Abstract
Overexpression of Rictor has been demonstrated to result in increased mTORC2 nucleation and activity leading to tumor growth and increased invasive characteristics in glioblastoma multiforme (GBM). However the mechanisms regulating Rictor expression in these tumors is not clearly understood. In this report, we demonstrate that Rictor is regulated at the level of mRNA translation via HSF1-induced HuR activity. HuR is shown to directly bind the 3′ UTR of the Rictor transcript and enhance translational efficiency. Moreover, we demonstrate that mTORC2/AKT signaling activates HSF1 resulting in a feed-forward cascade in which continued mTORC2 activity is able to drive Rictor expression. RNAi-mediated blockade of AKT, HSF1 or HuR is sufficient to downregulate Rictor and inhibit GBM growth and invasive characteristics in vitro and suppresses xenograft growth in mice. Modulation of AKT or HSF1 activity via the ectopic expression of mutant alleles support the ability of AKT to activate HSF1 and demonstrate continued HSF1/HuR/Rictor signaling in the context of AKT knockdown. We further show that constitutive overexpression of HuR is able to maintain Rictor expression under conditions of AKT or HSF1 loss. The expression of these components is also examined in patient GBM samples and correlative associations between the relative expression of these factors support the presence of these signaling relationships in GBM. These data support a role for a feed-forward loop mechanism by which mTORC2 activity stimulates Rictor translational efficiency via an AKT/HSF1/HuR signaling cascade resulting in enhanced mTORC2 activity in these tumors.
Keywords: mTORC2, AKT, HSF1, HuR, Rictor, translational regulation, glioblastoma
Introduction
The mechanistic target of rapamycin (mTOR) protein kinase integrates signal transduction networks coordinating cell growth, nutrient status, protein synthesis and autophagy 1. mTOR exists in two functionally distinct complexes, mTORC1 and mTORC2 2. The mTORC1 and mTORC2 kinase complexes both share mTOR, mLST8 (GβL), Deptor and Tti1/Tel2, however mTORC1 also specifically contains Raptor and PRAS40, while mTORC2 contains the subunits Rictor, mSIN1 and Protor1/2 3. The regulatory signals impinging on mTORC1 activity have been the focus of many studies, however by contrast, the mechanisms of mTORC2 regulation are not well understood. mTORC2 is activated by growth factor receptors and PI3K signaling 4. Association with the ribosome itself has additionally been demonstrated to result in mTORC2 activation 5, 6. mTORC2 has been shown to activate several downstream substrates, however its most well characterized function is the phosphorylation of serine 473 of AKT within the hydrophobic turn motif resulting in full activation of AKT 7. In GBM, the mutated epidermal growth factor receptor (EGFR) variant, EGFRvIII is known to activate mTORC2, in addition to PTEN loss 8-10. In an EGFR-PI3K driven Drosophila glial tumor model mTORC2 activity was required for GBM formation, and Rictor overexpression in a GEMM was sufficient to induce gliomagenesis 11, 12.
Rictor is a 200 kD protein initially identified as the defining component of mTORC2 and lacks any significant sequence homology between yeast and mammals 4, 13. Moreover, Rictor lacks any structural domains of known function but contains a C-terminus which is conserved among vertebrates. The degree of Rictor association with mTOR appears to be inversely correlated to Raptor expression and varies in different cell types 7, 13. Rictor is overexpressed in several cancers leading to hyperactive mTORC2 and has been shown to play a causal role in glioma formation 12, 14-17. Rictor expression has been demonstrated to be regulated transcriptionally and via protein degradation 18, 19, however recent studies have suggested that Rictor expression may also be regulated post-transcriptionally 20.
Here we describe a feed-forward cascade involving activation of AKT/HSF1 resulting in HSF1-induced HuR expression. Rictor is demonstrated to be a target of HuR leading to enhancement of Rictor mRNA translation and elevated mTORC2 activity. Rictor mRNA is demonstrated to be subject to translational control and shown that HuR binds to the Rictor 3′ UTR enhancing translational efficiency. Data is shown which demonstrate that HuR is a direct target of HSF1. Knockdown of AKT, HSF1 or HuR results in down-regulation of Rictor expression and impedes GBM growth, migration and invasive properties. Uncoupling HuR expression from its native promoter via viral expression maintained Rictor expression under conditions of AKT or HSF1 loss. Furthermore, an examination of clinical GBM specimens supports the proposed signaling relationships.
Results
Rictor is regulated at the level of mRNA translation
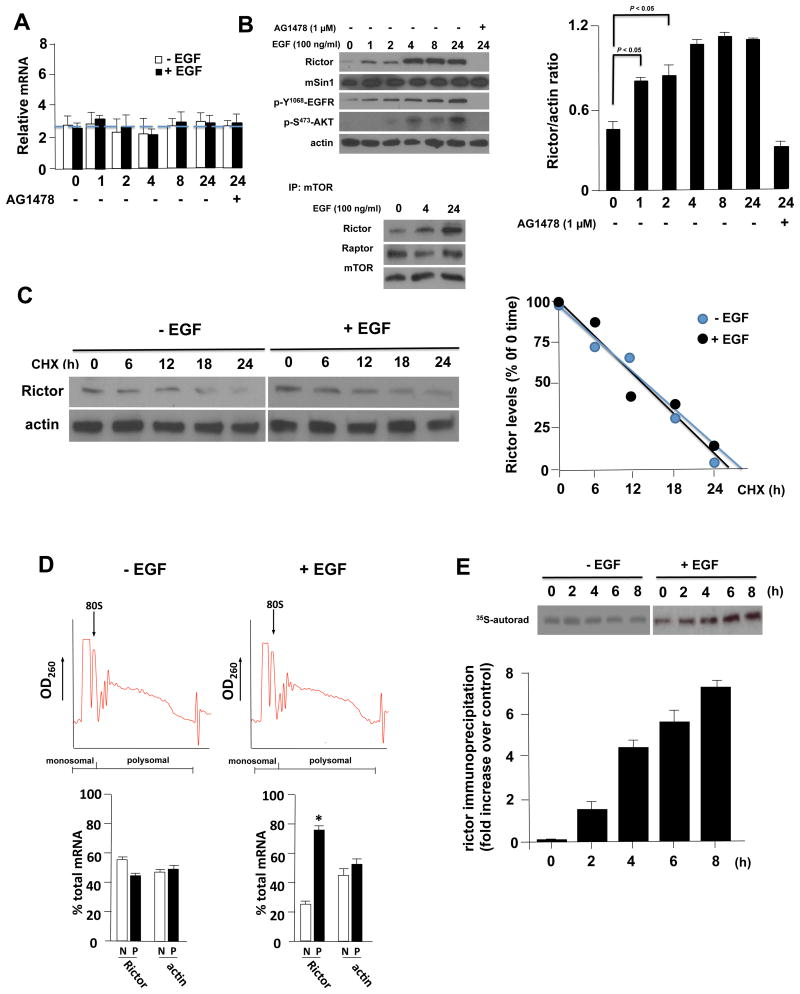
To begin investigating the mechanism(s) of Rictor expression in GBM we examined steady-state mRNA and protein levels in U138 GBM cells following stimulation. This line expresses low levels of Rictor and we reasoned that signaling which induces expression would be readily discernable. We treated cells with epidermal growth factor (EGF) and monitored mRNA and protein levels at time points following stimulation. As shown in figure 1A, EGF treatment did not alter the steady-state levels of Rictor mRNA, suggesting enhanced transcription or effects on mRNA stability do not regulate Rictor expression, however we did observe a marked increase in protein levels (Fig. 1B, top left panel). mTORC2 formation (Fig. 1B, lower left panel) and activity (p-S473-AKT levels, top left panel) were markedly induced concomitant with Rictor induction following EGF exposure. mSin1 levels appeared unaffected by EGF treatment. To examine the possibility that degradation of Rictor was inhibited by EGF exposure, we monitored the turnover of Rictor in the absence and presence of EGF. As shown in figure 1C, EGF treatment did not alter Rictor degradation. We also performed polysome analysis on the Rictor mRNA following EGF stimulation and as shown in figure 1D, Rictor mRNA displayed a marked shift to polysome-containing sucrose density gradient fractions consistent with an increase in the translational efficiency of this transcript and the accumulation of Rictor protein following EGF exposure. We then examined new Rictor protein synthesis in 35S-metabolically labeled U138 cells following EGF treatment. As shown in figure 1 E, Rictor protein synthesis increased ∼4-fold in EGF-treated cells by 4 hours relative to control non-treated cells. These data demonstrate that the Rictor mRNA is subject to translational regulation following EGF treatment.
Figure 1. Rictor mRNA is regulated at the level of translation.
