Abstract
Widely developed for clinical screening, electrocardiogram (ECG) recordings capture the cardiac electrical activity from the body surface. ECG analysis can therefore be a crucial first step to help diagnose, understand and predict cardiovascular disorders responsible for 30% of deaths worldwide. Computational techniques, and more specifically machine learning techniques and computational modelling are powerful tools for classification, clustering and simulation, and they have recently been applied to address the analysis of medical data, especially ECG data. This review describes the computational methods in use for ECG analysis, with a focus on machine learning and 3D computer simulations, as well as their accuracy, clinical implications and contributions to medical advances. The first section focuses on heartbeat classification and the techniques developed to extract and classify abnormal from regular beats. The second section focuses on patient diagnosis from whole recordings, applied to different diseases. The third section presents real-time diagnosis and applications to wearable devices. The fourth section highlights the recent field of personalized ECG computer simulations and their interpretation. Finally, the discussion section outlines the challenges of ECG analysis and provides a critical assessment of the methods presented. The computational methods reported in this review are a strong asset for medical discoveries and their translation to the clinical world may lead to promising advances.
Keywords: machine learning, computer simulations, electrocardiogram, interpretation and analysis, classification
1. Introduction
Cardiovascular disorders are a major burden worldwide, causing 30% of the deaths in the world according to the World Health Organization [1]. Therefore, early detection of the patients at risk, and a better understanding of the disease mechanisms are crucial to improve diagnosis and treatment. Widely used by clinicians as a routine modality in hospitals, electrocardiogram (ECG) recordings capture the propagation of the electrical signal in the heart from the body surface. Therefore, many cardiac structural or electrophysiological abnormalities have a signature on the ECG and their identification can help diagnose cardiac disorders. ECG recordings include different formats: Holter ECGs record the electrical activity of the heart over longer periods of time (several hours), whereas standard 12-lead ECGs provide information on cardiac activity from 12 different perspectives (leads) over several heartbeats. Manually studying large amounts of ECG data can be tedious and time-consuming. Therefore, there is a need for powerful computational methods to maximize the information extracted from comprehensive ECG datasets [2]. The variety of ECG formats and their clinical applications also call for a diversity of computational techniques to address this need.
In this review, we aim to describe the clinical applications and main machine learning methods currently used for ECG analysis to enable heartbeat and patient classification, and advanced computer simulation to explain cardiac ECG phenotypes. Although ECG signal processing techniques have been described before [3], here we review how machine learning methods have recently been used to automatically learn the dataset structure in order to make predictions. Learning can be supervised (training examples are given to the algorithm which learns the dataset relationships before testing them on unknown data), or unsupervised (the algorithm learns the data structure by itself). Classification of heartbeats [4–6] is probably the most developed application of machine learning to the ECG. It focuses on the detection of abnormal, irregular beats that may occur at unpredicted times and helps to detect arrhythmias. Other studies focus on patient classification [7–9], based on the overall behaviour of the ECG, to diagnose specific diseases. In addition, with the development of wearable devices and the need for real-time diagnosis, other challenges such as speed or memory requirements have emerged, requiring the adaptation of these methods for quick classification [10–12]. Analysing the ECG with machine learning methods is a promising approach but dealing with medical data for clinical applications raises some additional challenges, such as the lack of databases available for validation and the need to interpret ECG abnormalities at the organ and cellular level. 3D computer simulations are a powerful tool to address these issues. They allow personalized simulations of the ECG, allowing the interpretation of the ECG signals [13–15] as well as the generation of synthetic data for training and validation purposes.
2. Heartbeat classification
A heartbeat is defined as the sequence of electrical events happening in a whole cardiac cycle, from depolarization to repolarization. For a normal beat in sinus rhythm it includes the P wave, the QRS complex and the T wave. Heartbeat classification focuses on the automatic identification of beats of different nature, and can be useful for detecting ectopic beats or arrhythmic events. It is the most developed application of machine learning methods to ECG analysis, mostly because of the databases publically available for training and testing such as the MIT-BIH [16], composed of 48 half-hour excerpts of two-channel ambulatory ECG recordings and initially developed to evaluate arrhythmia detectors. Other databases are also available and widely used to develop these techniques such as those contained in Physionet's Physiobank, INCART, or the American Heart Association database [17] (table 1). The objective, performance and validation of the studies presented in the section below are summarized in table 2.
Table 1.
Summary table of the major databases used for classification of ECG signals.
| database | type of recordings | number of recordings | annotations |
|---|---|---|---|
| MIT-BIH Arrhythmiaa | — 30-min excerpts — 2-channel ambulatory ECG — 360 Hz |
48 | beat-by-beat annotations for each beat in each recording (approx. 110 000 annotations) |
| QT databasea | — 15-min. excerpts — 2-channel ECG — 250 Hz |
105 | — reference beat annotations — segmentation of waveforms (for 30 to 100 normal beats per recording) |
| American Heart Association ventricular arrhythmiaa | — 2-channel excerpts — analogue ambulatory ECG — 250 Hz |
80 for training—75 for testing | — 8 classes of recordings (level of ventricular ectopy) — final 30 min annotated beat-by-beat |
| INCARTa | — 30-min ECG — 12 leads — 275 Hz |
75 | — 175 000 beat annotations — 10 classes pathological diagnosis |
| UCI Machine Learning: Arrhythmia dataset | — 279 attributes (age, sex, height, waveforms description over 12 leads such as duration, amplitudes, areas) | 452 | 16 arrhythmia classes labelled |
| Long-Term-STa | — between 21 and 24 h — 2 or 3 ECG signals — 250 Hz |
86 | — annotated ST episode — QRS annotations — ST level measures |
aPhysioBank datasets [17] available at https://physionet.org/:
— gathers 60 databases (4TB) of physiological signals: cardiopulmonary, neural, other biomedical signals
— freely available
— healthy subjects and patients (sudden cardiac death, congestive heart failure, epilepsy, gait disorders, sleep apnoea, ageing)
Table 2.
Summary table of reviewed machine learning classification methods, along with their objective, dataset, performance and validation. AF, atrial fibrillation; ANN, artificial neural network; CNN, convolutional neural network; HMM, hidden Markov model; LBBB, left bundle branch block; LD, linear discriminant; LSTM, long short-term memory network; MLP, multilayer perceptron; PVC, premature ventricular contraction; RBBB, right bundle branch block; RNN, recurrent neural network; SVEB, supraventricular ectopic beat; SVM, support vector machine; VEB, ventricular ectopic beat.
| study | classes and focus | method | performance | validation |
|---|---|---|---|---|
| heartbeat classification | ||||
| Chazal et al. [18] | normal, VEB, SVEB fusion of normal and VEB, unknown | LD with QRS-based and time intervals features | SVEB: 75.9% (sensitivity) VEB: 77.7% (sensitivity) |
on 50 000 independent beats (MIT-BIH) |
| Llamedo & Martínez [4] | normal, VEB and SVEB classification | LD (RR intervals and wavelet transform features) with floating feature selection | 93% global accuracy | on independent MIT-BIH beats and INCART |
| Yeh et al. [19] | normal, LBBB, RBBB, PVC, atrial premature contractions | LD | 96.23% (global accuracy) | on 14 30-min excerpts (MIT-BIH) |
| Ubeyli [6] | normal, congestive heart failure, ventricular tachyarrhythmia, AF | SVM with error output correction code and discrete wavelet transform | 98.61% accuracy | on 360 independent beats (Physionet) |
| Melgani & Bazi [20] | normal, atrial premature beat, PVC, RBBB, LBBB, and paced beat | SVM optimized by particle swarm optimization | 89.72% accuracy | on 40 438 independent beats (from 20 patient records of MIT-BIH) |
| Asl et al. [21] | normal, PVC, AF, sick sinus syndrome, ventricular fibrillation, 2° heart block | SVM with heart rate variability features and discriminant analysis feature reduction | 99.16% accuracy | on 463 testing segments of MIT-BIH (average over 100 different runs) |
| Nasiri et al. [22] | normal, RBBB, LBBB, and paced beat | SVM with principal component analysis and genetic algorithm | 93.46% accuracy | 50% of MIT-BIH for testing |
| Ganeshkumar & Kumaraswamy [23] | normal, PVC, paced, atrial premature beat, LBBB and RBBB | random forest (30 trees) on 150 beats from MIT-BIH | 92.16% accuracy | not validated on independent dataset |
| de Oliveira et al. [24] | PVC detection | Bayesian network framework using channel fusion | 99.69% sensitivity | on QT database (25%—23 765 beats for testing) |
| Coast et al. [25] | VEB detection (over American Heart Association database) | HMM with states corresponding to ECG waveforms or intervals | 97.25% sensitivity | on 799 independent beats |
| Koski [26] | PVC detection | HMM and broken line approximation (30 states) | 100% accuracy | on only 4 beats |
| Andreao et al. [27] | PVC detection | HMM and rule-based system | 99.79% sensitivity | on 61 543 test beats from QT database |
| Niwas et al. [28] | normal, LBBB, RBBB, atrial premature beat, SVEB, PVC, AF, ventricular fibrillation, sick sinus syndrome, fusion of VEB and normal | ANN with heartbeat intervals and spectral entropy features | 99.02% accuracy | on 180 (unspecified) independent datasets |
| Inan et al. [29] | PVC detection | feed-forward MLP with wavelet transform and time intervals features | 96.82% accuracy | on 22 ECG recordings from MIT-BIH |
| Ubeyli [30] | normal, congestive heart failure, ventricular tachyarrhythmia, AF | RNN with Levenberg–Marquardt training algorithm and eigenvectors | 98.06% accuracy | on 360 beats from Physionet |
| Lagerholm et al. [31] | normal, LBBB, RBBB, atrial premature, aberrated atrial premature, nodal premature, SVEB, VEB, fusion normal and VEB, ventricular flutter, atrial escape, nodal escape, ventricular escape, unknown | Self-organizing networks with Hermite transform and RR intervals features | 1.5% of misclassification | not validated on independent database |
| Linh et al. [32] | normal, LBBB, RBBB, atrial premature, VEB, ventricular flatter wave, ventricular escape | TSK fuzzy network with Hermite transform | 96% accuracy | on 3668 beats from MIT-BIH |
| Ozbay et al. [33] | normal, sinus bradycardia, ventricular tachycardia, sinus arrhythmia, atrial premature contraction, paced, RBBB, LBBB, AF and atrial flutter | MLP with fuzzy clustering neural network architecture | 99.9% accuracy | on 5342 segments from 92 patients (MIT-BIH) |
| ECG recording analysis for patient diagnosis, monitoring and stratification | ||||
| Mincholé et al. [34] | ischaemic and non-ischaemic ST-segment changes | multivariate discriminant analysis with Wilk's Lambda minimization | 87.5% accuracy | cross validated estimation on LTST database |
| Faganeli & Jager [35] | patients with transient ischaemia episodes | decision trees with heart rate and Legendre polynomial coefficients features | 98.1% sensitivity, 85.2% specificity | bootstrap method |
| Rahman et al. [8] | hypertrophic cardiomyopathy detection on 12-lead ECG | SVM and random forest with 264 time intervals and waveforms amplitude features | precision of 0.84 | fivefold cross validation over 10 930 beats |
