Abstract
The covalent attachment of ubiquitin (Ub) or Ub chains to cellular proteins is a versatile post-translational modification involved in a variety of eukaryotic cellular events. Recently, the post-translational modification of Ub itself by phosphorylation has emerged as an important component of the Ub-signaling system. Specifically, Ub phosphorylation at serine-65 was shown to activate parkin-mediated mitochondrial quality control. However, the impact of phosphorylation on Ub structure and interactions is poorly understood. Here we investigate the recently reported structural changes in Ub upon serine-65 phosphorylation, namely, the equilibrium between a native-like and a novel, alternate conformer of phosphorylated Ub (pUb). We show that this equilibrium is pH-dependent, and the two pUb conformers are linked to the different charge states of the phosphate group. We examined pUb binding to a known Ub-receptor and found that the alternate conformer is binding incompetent. Furthermore, serine-65 phosphorylation affects the conformational equilibrium of K48-linked Ub dimers. Lastly, our crystal structure of S65D Ub and NMR data indicate that phosphomimetic mutations do not adequately reproduce the salient features of pUb. Our results suggest that the pH-dependence of the conformations and binding properties of phosphorylated Ub and polyUb could provide an additional level of modulation in Ub-mediated signaling.
Introduction
The covalent attachment of ubiquitin (Ub) or Ub chains (polyUb) to cellular proteins is one of the most versatile known post-translational modifications, with numerous signals being encoded by different types of Ub-Ub linkages1. Recently, Ub phosphorylation at S65 has come to light as a novel and important component in the PINK1 (PTEN-induced novel kinase protein 1)-Parkin pathway. Parkin is an E3 Ub ligase2,3 believed to have a significant role in mitochondrial quality control3–6, and defects in the PINK1-Parkin pathway have been linked to autosomal recessive forms of early-onset Parkinson’s Disease5,7–10. In its native conformation, parkin is autoinhibited11–16, and phosphorylation of both parkin and Ub by PINK1 at a conserved S65 appears to be necessary for complete activation of parkin’s E3 ligase activity17–22. In addition to S65, phosphorylation of Ub has been observed at several other residues23–26, suggesting that phosphorylated Ub (pUb) may play significant roles within the Ub-signaling system. The roles of Ub phosphorylation at the non-S65 sites and the respective kinases and phosphatases remain unknown. Ub modification by phosphorylation has thus emerged as an important regulatory mechanism of the Ub signaling system that requires rigorous exploration and characterization.
A recent study by Wauer et al. of S65 pUb27 found evidence of a novel, alternate conformer of Ub, designated here as pUbalt, which is in slow exchange (~2 per sec) with the “native-like” conformer, designated as pUbnat. The roles and origins of this novel conformer, however, are unclear. Strikingly, there is no evidence of such a conformer in phosphomimetic Ub variants, in which S65 is replaced with aspartate or glutamate, raising interesting questions about the origin of the pUbalt conformer. Since both phosphomimetic mutation and phosphorylation at S65 introduce a negative charge at the same position of Ub, we asked the question of what unique properties resulting from phosphorylation could result in the pUbalt conformer.
Here, we put forth and test two hypotheses to understand the origins of this conformer: (1) that it is an NMR artifact resulting from Ub oligomerization, and (2) that it results from a (−2) charge on the phosphate group. To test these hypotheses we analyzed temperature and pH-dependence of both pUb conformers, determined their spin-relaxation rates, and characterized and compared their ability to bind to a known Ub-receptor with the binding properties of wild-type Ub (UbWT) and phosphomimetic S65D Ub (UbS65D) variant. We also present the first crystal structure of UbS65D, which reveals the structural effects of introducing a single negative charge at position 65 in Ub. Given that phosphorylated Ub chains are found in cells28,29, understanding how Ub phosphorylation affects the structure of polyUb chains is critical to a complete understanding of the effect of this modification. Therefore, we also examined the effect of Ub phosphorylation on the conformations of Ub dimers linked via K48, the most abundant linkage in cells. Our results suggest that the singly- and doubly-deprotonated states of phosphorylated S65 have differential effect on Ub structure and interactions, and that phosphomimics are poor structural models for pUb.
Results
After purifying phosphorylated Ub (see Methods), we recorded a 1H-15N NMR correlation spectrum of the protein and observed the additional signals reported by Wauer et al.27 (Figure S1a). Shifts in NMR signals reflect a change in the electronic environment of nuclei which could reflect an altered 3D structure inside Ub but could also be caused by “external” factors such as intermolecular interactions. Note that the residues exhibiting strongly shifted alternate signals in pUb are located at or near the hydrophobic patch on Ub surface comprising residues L8, I44, and V70 (Figure S1f). Because a negative charge near the hydrophobic patch surface can promote interactions between a Ub-like domain and Ub30 (and by extension between two Ubs), which could subsequently affect chemical shifts of the residues at or near the interface, we tested whether the additional signals observed upon S65 phosphorylation could be a result of noncovalent Ub oligomerization upon the attachment of the phosphoryl group. To this end, we determined longitudinal (R1) and transverse (R2) 15N relaxation rates for backbone amides in pUb and compared the results for the “native-like” and the “alternate” NMR signals. The formation of a Ub dimer is expected to increase the R2 and decrease the R1 by approximately 1.6 fold31. Our results show essentially no difference between the two groups of signals (Fig. 1a and b) and therefore no evidence of oligomerization. This rules out Ub:Ub interactions as a possible cause of the observed pUbalt conformer. Furthermore, the similarity of the transverse relaxation rates for both Ub conformers allowed us to use intensities of the corresponding NMR signals in order to quantify the relative populations of pUbnat and pUbalt in the subsequent analysis.
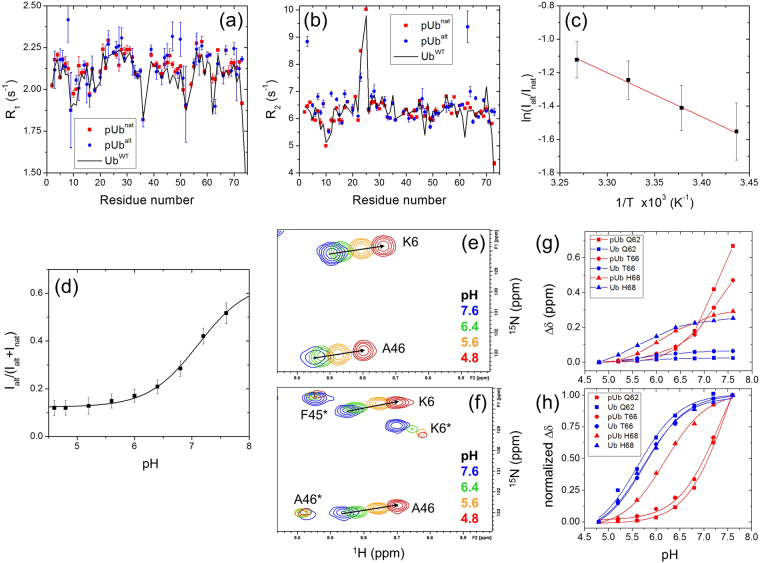
Figure 1.