(A) Steady-state Rictor mRNA levels in U138 cells in the absence or presence of EGF (100 ng/ml) for the indicated time points (h) (left panel). U138 cells were treated as indicated with EGF or in combination with the EGFR inhibitor AG1478 (5 μM; negative control). Mean + S.D. are shown, n = 3. (B) Rictor protein accumulation in U138 stimulated with EGF or both AG1478 treatment as indicated (top panel). Cell lysates were subjected to immunoblot analyses for the indicated proteins and blots were quantified by densitometry and results depicted in the right panel. Mean + S.D. are shown, n = 3. mTORC2 formation is enhanced by EGF stimulation (lower panel). U138 cells were treated with EGF and lysates immunoprecipitated with α-mTOR antibodies. Immunoprecipitates were immunoblotted for the indicated proteins. Experiments were repeated three times with similar results. (C) Rictor protein stability is not affected by EGF stimulation. U138 cells were subjected to cycloheximide (CHX)-chase experiments and Rictor protein levels examined at the indicated time points in the absence or presence of EGF (100 ng/ml) (left panel). Band intensities were quantified by densitometry and displayed graphically (right panel). (D) Polysome distribution of Rictor and actin mRNAs. U138 cells were treated with EGF (100 ng/ml, 8 h) and extracts subjected to sucrose density gradient centrifugation and divided into 11 1-ml fractions which were pooled into nonribosomal, monosomal fraction (N, white bars) and a polysomal fraction (P, black bars). Purified RNAs were used in real time quantitative rt-PCR analyses to determine the distributions of Rictor and actin mRNAs across the gradients. Polysome tracings are shown above values obtained from the rt-PCR analyses which are displayed graphically below. rt-PCR measurements were performed in quadruplicate and the mean and + S.D. are shown. (E) New Rictor protein synthesis in U138 following EGF stimulation (100 ng/ml). Cells were pulsed with 35S-methionine/cysteine for 1 h and chased with cold amino acids for the indicated time points. Rictor immunoprecipitates were analyzed by autoradiography and the fold increase new Rictor synthesis following stimulation relative to controls without EGF is shown. Mean +S.D. are shown, n =3.
HuR binds to the 3′ UTR of Rictor mRNA and promotes translation
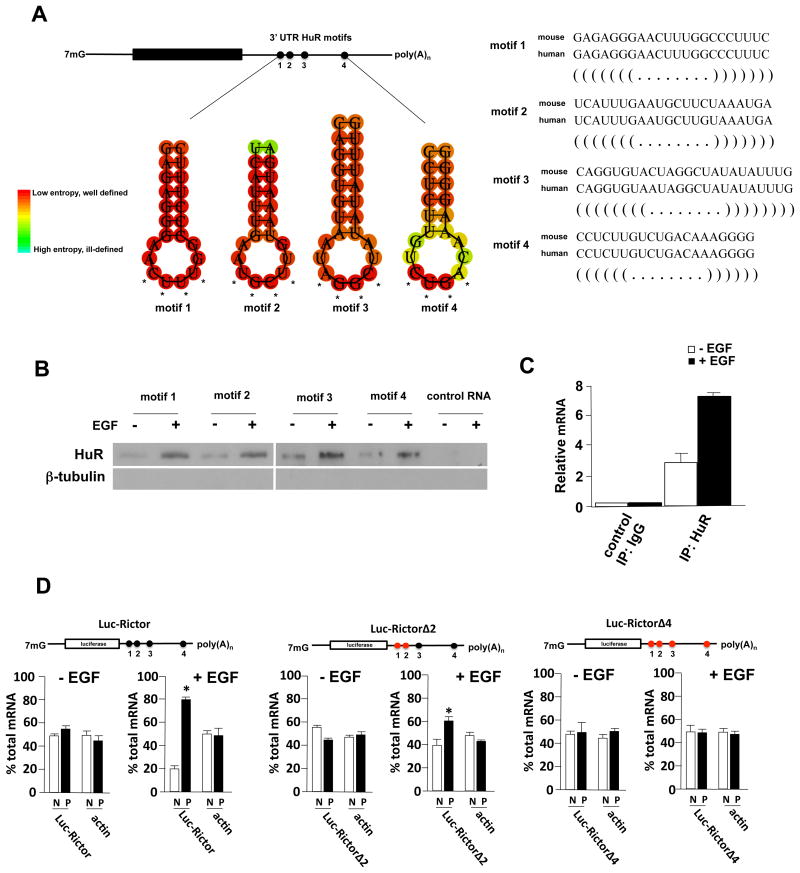
To understand the mechanism by which Rictor mRNA translation is regulated we examined the structure of the Rictor transcript. In a computational survey of RNA-binding protein motifs, we identified four consensus HuR binding sites located within the 3′ UTR of the Rictor mRNA which were conserved in human and mouse transcripts (Fig. 2A). These sites were found to be significantly more U-rich than AU-rich and adhered to the consensus sequences derived from studies by López de Silanes et al 21 and separately by Tenenbaum and coworkers 22 (Fig. 2B). As HuR is known to enhance mRNA stability and translation 23 we determined whether HuR would bind to these sequences. In an RNA pull-down assay, extracts from U138 cells treated without or with EGF, were mixed with biotinylated HuR binding site motif(s) RNA sequences as shown in Figure 2B. HuR was preferentially precipitated by each of the HuR binding site motifs (1-4) in a manner dependent on EGF stimulation. HuR was not detected in samples which were precipitated by a nonspecific control RNA. Similarly, in HuR immunoprecipitates we were able to detect Rictor 3′ UTR sequences by rt-PCR, which was enhanced in extracts treated with EGF (Figure 2C). To investigate whether these Rictor 3′ UTR HuR binding motifs were involved in regulating translational efficiency, we generated heterologous reporter mRNA transcripts in which the full-length Rictor 3′ UTR was fused to luciferase and the HuR binding motifs mutated (Figure 2D). The effects of these mutations on mRNA translation were subsequently assessed. As shown in figure 2D, the full-length Rictor 3′ UTR containing mRNAs were shifted to polysomal fractions by EGF stimulation consistent with enhanced HuR binding. However, mutating motifs 1 and 2 reduced the amount of reporter mRNA which was polysome associated and mutating all four HuR binding motifs completely abolished EGF-stimulated mRNA reporter polysome association. Taken together these data demonstrate that HuR binds to the Rictor 3′ UTR to stimulate translational efficiency.
Figure 2. HuR binds to the 3′ UTR of Rictor and stimulates translation.
(A) Sequence and structure of predicted HuR binding motifs within the Rictor 3′ UTR (left panel). Structural alignment of mouse and human HuR motifs within the Rictor 3′ UTRs (right panel). (B) Identification of HuR in RNA pull-down assays utilizing biotinylated HuR binding motifs as indicated. Biotinylated nonspecific RNA was used as a negative control. Cytoplasmic extracts of U138 cells treated in the absence or presence of EGF (100 ng/ml, 8 h) were incubated with biotinylated RNAs corresponding to HuR binding motifs 1-4 as shown and precipitated with streptavidin-Sepharose beads. Bound fractions were analyzed by immunoblotting for HuR and tubulin. Experiments were repeated three times with similar results. (C) HuR binds to Rictor 3′ UTR RNA containing HuR binding motifs 1-4 in cells and binding is stimulated by EGF (100 ng/ml, 8 h). Control IgG or anti-HuR antibody was used to immunoprecipitate (IP) lysates from U138 cells, and bound RNA was amplified by PCR of the Rictor 3′ UTR sequences. Relative amounts of Rictor 3′ UTR RNA are displayed graphically. Mean + S.D. are shown, n = 3. (D) Polysome analyses of Luc-Rictor 3′ UTR containing reporter mRNAs. U138 cells expressing the indicated reporter mRNAs, native Rictor 3′ UTR (Luc-Rictor), mutant 1-2 (Luc-RictorΔ2) in which HuR binding motifs 1 and 2 were mutated, and mutant 1-4 (Luc-RictorΔ4) in which all four HuR binding motifs were mutated (4 unpaired bases were changed to C within the loop structures of the predicted hairpin loop motifs, see Fig. 2A, asterisks), were treated with EGF (100 ng/ml, 8 h) and lysates subjected to polysome analyses as in Fig. 1C. Purified RNAs were used in real time quantitative rt-PCR analyses to determine the distributions of Luc-Rictor and actin mRNAs across the gradients. Values obtained from the rt-PCR analyses are displayed graphically below. rt-PCR measurements were performed in quadruplicate and the mean and + S.D. are shown.
