Abstract
Significance: Peroxiredoxins (Prxs) are thiol peroxidases with multiple functions in the antioxidant defense and redox signaling network of the cell. Our progressing understanding assigns both local and global significance to plant Prxs, which are grouped in four Prx types. In plants they are localized to the cytosol, mitochondrion, plastid, and nucleus. Antioxidant defense is fundamentally connected to redox signaling, cellular communication, and acclimation. The thiol–disulfide network is central part of the stress sensing and processing response and integrates information input with redox regulation.
Recent Advances: Prxs function both as redox sensory system within the network and redox-dependent interactors. The processes directly or indirectly targeted by Prxs include gene expression, post-transcriptional reactions, including translation, post-translational regulation, and switching or tuning of metabolic pathways, and other cell activities. The most advanced knowledge is available for the chloroplast 2-CysPrx wherein recently a solid interactome has been defined. An in silico analysis of protein structure and coexpression reinforces new insights into the 2-CysPrx functionality.
Critical Issues: Up to now, Prxs often have been investigated for local properties of enzyme activity. In vitro and ex vivo work with mutants will reveal the ability of Prxs to interfere with multiple cellular components, including crosstalk with Ca2+-linked signaling pathways, hormone signaling, and protein homeostasis.
Future Directions: Complementation of the Prxs knockout lines with variants that mimic specific states, namely devoid of peroxidase activity, lacking the oligomerization ability, resembling the hyperoxidized decamer, or with truncated C-terminus, should allow dissecting the roles as thiol peroxidase, oxidant, interaction partner, and chaperone. Antioxid. Redox Signal. 28, 609–624.
Keywords: : thiol peroxidase, signaling, post-translational modifications, redox network, 2-cysteine peroxiredoxin, interactome, chloroplast
Biochemical Features and Dynamics of Peroxiredoxins
The central chemical group in peroxiredoxins (Prxs) is the cysteinyl thiol with low pK value, the peroxidatic CysP. Together with plant glutathione peroxidases that in animals and Chlamydomonas function with a selenocysteine instead of the catalytic cysteine found in plants, Prxs constitute the thiol peroxidases of plant cells (23, 32). The thiol-based mechanism is independent of cofactors such as metal ions or prosthetic groups. Thus thiol peroxidases employ a simple nucleophilic attack mechanism in the primary catalytic reaction (46, 100).
Circumstantial evidence indicates that Prxs are ancient high-affinity enzymes for peroxide detoxification when atmospheric oxygen still was very low (31). After formation of the sulfenic acid derivative by reaction of CysP with the peroxide substrate, the CysP thiol is subjected to various redox modifications ranging from disulfide, sulfinic to sulfonic acid derivatives. The site of the resolving cysteine (CysR) and the quaternary interaction interfaces define the distinct forms of Prxs that have emerged during evolution and exhibit differential specificities toward peroxide substrates, distinct subcellular localization, and regulatory capabilities. The often highly abundant Prxs stabilize redox homeostasis, transmit redox information, and control cell metabolism, in particular, photosynthesis.
Recent research focused on the spatial and temporal occurrence of post-translational modifications (PTMs) of Prxs and their mechanistic coupling to redox reaction chains. Oxidations on thiols occur with increasing oxidation number starting with disulfide (S–S), sulfenic (–SOH), sulfinic (–SO2H) to sulfonic (–SO3H) acid. Except for the sulfonic acid, each of these modifications bears functional significance. Reduction is needed for regeneration (26, 106).
Disulfide formation in 2-CysPrx causes a major conformational switch because the C-terminus carrying the CysR residue has to locally unfold and move to the sulfenylated peroxidatic cysteine. This mechanism destabilizes the dimer–dimer interface resulting in loss of decamer integrity (121). This effect can be analyzed by size exclusion chromatography (SEC) wherein disulfide-bridged oxidized Arabidopsis 2-CysPrx separates as dimer, whereas the reduced form separates as decamer and dimer (62). The oxidized dimers may ease rereduction by NADPH-dependent thioredoxin reductase C (NTRC) or thioredoxins (Trxs) such as Trx-x, CDSP, or Trx-m (26, 96).
Hyperoxidation of the CysP to sulfinic acid in the H2O2 detoxification cycle occurs with a certain frequency of about 1 out of 250 turnovers for Arabidopsis thaliana 2-CysPrx when exposed to a constant H2O2 generation rate of 1 μM H2O2 min−1 from a glucose oxidase system (62). Hyperoxidation opens up additional possibilities for signaling because the hyperoxidized decamer adopts a more stable conformation than the reduced decamer and at least in vitro acts as chaperone preventing precipitation of citrate synthase and insulin at high temperature and reducing conditions, respectively (59). Moreover, S-nitrosylation (36, 104), phosphorylation (see Table 2), Cys-glutathionylation (84), and Lys-acetylation (83) are reported PTMs of Prxs, which, in turn, alter Prx properties. Thus activity and function of Prxs are integrated into multiple signaling pathways as discussed hereunder.
Table 2.
Phosphorylation Sites of Peroxiredoxins and Selected Redox Proteins
| Protein | Protein ID | p-site | Literature |
|---|---|---|---|
| AtPrxIIE | Q949 U7 | 80PDsTL84 | 7 |
| 129FVsKA133 | 103 | ||
| AtPrxIIF | Q9 M7T0 | 35EGtDI39 | 7 |
| 51ARsWD55 | 103 | ||
| 138EFyGD142 | 123 | ||
| 147HKsLG151 | 7 | ||
| AtPrxQ | Q9LU86 | 138DDsAS142 | 103 |
| At2-CysPrxA | Q96291 | 96KLsDY100 | 103 |
| 159RKsGG163 | 114 | ||
| 208HStIN212 | 103, 114 | ||
| 257KLsKEY262 | 103 | ||
| At2-CysPrxB | Q9C5R8 | 214QHsTI218 | 114 |
| 215HStIN219 | 103, 114 | ||
| 264KLsKEY269 | 103 | ||
| Mt2-CysPrxB | G7ZUV5 | 158RKsGG162 | 105 |
| 179SKsYG183 | 105 | ||
| AtCDSP32 | Q9SGS4 | 117ELsRT121 | 70 |
| 233SRsMS237 | 114 | ||
| 237SMsET241 | 99, 103, 114 | ||
| AtACHT1 | Q8LEK4 | 202VGsIS206 | 103 |
| AtTrx-m2 | Q9SEU8 | 178TSsLD182 | 37 |
| AtTrx-m4 | Q9SEU6 | 156IRsVP160 | 103 |
| 171KDsII175 | 103 | ||
| AttAPX | Q42593 | 79AAsDA83 | 70 |
| 213PPsPA217 | 103 | ||
| 371SDsMK375 | 103 | ||
| MttAPX | MEDTR3G088160 | 228PpsPA232 | 105 |
| 255AHtLG259 | 105 | ||
| 367DGsPN371 | 105 | ||
| AtsAPX | Q42592 | 234PPsPA238 | 78, 116, 117 |
| 237PAtHL241 | 116 | ||
| MtAPX1 | G7JRI5 | 2GKsYP6 | 105 |
| 5YPtVS9 | 105 | ||
| 47FDsKT51 | 105 | ||
| 211LLsDP215 | 105 | ||
| 241KLsEL245 | 105 | ||
| MtAPX4 | MEDTR5G064610 | 157GLsAA161 | 105 |
| AtGPX1 | P52032 | 176GPsTA180 | 123 |
| AtGPX2 | O04922 | 165GQAsS169 | 128 |
| 165GQASs169 | 103 | ||
| AtFD1 | O04090 | 25LRsLP29 | 70 |
| 40LKsTAR45 | 70 | ||
| 41KStAR45 | 70 |
Phosphorylation sites (p-site) with phosphorylated serine, threonine, and tyrosine (small letters s, t, y in bold) of peroxiredoxins, thioredoxins, ascorbate peroxidases, glutathione peroxidases, CDSP32, and ACHT1 revealed by employing databases mentioned on the next page (48, 127). The sequence containing the modified aminoacid (aa) was arbitrarily set 5–6 aa long. Aminoacid positions refer to primary sequence of the proteins as counted from N to C terminus with transit peptide included. Following proteins were queried: superoxide dismutase, PrxIIE, PrxIIF, PrxQ, 2-CysPrx A, 2-CysPrx B, Apx, ascorbate peroxidase, tApx, thylakoid ascorbate peroxidase, sApx, stromal ascorbate peroxidase; chloroplast Gpx, chloroplast; Trx members (Trx-f1, -f2, m1-4, -x, -y1, -y2,-z), Trx-like proteins (NTRC, CDSP32, ACHT 1–4 [atypical cysteine histidine-rich Trx], and APR 1–3 [protein disulfide isomerase-like proteins]), Fd, ferredoxin 1; FNR, ferredoxin-dependent NADPH reductase 1.
P-sites were found in the cited references.
Basically, there exist two common but in fact highly related classifications of Prxs. Prxs are assigned to six distinct groups based on their amino acid sequence, cysteine localization, interaction interface, and N-/C-terminal extensions. Classification and naming of Prxs changed along with novel discoveries, improved analytical methods, and also depending on the organism. Table 1 provides an overview on the Prx nomenclature. The basic classification follows that of (49). Additional information was retrieved from the Prx classification index (PREX) and concerned sequence and structural features (111). Types A, B, C, and D Prxs are present in plants, whereas types E and F are only found in some bacteria. Prxs are often classified in typical 2-cysteine (type A), typical 1-cysteine (type B), and atypical 2-cysteine Prxs, which cover most Prxs. The latter are separated into PrxQ (type C) and type II Prx (type D). Additional features distinguishing Prx types concern the regeneration and subunit composition and interaction mechanisms. The assignment considers the position of the peroxidatic nucleophilic cysteine (CysP) and the presence, position, or absence of a CysR capable of reacting with oxidized CysP (sulfenic acid). The minimal unit can be monomer or dimer. In type A Prx, dimers form intermolecular disulfide bridges (+CysR). Dimers of type B Prx lack the CysR and are linked noncovalently. Type C monomers carry a CysR and generate intramolecular disulfide bonds. Type D Prxs form intramolecular disulfide bonds with CysR as well and noncovalently linked dimers. Trxs, NTRC, glutaredoxin (Grx)/glutathione (GSH), or other electron donors mentioned in the text restore the catalytically active CysP in Prxs belonging to types A, C, and D. In the CysR-free type B Prx, the sulfenic acid immediately reacts with a thiol or another reductant such as ascorbate. The review by Dietz (31) covers many aspects of the Prx family in plants. A genome-wide search allowed us to define the minimum set of six Prxs expressed in all higher plants [31], namely one 1-CysPrx related to embryogenesis, one chloroplast PrxQ, one plastid 2-CysPrx, and one each of cytosolic, mitochondrial, and plastidic type II Prxs. Variation of this minimum set of Prxs frequently occurs with multiple isoforms such as two 2-CysPrxs and three cytosolic PrxIIB-D in A. thaliana, but two Prx II in poplar (PrxIIB, PrxIIC) and only one in rice (PrxIIB).
Table 1.
