Abstract
Aims: Circadian rhythms permeate all levels of biology to temporally regulate cell and whole-body physiology, although the cell-autonomous mechanism that confers ∼24-h periodicity is incompletely understood. Reports describing circadian oscillations of over-oxidized peroxiredoxin abundance have suggested that redox signaling plays an important role in the timekeeping mechanism. Here, we tested the functional contribution that redox state and primary metabolism make to mammalian cellular timekeeping.
Results: We found a circadian rhythm in flux through primary glucose metabolic pathways, indicating rhythmic NAD(P)H production. Using pharmacological and genetic perturbations, however, we found that timekeeping was insensitive to changes in glycolytic flux, whereas oxidative pentose phosphate pathway (PPP) inhibition and other chronic redox stressors primarily affected circadian gene expression amplitude, not periodicity. Finally, acute changes in redox state decreased PER2 protein stability, phase dependently, to alter the subsequent phase of oscillation.
Innovation: Circadian rhythms in primary cellular metabolism and redox state have been proposed to play a role in the cellular timekeeping mechanism. We present experimental data testing that hypothesis.
Conclusion: Circadian flux through primary metabolism is cell autonomous, driving rhythmic NAD(P)+ redox cofactor turnover and maintaining a redox balance that is permissive for circadian gene expression cycles. Redox homeostasis and PPP flux, but not glycolysis, are necessary to maintain clock amplitude, but neither redox nor glucose metabolism determines circadian period. Furthermore, cellular rhythms are sensitive to acute changes in redox balance, at least partly through regulation of PER protein. Redox and metabolic state are, thus, both inputs and outputs, but not state variables, of cellular circadian timekeeping. Antioxid. Redox Signal. 28, 507–520.
Keywords: : circadian rhythm, redox signaling, primary metabolism, clock gene, mammalian, pentose phosphate pathway
Introduction
Mammalian cells exhibit cell-autonomous ∼24-h rhythms in the organization of a broad range of biological processes, from metabolism (11) to redox balance (56), and from cell division (14) to DNA repair (67). At the whole-organism level, these individual cellular rhythms are synchronized by endocrine cues to facilitate anticipation of, and adaptation to, the environmental cycle of day and night (45). This is most evident in the daily cycle of human sleep and wake, but it extends to regulate most aspects of physiology and organismal function. In consequence, disruption of circadian rhythms, which occurs during shift work and social jetlag, is associated with an increased risk of chronic diseases such as type II diabetes, various cancers, and neurodegenerative disorders (33, 64). Understanding the cellular clock mechanism and how it achieves the temporal coordination of cell biology is, therefore, an important biomedical research objective.
Innovation.
Circadian rhythms in primary cellular metabolism and redox state have previously been proposed to play a role in the timekeeping mechanism. We tested this hypothesis, and found that although primary metabolism and redox signaling can function as both an input and an output of the cellular clockwork, they do not determine its period and are, thus, unlikely to constitute essential mechanistic timekeeping components. Importantly, we do find that PER2 stability is acutely sensitive to H2O2 transients, implicating this clock protein as a circadian effector of redox signaling.
Many of the individual “clock genes” and proteins that contribute to the fidelity of cellular timekeeping have been identified, and are believed to function in a delayed gene expression feedback loop whereby complexes containing the transcription factor BMAL1 activate the expression of period (1/2) and cryptochrome (1/2) genes, as well as other “clock-controlled genes,” such as the antioxidant regulator NRF2 (24, 54, 59). The encoded PER and CRY proteins form inhibitory complexes that eventually repress BMAL1-containing complex activity. The kinetics of this feedback inactivation are post-translationally regulated to give rise to persistent oscillations in the expression of clock and clock-controlled genes that occur with a period of ∼24 h. The precise cascade of molecular mechanisms that determines the period of these oscillations is incompletely understood (49), though it certainly involves the regulation of clock protein activity and stability by conserved kinases such as casein kinase 1δ/ɛ (43, 44). Various accessory feedback loops contribute to the fidelity and amplitude of circadian gene expression [e.g., (5, 42, 58)], whereas changes in circadian phase are effected by extracellular cues acting through several different signaling mechanisms (e.g., glucorticoid receptor, growth factor and G-protein receptor signaling) to stimulate acute changes in clock protein expression via intracellular signal transduction cascades (7, 8, 22, 65, 68).
Complicating this picture are recent observations that metabolic and redox pathways continue to exhibit circadian regulation in the absence of rhythmic clock gene expression (50, 51), suggesting the existence of an underlying post-transcriptional timekeeping mechanism [reviewed in ref. (56)]. Most notably, levels of redox co-factors and the abundance of over-oxidized peroxiredoxin (PRX-SO2/3) exhibit circadian regulation in isolated human and mouse erythrocytes (17, 51), which are naturally anucleate and, therefore, cannot be dependent on cycling clock gene transcription. Rhythms of PRX-SO2/3 and redox balance have been reported in vivo, as well as in cultured tissues and cells (27, 37), and also in other phylogenetic groups (4, 12, 32, 34, 46, 80). In addition, although PRX itself plays no timekeeping role (16), the activity of several clock proteins has been suggested to be redox sensitive (47, 57, 70, 81). Moreover, nuclear translocation of the antioxidant regulator NRF2 exhibits ultradian and circadian rhythms (54, 78), and SIRT1-mediated deacetylation of certain clock proteins is reported to be dependent on rhythmic availability of NAD+ (53, 58).
These observations suggest the possibility that redox-based oscillations may constitute a bona fide and conserved mechanism of the cellular clock in eukaryotes: a compelling hypothesis proposing that an as-yet uncharacterized post-translational oscillation in metabolism and redox balance reciprocally regulates clock gene expression to determine the period of the cellular circadian clock (60). Conversely, however, it is equally plausible that redox and metabolic rhythms are simply post-translational outputs of cellular clock function, and thus irrelevant to the timekeeping mechanism. A third possibility is that changes in metabolism and redox balance constitute a non-essential auxiliary cellular clock mechanism. In this case, they would be rhythmically regulated, with transient changes in metabolic activity and/or redox balance being competent to regulate the amplitude and phase of circadian gene expression, but not required to oscillate for the cellular timekeeping mechanism to function.
To distinguish between these three models, we used pharmacological and genetic approaches to perturb primary metabolic pathways and cellular redox balance, thereby assessing the causal relationships between cellular timekeeping and redox/metabolism. We found not only evidence for cell-autonomous circadian regulation of primary metabolism but also that inhibition of glycolysis alone had no major effect on the cellular clock. We further observed that the amplitude and phase of PER protein expression is sensitive to redox balance and oxidative pentose phosphate pathway (PPP) activity, but that the period of oscillation was essentially insensitive to these perturbations. Our data indicate that previously observed oscillations in cellular redox state are not required for the cellular clock to function, since the period of oscillation is robust against severe chronic oxidative as well as reductive stress.
Finally, we found that pulses of physiologically relevant concentrations of H2O2 elicited phase-dependent phase shifts, which strongly correlated with immediate changes in the expression level of the PER2 clock protein, providing a mechanistic basis for understanding how transient redox signals may contribute to cellular clock regulation under more physiological settings. We, therefore, conclude that redox signaling is not essential to the cellular timekeeping mechanism (i.e., it is not a state variable of the oscillation) but that it does function as both an input and an output of the circadian clockwork.
Results
Cell-autonomous rhythms of glucose catabolism drive rhythmic NAD(P)H generation
Circadian rhythms in the levels of several redox co-factors, primary metabolites, and the abundance of over-oxidized PRX have been observed in several cultured mammalian cells and tissues, as well as in vivo (17, 51, 53, 56). Here, we set out to test the relationship between circadian timekeeping, redox balance, and metabolic flux (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/ars), by using a primary cellular model: immortalized, but non-transformed, fibroblasts derived from the lung tissue of adult PER2::luciferase (LUC) mice (29, 75, 79). The attraction of this model is that fibroblasts contact-inhibit to form confluent, quiescent cell monolayers that express robust circadian rhythms in bioluminescence due to the fusion of the firefly LUC open reading frame at the 3′-end of the core clock gene Per2. This fusion allows us to longitudinally follow PER2::LUC activity, a well-characterized and faithful reporter of PER2 protein levels, and thus clock activity (29, 75, 79). Indeed, in this system, both PER2::LUC and PRX oxidation show overt circadian oscillations (Fig. 1A and Supplementary Fig. S2F). To validate key observations from this model, we also employed stable human and mouse cell lines expressing other well-established reporters of circadian transcriptional cycles. These are presented as Supplementary Figures.
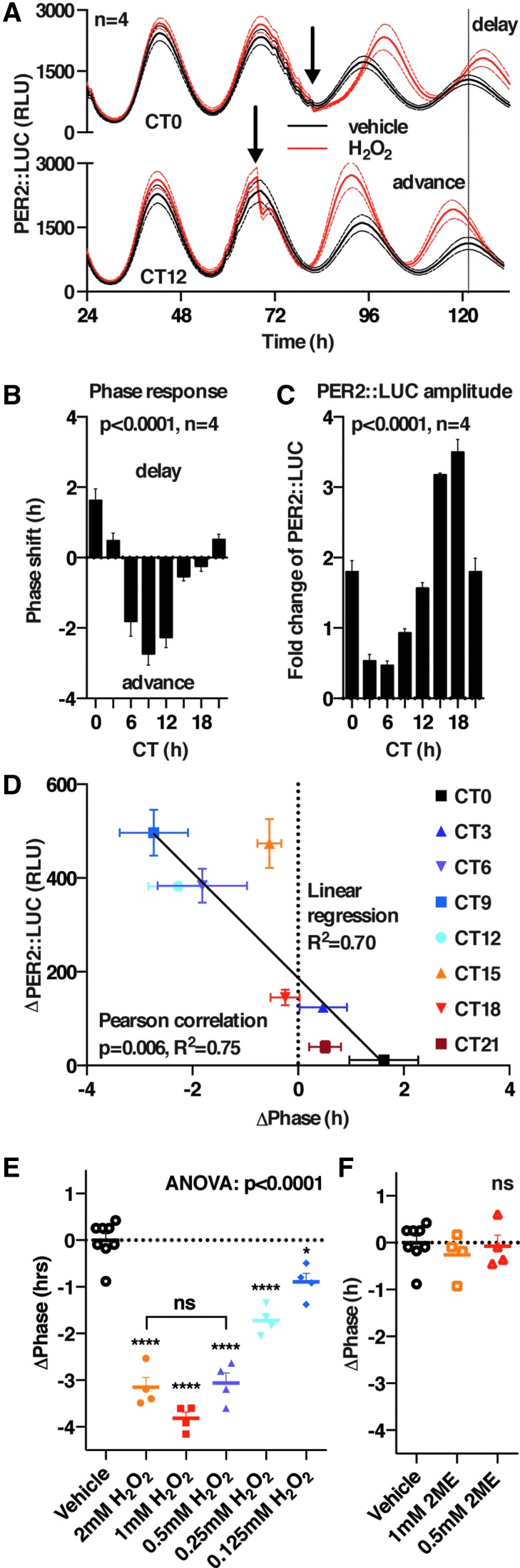
FIG. 1.
