Abstract
Introduction
In mammals, FADS2 catalyzes “front-end” Δ4-, Δ6-, and Δ8-desaturation of fatty acyl chains, whereas FADS1 has Δ5-desaturase activity. Eighteen and 20-carbon precursors to highly unsaturated n-3 and n-6 fatty acids are the usual substrates for FADS1 and FADS2. Our main objective was to characterize the metabolic fate of oleic acid (OA) due to action of FADS gene products.
Methods
MCF-7 cells were stably transformed with either FADS1 or FADS2 or empty vector. A series of dose-response experiments were conducted with albumin-bound fatty acid substrates (18:1n-9 and 20:1n-9) provided in concentrations up to 100 μM. Cells were harvested after 24 h, after which FAME were prepared and analyzed by GC-FID and covalent adduct chemical ionization tandem mass spectrometry (CACI-MS/MS).
Results
When stably transformed cells were incubated with 18:1n-9, FADS1 and control cells elongated 18:1n-9 → 20:1n-9 (11-20:1), while FADS2 cells Δ6 desaturated, elongated, and then Δ5 desaturated via FADS1 coded activity leading to Mead acid, 9-18:1 → 6,9-18:2 → 8,11-20:2 (20:2n-9) → 6,8,11-20:3 (20:3n-9). Surprisingly, FADS1 cells Δ7 desaturated 11-20:1 → 7,11-20:2, the latter detected at low levels in control and FADS2 cells. Our results imply three pathways operate on 18:1n-9: 1) 18:1n-9 → 18:2n-9 → 20:2n-9 → 20:3n-9; 2) 18:1n-9 → 20:1n-9 → 20:2n-9 → 20:3n-9 and 3) 18:1n-9 → 20:1n-9 → 7,11-20:2.
Conclusion
Alternative pathways for oleic acid metabolism exist depending on FADS2 or FADS1 activities, we present the first evidence of Δ7 desaturation via the FADS1 gene product.
Keywords: Oleic acid, fatty acid desaturases, Δ7 desaturation, Mead acid
Introduction
In mammals, the metabolism of omega-3, omega-6 and omega-9 unsaturated fatty acids (UFA) is carried out by one set of desaturation and elongation enzymes. Fatty acid desaturases (FADS) catalyze the site-specific introduction of cis double bonds into hydrocarbon chains, required for the biosynthesis of highly unsaturated fatty acids [1]. The FADS gene cluster comprises three genes - FADS1, FADS2, and FADS3 spanning 100 kb region on the long arm of chromosome 11 (11q12-q13.1) [2]. FADS2 is a multifunctional even numbered desaturase, shown to catalyze “front-end” Δ4-, Δ6-, and Δ8-desaturation of fatty acyl chains acting on at least ten substrates (one saturate, one monounsaturate and eight polyunsaturates), whereas FADS1 has only Δ5 desaturation activity [1, 3, 4]. The elongase gene (ELOVL) family in mammals comprises of seven members (ELOVL1-ELOVL7). Among the seven elongases, ELOVL2 and ELOVL5 are polyunsaturated fatty acid (PUFA)-specific, whereas ELOVL4 prefer saturated fatty acids (SFA) and very long-chain PUFA (C28-C38). ELOVL1, ELOVL3, ELOVL6 and ELOVL7 prefer SFA and monounsaturated fatty acids (MUFA) as substrates [5, 6].
Δ6 desaturation of 18-carbon substrates by FADS2 is the rate-limiting step in the biosynthesis of long chain polyunsaturated fatty acids (LCPUFA) under many but not all physiological conditions [1, 6]. Among three common substrates for FADS2, 18:3n-3 displays the greatest affinity, followed by 18:2n-6, and 18:1n-9 (oleic acid) [7]. Under typical physiological conditions, Mead acid (5,8,11-eicosatrienoic acid, 20:3n-9), a trienoic derivative of oleic acid, comprises less than 1% of total erythrocyte fatty acids [8]. In contrast, when essential fatty acid deficiency (EFAD) is experimentally induced, for instance in human umbilical epithelial cells, 20:3n-9 content rises to 5% of total fatty acids after 16 days [9]. Because cellular Mead acid content increases when omega-3 and omega-6 levels are depleted, the presence of elevated Mead acid levels and the triene-tetraene ratio (Mead acid/arachidonic acid; [20:3n-9]/[20:4n-6]) serves as a biomarker for EFAD [10, 11].
