MAIN TEXT
Most monogenic cases of obesity in humans have been linked to mutations in genes of the leptin-melanocortin pathway. Specifically, mutations in the Melanocortin-4 Receptor (MC4R), account for 3–5% of all severe obesity cases in humans1–3. Recently, adenylate cyclase 3 (ADCY3) mutations have been implicated in obesity4,5. ADCY3 is expressed at the primary cilia of neurons6, organelles that function as hubs for select signaling pathways. Mutations that disrupt the functions of primary cilia cause ciliopathies, rare recessive pleiotropic diseases, of which obesity is a cardinal manifestation7. We demonstrate that MC4R co-localizes with ADCY3 at the primary cilium of a subset of hypothalamic neurons, that obesity-associated MC4R mutations can impair ciliary localization and that inhibition of adenylyl-cyclase signaling at the primary cilia of these neurons increases body weight. These data point at impaired signaling from the primary cilia of MC4R neurons as a common pathway for genetic causes of obesity in humans.
The majority of mammalian cells, including neurons, possess a single, immotile primary cilium, an organelle that transduces select signals7,8. Defects in the genesis or function of primary cilia cause a range of overlapping human diseases, collectively termed ciliopathies7,9,10. Several ciliopathies, such as Bardet-Biedl syndrome and Alström syndrome, cause obesity11, and mutations in genes encoding ciliary proteins, such as CEP19 and ANKRD26, cause non-syndromic obesity in mice and humans12,13. While the mechanisms underlying a number of ciliopathy-associated phenotypes, such as polycystic kidney disease or retinal degeneration, have been at least partly elucidated, how ciliary dysfunction leads to obesity remains poorly understood7,11. Ubiquitous ablation of the primary cilia of neurons in adult mice causes an increase in food intake and obesity, suggesting that ciliopathy-associated obesity involves the post-developmental disruption of anorexigenic neuronal signals14. Recently, genetic and epigenetic studies have suggested a role for ADCY3 variations in human obesity4,15 and loss of function mutations in Adcy3 in mice leads to a severe obesity phenotype5. ADCY3, a member of the adenylyl cyclase family that mediate Gs signaling from G-Protein Coupled Receptors (GPCRs), is specifically expressed at the primary cilia of neurons6.
The melanocortin 4 Receptor (MC4R) is a Gs-coupled GPCR that transduces anorexigenic signals in the long-term regulation of energy homeostasis16. Heterozygous mutations in MC4R are the most common monogenic cause of severe obesity in humans and individuals with homozygous null mutations display severe, early-onset obesity1–3. Similar to humans, deletion of Mc4r in mice causes severe obesity17. MC4R is a central component of the melanocortin system, a hypothalamic network of neurons that integrates information about peripheral energy stores and that regulates food intake and energy expenditure18. Despite being a major target for the pharmacotherapy of obesity, nothing is known about the sub-cellular localization of MC4R.
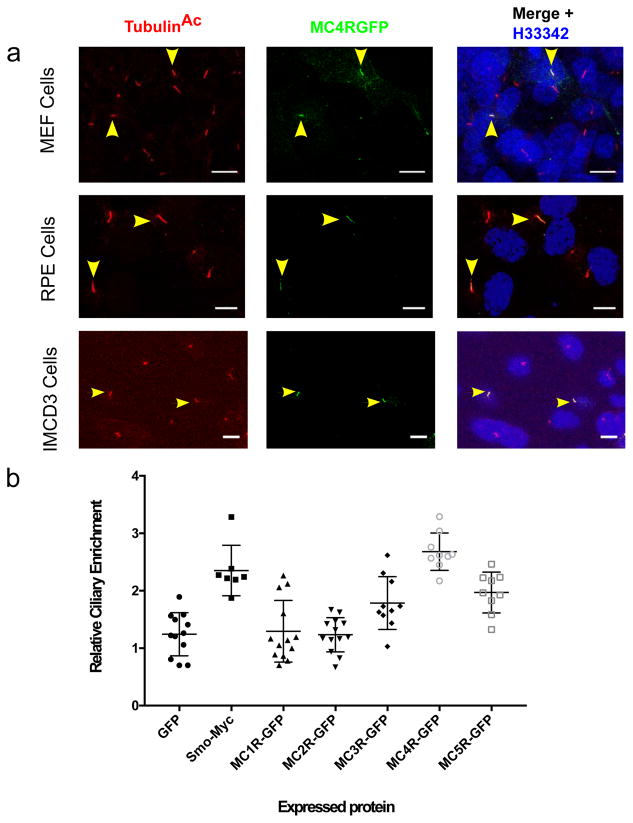
When expressed in un-ciliated heterologous cells, MC4R traffics to the cell membrane2. However, in ciliated cells such as mouse embryonic fibroblasts (MEFs), Retinal Pigment Epithelium (RPE), Inner Medullary Collecting Duct (IMCD3) cells, we find that a previously well-characterized, functional, C-terminally GFP-tagged MC4R (MC4R-GFP)19 localizes to primary cilia (Fig. 1A). In a quantitative assay, developed in IMCD3 cells, the ciliary enrichment of MC4R was comparable to that of Smoothened (SMO), a known cilium-enriched protein20,21 and was the strongest among members of the melanocortin receptor family (Fig. 1 B).