NMR characterization of the two conformers of pUb. (a,b) Longitudinal (R1) and transverse (R2) 15N relaxation rates for the pUbnat (red) and pUbalt (blue) signals of pUb, as well as for UbWT (black line). (c) Temperature dependence of the ratio of signal intensities, Ialt and Inat, of the pUbalt and pUbnat conformers, averaged over 16 residues (the symbols and error bars represent the mean and the standard deviation). The line represents a linear regression fit to the Boltzmann distribution of the relative populations of the two conformers, see Equation (1) (Methods). (d) The ratio of signal intensities of the two pUb conformers representing the relative population of pUbalt, as a function of pH. The ratio was averaged over 27 residues, the error bars represent standard deviations. The line represents the fit of these data to Equation (2) (see Methods). (e,f) Representative regions of the 1H-15N correlation spectra of UbWT (e) and pUb (f) showing signal shifts for select residues as a function of pH. The asterisks in F indicate pUbalt signals. (g,h) CSPs (g) and normalized CSPs (h) as a function of pH for the indicated residues in UbWT and pUbnat. The curves in (h) represent the results of fit of the normalized CSPs to Equation (3) (Methods). The pKa values extracted from the CSPs of Q62, T66, and H68 signals are 5.6, 5.8, and 5.7, respectively, in UbWT and 7.5, 7.3, and 6.2 for the signals of the same residues in pUbnat.
The 15N relaxation rates in pUb are generally similar to those in UbWT (Fig. 1a and b), both in the overall level and the residue-specific variations, indicating that the rotational diffusion (hence the shape) and the local backbone motions characteristic for Ub are preserved in pUb. Interestingly, several residues located near the phosphorylation site (most notably, I3 and K63) exhibited elevated R2 values in pUbalt, suggesting the presence of conformational exchange on the μs-ms time scale in this conformer. In addition, the 15N relaxation rates for L73 in pUbalt are markedly higher than in pUbnat and UbWT, consistent with the suggested retraction of the C-terminal tail which would render residues 72–73 less flexible.
The conformational equilibrium of S65 phosphorylated Ub is controlled by pH
Since the observed pUbalt and pUbnat NMR signals belong to distinct conformational states of pUb, we utilized the temperature dependence of the relative intensities of these signals in order to thermodynamically characterize the equilibrium between the corresponding states. As expected, the difference in the relative populations of pUbalt and pUbnat decreased with temperature. The linear dependence of the natural logarithm of the populations ratio versus 1/T in the temperature range from 291 to 306 K (Fig. 1c) allowed us to extract thermodynamic parameters of this equilibrium: ΔH = 22.0 ± 5.8 kJ/mol, ΔS = 62.5 ± 18.9 J/(mol·K), and ΔG = 3.32 ± 0.31 kJ/mol at 25 °C.
Because the covalently attached phosphate group can be singly or doubly deprotonated (having the (−1) or (−2) charge, respectively), we also hypothesized that the two observed Ub conformers could be coupled to the charge state of the phosphate group on S65. To test if the charge state of the phosphate controls the equilibrium between pUbnat and pUbalt, we prepared pUb in a citric acid-phosphate buffer at pH 7.6 and then gradually decreased the pH down to 4.6, recording NMR spectra at every titration point. We observed a systematic decrease in the intensity of the pUbalt signals over the course of the pH titration: these signals were comparable in intensity to the corresponding pUbnat signals at pH 7.6 but almost vanished at pH 4.6. This decrease of the pUbalt signals at low pH was not caused by a loss of the phosphoryl group, as evident from the absence of the signal of unphosphorylated S65 in the pUb spectrum at pH 4.8 (Figure S2c,d) indicating that the protein was still entirely in its phosphorylated state. The ratio of signal intensities, Ialt/(Ialt + Inat), reflecting the fraction of Ub molecules in the pUbalt conformation, exhibited a sigmoidal behavior as a function of pH (Fig. 1d) which could be fit to a single pKa value of 7.08 ± 0.13 (the individual values for different residues spread from 6.8 to 7.4). This pKa value is consistent with the pKa2 (6.9–7.2) of phosphoric acid32,33 (related to the equilibrium between dihydrogen and monohydrogen phosphate ions) and somewhat higher than the reported random-coil pKa value (≈6) of pSer34. Note that H68, the other Ub residue titratable in the pH range (4.6–7.6) studied here, has a pKa of 5.535 and therefore is not expected to have a significant effect on the pKa values determined above. This suggests that the observed pH dependence of the relative intensities of pUbalt and pUbnat signals (Fig. 1d) primarily reflects the change in the charge/protonation state of the phosphate group on S65.
Our observation of the pH-dependent conformational switch in pUb generally agrees with the report by Dong et al.36 that appeared online as this manuscript was in the process of submission. The general agreement of our pKa values with those derived in that publication using 31P NMR signals of pS65 corroborates our findings.
Interestingly, in addition to the changes in the relative signal intensities of the pUbnat and pUbalt conformers with pH, we observed pH-dependent shifts for many amides in pUb. Notably, the pUbalt signals showed significantly lesser shifts with pH than the pUbnat signals. To examine the effects of phosphorylation on the chemical shifts of pUb, we compared our results with a similar pH titration of UbWT. Both pUbnat and UbWT signals generally followed a linear trajectory on the 1H-15N map indicating that the 1H and 15N resonances are affected by pH proportionally (Figs 1e and f and S2a,b). We examined amide chemical shift perturbations (CSPs) as a function of pH for each residue in order to determine whether the observed effects were caused by the phosphate on S65 or by some other titratable group, such as H68. We found that the CSPs of the residues in the vicinity of H68 (e.g. K6, I44, A46, G47, Q49, L69) behaved similarly for both UbWT and pUbnat, and their pH dependence was consistent with the reported pKa of H68 (pKa≈5.535). However, residues closer to the phosphorylation site, e.g., Q62 and T66, showed far greater CSPs in pUb compared to UbWT (Figs 1g and S2a,b). In addition, their normalized CSPs in pUb showed a comparatively larger increase between pH 6.5 and 7.5, in the vicinity of pKa2 of phosphate (Fig. 1h), and could be fit with a single pKa value of 7.5 (Q62) and 7.3 (T66). Also the side chain amide signals of Q62 in pUb shifted significantly over the course of the pH titration (pKa = 7.5) whereas those of UbWT did not move (Figure S2a,b,e). Interestingly, these shifts were strikingly different for the Hε21 and Hε22 protons, both in the magnitude and the direction, suggesting differential contacts of the two protons with the surrounding atoms. The implications of this will be considered in the discussion section.
Do phosphomimetic mutations reproduce the effect of phosphorylation?
From the pH titration of pUb it became apparent that the (−2) charge of the phosphate group is necessary for the pUbalt conformer to be present. This led us to hypothesize that the phosphomimetic Ub variants (UbS65D and UbS65E) commonly used to mimic phosphorylation in vivo can only model the pUbnat conformer, owing to the fact that neither an aspartate nor glutamate residue is able to attain a (−2) charge. To test the ability of the phosphomimics to structurally model both conformers, and to test the effects of a single negative charge at S65 on Ub structure, we collected 1H-15N NMR spectra for both UbS65D and UbS65E. Surprisingly, both mutations resulted in very striking spectral perturbations (Figs 2a,b,d,e and S3e,f). However, the absence of any new signals in either phosphomimetic variant, in agreement with the previous observations27, indicates that no pUbalt–like conformer exists for UbS65D or UbS65E. Puzzlingly, of the three S65 modifications considered here, S65D resulted in much greater magnitudes of CSPs vs. UbWT (Fig. 2a–f). To understand whether the observed large CSPs were due to structural or charge-related effects, we determined the structure of UbS65D using X-ray crystallography.