Emerging biochemical evidence indicates a functional role for Mead acid. Its involvement in anti-inflammatory signaling pathways may partially compensate for the reduced serum levels of omega-3 and omega-6 fatty acids, both of which modulate the immune response [12]. Hamazaki et al. demonstrated that Mead acid reduces angiogenesis by inhibiting vascular endothelial growth factor-A (VEGF-A) function and attenuates the activity of osteoblasts [13, 14]. Therefore, Mead acid may promote cartilage synthesis, as further evidenced by elevated Mead acid concentrations in chondrocytes [15]. The anti-angiogenic activity of Mead acid may also serve to inhibit tumorigenesis in certain types of cancer [16]. Full elucidation of Mead acid synthesis and metabolism may therefore contribute to the development of novel immunological and cancer therapeutics.
MCF7 cells have no native capacity for the biosynthesis of LCPUFA from linoleic and α-linolenic acids because of the absence of Δ6-desaturase activity. Earlier, we showed restoration of this metabolic defect by transient transfection of FADS2 and both FADS1 and FADS2 competing for the same fatty acid substrate [17]. Here by using MCF7 cells stably expressing FADS1 or FADS2 we investigate the role of these enzymes in oleic acid metabolism.
Materials and Methods
Chemicals
Fatty acids (18:1n-9, 20:1n-9, and 22:1n-9) were purchased from Nu-Chek Prep (Elysian, MN). Solvents for lipid extraction are HPLC grade from Sigma-Aldrich (St. Louis, MO, US) or Burdick & Jackson (Muskegon, MI, US). Media and solution for cell culture were obtained from Life Technologies (NY), Corning (MA) and ThermoFisher Scientific (MA).
Cell culture
MCF-7 cells stably expressing FADS1 and FADS2 were created using the pcDNA3.1 expression vector system along with empty vector cells as control; details are available elsewhere [3]. Briefly, the antibiotic-resistant transformants were selected to generate pure (i.e. clonal) stably transformed FADS1 and FADS2 cells. Based on this selection method we expect 100% of the cells to contain FADS1 and FADS2. The cells were maintained on MEM-α media containing 10% (v ⁄ v) of heat-inactivated (30 min, 56°C) fetal bovine serum (FBS), 10 mM HEPES buffer and 0.5 mg/ml geneticin at 37°C with 5% CO2. Dosage studies were carried out by using 1×106 cells in 60×15 mm culture dishes and were grown for 48 h in 10% FBS to 80% confluence. Cells were incubated with bovine serum albumin (BSA)-bound fatty acid (18:1n-9 or 20:1n-9) substrates at concentrations of 0, 20, 50 and 100 μM for 24 hours (h). The BSA-bound fatty acid stock consists of 0.33 mM BSA plus 1 mM fatty acid substrate in10 ml PBS. For 100 μM treatment 0.5 ml is used from 10 ml stock. BSA is used to bound fatty acid substrates to be added exogenously to the cells, so zero dose does not contain BSA. While supplementing fatty acid substrate cells were grown in FBS free media. After 24 h, the cells were washed twice with PBS and harvested using trypsin for fatty acid analysis.
Fatty acid analysis
Fatty acid methyl esters (FAME) were prepared by using a modified one-step method [18], and quantitatively analyzed by a Hewlett Packard 5890 series II GC-FID with a BPX 70 column (20 m, 0.22 mm inner diameter, 0.25 m film); Hewlett Packard, Palo Alto, CA, U.S.A.), with H2 as a carrier gas. FAME were structurally identified by GC-covalent adduct chemical ionization tandem mass spectrometry (GC-CACI-MS/MS) as previously described [3]. An equal weight FAME mixture (462A; Nu-Chek Prep, Inc.) was used to calculate response factors on a daily basis [19], and peak area was normalized to 18:2n-6 and 20:3n-6 as in previous studies[4, 20]. Substrate availability alters substrate utilization. Treatment with n-9 fatty acids (FA) has effects on the n-7, n-9 FA families, as well as saturates (C16:0 and C18:0). Use of these FA for normalization produces data that are not clearly related to the hypothesis. While the n-6 and n-3 FA are products of FADS1 and FADS2 action, we did not add n-3 and n-6 substrates exogenously either via direct supplementation or via media as we used FBS free media. The n-3 FA are always present at minor levels even if not added exogenously from FBS while growing the cells. As the n-3 are in trace amounts, we were not able to use them for normalization. The n-6 FA that are present in these cells come from FBS prior to FA treatment and are in amounts which can be used for normalization. In our analysis neither 18:2n-6 nor 20:3n-6 did changed between FADS2, FADS1 and Control treatments, so we used them for normalization. Three biological replicates were used. GC analyses were performed in triplicate for each biological replicate. FAME were dissolved in heptane and stored at -20°C until analysis.
