Abstract
The sea lamprey (Petromyzon marinus) serves as a comparative model for reconstructing vertebrate evolution. To enable more informed analyses, we developed a new assembly of the lamprey germline genome that integrates several complementary datasets. Analysis of this highly contiguous (chromosome-scale) assembly reveals that both chromosomal and whole-genome duplications have played significant roles in the evolution of ancestral vertebrate and lamprey genomes, including chromosomes that carry the six lamprey HOX clusters. The assembly also contains several hundred genes that are reproducibly eliminated from somatic cells during early development in lamprey. Comparative analyses show that gnathostome (mouse) homologs of these genes are frequently marked by Polycomb Repressive Complexes (PRCs) in embryonic stem cells, suggesting overlaps in the regulatory logic of somatic DNA elimination and repressive/bivalent states that are regulated by early embryonic PRCs. This new assembly will enhance diverse studies that are informed by lampreys’ unique biology and evolutionary/comparative perspective.
The sea lamprey is a member of an ancient lineage that diverged from the vertebrate stem approximately 550 million years ago (MYA). By virtue of this deep evolutionary perspective, lamprey has served as a critical model for understanding the evolution of several conserved and derived features that are relevant to broad fields of biology and biomedicine. Studies have used lampreys to provide perspective on the evolution of developmental pathways that define vertebrate embryogenesis1,2, vertebrate nervous and neuroendocrine systems2,3, genome structure4, immunity5, clotting6 and others7. These studies reveal aspects of vertebrate biology that have been conserved over deep evolutionary time and reveal evolutionary modifications that gave rise to novel features that emerged within the jawed vertebrate lineage (gnathostomes). Lampreys also possess several features that are not observed in gnathostomes, which could represent either aspects of ancestral vertebrate biology that have not been conserved in the gnathostomes or features that arose since the divergence of the ancestral lineages that gave rise to lampreys and gnathostomes. These include the ability to achieve full functional recovery after complete spinal cord transection, deployment of evolutionarily independent yet functionally equivalent adaptive immune receptors, and the physical restructuring of the genome during development known as programmed genome rearrangement (PGR).
PGR results in the physical elimination of ~0.5 Gb of DNA from its ~2.3 Gb genome8–10. The elimination events that mediate PGR are initiated at the 7th embryonic cell division and are essentially complete by 3 days post fertilization11,12. As a result, lampreys are effectively chimeric, with germ cells possessing a full complement of genes and all other cell types possessing a smaller, reproducible, fraction of the germline genome. Previous analyses support the idea that the somatic genome lacks several genes that contribute to the development and maintenance of germ cells but are potentially deleterious if misexpressed in somatic lineages. However, our understanding of the mechanisms and consequences of PGR remains incomplete, as only a fraction of the germline genome has been sequenced to date.
In contrast to the germline genome, the somatically retained portions of the genome are relatively well characterized. Because PGR was not known to occur in lampreys prior to 20098, sequencing efforts focused on somatic tissues from which DNA or intact nuclei could be readily obtained (e.g. blood and liver)13. Sequencing of the sea lamprey somatic genome followed an approach that had proven successful for other vertebrate genomes prior to the advent of next generation sequencing technologies (Sanger sequencing of clone ends, fosmid ends and BAC ends). Due to the abundance of highly-identical interspersed repetitive elements and moderately high levels of polymorphism (approaching 1%), assembly of the somatic genome resulted in a consensus sequence that was substantially more fragmentary than other Sanger-based vertebrate assemblies14. Nonetheless, this initial assembly yielded significant improvements in our understanding of the evolution of vertebrate genomes and fundamental aspects of vertebrate neurobiology, immunity and development1–7.
Here we present the first assembly of the sea lamprey germline genome. Through extensive optimization of assembly pipelines, we identified a computational solution that allowed us to generate an assembly from next-generation sequence data (Illumina and Pacific Biosciences reads) that surpasses the existing Sanger-based somatic assembly. Analysis of the resulting assembly reveals several hundred genes that are eliminated from somatic tissues by PGR and sheds new light on the evolution of genes and functional elements in the wake of ancient large-scale duplication events.
RESULTS
Assembly and Annotation of the Sea Lamprey Genome
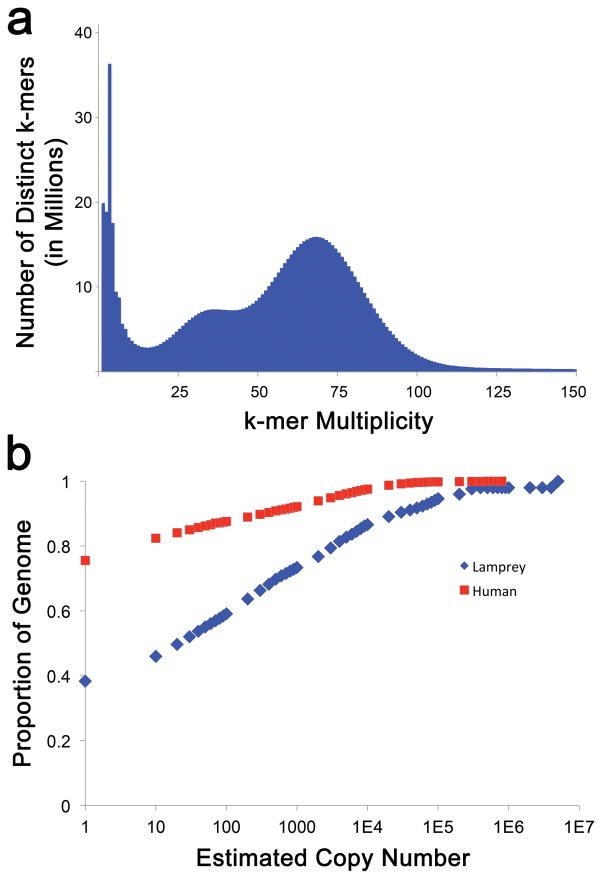
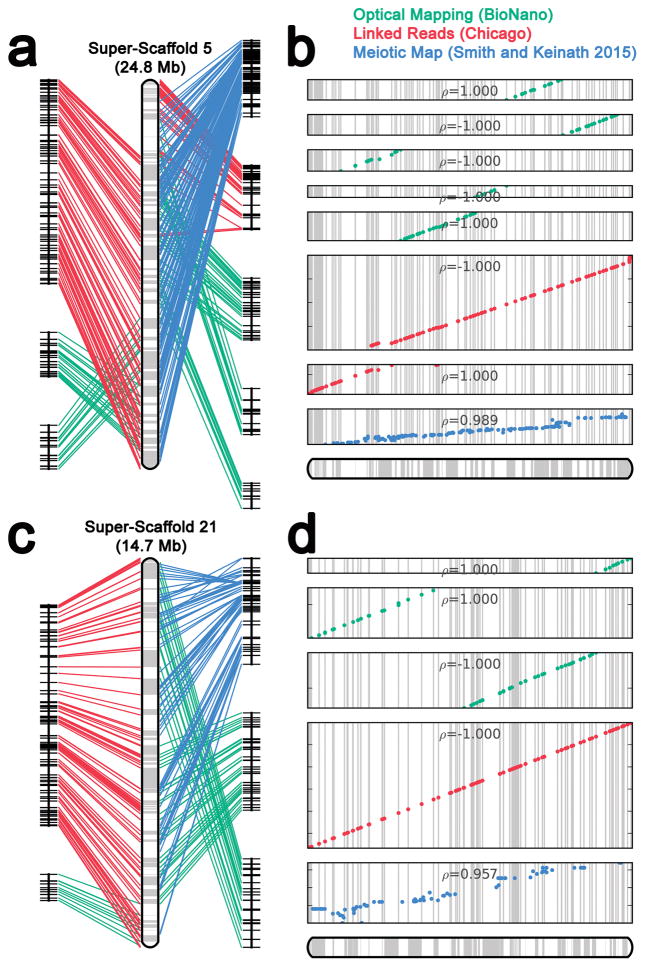
Several shotgun-sequencing and scaffolding datasets were generated in order to permit assembly of the lamprey germline genome (>100X sequence coverage in Illumina paired end reads, >300X physical coverage in 4kb Illumina mate pairs and >600X physical coverage in 40kb Illumina mate pairs). Previous analyses demonstrated that the lamprey genome is highly repetitive and initial analysis of Illumina shotgun sequence data confirmed that the repeat content of lamprey (~60% high-identity repeats) is substantially higher than that of human (Figure 1). To enable the development of a highly contiguous assembly, we also generated ~17X genome coverage in single molecule long-read data (Pacific Biosciences XL/C2 chemistry, N50 read length = 5424) and performed hybrid assembly using DBG2OLC15. This approach yielded an assembly with contiguity statistics (23,286 contigs, N50 = 164,585 bp) that rivaled a previously published Sanger-based assembly of the somatic genome13. To further improve the large-scale structure of this assembly, we integrated scaffolding data (~56X coverage in BioNano optical mapping: >150 kb molecules, and 325 million Chicago (Dovetail) linked read pairs: 2X152 bp), as well as published meiotic mapping data4. Linkages identified through these three independent datasets were cross-validated and integrated using AllMaps (Figure 2)16. This integrated scaffolding approach allowed us to further increase the contiguity of the assembly (12,077 contigs, N50 = 12 Mb, N50 contig number = 34). In total, 74.8% of the current germline genome assembly is anchored to one of 94 previously-defined linkage groups4 and >80% of the assembly is present in super-scaffolds that are 1 Mb or longer. Given that the sea lamprey has 99 pairs of chromosomes in its germline, this integrated assembly appears to approach chromosome-scale contiguity.
