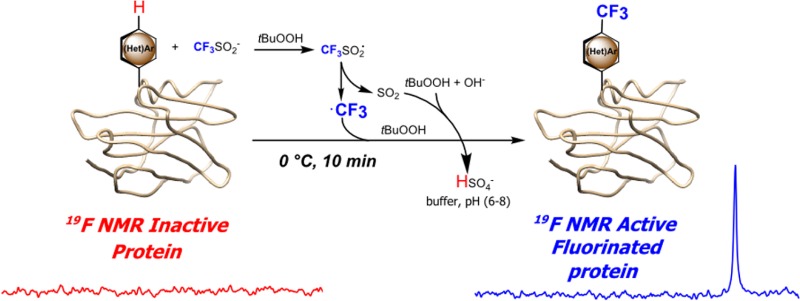
Abstract
The incorporation of fluorine can not only significantly facilitate the study of proteins but also potentially modulate their function. Though some biosynthetic methods allow global residue-replacement, post-translational fluorine incorporation would constitute a fast and efficient alternative. Here, we reveal a mild method for direct protein radical trifluoromethylation at native residues as a strategy for symmetric-multifluorine incorporation on mg scales with high recoveries. High selectivity toward tryptophan residues enhanced the utility of this direct trifluoromethylation technique allowing ready study of fluorinated protein constructs using 19F-NMR.
Although absent from nearly all-natural products, small organofluorine compounds have found widespread applications from materials,1 to clinical diagnostics (19F-MRI/18F-PET)2,3 and medicinal chemistry (where 30%4 of new small molecule drugs contain F to alter physicochemical properties5 and/or modulate affinity6). Incorporation of fluorine into biomolecules is more rare but potential applications in structural re-engineering7 and as reporter tags for “zero-background” 19F-NMR8,9 raise striking possibilities. For the latter, the benefits include: high sensitivity, extreme responsiveness to local environment, broad chemical shift range and low reactivity once incorporated.10 This has helped to elucidate structural, functional and dynamic11,12 interactions in receptor function landscapes13 and enzymic catalysis.14 Indeed, fluorine’s features can render it superior over other nuclei in, for example, in-cell NMR spectroscopy15 or protein-observed 19F-NMR (PrOF) protein–ligand binding.16−19
Apart from (semi)synthetic ligations,20 two main strategies are used for generating fluorinated proteins: incorporation of fluorinated amino acids using biosynthetic methods or chemical modification.21 The former is typically only residue-specific22 but can be achieved in a site-selective manner through sense or nonsense codon reassignment (e.g., for positioning of CF3-l-phenylalanine23). However, for operational simplicity and to avoid identified issues with the former,24 the latter modification method has often been preferred as more direct. It has relied largely on reaction of nucleophilic side chains with prosthetic groups bearing fluorine (typically cysteine and lysine) often attached via linkers or prosthetics25,26 that can perturb structure and create distance imprecision.24 A lack of other methods for installing fluorine into biomolecules has been highlighted as a restriction;24,27 due to low relative abundance and roles in interiors, aromatic amino acids are attractive targets. Therefore, direct (hetero)aromatic trifluoromethylation could be a powerful tool allowing (a) “zero/short linker” fluorine incorporation via (b) a higher sensitivity multiequivalent-F system.28,29 Here, we describe a novel method that exploits the chemoselectivity of radicals in protein chemistry to achieve this aim (Figure 1).
Figure 1.

Direct radical trifluoromethylation of protein substrates. ·CF3 radical targeting (hetero)aromatic residues can be generated from sodium trifluoromethanesulfinate (NaTFMS, Langlois’ reagent) under aqueous, oxidative conditions.
Direct C–CF3-bond trifluoromethylation presents a striking synthetic challenge in biomolecules. Most strategies for C–H functionalization have been applied only to small molecules using methods or conditions essentially incompatible with proteins (elevated temperatures,30 organic solvents31 or strong acids32). Among possible strategies, we identified radical-based approaches.33 We have demonstrated the benign nature of radicals for C–C-bond formation through use of a designed unnatural amino acid.34 This illustrated radical efficiency under ambient aqueous conditions and operational simplicity. We therefore considered whether a suitable ·CF3 precursor might be generated that could allow natural amino acid modification.
