Abstract
Background
Rhizoma Paridis (Chonglou) is a commonly used and precious traditional Chinese medicine. Paris polyphylla Smith var. yunnanensis (Franch.) Hand. -Mazz. and Paris polyphylla Smith var. chinensis (Franch.) Hara are the two main sources of Chonglou under the monograph of Rhizoma Paridis in Chinese Pharmacopoeia. In the local marketplace, however, this medicine is prone to be accidentally contaminated, deliberately substituted or admixed with other species that are similar to Rhizoma Paridis in shape and color. Consequently, these adulterations might compromise quality control and result in considerable health concerns for consumers. This study aims to develop a rapid and sensitive method for accurate identification of Rhizoma Paridis and its common adulterants.
Methods
DNA barcoding coupled with high resolution melting analysis was applied in this research to distinguish Rhizoma Paridis from its adulteration. The internal transcribed spacer 2 (ITS2) barcode was selected for HRM analysis to produce standard melting profile of the selected species. DNA of the tested herbal medicines was isolated and their melting profiles were generated and compared with the standard melting profile of P. polyphylla var. chinensis.
Results
The results indicate that the ITS2 molecular regions coupled with HRM analysis can effectively differentiate nine herbal species, including two authentic origins of Chonglou and their seven common adulterants. Ten herbal medicines labeled “Chonglou” obtained from a local market were collected and identified with our methods, and their sequence information was analyzed to validate the accuracy of HRM analysis.
Conclusions
DNA barcoding coupled with HRM analysis is a accurate, reliable, rapid, cost-effective and robust tool, which could contribute to the quality control of Rhizoma Paridis in the supply chain of the natural health product industry (NHP).
Electronic supplementary material
The online version of this article (10.1186/s13020-018-0162-4) contains supplementary material, which is available to authorized users.
Keywords: Authentication, Rhizoma Paridis, DNA barcoding, High resolution melting, Adulterations, Natural health products
Background
Rhizoma Paridis (Chonglou) is a precious traditional Chinese medicine. It originates from the dried rhizomes of both Paris polyphylla Smith var. yunnanensis (Franch.) Hand. -Mazz. and P. polyphylla Smith var. chinensis (Franch.) Hara, according to the Chinese Pharmacopoeia [1]. This medicine is most notably an ingredient of the hemostasis prescription ‘Yunnan Baiyao’ [2]. It plays an important clinical role in treating snake bites, fractures, parotitis, tumors, analgesia and traumatic bleeding. Recent pharmacological studies have indicated that Chonglou exhibits a variety of pharmacological activities, including anti-cancer, anti-inflammatory, antibacterial and immuno-regulatory activities [3]. Unfortunately, slow growth and over-harvesting for the past several years have led to a significant decline in Chonglou’s population and caused it to be listed as a vulnerable species by the IUCN (International Union for Conservation of Nature and Natural Resources). The lack of supply and subsequent high price can result in adulteration of Chonglou, including accidentally contamination, deliberately substitution or admixture with other species [4]. More specifically, Chonglou is often contaminated with several common adulterants: P. thibetica Franch, P. tengchongensis Y.H. Ji, C.J. Yang & Y.L. Huang, P. forrestii (Takht.) H. Li, P. mairei H. Lév, Tupistra spp., Trillium tschonoskii Maxim. and Polygonum paleaceum Wall. These adulterants are usually of poor quality and some might even be toxic. Contaminated with these plants can pose considerable health threat for the consumers [5]. It was reported that several Tupistra species can induce headache and vomiting [6]. Thus, Chonglou authenticity is not only an issue of medicinal quality but also of safety. A fast, reliable and robust method is urgently needed to be established for the herbal industry and regulatory agencies to identify Chonglou species from their adulterants.