Figure 1. Distribution of k-mer copy numbers in germline shotgun sequencing data.
a) The spectrum of error corrected 25-mers reveals a modal count of 68 and a second hump at half of this value, corresponding to allelic k-mers. k-mer multiplicity is defined as the number of times a k-mer was observed in the sequence dataset. b) Less than 40% of the lamprey genome can be represented by single-copy 25-mers, whereas >75% of the human genome can be represented by single-copy k-mers of this same length. The X-axis is plotted on a log scale to aid in visualization of patterns at lower estimated copy numbers.
Figure 2. Long-range scaffolding and assessment of long-range contiguity of lamprey super-scaffolds.
Data from three independent strategies were used to place contigs on larger chromosomal structures. Data from meiotic maps (blue), Dovetail maps (red) and optical maps (green) complement and extend one another. a) Information used to generate super-scaffold 5, b) Ordering of anchors along super-scaffold 5. c) Information used to generate super-scaffold 21, d) Ordering of anchors along super-scaffold 21. ρ = Pearson correlation coefficient based on the following numbers of markers, Panel a, top to bottom: n=18, 28, 14, 10, 34, 156, 78 and 162 independent scaffolding anchors; Panel b, top to bottom: n=10, 22, 36, 196 and 79 independent scaffolding anchors.
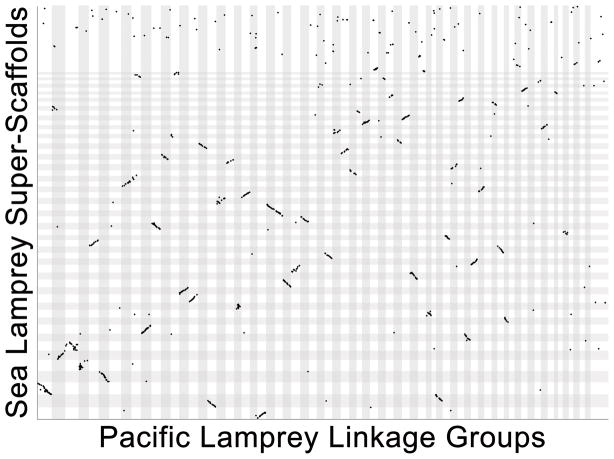
Our long-range scaffolding approach used three independent methods that extend and cross-validate one another (Figure 2) and we consider strong agreement among these three methods as evidence that the large-scale structure of the assembly accurately reflects the structure of P. marinus chromosomes. For many vertebrates, it is possible to independently assess long-range contiguity by measuring conservation of gene orders with closely related species. Highly contiguous assemblies are not yet available for any other jawless vertebrate, although an unanchored draft assembly does exist for the Arctic lamprey (Lethenteron camtschaticum: syn. Lethenteron japonicum)17. To provide perspective on the chromosomal structure of a closely related species, we developed a meiotic map for the Pacific lamprey (Entosphenus tridentatus). The species is a representative of a clade of lampreys (genera Entosphenus, Lethenteron and Lampetra) that diverged from the lineage represented by Petromyzon ~40 MYA18, and embryos of known parentage are available through ongoing hatchery efforts aimed at restoring the species to its native waterways in the Pacific Northwest19. Meiotic mapping was performed using restriction site associated DNA (RAD) sequencing of 94 F1 siblings generated from a controlled cross between two wild-captured individuals. The resulting meiotic map provides dense coverage of the genome and represents 83 linkage groups, covering 9956 cM with an average intermarker distance of 3.4 cM (Supplementary Table 1). Alignment of RAD markers to the sea lamprey genome identified 1733 homologous sequences, which show strong conservation of synteny and gene order (Figure 3, Supplementary Table 1). This broad conservation of gene order is considered strong evidence that the sea lamprey assembly and Pacific lamprey meiotic map accurately reflect the chromosomal structure of their respective species.
Figure 3. Alignment of the Pacific lamprey (E. tridentatus) meiotic map to assembled sea lamprey (P. marinus) super-scaffolds.
The relative position of homologous sequences is shown for sea lamprey (y-axis) and pacific lamprey (X-axis). A single homologous site (aligning RAD-seq read, Supplementary Table 1) is marked by a single dot. Chromosomes and linkage groups (LGs) are ordered from longest to shortest within species and individual chromosomes/LGs are highlighted by alternating dark and light shading. Groups of adjacent dots (regions showing conservation of synteny and gene order) appear as diagonal lines.
The repetitive nature of the lamprey genome presents challenges not only to its assembly, but also the identification of genes within assembled contigs. This is largely attributable to the interspersion of transposable coding sequences within and among the coding sequences of low-copy genes. To circumvent these issues we used a two-tiered approach to gene prediction. Annotation and identification of repetitive elements was performed using RepeatModeler and RepeatMasker20,21. The entire set of annotated repeats, published gene models and transcriptomic datasets10,13 were integrated to generate a conservative set of 18,205 gene predictions using MAKER22. After generating initial gene calls, a second round of gene predictions was generated that permitted extraction of gene models that include low-copy repetitive sequences, yielding another 2,745 gene models for a total of 20,950 MAKER gene models. In total, Maker was able to assign 18,367 of these gene models to a likely vertebrate homolog on the basis of multispecies blast alignments, which included the vast majority of single-copy orthologs expected for lamprey (Supplementary Note)23,24. An additional 2,583 genes (12%) could not be immediately assigned a homolog on the basis of multispecies alignments. While these may represent lamprey-specific genes, careful manual curation will likely be necessary to define their precise evolutionary origins. Such efforts will be enabled through the publicly available genome browser (see URLs). This annotation set was subsequently used to identify the location of 35382 lncRNA transcripts in 18857 lncRNA gene bodies (Supplementary Note, Supplementary Table 2 and Supplementary Figure 1). These and other annotation sets, including RNA sequencing and genome re-sequencing tracks are available through SIMRbase (see URLs).