mTORC2/AKT/HSF1 signaling stimulates HuR transcription in GBM
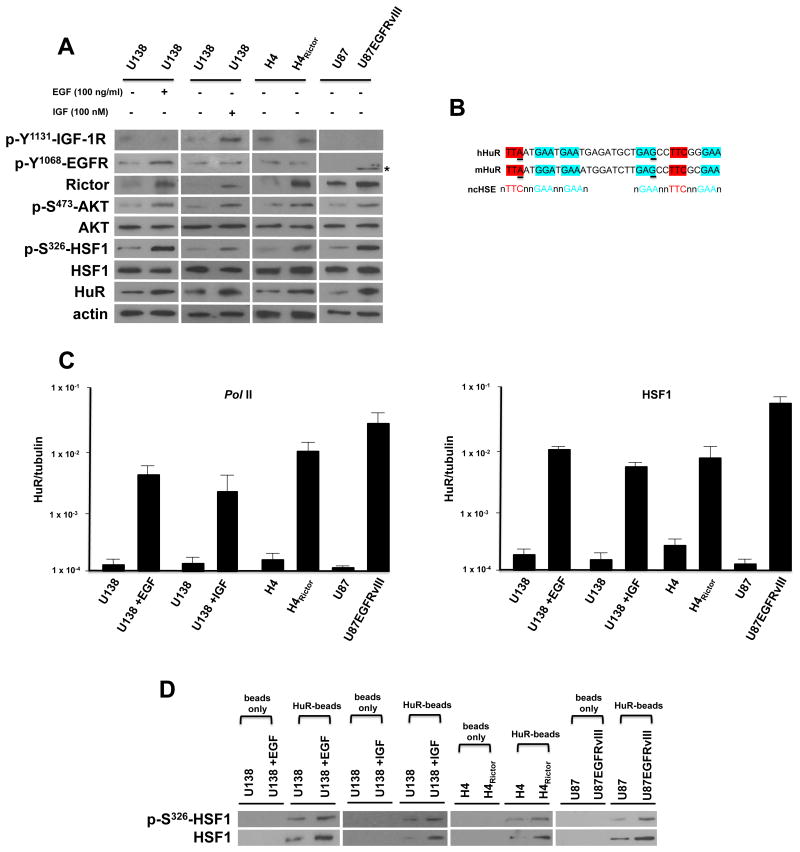
Overexpression of Rictor in glioblastoma cells has been demonstrated to increase the nucleation of mTORC2 resulting in elevated kinase activity 12, 16. We sought to identify a signaling mechanism linking the mTORC2/AKT axis to the regulation of HuR. AKT has been shown to directly activate HSF1 via phosphorylation of serine 326 24. Additionally, Chou et al have recently demonstrated that HSF1 appears to regulate β-catenin expression via effects on HuR in breast cancer cells 25. These relationships suggested that mTORC2/AKT signaling may activate HSF1/HuR leading to enhanced translation of Rictor which in turn, would enhance mTORC2 activity in a feed-forward loop promoting GBM proliferation and motility. We examined these cascades in three GBM cell line models of elevated mTORC2 activity. mTORC2 is known to be activated by EGF or IGF stimulation, overexpression of the mutated constitutively active EGFRvIII allele, as well as, by overexpression of Rictor 8, 12, 16, 26. As shown in Figure 3A, in U138 cells stimulated with EGF or IGF, phospho-S473-AKT, phospho-S326-HSF1, HuR and Rictor levels were enhanced following stimulation (see also Supplemental Figure S1A). In H4 cells in which Rictor is overexpressed 16, phospho-S473-AKT, phospho-S326-HSF1 and HuR expression was enhanced relative to the levels of these proteins in parental H4 cells. Similarly, in U87 cells overexpressing the mutant EGFRvIII allele, phospho-S473-AKT, phospho-S326-HSF1, HuR and Rictor levels were elevated compared to parental U87 cells. We confirmed that knockdown of another obligate component of mTORC2, mSin1, blocked AKT activity and the proposed downstream loop signaling following EGF exposure (Supplemental figure. S1B). While these data supported the notion of an mTORC2/phospho-S473-AKT/phospho-S326-HSF1/HuR/Rictor feed-forward cascade, the mechanism by which activated HSF1 leads to increases in HuR expression was unclear in GBM cells. To examine whether HuR was a direct transcriptional target of HSF1 we searched the HuR promoter for canonical or noncanonical heat shock elements (HSEs) 27. We identified tandem noncanonical HSEs beginning at position -475 within the HuR promoter and these sequences were conserved in the mouse HuR promoter (Figure 3B). Interestingly, Mendilo et al identified this region of the HuR promoter as a binding target of HSF1 in a ChIP-Seq study 28. To determine whether these HSEs were capable of mediating transcriptional responses directed by HSF1, we determined both RNA Pol II and HSF1 occupancy via ChIP assays followed by quantitative PCR. As shown in Figure 3C (left panel), Pol II content within the HuR promoter containing the tandem HSEs was increased in U138 cells following EGF or IGF stimulation. There was marked enhancement of Pol II content within the HuR promoter in H4Rictor - or U87EGFRvIII-overexpressing cells relative to parental cells. Similar results were obtained analyzing HSF1 occupancy of the HuR promoter as EGF or IGF stimulation, Rictor- or EGFRvIII-overexpression all resulted in increases in HSF1 occupancy (Figure 3C, right panel). The phosphorylation state of functionally bound HSF1 was assessed in a series of in vitro DNA-pull down assays (Figure 3D). Consistent with the chromatin immunoprecipitation experiments, EGF or IGF stimulation, Rictor- or EGFRvIII-overexpression resulted in higher levels of bound phospho-S326-HSF1 and total HSF1 compared to unstimulated U138 cells or parental H4 and U87 cells.
Figure 3. mTORC2 activation leads to HSF1-stimulated HuR transcription.
(A) Signaling effects of mTORC2 stimulation by EGF, IGF, Rictor or EGFRvIII overexpression. The GBM lines were treated with EGF (100 ng/ml, 8 h) or (IGF 100 nM, 8 h) as shown and lysates immunoblotted for the indicated proteins. Asterisk corresponds to the truncated phosphorylated Y1068 mutant EGFRvIII. (B) Sequence and alignment of tandem noncanonical heat-shock elements (HSEs) identified within the promoter region of HuR (-475 to -441). Previously identified noncanonical HSE (ncHSE) are shown below the human and mouse sequences. Underlined nucleotides differ from those noncanonical HSEs which have been described 28. (C) ChIP analyses of HuR promoter activity and HSF1 association in the indicated GBM cell lines. RNA polymerase II (Pol II) association with HuR promoter (left panel) and HSF1 association with HuR promoter (right panel) were determined and ChIP-quantitative PCR data are expressed as a ratio of HuR to tubulin. Mean and + S.D. are shown, n = 3. (D) Sepharose beads were conjugated with the ncHSEs HuR DNA (HuR-beads) or beads without linked DNA and incubated with nuclear extracts from the indicated cell lines. Following recovery by centrifugation and washing of the beads, bound material was analyzed by immunoblot for phospho-S326-HSF1 and total-HSF1. These experiments were performed twice with similar results.