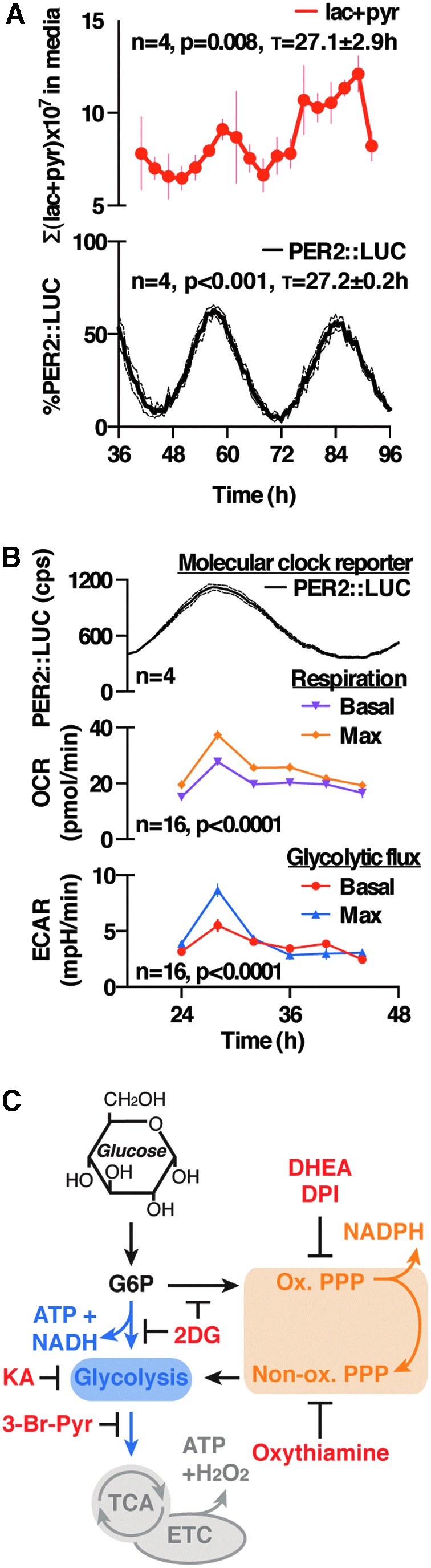
Cell autonomous circadian regulation of primary metabolism in mouse fibroblasts. (A) Liquid chromatography coupled to mass spectrometry analysis of perfused cell media reveals a significant variation (by cosinor analysis) in glycolytic end products (lac + pyr, lactate and pyruvate), peaking at the same time as PER2::LUC (molecular clock reporter) recorded from the same cultures simultaneously (mean ± SEM). (B) Extracellular flux analysis over a circadian cycle indicates a significant variation in primary metabolism, in the same phase as peak activity of PER2::LUC bioluminescence recorded from parallel cultures (mean ± SEM, one-way ANOVA). (C) Schematic of primary metabolic pathways, indicating the activity of the drugs used throughout this article. The validation of pathway inhibition by these inhibitors is shown in Supplementary Figure S2. ANOVA, analysis of variance; ECAR, extracellular acidification rate; ETC, electron transport chain; LUC, luciferase; OCR, oxygen consumption rate; PPP, pentose phosphate pathway; SEM, standard error of the mean; TCA, tricarboxylic acid cycle.
Cytosolic redox homeostasis is a dynamic equilibrium that is ultimately dependent on the rate of production of oxidizing reactive oxygen species (ROS) and reducing equivalents in the form of NADPH (and to a lesser extent NADH), which are primarily determined by rates of primary metabolic flux (23). Isotopic labeling of glucose is a sensitive method for the measurement of metabolic flux, since pathway fluxes can be interpolated from the abundance and label distribution of glycolytic end products, lactate and pyruvate (13, 18, 19, 25). In longitudinal assays of PER2::LUC fibroblasts, continuously perfused with media containing labeled glucose and luciferin, we detected a rhythm in total media lactate and pyruvate production (measured by liquid chromatography coupled to mass spectrometry [LC-MS] analysis) that peaked approximately at the same time as PER2 (Fig. 1A). This correlated with an increase in media acidification (extracellular acidification rate [ECAR]: reporting glycolytic rate) and respiration (oxygen consumption rate [OCR]: reporting respiratory rate) measured by extracellular flux analysis, indicating that primary metabolic flux is, indeed, clock regulated and peaks during what corresponds to the active phase in vivo (Fig. 1B), and in agreement with prior indications in the human U2OS cell line (2).
We then investigated whether circadian glucose catabolism is actively regulated in our PER2::LUC fibroblasts. At the beginning of glucose catabolism, there occurs a branch point that directs glucose-6-phosphate toward glycolysis (producing ATP and NADH), or the PPP, which produces reducing equivalents in the oxidative portion (NADPH) as well as biosynthetic intermediates during the non-oxidative portion (Fig. 1C). Using 13C-based flux analysis (Supplementary Fig. S1), we tested whether dynamic rerouting of metabolic flux occurred between glycolysis and the PPP over the circadian cycle.
We found no significant rhythm in the relative proportion of glucose catabolism between the two pathways (Supplementary Fig. S2A). This suggests that any rhythm in the NADP+:NADPH redox couples (51, 61) must occur passively, as a function of changes in co-factor oxidation and/or overall changes in glucose utilization. We also detected no significant rhythm in the ratio of lactate:pyruvate (Supplementary Fig. S2A), which is reflective of the free NAD+:NADH ratio (19), indicating that previously reported rhythms of NAD+:NADH may not represent the freely available pool. Critically, however, our observations indicate that the rate of production of NADH and NADPH shows a circadian rhythm in these cells, which is the greatest around the same phase as peak PER2 levels. This means that the generation of reductive potential is highest at the same time as maximal mitochondrial activity, based on our OCR measurements.
Glucose catabolism primarily determines the amplitude of circadian gene expression, not its period, via the PPP
We next asked whether rhythmic glucose catabolism and NAD(P)H production were, in fact, required for cellular timekeeping, or might instead be a consequence of it. We treated cells with 2-deoxyglucose (2DG), an anti-metabolite that is phosphorylated by hexokinase to produce 2DG-6-P, which dose dependently inhibits downstream flux through both glycolysis and the PPP (10) (Figs. 1C and 2D, and Supplementary Fig. S2B–E). We observed that 2DG elicited a profound and dose-dependent suppression of PER2::LUC rhythms (Fig. 2A, B, and E), whereas effects on period were small, inconsistent, and difficult to quantify reliably due to rapid damping. Oscillations recovered gradually on changing cells back into normal (2DG-free) media (Fig. 2A), indicating that the damping of PER2 rhythms was not due to cytotoxicity, with the slow recovery presumably being attributable to intracellular 2DG-6-P accumulation. We observed very similar results in additional cell lines (human U2OS cells and mouse NIH3T3 cells) expressing two different bioluminescent reporters for clock gene transcription (Per2:LUC and Bmal1:LUC, respectively) (Supplementary Fig. S3). Importantly, ATP levels in cells treated with 2DG for 3 days were not decreased (Fig. 2C) and actually increased at one concentration, as also occurs during Mg2+ depletion (28); and 2DG treatment had only a very modest effect on bioluminescence levels in cells expressing firefly LUC under the constitutive SV40 promoter (Fig. 2E and Supplementary Fig. S3B, E). In addition, we observed that depletion of glucose from the media did not phenocopy the effects of 2DG, instead eliciting a slight but significant increase in circadian period (Fig. 2F, G), as previously (40). We, therefore, conclude that 2DG treatment impacts clock gene amplitude through a different mechanism than glucose starvation, and it is not an artifact of LUC inhibition, ATP depletion, or general inhibition of gene expression.
FIG. 2.
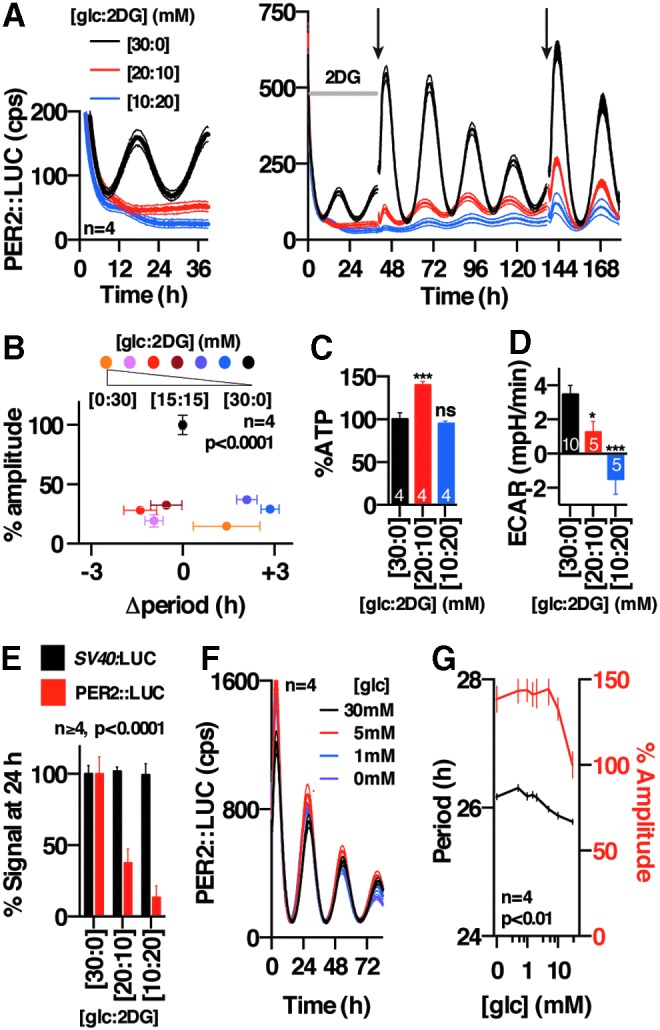
2DG reversibly dampens circadian rhythms in mouse fibroblasts. (A) 2DG immediately dampens PER2::LUC rhythms (left panel), which gradually recover on a change to 2DG-free media (black arrows, right panel); the ratio of glucose and 2DG concentrations, respectively, is indicated in square brackets. Mean ± SEM of representative experiments are plotted; statistical test: circadian fit versus straight line. (B) Effect of 2DG treatment on PER2::LUC amplitude is dose dependent, whereas the effect on period is not (mean ± SEM, one-way ANOVA for each is reported). (C) 2DG treatment does not significantly decrease cellular ATP levels after 72 h (mean ± SEM, replicate number, and Holm-Sidak p-values vs. control). (D) 2DG significantly reduces glycolytic flux (ECAR; mean ± SEM, replicate number, and Holm-Sidak p-values vs. control). (E) Comparison of bioluminescence from 2DG-treated PER2::LUC and SV40:LUC fibroblasts at 24 h (mean ± SEM, two-way ANOVA for 2DG vs. reporter interaction is reported). (F) Effect of glucose depletion on PER2::LUC rhythms (representative concentrations, mean ± SEM). (G) Effect of different glucose concentrations on circadian period and PER2::LUC bioluminescence at 24 h (one-way ANOVA p-values for each are reported). *p < 0.05, ****p < 0.0001, ns, p > 0.05. 2DG, 2-deoxyglucose.