Vertebrate Genome Evolution
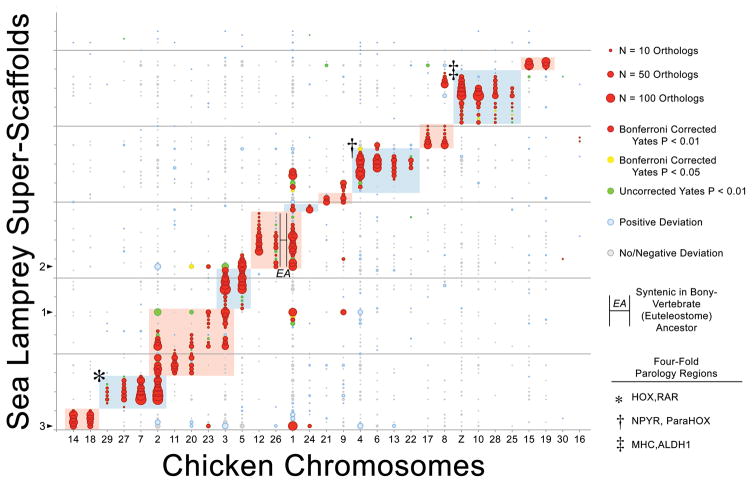
Lamprey occupies a critical phylogenetic position with respect to reconstructing ancestral karyotypes and inferring the timing and mode of duplication events that occurred in ancestral vertebrate and gnathostome lineages. Alignment to chicken25 and gar26 genomes (Supplementary Tables 3–5) permits reconstruction of ancestral orthology groups that are highly consistent with previous reconstructions that were based on the lamprey meiotic map4. Because these comparisons require resolution of homologies that are the product of duplication (i.e. 1:1 orthology is not expected) our operational definition of “orthology groups” is expanded to include higher-order relationships (see Smith and Keinath, 2015 for more detail)4. Inclusion of comparative mapping data from the recently published gar genome assembly provides further support for the observation that the majority of ancestral vertebrate chromosomes experienced a single large-scale duplication event in the ancestral vertebrate lineage (Figure 4, Supplementary Figure 2). Most ancestral orthology groups correspond to two derived chicken chromosomes (6/11 chicken/lamprey orthology groups identified here). Three other orthology groups possess four derived chromosomes suggesting that these groups have experienced an additional large-scale duplication: these include well defined four-fold orthology regions harboring HOX and MHC in one orthology group, NPYR and ParaHox clusters in a second, and RAR and ALDH1 loci in a third4 (Figure 4). Two remaining orthology groups present more complex ratios of ancestral:derived chromosomes. Notably though, comparative mapping with gar reveals that chicken chromosome 26 and a portion of chicken chromosome 1 were fused in the bony vertebrate (Euteleostome) ancestor approximately 450 MYA and subsequently experienced a derived fission in the chicken lineage. Other deviations from 1:2 or 1:4 are interpreted as the product of derived fission/fusion events that occurred during the first 150 MY following divergence of basal lamprey and gnathostome lineages, derived fission/fusion events in the lamprey lineage, or misassembled regions of the lamprey genome. While it is possible that the observed genome-wide patterns of conserved synteny could have arisen through two whole genome duplication events (the 2R hypothesis)27,28 accompanied by large numbers of chromosome losses29,30, a previously-proposed alternate scenario involving one whole genome duplication preceded by three distinct chromosome-scale duplication events requires fewer evolutionary steps and is consistent the data underlying all previous reconstructions4.
Figure 4. The distribution of conserved syntenies in chicken and lamprey reveals patterns of ancient large-scale duplication.
These patterns are consistent with those from the lamprey somatic genome assembly and reveal both chromosomal/segmental and whole genome duplications. Lamprey super-scaffolds are oriented along the y-axis and chicken chromosomes are oriented along the x-axis. Circles reflect counts of syntenic orthologs on the corresponding lamprey and chicken chromosomes, with the size of each circle being proportional to the number of orthologs on that pair. The color of each circle represents the degree to which the number of observed orthologs deviates from null expectations under a uniform distribution across an identical number of lamprey and chicken chromosomes with identical numbers of orthology-informative genes. Shaded regions of the plot designate homology groups that correspond to presumptive ancestral chromosomes. Syntenic groups that are linked by lines marked EA are predicted to correspond to a single chromosome in the Euteleostome ancestor, based one conserved synteny with spotted gar (Lepisosteus oculatus). The three largest super-scaffolds are marked with an arrow along the y-axis. The ordering of lamprey super-scaffolds along the y-axis is provided in Supplementary Table 4.
Lamprey HOX Clusters: Duplication and Divergence
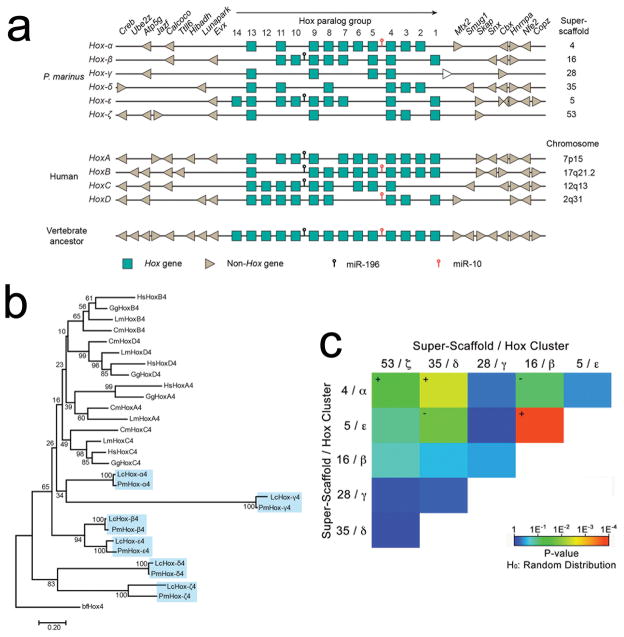
Historically, descriptions of genome duplications have relied heavily on the HOX gene clusters. This is partially due to their highly conserved organization with respect to gene order and orientation, which contributes to generation of coordinated patterns of axial expression (collinearity), associated with their roles in embryonic development. Assembly of the Arctic lamprey genome led to the tentative prediction of at least six, and possibly eight, HOX clusters, suggesting that the duplication history of at least the lamprey HOX-bearing chromosomes differs from that of the jawed vertebrates17. We identify 42 Hox genes in the sea lamprey, which all fall within six HOX clusters that are highly similar in content to the HOX clusters predicted in the Arctic lamprey (Figure 5A, Supplementary Figures 3–4). Additionally, we are able to place these in their broader chromosomal context, revealing that these six HOX clusters are embedded in larger chromosomal regions that share conserved synteny with the presumptive ancestral HOX-bearing chromosome (Figure 4).
Figure 5. Structure and Evolution of HOX clusters.
a) Six Hox clusters can be identified within the sea lamprey genome assembly. Lamprey cluster designations α through ζ follow the convention of Mehta et al17. Hox genes are represented as boxes, with the direction of their transcription indicated by the black arrow. Flanking non-Hox genes are depicted as arrowheads, which indicate their direction of transcription. The positions of known micro-RNAs are indicated. The four human Hox loci and the inferred ancestral vertebrate Hox locus40 are shown for comparison. The white arrow downstream of the lamprey Hox-γ cluster represents PMZ_0048273, an uncharacterized non-Hox gene. b) The evolutionary history was inferred using the Neighbor-Joining method41. The optimal tree with the sum of branch length = 9.68 is shown. The percentage of replicate trees in which the associated taxa clustered together (bootstrap test with 100 replicates) are shown next to the branches. c) Tests for enrichment of 2-copy duplicates among all pairs of Hox-bearing chromosomes (super-scaffolds). Colors correspond to the degree to which the counts of shared duplicates on each pair of chromosomes deviates from the expected value given an identical number of chromosomes and paralogs retained on each chromosome (Probability estimates were generated using two-tailed χ2 tests and a total of n=200 independent pairs of duplicated genes: see Supplementary Table 6). Plus and minus symbols indicate the direction of deviation from expected for chromosome pairs with P<0.01.
In principle, a number of duplication scenarios could potentially explain the existence of six paralogous HOX-bearing chromosomes. These include: 1) whole-genome duplication then triplication, or vice versa; 2) A gnathostome-like duplication history (either 2R accompanied by large numbers of chromosome losses29,30, or one whole genome duplication preceded by three chromosome-scale duplication events4) followed by a further round of whole genome duplication (yielding eight ancestral HOX clusters) and loss of two entire paralogous chromosomes; 3) A gnathostome-like duplication history followed by duplication of two individual chromosomes. Initial synteny comparisons between lamprey and gnathostome HOX loci revealed no clear orthology relationships, but show substantial similarities in the gene content of lamprey HOXε and HOXβ clusters. Notably, phylogenetic analyses of paralogy groups with ≥4 retained copies (HOX4, 8, 9, 11 and 13) also reveal no clear orthology between lamprey and gnathostome clusters, but reproducibly place members of HOX ε and β clusters in sister clades with high bootstrap support (Figure 5B, Supplementary Figures 5–9). Taken at face value this would seem to suggest that ε and β clusters diverged from one another more recently than other paralogous clusters, apparently lending support to scenario 3. Alternately, this might also reflect greater functional constraint with respect to the membership of these clusters.
