Abstract
In yeast, the U2 small nuclear RNA (snRNA) component of the spliceosome is targeted for additional post-transcriptional modifications in response to cellular stress. Uridines 56 and 93 are both modified to pseudouridines (Ψ) during nutrient deprivation, while U56 is also pseudouridylated during heat shock. Both positions are located within stem II, which must toggle between two mutually-exclusive structures during splicing. Stem IIa forms during spliceosome assembly, and stem IIc forms during the catalytic steps. We have studied how uridine 56 and 93 pseudouridylation impacts conformational switching of stem II. Using single molecule Förster resonance energy transfer (smFRET), we show that Ψ56 dampens conformational dynamics of stem II and stabilizes stem IIc. In contrast, Ψ93 increases dynamics of non-stem IIc conformations. Pseudouridylation impacts conformational switching of stem II by Mg2+ or the U2 protein Cus2; however, when Mg2+ and Cus2 are used in combination the impacts of pseudouridylation can be suppressed. These results show that stress-induced post-transcriptional modification of U56 and U93 alter snRNA conformational dynamics by distinct mechanisms and that protein and metal cofactors of the spliceosome alter how snRNAs respond to these modifications.
Keywords: Single-molecule FRET, snRNA, dynamics, spliceosome, pseudouridine
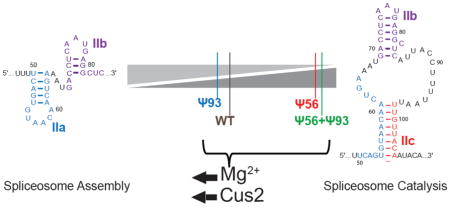
Graphical Abstract
INTRODUCTION
Post-transcriptional modifications can have far-reaching consequences on the structure and function of both coding and non-coding RNA transcripts. There are more than 100 distinct RNA modifications including pseudouridine, 2′-O-methyl-ribose and N1- and N6-methyladenosine [1–9]. Pseudouridine (Fig. 1a) is the most abundant and many sites of pseudouridylation are evolutionarily conserved [10]. Pseudouridine introduces a new hydrogen bond donor on the non-Watson Crick face of the nucleotide, which can allow for hydrogen bonding with the phosphodiester RNA backbone [11–13] or alter interactions with other molecules [14]. Additionally, pseudouridine’s carbon-carbon glycosidic linkage can increase base flexibility to permit optimal base pairing orientation and π-stacking with neighboring bases, both of which can stabilize RNA duplexes [11, 13]. Destabilization of RNA structures by pseudouridine incorporation into loop regions has also been observed [13], highlighting that pseudouridylation can have position and structure-specific effects. While many pseudouridines are constitutively incorporated into RNAs, others are inducible and appear in both non-coding and mRNAs under certain cellular conditions such as environmental or nutrient stress [2, 3, 15]. Even though pseudouridylation sites have been annotated in both Saccharomyces cerevisiae (yeast) and humans [2, 3], the impacts these modifications have on many RNA’s structures and functions are less well understood.
Figure 1.
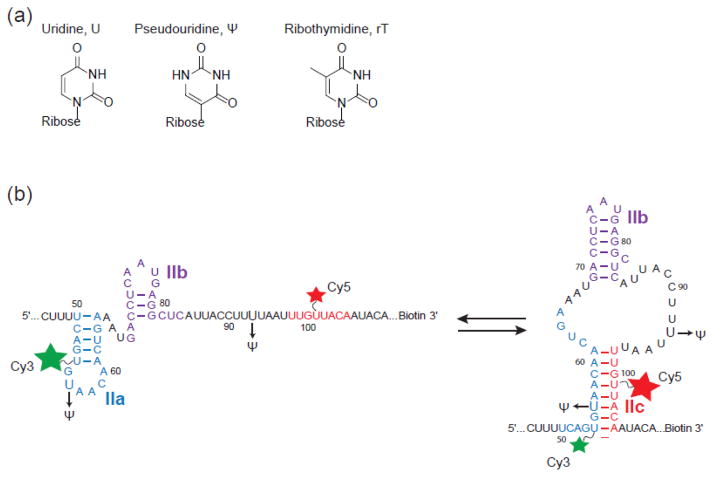
Schematic of U2 stem II model RNAs used in smFRET experiments. (a) Chemical structures of uridine, pseudouridine, ribothymidine. (b) U2 stem II fluctuates between IIa (left) and IIc (right) basepairing. The locations of fluorophore attachment (stars) and pseudouridine incorporation (Ψ) are shown. The green and red stars indicated the donor (Cy3) and acceptor (Cy5) FRET fluorophores, respectively. Basepairing schemes depict stem II conformations observed in the spliceosome by cryo-electron microscopy (cryo-EM) before activation (stem IIa) and after the first catalytic step (stem IIc) [39, 42].
The snRNA components of the spliceosome contain sites of both constitutive and inducible pseudouridylation [16]. The spliceosome is responsible for removing intronic sequences from pre-mRNAs, and the splicing machinery requires five distinct small nuclear ribonucleoproteins (snRNPs) for function [17]. Each snRNP (U1, U2, U4, U5, and U6) is composed of a uridine-rich snRNA and a host of protein cofactors. The U1, U2, and U5 snRNAs contain a number of constitutively incorporated pseudouridines [18] and these modifications have been shown to impact splicing activity [19–21]. Constitutive pseudouridylation of the U2 snRNA at positions 42 and 44 appears to be important for formation of a snRNA structure needed for pairing between the snRNA and branchsite (BS) [22]. Once paired, pseudouridylation at position 35 may help to stabilize this duplex [23–25]. Inducible pseudouridylation of the snRNAs may impart particular functions to the spliceosome that are necessary for certain cellular events. For example, pseudouridylation of the U6 snRNA is required for yeast filamentous growth but not at other times [26]. The functions of inducible pseudouridines in the U2 snRNA are less clear.
Under stress conditions, pseudouridines are induced at positions 56 and 93 of the U2 snRNA [15, 27]. Pseudouridine 56 (Ψ56) is incorporated by Pus7 during heat stress and to a lesser extent during nutrient deprivation [15]. In contrast, Ψ93 is incorporated by the H/ACA snR81 snoRNP via the environment-sensing TOR signaling pathway only during nutrient deprivation [15, 27]. While nutrient deprivation results in formation of both Ψ56 and Ψ93, their simultaneous presence in the same RNA molecule has not yet been confirmed.
