 (Fig 3H, bottom panel), suggesting that in B cells, EAT-2 can be recruited to SLAMF6 and is activated. To directly show the involvement of EAT2 in B cell survival, EAT-2 levels were knocked down in using siRNA against EAT2. siEAT2 or siControl treated B cells were cultured with naïve T cells for 48 hours. Reduction of about 40% in EAT-2 protein levels were detected in siEAT2 treated B cells (Supplementary Fig 2C). In this setting, SLAMF6 (Fig. 3I) and live B cells (Fig. 3J) levels showed a significant reduction of about 20%. This further suggests that EAT2 regulates SLAMF6 expression and B cell survival.
B cell survival is regulated by CD4+ T cells
Next, we wished to identify which T cell subpopulation, CD4+ or CD8+, is involved in the maintenance of naïve B cells. First, as seen in Supplementary Fig. 2D, CD4 T cells express higher levels of SLAMF6 than CD8 T cells. Next, wt, SAP−/−, or SLAMF6−/− CD4+ or CD8+ T cells were purified (Supplementary Fig. 2E) and co-cultured with B cells. As shown in Fig 4A, a greater number of conjugates of B cells with CD4+ T cells compared to CD8+ T cells were detected. In addition, only wt CD4+ T cells managed to support B cell survival (Fig. 4B) and SLAMF6 expression on B cells (Fig. 4C). Thus, CD4+ T cells regulate B cell survival in SAP and SLAMF6 dependent manner.
Figure 4. Naïve CD4+ T cells induce B cell survival regulation via MIF secretion.
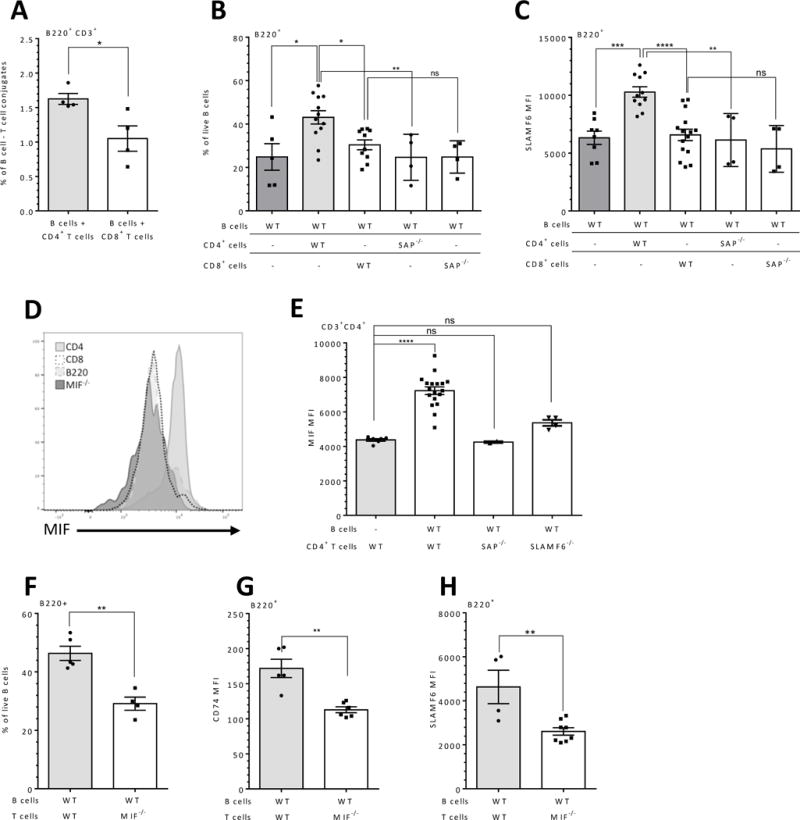
(A) Purified naïve wt B cells were cultured at a 1:1 ratio with 5×106 wt CD4+ or CD8+ T cells. After 24 hrs, the cells were analyzed for T/B cell conjugates by flow cytometry by double positive staining for B220+ and CD3+. Results are shown as the percentage of conjugates. (B-C) Naïve wt B splenocytes were cultured alone or at a 1:1 ratio with 5×106 wt or SAP−/− CD4+ T or CD8+ T cells. After 24 hrs, cells were analyzed by flow cytometry for Annexin V/7AAD N=4. (B), or SLAMF6 surface expression (C) on B cells. N=4. (D) MIF expression was analyzed 5 hrs following Monensin treatment by FACS staining. Histograms show MIF expression by CD4+T cells, CD8+ T cells, and B cells compared to MIF expression on MIF−/− splenocytes (background). (E) Naïve wt/SAP−/−/SLAMF6−/− T splenocytes were cultured alone or at a 1:1 ratio with 5×106 wt B cells. After 24 hrs, expression of MIF in CD3+CD4+ was analyzed by flow cytometry. N=4 (F-H) Naïve wt B splenocytes were cultured alone or at a 1:1 ratio with 5×106 wt or MIF−/− T cells. After 24 hrs, cells were analyzed by flow cytometry for Annexin V/7AAD (F), CD74 (G) or SLAMF6 (H) surface expression on B cells. N=3. Results are shown as percentage (F) or MFI (G-H). In all experiments, each dot represents a biological repeat. N represents the number of experiments. Bars showing SEM; ns p≥ 0.05, * p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Since CD74 expression is regulated by the SLAMF6 mediated T/B interaction, we wished to determine whether the SLAMF6/SAP dependent B and T interaction regulates the expression of CD74 ligand, MIF, in T cells. As shown in Fig. 4D, CD4+ T cells expressed higher levels of intracellular MIF compared to its levels in CD8+ T cells and B cells. Next, regulation of MIF expression levels in T cells by splenic B cells was determined. While B cells elevated MIF levels in wt CD4+ T cells in the co-culture setting, MIF levels in CD4+ T cells lacking SAP or SLAMF6 were not upregulated by B cells (Fig. 4E). This elevation was specific to the CD4+ T cells, and was not detected in the CD8+ population (Supplementary Fig. 2F).
