Abstract
Drug development continues to be costly and slow, with medications failing due to lack of efficacy or presence of toxicity. The promise of pharmacogenomic discovery includes tailoring therapeutics based on an individual's genetic makeup, rational drug development, and repurposing medications. Rapid growth of large research cohorts, linked to electronic health record (EHR) data, fuels discovery of new genetic variants predicting drug action, supports Mendelian randomization experiments to show drug efficacy, and suggests new indications for existing medications. New biomedical informatics and machine learning approaches advance the ability to interpret clinical information, enabling identification of complex phenotypes and subpopulations of patients. We review the recent history of use of “big data” from EHR-based cohorts and biobanks supporting these activities. Future studies using EHR data, other information sources, and new methods will promote a foundation for discovery to more rapidly advance precision medicine.
Introduction
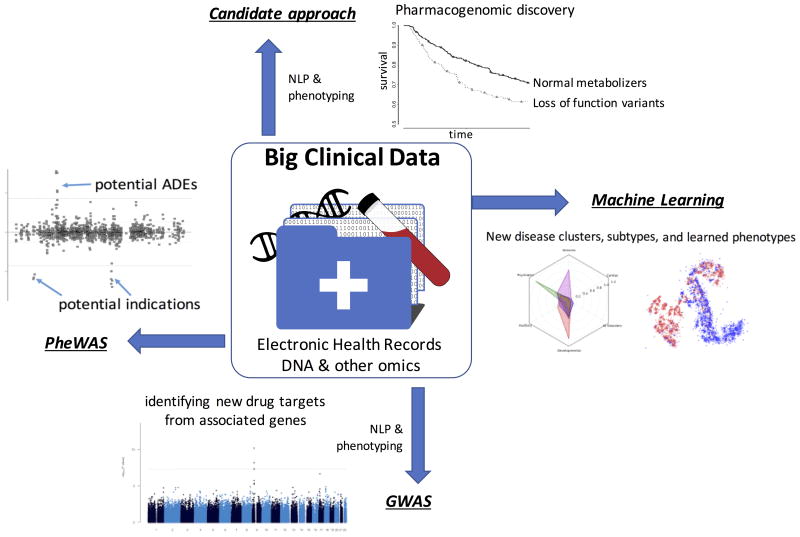
The completion of the Human Genome Project in 2003 ushered in a promise of a new era of personalized medicine. Envisioned were greater understandings of the genome to guide therapy for Mendelian diseases, an untangling of the basis of genetic influences underlying familial diseases, and the advancement of knowledge to lead to new therapeutics. While the impact of genetics on variable drug actions had been studied for decades before the Human Genome Project, the pace of discovery in the last 15 years has led to richer understandings of the workings of the genome and an uncovering of the genetic influences for drug responses as well as hundreds of traits and diseases, including both Mendelian and complex diseases. Recently, we are also seeing the advent of genetic-tailored therapies for common and rare diseases, and the development of new therapeutics based on genetic findings. A rate limiting step, however, has been the development of sufficiently large cohorts with the exposure, covariate and outcome phenotype data necessary for study. The majority of early studies focused on well-defined observational cohorts or randomized controlled trials. The growth of large national research cohorts and networks incorporating rich and broad phenotype data coupled to DNA biobanks is providing a transformational accelerant to discovery. Pioneered through several sites and networks such as the Electronic Medical Records and Genomics (eMERGE) network,1 electronic health record (EHR) data has proven a powerful “big data” tool for genomic discovery in the study of disease and therapeutics (Figure 1). Here, we review some of the contributions of routinely collected healthcare data, genomics, and large research cohorts to discover and prioritize potential drug targets, accelerate pharmacogenomics, predict side effects, uncover more precise understandings of diseases, repurpose medications, and mine data for unknown drug effects, with the ultimate goal of optimizing therapeutic efficacy while minimizing of adverse effects.
Figure 1. Electronic Health Records support genomic and pharmacogenomic discovery.
NLP=Natural Language Processing. PheWAS=Phenome-wide association study; GWAS=Genome-wide association study. Disease clusters adapted from Lingren et al.74 and Lasko et al.64
Genomics as a tool to prioritize drug targets
A growing body of evidence shows that genetic targets often suggest effective drug targets.2,3 Much of this evidence comes from genome-wide association studies (GWAS), which can survey millions of single nucleotide polymorphisms (SNPs) across the genome in a hypothesis-free approach. GWASs provide a systematic assessment of the impact of individual genetic variants for a given trait and have been responsible for discovery of >49,000 single nucleotide polymorphism (SNP)-trait associations through over 3000 publications as of this publication. An early example of the power of GWAS to find drug targets was in the analysis of low density lipoprotein (LDL) cholesterol levels in 2008, which demonstrated that common variants in HMGCR have small effects on LDL-cholesterol.4 HMGCR encodes HMG-CoA reductase, the target for the potent statin drug class. In 2014, Okada et al. reported a large GWAS of more than 100,000 cases and controls for rheumatoid arthritis (RA), identifying 101 loci associated with RA.5 These loci identified drug targets for 18 of the 27 approved RA drugs at the time and suggested several novel therapeutics. Similarly, the genetic variants associated with type 2 diabetes identify the drug targets for thiazolidinediones, sulfonylureas, glucagon-like peptide-1 (GLP-1) receptor agonists, and one of the newest class of antidiabetic drug classes, sodium-glucose cotransporter-2 (SGLT2) inhibitors.6 A prevailing theme in these GWAS findings is that these associations often find small effect sizes (often between common, likely non-functional SNPs and the disease) but represent drug targets with significant impact on the disease or trait.