Results
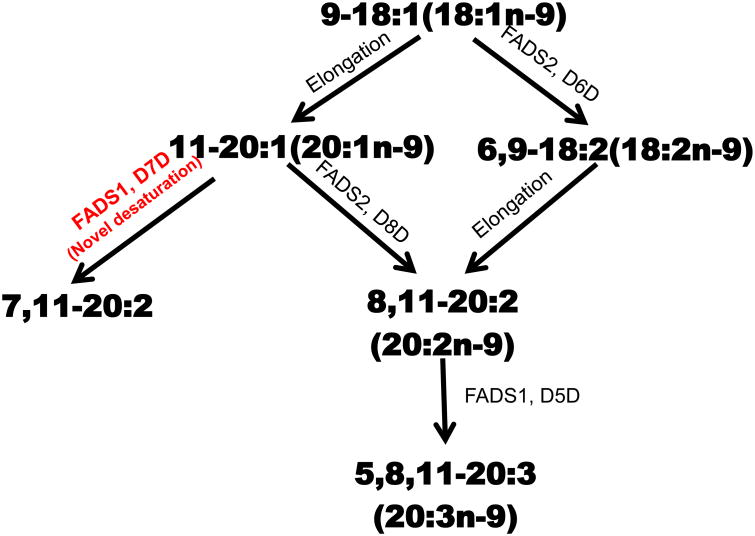
The fate of oleic acid (18:1n-9) was evaluated using stably transformed FADS1 and FADS2 MCF7 cells. Figure 1 shows three metabolic routes to two fates for 18:1n-9 (a) 18:1n-9 is desaturated via Δ6 desaturation (D6D) to 18:2n-9 followed by elongation to 8,11-20:2 and Δ5 desaturation (D5D) to 20:3n-9 (b) 18:1n-9 is elongated to 20:1n-9 followed by Δ8 desaturation (D8D) by FADS2 to 8,11-20:2. The 8,11-20:2 is then Δ5 desaturated to 20:3n-9 or (c) Δ7 desaturation (D7D) by FADS1 to 7,11-20:2.
Figure 1.
Oleic acid Pathway Summary. Metabolic fate of oleic acid (18:1n-9). Oleic acid is metabolized to 20:3n-9 via (a) Δ6-pathway (Δ6 desaturation (D6D) → elongation → Δ5 desaturation (D5D)) or (b) elongation/Δ8-pathway (elongation → Δ8 desaturation (D8D) → Δ5 desaturation (D5D)) to biosynthesize Mead acid (20:3n-9). (c) Elongation/Δ7-pathway (elongation → Δ7 desaturation (D7D)) to biosynthesize the novel 7,11-20:2.
Figure 2 shows chromatograms of control, FADS1 and FADS2 stable MCF7 cell treatments dosed at 0 μM or 100 μM of 18:1n-9 or 20:1n-9 substrates. When cells are treated with 100 μM of 18:1n-9, the elongation product 20:1n-9 is seen in control and FADS1 cells, whereas in FADS2 cells, 18:1n-9 is (a) desaturated via D6D to 18:2n-9, followed by elongation to 8,11-20:2 or (b) elongated to 20:1n-9 followed by D8D by FADS2 to 8,11-20:2. (c) FADS1 cells D7D 20:1n-9 to a novel product, 7,11-20:2. GC-MS analysis was performed to unambiguously identify methyl 20:2 (Figure 3). Figure 3: Panel A is the MS-1 spectrum showing peaks at m/z 376, 323, 291, and 273, corresponding to the [M+54], [MH]+, [M+54-32], and [M+54-32-18] ions, respectively, characteristic of a 20:2 FAME. Panel B is the MS-2 collisional dissociation spectrum of [M+54], yielding ions at m/z 278 and 208 corresponding, respectively, to the α and ω diagnostic ions for 7,11-20:2 and positively identifying this novel product. Panel C is the MS-2 collisional dissociation spectrum of [M+54], yielding ions at m/z 264 and 234 corresponding, respectively, to the α and ω diagnostic ions for 8,11-20:2.
Figure 2.