Various putative ·CF3 sources35 were tested using model amino acid substrates. Langlois’ reagent (NaTFMS) proved most promising via multiple redox initiation modes (e.g., transition-metal-free, photocatalytic) under mild conditions.36−38 Baran et al. identified that NaTFMS may be used to modify small molecule N-heterocycle caffeine when spiked into cell lysate, which has “important implications in the area of bioconjugation”.39 Advantageously, 19F-NMR allowed us ready reactivity screening of all abundant natural amino acids (SI Figure S1) and revealed selective reactivity of NaTFMS/TBHP39 with only aromatic amino acids and free cysteine ((hetero)arene-C–H or Cys-S–H trifluoromethylation) and none toward any other natural amino acids. 19F-NMR suggested near homogeneous product formation from tryptophan (Trp) and cysteine (Cys); low levels of products from His, Tyr and Phe, appeared as complex mixtures of apparent regioisomers.
Next, limited (<40%) conversion competition assays (Figure 2) using mixtures of equimolar Trp, Phe, Tyr, Cys, His, Met with NaTFMS/TBHP were used to assess residue-specific selectivity. These revealed preferential formation of the Trp-C2 isomer at a krel > ∼7.5-fold. This could be further increased to krel > ∼30-fold by lowering the pH to 6 (SI Table T2), potentially due to reduced reactivity of protonated His residues. The main side reaction was oxidative dimerization of Cys without trifluoromethylation. These observations suggested not only the cysteine disulfide is inert but in the absence of free Cys, strongly chemo- and regio- Trp-selective trifluoromethylation might be possible in proteins. This was confirmed by a similar competition reaction in the absence of Cys (SI Figure S3), along with His modification as minor (<1:24) side-reaction. Optimization of the reaction toward Trp revealed the key role of pH (SI Table T3). In weakly or nonbuffered aqueous conditions, rapid acidification was observed, consistent with a mechanism that leads to hydrogensulfate formation (Figure 1).40 Useful conversions to CF3-Trp (>50%) were obtained at a moderate pH ∼ 6 (Figure S4), optimal for most biomolecules.
Figure 2.

Limited-conversion competition assay of natural amino acids (0.03 mmol) revealed chemo- and regio-selectivity toward Trp with minor products (Cys and lower levels of Phe, His, Tyr isomers, determined by 19F-NMR, LC-MS). See Figure S3 for parallel reaction in absence of Cys.
Together, these results prompted investigation in more complex peptidic and protein substrates. First, reaction of N-acetyl-l-tryptophanamide confirmed reactivity and identical regio-selectivity to that seen for Trp. Second, as a simple model peptide, we chose melittin, a 26-residue peptide with a single Trp residue at Trp19. Use of excess NaTFMS/TBHP with methionine as an oxidative buffer allowed short reaction times (Figure 3). The exclusive site of modification at Trp19 was confirmed by LC-MS/MS analysis (SI Figure S3); Trp19 oxidation and dual Trp19 CF3-ylation were observed under prolonged conditions. 4-Hydroxy-TEMPO not only allowed ready termination of the reaction at varied time points but also confirmed the radical-mediated nature.
Figure 3.