With rapid advances in sequencing technologies, Herbgenomics provides an effective platform to support biological analyses of complex herbal products [7, 8]. It is now being widely applied to DNA-based herbal identification through the establishment of an herbal gene bank. DNA-based methods have gained popularity for unequivocal plant species identification because DNA molecules can be found in all tissues and they cannot be affected by environment or physiological factors [9–13]. DNA barcoding, a method that use the sequence of a standard region of DNA for the purpose of species identification, has been recognized as a powerful tool for the identification of medicinal plants and their adulterants at the species level [4, 14–17]. However, DNA sequencing of each individual sample is expensive, time consuming and cannot easily be applied to studies of large samples because of financial constraints [18]. High resolution melting (HRM) analysis is a closed-tube post-PCR analysis that can differentiate between samples even with a single base change in DNA sequences. This technique permits the genotyping and the quantification of adulterants and has been proved to be fast, reliable, low-cost in terms of consumables [19, 20]. Very recently, the Bar-HRM analysis has been introduced for species identification and authentication. The use of just Bar-HRM for species authentication and detection of adulteration in commercial products has been reported [21, 22]. To date, it has not been applied as a tool for the species authentication of Chonglou.
The goal of this research is to develop a rapid, scientifically rigorous and accurate method to identify and authenticate two types of Chonglou, namely P. polyphylla var. yunnanensis and P. polyphylla var. chinensis, from their adulterants. For that purpose, Bar-HRM analysis using universal DNA barcoding ITS2 region was conducted. The proposed methodology was further applied to differentiate ten herbal medicines labeled as Chonglou bought in market place. This will provide the first evidence for the utility of Bar-HRM analysis to test the authenticity of Chonglou sold in the local market place.
Methods
Reference plant material and DNA extraction
Thirty two fresh rhizome samples from nine species were collected from the Yunnan Province, People’s Republic of China, including P. polyphylla var. yunnanensis, P. polyphylla var. chinensis, P. thibetica, P. tengchongensis, P. forrestii, P. mairei, Tupistra spp., T. tschonoskii and P. paleaceum (Table 1). The rhizomes were gently washed and dried at 50 °C. All of the materials were authenticated by Professor Conglong Xia of the College of Pharmaceutical Science, Dali University. In addition, ten herbal medicines labeled “Chonglou” were purchased from a local market in the Yunnan Province. All voucher specimens and herbal medicines were deposited in the Herbarium of Dali University (DLU). Samples taken from dried rhizomes (30 mg) were rubbed for 2 min at a frequency of 30 r/s. Extraction proceeded by using the modified Plant Genomic DNA Extraction Kit (Tiangen Biotech Co., Ltd., Beijing, China). The DNA was diluted to 50 ng/μL and later stored at − 20 °C for further use.
Table 1.
The origin of the plant species used in the study and their GenBank accession numbers
| No. | Family | Botanical name | Voucher number | Genbank accessions number |
|---|---|---|---|---|
| 1 | Liliaceae | Paris polyphylla var. yunnanensis (Franch.) Hand. -Mazz. | YN0001MT01, YN0001MT02, YN0001MT03, YN0001MT04, YN0001MT05, YN0001MT06 | KX458130, KX458131, KX458132, KX458133, KX458134, KX458135 |
| 2 | Paris polyphylla var. chinensis (Franch.) H. Hara | YN0011MT01, YN0011MT02, YN0011MT03 | KX458140, KX458141, KX458142 | |
| 3 | Paris thibetica Franch. | YN0012MT01, YN0012MT02, YN0012MT03 | KX458136, KX458137, KX458138, KX458139 | |
| 4 | Paris tengchongensis Y.H. Ji, C.J. Yang & Y.L. Huang | YN0007MT01, YN0007MT02, YN0007MT03, YN0007MT04, YN0007MT05, YN0007MT06 | KX458121, KX458122, KX458123, KX458124, KX458125, KX458126 | |
| 5 | Paris forrestii (Takht.) H. Li | YN0009MT01, YN0009MT02, YN0009MT03 | KX458127, KX458128, KX458129 | |
| 6 | Paris mairei H. Lév | YN0005MT01, YN0005MT02, YN0005MT03 | KX458118, KX458119, KX458120 | |
| 7 | Tupistra spp. (not define) | YN0016MT03, YN0016MT04, YN0016MT05 | / | |
| 8 | Trillium tschonoskii Maxim. | YN0016MT10, YN0016MT11, YN0016MT12 | KX458143, KX458144, KX458145 | |
| 9 | Polygonaceae | Polygonum paleaceum Wall. | YN0016MT01, YN0016MT02 | KX458146, KX458147 |
HRM-PCR amplification
A conserved region of the ITS2 region of nuclear ribosomal DNA was used as the optimal region for the assay because of it has previously correctly identified the medicinal plants of the genus Paris [23]. DNA was amplified using the polymerase chain reaction (PCR). The real-time PCR mixture (25 μL) contained 50 ng of genomic DNA, 12.5 μL of 2 × HRM PCR master mix, 1 μL forward (5′-ATGCGATACTTGGTGTGAAT-3′) primer (10 μmol/L), and 1 μL reverse (5′-GACGCTTCTCCAGACTACAAT-3′) primer (10 μmol/L), and distilled water up to the final volume. Real-time PCR was performed in a Rotor-Gene Q MDx cycler (Qiagen, Hilden, Germany) under the following conditions: 94 °C for 3 min followed by 40 cycles of 94 °C for 30 s, 56 °C for 30 s and 72 °C for 45 s. The fluorescent data were acquired at the end of each extension step of the PCR cycle. HRM was performed as follows: pre-melt at 95 °C for 1.5 min, followed by a temperature raise from 65 to 95 °C with 0.1 °C degree increments and a 2 s hold time for each acquisition step.