Figure 1. MC4R localizes to the primary cilia in heterologous cells.
A) Representative confocal microscopy images of transiently transfected MEF, RPE and IMCD3 cells, transfected with MC4R-EGFP labeled for the cilia specific protein acetylated Tubulin (TubulinAc, red) and GFP (green), and nuclei with Hoechst 33342 (blue). MC4R-GFP localize to the primary cilium (yellow arrowheads). Scale bars represent 10 μm. (B) Relative ciliary enrichment of melanocortin receptor family members compared to ciliary enrichment of GFP (negative control) and Smoothen (postive control). Data are Mean±sem. Means were compared to GFP (n=12 cells, mean=1.24) and Dunnet’s multiple comparison test was applied. Smo-Myc: n=7 cells, mean= 2.35, p=0,0001; MC1RGFP: n=13 cells, mean=1.29, p=0.9995, MC2RGFGP: n=13 cells, mean=1.23, p=0.9999, MC3RGFP: n=10 cells, mean=1.24, p=0.014; MC4RGFP: n=9 cells, mean=2.68, p=0.0001, MC5RGFP: n=9 cells, mean=1.96, p=0.0008.
We set out to determine if, and to what extent, MC4R localizes to primary cilia in vivo in mice. Most of the anorexigenic activity of MC4R is due to its function in a subset of Single Minded 1 (SIM1)-expressing neurons of the paraventricular nucleus of the hypothalamus (PVN)22 and all MC4R expressing neurons in the PVN express SIM123. Using a transgenic mouse line in which GFP is expressed in all SIM1-expressing neurons, we first investigated whether SIM1 expressing PVN neurons are ciliated. We find that an Adenylate-Cyclase 3 (ADCY3)-positive primary cilium was found at a majority of SIM1-expressing neurons of the PVN (Supplementary Fig. 1).
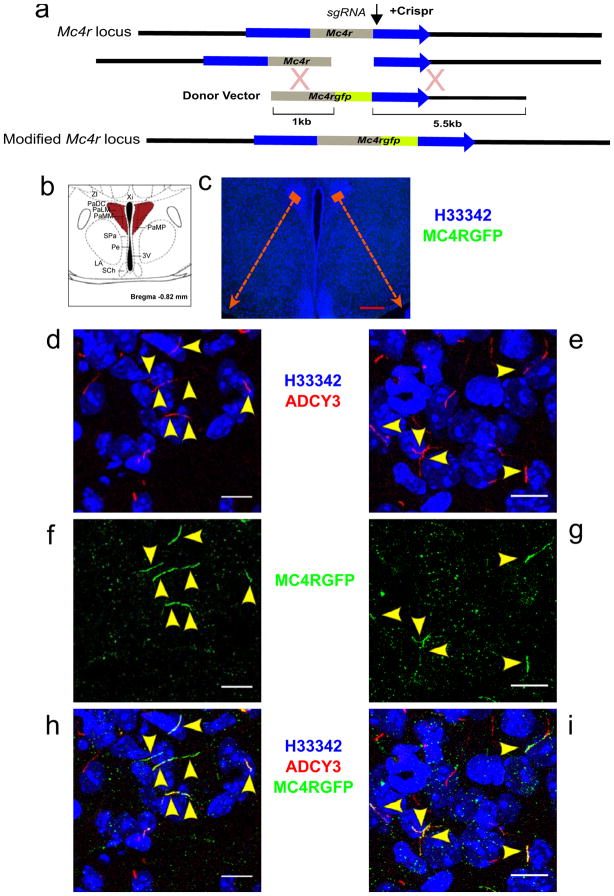
Previous attempts to determine the subcellular localization of MC4R in vivo in mice have been unsuccessful due to the small number of neurons in which it is expressed, its low abundance and the lack of tractable antibodies. Using Cas9-mediated recombination in mouse zygotes, we inserted a GFP tag in frame at the C-terminus of the endogenous mouse Mc4r locus (Fig. 2A). MC4R-GFP/+ knock-in mice do not have an obvious energy metabolism phenotype and are fertile, suggesting that the C-terminal GFP does not significantly impair the trafficking or function of MC4R in these mice. Confocal imaging of the PVN of these mice demonstrates that MC4R co-localizes with ADCY3 to the primary cilia of a subset of PVN neurons in vivo (Fig. 2, B–I).
Figure 2. MC4R localizes specifically to the primary cilia of a subset of PVN neurons in vivo.