To gain further perspective on the duplication history of lamprey HOX clusters, we extended analyses to compare the chromosome-wide distribution of 2-copy paralogs on all HOX-bearing chromosomes. Because post-duplication patterns of conserved synteny are strongly driven by paralog loss, we reasoned that more recent duplication events should yield pairs of chromosomes that share more 2-copy duplications, exclusive of all other paralogous chromosomes (the latter of which would have experienced more extensive loss of redundant paralogs over time). Two pairs of chromosomes were observed to share more duplicates relative to all other pairwise combinations of HOX-bearing chromosomes. The strongest enrichment of 2-copy paralogs was observed between super-scaffolds 5 and 16 (χ2=14.22, P=1.6E−4, df=1, Figure 5, Supplementary Table 6), which carry the HOX ε and β clusters. In conjunction with the internal structure of HOX clusters and consistent phylogenetic clustering of ε and β Hox members, we interpret this as indicating that the ε– and β– bearing chromosomes trace their ancestry to a chromosome-scale duplication event that occurred substantially more recently than the genome/chromosome-scale duplication events that define all other pairwise contrasts, perhaps within the last 200–300 MY. Only one other pair of chromosomes shows significant enrichment of 2-copy paralogs relative to all other contrasts. The chromosomes bearing HOX α and δ clusters are enriched in shared 2-copy paralogs (χ2=8.41, P=3.7E−3, df=1, Figure 5, Supplementary Table 6), although α and δ HOX members show no consistent pattern of clustering within gene trees. This difference could be interpreted as indicating that these two chromosomes are the product of a slightly older duplication event, or alternately it might reflect differential constraints relative to the retention of duplicates by individual pairs of paralogous chromosomes. However, it is unclear what processes might constrain the evolution of one pair of paralogous chromosomes relative to all others.
Programmed Genome Rearrangement
Identification of eliminated DNA
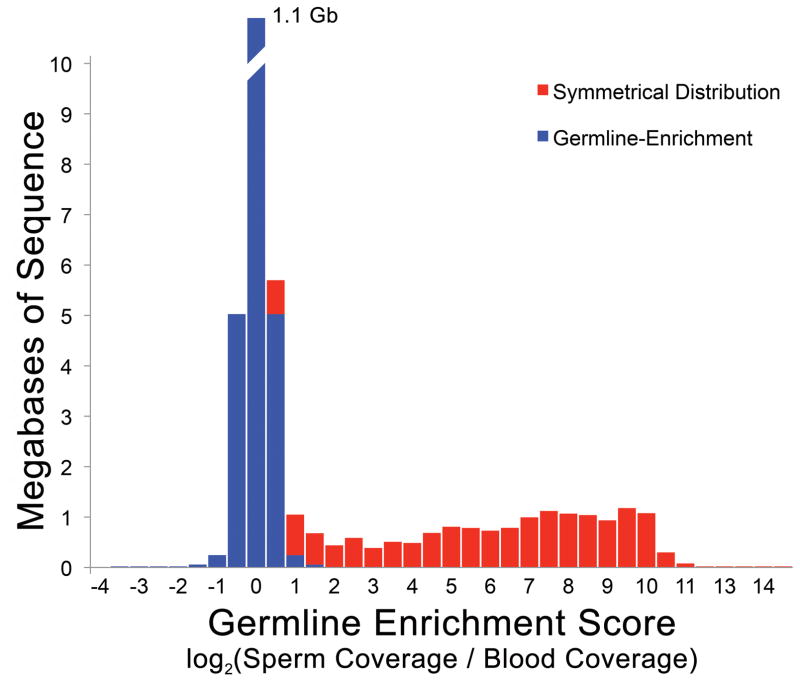
In lampreys approximately 20% of zygotically inherited DNA is eliminated from somatic cell lineages during early embryogenesis, being retained only by the germline8,10,31. To identify germline-enriched (i.e. somatically-eliminated) regions, we generated whole genome shotgun sequence data for both sperm (73X coverage) and blood (80X coverage) DNAs that were isolated from the same individual. Analysis of read counts identified 1077 super-scaffolds with enrichment scores [log2(standardized sperm coverage/blood coverage)] exceeding two, over more than 80% of the scaffold (Figure 6, Supplementary Table 7). These presumptively germline-specific regions covered ~13 Mb of the genome assembly and contain 356 annotated protein coding genes. The distribution of enrichment scores also suggests that other regions with lower enrichment scores are likely to be impacted by PGR. To further evaluate our predictions, we designed primers for the 96 longest super-scaffolds with enrichment scores of two or higher. In total, primers from 90 (94%) of these scaffolds yielded specific amplification in testes relative to blood, confirming that they are deleted during PGR (Supplementary Table 8).
Figure 6. Germline Enrichment of Single/Low-Copy DNA Sequences.
Comparative sequencing reveals germline enrichment of several single/low-copy intervals. The distribution of coverage ratios reveals a long tail corresponding to segments with higher sequence coverage in sperm relative to blood. This excess is highlighted in red, assuming a symmetrical distribution of enrichment scores for non-eliminated regions and an absence of somatic-specific sequence.
Notably, the estimates above only account for single copy DNA of sufficient complexity to yield unique alignments. Eliminated sequences with retained paralogs or that contain low copy repetitive elements are expected to show relatively lower enrichment scores. To gain further insight into elimination of repetitive DNA, we performed similar analyses targeting repetitive sequences (Supplementary Note). These analyses identify an additional 102 Mb of eliminated sequence that can be directly assigned to assemblable repetitive sequences and indicate remaining fractions of the germline-specific subgenome likely consist of arrays of short or incomplex/simple repetitive sequence that are less amenable to sequencing, mapping or assembly (Supplementary Note and Supplementary Figure 10).
Function of PGR
It has been proposed that PGR serves to prevent the expression of genes with beneficial functions in the germline and deleterious functions in soma (e.g. oncogenesis, aging)8,10,12. To gain further insight into the functions of eliminated genes and the underlying evolutionary logic of PGR, we asked whether human homologs of eliminated genes are enriched for defined functional categories. In interpreting these ontology enrichment studies, it is important to recognize that these analyses define a single human or mouse ortholog for each lamprey gene. While this scenario does not accurately reflect duplication events that have structured lamprey and gnathostomes, or divergence in gene functions over more than 500 MY of independent evolution, they are expected to provide some (albeit conservative) perspective on the likely function of lamprey genes. Despite this deep divergence, ontology analyses revealed enrichment for several categories, including pathways related to oncogenesis, including: regulation of cell division, epithelial migration, adhesion, and cell fate commitment (Supplementary Table 9, Supplementary Note).
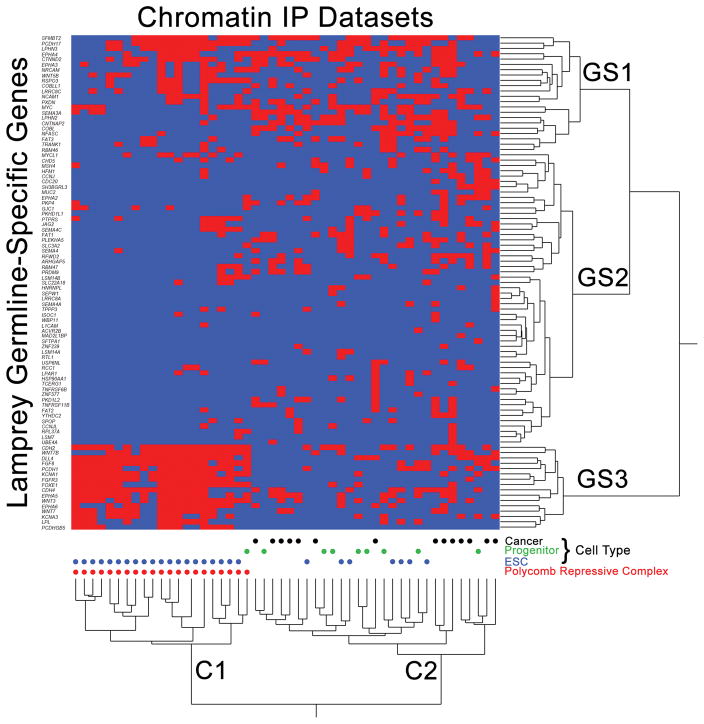
While ontology analyses provide some insight into the likely functions of eliminated genes, it is important to recognize that curated ontology databases do not capture all of the biological functions that are encoded in the genome. To gain additional insight into the functional consequences of PGR, we searched for enrichment of eliminated orthologs among 645 chromatin immunoprecipitation (ChIP) experiments (ChEA 2016)32,33 (Supplementary Table 10). To identify subcategories of enriched ChIP datasets, we performed 2-way hierarchical clustering of presence/absence calls from the top 50 enriched ChIP datasets. These analyses revealed two distinct categories of lamprey genes and ChIP experiments (Figure 7). One cluster (Figure 7, C1) corresponds to the binding sites of polycomb repressive complex (PRC) genes in mouse embryonic stem cells, apparently indicating that that these genes may be marked by bivalent promoters in embryonic stem cells (ESCs), then presumably released from silencing in germline at later developmental stages. To test this idea, we more closely examined a cluster of genes that was highly enriched within C1 ChIP experiments (GS3). Notably, all of these genes were previously found to be marked by bivalent (poised) promoters in murine ESCs and primordial germ cells34 (poised in ESCs: 16/16, χ2=77.0, P=8.8E−19, df=1: poised in PGCs: 15/16, χ2=47.3, P=3.1E−12, df=1). A second cluster of eliminated genes (GS1) also showed strong enrichment for these two functional categories (poised in ESCs: 14/22, χ2=34.6, P=2.0E−9, df=1; poised in PGCs: 14/22, χ2=23.2, P=7.5E−7, df=1).
