Abstract
In 2020, 146,063 deaths due to pancreatic cancer are estimated to occur in Europe and the United States combined. To identify common susceptibility alleles, we performed the largest pancreatic cancer GWAS to date, including 9040 patients and 12,496 controls of European ancestry from the Pancreatic Cancer Cohort Consortium (PanScan) and the Pancreatic Cancer Case-Control Consortium (PanC4). Here, we find significant evidence of a novel association at rs78417682 (7p12/TNS3, P = 4.35 × 10−8). Replication of 10 promising signals in up to 2737 patients and 4752 controls from the PANcreatic Disease ReseArch (PANDoRA) consortium yields new genome-wide significant loci: rs13303010 at 1p36.33 (NOC2L, P = 8.36 × 10−14), rs2941471 at 8q21.11 (HNF4G, P = 6.60 × 10−10), rs4795218 at 17q12 (HNF1B, P = 1.32 × 10−8), and rs1517037 at 18q21.32 (GRP, P = 3.28 × 10−8). rs78417682 is not statistically significantly associated with pancreatic cancer in PANDoRA. Expression quantitative trait locus analysis in three independent pancreatic data sets provides molecular support of NOC2L as a pancreatic cancer susceptibility gene.
Genetic variants associated with susceptibility to pancreatic cancer have been identified using genome wide association studies (GWAS). Here, the authors combine data from over 9000 patients and perform a meta-analysis to identify five novel loci linked to pancreatic cancer.
Introduction
Pancreatic cancer is currently the third leading cause of cancer-related deaths in the United States and the fifth leading cause in Europe1,2, and is predicted to become the second leading cause of cancer-related deaths in the United States by 20303,4. Incidence rates of pancreatic cancer have also gradually increased1. Genetic susceptibility plays an important role in pancreatic cancer risk through mutations in known genes for hereditary cancer or hereditary pancreatitis5–11, and common genetic variants identified through genome-wide association studies (GWAS)12–16.
With the aim of identifying additional common pancreatic cancer risk loci, the Pancreatic Cancer Cohort Consortium (PanScan: https://epi.grants.cancer.gov/PanScan/) and the Pancreatic Cancer Case-Control Consortium (PanC4: http://www.panc4.org/) have performed GWAS of pancreatic ductal adenocarcinoma (PDAC) in populations of European ancestry. These studies, namely PanScan I12, PanScan II13, PanScan III14,15, and PanC416, have led to the identification of 13 genomic loci carrying 17 independent pancreatic cancer risk signals on chromosomes 1q32.1 (two independent signals in NR5A2), 2p14 (ETAA1), 3q28 (TP63), 5p15.33 (three independent risk loci in the CLPTM1L-TERT gene region), 7p14.1 (SUGCT), 7q23.2 (LINC-PINT), 8q24.1 (two independent risk loci in the MYC-PVT1 gene region), 9q34.2 (ABO), 13q12.2 (PDX1), 13q22.1 (non-genic), 16q23.1 (BCAR1), 17q24.3 (LINC00673), and 22q12.1 (ZNRF3)12–16. A fourth independent risk locus at 5p15.33 (TERT) was identified through a candidate gene analysis by the PANcreatic Disease ReseArch (PANDoRA) case–control consortium17,18. GWAS in populations from China19 and Japan20 have identified eight additional GWAS significant pancreatic cancer risk loci on chromosomes 5p13.1 (DAB2), 6p25.3 (FOXQ1), 7q36.2 (DPP6), 12p11.21 (BICD1), 10q26.11 (PRLHR), 21q21.3 (BACH1), 21q22.3 (TFF1), and 22q13.32 (FAM19A5)19. The overlap among loci identified in the European and Asian ancestry scans with current sample sizes is limited14,16,21.
Here we report the findings of the largest Pancreatic Cancer GWAS study to date. Five novel regions of association were identified. A locus at rs78417682 (7p12/TNS3, P = 4.35 × 10−8) was identified in meta-analysis of the PanScanI/II, PanScanIII, and PanC4 data. Four additional loci, rs13303010 at 1p36.33 (NOC2L, P = 8.36 × 10−14), rs2941471 at 8q21.11 (HNF4G, P = 6.60 × 10−10), rs4795218 at 17q12 (HNF1B, P = 1.32 × 10−8), and rs1517037 at 18q21.32 (GRP, P = 3.28 × 10−8) were identified after replication in additional cases and controls from the PANDoRA consortium.
Results
Association analysis
In the current study, we performed the largest association analysis of pancreatic cancer risk to date, including 9040 individuals diagnosed with pancreatic cancer and 12,496 control individuals of European ancestry (Supplementary Table 1) from four GWAS studies (PanScan I, PanScan II, PanScan III, and PanC4). These individuals were previously genotyped on the Illumina HumanHap550, 610-Quad, OmniExpress, and OmiExpressExome arrays, respectively12–16. Because of the extensive overlap of variants on the arrays, data from PanScan I and PanScan II were analyzed jointly, while PanScan III and PanC4 were each analyzed separately. Imputation was performed using the 1000 G (Phase 3, v1) reference data set22. After quality control 11,381,182 variants were analyzed for 21,536 individuals (7167 in PanScan I+II, 6785 in PanScan III, and 7584 in PanC4). A quantile–quantile plot (Supplementary Figure 1) showed little evidence of systematic inflation (λ = 1.002 for PanScan I+II, λ = 1.051 for PanScan III, λ = 1.025 for PanC4, and λ = 1.05 for the meta-analysis).
In a fixed-effect meta-analysis of PanScan I+II, PanScan III, and PanC4, we observed robust associations at our previously identified susceptibility loci in individuals of European ancestry12–16 (Supplementary Table 2). We also noted one novel locus that met the genome-wide significance threshold (P < 5 × 10−8: Wald test) at chromosome 7p12 in the TNS3 gene, and nine additional promising loci (P < 1 × 10−6: Wald test). These 10 loci were carried forward to an independent replication in up to 2737 pancreatic cancer cases and 4752 control individuals from the PANcreatic Disease ReseArch (PANDoRA) consortium23. In a combined meta-analysis of up to 11,537 pancreatic cancer cases and 17,107 control individuals from PanScan I+II, PanScan III, PanC4, and PANDoRA, we identified three additional loci of genome-wide significance: rs13303010 at 1p36.33 (NOC2L, odds ratio (OR) = 1.26; 95% confidence interval (CI) 1.19–1.35, P = 8.36 × 10−14: Wald test), rs2941471 at 8q21.11 (HNF4G, OR = 0.89, 95% CI 0.85–0.93, P = 6.60 × 10−10: Wald test), and rs4795218 at 17q12 (HNF1B, OR = 0.88, 95% CI 0.84–0.92, P = 1.32 × 10−8: Wald test). A locus that was previously reported to be suggestive in the PanC4 study at 18q21.32 in the GRP gene16 also surpassed the significance threshold (rs1517037, OR = 0.86, 95% CI 0.80–0.91, P = 3.28 × 10−8: Wald test) in our meta-analysis. (Table 1 and Fig. 1). The single-nucleotide polymorphism (SNP) at 7p12 in TNS3 (rs73328514) was not significantly associated with pancreatic cancer in PANDoRA (OR = 0.94, PPANDoRA = 0.31; OR = 0.85, PCombined = 1.35 × 10−7: Wald test).
Table 1.