Overview of Prx Subfamilies in Plants, Mammals, and Yeast
| Plant | Mammal | ||||||
|---|---|---|---|---|---|---|---|
| Type | Arabidopsis thaliana(1) | Oryza sativa(1) | Beta vulgaris(2) | Homo sapiens(1) | Mus musculus(1) | Rattus norvegicus(1) | Yeast Saccharomyces cerevisiae(1) |
| A | 2-CysPrxA (cl) Q96291 | 2-CysPrxA (cl) Q6ER94 | 2-CysPrx (cl) XM_010685313.1 | Prdx(ct) Q06830 | Prdx1(ct) P35700 | Prdx1(ct) Q63716 | Prx1, Tsa1 (ct) P34760 |
| 2-CysPrxB (cl) Q9C5R8 | — | — | Prdx2(ct) P32119 | Prdx2(ct) Q61171 | Prdx2(ct) P35704 | Prx2, Tsa2 (ct) Q04120 | |
| Prdx3(mt) P30048 | Prdx3(mt*) P20108 | Prdx3(mt*) Q9Z0 V6 | |||||
| Prdx4(ct) Q13162 | Prdx4(ct*) O08807 | Prdx4 (ct*) Q9Z0 V5 | |||||
| B | 1-CysPrx (ct, n) O04005 | 1-CysPrx (ct, n*) P0C5C9 | 1-CysPrx (ct, n) XM_010669313.1 | Prdx6(ct) P30041 | Prdx6(ct) O08709 | Prdx6(ct) O35244 | mTpx (mt) P34227 |
| C | PrxQ (cl) Q9LU86 | PrxQ (cl) P0C5D5 | PrxQ (cl) XM_010689254.1 | — | — | — | nTpx (n) P40553 |
| D | PrxIIA (—) Q7G959 | — | — | Prdx5(ct,mt,pr) P30044 | Prdx5 (ct,mt,pr) P99029 | Prdx5 (ct,mt,pr) Q9R063 | cTpx3 (ct) P38013 |
| PrxIIB (ct) Q9XEX2 | PrxIIB (ct) Q9FR35 | PrxIIB (ct) XM_010693075.1 | |||||
| PrxIIC (ct) Q9SRZ4 | — | — | |||||
| PrxIID (ct) O22711 | — | — | |||||
| PrxIIE (cl) Q949 U7 | PrxIIE1 (cl*) Q69TY4 | PrxIIE (cl) XM_010685588.1 | |||||
| PrxIIF (mt) Q9 M7T0 | PrxIIE2 (cl*) Q7F8S5 | — | |||||
| PrxIIF (mt*) Q9SDD6 | PrxIIF (mt) XM_010696978.1 | ||||||
Based on Hofmann et al. (49), Prxs were sorted with regard to their respective biochemical property of peroxidative cysteines and structure. In addition to the two plant model organisms, the newly sequenced genome of Beta vulgaris was included as additional example (51). For further information concerning subcellular localization, function, biochemical properties, and crystallographic data of annotated proteins, see the respective Uniprot entry (1) or reference sequence for B. vulgaris (2). Subcellular localization is given in parentheses as follows: nucleus = n, cytosol = ct, chloroplast = cl, mitochondria = mt, endoplasmatic reticulum = er, peroxisome = pr, not known = —. An * indicates manual assertion inferred from sequence similarity.
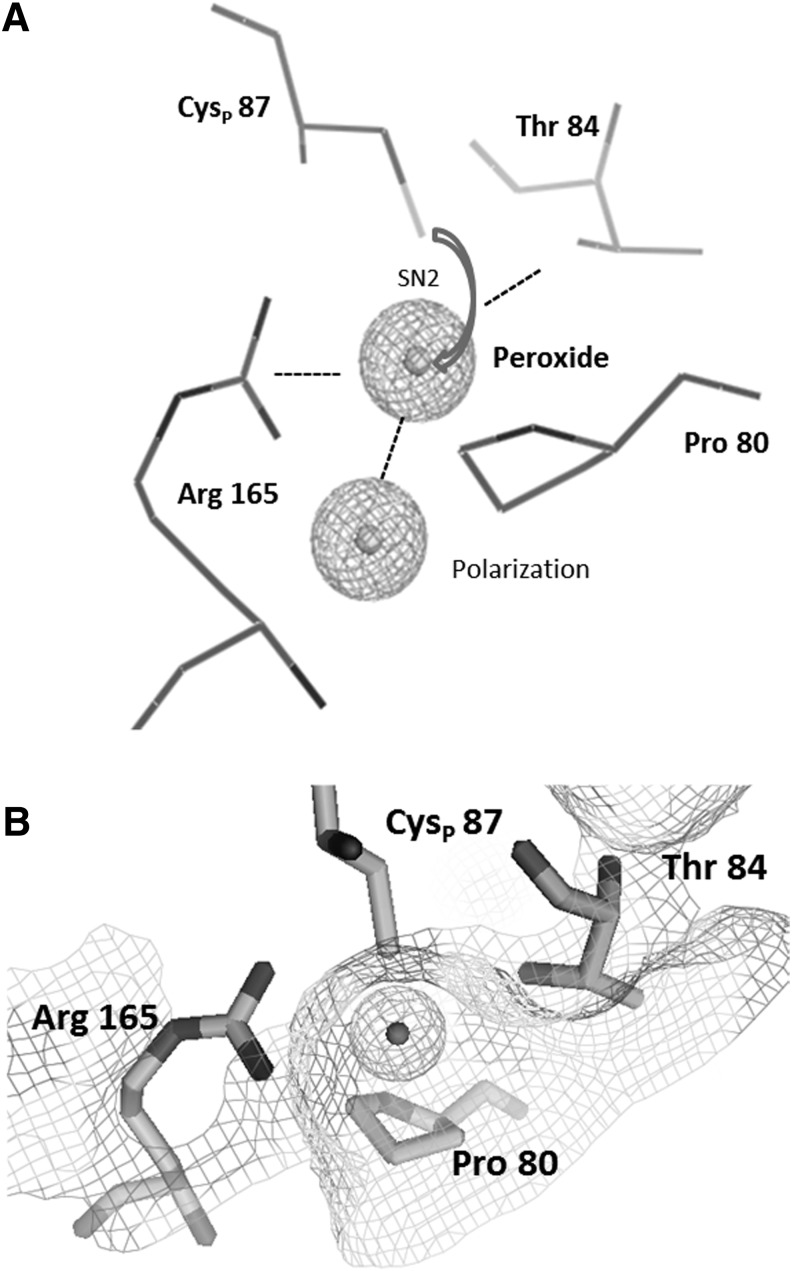
All Prxs and glutathione peroxidases (Gpxs) have a highly conserved structure based on the typical Trx-like fold. Based on this observation, the hypothesis emerged that all Prxs have a common Trx ancestor. The proper Trx-like fold is built from a βαβββα structure (97). The expanded structure of Prxs consists of seven-stranded β-sheets surrounded by five α-helices in β1-β2-α1-β3-α2-β4-α3-β5-β6-α4-β7-α5 topology. The properties of the catalytic center composed of the CysP in its conserved environment were analyzed by crystallography, bioinformatics, and activity assays. These amino acids are arranged in a reaction pocket between sheets β3 and β6, helix α2, and the loop connecting α2 and β3 (81). This position is conserved whereas the resolving Cys is variably positioned in helices α2, α3, and α5 or in the C-terminal region for intermolecular resolving reaction (91). The physicochemical properties of the specified amino acid residues combined with a nucleophilic displacement reaction (SN2) provide a framework for the enzymatic mechanism (Fig. 1). The Arg and Thr/Ser together with the Cys thiolate support the binding of peroxides. The oxygen–thiolate interaction polarizes the terminal oxygen to facilitate the Cys–S–O bond formation. The hydrophobic Pro protects the highly active cysteine site from exposure to the solvent and promotes the high peroxide specificity. Amino acid sequence comparison between At2-CysPrx, PsPrxIIF, and BtPrx3 underlines a conserved positioning for flanking amino acids and peroxidative cysteine in different Prxs and across species such as BtPrx3 102PXXXTXXCX…XR185, PsPrxIIF 80PXXXTXXCX…XR165, and At2-CysPrx 162PXXXTXXCX…XR195. Around 83% of all CysP display a pKa between 5.1 and 6.3 and thus are present in the thiolate form (Cys-S−) at physiological pH of, for example, 7.2 in the cytosol (93). The nucleophilic characteristics of the catalytic center originating from deprotonation at low-pH value (low pKa) generates a rather efficient peroxide detoxification microenvironment with high substrate affinity in the low micromolar range (39, 93). However, the turnover number is low with 10–250 reactions/min. The catalytic efficiency of Prxs is in the range of 105–108 M−1 s−1 and depends strongly on the thiol regeneration mechanism (96). The variation by three orders of magnitudes points to diversification of Prxs across all kingdoms. As shown for AhpC in Salmonella (86) and Prdx2 in human systems (66), an efficiency of 107 M−1 s−1 for H2O2 is similar to catalase (107 M−1 s−1) or selenocysteine-based Gpx (108 M−1 s−1) (122).
FIG. 1.
Peroxide binding pocket with conserved amino acids. (A) Graphical presentation of the catalytic center of the mitochondrial PrxIIF of pea. The template was accessed from RCSB PDB with the model of 2PWJ (2007) at 2.8 Å resolution. The conserved amino acids CysP 87, Arg 165, Thr 84, and Pro 80 are depicted using PyMOL (29). The peroxyl moiety was placed in appropriate position in the catalytic reaction pocket to simulate peroxide binding. (B) Potential surface mesh is laid on the active site residues and the terminal oxygen of substrate.
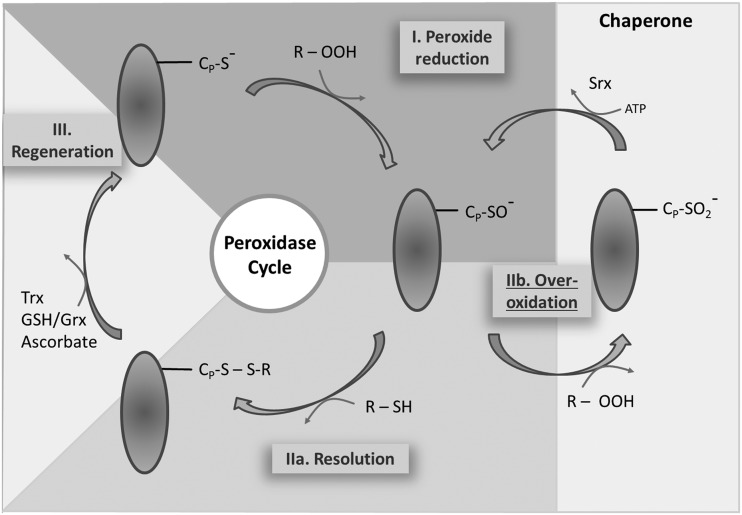
The thiol peroxidase cycle consists of three steps: the reduction of the peroxide substrate and generation of the sulfenic acid form of CysP, the resolution of the sulfenic acid to release water, and the regeneration of the thiol by electron transmitters (Fig. 2). (I) After binding of the peroxide to CysP, the oxygen–oxygen bond of the peroxide is disrupted and the corresponding alcohol from alkylhydroperoxide, water from H2O2, and nitrite from peroxinitrite are released. The active thiol group of the CysP is converted to a sulfenic acid (–SOH) group. (IIa) The CysR on the same or a different molecule attacks the sulfenic acid and forms an inter- or intramolecular disulfide bond (Cys–S–S–Cys). The disulfide bonded Prx adopts the inactive form. (IIb) Peroxide competes with the CysR for the sulfenic acid group. A second oxidation can take place before the resolving step occurs. This is considered as a regulatory mechanism that allows for inactivation and local peroxide accumulation in line with the flood gate model. The sulfinic acid is described as hyperoxidized form of Prxs. Its conversion to the active thiol peroxidase requires an additional regeneration step by sulfiredoxin (Srx) (94). (III) In the final step, the disulfide is reduced by specific redox transmitters such as Trxs, NTRC, Grx in combination with GSH, and ascorbate (reviewed in Ref. 31).
FIG. 2.
Schematics of the peroxidase cycle and hyperoxidation cycle of Prxs. (I) Reduced Prx scavenges peroxides (H2O2) releasing water. The peroxidatic cysteine (CysP) forms a sulfenic acid derivative. (IIa) In the resolving step, a second cysteine (CysR) attacks the sulfenic acid to form an inter- or intramolecular disulfide. (IIb) The reaction with an additional peroxide may hyperoxidize CysP to the sulfinic acid form. Hyperoxidation of 2-CysPrx can be retroreduced by the ATP-dependent sulfiredoxin, returning sulfenic forms into the cycle. (III) The regeneration step is catalyzed by a thiol reductant-like NTRC or Trx, reducing the catalytic and resolving cysteine and converting the Prxs to an active thiol peroxidase.