| Bailón et al. [36] | diagnosis of coronary artery disease | multivariate discriminant analysis with repolarization, depolarization and heart rate variability features | 94% sensitivity, 92% specificity | cross validated estimation (leave one out) |
| Kawazoe et al. [37] | risk of ventricular fibrillation in Brugada syndrome patients | logistic regression with syncope, R–J interval, QRS duration, and Tpeak–Tend dispersion as features | 97.1% sensitivity, 63.0% specificity | leave-one-out cross validation over 143 patients |
| Pourbabaee & Lucas [7] | paroxysmal AF episodes detection | MLP with time interval and morphological waveform features | 87.5% accuracy | over 16 recordings from 2001 Computing in Cardiology challenge |
| Colloca et al. [38] | AF episodes detection | SVM optimized with grid-search | 85.45% accuracy with 100% sensitivity to AF | Series 200 of the MIT-BIH Arrhythmia (with 8 AF subjects) |
| Asgari et al. [39] | AF episodes detection | SVM with stationary wavelet transform | 97.0% sensitivity | twofold stratified cross validation on MIT-BIH |
| Acharya et al. [40] | ischaemic/dilated cardiomyopathy, complete heart block, sick sinus syndrome, AF, ectopics, normal | ANN with fuzzy equivalence | 85–95% accuracy | on 66 testing samples |
| Zheng et al. [41] | congestive heart failure (from 2-lead ECGs) | CNN | 94.7% accuracy | 10-fold cross validation over 15 subjects |
| Kannathal et al. [9] | normal, abnormal (PVC, RBBB, LBBB, paced), life threatening (sick sinus syndrome, ischaemic heart disease, ventricular fibrillation) | ANN with radial basis function | 99% accuracy | on 200 independent testing patients |
| Zhang et al. [12] | asystole, extreme bradycardia, extreme tachycardia, ventricular tachycardia or ventricular flutter/fibrillation arrhythmia | SVM with genetic algorithm | 93% true positive rate | fivefold cross validation over 750 recordings (2015 Physionet Challenge) |
| real-time episodes' detection and wearable devices | ||||
| Kiranyaz et al. [10] | VEB and SVEB detection | 1D patient-specific CNN | 98.6% accuracy | on 41 766 testing beats from MIT-BIH |
| Chauhan & Vig [42] | PVC, atrial premature contraction, paced, ventricular couplet | deep LSTM network | 0.975 precision, 0.9645 F-score | testing set of unknown size |
| Jeon et al. [43] | normal beats, AF, myocardial ischaemia classification | SVM on ARM processor | 95.1% sensitivity | 10-fold cross validation over MIT-BIH AF, 2001 and 2004 CinC challenge and STT database |
| Leutheuser et al. [44] | normal and abnormal (all MIT-BIH labels that are not normal), on Android devices | decision tree classifier with heartbeat features | 89.6% accuracy | not validated on independent dataset |
| Oresko et al. [11] | normal, RBBB, PVC, paced or fusion of paced and normal beat detection on smartphone | feed-forward MLP with QRS morphological beat pattern | 99% accuracy for normal—81% accuracy for fusion | threefold cross validation over 5421 beats (MIT-BIH) |
| Oster et al. [45] | normal and SVEB, fusion and VEB, and unknown | switching Kalman filters with X-factor mode | 99.2% F1-score | independent validation on INCART |
2.1. Clinical objectives and ECG data
The MIT-BIH arrhythmia database considers 15 heartbeat classes, which have been also used in other studies [18]. Due to this variety of heartbeat label sets, the classification objectives of the different studies may be different, making their performances harder to compare. Some studies focus on binary classification to distinguish between normal and abnormal beats [26,46], normal and premature ventricular beats (PVBs) [24,29] or normal and diseased beats [47]. Other works follow the classification recommendations of labelling rules such as the AAMI guidelines (normal, ventricular, supraventricular, fusion of normal and ventricular, and unknown beats [5]). However, most of these techniques report their classification performances in the same metrics, facilitating their comparison. Accuracy (%) measures the amount of correctly classified samples compared to the total number of samples classified. Sensitivity (resp. specificity) (%), or true positive (resp. negative) rate, measures the amount of positive (resp. negative) samples correctly classified.
Heartbeat classification can also be performed in recordings of different length, such as standard 12-lead ECGs, lasting for several seconds, or Holter ECGs, recorded for several hours. Longer recordings allow the analysis of the ECG over time and the identification of time-dependent abnormalities, such as changes in the beat morphologies with time or changes in heart rate. In addition to the length of the recording, the number of ECG leads may differ and various methods are proposed to handle multi-channel data. Some studies focus only on single lead data [48]. Others combine features computed over several different leads in a single feature vector [18]. Another approach is to combine the output of classifiers when applied over the different leads following a voting approach. For example, in Zhang & Luo's work [49], the outputs of several support vector machine (SVM) classifiers were merged based on the decision of each classifier.
2.2. Feature extraction and dimensionality reduction
Most machine learning classification techniques require the definition of a feature vector to describe the ECG beat and the training of a classifier. Each heartbeat is composed of multiple waves describing different events of the cardiac cycle (P-wave, QRS complex, T-wave) (figure 1). Morphological features, such as slopes, peaks, amplitudes [18,54], describe the shape of the ECG waveforms. They may be able to capture changes in the heart rhythm, such as sinus rhythm versus fibrillation, in which the complexes exhibit different morphologies. Some works focus on time interval features to characterize the dynamics of ECG phenomena such as QRS duration, QT interval or heart rate, defined as the number of beats per unit of time [22,23,31,46,55]. Morphological features include the coefficients of the Hermite transform, the wavelet transform or the discrete cosine transform [29,32] that aim to model the ECG beat instead of extracting features from the raw data. In most studies, a combination of these features is used to characterize the ECG signal. Other works such as Llamedo et al. [56] combine the signal's multiple channels before performing feature extraction. They investigated several strategies such as combining features from the wavelet transform (a time–frequency representation of the signal by mathematical functions called wavelets) from all the leads, computing the wavelet transform from the vectocardiogram leads, or computing the features from the two first principal component of the ECG leads' principal component analysis (PCA).
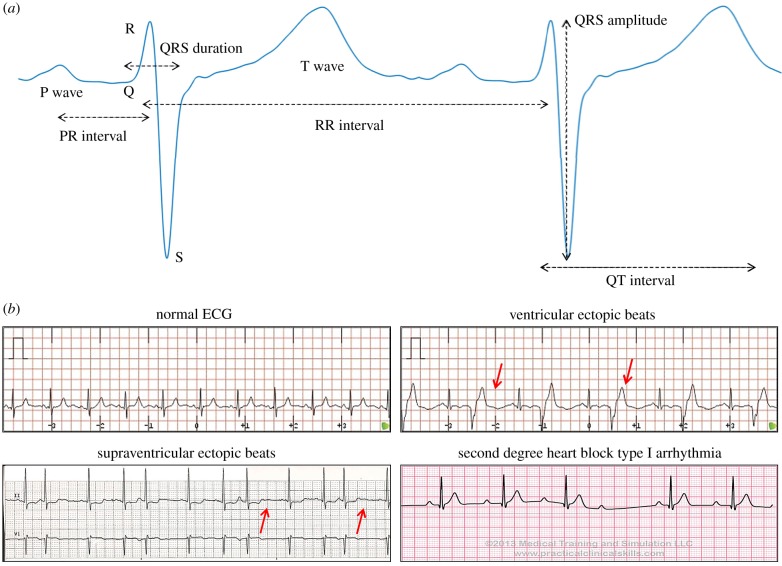
Figure 1.
Example of data available for the analysis of ECG signals. (a) ECG waveforms (P, QRS and T waves) and standard features extracted from an ECG beat. The RR interval is measured as the peak-to-peak interval between two consecutive QRS complexes, the PR interval is defined as the duration from the beginning of the P wave to the beginning of the QRS complex, the QRS duration (or width) is the duration between the start and the end of the QRS complex, the QRS amplitude is defined as the absolute value of the difference between the minimum and the maximum of the QRS complex, and the QT interval is measured as the time between the beginning of the Q wave and the end of the T wave. (b) Examples of different ECG waveforms: normal ECG [50], ventricular ectopic beats [51], supraventricular ectopic beats [52] and second degree heart block arrhythmia [53]. (Online version in colour.)
One of the challenges is that the ratio between the amount of available training data and the number of extracted features is too small, which may lead to overfitting. The number of features must therefore be reduced for good generalization and performance and two main techniques are usually used: dimensionality reduction and feature selection. Dimensionality reduction aims to reduce the size of the space in which the data are represented by computing a reduced number of dimensions that contain most of the information of the dataset. As many features can be extracted from ECG signals, dimensionality reduction algorithms are often performed before running the classifier. Examples of dimensionality reduction techniques include PCA (linear or nonlinear) [22] or linear discriminant analysis [18]. Feature selection selects only a small subset of the most significant features in the classification. For this purpose, some studies include an optimization step testing different feature combinations (via an optimization algorithm such as genetic algorithm or particle swarm optimization, or statistical distribution analysis such as Gini's index) and retrieving only the relevant features for further analysis [20,22,57]. For example, Mar et al. [58] performed classification between normal, ventricular, premature ventricular and fusion beats based on the idea of Llamedo & Martínez [4] to use the sequential forward floating search (SFFS) feature selection procedure. This improved the classification accuracy of the multi-layer perceptron (MLP) classifier from 79% using 71 features to 90% with only nine features.
Feature quality and robustness is a challenge as poor quality features resulting from low quality delineators, filtering or approximations may lead to low performance and generalization properties despite powerful classification algorithms. Solutions to tackle this issue have been proposed such as in Llamedo & Martínez [4]. In their study they introduced the use of robust surrogates of typical features, using for example directly the wavelet transform signal instead of the QRS width to reduce the effect of delineation errors.
2.3. Machine learning methods for heartbeat classification
In terms of heartbeat classification accuracy, all the machine learning methods reported below present similar good performances (approx. 95–99%). From the clinical viewpoint, two important benefits can be highlighted. Firstly, the outcomes of random forests and linear techniques, contrary to SVMs or neural networks, are clinically interpretable, providing the opportunity to discover new biomarkers and enhance their importance in discriminating specific types of heartbeats. Secondly, neural network and Bayesian models may allow the analysis of the ECG without any preprocessing of the signal, which avoids the need for prior information on the biomarkers and may help discover new knowledge.
2.3.1. Linear and quadratic discriminants
De Chazal et al. [18] implemented linear discriminants (LD) with weighted likelihood to classify 50 000 heartbeats in five classes (normal, ventricular ectopic, supraventricular ectopic, fusion of a normal and ventricular ectopic, or unknown beat type) from 22 recordings of the MIT-BIH arrhythmia database using QRS-based and time intervals features. The LD technique computes mean and covariance for the training data in order to maximize the likelihood. Posterior probabilities are then computed to output the final classification labelling. Their study reached a sensitivity of 75.9% (positive predictivity 38.5%, false positive rate 4.7%) for supraventricular ectopic beats, and a sensitivity of 77.7% (positive predictivity 81.9%, false positive rate 1.2%) for ventricular ectopic beats (VEBs). Similarly, another work by Llamedo & Martínez [4] used LD and quadratic discriminants to perform the classification of normal, ventricular and supraventricular beats. They extracted features from the RR interval and characterized the morphology of the ECG by discrete wavelet transform. A global accuracy of 93% (84% sensitivity and 75% positive predictivity) was achieved on the MIT-BIH arrhythmia database as well as on the MIT-BIH supraventricular arrhythmia and INCART databases. Similar work was conducted by Yeh et al. [19], using LD for classification between normal and five different classes of abnormal beats for arrhythmia diagnosis. Evaluated on the recordings from the MIT-BIH database, their method reported correct detection between 84.68% and 98.97% for the five classes studied.