Novel pancreatic cancer susceptibility loci
| Chra SNP Positionb gene | Effect allele (minor)/reference allele | Statistic | PanScan I/II 3535 cases and 3642 controls | PanScan III 1582 cases and 5203 controls | PanC4 3933 cases and 3651 controls | ALL GWAS 9040 cases and 12,496 controls | PANDoRA 2497 cases and 4611 controls | GWAS+PANDoRa 11,537 cases and 17,107 controls |
|---|---|---|---|---|---|---|---|---|
| 1p36.33 rs13303010 894,573 NOC2L | G/A | MAFc cases;controls | 0.14; 0.13 | 0.12; 0.10 | 0.13; 0.11 | 0.14; 0.10 | – | |
| Infod | 0.42 | g | g | g | – | |||
| OR (CI) | 1.15 (1.01–1.26) | 1.22 (1.09–1.33) | 1.16 (1.07–1.24) | 1.20 (1.12–1.29) | 1.45 (1.33–1.57) | 1.26 (1.19–1.35) | ||
| P value | 3.64 × 10−2 | 1.48 × 10−3 | 9.54 × 10−4 | 7.30 × 10−7 | 6.00 × 10−10 | 8.36 × 10−14 | ||
| Heterogeneity P valuee | 6.49 × 10−1 | 4.57 × 10−2 | ||||||
| 7p12 rs73,328,514 47488569 TNS3 | T/A | MAF cases; controls | 0.09; 0.11 | 0.10; 0.12 | 0.10; 0.12 | 0.10; 0.11 | – | |
| Info | 0.93 | 0.97 | 0.97 | g | – | |||
| OR (CI) | 0.80 (0.71–0.89) | 0.88 (0.76–1.02) | 0.82 (0.74–0.92) | 0.83 (0.77–0.88) | 0.94 (0.83–1.06) | 0.85 (0.80–0.90) | ||
| P value | 8.38 × 10−5 | 9.31 × 10−2 | 3.61 × 10−4 | 4.35 × 10−8 | 3.08 × 10−1 | 1.35 × 10−7 | ||
| Heterogeneity P value | 5.98 × 10−1 | 2.35 × 10−1 | ||||||
| 8q21.11 rs2941471 76,470,404 HNF4G | G/A | MAF cases; controls | 0.40; 0.43 | 0.41; 0.42 | 0.41; 0.43 | 0.40; 0.43 | ||
| Info | 1.0 | 1.0 | 1.0 | g | ||||
| OR (CI) | 0.87 (0.79–0.94) | 0.91 (0.80–1.01) | 0.89 (0.82–0.96) | 0.89 (0.86–0.94) | 0.87 (0.79–0.94) | 0.89 (0.85–0.93) | ||
| P value | 2.39 × 10−4 | 8.30 × 10−2 | 2.19 × 10−3 | 4.73 × 10−7 | 2.42 × 10−4 | 6.60 × 10−10 | ||
| Heterogeneity P value | 7.73 × 10−1 | 7.87 × 10−1 | ||||||
| 17q12 rs4795218 36,078,510 HNF1B | A/G | MAF cases; controls | 0.20; 0.23 | 0.22; 0.23 | 0.21; 0.23 | 0.21; 0.23 | ||
| Info | 0.96 | 0.96 | 0.95 | g | ||||
| OR (CI) | 0.87 (0.80–0.95) | 0.88 (0.78–0.98) | 0.88 (0.81–0.95) | 0.88 (0.82–0.93) | 0.90 (0.82–0.98) | 0.88 (0.84–0.92) | ||
| P value | 1.12 × 10−3 | 2.29 × 10−2 | 1.11 × 10−3 | 2.73 × 10−7 | 1.38 × 10−2 | 1.32 × 10−8 | ||
| Heterogeneity P value | 9.96 × 10−1 | 9.78 × 10−1 | ||||||
| 18q21.32 rs1517037 56,878,274 GRP | T/C | MAF cases; controls | 0.16; 0.19 | 0.17; 0.19 | 0.17; 0.18 | 0.17; 0.19 | ||
| Info | g | g | g | g | – | |||
| OR (CI) | 0.82 (0.75–0.89) | 0.92 (0.82–1.04) | 0.90 (0.83–0.98) | 0.87 (0.82–0.93) | 0.87 (0.79–0.97) | 0.86 (0.80–0.91) | ||
| P value | 7.56 × 10−6 | 1.90 × 10−1 | 1.64 × 10−2 | 8.81 × 10−7 | 1.17 × 10−2 | 3.28 × 10−8 | ||
| Heterogeneity P value | 1.87 × 10-1 | 7.73 × 10-2 | 1.03 × 10-1 |
a Cytogenetic regions according to NCBI Human Genome Build 37
b SNP position according to NCBI Human Genome Build 37
c Minor allele frequency
d Quality of imputation metric. See online methods for more detail. If a SNP is genotyped and not imputed, a “g” is reported
e P value from test of heterogeneity
Fig. 1.
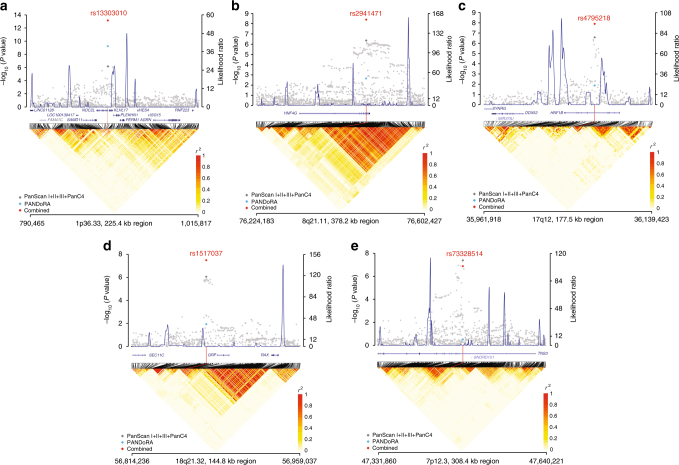
Association results, recombination hotspots, and LD plots for new pancreatic cancer susceptibility regions. The top half of each panel shows the association results for the meta-analysis of PanScan I+II, PanScan III, and PanC4 (gray diamonds). The results for the replication of the marker SNP at each locus are shown for PANDoRA (light blue diamonds) and the combined meta-analysis results (red diamonds). Overlaid are likelihood ratio statistics estimating putative recombination hotspots across the region based on the inference using the CEU 1000 G Phase 3 data. Genomic coordinates are plotted on the x axis (Genome build hg19), P values for the association analysis are shown on the left y axis, and recombination hotspot likelihood ratio on the right y axis. The bottom half of each panel shows LD heat maps based on r2 values from the 1000 G Phase 3 CEU population for all variants included in the analysis. Shown are results for chromosomes 1p36.33 (a), 8q21.11 (b), 17q12 (c), 18q21.32 (d), and 7p12 (e)
The marker SNP at 1p36.33 (rs13303010) maps to the first intron of the NOC2L gene, which encodes the nucleolar complex protein 2 homolog (NOC2-like protein, also known as novel INHAT repressor), an inhibitor of histone acetyltransferase (HAT) activity and transcriptional repressor24. This protein also directly binds to p53, stabilizing an interaction between the mitotic kinase Aurora B and p53, resulting in inhibition of p53-mediated transcriptional activation24,25. Likewise, NOC2-like protein binds and inhibits transcriptional activity of the closely related tumor suppressor protein, p63 (TAp63)26. Notably, we have previously identified a pancreatic cancer risk locus intronic to the TP63 gene16.
At chromosome 8q21.11, the newly associated SNP (rs2941471) is intronic to HNF4G, which encodes hepatocyte nuclear factor 4 gamma, a transcription factor (TF) of the nuclear receptor superfamily. Mice lacking Hnf4g have higher numbers of pancreatic β-cells, increased glucose-induced insulin secretion, and improved glucose tolerance27. Of the multiple GWAS that have reported association signals at this locus for other conditions, the variant with the highest linkage disequilibrium (LD) with our pancreatic cancer-associated variant, rs2941471, has been significantly associated with variations in serum urate concentrations (rs294148428, r2 = 0.56, in 1000 G EUR). Interestingly, we have previously shown that urate levels are associated with pancreatic cancer risk29. Additional variants in the HNF4G gene region, including those significantly associated with body mass index (BMI)30, obesity31, and breast cancer32 are less correlated with rs2941471 (r2 = 0.02–0.12).
The signal at 17q12 (rs4795218) maps to the fourth intron of HNF1B, encoding another member of the hepatocyte nuclear factor family. HNF1B plays an important role in pancreatic development, acting in a transcriptional network that controls the differentiation of multipotent progenitor cells to acinar, ductal, and endocrine cells33,34. Mutations in HNF1B account for a small percentage (1–2%) of maturity onset diabetes of the young (MODY)35. In addition, variants in the HNF1B gene region that are modestly linked with rs4795218 have previously been associated with the development of prostate cancer (rs4794758, r2 = 0.59 in 1000 G EUR)36. Although additional variants in this region have been associated with other cancers including prostate36, endometrial37, and testicular38 cancers, they do not appear to mark the same signal (rs4430796, rs11263763, rs7501939, respectively, r2 with rs4795218 ≤ 0.005 in 1000 G EUR).