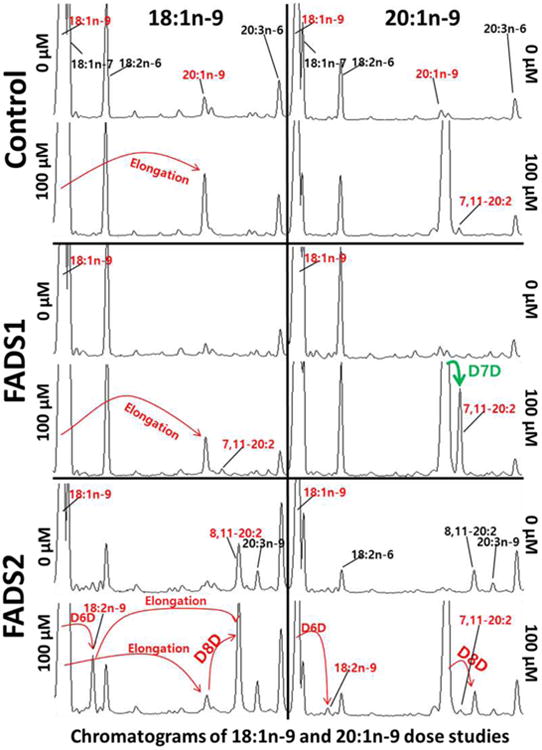
Chromatograms of 18:1n-9 and 20:1n-9 dose studies using stable FADS1 and FADS2 MCF7 cells. The 18:1n-9 dose caused 18:1n-9 elongation to 20:1n-9 in control and FADS1 cells. (a) The FADS2 cells Δ6-desaturate (D6D) 18:1n-9 to 18:2n-9 and then elongate to 20:2n-9 or (b) Δ-8-desaturate (D8D) 20:1n-9 to 8,11- 20:2. In addition, (c) the FADS1cells Δ7-desaturate (D7D) 20:1n-9 to 7,11- 20:2.
Figure 3.
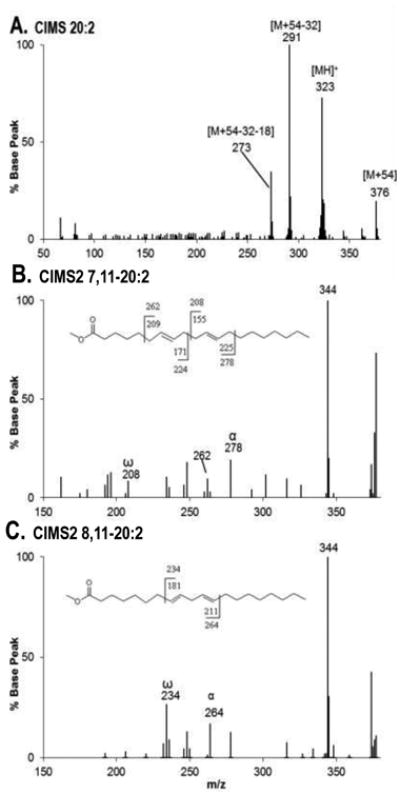
Data Showing CACI-MS1 and CACI-MS2 spectra of 20:2 FAME. A: CACI-MS1 spectrum showing m/z 376, 323, 291, and 273 characteristic ions of 20:2 FAME. B: CACI-MS/MS spectrum of the 20:2 FAME showing FADS1 action. Novel product 7,11-20:2 was positively identified based on the detection of diagnostic ions m/z 278 and 208. C: CACI-MS/MS spectrum of the 20:2 FAME showing FADS2 action. 8,11-20:2 was positively identified based on the detection of diagnostic ions m/z 264 and 234.
Figure 4 shows MCF7 cells stably expressing FADS1, FADS2, and control (empty vector) incubations with various concentrations (0, 20, 50 and 100 μM) of 18:1n-9 or 20:1n-9 substrates. When cells treated with 18:1n-9 control and FADS1 cells synthesize 20:1n-9 via chain elongation. In contrast, FADS2 cells accumulate D6D product 18:2n-9, which increased with increasing concentration of 18:1n-9. Increasing precursor 18:1n-9 not only increased product 18:2n-9 but also increased 8,11-20:2 by several fold. Similarly, when cells were treated with 20:1n-9, FADS1 cells accumulate D7D product 7,11-20:2, which steadily increased with increasing concentrations. No obvious changes were seen in control cells, whereas, in FADS2 cells 8,11-20:2 increased at concentrations 20 and 50 μM. Based on the unexpected observation of Δ7 desaturation of 20:1n-9, we performed similar experiments with 22:1n-9 (erucic acid). No desaturation product was observed for 22:1n-9 upon incubation with FADS1, FADS2, or control cells (data not shown). When cells were treated with oleic acid, the percent conversion to all downstream FA by FADS2 was 20% and by FADS1 was 3%.
Figure 4.