The Rotor-Gene Q software (Qiagen, Hilden, Germany) was used to genotype the different varieties and the herbal products. The negative derivative of the fluorescence (F) over temperature (T) (dF/dT) curve displayed the melting temperature (Tm), and the normalized raw curve depicted the decreasing fluorescence against increasing temperature. Pre- and post-melt normalization regions were set to define the temperature boundaries of the normalized and difference plots and to generate normalized melting curves of the normalized and difference plots [24]. P. polyphylla var. chinensis was also used as reference species. To assess the discriminative power of Bar-HRM analysis, samples from both the fresh plants and dry tissue were used for PCR amplification and HRM analysis.
Sequencing and data analysis
To verify the HRM analysis results, all the PCR products were directly sequenced in two directions using the automated ABI 3730 XL sequencer (Applied Biosystems, Foster City, CA, USA). Sequence results were submitted to the GenBank database (accession numbers were listed in Table 1), assembled with CodonCode Aligner 5.1.5 (CodonCode Co., USA) and aligned using Clustal W. The neighbor-joining (NJ) trees were calculated and constructed using the MEGA 5.05 software with the Bootstrap method (1000 resembling) and the K2P model [25].
The Minimum Standards of Reporting Checklist contains details of the experimental design, and statistics, and resources used in this study (Additional file 1).
Results
Barcoding of PCR products by HRM analysis
The amplicons of ITS2 were analyzed with HRM to define their Tms. There exist some differences in ITS2 region among Chonglou and their adulterate species reflected by their Tm values (Table 2). Although the Tms value of Paris genus were close to each other, each species still could be clearly discerned by the plots of the normalized melting curves (Fig. 1). Average genotype confidence percentages (GCPs) were also calculated and a cut-off value of 90% was used to assign a genotype for each region. The GCPs from the HRM analyses of the ITS2 region of nine species are listed in Table 3. As shown in Table 3, all species were sufficiently and confidently identified with a value of 100%. The highest GCP (81.28%) was found between the P. mairei and P. polyphylla var. chinensis species, while the lowest (0.0) was between Paris and the other genera. The melting profiles of the ITS2 amplicons from the nine species were assessed by plotting two different curves: the derivate melt curve (Fig. 1a) and the difference plot melt curve (Fig. 1b). The normalized HRM curves with the barcode marker ITS2 showed that all the nine species could be easily and visually distinguished.
Table 2.
The values of the melting temperatures of nine species measured by HRM analysis using ITS2 as marker
| No. | Botanical name | Examin | Tm (°C) |
|---|---|---|---|
| 1 | Paris polyphylla var. yunnanensis (Franch.) Hand. -Mazz. | 6 | 87.31 ± 0.07 |
| 2 | Paris polyphylla var. chinensis (Franch.) H. Hara | 3 | 86.88 ± 0.02 |
| 3 | Paris thibetica Franch. | 3 | 86.84 ± 0.02 |
| 4 | Paris tengchongensis Y.H. Ji, C.J. Yang & Y.L. Huang | 6 | 87.55 ± 0.04 |
| 5 | Paris forrestii (Takht.) H. Li | 3 | 86.68 ± 0.04 |
| 6 | Paris mairei H. Lév | 3 | 87.37 ± 0.03 |
| 7 | Tupistra spp. (not define) | 3 | 92.66 ± 0.08 |
| 8 | Trillium tschonoskii Maxim. | 3 | 88.91 ± 0.17 |
| 9 | Polygonum paleaceum Wall. | 2 | 91.02 ± 0.02 |
Fig. 1.