The two novel risk loci in genes of the hepatocyte nuclear factor family are intriguing in light of our previously published suggestive evidence of association with other members of this family, including a locus at 12q24.31 in the HNF1A gene (rs1182933, OR = 1.11, P = 3.49 × 10−7: Wald test) and a locus on 20q13.11 ~20 kb downstream of the HNF4A gene (rs6073450, OR = 1.09, P = 4.55 × 10−6: Wald test; Supplementary Table 3)16. Members of this family of TFs play important roles in pancreatic development and regulate specific gene expression programs in pancreatic acini, pancreatic islets, and hepatocytes in adults39–42. Importantly, HNF1A appears to be a critical member of a signaling network that maintains homeostasis in the adult pancreas40,43. We have also previously shown that HNF1A may be a tumor suppressor gene in the pancreas44,45. Inherited mutations in several genes of this family cause pancreatic beta cell dysfunction resulting in MODY: HNF4A (MODY 1), HNF1A (MODY 3), and HNF1B (MODY 5)35. Common variants in or close to some of these genes have also been significantly associated with type 2 diabetes (T2D) and body mass index (BMI)/obesity, both known epidemiologic risk factors for pancreatic cancer46. However, the low LD between those signals on 8q21.11/HNF4G (PDAC-rs2941471 and BMI-rs1740581947, r2 = 0.05), 17q12/HNF1B (PDAC-rs4795218 and T2D-rs443079648, r2 = 0.0007), and 12q24.31/HNF1A (PDAC-rs7310409 and T2D-rs1242735348, r2 = 0.18) indicates that the underlying functional mechanism for the pancreatic cancer GWAS signals may differ from those for adult-onset T2D and BMI.
At 18q21.32, the signal marked by rs1517037 is ~10 kb upstream of the GRP gene, which encodes a member of the bombesin-like family of gastrin-releasing peptides that stimulates the release of gastrointestinal hormones, including amylase49, a marker of acute pancreatitis. Correlated variants at this locus (r2 = 0.82–1.00) are associated with inflammatory bowel disease (IBD)50 and BMI47.
The locus at 7p12 is marked by an intronic SNP (rs73328514) in TNS3. This gene encodes Tensin-3, a member of a family of focal adhesion-associated proteins (Tensin-1 through Tensin-4) that regulate cell adhesion and migration51 and may play a role in metastasis52.
We identified suggestive evidence for additional risk loci in the meta-analysis of PanScan and PanC4 data; at 9q31.1 in the SMC2 gene (rs2417487, P = 1.49 × 10−7: Wald test), at 4q31.22 near EDNRA (rs6537481, P = 1.15 × 10−7: Wald test), and at 16q24.1 near LINC01081/LINC00917 (rs7200646, P = 1.39 × 10−7: Wald test) but these were not significantly associated in PANDoRA (P = 0.38, 0.91 and 0.93, respectively: Wald tests; Supplementary Table 4). SMC2 encodes a subunit of condensin and is necessary for chromosome organization, cell division, and DNA repair53. EDNRA encodes the endothelin-1 receptor and has been associated with pancreatic cancer prognosis54. The locus on 16q24.1 lies ~200–300 kb upstream of a cluster of genes of the forkhead family of TFs (FOXF1, FOXC2, and FOXL1), known for their roles in development, cell proliferation, and several diseases, including cancer55.
We further estimated a polygenetic risk score (PRS) for pancreatic cancer using the 22 independent genome-wide significant risk SNPs in the Caucasian population12–16. The OR for pancreatic cancer among individuals above the 90th percentile the risk distribution was 2.20 (95% CI 1.83–2.65) compared with those with a PRS in the 40–60th percentile (Supplementary Table 5). We also assessed eight pancreatic cancer risk loci identified in Chinese and Japanese individuals in our data and noted one nominally significant locus in the combined PanScan and PanC4 results (6p25.3, rs9502893, OR = 0.94, 95% CI 0.92–0.97, P = 0.009: Wald test; Supplementary Table 6).
Pathway enrichment analysis
Pathway enrichment analysis for genes in currently known pancreatic cancer risk loci was performed using gene set enrichment analysis (GeneCodis; http://genecodis.cnb.csic.es/analysis)56 and Data-Driven Expression Prioritized Integration for Complex Traits (DEPICT; https://data.broadinstitute.org/mpg/depict/)57. The most significant enrichment was seen for the terms “Maturity onset diabetes of the young” (Kyoto Encyclopedia of Genes and Genomes (KEGG), P = 5.5 × 10−9, Hypergeometric distribution test), “Sequence-specific DNA-binding transcription factor activity” (GO Molecular Function, P = 3.1 × 10−4), “Cellular response to UV” (GO Biological Process, P = 4.2 × 10−4, Hypergeometric distribution test) as well as multiple gastrointestinal tissues (DEPICT, P = 5.1 × 10−5−0.004, Welch’s t-test; Supplementary Tables 7 and 8).
Expression analysis
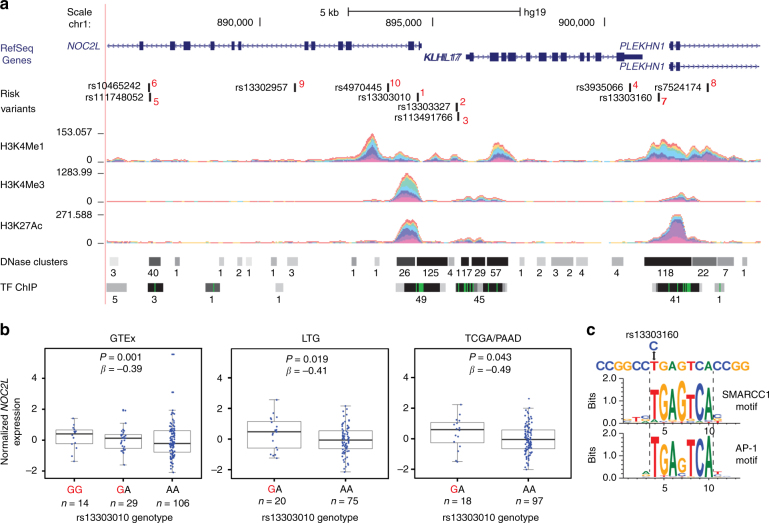
To begin unraveling the functional consequences of the newly discovered risk alleles, we performed expression quantitative trait locus (eQTL) analyses in three independent pancreatic tissue sample sets. We first assessed eQTLs in the publicly available Genotype-Tissue Expression (GTEx) project data for 149 histologically normal pancreatic tissue samples (including genes in a 1MB window centered on the marker SNP at each locus). Nominally significant eQTLs (P < 0.05) from this analysis (Supplementary Table 9, Supplementary Figure 2) were then carried forward to replication in two additional sample sets: (1) 95 histologically normal pancreatic samples (Laboratory of Translational Genomics, Laboratory of Translational Genomics (LTG) set58) and (2) 115 pancreatic tumors (The Cancer Genome Atlas, TCGA, Pancreatic Adenocarcinoma, PAAD, samples58; Table 2). The most notable eQTL in this analysis was seen for 1p36.33, where the risk-increasing allele at rs13303010 was associated with higher NOC2L expression in all three data sets (GTEx: P = 0.01, β = 0.39; LTG: P = 0.019, β = 0.41; TCGA: P = 0.043, β=0.49: T-statistic; Fig. 2b). An additional eQTL for a nearby gene, KLHL17, was significant in GTEx (P = 2.1 × 10−5, β = −0.42: T-statistic) but not in LTG (P = 0.131, β = −0.32: T-statistic) or TCGA (P = 0.654, β = −0.11: T-statistic). At 8q21.11, the risk allele (rs2941471-A) was associated with higher expression of HNF4G in GTEx (P = 0.038, β = 0.15: T-statistic) and LTG (P = 0.024, β = 0.28: T-statistic) samples, but not in TCGA (P = 0.80, β = −0.029: T-statistic).