We next asked whether the effect of 2DG treatment might be attributable to its action on the glycolytic pathway or PPP. Genetic knockouts of glycolytic and PPP enzymes are cytotoxic and so to accomplish this we employed transient RNAi knockdown of either glucose-6-phosphate dehydrogenase (G6pdh) or phosphoglucose isomerase (Gpi), the enzymatic gatekeepers of the PPP and glycolytic pathways, respectively. Although cellular levels for both proteins were reduced by ∼50%, we observed that only knockdown of G6pdh elicited a significant reduction in PER2 levels, whereas Gpi knockdown did not (Fig. 3). This was accompanied by a significant but small shortening of period by 0.6 ± 0.1 h (t-test, p = 0.001), which we do not consider to be noteworthy (see the section “Materials and Methods”).
FIG. 3.
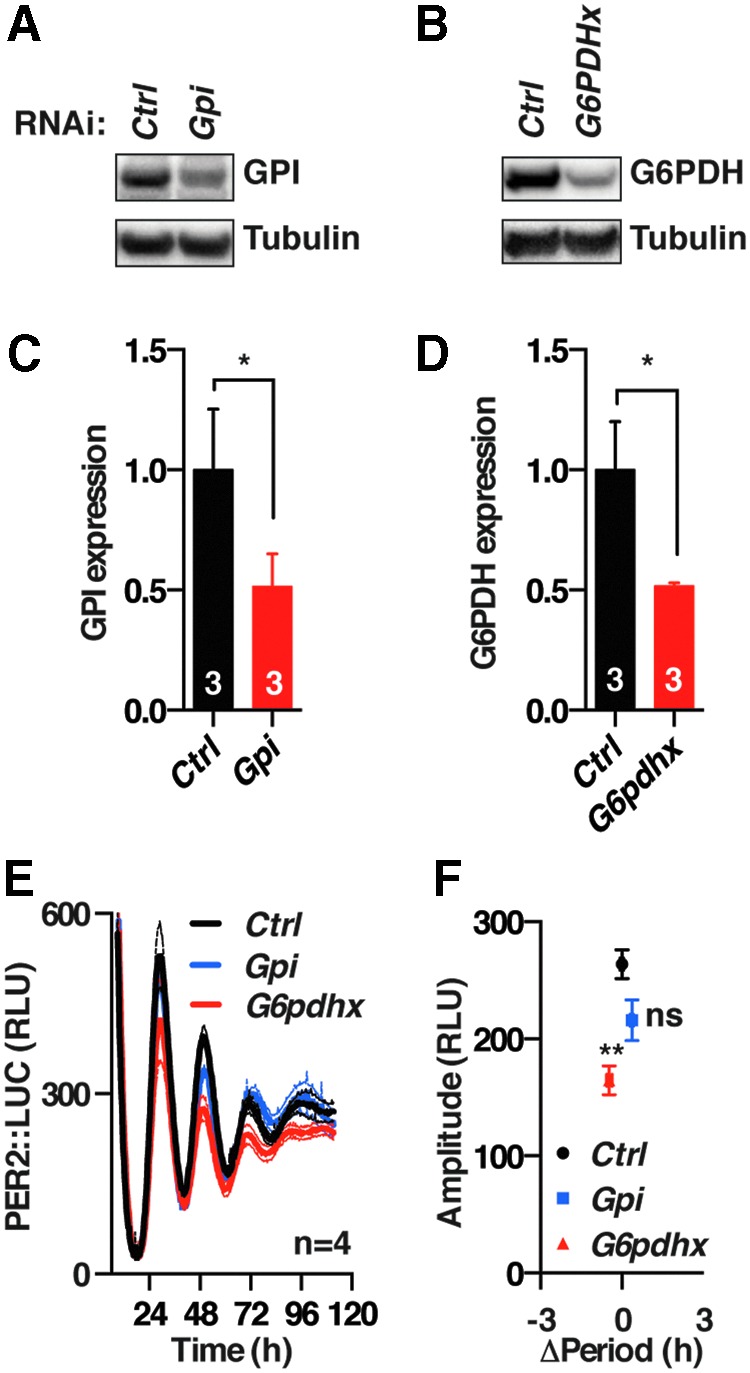
G6PDH activity regulates the amplitude of circadian rhythms in mouse fibroblasts. Western blot analysis (A, B) and its quantification (C, D) from cells silenced for Gpi or G6pdh (mean ± SEM, Student's t-test). (E) Effect of RNAi silencing of Gpi and G6pdh on PER2::LUC rhythms (mean ± SEM). (F) Silencing of G6PDH but not of GPI results in decreased clock protein amplitude, measured at the second peak of bioluminescence (mean ± SEM, Student's t-test). *p < 0.05; **p < 0.01, ns, p > 0.05. G6PDH, glucose-6-phosphate dehydrogenase; GPI, phosphoglucose isomerase.
The activity and abundance of essential metabolic enzymes is typically much greater than cellular demand, explaining why reduced enzyme levels might not elicit as strong an effect as 2DG treatment, and we, therefore, sought to test whether the greater sensitivity of PER2 oscillations to PPP over glycolytic activity, indicated by genetic knockdown, could be validated by using pharmacological approaches. We found that inhibitors of the oxidative PPP (flavoprotein inhibitor diphenyleneiodonium [DPI; (62)] and G6PDH inhibitor dehydroepiandrosterone [DHEA; (31)] consistently phenocopied the effect of 2DG in all three cellular models (Fig. 4A–E and Supplementary Fig. S4A, B, F, and G), whereas selective inhibition of the non-oxidative PPP (oxythiamine; Fig. 4F, G, and Supplementary Fig. S4C–E) did not. Since DPI and DHEA belong to different classes of drugs and inhibit PPP flux by different mechanisms, our results suggest that PPP flux can regulate the level of cellular clock gene expression. In contrast, we found that selective, established inhibitors of different glycolytic enzymes (GAPDH inhibitor koningic acid [KA; (66)] and pyruvate analog 3-bromopyruvate (55) did not elicit a decrease in the amplitude of clock gene expression and evoked small, inconsistent effects on circadian period in the three different and independent cellular models that were employed (Fig. 5 and Supplementary Fig. S5). Importantly, the action and selectivity of the glycolytic versus PPP flux inhibitors was confirmed by LC-MS-based metabolic tracing (Supplementary Fig. S2B–E), where we confirmed major effects of each glycolytic and PPP inhibitor on flux through the PPP and glycolysis, respectively, as well as effects on NADH availability and glucose utilization (Supplementary Fig. S2B–E).
FIG. 4.
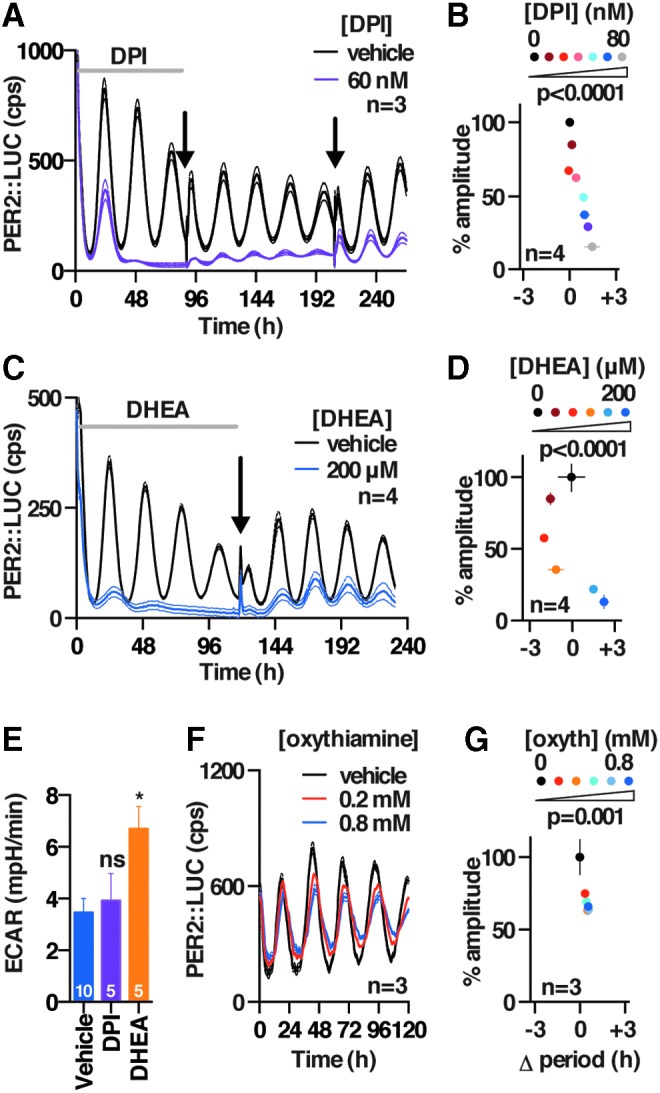
PPP inhibition reversibly dampens circadian rhythms in mouse fibroblasts. Effect of DPI (flavoprotein inhibitor) (A, B) and DHEA (G6PDH inhibitor) (C, D) on PER2::LUC rhythms. Left panels show traces of representative concentrations (mean ± SEM). Black arrows indicate change to drug-free media. Right panel shows dose-dependent effects of drug treatments (mean ± SEM, one-way ANOVA p-value for each parameter is reported). (E) DPI and DHEA do not reduce glycolytic flux (replicate numbers and Holm-Sidak p-values are reported). (F) Effect of oxythiamine (oxyth, transketolase inhibitor) on PER2::LUC rhythms (representative mean ± SEM). (G) Effect of oxythiamine treatment on PER2::LUC amplitude and period (one-way ANOVA p-value for each parameter is reported). *p < 0.05, ns, p > 0.05. DHEA, dehydroepiandrosterone; DPI, diphenyleneiodonium.