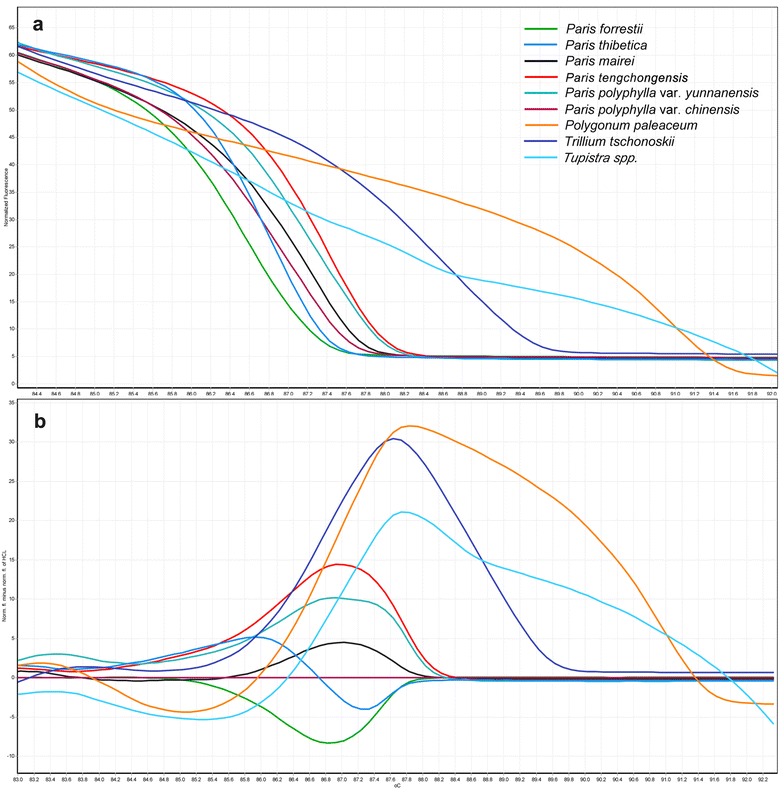
a The normalized melting profiles of nine plant species obtained by HRM analysis with ITS2 as the universal marker. b The difference graphs of nine species using Paris polyphylla var. chinensis as a reference genotype. A color-code table listing the species was used
Table 3.
The results of average genotype confidence percentages from the HRM analysis of nine species
| Taxon | P. forrestii | P. tengchongensis | P. mairei | P. polyphylla var. chinensis | P. polyphylla var. yunnanensis | P. thibetica | Trillium tschonoskii | Polygonum paleaceum | Tupistra spp. |
|---|---|---|---|---|---|---|---|---|---|
| P. forrestii | 100 | ||||||||
| P. tengchongensis | 0.13 | 100 | |||||||
| P. mairei | 19.11 | 16.85 | 100 | ||||||
| P. polyphylla var. chinensis | 48.71 | 4.55 | 81.28 | 100 | |||||
| P. polyphylla var. yunnanensis | 1.24 | 72.1 | 44.33 | 18.75 | 100 | ||||
| P. thibetica | 28.57 | 3.32 | 47.6 | 63.51 | 16.66 | 100 | |||
| Trillium tschonoskii | 0 | 0.06 | 0 | 0 | 0.01 | 0 | 100 | ||
| Polygonum paleaceum | 0 | 0 | 0 | 0 | 0 | 0 | 0.04 | 100 | |
| Tupistra spp. | 0 | 0.02 | 0.02 | 0.01 | 0.02 | 0 | 0.94 | 0.6 | 100 |
The highest GCP was found between the P. mairei and P. polyphylla var. chinensis, which are presented in italics
Species authentication using DNA sequencing
To validate the results of HRM analysis, amplicon sequences of all samples from the nine reference plant species were sequenced. Twenty-seven ITS2 sequences from eight species were successfully obtained, except for Tupistra spp. These sequences were used to construct phylogenetic tree [neighbor-joining (NJ)]. Eight species can be clearly distinguished by the NJ tree (Fig. 2), namely P. polyphylla (var. yunnanensis and var. chinensis), P. thibetica, P. tengchongensis, P. forrestii, P. mairei, T. tschonoskii and P. paleaceum. The genetic relationships reflected by the NJ tree were in good agreement with the Bar-HRM results. The sequence of each amplicon and the alignment among eight species are shown in Fig. 3. Sequencing results revealed that the differences in the melting curves maybe caused by deletions, insertions and a number of SNPs in the ITS2 region. In brief, the variations of the eight species in ITS2 region are capable to produce differences in the HRM analysis profile.