Perhaps the first major prospective example of a primarily genomic discovery leading to a new therapeutic drug class was the recent development of proprotein convertase subtilisin/kexin type 9 (PCSK9) inhibitors, for which there are now two drugs available in the US. By sequencing PCSK9 in individuals with very low levels of LDL in the multiethnic Dallas Heart Study, Cohen et al. found two loss of function (LOF) variants in individuals of African ancestry that resulted in a 40% decrease in LDL cholesterol levels.7 They were later able to show individuals with LOF variants in PCSK9 had a 88% reduction in cardiovascular disease in African Americans.8 Missense variants associated with lesser changes in LDL discovered in European Americans were also associated with reduced cardiovascular disease. Following these promising genetic findings, monoclonal antibodies have been developed against PCSK9 and were approved in 2015. Randomized controlled trials have shown that these PCSK9 inhibitors further reduce LDL cholesterol levels and cardiovascular disease when combined with statin therapy.9
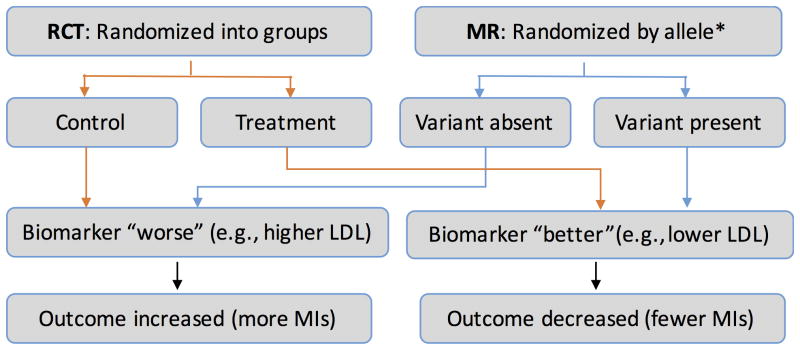
The studies of PCKS9 follow the pattern of Mendelian randomization (MR) studies (Figure 2). MR is a technique used to provide evidence for the causality of a biomarker on a disease state in conditions in which randomized controlled trials are difficult or too expensive to pursue. For example, LDL and high-density lipoprotein (HDL) levels have long been associated with myocardial infarction in observational cohorts, but it was unclear whether they are markers for the disease process or causal for the outcome; perhaps LDL and HDL levels are indicators for diet, activity level, or other unknown factors that contribute to the pathogenesis of disease. It is essentially impossible (and possibly unethical) to perform the definitive study, a randomized control trial that alters participants' LDL or HDL levels. However, a number of genetic variants have been found that alter LDL and HDL levels. Since alleles randomly distribute at meiosis, studying the impact of biomarker-influencing alleles provides a naturally occurring randomization of the risk factor. Genetic variants are generally not associated with behavioral, social, and some physiological factors – reducing confounding. Thus, by studying the impact on the clinical outcome of the variants associated with the biomarker, one can assess causality of the biomarker to the outcome. MR has proven a powerful tool in recent years. MR studies have demonstrated clear associations between LDL and triglyceride levels and cardiovascular disease while casting doubt on the role of HDL in protecting against cardiovascular disease.10,11 The latter is particular interesting as cholesteryl ester transfer protein (CETP) inhibitors, medications effective at raising HDL, have so far not been successful at reducing cardiovascular events12 with the exception of anacetrapib whose cardiovascular event reduction could be through the reduction of non-HDL cholesterol.13 MR has also cast doubt on the causality of C-reactive protein in heart disease risk, 14 decreasing enthusiasm for the development of therapeutics targeting C-reactive protein levels.
Figure 2. Mendelian Randomization (MR) vs. Randomized Controlled Trials (RCT).
MI=myocardial infarction. LDL=low density lipoprotein levels. *Allele could be a single SNP or group of SNPs (e.g., genetic risk score).
Another example of MR demonstrating a clinical effect is seen with ezetimibe, which lowers LDL cholesterol by inhibiting Niemann-Pick C1-like protein 1 (encoded by NPC1L1). Ezetimibe was introduced as a new class of LDL cholesterol medication in 2002 as an alternate to or adjuvant with statin therapy. However, its efficacy in reducing cardiovascular disease had been in doubt15 until a MR study demonstrated that individuals with LOF variants in NPC1L1 had both reduced LDL-cholesterol levels and a 53% reduction in cardiovascular disease.16 The reduction in cardiovascular events was demonstrated through study of 113,094 individuals with genotyping for these LOF variants. Importantly, 21,131 of those individuals were identified in an EHR-linked biobank with extant genotyping whose cases and controls were found within about 3 weeks, demonstrating the potential power for reuse of clinical datasets for discovery. Soon after this study was published, the IMPROVE-IT randomized controlled trial of ezetimibe + simvastatin vs. simvastatin alone demonstrated a small cardiovascular benefit with the addition of ezetimibe.17
Recent exome array and exome sequencing studies using large EHR-linked populations have elucidated other new targets for lipid disease. Stitziel et al. studied 193,638 individuals (23,576 from EHR cohorts) with exome array data for rare variants associated with cardiovascular disease, plasma lipids, blood pressure, and type 2 diabetes.18 They identified novel rare missense variants in SVEP1, ANGPTL4 (which inhibits lipoprotein lipase, or LPL), and LPL associated with cardiovascular disease. The LOF ANGPTL4 variants were associated with reduced triglyceride levels as well as protection from coronary artery disease. Similarly, the DiscovEHR cohort at Geisinger Health System is proving a powerful discovery resource, currently with EHR-linked DNA collected on more than 100,000 participants. An analysis more than 50,000 participants with whole exome sequence data from Geisinger and Regeneron investigators also found that the rare ANGPTL4 E40K variant (rs116843064) found in the Stitziel study was associated with reduced triglyceride levels, increased HDL levels, and lower risk of coronary artery disease.19 This group further demonstrated that monoclonal antibody inhibition of Angptl4 in mice reduced triglyceride levels. A similar theme is evolving with other lipid traits, leveraging use of healthcare-derived data: rare variant discovery suggesting new drug targets and in some cases leading to development of new candidate therapeutics.20–22
MR can also be used to evaluate expected safety profiles of a given medication. An example comes from PCSK9 inhibitors. Given the risk of type 2 diabetes with statins, Schmidt et al. performed a MR study using PCSK9 functional variants, looking at replication of known cholesterol effects and evaluating for a potential effect on type 2 diabetes. 23 Their meta-analysis (including EHR samples) replicated known LDL-cholesterol lowering effects and showed a potential risk of increased glucose and type 2 diabetes.
Accelerating pharmacogenetic discovery with EHRs
Pharmacogenetics focuses on the discovery of genetic variants that alter medication response, through alteration of effective drug levels via changes in absorption, distribution, metabolism, or excretion of medication, differences in effect, such as variants in drug receptors, or via off-target effects, such as drug hypersensitivity examples. One early pharmacogenomic discovery was a description of TPMT genotypes and activity.24 Since the sentinel studies identifying the impact of genetic variants on enzyme function and thus drug response, GWAS have become a powerful tool for the elucidation of the genetic basis of diseases and traits. However, the vast majority of GWAS to date have explored disease outcomes or phenotypic traits, with only ∼9% performed on pharmacogenomic traits, with the majority of these focused on anticoagulants and antiplatelet therapies, statins, chemotherapy, and psychiatric medications.25 Several challenges may hinder the collection of large cohorts with the information required for pharmacogenomic discovery. As with all GWAS, large sample sizes are needed and many of the events are rare or may be largely unpredictable, requiring longitudinal follow-up of large populations to experience. Traditional prospective population cohorts are often not large enough to note very rare adverse events (such as Stevens-Johnson Syndrome or heparin induced thrombocytopenia) or lack the regular and comprehensive collection of medication exposures and diverse outcomes necessary to evaluate more common drug effects (e.g., a response to blood pressure medication in a psychiatric cohort). In addition, retrospective assessment of outcomes with high morbidity and mortality may be difficult to ascertain.