Prx Properties of Typical 1-CysPrx and 2-CysPrx
1-cysteine peroxiredoxin
Typical 1-cysteine peroxiredoxins (1-CysPrxs) belong to the B-type class and have a homodimer interface like A-type Prxs. 1-CysPrxs carry the CysP but lack the CysR. The sulfenic acid is resolved by other interaction partners. Information on plant 1-CysPrxs is still scarce. They are preferentially expressed in seeds wherein, for example, in barley they are found in the nucleus of aleurone and scutellum cells (113). Owing to its mostly confined expression in seeds, 1-CysPrx has mostly moved out of focus of current redox research. In 2009, Pulido and coworkers described the NTR/Trx system as electron donor for regeneration of 1-CysPrx (95). Enzyme assays with heterologously expressed wheat 1-CysPrx showed NTR-dependent peroxidase activity as well as localization in the nucleus as shown by immunolabeling. With 70 kDa, the molecular mass of NTR dimers is too big to diffuse through nuclear pores and thus the translocation mechanism to the nucleus remains elusive in the light of the missing nuclear localization sequence (95). It may be speculated that 1-CysPrx is taken piggyback by other cargo. Oxidized 1-CysPrx can be regenerated with ascorbate as electron donor. The efficiency appears to be low compared with that with the Trx system both for yeast mTpx and rat Prdx6. The antioxidant role of erythroascorbate remains a matter of debate despite the fact that the kcat/KM values for erythroascorbate with 104 M−1s−1 were one order of magnitude below that of Trx with 105 M−1s−1 (72). A recent report compared kinetic reactivity of the 1-CysPrx sulfenic acid with ascorbate across many species (3). The catalytic efficiencies kcat/KM for H2O2 decomposition were similar for yeast, bacteria, plant, and human 1-CysPrx with values of around 103 M−1s−1. The significance of the ascorbate-dependent regeneration in plants is still difficult to judge. Its contribution likely depends on the relative concentration of the various reductants, because in addition to Trx also GSH and glutathione-S-transferase and glutaredoxin, respectively (GST/GSH, Grx/GSH), regenerate 1-CysPrx at least in mammalia (64) and yeast (87). The link of 1-CysPrx to various regeneration systems may increase the viability of seeds facing harsh environmental conditions. Indeed Arabidopsis seeds overexpressing the PER1 homologue of Nelumbo nucifera (24) germinated better by 25%–50% than wild type (WT) in controlled deterioration treatments. The seedlings accumulated fivefold more biomass when grown on ABA for 10 days. These observations and increased germination at 50°C underpin the crucial role of 1-CysPrx in the redox network of seeds.
2-cysteine peroxiredoxin
Typical 2-CysPrxs interact through an obligate monomer–monomer interface orientated in parallel to the β-sheet bundles (B-type interface) in the Trx fold. In addition, intermolecular thiol–disulfide bonds are formed in the homodimer (46). 2-CysPrx is a highly abundant protein in the chloroplast stroma (100–120 μM in total for both 2-CysPrx A and B in A. thaliana) and likely involved in redox regulation in both the stroma and near the thylakoids (58, 89). Immunolabeling of 2-CysPrx with gold nanoparticles-conjugated antibodies revealed a location vicinal to thylakoid membranes of barley chloroplasts for 72% of all counted particles. Control analyses of etioplasts resulted in less (32%) label in proximity to the prolammelar body. Preimmune serum gave no such label in chloroplasts (58). Muthuramalingam et al. (77) detected 2-CysPrx in isolated photosystem II-enriched particles. Cerveau et al. (21) analyzed carefully washed membranes obtained from control or water-stressed A. thaliana WT and srx mutants and failed to detect 2-CysPrx in the thylakoid fraction. These controversial observations indicate that 2-CysPrx possibly associates with the thylakoids in a dynamic and conformational state-dependent manner and that this association is lost during extensive washing.
2-CysPrx adopts five different quaternary conformations. The reduced dimer, the reduced decamer, and the oxidized dimer take part in the peroxidase cycle already described. The hyperoxidized decamer and the hyperoxidized hyperaggregate realize the chaperone function of 2-CysPrx (60). Site-directed mutated variants of Arabidopsis 2-CysPrx revealed variable chaperone activity with lowest activity of a C-terminally truncated version and a disulfide-bond mimicking variant and highest activity for the hyperoxidation-state-mimicking variant C54D (59). 2-CysPrx, together with PrxIIE and PrxQ, is part of the complex thiol–disulfide network of the chloroplast, functions as redox sensors, and participates in regulation and maintenance of metabolic homeostasis (Fig. 3) (30). Owing to its high affinity and reactivity, 2-CysPrx may be considered as primary reactive oxygen species (ROS) sensor. In line with this hypothesis, 2-CysPrx was among the most sensitive targets of oxidation by H2O2 in vitro and methylviologen treatment in vivo (76). Regeneration is achieved by, for example, NTRC and Trx-x, but also by Trx-f1 and thereby is connected to the electron input elements ferredoxin (Fd) and NADPH (26, 90). The peroxidase activity is limited by the slow regeneration reaction. Increasing ROS turnover may disturb this process by hyperoxidizing the catalytic cysteines after ∼250 peroxidase cycles (56, 62). Then the alternative ATP-dependent regeneration system of Srx regenerates the active 2-CysPrx in a very slow reaction with about 1 min−1. Initially, Srx was thought to be specific for 2-CysPrx in the chloroplast until Iglesias-Baena et al. (53) proved the dual targeting of Srx also to the mitochondrium. The data revealed that Srx reduces hyperoxidized PrxIIF in the mitochondrion.
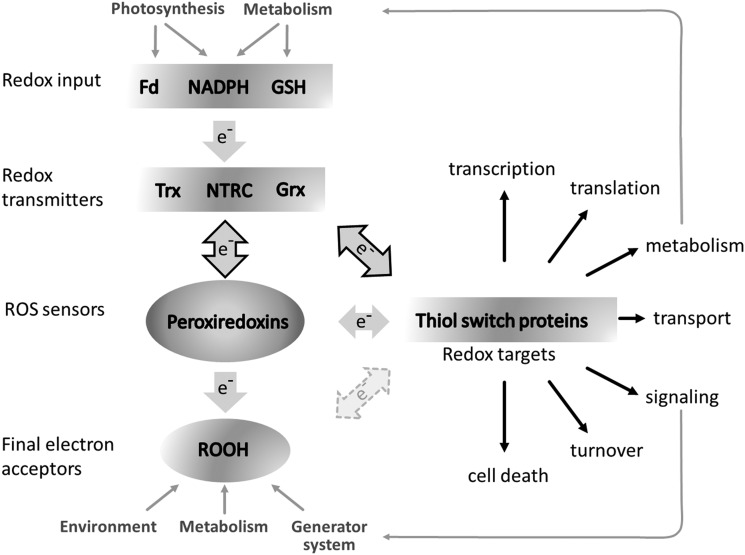
FIG. 3.
Role of Prxs in the thiol-dependent redox network. Environmental fluctuations and metabolic disequilibria generate ROS that drain electrons from Prxs that may be considered as redox sensors. Oxidized thiol peroxidases such as Prxs are reduced by redox transmitters, which receive electrons from redox input elements such as NADPH and ferredoxin (Fd). The redox transmitters (Trx, Grx, NTRC, and ACHT) control the redox state of redox targets that act as thiol switches, regulate virtually all cellular processes, and modulate signaling that feeds back to the reducing site and the generator systems. Prxs and ROS may also directly oxidize target proteins by proximity-based oxidation. NTRC, NADPH-dependent thioredoxin reductase C; ROS, reactive oxygen species.
The occurrence and functional link between oligomerization, hyperoxidation, and chaperoning still need to be further investigated for structural and functional features. Noichri and coworkers (82) suggested that stacked reduced decamers act as precursors for stacked hyperoxidized aggregates in Saccharomyces cerevisiae. Two different high molecular weight (HMW) fractions were identified by SEC and native polyacrylaminde gel electrophoresis (PAGE) or SDS-PAGE, depending on the redox state. The occurrence of hyperoxidized 2-CysPrx follows a circadian rhythm in eukaryotic cells. The circadian hyperoxidation does not depend on transcriptional circuits and correlates with release of H2O2 from mitochondria in yeast (101). It should be noted that the fraction of hyperoxidation is rather small with an estimated share of up to 1.6% of total 2-CysPrx in erythrocytes of WT mice (25). The circadian oscillation of 2-CysPrx hyperoxidation also occurs in A. thaliana and other plants (34). But again the amount subjected to hyperoxidation is low (21). For the time being, the circadian appearance of hyperoxidized 2-CysPrx is considered as indicator for daily alterations in cell redox milieu possibly linked to specific metabolic activities such as FeS center assembly. A major accumulation of hyperoxidized 2-CysPrx was also not seen under temperature or water stress (21). The absence of HMW complexes was interpreted as indicator that the chaperone function might be less important than previously proposed. In line with this suggestion, hyperoxidized 2-CysPrx could neither be detected in nonreducing gels nor in Western blots using sulfinic/sulfonic antibody. Srx and NTRC transcript levels coding for the main regulators of 2-CysPrx redox state were unaffected under stress (94).
Apparently the system consisting of 2-CysPrxs, NTRC, and Srx guarantees a rather stable redox state of 2-CysPrx under most conditions. Interestingly, hyperoxidized 2-CysPrx was found in plants lacking NTRC. 2-CysPrx can turn hyperoxidized only if the peroxidase cycle is running. Therefore, the hyperoxidation in ntrc background proves that 2-CysPrx receives sufficient electrons by NTRC-independent components such as Trx-x, CDSP32, Trx-f, and Trx-m to sustain the peroxidase cycle. While questioning the significance of circadian oscillations in hyperoxidation, the authors suggest a light-dependent regulation of 2-CysPrx hyperoxidation dependent on photosynthetic electron transport (PET) and the Fd/FTR/Trx system (21). Alternatively, the small fraction of hyperoxidized 2-CysPrx might realize a gain of function by specific interaction with targets apart from general chaperoning.
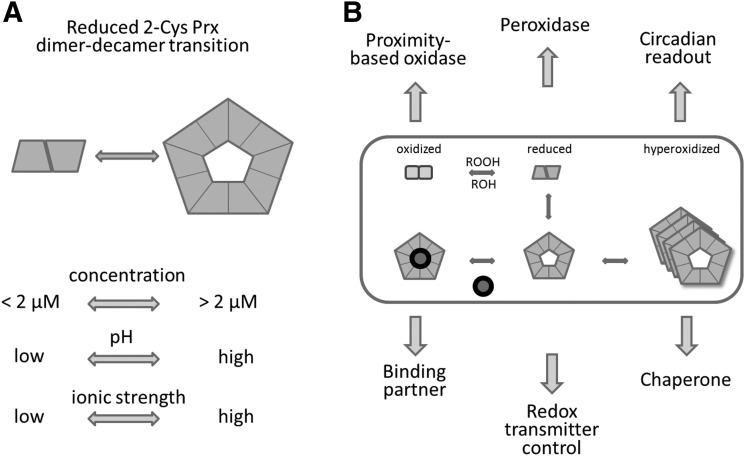
A peculiar feature of reduced 2-CysPrx is its dynamic transition from dimer to decamer. Several physicochemical parameters influence this transition and the stability of the reduced decamer, in particular pH, ionic strength, redox potential, and its own concentration (12, 62, 75) (Fig. 4A). Below the critical transition concentration (CTC), reduced 2-CysPrx exclusively adopts the dimeric conformation. With extremely high cooperativity, reduced dimers assemble to decamers above the CTC similar to the abrupt transition of free lipids to lipid micelles above the critical micelle concentration. SEC, isothermal titration microcalorimetry (ITC), and dynamic light scattering defined the CTC with about 2.2 μM at pH 8 and 4.2 μM at pH 7.2. The pH dependency might possibly be linked to a histidine in the dimer–dimer interface, which is protonated at lower pH. His protonation may decrease the ability to form decamers.
FIG. 4.
Multifunctionality of 2-CysPrx in plants. (A) Reduced 2-CysPrx adopts a dimer–decamer equilibrium. Several physicochemical parameters affect this equilibrium: the 2-CysPrx concentration, the pH, and the ionic strength. The critical transition concentration defines the minimum concentration for effective decamer association. Low pH and low ionic strength favor the dimer, whereas high pH and ionic strength support oligomerization. (B) 2-CysPrx is involved in multiple cellular processes based on its redox-dependent conformational state, namely as thiol peroxidase, oxidant of redox transmitters in the redox network, proximity-based oxidase, chaperone, protein interactor, and also as redox indicator in the circadian rhythm.
Stromal proteins interfere with the CTC, for example, the cyclophilin 20-3 (Cyp20-3), a peptidyl-prolyl-cis/trans isomerase that regulates cysteine synthesis (85). In contrast, titration of 2-CysPrx in 200 μg/ml stromal protein extract significantly decreased the CTC to 0.6 μM (62). Apparently complex and dynamic mechanisms regulate the dimer–decamer equilibrium, wherein a relatively small portion of dimers interact with several partners, or the binding partners of the decamer affect the equilibrium between dimer and decamer. The schematic depicted in Figure 4B summarizes the conformational states and their proposed or proven functions.