Some studies propose more elaborate ways of using these classifiers that may lead to patient-specific techniques and allow expert assistance. For example, de Chazal & Reilly [5] proposed a patient-adaptable system in which a classifier is trained to annotate the first beats of a recording, then checked by a clinical expert. This patient-specific adaptation was then added to a global classifier. This technique was shown to increase the performance of the classifier by 10% (accuracy of 94%) for the classification of heartbeats according to the AAMI classes. Another work by Llamedo & Martínez [59] also combined a global classifier (linear discriminant analysis) and a patient-specific step (an expectation–maximization clustering algorithm). This led to an automatic patient-adaptable technique which can also incorporate the input of a cardiologist (semi-automatic) when the clustering requires guidance from expert annotation because of interpatient variability.
2.3.2. Support vector machines
SVMs are very popular class of machine learning algorithms because of their good classification and generalization properties. It is a supervised learning method which learns the best separating hyperplane to maximize the margin between two classes in the feature space (figure 2a). This decision boundary is then used to classify unknown testing data [60]. SVM application to ECG beat classification and its optimization have been widely studied. For example, Ubeyli [6] applied SVM with error output correction code to classify heartbeats from four classes (normal, congestive heart failure, ventricular tachyarrhythmia and atrial fibrillation (AF)) from the Physionet database. Discrete wavelet transform was used to preprocess the data and extract features. They reached an accuracy of 98.61% (sensitivity of 98.89%) on a testing set of 360 beats.
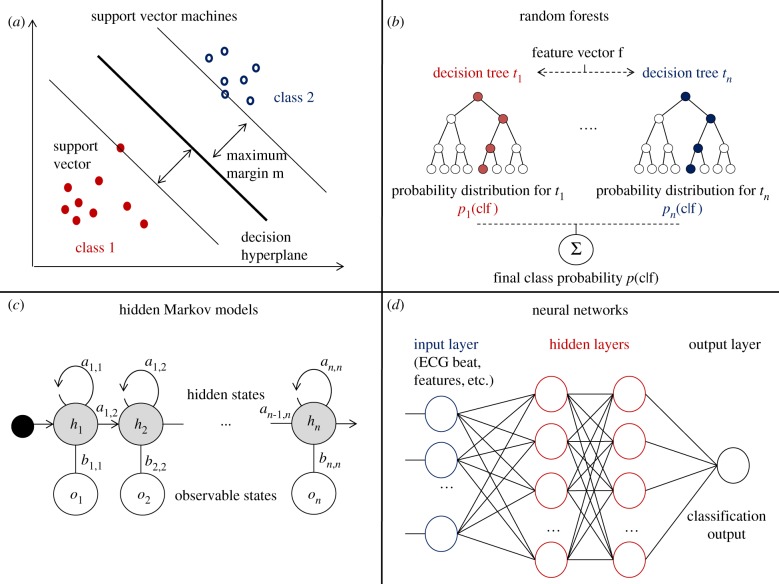
Figure 2.
Main machine learning methods used for ECG classification. (a) Support vector machine binary classification by maximization of the margin m. (b) Random forest classification using n decision trees. (c) Hidden Markov model with n states, transition matrix (ai,j) and emission matrix (bi,j). (d) Neural network with two hidden layers. (Online version in colour.)
SVM optimization techniques have been investigated to improve the choice of features and parameters, reduce overfitting and speed-up the classification. For example, Melgani & Bazi [20] performed arrhythmia classification with SVM optimized by particle swarm optimization (PSO). PSO reduced the number of features from 303 to 46 and reached an overall accuracy of 89.72% in detecting five heartbeat classes (normal, atrial premature beat, ventricular premature beat, right bundle branch block, left bundle branch block and paced beat) from the MIT-BIH database. Another work by Asl et al. [21] focused on the classification of heartbeats in six classes for arrhythmia detection (normal, premature ventricular contraction (PVC), AF, sick sinus syndrome, ventricular fibrillation and second degree heart block). They used 1367 ECG segments from the MIT-BIH database and designed feature vectors based on heart rate variability. They reached 99.16% accuracy with discriminant analysis feature reduction (15 to five features). In another study, Nasiri et al. [22] used PCA and genetic algorithm to determine the best parameters to tune the SVM algorithm and to perform feature reduction to remove the features that may lower the accuracy. They reached an accuracy of 93.46% for classifying between normal, right and left bundle branch block, and paced beat on the MIT-BIH arrhythmia database with genetic algorithm. Similar applications of SVM algorithms to ECG beat classification were implemented by Li et al. [61], Rabee & Barhumi [62], or Mehta & Lingayat [48]
Methods based on SVM and optimized SVM therefore reach high accuracies ranging from 90% to 99% in multi-class classification. A drawback of SVM classification for clinical applications is its lack of interpretability in order to evaluate each feature's influence and extract relevant discriminant biomarkers.
2.3.3. Random forests
The random forest method [63] is an ensemble learning technique combining the classification outputs of the decision trees that compose the forest (figure 2b). A useful property of random forests is their ability to rank the variables according to their importance in the classification and therefore allow feature selection to avoid overfitting. Ganeshkumar & Kumaraswamy [23] investigated arrhythmia detection by identifying six heartbeat classes (normal, PVC, paced, atrial premature beat, and left and right bundle branch block) using random forest. They reached an accuracy of 92.16% on 150 beats extracted from the MIT-BIH database with 30 trees but their method was not validated over an independent testing set. In a recent study, Rahman et al. [8] compared the heartbeats of 12-lead ECG from 1000 hypertrophic cardiomyopathy (HCM) patients to a group of ischaemic and non-ischaemic cardiomyopathy patients. Their implementation used a random forest algorithm with 500 trees and 264 features (after feature selection) obtained from ECG waveforms morphology and duration. They reached an accuracy of 89% validated with fivefold cross validation. Similar random forest classifiers for ECG classification were developed by Sathish & Vimal [64], or Emanet [65].
2.3.4. Bayesian networks
Bayesian networks are probabilistic graphical models representing variables and their probabilistic relationships. Following the successful application of Bayesian frameworks techniques to ECG segmentation [66] and denoising [67], de Oliveira et al. [24] introduced the first approach of ECG beat classification using Bayesian networks. Their work focused on PVBs detection by the implementation of a Bayesian network framework using channel fusion. They reached a sensitivity of 99.69% (positive predictivity 98.79%). Validation of their method was performed by splitting the labelled heartbeats from the QT database into 75% as training set and 25% as test set. Similarly, Oster et al. [45] developed a model-based filtering approach using switching Kalman filters to classify ventricular beats. In addition to the classification performance, their approach presented the ability to detect unknown beat morphologies. They reached an accuracy of 97.3% (positive predictivity 99.96%) for ventricular beats classification on the MIT-BIH and INCART databases.
A simple type of Bayesian networks are hidden Markov models (HMM) which represent the system of study as a set of hidden states and transition probabilities where the sequence of hidden states is estimated from the observations [68] (figure 2c). HMM do not require prior knowledge and the estimation of the parameters is automated. This technique is popular for sequence modelling and temporal pattern analysis. Hence, several studies focus on the application of HMM to ECG modelling and prediction. Coast et al. [25] investigated the use of HMM for classification and modelling of normal and VEBs from the American Heart Association database (six 30-min ECG recordings). Each ECG waveform and duration was represented by a HMM state. Their method reached a sensitivity of 97.25% (specificity of 85.67% over 799 VEB beats). Similarly, Koski [26] performed ECG segmentation and classification between normal and PVBs signals using HMM. They estimated the ECG signal by broken line approximation. The classifier was composed of 20 to 30 states. They reported an accuracy of 100% on the very small testing set used (four testing beats). Later, Andreao et al. [27] performed modelling and classification of PVC beats using 59 recordings from the QT database. They combined an HMM and a rule-based system for beat identification (based on the beat prematurity and enlarged QRS morphology criteria). They reached a sensitivity of 99.79% (specificity of 99.96%) for PVB detection over the testing set. Most of these techniques based on HMM also integrate and estimate the parameters of the delineation of ECG waveforms in the model, avoiding the need for a separate segmentation step.
2.3.5. Neural networks
In the last 15 years, neural networks [69] have been very popular in ECG classification. In this family of machine learning algorithms inspired by biological processes, the interconnected neurons of the system learn the structure of the data from training examples (figure 2d). Neural networks are powerful for their ability to detect patterns and extract data structure without expert knowledge. An example of neural network application to ECG heartbeat classification was presented by Niwas et al. [28]. They implemented an artificial neural network (ANN) trained on feature vectors composed of heartbeat intervals and spectral entropy. They reached an accuracy of 99.02% in the classification of testing heartbeats in 10 classes from the MIT-BIH database. Similarly, Inan et al. [29] studied the detection of PVC beats. They used morphological features derived from the wavelet transform and time intervals features as input to a feed-forward MLP neural network with a single hidden layer. They obtained 96.82% accuracy for PVC beat classification on 22 ECG recordings from the MIT BIH database.
In another study, Ubeyli et al. [30] used a more complex neural network architecture, recurrent neural networks (RNN), which differ from standard ANN by the presence of direct cycles in the neurons architecture. This allows the representation of dynamic temporal processes. They trained the RNN with Levenberg–Marquardt training algorithm and eigenvector based features. They tackled the classification of heartbeats in four classes on 720 heartbeats from the Physionet database and reached an accuracy of 98.06%. Another study by Lagerholm et al. [31] presented a clustering method for arrhythmia over 48 recordings of the MIT-BIH database. They preprocessed the signal with the Hermite transform which provided a better robustness to noise. The Hermite transform and the RR interval defined the feature vector. They implemented self-organizing networks (SON) to perform clustering in 25 groups and reach 1.5% of misclassification. SON also conserves the neighbouring structure of the data, representing similar clusters physically close on the map, and therefore facilitating the interpretation of the results by the clinicians. Similarly, Linh et al. [32] used the Hermite transform of QRS complexes coupled to a TSK fuzzy network, an association of neural network with logical rules, to classify six types of heart rhythms (premature ventricular ectopic, left bundle branch block, right bundle branch block, atrial premature beat, ventricular flatter wave, and ventricular escape beat) for arrhythmia. They reached 96% accuracy in detecting these heartbeat classes, although no validation dataset is described in the paper. In addition, their method was less sensitive to morphological variations of the ECG and handled heartbeat variability. Another method implementing fuzzy techniques was developed by Ozbay et al. [33]. Their fuzzy clustering neural network architecture achieved 99.9% accuracy in arrhythmia detection (classifying normal beats, sinus bradycardia, ventricular tachycardia, sinus arrhythmia, atrial premature contraction, paced beats, right and left bundle branch block, AF and atrial flutter) on 5342 segments from the MIT/BIH database. Similar works on neural networks were conducted by Jadhav et al. [70], Meau et al. [71], Dokur & Olmez [72] or Das et al. [73].
3. ECG recording analysis for patient diagnosis, monitoring and stratification
Rather than classifying heartbeats, some studies focus on diagnosing patients based on their ECG (as summarized in table 2). Patient classification and diagnosis requires analysing the ECG recordings as a whole (time changes, various beat morphologies) rather than analysing a single isolated beat. The machine learning methods described above can be adapted to this task. Clinical applications include risk stratification or disease monitoring, and these studies may be able to provide insight into the structure of diseased populations thanks to clustering techniques, highlight which biomarkers are significant to distinguish between disorders, provide automatic diagnosis (or semi-automatic, taking into account expert input), and analyse ECG changes over time, which may be tedious to perform visually. The paragraphs below highlight some of the clinical areas that have benefited from the use of these machine learning methods to patient classification.