Figure 7. Enrichment analysis provides insight into the function of germline specific sequences.
Homologs of eliminated genes show strong overlap for the binding targets of polycomb repressive complexes in mouse embryonic stem cells (ESCs) and the binding sites of transcription factors in multipotent progenitor lineages and cancer cells (from ChEA 2016)32. Red cells denote ChIP experiments (x-axis) that identify peaks overlapping orthologs of lamprey genes (y-axis). ChIP enrichment statistics and ordering along the x-axis are provided in Supplementary Table 9. Labels GS1, GS2 and GS3 denote three primary clusters of germline-specific genes, C1 and C2 denote two primary clusters of ChIP experiments.
Other enriched ChIP experiments (C2) correspond primarily to the binding targets of transcriptional modifiers in embryonic stem cells (N = 7), embryonic progenitor lineages (N = 7) and transcriptional activators in cancer (N = 15; Figure 7). Notably, all but one (PCDHGB5) of the genes detected in C1 are present in one or more experiments in C2. Overall, comparisons with ChIP analyses performed in non-eliminating species lends further support to the idea that PGR acts to prevent misexpression of “germline” genes and suggests that misexpression of orthologous genes may be directly contributing to oncogenesis in a diverse range of cancers. Moreover, these comparative analyses provide new insight into the regulatory functions of PGR by revealing overlap between early gene silencing events that are achieved by PGR and those that are mediated by the PRC during differentiation of germline and soma.
DISCUSSION
The lamprey genome presents an interesting target for sequencing due to its phylogenetic position and unique genome biology, yet a particularly challenging target given its high repeat content and divergence from other species with highly contiguous assemblies. In an attempt to resolve this complexity, we leveraged several complementary technologies to generate a highly contiguous assembly that approaches the scale of entire chromosomes. Moreover, we were able to validate the chromosome-scale contiguity of our assembly by generating a dense meiotic map for a related species. The high contiguity of our assembly provides critical context for understanding the evolution of gene content and genome structure in vertebrates. Here we highlighted the utility of this assembly in addressing fundamental questions related to understanding changes in large-scale structure of vertebrate genomes, specifically: reconstructing the deep evolutionary origins of vertebrate chromosomes and understanding how PGR mediates genetic conflicts between germline and somatic tissues.
Our improved assembly permits robust resolution of a complement of ancestral chromosomes that existed before the divergence of ancestral gnathostome and agnathan lineages, and prior to whole genome duplication(s) within the shared ancestral lineage of all extant vertebrates. These reconstructions largely validate previous analyses that were performed using meiotic mapping data, but provide improved resolution of ancestral homology groups. Analyses also lend further support to the idea that chromosome-scale duplication events may have been more common over the course of vertebrate ancestry than has been appreciated from the analysis of bony vertebrate genomes. Parallel lines of evidence supporting a relatively recent duplication having given raise to lamprey HOX ε and β-bearing chromosomes further highlights the potential for large-scale duplication outside of the context of whole genome duplication. It appears that two features of lamprey biology might favor the fixation of chromosomal duplications. First, lampreys possess a large number of small chromosomes and consequently chromosomal duplications will generally impact fewer genes than similar events in human. Duplication events (in addition to a single presumptive whole-genome duplication) appear to have impacted other groups of lamprey chromosomes, though not all (Supplementary Figure 11). Second, individuals are highly fecund (~100,000 eggs per female) and therefore a single mutant can introduce thousands of carriers (including stable carriers) into a population4,35–37. While it is likely that the reproductive biology and distribution of chromosome sizes has fluctuated over the course of vertebrate evolution, available evidence suggests that lampreys have possessed similar karyotypes and reproductive biologies for hundreds of millions of years. As such, extant lampreys may represent a better model for conceptualizing phases of evolution where ancestral vertebrates were characterized by higher fecundity and larger numbers of relatively gene poor microchromosomes, in addition to providing phylogenetic perspective on early stages of vertebrate genome evolution.
The assembly also identifies a large number of genes that are reproducibly eliminated via PGR. These genes reveal a strong overlap in the targets of PGR-mediated elimination and the targets silencing via PRC proteins in embryonic stem cells. The PRC is a deeply conserved complex that plays roles in gene silencing related to the maintenance of stem cell identity, silencing of oncogene expression and X-chromosome inactivation, among other functions38,39. These well-defined functions of PRC mirror several aspects of PGR, particularly in that both act to achieve strong transcriptional silencing both appear to target an overlapping subset of proto-oncogenes. It is interesting to speculate that the overlapping targets of PGR and PRC may indicate that the two mechanisms share common underlying mechanisms. However, it is notable that PRC repression is strongly associated with the deposition and binding to tri-methylated lysine 27 of histone H3 (H3K27me3), whereas previous studies have shown that this mark is absent prior to the onset of PGR in lamprey embryos11. It therefore appears that PGR acts to (in part) regulate a subset of germline-expressed targets of PRC and that it may work upstream of PRC in lamprey embryos.
The analyses presented here address a focused set of topics that are specifically related to understanding the evolution and development of genome structure in lamprey and other vertebrates. We anticipate that this assembly will substantially improve our ability to use lamprey as a comparative evolutionary model. Because sequences are anchored to their broader chromosomal structure, the current assembly should enhance the ability to reconstruct the deep evolutionary history of the vast majority of genes within vertebrate genomes, and perform robust tests of hypotheses related to historical patterns of duplication and divergence. Moreover, the availability of a highly contiguous assembly for an agnathan species should aid in the development and analysis of other genome assemblies from this highly informative vertebrate lineage.
MATERIALS and METHODS
Sequencing
Fragment libraries were prepared by Covaris shearing of sperm genomic DNA isolated from a single individual and size selected to achieve average insert sizes of ~205 and 231 bp. These libraries were sequenced on the Illumina HiSeq2000 platform. Two separate 4kb mate pair libraries were generated. One 4kb library was prepared and sequenced by the Genomic Services Laboratory at HudsonAlpha (Huntsville, AL) and another was prepared and sequenced using the standard Illumina mate-pair kit. Two 4kb libraries were prepared and sequenced by Lucigen (Middleton, WI). Long reads were prepared by the University of Florida Interdisciplinary Center for Biotechnology Research (Gainesville, FL) and sequenced using Pacific Biosciences (Menlo Park, CA) XL/C2 chemistry on a Single Molecule, Real-Time (SMRT) Sequencing platform.
Hybrid Assembly
Hybrid assembly of Illumina fragment reads and Pacific Biosciences single molecule reads was performed using the programs SparseAssembler42 and DBG2OLC15. First 159Gb of the high quality paired end reads were used to construct short but accurate de Bruijn graph contigs using programs SparseAssembler42 with k-mer size 51 and a skip length of 15. The program DBG2OLC15 was then used to map short contigs to PacBio SMRT sequencing reads and generate a hybrid assembly. Each PacBio read was compressed using high quality short read contigs and aligned to all other reads for structural error correction wherein chimeric PacBio reads are identified and trimmed. A read overlap-based assembly graph was generated and unbranched linear regions of the graph were output as the initial assembly backbones. Consensus sequences for the backbones were generated by joining overlapped raw sequencing reads and short read contigs. In practice, many regions of the initial consensus sequences can be erroneous due to the high error rates of the PacBio reads. In order to polish each backbone, all related PacBio reads and contigs are first collected and realigned using Sparc43 to calculate the most likely consensus sequence for the genome.
Scaffolding
Scaffolding of the hybrid assembly was performed using SSPACE 2.044 to incorporate mate pair data, followed by ALLMAPS version 0.5.316 to incorporate optical mapping (BioNano), linked-read (Dovetail) and previously-published meiotic mapping data4. Scaffolding by SSPACE imposed a stringent scaffolding threshold requiring 5 or more consistent linkages to support scaffolding of any pair of contigs. Scaffolding via ALLMAPS was implemented with default parameters and with equal weights assigned to all three types of mapping data with initial anchoring to meiotic maps. For scaffolds without linkage mapping data, additional ALLMAPS runs were performed using the remaining datasets. Conflicts among the three mapping methods were resolved by majority rule or by manually breaking contigs that could not be placed by majority rule.