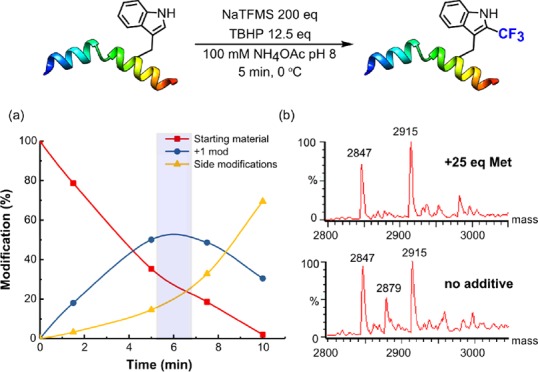
Reaction of melittin (MWcalc= 2847) with NaTFMS/TBHP. (a) Relative conversions from LC-MS with the optimal reaction time highlighted in blue. (b) Deconvoluted LC-MS spectra of reaction mixtures after 5 min ± Met. Undesired oxidation (+32) is curtailed by sacrificial reductant giving melittin-TrpCF3 (+68, MWcalc = 2915, MWfound = 2915).
As a natural protein substrate, we chose model hemoprotein–horse heart myoglobin (Mb). Mb contains two buried Trp (Trp7, Trp14) in the presence of several other potentially reactive, more accessible residues (SI Figure S6). Optimization of conditions (pH, temperature, reactants loading, SI Table T4, SI Figure S7) allowed modulation of reactivity by CF3 copy-number. In this way, at pH 6 (100 mM ammonium acetate), 0 °C, using 200 equiv NaTFMS after 10 min we were able to create myoglobin bearing primarily one or two CF3 groups (Figure 4a). Higher equivalents of Langlois’ reagent allowed further shortening of reaction times, without apparent adverse effects on protein (SI Table T4, entries 5,6). In contrast to literature observations,39 we observed no significant influence of Zn2+ as an additive. Control of pH also minimized concomitant oxidation (SI Figure S8). Tryptic digest-LCMS/MS revealed that Trp7 and Trp14 were the dominant sites of modification (Figure 3b and SI Figure S9) despite their buried nature, highlighting the low steric hindrance of ·CF3, even in these congested environments. This accessibility to buried sites complements existing nucleophilic, prosthetic-mediated methods, which are more applicable to exposed sites, requiring partial denaturation to access buried sites.41−43 Similarly, accessibility is needed for standard labeling of Cys with fluorinated tags. Importantly, CD spectroscopy revealed that trifluoromethylation had negligible influence on the structure of Mb-TrpCF3 protein (SI Figure S10).
Figure 4.
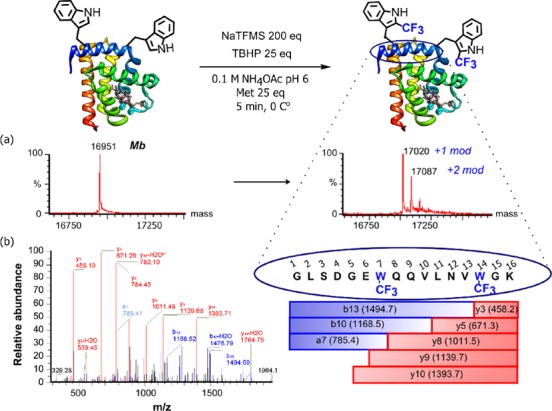
Direct trifluoromethylation of proteins (a) myoglobin (Mb, MWcalc = 16951.5) deconvoluted LC-MS spectrum showing singly- (MWcalc = 17019, MWfound = 17020) and doubly- modified (MWcalc = 17087, MWfound = 17087), (b) tryptic-LC-MS/MS confirmed site-selectivity at Trp7 and Trp14.
Next, we tested the prospective utility of our developed direct trifluoromethylation as a tool for enabling protein 19F-NMR. Pleasingly, mg-scale reactions allowed straightforward “labeling” of Mb with CF3 with ∼65–80% recovery. As a proof-of-principle, we used 19F-NMR to directly probe sites of modification (Figure 5a). The native protein spectra of modified Mb revealed two major 19F-peaks. To elucidate their origin, protein was digested with trypsin (to free residues from their microenvironment and remove nonequivalence); the resonances collapsed to δF −58.8 ppm, in good agreement to that for 2-CF3-Trp in model Trp-CF3bis-amide (δF −58.76). This was further reinforced by denaturative 19F-NMR structural analyses:44 titration with urea induced a gradual change in chemical shift (Figure 5b) that could be explained by a known progressive loss of secondary structure in Mb under such conditions.45 Treatment with 10 M urea resulted in full coalescence to δ-58.74 ppm (Figure 5a, full spectrum Figure S11).46,47
Figure 5.