3.1. Pro-arrhythmic ventricular diseases
Several works approach patient diagnosis by identifying abnormal excerpts or events in the recordings. For example, ischaemia is a disease condition normally manifested in ambulatory recordings by transient deviations of the ST segment voltage. Therefore, the automatic identification of ST deviations due to ischaemia and due to non-ischaemic events (such as or axis shifts, and heart rate or conduction changes) has been widely investigated. In particular, Mincholé et al. [34] automatically derived heart rate related depolarization and repolarization indices to discriminate between ST-segment changes due to ischaemia and non-ischaemic ST episodes due to heart rate. They reached an accuracy of 87.5% on the LTST database. They applied a multivariate discriminant analysis using the Wilk's lambda minimization as the criteria for inclusion and removal of features.
On the same database, Faganeli & Jager [35] developed a decision-tree based classification to distinguish between ischaemic and non-ischaemic heart rate-related ST-segment episodes. They evaluated their performance by bootstrap method and reached a sensitivity of 98.1% and specificity of 85.2% with features from heart rate and Legendre orthonormal polynomial coefficients. Another work by Bailón et al. [36] aimed at diagnosing coronary heart disease based on exercise ECG indexes. They developed an automated method to extract repolarization, depolarization and heart rate variability (HRV) indexes from noisy exercise recordings. A multivariate discriminant analysis then classified patients into two classes: ischaemic and low risk patients based on independent set of indexes. HRV indexes provided the best results with a sensitivity of 94% and specificity of 92% to classify 65 ischaemic and 40 low-risk cases. Another disease condition investigated is Brugada syndrome, which leads to a high risk of sudden cardiac death because of episodes of ventricular fibrillation (VF) in patients with no structural heart disease [74]. Kawazoe et al. [37] investigated risk of VF in patients with Brugada syndrome thanks to a logistic regression model, cross validated over a database of 143 patients (35 with VF, 108 without). Syncope episodes, R–J interval in lead V1, QRS duration in lead V6, and Tpeak–Tend dispersion were the best discriminating features identified by logistic regression. They led to a sensitivity of 97.1% and a specificity of 63.0% by leave-one-out cross validation.
The study by Rahman et al. [8], mentioned earlier, presented a patient classifier to detect patients affected by HCM based on standard 12-lead ECG. They classified a patient as HCM if the majority of the beats show HCM beat morphology. Their dataset was composed of 221 HCM patients and 541 control (non-HCM patients, but with implemented cardiac defibrillator). Two hundred and sixty-four standard ECG features, such as time intervals and waveforms amplitude, were extracted by feature selection (information gain criterion), and used to perform SVM and random forest classifications with fivefold cross validation. They reached similar performance results with the two classifiers, with a precision of 0.84 (0.89 sensitivity, 0.93 specificity).
Acharya et al. [40] were interested in diagnosing several groups of cardiac disorders based on ECG signals: ischaemic or dilated cardiomyopathy, complete heart block, sick sinus syndrome, AF or ectopics, and normal beats. They developed an ANN coupled with a fuzzy equivalence relation and reached an accuracy of 85–95% using four heart rate variability features. In another work, Zheng et al. [41] applied convolutional neural network (CNN) to time-series classification using a multi-channel technique. They aimed to classify congestive heart failure from two-lead ECG of 15 subjects by extracting sub-sequences of the signal. The model learned features from the ECG throughout the layers of the CNN and reached an accuracy of 94.7% for congestive heart failure detection on this dataset.
3.2. Atrial fibrillation
AF is a condition investigated by many studies as its irregular and repetitive episodes can lead to heart failure, stroke and double the risk of mortality [75]. In Pourbabaee & Lucas [7], time interval and morphological features were extracted from the three main ECG waveforms (P wave, QRS and T wave) of 25 patients with paroxysmal AF episodes and 25 healthy subjects from the 2001 Computers in Cardiology Challenge database. The MLP implemented classified accurately 87% of the testing dataset, and suggested the importance of QRS-based features in the classification after testing the influence of each set of features. Similarly in [38], Colloca et al. performed detection of atrial fibrillation episodes via SVMs, evaluated on the MIT-BIH database with a specificity of 99.72% on the testing set. In another study by Asgari et al. [39], stationary wavelet transform and SVM were designed to identify AF events with sensitivity and specificity of 97.0% and 97.1% respectively. More detailed techniques for AF management and detection can be found in [76].
3.3. Long-term patient monitoring
ECG analysis can also be used for long-term monitoring of patients in clinical care, to detect abnormal rhythmic events that may occur suddenly. For example in Kannathal et al. [9], three degrees of disease severity (normal, abnormal, life threatening) were predicted from the ECG of patients in an intensive care unit (ICU) using ANNs (600 training patients, 200 testing patients). This reached an accuracy of 99% with RBF (radial basis function) networks. Similarly in Zhang et al. [12], a combination of genetic algorithm and SVM was used to detect false critical arrhythmia alarms in ICUs with a true-positive rate of 93% (true-negative rate of 94%).
In this context, an example of successful integration of computational techniques in the clinical environment is the recent collaboration between Microsoft and the Cleveland Clinic [77], focused on the analysis of data from the ICU (clinical data, medical records, etc.). They aimed to monitor and identify high-risk patients, using machine learning and advanced data analytics (Azure Machine Learning).
4. Real-time episode detection and wearable devices
The integration of classification techniques in clinical settings requires the detection of ECG abnormalities in real-time to be used in the hospital environment at bedside, or on wearable devices. This involves the development of classification algorithms with low complexity and low memory requirements, which add different challenges to these techniques (summarized in table 2).
4.1. Real-time diagnosis
Real-time analysis and classification of ECG signals find clinical applications in the detection of sudden abnormal heart rhythms (more than in the diagnosis of long-term diseases), especially in ICUs where the real-time monitoring of patients is crucial. In addition, future progress could see memory networks learn to predict severe events in real time, allowing clinicians to take action before fatal arrhythmias occur.
The standard machine learning methods presented earlier may perform real-time classification if optimized to deal with a large influx of incoming data. However, most of them require the extraction of handcrafted features before classification. In addition to the already mentioned risk of poor feature quality due to inaccurate delineation of the ECG waves or an imprecise extraction of ECG features, these methods may struggle to run in real time. Recently, powerful tools have been developed in the field of deep learning, helping real-time classification of very complex signals by the design of more complex and deep networks. Deep learning is a field of machine learning in which several hidden layers of computation are added to neural networks (learning algorithms inspired by biological brain networks) in order to model more complex behaviours and data structures. Popular applications of these techniques are image recognition and generation, and speech and time-series classification [78,79]. In Kiranyaz et al. [10], a 1D CNN merged feature extraction and classification in one step to develop a personalized patient classifier that can be used in real time, once trained, to classify longer recordings, on wearable devices for instance. They reached an accuracy of 98.6% (95% sensitivity, 98.1% specificity) on 24 test recordings of the MIT-BIH database in classifying ventricular and supraventricular ectopic beats. Presented earlier, Zheng et al. [41] applied CNNs to the classification of time-series data using a multi-channel technique. One of their applications was the classification of congestive heart failure in two-lead ECGs from 15 subjects by extracting sub-sequences of the signal. The model learnt directly the features from these time series throughout the layers of the CNN and reached an accuracy of 94.7% on this dataset. In Chauhan & Vig [42], a deep long short-term memory (LSTM) network was implemented to tackle the classification of various types of ECG beats (PVC, atrial premature contraction, paced beats and ventricular couplet) from the MIT-BIH database. An advantage of LSTM networks is their ability to take a raw ECG signal as input without any preprocessing and automatically discover key features thanks to their ability to ‘remember’ past events. In this study, the LSTM network ran in a short amount of time once trained (testing time of 0.5 s for a 20-min ECG signal on a 16 core CPU machine) making it suitable for time-constrained applications like real-time classification.
4.2. Application to wearable devices
Wearable devices for ECG monitoring have a key impact on the recent effort to move the clinic to the home, especially in the case of monitoring elderly patients or long-term diseases (figure 3). Their development aims to reduce the costs for prevention and monitoring, by freeing expert time and space in clinics. Current technologies are progressing towards this goal but challenges remain, such as portability (battery, computational costs) or reliability of the abnormality detection.
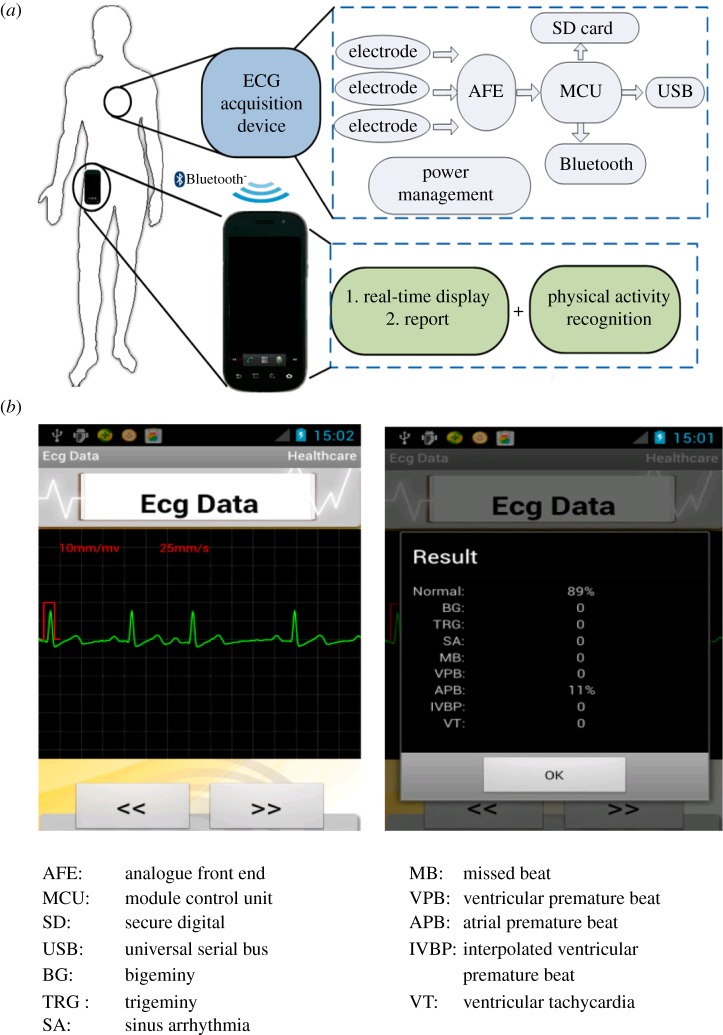
Figure 3.
Example of an ECG classification system on wearable device showing the methodology (a), and output (b). From Miao et al. [80]. Panel a details the ECG acquisition sensor implanted in the wearable device: the signal, recorded by electrodes, is amplified and filtered by the AFE module, converted to digital signal by the MCU module and recorded on the SD card, transmitted to the USB port or sent directly to the phone via Bluetooth. Panel b describes the output panel provided to the user: a screenshot of the ECG excerpt (left), and a summary of the normal and abnormal beats recorded (right).
Additional challenges arise for classification methods, such as the need for rapid and real-time analysis of ECG signals to avoid the storage of large amounts of data, and the ability to automatically handle noisy data, as wearable devices may generate data more affected by movement, noise or changes in heart rate than those generated in standard clinical equipment.