Fig. 2.
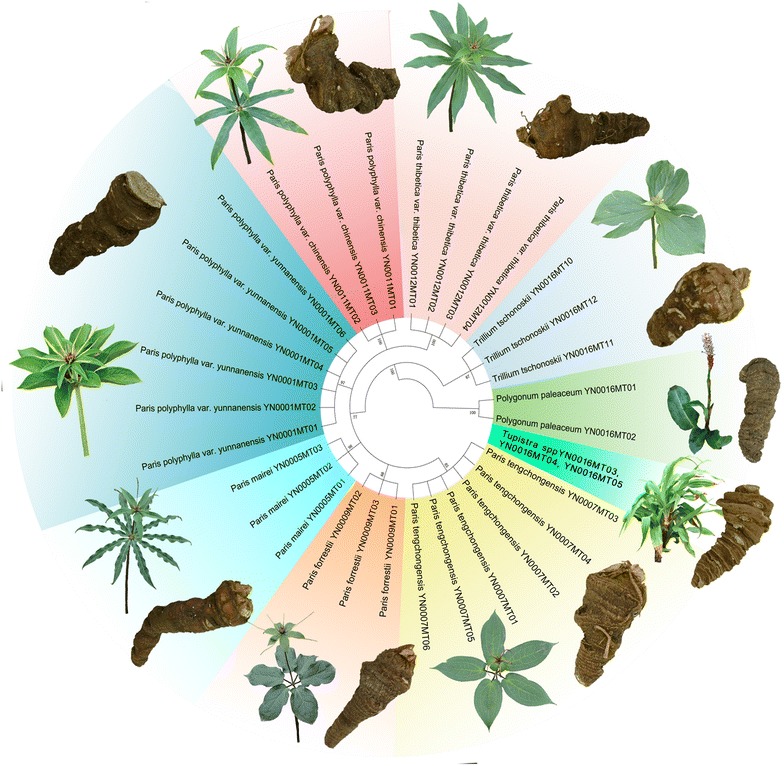
The phylogenetic tree of Rhizoma Paridis and its adulterants constructed from the ITS2 sequences using the N-J method (Bootstrap scores ≥ 50%). Representatives of the species illustrate the morphological variation in Paris and related species
Fig. 3.