Interestingly, both Ψ56 and Ψ93 are found within the conformationally dynamic stem II region which adopts two mutually exclusive structures during splicing (Fig. 1b) [28, 29]. Stem IIa is required for spliceosome assembly and likely forms transiently while the spliceosome active site rearranges after 5′ splice site (SS) cleavage and before exon ligation [30]. In contrast, stem IIc is present during these catalytic steps [31, 32]. Thus, distinct conformations of stem II can promote or inhibit spliceosome assembly or catalysis [32, 33]. Recently, smFRET has been used to study stem II dynamics in the absence of the spliceosome [34]. This work revealed that this region of the snRNA is intrinsically dynamic and that other, trans-acting factors can influence these dynamics. Specifically, Mg2+ and the U2 accessory protein Cus2 [35] both were able to switch stem II away from a predominant IIc conformation and towards IIa. Given the role of stem II in splicing and its intrinsic conformational flexibility, the discovery of inducible pseudouridines in this region suggests that post-transcriptional modification may directly impact either spliceosome assembly or catalysis by altering stem II dynamics. Indeed, Ψ93 appears to influence exon ligation in yeast when non-consensus 3′ SS are present [15].
To gain further insight into the consequences of pseudouridylation of the U2 snRNA, we have used smFRET to directly study the structure and dynamics of a stem II model RNA in the presence and absence of these modifications. Both modifications alter the dynamics of the model RNA and the relative abundance of stem IIc relative to other RNA structures. However, the influences of specific modifications are distinct: Ψ56 stabilizes stem IIc while Ψ93 increases the conformational dynamics of RNAs in non-stem IIc conformations. Moreover, both modifications impact how stem II responds to protein and metal ion cofactors. Together these data reveal how RNA post-transcriptional modifications can influence a conformational switch found within the U2 snRNA.
RESULTS
Our laboratory has previously used smFRET to study stem II conformational dynamics [34]. In those studies, a model RNA containing stem II was prepared by splinted ligation of two RNA fragments containing Cy3 and Cy5 fluorophores attached to uridines 56 and 101, respectively. The model RNA reversibly switched between multiple conformations identified as stem IIc (high FRET) and a set of conformations consistent with stem IIa pairing (mid-FRET). However, Cy3 incorporation at uridine 56 precludes analysis of the impact of pseudouridylation at this same position. Therefore, we prepared a new stem II smFRET reporter in which Cy3 was attached to uridine 54 to allow for inclusion of Ψ56 (Fig. 1b). As with the previous reporter, we predicted that this RNA would exhibit high FRET upon stem IIc formation and a mid-FRET signal due to stem IIa conformers.
The new stem II smFRET reporter with Cy3 at position 54 reproduced the behaviors observed with the previous RNA. Time trajectories from Cy3 and Cy5 fluorophores located on individual RNA molecules showed anti-correlated changes in fluorescence intensity (Fig. 2a). These values were used to determine FRET efficiency values (EFRET) which confirmed the expected transitions between high and medium FRET (Fig. 2b). When EFRET trajectories from many molecules were combined, they showed a sharp peak near a high EFRET of 0.96 and a broader distribution of EFRET from ~0.5–0.9 (Fig. 3a). Based on our previous work, we predict that the high EFRET peak corresponds to stem IIc formation while the broad distribution of EFRET values represents a set of conformations that form upon stem IIa pairing. In addition, the new smFRET reporter replicated the destabilization of stem IIc upon addition of Mg2+ that was previously observed (Fig 3a, b). Movement of the fluorophore from uridine 56 to uridine 54 resulted in a slight stabilization of stem IIc in the presence of Mg2+ since some high EFRET was still observed at 10 mM Mg2+, unlike with the previous reporter. Stem IIa also appeared to be more homogeneous in the presence of Mg2+ with the new reporter since we now observed a much sharper peak centered near EFRET of 0.75 (Fig. 3a, b) rather than the broad distribution noted earlier at 0.54. Similar data were obtained when the new reporter was incubated with the Cus2 protein, which also results in destabilization of stem IIc and accumulation of stem IIa (Fig. 4a). Since the new smFRET reporter recapitulated the overall features of reversible and spontaneous conformational switching of stem II as well as the RNA’s response to Mg2+ and Cus2, we next determined how pseudouridine influenced these behaviors.
Figure 2.
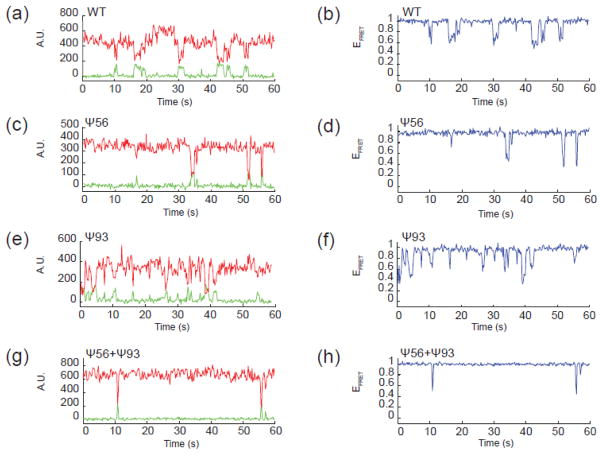
Representative single molecule fluorescence data for U2 stem II model RNAs. (a, c, e, g) Fluorescence intensity data collected from a single RNA molecule by excitation of the Cy3 FRET donor and by simultaneously monitoring Cy3 (green) and Cy5 (red) fluorescence emission and reported in arbitrary fluorescence units (A.U.). Anti-correlated changes in Cy3 and Cy5 fluorescence intensity are observed for both the WT RNA (a) and each variant containing the indicated Ψ bases (c, e, g). (b, d, f, h) EFRET values calculated from raw fluorescence data shown in (a, c, e, g) for each RNA molecule.
Figure 3.
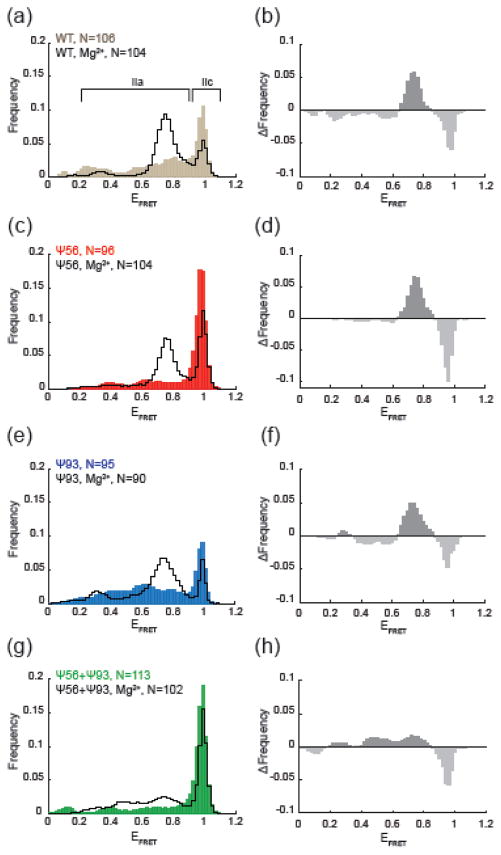
Influence of pseudouridylation on Mg2+-dependent conformational switching. (a, c, e, g) Histograms of EFRET values obtained from the indicated number (N) of single RNA molecules in the absence (filled, colored bars) or presence of 10 mM Mg2+ (black lines). (b,d,f,h) Changes in EFRET due to Mg2+ addition for each of the indicated RNAs. Each plot represents the result of subtraction of the histogram obtained in the absence of Mg2+ from the histogram obtained in the presence of Mg2+.