Machine learning methods (presented in §2) must tackle challenges of speed and memory requirements in order to be embedded on these portable devices. In [43], a portable device was designed to perform classification of normal beats, atrial fibrillation, and myocardial ischaemia in real time and with a sensitivity of 95.1% and specificity of 95.9% using an SVM algorithm. Similarly, Leutheuser et al. [44], compared several techniques for arrhythmia detection from ECG signals on Android mobile devices evaluated on the MIT-BIH databases and stressed the importance of balancing computational costs and memory demand in the design of such techniques. In 2010, Oresko et al. [11] implemented a smartphone-based wearable platform to perform real-time ECG acquisition and beat classification for cardiovascular disorder detection. They based the classification step on the design of a feed-forward MLP neural network and use the original QRS morphological beat as input to the classifier. This reduced the amount of preprocessing work and the method reaches a prediction accuracy greater than 90% to detect right bundle branch block, PVC, paced or normal beats.
An additional challenge for remote monitoring is the handling of unknown beat morphologies that the algorithm may encounter that could lead to false alarms. These should be reduced as much as possible on wearable devices when the patient is at home to avoid unnecessary alarming messages, or in the ICU to optimize the nurses' time. Mentioned earlier, Oster et al. [45] tackled this issue by implementing switching Kalman filters with an additional X-factor mode to account for unseen beat morphologies. The analysis of the quality of these unknown beats confirmed that 639 out of 954 (approx. 2/3) of these beats were of poor or medium quality and should therefore be discarded for monitoring and diagnosis. Additional examples of such methods can be found in the electronic supplementary material.
AliveCor [81] is a striking example of success in translating these computational methods to the clinic. The wearable technology is integrated in a smartphone application and records ECG and blood pressure data from patients. These measurements are then analysed with a machine learning algorithm to help detect AF. AliveCor received clearance from the Food and Drug Administration [82]. It is now widely used in the clinic for ECG monitoring. A further collaboration with the Mayo Clinic plans to develop the technology to discover hidden physiological signals from ECG data [83].
5. ECG computer simulations
One of the main challenges emerging from the summary of the classification studies presented earlier is the generalization to larger and different databases. Indeed, a limited number of databases and recordings are available for testing and validation, which may explain the limited number of heartbeat types and cardiac conditions investigated by these studies. Very few works actually explore the generalization performance of their techniques to other databases as in Llamedo & Martínez [4] who reported and tackled this issue by evaluating their method over three different databases to prove good generalization properties. The 3D computer simulations techniques presented in this section may be very helpful in addressing this problem given their ability to generate synthetic data that could then be used as validation datasets for the classification studies. In addition, computer simulations using anatomically-based multiscale models of the electrical activity of the heart can help to interpret ECG findings from machine learning studies. Computer modelling studies can therefore find clinical applications in both the generation of synthetic ECG data and the interpretation of ECG abnormalities by linking structural or electrophysiological changes to ECG abnormalities. The drive towards personalized simulations also provides a unique opportunity for research, and clinical benefit is directly seen in therapy testing, and reduction of invasive techniques.
In 2013, Sovilj et al. [84] presented simulations of the ECG using an ellipsoid 3D model of a heart embedded in a torso. They show the effect of simulated myocardial infarctions at various locations on ECG changes, specifically in the ST segment. Another study by Bacharova et al. [85] used computer simulations to investigate the influence of left ventricular (LV) mass on QRS, and specifically increased QRS amplitude, in the context of LV hypertrophy. In a more recent work, Bacharova et al. [86] investigated the effect of slow ventricular activation on the QRS complex and showed how alteration of electrical properties may mimic ECG morphologies associated with anatomical abnormalities. These approaches have the benefit of speed and low memory requirements, but they do not take into account a realistic anatomy of the heart.
Simulations using image-based anatomical models have also been used extensively to investigate the ionic and structural basis of ECG (figure 4). Substantial work by Potse and colleagues has used several approaches for investigations of the underlying basis of ECG changes in disease, from ST elevation in ischaemia to personalized models of heart failure patients [88–90]. Furthermore, in [91], Chen et al. used an MRI-based computer simulation model from patients with acute myocardial infarction to show that instabilities in the QT interval may predict ventricular tachycardia onset. Similarly, Wilhelms et al. [92] applied realistic detailed 3D computer simulations to better understand the mechanisms underlying shifts in the ECG ST segment in acute cardiac ischaemia.
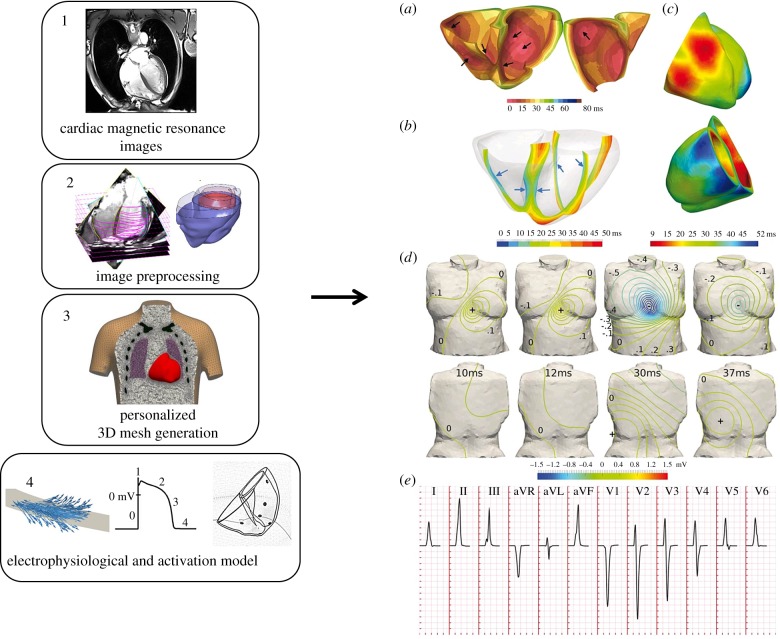
Figure 4.
Computational pipeline for ECG simulation from magnetic resonance images through 3D meshes and cellular electrophysiological models (left, from Zacur et al. [87]) and the obtained simulation of the cardiac electrical activity (right, from Cardone-Noott et al. [13]). Personalized cardiac magnetic resonance images (1) are segmented and preprocessed (2). From this information, surface and volumetric meshes are generated (3) and an electrophysiological model defining the electrical activity in the cells is implemented (4). The obtained simulated electrical conduction (right panel) can then be investigated.
The simulation of the ECG through 3D computer simulations also finds applications in understanding the effect of drugs on the ECG [15,93]. Sebastian et al. [94] investigated the effect of dofetilide, a drug affecting the IKr current on a patient-specific model simulating the pseudo-ECG. They observed a prolongation of the QT interval of 100 ms with a total IKr block, in line with a prolongation of the APD90 in cardiac cells. Zemzemi et al. [15] investigated the influence of drugs on the ECG signal using 3D computer simulations, allowing drug testing in silico rather than in vivo. They simulated channel conductance block for the hERG and fast sodium channels and observed QT prolongation for 50% hERG block (6%) as well as QRS and QT prolongation for 50% fast sodium current block (12% and 5% respectively).
Another work by Cardone-Noott et al. [13] investigated the effect of changes in conductivities on the ECG morphology and how the variability in the activation sequence relates to changes in QRS biomarkers, by designing a 3D simulation pipeline with a human volumetric mesh and activation model based on the cellular O'Hara–Rudy model.
Phenomenological approaches to modelling and simulation of electrical propagation have also been proposed using fast algorithms based on graph based theory [95,96]. This may facilitate both simulations of the ECG as well as parameter inference from the ECG, from clinical datasets, by taking into account parameter uncertainty such as in Wallman et al. [97] and Konukoglu et al. [98].
In addition, abnormalities in human atrial electrophysiology and their consequences on ECG patterns have also been investigated building on substantial work on multiscale modelling of atrial dynamics. Geometrical atria models have been described and simple models represent the shape of the atria as a 2D or 3D folded sheet [99,100]. As in the case of ventricular models, these simple models are not derived from realistic imaging data but are based on a series of assumptions. The characteristics of the tissue can be easily modelled such as homogeneous tissue, anisotropy [100], etc., and these simple models allow the investigation of essential questions regarding atria behaviour such as the anatomy. Vigmond et al. [100] for example showed the importance of superior vena cava and pulmonary vein openings in rotors by using a model of the atria built with two spheres containing holes as anatomical elements. However, despite being very useful because of their simplicity, these models do not possess the anatomical details that would be obtained with imaging data.
As in ventricular models, another type of model therefore emerged, using imaging data (from MRI or CT scans) [101–103], including realistic anatomical elements of the whole atria, such as sinoatrial node, left and right atrium appendage, Bachmann bundle, etc. [104].
As in [105], these models allow the simulation of the body surface ECG in normal and arrhythmic cases to investigate P wave abnormalities in the ECG and their correlation with atrial fibrillation dynamics and propensity.
Computer simulation models of whole ventricles and atria therefore allow the modelling of the electrical propagation throughout the heart from ionic dynamics to the ECG. ECG computer simulations pipelines are powerful tools to provide a deeper understanding of the impact of cardiac diseases and treatments on the ECG. Furthermore, with advances in high performance computing and development of fast simulation methods, they can bring the generation of synthetic ECG data under various conditions, to improve training of machine learning classifiers and the development of new ECG features.
6. Discussion
The electrocardiogram is cheap, non-invasive and widely used in clinical practice. As a recording of the body surface electrical activity, it provides information about heart rhythm abnormalities and helps detect diseases. However, visual inspection of the ECG provides discrete clinically interpreted features which cannot objectively capture the diversity of ECG abnormalities and morphologies. This is why computational methods are required, as they can make sense of multivariate complex datasets and detect differences that might be challenging for the human eye. However, analysing ECG data presents many challenges. Indeed, most large clinical studies still record ECG on paper print-outs, requiring manual digitization before computational techniques can be applied. Digital ECG clinical acquisition is still to be implanted in many hospitals. In addition, many ECG databases are not publicly available, gather low numbers of patients, and require extensive signal preprocessing techniques to denoise the recordings for computational analysis. As a consequence, most studies in the literature and reviewed here focus on large, publicly available databases of ECG recordings. Some of these databases such as the widely used MIT-BIH were originally analogue ECG recordings on tapes that were then digitized. Some frequency-domain artefacts were identified, although they should not pose problems for beat classification or wave delineation [106]. Many methods are trained and tested on the same databases, despite ensuring separated training and testing sets, which may limit the generalization of these techniques to different clinical databases or other cardiac disorders. Availability of large clinical ECG datasets is therefore required for technical developments in machine learning application to ECG analysis and classification.
These challenges will also apply to other recording techniques such as body surface mapping (BSM) which samples multiple points around the chest to provide a more detailed mapping of the body surface electrical activity than the standard 12-lead ECG. It has been shown to be more accurate than the ECG in detecting transient episodes like myocardial ischaemia [107]. It is a rich source of data but is, by far, less commonly used and accessible than standard ECG. Intracardiac recordings also present an alternative to measure the electrical activity in the heart, but they are invasive, and could therefore not be obtained for large numbers of patients, thus showing less interest for its analysis with machine learning techniques.
The clinical integration of these techniques also presents limitations. First, the gold standard for electrocardiographic abnormality detection remains expert annotations, and there is no reference dataset to compare all these studies, limiting the analysis of their performances for clinical use. However, the use of these techniques provides automaticity and consistency in the analysis of large datasets, which are needed more and more when manual classification cannot be performed. More importantly, the benefit of computer techniques goes beyond these classification tasks where computational methods try to reproduce expert judgement, and can help uncover new knowledge and discover new biomarkers by unravelling structures in the data that were unknown before.