To directly determine whether MIF plays a role in this crosstalk, murine wt splenic B cells were cultured alone or with naïve wt or MIF-deficient T cells (27) for 24 hrs. FACS analysis revealed a reduced B cell survival (Fig. 4F), CD74 cell surface expression (Fig. 4G), and SLAMF6 (Fig. 4H) on B cells incubated with MIF-deficient T cells. These results imply that MIF secreted from T cells is essential for B cell survival in a SALMF6 and CD74 dependent manner. CD74 expression on B cells cultured with T cells deficient in SAP, SLAMF6 or MIF, did not induce an upregulation of CD74 expression on B cells, suggesting that all these molecules contribute to the T cell support of B cell survival (Supplementary Fig. 2G).
Following immunization or infection, binding of CD40L on the T cells to CD40 on B cells promotes B cell proliferation, germinal center development and the differentiation of B cells into antibody-secreting plasma cells (36). It was previously shown that SAP downregulates the expression of CD40L on T cells, as SAP−/− T cells exhibit higher levels of CD40L (23). We therefore determined CD40L expression on naïve T cells in the unstimulated co-culture setting. WT/SLAMF6−/−/SAP−/− CD4+ T cells were cultured alone or with B cells, and CD40L was analyzed on the T cells. As described previously, SAP−/− CD4+ T cells exhibited elevated levels of CD40L compared to wt cells (Supplementary Fig. 2H). However, the presence of B cells in the co-culture had no effect on CD40L cell surface expression. Furthermore, comparable levels of CD40L expression were observed on wt CD4+/CD8+ or SLAMF6−/− CD4+ T cells (Supplementary Fig. 2I). Thus, CD40L does not play a part in the naïve T/B interaction.
SAP and SLAMF6 regulate B cell survival in vivo
To determine the in vivo role of SAP and SLAMF6 in naïve T/B interactions, and regulation of B cell survival, purified wt splenic B cells were adoptively transferred together with purified wt or SAP−/− splenic T cells into lymphocyte-deficient RAG1−/− recipients, which lack mature B and T cells. The mice were sacrificed 24 hrs after the cell transfer. CD74 (Fig. 5A) and SLAMF6 (Fig. 5B) cell surface expression levels were significantly lower on B cells co-transferred with SAP deficient naïve T cells, compared to their levels in the presence of wt T cells. In addition, the percentage of the live B cell population was downregulated when B cells were transferred together with SAP deficient T cells (Fig. 5C). Moreover, to directly show the role of CD4+ T cells in vivo, wt naïve B cells were adoptively transferred into RAG1−/− alone or with WT CD8+ T cells, and WT or SAP−/− CD4+ T cells. As seen in Fig. 5D, only wt CD4+ supported B cell survival.
Figure 5. SAP mediated signaling in T cells regulates CD74 expression and B cell survival in vivo, and anti-CD4 downregulates SLAMF6 expression in vivo.
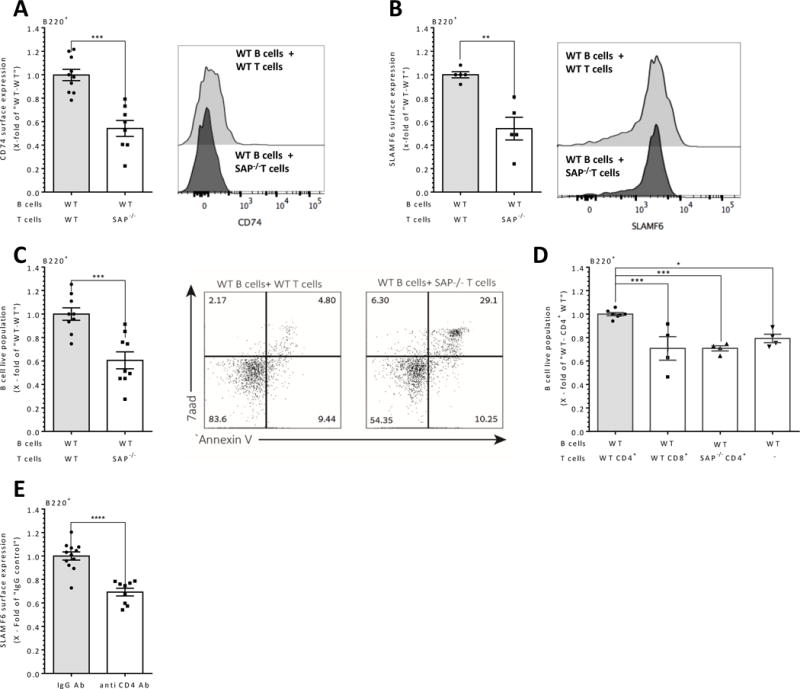
(A-D) Naïve wt B cells were injected into the tail vein of RAG1−/− mice together with 5×106 WT or SAP−/− T cells. After 24 hrs, splenocytes of RAG1−/− recipients were harvested and analyzed by flow cytometry for CD74. N=4. (A), right panel shows representative histograms of CD74 expression), SLAMF6. (B), right panel shows representative histograms of SLAMF6 expression) and Annexin V/7-AAD. N=3._ (C), right panel shows representative dot plots of Annexin V/7AAD). All results are shown as x- fold of “WT B cells and WT T cells”. (D) Naïve wt B cells were injected into the tail vein of RAG1−/− mice together with 5×106 WT or SAP−/− CD4+ or CD8+ T cells. After 24 hrs, splenocytes of RAG1−/− recipients were harvested and analyzed by flow cytometry for B cell survival. Results are shown as x-fold of “WT B cells and WT CD4+ T cells”. N=3. (E) WT mice were injected i.v. to the tail vein with 150 μg anti-CD4 neutralizing Ab or with PBS alone as control. After 4 days following the injection, PB was collected from the tail vein and analyzed for SLAMF6 expression on B cells. N=3. Results are shown as fold change relative to the control group, bars indicate SEM. In all results, each dot represents a biological repeat. N represents the number of experiments. **p<0.01, ***p<0.001, ***p<0.001, ****p<0.0001.