A) Strategy used to target the mouse Mc4r locus by Crispr/Cas9. B) Schematic representation of hypothalamic region studied in C–I. C) Coronal section of the PVN of the hypothalamus of a heterozygous MC4R-GFP mouse immunostained for GFP and Adcy3. Note that no immunofluorescence is detectable at this resolution. D–I) Maximal intensity projections of confocal sections through two PVN regions indicated in C reveals MC4R-GFP (green) co-localized with neuronal primary cilia (Adcy3, red) in a subset of PVN neurons. Co-localization of MC4R-GFP with cilia has been observed in the PVN of over 10 mice (male and female) derived from two independent founders. Red scale bars represent 200 μm, White scale bars represent 10 μm.
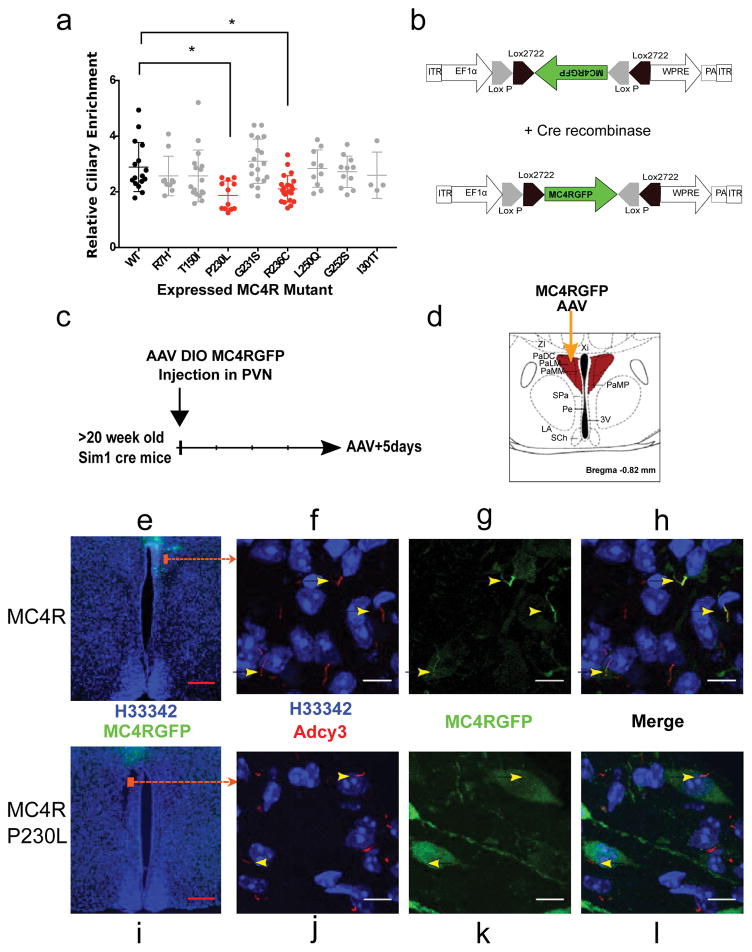
If MC4R localization to the primary cilia is essential for its function, then human obesity-causing mutations in MC4R may impair its function by compromising its ciliary localization. Heterozygous MC4R mutations are the most common genetic cause of severe childhood obesity1. Over fifty different obesity-associated mutations in MC4R have been described24 (Supplementary Fig. 2). Functional assessment of the effects of these mutations in non-ciliated cells has revealed that many of these mutations disrupt trafficking of the receptor to the membrane or impair ligand activation (Supplementary Fig. 2). In non-ciliated HEK293 cells, eight obesity-associated MC4R mutant proteins (p.(Arg7His), p.(Thr150Ile), p.(Pro230Leu), p.(Gly231Ser), p.(Arg236Cys), p.(Leu250Gln), p.(Gly252Ser), p.(Ile130Thr) ) traffic normally to the cell membrane and respond normally to α-MSH activation2,24–27. To determine whether any of these mutations alter ciliary localization of MC4R, we quantified their ciliary enrichment in IMCD3 cells (Fig. 3A). We found that P230L and R236C significantly decreased MC4R ciliary localization. Interestingly, these two mutations are located in the third intracellular domain of MC4R (Supplementary Fig. 2), a domain previously implicated in ciliary localization of other GPCRs28.
Figure 3. A subset of human obesity-associated mutations selectively impair ciliary localization of MC4R.
A) Relative ciliary enrichment of eight obesity-associated mutant MC4R compared to the wild-type (WT) receptor. Data are Mean±sem. Means were compared to MC4RWT (n=16 cells, mean=2.89) and Dunnet’s multiple comparison test was applied. MC4RR7H: n=10 cells, mean= 2.57, p=0.28; MC4RT150I: n=17 cells, mean=2.57, p=0.21; MC4RP230L: n=12, mean=1.87, p=0.0004, MC4RG231S: n=18, mean=3.10, p=0.39, MC4RR236C: n=22, mean=2.1, p=0.0013, MC4RL250Q: n=10, mean=2.83, p=0.85; MC4RG252S: n=11, mean=2.72, p=0.54; MC4RI301T: n=4, mean=2.59, p=0.47. B) Design of DIO AAV expressing MC4RGFP and MC4RP230LGFP. C) Experimental protocol D) Schematic representation of injection site. E, I) Coronal section of the PVN of the hypothalamus of Sim1 cre mice injected with the MC4RGFP DIO AAV (E) and the MC4RP230LGFP DIOAAV (I) respectively. (F–H and J–L) Maximal intensity confocal projection of sections through the PVN regions indicated in E and I respectively reveals co-localization of MC4RGFP (F–H) but not MC4RP230LGFP (J–L) with neuronal primary cilia in PVN neurons. Expressions of MC4R-GFP (WT vs P230L) have been observed in the PVN of over 6 mice (male and female). Red scale bars represent 200 μm, White scale bars represent 10 μm.