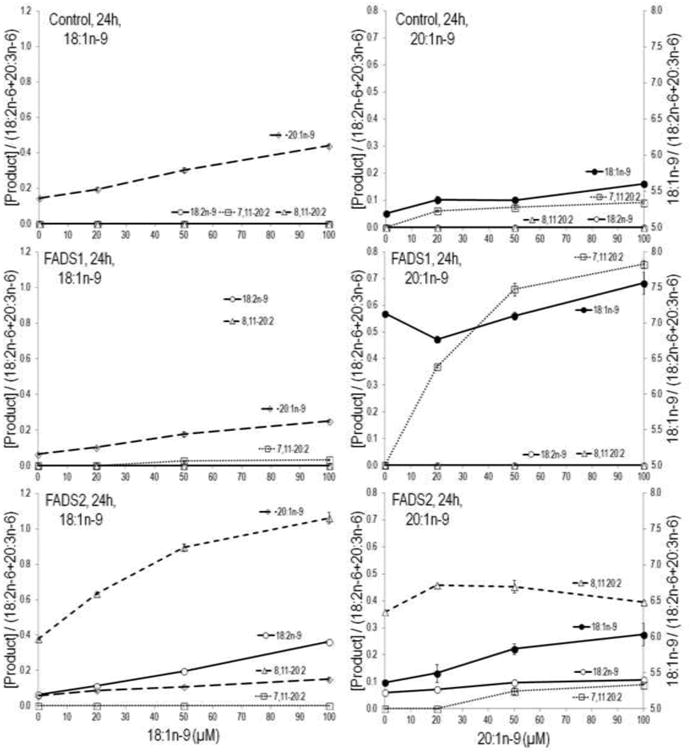
Stably transformed FADS1, FADS2, and control (empty vector) MCF7 cells, incubated with various concentrations (0, 20, 50 and 100 μM) of 18:1n-9 or 20:1n-9 substrates. The FADS1 and control cells elongate 18:1n-9 to 20:1n-9. The FADS2 cells Δ6 desaturate (D6D) 18:1n-9 to 18:2n-9. Increasing precursor 18:1n-9 not only increased product 18:2n-9 but also increased 8,11-20:2 by several folds. In addition, FADS1 cells accumulated Δ7 desaturation (D7D) product 7,11-20:2, whereas, FADS2 cells accumulated the Δ8 desaturation (D8D) product 8,11-20:2 which increased at substrate concentrations 20 and 50 μM.
When incubated with 11-20:1, the novel 7,11-20:2 product was also detected in control and FADS2 cells (Figure 2 and Figures 4, top and bottom right panels). Though MCF7 cells have no native FADS2 activity, they do have FADS1 activity. These data demonstrate that 7,11-20:2 appears in cells when substrate 11-20:1 is provided even in cells with FADS2 activity. In 18:1n-9 treated cells, 7,11-20:2 is detected only in FADS1 but not control or FADS2 cells, consistent with 9-18:1 → 11-20:1 → 7,11-20:2 and the last step catalyzed by FADS1 (Figures 2 and 4).
Discussion
Oleic acid (18:1n-9) exists in various animal and vegetable fats and oils and serves as a precursor for the synthesis of Mead acid (20:3n-9). In the present study, by using MCF7 cells stably transformed with FADS2 and FADS1 we investigated the pathways leading to the biosynthesis of Mead acid. Conversion of oleic acid to Mead acid has been shown earlier by using siRNA-mediated knockdown assays [21] to proceed via two pathways. (a) ‘classical’ Δ6-pathway (Δ6 desaturation → elongation → Δ5 desaturation), or (b) ‘alternative’ elongation/Δ8-pathway (elongation → Δ8 desaturation → Δ5 desaturation) Supplementary Figure 1. In classical Δ6-pathway, similar to the PUFA pathway FADS2 catalyzes the rate limiting Δ6 desaturation step to form 18:2n-9, which is followed by elongation reaction catalyzed by ELOVL5 and Δ5 desaturation to yield 20:3n-9. However, in the alternative pathway 18:1n-9 first is elongated to 20:1n-9. This step is known to be catalyzed by ELOVL3, ELOVL5 or ELOVL7 [5, 21, 22]. The fate of 20:1n-9 then follows Δ8 desaturation (FADS2) → Δ5 desaturation (FADS1) to yield Mead acid. Interestingly, our stable FADS1 cells D7D 20:1n-9 to 7,11-20:2.
Desaturases and elongases are well known to compete for the same substrates. For example, 18:2n-6 and 18:3n-3 EFA competes to access FADS2 (Δ6-desaturase activity). Earlier we showed that FADS1 and FADS2 compete for the same substrates [17]. In MCF7 cells with substantial absence of FADS2 (Δ8 desaturation activity), the D5D (FADS1) operates on 20:2n-6 and 20=3n-3 to yield 5,11,14-20=3 and 5,11,14,17-20=4, respectively [17]. Similarly, here we show FADS1 acting on 20:1n-9 to yield 7,11-20:2, demonstrating that the metabolic fate of 20:1n-9 depends on FADS1 and FADS2 activities.