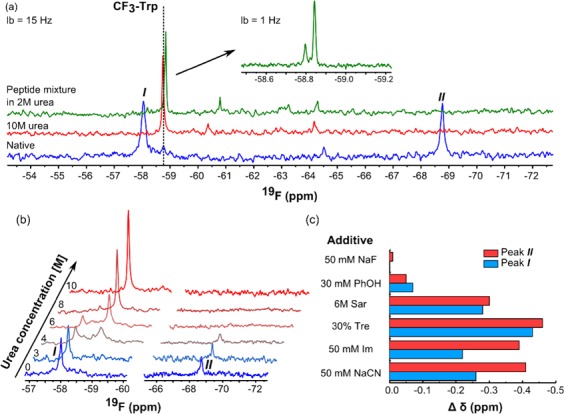
19F-NMR analyses enabled by direct trifluoromethylation. (a) 19F-NMR of CF3Trp-Mb (700 μM in 100 mM NH4OAc, pH 8, blue), denatured by 10 M urea (300 μM, red) and peptide mixture from tryptic digest (from 2 mM protein, green). Dashes indicate chemical shift for small molecule model. Inset at higher field with individual Trp resolved. (b) Urea titration by 19F-NMR (protein 200–400 μM). (c) Chemical shift differences from ligands in PrOF, protein concentration (100–300 μM).
We extended essentially the same protocol to other model proteins with different folds and Trp copy number: pantothenate synthetase (PanC with single Trp306) and more demanding enzyme lysozyme (Ly), which possesses six potentially reactive Trp (Trp28, Trp62, Trp63, Trp108, Trp111 and Trp123) and eight Cys engaged within each other as disulfides. Both tryptic-LC-MS/MS and 19F-NMR (SI Figures S12–17) not only confirmed Trp residues as primary modification sites in both proteins but also revealed no reaction of Cys residues, consistent with our prior observations. Thermal denaturation also gave rise to resonance coalesence, as for Mb (SI Figure S19). We were also able to determine that trifluoromethylation had negligible influence upon both the structure of Ly (via CD, SI Figure S20) but also upon its enzymatic function (SI Figure 22, SI Table T5), which was essentially unaltered. We were also able to use this fully active CF3-ylated Ly in quantitative 19F-NMR PrOF16−19 (SI Figure 21) to directly determine the binding of known alkaloid inhibitor ligand berberine (Kd 20 μM, consistent with values determined by complementary methods48). The potential of our direct trifluoromethylation method was also demonstrated in more complex structural experiments for different protein–ligand states studied by PrOF.16−19 This allowed observation of displacement of water bound at hemin iron (which we confirmed by UV–vis49 was retained after CF3-ylation, Figure S23) by known ligands cyanide, fluoride, imidazole (Im)50 (Figure 5c, full spectra Figure S24).16 We also tested effects of common osmo/cryo- protectants (sarcosine – Sar, trehalose – Tre)51 and putative ligand phenol.52 CN–, Im caused clear upfield shifts, consistent with being known specific ligands.47 Both Trp residues are located within the same helix; both signals reported changes, yet these were more pronounced for more-tightly packed Trp14 (Figure S26). Interestingly, known ligand F– did not trigger a change in chemical shift suggesting that this small ligand has a negligible impact on overall protein conformation. Conversely, Tre and Sar caused globally observed changes, consistent with nonspecific interactions or changes in solute properties, in agreement with the mode of actions of these compounds.53 Phenol, which inhibits the dehaloperoxidase activity of myoglobins,52 showed only small changes implying only nonspecific binding, likely not at hemin (Figure S25).