Table 2.
Expression quantitative trait loci (eQTLs) for marker SNPs on chromosomes 1p36.33 and 8q21.11 in histologically normal pancreatic tissue samples from GTeX (n = 149) and LTG (n = 95) as well as pancreatic tumor samples from TCGA (PDAC, n = 115)
| Chr1p36.33: eQTLs for rs13303010 | ||||||
|---|---|---|---|---|---|---|
| GTeX pancreas | LTG pancreas | TCGA PDAC | ||||
| Gene name | P value | Effect size* | P value | Effect size* | P value | Effect size* |
| KLHL17 | 2.10 × 10-5 | −0.42 | 0.131 | −0.32 | 0.654 | −0.11 |
| NOC2L | 0.001 | 0.39 | 0.019 | 0.41 | 0.043 | 0.49 |
| SAMD11 | 0.023 | −0.26 | 0.500 | 0.14 | 0.397 | −0.18 |
| DVL1 | 0.042 | −0.14 | 0.280 | 0.18 | 0.085 | −0.37 |
| Chr8q21.11: eQTLs for rs2941471 | ||||||
|---|---|---|---|---|---|---|
| GTeX pancreas | LTG pancreas | TCGA PDAC | ||||
| Gene name | P value | Effect size* | P value | Effect size* | P value | Effect size* |
| HNF4G | 0.038 | 0.15 | 0.024 | 0.28 | 0.803 | −0.029 |
Expression QTLs were assessed in GTeX pancreatic tissue samples for all RefSeq genes within a 1 MB region centered on the marker SNP at each locus. Nominally significant findings were attempted for replication in the LTG and TCGA pancreatic tissue samples. *Effect size is the estimated eQTL effect size or beta (β) and its direction is shown for the risk increasing allele at each locus
Fig. 2.
Functional analysis of the 1p36.33 risk locus. a The set of most likely functional variants at 1p36.33 and their P value rank (1–10, in red) is shown as well as overlapping RefSeq genes on chr1: 885,555-904,522 (NCBI GRCh37/Hg19). ENCODE data for histone modification marks (H3K4me1, H3K4me3, H3K27Ac) are indicated by colored density plots. Open chromatin (DNase hypersensitivity regions, DNase clusters) and binding of transcription factors (TF ChIP) are indicated by horizontal bars. The numbers next to each bar indicate the number of cell lines with DNase clusters, or the number of different transcription factors bound across all tested cell lines. The panel is adapted from the UCSC Genome Browser. b Expression QTLs in histologically normal autopsy-derived pancreatic tissues (n = 149) from the GTEx consortium (GTEx), the Laboratory of Translational Genomics histologically normal adjacent-to-tumor pancreatic tissue set (LTG, n = 95), and the TCGA pancreatic cancer tissue set (TCGA/PAAD, n = 115). Normalized NOC2L expression is shown on the y axis and genotypes at the marker SNP at 1p36.33 on the x axis. Risk-increasing alleles are marked in red. Note that no samples in the LTG and TCGA/PAAD sets were of the minor homozygous risk genotype (GG). The box-and-whisker plots show the median (horizontal middle line within each box), interquartile range (top and bottom horizontal lines of each box), and 1.5 times the IQR (whiskers). c Analysis of the effects of 1p36.33 variants on transcription factor motifs for rs13303160 (r2 = 0.93 with rs13303010 in 1000 G EUR). The risk allele (C) at this marker alters predicted DNA-binding motifs for SMARCC1 and AP-1 proteins
At 1p36.33/NOC2L, we analyzed the set of variants most likely to be functional (LR > 1:100, n = 10) for overlap with transcriptionally active chromatin and effects on predicted TF-binding sites. The most notable variant in this analysis was rs13303160 (r2 = 0.93 with rs13303010) that overlaps open chromatin and prominent histone modification marks in ENCODE data (Fig. 2a, Supplementary Table 10) and where the risk allele is predicted to strongly disrupt TF-binding motifs for SMARCC1 (also known as BAF155) and several AP-1 proteins (Fig. 2c, Supplementary Table 11). These analyses suggest that altered SMARCC1 or AP-1 binding at rs13303160 may lead to higher levels of NOC2L mRNA. An increase in NOC2L protein would be expected to result in lower levels of histone acetylation and repression of p53 and p63 transcriptional activity24–26,59. We also assessed differential expression of NOC2L (and additional genes at the five novel loci) in pancreatic tumors44 and noted a 3.98-fold (P = 9.69 × 10−10: EdgeR, Exact test) increased expression in pancreatic cancer cell lines (n = 9) as compared with histologically normal pancreatic tissue samples (n = 10; Supplementary Table 12).
Discussion
This study demonstrates the power of large-scale collaborations in identifying new risk loci for pancreatic cancer, a deadly disease that presents challenges in accruing large sample sets for genetic studies. We herein add to the number of pancreatic cancer risk loci in or close to genes involved in MODY. As these genes also play roles in pancreatic development and acinar homeostasis, they may help explain underlying mechanisms at these loci. However, due to the low LD with BMI and T2D GWAS variants, the underlying mechanisms may differ between these epidemiologically and pathophysiologically associated conditions. We also describe potential functional underpinnings of risk loci, in particular for the locus on chromosome 1p36.33 in NOC2L, that require further detailed investigation.
Methods
Study participants
Participants were drawn from the Pancreatic Cancer Cohort Consortium (PanScan) and the Pancreatic Cancer Case-Control Consortium (PanC4) and individuals were included from 16 cohort and 13 case–control studies genotyped in four previous GWAS phases, namely PanScan I, PanScan II, PanScan III, and PanC412–14,16. Samples from the PANDoRA case–control consortium were used for replication23. The details on cases (individuals with PDAC) and controls have been previously described12–14,16.
All studies obtained informed consent from study participants and Institutional Review Board (IRB) approvals including IRB certifications permitting data sharing in accordance with the NIH Policy for Sharing of Data Obtained in NIH Supported or Conducted GWAS. The PanScan and PanC4 GWAS data are available through dbGAP (accession numbers phs000206.v5.p3 and phs000648.v1.p1, respectively).
Genotyping, imputation, and association analysis
Genotyping for PanScan was performed at the Cancer Genomics Research Laboratory (CGR) of the National Cancer Institute (NCI) of the National Institutes of Health (NIH) using the Illumina HumanHap series arrays (Illumina HumanHap550 Infinium II, Human 610-Quad) for PanScan I and II, respectively, and the Illumina Omni series arrays (OmniExpress, Omni1M, Omni2.5, and Omni5M) for PanScan III12–14. Genotyping for the PanC4 GWAS was performed at the Johns Hopkins Center for Inherited Disease Research (CIDR) using the Illumina HumanOmniExpressExome-8v1 array. Imputation was performed using the 1000 Genomes (1000 G) Phase 3, Release 1 reference data set22 and IMPUTE2 (http://mathgen.stats.ox.ac.uk/impute/impute_v2.html)60 as previously described14,18. Because of the large overlap of variants on genotyping arrays for PanScan I and II, these data sets were imputed and analyzed together. The PanScan III and PanC4 GWAS data sets were each imputed and analyzed separately. For quality control, variants were excluded based on (1) completion rate <90%; (2) MAF <0.01; and (3) low-quality imputation score (IMPUTE2 INFO score <0.3). After quality control, 11,381,182 SNPs genotyped or imputed in 5107 pancreatic cancer patients and 8845 controls of European ancestry were included in the analysis for PanScan I-III and 3933 cases and 3651 controls for PanC4 (Supplementary Table 1). The association analysis was performed using SNPTEST (http://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html)61 based on probabilistic genotype values from IMPUTE260, with parallel covariate adjustments: study, geographical region, age, sex, and principal components (PCA) of population substructure as were used in PanScan12–14 and study, age, sex, and PCA population substructure as were used in PanC416. The score statistic of the log additive genetic association magnitude was used. Summary statistics from PanScan I and II, PanScan III, and PanC4 were used for a meta-analysis using the fixed-effects inverse-variance method based on β estimates and SEs (http://csg.sph.umich.edu/abecasis/metal/). Heterogeneity was not observed for the SNPs identified as GWAS significant or suggestive in the combined study (Pheterogeneity ≥ 0.32). IMPUTE2 information scores were above 90% for SNPs (P < 1 × 10−6), except for rs13303010 in the PanScan I+II data (INFO = 0.42; Table 1). The estimated inflation of the test statistic, λ, was 1.002 for PanScan I+II, 1.051 for PanScan III, and 1.024 for PanC462.