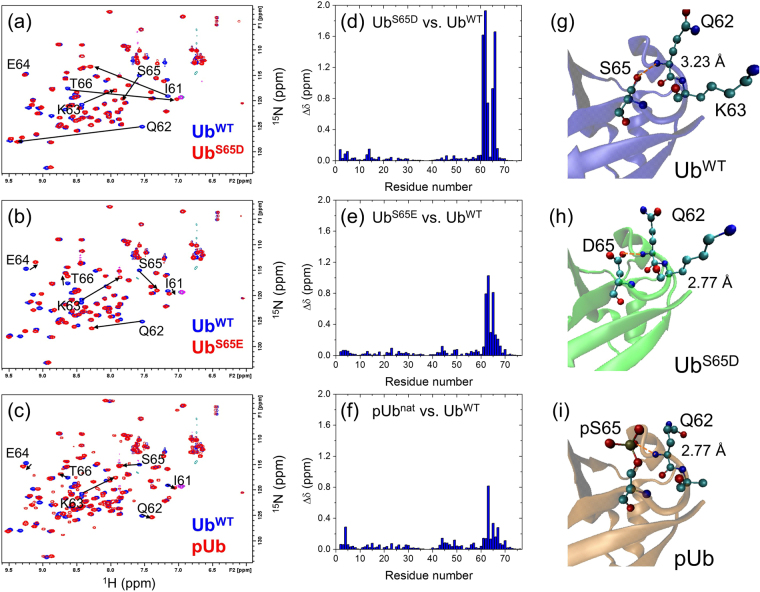
Figure 2.
NMR and structural comparison of pUb and phosphomimic Ub variants with UbWT. (a–c) Overlay of 1H-15N correlation spectra of (a) UbS65D, (b) UbS65E, and (c) pUb (all colored red) with the spectrum of UbWT (blue). Signals belonging to several residues around the modification site are indicated, their shifts are marked with arrows. (d and e) Spectral differences seen in panels (a–c) quantified as amide chemical shift perturbations in (d) UbS65D, (e) UbS65E, and (f) pUb versus UbWT as a function of residue number. (g–i) Comparison of the crystal structures of (g) UbWT (PDB ID: 1UBQ), (h) UbS65D (this work), and (i) pUbnat (PDB ID: 4WZP); zoom on the area containing residue 65. The numbers indicate the distance (shown by the orange dashed line) between the backbone amide nitrogen of Q62 and the closest to it oxygen of the side chain of S65, D65, or pS65, respectively.
The structure of UbS65D was resolved to 1.2 Å. The two-molecule unit cell is shown in Figure S3a. Indexing and refinement statistics are shown in Table S1. The UbS65D structure shows little overall deviation from UbWT (PDB ID: 1UBQ) (Figure S3b). With the flexible C-terminal tail included, the UbS65D and UbWT structures superimpose with a backbone RMSD of 0.56 Å for chain A of the two-chain asymmetric unit and 0.94 Å for chain B. Removing the flexible-tail residues R74, G75, and G76 decreases RMSD to 0.50 Å for chain A, and 0.69 Å for chain B. The backbone structures of UbS65D and UbWT are very similar, even in the loop containing S65 (Figs 2g,h and S3d). There are minor positional differences in the side chains of residues 62 to 65. Elsewhere in the molecule, the greatest structural deviation is in the C-terminal tail, which is likely a result of the tail’s flexibility, as well as in the β1/β2 loop comprising residues T7 to G10. An examination of the polar contacts between Q62 and S/D65 showed noticeable differences in the backbone environment of Q62 in the mutant compared to UbWT. The side chain oxygen (Oδ2) of D65 in UbS65D is 0.44–0.47 Å closer to the backbone amide nitrogen of Q62 than the S65 oxygen in UbWT (Fig. 2g,h). At the same time, it is 0.94–0.95 Å farther from the backbone carbonyl oxygen of Q62.
UbS65D is also structurally similar to the crystallized pUbnat conformer of pUb (PDB ID: 4WZP, Figs 2i and S3d)27. The backbone RMSD is 0.63 Å (chain A, tail residues not included). In the pUb structure, the closest phosphate oxygen to the amide group of Q62 is at about the same distance as the D65 side chain oxygen in UbS65D (Fig. 2i). Furthermore, for all three Ub variants considered here (UbWT, UbS65D, and pUbnat) the crystal structures predict a polar/hydrogen-bond contact between the side chain oxygen of residue 65 and the backbone amide nitrogen of Q62, as well as a hydrogen bond between the backbone amide nitrogen of residue 65 and the carbonyl oxygen of Q62. However, the chemical shift differences between UbS65D and pUb are nonetheless very striking (Figure S3f), suggesting that the phosphomimic does not fully reproduce the chemical environment of the pUbnat conformer in solution.
The effect of S65 phosphorylation on the ligand-binding properties of ubiquitin
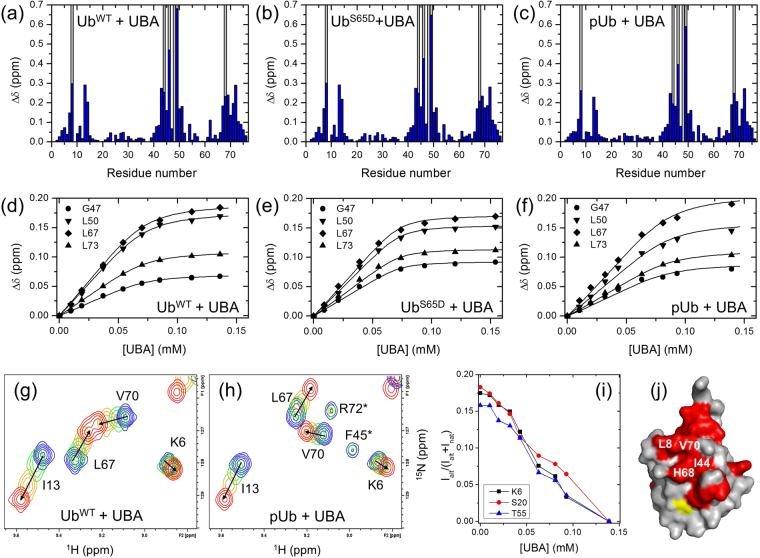
We next tested the ability of pUb to bind to a well-characterized Ub-binding partner and one of the strongest Ub-binding domains37, the ubiquitin-associated domain (UBA) of human ubiquilin−1/hPLIC138 involved in regulation of Ub-mediated proteasomal degradation of proteins39. We hypothesized that the structural perturbations in pUb due to the pUbalt conformer would impair UBA binding. To determine binding affinity, we titrated 15N-labeled pUb with unlabeled UBA and monitored changes in the chemical shifts of the pUb signals. By monitoring the pUbnat signals we found that pUb binds UBA with a comparable affinity to WT Ub (Fig. 3a–f, Table S2). By contrast, the pUbalt signals barely shifted upon the addition of UBA (Fig. 3g and h) but instead uniformly decreased in intensity, to an even greater extent than upon reduction of pH (Figs 3i and S4). This suggests that the pUbalt conformer is not involved in UBA binding, and the observed disappearance of the pUbalt signals reflects a shift in the conformational equilibrium between pUbalt and pUbnat upon UBA binding to the latter. We also characterized the ability of the phosphomimetic UbS65D to bind the UBA, in order to test how well UbS65D can mimic pUb functionally. We found that UbS65D has a somewhat enhanced affinity for UBA compared to pUb and even UbWT: the derived dissociation constant values were Kd = 4.9 ± 1.2 μM (UbWT), 1.8 ± 0.5 μM (UbS65D), and 8.0 ± 2.5 μM (pUbnat) (Fig. 3d–f, Table S2). For all three Ub variants, UbWT, UbS65D, and pUb, strong signal attenuations were observed for residues L8, I44, A46, K48, Q49, and H68 (Fig. 3a–c), indicating an intermediate or slow exchange on the NMR chemical shift timescale, as reported previously37,38. This suggests that the canonical ligand-binding interface remains intact in the pUbnat conformer of the phosphorylated Ub (Fig. 3j).