Use of EHRs may be a particularly advantageous tool for assessing drug effects given their continuous and prospective longitudinal assessment of clinically relevant outcomes and medication exposures (Table 1). Moreover, use of EHR data for pharmacogenomic study has an added benefit in representing the “real world” conditions of patient medication use, comorbid conditions, secular treatment trends, and interacting medications that may better represent the clinical effect of drug genome interactions. EHR-based candidate gene studies and GWAS have proven fruitful for replication and discovery of pharmacogenomic phenotypes. Delaney et al. replicated the association between cardiovascular risk and CYP2C19*2 and ABCB1 in patients receiving clopidogrel; the effect size in this study was nearly identical to prior efforts.26 Similarly, associations between variants in CYP2C9, VKORC1 and CYP4F2 and steady state warfarin dose in European ancestry individuals have been replicated in EHR data sets.27 EHRs have been used for pharmacogenetic traits have identified a number of other pharmacogenetic associations, some of which are summarized in Table 1. An analysis of a project at Vanderbilt that studied 31 EHR-defined drug phenotypes found that use of the EHRs decreased cost 72% per subject, shortened study time, and more efficiently leveraged valuable patient data.28 Furthermore, data from 90% of the individuals were used in more than one study, suggesting the strong reuse potential for these data.
Table 1. Selected EHR-based genomic studies predicting drug effects.
All reported results were significant in their respective studies.
| Phenotype | Type | Cases | Gene/SNP |
|---|---|---|---|
| Replications of pharmacogenetic effects using EHR biobanks | |||
| Warfarin-stable dose27 | Candidate | 1167 | VKORC1, CYP2C9, CYP4F2, CALU variants |
| Warfarin-stable dose (pediatrics)81 | Candidate | 100 | CYP2C9, VKORC1 variants |
| Clopidogrel efficacy26 | Candidate | 225 | CYP2C19*2, ABCB1 rs1045642 |
| Tacrolimus stable dose82 | Candidate | 446 | CYP3A5 rs776746 |
| Pharmacogenetic discoveries using EHR biobanks | |||
| ACEI-induced cough30 | GWAS | 1,346 | KCNIP4 rs145489027 |
| Serum creatinine during vancomycin therapy | GWAS | 745 | GJA1 rs2789047 |
| Anthracycline induced cardiomyopathy83 | GWAS | 385 | rs7542939 (near PRDM2) |
| Warfarin-related bleeds84 | Candidate | 249 | CYP2C9*3 |
| Statin LDL reduction85 | Candidate | PCSK9 R46L | |
| Heparin-induced thrombocytopenia | GWAS | 73 | GPR65 rs10782473 |
| EHR data for drug target discovery | |||
| Drug targets for RA (includes EHR)5 | GWAS | 103,638 | 101 risk loci; 98 gene candidates suggesting |
| Statin LDL-lowering effect (includes EHR)32 | GWAS | 18,596 | LDL-lowering effect of statins mediated by LPA, APOE, SLCO1B1, and a SORT1/CELSR2/PSRC1 loci |
| Triglyceride levels and cardiovascular disease (EHR only)19 | Exome sequencing | 42,930 | ANGPTL4 |
| NPC1L1 LOF variants on LDL and cardiovascular disease (includes EHR)16 | NPC1L1 sequencing; exome array | NPC1L1 (ezetimibe target) lowers LDL and protects against CV disease | |
| PheWAS-based drug effect discovery | |||
| Effects of TYK2 partial LOF variants40 | PheWAS | 29,377 | Potential indications for RA and Psoriasis; no potential adverse events associated |
| Analysis of PheWAS Catalog for known drug targets | PheWAS | 13,835 | 127 replicated drug/indication pairs; 2,583 drug-indication pairs suggested |
Given the nuances of defining exposure, covariate, and outcome phenotypes for pharmacogenomic studies, sharing of data extraction methods, validation across different sites and EHR systems, and codification of variable definitions are necessary for efficient progress in the field. Networks such as eMERGE Network have established a strong track record for accurate identification of disease and drug response phenotypes from the EHRs, with median positive predictive values (PPV) of published algorithms >95%.29 Typically these algorithms involve the combination of billing codes, medication data, lab values, and text mining to achieve these results. However, most of these were disease or trait focused algorithms; algorithms for pharmacogenomics often require sequencing of medical events and medical exposures in time. Research has shown that many pharmacogenetic traits can be assessed with accurate performance without requiring manual review, just as for disease algorithms. Examples include angiotensin converting enzyme inhibitor induced cough,30 warfarin27 or vancomycin31 dose, and statin effect of LDL-cholesterol lowering.32,33 A common theme across these pharmacologic phenotypes is that they involve chronically administered medications (alleviating the need to accurately determine a medication stop date), inpatient administrations with clear documentations of medication exposures, or a clinical event that is relatively unambiguous (e.g., drug level or physician-asserted adverse event). Other pharmacogenomic algorithms sometimes require manual review to confirm true positive cases. For example, an algorithm for drug-induced liver injury, in which algorithm sensitivity was intentionally favored over positive predictive value in order to capture all events, found a PPV of 20%,34 and the clopidogrel-MACE phenotyping algorithm referenced earlier had a 44% PPV rate for cases, primarily due to difficulty automatically ascertaining the stop date for clopidogrel.26
Using EHR data for pharmacogenomic studies is not without bias. As noted above, assessing accurate medication start and stop dates can be challenging, and for most health systems, the medication exposures are based on prescribing records instead of pharmacy fill records or pill counts and thus can overrepresent the medications people actually take. Inaccuracies in understanding medication start and stop dates can lead to challenges determining if an event occurred during the medication exposure or not or with interacting medications or not. Healthcare events can occur at outside providers, leading to cases misclassified as controls. Use of over-the-counter (OTC) medications is not rigorously documented in the EHR, precluding assessment of these medications or of drug-drug interactions with OTC drugs. The vast majority of these biases for pharmacogenetic studies will bias toward a null result, reducing the likelihood of false positive associations but hindering the identification of potentially important drug gene interactions.