To further determine whether the P230L mutation alters ciliary localization in vivo, we injected AAVs that express MC4R-P230L-GFP and MC4R-GFP in a Cre-dependent fashion into Sim1-Cre transgenic mice (Fig. 3 B–D). The human wild-type MC4R-GFP localized to primary cilia of Sim1-expressing PVN neurons (Fig. 3 E–H) confirming that the human receptor also traffics to the cilia in vivo. In contrast, MC4R-P230L-GFP failed to co-localize with ADCY3 to primary cilia (Fig. 3 I–L). Together, these results suggest that MC4R mutations may cause human obesity by altering the ciliary localization of the receptor.
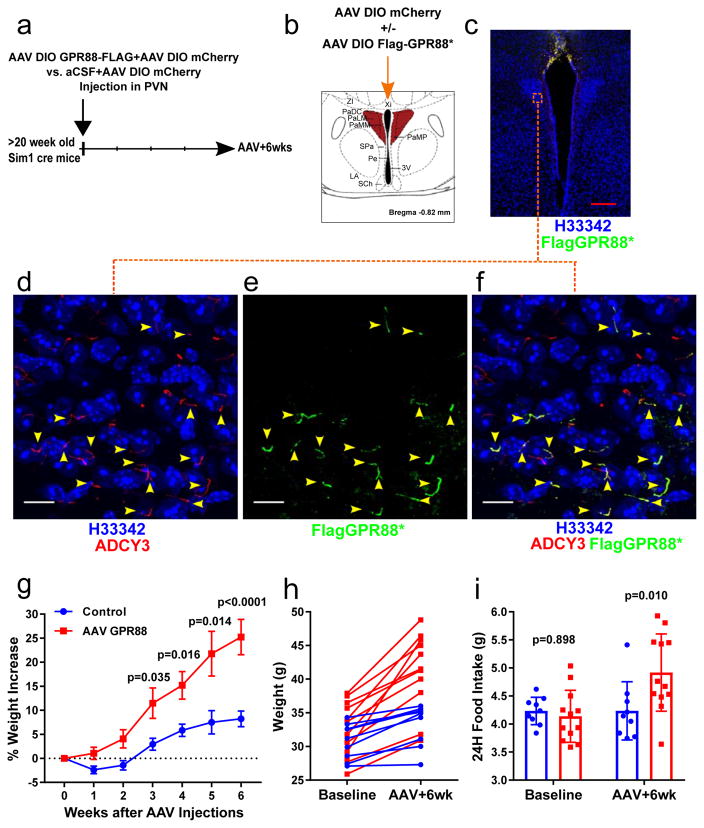
If MC4R and ADCY3 function at the primary cilia to regulate body weight, we predicted that specific inhibition of Adenylyl cyclase at the primary cilia of MC4R expressing neurons should be sufficient to cause obesity. Specific inhibition of Adenylyl cyclase at primary cilia of neuron can be achieved by expression of a constitutively active version of the cilia specific Gi-protein coupled receptor GPR88 (GPR88p.(Gly283His) or GPR88*)29. GPR88* was delivered to Sim1 expressing neurons of the PVN using the same approach used for the hMC4RGFP DIO-AAV (Fig 3) but using high level of virus delivered at the midline to ensure large coverage of PVN neurons (Supplementary Figure 3). As visualization of cilia expression requires confocal imaging, a DIO-AAV expressing mCherry was co-injected with the DIO-AAV expressing a Flag tagged version of GPR88* to facilitate verification of the accuracy of the targeting and the coverage of the PVN at the end of the experiment in each mouse (Supplementary Fig. 3). Weight-paired littermate mice injected only with the mCherry DIO-AAV were used as controls. Following AAV injections, mice in which Flag-GPR88* was expressed at the primary cilia of Sim1 expressing PVN neurons increased their food intake and gained significant weight compared to controls (Figure 4) demonstrating that Adenylyl cyclase signaling at the primary cilia of these neurons is essential for the regulation of body weight.
Figure 4. inhibition of Adenylyl cyclase at the primary cilia of Sim1 PNV neurons is sufficient to cause weight gain.