Figure 4.
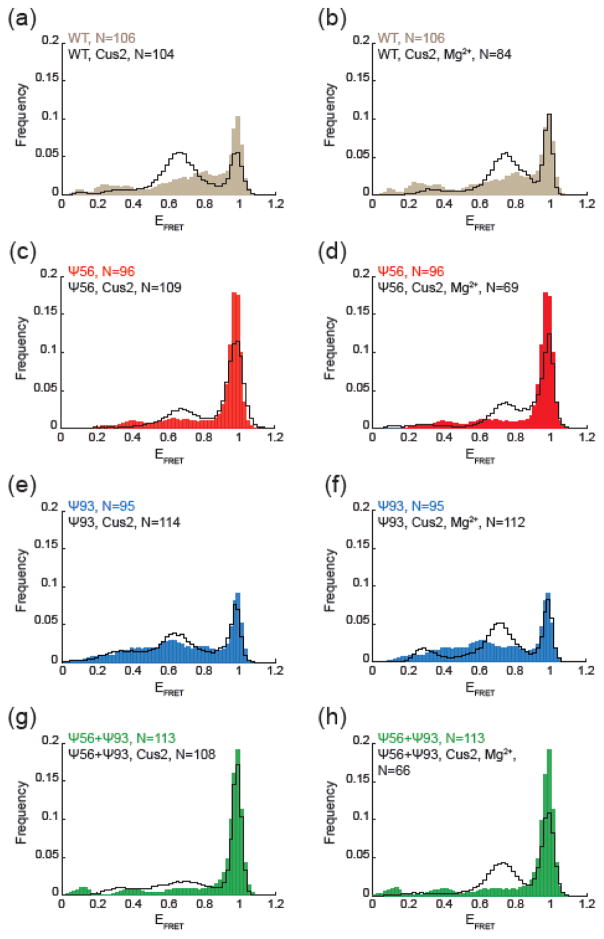
Influence of pseudouridylation on Cus2-dependent conformational switching. (a, c, e, g) Histograms of EFRET values obtained from the indicated number (N) of single RNA molecules in the absence (filled, colored bars) or presence of 6 μM Cus2 (black lines). (b, d, f, h) Histograms of EFRET values obtained from the indicated number (N) of single RNA molecules in the absence (filled, colored bars) or presence of both 6 μM Cus2 and 10 mM Mg2+ (black lines).
Pseudouridylation Changes Stem II Dynamics
We next prepared smFRET reporter RNAs by splinted ligation of synthetic oligonucleotides containing pseudouridine at position 56 (Ψ56), position 93 (Ψ93), or at both positions (Ψ56+Ψ93). Analysis of the fluorescence trajectories of single molecules of each RNA variant indicated that pseudouridylation was impacting stem II dynamics. In RNAs containing Ψ56, we observed fewer transitions of the RNA out of the high EFRET, stem IIc state suggesting a suppression of dynamics and stabilization of this conformation (Fig 2c, d). Histogram analysis confirmed this result and showed a sharp peak at high EFRET (Fig. 3c). Quantification of the EFRET data showed that this peak resulted from a 1.7-fold increase in the fraction of high EFRET observations (those >0.9) in comparison to the wild type (WT) reporter.
By comparison, Ψ93 incorporation did not cause a similar increase in stem IIc. Rather, Ψ93 RNAs show increased and more varied transitions between EFRET states in comparison to the WT reporter (Fig. 2e, f). Histogram analysis confirmed broadening of EFRET observations <0.9 as well as accumulation of EFRET values near 0.4 and 0.6 relative to other RNAs (Fig. 3e). These increased numbers of observations at mid-EFRET values are accompanied by a ~20% decrease in the fraction of EFRET observations >0.9, indicating a slightly lower abundance of stem IIc in the Ψ93 RNA. When Ψ56 and Ψ93 were simultaneously present in the same molecule, the impact of Ψ56 was dominant and few transitions out of a long-lived high EFRET state were observed (Fig. 2g, h; Fig. 3g). Stabilization of high EFRET by Ψ56 likely prevented observation of increased dynamics in the low- to mid-EFRET range due to Ψ93.
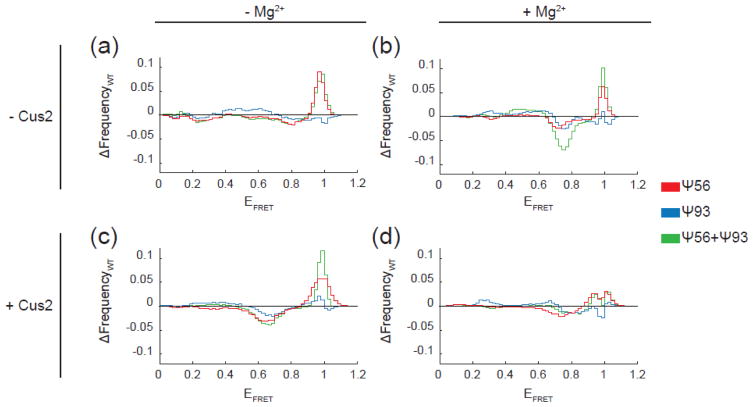
To more easily visualize the specific impacts of Ψ56 and Ψ93, we constructed EFRET difference histograms by subtracting the WT histogram from those obtained after pseudouridine incorporation (Fig. 5a). In these difference histograms, positive peaks represent EFRET states stabilized or increasingly populated by pseudouridine incorporation and negative peaks represent EFRET states repressed by pseudouridine. The difference histogram clearly shows an increase in high EFRET observations due to Ψ56 even when Ψ93 is also present (Fig. 5a, red and green lines). This is consistent with Ψ56 stabilizing stem IIc, which includes Ψ56 paired to A103. Ψ93 shows little change in the high EFRET peak but instead shows a small but very broadly distributed increase in EFRET observations from 0.3–0.7 (Fig. 5a, blue line). Since Ψ93 is not predicted to be stably base-paired in either stem IIa or IIc, this suggests that Ψ93 may more subtly influence stem II dynamics, particularly once the RNA has transitioned out of stem IIc.
Figure 5.
The impact of pseudouridylation on stem II switching in comparison to the unmodified RNA. Each plot represents the result of subtraction of the histogram obtained from the WT RNA under the indicated condition from the histogram obtained using RNA containing one or more pseudouridines (see Fig. 3 and 4). Signals above zero indicate an increase in observances of those particular EFRET values due to pseudouridine incorporation, while signals below zero indicate a reduction in observances of those EFRET values.