Repurposing existing medications and predicting side effects through phenome-wide approaches
Phenome-wide association studies (PheWAS) provide a way to serially test an independent variable against a comprehensive range of phenotype outcomes. The first PheWAS was performed in EHR data using genetic variants,35 but PheWAS has also been performed using observational cohort data.36 PheWAS has been used to replicate hundreds of known SNP-phenotype associations.37–39 Using the hypothesis established above that genomic associations can suggest effective drug targets (and their effects), PheWAS can provide an approach by which one can simultaneously assess both potential drug indications and their on-target side effects. Diogo et al. found functional variants in TYK2 associated with rheumatoid arthritis (RA), and then performed a PheWAS on these variants to look for other potential indications and adverse effects for medications targeting TYK2 used in RA.40 Their PheWAS of these partial LOF variants supported the potential for blockade of TYK2 as a treatment (as noted by odds ratios <1) for RA, inflammatory bowel disease, and systemic lupus erythematosus, and did not identify significant adverse events (as would be noted by odds ratios>1 and significant p-values). Rastegar-Mojarad et al.41 applied a similar approach using published SNP-phenotype associations in the PheWAS catalog to test this hypothesis on a large scale using common variants in genes that represent current drug targets as identified in DrugBank. They found evidence for 127 drug-indication pairs and identified 2583 potential novel drug-disease associations.
Following this paradigm, another potential avenue for genetics is the repurposing of existing medications with known safety profiles for new indications. Such use cases may be especially useful for special populations such as obstetrics, geriatrics, or pediatrics, for which development and testing of new medications can be more challenging, and for minority populations who have historically not been well represented in phase I-III clinical trials. Leveraging the known drug targets of existing US Food and Drug Administration (FDA)-approved medications could identify additional indications through PheWAS of variants in the drug target genes. In this repurposing model, new indications are recognized by having the same direction of effect for a given genetic variant in the target gene as existing indications; potential side effects are noted by having in opposite direction of effect for a given variant as the existing indications (Figure 1, PheWAS component). Thus, for this approach, it is not necessary to find LOF variants in a gene; rather the known indications provide orientation to potential new indications or side effects using all available common variants in the gene. Thus, the vast catalog of EHR-associated GWAS data could aid in this sort of discovery, and an opportunity to develop or extend EHR-linked cohorts with populations underrepresented in biomedical research, including minorities, the elderly, children, and pregnant women. Limitations of this approach include having sufficiently large samples sizes and finding variants clearly associated with the drug target, since many common variants in GWAS studies are not necessarily clearly linked to expression or function of their nearest gene.
Discovering of drug effects using large-scale clinical data alone
While much of the discovery discussed above has involved the study of genomic data linked to EHRs, a number of investigators have also shown the utility of EHR data by itself to identify potential adverse or therapeutic drug effects and drug-drug interactions. Brownstein et al. demonstrated a two standard deviation rise in myocardial infarctions in their hospital systems 8 months after the introduction of rofecoxib that resolved within one month of rofecoxib's withdrawal from the market.42 LePendu et al. applied natural language processing to large scale EHR data to find known drug adverse events.43 Of note, in the retrospective analysis, their approach also identified a significant association between rofecoxib and myocardial infarction using data from before this safety signal was identified in a clinical trial. A similar approach was used to show that the claudication drug cilostazol may be safe in patients with congestive heart failure, a listed contraindication.44 Tatonetti at al. evaluated the combination of paroxetine and pravastatin on glucose levels, 45 showing the hyperglycemic effect originally discovered from the FDA's Adverse Event Reporting System (AERS) database was found in a small population in their EHR. They replicated the effect in two other EHRs.
Identification of new therapeutic effects can lead to drug repurposing. Xu et al. evaluated the effect of metformin on cancer mortality in two health systems.46 Their results demonstrated, as have others using administrative data,47 improved survival in the metformin treated group for many cancers within both healthcare systems. Statins have also been a popular target for population studies for potential repurposing. A study of the Danish population suggested an improvement in cancer survival with statin therapy.48 However, a recent randomized controlled trial evaluating the use of pravastatin in small cell lung cancer did not find a mortality benefit,49 and another recent study using Surveillance, Epidemiology, and End Results (SEER) showed no effect in a broader population when using methods selecting only users initiated on statins after developing cancer.50 This latter study highlights some of the challenges when evaluating retrospective clinical data for new drug effects; the effect on cancer survival was only seen when an immortal bias was introduced by the selection criteria. A recent population study using Canadian claims data suggested statin use may improve all-cause and lung-related mortality in patients with COPD.51
Unlike genetic studies, which have the benefit of an exposure (the genotype) that is not influenced by clinical care, use of clinical data for drug effect studies must be carefully considered for biases. Individuals take medications for a given indication; often their alternatives are often not chosen with equipoise, such as with statins, which are clearly the first line therapy for hyperlipidemia. Moreover, one could choose a medication specifically because of the potentially “additional” indication in question. For instance, physicians may preferentially choose metformin in a patient with cancer and mildly elevated glucose values or otherwise well-controlled diabetes, leading to indication bias. Statistical approaches such as propensity score adjustments or penalized regression models using dense phenotype models may help counter some of these confounders but may not be able to adjust for all factors. One must also be wary of immortal time biases and selection biases if methods employed require presence of an exposure in one of the analyses groups not present at time zero. Careful attention to the design of phenotype algorithm in prospectively captured data sets (such as EHR and claims data) can ameliorate this bias.50
Machine learning approaches applied to large data sets
As available clinical data sets and their size have increased, there has been a rise in use of novel methods to explore these data. Machine learning methods have been applied to clinical data sets for more than a decade but typically for very focused problems, such as to improve identification of a case status for a defined problem (e.g., detection of tobacco exposure52,53, extracting elements of diabetic foot exams,54 or identifying clinical diseases55,56) or to improve general purpose tools such as natural language processing algorithms.57,58 These approaches typically have been hampered by their requirement for well-labeled training sets – referred to as supervised machine learning methods. The supervised machine learning approach has been used to aid in identifying phenotypes from clinical data records, particularly for disease phenotypes.59,60 The creation of these training sets requires costly annotation by clinical or domain experts. Supervised machine learning approaches have also been used to find drug adverse events,61,62 although to date such approaches typically have not achieved the same level of performance as with disease phenotype models, a recognition of the inherent challenges in defining drug efficacy and adverse events in EHR data sets mentioned above.