In conclusion, we have demonstrated direct trifluoromethylation of natural residues in proteins. Tuned radical chemistry allows fast (<10 min) and chemo-selective modification of Trp with a useful but not absolute selectivity over other putative residues in peptidic systems and proteins. This method appears to provide a selective and direct protocol for trifluoromethylation of proteins. Methods exploiting photochemical generation54,55 or the use of electrophilic alkylation chemistry56 suggest lower and/or differing selectivities. Interestingly, in the context of the known ambiguity of ·CF3 toward heteroarenes,57 this selectivity seems to exploit electrophilic polarity; our preliminary results using known precursors to nucleophilic ·CF2Me under essentially identical conditions failed. Trp vs His selectivity is increased at pH 6 (Table T2), also consistent with lower reactivity of electrophilic ·CF3 toward protonated His. It allows direct access to 19F-NMR experiments without the requirement of additional mutation of protein substrates and use of linkers, as is currently the case.58 In particular, we showed through fractional CF3-“labeling” 19F-NMR spectra can be obtained directly with no significant line broadening.59 Although Trp residues are rare, their frequent role and in proximity to protein–ligand/protein interactions and presence at hydrophobic interfaces makes them useful probes in PrOF assays.60−63 We speculate too that the reactivity observed here with typically inaccessible sites reflects both the lack of bulk (no ‘linker’/ “zero size”) and prior observations that biphasic/interfacial regions enhance small molecule radical reactions.40 It therefore complements other Trp-modification, such as the use of Rh-carbenoids that work best with surface residues and currently allow more generality in modifying group than the method we present here.42 Other applications can also be envisaged: tuning of intrinsic Trp fluorescence64 or physicochemical effects via fluorination (e.g., “Teflon” proteins65). We observed even low levels of fluorine incorporation altered fluorescence spectra (SI Figure S27) and changed chromatographic behaviors (SI Figure S28).
Acknowledgments
Funded under the EU Horizon 2020 programme, Marie Skłodowska-Curie grant agreement No. 675071, and by a Croucher Foundation Fellowship.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.7b10230.
Experimental and characterization data (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Leclerc N.; Chavez P.; Ibraikulov O. A.; Heiser T.; Leveque P. Polymers 2016, 8, 11. 10.3390/polym8010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirotta I.; Dichiarante V.; Pigliacelli C.; Cavallo G.; Terraneo G.; Bombelli F.; Metrangolo P.; Resnati G. Chem. Rev. 2015, 115, 1106. 10.1021/cr500286d. [DOI] [PubMed] [Google Scholar]
- Preshlock S.; Tredwell M.; Gouverneur V. Chem. Rev. 2016, 116, 719. 10.1021/acs.chemrev.5b00493. [DOI] [PubMed] [Google Scholar]
- Zhou Y.; Wang J.; Gu Z.; Wang S.; Zhu W.; Acena J. L.; Soloshonok V. A.; Izawa K.; Liu H. Chem. Rev. 2016, 116, 422. 10.1021/acs.chemrev.5b00392. [DOI] [PubMed] [Google Scholar]
- Purser S.; Moore P. R.; Swallow S.; Gouverneur V. Chem. Soc. Rev. 2008, 37, 320. 10.1039/B610213C. [DOI] [PubMed] [Google Scholar]
- Muller K.; Faeh C.; Diederich F. Science 2007, 317, 1881. 10.1126/science.1131943. [DOI] [PubMed] [Google Scholar]