Meiotic Mapping E. tridentatus
A meiotic map was generated for E. tridentatus using a single outbred adult pair collected from Willamette Falls (Oregon City, OR, USA) and from which larvae were artificially propagated in May 2013 at the USGS Columbia River Cook Laboratory (Cook, WA, USA) and reared for 2 weeks until they were sacrificed after hatching at around ~10 mm in total length. Restriction site–associated DNA sequencing (RAD-seq; Miller et al. 2007) Illumina sequencing libraries were prepared using a modified version of a previously published protocol (Miller et al. 2012). A total of 250 ng of DNA from each sample was added to a 100 μL restriction digest using the Sbf1 restriction enzyme (New England Biolabs, Ipswich, MA, USA). Each sample was then tagged by ligation of one of 96 unique barcoded adapters (P1 adapter) to the Sbf1 site. Once barcoded, the samples were mixed together into three libraries of 96 individuals per library, and approximately 4 μg of each was sheared using a Bioruptor UCD-300 instrument (Diagenode, Denville, NJ, USA). Following sonication, each library was concentrated using the Qiagen MinElute PCR purification kit (Qiagen) in preparation for size selection by agarose gel electrophoresis. Prior to sequencing RAD-seq libraries were quantified by qPCR and Illumina library quantification standards (Kappa Biosystems Inc, Woburn, MA, USA) on an ABI 7900HT Sequence Detection System (Life Technologies). Libraries were sequenced with single-end 100-bp reads on an Illumina HiSeq2000 sequencer (Illumina Inc., San Diego, CA, USA). Genotypes from 94 individuals with the greatest marker densities were used to reconstruct a consensus meiotic map from maternal and paternal meiosis. Maximum likelihood mapping and manual curation were performed using the JoinMap software package with default parameters for an outbred crossing design, except that the number of optimization rounds was increased to ten in order to better optimize the internal ordering of markers45,46.
Annotation
Identification of Repetitive Elements
Repeats were identified within assembled scaffolds using RepeatModeler20 and annotated using RepeatMasker version open-4.0.521 (see URLs) and a library of vertebrate repeats from repbase (repeatmaskerlibraries-20140131).
Identification of Coding Sequences
Genome annotations were produced using the MAKER47–49 genome annotation pipeline, which supports re-annotation using pre-existing gene models as input. Previous Petromyzon marinus gene models (WUGSC 7.0/petMar2 assembly)50 were mapped against the new genome assembly into GFF3 format and were used as prior model input to MAKER for re-annotation. Snap51 and Augustus52,53 were also used with MAKER and were trained using the pre-existing lamprey gene models. Additional input to MAKER included previously-published mRNA-seq reads derived from lamprey embryos and testes10,12,13 and assembled using Trinity54, as well as mRNA-seq reads (NexSeq 75–100 bp paired-end) were derived from whole embryos and dissected heads at Tahara stage 20, as well as dissected embryonic dorsal neural tubes at Tahara stage 18, 20 and 21. The following protein datasets were also used: Ciona intestinalis (sea squirt)55, Lottia gigantea (limpet)56, Nematostella vectensis (sea anemone)57, Takifugu rubripes (pufferfish)58, Branchiostoma floridae (lancelet)59, Callorhinchus milii (elephant shark)60, Xenopus tropicalis (western clawed frog)61, Drosophila melanogaster (fruit fly)62, Homo sapiens (human)63,64, Mus musculus (mouse)65, Danio rerio (zebrafish)66, Hydra magnipapillata67, Trichoplax adhaerens68, and the Uniprot/Swiss-Prot protein database69,70. Protein domains were identified in final gene models using the InterProScan domain identification pipeline71–73, and putative gene functions were assigned using BLASTP74 identified homology to the Uniprot/Swiss-Prot protein database.
lncRNA annotation
Putative lncRNAs were predicted from RNA-Seq reads obtained from brain, heart, kidney, and ovary/testis sampled from two ripe adult individuals (one female, one male). In total, 8 libraries were produced using the Illumina stranded TruSeq mRNA kit (Illumina Inc.). Sequencing (single-end, directional 100 bp) was performed on a HiSeq 2000. The resulting reads were mapped to the germline genome assembly using GSNAP (v2017-04-24)75; the resulting bam files were then assembled into transcript models using StringTie (v1.3.3b)76. The following parameters were optimized in order to maximize the number of predicted lncRNAs and reduce the number of assembly artifacts: 1) Minimum isoform abundance of the predicted transcripts as a fraction of the most abundant transcript assembled at a given locus: lower abundance transcripts are often artifacts of incompletely spliced precursor of processed transcripts; 2) minimum read coverage allowed for the predicted transcripts; 3) minimum locus gap separation value: reads that are mapped closer than 10 bp distance are merged together in the same processing bundle; 4) smallest anchor length: junctions that do not have spliced reads that align across them with at least 10 bases on both sides are filtered out; 5) minimum length allowed for the predicted transcripts (200 bp); 6) minimum number of spliced reads that align across a junction (i.e. junction coverage); 7) removal of monoexonic transcripts. The resulting transcriptomes from each library were then merged into a single GTF file (--merge option in StringTie).
Transcripts overlapping (in sense) exons of the protein coding annotated genes were removed using the script FEELnc_filter.pl77. The filtered gene models file was then used to compute the Coding Potential Score (CPS) for each of the candidate non-coding transcript with the script FEELnc_codpot.pl77. In the absence of a species-specific lncRNA set, as is the case for P. marinus, the implemented machine-learning strategy requires to simulate non-coding RNA sequences to train the model by shuffling the set of mRNAs while preserving their 7-mer frequencies. This approach is based on the hypothesis that at least some lncRNAs are derived from “debris” of protein-coding genes78. The simulated data were then used to calculate the CPS cutoff separating coding (mRNAs) from non-coding (lncRNAs) using 10 fold cross-validation on the input training files in order to extract the CPS that maximizes both sensitivity and specificity.
Analysis of Conserved Synteny
Analyses of conserved synteny were performed as previously described4. Briefly, predicted protein sequences from the lamprey genome were aligned to proteins from Gar (LepOcu1: GCA_000242695.1) and Chicken (Galgal4: GCA_000002315.2) genome assemblies79. All alignments with bitscore ≥100 and ≥90% of the best match (within a species) were considered putative orthologs of each lamprey, chicken or gar gene. Groups of orthologs were filtered to remove those with more than 6 members in any given species. Enrichment of orthologs on chromosomes or chromosomal segments was assessed using χ2 tests, incorporating Yates’ correction for continuity and Bonferroni corrections for multiple testing as previously described4.
Identification and Characterization of Germline-Specific/Enriched Sequences
Single-Copy Genes
To identify germline-specific regions, we separately aligned paired end reads from blood and sperm DNA to the germline genome assembly using BWA-MEM (v.0.7.10)80 with default parameters and filtered to exclude unmapped reads and supplementary alignments (samtools v.1.2 with option: view -F2308)81. Initial coverage analyses was implemented using bedtools v2.23.082 and revealed that the modal coverage of reads from sperm DNA was slightly lower than the coverage of reads from blood, ~73X and ~80X, respectively, but contained a larger amount of low-copy DNA (Supplementary Figure 12). To identify germline-enriched intervals, data were filtered to remove regions with coverage both from sperm and blood of < 10 (underrepresented regions: computed with genomecov -bga, bedtools v2.23.0) and also regions with coverage exceeding three times the modal value in sperm or blood (high-copy regions). The remaining data were processed to generate coverage ratios for discreet intervals containing 1,000 bp (or >500 bp at contig ends) of approximately single-copy sequence. Identification of contiguous intervals and re-estimation of coverage ratios was performed using DNAcopy version 1.42.083 after removing trailing windows that were less that 500bp in length. Ontology analyses used naming assignments that were generated using multispecies blast alignments via MAKER47–49 and were performed using Enrichr33.
Repetitive Sequences
High-identity repetitive elements were assembled de-novo from k-mers (K=31) that were abundant in sperm and blood reads, with k-mer counting via Jellyfish version 2.2.484 and assembly using Velvet version 1.2.1085. Copy number thresholds for abundant k-mers set at 3X modal copy numbers for 31-mers: 165 for sperm and 180 for blood. Abundant k-mers from sperm and blood were combined and used as a single-end reads for Velvet running with 29-mers. These analyses resulted in de novo repeat library with 130,632 sequences (overall length ~11Mb with individual contigs lengths range from 57 bases to 15.5 kb). These repeats were annotated using RepeatMasker version open-4.0.521 (see URLs) and repeat libraries generated for the germline assembly and from Repbase (repeatmaskerlibraries-20140131: “vertebrate repeats”).