The recent interest in computational ECG analysis is twofold. For the computational field, the challenge and diversity offered by ECG recordings provide a rich environment to develop new methods. From the clinical perspective, these methods provide a new horizon on how ECGs can be analysed, developing novel biomarkers to diagnose cardiac diseases. Progress in this field is recent, which explains why several studies investigate the behaviours of different machine learning methods on ECG data. The fact that the ECG signal can be analysed by so many techniques actually provides a wide range of options depending on the goal and requirements of the study. These techniques were recently introduced in the clinical environment, and growing interest has been shown from the clinical field [2,108]. However, translating novel techniques to the clinic requires both technology developments and also addressing practical challenges such as regulation approval, and inclusion in clinical protocols, which require intersectorial collaborations [109]. The novel approaches for ECG analysis reviewed here, in synergy with other modalities such as intracardiac electrical recordings or imaging techniques, have the potential to improve patient risk stratification and precision medicine.
7. Conclusion
This review shows that computational techniques have been widely developed to analyse ECG signals and are strong candidates to help clinical advances by providing a better understanding of medical challenges. Machine learning techniques provide accurate and automatic classifications of heartbeats to detect arrhythmias or unexpected changes in heart morphology. They also help in automatic disease diagnosis, monitoring and stratification by handling long ECG recordings for which visual and manual inspections can be tedious and time consuming. Their adaptability to real-time requirements and embedding on wearable devices ensures an efficient and reliable monitoring of the ECG activity in hospital settings or at home. Finally, 3D computer simulations are powerful tools to interpret the ECG and they may soon become invaluable by generating large datasets of synthetic data for the training of machine learning classifiers. Despite the many challenges they face and the novelty of their introduction to clinical practice, these computational methods are becoming a powerful tool for medical advances and their integration in clinical settings should help improve patient care.
Supplementary Material
Data accessibility
This article has no additional data.
Authors' contributions
A.L. designed and wrote the paper and carried out the literature review of the field. A.M., J.P.M., P.L. and B.R. provided advice on the shape of the review and input on the manuscript. All authors gave final approval for publication.
Competing interests
We have no competing interests.
Funding
A.L. is supported by a scholarship provided by the British Heart Foundation Centre of Research Excellence. A.M. and B.R. are supported by B.R.'s Wellcome Trust Senior Research Fellowship in Basic Biomedical Sciences, the CompBiomed project (grant agreement no 675451) and the NC3R Infrastructure for Impact award (NC/P001076/1). The project has been also funded by DPI2016-75458-R, from MINECO, Spain and ‘Grupo Consolidado BSICoS’ from DGA, Aragón, Spain.
References
- 1.Institute of Medicine (US) Committee on Preventing the Global Epidemic of Cardiovascular Disease: Meeting the Challenges in Developing Countries. 2010. Epidemiology of cardiovascular disease. In Promoting cardiovascular health in the developing world: a critical challenge to achieve global health (eds Fuster V, Kelly BB). Washington, DC: National Academies Press; See https://www.ncbi.nlm.nih.gov/books/NBK45688/ [PubMed] [Google Scholar]
- 2.Obermeyer Z, Emanuel EJ. 2016. Predicting the future—big data, machine learning, and clinical medicine. N. Engl. J. Med. 375, 1216–1219. ( 10.1056/NEJMp1606181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sörnmo L, Laguna P. 2005. Bioelectrical signal processing in cardiac and neurological applications (ed. LS Laguna). Burlington: Academic Press; See http://www.sciencedirect.com/science/article/pii/B9780124375529500015 (accessed 10 October 2014) [Google Scholar]
- 4.Llamedo M, Martinez JP. 2011. Heartbeat classification using feature selection driven by database generalization criteria. IEEE Trans. Biomed. Eng. 58, 616–625. ( 10.1109/TBME.2010.2068048) [DOI] [PubMed] [Google Scholar]
- 5.de Chazal P, Reilly RB. 2006. A patient-adapting heartbeat classifier using ECG morphology and heartbeat interval features. IEEE Trans. Biomed. Eng. 53, 2535–2543. ( 10.1109/TBME.2006.883802) [DOI] [PubMed] [Google Scholar]
- 6.Übeyli ED. 2007. ECG beats classification using multiclass support vector machines with error correcting output codes. Digit. Signal Process. 17, 675–684. ( 10.1016/j.dsp.2006.11.009) [DOI] [Google Scholar]
- 7.Pourbabaee B, Lucas C. 2008. Automatic detection and prediction of paroxysmal atrial fibrillation based on analyzing ECG signal feature classification methods. In 2008 Cairo International Biomedical Engineering Conference pp. 1–4, Cairo, Egypt, 18-20 December. Piscataway, NJ: IEEE; ( 10.1109/CIBEC.2008.4786068) [DOI] [Google Scholar]
- 8.Rahman QA, Tereshchenko LG, Kongkatong M, Abraham T, Abraham MR, Shatkay H. 2015. Utilizing ECG-based heartbeat classification for hypertrophic cardiomyopathy identification. IEEE Trans. Nanobioscience. 14, 1 ( 10.1109/TNB.2015.2407291) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kannathal N, Acharya UR, Lim CM, Sadasivan P, Krishnan S. 2003. Classification of cardiac patient states using artificial neural networks. Exp. Clin. Cardiol. 8, 206–211. [PMC free article] [PubMed] [Google Scholar]
- 10.Kiranyaz S, Ince T, Gabbouj M. 2016. Real-time patient-specific ECG classification by 1-D convolutional neural networks. IEEE Trans. Biomed. Eng. 63, 664–675. ( 10.1109/TBME.2015.2468589) [DOI] [PubMed] [Google Scholar]
- 11.Oresko JJ, Jin Z, Cheng J, Huang S, Sun Y, Duschl H, Cheng AC. 2010. A wearable smartphone-based platform for real-time cardiovascular disease detection via electrocardiogram processing. IEEE Trans. Inf. Technol. Biomed. 14, 734–740. ( 10.1109/TITB.2010.2047865) [DOI] [PubMed] [Google Scholar]
- 12.Zhang Q, Chen X, Fang Z, Xia S. 2016. False arrhythmia alarm reduction in the intensive care unit using data fusion and machine learning. In 2016 IEEE-EMBS International Conference on Biomedical and Health Informatics (BHI), pp. 232–235, 24-27 February, Las Vegas, NV. Piscataway, NJ: IEEE; ( 10.1109/BHI.2016.7455877) [DOI] [Google Scholar]
- 13.Cardone-Noott L, Bueno-Orovio A, Mincholé A, Zemzemi N, Rodriguez B. 2016. Human ventricular activation sequence and the simulation of the electrocardiographic QRS complex and its variability in healthy and intraventricular block conditions. Europace 18(Suppl. 4), iv4–iv15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vadakkumpadan F, Rantner LJ, Tice B, Boyle P, Prassl AJ, Vigmond E, Plank G, Trayanova N. 2009. Image-based models of cardiac structure with applications in arrhythmia and defibrillation studies. J. Electrocardiol. 42, 157.e1–157.e10. ( 10.1016/j.jelectrocard.2008.12.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zemzemi N, Bernabeu MO, Saiz J, Cooper J, Pathmanathan P, Mirams GR, Pitt-Francis J, Rodriguez B. 2013. Computational assessment of drug-induced effects on the electrocardiogram: from ion channel to body surface potentials. Br. J. Pharmacol. 168, 718–733. ( 10.1111/j.1476-5381.2012.02200.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moody GB, Mark RG. 2001. The impact of the MIT-BIH arrhythmia database. IEEE Eng. Med. Biol. Mag. 20, 45–50. ( 10.1109/51.932724) [DOI] [PubMed] [Google Scholar]
- 17.Goldberger AL, et al. 2000. PhysioBank, PhysioToolkit, and PhysioNet components of a new research resource for complex physiologic signals. Circulation 101, e215–e220. ( 10.1161/01.CIR.101.23.e215) [DOI] [PubMed] [Google Scholar]
- 18.de Chazal P, O'Dwyer M, Reilly RB. 2004. Automatic classification of heartbeats using ECG morphology and heartbeat interval features. IEEE Trans. Biomed. Eng. 51, 1196–1206. ( 10.1109/TBME.2004.827359) [DOI] [PubMed] [Google Scholar]
- 19.Yeh Y-C, Wang W-J, Chiou CW. 2009. Cardiac arrhythmia diagnosis method using linear discriminant analysis on ECG signals. Measurement 42, 778–789. ( 10.1016/j.measurement.2009.01.004) [DOI] [Google Scholar]
- 20.Melgani F, Bazi Y. 2008. Classification of electrocardiogram signals with support vector machines and particle swarm optimization. IEEE Trans. Inf. Technol. Biomed. 12, 667–677. ( 10.1109/TITB.2008.923147) [DOI] [PubMed] [Google Scholar]
- 21.Asl BM, Setarehdan SK, Mohebbi M. 2008. Support vector machine-based arrhythmia classification using reduced features of heart rate variability signal. Artif. Intell. Med. 44, 51–64. ( 10.1016/j.artmed.2008.04.007) [DOI] [PubMed] [Google Scholar]
- 22.Nasiri JA, Naghibzadeh M, Yazdi HS, Naghibzadeh B. 2009. ECG arrhythmia classification with support vector machines and genetic algorithm. In Third UKSim European Symposium on Computer Modeling and Simulation, 2009 EMS ’09 pp. 187–192. 25-27 November, Athens, Greece. Piscataway, NJ: IEEE; ( 10.1109/EMS.2009.39) [DOI] [Google Scholar]
- 23.Ganeshkumar R, Kumaraswamy YS. 2012. Investigating cardiac arrhythmia in ECG using random forest classification. Int. J. Comput. Appl. 37, 31–34. ( 10.5120/4599-6557) [DOI] [Google Scholar]
- 24.de Oliveira LSC, Andreão RV, Sarcinelli-Filho M. 2010. The use of Bayesian networks for heart beat classification. Adv. Exp. Med. Biol. 657, 217–231. ( 10.1007/978-0-387-79100-5_12) [DOI] [PubMed] [Google Scholar]
- 25.Coast DA, Stern RM, Cano GG, Briller SA. 1990. An approach to cardiac arrhythmia analysis using hidden Markov models. IEEE Trans. Biomed. Eng. 37, 826–836. ( 10.1109/10.58593) [DOI] [PubMed] [Google Scholar]
- 26.Koski A. 1996. Modelling ECG signals with hidden Markov models. Artif. Intell. Med. 8, 453–471. ( 10.1016/S0933-3657(96)00352-1) [DOI] [PubMed] [Google Scholar]
- 27.Andreão RV, Dorizzi B, Boudy J. 2006. ECG signal analysis through hidden Markov models. IEEE Trans. Biomed. Eng. 53, 1541–1549. ( 10.1109/TBME.2006.877103) [DOI] [PubMed] [Google Scholar]
- 28.Issac Niwas S, Shantha Selva Kumari R, Sadasivam V. 2005. Artificial neural network based automatic cardiac abnormalities classification. In Sixth International Conference on Computational Intelligence and Multimedia Applications, 2005, pp. 41–46. 16-18 August, Las Vegas, NV. Piscataway, NJ: IEEE; ( 10.1109/ICCIMA.2005.13) [DOI] [Google Scholar]
- 29.Inan OT, Giovangrandi L, Kovacs GTA. 2006. Robust neural-network-based classification of premature ventricular contractions using wavelet transform and timing interval features. IEEE Trans. Biomed. Eng. 53, 2507–2515. ( 10.1109/TBME.2006.880879) [DOI] [PubMed] [Google Scholar]
- 30.Übeyli ED. 2009. Combining recurrent neural networks with eigenvector methods for classification of ECG beats. Digit. Signal Process. 19, 320–329. ( 10.1016/j.dsp.2008.09.002) [DOI] [Google Scholar]