- Marsh E. N. Acc. Chem. Res. 2014, 47, 2878. 10.1021/ar500125m. [DOI] [PubMed] [Google Scholar]
- Danielson M. A.; Falke J. J. Annu. Rev. Biophys. Biomol. Struct. 1996, 25, 163. 10.1146/annurev.bb.25.060196.001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.; Zhang Z. T.; Jiang B.; Zhang X.; Li C.; Liu M. Anal. Bioanal. Chem. 2014, 406, 2279. 10.1007/s00216-013-7518-5. [DOI] [PubMed] [Google Scholar]
- Yu J. X.; Hallac R. R.; Chiguru S.; Mason R. P. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 70, 25. 10.1016/j.pnmrs.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung R. L. C.; Robinson M. D. M.; Ajabali A. A.; Karunanithy G.; Lyons B.; Raj R.; Raoufmoghaddam S.; Mohammed S.; Claridge T. D. W.; Baldwin A. J.; Davis B. G. J. Am. Chem. Soc. 2017, 139, 5277. 10.1021/jacs.6b11040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manglik A.; Kim T. H.; Masureel M.; Altenbach C.; Yang Z. Y.; Hilger D.; Lerch M. T.; Kobilka T. S.; Thian F. S.; Hubbell W. L.; Prosser R. S.; Kobilka B. K. Cell 2015, 162, 1431. 10.1016/j.cell.2015.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L.; Van Eps N.; Zimmer M.; Ernst O. P.; Prosser R. S. Nature 2016, 533, 265. 10.1038/nature17668. [DOI] [PubMed] [Google Scholar]
- Kim T. H.; Mehrabi P.; Ren Z.; Sljoka A.; Ing C.; Bezginov A.; Ye L.; Pomes R.; Prosser R. S.; Pai E. F. Science 2017, 355, eaag2355. 10.1126/science.aag2355. [DOI] [PubMed] [Google Scholar]
- Ye Y.; Liu X.; Chen Y.; Xu G.; Wu Q.; Zhang Z.; Yao C.; Liu M.; Li C. Chem. - Eur. J. 2015, 21, 8686. 10.1002/chem.201500279. [DOI] [PubMed] [Google Scholar]
- Gee C. T.; Arntson K. E.; Urick A. K.; Mishra N. K.; Hawk L. M.; Wisniewski A. J.; Pomerantz W. C. Nat. Protoc. 2016, 11, 1414. 10.1038/nprot.2016.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urick A. K.; Calle L. P.; Espinosa J. F.; Hu H.; Pomerantz W. C. ACS Chem. Biol. 2016, 11, 3154. 10.1021/acschembio.6b00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arntson K. E.; Pomerantz W. C. J. Med. Chem. 2016, 59, 5158. 10.1021/acs.jmedchem.5b01447. [DOI] [PubMed] [Google Scholar]
- Urick A. K.; Hawk L. M.; Cassel M. K.; Mishra N. K.; Liu S.; Adhikari N.; Zhang W.; dos Santos C. O.; Hall J. L.; Pomerantz W. C. ACS Chem. Biol. 2015, 10, 2246. 10.1021/acschembio.5b00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malins L. R.; Payne R. J. Curr. Opin. Chem. Biol. 2014, 22, 70. 10.1016/j.cbpa.2014.09.021. [DOI] [PubMed] [Google Scholar]
- Frieden C.; Hoeltzli S.; Bann J. Methods Enzymol. 2004, 380, 400. 10.1016/S0076-6879(04)80018-1. [DOI] [PubMed] [Google Scholar]
- Marsh E. N.; Suzuki Y. ACS Chem. Biol. 2014, 9, 1242. 10.1021/cb500111u. [DOI] [PubMed] [Google Scholar]
- Hammill J. T.; Miyake-Stoner S.; Hazen J. L.; Jackson J. C.; Mehl R. A. Nat. Protoc. 2007, 2, 2601. 10.1038/nprot.2007.379. [DOI] [PubMed] [Google Scholar]
- Kitevski-LeBlanc J. L.; Prosser R. S. Prog. Nucl. Magn. Reson. Spectrosc. 2012, 62, 1. 10.1016/j.pnmrs.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Gerig J. T. Prog. Nucl. Magn. Reson. Spectrosc. 1994, 26, 293. 10.1016/0079-6565(94)80009-X. [DOI] [Google Scholar]
- Basle E.; Joubert N.; Pucheault M. Chem. Biol. 2010, 17, 213. 10.1016/j.chembiol.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Tian H.; Furstenberg A.; Huber T. Chem. Rev. 2017, 117, 186. 10.1021/acs.chemrev.6b00084. [DOI] [PubMed] [Google Scholar]
- Rapidly rotating CF3 produces strong, narrow NMR signal.