Pseudouridylation Dampens Mg2+-Dependent Conformational Switching
We next tested each reporter RNA for the ability to undergo conformational switching in the presence of Mg2+. RNAs containing Ψ56 or Ψ93 still switched out of IIc and towards IIa in the presence of Mg2+ (Fig. 3a, e). Difference histograms comparing each RNA in the absence and presence of Mg2+ showed the loss of high EFRET and gain of mid-EFRET signals consistent with the switch (Fig. 3b, d, f). Ψ56 stabilization of stem IIc persisted in the presence of Mg2+ since we observed an increased abundance of high EFRET signals relative to WT (Fig. 3a vs. Fig. 3c and Fig. 5b). Ψ93 also did not prevent accumulation of stem IIa in response to Mg2+ (Fig. 3e and Fig. 5b). Surprisingly, while both Ψ56 and Ψ93 permitted Mg2+ dependent switching in isolation, their combinatorial effect is not additive. The combination of Ψ56 and Ψ93 resulted in RNAs with a much dampened response to Mg2+ that maintained a predominant stem IIc conformation with only a smaller increase in stem IIa EFRET signals relative to WT (Fig. 3g,h and Fig. 5b).
We examined RNAs containing substitutions at positions 56 and 93 further by introduction of ribothymidine (rT). This nucleotide lacks the hydrogen bond donor present at the non-Watson Crick face of pseudouridine; however, it introduces steric bulk at this same position due to the methyl group (Fig. 1a). RNAs containing only rT56 or rT93 behaved similarly to their pseudouridine counterparts both in the presence or absence of Mg2+ (Sup. Fig. 1a, b). This indicates that N1 hydrogen bonding is not required for Ψ56 or Ψ93 to alter stem II structural dynamics. However, the RNA containing both rT56 and rT93 behaved similarly to the WT RNA under both conditions and failed to induce stem IIc stabilization or resistance to Mg2+ (Sup. Fig. 1c). This indicates that stabilization of stem IIc in the Ψ56+Ψ93 smFRET reporter RNA requires the presence of pseudouridine.
Cus2 and Mg2+ Together Suppress the Impact of Pseudouridylation on Stem II
To examine if pseudouridylation impacted the ability of the U2 protein Cus2 to trigger conformational switching as may occur during spliceosome assembly, we carried out smFRET experiments either in the presence of Cus2 alone or in combination with Mg2+. Electrophoretic mobility shift assays (EMSAs) confirmed that Cus2 interacted and shifted WT as well as each pseudouridylated RNA similarly in the presence or absence of Mg2+ (Sup. Fig. 2). As previously noted [34, 35], Cus2 binding of stem II results in two slower-migrating bands by EMSA, which is possibly related to the structural dynamics of stem II itself under non-denaturing conditions.
In our smFRET assay, addition of Cus2 caused a decrease in stem IIc EFRET signals and an increase in signals at lower EFRET (Fig. 4a–d). Protein-dependent conformational switching followed the same trends observed for Mg2+-dependent switching: Ψ56 RNAs in stem IIc were more resistant to switching than WT, Ψ93 RNAs showed similar switching to WT, and the combination of Ψ56 and Ψ93 resulted in RNAs with stabilized stem IIc and minimal sensitivity to Cus2 (Fig. 5c). In all cases, the peaks that appeared in the presence of Cus2 were slightly shifted towards lower EFRET values than those that appeared in response to Mg2+ (~0.65 vs. ~0.75). This is consistent with previous observations using a different stem II reporter that suggested distinct effects of protein and metal ions on stem II structure [34].
When Mg2+ and Cus2 were used in combination, all RNAs (including Ψ56+Ψ93) were able to depopulate the stem IIc conformation and accumulate stem IIa (Fig. 4e–h). In fact, the resulting distribution of EFRET states under these conditions most closely resembled that observed with the WT RNA for each of the reporters, regardless of the pseudouridylation state (Fig. 5d). Together our smFRET data indicate that while pseudouridylation has distinct effects on conformational dynamics of the stem II region of U2 (Fig. 3), these effects can be suppressed by other factors such as the combination of Mg2+ and snRNP proteins.
RNase T1 Probing of Pseudouridine-Containing Stem II RNAs
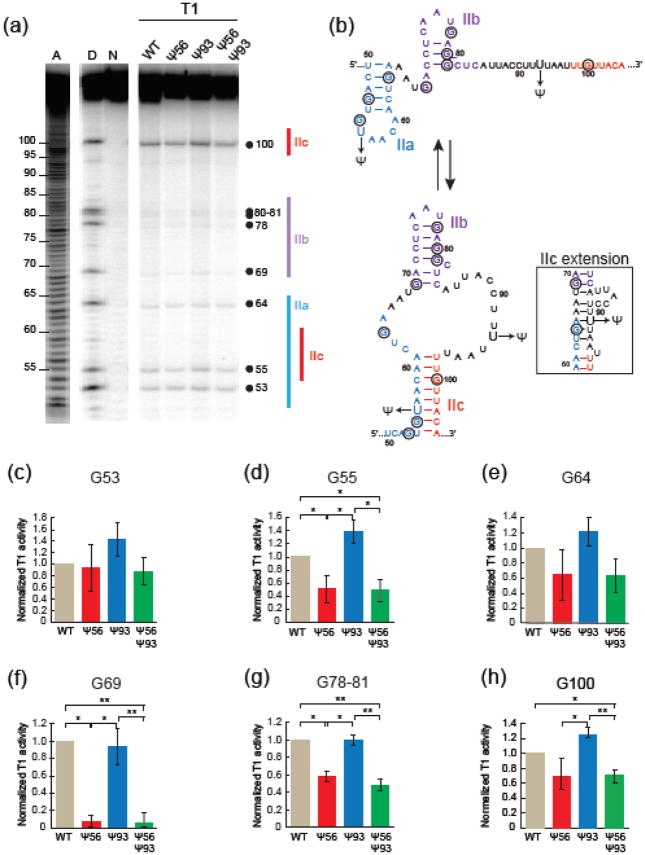
Given the sensitive nature of stem II to structural perturbation, it is possible that the fluorophores used to study FRET also are changing the RNA dynamics. To examine the impact of pseudouridylation on stem II conformation in solution and without fluorophore attachment, we structurally probed each RNA using RNase T1 (Fig. 6a). RNase T1 preferentially cleaves RNA 3′ of single stranded guanosines (G) [36]. We have previously shown that cleavage of stem II at G100 correlates well with RNA structure since this nucleotide is unpaired in stem IIa but paired in stem IIc (Fig. 6b) [34]. We analyzed cleavage at this position in addition to the eight other guanosines present in the WT, Ψ56, Ψ93, and Ψ56+Ψ93 RNAs (Fig. 6c–h). G55 is also predicted to be paired in stem IIc but not IIa and exhibit reduced cleavage upon stem IIc formation. Consistent with this and with the smFRET results (Fig. 2), G55 showed reduced cleavage in the Ψ56 and Ψ56+Ψ93 RNAs in comparison with WT (Fig. 6d). Ψ93, on the other hand, more closely resembled WT in both this and the smFRET assays. Similar results were obtained at G100, with Ψ56+Ψ93 showing reduced cleavage in comparison with WT and Ψ93. Cleavage at G100 also supported more stem IIc formation in the Ψ56 RNA in comparison to the Ψ93 RNA, although cleavage at this position in Ψ56 and WT was more comparable.