The Prx Interactome
2-CysPrxs, such as PrxIIE and PrxQ, are part of the chloroplast redox-regulatory network and are targeted by ROS as oxidants and electron transmitters as reductants (Fig. 3). Chloroplast Prxs and Gpx are preferentially reduced by some members of the whole chloroplast Trx family, consisting of Trx-f1, -f2, m1-4, -x, -y1, -y2,-z, 9 Trx-like proteins (NTRC, CDSP32, ACHT 1–4 [atypical cysteine histidine-rich Trx], and APR-1–3 [protein disulfide isomerase-like proteins]). The thiol peroxidases and two Asc-dependent peroxidases (thylakoid Apx and stromal Apx) reveal distinct suborganellar localization in the plastids (14, 61, 71).
The stromal 2-CysPrx is part of an intricate protein–protein interaction network. NTRC and Trx-x, together with CDSP32, are described as the predominant reductants of oxidized 2-CysPrx in vitro and in vivo and regenerate the active thiol peroxidase (Fig. 2) (14, 17). Reduced Cyp20-3 interacts with reduced 2-CysPrx as shown by fluorescence resonance energy transfer in protoplasts, ITC, and in vitro binding assays (62, 77).
Cyp20-3 binds 12-oxophytodienoic acid (OPDA), the precursor of jasmonates, with high affinity (Kd = 196 nM) (85). OPDA-liganded Cyp20-3 activates the chloroplast cysteine synthase complex consisting of serine acetyltransferase 2.1 (Serat) and O-acetylserine thiol lyase-B (OASTL-B). However, Cyp20-3 may not be necessary for chloroplast cysteine synthesis because formation of the cysteine synthase complexes occurs spontaneously (ΔG = −33 kcal mol−1) (112). In addition, only about 20% of cellular GSH synthesis derives from chloroplast Serat 2.1/OASTL-B, whereas 80% originate from mitochondrial and cytosolic cysteine synthase (112). Nonetheless, Cyp20-3 is regarded as a mediator of stress-dependent redox responses. The interaction between 2-CysPrx and Cyp20-3 occurs under reducing conditions and is suggested to compete with the activation of the cysteine synthase depending on the binding of OPDA and a potential conformational shift. This mechanism would allow for a negative regulatory feedback, slowing down stress-induced cysteine synthesis after re-establishing redox homeostasis.
Dangoor et al. (27) described an interaction of 2-CysPrx with ACHT1 as part of the chloroplast redox-regulatory network. 2-CysPrx is regarded as oxidant of ACHT1, which is suggested to regulate metabolism and acclimation processes, for example, during light–dark transitions of photosynthesis. Furthermore, ACHT4 also interacts with 2-CysPrx using a similar mechanism like ACHT1. The ADP-glucose pyrophosphorylase is an essential enzyme of starch synthesis. Its small subunit is implicated in the redox network as downstream target of ACHT4 (35). During dark–light transitions, the fast reoxidation of ACHT4 depends on 2-CysPrx. 2-CysPrx likely functions as oxidant to switch off starch synthesis in the dark similar to the proposed hypothesis that 2-CysPrx acts as a local redox sink transferring electrons to ROS to shut down photosynthesis. There is a need to kinetically resolve the redox state of components of the thiol–disulfide redox regulatory network in WT and mutants to better define the role of 2-CysPrx in rapid redox regulation (45).
Recently, an ex vivo approach based on coimmunoprecipitation (CoIP) and mass spectrometry allowed for identifying a set of 2-CysPrx interactors (20). The strength of this study rested on the comparison of CoIP proteomes of WT and 2-CysPrxA/B-deficient A. thaliana. Polypeptides recovered with anti-2CysPrx antibody from 2cysprxA/B-CoIPs were considered as false positives and subtracted from the polypeptides detected in WT CoIPs. This approach identified 67 interacting polypeptides from the chloroplast. The 91 interactions with polypeptides from extraplastidic compartments may originate from lost compartmentation during extraction. Fructose-1,6-bisphosphatase, ACHT2, and CDSP32 were among the known interactors. Surprisingly, Trx, NTRC, and Cyp20-3 were not found possibly because of the weak nature of interactions.
Interestingly, binding to other cyclophilins such as Cyp37/38 and CLP protease supports a role of 2-CysPrx protein in protein folding or regulation. The interactions with, for example, protochlorophyllide reductase B, 5-adenylylsulfate reductase (APR), GSH reductase, and three ferredoxin (Fd)-dependent proteins, namely Fd-sulfite reductase, Fd-nitrite reductase, and Fd-glutamate synthase reinforce the suggested function of 2-CysPrx in the redox-regulatory network of the chloroplast (20). Future work will have to address the significance of the interactions for interactor function and chloroplast metabolism.
A prerequisite for protein interactions are partner-specific surface-exposed residues, which form the interface either permanently or become surface exposed after conformational changes induced by interaction or variation of the milieu. A 2-CysPrx site with binding characteristics is the flexible C-terminus that is involved in binding the Ser protease Omi/HtrA2 and embraces Srx1 during the retroreduction of the hyperoxidized sulfinylated 2-CysPrx (50, 56). As shown for the interaction of At2-CysPrx with NTRC, the deletion of the C-terminus decreases the affinity for NTRC and alters the peroxidase and chaperone activities (59). The best studied and closest sequence homologues of At2-CysPrx likely are human Prdx1 and Prdx2. Analysis of their interactomes allowed the identification of the sequences CXXC, PXXP, and LXXL as motifs found in either Prdx1- or Prdx2-binding partners (15).
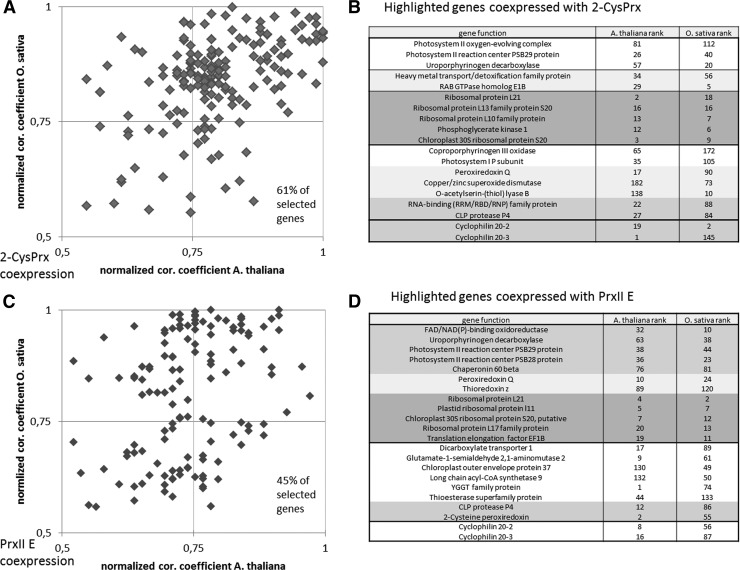
Coexpression frequently allows pinpointing to functional relationships. Transcriptional coexpression with Prxs has been reported by Muthuramalingam et al. (77) and Dietz (31) and identified Cyp20-3 as best hit for coexpression with values of 0.97–0.99. Here we extended this analysis to include the transcriptome of Oryza sativa cv. japonica (Rice Oligonucleotide Array Database [ www.ricearray.org]) in comparison with A. thaliana (taken from ATTED-II [http://atted.jp]). The obtained sets of highly coexpressed genes showed a significant match between Arabidopsis and rice for 2-CysPrx or PrxIIE. Sets of 61% for 2-CysPrx and 45% for PrxIIE of the identified Arabidopsis genes were also highly coregulated in rice (correlation coefficient >0.5) (Fig. 5). Strong connections to photosynthesis, antioxidant partners, and ribosomal activity were seen in both species. The comparison is still constrained by the incomplete annotation of the rice genome, possibly causing an underestimation of the coregulated genes, but also limiting the differentiation of the Arabidopsis and rice response.
FIG. 5.
Transcripts coexpressed with 2-CysPrx and PrxIIE in Arabidopsis thaliana and Oryza sativa. The correlation coefficients for O. sativa and A. thaliana were obtained from the ATTED-II and the Rice Oligonucleotide Array Database (6, 19). Correlation coefficients were normalized and ranked with 1 for the transcript with maximum level of coregulation and higher numbers according to the position in the list. The query list comprised the Arabidopsis genes with a correlation >0.5. The homologous genes were searched in rice, the degree of coexpression with 2-CysPrx and PrxIIE was determined and plotted against the corresponding coefficient of Arabidopsis. (A) The transcripts in the area with high correlation of >0.5 in both rice and Arabidopsis represent 61% of total analyzed genes, indicating a high level of similarity in coregulation in both species. (C) For PrxIIE, 45% of the selected genes ranged in the area of high coexpression in both species. A selection of interesting genes from this group is given for 2-CysPrx (B) and PrxIIE (D)
Post-Translational Effects
The cysteine oxidation is the prime PTM on Prxs. The robust Trx-fold, conserved motifs, and flexible structures mediate conformational changes along with redox-dependent thiol modification. Apart from their thiol peroxidase function, Prxs can be considered as a prototypic thiol switch protein that is based on present-day knowledge evolved early during evolution. The dithiol–disulfide transition modifies conformation and function. Nowadays, the thiol switch mechanism is recognized for its broad significance in plants and other organisms with oxygenic metabolism (110). Such thiol switch proteins play fundamental roles in regulation and signaling of eukaryotes and prokaryotes, including mitochondria (4, 5, 40, 44, 79). ROS are generated at low rates by metabolism and, for example, by NADPH oxidases under normal conditions. They are detoxified by antioxidant enzymes, including thiol peroxidases (32). Thiol peroxidases are regenerated by redox transmitters, which then may oxidize other thiol switch proteins. Accumulating ROS under stress in addition may target sensitive thiols of other proteins (76).
Reversibility of disulfide formation, sulfenylation, and possibly sulfinylation contrasts sulfonylation and oxidative modifications of other amino acid residues, which often mark proteins for degradation (10, 28). Thus it was a very important finding that the hyperoxidized sulfinic acid forms of 2-CysPrx and PrxIIF can be regenerated by Srx (16, 53, 118). The specificity or promiscuity of Srx in reduction of sulfinylated proteins still needs to be scrutinized because hyperoxidation of 1-CysPrx may be irreversible (118). These authors state that hyperoxidized 1-CysPrx but also glyceraldehyde-3-phosphate dehydrogenase might be degraded because neither of them is a target of Srx.
Redox-dependent inhibition of Prx peroxidase activity triggers alternative functions and signaling pathways. Hyperoxidized 2-CysPrx prevents heat-induced denaturation of citrate synthase in vitro (54, 59). Overexpression of 2-CysPrx suppresses aggregation of cyclophilin 20-3 coexpressed in protoplasts in vivo (77). Combining redox-dependent peroxidase and chaperone function in a highly abundant protein provides the opportunity to operate a molecular redox switch in response to oxidative stress. The functional switch from peroxidase to chaperone is part of the floodgate model. Hyperoxidation of Prxs might shut down the detoxification in the thiol peroxidase branch and temporally and spatially enhance the ROS-dependent signaling (121). Analysis of 2-cysprx A/B knockout mutants revealed enhanced photo-oxidative damage and higher H2O2 levels in high light. Considering its high abundance in chloroplasts and its association with the thylakoids, the 2-CysPrx might act as a local redox-controlled switch to allow for the release of ROS signals from the photosynthetic electron transport chain. In turn, ROS release may change chloroplast processes but also initiate retrograde control of extrachloroplast processes. However, regulation of ROS-responsive marker genes such as ZAT12, BAP1, HSFA2, or OXI1 was unaltered in 2cysprxA/B compared with WT and only responded after introducing the additional knockout in the thylakoid ascorbate peroxidase (tApx) gene. This might indicate a synergistic floodgate control by thiol and ascorbate peroxidases and mutual compensation of the lost other enzyme (9).
PTMs of Prxs by glutathionylation, nitrosylation, acetylation, or phosphorylation partly mimic cysteine hyperoxidation and thus oligomerization, and realize oxidation-independent regulation and stress responses. The tripeptide GSH is present at millimolar concentrations in plasmatic compartments and functions as redox–thiol buffer, electron donor, substrate for phytochelatin synthesis, and in S-glutathionylation (80). S-glutathionylation is necessary for redox regeneration of 1-CysPrx and type II Prxs, which then depends on GST or Grx-coupled cysteine reduction.