A Polygenic Risk Score (PRS) was constructed for each individual by summing the number of risk alleles carried for all established pancreatic cancer risk loci identified by GWAS, weighted by their estimated effect size. Individuals were grouped by percentiles, and the association of the PRS (as percentile groupings) with pancreatic cancer was estimated using logistic regression.
DEPICT analysis was used to prioritize causal genes at currently known pancreatic cancer risk loci12–16 identify gene sets enriched across risk loci, and tissues in which genes at risk loci are highly expressed57. No genes or SNP–gene pairs were significant at false discovery rate (FDR) < 0.05 (data not shown) but significant tissue enrichment is shown in Supplementary Table 7. Additional gene set enrichment analysis was performed using GeneCodis3 (http://genecodis.cnb.csic.es/). Genes (n = 65) were located in the currently known pancreatic cancer risk loci identified in subjects of European descent (for genes located +/−100 kb from the most significant SNP at the 22 risk loci) based on KEGG, Gene Ontology (Biological Process and Molecular Function) annotations using GeneCodis3 with reporting of FDR-corrected hypergeometric P values (Supplementary Table 8)56.
Replication
Ten promising signals (P < 10−6) were selected for replication in samples from the PANDoRA consortium23. Genotyping was performed by custom TaqMan genotyping assays (Applied Biosystems) at the German Cancer Research Center (DKFZ) in Heidelberg, Germany, for 2770 pancreatic cancer patients and 5178 controls, of which 2737 cases and 4753 controls had complete age and clinical data and did not overlap with other study individuals. Duplicate quality-control samples (n = 607 pairs) showed 99.48% genotype concordance. SNP quality metrics were performed for each SNP by plate; plates with <80% genotype completion rates were dropped from the analysis. Individuals were excluded if they were missing data on two or more SNPs after excluding SNPs on plates with low genotype completion rates. The association analysis for PANDoRA was adjusted for age and study in the same manner as previously described14–16. Heterogeneity between studies was assessed using the Cochran’s Q-test. Association analysis was also performed for the set of variants previously replicated in PANDoRA as part of the PanScan III14 and PanC416 GWAS studies (Supplementary Table 3).
Using SequenceLDhot, recombination hotspots for association plots were generated as previously described12–14. Recombination hotspot inference was performed using the 1000 G CEU samples (n = 99). The LD heatmap was prepared using the 1000 G Phase 3 CEU data, and the snp.plotter R software package63.
eQTL analysis
The publicly available GTEx data64 (http://www.gtexportal.org/; version 6) were used to assess eQTLs in pancreatic tissue samples (n = 149). RefSeq genes located within +/−500 kb of the marker SNP for each GWAS significant locus were assessed for cis-eQTL effects. Nominally significant eQTLs from this analysis (P < 0.05) were then taken forward to further analysis in two additional pancreatic tissue sample sets58: (1) the LTG and (2) The Cancer Genome Atlas (TCGA) pancreatic adenocarcinoma (PAAD) samples.
The LTG set included 95 histologically normal pancreatic tissue samples from participants of European ancestry collected at three participating sites: Mayo Clinic in Rochester, MN (45 samples, adjacent to tumor); Memorial Sloan Kettering Cancer Center in New York City, NY (34 samples, adjacent to tumor); and Penn State College of Medicine, Hershey, PA (16 samples, from tissue donors via the Gift of Life Donor Program, Philadelphia, PA) as previously described58. Samples were confirmed to be non-tumorous with ≥80% epithelial component by histological review and macro-dissected when needed. The project was approved by the Institutional Review Board of each participating institution as well as the NIH.
In short, RNA (RIN >7.5) isolated from fresh frozen histologically normal pancreatic tissue samples (LTG samples) with the Ambion mirVana kit was poly-A-enriched and subjected to massively parallel paired-end sequencing (Illumina’s HiSeq2000/TruSeq v3 sequencing). MapSplice was used to align reads and RSEM (v1.2.14) for gene expression quantification (TPM)65,66 using the hg19/GRCh37-based UCSC “RefSeq” track for gene annotation. DNA for genotyping was isolated from blood (Mayo Clinic samples), histologically normal fresh frozen pancreatic tissue samples (Penn State samples), or histologically normal fresh frozen spleen or duodenum tissue samples (MSKCC samples) using the Gentra Puregene Tissue Kit (Qiagen). DNA samples were genotyped on the Illumina OmniExpress or Omni1M arrays at the CGR of the Division of Cancer Epidemiology and Genetics, NCI, NIH. After quality control, genotypes were imputed using the 1000 G (Phase 1, v3) imputation reference data set and IMPUTE 267. Pre-imputation exclusion filters of Hardy Weinberg Equilibrium P < 1 × 10−6, minor allele frequency (MAF) <0.01, genotype missing rate >0.05, A/T and G/C pairs on ambiguous DNA strand (MAF > 0.45), and significantly different allele frequency between sample data and the 1000 G reference data (P < 7 × 10−8: Fisher’s exact test) were used. Post-imputation variants (single-nucleotide variants (SNP) and small insertion-deletion (indel) polymorphisms) with MAF < 0.05 or imputation quality scores (INFO score) <0.5 were removed from the final analysis58.
The second sample set included RNA-sequencing (RNA-seq) and genotype data from tumor-derived pancreatic samples obtained from TCGA PAAD data set by permission through the TCGA Data Access Committee. We excluded samples of non-European ancestry, with history of neo-adjuvant therapy prior to surgery, or with histological subtypes other than PDAC, leaving a total of 115 tumor samples for analysis58. TCGA mRNA-seq data (level 1 read data, generated using Illumina’s HiSeq2000) for pancreatic cancer samples (TCGA PAAD) were processed in the same manner as the histologically normal LTG samples described above. Blood-derived DNA samples for TCGA PAAD samples were genotyped on Affymetrix 6.0 arrays and processed in the same manner as for the LTG samples58.
The eQTL analysis was performed separately in histologically normal (LTG) and tumor-derived (TCGA PAAD) pancreatic samples using the Matrix eQTL (http://www.bios.unc.edu/research/genomic_software/Matrix_eQTL/) software package68. We tested associations between genotyped and imputed SNPs and the expression of genes evaluated by mRNA-sequencing after upper quantile normalization within samples and normal quantile transformation for each gene across samples by regressing the imputed dosage of the minor allele for each variant against normalized gene expression values68. Linear models were adjusted for age, sex, study, and the top five principal components (PCs) each for genotypes and gene expression to account for possible measured or hidden confounders58. The T-statistics from the linear regression is reported. For the tumor samples, we further adjusted for tumor stage and sequence-based tumor purity as per information provided by TCGA.
Bioinformatic analysis of functional potential
Variants at the new risk loci were assessed for potential functionality by examining their location in open (DNase Hypersensitivity Regions, DHS) and active chromatin (as per promoter and enhancer histone modification marks) in the ENCODE data. For this, we used HaploReg 4.1 (http://www.broadinstitute.org/mammals/haploreg/haploreg.php) and the UCSC Genome Browser (http://genome.ucsc.edu/). Variants correlated with the most significant variant at each locus at r2 > 0.7 in 1000 G EUR populations were included, except for 1p36.33/NOC2L where P value LR > 1:100 was used.