Recently, the availability of newer machine learning methods combined with very large data sets has given rise to “deep learning” methods that can learn distinguishing features from a dataset without supervision. These approaches have garnered substantial interest from academia as well as industry which may be better equipped to support the large-scale computing needs required to employ deep learning methods on unstructured information. One of the early examples of these methods was a demonstration project by Google that was able to learn patterns such as human faces, cats, flowers, wine, pizza, and many other characteristics from images selected from YouTube videos.63 These methods are also being applied to clinical studies. Lasko et al. used deep learning methods to learn features from time-compressed curves of uric acid laboratory values to differentiate tumor lysis syndrome from gout with an area under the receiver operator characteristic curve (AUC) of 0.97.64 A team of Google investigators applied deep learning to more than 128,000 images of patients with diabetic retinopathy and controls without diabetic retinopathy.65 Their algorithms were able to identify diabetic retinopathy with similar performance to ophthalmologists with an AUC of 0.99. Similarly, a research group from Stanford was able to identify melanomas with similar performance to trained pathologists using deep learning applied to a training set of 129,450 pathology images.66
Following the promises of machine learning successes applied to real-world data such as these and others within computer science domains, a number of groups are exploring accumulation of real world evidence to aid in prescribing decisions.67 The FDA has demonstrated its commitment to use of real-world data for safety via the Sentinel Initiative68 and, more recently, the Information Exchange and Data Transformation (INFORMED) Initiative,69 which seeks to create a big data environment to advance cancer care. In addition, a number of companies are also exploring applications of machine learning to health, including International Business Machine70–72 and Alphabet companies (which were involved in some of the studies mentioned above).
Big data approaches may also help identify subtypes of disease, which could suggest differential treatment and prognoses. Li et al. used EHR disease data (from billing codes) to identify different clusters of patients with type 2 diabetes, which associated with different genetic variants, and included clusters with differential risks of cardiovascular disease, metabolic factors, and cancers. Doshi-Velez et al. applied hierarchical classifications to identify subtypes of autism, finding disease-associated clusters of phenotypes also presented at different ages and different disease trajectories.73 These autism subclusters were replicated at two other hospital EHR systems.74 Nonnegative tensor factorization has also been used to find novel clusters of diseases with medications when evaluating EHR data.75 Collectively, these approaches to defining subtypes of diseases with differential comorbid disease risk and different prognoses could suggest future implications for differential treatment using big data approaches. However, at the current time, the prognostic value of these computationally defined phenotypes for either prognosis or tailoring medical therapy is unknown.
Discussion
The identification of new, effective, and safe therapeutic targets remains challenging and expensive, with 90% of most novel therapeutics entering clinical trials failing efficacy or safety trials.76 In addition, many adverse drug reactions are unpredictable and costly, amounting to an estimated $4 billion in the US.77 A growing body of research is demonstrating the efficacy of genomic approaches linked to EHR data to prioritize drug targets for diseases, replicate and discover clinically relevant genetic variants that influence drug response, and rapidly add samples to large consortia for experiments using techniques such as MR, GWAS and PheWAS.
A major challenge to discovery is having large richly phenotypes populations available for research. Demonstrating the power of very large numbers, a recent GWAS of height on over 700,000 individuals discovered associations with 83 rare variants with effect sizes up to ten times larger than typically seen with common variants.78 In contrast, most of the research using EHR data has been applied to single healthcare systems, which in addition to limiting sample size, allows for certain biases based on coding and clinical practice variations, geographic location, and population architecture (especially with genetic studies). A current standard for genomic studies is replication in an independent cohort, or use of large meta-analyses that combine data from many independent cohorts. Legal and privacy concerns hinder sharing of clinical data. However, networks such as eMERGE network have pioneered multisite studies using EHR data linked to genetic information, including many of the EHR-based genetic studies referenced in this paper. The growing adoption of common data models79 for EHR data, including the data models put forth by the Observational Health Data Sciences and Informatics (OHDSI) network80 and the Patient-Centered Clinical Research Network (PCORnet),81 are proving effective at aggregating disparate EHR datasets.
In the future, clinical data studies will integrate EHR data with multiple other big data sources. Currently, most of the US prescription medication data is amalgamated at SureScripts, which has built a e-prescription network with most of US retail pharmacies and pharmacy benefit managers, and some other countries have national prescribing records. Incorporation of data from pharmacies would allow a more comprehensive picture of a patient exposure to medications and allow for more accurate construction of timelines of medication exposures and clinical events. In addition, integration with patient portals may aid discovery of drug adverse events. Patient generated messages to their physicians about drugs they are taking also contain information about adverse events they experience, which may not be otherwise documented in a clinical visit. Patient portals could also be actively used for dynamic feedback from patients as they are taking medications. Questionnaires could be sent to patients allowing for the collection of how they are responding to medication and whether or not they have had an adverse event. When coupled to technologies such natural language processing or intelligent form completion, a structured representation of medication exposures and response could be developed for future data mining. These sorts of collections of multiple complementary data sets could enable richer and more complete discovery than current limitations relying on a single resource such as the electronic health records – as well as benefiting clinical care.
Translation of discoveries personalizing drug therapy to clinical impact will require new types of clinical decision support implemented within EHRs. The incorporation of omic information into care – from the “simple” translation of particular variants into a recommended pharmacologic pathway to machine learning approaches operating on potentially millions of variables – are beyond what can be expected from a provider. Early decision support incorporating single or multiple variants have been implemented for pharmacogenetic prescribing at a variety of sites and within a variety of EHR systems, as propagated by NIH-funded networks such as the Electronic Medical Records and Genomics (eMERGE) and the Implementing Genomics Into Practice (IGNITE) networks.1,82–87 These systems provide the beginnings of automating genomic decision support at the point of care and have proven effective at altering prescribing behaviors and improving outcomes.87,88 To support machine learning approaches in clinical care, the new models of clinical decision support will be needed that reason on a much higher density of information, potentially modeled and computed outside the EHR in special omic resources.
Newer national cohorts have the promise of dramatically scaling “big data” discoveries to accelerate pharmaceutical development, repositioning, and adverse event prediction (Table 2). The Million Veteran Program has currently recruited over 600,000 veterans with a goal to reach at least 1 million individuals, each with linkage to the VA's longitudinal EHR system, which also includes pharmacy data.89 Other large cohort studies such as the China Kadoorie Biobank90 and the UK Biobank91 also leverage routinely-collected healthcare data. Similarly, the All of Us Research Program (formerly known as the Precision Medicine Initiative Cohort Program) has as its goal a collection of 1 million or more individuals who agree to be recontacted, will complete health surveys, and will share biospecimens and EHR data.92 Collectively, these cohorts envision an international set of millions of participants with molecular data (including genomics) linked to dense EHR data and other phenomic data that will be accessible to many researchers. The integration of medication and clinical data in a way that preserves chronicity will enable pharmacologic and genetic discovery with the sample sizes and phenotypic density necessary to advance discovery of medication effects, to prioritize potential new drug targets, and to accelerate drug repurposing. Perhaps paradoxically, it will be through the collection of very large research cohorts that we will significantly advance the care of individual patients.
Table 2. Large cohorts leveraging clinical data for genomics research.