Under conditions of GSSG accumulation, the mass of PsTrx-h2 increases to 14,361.1 Da corresponding to addition of 305.5 Da of GSH to 14,056.9 Da (42). Another example of reversible glutathionylation occurs with chloroplast glyceraldehyde-3-phosphate dehydrogenase and NADP-dependent malate dehydrogenase (13, 69). The GSH/GSSG pool participates in redox homeostasis and controls various signaling pathways. Reversible S-glutathionylation introduces a negative charge and a bulky tripeptide and protects thiols from hyperoxidation.
Peroxynitrite (ONOO−) serves as substrate for 2-CysPrx and PrxIIE (8, 107). Biotic and abiotic stress conditions provoke nitrosoglutathione (GSNO) accumulation, which is a storage pool for NO. NO is a radical and participates in cell signal transduction (38). A proteome analysis identified >1000 S-nitrosylated targets in A. thaliana and likewise in Kalanchoe pinnata, Brassica juncea, and Citrus aurantium. These proteins are involved in major cellular activities such as primary and secondary metabolism, photosynthesis, genetic information processing, cellular architecture, and response to biotic and abiotic stresses (8). As part of detoxification networks, PrxIIE and PrxIIB (cytosol), PrxIIF (mitochondria), and GR and Gpx are targets of S-nitrosylation. These and other nitrosylated polypeptides were inventoried by MS/MS analysis after addition of GSNO to Arabidopsis cell cultures or exposing leaves to 1 ppm NO via the gaseous phase (63). Proof of physiological significance is scarce. S-nitrosylation of PrxIIE is suggested to ease ONOO−-dependent tyrosine nitration as signaling event in pathogen defense (104). Plant Prxs are Lys acetylated as reported for Os1-CysPrx (47) and for Os2-CysPrx (124). Acetylation sites are KSGGLGDLK(ac)TK, GLFIIDK(ac)HSTINNLAIGR, and SISK(ac)GVLIPDQGIALR in Os2-CysPrx. Lys acetylation in mammalian astrocytes was connected to the oxidation state of Cys residues because many acetylation sites are detected in the vicinity of oxidized cysteines in cell lines depleted of most of their GSH (88). Human Prdx2 is subjected to N-terminal acetylation, which protects the catalytic cysteine from hyperoxidation to irreversible sulfonic acid, retaining peroxidase activity by Srx regeneration (109). The physiological role of acetylation awaits investigation in plants.
More is known about the physiological role of plant protein phosphorylation (98, 102, 115, 126). A recent analysis of the phosphoproteome of grapevine (67) confirmed phosphorylation of Vv-Apx (Vitis vinifera) in addition to the cytoplasmic VvPrxIIB in response to phytoplasma infection. A similar pattern was found in A. thaliana after wounding and in Medicago truncatula after rhizobial infection (52, 105). To address the consequences of kinase inhibition, Xu et al. (125) observed reduction in the drought-induced H2O2 production and a decrease in the drought-enhanced activities of antioxidant enzymes such as SOD, Apx, and Gpx in maize. Queries to the online databases IPTM net (http://research.bioinformatics.udel.edu/iptmnet), P3DB (www.p3db.org), and PhosPhat 4.0 (http://phosphat.uni-hohenheim.de) retrieve serine/threonine protein kinases involved in modifying redox proteins. Of the redox proteins mentioned in this review, PrxIIE, PrxIIF, PrxQ, 2-CysPrxA, 2-CysPrxB, CDSP32, ACHT 1, Trx-m4, Apx, and Gpx are found to be phosphorylated among others (Table 2). The table provides detailed information on the identified phosphorylated peptides of the redox proteins.
Interestingly, At2-CysPrx and the mammalian 2-CysPrx type Prdx1 have the C-terminal SKEYF site in common, which is phosphorylated at the Ser in A. thaliana and at the Tyr in mouse, rat, and human (119). In vitro phosphorylation of Tyr in the respective site of human Prdx1 decreases the peroxidase activity by 80% (119). According to their proposed model, a local rise in peroxide concentration leads to MAPK activation, and phosphorylation of peroxiredoxin, in turn, allows for further signaling. Whether this model is applicable to plant 2-CysPrxs and other redox proteins is not known. Altogether, these results indicate a relationship between redox regulation, PTMs, and signaling. The PTM of redox proteins and their implications in planta still needs to be explored.
Membrane Interaction
The C-terminus of 2-CysPrx bears the motif SKEYF, which is conserved among plant 2-CysPrx but absent in its prokaryotic homologue, the alkyl hydroperoxide reductase C (AhpC). In addition to the feature of SKEYF phosphorylation mentioned in the previous section, the SKEYF motif is also present in the C-terminal sequence 186NVDDSKEYFSKHN198 of the human 2-Cys Prx, Prdx2, where it mediates its membrane attachment (22). Therefore, as shown for barley 2-CysPrx, the C-terminus that is essential for binding to NTRC (59) likely plays a role in 2-CysPrx membrane association, in regulation of its peroxidase activity (see previous section), or both. The SKEYF-containing sequence 184EKSMKPDPKLSKEYFSAI201 is computationally predicted to interact with membranes (Table 3). A different approach to investigate the possibility of peripheral membrane association of 2-CysPrx considers electrostatic interactions between the decamer and the membrane (68). Here it is striking that the outer pentagon sites of 2-CysPrx bear Lys-rich C-termini and the inner doughnut sites are enriched in positively charged residues. Yet, it is unknown whether these features play a role in realizing the putative amphitropic character of 2-CysPrx, wherein barley 2-CysPrx is found both in the soluble plastid proteome and associated with the thylakoid (58). Amphitropic proteins reversibly interact with the membrane by lipid-binding regions containing positively charged and hydrophobic amino acids residing in coil-helical regions (18, 55). Hydrophobic faces are also often involved in membrane interactions (41) and the analysis shown in Table 3 revealed a hydrophobic face sequence 162LQALQYIQENPDEVCPAG179. The hydrophobic face has negatively charged residues opposed to their hydrophobic domains, which could play a role in At-2CysPrx membrane association as proposed for the human Prdx2 membrane interaction model (92). Protons or Ca2+ ions may neutralize the negatively charged hydrophobic faces to imbed 2-CysPrx in the membrane.
Table 3.
At2-CysPrx Sequences with α-Helical Characteristics, Analyzed with HELIQUEST
| Name | Sequence | z | <H> | <μH> | D |
|---|---|---|---|---|---|
| At2-CysPrx 162–179 | LQALQYIQENPDEVCPAG | −3 | 0.426 | 0.337 | 0.67a |
| At2-CysPrx 184–201 | EKSMKPDPKLSKEYFSAI | 1 | 0.172 | 0.418 | 0.72 |
The analysis with the HELIQUEST program used the default settings. Regions of At2-CysPrx were selected with minimum 50% α-helical character, but no β-strand content and no dimer–dimer interaction site (120). Secondary structure features of At2-CysPrx are based on the Phyre 2 (www.sbg.bio.ic.ac.uk/phyre2) alignment with human peroxiredoxin as template (1qmv, 62% identity, 100% confidence). Sequences with putative lipid-binding helices and hydrophobic faces (a) are presented. According to the discrimination factor equation D = 0.944 (<μH>) + 0.33 (z), possible lipid-binding helices exist, when the calculated discrimination factor D lies between 0.68 and 1.33 (41).
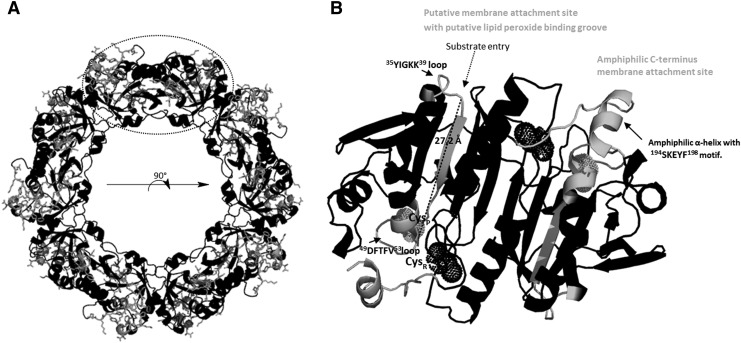
The Aramemnon online tool (http://aramemnon.uni-koeln.de/request.ep) was then used to explore the lipid-binding characteristics of 2-CysPrx (108). A lipid anchor modification was not predicted, but a weak transmembrane helical region was assigned to the 37GKKYVILFFYPLDFTFVCPTE57 sequence. Support for this sequence motif as putative site of membrane binding comes from the observation that the contained ILFF motif is present in membrane-associated proteins such as membrane-associated protein (At1 g80910), Gibberellin receptor (At3 g05120), and GDSL lipase (At1 g54790). The ILFF motif is also present in the phosphatidic acid-binding site of Raf 1, A-Raf, and B-Raf (2, 43). In the human 1-CysPrx Prdx6 that has both lipase and peroxidase activities, the ILF motif is present in its α/β hydrolase fold (65). Of the three putative membrane interaction sites, the 184EKSMKPDPKLSKEYFSAI201 and 37GKKYVILFFYPLDFTFVCPTE57 sites are rich in Lys content and are present on both halves of the decameric donut (Fig. 6). The latter site is suggested to contain a binding groove for lipid peroxide substrates. The site starting with the 35YIGKK39 loop and ending with the 49DFTFV53 loop contains the CysP, is highly hydrophobic, and is proposed to have a length of ∼27Å and a width spanning from 4 to 12Å resembling the lengths of known lipid-binding proteins such as β-lactoglobulin (PDB ID: 1EXS) or hPrdx6 (PDB ID: 1PRX) (Fig. 6B).
FIG. 6.
Putative membrane attachment sites of At-2-CysPrx. (A) 2-CysPrx pentadimer with membrane attachment sites highlighted in gray and residing residues shown as stick models. Each subunit contains two membrane attachment sites. The 3D model was constructed with Phyre 2 (www.sbg.bio.ic.ac.uk/∼phyre) and PyMOL (29). hPrx2 (PDB ID 1qmv, 62% identity) was used as template (57). (B) 90° turn of 2-CysPrx pentadimer as indicated with viewpoint on the indicated dimer (dotted circle) after rotation. Each monomer is suggested to mediate membrane attachment of one homodimer via two sites (highlighted in gray), the C-terminus and the loop surrounding the substrate entry site. Stick models of residues visible in (A) are not shown in (B) in order to ease the perspective on the lipid peroxide-binding groove (length as dotted line in Ångstrom [Å]). The peroxide-binding groove starts (amino acids counted from the N-terminus) at the positively charged Lys-rich loop near the entrance (aa 35–39), proceeds to the 42ILF44-motif of the β-strand, and ends at the 49DFTFV53 loop close to the CysP (dotted spheres) of the cavity. The resolving Cys of the adjacent monomer is shown as dotted black sphere.
The Ca2+-associated protein calpromotin nowadays called Prdx2 in mammals interacts with integral membrane proteins such as the gardos channel (Uniprot entry O15554), stomatin (Uniprot entry P27105), and band 3 (Uniprot entry P02730) (68, 73, 74). In contrast to Cha et al. (22), Matte et al. (68) proposed that the N-terminus of cytosolic band 3 (Cdb3) is crucial for Prx2 membrane association, whereas Plishker et al. (92) assume that the redox milieu and Ca2+ are essential for the switch from the soluble to the membrane-bound state. Alternatively, binding proteins may mediate thylakoid attachment of 2-CysPrx. Muthuramalingam et al. (77) detected 2-CysPrx in photosystem II-enriched fractions and were able to coimmunoprecipitate D1 protein and 2-CysPrx. Prdx2 is highly similar to At2-CysPrx. Therefore, one may ask whether the binding epitopes of the mentioned proteins can be found in At2-CysPrx. A protein Blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) search of the A. thaliana uniprot databank with the gardos channel retrieved two pore domain potassium channels, the vacuolar AtTPK1 (At5 g55630) and the plasmamembrane AtTPK4 (At1 g02510). They share the GYGD consensus motif with the gardos channel and are Ca2+-activated K+ channels but reside in a different compartment than the plant 2-CysPrx. The A. thaliana genome contains genes encoding stomatin-like proteins (33). Future studies focusing on the interaction of 2-CysPrx with integral membrane proteins will be of great interest.