Figure 3.
NMR titration analysis of binding of the UBA domain of ubiquilin-1 to the Ub variants studied here. (a–c) Spectral perturbations at the titration endpoint for each residue and (d–f) the representative titration curves for UbWT, UbS65D, and pUb. Blue bars in (a–c) show the magnitude of amide CSPs as a function of residue number in Ub, while the grey bars indicate residues exhibiting strong signal attenuations during titration (>75%). The curves in (d–f) correspond to a global fit of all analyzed residues to a single-site binding model (see Table S2). (g,h) Fragments of the 1H-15N correlation spectra of UbWT (g) and pUb (h) illustrating NMR signal behavior in the course of UBA titration, colored from blue (free Ub or pUb) to red (titration endpoint). (i) The reduction in the relative intensity of the pUbalt signals upon addition of UBA. (j) Map on the surface of Ub of the consensus residues (painted red) exhibiting strong CSPs and/or signal attenuations upon UBA binding. Hydrophobic patch residues are indicated. S65 is painted yellow.
We also examined the effect of S65 phosphorylation on Ub binding to K48-specific deubiquitinase OTUB1, which interacts with the hydrophobic patch surface on Ub or Ub240. The addition of OTUB1 to 15N-labeled pUb resulted in significant attenuation of pUbnat signals (Figure S5c), consistent with the expected 5–6 fold increase in the molecular mass upon pUb binding to a 31 kDa protein. The residue-specific shifts and attenuations of pUbnat NMR signals map the OTUB1-interaction surface to the hydrophobic patch surface of pUb (Figure S5a,c,e). These results are consistent with our previous observations for OTUB1 binding to unphosphorylated Ub and Ub240 and with the structural data indicating that S65 is not directly involved in interactions with OTUB141,42. By contrast, only minimal shifts and signal attenuations were detected for pUbalt, suggesting that this conformer does not interact with OTUB1. The inability of pUbalt to bind OTUB1 is in line with the observed inhibitory effect of S65 phosphorylation on polyUb disassembly by this enzyme27.
Phosphorylation of S65 perturbs polyubiquitin conformations
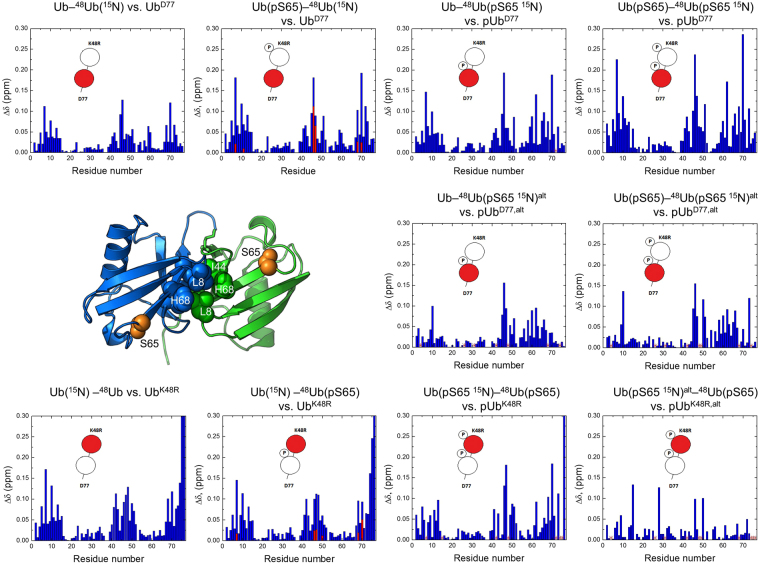
Given that phosphorylated polyUb chains are found in cells28,29 and PINK1 has been identified as a polyUb kinase27, understanding how S65 phosphorylation affects interdomain interactions and conformational properties of polyUb chains is critical to a complete understanding of the consequences of this modification. Since the pUbalt conformer turned out to be binding incompetent with respect to an intermolecular interaction, we next examined how it would behave in an intramolecular setting, when the binding partner is another Ub within the same chain. Because linkage via K48 is the most abundant in cells43, and K48-linked Ub chains act as signals for proteasomal degradation44, we examined the effect of S65 phosphorylation on K48-linked Ub dimers. Specifically, we tested whether the Ub:Ub interface, a hallmark of the K48-linked Ub dimer44–46, is affected by the phosphorylation. For this we synthesized K48-linked dimers with S65 phosphorylated in one or both Ub units. Figure 4 contains schematic diagrams of every phosphorylated Ub dimer that was synthesized. Two versions of each dimer were made: one with 15N-labeled proximal Ub, and one with 15N-labeled distal Ub (the labeled unit is shown as a red circle in Fig. 4). To denote these dimers, we will use the Ub chain nomenclature introduced by Nakasone et al.47, in which Ub units are written in the distal to proximal direction (left to right). We append this nomenclature by the addition of a (pS65) to indicate the phosphorylated Ub. For example, Ub(15N)–48Ub(pS65) denotes a K48-linked dimer 15N-labeled on the distal Ub and phosphorylated on the proximal Ub. For brevity, we will omit the K48R and D77 designations in our notations, unless necessary, as all our Ub dimers possess these mutations in the distal and proximal units, respectively.
Figure 4.
NMR analysis of the effect of Ub phosphorylation on the spectra of K48-linked Ub dimer. Each CSP plot shown here compares the spectra of a given Ub unit in the dimer with the respective monomeric Ub variant. Also shown are cartoon representations of each dimer, where phosphorylated Ub unit is marked with a “P” and the 15N-labeled unit (which acts as a reporter in each case) is colored red. Following the established nomenclature in the Ub field44, the Ub unit containing free C terminus is called “proximal” while the other Ub, conjugated to it through its C-terminal G76, is called “distal”. The proximal Ub has D77 as the C-terminal residue while the distal Ub contains a K48R mutation. The red bars on the plots for Ub(15N)–48Ub(pS65) and Ub(pS65)–48Ub(15N) indicate CSPs for the additional signals. Red asterisks indicate signals that could not be confidently assigned due to signal overlap. Also shown is the 3-D structure of the closed state of K48-linked Ub dimer (PDB ID: 1AAR); the hydrophobic patch residues L8, I44, V70 as well as H68 of the distal (blue) and proximal (green) Ubs are shown as spheres, the site of phosphorylation, S65, on each Ub is shown as orange spheres.
The first two Ub dimers we tested contained a single phosphate group, and were 15N-labeled on the unphosphorylated Ub unit: Ub(15N)–48Ub(pS65) and Ub(pS65)–48Ub(15N). For both dimers, the CSP patterns at pH 6.8 resemble those in the corresponding unphosphorylated dimers (Fig. 4), indicating the formation of the Ub:Ub interface45. The CSPs for the residues forming the interface were somewhat stronger in the proximal Ub and weaker in the distal Ub compared to those in the unphosphorylated dimers (see also Figures S6c and S7c). In both cases the amide signals shifted in the same direction as in the unphosphorylated dimer (Figures S6d and S7d), suggesting similar interatomic contacts. Furthermore, only negligible CSPs were observed at pH 4.6, indicating absence of the Ub:Ub interface at low pH (Figure S8). This behavior is in full agreement with the pH-dependent conformational switch reported for the unphosphorylated dimer45,48.