Candidate functional variants at 1p36.33 were selected by comparing the likelihood of each variant from the association analysis with the likelihood of the most significant variant. Ten variants had likelihood ratios, LR > 1:100 relative to the most significant SNP: rs13303010 (P value rank 1), rs13303327 (rank 2), rs113491766 (rank 3), rs3935066 (rank 4), rs111748052 (rank 5), rs10465242 (rank 6), rs13303160 (rank 7), rs7524174 (rank 8), rs13302957 (rank 9), and rs4970445 (rank 10). They were all highly correlated with rs13303010 (r2 = 0.52–1.00, 1000 G EUR data). These 10 variants were considered the set of variants most likely to contain the functional variant(s) at 1p36.33. Possible allelic effects of these top 10 variants on TF-binding motifs were determined using PrEdict Regulatory Functional Effect of SNPs by Approximate P value Estimation (PERFECTOS-APE; http://opera.autosome.ru/perfectosape/) analysis that determines the probability of a TF motif (using position weight matrices, from HOCOMOCO-10, JASPAR, HT-SELEX, SwissRegulon, and HOMER databases) in the DNA sequence overlapping each variant. The fold change in probability of there being a TFBS present for each allele of a variant is then calculated69. Two dbSNP variants at 1p36.33 with the same bp location, rs111748052 (−/ATTTT) and rs10465241 (C/T), may be two independent variants (as indicated in dbSNP), or a single tri-allelic marker (alleles are shown as C/CATTTT/T in 1000 G). As PERFECTOS-APE does not analyze indel variants, we analyzed the two indels among the 10 potential functional variants, rs111748052 and rs113491766, using a different program, sequence TF Affinity Prediction. This program calculates the total affinity of a sequence for a TF (as given by TRANSFAC and JASPAR databases) on the basis of a biophysical model of the binding energies between a TF and DNA70. The probability for a given TFBS for each variant of the indel was then compared as in PERFECTOS-APE to determine the fold change effect of the indel on the presence of the TFBS.
Gene expression analysis
Gene expression was assessed for genes that are closest to the reported variants at chromosomes 1p36.33: NOC2L, KLHL17, and PLEKHN1; 7p12: TNS3; 8q21.11: HNF4G; 17q12: HNF1B; 18q21.32: GRP, as well as two additional genes at 1p36.33 exhibiting nominally significant eQTLs in GTEx (1p36.33/SAMD11/DVL1). We assessed differential expression of these genes in pancreatic tumor samples (PDAC, n = 8), histologically normal (non-malignant) pancreatic tissue samples (n = 10), and pancreatic cell lines (n = 9) by RNA-seq as described previously44. We compared gene expression in tumors (T) and cell lines (C) to histologically normal pancreatic tissue samples (N) by EdgeR analysis. P values for differential expression in tumor vs. normal (TvN) and Cell lines vs. normal (CvN) represent an exact statistic using the normalized pseudo-counts and tagwise dispersion estimates per gene.
Data availability
The PanScan and PanC4 genome-wide association data that support the findings of this study are available through dbGAP (accession numbers phs000206.v5.p3 and phs000648.v1.p1, respectively).
Electronic supplementary material
Acknowledgements
This work was supported by RO1 CA154823, the Lustgarten Foundation, and federal funds from the NCI, US NIH under contract number HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services, and mention of trade names, commercial products, or organizations does not imply endorsement by the US government. Genotyping Services were provided by the CIDR and the NCI’s CGR. CIDR is fully funded through a federal contract from the NIH to the Johns Hopkins University, contract number HHSN268201100011I. The IARC/Central Europe study was supported by a grant from the US NCI at the NIH (R03 CA123546-02) and grants from the Ministry of Health of the Czech Republic (NR 9029-4/2006, NR9422-3, NR9998-3, and MH CZ-DRO-MMCI 00209805). The work at Johns Hopkins University was supported by the NCI Grants P50CA062924 and R01CA97075. Additional support was provided by, Susan Wojcicki, and Dennis Troper, and the Sol Goldman Pancreas Cancer Research Center. The Mayo Clinic Biospecimen Resource for Pancreas Research study is supported by the Mayo Clinic SPORE in Pancreatic Cancer (P50 CA102701). The Memorial Sloan Kettering Cancer Center Pancreatic Tumor Registry is supported by P30CA008748, the Geoffrey Beene Foundation, the Arnold and Arlene Goldstein Family, Foundation, and the Society of MSKCC. The PACIFIC Study was supported by RO1CA102765, Kaiser Permanente, and Group Health Cooperative. The Queensland Pancreatic Cancer Study was supported by a grant from the National Health and Medical Research Council of Australia (NHMRC; Grant number 442302). R.E.N. is supported by a NHMRC Senior Research Fellowship (#1060183). The UCSF pancreas study was supported by NIH-NCI grants (R01CA1009767, R01CA109767-S1, and R0CA059706) and the Joan Rombauer Pancreatic Cancer Fund. Collection of cancer incidence data was supported by the California Department of Public Health as part of the statewide cancer reporting program; the NCI’s SEER Program under contract HSN261201000140C awarded to CPIC; and the CDC’s National Program of Cancer Registries, under agreement #U58DP003862-01 awarded to the California Department of Public Health. The Yale (CT) pancreas cancer study is supported by NCI at the U.S. NIH, grant 5R01CA098870. The cooperation of 30 Connecticut hospitals, including Stamford Hospital, in allowing patient access is gratefully acknowledged. The Connecticut Pancreas Cancer Study was approved by the State of Connecticut Department of Public Health Human Investigation Committee. Certain data used in that study were obtained from the Connecticut Tumor Registry in the Connecticut Department of Public Health. The authors assume full responsibility for analyses and interpretation of these data. Studies included in PANDoRA were partly funded by the Czech Science Foundation (No. P301/12/1734), the Internal Grant Agency of the Czech Ministry of Health (IGA NT 13 263); the Baden-Württemberg State Ministry of Research, Science and Arts (Professor H. Brenner), the Heidelberger EPZ-Pancobank (Professor M.W. Büchler and team: Professor T. Hackert, Dr. N. A. Giese, Dr. Ch. Tjaden, E. Soyka, M. Meinhardt; Heidelberger. Stiftung Chirurgie and BMBF grant 01GS08114), the BMBH (Professor P. Schirmacher; BMBF grant 01EY1101), the “5 × 1000” voluntary contribution of the Italian Government, the Italian Ministry of Health (RC1203GA57, RC1303GA53, RC1303GA54, and RC1303GA50), the Italian Association for Research on Cancer (Professor A. Scarpa; AIRC n. 12182), the Italian Ministry of Research (Professor A. Scarpa; FIRB - RBAP10AHJB), the Italian FIMP-Ministry of Health (Professor A. Scarpa; 12 CUP_J33G13000210001), and by the National Institute for Health Research Liverpool Pancreas Biomedical Research Unit, UK. We would like to acknowledge the contribution of Dr. Frederike Dijk and Professor Oliver Busch (Academic Medical Center, Amsterdam, the Netherlands). Assistance with genotype data quality control was provided by Cecelia Laurie and Cathy Laurie at the University of Washington Genetic Analysis Center. The American Cancer Society (ACS) funds the creation, maintenance, and updating of the Cancer Prevention Study II cohort. Cancer incidence data for CLUE were provided by the Maryland Cancer Registry, Center for Cancer Surveillance and Control, Department of Health and Mental Hygiene, 201 W. Preston Street, Room 400, Baltimore, MD 21201, http://phpa.dhmh.maryland.gov/cancer, 410-767-4055. We acknowledge the State of Maryland, the Maryland Cigarette Restitution Fund, and the National Program of Cancer Registries of the Centers for Disease Control and Prevention for the funds that support the collection and availability of the cancer registry data. We thank all the CLUE participants. The Melbourne Collaborative Cohort Study (MCCS) recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC grants 209057 and 396414 and by the infrastructure provided by Cancer Council Victoria. Cases and their vital status were ascertained through the Victorian Cancer Registry and the Australian Institute of Health and Welfare, including the National Death Index and the Australian Cancer Database. The NYU study (AZJ and AAA) was funded by NIH R01 CA098661, UM1 