Figure 6.
Structural probing of U2 stem II RNAs by RNase T1 protection assay. (a) Denaturing polyacrylamide gel of 5′-[P32]-labeled stem II RNAs incubated with RNase T1. The lanes labeled “A”, “D”, and “N” are standards representing a RNA ladder generated by alkaline hydrolysis, cleavage carried out under RNA denaturing conditions to identify the position of each guanosine, and the native intact RNA, respectively. The position of each guanosine and stem II structure are noted beside the gel. (b) 2D RNA representation of stem IIa and IIc with guanosine nucleotides circled. RNase T1 preferentially targets single-stranded guanosines. An alternative stem IIc pairing conformation, the IIc extension, is shown in the inset. (c–h) Quantification of cleavage at each guanosine within the stem II RNA. Each bar represents the average from four replicates, while error bars represent ±S.D. Data were normalized to the total band intensity in each lane as well as to the extent of cleavage of the WT RNA in each replicate. The F-test was used to determine the variance between data sets, and the t-test was then used to calculate the p-values. For clarity, comparisons which did not display significant differences in cleavage are not indicated by brackets. p ≥ 0.05; *, 0.01<p<0.05; **, 0.001<p<0.01.
While G53 is predicted to be paired in stem IIa and unpaired in stem IIc, we did not detect significant differences in cleavage at this position (Fig. 6c). Surprisingly, pseudouridine incorporation also changed cleavage patterns within stem IIb. G69 and G78-81 showed differences in cleavage, again with Ψ56 and Ψ56+Ψ93 showing reduced cleavage in comparison to WT and with Ψ93 showing similar cleavage to WT (Fig. 6f, g). This supports the conclusion that Ψ93 results in RNAs with similar dynamic and structural characteristics as WT, consistent with the transitions observed by smFRET. We previously described how subtle changes in stem IIb influences stem IIa/IIc dynamics [34]. These cleavage assays indicate that pseudouridylation in stem IIa/IIc can also alter stem IIb structure providing further evidence for an intimate connection between each stem II subunit.
We also did not detect any significant differences in cleavage at G64. One possible explanation for this could be that G64 is in a similar environment and paired within each RNA. Together with the other cleavage data a possible explanation for this could be formation of an alternate stem IIc basepairing structure (the stem IIc extension, Fig. 6b) that has been previously proposed [15]. This structure would explain the similar cleavage of G64 in each RNA, independent of stem IIa or IIc formation. The stem IIc extension may also explain reduced cleavage of G55, G69, G78-81, and G100 in Ψ56 and Ψ56+Ψ93 RNAs since stem IIc formation would incorporate these nucleotides into an extended RNA duplex. It is interesting to note that this structure appears to be stabilized by Ψ56 incorporation but not Ψ93, even though the latter is part of the stemIIc extension. This suggests that stabilization of the base of stem IIc could be necessary for formation of the stem IIc extension. Together the T1 cleavage data support the smFRET results and indicate that Ψ56 or Ψ56+Ψ93 incorporation results in stabilization of an RNA structure consistent with stem IIc, while the Ψ93-containing RNAs exhibit structures and dynamics similar to WT.
Induction of Ψ 93 in Yeast Does Not Inactivate Spliceosomes with Altered Assembly or Activation Pathways
It has previously been shown that a modified pseudouridine synthase guide RNA (snR81MUT) can be used to induce Ψ93 formation in U2 in yeast cells under normal growth conditions [15]. One consequence of this is a decrease in usage of a non-consensus 3′ SS in a ACT1-CUP1 splicing reporter. In our experiments, Ψ93 appears to change stability and dynamics of stem IIa leading to a broad distribution of low- to mid-FRET states. We wondered if this could also result in a change in splicing in the presence of spliceosome mutations that impact steps associated with stem IIa formation or toggling.
We first combined the snR81MUT plasmid with a yeast strain in which Cus2 (which is not essential) had been deleted. We predicted that changes in stem IIa structure due to snR81MUT-guided Ψ93 formation could result in defects in spliceosome assembly or BS usage in the absence of Cus2 since this protein may help stabilize stem IIa at this stage. The Cus2Δ strains transformed with either the snR81MUT or control plasmids were viable and showed no evidence of proliferative defects under normal growth conditions (data not shown). We then examined the Cus2Δ/snR81MUT strain for changes in splicing activity using a variety of ACT1-CUP1 branchsite-usage reporters, which allow in vivo splicing activity to be monitored using a Cu2+ tolerance growth assay [40]. We detected no changes in growth for any of the Cus2Δ/snR81MUT strains with any of the tested reporters relative to the control strains (Sup. Fig. 3).
We also predicted that the Prp2Q548N allele (which causes a defect in spliceosome activation and a cold sensitive phenotype [37]) may result in genetic interactions with snR81MUT since stem IIa to IIc switching occurs during or after Prp2 function [38, 39] and may be necessary for active site folding. We combined the snR81MUT plasmid with yeast strains harboring Prp2WT or the Prp2Q548N allele and detected no changes in growth of the Prp2Q548N/snR81MUT strain relative to controls at either permissive or restrictive temperatures (Sup. Fig. 4). Together the data suggest that induction of Ψ93 either does not influence assembly or activation steps in splicing or does not introduce a new, rate-limiting step in vivo under these conditions. It is possible that Mg2+, Cus2, or other cellular factors function to dampen the impact of Ψ93 in cells during these steps.
DISCUSSION
In this study we have analyzed the impacts of stress-induced pseudouridylation on the dynamics of the stem II region of the U2 snRNA. The discovery of post-transcriptional RNA modifications within stem II is intriguing since this region of the RNA must toggle between at least two alternate structures (stem IIa and IIc) to facilitate spliceosome assembly and catalysis [31, 32]. Both stem IIa and IIc have been observed in recent cryo-EM structures of the spliceosome captured at different stages of the splicing reaction [38, 39, 41–44]. Changes in how stem II interconverts between IIa and IIc could therefore modulate multiple steps in pre-mRNA splicing. Our results show that pseudouridylation alters the intrinsic dynamics present within a RNA model of U2 stem II and support the notion that post-transcriptional modifications can change snRNA structural rearrangements.