For downstream analyses we used a set of model repeats representing the union of de novo repeats, those identified within assembled genomic sequences via RepeatModeler20 and an updated assembly of the previously-identified Germ1 element8. Enrichment analyses were performed by separately aligning paired end reads from blood and sperm DNA to the repeat dataset. As with single-copy sequence, alignments were pre-filtered to exclude unmapped reads and supplementary alignments. The remaining data were processed to generate average coverage ratios for intervals of approximately 100bp.
Manual curation of HOX Clusters
Manual curation of gene models was carried out using Apollo86 implemented in JBrowse87. Indels in the assembly were identified and corrected by comparison with RNAseq and genomic DNA re-sequencing data. Gene predictions from Maker were refined based on whole embryo RNA-seq data from multiple developmental stages and homology with gene sequences from other vertebrates.
In addition to the 42 clustered Hox genes in the genome assembly, 6 further Hox genes were predicted that did not fall within the 6 HOX clusters. To investigate these genes further, the genomic scaffolds harboring these gene loci were extracted and used as queries for alignment against the assembly by BLAST88. Five of these gene loci (homologs of hoxA3, D8, C9, B13 and B13a) were found to align with high sequence similarity (>97% identity) across long stretches of their sequence (>4kb, containing predicted Hox coding sequence and flanking, non-coding sequence) to loci of individual members of the 42 clustered lamprey Hox genes (Supplementary Table 13). These loci could represent either recent duplications of Hox loci or could be assembly artifacts arising from the relatively high heterozygosity of the lamprey genome. Based on their exceptionally high levels of coding and non-coding sequence similarity to clustered Hox loci, we infer that these 5 loci are assembly artifacts due to polymorphism and that they do not represent additional singleton Hox genes in the lamprey genome. The 6th predicted singleton Hox gene shows equal levels of homology to ANTP-class homeobox genes of both Hox and non-Hox families, suggesting it is a derived ANTP-class homeobox gene and not necessarily a Hox gene.
Phylogenetic analysis of Hox genes
Phylogenetic analysis was performed on Hox paralog groups with 4 or more members in sea lamprey: groups 4, 8, 9, 11 and 13. For each paralog group, predicted Sea lamprey Hox protein sequences were aligned against homologs from other vertebrate species and amphioxus, retrieved from Genbank. Our approach was informed by the experiences detailed by Kuraku et al89, Qiu et al90, Mehta et al17 and Manousaki et al91. In selecting jawed vertebrate taxa for these analyses, we avoided teleost fish and Xenopus laevis as these lineages have undergone additional genome duplication events, which can lead to their co-orthologous genes/proteins being more derived than those from non-duplicated lineages. Thus, we opted for Elephant shark (C. milii) and coelacanth (L. menadoensis) as Chondrichthian and ‘basal’ Sarcopterygian representatives respectively, both of which having slowly evolving protein-coding genes and well characterized Hox gene complements92,93. Urochordates are the sister group of vertebrates but the divergent nature of their Hox genes led us to favor the cephalochordate amphioxus as a source for outgroup sequences in our analyses. We chose to perform protein alignments rather than DNA alignments due to the high coding GC content in lamprey, which can result in artifactual clustering of lamprey genes in DNA trees. Nevertheless, the unique pattern of amino-acid composition in lamprey proteins is an unavoidable complicating factor that impinges on their phylogenetic analysis and can lead to artifactual clustering of lamprey proteins, as described in Qiu et al90. The MEGA741 software suite was used for sequence alignment, best-fit substitution model evaluation and phylogeny reconstruction. Protein alignments were performed with full available length protein sequences using MUSCLE41. Best-fit substitution models were evaluated and chosen for each alignment. Maximum likelihood, neighbor joining and maximum parsimony approaches were used for phylogenetic analysis, with 100 bootstrap replicates generated for node support. For each method, all positions in the alignment containing gaps and missing data were eliminated.
Code Availability
Custom code (DifCover) is available on GitHub (see URLs)
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM104123 to JJS, the Stowers Institute under award number SIMR-1001 to HJP, MEC, LMW, SR and RK and the Bonneville Power Administration to JEH and SRN. EEE is an investigator of the Howard Hughes Medical Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Support and resources from the Center for High Performance Computing at the University of Utah are gratefully acknowledged. Additional computational support was provided by The University of Kentucky High Performance Computing complex.
Footnotes
AUTHOR CONTRIBUTIONS
JJS, REK, CTA and GE conceived of the study. JJS, NT, CY, CH, MCK, HJP, MEC, JEH, SRN, VAT, CW, CS, HK, FL, LMW, SR, CB, EEE, DH, TS-S, MY and REK contributed analyses. JJS, NT, MCK, HJP and REK wrote the manuscript.
COMPETING FINANCIAL INTERESTS STATEMENT
EEE is on the scientific advisory board (SAB) of DNAnexus, Inc.
RESEARCH ANIMALS
This study complied with all relevant ethical guidelines and was performed under protocol number 2011-0848 (University of Kentucky Institutional Animal Care and Use Committee).
URLs
SIMRbase /Lamprey Genome Browser:
https://genomes.stowers.org/organism/Petromyzon/marinus
DifCover: https://github.com/timnat/DifCover
RepeatMasker: http://www.repeatmasker.org
Original data pertaining to the Chicago assembly (Dovetail) and Hox cluster curation can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/LIBPB-1215.
DATA AVAILABILITY
Accession Numbers
Genome Assembly: (NCBI Genome: PIZI00000000.1, BioProject: PRJNA357048). Raw sequence data used for genome assembly (NCBI SRA: SRR5503831-43). Re-sequencing data for detection of eliminated segments (NCBI SRA: SRR5535434-5). Previously published RNAseq data were used for annotation (NCBI SRA: SRX110029.2 - SRX110035.2)13, (NCBI SRA: SRX1483277 - SRX1483282)12 and (NCBI SRA: SRX104180)10.
References
- 1.Parker HJ, Bronner ME, Krumlauf R. A Hox regulatory network of hindbrain segmentation is conserved to the base of vertebrates. Nature. 2014;514:490–493. doi: 10.1038/nature13723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green SA, Simoes-Costa M, Bronner ME. Evolution of vertebrates as viewed from the crest. Nature. 2015;520:474–482. doi: 10.1038/nature14436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sower SA, et al. Emergence of an Ancestral Glycoprotein Hormone in the Pituitary of the Sea Lamprey, a Basal Vertebrate. Endocrinology. 2015;156:3026–3037. doi: 10.1210/en.2014-1797. [DOI] [PubMed] [Google Scholar]
- 4.Smith JJ, Keinath MC. The sea lamprey meiotic map improves resolution of ancient vertebrate genome duplications. Genome Res. 2015;25:1081–1090. doi: 10.1101/gr.184135.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das S, et al. Evolution of two prototypic T cell lineages. Cell Immunol. 2015;296:87–94. doi: 10.1016/j.cellimm.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doolittle RF. Bioinformatic Characterization of Genes and Proteins Involved in Blood Clotting in Lampreys. J Mol Evol. 2015;81:121–130. doi: 10.1007/s00239-015-9701-0. [DOI] [PubMed] [Google Scholar]
- 7.McCauley DW, Docker MF, Wyhard S, Li W. Lampreys as Diverse Model Organisms in the Genomics Era. BioScience. 2015;65:1046–1056. doi: 10.1093/biosci/biv139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith JJ, Antonacci F, Eichler EE, Amemiya CT. Programmed loss of millions of base pairs from a vertebrate genome. Proc Natl Acad Sci USA. 2009;106:11212–11217. doi: 10.1073/pnas.0902358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith JJ, Stuart AB, Sauka-Spengler T, Clifton SW, Amemiya CT. Development and analysis of a germline BAC resource for the sea lamprey, a vertebrate that undergoes substantial chromatin diminution. Chromosoma. 2010;119:381–389. doi: 10.1007/s00412-010-0263-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith JJ, Baker C, Eichler EE, Amemiya CT. Genetic consequences of programmed genome rearrangement. Curr Biol. 2012;22:1524–1529. doi: 10.1016/j.cub.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Timoshevskiy VA, Herdy JR, Keinath MC, Smith JJ. Cellular and Molecular Features of Developmentally Programmed Genome Rearrangement in a Vertebrate (Sea Lamprey: Petromyzon marinus) PLoS Genet. 2016;12:e1006103. doi: 10.1371/journal.pgen.1006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryant SR, Herdy JR, Amemiya CT, Smith JJ. Characterization of Somatically-Eliminated Genes During Development: Lamprey (Petromyzon marinus) Mol Biol Evol. 2016;33:2337–2344. doi: 10.1093/molbev/msw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith JJ, et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. 2013;45:415–421. doi: 10.1038/ng.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Speir ML, et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 2016;44:D717–725. doi: 10.1093/nar/gkv1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye C, Hill CM, Wu S, Ruan J, Ma ZS. DBG2OLC: Efficient Assembly of Large Genomes Using Long Erroneous Reads of the Third Generation Sequencing Technologies. Scientific reports. 2016;6:31900. doi: 10.1038/srep31900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang H, et al. ALLMAPS: robust scaffold ordering based on multiple maps. Genome Biol. 2015;16:3. doi: 10.1186/s13059-014-0573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehta TK, et al. Evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum) Proc Natl Acad Sci U S A. 2013;110:16044–16049. doi: 10.1073/pnas.1315760110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuraku S, Kuratani S. Time scale for cyclostome evolution inferred with a phylogenetic diagnosis of hagfish and lamprey cDNA sequences. Zoolog Sci. 2006;23:1053–1064. doi: 10.2108/zsj.23.1053. [DOI] [PubMed] [Google Scholar]