Limited to cohorts exceeding 100,000 individuals with biosamples. Sizes reported are as of 9/2017.
| Biobank | Region | Start Year | Size | Website |
|---|---|---|---|---|
| eMERGE | U.S. | 2007 | 105,325 | gwas.net |
| BioVU | U.S. | 2007 | >247,000 | victr.vanderbilt.edu/pub/biovu |
| UK Biobank | U.K. | 2006 | 512,000 | ukbiobank.ac.uk |
| Million Veteran Program | U.S. | 2011 | >580,000 Goal: 1 million |
www.research.va.gov/MVP/default.cfm |
| Kaiser Permanente Biobank | U.S. | 2009 | 240,000 | www.rpgeh.kaiser.org |
| China Kadoorie Biobank | China | 2004 | 510,000 | ckbiobank.org |
| All of Us Research Program | U.S. | 2017 | Goal: 1 million or more | joinallofus.org |
| Taiwan Biobank | Taiwan | 2005 | 86,695 Goal: 200,000 |
www.twbiobank.org.tw |
| Geisinger MyCode | U.S. | 2007 | >150,000 |
eMERGE: The Electronic Medical Records and Genomics Network
Acknowledgments
Precision Medicine Initiative, PMI, and All of Us are service marks of the U.S. Department of Health and Human Services.
Funding: The authors would like to acknowledge funding by NIH through R01 LM 010685, P50 GM115305, U01 HG008672, U01 HG007253, R01 HL133786 and U2C OD023196.
Footnotes
Conflict of Interest: The authors have no competing interests as defined by the American Society for Clinical Pharmacology and Therapeutics, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
References
- 1.Gottesman O, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med Off J Am Coll Med Genet. 2013;15:761–771. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanseau P, et al. Use of genome-wide association studies for drug repositioning. Nat Biotechnol. 2012;30:317–320. doi: 10.1038/nbt.2151. [DOI] [PubMed] [Google Scholar]
- 3.Nelson MR, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47:856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 4.Kathiresan S, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okada Y, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Florez JC. Mining the Genome for Therapeutic Targets. Diabetes. 2017;66:1770–1778. doi: 10.2337/dbi16-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen J, et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 8.Cohen JC, Boerwinkle E, Mosley THJ, Hobbs HH. Sequence Variations in PCSK9, Low LDL, and Protection against Coronary Heart Disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 9.Sabatine MS, et al. Evolocumab and Clinical Outcomes in Patients with Cardiovascular Disease. N Engl J Med. 2017;376:1713–1722. doi: 10.1056/NEJMoa1615664. [DOI] [PubMed] [Google Scholar]
- 10.Holmes MV, et al. Mendelian randomization of blood lipids for coronary heart disease. Eur Heart J. 2014 doi: 10.1093/eurheartj/eht571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voight BF, et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. The Lancet. 2012;380:572–580. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohammadpour AH, Akhlaghi F. Future of cholesteryl ester transfer protein (CETP) inhibitors: a pharmacological perspective. Clin Pharmacokinet. 2013;52:615–626. doi: 10.1007/s40262-013-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Group, T. H.-R. C. Effects of Anacetrapib in Patients with Atherosclerotic Vascular Disease. N Engl J Med. 2017;0 doi: 10.1056/NEJMoa1706444. null. [DOI] [PubMed] [Google Scholar]
- 14.Elliott P, et al. Genetic Loci associated with C-reactive protein levels and risk of coronary heart disease. JAMA J Am Med Assoc. 2009;302:37–48. doi: 10.1001/jama.2009.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kastelein JJP, et al. Simvastatin with or without Ezetimibe in Familial Hypercholesterolemia. N Engl J Med. 2008;358:1431–1443. doi: 10.1056/NEJMoa0800742. [DOI] [PubMed] [Google Scholar]
- 16.Myocardial Infarction Genetics Consortium Investigators et al. Inactivating mutations in NPC1L1 and protection from coronary heart disease. N Engl J Med. 2014;371:2072–2082. doi: 10.1056/NEJMoa1405386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cannon CP, et al. Ezetimibe Added to Statin Therapy after Acute Coronary Syndromes. N Engl J Med. 2015;372:2387–2397. doi: 10.1056/NEJMoa1410489. [DOI] [PubMed] [Google Scholar]
- 18.Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators et al. Coding Variation in ANGPTL4, LPL, and SVEP1 and the Risk of Coronary Disease. N Engl J Med. 2016;374:1134–1144. doi: 10.1056/NEJMoa1507652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dewey FE, et al. Inactivating Variants in ANGPTL4 and Risk of Coronary Artery Disease. N Engl J Med. 2016 doi: 10.1056/NEJMoa1510926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nioi P, et al. Variant ASGR1 Associated with a Reduced Risk of Coronary Artery Disease. N Engl J Med. 2016;374:2131–2141. doi: 10.1056/NEJMoa1508419. [DOI] [PubMed] [Google Scholar]
- 21.Dewey FE, et al. Genetic and Pharmacologic Inactivation of ANGPTL3 and Cardiovascular Disease. N Engl J Med. 2017;377:211–221. doi: 10.1056/NEJMoa1612790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham MJ, et al. Cardiovascular and Metabolic Effects of ANGPTL3 Antisense Oligonucleotides. N Engl J Med. 2017;377:222–232. doi: 10.1056/NEJMoa1701329. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt AF, et al. PCSK9 genetic variants and risk of type 2 diabetes: a mendelian randomisation study. Lancet Diabetes Endocrinol. 2017;5:97–105. doi: 10.1016/S2213-8587(16)30396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinshilboum RM, Sladek SL. Mercaptopurine pharmacogenetics: Monogenic inheritance of erythrocyte thiopurine methyltransferase activity. Am J Hum Genet. 1980;32:651–662. [PMC free article] [PubMed] [Google Scholar]
- 25.Giacomini KM, et al. Genome-wide association studies of drug response and toxicity: an opportunity for genome medicine. Nat Rev Drug Discov. 2017;16:70–70. doi: 10.1038/nrd.2016.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delaney JT, et al. Predicting clopidogrel response using DNA samples linked to an electronic health record. Clin Pharmacol Ther. 2012;91:257–263. doi: 10.1038/clpt.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramirez AH, et al. Predicting warfarin dosage in European-Americans and African-Americans using DNA samples linked to an electronic health record. Pharmacogenomics. 2012;13:407–418. doi: 10.2217/pgs.11.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowton E, et al. Biobanks and electronic medical records: enabling cost-effective research. Sci Transl Med. 2014;6:234cm3. doi: 10.1126/scitranslmed.3008604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirby JC, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc JAMIA. 2016;23:1046–1052. doi: 10.1093/jamia/ocv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mosley JD, et al. A genome-wide association study identifies variants in KCNIP4 associated with ACE inhibitor-induced cough. Pharmacogenomics J. 2015 doi: 10.1038/tpj.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Driest SL, et al. Genome-Wide Association Study of Serum Creatinine Levels during Vancomycin Therapy. PloS One. 2015;10:e0127791. doi: 10.1371/journal.pone.0127791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Postmus I, et al. Pharmacogenetic meta-analysis of genome-wide association studies of LDL cholesterol response to statins. Nat Commun. 2014;5:5068. doi: 10.1038/ncomms6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei WQ, et al. Characterization of Statin Dose Response in Electronic Medical Records. Clin Pharmacol Ther. 2013 doi: 10.1038/clpt.2013.