FIG. 5.
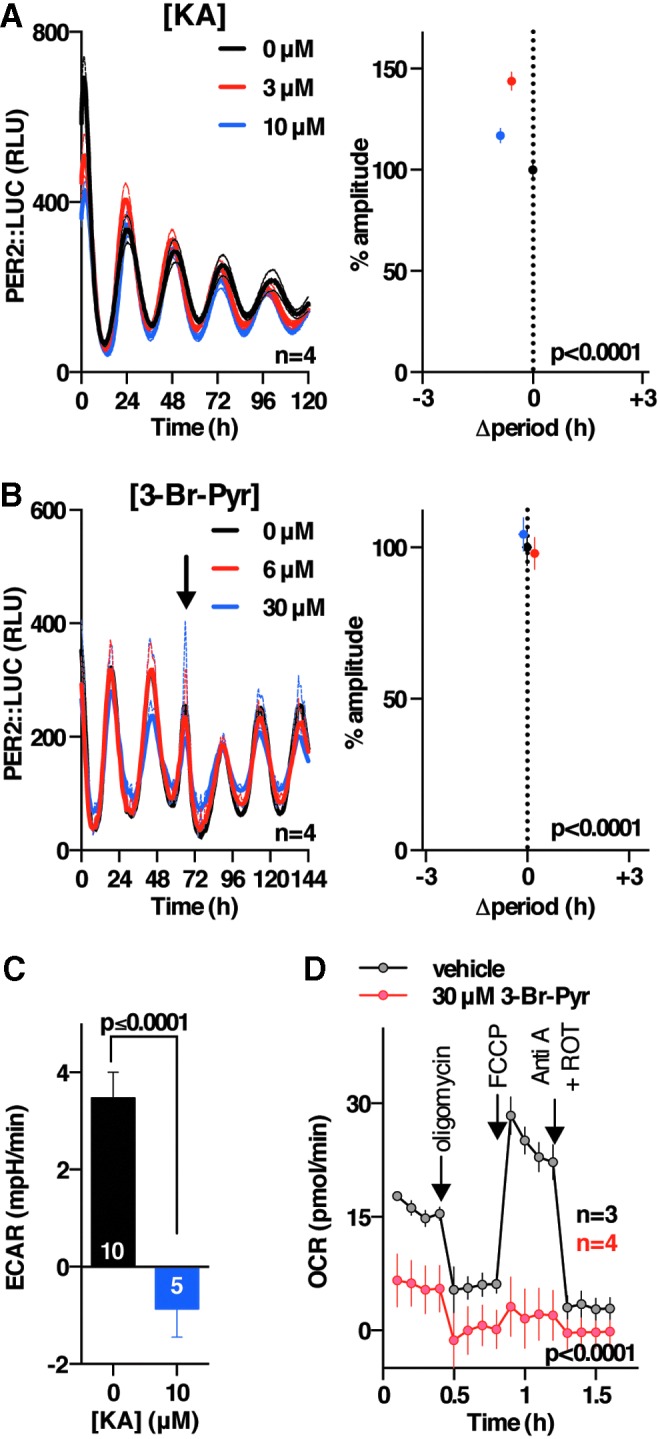
Circadian rhythms in mouse fibroblasts are insensitive to glycolytic inhibition. (A). Left: effect of KA (GAPDH inhibitor) on PER2::LUC rhythms (mean ± SEM). Right: effect of KA treatment on PER2::LUC amplitude and period (mean ± SEM, one-way ANOVA p-value for each parameter is reported). (B). Left: effect of 3-Br-Pyr (pyruvate analogue preventing flux through glycolysis) on PER2::LUC rhythms (representative mean ± SEM). Right: effect of 3-Br-Pyr treatment on PER2::LUC amplitude and period (mean ± SEM, one-way ANOVA p-value for each parameter is reported). (C) Effect of KA treatment on glycolytic flux (unpaired t-test p-value and replicate number are reported). (D) Effect of 3-Br-Pyr treatment on respiration (two-way ANOVA p-value and replicate number). 3-Br-Pyr, 3-bromopyruvate; Anti A, antimycin A; KA, koningic acid; ROT, rotenone.
Thus, genetic and pharmacological evidence suggests that the amplitude of circadian gene expression oscillations is differentially sensitive to oxidative PPP compared with glycolytic activity. Critically, the effect of severe and chronic metabolic inhibition on circadian period was very modest and also inconsistent between our different cellular models. This is hard to reconcile with models in which metabolic/redox oscillations play essential and conserved roles within the circadian timekeeping mechanism, but it does suggest that PPP activity may have an accessory role in regulating clock amplitude. We, therefore, wondered how PPP inhibition might impact circadian gene expression cycles.
Redox balance primarily affects clock gene expression amplitude, not circadian period
A potential consequence of a sustained reduction in flux through the oxidative PPP flux is a reduction in the rate with which NADPH can be regenerated from NADP+. In turn, this would be expected to induce a sustained oxidation of cellular redox balance. Correspondingly, we found that 24 h treatment with 2DG significantly increased the cellular NADP:NADPH ratio (Supplementary Fig. S6A), indicating a more oxidized cellular environment; and that reducing reagent 2-mercaptoethanol (2ME) transiently attenuated the effect of 2DG on PER2 amplitude (Supplementary Fig. S6B). To test whether chronic perturbation of cellular redox homeostasis might constitute the means whereby PPP inhibition impacts the amplitude of circadian gene expression, we incubated cells with a range of concentrations of 2ME or glucose oxidase (GOX, catalyzing the oxidation of glucose to form d-glucono-1,5-lactone and the continuous production of H2O2). We observed that the reducing treatment elicited a dose-dependent and reversible reduction in the amplitude of circadian gene expression (Fig. 6A top, B and Supplementary Fig. S6C), whereas the oxidizing treatment initially increased and then reduced the amplitude of clock gene oscillations (Fig. 6A, C). We further noted that there was a significant interaction between circadian amplitude and the circadian phase at which the redox treatment occurred: Cells were less sensitive to 2ME treatment that began at the trough of PER2 expression (CT0) than at its peak (CT12). Conversely, treatments with all but the highest levels of GOX evoked greater increases in the subsequent amplitude of PER2::LUC oscillations, when added at CT0, than was observed when treatments commenced 12 h later. Although we did see a significant interaction between the period of oscillation and phase of treatment with both 2ME and GOX (p < 0.0001, two-way analysis of variance), the effect size was very small (<7% control) compared with the effect on amplitude. GOX and 2ME treatment, respectively, impose chronic oxidative and reductive stresses, which are well outside the normal range of cellular redox homeostasis. Our observation, therefore, suggests that the period of oscillation is quite robust against these “sledgehammer” treatments.
FIG. 6.
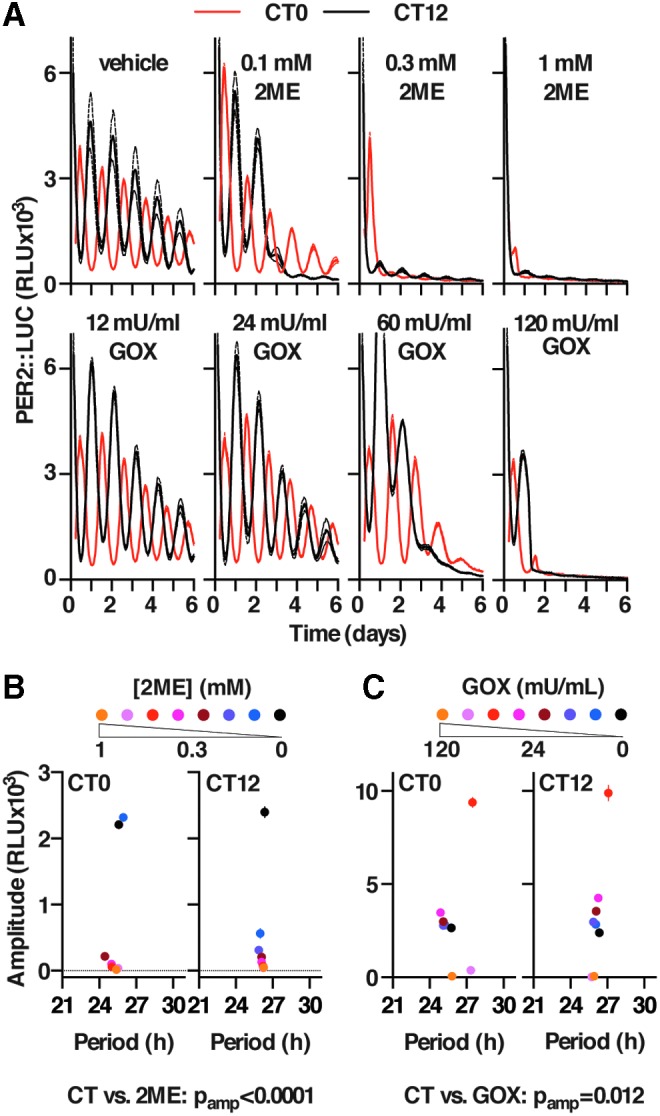
Redox balance regulates the amplitude of circadian rhythms in mouse fibroblasts. (A) Effect of chronic GOX and 2ME treatment on PER2::LUC rhythms, with treatment beginning at two opposite circadian phases (CT, CT0 or CT12) (representative mean ± SEM are shown [n = 4]). (B) Effect of 2ME and (C) GOX treatment on PER2::LUC amplitude and period (two-way ANOVA p-values for amplitude vs. concentration effect [n = 4]). 2ME, 2-mercaptoethanol; CT, circadian time; GOX, glucose oxidase.