Final Remarks
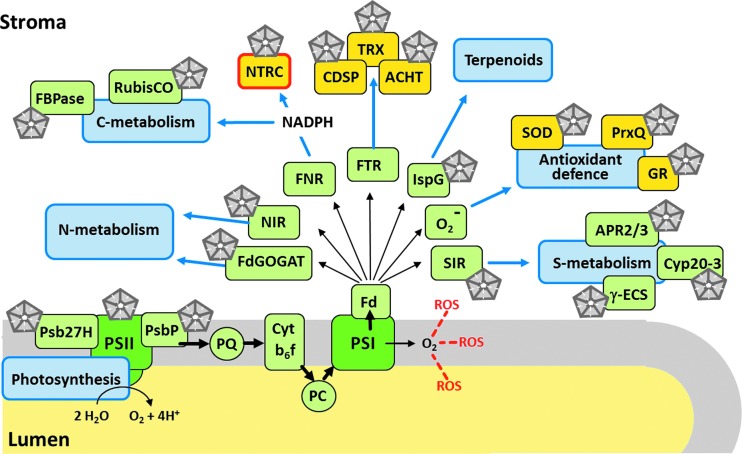
Research on plant Prxs began with the identification of 2-CysPrx in the chloroplast and 1-CysPrx in the nucleus 20 years ago (11, 113). Since then, the biochemical features, cell biological mechanisms, and physiological implications of different plant Prxs have been elucidated from various perspectives, but the most significant advancement has been achieved for the chloroplast 2-CysPrx either by targeted investigation or by coincidental findings, for example, by identifying NTRC as most efficient but not exclusive electron donor of 2-CysPrx. Since the last comprehensive review (31), significant progress has been achieved in identifying the interactome as selectively displayed in Figure 7 for 2-CysPrx (20). Key components of the major metabolic pathways that consume reductive power have been proven or are suggested to interact with 2-CysPrx. This concerns photosynthetic light reactions, carbon metabolism, nitrogen and sulfur metabolism, antioxidant defense, and secondary compound synthesis. In this context, it is intriguing that ferredoxin-dependent proteins constitute a major fraction of interacting partners. Important interactions with proteins or membranes need to be confirmed and explored with respect to binding selectivity for redox-dependent conformational states. The successful proteomics analysis indicates that there is a need to conduct metabolomics and transcriptomics analyses to reach the global assessment of the physiological significance of the different Prxs. This includes the interference with Ca2+ and hormone signaling. Most importantly we need complementation of the 2-CysPrx knockout line with variants that mimic specific states of 2-CysPrx, namely devoid of peroxidase activity, lacking the oligomerization ability, resembling the hyperoxidized decamer, or with truncated C-terminus. It will be interesting to find out which features of the knockout phenotype can be complemented by each of the variants and this approach should allow dissecting the roles as thiol peroxidase, oxidant, interaction partner, and chaperone.
FIG. 7.
Assignment of the 2-CysPrx interacting partners to selected metabolic pathways of the chloroplast. The figure is based on diverse findings, in particular, the interactome article by Cerveau et al. (20) and focuses on the photosynthetic electron transport and the associated ferredoxin (Fd), which distributes electron via diverse Fd-dependent proteins to various reductive pathways. The pentagon icon indicates interaction of 2-CysPrx with the proteins. See text for further discussion. ACHT, atypical chloroplast thioredoxin; APR, adenylylsulfate reductase; Cyp20-3, cyclophilin 20-3; γ-ECS, gamma glutamyl cysteine synthase; FBPase, fructose-1,6-bisphosphatase; Fd-GOGAT, ferredoxin-dependent glutamate synthase; FNR, ferredoxin-dependent NADPH reductase; FTR, ferredoxin-dependent thioredoxin reductase; GR, glutathione reductase; IspG, ferredoxin-dependent 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase; NIR, ferredoxin-dependent nitrite reductase; PC, plastocyanine; PQ, plastoquinone; Psb, photosystem II; PSI/II, photosystem I/II; RubisCO, ribulose-1,5-bisphosphatase; SIR, ferredoxin-dependent sulfite reductase; SOD, superoxide dismutase.
Abbreviations Used
- 2-CysPrx
2-cysteine peroxiredoxin
- APR
adenylylsulfate reductase
- Apx
ascorbate peroxidase
- CoIP
coimmuno precipitation
- CTC
critical transition concentration
- Cyp20-3
cyclophilin 20–3
- Fd
ferredoxin
- Gpx
glutathione peroxidase
- GSH/GSSG
reduced/oxidized glutathione
- GSNO
nitrosoglutathione
- GST
glutathione-S-transferase
- HMW
high molecular weight
- ITC
isothermal titration microcalorimetry
- NTRC
NADPH-dependent thioredoxin reductase C
- OPDA
oxophytodienoic acid
- PAGE
polyacrylamide gel electrophoresis
- PET
photosynthetic electron transport
- Prx/Prdx
peroxiredoxin
- PTMs
post-translational modifications
- ROS
reactive oxygen species
- sApx
stromal ascorbate peroxidase
- SEC
size exclusion chromatography
- SOD
superoxide dismutase
- Srx
sulfiredoxin
- tApx
thylakoid-bound peroxidase
- Trx
thioredoxin
Acknowledgment
The work in this article was supported by the Deutsche Forschungsgemeinschaft DI346/14 and SPP1710.
References
- 1.Altschul SF, Gish W, Miller W, Myers EW, and Lipman DJ. Basic local alignment search tool. J Mol Biol 215: 403–410, 1990 [DOI] [PubMed] [Google Scholar]
- 2.Andresen BT, Rizzo MA, Shome K, and Romero G. The role of phosphatidic acid in the regulation of the Ras/MEK/Erk signaling cascade. FEBS Lett 531: 65–68, 2002 [DOI] [PubMed] [Google Scholar]
- 3.Anschau V, Ferrer-Sueta G, Portillo-Ledesma S, and Netto LES. Kinetic Characterization of Sulfenic Acid Reduction in 1-Cys Peroxiredoxins by Ascorbate. In Annual Meeting of the Society for Free Radical Biology and Medicine (SFRBM). Soc Free Radic Biol Med 76: S138, 2014 [Google Scholar]
- 4.Antelmann H. Enzyme regulation: a thiol switch opens the gate. Nat Chem Biol 11: 4–5, 2015 [DOI] [PubMed] [Google Scholar]
- 5.Antelmann H. and Helmann JD. Thiol-based redox switches and gene regulation. Antioxid Redox Signal 14: 1049–1063, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aoki Y, Okamura Y, Tadaka S, Kinoshita K, and Obayashi T. ATTED-II in 2016: a plant coexpression database towards lineage-specific coexpression. Plant Cell Physiol 57: e5, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aryal UK, Krochko JE, and Ross AR. Identification of phosphoproteins in Arabidopsis thaliana leaves using polyethylene glycol fractionation, immobilized metal-ion affinity chromatography, two-dimensional gel electrophoresis and mass spectrometry. J Proteome Res 11: 425–437, 2011 [DOI] [PubMed] [Google Scholar]
- 8.Astier J, Kulik A, Koen E, Besson-Bard A, Bourque S, Jeandroz S, Lamotte O, and Wendehenne D. Protein S-nitrosylation: what's going on in plants?. Free Radic Biol Med 53: 1101–1110, 2012 [DOI] [PubMed] [Google Scholar]
- 9.Awad J, Stotz HU, Fekete A, Krischke M, Engert C, Havaux M, Berger S, and Mueller MJ. 2-cysteine peroxiredoxins and thylakoid ascorbate peroxidase create a water-water cycle that is essential to protect the photosynthetic apparatus under high light stress conditions. Plant Physiol 167: 1592–1603, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bader N. and Grune T. Protein oxidation and proteolysis. Biol Chem 387: 1351–1355, 2006 [DOI] [PubMed] [Google Scholar]
- 11.Baier M. and Dietz KJ. Primary structure and expression of plant homologues of animal and fungal thioredoxin-dependent peroxide reductases and bacterial alkyl hydroperoxide reductases. Plant Mol Biol 31: 553–564, 1996 [DOI] [PubMed] [Google Scholar]
- 12.Barranco-Medina S, Krell T, Bernier-Villamor L, Sevilla F, Lázaro JJ, and Dietz KJ. Hexameric oligomerization of mitochondrial peroxiredoxin PrxIIF and formation of an ultrahigh affinity complex with its electron donor thioredoxin Trx-o. J Exp Bot 59: 3259–3269, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bedhomme M, Adamo M, Marchand CH, Couturier J, Rouhier N, Lemaire SD, Zaffagnini M, and Trost P. Glutathionylation of cytosolic glyceraldehyde-3-phosphate dehydrogenase from the model plant Arabidopsis thaliana is reversed by both glutaredoxins and thioredoxins in vitro. Biochem J 445: 337–347, 2012 [DOI] [PubMed] [Google Scholar]
- 14.Bernal-Bayard P, Ojeda V, Hervás M, Cejudo FJ, Navarro JA, Velázquez-Campoy A, and Pérez-Ruiz JM. Molecular recognition in the interaction of chloroplast 2-Cys peroxiredoxin with NADPH-thioredoxin reductase C (NTRC) and thioredoxin x. FEBS Lett 588: 4342–4347, 2014 [DOI] [PubMed] [Google Scholar]
- 15.Bertoldi M. Human peroxiredoxins 1 and 2 and their interacting protein partners; through structure toward functions of biological complexes. Protein Peptide Lett 23: 69–77, 2016 [DOI] [PubMed] [Google Scholar]
- 16.Biteau B, Labarre J, and Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature 425: 980–984, 2003 [DOI] [PubMed] [Google Scholar]
- 17.Broin M, Cuiné S, Eymery F, and Rey P. The plastidic 2-cysteine peroxiredoxin is a target for a thioredoxin involved in the protection of the photosynthetic apparatus against oxidative damage. Plant Cell 14: 1417–1432, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burn P. Talking point Amphitropic proteins: a new class of membrane proteins. Trends Biochem Sci 13: 79–83, 1988 [DOI] [PubMed] [Google Scholar]
- 19.Cao P, Jung KH, Choi D, Hwang D, Zhu J, and Ronald PC. The rice oligonucleotide array database: an atlas of rice gene expression. Rice 5: 17, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerveau D, Kraut A, Stotz HU, Mueller MJ, Couté Y, Rey P. Characterization of the Arabidopsis thaliana 2-Cys peroxiredoxin interactome. Plant Sci 252: 30–41, 2016 [DOI] [PubMed] [Google Scholar]
- 21.Cerveau D, Ouahrani D, Marok MA, Blanchard L, and Rey P. Physiological relevance of plant 2-Cys peroxiredoxin overoxidation level and oligomerization status. Plant Cell Environ 39: 103–119, 2016 [DOI] [PubMed] [Google Scholar]
- 22.Cha MK, Yun CH, and Kim IH. Interaction of human thiol-specific antioxidant protein 1 with erythrocyte plasma membrane. Biochemistry 39: 6944–6950, 2000 [DOI] [PubMed] [Google Scholar]
- 23.Chae HZ, Chung SJ, and Rhee SG. Thioredoxin-dependent peroxide reductase from yeast. J Biol Chem 269: 27670–27678, 1994 [PubMed] [Google Scholar]
- 24.Chen HH, Chu P, Zhou YL, Ding Y, Li Y, Liu J, Jiang LW, and Huang SZ. Ectopic expression of NnPER1, a Nelumbo nucifera 1-cysteine peroxiredoxin antioxidant, enhances seed longevity and stress tolerance in Arabidopsis. Plant J 88: 608–619, 2016 [DOI] [PubMed] [Google Scholar]
- 25.Cho CS, Yoon HJ, Kim JY, Woo HA, and Rhee SG. Circadian rhythm of hyperoxidized peroxiredoxin II is determined by hemoglobin autoxidation and the 20S proteasome in red blood cells. Proc Natl Acad Sci USA 111: 12043–12048, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collin V, Issakidis-Bourguet E, Marchand C, Hirasawa M, Lancelin JM, Knaff DB, and Miginiac-Maslow M. The Arabidopsis plastidial thioredoxins New functions and new insights into specificity. J Biol Chem 278: 23747–23752, 2003 [DOI] [PubMed] [Google Scholar]
- 27.Dangoor I, Peled-Zehavi H, Wittenberg G, and Danon A. A chloroplast light-regulated oxidative sensor for moderate light intensity in Arabidopsis. Plant Cell 24: 1894–1906, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dat J, Vandenabeele S, Vranová E, Van Montagu M, Inzé D, and Van Breusegem F. Dual action of the active oxygen species during plant stress responses. Cell Mol Life Sci 57: 779–795, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeLano WL. The PyMOL molecular graphics system, 2002. DeLano Scientific, Palo Alto, CA: www.pymol.org (accessed July5, 2017) [Google Scholar]
- 30.Dietz KJ. Redox signal integration: from stimulus to networks and genes. Physiol Plantarum 133: 459–468, 2008 [DOI] [PubMed] [Google Scholar]