- 31.Lagerholm M, Peterson C, Braccini G, Edenbrandt L, Sornmo L. 2000. Clustering ECG complexes using Hermite functions and self-organizing maps. IEEE Trans. Biomed. Eng. 47, 838–848. ( 10.1109/10.846677) [DOI] [PubMed] [Google Scholar]
- 32.Linh TH, Osowski S, Stodolski M. 2003. On-line heart beat recognition using Hermite polynomials and neuro-fuzzy network. IEEE Trans. Instrum. Meas. 52, 1224–1231. ( 10.1109/TIM.2003.816841) [DOI] [Google Scholar]
- 33.Ozbay Y, Ceylan R, Karlik B. 2006. A fuzzy clustering neural network architecture for classification of ECG arrhythmias. Comput. Biol. Med. 36, 376–388. ( 10.1016/j.compbiomed.2005.01.006) [DOI] [PubMed] [Google Scholar]
- 34.Mincholé A, Jager F, Laguna P. 2010. Discrimination between ischemic and artifactual ST segment events in Holter recordings. Biomed. Signal Process. Control. 5, 21–31. ( 10.1016/j.bspc.2009.09.001) [DOI] [Google Scholar]
- 35.Faganeli J, Jager F. 2010. Automatic classification of transient ischaemic and transient non-ischaemic heart-rate related ST segment deviation episodes in ambulatory ECG records. Physiol. Meas. 31, 323 ( 10.1088/0967-3334/31/3/004) [DOI] [PubMed] [Google Scholar]
- 36.Bailón R, Mateo J, Olmos S, Serrano P, García J, del Río A, Ferreria IJ, Laguna P. 2003. Coronary artery disease diagnosis based on exercise electrocardiogram indexes from repolarisation, depolarisation and heart rate variability. Med. Biol. Eng. Comput. 41, 561–571. ( 10.1007/BF02345319) [DOI] [PubMed] [Google Scholar]
- 37.Kawazoe H, et al. 2016. Risk stratification of ventricular fibrillation in Brugada syndrome using noninvasive scoring methods. Heart Rhythm 13, 1947–1954. ( 10.1016/j.hrthm.2016.07.009) [DOI] [PubMed] [Google Scholar]
- 38.Colloca R, Johnson AE, Mainardi L, Clifford GD. 2013. A support vector machine approach for reliable detection of atrial fibrillation events. Comput. Cardiol. 40, 1047–1050. [Google Scholar]
- 39.Asgari S, Mehrnia A, Moussavi M. 2015. Automatic detection of atrial fibrillation using stationary wavelet transform and support vector machine. Comput. Biol. Med. 60, 132–142. ( 10.1016/j.compbiomed.2015.03.005) [DOI] [PubMed] [Google Scholar]
- 40.Rajendra Acharya U, Subbanna Bhat P, Iyengar SS, Rao A, Dua S. 2003. Classification of heart rate data using artificial neural network and fuzzy equivalence relation. Pattern Recogn 36, 61–68. ( 10.1016/S0031-3203(02)00063-8) [DOI] [Google Scholar]
- 41.Zheng Y, Liu Q, Chen E, Ge Y, Zhao JL. 2014. Time series classification using multi-channels deep convolutional neural networks. In Web-age information management (eds Li F, Li G, Hwang S, Yao B, Zhang Z), pp. 298–310. Springer International Publishing; See http://link.springer.com/chapter/10.1007/978-3-319-08010-9_33. (accessed 6 January 2015) [Google Scholar]
- 42.Chauhan S, Vig L. 2015. Anomaly detection in ECG time signals via deep long short-term memory networks In 2015 IEEE International Conference on Data Science and Advanced Analytics (DSAA), pp. 1–7. 19–21 October, Paris, France. Piscataway, NJ: IEEE ( 10.1109/DSAA.2015.7344872) [DOI] [Google Scholar]
- 43.Jeon T, Kim B, Jeon M, Lee B-G. 2014. Implementation of a portable device for real-time ECG signal analysis. Biomed. Eng. Online 13, 160 ( 10.1186/1475-925X-13-160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leutheuser H, et al. 2014. Comparison of real-time classification systems for arrhythmia detection on Android-based mobile devices. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2014, 2690–2693. [DOI] [PubMed] [Google Scholar]
- 45.Oster J, Behar J, Sayadi O, Nemati S, Johnson AEW, Clifford GD. 2015. Semisupervised ECG ventricular beat classification with novelty detection based on switching Kalman filters. IEEE Trans. Biomed. Eng. 62, 2125–2134. ( 10.1109/TBME.2015.2402236) [DOI] [PubMed] [Google Scholar]
- 46.Gao D, Madden M, Chambers D, Lyons G. 2005. Bayesian ANN classifier for ECG arrhythmia diagnostic system: a comparison study In 2005 IEEE International Joint Conference on Neural Networks, 2005 IJCNN ’05 Proceedings, vol. 4, pp. 2383–2388. [Google Scholar]
- 47.Ouyang N, Yamauchi K, Ikeda M. 1998. Training a NN with ECG to diagnose the hypertrophic portions of HCM In The 1998 IEEE International Joint Conference on Neural Networks Proceedings, 1998 IEEE World Congress on Computational Intelligence, vol. 1 pp. 306–309. 4-9 May, Anchorage, AK. Piscataway, NJ: IEEE ( 10.1109/IJCNN.1998.682282) [DOI] [Google Scholar]
- 48.Mehta SS, Lingayat NS.. 2008. Detection of QRS complexes in electrocardiogram using support vector machine. J. Med. Eng. Technol. 32, 206–215. ( 10.1080/03091900701507183) [DOI] [PubMed] [Google Scholar]
- 49.Zhang Z, Luo X. 2014. Heartbeat classification using decision level fusion. Biomed. Eng. Lett. 4, 388–395. ( 10.1007/s13534-014-0158-7) [DOI] [Google Scholar]
- 50. Cardionetics Ltd. Sinus rhythm. See https://www.cardionetics.com/sinus-rhythm. (accessed 10 August 2017)
- 51. Cardionetics Ltd. Ventricular ectopic beats. See https://www.cardionetics.com/ventricular-ectopic-beats . (accessed 10 August 2017)
- 52. CardiacMonitoring.com. Supraventricular ectopic beat. See http://cardiacmonitoring.com/ecg-arrhythmia-guide/supraventricular-arrhythmias/supraventricular-ectopic-beat/ (accessed 10 August 2017).
- 53.Medical Training and Simulation LLC. Practical clinical skills. See https://www.practicalclinicalskills.com. (accessed 10 August 2017).
- 54.Bortolan G, Degani R, Willems JL. 1990. Neural networks for ECG classification In Computers in Cardiology 1990, Proceedings, pp. 269–272. 23-26 September, Chicago, IL. Piscataway, NJ: IEEE ( 10.1109/CIC.1990.144212) [DOI] [Google Scholar]
- 55.Gharaviri A, Dehghan F, Teshnelab M, Moghaddam HA. 2008. Comparison of neural network, ANFIS, and SVM classifiers for PVC arrhythmia detection In 2008 International Conference on Machine Learning and Cybernetics, pp. 750–755. 12-15 July, China. Piscataway, NJ: IEEE ( 10.1109/ICMLC.2008.4620504) [DOI] [Google Scholar]
- 56.Llamedo M, Khawaja A, Martinez JP. 2012. Cross-database evaluation of a multilead heartbeat classifier. IEEE Trans. Inf. Technol. Biomed. 16, 658–664. ( 10.1109/TITB.2012.2193408) [DOI] [PubMed] [Google Scholar]
- 57.Doughty E., Goldfeder R.2013. Characterizing and diagnosing hypertrophic cardiomyopathy from ECG data. Stanford University. See http://cs229.stanford.edu/proj2013/DoughtyGoldfeder-CharacterizingAndDiagnosingHypertrophicCardiomyopathyFromECGData.pdf. (accessed 15 October 2014)
- 58.Mar T, Zaunseder S, Martínez JP, Llamedo M, Poll R. 2011. Optimization of ECG classification by means of feature selection. IEEE Trans. Biomed. Eng. 58, 2168–2177. ( 10.1109/TBME.2011.2113395) [DOI] [PubMed] [Google Scholar]
- 59.Llamedo M, Martinez JP. 2012. An automatic patient-adapted ECG heartbeat classifier allowing expert assistance. IEEE Trans. Biomed. Eng. 59, 2312–2320. ( 10.1109/TBME.2012.2202662) [DOI] [PubMed] [Google Scholar]
- 60.Cortes C, Vapnik V. 1995. Support-vector networks. Mach. Learn. 20, 273–297. ( 10.1007/BF00994018) [DOI] [Google Scholar]
- 61.Li Q, Rajagopalan C, Clifford GD. 2014. A machine learning approach to multi-level ECG signal quality classification. Comput. Methods Programs Biomed. 117, 435–447. ( 10.1016/j.cmpb.2014.09.002) [DOI] [PubMed] [Google Scholar]
- 62.Rabee A, Barhumi I. 2012. ECG signal classification using support vector machine based on wavelet multiresolution analysis In 2012 11th International Conference on Information Science, Signal Processing and their Applications (ISSPA), pp. 1319–1323. 2-5 July, Montreal, QC. Piscataway, NJ: IEEE ( 10.1109/ISSPA.2012.6310497) [DOI] [Google Scholar]
- 63.Breiman L. 2001. Random forests. Mach. Learn. 45, 5–32. ( 10.1023/A:1010933404324) [DOI] [Google Scholar]
- 64.Sathish B, Vimal C. 2010. Random forest classifier based ECG arrhythmia classification. Int J. Heal. Inf. Syst. Inf. 5, 1–10. ( 10.4018/jhisi.2010040101) [DOI] [Google Scholar]
- 65.Emanet N. 2009. ECG beat classification by using discrete wavelet transform and Random Forest algorithm In Fifth International Conference on Soft Computing, Computing with Words and Perceptions in System Analysis, Decision and Control, 2009 ICSCCW 2009, pp. 1–4. 2-4 September, Famagusta, Cyprus. Piscataway, NJ: IEEE ( 10.1109/ICSCCW.2009.5379457) [DOI] [Google Scholar]
- 66.Sayadi O, Shamsollahi MB.. 2009. A model-based Bayesian framework for ECG beat segmentation. Physiol. Meas. 30, 335 ( 10.1088/0967-3334/30/3/008) [DOI] [PubMed] [Google Scholar]
- 67.Sameni R, Shamsollahi MB, Jutten C, Clifford GD. 2007. A nonlinear Bayesian filtering framework for ECG denoising. IEEE Trans. Biomed. Eng. 54, 2172–2185. ( 10.1109/TBME.2007.897817) [DOI] [PubMed] [Google Scholar]
- 68.Rabiner L. 1989. A tutorial on hidden Markov models and selected applications in speech recognition. Proc. IEEE 77, 257–286. ( 10.1109/5.18626) [DOI] [Google Scholar]
- 69.Bishop CM. 1995. Neural networks for pattern recognition. New York, NY: Oxford University Press, Inc. [Google Scholar]
- 70.Jadhav SM, Nalbalwar SL, Ghatol A. 2010. Artificial Neural Network based cardiac arrhythmia classification using ECG signal data In Electronics and Information Engineering (ICEIE), 2010 International Conference On, pp. V1-228–V1-231. 1-3 August, Kyoto, Japan. Piscataway, NJ: IEEE ( 10.1109/ICEIE.2010.5559887) [DOI] [Google Scholar]
- 71.Meau YP, Ibrahim F, Narainasamy SAL, Omar R. 2006. Intelligent classification of electrocardiogram (ECG) signal using extended Kalman Filter (EKF) based neuro fuzzy system. Comput. Methods Programs Biomed. 82, 157–168. ( 10.1016/j.cmpb.2006.03.003) [DOI] [PubMed] [Google Scholar]
- 72.Dokur Z, Ölmez T. 2001. ECG beat classification by a novel hybrid neural network. Comput. Methods Programs Biomed. 66, 167–181. ( 10.1016/S0169-2607(00)00133-4) [DOI] [PubMed] [Google Scholar]
- 73.Das MK, Ari S, Das MK, Ari S.. 2014. ECG beats classification using mixture of features. Int. Sch. Res. Not. 2014, e178436 ( 10.1155/2014/178436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brugada P, Brugada J. 1992. Right bundle branch block, persistent ST segment elevation and sudden cardiac death: a distinct clinical and electrocardiographic syndrome. A multicenter report. J. Am. Coll. Cardiol. 20, 1391–1396. ( 10.1016/0735-1097(92)90253-J) [DOI] [PubMed] [Google Scholar]
- 75.Alonso A, Agarwal SK, Soliman EZ, Ambrose M, Chamberlain AM, Prineas RJ, FolsoM AR. 2009. Incidence of atrial fibrillation in whites and African-Americans: the Atherosclerosis Risk in Communities (ARIC) study. Am. Heart J. 158, 111–117. ( 10.1016/j.ahj.2009.05.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mainardi L, Sörnmo L, Cerutti S. 2008. Understanding atrial fibrillation: the signal processing contribution, part I. Synth. Lect. Biomed. Eng. 3, 1–129. [Google Scholar]
- 77.Cortana Intelligence and ML Blog Team. 2016. Cleveland Clinic to identify at-risk patients in ICU using Cortana Intelligence. Available at: https://blogs.technet.microsoft.com/machinelearning/2016/09/26/cleveland-clinic-to-identify-at-risk-patients-in-icu-using-cortana-intelligence-suite/ (accessed 1 December 2017).