To our surprise, several residues in the 15N-labeled Ub units exhibited new signals resulting from phosphorylation of the opposite Ub unit in the dimer, indicating that these residues sense two different environments. We denote these as “additional” signals. The NMR spectra allowed us to rule out contamination of our samples with either unreacted Ub monomer or unphosphorylated dimer (Figures S6a,b and S7a,b), thus confirming that the new signals are genuine to the phosphorylated dimer. Interestingly, the residues exhibiting these additional signals cluster around the Ub:Ub interface (Figures S6f and S7f). We noted that these signals were much closer to the signals from monomeric Ub than to Ub dimer, and in many cases we observed a linear movement, from the monomer signal to the additional signal to the dimer signal (Figures S6e and S7e). We therefore hypothesize that the additional signals correspond to those dimers where the phosphorylated Ub unit is in the pUbalt state. Judging by the smaller signal shifts, these dimers have a weaker interface. In line with our hypothesis, the additional signals were absent at pH 4.6, where pUbalt is only weakly populated (Fig. 1d), although this could also reflect the disappearance of the Ub:Ub interface at low pH45,48. Overall, we conclude that while the Ub:Ub interface is generally preserved in the singly phosphorylated Ub dimers, even a single phosphate group on one Ub unit is enough to perturb the conformational states of the K48-linked Ub dimer.
We next examined three additional dimers: Ub–48Ub(pS65, 15N), Ub(pS65)–48Ub(pS65, 15N), and Ub(pS65, 15N)–48Ub(pS65). The latter two are phosphorylated on both Ub units. For each dimer, CSPs were determined for both pUbnat and pUbalt conformers (Fig. 4). Our findings can be summarized as follows. First, despite the relatively tight45,46 interface between the two Ub units, the pUbalt conformer was still present in both the proximal and distal units. Second, the largest CSPs for each phosphorylated dimer were found in the same residues as for the unphosphorylated dimers, indicating that the Ub:Ub interface is generally preserved even when both Ubs are phosphorylated. Third, the magnitude of the CSPs in the proximal Ub appears to increase with each additional phosphate (Fig. 4) suggesting strengthening of the interface. Fourth, the signal shifts for the pUbalt conformer upon dimer formation were overall significantly smaller than those for the pUbnat conformer, suggesting that the pUbalt conformer does not participate in the Ub:Ub interface or at least plays a lesser role in forming it (Fig. 4). Lastly, when using the UbD77 mutant (phosphorylated at S65) as a reference for the proximal Ub in the dimers, we observed that the additional Asp (D77) at Ub C terminus slightly stabilized the Ubalt conformer (Figure S9a), possibly through electrostatic interactions of the additional negative charge on the C terminus with the positively charged residues surrounding the β5 strand.
Discussion
Our study confirmed that the alternate NMR signals (pUbalt) caused by phosphorylation of S65 are not an artifact of Ub:Ub binding but in fact reflect the internal conformational equilibrium between the native-like and an alternate states of pUb. Consistent with the previous report27, phosphomimetic mutations do not result in the novel conformation of pUb. Furthermore, they do not mimic the chemical shifts observed for pUb, even for the pUbnat conformer (Figure S3f). This raises the question: what differences between UbS65D/E and pUb are responsible for the presence of the pUbalt conformer? The dramatic reduction of the pUbalt conformer signals at low pH provides a clue, suggesting that the protonation state of the phosphate group regulates the equilibrium between the two pUb conformers. At pH 4.6, the phosphate group is predominantly singly deprotonated, carrying the (−1) charge. As the pH increases toward the pKa2 of phosphate, the fraction of the doubly-deprotonated phosphate groups, with the (−2) charge, increases, and so does the fraction of pUbalt. Thus the presence of this conformer appears to be coupled to the protonation/charge state of the phosphate on S65.
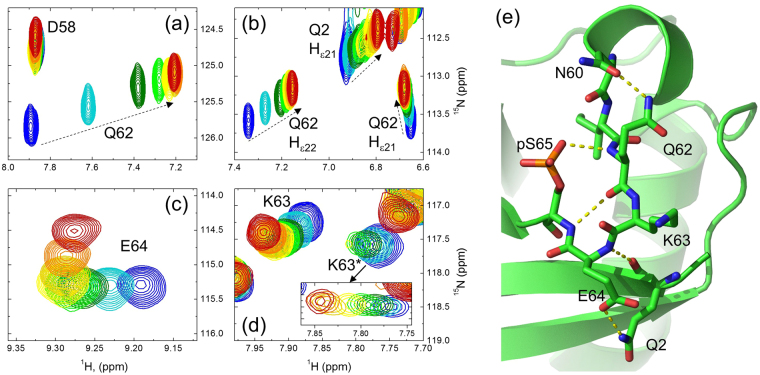
This in turn raises an intriguing question regarding the structural mechanism that relates protonation/deprotonation of pS65, expected to be ultrafast for a solvent-exposed phosphate group (e.g.49,50), to the slow (ca. 2 per sec) exchange between the pUbnat and pUbalt conformers. One can speculate that hydrogen bonds between the phosphate group and surrounding atoms in Ub might slow down the proton transfer between the phosphate and water. In fact, the crystal structure of pUbnat suggests a hydrogen bond between a phosphate oxygen of pS65 and the backbone amide nitrogen (NH) of Q62, and indeed, our NMR study revealed a strong pH-dependent shift (0.66 ppm in 1H and 0.65 ppm in 15N) of the Q62 amide signal in pUb but not in UbWT (see Figs 5a and S2a,b). Interestingly, we also found that the two side-chain amide hydrogens of Q62 in pUb behave differently with pH. Specifically, the signal of the Hε22 proton showed a dramatic shift (0.19 ppm) over the course of our pH titration, while the Hε21 signal shifted much less (see Fig. 5b). Both signals were virtually stationary in the pH titration of UbWT (Figure S2b), indicating that the shifts detected in pUb arise from titration of the phosphate. The differential behavior of the NMR signals of the two hydrogens suggests a specific hydrogen-bonding pattern involving Q62. Indeed, the crystal structure of pUbnat suggests a hydrogen bond between the side chain Nε2 atom of Q62 and the side chain Oδ1 oxygen of N60, mediated by one of the amide protons (likely Hε22) of Q62 (Fig. 5e). Of note, this hydrogen bond is not present in UbWT. Together with the hydrogen bonds between the phosphate group and NH of Q62, between NH of S65 and C’O of Q62, as well as between the side chain oxygen of E64 and the Nε2 nitrogen of Q2 (not present in UbWT or UbS65D), these interactions form a network of hydrogen bonds involving pS65 (Fig. 5e). This suggests an intriguing possibility, yet to be tested, that these interactions might play a role in slowing down and relaying the protonation/deprotonation of the pS65 phosphate group to the transitions between the pUbnat and pUbalt conformers.
Figure 5.