Redox signals regulate the cellular clock via PER2 stability
The phase-dependent effects of reducing or oxidizing the cellular redox environment indicated that redox signaling might play a bona fide role in regulating clock protein activity, as has previously been proposed in other model organisms such as Neurospora crassa (80). We tested this hypothesis by using H2O2, the putative second messenger in redox signaling relays (23, 35). Cells were treated with vehicle or a bolus of H2O2, every 3 h over a complete circadian cycle, and the effects on the subsequent rhythm were analyzed (Fig. 7 and Supplementary Figs. S7 and S8). We observed no consistent changes in circadian period, whereas we did observe phase shifts that conformed to a classic circadian phase response curve, with delays of ∼2 h occurring around CT12 (peak PER2) and advances of ∼2 h occurring over a smaller window around CT0 (Fig. 7B). Although we also observed a phase-dependent and significant change in amplitude (Fig. 7C), we observed no correlation between the phase shifts and the subsequent amplitude of PER2 expression (R2 = 0.05, data not shown). Interestingly, a strong correlation was observed between the acute decrease in total PER2::LUC levels occurring immediately after H2O2 addition and the magnitude and direction of the phase shift elicited (R2 = 0.75) (Fig. 7D). The phase shift occurred in a dose-dependent manner, with maximum effect occurring at ≥0.5 mM H2O2, and it remained significant at physiologically relevant concentrations of H2O2 (Fig. 7E); whereas control treatments with 2ME elicited no such effect (Fig. 7F). These results suggest a possible physiological role for H2O2 signaling: to relay extracellular (e.g., growth factor) or intracellular (e.g., mitochondrial metabolic) cues to the cellular clock and alter its phase, since PER2 is a bona fide state variable of the cellular clockwork (21).
FIG. 7.
Acute H2O2 treatment elicits phase-dependent PER2 degradation and circadian phase shifts in mouse fibroblasts. (A) Effect of an acute H2O2 bolus (1 mM final, black arrow) on PER2::LUC rhythms (representative mean ± SEM are shown). (B) Phase response curve to H2O2 treatment at indicated CT (mean ± SEM, one-way ANOVA p-value). (C) Effect of H2O2 bolus at indicated CT on subsequent peak PER2::LUC (mean ± SEM, one-way ANOVA p-value, and replicate numbers are reported). (D) Correlation between the immediate decrease in PER2::LUC bioluminescence after the H2O2 bolus and the subsequent circadian phase shift (mean ± SEM, correlation statistics). (E) Phase shift induced by an acute H2O2 bolus at CT12 occurs at physiologically relevant H2O2 concentrations and is dose dependent up to 1 mM (mean ± SEM, one-way ANOVA p-value). (F) No phase shift is induced by an acute 2ME bolus at CT12 (mean ± SEM, one-way ANOVA p-value).
Acute changes in the expression of PER2 protein abundance are an accepted mechanism by which circadian phase shifts occur (both in cells and in vivo) (1, 6, 8, 9, 21, 72), and our data, thus, suggest PER2 to be a functional circadian effector of redox signaling in mammalian cells. To confirm that the drop in LUC signal was a true report of decreased PER2 protein levels, we used immortalized fibroblasts isolated from knock-in transgenic mice expressing the PER2::VENUS fusion protein (73), as the VENUS-tag allows for clear quantification of PER2 by Western blot using an anti-GFP antibody. At the peak of PER2 expression (i.e., CT12), we treated the cells with H2O2 or the disulfide-inducing reagent diamide and analyzed protein levels by immunoblotting. Similar to the results of our longitudinal assays (Fig. 7A and Supplementary Fig. S7), we observed that PER2 levels were significantly reduced after an acute oxidative insult (Fig. 8A). This reduction was dose dependent and correlated well with the phase shift reported in Figure 7E.
FIG. 8.
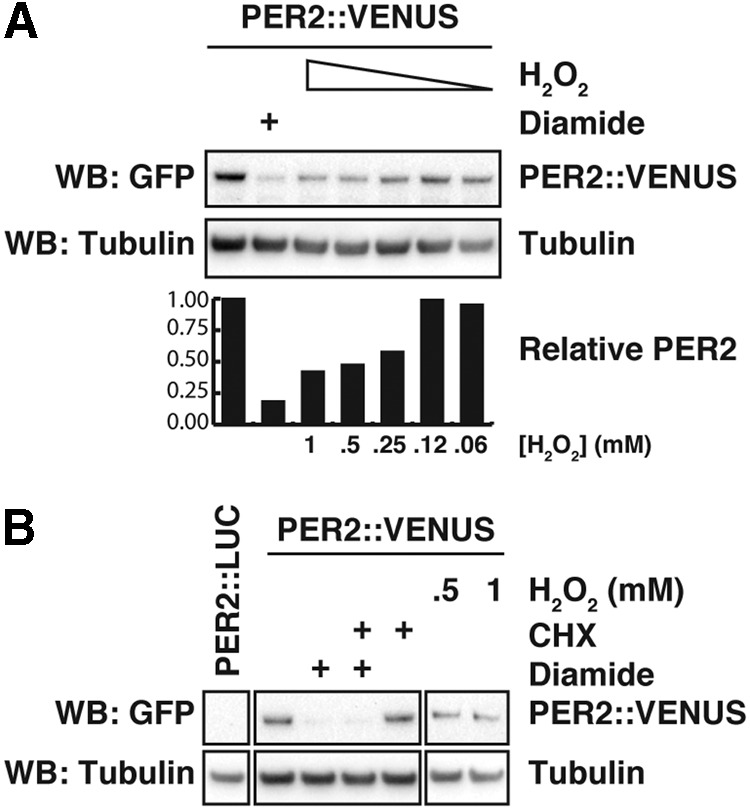
Acute H2O2 and diamide treatment induce rapid PER2 degradation. (A) Western blot analysis of cell lysates of PER2::VENUS cells treated with H2O2 (60 μM–1 mM) or diamide (250 μM) for 15 min at the expected peak of PER2 expression. The graph depicts the PER2::VENUS levels relative to tubulin expression. (B) Experiment as in (A). Effects of acute oxidative stress (H2O2 or 250 μM diamide) for 15 min, at the normal PER2 peak, on PER2 stability, in the presence or absence of the inhibitor of protein synthesis CHX (10 μM). A similar experiment assessing PER2::LUC bioluminescence can be found in Supplementary Figure S9. CHX, cycloheximide.
The observed reduction of PER2 protein levels could be either the effect of a temporary inhibition of translation or the result of increased PER2 protein degradation. We therefore compared the rate of PER2 decay on redox stress with that of cells treated with a saturating concentration of translation inhibitor cycloheximide (CHX) and found that both H2O2 and diamide induced a greater decrease in PER2 levels than CHX alone, indicating that PER2 degradation is increased on an oxidative insult (Fig. 8B). This result was recapitulated with PER2::LUC fibroblasts, where we observed that H2O2 decreased PER2 acutely, even in the presence of saturating CHX (Supplementary Fig. S9).
Discussion
We investigated the causal relationships between redox metabolism and circadian gene expression cycles in cultured mammalian cells. In keeping with previous observations, we observed compelling evidence that total glucose catabolism is circadian regulated (2, 30, 38), meaning that the rate of glycolytic NADH production and PPP-mediated NADPH production is also rhythmic. We found no evidence for active cell-autonomous circadian redistribution of metabolic flux between the NADH- and ATP-producing glycolytic pathway and the NADPH-producing PPP, suggesting that this cannot be a general feature of the circadian clock in mammalian cells. It seems more likely that, in the absence of oxidative or reductive stress, any rhythm in steady-state redox co-factor levels arises passively, as a consequence of rhythms in overall glucose catabolism and/or cofactor consumption. We should mention, however, that despite our repeated efforts, we have been unable to find evidence for overt circadian regulation of glutathione redox potential, the NAD+/NADH ratio, or the NADP+/NADPH ratio in any of the cell culture models employed in this study. This indicates that although PRX-SO2/3 rhythms may result from a cell-autonomous oscillation in cellular ROS production and/or redox potential, the oscillation's amplitude lies beneath the threshold of detection in our cell models, it does not appear to have any major role in the determination of circadian period, and it is, therefore, unlikely to constitute an essential timekeeping mechanism. Indeed, it is plausible that PRX-SO2/3 rhythms result from the rhythmic proteasomal degradation of over-oxidized PRX, as has been suggested in mouse erythrocytes (13).
Our data indicate that circadian gene expression is not affected by circadian oscillations in (NADH-generating) glycolytic flux, whereas chronic PPP inhibition and perturbation of redox balance primarily determine the amplitude of oscillation and not its period. This again suggests that circadian oscillations in redox homeostasis and primary metabolism do not play any essential role in the cellular timekeeping mechanism itself, whereas there exists a permissive range of PPP activity and redox state, beyond which clock gene expression rhythms cannot be sustained. We observed that the clock in U2OS cells, in particular, was profoundly resilient to reductive stress, with concentrations of 0.5 mM 2ME eliciting minimal effects on the amplitude and period of circadian rhythms in transcription (Supplementary Fig. S6D). We also observed no effect on circadian rhythms of the NRF2 activator sulforaphane or NRF2 RNAi, nor did NRF2 levels change consistently during PPP inhibition (Supplementary Fig. S10) (54). Again, a timekeeping mechanism that is effectively insensitive to chronic reductive stress is not readily compatible with redox oscillations playing some essential and conserved role in the cellular clockwork.