A) Experimental protocol (See also supplementary Figure 3). B) Midline stereotaxic injections of AAV DIO Flag-GPR88* + AAV DIO mCherry or AAV DIO mCherry were performed in male Sim1 cre mice. C) Coronal section of the PVN of the hypothalamus of a Sim1 cre mouse injected with the AAV DIO Flag-GPR88* + AAV DIO mCherry. D–F) Maximal intensity projections of confocal sections through the PVN region indicated by a red square in C. Arrows indicate cilia expressing GPR88. Scale bars represent 10 μm. G) Percent weight changes (mean ± SE) of Sim1cre mice following midline PVN injection of AAV DIO GPR88(G283H)+ AAV DIO mCherry (n=12) or AAV DIO mCherry (n=9). Mice were paired at baseline by body weights and litters. Repeated measures of two-way ANOVA followed by Sidak’s multiple comparisons test were performed (treatment F(1, 19) = 8.898, P = 0.0076; time F(5, 95) = 49.07, P < 0.0001; interaction F(5,95) = 8.789, P < 0.0001; p values from Sidak’s multiple comparisons test are shown in the figure). H) Individual weight changes of Sim1cre mice in G) (each line represents one mouse). I) Food intake at baseline and 6 weeks after AAV injections (mean± SD). Repeated measures of two-way ANOVA followed by Sidak’s multiple comparisons test were performed (treatment F(1, 19) = 2.41, P = 0.1370; time F(1, 19) = 9.328, P = 0.0065; interaction F(1, 19) = 9.196, P = 0.0068; p values from Sidak’s multiple comparisons test are shown in the figure). Red scale bars represent 200 μm, White scale bars represent 10 μm.
Combined, our data suggest that impaired signaling from the primary cilia of MC4R expressing neurons is a common pathway for syndromic and non-syndromic causes of monogenic obesity in humans. Our data do not indicate, however, that the primary cilia is necessary for Gs coupling and ADCY activation by MC4R since these occur in non ciliated cells. Rather our data suggest that this signaling has to occur at the primary cilia since impairing localization of MC4R at the primary cilia or inhibiting ADCY at the primary cilia impairs regulation of body weight. This functional link between MC4R and ciliopathy-associated obesity parallels findings underlying other human ciliopathy associated phenotypes. For example, syndromic and non-syndromic polycystic kidney disease is linked to impaired function of Polycytin 1 and 2, proteins expressed at the primary cilia of renal tubular cells, while impaired function of Rhodopsin (RHO) in the anterior segment of retinal cells, a specialized primary cilia, is a common pathway for both common and ciliopathy associated retinal phenotypes7.
Our findings also provide important new insights into the sub-cellular basis underlying the relationship between short-term regulation of food intake and long-term regulation of energy homeostasis. PVN MC4R expressing neurons are part of a neuronal circuitry implicated in short term control of feeding as they receive synaptic gabaergic and glutamatergic inputs in particular from the arcuate nucleus of the hypothalamus30. MC4R itself, however, controls long-term energy homeostasis as evidenced by the phenotype of MC4R deficient mice or humans and both MC4R ligands have a slower effect on food intake regulation. In strong support of this model, a recent report has elegantly established that the PVN MC4R expressing neurons receive fast-acting food intake regulating synaptic inputs from the ARC that are post-synaptically modulated by MC4R through its slower acting neuropeptide ligands αMSH and AGRP30.
Our finding that MC4R localizes to the primary cilia of MC4R PVN neurons provides for a sub-cellular compartmentalization of the slower signaling by the endogenous MC4R ligands, allowing for an independent control of long-term energy homeostasis, in neurons also implicated in the short-term regulation of food intake.
ONLINE METHODS
Studies in cell lines
Expression plasmids
hMC1R-GFP, hMC2R-GFP, hMC3R-GFP and hMC5R-GFP expression constructs were constructed as previously described for hMC4R-GFP18, as have the hMC4R mutant constructs used25.
Ciliary expression of MC4R in cultured cells
Cell lines [IMCD3 (ATCC CRL2123), MEF (NIH/3T3 ATCC CRL-1658) and RPE (ATCC CRL-4000)] were maintained at 37°C and 5% CO2 and cultured in 50% DMEM/50% Ham’s F-12 nutrient mix (CCF, UCSF, San Francisco, CA), 10% FBS, and 2mM Glutamax (GIBCO, CA) (IMCD3) or DMEM (CCF) with 10% FBS (MEF). Effectene (QIAGEN, Chatsworth, CA) was used for transfections according to the manufacturer’s protocol. Upon reaching confluency, cells were cultured in Opti-MEM (Life Technologies) for 24h to induce ciliation. Plasmid-transfected cells were fixed and stained prior to imaging by confocal microscopy. For immunofluorescence staining, cells were fixed with 4% PFA for 20 min at 4°C, permeabilized in binding buffer (10% BSA, 2% goat serum and 0.02% NaAzide in PBS) with 0.3% Triton-X100 (Sigma), and blocked in binding buffer with 3% goat serum. Primary antibodies were added in binding buffer and incubated overnight at 4°C. Subsequently, cells were washed in PBS followed by a 1h incubation with secondary antibodies and 5 min incubation with Hoechst33342 at room temperature and mounted in Prolong Gold (Life Technologies). GFP was detected by chicken polyclonal anti-GFP (abcam, ab13970). The Flag epitope was detected by mouse monoclonal anti-FlagM2 (Sigma, F1804). Primary cilia were detected by mouse monoclonal anti-Acetylated tubulin (Sigma, T7451), rabbit anti-Arl13b (gift of Tamara Caspary), or rabbit anti-Adcy3 (Santa Cruz Biotechnology, sc-588). Secondary antibody: goat anti-chicken Alexa fluor 488 (Invitrogen, A11039), goat anti-mouse Alexa fluor 555 (Invitrogen, A21424), goat anti-rabbit Alexa fluor 555 (Invitrogen, A21429), or goat anti-mouse Alexa fluor 633 (Invitrogen, A21052). Nuclei were labeled by DAPI, To-Pro3 (Invitrogen) or Hoechst33342 (Invitrogen).