Differential behavior of NMR signals with pH relates to the hydrogen bonding network around the phosphorylation site. (a–d) Shifts of 1H-15N NMR signals of the backbone amides of (a) Q62, (c) E64, (d) K63, and (b) the side chain amides of Q62 and Q2 upon gradual change in pH from 7.6 (blue) to 4.8 (red). Both pUbnat and pUbalt signals of K63 are seen in (d). The inset depicts the movement of the K63 pUbalt signal (K63*); the intensities were artificially scaled up as the pH decreased in order to follow the vanishing signal. To guide the eye, the dashed arrows in (a and b) are drawn in the direction of signal shifts as the pH decreases. (e) Fragment of the crystal structure of pUbnat (PDB ID: 4WZP) showing the relevant residues (in stick representation) surrounding the phosphorylation site and the PyMol-predicted polar/hydrogen bonds (yellow dots) between them.
Interestingly, the pUbalt conformer did not completely disappear at low pH but plateaued at ca. 10% of the total protein (Fig. 1d). This suggests that other factors contribute to its stabilization at low pH. In this regard we noticed that pUbnat amide signals of the residues close to pS65 (most prominently, K63 and E64) did not shift linearly with pH but instead changed the direction of the shift below pH 5.6 (Fig. 5c and d). This could reflect protonation of the neighboring ionizable groups (e.g., E64, H68) and/or some local structural rearrangement in pUbnat. Note that such behavior is not present in the UbWT spectra.
Given the role of the protonation state of the phosphate in controlling the conformational equilibrium of pUb, the inability of aspartate and glutamate to adopt a (−2) charge on their side chain explains the observed structural and spectroscopic differences between phosphomimetic and phosphorylated Ub and suggests that phosphomimetic Ub is a poor structural model for pUb. It should nonetheless be noted that even a (−1) charge on residue 65 in Ub is sufficient to cause large CSPs (Fig. 2d and e), indicating that the charge at this position significantly affects the chemical environment of the backbone amides of the neighboring residues.
Many studies have used such phosphomimetic mutations to simulate phosphorylation of parkin17,18,29,51 and mono-17,18,51,52 and polyUb29. While these variants have generally been sufficient to model Ub phosphorylation in several cases, differences between UbS65D/E and pUb have been reported27,53, matching our observations. Furthermore, although UbS65D/E mutants have proven to be adequate surrogates for S65 pUb in its role as a parkin activator17,18,29, it is unclear whether these phosphomimics will be adequate in studies of other potential roles of pUb, particularly those that involve the pUbalt conformer, as well as for other phosphorylation sites on Ub.
The canonical ligand-binding surface of Ub does not appear to be disrupted either by phosphorylation (in case of the pUbnat conformer) or by phosphomimetic mutations of S65, as demonstrated by our experiments with the UBA of ubiquilin-1. However, our detailed analysis of the behavior of the NMR signals of the two pUb conformers upon addition of UBA suggests that pUbalt is incapable of binding to a ligand like ubiquilin-1 UBA, and it is the pUbnat conformer that binds the UBA. Similar conclusions can be drawn from our analysis of OTUB1 binding to pUb. From a structural perspective, the β5-strand slippage by two residues in pUbalt proposed in27 and recently shown by NMR36 disrupts the canonical Ub hydrophobic surface patch by shifting V70 to where H68 is in UbWT and by placing a positively charged side chain of R72 at the location of V70 in UbWT (Figure S10). Overall, such a shift would amount to mutating V70 to R, H68 to V, and possibly T66 to H. From this point of view the inability of pUbalt to bind UBA is in line with the fact that the V70R mutation in Ub is not tolerated by yeast cells54. This also suggests that the pUbalt conformer might be incapable of binding to other Ub-receptors that target the L8-I44-V70 hydrophobic patch of Ub, which could potentially impact pUb recognition. The weak “additional” signals detected here in K48-linked Ub dimers and reflecting a weaker interface between Ub and pUbalt seem to support these predictions. Our data indicate that the presence of the pUbalt conformer did not significantly impact the overall strength of UBA binding, likely due to the dynamic equilibrium between pUbalt and pUbnat. Indeed, our binding assays show that UBA binding to pUb stabilizes the pUbnat conformer. Published structures of pUb in complex with parkin55,56 do not show any evidence of the pUbalt conformer. However, although the latter conformer has not yet been found to be relevant for parkin activation, it may perhaps be relevant for other interactions. It remains to be seen whether a ligand exists that can specifically recognize and stabilize the pUbalt conformer of pUb as suggested recently57.
Our Ub dimer studies also provide insights into the impact of phosphorylation on the structure of Ub chains. K48-linked Ub dimers exist in solution in a dynamic equilibrium of multiple conformational states, including the “closed” state (wherein the hydrophobic patch residues form the “canonical” noncovalent Ub:Ub interface) and various “open” states, some of which are pre-organized for receptor recognition48. The CSPs (dimer vs. monoUb) reflect this equilibrium45 and therefore are uniquely suited for monitoring changes in the conformational ensemble of K48-linked Ub dimers. We found that the Ub:Ub interface characteristic of K48-linked chains at neutral pH is generally preserved upon phosphorylation of S65 and disappears at low pH (Figure S8), as in the unphosporylated dimer45. However, the additional NMR signals detected in the unphosphorylated Ub units of K48-linked dimers show that the conformational changes in the phosphorylated Ub unit are acutely sensed by the opposite Ub unit in the chain. The smaller magnitude of the CSPs (Fig. 4) associated with these signals suggests a weaker interface, i.e. a less populated closed state compared to the unphosphorylated K48-linked dimer. We hypothesize that this reflects those dimers where the phosphorylated Ub unit is in the pUbalt conformer. Moreover, for the dimers phosphorylated on both Ub units we did observe an additional (third) signal for G47 (Figure S9b). Taken together, our results indicate that, through the pUbalt state, Ub phosphorylation at S65 affects not only Ub-receptor interactions, but also the conformations that K48-linked Ub chains adopt in solution, and might serve as an additional mechanism of regulation in polyUb-mediated signaling pathways.
Ub phosphorylation at S65 presents and interesting and potentially important example that not only the negative sign but also the actual value of the charge/protonation state of a phosphorylated side chain can result in different structural and recognition properties of a protein. This can have implications for phosphorylation of other proteins. While the biological role of the pUbalt conformer of pUb remains elusive, it is clear, however, that a phosphomimetic mutation is not a foolproof model for phosphorylated protein, and using phosphomimics to study the effects of phosphorylation in vitro and in vivo should be done with care.
Materials and Methods
UbWT and both phosphomimetic variants (UbS65D, UbS65E) were grown and purified using the standard Ub purification protocol45,58. Ub used for NMR experiments was grown in 15N-enriched minimal media. PINK1 was grown and purified as described previously19, although Rosetta cells were used in place of BL21-RP. The UBA domain of human ubiquilin-1 was expressed and purified as described38. The UBA construct used in this study contained a V545Y mutation in order to enable its concentration determination through absorbance at 280 nm. Human OTUB1 was expressed and purified as described40.
The protocol for Ub phosphorylation was adapted from19,27. To generate pUb, 2 mM Ub were incubated with 32 μM maltose-binding protein-tagged-TcPINK1 in a 0.30 mL volume. The reaction buffer contained 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, and 2 mM DTT. 10 mM ATP was added to start the reaction. The reaction was incubated at room temperature for 3 hours. ESI-MS was used to confirm phosphorylation of Ub (Figure S1b,c). Mass spectrometry was performed on a JEOL AccuTOF-CS mass spectrometer in positive electrospray ionization mode.