AKT, HSF1 and HuR are required for Rictor expression and their loss inhibits GBM cell line growth, motility and invasiveness
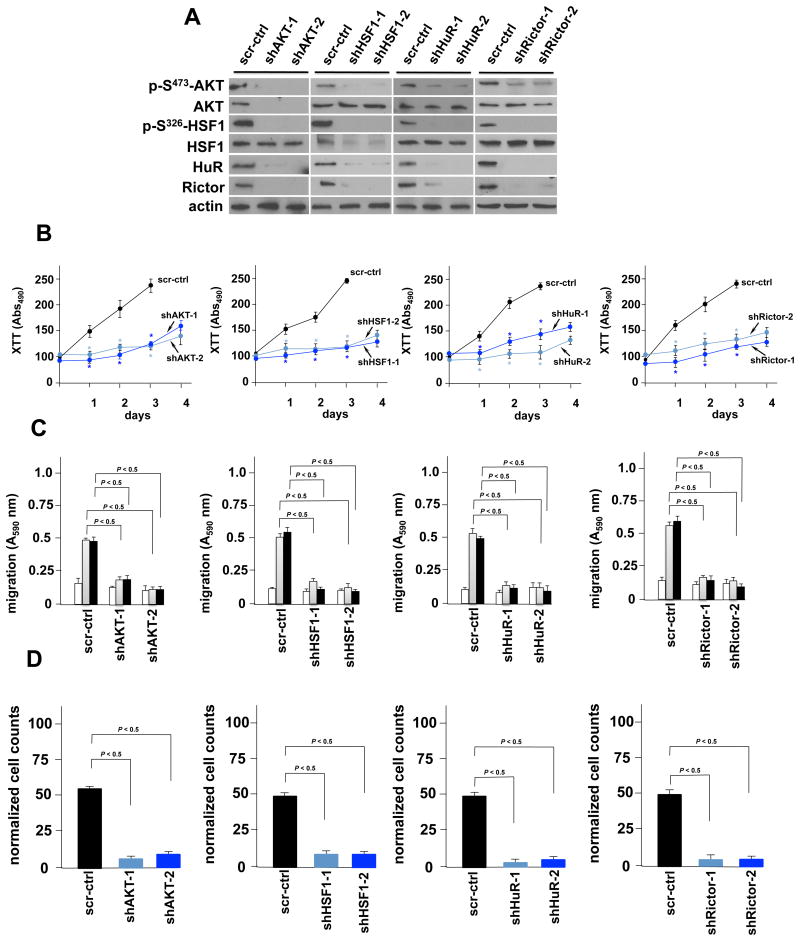
We next investigated whether loss of AKT, HSF1 or HuR altered Rictor and mTORC2 activity. DBTRG-05MG cells, which harbor elevated mTORC2 activity (B. Holmes and J. Gera, unpublished results), were stably transduced with lentiviral vectors expressing shRNAs targeting AKT, HSF1, HuR, Rictor or a control scrambled sequence non-targeting control. Cells expressing the shRNAs were immunoblotted for the signaling constituents predicted in the feed-forward loop. As shown in Figure 4A (see also Supplemental Figure S2), knockdown of AKT resulted in a decline of phospho-S326-HSF1, HuR and Rictor consistent with a pathway involving a concerted AKT/phospho-S326-HSF1/HuR/Rictor cascade. Knockdown of HSF1 resulted in down-regulation of HuR, Rictor and phospho-S473-AKT levels, while knockdown of HuR led to reduced expression of Rictor, phospho-S473-AKT and phospho-S326-HSF1. Knockdown of Rictor abrogated mTORC2 activity and resulted in reduced phospho-S473-AKT levels, consistent with previous results 16, but also reduced phospho-S326-HSF1 and HuR expression. We additionally determined the effects of knockdown of each of these signaling components on growth, migration and invasive characteristics in the lines. As shown in Figure 4B, growth of AKT, HSF1, HuR and Rictor shRNA-expressing lines was markedly reduced compared to control non-targeting shRNA-expressing cells. The migratory capacity (Figure 4C) was also significantly reduced in these knockdown lines as loss of AKT, HSF1, HuR and Rictor impeded the ability of cells to migrate on either vitronectin or fibronectin in Boyden chambers. The ability of AKT, HSF1, HuR or Rictor knockdown cells to migrate was reduced by ∼ 60-70% relative to control scrambled non-targeting shRNA expressing cells. As shown in Figure 4D, AKT, HSF1, HuR and Rictor knockdown cells were also inhibited in their ability to invade Matrigel relative to control cells.
Figure 4. AKT, HSF1 and HuR are required for Rictor expression and their blockade inhibits GBM cell line growth, motility and invasiveness.
(A) shRNA-mediated knockdown of AKT, HSF1, HuR and Rictor in DBTRG-05MG GBM cells. Cells expressing the indicated shRNA or nontargeting shRNA (negative control, scr; scrambled sequence) were immunoblotted for the indicated proteins. (B) Effects of AKT, HSF1, HuR and Rictor knockdown on cell growth in as indicated. Control (scr-ctrl) U87EGFRvIII cells expressed a nontargeting-scrambled shRNA. (*, P < 0.05). Mean ± S.D. are shown, n = 3. (C) Migration of control (scr-ctrl) or AKT, HSF1, HuR or Rictor shRNA-expressing knockdown clones. Cells were seeded into Boyden chambers and allowed to migrate towards BSA (white bars), vitronectin (grey bars) or fibronectin (black bars). Data represent mean +S.D. of three independent experiments. (D) Invasive potential of control or AKT, HSF1, HuR or Rictor shRNA-expressing knockdown cells migrating through matrigel. Data represent mean +S.D. of three independent experiments.
Effects of modulating AKT or HSF1 activity on HuR/Rictor signaling in GBM
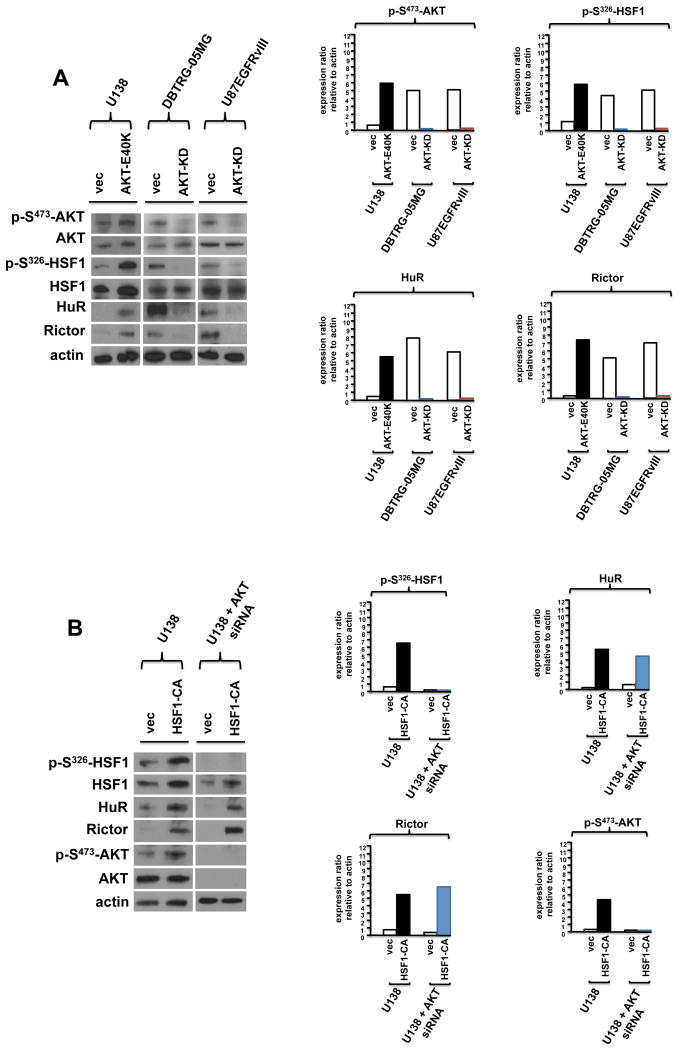
We next sought to investigate the effects of modulating the activities of AKT and HSF1 via genetic approaches on the proposed feed-forward signaling loop. A constitutively active allele of AKT was stably expressed in U138 (AKT-E40K), while a dominant negative AKT-KD (AKT-K179M) was expressed in DBTRG-05MG and U87EGFRvIII cells. As shown in figure 5A, expression of the constitutively active AKT resulted in a marked increase in phospho-S326 HSF1, HuR and Rictor levels as compared to control cells containing the empty vector. Conversely, in DBTRG-05MG and U87EGFRvIII expressing the dominant negative AKT, substantially reduced levels of phospho-S326 HSF1, HuR and Rictor were observed. We also examined the effects of a constitutively active HSF1 mutant (HSF1-CA) when ectopically expressed in U138 cells. As shown in figure 5B, cells expressing this HSF1 mutant demonstrated elevated levels of HuR and Rictor (upper panel). Moreover, when U138 expressing the constitutively active HSF1 allele were subjected to AKT knockdown via siRNA treatment, these cells maintained elevated levels of phospho-S326 HSF1, HuR and Rictor relative to control cell expressing vector alone. We further examined the translational state of Rictor mRNAs via polysome analyses from control (vector only) and HSF1-CA expressing cells treated with AKT siRNAs, and as shown in figure 5B (lower panel), a majority of the Rictor mRNA was associated non-ribosomal/monosomal material from control cells, whereas, in HSF1-CA expressing cells a marked shift in Rictor mRNA translational state was observed with most of the transcripts associated with polysomes.
Figure 5.
Modulation of AKT or HSF1 activity influences downstream signaling components of the feedforward loop. (A) U138, DBTRG-05MG or U87EGFRvIII lines stably expressing the constitutively active AKT (AKT-E40K) or dominant negative AKT-KD (AKT-K179M) alleles or empty vector control constructs as shown were immunoblotted for the indicated proteins (left panel). Protein expression levels were quantified and shown in the right panel. Experiments were repeated twice with similar results. (B) U138 cells stably expressing a constitutively active HSF1 mutant (HSF1-CA) were treated with siRNAs targeting AKT as indicated. Extracts were subsequently immunoblotted for the proteins shown (left panel). Protein expression was quantified and displayed graphically in the right panel. Experiments were repeated twice with similar results.