- Kim H. J.; Howell S. C.; Van Horn W. D.; Jeon Y. H.; Sanders C. R. Prog. Nucl. Magn. Reson. Spectrosc. 2009, 55, 335. 10.1016/j.pnmrs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natte K.; Jagadeesh R. V.; He L.; Rabeah J.; Chen J.; Taeschler C.; Ellinger S.; Zaragoza F.; Neumann H.; Brückner A.; Beller M. Angew. Chem., Int. Ed. 2016, 55, 2782. 10.1002/anie.201511131. [DOI] [PubMed] [Google Scholar]
- Nagib D. A.; MacMillan D. W. Nature 2011, 480, 224. 10.1038/nature10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.; Li Z.; Kan J.; Huang S.; Su W.; Li Y. Nat. Commun. 2017, 8, 14353. 10.1038/ncomms14353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M.; Lo J. C.; Edwards J. T.; Baran P. S. J. Am. Chem. Soc. 2016, 138, 12692. 10.1021/jacs.6b08856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller M. F.; Fung M. K.; Wright T. H.; Katz M. E.; Kent D. V. Science 2016, 354, 225. 10.1126/science.aaf5466. [DOI] [PubMed] [Google Scholar]
- Jiang Y.; Yu H.; Fu Y.; Liu L. Sci. China: Chem. 2015, 58, 673. 10.1007/s11426-014-5178-8. [DOI] [Google Scholar]
- Lefebvre Q. Synlett 2016, 28, 19. 10.1055/s-0036-1588643. [DOI] [Google Scholar]
- Li L.; Mu X.; Liu W.; Wang Y.; Mi Z.; Li C. J. J. Am. Chem. Soc. 2016, 138, 5809. 10.1021/jacs.6b02782. [DOI] [PubMed] [Google Scholar]
- Wang D.; Deng G.; Chen S.; Gong H. Green Chem. 2016, 18, 5967. 10.1039/C6GC02000C. [DOI] [Google Scholar]
- Fujiwara Y.; Dixon J. A.; O’Hara F.; Funder E. D.; Dixon D. D.; Rodriguez R. A.; Baxter R. D.; Herle B.; Sach N.; Collins M. R.; Ishihara Y.; Baran P. S. Nature 2012, 492, 95. 10.1038/nature11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y.; Brueckl T.; Baxter R.; Fujiwara Y.; Seiple I.; Su S.; Blackmond D.; Baran P. S. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 14411. 10.1073/pnas.1109059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seim K. L.; Obermeyer A. C.; Francis M. B. J. Am. Chem. Soc. 2011, 133, 16970. 10.1021/ja206324q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antos J. M.; McFarland J. M.; Iavarone A. T.; Francis M. B. J. Am. Chem. Soc. 2009, 131, 6301. 10.1021/ja900094h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y.; Ishiyama T.; Sasaki D.; Abe J.; Sohma Y.; Oisaki K.; Kanai M. J. Am. Chem. Soc. 2016, 138, 10798. 10.1021/jacs.6b06692. [DOI] [PubMed] [Google Scholar]
- Lian C.; Le H.; Montez B.; Patterson J.; Harrell S.; Laws D.; Matsumura I.; Pearson J.; Oldfield E. Biochemistry 1994, 33, 5238. 10.1021/bi00183a029. [DOI] [PubMed] [Google Scholar]
- Muthuselvi L.; Miller R.; Dhathathreyan A. Chem. Phys. Lett. 2008, 465, 126. 10.1016/j.cplett.2008.09.037. [DOI] [Google Scholar]
- We tentatively assign I as Trp7 and II as Trp14 by comparison with universally labelled [4F]-Trp-Mb despite highly dispersed peaks in those studies. See the following ref:
- Pearson J. G.; Montez B.; Le H. B.; Oldfield E.; Chien E. Y. T.; Sligar S. G. Biochemistry 1997, 36, 3590. 10.1021/bi961664h. [DOI] [PubMed] [Google Scholar]