RNA dynamics can be altered by a number of factors including mutations to the nucleotide sequence, buffer conditions, and temperature in addition to the post-transcriptional modifications studied here. It is also possible that the fluorophores which enable the smFRET experiments have themselves perturbed these intrinsic dynamics. Structural probing is consistent with our smFRET observations that Ψ56 and Ψ56+Ψ93-containing RNAs increase the abundance of stem IIc while the Ψ93-containing RNA more closely resembles the unmodified WT. Further, the ability of Cus2 to associate with each RNA in bulk EMSA assays supports its alteration of stem II smFRET despite Ψ56 and Ψ93 incorporation. Our combined results reveal how pseudouridylation changes the relative behaviors of each modified RNA.
It was previously hypothesized that Ψ56 and Ψ93 could both stabilize stem IIc and in combination inhibit splicing by preventing toggling to stem IIa [15]. Our data support stem IIc stabilization by Ψ56 but not by Ψ93, which instead appears to increase dynamics of non-stem IIc conformations. When RNAs simultaneously contained both Ψ56 and Ψ93, toggling to stem IIa was still permitted in the presence of Mg2+ and Cus2. Together these results suggest that Ψ56 and Ψ93 may differentially impact stem II behavior within the spliceosome and that their simultaneous presence in stem II may not necessarily lead to prevention of stem II toggling or splicing inhibition.
The pathways and conditions under which the U2 snRNA becomes inducibly pseudouridylated at Ψ56 and Ψ93 are distinct. Both heat shock and nutrient deprivation result in Ψ56 incorporation via Pus7, while only nutrient stress has been shown to result in Ψ93 incorporation [15, 27]. Difficulties in isolating specific phenotypes associated with Ψ56 and Ψ93 incorporation in vivo have complicated analysis of the consequences of these modifications. We observed pronounced stabilization of stem IIc after Ψ56 incorporation. Given that the calculated Tm for stem IIc is near room temperature (~20°C, determined using OligoCalc, [45]), Ψ56 may prove beneficial for maintaining stem IIc integrity at elevated temperatures experienced during heat shock. How Ψ56 functions during nutrient deprivation is less clear since stem IIc stabilization could inhibit spliceosome assembly and limit cell growth or promote the catalytic steps of splicing and facilitate gene expression.
Stabilization of stem IIc by Ψ56 is consistent with pseudouridine functioning to increase duplex stability since Ψ56 is paired within stem IIc itself. Stabilization due to Ψ56 could arise from a number of factors in addition to N1 hydrogen bonding, including changes in the glycosidic bond which alters base orientation and duplex stability [11, 13] or facilitating formation of the stem IIc extension. We cannot exclude the possibility that Ψ56 incorporation contributes to stem IIa destabilization, since pseudouridine incorporation into loops has been proposed to alter loop flexibility [13]. Interestingly, an increase in stem IIc was also observed when position 56 was modified with rT. This suggests that presence of a new hydrogen bond donor or C-C glycosidic bond at Ψ56 is not obligatory for increasing the abundance of stem IIc and presence of steric bulk at this position can produce the same outcome via a different mechanism.
In cryo-EM structures of C and C* spliceosomes, position 56 is paired within stem IIc and was modeled [41–43]. While no protein contacts were made directly to uridine 56, stem IIc is located adjacent to several spliceosome proteins including the Sm proteins of U2 and Prp17. It is intriguing to conjecture that these or other proteins could function similarly to rT56 and Ψ56 by making transient contact with stem IIc in order to stabilize the structure during spliceosome rearrangements.
Ψ93’s ability to induce conformational flexibility suggests that Ψ93 may interfere with transient IIa formation by permitting other, off-pathway structures to form once IIc has unwound. Inefficient stem IIc→IIa→IIc switching by the spliceosome as the active site rearranges after 5′ SS cleavage could account for Ψ93’s mild inhibition of exon ligation [15]. Improved methods for evaluating splicing defects under non-optimal growth conditions such as nutrient deprivation may facilitate detection of additional phenotypes due to this and other snRNA modifications.
It is now clear that many cellular RNAs are post-transcriptionally modified [1–9]; however, the functional relevance of only a small subset of these modifications has been elucidated. Our work highlights some of the challenges in understanding the biological consequences of RNA modification. Even though stem II RNA dynamics were altered due to pseudouridine incorporation, these changes were suppressed by both Mg2+ and Cus2 and most strongly when both were present simultaneously. This suggests that spliceosome proteins and cellular ions can buffer splicing from the consequences of pseudouridylation. Buffering may be in place for other RNA modifications on other transcripts as well. Some RNA modifications may become functionally relevant inside cells only in the presence or absence of additional trans-acting proteins or small molecules.
MATERIAL AND METHODS
Preparation of Stem II Model RNAs
The stem II model RNAs used in this study were generated by splinted ligation of two chemically synthesized RNA oligos from Integrated DNA Technologies (Coralville, IA) as described in Rodgers et al. [34] using a DNA splint with the sequence 5′-AAAAGGTAATGAGCCTCATTGAGGTCATTTCA-3′. The RNA oligos represented the yeast U2 snRNA nt 31–78 (5′ piece) and nt 79–126 (3′ piece) and were synthesized with modified nucleotides at the desired positions (Sup. Table 1).
For smFRET experiments, the 5′ and 3′ RNA oligos were labeled with Cy3-NHS or Cy5-NHS (GE Healthcare Life Sciences, Pittsburgh, PA), respectively, on amino allyl uridines introduced into positions 54 and 101 prior to splinted ligation. Labeling reactions were carried out overnight in 100 mM NaHCO3 pH 8.5 in the dark at room temperature. Excess dye was removed using a G-25 MicroSpin column (GE Healthcare Life Sciences, Pittsburgh, PA) and labeled RNA was further purified via denaturing, polyacrylamide gel electrophoresis (PAGE). Labeled RNAs were extracted from the gel overnight in RNA extraction buffer (0.3 M NaOAc pH 5.2, 1 mM EDTA, 10% v/v phenol pH 4.5), in the dark, and at room temperature. This was followed by ethanol precipitation and resuspension of the labeled RNAs in nuclease-free water (Ambion®, Thermo Fisher Scientific, Rockford, IL).
For model RNAs analyzed by RNase T1 digestion, the 5′ oligo was labeled with [32P] using γ-[32P]-labeled ATP (Perkin Elmer) and polynucleotide kinase (New England Biolabs) prior to ligation, which was then carried out using a ratio of 1:2:1 5′ oligo:DNA splint:3′ oligo.
smFRET Data Collection and Analysis
Biotinylated RNA molecules were immobilized via streptavidin on passivated slides as previously described [34, 46] except that 25.4×76.2×1 mm quartz slides and 24×60 mm type 0 coverslips (Thermo Fisher Scientific, Gold Seal™) were used. Molecules were imaged in 1x imaging buffer (20 mM HEPES pH 7.9, 125 mM KCl, 5% v/v glycerol, 0.1% v/v Triton X-100) containing either 1 mM EDTA or 10 mM MgCl2, a glucose oxidase/catalase oxygen scavenging system, and 2 mM Trolox as previously described [34].