Microscopes
Imaging of transfected immortalized cells was performed on a Zeiss Upright Axioscope 2 Plus Fluorescence Microscope and/or on a Leica SL, a Leica SP5 or a Zeiss LSM 780 confocal microscope.
Quantification of ciliary localization in cultured cells
A 3-plane Z-stack of transiently transfected IMCD3 cells was acquired on an Olympus IX-70 microscope, and best focus of average was recorded using Metamorph software (Molecular Devices, Sunnyvale, CA). Relative ciliary enrichment was calculated using Matlab Software as the ratio between the green chanel pixel intensity of GFP-chimera expression at the primary cilium versus pixel intensity of the cell body, wherein the cilium was defined by acetylated tubulin staining recorded in the red channel.
In vivo studies in mice
All animal procedures were approved by the Institutional Animal Care and Use Committee of the University of California, San Francisco. Zygote injection and implantation was performed at the transgenic core of the Gladstone Institute.
Generation of Mc4r-GFP knock-in mice
Super-ovulated female FVB/N mice (4 weeks old) were mated to FVB/N stud males. Fertilized zygotes were collected from oviducts and injected with (1) Cas9 protein (50 ng/ul), (2) a donor vector (20 ng/ul) consisting of 1kb of 5’flanking sequence (i.e. the MC4R coding sequence) followed by GFP (cloned in frame) and 5.5 kb of 3’flanking sequence and (3) a sgRNA (25 ng/ul) of which the guide sequence (see supplementary table) was designed to target nucleotides immediately downstream the MC4R stop codon in a short region that was not present in the donor vector into pronucleus. Injected zygotes were implanted into oviducts of pseudopregnant CD1 female mice. Pups were genotyped for insertions at the correct loci by PCR. Tissue specific expression of Mc4r-GFP was verified by qPCR. Imaging experiments were done in F2–F5 mice from two different founders.
Origin of the other mouse lines used
Mice expressing Cre under the control of the Sim1 promoter, Tg(Sim1-cre)1Lowl, were obtained from Jackson Laboratories (Bar Harbor, ME). Sim1-GFP mice, Tg(Sim1-EGFP)AX55Gsat, were obtained from the Mutant Mouse Regional Resource Center (Davis, CA).
Mice were housed in a barrier facility and maintained on a 12:12 light cycle (on: 0700-1900) at an ambient temperature of 23±2°C and relative humidity 50–70%. Mice were fed with rodent diet 5058 (Lab Diet) and group-housed up to 5. Experiments were performed with weight matched littermates.
Generation and injection of AAVs
AAV DIO MC4RGFP, AAV DIO P230LMC4RGFP and AAV DIO GPR88* plasmids were generated by replacing hChR2(H134R)-EYFP in pAAV-Ef1a-DIO- hChR2(H134R)-EYFP-WPRE-pA (obtained from K. Desseiroth, Stanford University) with hMC4RGFP, P230LMC4RGFP or GPR88(G283H) respectively. AAV DJ were prepared and titrated by the Stanford Neuroscience Viral Core which also provided the stock mCherry DIO-AAV ( GVVC-AAV-14).
DIO AAV were injected in the PVN of Tg(Sim1-cre)1Lowl mice to anatomically and genetically restrict expression to Sim1 expressing PVN neurons. For experiments presented in Figure 3, 0.2 ul of AAV DIO MC4RGFP or AAV DIO P230LMC4RGFP were stereotaxically injected unilaterally in the PVN (coordinates: AP=−0.8, ML=0.2, DV=−5.2). Mice were sacrificed 7 days after the injections. For experiments presented in Figure 4 (and supplementary Figure 3), AAV DIO mCherry +/− AAV DIO Flag-GPR88* were stereotaxically injected in 1ul at the midline just above the third ventricle (coordinates: AP=−0.8, ML=0.0, DV=−5.2). Weight was measured for 2 months, after which mice were sacrificed to confirm the site of injection. Mice with missed injections were excluded prior to data analysis. mCherry expression was assessed in all mice by widefield microscopy to verify the accuracy and extent of the AAV infection and GPR88* expression since in mice injected with both AAV DIO mCherry and AAV DIO Flag-GPR88* infected neurons were infected with both viruses (supplementary figure 3).