To further investigate the role of CD4+ T cells in the maintenance of naïve B cells, CD4+ cells were depleted by injection of anti-CD4 antibody (Supplementary Fig. 2J) (37). After 4 days, spleens were harvested and analyzed for cell surface markers. SLAMF6 levels were significantly reduced on B cells derived from anti-CD4 treated mice compared to control injected mice (Fig. 5E), suggesting a role for SLAMF6 expression on CD4+ cells in B cell maintenance. CD74 expression levels were not altered at this time point (data not shown). These results show that naïve CD4+ T cells are required in vivo for the maintenance of B cells in an SAP-dependent manner.
The role of SAP and SLAMF6 in human B cell survival
We next wished to determine whether a similar SAP dependent T cell-induced survival pathway exists in human B cells; to this end, SAP expression in healthy human PB T cells was knocked-down by SAP siRNA (Supplementary Fig. 3A). SAP deficient T cells were then co-cultured with untreated human PB B cells, and their SLAMF6 expression levels were analyzed. As shown in Fig. 6A and Supplementary Fig 3B, lower SLAMF6 cell surface expression levels were detected on B cells cultured with SAP deficient T cells compared to B cells cultured with T cells treated with control siRNA. CD74 mRNA (24 h; Fig. 6B) and protein (48 h; Fig. 6C and Supplementary Fig 3C) levels were then analyzed in B cells incubated with T cells deficient in SAP. CD74 message and protein levels were significantly reduced in healthy human B cells co-cultured with SAP-deficient T cells, compared to their levels when incubated with T cells treated with a control siRNA.
Figure 6. SAP dependent B/T cell crosstalk regulates the expression of CD74, SLAMF6 and B cell survival in human cells.
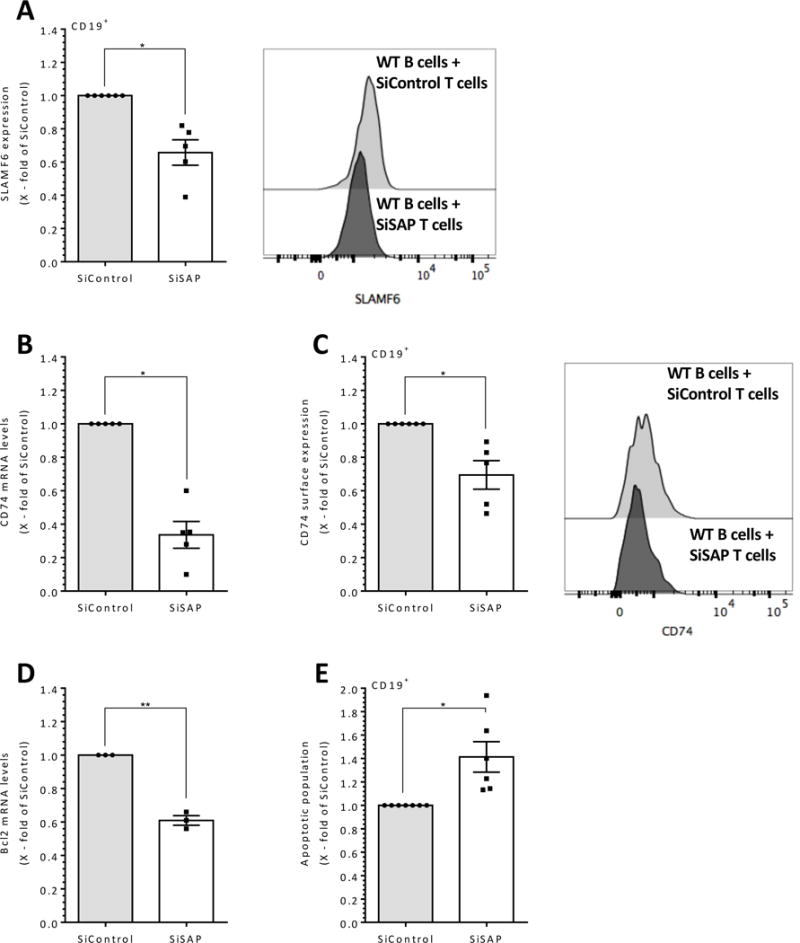
(A-D) Healthy human PB B cells were co-cultured with healthy human PB T cells that were treated with control scrambled or SAP siRNA. Following 24 hrs of incubation, RNA was purified, and mRNA was analyzed by qRT-PCR for CD74 mRNA (B) and Bcl2 (D). After 48 hrs, cell surface expression of SLAMF6 (A, right panel shows representative SLAMF6 histograms, N=3), CD74 (C, right panel shows representative CD74 histograms), and Annexin V (E), on CD19+ positive population was determined by FACS. Results are shown as fold change relative to the si-control group; bars indicate SEM. Each dot represents a biological repeat. N represents the number of experiments. * p<0.05, **p<0.01, ***p<0.001
Next, we determined whether reduced SAP, SLAMF6 and CD74 levels lead to cell death in human B cells. Incubation of B cells with SAP knockdown T cells significantly reduced their Bcl-2 mRNA levels (Fig. 6D) and induced their death (Fig. 6E). Collectively, these results suggest that SAP plays a dominant role in T cell induction of peripheral human B cell survival by regulating surface CD74 and SLAMF6 expression.