The phosphorylated Ub was separated from unphosphorylated protein using a SPHP high performance cation exchange column (GE Healthcare). Column equilibration and wash buffer was 50 mM ammonium acetate (pH 4.5). Elution buffer was wash buffer plus 1 M NaCl. The phosphorylated protein eluted at 10% elution buffer. Purity was confirmed by ESI-MS and phosphorylation at the S65 site was confirmed by matching the NMR spectrum of the protein to published spectra27.
K48-linked Ub dimers were prepared enzymatically (see also refs45,58). First, the corresponding Ubs were phosphorylated as monomers and purified using the protocol detailed above. The formation of conjugates longer than dimers was blocked using chain terminating mutations, namely, a K48R mutation in the distal Ub and an addition of D77 at the C-terminus of the proximal Ub. Both UbK48R and UbD77 monomers were mixed in a 1:1 ratio for the dimerization reaction. The reaction contained 2 mM total Ub, 20 μM E2-25K, 1 μM human E1, 2 mM ATP, 5 mM MgCl2, 10 mM creatine phosphate, 0.6 U/mL inorganic phosphate, 0.6 U/mL creatine phosphokinase, and 3 mM TCEP in 50 mM Tris, pH 8.0. Dimers were separated from unreacted monomers using high-performance cation chromatography followed by size exclusion chromatography.
Residues used for the analysis of pH titration of pUb were selected based on several criteria: (1) NMR signals that overlapped partially or entirely with each other were excluded. (2) Signals that could not be unambiguously assigned due to attenuation or migration were excluded. (3) Signals not present in the alternate conformer were excluded.
NMR experiments were performed on a Bruker Avance–III 600 MHz spectrometer equipped with the TCI cryoprobe. All experiments were performed at 23 °C except for temperature dependence studies. The buffer for all NMR samples (except for pH titration) contained 20 mM sodium phosphate, 0.02% NaN3 and 7% D2O, the pH was 6.8. Total correlation spectroscopy (TOCSY) and nuclear Overhauser effect spectroscopy (NOESY) were used for signal assignments of UbS65D and UbS65E. NMR signal assignments for both pUb conformers were from reference27; we corrected assignments for G53 and G75. 1H-15N SOFAST-HMQC was used for most experiments, while HSQC was used for analysis of the temperature dependence. 15N relaxation experiments were performed as described59. NMR data were processed using TopSpin (Bruker Inc.) and analyzed using Sparky60 and in-house Matlab programs. Amide chemical shift perturbations (CSPs) for each residue were quantified as
where ΔδH and ΔδN are shifts of the 1H and 15N resonances, respectively. The fraction of pUb molecules in the pUbalt conformation was determined as
where Ialt and Inat are signal intensities for a given residue corresponding to pUbalt and pUbnat conformations, respectively.
The thermodynamic parameters, ΔH and ΔS, for the equilibrium between the two pUb conformers were obtained using a linear regression analysis of the dependence of their relative populations,
| 1 |
on the reciprocal temperature, according to the Boltzmann distribution.
The pH dependence of the fraction of pUb molecules in the pUbalt conformation was fit to the following equation reflecting the acid-base equilibrium:
| 2 |
The shifts in NMR signals of Ub and pUbnat as a function of pH were quantified as CSPs, and the normalized CSPs were fit to the following equation:
| 3 |
where Δδ(pH) for each residue was computed as the CSP at a given pH value versus the signal position at pH 4.8.
The dissociation constant, Kd, for UBA binding was obtained by fitting the measured CSP values (Δδ) in Ub or Ub variants to a single-site binding model61:
| 4 |
where Δδmax is the CSP at saturation and [P]t and [L]t are the total concentrations of the protein (Ub) and the ligand (UBA), respectively. Kd and Δδmax were treated as global fitting parameters using an in-house Matlab program Kdfit61.
Crystallization conditions for UbS65D were 0.1 M Tris pH 8.0, 0.2 M MgCl2, and 25% polyethylene glycol (PEG) 3350. Protein concentration was 4.23 mM and protein in 20 mM Hepes buffer was mixed 1:1 with crystallization buffer. Indexing of X-ray diffraction data for UbS65D was performed with Mosflm62. Phenix63 was used for molecular replacement. The crystal structure was refined using CCP4i64 and Coot65 software. Visual Molecular Dynamics66 was used for structural alignment of proteins, as well as for generation of figures.
Data Availability
The atom coordinates of UbS65D have been deposited with the Protein Data Bank (accession code: PDB ID: 5W46).
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
We thank Miratul Muqit for the PINK1 plasmid, Karina Herrera-Guzman and Mark Nakasone for the UBA material, Paul Paukstelis for help and advice with X-ray data analysis and structure refinement, and Christina Camara for help with NMR signal assignments. This research was supported in part by a grant to the University of Maryland from the Howard Hughes Medical Institute Undergraduate Science Education Program (Undergraduate Research Fellowship for Y.K.) and by NIH grant GM065334 to D.F.
Author Contributions
Y.K. and D.F. conceived and designed the experiments; Y.K., D.F. and M.-Y.L. performed NMR studies, R.H.S. collected diffraction data and performed initial analysis; Y.K. and D.F. analyzed the data and wrote the paper.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-20860-w.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fushman D, Wilkinson KD. Structure and recognition of polyubiquitin chains of different lengths and linkage. F1000 Biol Rep. 2011;3:26. doi: 10.3410/B3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shimura H, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Gen. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 3.Wenzel DM, Klevit RE. Following Ariadne’s thread: a new perspective on RBR ubiquitin ligases. BMC Biol. 2012;10:24. doi: 10.1186/1741-7007-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corti O, Lesage S, Brice A. What genetics tells us about the causes and mechanisms of Parkinson’s disease. Physiol Rev. 2011;91:1161–1218. doi: 10.1152/physrev.00022.2010. [DOI] [PubMed] [Google Scholar]
- 6.Narendra, D., Walker, J. E. & Youle, R. Mitochondrial Quality Control Mediated by PINK1 and Parkin: Links to Parkinsonism. Cold Spring Harbor Perspectives in Biology4, 10.1101/cshperspect.a011338 (2012). [DOI] [PMC free article] [PubMed]
- 7.Sun M, et al. Influence of heterozygosity for Parkin mutation on onset age in familial Parkinson disease The GenePD study. Arch Neurol. 2006;63:826–832. doi: 10.1001/archneur.63.6.826. [DOI] [PubMed] [Google Scholar]
- 8.Kitada T, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 9.Valente EM, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, et al. Risk of Parkinson disease in carriers of Parkin mutations - Estimation using the kin-cohort method. Arch Neurol. 2008;65:467–474. doi: 10.1001/archneur.65.4.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wauer T, Komander D. Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 2013;32:2099–2112. doi: 10.1038/emboj.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trempe JF, et al. Structure of Parkin Reveals Mechanisms for Ubiquitin Ligase Activation. Science. 2013;340:1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- 13.Matsuda N, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong H, et al. Parkin, PINK1, and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J Clin Invest. 2009;119:650–660. doi: 10.1172/JCI37617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaugule VK, et al. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 2011;30:2853–2867. doi: 10.1038/emboj.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riley BE, et al. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat Commun. 2013;4:1962. doi: 10.1038/ncomms2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koyano F, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 18.Kane LA, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kazlauskaite A, et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser(65) Biochem J. 2014;460:127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okatsu K, et al. PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nat Commun. 2012;3:1016. doi: 10.1038/ncomms2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondapalli C, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biology. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shiba-Fukushima, K. et al. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep2, 10.1038/srep01002 (2012). [DOI] [PMC free article] [PubMed]
- 23.Lee HJ, et al. Quantitative analysis of phosphopeptides in search of the disease biomarker from the hepatocellular carcinoma specimen. Proteomics. 2009;9:3395–3408. doi: 10.1002/pmic.200800943. [DOI] [PubMed] [Google Scholar]
- 24.Bennetzen MV, et al. Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol Cell Proteomics. 2010;9:1314–1323. doi: 10.1074/mcp.M900616-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu TL, et al. Survey of tyrosine kinase signaling reveals ROS kinase fusions in human cholangiocarcinoma. PLoS One. 2011;6:e15640. doi: 10.1371/journal.pone.0015640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malik R, et al. Quantitative analysis of the human spindle phosphoproteome at distinct mitotic stages. J Proteome Res. 2009;8:4553–4563. doi: 10.1021/pr9003773. [DOI] [PubMed] [Google Scholar]
- 27.Wauer, T. et al. Ubiquitin Ser 65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J (2014). [DOI] [PMC free article] [PubMed]
- 28.Ordureau A, et al. Quantitative Proteomics Reveal a Feedforward Mechanism for Mitochondrial PARKIN Translocation and Ubiquitin Chain Synthesis. Mol Cell. 2014;56:360–375. doi: 10.1016/j.molcel.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okatsu K, et al. Phosphorylated ubiquitin chain is the genuine Parkin receptor. J Cell Biol. 2015;209:111–128. doi: 10.1083/jcb.201410050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nowicka U, et al. DNA-Damage-Inducible 1 Protein (Ddi1) Contains an Uncharacteristic Ubiquitin-like Domain that Binds Ubiquitin. Structure. 2015;23:542–557. doi: 10.1016/j.str.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fushman D, Varadan R, Assfalg M, Walker O. Determining domain orientation in macromolecules by using spin-relaxation and residual dipolar coupling measurements. Progr NMR Spetcrosc. 2004;44:189–214. doi: 10.1016/j.pnmrs.2004.02.001. [DOI] [Google Scholar]
- 32.Haynes, W. CRC Handbook of Chemistry and Physics. 97 edn, (CRC Press, 2016).