Because PPP inhibition and chronic redox disruption phenocopied the effect of 2DG on clock gene expression rhythms in three independent mammalian cell lines expressing three different bioluminescent reporters, we think it plausible that PPP/redox regulation of the amplitude of clock gene expression rhythms may reflect a general feature of how redox signals are integrated into the mammalian cellular circadian clockwork. We do note, however, that our findings using three different PPP inhibitors (2DG, DPI, DHEA) differ markedly from the reported effect of the niacin antagonist, 6-aminonicotinamide (6AN), which increases circadian period in (transformed) U2OS cells when employed at millimolar concentrations (61). 6AN becomes incorporated into NAD+ and NADP+ antimetabolites (6ANAD and 6ANADP), and thereby inhibits a wide range of NAD(P)+-dependent cellular processes (74). Unlike in U2OS cells however, we found that 6AN did not elicit period lengthening of PER2::LUC rhythms in non-transformed mouse fibroblasts (Supplementary Fig. S11). It is, therefore, possible either that the clock in these transformed cells, which rely heavily on Warburg metabolism (41), is more sensitive to 6AN or that the action of 6AN on the circadian clock is not specific to inhibition of the PPP.
In (non-transformed) mouse fibroblasts, we observed a phase-dependent and opposite sensitivity to reductive and oxidative stress in terms of the subsequent amplitude of clock gene expression rhythms. Specifically, cells were more tolerant to oxidative stress around the peak of PER2 expression (CT12), when combined glycolytic and PPP flux was greatest, and more tolerant to reductive stress around the nadir of PER2 expression, when the lowest rates of carbohydrate metabolism were observed. This establishes a functional consequence for circadian regulation of primary metabolism and redox balance that could have interesting chronotherapeutic applications (52). For example, in the context of pro-oxidant chemotherapies, one might anticipate that milder side effects will be observed in non-tumor cells (expressing normal circadian rhythms) when treatments coincide with the peak of PER2 expression, that is, occurring around anticipated dawn in humans (3, 20).
Circadian rhythms in the abundance of over-oxidized PRXs have been interpreted to indicate the existence of an underlying “redox oscillator” (60, 76). Our data strongly support the interpretation that redox signaling can elicit phase shifts, that NAD(P)H generation is rhythmic, and that the cellular redox environment appears to regulate the amplitude of circadian gene expression. However, our observations also clearly indicate that cellular timekeeping is remarkably robust against perturbation of redox balance in terms of the period of oscillation. We therefore conclude that our observations are most consistent with the third model that we introduced: that any circadian variations in redox balance are a cellular clock output, likely to result at least partially from rhythmic flux through primary metabolic pathways, but are not essential to cellular timekeeping. Moreover, redox signaling is competent to modulate the level of PER protein expression and thus, acute changes in redox balance can elicit phase-dependent circadian phase shifts. If our interpretation is correct, this would establish redox balance as an auxiliary feedback loop, akin to a zeitnehmer (63), a cellular system that is rhythmically regulated and can regulate the rhythm but is not essential to the timekeeping mechanism itself. Future work will need to test the potential physiological relevance of our findings, and the molecular mechanism that facilitates such rapid PER2 degradation in response to acute increases in intracellular H2O2.
Materials and Methods
Mammalian cell culture
Primary fibroblasts isolated from lung tissue (71) of adult PER2::LUC male mice [kindly provided by J. S. Takahashi (UT Southwestern, United States) (79)] and PER2::VENUS fibroblasts (kindly provided by M. Hastings [MRC LMB, United Kingdom (73)] were cultured as previously described (48) and immortalized by serial passage (77). U2OS and NIH3T3 cells were obtained from ATCC and used to generate transcriptional LUC-reporter cell lines as previously described (15, 48), and they were not passaged above 30 times. All cells were confirmed as being free of mycoplasma.
Silencing experiments
Cells were seeded in 24-well plates at a density of 1 × 105/well and transfected with HiPerFect transfection reagent (Qiagen) by using the manufacturer's recommended reverse transfection protocol. Media were changed after 20 h, and cells were allowed to grow to confluence for a further 48 h. Cells were then synchronized by using 100 nM dexamethasone for 2 h, changed into air medium and bioluminescence was recorded as described later. We observed more rapid damping and increased baseline bioluminescence in recently transfected cells using this protocol than in control cells of other experiments, which was controlled for in our analyses. In light of prior observations, this was most likely associated with an increased rate of intercellular desynchronization (48, 75). To assay for protein expression, parallel plates were lysed (in triplicate) for immunoblotting 96 h after transfection.
Bioluminescence recordings
Bioluminescence recordings under static conditions were performed in 20 mM HEPES-buffered air medium (48) supplemented with 2% B-27 (Life Technologies; 50 × ), 0.3 or 1 mM luciferin (Biosynth AG; 0.3 mM for U2OS cells and 1 mM for fibroblasts), 1 × glutamax (Life Technologies), 100 U/ml penicillin/100 μg/ml streptomycin, and 10% FetalClone™ II or III serum (HyClone™; II for U2OS and III for fibroblasts). Osmolality was adjusted to 350 mOsm with NaCl. Before each experiment, cells were synchronized by using temperature cycles (12 h 32°C–12 h 37°C) for at least 3 days. Experiments were begun immediately after a medium change from culture medium to air medium, with or without drug as indicated (at 0 h). Vehicle treatments were performed with water, dimethyl sulfoxide (DMSO), or ethanol as appropriate, with the final concentration of solvent never being more than 0.3% DMSO or 0.1% ethanol. Bioluminescence activity was measured in a Lumicycle (Actimetrics), an LB962 plate reader (Berthold Technologies), or an ALLIGATOR (Cairn Research). Lumicycle and plate reader measurements are reported as counts per second, whereas ALLIGATOR measurements are reported as relative luminescence units. For technical reasons, the bioluminescence data presented in Figure 1A and Supplementary Figure S2A were normalized to the first peak of PER2::LUC bioluminescence to account for differences in the efficiency of light detection, resulting from variable distances of microfluidic devices from the camera lens.
Metabolic labeling strategy
Classically, either singly or doubly 13C-labeled glucose has been used to investigate flux through the PPP (13, 25). Glucose passing though the PPP loses its first carbon molecule as CO2 during the oxidative phase, after which rearrangements in the non-oxidative PPP produce differently labeled metabolites to those from glucose that has passed solely through glycolysis, allowing for calculation of PPP flux. However, 13C may also be present at additional glucose carbons due to its natural abundance (∼1%) or due to impurities from the enrichment process. This has the potential to introduce error into calculations of the relative fluxes between glycolysis and the PPP by using either the singly or doubly labeled method. To avoid this problem, we utilized 1,2,3-13C-labeled glucose as our labeled substrate. Metabolism of 1,2,3-13C glucose through either glycolysis or the PPP produces fully 13C-labeled or completely unlabeled (12C) 3-carbon metabolites (lactate and pyruvate, Supplementary Fig. S1), which may serve as substrates for mitochondrial respiration or are exported into the cell culture media. 13C and 12C lactate/pyruvate are produced in different ratios depending on which metabolic pathways they have passed through: Flux through glycolysis produces a 1:1 ratio of 13C-labeled to 12C-unlabeled product, whereas flux through the PPP produces a ratio of 2:3 (Supplementary Fig. S1). Analysis of the ratios of these two metabolites allows for accurate determination of the relative metabolic fluxes between the two pathways. This method is not affected by three-carbon metabolites produced by other pathways as a result of impurities in the labeled glucose, as these will be singly or doubly labeled, allowing for more accurate determination of primary metabolic flux. For analysis of perfusion time courses, we only consider samples that were collected after the levels of 1,2,3-13C-glucose in the perfusate had attained a stable baseline, to avoid any potential confounding effects that might otherwise be introduced by unlabeled glucose from the media in which the cells were previously cultured.
Perfusion recordings
Recordings under perfusion were carried out by using cells seeded in micro-slide 0.6 (Ibidi). Immediately before recording, cells were changed into bicarbonate-buffered Dulbecco's modified Eagle medium (DMEM) containing 1 g/L d-glucose and supplemented with 2% Fetalclone III™ serum, B-27, glutaMAX, luciferin, penicillin, and streptomycin as for air medium. Cultures were then placed at 37°C, 5% CO2 in an ALLIGATOR and bioluminescence was recorded while cells were under continuous media perfusion at a rate of 50 μl/h. Media were kept at room temperature within 20 ml BD Plastipak syringes and were driven across the cells by using an NE-1600 programmable syringe pump. Out-flow media were collected on ice and, subsequently, snap frozen every 4 h.
Pharmacological perturbations
KA was supplied by 2BScientific. All other drugs and chemicals were used in concentrations as indicated, and they were from Sigma. DPI and carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP) were dissolved in DMSO, KA in EtOH, DHEA in MeOH, GOX in phosphate-buffered saline, and 6AN and oxythiamine in air medium. All other drugs were dissolved in H2O. We found that NIH3T3 fibroblast cultures required 1 mM mannose to remain viable in the presence of 2DG [presumably to provide a substrate for protein glycosylation (39)]. Similarly, we found that U2OS cells required 10 mM pyruvate to remain viable in the presence of KA [presumably providing a substrate for mitochondrial respiration to maintain ATP production (26)]. The 3T3 and U2OS experiments and controls reported in Supplementary Figure S3 and S5 only were, therefore, performed with either mannose or pyruvate supplementation, respectively. The non-transformed fibroblasts that were used for experiments reported in main figures were not so supplemented in any case.
ATP measurements
ATP measurements were performed as previously described (28).
Extracellular flux analysis
Measurements of ECAR (a reliable proxy for glycolytic rate) and rate of OCR (which correlates with cellular respiration rate) were performed with a Seahorse XFe96 analyzer (Agilent) by using the Seahorse XF Cell Mito Stress test and Glycolysis Stress Test Kits (Agilent). Cells were seeded in Seahorse XF96 cell culture microplates, grown to confluence, subjected to temperature cycles for 72 h, given a media change, and incubated at constant 37°C. Immediately before each measurement, a plate was removed from the incubator; media were then changed to Seahorse XF Media (Agilent) and warmed to 37°C on a 37°C isothermal pad. Seahorse XF Media were supplemented with 1% glutaMAX, and also 11 mM glucose and 200 μM pyruvate for those cells used with the Mito Stress Test. Plates were calibrated with Seahorse XF Calibrant (Agilent) before every test, as per the manufacturer's instructions. Basal respiration and glycolysis is determined before any drug treatment in cells but in the presence of glucose. Maximal respiration is determined after treatment with the proton ionophore FCCP. Maximal glycolytic capacity is determined after the addition of ATP synthase inhibitor oligomycin.