- Jash C.; Kumar G. S. RSC Adv. 2014, 4, 12514. 10.1039/c3ra46053c. [DOI] [Google Scholar]
- Richards M. P. Antioxid. Redox Signaling 2013, 18, 2342. 10.1089/ars.2012.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W. C.; Gonzalez G.; Wittenberg J. B.; Hille R.; Dakappagari N.; Jacob A.; Gonzalez L. A.; Gilles-Gonzalez M. A. Chem. Biol. 1996, 3, 841. 10.1016/S1074-5521(96)90070-8. [DOI] [PubMed] [Google Scholar]
- Olsson C.; Jansson H.; Swenson J. J. Phys. Chem. B 2016, 120, 4723. 10.1021/acs.jpcb.6b02517. [DOI] [PubMed] [Google Scholar]
- Huang X.; Wang C. X.; Celeste L. R.; Lovelace L. L.; Sun S. F.; Dawson J. H.; Lebioda L. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun. 2012, 68, 1465. 10.1107/S1744309112045514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S.; Ali S. A.; Islam A.; Hassan M. I.; Ahmad F. Arch. Biochem. Biophys. 2016, 591, 7. 10.1016/j.abb.2015.11.035. [DOI] [PubMed] [Google Scholar]
- Labroo V. M.; Labroo R. B.; Cohen L. A. Tetrahedron Lett. 1990, 31, 5705. 10.1016/S0040-4039(00)97937-1. [DOI] [Google Scholar]
- Cheng M.; Zhang B.; Cui W.; Gross M. L. Angew. Chem., Int. Ed. 2017, 56, 14007. 10.1002/anie.201706697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone S.; Kieltsch I.; Flögel O.; Lelais G.; Togni A.; Seebach D. Helv. Chim. Acta 2008, 91, 2035. 10.1002/hlca.200890217. [DOI] [Google Scholar]
- O’Hara F.; Blackmond D. G.; Baran P. S. J. Am. Chem. Soc. 2013, 135, 12122. 10.1021/ja406223k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abboud M. I.; Hinchliffe P.; Brem J.; Macsics R.; Pfeffer I.; Makena A.; Umland K. D.; Rydzik A. M.; Li G. B.; Spencer J.; Claridge T. D.; Schofield C. J. Angew. Chem., Int. Ed. 2017, 56, 3862. 10.1002/anie.201612185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitevski-LeBlanc J. L.; Evanics F.; Prosser R. S. J. Biomol. NMR 2010, 48, 113. 10.1007/s10858-010-9443-7. [DOI] [PubMed] [Google Scholar]
- Chilkoti A.; Tan P. H.; Stayton P. S. Proc. Natl. Acad. Sci. U. S. A. 1995, 92, 1754. 10.1073/pnas.92.5.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert M.; Nakamura A.; Wang Y. S.; Eiler D.; Söll D.; Guo L. T. Nucleic Acids Res. 2015, 43, 11061. 10.1093/nar/gkv1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H.; Yang X.; Lu Z.; Liu N.; Chen Y. PLoS One 2014, 9, e103792. 10.1371/journal.pone.0103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A.; Tsukada T.; Auer S.; Furuta T.; Wada M.; Koivula A.; Igarashi K.; Samejima M. J. Biol. Chem. 2013, 288, 13503. 10.1074/jbc.M113.452623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz J. R.Principles of fluorescence spectroscopy; 3rd ed.; Springer: New York, NY, 2006. [Google Scholar]
- Budisa N.; Pipitone O.; Siwanowicz I.; Rubini M.; Pal P. P.; Holak T. A.; Gelmi M. L. Chem. Biodiversity 2004, 1, 1465. 10.1002/cbdv.200490107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.