To further examine the role of SLAMF6 in B cells, SLAMF6 expression in healthy human PB T cells was knocked-down by SLAMF6 siRNA (Supplementary Fig. 3D). SLAMF6 deficient T cells were then co-cultured with human PB B cells. CD74 surface levels were analyzed after 48 hrs. As shown in Fig. 7A and Supplementary Fig 3E, lower levels of surface CD74 were detected on B cells incubated with SLAMF6 knockdown T cells. In addition, healthy total PB cells (T and B cells) were incubated in the presence of either SLAMF6 neutralizing (32) or control IgG antibodies. Reduced T/B cell interactions through SLAMF6 resulted in lower CD74 cell surface levels on B cells when compared to IgG-treated cells (Fig. 7B). Furthermore, blocking SLAMF6 reduced Bcl-2 protein levels (Fig. 7C). These results suggest that SLAMF6 regulates CD74 cell surface expression and human B cell survival.
Figure 7. The role of SLAMF6 in CD74 regulation on human B cells in vitro and in vivo.
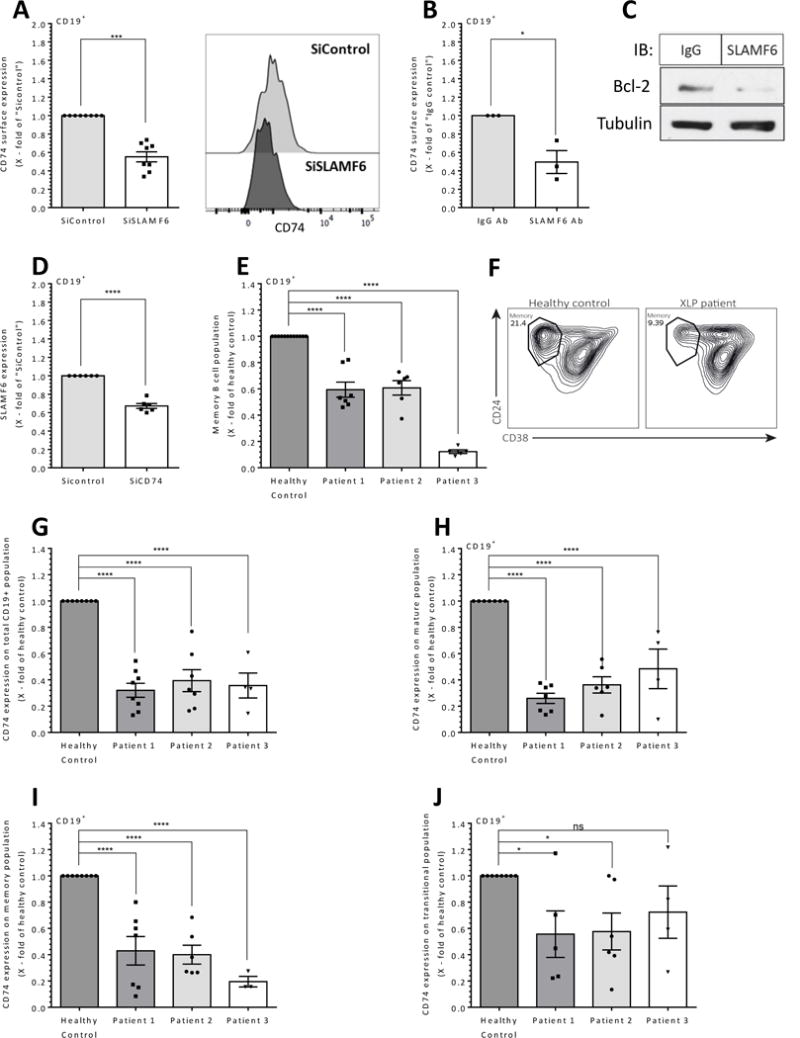
(A) Healthy human B cells were co-cultured with healthy human PB T cells which were treated with SLAMF6 or control siRNA. After 48 hrs, CD74 surface expression was analyzed by FACS. Right panel shows representative CD74 histograms. N=3 (B-C) Purified PB B and T cells were incubated in the presence of anti-SLAMF6 or control IgG antibodies. After 24 hrs, (B) CD74 surface expression was analyzed by flow cytometry (C) Purified B cells were lysed and analyzed for Bcl2 and tubulin by western blot analysis (N=3). (D) PB B cells were treated with control scrambled or CD74 siRNA and co-cultured with non-treated healthy PB T cells. After 48 hrs, the cells were analyzed by flow cytometry for SLAMF6. N=3. Results are shown as fold change relative to the si-control group; bars indicate SD. Each dot represents one sample. N represents the number of experiments.* p<0.05, ***p<0.001, ****p<0.0001
(E-F) Lymphocytes were isolated from PB derived from XLP and healthy patients and stained for B cell populations. Results are shown as fold change relative to the control group; bars indicate SD. Each repeat represents a different blood sample from the same patient. (E) Memory B cells (Patient 1, n=8; Patient 2, n=6; Patient 3, n=5). (F) Representative contour plots of B cell population in healthy control and in XLP patients. (G-J) CD74 expression was analyzed on B cell populations derived from subjects with XLP and compared to healthy controls. CD74 surface expression on the total CD19+ population (Patient 1, n=8; Patient 2, n=7; Patient 3, n=4) (G), mature B cell population (Patient 1, n=7; Patient 2, n=6; Patient 3, n=3) (H), memory B cell population (patient 1, n=7; Patient 2, n=6; Patient 3, n=3) (I), and transitional B cell population (Patient 1, n=5; Patient 2, n=6; Patient 3, n=3) (J). ns p> 0.05, * p<0.05, **p<0.01, ***p<0.001.