Constitutive HuR expression prevents loss of Rictor under conditions of AKT or HSF1 knockdown
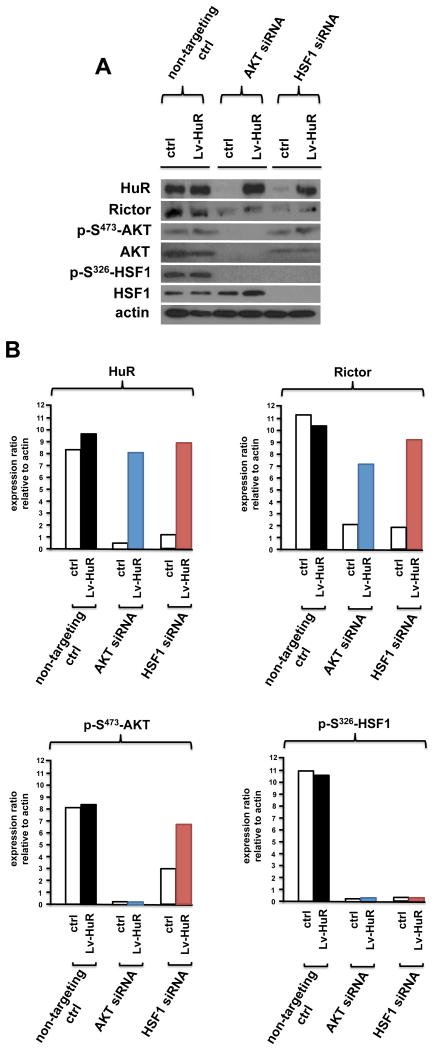
To gain further insight as to whether an mTORC2/phospho-S473-AKT/phospho-S326-HSF1/HuR/Rictor signaling cascade was operative in GBM, we investigated whether HuR expression driven from a viral vector would prevent Rictor loss under conditions of AKT or HSF1 knockdown. DBTRG-05MG cells were stably transduced with a lentiviral construct which expresses HuR and cells treated with siRNAs targeting AKT, HSF1 or control scrambled non-targeting siRNAs. As shown in figure 6A, viral driven expression of HuR (Lv-HuR) maintained Rictor abundance under conditions of AKT or HSF1 loss. Treatment of cells constitutively expressing viral driven HuR with siRNAs targeting AKT, abolished phospho-S473-AKT and total AKT levels, as well as, phospho-S326-HSF1 levels, but did not result in reduced Rictor expression (see Figure 6B). Similarly, knockdown of HSF1 in cells expressing viral driven HuR resulted in inhibition of phospho-S326-HSF1 and total HSF1, however Rictor expression was maintained. Taken together these data are consistent with a feed-forward loop in which mTORC2 activity leads to signaling through an AKT/HSF1/HuR/Rictor cascade in GBM cells.
Figure 6. Overexpression of HuR prevents loss of Rictor expression under conditions of AKT or HSF1 loss.
(A) Viral vector driven expression of HuR prevents inhibition of Rictor expression in DBTRG-05MG cells treated with non-targeting (scr-ctrl), AKT-, or HSF1-targeting siRNAs. Lysates were subsequently immunoblotted for the indicated proteins. (B) Quantification of HuR, Rictor, p-S473-AKT and p-S326-HSF1 protein levels from experiments in (A) by densitometry. Experiments were repeated twice with similar results.
Alterations in mTORC2/AKT/HSF1/HuR signaling affect tumor xenograft growth and Rictor mRNA translation
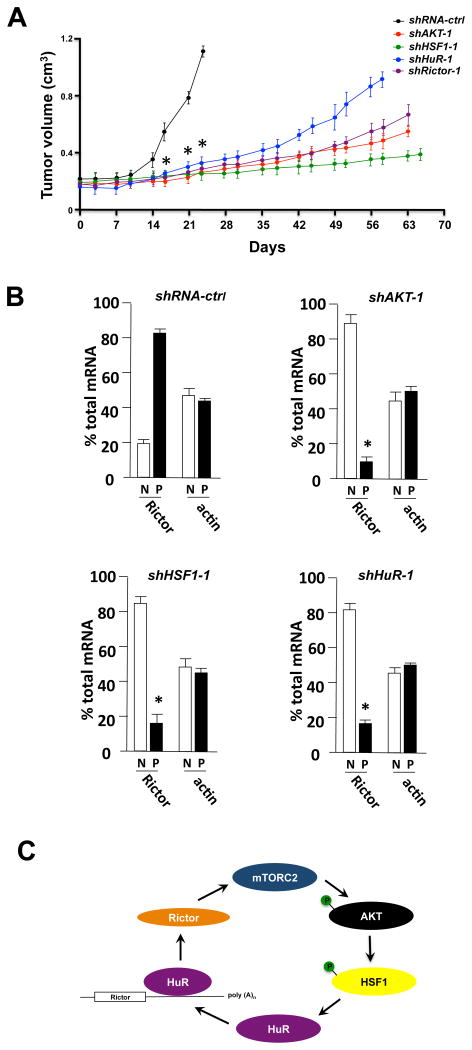
To determine if shRNA mediated knockdown of AKT, HSF1, or HuR would affect the in vivo growth rates of murine xenografts, we subcutaneously implanted DBTRG-05MG cells expressing these shRNAs into SCID mice and monitored tumor growth. As shown in figure 7A, cells expressing non-targeting shRNAs exhibited rapid growth with a latency period of 14 days. Conversely, tumors expressing shRNAs targeting AKT, HSF1, or HuR grew significantly slower and with longer latency periods (*, P < 0.05; latency periods; shRNA-AKT, 28 days; shRNA-HSF1, 30 days; shRNA-HuR, 23 days). We also expressed shRNAs targeting Rictor in DBTRG-05MG cells and knockdown resulted in significantly slower growth and longer latency period (shRNA-Rictor, 23 days), confirming our previous results in other GBM cell lines 16. Tumors from mice at autopsy were subjected to polysome analyses to determine the relative translational state of the Rictor mRNA. As shown in Figure 7B, in tumors expressing the non-targeting control shRNA 80% of Rictor mRNA was polysome associated and well translated, however in tumors expressing AKT, HSF1 or HuR shRNAs, Rictor mRNA shifted markedly to non-ribosomal/monosomal fractions indicating reduced translational efficiency and consistent with the reduced growth rates of these tumors.
Figure 7. Knockdown of AKT, HSF1 or HuR inhibit GBM growth in vivo.
(A) DBTRG-05MG cells expressing shRNA targeting AKT, HSF1, HuR, Rictor or non-targeting (ctrl) were monitored for tumor growth for up to 70 d. (n = 4-5 per group; *, P < 0.05). (B) Polysome analyses were performed on cells from tumors harvested at autopsy (as in Figure 1D) for the cells expressing the indicated shRNAs. The distribution of Rictor and actin mRNAs across the gradients was determined and quantified via rt-PCR as before (*, P < 0.05). (C) Proposed model of feed-forward regulation of Rictor expression and mTORC2 activity via AKT/HSF1/HuR signaling.
mTORC2/AKT/HSF1/HuR/Rictor signaling in GBM patients
To assess whether these signaling relationships were valid in clinical GBM samples we analyzed an independent set of 34 flash-frozen GBM and 5 normal brain samples. Each tumor sample was confirmed histologically, tumor extracts prepared, and the total relative abundance of phospho-S473-AKT, phospho-S326-HSF1, HuR and Rictor determined by Western analyses. These data are summarized in table 1 and supplemental figure S5. As shown, elevated mTORC2 activity, as determined by immunoblotting for phospho-S473-AKT levels, was observed in 22 of 34 tumor samples (65%, P < 0.05) consistent with the degree of hyperactivity previously observed 8, 16. Phospho-S326-HSF1, HuR and Rictor expression was elevated in 74% (25 of 34, P < 0.05), 62% (21 of 34, P < 0.05) and 74 % (25 of 34, P < 0.05) of samples, respectively. Significant direct correlations were observed between samples harboring elevated phospho-S473-AKT and increased phospho-S326-HSF1, and those containing elevated phospho-S326-HSF1 and increased HuR levels (P values less then 0.05 for both correlations). A highly significant direct correlation was found between elevated HuR containing samples and those tumors expressing high levels of Rictor (P < 0.01). These data strongly support the feed-forward signaling pathway observed in the GBM cell line experiments and provide evidence of these signaling relationships in patient samples.
Table 1.