In 2001, Destaillats et al. [23] reported that seed oils of two Taxaceae species (Taxus chinensis and T. baccata) contain a ∆7-PUFA dihomotaxoleic (DHT; cis-7,cis-11 20:2) acid. The T. chinensis and T. baccata species contain taxoleic acid (TA; all-cis 5,9-18:2) at 16.4 and 10.6% and dihomotaxoleic at 0.13 and 0.06% of total fatty acids, respectively. The presence of 5,9-18:2 and 7,11-20:2 in these two species seed oils led Destaillats et al. [23] to propose that DHT could be the elongation product of TA. The 7,11-20:2 is also seen in the hardshell clam (Mercenaria mercenaria) [24]. Here we provide the first molecular evidence showing that FADS1 introduces a double bond at the Δ7 position leading to the biosynthesis of 7,11-20:2 (DHT).
The all-cis Δ7 monounsaturated and polyunsaturated fatty acids have been previously reported in numerous samples. In human milk lipids, Precht and Molkentin [25] reported existence of cis-7 16:1. In plant species like Arabidopsis thaliana, Spinacia oleracea, and Brassica napus which are widely known as 16:3 plants, Δ7-methylene interrupted hexadeca-7,10,13-trienoic acid is one of the most abundant fatty acids comprising up to 30% of the total [26, 27]. The dienoic 7,11-18:2 was found in the sponge Haliclona cinerea from the Black Sea [28]. By using labelling studies Zhukova has shown presence of non-methylene-interrupted dienoic 22:2 (7,13) and 22:2 (7,15) in the bivalve mollusk Scapharca broughtoni. Similarly, Kawashima et al. [29] identified non-methylene-interrupted dienoic fatty acids in Cellana grata. The 20 carbon 7,13-20:2 and 7,10,13-20:3 were found in liver lipids of rats raised on a fat free diet [30]. The 7,10,13-20:3 and 7,10,13,16-20:4 (20:4n-4) were also found in Bathymodiolus japonicus and Bathymodiolus platifrons two cold-seep mussels, which host methane-oxidizing bacteria [31].
In conclusion, we provide the first molecular evidence of Δ7desaturation via the FADS1 gene product. An alternative pathway for oleic acid metabolism exists, and its metabolic significance depends on FADS1 and FADS2 activities. Production of 7,11-20:2 is found when oleic acid's elongation product, 11-20:1 is sufficient.
Supplementary Material
Highlights.
To date FADS1 is known to catalyze only 5 desaturation.
We show that FADS1 catalyzes 7 desaturation of 11-20:1 to yield 7,11-20:2
In 20:1n-9 supplemented cells 7,11-20:2 is found at low concentration in basal MCF7 cells
Acknowledgments
This work was supported by NIH grant R01 AT007003 from the National Center for Complementary and Integrative Health (NCCIH) and the Office of Dietary Supplements (ODS). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Abbreviations
- FADS
fatty acid desaturases
- UFA
unsaturated fatty acids
- ELOVL
elongase gene
- PUFA
polyunsaturated fatty acid
- MUFA
monounsaturated fatty acids
- SFA
saturated fatty acids
- LCPUFA
long chain polyunsaturated fatty acids
- EFAD
essential fatty acid deficiency
- VEGF-A
vascular endothelial growth factor-A
- FBS
fetal bovine serum
- FAME
Fatty acid methyl esters
- GC-CACI-MS/MS
GC-covalent adduct chemical ionization tandem mass spectrometry
- D6D
Δ6 desaturation
- D5D
Δ5 desaturation
- D8D
Δ8 desaturation
- D7D
Δ7 desaturation
- DHT
dihomotaxoleic
- TA
taxoleic acid
Footnotes
Author Contributions: J.T.B., K.S.D.K., HP. designed research; HP., M.G.E., K.V.L executed the research; J.T.B. and K.S.D.K. contributed new reagents/analytic tools; HP., M.G.E., K.V.L, P.L analyzed and interpreted the data; and J.T.B., K.S.D.K., H.P. wrote the first draft and all authors approved the final draft.