Finally, to determine whether CD74 auto-regulates SLAMF6 expression, CD74 expression was downregulated in healthy human PB B cells by CD74 siRNA (Supplementary Fig. 3F). Reduced expression levels of SLAMF6 were detected on B cells with knockdown of CD74 (Fig 7D).
SAP and SLAMF6 regulate survival of XLP B cells
XLP is a severe immunodeficiency associated with mutations of the SH2D1A gene that encodes SAP. Although, the absolute number of total B cells in XLP patients is normal (38), these patients exhibit a marked reduction in their PB memory B cell population (39-41), and Fig. 7 E-F.
To determine whether the lack of SAP in these patients regulates B cell maintenance through the control of CD74 expression, CD74 expression was analyzed in XLP patient- derived PB B cells from three patients. CD74 cell surface expression on total PB B cells (CD19+ cells; Fig. 7G), naïve (CD19+CD24lowCD38int; Fig. 7H), and memory (CD19+CD24hiCD38low; Fig. 7I) PB B cells derived from the XLP patients was significantly reduced compared to its levels on wt human B cells. Almost no change in CD74 expression was observed on the transitional subpopulation (CD19+ CD24hiCD38hi; Fig. 7J). The reduced expression levels of CD74 in the naïve state of the B cells suggests that SAP expression in T cells regulates B cell survival from the naïve stage.
Discussion
Peripheral B-cell numbers are tightly regulated by various homeostatic mechanisms in order to maintain a functional and selective humoral immune response. Our current study provides evidence that naïve mature B cell survival is not only an intrinsic process, but its regulation requires CD4+ T cells in a SLAM/SAP dependent manner. SLAMs and SAP mediate many immunological processes, including the interaction of CD4+ T cell with B cells in the germinal centers (GC), whereas SAP−/− CD4+ T cells are unable to form long-term conjugates with cognate B cells, leading to perturbed GC formation (22-24). Our study followed an antigen-independent interaction between naïve B and T cells. Our results reveal a novel role for SAP and SLAMF6 in regulation of mature naive B cell survival. T cells lacking SAP cannot support the SLAMF6-induced B cell survival pathway. The two cell types can interact through SLAMF6, but the absence of SAP leads to lower CD74 expression on B cells and their reduced survival.
The adaptor molecule SAP controls signal transduction pathways downstream of the SLAM family receptors, and is a key regulator of normal immune function in T, NK, and NKT cells (15, 18). However, B cells do not express SAP (19), and EAT2 was described as a functional homologue in these cells (20, 21). Our results show that EAT-2 protein is expressed in B cells. Activation of SLAMF6 induces EAT-2 binding to SLAMF6, resulting in EAT-2 phosphorylation.
It was previously shown that the lymphocyte populations are grossly normal in SAP deficient mice, although occasional mutant animals exhibit a lower percentage of B cells in the spleen (25). The fact that the steady state levels of B cells are not dramatically altered in the SAP−/− mice might be due to redundancy and constant replenishment of the B population, and from the support provided by other cells in the environment.
The SLAMF6-induced cascade auto-regulates its own expression. Abrogation of stimulation via SLAMF6 and signaling through SAP leads to downregulation of SLAMF6 expression on B cells but not on T cells. The lack of SAP in T cells results in reduced SLAMF6 expression levels on B cells due to the perturbed signaling cascade induced by the T/B interaction in B cells. SLAMF6 elevates CD74 expression levels, which increases the survival potential of naïve B cells. Beyond its role in B cell survival, CD74 regulates SLAMF6 expression levels.
CD4+ T cells are the main players in supporting B cell survival. In vivo depletion of CD4+ T cells in mice significantly decreased SLAMF6 expression. CD4+ T cells express high levels of MIF, a ligand of CD74. T-B crosstalk induces the expression of MIF in the CD4+ T population via SAP and SLAMF6. CD4+ T cells can support the survival of B cells by secretion of MIF. Deficiency of MIF has an incomplete effect on B cell survival. This could be explained by the existence of an additional CD74 ligand. Another ligand of CD74 is MIF2, also termed D-dopachrome tautomerase (D-DT) (42). MIF1 (a.k.a MIF) and MIF2 possess overlapping functions and expression patterns (43). It is possible that MIF2 might play an additional role in the T cell-mediated maintenance of naïve B cells. MIF1 and MIF2 binding to CD74 induces a survival cascade involving NF-κB activation and Bcl-2 expression (5).
Collectively, our results suggest a model whereby B cells can manipulate T cells signaling via SLAMF6 and its adaptor SAP to maintain their full survival potential. Additional SLAM family members have been implicated in survival of specific B cell populations; SLAMF5 (CD84) induces survival of CLL cells, which upregulate its expression in a CD74/MIF dependent manner (12, 33). SLAMF1 (CD150) regulates apoptosis, proliferation and differentiation and IgG synthesis of B cells (44, 45). Analysis of these SLAM members shows that while they might have some role in the T/B interaction described here, it is less significant than that of SLAMF6.
XLP is a severe immunodeficiency associated with mutations of the gene that encodes SAP. Although the absolute number of total B cells in XLP patients is normal (38), these patients exhibit a marked reduction in their PB memory B cell population (39-41). However, previous analysis of the B cells in XLP patients revealed an increase in the circulating immature/transitional B cells (46). Our results suggest that the elevation of the immature population may result from a constant death of the mature B population that needs to be constantly replenished.