- 31.Dietz KJ. Peroxiredoxins in plants and cyanobacteria. Antioxid Redox Signal 15: 1129–1159, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dietz KJ. Thiol-Based Peroxidases and Ascorbate Peroxidases: Why Plants Rely on Multiple Peroxidase Systems in the Photosynthesizing Chloroplast?. Mol Cells 39: 20–25, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunkley TP, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, and Watson RB. Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci USA 103: 6518–6523, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, Maywood ES, Hastings MH, Baliga NS, Merrow M, Millar AJ, Johnson CH, Kyriacou CP, O'Neill JS, and Reddy AB. Peroxiredoxins are conserved markers of circadian rhythms. Nature 485: 459–464, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eliyahu E, Rog I, Inbal D, and Danon A. ACHT4-driven oxidation of APS1 attenuates starch synthesis under low light intensity in Arabidopsis plants. Proc Natl Acad Sci USA 112: 12876–12881, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engelman R, Weisman-Shomer P, Ziv T, Xu J, Arnér ES, and Benhar M. Multilevel regulation of 2-Cys peroxiredoxin reaction cycle by S-nitrosylation. J Biol Chem 288: 11312–11324, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Engelsberger WR. and Schulze WX. Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen–starved Arabidopsis seedlings. Plant J 69: 978–995, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Espunya MC, De Michele R, Gómez-Cadenas A, and Martínez MC. S-Nitrosoglutathione is a component of wound-and salicylic acid-induced systemic responses in Arabidopsis thaliana. J Exp Bot 63: 3219–3227, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferrer-Sueta G, Manta B, Botti H, Radi R, Trujillo M, and Denicola A. Factors affecting protein thiol reactivity and specificity in peroxide reduction. Chem Res Toxicol 24: 434–450, 2011 [DOI] [PubMed] [Google Scholar]
- 40.Foyer CH. and Noctor G. Ascorbate and glutathione: the heart of the redox hub. Plant Physiol 155: 2–18, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gautier R, Douguet D, Antonny B, and Drin G. HELIQUEST: a web server to screen sequences with specific α-helical properties. Bioinformatics 24, 2101–2102, 2008 [DOI] [PubMed] [Google Scholar]
- 42.Gelhaye E, Rouhier N, Gérard J, Jolivet Y, Gualberto J, Navrot N, Ohlsson PI, Wingsle G, Hirasawa M, Knaff DB, Wang H, Dizengremel P, Meyer Y, and Jacquot JP. A specific form of thioredoxin h occurs in plant mitochondria and regulates the alternative oxidase. Proc Natl Acad Sci U S A 101: 14545–14550, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosh S, Moore S, Bell RM, and Dush M. Functional analysis of a phosphatidic acid binding domain in human Raf-1 Kinase mutations in the phosphatidate binding domain lead to tail and trunk abnormalities in developing zebrafish embryos. J Biol Chem 278: 45690–45696, 2003 [DOI] [PubMed] [Google Scholar]
- 44.Groitl B. and Jakob U. Thiol-based redox switches. Biochim Biophys Acta 1844: 1335–1343, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gutscher M, Sobotta MC, Wabnitz GH, Ballikaya S, Meyer AJ, Samstag Y, and Dick TP. Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J Biol Chem 284: 31532–31540, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hall A, Karplus PA, and Poole LB. Typical 2-Cys peroxiredoxins–structures, mechanisms and functions. FEBS J 276: 2469–2477, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He D, Wang Q, Li M, Damaris RN, Yi X, Cheng Z, and Yang P. Global proteome analyses of lysine acetylation and succinylation reveal the widespread involvement of both modification in metabolism in the embryo of germinating rice seed. J Proteome Res 15: 879–890, 2016 [DOI] [PubMed] [Google Scholar]
- 48.Heazlewood JL, Durek P, Hummel J, Selbig J, Weckwerth W, Walther D, and Schulze WX. PhosPhAt: a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res 36(Suppl 1): D1015–D1021, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hofmann B, Hecht HJ, and Flohé L. Peroxiredoxins. Biol Chem 383: 347–364, 2002 [DOI] [PubMed] [Google Scholar]
- 50.Hong SK, Cha MK, and Kim IH. Specific protein interaction of human Pag with Omi/HtrA2 and the activation of the protease activity of Omi/HtrA2. Free Radic Biol Med 40: 275–284, 2006 [DOI] [PubMed] [Google Scholar]
- 51.Hossain MS, ElSayed AI, Moore M, and Dietz KJ. Redox and Reactive Oxygen Species Network in Acclimation for Salinity Tolerance in Sugar Beet. J Exp Bot 68: 1283–1298, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang C, Verrillo F, Renzone G, Arena S, Rocco M, Scaloni A, and Marra M. Response to biotic and oxidative stress in Arabidopsis thaliana: analysis of variably phosphorylated proteins. J Proteomics 74: 1934–1949, 2011 [DOI] [PubMed] [Google Scholar]
- 53.Iglesias-Baena I, Barranco-Medina S, Sevilla F, and Lázaro JJ. The dual-targeted plant sulfiredoxin retroreduces the sulfinic form of atypical mitochondrial peroxiredoxin. Plant Physiol 155: 944–955, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jang HH, Lee KO, Chi YH, Jung BG, Park SK, Park JH, Lee JR, Lee SS, Moon JC, Yun JW, Choi YO, Kim WY, Kang JS, Cheong GW, Yun DJ, Rhee SG, Cho MJ, and Lee SY. Two enzymes in one: two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell 117: 625–635, 2004 [DOI] [PubMed] [Google Scholar]
- 55.Johnson JE. and Cornell RB. Amphitropic proteins: regulation by reversible membrane interactions (review). Mol Membr Biol 16: 217–235, 1999 [DOI] [PubMed] [Google Scholar]
- 56.Jönsson TJ, Johnson LC, and Lowther WT. Structure of the sulphiredoxin–peroxiredoxin complex reveals an essential repair embrace. Nature 451: 98–101, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kelley LA, Mezulis S, Yates CM, Wass MN, and Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.König J, Baier M, Horling F, Kahmann U, Harris G, Schürmann P, and Dietz KJ. The plant-specific function of 2-Cys peroxiredoxin-mediated detoxification of peroxides in the redox-hierarchy of photosynthetic electron flux. Proc Natl Acad Sci USA 99: 5738–5743, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.König J, Galliardt H, Jütte P, Schäper S, Dittmann L, and Dietz KJ. The conformational bases for the two functionalities of 2-cysteine peroxiredoxins as peroxidase and chaperone. J Exp Bot 64: 3483–3497, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.König J, Lotte K, Plessow R, Brockhinke A, Baier M, and Dietz KJ. Reaction mechanism of plant 2-Cys peroxiredoxin role of the C terminus and the quaternary structure. J Biol Chem 278: 24409–24420, 2003 [DOI] [PubMed] [Google Scholar]
- 61.Lemaire SD, Michelet L, Zaffagnini M, Massot V, and Issakidis-Bourguet E. Thioredoxins in chloroplasts. Curr Genet 51: 343–365, 2007 [DOI] [PubMed] [Google Scholar]
- 62.Liebthal M, Strüve M, Li X, Hertle Y, Maynard D, Hellweg T, Viehhauser A, and Dietz KJ. Redox-dependent conformational dynamics of decameric 2-cysteine peroxiredoxin and its interaction with cyclophilin 20–23. Plant Cell Physiol 57: 1415–1425, 2016 [DOI] [PubMed] [Google Scholar]
- 63.Lindermayr C, Saalbach G, and Durner J. Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol 137: 921–930, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Manevich Y, Feinstein SI, and Fisher AB. Activation of the antioxidant enzyme 1-CYS peroxiredoxin requires glutathionylation mediated by heterodimerization with πGST. Proc Natl Acad Sci U S A 101: 3780–3785, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Manevich Y, Reddy KS, Shuvaeva T, Feinstein SI, and Fisher AB. Structure and phospholipase function of peroxiredoxin 6: identification of the catalytic triad and its role in phospholipid substrate binding. J Lipid Res 48: 2306–2318, 2007 [DOI] [PubMed] [Google Scholar]
- 66.Manta B, Hugo M, Ortiz C, Ferrer-Sueta G, Trujillo M, and Denicola A. The peroxidase and peroxynitrite reductase activity of human erythrocyte peroxiredoxin 2. Arch Biochem Biophys 484: 146–154, 2009 [DOI] [PubMed] [Google Scholar]
- 67.Margaria P, Abbà S, and Palmano S. Novel aspects of grapevine response to phytoplasma infection investigated by a proteomic and phospho-proteomic approach with data integration into functional networks. BMC Genomics 14: 1, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matte A, Bertoldi M, Mohandas N, An X, Bugatti A, Brunati AM, Rusnati M, Tibaldi E, Siciliano A, Turrini F, Perrotta S, and De Franceschi L. Membrane association of peroxiredoxin-2 in red cells is mediated by the N-terminal cytoplasmic domain of band 3. Free Radic Biol Med 55: 27–35, 2013 [DOI] [PubMed] [Google Scholar]
- 69.Michelet L, Zaffagnini M, Marchand C, Collin V, Decottignies P, Tsan P, Lancelin JM, Trost P, Miginiac-Maslow M, Noctor G, and Lemaire SD. Glutathionylation of chloroplast thioredoxin f is a redox signaling mechanism in plants. Proc Natl Acad Sci U S A 102: 16478–16483, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mithoe SC, Boersema PJ, Berke L, Snel B, Heck AJ, and Menke FL. Targeted quantitative phosphoproteomics approach for the detection of phospho-tyrosine signaling in plants. J Proteome Res 11: 438–448, 2012 [DOI] [PubMed] [Google Scholar]
- 71.Mock HP, and Dietz KJ. Redox proteomics for the assessment of redox-related posttranslational regulation in plants. Biochim Biophys Acta 1864: 967–973, 2016 [DOI] [PubMed] [Google Scholar]
- 72.Monteiro G, Horta BB, Pimenta DC, Augusto O, and Netto LE. Reduction of 1-Cys peroxiredoxins by ascorbate changes the thiol-specific antioxidant paradigm, revealing another function of vitamin C. Proc Natl Acad Sci USA 104: 4886–4891, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moore RB. and Shriver SK. Protein 7.2 b of human erythrocyte membranes binds to calpromotin. Biochem Biophys Res Commun 232, 294–297, 1997 [DOI] [PubMed] [Google Scholar]
- 74.Moore RB, Mankad MV, Shriver SK, Mankad VN, and Plishker GA. Reconstitution of Ca (2+)-dependent K+ transport in erythrocyte membrane vesicles requires a cytoplasmic protein. J Biol Chem 266, 18964–18968, 1991 [PubMed] [Google Scholar]
- 75.Morais MA, Giuseppe PO, Souza TA, Alegria TG, Oliveira MA, Netto LE, and Murakami MT. How pH modulates the dimer-decamer interconversion of 2-Cys peroxiredoxins from the Prx1 subfamily. J Biol Chem 290: 8582–8590, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Muthuramalingam M, Matros A, Scheibe R, Mock HP, and Dietz KJ. The hydrogen peroxide-sensitive proteome of the chloroplast in vitro and in vivo. Front Plant Sci 4: 54, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Muthuramalingam M, Seidel T, Laxa M, De Miranda SMN, Gärtner F, Ströher E, Kandlbinder A, and Dietz KJ. Multiple redox and non-redox interactions define 2-Cys peroxiredoxin as a regulatory hub in the chloroplast. Mol Plant 2: 1273–1288, 2009 [DOI] [PubMed] [Google Scholar]
- 78.Nakagami H, Sugiyama N, Mochida K, Daudi A, Yoshida Y, Toyoda T, Tomita M, Ishihama Y, and Shirasu K. Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol 153: 1161–1174, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nietzel T, Mostertz J, Hochgräfe F, and Schwarzländer M. Redox regulation of mitochondrial proteins and proteomes by cysteine thiol switches. Mitochondrion 33: 72–83, 2016 [DOI] [PubMed] [Google Scholar]
- 80.Noctor G, Mhamdi A, Chaouch S, Han YI, Neukermans J, Marquez-Garcia B, Queval G, and Foyer CH. Glutathione in plants: an integrated overview. Plant Cell Environ 35: 454–484, 2012 [DOI] [PubMed] [Google Scholar]
- 81.Noguera-Mazon V, Krimm I, Walker O, and Lancelin JM. Protein–protein interactions within peroxiredoxin systems. Photosynthesis Res 89: 277–290, 2006 [DOI] [PubMed] [Google Scholar]