Conflict of interest: All authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Park WJ, Kothapalli KS, Lawrence P, Tyburczy C, Brenna JT. An alternate pathway tolong-chain polyunsaturates: the FADS2 gene product Delta8-desaturates 20:2n-6 and 20:3n-3. J Lipid Res. 2009;50:1195–1202. doi: 10.1194/jlr.M800630-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marquardt A, Stohr H, White K, Weber BH. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 2000;66:175–183. doi: 10.1006/geno.2000.6196. [DOI] [PubMed] [Google Scholar]
- 3.Park HG, Park WJ, Kothapalli KS, Brenna JT. The fatty acid desaturase 2 (FADS2) gene product catalyzes Delta4 desaturation to yield n-3 docosahexaenoic acid and n-6 docosapentaenoic acid in human cells. FASEB J. 2015;29:3911–3919. doi: 10.1096/fj.15-271783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park HG, Kothapalli KS, Park WJ, DeAllie C, Liu L, Liang A, Lawrence P, Brenna JT. Palmitic acid (16:0) competes with omega-6 linoleic and omega-3 a-linolenic acids for FADS2 mediated Delta6-desaturation. Biochim Biophys Acta. 2016;1861:91–97. doi: 10.1016/j.bbalip.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guillou H, Zadravec D, Martin PG, Jacobsson A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog Lipid Res. 2010;49:186–199. doi: 10.1016/j.plipres.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Zhang JY, Kothapalli KS, Brenna JT. Desaturase and elongase-limiting endogenous long-chain polyunsaturated fatty acid biosynthesis. Curr Opin Clin Nutr Metab Care. 2016;19:103–110. doi: 10.1097/MCO.0000000000000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenner RR. The oxidative desaturation of unsaturated fatty acids in animals. Mol Cell Biochem. 1974;3:41–52. doi: 10.1007/BF01660076. [DOI] [PubMed] [Google Scholar]
- 8.Fokkema MR, Smit EN, Martini IA, Woltil HA, Boersma ER, Muskiet FA. Assessment of essential fatty acid and omega3-fatty acid status by measurement of erythrocyte 20:3omega9 (Mead acid), 22:5omega6/20:4omega6 and 22:5omega6/22:6omega3. Prostaglandins Leukot Essent Fatty Acids. 2002;67:345–356. doi: 10.1054/plef.2002.0440. [DOI] [PubMed] [Google Scholar]
- 9.Lerner R, Lindstrom P, Berg A, Johansson E, Rosendahl K, Palmblad J. Development and characterization of essential fatty acid deficiency in human endothelial cells in culture. Proc Natl Acad Sci U S A. 1995;92:1147–1151. doi: 10.1073/pnas.92.4.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siguel EN, Chee KM, Gong JX, Schaefer EJ. Criteria for essential fatty acid deficiency in plasma as assessed by capillary column gas-liquid chromatography. Clin Chem. 1987;33:1869–1873. [PubMed] [Google Scholar]
- 11.Holman RT. The ratio of trienoic: tetraenoic acids in tissue lipids as a measure of essentialfatty acid requirement. J Nutr. 1960;70:405–410. doi: 10.1093/jn/70.3.405. [DOI] [PubMed] [Google Scholar]
- 12.Hsu LC, Wen WH, Chen HM, Lin HT, Chiu CM, Wu HC. Evaluation of the Anti-Inflammatory Activities of 5,8,11-cis-Eicosatrienoic Acid. Food and Nutrition Sciences. 2013;4:113–119. [Google Scholar]
- 13.Hamazaki T, Nagasawa T, Hamazaki K, Itomura M. Inhibitory effect of 5,8,11-eicosatrienoic acid on angiogenesis. Prostaglandins Leukot Essent Fatty Acids. 2012;86:221–224. doi: 10.1016/j.plefa.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Hamazaki T, Suzuki N, Widyowati R, Miyahara T, Kadota S, Ochiai H, Hamazaki K. The depressive effects of 5,8,11-eicosatrienoic Acid (20:3n-9) on osteoblasts. Lipids. 2009;44:97–102. doi: 10.1007/s11745-008-3252-8. [DOI] [PubMed] [Google Scholar]
- 15.Adkisson HDt, Risener FS, Jr, Zarrinkar PP, Walla MD, Christie WW, Wuthier RE. Unique fatty acid composition of normal cartilage: discovery of high levels of n-9 eicosatrienoic acid and low levels of n-6 polyunsaturated fatty acids. FASEB J. 1991;5:344–353. doi: 10.1096/fasebj.5.3.2001795. [DOI] [PubMed] [Google Scholar]
- 16.Kinoshita Y, Yoshizawa K, Hamazaki K, Emoto Y, Yuri T, Yuki M, Shikata N, Kawashima H, Tsubura A. Mead acid inhibits the growth of KPL-1 human breast cancer cells in vitro and in vivo. Oncol Rep. 2014;32:1385–1394. doi: 10.3892/or.2014.3390. [DOI] [PubMed] [Google Scholar]