Mouse Metabolism Studies
For experiments presented in Figure 4 (and supplementary Figure 3), mice were single housed after AAV injections. Weight was measured for 2 months. Food intake was measured by CLAMS (Columbus Instruments, Columbus, OH) at baseline and 6 weeks after AAV injections. Mice were tested over 96 continuous hours, and the data from the middle 48 hours were analyzed.
Immunofluorescence studies of mouse hypothalamus
Mice were perfused trans-cardially with PBS followed by 4% paraformaldehyde fixation solution. Brains were dissected and post-fixed in fixation solution at 4°C overnight, soaked in 30% sucrose solution overnight, embedded in O.C.T. (Tissue-Tek, Sakura Finetek USA, INC., Torrance, CA), frozen, and cut into 20–35 μm coronal sections. After washing, sections were blocked for 1 hr (3% normal goat serum in PBS, 0.4% Triton X-100, 0.2% sodium azide) followed by incubation with primary antibody: chicken anti GFP (abcam, ab13970), rabbit anti-Adcy3 (Santa Cruz Biotechnology, sc-588) or mouse anti FLAG M1 (Sigma, F3040) overnight at 4°C. Sections were extensively washed in PBS, and then incubated with secondary antibody: goat anti-chicken Alexa fluor 488 (Invitrogen, A11039), goat anti-mouse Alexa fluor 488 (Invitrogen, A11001), or goat anti-rabbit Alexa fluor 633 (Invitrogen, A21071).
Image capture and processing
Widefield images were generated using a Zeiss ApoTome microscope. Confocal images were generated using a Zeiss LSM 780 confocal microscope. In confocal images, MC4R-GFP was labeled with Alexa 488, and the neuronal cilia marker Adcy3 was labeled with Alexa 633. For Alexa 488, the detector range was set from 490–534 nm. For Alexa 633, the detector range was set from 600–750 nm. Images were processed with Fiji. Maximal intensity Z projections are from at least 20 slices over 10 μm.
In confocal images, GPR88-FLAG was labeled with Alexa 488, and the neuronal cilia marker Adcy3 was labeled with Alexa 633. For Alexa 488, the detector range was set from 490–534 nm. For Alexa 633, the detector range was set from 600–750 nm. mCherry was detected by direct fluorescence. Images were processed with Fiji. Maximal intensity Z projections are from at least 40 slices over 20 μm.
Statistics
Sample sizes were chosen based upon the estimated effect size drawn from previous publications7 and from the performed experiments. Data distribution were assumed to be normal but this was not formally tested. All test used are indicated in the figures. We analyzed all data using Prism 7.0 (GraphPad Software).
Supplementary Material
Acknowledgments
This research was supported by the UCSF DRC NIH P30DK063720 and the UCSF NORC NIH P30DK098722; by an AMC Graduate School PhD Scholarship to J.E.S.; NIH R01AR05439 & NIH R01GM095941, a Burroughs Wellcome Fund, the David & Lucile F. Packard Foundation to J.F.R; NIH R01DK60450 to C.V.; NIH RO1DK106404 to C.V. and J.F.R.;NIH RO1DA012864 and NIH RO1DA010711 to M.V. and a New Frontier Research Award through the UCSF Program for Breakthrough Biomedical Research to C.V., J.R, and M.V.
Footnotes
AUTHOR CONTRIBUTIONS:
C.V and J.F.R supervised the research. J.E.S, Y.W, C.V. and J.F.R. conceived and designed experiments, performed experiments, performed statistical analysis, analyzed the data and wrote the paper. S.Z. Performed experiments. A.B. Performed experiments and analyzed data relevant to Figure 4. B.E. Conceived and performed experiments and anlayzed data relevant to figure 2. A.M and M.V contributed regents and expertise relevant to Figure 4.
COMPETING FINANCIAL INTERESTS:
The authors declare no competing financial interests.
DATA AVAILABILITY:
All the data supporting the findings of this study are available from the corresponding author upon request.