Together, our results suggest that T cells have the ability to regulate peripheral B cell survival through SLAMF6/SAP. This pathway, in turn, increases MIF secretion which activates CD74. This consequently enhances cell survival and SLAMF6 expression, thereby forming a positive feedback loop supporting B cell maintenance (Supplementary Fig. 4).
Supplementary Material
Key points.
Naïve CD4+ T cells support B cell survival in vitro and in vivo.
The T cell dependent survival control is mediated through SLAMF6/SAP, resulting in upregulation of CD74 expression by B cells.
The lack of SAP in XLP patients perturbs B cell maintenance through the control of CD74 expression from the mature naïve B cell stage.
Acknowledgments
The authors wish to thank members of the Shachar lab for fruitful discussion and support. We would like also to thank Dr. Ziv Shulman for his suggestions and help with reagents. I.S. is the incumbent of the Dr. Morton and Ann Kleiman Professorial Chair.
Footnotes
This research was supported in part by the Binational Science Foundation (BSF), the Cooperation Program in Cancer Research of the Deutsches Krebsforschungszentrum (DKFZ) and Israel’s Ministry of Science, Technology and Space (MOST), Quinquin foundation and Rubenstein charitable foundation.
The authors declare no competing financial interests
Authors’ contribution
L. R. - designed research, performed research, analyzed data and wrote the paper.
S.C. - designed research, performed research, analyzed data and wrote the paper.
M. K. - designed research, performed research and analyzed data.
E.B. - designed research, performed research, analyzed data.
L. R. - designed research, performed research, analyzed data.
H. L. - designed research, performed research, analyzed data.
A.B. - designed research, performed research, analyzed data.
Z.P.- designed research, performed research, analyzed data.
R. B. - provided essential reagent and wrote the paper.
P.S.- provided essential reagent and wrote the paper.
S. B.-H.- designed research, analyzed data.
I. S.- designed research, analyzed data and wrote the paper.
The authors have declared that no conflict of interest exists.
References
- 1.Kraus M, Alimzhanov MB, Rajewsky N, Rajewsky K. Survival of resting mature B lymphocytes depends on BCR signaling via the Ig alpha/beta heterodimer. Cell. 2004;117:787–800. doi: 10.1016/j.cell.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 2.Srinivasan L, Sasaki Y, Calado DP, Zhang B, Paik JH, DePinho RA, Kutok JL, Kearney JF, Otipoby KL, Rajewsky K. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 2009;139:573–586. doi: 10.1016/j.cell.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackay F, Figgett WA, Saulep D, Lepage M, Hibbs ML. B cell stage and context dependent requirements for survival signals from BAFF and the B cell receptor. Immunol Rev. 2010;237:205–225. doi: 10.1111/j.1600-065X.2010.00944.x. [DOI] [PubMed] [Google Scholar]
- 4.Shachar I, Haran M. The secret second life of an innocent chaperone: the story of CD74 and B cell/chronic lymphocytic leukemia cell survival. Leuk Lymphoma. 2011;52:1446–1454. doi: 10.3109/10428194.2011.565437. [DOI] [PubMed] [Google Scholar]
- 5.Bucala R, Shachar I. The integral role of CD74 in antigen presentation, MIF signal transduction, and B cell survival and homeostasis. Mini Rev Med Chem. 2014;14:1132–1138. doi: 10.2174/1389557515666150203144111. [DOI] [PubMed] [Google Scholar]
- 6.Stumptner-Cuvelette P, Benaroch P. Multiple roles of the invariant chain in MHC class II function. Biochimica et biophysica acta. 2002;1542:1–13. doi: 10.1016/s0167-4889(01)00166-5. [DOI] [PubMed] [Google Scholar]
- 7.Ong GL, Goldenberg DM, Hansen HJ, Mattes MJ. Cell surface expression and metabolism of major histocompatibility complex class II invariant chain (CD74) by diverse cell lines. Immunology. 1999;98:296–302. doi: 10.1046/j.1365-2567.1999.00868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leng L, Metz CN, Fang Y, Xu J, Donnelly S, Baugh J, Delohery T, Chen Y, Mitchell RA, Bucala R. MIF Signal Transduction Initiated by Binding to CD74. J Exp Med. 2003;197:1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Starlets D, Gore Y, Binsky I, Haran M, Harpaz N, Shvidel L, Becker-Herman S, Berrebi A, Shachar I. Cell Surface CD74 initiates a signaling cascade leading to cell proliferation and survival. Blood. 2006;107:4807–4816. doi: 10.1182/blood-2005-11-4334. [DOI] [PubMed] [Google Scholar]
- 10.Gore Y, Starlets D, Maharshak N, Becker-Herman S, Kaneyuki U, Leng L, Bucala R, Shachar I. Macrophage migration inhibitory factor (MIF) induces B cell survival by activation of a CD74/CD44 receptor complex. J Biol Chem. 2008;283:2784–2792. doi: 10.1074/jbc.M703265200. [DOI] [PubMed] [Google Scholar]