- 78.Lecun Y, Bengio Y. 1998. Convolutional networks for images, speech, and time series. In The handbook of brain theory and neural networks (ed. MA Arbib), pp. 255–258. Cambridge, MA: MIT Press.
- 79.Hochreiter S, Schmidhuber J. 1997. Long short-term memory. Neural Comput. 9, 1735–1780. ( 10.1162/neco.1997.9.8.1735) [DOI] [PubMed] [Google Scholar]
- 80.Miao F, Cheng Y, He Y, He Q, Li Y. 2015. A wearable context-aware ECG monitoring system integrated with built-in kinematic sensors of the smartphone. Sensors 15, 11 465–11 484. ( 10.3390/s150511465) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lau JK, Lowres N, Neubeck L, Brieger DB, Sy RW, Galloway CD, Albert DE, Freedman SB. 2013. iPhone ECG application for community screening to detect silent atrial fibrillation: a novel technology to prevent stroke. Int. J. Cardiol. 165, 193–194. ( 10.1016/j.ijcard.2013.01.220) [DOI] [PubMed] [Google Scholar]
- 82.AliveCor's heart monitor for iPhone receives FDA clearance. See https://www.alivecor.com/press/press_release/alivecors-heart-monitor-for-iphone-receives-fda-clearance/ (accessed 1 December 2017).
- 83.PR Newswire Association LLC. AliveCor and Mayo Clinic announce collaboration to identify hidden health signals in humans. See https://www.prnewswire.com/news-releases/alivecor-and-mayo-clinic-announce-collaboration-to-identify-hidden-health-signals-in-humans-300349847.html (accessed 1 December 2017).
- 84.Sovilj S, Magjarević R, Lovell NH, Dokos S. 2013. A simplified 3D model of whole heart electrical activity and 12-lead ECG generation. Comput. Math. Methods Med. 2013, 1–10. ( 10.1155/2013/134208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bacharova L, Szathmary V, Potse M, Mateasik A. 2012. Computer simulation of ECG manifestations of left ventricular electrical remodeling. J. Electrocardiol. 45, 630–634. ( 10.1016/j.jelectrocard.2012.07.009) [DOI] [PubMed] [Google Scholar]
- 86.Bacharova L, Szathmary V, Svehlikova J, Mateasik A, Tysler M. 2016. QRS complex waveform indicators of ventricular activation slowing: simulation studies. J. Electrocardiol. 49, 790–793. ( 10.1016/j.jelectrocard.2016.07.032) [DOI] [PubMed] [Google Scholar]
- 87.Zacur E, Minchole A, Villard B, Carapella V, Ariga R, Rodriguez B, Grau V. 2017. MRI-based heart and torso personalization for computer modeling and simulation of cardiac electrophysiology. In Imaging for patient-customized simulations and systems for point-of-care ultrasound (ed. Cardoso M. et al.), pp. 61–70. BIVPCS 2017, POCUS 2017. Lecture Notes in Computer Science, vol. 10549 Cham, Switzerland: Springer. [Google Scholar]
- 88.Potse M, Coronel R, Falcao S, LeBlanc A-R, Vinet A. 2007. The effect of lesion size and tissue remodeling on ST deviation in partial-thickness ischemia. Heart Rhythm 4, 200–206. ( 10.1016/j.hrthm.2006.10.022) [DOI] [PubMed] [Google Scholar]
- 89.Sánchez C, D'Ambrosio G, Maffessanti F, Caiani EG, Prinzen FW, Krause R, Auricchio A, Potse M. 2017. Sensitivity analysis of ventricular activation and electrocardiogram in tailored models of heart-failure patients. Med. Biol. Eng. Comput. ( 10.1007/s11517-017-1696-9) [DOI] [PubMed] [Google Scholar]
- 90.Potse M, et al. 2014. Patient-specific modelling of cardiac electrophysiology in heart-failure patients. EP Europace 16(Suppl. 4), iv56–iv61. ( 10.1093/europace/euu257) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen X, Hu Y, Fetics BJ, Berger RD, Trayanova NA. 2011. Unstable QT interval dynamics precedes VT onset in patients with acute myocardial infarction: a novel approach to detect instability in QT interval dynamics from clinical ECG. Circ. Arrhythm Electrophysiol. 4, 858–866. ( 10.1161/CIRCEP.110.961763) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wilhelms M, Dössel O, Seemann G. 2011. In silico investigation of electrically silent acute cardiac ischemia in the human ventricles. IEEE Trans. Biomed. Eng. 58, 2961–2964. ( 10.1109/TBME.2011.2159381) [DOI] [PubMed] [Google Scholar]
- 93.Benson AP, Al-Owais M, Holden AV. 2011. Quantitative prediction of the arrhythmogenic effects of de novo hERG mutations in computational models of human ventricular tissues. Eur. Biophys. J. 40, 627–639. ( 10.1007/s00249-010-0663-2) [DOI] [PubMed] [Google Scholar]
- 94.Sebastian R, Heidenreich E, Dux-Santoy L, Rodriguez JF, Ferrero JM, Saiz J. 2010. Modeling drug effects on personalized 3D models of the heart: a simulation study. In Statistical atlases and computational models of the heart, pp. 222–231. Berlin, Germany: Springer; See https://link.springer.com/chapter/10.1007/978-3-642-15835-3_23. [Google Scholar]
- 95.van Dam PM van, Oostendorp TF, van Oosterom A. 2009. Application of the fastest route algorithm in the interactive simulation of the effect of local ischemia on the ECG. Med. Biol. Eng. Comput. 47, 11–20. ( 10.1007/s11517-008-0391-2) [DOI] [PubMed] [Google Scholar]
- 96.Wallman M, Smith N, Rodriguez B. 2011. Estimation of activation times in cardiac tissue using graph based methods. In Functional imaging and modeling of the heart, pp. 71–79. Berlin, Germany: Springer; See https://link.springer.com/chapter/10.1007/978-3-642-21028-0_9 [Google Scholar]
- 97.Wallman M, Smith NP, Rodriguez B. 2014. Computational methods to reduce uncertainty in the estimation of cardiac conduction properties from electroanatomical recordings. Med. Image Anal. 18, 228–240. ( 10.1016/j.media.2013.10.006) [DOI] [PubMed] [Google Scholar]
- 98.Konukoglu E, et al. 2011. Efficient probabilistic model personalization integrating uncertainty on data and parameters: application to eikonal-diffusion models in cardiac electrophysiology. Prog. Biophys. Mol. Biol. 107, 134–146. ( 10.1016/j.pbiomolbio.2011.07.002) [DOI] [PubMed] [Google Scholar]
- 99.Dokos S, Cloherty SL, Lovell NH. 2007. Computational model of atrial electrical activation and propagation. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2007, 908–911. [DOI] [PubMed] [Google Scholar]
- 100.Vigmond EJ, Ruckdeschel R, Trayanova N. 2001. Reentry in a morphologically realistic atrial model. J. Cardiovasc. Electrophysiol. 12, 1046–1054. ( 10.1046/j.1540-8167.2001.01046.x) [DOI] [PubMed] [Google Scholar]
- 101.Adeniran I, MacIver DH, Garratt CJ, Ye J, Hancox JC, Zhang H. 2015. Effects of persistent atrial fibrillation-induced electrical remodeling on atrial electro-mechanics—insights from a 3D model of the human atria. PLoS ONE 10, e0142397 ( 10.1371/journal.pone.0142397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Krueger MW, et al. 2013. Personalization of atrial anatomy and electrophysiology as a basis for clinical modeling of radio-frequency ablation of atrial fibrillation. IEEE Trans. Med. Imaging 32, 73–84. ( 10.1109/TMI.2012.2201948) [DOI] [PubMed] [Google Scholar]
- 103.Seemann G, Höper C, Sachse FB, Dössel O, Holden AV, Zhang H. 2006. Heterogeneous three-dimensional anatomical and electrophysiological model of human atria. Phil. Trans. R. Soc. A 364, 1465–1481. ( 10.1098/rsta.2006.1781) [DOI] [PubMed] [Google Scholar]
- 104.Colman MA, Aslanidi OV, Stott J, Holden AV, Zhang H. 2011. Correlation between P-wave morphology and origin of atrial focal tachycardia—insights from realistic models of the human atria and torso. IEEE Trans. Biomed. Eng. 58, 2952–2955. ( 10.1109/TBME.2011.2161305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Aslanidi OV, Colman MA, Stott J, Dobrzynski H, Boyett MR, Holden AV, Zhang H. et al. 2011. 3D virtual human atria: a computational platform for studying clinical atrial fibrillation. Prog. Biophys. Mol. Biol. 107, 156–168. ( 10.1016/j.pbiomolbio.2011.06.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Physionet.org. Introduction. MIT-BIH Arrhythmia Database Directory. See https://physionet.org/physiobank/database/html/mitdbdir/intro.htm. (accessed 30 October 2017)
- 107.Robinson MR, Curzen N. 2009. Electrocardiographic body surface mapping: potential tool for the detection of transient myocardial ischemia in the 21st century? Ann. Noninvasive Electrocardiol. 14, 201–210. ( 10.1111/j.1542-474X.2009.00284.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ambale-Venkatesh B, et al. 2017. Cardiovascular event prediction by machine learning: the multi-ethnic study of atherosclerosis. Circ. Res. 121, 1092–1101. ( 10.1161/CIRCRESAHA.117.311312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rodriguez B, et al. 2016. Human-based approaches to pharmacology and cardiology: an interdisciplinary and intersectorial workshop. Europace 18, 1287–1298. ( 10.1093/europace/euv320) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.