Relative protein levels of phospho-S473AKT, phospho-S326-HSF1, HuR, and Rictor in normal and glioblastoma samples.
| Samples | pS473-AKT expression § | pS326-HSF1 expression # | HuR expression¥ | Rictor expression≠ |
|---|---|---|---|---|
| Normal | ||||
| 1 | 1.1 | 1.6 | 1.9 | 1.5 |
| 2 | 1.7 | 2.1 | 1.3 | 1.1 |
| 3 | 1.2 | 1.3 | 1.4 | 1.4 |
| 4 | 1.6 | 1.4 | 1.8 | 1.9 |
| 5 | 1.5 | 1.8 | 1.7 | 1.2 |
| GBM | ||||
| 1 | 54.3*++ | 26.3ˆ++ | 37.5″++ | 65.9«++ |
| 2 | 15.7*+ | 19.7ˆ+ | 8.1 | 10.2«+ |
| 3 | 26.9*++ | 17.3ˆ++ | -‡ | 49.4«++ |
| 4 | 36.2*++ | 43.8ˆ++ | 9.62 | 54.1«++ |
| 5 | 48.4*++ | 34.3ˆ++ | 42.7″++ | 26.8«++ |
| 6 | 3.5 | 0.7 | 2.7 | 1.8 |
| 7 | 26.9*++ | 16.4ˆ++ | 53.1″++ | 29.3«++ |
| 8 | 5.5 | 2.4 | 3.9 | 6.9 |
| 9 | 32.4*++ | 45.9ˆ++ | 57.3″++ | 48.2«++ |
| 10 | 23.6*++ | 29.7ˆ++ | 42.8″++ | 37.4«++ |
| 11 | 17.6*+ | 11.3ˆ+ | 13.1″+ | 9.7«+ |
| 12 | 39.1*++ | 56.2ˆ++ | 64.5″++ | 25.8«++ |
| 13 | 4.3 | 13.5ˆ+ | 8.2 | 3.4 |
| 14 | 32.7*++ | 26.9ˆ++ | 29.2″++ | 41.9«++ |
| 15 | 56.8*++ | 32.8ˆ++ | 46.9″++ | 41.0«++ |
| 16 | 3.8 | -† | 4.2 | 2.1 |
| 17 | 7.8 | 10.3ˆ+ | 11.3″+ | 7.5 |
| 18 | 42.7*++ | 38.6ˆ++ | 52.6″++ | 31.1«++ |
| 19 | 27.9*++ | 29.4ˆ++ | 24.8″++ | 30.4«++ |
| 20 | 43.6*++ | 48.1ˆ++ | 39.7″++ | 37.9«++ |
| 21 | 6.7 | 5.3 | 4.6 | 3.6 |
| 22 | 46.1*++ | 26.7ˆ++ | 38.6″++ | 42.8«++ |
| 23 | 7.2 | 5.4 | -‡ | 2.4 |
| 24 | 78.4*++ | 69.4ˆ++ | 52.8″++ | 34.8«++ |
| 25 | 53.2*++ | 66.8ˆ++ | 29.8″++ | 53.1«++ |
| 26 | 69.2*++ | 13.4ˆ++ | 42.5″++ | 16.4«+ |
| 27 | 7.2 | 5.2 | 3.7 | 2.6 |
| 28 | 76.3*++ | 84.3ˆ++ | 63.8″++ | 58.4«++ |
| 29 | 7.0 | 2.4 | 6.4 | 5.8 |
| 30 | 16.2*+ | 14.7ˆ+ | 9.1 | 12.5«+ |
| 31 | 43.8*++ | 38.6ˆ++ | 5.5 | 49.2«++ |
| 32 | 62.5*++ | 39.4ˆ++ | 46.4ˆ++ | 58.6ˆ++ |
| 33 | 28.4*++ | 51.3ˆ++ | 33.5″++ | 32.9«++ |
| 34 | 48.1*++ | 50.7ˆ++ | 43.7″++ | 29.6«++ |
Note: Five normal brain and 34 independent quick-frozen GBM samples were assessed for phosphorylated AKT, phosphorylated HSF1, HuR and Rictor levels by Western analyses as described in Experimental Procedures and quantified by densitometry. 22 of 34 tumor samples (65%) had markedly higher mTORC2 activity as determined by monitoring expression levels of phospho-S473-AKT relative to normal brain. HSF1 activity was determined by monitoring phospho-Ser326-HSF1.
phospho-S473-AKT expression > 2-fold above mean of normal brain.
Markedly increased mTORC2 activity; ++ > 20-fold increase above mean of normal brain (dark gray shaded row); + > 10-fold increase above mean of normal brain (light gray shaded row).
phospho-S326-HSF1 expression > 2-fold above mean of normal brain.
Markedly increased phospho-S326-HSF1 expression; ++ > 20-fold increase above mean of normal brain + > 10-fold increase above mean of normal brain
Undetectable phospho-Ser326-HSF1.
HuR expression > 2-fold above mean of normal brain
Markedly increased HuR expression; ++ > 20-fold increase above mean of normal brain + > 10-fold increase above mean of normal brain
Undetectable HuR expression.
Rictor expression > 2-fold above mean of normal brain
Markedly increased Rictor expression; ++ > 20-fold increase above mean of normal brain + > 10-fold increase above mean of normal brain.
Discussion
Our current understanding suggests that mTORC2 can be regulated by RTKs, ribosomes, TSC1-TSC2, Rac1, and the expression levels of Rictor 2, 4. Very recent data suggests that the polyubiquitination status of GβL may additionally regulate the homeostasis of mTORC2 formation and activation 29. Our data suggest that mTORC2 is also subject to autoregulation via feed-forward signaling through an AKT/HSF1/HuR/Rictor cascade in GBM (see Figure 7C).
Although Rictor is necessary for the stability and activity of mTORC2 and reports have described the role of phosphorylation and acetylation events in these processes, little is known regarding control of Rictor expression via post-transcriptional control mechanisms 4, 30. The 3′ UTR of Rictor is relatively long suggesting that it contains post-transcriptional regulatory sequences. Furthermore, the 3′ UTR contains several segments of AU-or U rich cis-regulatory sequences implicated in mRNA turnover and translational control 31, 32. Our analysis identified several HuR binding sites (see figure 2A) and additional regulatory domains are likely to be present in such a large 3′ UTR, possibly controlling mRNA stability under particular conditions. Our data supporting the ability of the Rictor transcript to be subject to translational control is reinforced by recent data from Yasuda and colleagues, who investigated the role of Mdm20 in actin remodeling via Rictor-mediated mTORC2 activity 20. Mdm20 was suggested to regulate the expression of Rictor at the level of de novo protein synthesis.
Our study implicates HSF1/HuR signaling in the regulation of Rictor expression and mTORC2 activity. Recent data support an HSF1/HuR cascade in the regulation of β-catenin levels in breast cancers 25. Gabai et al, also observed that HSF1 controls the expression of HIF-1α via effects on HuR 33. HSF1's affects on HuR were determined to be at the level of transcription. We identified tandem non-canonical HSEs within the promoter of HuR and showed that these sites were capable of association with activated HSF1 and promoted HuR expression as determined by increased Pol II occupancy (see Figure 3). These sequences were also identified as HSF1 binding sites in a high-resolution survey of HSF1 genome occupancy in breast cancer cells 28. These data support the notion that HuR is a direct target of activated HSF1 in GBM.
One prediction of the model presented here is that modulation of a single pathway component should similarly affect other components of the signaling loop. In agreement with this notion, the genetic inhibition of mSin1, AKT, HSF1, HuR or Rictor expression individually, resulted in the down-regulation of all other components within the signaling loop (see Supplemental figure S1B and Figures 4A and 5A). Similarly, enhancement of an individual components activity resulted in a concomitant increase in downstream signaling loop components activity or expression. Growth factor-stimulated mTORC2, ectopic expression of Rictor or constitutively activated alleles of AKT or HSF1 led to marked increases in downstream components activity or expression (see Figure 3A and Figures 5A & B). The capacity of a single genetic ablation of a pathway component to influence the entire signaling cascade suggests that targeting a single component may yield enhanced therapeutic effectiveness.
All of the components of the signaling cascade we have delineated have established roles in GBM growth and migration. Rictor/mTORC2 have important roles regulating these functions and additionally regulate the metabolic reprograming of these tumors 2, 16. AKT and HSF1 have been shown to mediate glioma growth and survival 34, 35, while HuR appears to be overexpressed in GBM and its dysregulation results in the enhanced translation of mRNAs promoting growth, motility and drug resistance 36. Feed-forward signaling loops are well represented in cell signaling networks and provide a positive mechanism for reinforcing and stimulating network activity. Many of the downstream effectors of the feed-forward loop components described here have overlapping targets and the coordinated activation of this loop may serve to stimulate the expression of common targets at multiple levels of gene regulation. These results contribute to a better understanding of the signaling mechanisms regulating Rictor expression and mTORC2 activity in GBM.