Data were collected at a frame rate of 200 ms on a prism-based total internal reflection fluorescence microscope (TIRF) with laser excitation at 532 nm (15 mW) and images recorded on a EM-CCD camera (Andor) as described [34]. Data analysis was performed using custom Matlab software (Mathworks, Framingham, MA) scripts as previously described [34].
Cus2 Expression, Purification, and Binding Assays
Yeast Cus2 was recombinantly expressed and purified from E. coli as previously described [34, 35]. EMSA assays were carried out in triplicate using [32P]-labeled RNAs as previously described [34] except that EDTA was omitted from samples containing Mg2+.
RNase T1 Structural Probing
Structural probing was carried out following methods described by Wroblewska and Olejniczak [49]. Each reaction contained 9 ng of stem II RNA and 1 μg of yeast total RNA (Sigma-Aldrich, St. Louis, MO) diluted to 5 μl with RNase-free water. 5x buffer (2μL; 250 mM Tris pH 7.5, 10 mM EDTA) was then added. T1 RNase (3 μl; 0.03 U/μl) was added to each sample and incubated at 37°C for 3 min. T1 Stop buffer (10 μL; 8M urea, 40 mM EDTA, 50% deionized formamide, 0.02% w/v bromophenol blue, 0.02% w/v xylene cyanol) was then added. The U2 stem II T1 RNase ladder showing cleavage at every guanine was obtained from 9 μg of stem II RNA and 1 μg of yeast total RNA in 50 mM sodium acetate pH 4.3, 7M Urea buffer by digestion with 1U of T1 RNase at 55°C for 10 min before stopping the reaction by addition of equal volume of Stop buffer. An RNA ladder was produced by incubating 9 ng of stem II RNA and 1 μg of yeast total RNA with 50 mM sodium acetate pH 9.7 for 2 min at 90°C before adding an equal volume of Stop buffer. Samples and ladders were then analyzed by denaturing 10% PAGE and the gel was dried before the bands were imaged using a phosphorscreen. Band intensities were quantified using ImageQuant software (GE Healthcare Life Sciences, Pittsburgh, PA).
Yeast Growth Assays
Yeast strains containing either Prp2WT (yTJC267) or Prp2Q548N (yTJC278) have been reported previously [47]. Cus2 was deleted in strain 46α (MAT α cup1Δ ura3 his3 trp1 lys2 ade2 leu2; gift of David Brow, U. Wisconsin-Madison) by integration of a hygromycin resistance cassette [48] and confirmed by PCR to create strain yAAH1648. URA-marked plasmids for expression of the WT snR81 snoRNA (snR81WT), the U101A/U105A mutant for induction of Ψ93 (snR81MUT), or the empty yEPlac195 plasmid were obtained from Yi-Tao Yu (U. Rochester Medical Center) [15]. Yeast transformation and growth were carried out using standard methods [49].
ACT1-CUP1 growth assays were carried out by transforming strain 46α or yAAH1648 with the URA-marked plasmids encoding snR81 (or the empty plasmid control) and LEU-marked plasmids encoding the noted reporter pre-mRNA. Yeast were grown to mid-log phase in –URA/-LEU dropout media to maintain selection for the plasmids, adjusted to OD600 of 0.5 and equal volumes were spotted with a multi-blot replicator (VP407AH, V&P Scientific Inc, San Diego, CA) onto –URA/-LEU dropout plates containing 0, 0.025, 0.05, 0.075, 0.1, 0.15, 0.2, 0.25, 0.3, 0.35, 0.4, 0.45, 0.5, 0.6, 0.7, 0.8, 0.9, 1.0, 1.1, 1.2, 1.3, 1.4, 1.5, 1.6, 1.7, 1.8, 1.9, 2.0, 2.25, or 2.5 mM CuSO4. Plates were scored after 3 days growth at 30°C.
Temperature sensitivity assays were carried out by first transforming strain yTJC267 or yTJC278 with the URA-marked plasmids encoding snR81 (or the empty plasmid control). Yeast were grown to mid-log phase in –URA dropout media, adjusted to OD600 of 0.5, and equal volumes spotted onto –URA dropout plates at 1x, 10−1, 10−2, and 10−3 cell density. Plates were incubated at the indicated temperature and scored after 3 days of growth at 23°C, 30°C, and 37°C or after 10 days of growth at 16°C.
Supplementary Material
HIGHLIGHTS.
Pseudouridylation changes the conformational dynamics of U2 snRNA stem II
Pseudouridines induced under different conditions alter dynamics in distinct ways
Cofactors of the spliceosome suppress the impact of pseudouridylation
Acknowledgments
We thank Yi-Tao Yu (U. Rochester Medical Center) for sharing the snR81 plasmids. This work was supported by the National Institutes of Health (R00 GM086471, R01 GM112735), a Shaw Scientist Award, a Beckman Young Investigator Award, and startup funding from the University of Wisconsin-Madison, Wisconsin Alumni Research Foundation (WARF), and the Department of Biochemistry.