Metabolic labeling sample preparation
Cells were perfused (as described earlier) with 1,2,3-13C glucose substituted in place of unlabeled glucose. Outflow media were snap frozen. Subsequently, samples were defrosted and centrifuged in a bench-top centrifuge at 4°C, at maximum speed for 5 min. Fifty microliters of the resulting supernatant was resuspended in 750 μl extraction buffer (375 μl methanol, 225 μl acetonitrile +150 μl H2O) and then mixed at 1000 rpm for 15 min at 4°C in a ThermoMixer (Eppendorf). Samples were then centrifuged again at 4°C, at maximum speed in a bench-top centrifuge for 10 min, and the resulting supernatant was transferred to a new vial and stored at −80°C until analysis by LC-MS.
Cell culture medium analysis by LC-MS
The LC-MS analysis was performed on a Dionex U3000 UHPLC system coupled to a Q Exactive™ Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Scientific). The liquid chromatography system was fitted with a SeQuant ZIC-pHILIC column (150 mm × 2.1 mm. 3.5 μm) and guard column (20 mm × 2.1 mm 3.5 μm) from Merck Millipore. The mobile phase was composed of 20 mM ammonium carbonate and 0.1% ammonium hydroxide in water (solvent A), and acetonitrile (solvent B). The flow rate was set at 180 μl/min with the following gradient: 0 min 70% B, 1 min 70% B, 16 min 38% B, 16.5 min 70% B, and a hold at 70% B for 8.5 min. The operator was blinded to the samples, which were randomized to avoid bias due to machine drift. The acquired spectra were analyzed using XCalibur Qual Browser and XCalibur Quan Browser software (Thermo Scientific) by referencing to commercially available compounds and an internal library of compounds.
NADP+:NADPH ratio
Assays to determine NADP+:NADPH ratio were performed by using the NADP+/NADPH-Glo(TM) Assay (Promega) as per the manufacturer's instructions; 250 μM diamide was used as a positive control.
Data analysis
Bioluminescence data were analyzed with Prism Graphpad 6. Baseline subtraction was performed to detrend traces (24-h moving average or polynomial line fitting). Period, phase, and amplitude were determined by least-square fitting to a circadian damped cos wave with a linear baseline:
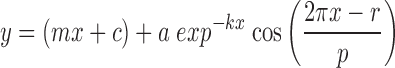 |
where m is the gradient of the baseline, c is the y offset, k describes the damping rate, a is the amplitude, r is the phase, and p is the period. The first 24 h of data were omitted from analysis to exclude the transient effects in clock gene expression that frequently occur on a media change. When indicated, the cos wave fit was compared with the straight-line fit; p-value indicates rejection of the straight line.
Recent work has revealed that luciferin itself (non-specifically) lengthens the circadian period by ∼1 h at high concentrations (≤2.5 mM) (29). To distinguish biologically relevant from statistically significant but non-specific effects on the cellular clock, informed by the excellent paper from the Ueda lab (36), we define a biologically relevant effect on period as being >5% and >3 standard deviations different than controls. In practice, this means that only changes in a period of >1 h are considered noteworthy.
Western blotting
Western blots were performed according to standard protocols, using NuPAGE™ Novex™ 4–12% Bis-Tris Protein Gels (ThermoFisher). Transfer to nitrocellulose membranes was achieved by using the iBlot dry blotting system (ThermoFisher). Blots were incubated with blocking buffer consisting of 0.125% bovine serum albumin (Sigma) and 0.125% milk powder (Marvel). PER2::VENUS was detected by using anti-GFP antibody (Sigma), G6PDH (Abcam; ab993), GPI (Abcam; 66340), NRF2 (Santa Cruz; sc-722), actin (Santa Cruz; sc-47778), and PRX-SO2/3 (Abcam; ab16830). Western blots were imaged on a Biorad chemidoc and quantifications were performed in FIJI (69), measuring integrated density of the bands. Protein expression was expressed relative to tubulin expression or Coomaasie stain of the gel, as indicated. Full annotated images for all blots are presented in Supplementary Figures S12 to S15.
Supplementary Material
Abbreviations Used
- 2DG
2-deoxyglucose
- 2ME
2-mercaptoethanol
- 3-Br-Pyr
3-bromopyruvate
- 6AN
6-aminonicotinamide
- ANOVA
analysis of variance
- Anti A
antimycin A
- CHX
cycloheximide
- CT
circadian time
- DHEA
dehydroepiandrosterone
- DMSO
dimethyl sulfoxide
- DPI
diphenyleneiodonium
- ECAR
extracellular acidification rate
- ETC
electron transport chain
- FCCP
carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone
- G6PDH
glucose-6-phosphate dehydrogenase
- Glc
glucose
- GOX
glucose oxidase
- GPI
phosphoglucose isomerase
- KA
koningic acid
- LC-MS
liquid chromatography coupled to mass spectrometry
- LUC
luciferase
- OCR
oxygen consumption rate
- PPP
pentose phosphate pathway
- PRX
peroxiredoxin
- ROS
reactive oxygen species
- ROT
rotenone
- SEM
standard error of the mean
- SFN
sulforaphane
- TCA
tricarboxylic acid cycle
Acknowledgments
The authors thank Alessandra Stangherlin, Rachel Edgar, and David Wong for valuable discussions. They would also like to thank all their reviewers for their constructive comments. M.P. was supported by the Dutch Cancer Foundation (KWF, BUIT-2014-6637) and EMBO (ALTF-654-2014). J.S.O. was supported by the Medical Research Council (MC_UP_1201/4) and the Wellcome Trust (093734/Z/10/Z).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Albrecht U, Albrecht U, Zheng B, Larkin D, Sun ZS, and Lee CC. mPer1 and mPer2 are essential for normal resetting of the circadian clock. J Biol Rhythms 16: 100–104, 2001 [DOI] [PubMed] [Google Scholar]
- 2.Altman BJ, Hsieh AL. Sengupta A, Krishnanaiah SY, Stine ZE, Walton ZE, et al. . MYC disrupts the circadian clock and metabolism in cancer cells. Cell Metab 22: 1009–1019, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archer SN, Laing EE, Moller-Levet CS, van der Veen DR, Bucca G, Lazar AS, et al. . Mistimed sleep disrupts circadian regulation of the human transcriptome. Proc Natl Acad Sci U S A 111: E682–E691, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, et al. . SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134: 317–328, 2008 [DOI] [PubMed] [Google Scholar]
- 5.Asher G. and Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab 13: 125–137, 2011 [DOI] [PubMed] [Google Scholar]
- 6.Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, and Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30: 525–536, 2001 [DOI] [PubMed] [Google Scholar]
- 7.Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, et al. . Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289: 2344–2347, 2000 [DOI] [PubMed] [Google Scholar]
- 8.Balsalobre A, Damiola F, and Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93: 929–937, 1998 [DOI] [PubMed] [Google Scholar]
- 9.Balsalobre A, Marcacci L, and Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured rat-1 fibroblasts. Curr Biol 10: 1291–1294, 2000 [DOI] [PubMed] [Google Scholar]
- 10.Barban S. and Schulze H. The effects of 2-deoxyglucose on the growth and metabolism of cultured human cells. J Biol Chem 236: 1887–1890, 1961 [PubMed] [Google Scholar]
- 11.Bass J. Circadian topology of metabolism. Nature 491: 348–356, 2012 [DOI] [PubMed] [Google Scholar]
- 12.Belden WJ, Larrondo LF, Froehlich AC, Shi M, Chen CH, Loros JJ, et al. . The band mutation in Neurospora crassa is a dominant allele of ras-1 implicating RAS signaling in circadian output. Genes Dev 21: 1494–1505, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brekke EMF, Walls AB, Schousboe A, Waagepetersen HS, and Sonnewald U. Quantitative importance of the pentose phosphate pathway determined by incorporation of 13C from [2-13C]- and [3-13C]glucose into TCA cycle intermediates and neurotransmitter amino acids in functionally intact neurons. J Cereb Blood Flow Metab 32: 1788–1799, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown SA. Circadian clock-mediated control of stem cell division and differentiation: beyond night and day. Development 141: 3105–3111, 2014 [DOI] [PubMed] [Google Scholar]
- 15.Burke TM, Markwald RR, Mchill AW, Chinoy ED, Snider JA, Bessman SC, et al. . Effects of caffeine on the human circadian clock in vivo and in vitro. Sci Transl Med 7: 1–8, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Causton HC, Feeney KA, Ziegler CA, and O'Neill JS. Metabolic cycles in yeast share features conserved among circadian rhythms. Curr Biol 25: 1056–1062, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho C-S, Yoon HJ, Kim JY, Woo HA, and Rhee SG. Circadian rhythm of hyperoxidized peroxiredoxin II is determined by hemoglobin autoxidation and the 20S proteasome in red blood cells. Proc Natl Acad Sci U S A 111: 12043–12048, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chokkathukalam A, Kim D-H, Barrett MP, Breitling R, and Creek DJ. Stable isotope-labeling studies in metabolomics: new insights into structure and dynamics of metabolic networks. Bioanalysis 6: 511–524, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen CE, Karlsson M, Winther JR, Jensen PR, and Lerche MH. Non-invasive in-cell determination of free cytosolic [NAD+]/[NADH] ratios using hyperpolarized glucose show large variations in metabolic phenotypes. J Biol Chem 289: 2344–2352, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuesta M, Cermakian N, and Boivin DB. Glucocorticoids entrain molecular clock components in human peripheral cells. FASEB J 29: 1360–1370, 2014 [DOI] [PubMed] [Google Scholar]
- 21.D'Alessandro M, Beesley S, Kim JK, Chen R, Abich E, Cheng W, et al. . A tunable artificial circadian clock in clock-defective mice. Nat Commun 6: 8587, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Damiola F, Le Minli N, Preitner N, Kornmann B, Fleury-Olela F, and Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14: 2950–2961, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickinson BC. and Chang CJ. Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat Chem Biol 7: 504–511, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunlap JC. Molecular bases for circadian clocks. Cell 96: 271–290, 1999 [DOI] [PubMed] [Google Scholar]
- 25.Dusick JR, Glenn TC, Lee WP, Vespa PM, Kelly DF, Lee SM, et al. . Increased pentose phosphate pathway flux after clinical traumatic brain injury: a [1,2-13C2]glucose labeling study in humans. J Cereb Blood Flow Metab 27: 1593–1602, 2007 [DOI] [PubMed] [Google Scholar]
- 26.Eberhart K, Renner K, Ritter I, Kastenberger M, Singer K, Hellerbrand C, et al. . Low doses of 2-deoxy-glucose sensitize acute lymphoblastic leukemia cells to glucocorticoid-induced apoptosis. Leukemia 23: 2167–2170, 2009 [DOI] [PubMed] [Google Scholar]