REFERENCES FOR MAIN TEXT
- 1.Lubrano-Berthelier C. Melanocortin 4 Receptor Mutations in a Large Cohort of Severely Obese Adults: Prevalence, Functional Classification, Genotype-Phenotype Relationship, and Lack of Association with Binge Eating. Journal of Clinical Endocrinology & Metabolism. 2006;91:1811–1818. doi: 10.1210/jc.2005-1411. [DOI] [PubMed] [Google Scholar]
- 2.Vaisse C, et al. Melanocortin-4 receptor mutations are a frequent and heterogeneous cause of morbid obesity. J Clin Invest. 2000;106:253–62. doi: 10.1172/JCI9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vaisse C, Clement K, Guy-Grand B, Froguel P. A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat Genet. 1998;20:113–4. doi: 10.1038/2407. [DOI] [PubMed] [Google Scholar]
- 4.Stergiakouli E, et al. Genome-wide association study of height-adjusted BMI in childhood identifies functional variant in ADCY3. Obesity (Silver Spring) 2014;22:2252–9. doi: 10.1002/oby.20840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z, et al. Adult type 3 adenylyl cyclase-deficient mice are obese. PLoS One. 2009;4:e6979. doi: 10.1371/journal.pone.0006979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop GA, Berbari NF, Lewis J, Mykytyn K. Type III adenylyl cyclase localizes to primary cilia throughout the adult mouse brain. J Comp Neurol. 2007;505:562–71. doi: 10.1002/cne.21510. [DOI] [PubMed] [Google Scholar]
- 7.Reiter JF, Leroux MR. Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol. 2017;18:533–547. doi: 10.1038/nrm.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–44. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green JA, Mykytyn K. Neuronal ciliary signaling in homeostasis and disease. Cell Mol Life Sci. 2010;67:3287–97. doi: 10.1007/s00018-010-0425-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–43. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaisse C, Reiter JF, Berbari NF. Cilia and Obesity. Cold Spring Harb Perspect Biol. 2017;9 doi: 10.1101/cshperspect.a028217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shalata A, et al. Morbid obesity resulting from inactivation of the ciliary protein CEP19 in humans and mice. Am J Hum Genet. 2013;93:1061–71. doi: 10.1016/j.ajhg.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acs P, et al. A novel form of ciliopathy underlies hyperphagia and obesity in Ankrd26 knockout mice. Brain Struct Funct. 2014 doi: 10.1007/s00429-014-0741-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davenport J, et al. Disruption of Intraflagellar Transport in Adult Mice Leads to Obesity and Slow-Onset Cystic Kidney Disease. Current Biology. 2007;17:1586–1594. doi: 10.1016/j.cub.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu L, Shen C, Seed Ahmed M, Ostenson CG, Gu HF. Adenylate cyclase 3: a new target for anti-obesity drug development. Obes Rev. 2016;17:907–14. doi: 10.1111/obr.12430. [DOI] [PubMed] [Google Scholar]
- 16.Krashes MJ, Lowell BB, Garfield AS. Melanocortin-4 receptor-regulated energy homeostasis. Nat Neurosci. 2016;19:206–19. doi: 10.1038/nn.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huszar D, et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 1997;88:131–41. doi: 10.1016/s0092-8674(00)81865-6. [DOI] [PubMed] [Google Scholar]
- 18.Morton GJ, Meek TH, Schwartz MW. Neurobiology of food intake in health and disease. Nat Rev Neurosci. 2014;15:367–78. doi: 10.1038/nrn3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lubrano-Berthelier C, et al. Intracellular retention is a common characteristic of childhood obesity-associated MC4R mutations. Hum Mol Genet. 2003;12:145–153. doi: 10.1093/hmg/ddg016. [DOI] [PubMed] [Google Scholar]
- 20.Aanstad P, et al. The extracellular domain of Smoothened regulates ciliary localization and is required for high-level Hh signaling. Curr Biol. 2009;19:1034–9. doi: 10.1016/j.cub.2009.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corbit KC, et al. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 22.Balthasar N, et al. Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell. 2005;123:493–505. doi: 10.1016/j.cell.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 23.Garfield AS, et al. A neural basis for melanocortin-4 receptor-regulated appetite. Nat Neurosci. 2015;18:863–71. doi: 10.1038/nn.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bromberg Y, Overton J, Vaisse C, Leibel RL, Rost B. In silico mutagenesis: a case study of the melanocortin 4 receptor. FASEB J. 2009;23:3059–69. doi: 10.1096/fj.08-127530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calton M, et al. Association of functionally significant Melanocortin-4 but not Melanocortin-3 receptor mutations with severe adult obesity in a large North-American case control study. Human Molecular Genetics. 2008 doi: 10.1093/hmg/ddn431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ersoy BA, et al. Mechanism of N-terminal modulation of activity at the melanocortin-4 receptor GPCR. Nat Chem Biol. 2012;8:725–30. doi: 10.1038/nchembio.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinney A, et al. Melanocortin-4 receptor gene: case-control study and transmission disequilibrium test confirm that functionally relevant mutations are compatible with a major gene effect for extreme obesity. J Clin Endocrinol Metab. 2003;88:4258–67. doi: 10.1210/jc.2003-030233. [DOI] [PubMed] [Google Scholar]
- 28.Berbari N, Johnson AD, Lewis J, Askwith C, Mykytyn K. Identification of ciliary localization sequences within the third intracellular loop of G protein-coupled receptors. Mol Biol Cell. 2008;19:1540–7. doi: 10.1091/mbc.E07-09-0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marley A, Choy RW, von Zastrow M. GPR88 reveals a discrete function of primary cilia as selective insulators of GPCR cross-talk. PLoS One. 2013;8:e70857. doi: 10.1371/journal.pone.0070857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fenselau H, et al. A rapidly acting glutamatergic ARC-->PVH satiety circuit postsynaptically regulated by alpha-MSH. Nat Neurosci. 2017;20:42–51. doi: 10.1038/nn.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.