ABBREVIATIONS
- BS
branchsite
- Ψ
pseudouridine
- rT
ribothymidine
- smFRET
single molecule Förster resonance energy transfer
- snRNA
small nuclear ribonucleic acid
- snRNP
small nuclear ribonucleoprotein
- SS
splice site
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, et al. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–6. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–62. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu Y, Pirnie SP, Carmichael GG. High-throughput and site-specific identification of 2′-O methylation sites using ribose oxidation sequencing (RibOxi-seq) RNA. 2017 doi: 10.1261/rna.061549.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai Q, Moshitch-Moshkovitz S, Han D, Kol N, Amariglio N, Rechavi G, et al. Nm-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat Methods. 2017 doi: 10.1038/nmeth.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–46. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12:767–72. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–4. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441–6. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Ma S, Yi C. Pseudouridine: the fifth RNA nucleotide with renewed interests. Curr Opin Chem Biol. 2016;33:108–16. doi: 10.1016/j.cbpa.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23:5020–6. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newby MI, Greenbaum NL. Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proc Natl Acad Sci U S A. 2002;99:12697–702. doi: 10.1073/pnas.202477199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kierzek E, Malgowska M, Lisowiec J, Turner DH, Gdaniec Z, Kierzek R. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 2014;42:3492–501. doi: 10.1093/nar/gkt1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaidyanathan PP, AlSadhan I, Merriman DK, Al-Hashimi H, Herschlag D. Pseudouridine and N-6 methyladenosine modifications weaken PUF protein/RNA interactions. RNA. 2017 doi: 10.1261/rna.060053.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu G, Xiao M, Yang C, Yu YT. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J. 2011;30:79–89. doi: 10.1038/emboj.2010.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu AT, Ge J, Yu YT. Pseudouridines in spliceosomal snRNAs. Protein & Cell. 2011;2:712–25. doi: 10.1007/s13238-011-1087-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wahl MC, Will CL, Lührmann R. The Spliceosome: Design Principles of a Dynamic RNP Machine. Cell. 2009;136:701–18. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Karijolich J, Yu YT. Spliceosomal snRNA modifications and their function. RNA Biol. 2010;7:192–204. doi: 10.4161/rna.7.2.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall KB, McLaughlin LW. Properties of a U1/mRNA 5′ splice site duplex containing pseudouridine as measured by thermodynamic and NMR methods. Biochemistry. 1991;30:1795–801. doi: 10.1021/bi00221a010. [DOI] [PubMed] [Google Scholar]
- 20.Yu YT, Shu MD, Steitz JA. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 1998;17:5783–95. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao X, Yu YT. Incorporation of 5-fluorouracil into U2 snRNA blocks pseudouridylation and pre-mRNA splicing in vivo. Nucleic Acids Res. 2007;35:550–8. doi: 10.1093/nar/gkl1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu G, Adachi H, Ge J, Stephenson D, Query CC, Yu YT. Pseudouridines in U2 snRNA stimulate the ATPase activity of Prp5 during spliceosome assembly. EMBO J. 2016;35:654–67. doi: 10.15252/embj.201593113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newby MI, Greenbaum NL. A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. RNA. 2001;7:833–45. doi: 10.1017/s1355838201002308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newby MI, Greenbaum NL. Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat Struct Biol. 2002;9:958–65. doi: 10.1038/nsb873. [DOI] [PubMed] [Google Scholar]
- 25.Yang C, McPheeters DS, Yu YT. Psi35 in the branch site recognition region of U2 small nuclear RNA is important for pre-mRNA splicing in Saccharomyces cerevisiae. J Biol Chem. 2005;280:6655–62. doi: 10.1074/jbc.M413288200. [DOI] [PubMed] [Google Scholar]
- 26.Basak A, Query CC. A pseudouridine residue in the spliceosome core is part of the filamentous growth program in yeast. Cell Rep. 2014;8:966–73. doi: 10.1016/j.celrep.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu G, Radwan MK, Xiao M, Adachi H, Fan J, Yu YT. The TOR signaling pathway regulates starvation-induced pseudouridylation of yeast U2 snRNA. RNA. 2016;22:1146–52. doi: 10.1261/rna.056796.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller EB, Noon WA. Intron splicing: a conserved internal signal in introns of Drosophila pre-mRNAs. Nucleic Acids Res. 1985;13:4971–81. doi: 10.1093/nar/13.13.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ares M, Jr, Igel AH. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev. 1990;4:2132–45. doi: 10.1101/gad.4.12a.2132. [DOI] [PubMed] [Google Scholar]
- 30.Zavanelli MI, Ares M., Jr Efficient association of U2 snRNPs with pre-mRNA requires an essential U2 RNA structural element. Genes Dev. 1991;5:2521–33. doi: 10.1101/gad.5.12b.2521. [DOI] [PubMed] [Google Scholar]
- 31.Perriman RJ, Ares M., Jr Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev. 2007;21:811–20. doi: 10.1101/gad.1524307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hilliker AK, Mefford MA, Staley JP. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev. 2007;21:821–34. doi: 10.1101/gad.1536107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zavanelli MI, Britton JS, Igel AH, Ares M., Jr Mutations in an essential U2 small nuclear RNA structure cause cold-sensitive U2 small nuclear ribonucleoprotein function by favoring competing alternative U2 RNA structures. Mol Cell Biol. 1994;14:1689–97. doi: 10.1128/mcb.14.3.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodgers ML, Tretbar US, Dehaven A, Alwan AA, Luo G, Mast HM, et al. Conformational dynamics of stem II of the U2 snRNA. RNA. 2016;22:225–36. doi: 10.1261/rna.052233.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan D, Perriman R, Igel H, Howe KJ, Neville M, Ares M., Jr CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol Cell Biol. 1998;18:5000–9. doi: 10.1128/mcb.18.9.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ehresmann C, Baudin F, Mougel M, Romby P, Ebel JP, Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–28. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wlodaver AM, Staley JP. The DExD/H-box ATPase Prp2p destabilizes and proofreads the catalytic RNA core of the spliceosome. RNA. 2014;20:282–94. doi: 10.1261/rna.042598.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan C, Wan R, Bai R, Huang G, Shi Y. Structure of a yeast activated spliceosome at 3.5 Å resolution. Science. 2016;353:904–11. doi: 10.1126/science.aag0291. [DOI] [PubMed] [Google Scholar]
- 39.Rauhut R, Fabrizio P, Dybkov O, Hartmuth K, Pena V, Chari A, et al. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science. 2016;353:1399–405. doi: 10.1126/science.aag1906. [DOI] [PubMed] [Google Scholar]
- 40.Lesser CF, Guthrie C. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993;133:851–63. doi: 10.1093/genetics/133.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wan R, Yan C, Bai R, Huang G, Shi Y. Structure of a yeast catalytic step I spliceosome at 3.4 Å resolution. Science. 2016;353:895–904. doi: 10.1126/science.aag2235. [DOI] [PubMed] [Google Scholar]
- 42.Galej WP, Wilkinson ME, Fica SM, Oubridge C, Newman AJ, Nagai K. Cryo-EM structure of the spliceosome immediately after branching. Nature. 2016;537:197–201. doi: 10.1038/nature19316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fica SM, Oubridge C, Galej WP, Wilkinson ME, Bai XC, Newman AJ, et al. Structure of a spliceosome remodelled for exon ligation. Nature. 2017 doi: 10.1038/nature21078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Yan C, Hang J, Finci LI, Lei J, Shi Y. An Atomic Structure of the Human Spliceosome. Cell. 2017;169:918–29. e14. doi: 10.1016/j.cell.2017.04.033. [DOI] [PubMed] [Google Scholar]
- 45.Kibbe WA. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 2007;35:W43–6. doi: 10.1093/nar/gkm234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crawford DJ, Hoskins AA, Friedman LJ, Gelles J, Moore MJ. Single-molecule colocalization FRET evidence that spliceosome activation precedes stable approach of 5′ splice site and branch site. Proc Natl Acad Sci U S A. 2013;110:6783–8. doi: 10.1073/pnas.1219305110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carrocci TJ, Zoerner DM, Paulson JC, Hoskins AA. SF3b1 mutations associated with myelodysplastic syndromes alter the fidelity of branchsite selection in yeast. Nucleic Acids Res. 2017;45:4837–52. doi: 10.1093/nar/gkw1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–53. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 49.Wroblewska Z, Olejniczak M. Hfq assists small RNAs in binding to the coding sequence of ompD mRNA and in rearranging its structure. RNA. 2016;22:979–994. doi: 10.1261/rna.055251.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amberg DC. In: Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. 2005. Burke D, Strathern JN, Burke D, editors. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.