- 19.Lampman R, et al. In: Jawless Fishes of the World. Orlov A, Beamish R, editors. Vol. 2. Cambridge Scholars Publishing; 2016. pp. 160–195. Ch 22. [Google Scholar]
- 20.Smit AFA, Hubley R. RepeatModeler Open-1.0. 2015. [Google Scholar]
- 21.Smit AFA, Hubley R, Green P. RepeatMasker Open-4.0. 2015. [Google Scholar]
- 22.Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011;12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 24.Hara Y, et al. Optimizing and benchmarking de novo transcriptome sequencing: from library preparation to assembly evaluation. BMC Genomics. 2015;16:977. doi: 10.1186/s12864-015-2007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warren WC, et al. A New Chicken Genome Assembly Provides Insight into Avian Genome Structure. G3 (Bethesda) 2017;7:109–117. doi: 10.1534/g3.116.035923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braasch I, et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet. 2016;48:427–437. doi: 10.1038/ng.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohno S. Evolution by Gene Duplication. Springer-Verlag; 1970. [Google Scholar]
- 28.Holland PW, Garcia-Fernandez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Dev Suppl. 1994:125–133. [PubMed] [Google Scholar]
- 29.Nakatani Y, Takeda H, Kohara Y, Morishita S. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 2007;17:1254–1265. doi: 10.1101/gr.6316407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murat F, Van de Peer Y, Salse J. Decoding plant and animal genome plasticity from differential paleo-evolutionary patterns and processes. Genome biology and evolution. 2012;4:917–928. doi: 10.1093/gbe/evs066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timoshevskiy VA, Lampman RT, Hess JE, Porter LL, Smith JJ. Deep ancestry of programmed genome rearrangement in lampreys. Dev Biol. 2017;429:31–34. doi: 10.1016/j.ydbio.2017.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lachmann A, et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sachs M, et al. Bivalent chromatin marks developmental regulatory genes in the mouse embryonic germline in vivo. Cell Rep. 2013;3:1777–1784. doi: 10.1016/j.celrep.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore CM, Best RG. Chromosomal Genetic Disease: Structural Aberrations. eLS; 2001. [Google Scholar]
- 36.Hardisty MW. Fecundity and Speciation in Lampreys. Evolution. 1963;17:17–22. [Google Scholar]
- 37.Hardisty MW, Cosh J. Primordial germ cells and fecundity. Nature. 1966;210:1370–1371. doi: 10.1038/2101370a0. [DOI] [PubMed] [Google Scholar]
- 38.Grossniklaus U, Paro R. Transcriptional silencing by polycomb-group proteins. Cold Spring Harb Perspect Biol. 2014;6:a019331. doi: 10.1101/cshperspect.a019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aloia L, Di Stefano B, Di Croce L. Polycomb complexes in stem cells and embryonic development. Development. 2013;140:2525–2534. doi: 10.1242/dev.091553. [DOI] [PubMed] [Google Scholar]
- 40.Acemel RD, et al. A single three-dimensional chromatin compartment in amphioxus indicates a stepwise evolution of vertebrate Hox bimodal regulation. Nat Genet. 2016;48:336–341. doi: 10.1038/ng.3497. [DOI] [PubMed] [Google Scholar]
- 41.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye C, Ma ZS, Cannon CH, Pop M, Yu DW. Exploiting sparseness in de novo genome assembly. BMC Bioinformatics. 2012;13(Suppl 6):S1. doi: 10.1186/1471-2105-13-S6-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye C, Ma ZS. Sparc: a sparsity-based consensus algorithm for long erroneous sequencing reads. PeerJ. 2016;4:e2016. doi: 10.7717/peerj.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 45.Stam P. Construction of Integrated Genetic-Linkage Maps by Means of a New Computer Package - Joinmap. Plant J. 1993;3:739–744. doi: 10.1111/J.1365-313x.1993.00739.X. [DOI] [Google Scholar]
- 46.Van Ooijen JW. Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet Res (Camb) 2011;93:343–349. doi: 10.1017/S0016672311000279. [DOI] [PubMed] [Google Scholar]
- 47.Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011;12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campbell MS, et al. MAKER-P: A Tool Kit for the Rapid Creation, Management, and Quality Control of Plant Genome Annotations. Plant Physiology. 2014;164:513–524. doi: 10.1104/pp.113.230144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cantarel BL, et al. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hwang JY, Smith S, Myung K. The Rad1-Rad10 complex promotes the production of gross chromosomal rearrangements from spontaneous DNA damage in Saccharomyces cerevisiae. Genetics. 2005;169:1927–1937. doi: 10.1534/genetics.104.039768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19:ii215–225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 53.Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637– 644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- 54.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature biotechnology. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dehal P, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 56.Simakov O, et al. Insights into bilaterian evolution from three spiralian genomes. Nature. 2013;493:526–531. doi: 10.1038/nature11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Putnam NH, et al. Sea Anemone Genome Reveals Ancestral Eumetazoan Gene Repertoire and Genomic Organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 58.Aparicio S, et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- 59.Putnam NH, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 60.Venkatesh B, et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature. 2014;505:174–179. doi: 10.1038/nature12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hellsten U, et al. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. 8392 [pii] [DOI] [PubMed] [Google Scholar]
- 63.Venter JC, et al. The Sequence of the Human Genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 64.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 65.Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 66.Howe K, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chapman JA, et al. The dynamic genome of Hydra. Nature. 2010;464:592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Srivastava M, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- 69.Consortium TU. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Research. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucl Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.The InterPro C et al. InterPro: An integrated documentation resource for protein families, domains and functional sites. Brief Bioinform. 2002;3:225–235. doi: 10.1093/bib/3.3.225. [DOI] [PubMed] [Google Scholar]
- 72.Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quevillon E, et al. InterProScan: protein domains identifier. Nucl Acids Res. 2005;33:W116–120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ. Basic Local Alignment Search Tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 75.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pertea M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wucher V, et al. FEELnc: a tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res. 2017;45:e57. doi: 10.1093/nar/gkw1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duret L, Chureau C, Samain S, Weissenbach J, Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312:1653–1655. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- 79.Yates A, et al. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 84.Marcais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee E, et al. Web Apollo: a web-based genomic annotation editing platform. Genome Biol. 2013;14:R93. doi: 10.1186/gb-2013-14-8-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH. JBrowse: a next-generation genome browser. Genome Res. 2009;19:1630–1638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kuraku S, Meyer A, Kuratani S. Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Mol Biol Evol. 2009;26:47–59. doi: 10.1093/molbev/msn222. [DOI] [PubMed] [Google Scholar]
- 90.Qiu H, Hildebrand F, Kuraku S, Meyer A. Unresolved orthology and peculiar coding sequence properties of lamprey genes: the KCNA gene family as test case. BMC Genomics. 2011;12:325. doi: 10.1186/1471-2164-12-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Manousaki T, et al. In: Jawless fishes of the world. Orlov A, Beamish R, editors. Vol. 1. Cambridge Scholars; 2016. pp. 2–16. [Google Scholar]
- 92.Ravi V, et al. Elephant shark (Callorhinchus milii) provides insights into the evolution of Hox gene clusters in gnathostomes. Proc Natl Acad Sci U S A. 2009;106:16327–16332. doi: 10.1073/pnas.0907914106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Amemiya CT, et al. Complete HOX cluster characterization of the coelacanth provides further evidence for slow evolution of its genome. Proc Natl Acad Sci USA. 2010;107:3622–3627. doi: 10.1073/pnas.0914312107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.