CA182934 and center grants P30 CA016087 and P30 ES000260. The PANKRAS II Study in Spain was supported by research grants from Instituto de Salud Carlos III-FEDER, Spain: Fondo de Investigaciones Sanitarias (FIS; #PI13/00082 and #PI15/01573) and Red Temática de Investigación Cooperativa en Cáncer, Spain (#RD12/0036/0050); and European Cooperation in Science and Technology (COST Action #BM1204: EU_Pancreas), Ministerio de Ciencia y Tecnología (CICYT SAF 2000-0097), Fondo de Investigación Sanitaria (95/0017), Madrid, Spain; Generalitat de Catalunya (CIRIT—SGR); “Red temática de investigación cooperativa de centros en Cáncer” (C03/10), “Red temática de investigación cooperativa de centros en Epidemiología y salud pública” (C03/09), and CIBER de Epidemiología (CIBERESP), Madrid. The Physicians’ Health Study was supported by research grants CA-097193, CA-34944, CA-40360, HL-26490, and HL-34595 from the NIH, Bethesda, MD, USA. The Women’s Health Study was supported by research grants CA-047988, HL-043851, HL-080467, and HL-099355 from the NIH, Bethesda, MD, USA. Health Professionals Follow-up Study is supported by NIH grant UM1 CA167552 from the NCI, Bethesda, MD, USA. Nurses’ Health Study is supported by NIH grants UM1 CA186107, P01 CA87969, and R01 CA49449 from the NCI, Bethesda, MD, USA. Additional support from the Hale Center for Pancreatic Cancer Research, U01 CA21017 from the NCI, Bethesda, MD, USA, and the United States Department of Defense CA130288, Lustgarten Foundation, Pancreatic Cancer Action Network, Noble Effort Fund, Peter R. Leavitt Family Fund, Wexler Family Fund, and Promises for Purple to B.M. Wolpin is acknowledged. The WHI program is funded by the National Heart, Lung, and Blood Institute, NIH, U.S. Department of Health and Human Services through contracts HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C, and HHSN268201600004C. The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found at http://www.whi.org/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Long%20List.pdf. We thank Laurie Burdett, Aurelie Vogt, Belynda Hicks, Amy Hutchinson, Meredith Yeager, and other staff at the NCI’s Division of Epidemiology and Genetics (DECG) CGR for GWAS genotyping. We also thank Bao Tran, Jyoti Shetty, and other members of the NCI Center for Cancer Research (CCR) Sequencing Facility for sequencing RNA from histologically normal pancreatic tissue samples (LTG samples). This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the NIH, Bethesda, MD, USA (http://biowulf.nih.gov). The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the NIH, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the pancreatic tissue data from the GTEx Portal on 05/04/17. The results published here are in part based upon data generated by The Cancer Genome Atlas (TCGA) managed by the NCI and NHGRI. Information about TCGA can be found at http://cancergenome.nih.gov/. We acknowledge the clinical contributors that provided PDAC samples and the data producers of RNA-seq and GWAS genotype data from TCGA Research Network. The data set used for the analyses described in this manuscript was obtained by formal permission through the TCGA Data Access Committee (DAC).
Author contributions
A.P.K., R.S.S., L.T.A., B.M.W., G.M.P., and H.A.R. organized and designed the study. A.P.K., L.T.A., F.C., J.W.H., A.J., J.Z., and M.B. organized and supervised the genotyping of samples. E.J.C., E.M., O.O., M.Z., A.Blackford. L.T.A., F.C., P.K., J.W.H., A.J., J.Z., F.Chen, and A.P.K. designed and conducted the statistical analysis. A.P.K. and L.T.A. drafted the first version of the manuscript. A.P.K., B.M.W., H.A.R., R.Z.S.-S., F.C., D.A., G.A., A.A.A., A.B., W.R.B., L.B.-F., S.I.B., A.Borgida, P.M.B., L.B., P.B., H.B., B.B.-d.-M., J.B., D.C., G.C., G.C., G.M.C., K.G.C., C.C.C., S.C., M.C., F.D., E.J.D., L.F., C.F., N.F., S.G., J.M.M.G., M.Gazouli, G.G.G., E.G., M.G., G.E.G., P.J.G., T.H., C.H., P.H., M.H., P.H., K.J.H., J.H., I.H., E.A.H., R.H., R.J.H., E.J.J., K.J., V.J., R.K., K.-T.K., E.A.K., M.K., C.K., M.H.K., J.K., R.J.K., D.L., S.L., R.T.L., I.-M.L., L.L., L.Lu., N.M., A.M., S.M., R.L.M., B.M.-D., R.E.N,. J.P.N., A.L.O., I.O., C.P., A.V.P., U.P., R.P., M.P., F.X.R., N.R., G.S., H.D.S., G.S., X.-O.S., D.S., J.P.S., P.S., M.S., R.T.-W., F.T., M.D.T., G.S.T., S.K.V., Y.V., K.V., P.V., J.W.-W., Z.W., N.W., E.W., H.Y., K.Y., A.Z.-J., W.Z., P.K., D.L., S.C., O.O., G.M.P., and L.T.A. conducted the epidemiological studies and contributed samples to the GWAS, expression and/or follow-up genotyping. All authors contributed to the writing of the manuscript.
Competing Interests
The authors declare no competing financial interests.
Footnotes
Alison P. Klein, Brian M. Wolpin, Harvey A. Risch and Rachael Z. Stolzenberg-Solomon contributed equally to this work. Donghui Li, Stephen Chanock, Ofure Obazee, Gloria M. Petersen and Laufey T. Amundadottir jointly supervised this work.
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41467-018-02942-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Alison P. Klein, Email: aklein1@jhmi.edu
Laufey T. Amundadottir, Email: amundadottirl@mail.nih.gov
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. Ca. Cancer J. Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay, J. et al. http://globocan.iarc.fr (IARC, Lyon, France, 2013).
- 3.Malvezzi M, Bertuccio P, Levi F, La Vecchia C, Negri E. European cancer mortality predictions for the year 2014. Ann. Oncol. 2014;25:1650–1656. doi: 10.1093/annonc/mdu138. [DOI] [PubMed] [Google Scholar]
- 4.Rahib L, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein AM, et al. High-risk melanoma susceptibility genes and pancreatic cancer, neural system tumors, and uveal melanoma across GenoMEL. Cancer Res. 2006;66:9818–9828. doi: 10.1158/0008-5472.CAN-06-0494. [DOI] [PubMed] [Google Scholar]
- 6.Jones S, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kastrinos F, et al. Risk of pancreatic cancer in families with Lynch syndrome. JAMA. 2009;302:1790–1795. doi: 10.1001/jama.2009.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowenfels AB, et al. Hereditary pancreatitis and the risk of pancreatic cancer. International Hereditary Pancreatitis Study Group. J. Natl Cancer Inst. 1997;89:442–446. doi: 10.1093/jnci/89.6.442. [DOI] [PubMed] [Google Scholar]
- 9.Roberts NJ, et al. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov. 2012;2:41–46. doi: 10.1158/2159-8290.CD-11-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goggins M, et al. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360–5364. [PubMed] [Google Scholar]
- 11.Giardiello FM, et al. Increased risk of cancer in the Peutz-Jeghers syndrome. N. Engl. J. Med. 1987;316:1511–1514. doi: 10.1056/NEJM198706113162404. [DOI] [PubMed] [Google Scholar]
- 12.Amundadottir L, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat. Genet. 2009;41:986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petersen GM, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet. 2010;42:224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolpin BM, et al. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat. Genet. 2014;46:994–1000. doi: 10.1038/ng.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang, M. et al. Three new pancreatic cancer susceptibility signals identified on chromosomes 1q32.1, 5p15.33 and 8q24.21. Oncotarget, 10.18632/oncotarget.11041 (2016). [DOI] [PMC free article] [PubMed]
- 16.Childs EJ, et al. Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nat. Genet. 2015;47:911–916. doi: 10.1038/ng.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campa, D. et al. TERT gene harbors multiple variants associated with pancreatic cancer susceptibility. Int. J. Cancer10.1002/ijc.29590 (2015). [DOI] [PMC free article] [PubMed]
- 18.Wang Z, et al. Imputation and subset-based association analysis across different cancer types identifies multiple independent risk loci in the TERT-CLPTM1L region on chromosome 5p15.33. Hum. Mol. Genet. 2014;23:6616–6633. doi: 10.1093/hmg/ddu363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu C, et al. Genome-wide association study identifies five loci associated with susceptibility to pancreatic cancer in Chinese populations. Nat. Genet. 2012;44:62–66. doi: 10.1038/ng.1020. [DOI] [PubMed] [Google Scholar]