- 33.Nelson, D. L. & Cox, M. M. Lehninger Principles of Biochemistry. 1328pp (Freeman W.H. & Company, 2017).
- 34.Bienkiewicz EA, Lumb KJ. Random-coil chemical shifts of phosphorylated amino acids. J Biomol NMR. 1999;15:203–206. doi: 10.1023/A:1008375029746. [DOI] [PubMed] [Google Scholar]
- 35.Fujiwara K, et al. Structure of the ubiquitin-interacting motif of S5a bound to the ubiquitin-like domain of HR23B. J Biol Chem. 2004;279:4760–4767. doi: 10.1074/jbc.M309448200. [DOI] [PubMed] [Google Scholar]
- 36.Dong X, et al. Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch. Proc Natl Acad Sci USA. 2017;114:6770–6775. doi: 10.1073/pnas.1705718114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 38.Zhang D, Raasi S, Fushman D. Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J Mol Biol. 2008;377:162–180. doi: 10.1016/j.jmb.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kleijnen MF, et al. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol Cell. 2000;6:409–419. doi: 10.1016/S1097-2765(00)00040-X. [DOI] [PubMed] [Google Scholar]
- 40.Wang T, et al. Evidence for bidentate substrate binding as the basis for the K48 linkage specificity of otubain 1. J Mol Biol. 2009;386:1011–1023. doi: 10.1016/j.jmb.2008.12.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiener R, Zhang X, Wang T, Wolberger C. The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature. 2012;483:618–622. doi: 10.1038/nature10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Juang YC, et al. OTUB1 co-opts Lys48-linked ubiquitin recognition to suppress E2 enzyme function. Mol Cell. 2012;45:384–397. doi: 10.1016/j.molcel.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 45.Varadan R, Walker O, Pickart C, Fushman D. Structural properties of polyubiquitin chains in solution. J Mol Biol. 2002;324:637–647. doi: 10.1016/S0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- 46.Cook WJ, Jeffrey LC, Carson M, Chen ZJ, Pickart CM. Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2) J Biol Chem. 1992;267:16467–16471. doi: 10.2210/pdb1aar/pdb. [DOI] [PubMed] [Google Scholar]
- 47.Nakasone MA, Livnat-Levanon N, Glickman MH, Cohen RE, Fushman D. Mixed-linkage ubiquitin chains send mixed messages. Structure. 2013;21:727–740. doi: 10.1016/j.str.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berlin K, et al. Recovering a Representative Conformational Ensemble from Underdetermined Macromolecular Structural Data. J Am Chem Soc. 2013;135:16595–16609. doi: 10.1021/ja4083717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eigen M. Proton transfer, acid-base catalysis, and enzymatic hydrolysis. Angew Chem Int Ed. 1964;3:1–19. doi: 10.1002/anie.196400011. [DOI] [Google Scholar]
- 50.Fersht, A. Structure and function in protein science. 162–163 (W. H. Freeman, 2003).
- 51.Yamano K, et al. Site-specific Interaction Mapping of Phosphorylated Ubiquitin to Uncover Parkin Activation. J Biol Chem. 2015;290:25199–25211. doi: 10.1074/jbc.M115.671446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walinda E, Morimoto D, Sugase K, Shirakawa M. Dual Function of Phosphoubiquitin in E3 Activation of Parkin. J Biol Chem. 2016;291:16879–16891. doi: 10.1074/jbc.M116.728600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herhaus L, Dikic I. Expanding the ubiquitin code through post-translational modification. EMBO Rep. 2015;16:1071–1083. doi: 10.15252/embr.201540891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roscoe BP, Thayer KM, Zeldovich KB, Fushman D, Bolon DNA. Analyses of the Effects of All Ubiquitin Point Mutants on Yeast Growth Rate. J Mol Biol. 2013;425:1363–1377. doi: 10.1016/j.jmb.2013.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wauer T, Simicek M, Schubert A, Komander D. Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature. 2015;524:370–374. doi: 10.1038/nature14879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar A, et al. Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity. Nat Struct Mol Biol. 2017;24:475–483. doi: 10.1038/nsmb.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schubert AF, et al. Structure of PINK1 in complex with its substrate ubiquitin. Nature. 2017;552:51–56. doi: 10.1038/nature24645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pickart CM, Raasi S. Controlled synthesis of polyubiquitin chains. Meth Enzymol. 2005;399:21–36. doi: 10.1016/S0076-6879(05)99002-2. [DOI] [PubMed] [Google Scholar]
- 59.Hall JB, Fushman D. Characterization of the overall and local dynamics of a protein with intermediate rotational anisotropy: Differentiating between conformational exchange and anisotropic diffusion in the B3 domain of protein G. J Biomol NMR. 2003;27:261–275. doi: 10.1023/A:1025467918856. [DOI] [PubMed] [Google Scholar]
- 60.Goddard, T. & Kneller, D. G. (University of California, San Fransisco).
- 61.Varadan R, et al. Solution conformation of Lys(63)-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem. 2004;279:7055–7063. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- 62.Battye TGG, Kontogiannis L, Johnson O, Powell HR, Leslie AGW. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Cryst D. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Cryst D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Cryst D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(33–38):27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atom coordinates of UbS65D have been deposited with the Protein Data Bank (accession code: PDB ID: 5W46).
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.