- 27.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, et al. . Peroxiredoxins are conserved markers of circadian rhythms. Nature 485: 459–464, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feeney KA, Hansen LLL, Putker M, Olivares-Yañez C, Day J, Eades LJ, et al. . Daily magnesium fluxes regulate cellular timekeeping and energy balance. Nature 532: 375–379, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feeney KA, Putker M, Brancaccio M, and ONeill JS. In-depth characterization of firefly luciferase as a reporter of circadian gene expression in mammalian cells. J Biol Rhythms 31: 540–550, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feneberg R. and Lemmer B. Circadian rhythm of glucose uptake in cultures of skeletal muscle cells and adipocytes in wistar-kyoto, wistar, goto-kakizaki, and spontaneously hypertensive rats. Chronobiol Int 21: 521–538, 2004 [DOI] [PubMed] [Google Scholar]
- 31.Gordon G, Mackow MC, and Levy HR. On the mechanism of interaction of steroids with human glucose 6-phosphate dehydrogenase. Arch Biochem Biophys 318: 25–29, 1995 [DOI] [PubMed] [Google Scholar]
- 32.Gyöngyösi N, Nagy D, Makara K, Ella K, and Káldi K. Reactive oxygen species can modulate circadian phase and period in Neurospora crassa. Free Radic Biol Med 58: 134–143, 2013 [DOI] [PubMed] [Google Scholar]
- 33.Hastings MH, Reddy AB, and Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci 4: 649–661, 2003 [DOI] [PubMed] [Google Scholar]
- 34.Hirayama J, Cho S, and Sassone-Corsi P. Circadian control by the reduction/oxidation pathway: catalase represses light-dependent clock gene expression in the zebrafish. Proc Natl Acad Sci U S A 104: 15747–15752, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holmström KM. and Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol 15: 411–421, 2014 [DOI] [PubMed] [Google Scholar]
- 36.Isojima Y, Nakajima M, Ukai H, Fujishima H, Yamada RG, Masumoto K, et al. . CKIepsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci U S A 106: 15744–15749, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kil IS, Lee SK, Ryu KW, Woo HA, Hu MC Bae SH, et al. . Feedback control of adrenal steroidogenesis via H2O2-dependent, reversible inactivation of peroxiredoxin III in mitochondria. Mol Cell 46: 584–594, 2012 [DOI] [PubMed] [Google Scholar]
- 38.Krishnaiah SY, Wu G, Altman BJ, Growe J, Rhoades SD, Coldren F, et al. . Clock regulation of metabolites reveals coupling between transcription and metabolism. Cell Metab 25: 961–974.e4, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurtoglu M, Gao N, Shang J, Maher JC, Lehrman MA, Wangpaichitr M, et al. . Under normoxia, 2-deoxy-d-glucose elicits cell death in select tumor types not by inhibition of glycolysis but by interfering with N-linked glycosylation. Mol Cancer Ther 6: 3049–3058, 2007 [DOI] [PubMed] [Google Scholar]
- 40.Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, et al. . AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326: 437–440, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liberti MV. and Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci 41: 211–218, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, and Kay SA. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet 4: e1000023, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, et al. . Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288: 483–492, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meng Q-JJ, Logunova L, Maywood ES, Gallego M, Lebiecki J, Brown TM, et al. . Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron 58: 78–88, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohawk JA, Green CB, and Takahashi JS. Central and Peripheral circadian clocks in mammals. Annu Rev Neurosci 35: 445–462, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, et al. . The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134: 329–340, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nangle SN, Rosensweig C, Koike N, Tei H, Takahashi JS, and Carla B. Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex. Elife 3: e03674, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Neill JS. and Hastings MH. Increased coherence of circadian rhythms in mature fibroblast cultures. J Biol Rhythms 23: 483–488, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Neill JS, Maywood ES, and Hastings MH. Cellular mechanisms of circadian pacemaking: beyond transcriptional loops. Handb Exp Pharmacol 67–103, 2013 [DOI] [PubMed] [Google Scholar]
- 50.O'Neill JS, Ooijen GV, Dixon LE, Troein C, Corellou F, Bouget F-Y, et al. . Circadian rhythms persist without transcription in a eukaryote. Nature 469: 554–558, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Neill JS. and Reddy AB. Circadian clocks in human red blood cells. Nature 469: 498–503, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ortiz-Tudela E, Innominato PF, Rol MA, Lévi F, and Madrid JA. Relevance of internal time and circadian robustness for cancer patients. BMC Cancer 16: 285, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peek CB, Affinati AH, Ramsey KM, Kuo H-Y, Yu W, Sena LA, et al. . Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science 342: 1243417, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pekovic-Vaughan V, Gibbs J, Yoshitane H, Yang N, Pathiranage D, Guo B, et al. . The circadian clock regulates rhythmic activation of the NRF2/glutathionemediated antioxidant defense pathway to modulate pulmonary fibrosis. Genes Dev 28: 548–560, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pietzke M, Zasada C, Mudrich S, and Kempa S. Decoding the dynamics of cellular metabolism and the action of 3-bromopyruvate and 2-deoxyglucose using pulsed stable isotope-resolved metabolomics. Cancer Metab 2: 9, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Putker M. and O'Neill JS. Reciprocal control of the circadian clock and cellular redox state—a critical appraisal. Mol Cells 39: 6–19, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, et al. . Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol 14: 1207–1213, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramsey KM, Yoshino J, Brace SC, Abrassart D, Kobayashi Y, Mercheva B, et al. . Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324: 651–654, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reppert SM. and Weaver DR. Coordination of circadian timing in mammals. Nature 418: 935–941, 2002 [DOI] [PubMed] [Google Scholar]
- 60.Rey G. and Reddy AB. Interplay between cellular redox oscillations and circadian clocks. Diabetes Obes Metab 17: 55–64, 2015 [DOI] [PubMed] [Google Scholar]
- 61.Rey G, Valekunja UK, Feeney KA, Wulund L, Milev NB, Stangherlin A, et al. . The pentose phosphate pathway regulates the circadian clock. Cell Metab 24: 1–12, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Riganti C, Gazzano E, Polimeni M, Costamagna C, Bosia A, and Ghigo D. Diphenyleneiodonium inhibits the cell redox metabolism and induces oxidative stress. J Biol Chem 279: 47726–47731, 2004 [DOI] [PubMed] [Google Scholar]
- 63.Roenneberg T. and Merrow M. Life before the clock: modeling circadian evolution. J Biol Rhythms 17: 495–505, 2002 [DOI] [PubMed] [Google Scholar]
- 64.Roenneberg T. and Merrow M. The circadian clock and human health. Curr Biol 26: R432–R443, 2016 [DOI] [PubMed] [Google Scholar]
- 65.Saini C, Morf J, Stratmann M, Gos P, and Schibler U. Simulated body temperature rhythms reveal the phase-shifting behavior and plasticity of mammalian circadian oscillators. Genes Dev 26: 567–580, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sakai K, Hasumi K, and Endo A. Inactivation of rabbit muscle glyceraldehyde-3-phosphate dehydrogenase by koningic acid. Biochim Biophys Acta 952: 297–303, 1988 [DOI] [PubMed] [Google Scholar]
- 67.Sancar A, Lindsey-Boltz LA, Kang T-H, Reardon JT, Lee JH, and Ozturk N. Circadian clock control of the cellular response to DNA damage. FEBS Lett 584: 2618–2625, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schibler U, Gotic I, Saini C, Gos P, Curie T, Emmenegger Y, et al. . Clock-talk: interactions between central and peripheral circadian oscillators in mammals. Cold Spring Harb Symp Quant Biol 80: 223–232, 2016 [DOI] [PubMed] [Google Scholar]
- 69.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. . Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmalen I, Reischl S, Wallach T, Klemz R, Grudziecki A, Prabu JR, et al. . Interaction of circadian clock proteins CRY1 and PER2 Is modulated by zinc binding and disulfide bond formation. Cell 157: 1203–1215, 2014 [DOI] [PubMed] [Google Scholar]
- 71.Seluanov A, Vaidya A, and Gorbunova V. Establishing primary adult fibroblast cultures from rodents. J Vis Exp pii: 2033, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, et al. . Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell 91: 1043–1053, 1997 [DOI] [PubMed] [Google Scholar]
- 73.Smyllie NJ, Pilorz V, Boyd J, Meng Q-J, Saer B, Chesham JE, et al. . Visualizing and quantifying intracellular behavior and abundance of the core circadian clock protein PERIOD2. Curr Biol 26: 1–7, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Street JC, Mahmood U, Ballon D, Alfieri AA, and Koutcher JA. 13C and 13P NMR investigation of effect of 6-aminonicotinamide on metabolism of RIF-1 tumor cells in vitro. J Biol Chem 271: 4113–4119, 1996 [DOI] [PubMed] [Google Scholar]
- 75.Welsh DK, Yoo S-H, Liu AC, Takahashi JS, and Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol 14: 2289–2295, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu L. and Reddy AB. Rethinking the clockwork: redox cycles and non-transcriptional control of circadian rhythms. Biochem Soc Trans 42: 1–10, 2014 [DOI] [PubMed] [Google Scholar]
- 77.Xu J. Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr Protoc Mol Biol Chapter 28: 1–8, 2005 [DOI] [PubMed] [Google Scholar]
- 78.Xue M, Momiji H, Rabbani N, Barker G, Bretschneider T, Shmygol A, et al. . Frequency modulated translocational oscillations of Nrf2 mediate the antioxidant response element cytoprotective transcriptional response. Antioxid Redox Signal 23: 613–629, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yoo S-H, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, et al. . PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A 101: 5339–5346, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yoshida Y, Iigusa H, Wang N, and Hasunuma K. Cross-talk between the cellular redox state and the circadian system in Neurospora. PLoS One 6: e28227, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yoshii K, Ishijima S, and Sagami I. Effects of NAD(P)H and its derivatives on the DNA-binding activity of NPAS2, a mammalian circadian transcription factor. Biochem Biophys Res Commun 437: 386–391, 2013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.