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Overby CL, et al. A collaborative approach to developing an electronic health record phenotyping algorithm for drug-induced liver injury. J Am Med Inform Assoc JAMIA. 2013;20:e243–e252. doi: 10.1136/amiajnl-2013-001930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denny JC, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pendergrass SA, et al. The use of phenome-wide association studies (PheWAS) for exploration of novel genotype-phenotype relationships and pleiotropy discovery. Genet Epidemiol. 2011;35:410–422. doi: 10.1002/gepi.20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denny JC, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31:1102–1111. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karnes JH, et al. Phenome-wide scanning identifies multiple diseases and disease severity phenotypes associated with HLA variants. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aai8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bush WS, Oetjens MT, Crawford DC. Unravelling the human genome-phenome relationship using phenome-wide association studies. Nat Rev Genet. 2016;17:129–145. doi: 10.1038/nrg.2015.36. [DOI] [PubMed] [Google Scholar]
- 40.Diogo D, et al. TYK2 protein-coding variants protect against rheumatoid arthritis and autoimmunity, with no evidence of major pleiotropic effects on non-autoimmune complex traits. PloS One. 2015;10:e0122271. doi: 10.1371/journal.pone.0122271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rastegar-Mojarad M, Ye Z, Kolesar JM, Hebbring SJ, Lin SM. Opportunities for drug repositioning from phenome-wide association studies. Nat Biotechnol. 2015;33:342–345. doi: 10.1038/nbt.3183. [DOI] [PubMed] [Google Scholar]
- 42.Brownstein JS, Sordo M, Kohane IS, Mandl KD. The Tell-Tale Heart: Population-Based Surveillance Reveals an Association of Rofecoxib and Celecoxib with Myocardial Infarction. PLOS ONE. 2007;2:e840. doi: 10.1371/journal.pone.0000840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LePendu P, et al. Pharmacovigilance Using Clinical Notes. Clin Pharmacol Ther. 2013 doi: 10.1038/clpt.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leeper NJ, et al. Practice-Based Evidence: Profiling the Safety of Cilostazol by Text-Mining of Clinical Notes. PLoS ONE. 2013;8:e63499. doi: 10.1371/journal.pone.0063499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tatonetti NP, et al. Detecting Drug Interactions From Adverse-Event Reports: Interaction Between Paroxetine and Pravastatin Increases Blood Glucose Levels. Clin Pharmacol Ther. 2011 doi: 10.1038/clpt.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu H, et al. Validating drug repurposing signals using electronic health records: a case study of metformin associated with reduced cancer mortality. J Am Med Inform Assoc JAMIA. 2014 doi: 10.1136/amiajnl-2014-002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang P, Li H, Tan X, Chen L, Wang S. Association of metformin use with cancer incidence and mortality: A meta-analysis. Cancer Epidemiol. 2013;37:207–218. doi: 10.1016/j.canep.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 48.Nielsen SF, Nordestgaard BG, Bojesen SE. Statin use and reduced cancer-related mortality. N Engl J Med. 2012;367:1792–1802. doi: 10.1056/NEJMoa1201735. [DOI] [PubMed] [Google Scholar]
- 49.Seckl MJ, et al. Multicenter, Phase III, Randomized, Double-Blind, Placebo-Controlled Trial of Pravastatin Added to First-Line Standard Chemotherapy in Small-Cell Lung Cancer (LUNGSTAR) J Clin Oncol Off J Am Soc Clin Oncol. 2017;35:1506–1514. doi: 10.1200/JCO.2016.69.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emilsson L, et al. Examining Bias in Studies of Statin Treatment and Survival in Patients With Cancer. JAMA Oncol. 2017 doi: 10.1001/jamaoncol.2017.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raymakers AJN, Sadatsafavi M, Sin DD, Vera MAD, Lynd LD. The Impact of Statin Drug Use on All-Cause Mortality in Patients With COPD. CHEST. 2017;152:486–493. doi: 10.1016/j.chest.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 52.Savova GK, Ogren PV, Duffy PH, Buntrock JD, Chute CG. Mayo Clinic NLP System for Patient Smoking Status Identification. J Am Med Inf Assoc. 2008;15:25–28. doi: 10.1197/jamia.M2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohen AM. Five-way Smoking Status Classification Using Text Hot-Spot Identification and Error-correcting Output Codes. J Am Med Inf Assoc. 2008;15:32–35. doi: 10.1197/jamia.M2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pakhomov SVS, Hanson PL, Bjornsen SS, Smith SA. Automatic classification of foot examination findings using clinical notes and machine learning. J Am Med Inform Assoc JAMIA. 2008;15:198–202. doi: 10.1197/jamia.M2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang M, et al. A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries. J Am Med Inform Assoc JAMIA. 2011;18:601–606. doi: 10.1136/amiajnl-2011-000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pakhomov SV, Buntrock JD, Chute CG. Automating the assignment of diagnosis codes to patient encounters using example-based and machine learning techniques. J Am Med Inf Assoc. 2006;13:516–25. doi: 10.1197/jamia.M2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Savova GK, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc JAMIA. 2010;17:507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y, Lasko TA, Mei Q, Denny JC, Xu H. A study of active learning methods for named entity recognition in clinical text. J Biomed Inform. 2015;58:11–18. doi: 10.1016/j.jbi.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carroll RJ, Eyler AE, Denny JC. Naïve electronic health record phenotype identification for rheumatoid arthritis. AMIA Annu Symp Proc. 2011;2011:189–196. [PMC free article] [PubMed] [Google Scholar]
- 60.Liao KP, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res. 2010;62:1120–1127. doi: 10.1002/acr.20184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin C, et al. Automatic identification of methotrexate-induced liver toxicity in patients with rheumatoid arthritis from the electronic medical record. J Am Med Inform Assoc JAMIA. 2014 doi: 10.1136/amiajnl-2014-002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin C, et al. Automatic prediction of rheumatoid arthritis disease activity from the electronic medical records. PloS One. 2013;8:e69932. doi: 10.1371/journal.pone.0069932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Le QV, et al. Building high-level features using large scale unsupervised learning. ArXiv11126209 Cs. 2011 [Google Scholar]
- 64.Lasko TA, Denny JC, Levy MA. Computational phenotype discovery using unsupervised feature learning over noisy, sparse, and irregular clinical data. PloS One. 2013;8:e66341. doi: 10.1371/journal.pone.0066341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gulshan V, et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 66.Esteva A, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khozin S, Blumenthal GM, Pazdur R. Real-world Data for Clinical Evidence Generation in Oncology. JNCI J Natl Cancer Inst. 2017;109 doi: 10.1093/jnci/djx187. [DOI] [PubMed] [Google Scholar]
- 68. [Accessed: 11th November 2017];FDA's Sentinel Initiative. Available at: https://www.fda.gov/Safety/FDAsSentinelInitiative/ucm2007250.htm.