- 11.Binsky I, Haran M, Starlets D, Gore Y, Lantner F, Harpaz N, Leng L, Goldenberg DM, Shvidel L, Berrebi A, Bucala R, Shachar I. IL-8 secreted in a macrophage migration-inhibitory factor- and CD74-dependent manner regulates B cell chronic lymphocytic leukemia survival. Proc Natl Acad Sci U S A. 2007;104:13408–13413. doi: 10.1073/pnas.0701553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Binsky-Ehrenreich I, Marom A, Sobotta MC, Shvidel L, Berrebi A, Hazan- Halevy I, Kay S, Aloshin A, Sagi I, Goldenberg DM, Leng L, Bucala R, Herishanu Y, Haran M, Shachar I. CD84 is a survival receptor for CLL cells. Oncogene. 2014;33:1006–1016. doi: 10.1038/onc.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calpe S, Wang NH, Romero X, Berger SB, Lanyi A, Engel P, Terhorst C. The SLAM and SAP gene families control innate and adaptive immune responses. Advances in Immunology. 2008;97:177–250. doi: 10.1016/S0065-2776(08)00004-7. Vol 97. [DOI] [PubMed] [Google Scholar]
- 14.Cannons JL, Tangye SG, Schwartzberg PL. SLAM family receptors and SAP adaptors in immunity. Annu Rev Immunol. 2011;29:665–705. doi: 10.1146/annurev-immunol-030409-101302. [DOI] [PubMed] [Google Scholar]
- 15.Ma CS, Nichols KE, Tangye SG. Regulation of cellular and humoral immune responses by the SLAM and SAP families of molecules. Annu Rev Immunol. 2007;25:337–379. doi: 10.1146/annurev.immunol.25.022106.141651. [DOI] [PubMed] [Google Scholar]
- 16.Engel P, Eck MJ, Terhorst C. The SAP and SLAM families in immune responses and X-linked lymphoproliferative disease. Nature Reviews Immunology. 2003;3:813–821. doi: 10.1038/nri1202. [DOI] [PubMed] [Google Scholar]
- 17.Latour S, Veillette A. The SAP family of adaptors in immune regulation. Seminars in Immunology. 2004;16:409–419. doi: 10.1016/j.smim.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 18.Sayos J, Wu C, Morra M, Wang N, Zhang X, Allen D, van Schaik S, Notarangelo L, Geha R, Roncarolo MG, Oettgen H, De Vries JE, Aversa G, Terhorst C. The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature. 1998;395:462–469. doi: 10.1038/26683. [DOI] [PubMed] [Google Scholar]
- 19.Detre C, Yigit B, Keszei M, Castro W, Magelky EM, Terhorst C. SAP modulates B cell functions in a genetic background-dependent manner. Immunol Lett. 2013;153:15–21. doi: 10.1016/j.imlet.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tangye SG, van de Weerdt BCM, Avery DT, Hodgkin PD. CD84 is up-regulated on a major population of human memory B cells and recruits the SH2 domain containing proteins SAP and EAT-2. European Journal of Immunology. 2002;32:1640–1649. doi: 10.1002/1521-4141(200206)32:6<1640::AID-IMMU1640>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 21.Veillette A. Immune regulation by SLAM family receptors and SAP-related adaptors. Nature Reviews Immunology. 2006;6:56–66. doi: 10.1038/nri1761. [DOI] [PubMed] [Google Scholar]
- 22.Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN. SAP- controlled T-B cell interactions underlie germinal centre formation. Nature. 2008;455:764–769. doi: 10.1038/nature07345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cannons JL, Yu LJ, Jankovic D, Crotty S, Horai R, Kirby M, Anderson S, Cheever AW, Sher A, Schwartzberg PL. SAP regulates T cell-mediated help for humoral immunity by a mechanism distinct from cytokine regulation. J Exp Med. 2006;203:1551–1565. doi: 10.1084/jem.20052097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cannons JL, Qi H, Lu KT, Dutta M, Gomez-Rodriguez J, Cheng J, Wakeland EK, Germain RN, Schwartzberg PL. Optimal Germinal Center Responses Require a Multistage T Cell:B Cell Adhesion Process Involving Integrins, SLAM- Associated Protein, and CD84. Immunity. 2010;32:253–265. doi: 10.1016/j.immuni.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Czar MJ, Kersh EN, Mijares LA, Lanier G, Lewis J, Yap G, Chen A, Sher A, Duckett CS, Ahmed R, Schwartzberg PL. Altered lymphocyte responses and cytokine production in mice deficient in the X-linked lymphoproliferative disease gene SH2D1A/DSHP/SAP. Proc Natl Acad Sci U S A. 2001;98:7449–7454. doi: 10.1073/pnas.131193098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elliott EA, Drake JR, Amigorena S, Elsemore J, Webster P, Mellman I, Flavell RA. The invariant chain is required for intracellular transport and function of major histocompatibility complex class II molecules. J Exp Med. 1994;179:681–694. doi: 10.1084/jem.179.2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fingerle-Rowson G, Petrenko O, Metz CN, Forsthuber TG, Mitchell R, Huss R, Moll U, Muller W, Bucala R. The p53-dependent effects of macrophage migration inhibitory factor revealed by gene targeting. Proc Natl Acad Sci U S A. 2003;100:9354–9359. doi: 10.1073/pnas.1533295100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao F, Cannons JL, Dutta M, Griffiths GM, Schwartzberg PL. Positive and negative signaling through SLAM receptors regulate synapse organization and thresholds of cytolysis. Immunity. 2012;36:1003–1016. doi: 10.1016/j.immuni.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang N, Satoskar A, Faubion W, Howie D, Okamoto S, Feske S, Gullo C, Clarke K, Sosa MR, Sharpe AH, Terhorst C. The cell surface receptor SLAM controls T cell and macrophage functions. J Exp Med. 2004;199:1255–1264. doi: 10.1084/jem.20031835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.George TC, Fanning SL, Fitzgerald-Bocarsly P, Medeiros RB, Highfill S, Shimizu Y, Hall BE, Frost K, Basiji D, Ortyn WE, Morrissey PJ, Lynch DH. Quantitative measurement of nuclear translocation events using similarity analysis of multispectral cellular images obtained in flow. J Immunol Methods. 2006;311:117–129. doi: 10.1016/j.jim.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 31.Zelman-Toister E, Bakos E, Cohen S, Zigmond E, Shezen E, Grabovsky V, Sagiv A, Hart G, Kaushansky N, Ben-Nun A, Maharshak N, Sonnenberg A, Alon R, Becker-Herman S, Shachar I. CD151 Regulates T-Cell Migration in Health and Inflammatory Bowel Disease. Inflamm Bowel Dis. 2016;22:257–267. doi: 10.1097/MIB.0000000000000621. [DOI] [PubMed] [Google Scholar]