Materials & Methods
Cell Lines, GBM samples, Transfections and Viral Transductions
All GBM cell lines were obtained from ATCC except U87EGFRvIII, which was kindly provided by Dr. Paul Mischel (LICR-UCSD). Lines were routinely tested to confirm the absence of mycoplasma and authenticated by STR profiling (ATCC). Flash-frozen normal brain and glioblastoma samples were obtained from the Cooperative Human Tissue Network under an approved Institutional Review Board protocol and informed consent obtained from each individual. Each sample was histopathologically reviewed and those containing greater than 95% tumor were utilized. Samples were homogenized in RIPA buffer using a Polytron homogenizer (Fisher, Pittsburgh, PA) followed by sonication to generate extracts for protein and RNA analysis. DNA transfections were performed using Effectene transfection reagent according to the manufacturer (Qiagen). For siRNA knockdowns, DBTRG-05MG, U87 or U87EGFRvIII cells were transfected with 10 nmol/L siRNA pools targeting AKT1, HSF1, HuR, Rictor, mSIN1 or a non-targeting scrambled control sequence. ON-TARGETplusSMARTpool siRNAs were obtained from GE Dharmacon and transfected using Lipofectamine RNAimax (Life Technologies). Lentiviral shRNA production and infection was performed as described 16.
Constructs and Reagents
The luciferase-human Rictor 3′ UTR constructs were generated by insertion of the full-length human Rictor 3′ UTR into the XbaI site of pGL3-promoter (Promega). Mutagenesis was performed using the QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent Technologies) with the appropriate mutagenic primers. TRC pLKO.1 library constructs expressing multiple shRNA-targeting AKT1, HSF1, HuR, Rictor or non-targeting controls were from GE Dharmacon. The constructs utilized had the following TRC designations: shAKT-1, TRCN0000010162; shAKT-2, TRCN0000010163; shHSF1-1, TRCN0000007480; shHSF1-2, TRCN0000007481; shHuR-1, TRCN0000017273; shHuR-2, TRCN0000017274; shRictor-1, TRCN0000074288; shRictor-2, TRCN0000074289; shmSin1, TRCN0000003150. The constitutively active AKT-E40K and dominant negative AKT-KD (AKT-K179M) constructs have been previously described 37, 38. The constitutively active HSF1 construct (HSF1-CA) was originally described by Voellmy and co-workers 39 and was kindly provided by Dr. Eugene Kandel (Roswell Park Cancer Institute). The HuR overexpression construct contained the full-length human HuR ORF cloned into pLenti-C-Myc-DDK-P2A-Puro (Lv-HuR) and was from Origene. Antibodies to the following proteins were used: phospho-S473-AKT (#9271, CST), AKT (#9272, CST), Rictor (#A300-459A, Bethyl Laboratories), actin (#ab3280, Abcam), phospho-Y1068-EGFR (#2234, CST), β-tubulin (#2146, CST), phospho-S326-HSF1 (ADI-SPA-902-D, Enzo Life Sciences), HSF1 (ADI-SPA-901-D, Enzo Life Sciences), HuR (07-468, EMD Millipore), RNA Pol II (#39097, Active Motif) and mSin1 (#07-2276, EMD Millipore). AG1478 was obtained from Selleckchem. EGF was from Life Technologies and all other reagents were from Sigma.
Polysome Analysis
Separation of polysomes was performed as described 40. Briefly, cell extracts were prepared and layered onto 15% to 50% sucrose gradients and spun at 38,000 rpm for 2 h at 4°C in a SW40 rotor (Beckman Instruments). Gradients were fractionated using a gradient fractionator system (Brandel Instruments) using a flow rate of 3 mL/min. The polysome profile of the gradients was monitored via UV absorbance at 260 nm. RNA was isolated and pooled into nonribosomal/monosomal and polysomal fractions. RNAs (100 ng) were subsequently used in quantitative reverse transcriptase-PCR analyses.
Metabolic labeling, Immunoblotting and Quantitative real time PCR
U138 cells were pulsed-labeled with [35S]methionine/cysteine at a final concentration of 100 μCi/ml. Cells were harvested following the indicated treatments and lysates prepared in RIPA buffer. Rictor protein was immunoprecipitated overnight with α-Rictor antibody and collected with protein G-Sepharose (GE Healthcare). The immunoprecipitate was washed four times and resuspended in SDS sample buffer and separated by SDS-PAGE. Gels were dried and visualized using a phosphorimager. Collectively, immunoprecipitation controls included equal numbers of cells plated per flask and equal amounts of quantitated total protein lysate per sample with equivalent amounts of antibody. Western blotting and quantitative real time PCR was performed as described 38.
Co-immunoprecipitations, Chromatin immunoprecipitations, RNA and DNA in vitro pull-down assays
mTOR-Rictor co-immunoprecipitations were performed as described utilizing 0.3% CHAPS-buffer to maintain mTORC2 complex integrity during lysis 7. Chromatin immunoprecipitation (ChIP) assays were performed as previously described 41. For RNA-pull down assays 38, extracts were prepared and biotinylated RNA oligonucleotides containing HuR binding site motif(s) added. The protein and biotinylated RNA complexes were recovered and the complexes were washed five times and resolved by gel electrophoresis. In vitro DNA-pull down assays were performed as described 41 using a double-stranded oligonucleotide containing the human HuR noncanonical HSE attached to streptavidin-Sepharose beads via a 5′ biotinylated plus strand according to the manufacture's recommendation (Invitrogen). Beads with bound proteins were analyzed by SDS-PAGE followed by immunoblotting.
Cell proliferation and migration assays
Cells growth was determined via XTT assays (Roche). Cell migration assays were performed using modified Boyden chambers (Chemicon) as previously described 42. For invasion assays through Matrigel, 2 × 104 cells were placed into the top well of Boyden chambers containing growth factor-reduced Matrigel extracellular basement membrane over a polyethylene terephthalate membrane (8-mm pores; BD Biosciences). Following 24 h culture, Matrigel was removed and invaded cells were fixed and stained. Cells adhering to the bottom of the membrane were counted.
Xenograft studies
All experiments were performed under an approved Institutional Animal Care and Use Committee protocol. Xenografts of shRNA-expressing cell lines were injected s.c. into the flanks of 4-6 week old female C.B.-17-scid (Taconic) mice as previously described 16. Two shAKT-1 and one shHuR-1 mice were excluded as they did not establish tumors. Sample sizes were chosen based on similar well-characterized experiments to ensure adequate power to detect a pre-specified effect size. Mice were randomly assigned to groups and the investigator blinded to assignments until final tumor analyses. Tumors were measured every 3-4 days and volumes calculated using the formula length × width × height × 0.5236. Tumors were harvested at autopsy for polysome analyses.
Statistical analysis
Statistical analyses were performed using unpaired Student's t tests and ANOVA models using Systat 13 (Systat Software, Chicago, IL). P values of less the 0.05 were considered significant. Significance in group comparisons was determined using a one-way analysis of variance and data generated showed normal distribution with similar variances, and analysis was completed assuming equal variances. To assess correlations of molecular markers in glioblastomas Spearman's rank correlation was used.
Supplementary Material
Acknowledgments
We thank Drs. Eugene Kandel, Jacob Fleischmann, Norimoto Yanagawa, Sanjai Sharma and Paul Mischel for cell lines and reagents. We also thank Dr. Alan Lichtenstein for comments on the manuscript and Jheralyn Martin and Janine Masri for technical assistance. This work was supported, in whole or in part, by VA MERIT I01BX002665 and NIH R01CA109312 grants.