- 69.Khozin S, Kim G, Pazdur R. Regulatory watch: From big data to smart data: FDA's INFORMED initiative. Nat Rev Drug Discov. 2017;16:nrd.201726. doi: 10.1038/nrd.2017.26. [DOI] [PubMed] [Google Scholar]
- 70.IBM's Watson can improve cancer treatment through better gene targeting. [Accessed: 11th November 2017];ScienceDaily. Available at: https://www.sciencedaily.com/releases/2017/08/170807110400.htm.
- 71.MD Anderson Cancer Center's IBM Watson project fails, and so did the journalism related to it. HealthNewsReview.org [Google Scholar]
- 72.Codella NCF, et al. Deep learning ensembles for melanoma recognition in dermoscopy images. IBM J Res Dev. 2017;61:5:1–5:15. [Google Scholar]
- 73.Doshi-Velez F, Ge Y, Kohane I. Comorbidity Clusters in Autism Spectrum Disorders: An Electronic Health Record Time-Series Analysis. Pediatrics peds. 2013:2013–0819. doi: 10.1542/peds.2013-0819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lingren T, et al. Electronic Health Record Based Algorithm to Identify Patients with Autism Spectrum Disorder. PloS One. 2016;11:e0159621. doi: 10.1371/journal.pone.0159621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ho JC, et al. Limestone: High-throughput candidate phenotype generation via tensor factorization. J Biomed Inform. 2014;52:199–211. doi: 10.1016/j.jbi.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat Rev Drug Discov. 2013;12:581–594. doi: 10.1038/nrd4051. [DOI] [PubMed] [Google Scholar]
- 77.Plumpton CO, Roberts D, Pirmohamed M, Hughes DA. A Systematic Review of Economic Evaluations of Pharmacogenetic Testing for Prevention of Adverse Drug Reactions. PharmacoEconomics. 2016;34:771–793. doi: 10.1007/s40273-016-0397-9. [DOI] [PubMed] [Google Scholar]
- 78.Marouli E, et al. Rare and low-frequency coding variants alter human adult height. Nature. 2017;542:186–190. doi: 10.1038/nature21039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rosenbloom ST, Carroll RJ, Warner JL, Matheny ME, Denny JC. Representing Knowledge Consistently Across Health Systems. Yearb Med Inform. 2017;26:139–147. doi: 10.15265/IY-2017-018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hripcsak G, et al. Characterizing treatment pathways at scale using the OHDSI network. Proc Natl Acad Sci. 2016;113:7329–7336. doi: 10.1073/pnas.1510502113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Collins FS, Hudson KL, Briggs JP, Lauer MS. PCORnet: turning a dream into reality. J Am Med Inform Assoc JAMIA. 2014;21:576–577. doi: 10.1136/amiajnl-2014-002864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.O'Donnell PH, et al. The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clin Pharmacol Ther. 2012;92:446–449. doi: 10.1038/clpt.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pulley JM, et al. Operational Implementation of Prospective Genotyping for Personalized Medicine: The Design of the Vanderbilt PREDICT Project. Clin Pharmacol Ther. 2012;92:87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weitzel KW, et al. Clinical pharmacogenetics implementation: Approaches, successes, and challenges. Am J Med Genet C Semin Med Genet. 2014;166:56–67. doi: 10.1002/ajmg.c.31390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Larson EA, Wilke RA. Integration of Genomics in Primary Care. Am J Med. 2015 doi: 10.1016/j.amjmed.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 86.Weitzel KW, et al. The IGNITE network: a model for genomic medicine implementation and research. BMC Med Genomics. 2016;9:1. doi: 10.1186/s12920-015-0162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cavallari LH, et al. Multisite Investigation of Outcomes With Implementation of CYP2C19 Genotype-Guided Antiplatelet Therapy After Percutaneous Coronary Intervention. JACC Cardiovasc Interv. 2017;3357 doi: 10.1016/j.jcin.2017.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Peterson JF, et al. Physician response to implementation of genotype-tailored antiplatelet therapy. Clin Pharmacol Ther. 2015 doi: 10.1002/cpt.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gaziano JM, et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–223. doi: 10.1016/j.jclinepi.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 90.Chen Z, et al. China Kadoorie Biobank of 0.5 million people: survey methods, baseline characteristics and long-term follow-up. Int J Epidemiol. 2011;40:1652–1666. doi: 10.1093/ije/dyr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sudlow C, et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Precision Medicine Initiative (PMI) Working Group Report to the Advisory Committee to the Director, NIH. The Precision Medicine Initiative Cohort Program – Building a Research Foundation for 21st Century Medicine [Google Scholar]
- 93.Vear SI, et al. The impact of age and CYP2C9 and VKORC1 variants on stable warfarin dose in the paediatric population. Br J Haematol. 2014;165:832–835. doi: 10.1111/bjh.12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Birdwell KA, et al. The use of a DNA biobank linked to electronic medical records to characterize pharmacogenomic predictors of tacrolimus dose requirement in kidney transplant recipients. Pharmacogenet Genomics. 2012;22:32–42. doi: 10.1097/FPC.0b013e32834e1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wells QS, et al. Genome-wide association and pathway analysis of left ventricular function after anthracycline exposure in adults. Pharmacogenet Genomics. 2017;27:247–254. doi: 10.1097/FPC.0000000000000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kawai VK, et al. Genotype and risk of major bleeding during warfarin treatment. Pharmacogenomics. 2014;15:1973–1983. doi: 10.2217/pgs.14.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Feng Q, et al. The effect of genetic variation in PCSK9 on the LDL-cholesterol response to statin therapy. Pharmacogenomics J. 2016 doi: 10.1038/tpj.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]