- 32.Uzana R, Eisenberg G, Sagi Y, Frankenburg S, Merims S, Amariglio N, Yefenof E, Peretz T, Machlenkin A, Lotem M. Trogocytosis is a gateway to characterize functional diversity in melanoma-specific CD8+ T cell clones. J Immunol. 2012;188:632–640. doi: 10.4049/jimmunol.1101429. [DOI] [PubMed] [Google Scholar]
- 33.Marom A, Barak AF, Kramer MP, Lewinsky H, Binsky-Ehrenreich I, Cohen S, Tsitsou-Kampeli A, Kalchenko V, Kuznetsov Y, Mirkin V, Dezorella N, Shapiro M, Schwartzberg PL, Cohen Y, Shvidel L, Haran M, Becker-Herman S, Herishanu Y, Shachar I. CD84 mediates CLL-microenvironment interactions. Oncogene. 2017;36:628–638. doi: 10.1038/onc.2016.238. [DOI] [PubMed] [Google Scholar]
- 34.Kageyama R, Cannons JL, Zhao F, Yusuf I, Lao C, Locci M, Schwartzberg PL, Crotty S. The receptor Ly108 functions as a SAP adaptor-dependent on-off switch for T cell help to B cells and NKT cell development. Immunity. 2012;36:986–1002. doi: 10.1016/j.immuni.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morra M, Lu J, Poy F, Martin M, Sayos J, Calpe S, Gullo C, Howie D, Rietdijk S, Thompson A, Coyle AJ, Denny C, Yaffe MB, Engel P, Eck MJ, Terhorst C. Structural basis for the interaction of the free SH2 domain EAT-2 with SLAM receptors in hematopoietic cells. Embo Journal. 2001;20:5840–5852. doi: 10.1093/emboj/20.21.5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quezada SA, Jarvinen LZ, Lind EF, Noelle RJ. CD40/CD154 interactions at the interface of tolerance and immunity. Annu Rev Immunol. 2004;22:307–328. doi: 10.1146/annurev.immunol.22.012703.104533. [DOI] [PubMed] [Google Scholar]
- 37.Arora S, McDonald RA, Toews GB, Huffnagle GB. Effect of a CD4- depleting antibody on the development of Cryptococcus neoformans-induced allergic bronchopulmonary mycosis in mice. Infect Immun. 2006;74:4339–4348. doi: 10.1128/IAI.01989-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindsten T, Seeley JK, Ballow M, Sakamoto K, St Onge S, Yetz J, Aman P, Purtilo DT. Immune deficiency in the X-linked lymphoproliferative syndrome. II. Immunoregulatory T cell defects. J Immunol. 1982;129:2536–2540. [PubMed] [Google Scholar]
- 39.Ma CS, Hare NJ, Nichols KE, Dupre L, Andolfi G, Roncarolo MG, Adelstein S, Hodgkin PD, Tangye SG. Impaired humoral immunity in X-linked lymphoproliferative disease is associated with defective IL-10 production by CD4+ T cells. J Clin Invest. 2005;115:1049–1059. doi: 10.1172/JCI23139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malbran A, Belmonte L, Ruibal-Ares B, Bare P, Massud I, Parodi C, Felippo M, Hodinka R, Haines K, Nichols KE, de Bracco MM. Loss of circulating CD27+ memory B cells and CCR4+ T cells occurring in association with elevated EBV loads in XLP patients surviving primary EBV infection. Blood. 2004;103:1625–1631. doi: 10.1182/blood-2003-07-2525. [DOI] [PubMed] [Google Scholar]
- 41.Ma CS, Pittaluga S, Avery DT, Hare NJ, Maric I, Klion AD, Nichols KE, Tangye SG. Selective generation of functional somatically mutated IgM+CD27+, but not Ig isotype-switched, memory B cells in X-linked lymphoproliferative disease. J Clin Invest. 2006;116:322–333. doi: 10.1172/JCI25720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merk M, Zierow S, Leng L, Das R, Du X, Schulte W, Fan J, Lue H, Chen Y, Xiong H, Chagnon F, Bernhagen J, Lolis E, Mor G, Lesur O, Bucala R. The D- dopachrome tautomerase (DDT) gene product is a cytokine and functional homolog of macrophage migration inhibitory factor (MIF) Proc Natl Acad Sci U S A. 2011;108:E577–585. doi: 10.1073/pnas.1102941108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merk M, Mitchell RA, Endres S, Bucala R. D-dopachrome tautomerase (D-DT or MIF-2): Doubling the MIF cytokine family. Cytokine. 2012;59:10–17. doi: 10.1016/j.cyto.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 45.Bhat R, Eissmann P, Endt J, Hoffmann S, Watzl C. Fine-tuning of immune responses by SLAM-related receptors. J Leukoc Biol. 2006;79:417–424. doi: 10.1189/jlb.0905537. [DOI] [PubMed] [Google Scholar]
- 46.Cuss AK, Avery DT, Cannons JL, Yu LJ, Nichols KE, Shaw PJ, Tangye SG. Expansion of functionally immature transitional B cells is associated with human-immunodeficient states characterized by impaired humoral immunity. J Immunol. 2006;176:1506–1516. doi: 10.4049/jimmunol.176.3.1506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