- 20.Low SK, et al. Genome-wide association study of pancreatic cancer in Japanese population. PLoS ONE. 2010;5:e11824. doi: 10.1371/journal.pone.0011824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campa D, et al. Lack of replication of seven pancreatic cancer susceptibility loci identified in two Asian populations. Cancer Epidemiol. Biomark. Prev. 2013;22:320–323. doi: 10.1158/1055-9965.EPI-12-1182. [DOI] [PubMed] [Google Scholar]
- 22.Consortium TGP. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campa D, et al. Dig. Liver Dis. 2013. Genetic susceptibility to pancreatic cancer and its functional characterisation: the PANcreatic Disease ReseArch (PANDoRA) consortium; pp. 95–99. [DOI] [PubMed] [Google Scholar]
- 24.Hublitz P, et al. NIR is a novel INHAT repressor that modulates the transcriptional activity of p53. Genes Dev. 2005;19:2912–2924. doi: 10.1101/gad.351205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu L, Ma CA, Zhao Y, Jain A. Aurora B interacts with NIR-p53, leading to p53 phosphorylation in its DNA-binding domain and subsequent functional suppression. J. Biol. Chem. 2011;286:2236–2244. doi: 10.1074/jbc.M110.174755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heyne K, et al. NIR, an inhibitor of histone acetyltransferases, regulates transcription factor TAp63 and is controlled by the cell cycle. Nucleic Acids Res. 2010;38:3159–3171. doi: 10.1093/nar/gkq016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baraille F, et al. Glucose tolerance is improved in mice invalidated for the nuclear receptor HNF-4gamma: a critical role for enteroendocrine cell lineage. Diabetes. 2015;64:2744–2756. doi: 10.2337/db14-0993. [DOI] [PubMed] [Google Scholar]
- 28.Kottgen A, et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat. Genet. 2013;45:145–154. doi: 10.1038/ng.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayers JR, et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat. Med. 2014;20:1193–1198. doi: 10.1038/nm.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berndt SI, et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat. Genet. 2013;45:501–512. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michailidou K, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 2013;45:353–361. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haumaitre C, et al. Lack of TCF2/vHNF1 in mice leads to pancreas agenesis. Proc. Natl Acad. Sci. USA. 2005;102:1490–1495. doi: 10.1073/pnas.0405776102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Solar M, et al. Pancreatic exocrine duct cells give rise to insulin-producing beta cells during embryogenesis but not after birth. Dev. Cell. 2009;17:849–860. doi: 10.1016/j.devcel.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 35.Fajans SS, Bell GI. MODY: history, genetics, pathophysiology, and clinical decision making. Diabetes Care. 2011;34:1878–1884. doi: 10.2337/dc11-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berndt SI, et al. Large-scale fine mapping of the HNF1B locus and prostate cancer risk. Hum. Mol. Genet. 2011;20:3322–3329. doi: 10.1093/hmg/ddr213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Painter JN, et al. Fine-mapping of the HNF1B multicancer locus identifies candidate variants that mediate endometrial cancer risk. Hum. Mol. Genet. 2015;24:1478–1492. doi: 10.1093/hmg/ddu552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kristiansen W, et al. Two new loci and gene sets related to sex determination and cancer progression are associated with susceptibility to testicular germ cell tumor. Hum. Mol. Genet. 2015;24:4138–4146. doi: 10.1093/hmg/ddv129. [DOI] [PubMed] [Google Scholar]
- 39.Odom DT, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molero X, et al. Gene expression dynamics after murine pancreatitis unveils novel roles for Hnf1alpha in acinar cell homeostasis. Gut. 2012;61:1187–1196. doi: 10.1136/gutjnl-2011-300360. [DOI] [PubMed] [Google Scholar]
- 41.El-Khairi R, Vallier L. The role of hepatocyte nuclear factor 1beta in disease and development. Diabetes Obes. Metab. 2016;18 Suppl 1:23–32. doi: 10.1111/dom.12715. [DOI] [PubMed] [Google Scholar]
- 42.Ktistaki E, Talianidis I. Modulation of hepatic gene expression by hepatocyte nuclear factor 1. Science. 1997;277:109–112. doi: 10.1126/science.277.5322.109. [DOI] [PubMed] [Google Scholar]
- 43.Boj SF, Parrizas M, Maestro MA, Ferrer J. A transcription factor regulatory circuit in differentiated pancreatic cells. Proc. Natl Acad. Sci. USA. 2001;98:14481–14486. doi: 10.1073/pnas.241349398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoskins JW, et al. Transcriptome analysis of pancreatic cancer reveals a tumor suppressor function for HNF1A. Carcinogenesis. 2014;35:2670–2678. doi: 10.1093/carcin/bgu193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo Z, et al. Hepatocyte nuclear factor 1A (HNF1A) as a possible tumor suppressor in pancreatic cancer. PLoS ONE. 2015;10:e0121082. doi: 10.1371/journal.pone.0121082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stolzenberg-Solomon RZ, Amundadottir LT. Epidemiology and inherited predisposition for sporadic pancreatic adenocarcinoma. Hematol. Oncol. Clin. North Am. 2015;29:619–640. doi: 10.1016/j.hoc.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Replication DIG, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park HS, Kwon HY, Lee YL, Chey WY, Park HJ. Role of GRPergic neurons in secretin-evoked exocrine secretion in isolated rat pancreas. Am. J. Physiol. Gastrointest. Liver Physiol. 2000;278:G557–G562. doi: 10.1152/ajpgi.2000.278.4.G557. [DOI] [PubMed] [Google Scholar]
- 50.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blangy A. Tensins are versatile regulators of Rho GTPase signalling and cell adhesion. Biol. Cell. 2017;109:115–126. doi: 10.1111/boc.201600053. [DOI] [PubMed] [Google Scholar]
- 52.Qian X, et al. The Tensin-3 protein, including its SH2 domain, is phosphorylated by Src and contributes to tumorigenesis and metastasis. Cancer Cell. 2009;16:246–258. doi: 10.1016/j.ccr.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirano T. Condensin-based chromosome organization from bacteria to vertebrates. Cell. 2016;164:847–857. doi: 10.1016/j.cell.2016.01.033. [DOI] [PubMed] [Google Scholar]
- 54.Rachidi SM, Qin T, Sun S, Zheng WJ, Li Z. Molecular profiling of multiple human cancers defines an inflammatory cancer-associated molecular pattern and uncovers KPNA2 as a uniform poor prognostic cancer marker. PLoS ONE. 2013;8:e57911. doi: 10.1371/journal.pone.0057911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Golson ML, Kaestner KH. Fox transcription factors: from development to disease. Development. 2016;143:4558–4570. doi: 10.1242/dev.112672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012;40:W478–W483. doi: 10.1093/nar/gks402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pers TH, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, M. et al. Characterizing cis-regulatory variation in the transcriptome of histologically normal and tumor-derived pancreatic tissues Gut 0:1–13 (2017). [DOI] [PMC free article] [PubMed]
- 59.Lopez-Bergami P, Lau E, Ronai Z. Emerging roles of ATF2 and the dynamic AP1 network in cancer. Nat. Rev. Cancer. 2010;10:65–76. doi: 10.1038/nrc2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 62.de Bakker PI, et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luna A, Nicodemus KK. snp.plotter: an R-based SNP/haplotype association and linkage disequilibrium plotting package. Bioinformatics. 2007;23:774–776. doi: 10.1093/bioinformatics/btl657. [DOI] [PubMed] [Google Scholar]
- 64.Consortium G. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang K, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38:e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 68.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vorontsov IE, et al. PERFECTOS-APE - predicting regulatory functional effect of SNPs by approximate P-value estimation. Bioinformatics. 2015;1:102–108. [Google Scholar]
- 70.Thomas-Chollier M, et al. Transcription factor binding predictions using TRAP for the analysis of ChIP-seq data and regulatory SNPs. Nat. Protoc. 2011;6:1860–1869. doi: 10.1038/nprot.2011.409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The PanScan and PanC4 genome-wide association data that support the findings of this study are available through dbGAP (accession numbers phs000206.v5.p3 and phs000648.v1.p1, respectively).