- 17.Park WJ, Kothapalli KS, Lawrence P, Brenna JT. FADS2 function loss at the cancer hotspot 11q13 locus diverts lipid signaling precursor synthesis to unusual eicosanoid fatty acids. PLoS One. 2011;6:e28186. doi: 10.1371/journal.pone.0028186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garces R, Mancha M. One-step lipid extraction and fatty acid methyl esters preparation from fresh plant tissues. Anal Biochem. 1993;211:139–143. doi: 10.1006/abio.1993.1244. [DOI] [PubMed] [Google Scholar]
- 19.Diau GY, Hsieh AT, Sarkadi-Nagy EA, Wijendran V, Nathanielsz PW, Brenna JT. The influence of long chain polyunsaturate supplementation on docosahexaenoic acid and arachidonic acid in baboon neonate central nervous system. BMC Med. 2005;3:11. doi: 10.1186/1741-7015-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park HG, Lawrence P, Engel MG, Kothapalli K, Brenna JT. Metabolic fate of docosahexaenoic acid (DHA; 22:6n-3) in human cells: direct retroconversion of DHA to eicosapentaenoic acid (20:5n-3) dominates over elongation to tetracosahexaenoic acid (24:6n-3) FEBS Lett. 2016;590:3188–3194. doi: 10.1002/1873-3468.12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ichi I, Kono N, Arita Y, Haga S, Arisawa K, Yamano M, Nagase M, Fujiwara Y, Arai H. Identification of genes and pathways involved in the synthesis of Mead acid (20:3n-9), an indicator of essential fatty acid deficiency. Biochim Biophys Acta. 2014;1841:204–213. doi: 10.1016/j.bbalip.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 22.Naganuma T, Sato Y, Sassa T, Ohno Y, Kihara A. Biochemical characterization of the very long-chain fatty acid elongase ELOVL7. FEBS Lett. 2011;585:3337–3341. doi: 10.1016/j.febslet.2011.09.024. [DOI] [PubMed] [Google Scholar]
- 23.Destaillats F, Wolff RL, Angers P. A new delta 7-polyunsaturated fatty acid in Taxus spp. Seed lipids, dihomotaxoleic (7,11-20:2) acid. Lipids. 2001;36:319–321. doi: 10.1007/s11745-001-0724-6. [DOI] [PubMed] [Google Scholar]
- 24.Klingensmith JS. Distribution of methylene and nonmethylene-interrupted dienoic fatty acids in polar lipids and triacylglycerols of selected tissues of the hardshell clam (Mercenaria mercenaria) Lipids. 1982;17:976–981. doi: 10.1007/BF02534595. [DOI] [PubMed] [Google Scholar]
- 25.Precht D, Molkentin J. Identification and quantitation of cis/trans C16:1and C17:1 fatty acid positional isomers in Germanhuman milk lipids by thin-layer chromatographyand gas chromatography/mass spectrometry. Eur J Lipid Sci Technol. 2000;102:102–113. [Google Scholar]
- 26.Heilmann I, Mekhedov S, King B, Browse J, Shanklin J. Identification of the Arabidopsis palmitoyl-monogalactosyldiacylglycerol delta7-desaturase gene FAD5, and effects of plastidial retargeting of Arabidopsis desaturases on the fad5 mutant phenotype. Plant Physiol. 2004;136:4237–4245. doi: 10.1104/pp.104.052951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Browse J, Warwick N, Somerville CR, Slack CR. Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the ‘16:3’ plant Arabidopsis thaliana. Biochem J. 1986;235:25–31. doi: 10.1042/bj2350025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joh YG, Elenkov IJ, Stefanov KL, Popov SS, Dobson G, Christie WW. Novel di-, tri-, and tetraenoic fatty acids with bis-methylene-interrupted double-bond systems from the sponge Haliclona cinerea. Lipids. 1997;32:13–17. doi: 10.1007/s11745-997-0003-6. [DOI] [PubMed] [Google Scholar]
- 29.Kawashima H, Ohnishi M, Ogawa S, Matsui K. Unusual fatty acid isomers of triacylglycerols and polar lipids in female limpet gonads of Cellana grata. Lipids. 2008;43:559–567. doi: 10.1007/s11745-008-3179-0. [DOI] [PubMed] [Google Scholar]
- 30.Schmitz B, Murawski U, Pfluger M, Egge H. Positional isomers of unsaturated fatty acids in rat liver lipids. Lipids. 1977;12:307–313. doi: 10.1007/BF02533353. [DOI] [PubMed] [Google Scholar]
- 31.Saito H. Unusual novel n-4 polyunsaturated fatty acids in cold-seep mussels (Bathymodiolus japonicus and Bathymodiolus platifrons), originating from symbiotic methanotrophic bacteria. J Chromatogr A. 2008;1200:242–254. doi: 10.1016/j.chroma.2008.05.094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.