Footnotes
Conflict of interest: The authors declare no competing financial interests
Author Contributions: Conceived and designed the experiments: BH, RNN, JG. Performed the experiments: BH, ABS, RSF, KAL, TB. Analyzed the data: BH, RNN, JG. Wrote the paper: BH, RNN, JG.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu SH, Bi JF, Cloughesy T, Cavenee WK, Mischel PS. Emerging function of mTORC2 as a core regulator in glioblastoma: metabolic reprogramming and drug resistance. Cancer biology & medicine. 2014;11:255–263. doi: 10.7497/j.issn.2095-3941.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cornu M, Albert V, Hall MN. mTOR in aging, metabolism, and cancer. Current opinion in genetics & development. 2013;23:53–62. doi: 10.1016/j.gde.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Oh WJ, Jacinto E. mTOR complex 2 signaling and functions. Cell cycle (Georgetown, Tex) 2011;10:2305–2316. doi: 10.4161/cc.10.14.16586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oh WJ, Wu CC, Kim SJ, Facchinetti V, Julien LA, Finlan M, et al. mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. The EMBO journal. 2010;29:3939–3951. doi: 10.1038/emboj.2010.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 7.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science (New York, NY) 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka K, Babic I, Nathanson D, Akhavan D, Guo D, Gini B, et al. Oncogenic EGFR signaling activates an mTORC2-NF-kappaB pathway that promotes chemotherapy resistance. Cancer discovery. 2011;1:524–538. doi: 10.1158/2159-8290.CD-11-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhattacharya K, Maiti S, Mandal C. PTEN negatively regulates mTORC2 formation and signaling in grade IV glioma via Rictor hyperphosphorylation at Thr1135 and direct the mode of action of an mTORC1/2 inhibitor. Oncogenesis. 2016;5:e227. doi: 10.1038/oncsis.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guertin DA, Stevens DM, Saitoh M, Kinkel S, Crosby K, Sheen JH, et al. mTOR complex 2 is required for the development of prostate cancer induced by Pten loss in mice. Cancer cell. 2009;15:148–159. doi: 10.1016/j.ccr.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Read RD, Fenton TR, Gomez GG, Wykosky J, Vandenberg SR, Babic I, et al. A kinome-wide RNAi screen in Drosophila Glia reveals that the RIO kinases mediate cell proliferation and survival through TORC2-Akt signaling in glioblastoma. PLoS genetics. 2013;9:e1003253. doi: 10.1371/journal.pgen.1003253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bashir T, Cloninger C, Artinian N, Anderson L, Bernath A, Holmes B, et al. Conditional astroglial Rictor overexpression induces malignant glioma in mice. PloS one. 2012;7:e47741. doi: 10.1371/journal.pone.0047741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Current biology : CB. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 14.Cheng H, Zou Y, Ross JS, Wang K, Liu X, Halmos B, et al. RICTOR Amplification Defines a Novel Subset of Patients with Lung Cancer Who May Benefit from Treatment with mTORC1/2 Inhibitors. Cancer discovery. 2015;5:1262–1270. doi: 10.1158/2159-8290.CD-14-0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Driscoll DR, Karim SA, Sano M, Gay DM, Jacob W, Yu J, et al. mTORC2 Signaling Drives the Development and Progression of Pancreatic Cancer. Cancer research. 2016;76:6911–6923. doi: 10.1158/0008-5472.CAN-16-0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masri J, Bernath A, Martin J, Jo OD, Vartanian R, Funk A, et al. mTORC2 activity is elevated in gliomas and promotes growth and cell motility via overexpression of rictor. Cancer research. 2007;67:11712–11720. doi: 10.1158/0008-5472.CAN-07-2223. [DOI] [PubMed] [Google Scholar]
- 17.Gibault L, Ferreira C, Perot G, Audebourg A, Chibon F, Bonnin S, et al. From PTEN loss of expression to RICTOR role in smooth muscle differentiation: complex involvement of the mTOR pathway in leiomyosarcomas and pleomorphic sarcomas. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2012;25:197–211. doi: 10.1038/modpathol.2011.163. [DOI] [PubMed] [Google Scholar]
- 18.Chen CC, Jeon SM, Bhaskar PT, Nogueira V, Sundararajan D, Tonic I, et al. FoxOs inhibit mTORC1 and activate Akt by inducing the expression of Sestrin3 and Rictor. Developmental cell. 2010;18:592–604. doi: 10.1016/j.devcel.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koo J, Wu X, Mao Z, Khuri FR, Sun SY. Rictor Undergoes Glycogen Synthase Kinase 3 (GSK3)-dependent, FBXW7-mediated Ubiquitination and Proteasomal Degradation. The Journal of biological chemistry. 2015;290:14120–14129. doi: 10.1074/jbc.M114.633057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yasuda K, Takahashi M, Mori N. Mdm20 Modulates Actin Remodeling through the mTORC2 Pathway via Its Effect on Rictor Expression. PloS one. 2015;10:e0142943. doi: 10.1371/journal.pone.0142943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tenenbaum SA, Lager PJ, Carson CC, Keene JD. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods (San Diego, Calif) 2002;26:191–198. doi: 10.1016/S1046-2023(02)00022-1. [DOI] [PubMed] [Google Scholar]
- 23.Srikantan S, Gorospe M. HuR function in disease. Frontiers in bioscience (Landmark edition) 2012;17:189–205. doi: 10.2741/3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carpenter RL, Paw I, Dewhirst MW, Lo HW. Akt phosphorylates and activates HSF-1 independent of heat shock, leading to Slug overexpression and epithelial-mesenchymal transition (EMT) of HER2-overexpressing breast cancer cells. Oncogene. 2015;34:546–557. doi: 10.1038/onc.2013.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou SD, Murshid A, Eguchi T, Gong J, Calderwood SK. HSF1 regulation of beta-catenin in mammary cancer cells through control of HuR/elavL1 expression. Oncogene. 2015;34:2178–2188. doi: 10.1038/onc.2014.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tandon M, Chen Z, Pratap J. Runx2 activates PI3K/Akt signaling via mTORC2 regulation in invasive breast cancer cells. Breast cancer research : BCR. 2014;16:R16. doi: 10.1186/bcr3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leach MD, Farrer RA, Tan K, Miao Z, Walker LA, Cuomo CA, et al. Hsf1 and Hsp90 orchestrate temperature-dependent global transcriptional remodelling and chromatin architecture in Candida albicans. Nature communications. 2016;7:11704. doi: 10.1038/ncomms11704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendillo ML, Santagata S, Koeva M, Bell GW, Hu R, Tamimi RM, et al. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150:549–562. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang B, Jie Z, Joo D, Ordureau A, Liu P, Gan W, et al. TRAF2 and OTUD7B govern a ubiquitin-dependent switch that regulates mTORC2 signalling. Nature. 2017;545:365–369. doi: 10.1038/nature22344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masui K, Shibata N, Cavenee WK, Mischel PS. mTORC2 activity in brain cancer: Extracellular nutrients are required to maintain oncogenic signaling. BioEssays : news and reviews in molecular, cellular and developmental biology. 2016;38:839–844. doi: 10.1002/bies.201600026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic acids research. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobson A, Peltz SW. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annual review of biochemistry. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 33.Gabai VL, Meng L, Kim G, Mills TA, Benjamin IJ, Sherman MY. Heat shock transcription factor Hsf1 is involved in tumor progression via regulation of hypoxia-inducible factor 1 and RNA-binding protein HuR. Molecular and cellular biology. 2012;32:929–940. doi: 10.1128/MCB.05921-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riemenschneider MJ, Betensky RA, Pasedag SM, Louis DN. AKT activation in human glioblastomas enhances proliferation via TSC2 and S6 kinase signaling. Cancer research. 2006;66:5618–5623. doi: 10.1158/0008-5472.CAN-06-0364. [DOI] [PubMed] [Google Scholar]
- 35.Dai C, Santagata S, Tang Z, Shi J, Cao J, Kwon H, et al. Loss of tumor suppressor NF1 activates HSF1 to promote carcinogenesis. The Journal of clinical investigation. 2012;122:3742–3754. doi: 10.1172/JCI62727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nabors LB, Gillespie GY, Harkins L, King PH. HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3′ untranslated regions of cytokine and angiogenic factor mRNAs. Cancer research. 2001;61:2154–2161. [PubMed] [Google Scholar]
- 37.Hsu JH, Shi Y, Hu L, Fisher M, Franke TF, Lichtenstein A. Role of the AKT kinase in expansion of multiple myeloma clones: effects on cytokine-dependent proliferative and survival responses. Oncogene. 2002;21:1391–1400. doi: 10.1038/sj.onc.1205194. [DOI] [PubMed] [Google Scholar]
- 38.Jo OD, Martin J, Bernath A, Masri J, Lichtenstein A, Gera J. Heterogeneous nuclear ribonucleoprotein A1 regulates cyclin D1 and c-myc internal ribosome entry site function through Akt signaling. The Journal of biological chemistry. 2008;283:23274–23287. doi: 10.1074/jbc.M801185200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuo J, Rungger D, Voellmy R. Multiple layers of regulation of human heat shock transcription factor 1. Molecular and cellular biology. 1995;15:4319–4330. doi: 10.1128/mcb.15.8.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cloninger C, Bernath A, Bashir T, Holmes B, Artinian N, Ruegg T, et al. Inhibition of SAPK2/p38 enhances sensitivity to mTORC1 inhibition by blocking IRES-mediated translation initiation in glioblastoma. Molecular cancer therapeutics. 2011;10:2244–2256. doi: 10.1158/1535-7163.MCT-11-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vartanian R, Masri J, Martin J, Cloninger C, Holmes B, Artinian N, et al. AP-1 regulates cyclin D1 and c-MYC transcription in an AKT-dependent manner in response to mTOR inhibition: role of AIP4/Itch-mediated JUNB degradation. Molecular cancer research : MCR. 2011;9:115–130. doi: 10.1158/1541-7786.MCR-10-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holmes B, Lee J, Landon KA, Benavides-Serrato A, Bashir T, Jung ME, et al. Mechanistic Target of Rapamycin (mTOR) Inhibition Synergizes with Reduced Internal Ribosome Entry Site (IRES)-mediated Translation of Cyclin D1 and c-MYC mRNAs to Treat Glioblastoma. The Journal of biological chemistry. 2016;291:14146–14159. doi: 10.1074/jbc.M116.726927. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.