- 82.Noichri Y, Palais G, Ruby V, D'Autreaux B, Delaunay-Moisan A, Nyström T, Molin M, and Toledano MB. In vivo parameters influencing 2-CysPrx oligomerization: the role of enzyme sulfinylation. Redox Biol 6: 326–333, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pan Y, Jin JH, Yu Y, and Wang J. Significant enhancement of hPrx1 chaperone activity through lysine acetylation. Chembiochem 15: 1773–1776, 2014 [DOI] [PubMed] [Google Scholar]
- 84.Park JW, Piszczek G, Rhee SG, and Chock PB. Glutathionylation of peroxiredoxin I induces decamer to dimers dissociation with concomitant loss of chaperone activity. Biochemistry 50: 3204–3210, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Park SW, Li W, Viehhauser A, He B, Kim S, Nilsson AK, Andersson MX, Kittle JD, Ambavaram MM, Luan S, Esker AR, Tholl D, Cimini D, Ellerstrom M, Coaker G, Mitchell TK, Pereira A, Dietz KJ, and Lawrence CB. Cyclophilin 20-3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc Natl Acad Sci USA 110: 9559–9564, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Parsonage D, Youngblood DS, Sarma GN, Wood ZA, Karplus PA, and Poole LB. Analysis of the Link between Enzymatic Activity and Oligomeric State in AhpC, a Bacterial Peroxiredoxin†. Biochemistry 44: 10583–10592, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pedrajas JR, McDonagh B, Hernández-Torres F, Miranda-Vizuete A, González-Ojeda R, Martínez-Galisteo E, Padilla CA, and Bárcena JA. Glutathione is the resolving thiol for thioredoxin peroxidase activity of 1-Cys peroxiredoxin without being consumed during the catalytic cycle. Antioxid Redox Signal 24: 115–128, 2016 [DOI] [PubMed] [Google Scholar]
- 88.Pehar M, Ball LE, Sharma DR, Harlan BA, Comte-Walters S, Neely BA, and Vargas MR. Changes in protein expression and lysine acetylation induced by decreased glutathione levels in astrocytes. Mol Cell Proteomics 15: 493–505, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peltier JB, Cai Y, Sun Q, Zabrouskov V, Giacomelli L, Rudella A, Ytterberg AJ, Rutschow H, and van Wijk KJ. The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol Cell Proteomics 5: 114–133, 2006 [DOI] [PubMed] [Google Scholar]
- 90.Pérez-Ruiz JM, Spínola MC, Kirchsteiger K, Moreno J, Sahrawy M, and Cejudo FJ. Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell 18: 2356–2368, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Perkins A, Nelson KJ, Parsonage D, Poole LB, and Karplus PA. Peroxiredoxins: guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem Sci 40: 435–445, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Plishker GA, Chevalier D, Seinsoth L, and Moore RB. Calcium-activated potassium transport and high molecular weight forms of calpromotin. J Biol Chem 267: 21839–21843, 1992 [PubMed] [Google Scholar]
- 93.Poole LB. The basics of thiols and cysteines in redox biology and chemistry. Free Radic Biol Med 80: 148–157, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Puerto-Galán L, Pérez-Ruiz JM, Guinea M, and Cejudo FJ. The contribution of NADPH thioredoxin reductase C (NTRC) and sulfiredoxin to 2-Cys peroxiredoxin overoxidation in Arabidopsis thaliana chloroplasts. J Exp Bot 66: 2957–2966, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pulido P, Cazalis R, and Cejudo FJ. An antioxidant redox system in the nucleus of wheat seed cells suffering oxidative stress. Plant J 57: 132–145, 2009 [DOI] [PubMed] [Google Scholar]
- 96.Pulido P, Spínola MC, Kirchsteiger K, Guinea M, Pascual MB, Sahrawy M, Sandalio LM, Dietz KJ, González M, and Cejudo FJ. Functional analysis of the pathways for 2-Cys peroxiredoxin reduction in Arabidopsis thaliana chloroplasts. J Exp Bot 61: 4043–4054, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Qi Y. and Grishin NV. Structural classification of thioredoxin-like fold proteins. Proteins 58: 376–388, 2005 [DOI] [PubMed] [Google Scholar]
- 98.Reiland S, Finazzi G, Endler A, Willig A, Baerenfaller K, Grossmann J, and Baginsky S. Comparative phosphoproteome profiling reveals a function of the STN8 kinase in fine-tuning of cyclic electron flow (CEF). Proc Natl Acad Sci USA 108: 12955–12960, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, and Baginsky S. Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol 150: 889–903, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rhee SG. Overview on Peroxiredoxin. Mol Cells 39: 1–5, 2016a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rhee SG. and Kil IS. Mitochondrial H2O2 signaling is controlled by the concerted action of peroxiredoxin III and sulfiredoxin: linking mitochondrial function to circadian rhythm. Free Radic Biol Med 99: 120–127, 2016 [DOI] [PubMed] [Google Scholar]
- 102.Rochaix JD. Redox regulation of thylakoid protein kinases and photosynthetic gene expression. Antioxid Redox Signal 18: 2184–2201, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roitinger E, Hofer M, Köcher T, Pichler P, Novatchkova M, Yang J, Schlögelhofer P, and Mechtler K. Quantitative phosphoproteomics of the ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia-mutated and rad3-related (ATR) dependent DNA damage response in Arabidopsis thaliana. Mol Cell Proteomics 14: 556–571, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Romero-Puertas MC, Laxa M, Mattè A, Zaninotto F, Finkemeier I, Jones AM, Perazzolli M, Vandelle E, Dietz KJ, and Delledonne M. S-nitrosylation of peroxiredoxin II E promotes peroxynitrite-mediated tyrosine nitration. Plant Cell 19: 4120–4130, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rose CM, Venkateshwaran M, Volkening JD, Grimsrud PA, Maeda J, Bailey DJ, and Westphall MS. Rapid phosphoproteomic and transcriptomic changes in the rhizobia-legume symbiosis. Mol Cell Proteomics 11: 724–744, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rouhier N, Gelhaye E, Sautiere PE, Brun A, Laurent P, Tagu D, Gerard J, de Fay E, Meyer Y, and Jacquot JP. Isolation and characterization of a new peroxiredoxin from poplar sieve tubes that uses either glutaredoxin or thioredoxin as a proton donor. Plant Physiol 127: 1299–1309, 2001 [PMC free article] [PubMed] [Google Scholar]
- 107.Sakamoto A, Tsukamoto S, Yamamoto H, Ueda-Hashimoto M, Takahashi M, Suzuki H, and Morikawa H. Functional complementation in yeast reveals a protective role of chloroplast 2-Cys peroxiredoxin against reactive nitrogen species. Plant J 33: 841–851, 2003 [DOI] [PubMed] [Google Scholar]
- 108.Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, and Kunze R. ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol 131: 16–26, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Seo JH, Lim JC, Lee DY, Kim KS, Piszczek G, Nam HW, Kim YS, Ahn T, Yun CH, Kim K, Chock PB, and Chae HS. Novel protective mechanism against irreversible hyperoxidation of peroxiredoxin Nα-terminal acetylation of human peroxiredoxin II. J Biol Chem 284: 13455–13465, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sevilla F, Camejo D, Ortiz-Espín A, Calderón A, Lázaro JJ, and Jiménez A. The thioredoxin/peroxiredoxin/sulfiredoxin system: current overview on its redox function in plants and regulation by reactive oxygen and nitrogen species. J Exp Bot 66: 2945–2955, 2015 [DOI] [PubMed] [Google Scholar]
- 111.Soito L, Williamson C, Knutson ST, Fetrow JS, Poole LB, and Nelson KJ. PREX: PeroxiRedoxin classification indEX, a database of subfamily assignments across the diverse peroxiredoxin family. Nucleic Acids Res 39(Suppl 1): D332–D337, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Speiser A, Haberland S, Watanabe M, Wirtz M, Dietz KJ, Saito K, and Hell R. The significance of cysteine synthesis for acclimation to high light conditions. Front Plant Sci 5: 776, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stacy RA, Munthe E, Steinum T, Sharma B, and Aalen RB. A peroxiredoxin antioxidant is encoded by a dormancy-related gene, Per1, expressed during late development in the aleurone and embryo of barley grains. Plant Mol Biol 31: 1205–1216, 1996 [DOI] [PubMed] [Google Scholar]
- 114.Umezawa T, Sugiyama N, Takahashi F, Anderson JC, Ishihama Y, Peck SC, and Shinozaki K. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci Signal 6: rs8, 2013 [DOI] [PubMed] [Google Scholar]
- 115.van Wijk KJ, Friso G, Walther D, and Schulze WX. Meta-analysis of Arabidopsis thaliana phospho-proteomics data reveals compartmentalization of phosphorylation motifs. Plant Cell 26: 2367–2389, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang P, Xue L, Batelli G, Lee S, Hou YJ, Van Oosten MJ, Zhang H, Tao WA, and Zhu JK. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc Natl Acad Sci USA 110: 11205–11210, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang X, Bian Y, Cheng K, Zou H, Sun SSM, and He JX. A comprehensive differential proteomic study of nitrate deprivation in Arabidopsis reveals complex regulatory networks of plant nitrogen responses. J Proteome Res 11: 2301–2315, 2012 [DOI] [PubMed] [Google Scholar]
- 118.Woo HA, Jeong W, Chang TS, Park KJ, Park SJ, Yang JS, and Rhee SG. Reduction of cysteine sulfinic acid by sulfiredoxin is specific to 2-cys peroxiredoxins. J Biol Chem 280: 3125–3128, 2005 [DOI] [PubMed] [Google Scholar]
- 119.Woo HA, Yim SH, Shin DH, Kang D, Yu DY, and Rhee SG. Inactivation of peroxiredoxin I by phosphorylation allows localized H2O2 accumulation for cell signaling. Cell 140: 517–528, 2010 [DOI] [PubMed] [Google Scholar]
- 120.Wood ZA, Poole LB, Hantgan RR, and Karplus PA. Dimers to Doughnuts: Redox-Sensitive Oligomerization of 2-Cysteine Peroxiredoxins†. Biochemistry 41: 5493–5504, 2002 [DOI] [PubMed] [Google Scholar]
- 121.Wood ZA, Poole LB, and Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science 300: 650–653, 2003a [DOI] [PubMed] [Google Scholar]
- 122.Wood ZA, Schröder E, Harris JR, and Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci 28: 32–40, 2003b [DOI] [PubMed] [Google Scholar]
- 123.Wu XN, Rodriguez CS, Pertl-Obermeyer H, Obermeyer G, and Schulze WX. Sucrose-induced receptor kinase SIRK1 regulates a plasma membrane aquaporin in Arabidopsis. Mol Cell Proteomics 12: 2856–2873, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xiong Y, Zhang K, Cheng Z, Wang GL, and Liu W. Data for global lysine-acetylation analysis in rice (Oryza sativa). Data Brief 7: 411–417, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xu S, Ding H, Su F, Zhang A, and Jiang M. Involvement of Protein Phosphorylation in Water Stress-induced Antioxidant Defense in Maize Leaves. J Integr Plant Biol 51: 654–662, 2009 [DOI] [PubMed] [Google Scholar]
- 126.Yang Z, Guo G, Zhang M, Liu CY, Hu Q, Lam H, and Li N. Stable isotope metabolic labeling-based quantitative phosphoproteomic analysis of Arabidopsis mutants reveals ethylene-regulated time-dependent phosphoproteins and putative substrates of constitutive triple response 1 kinase. Mol Cell Proteomics 12: 3559–3582, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yao Q, Ge H, Wu S, Zhang N, Chen W, Xu C, Gao J, Thelen JJ, and Xu D. P3DB 3.0: from plant phosphorylation sites to protein networks. Nucleic Acids Res 42: D1206–D1213, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang H, Zhou H, Berke L, Heck AJ, Mohammed S, Scheres B, and Menke F. L. Quantitative phosphoproteomics after auxin-stimulated lateral root induction identifies an SNX1 protein phosphorylation site required for growth. Mol Cell Proteomics 12: 1158–1169, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]