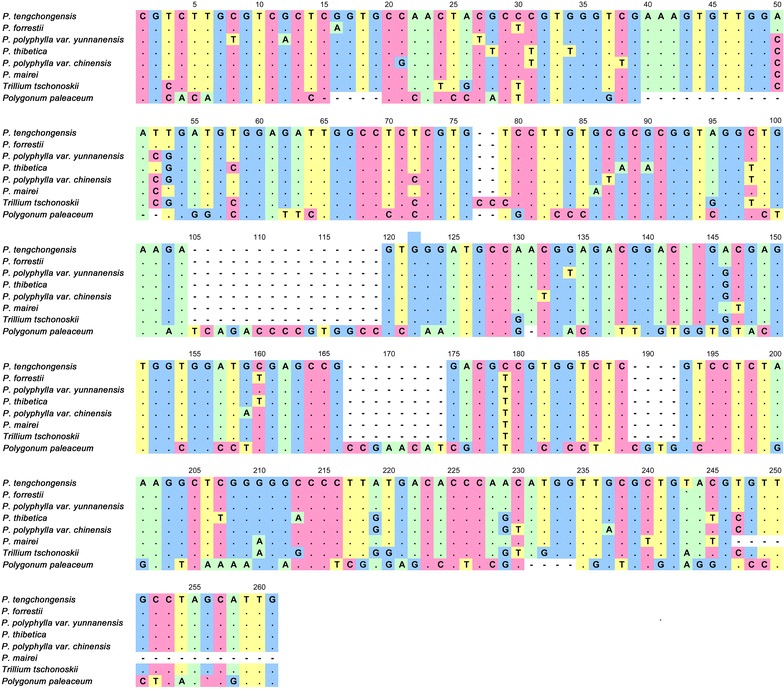
The DNA sequence alignment analysis of nine plant species showing differences in the DNA level alignment confirmed the Bar-HRM results
Application of Bar-HRM to commercial herbal medicine
As mentioned previously, Chonglou sold in the local market places can be confused with certain other species that are similar in shape and color with the authentic Chonglou. To evaluate the accuracy of Bar-HRM method and to assure the quality of Chonglou in local market, the proposed methodology was further applied to authenticate ten batches of samples labelled as “Chonglou”. The HRM curves of nine reference plant species and ten samples are shown in Fig. 4. Tm values and GCPs of ten samples obtained from the HRM analysis are shown in Tables 4 and 5. As shown in Fig. 4 and Tables 4 and 5, the Tms of five Chonglou samples (Com-1, Com-2, Com-3, Com-8, and Com-10) ranged from 87.29 to 87.33 °C, which agreed with the Tm of P. polyphylla var. yunnanensis, and the curves overlapped completely. These results demonstrated that all these five samples were P. polyphylla var. yunnanensis. Among them, three samples (Com-4, Com-5, and Com-6) were identified as P. tengchongensis. Com-7 sharing a similar plot with T. tschonoskii. Com-9 was identified as P. mairei. These results indicate that Bar-HRM analysis can be effectively used to differentiate Chonglou from its adulterants.
Fig. 4.
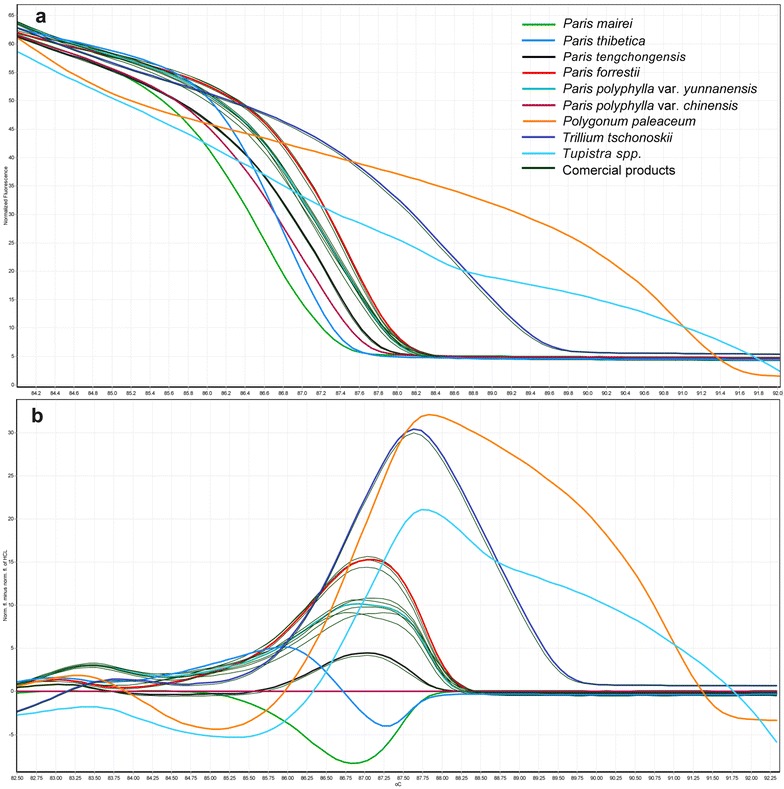
The Bar-HRM analysis of nine plant species and ten commercial Chonglou products using HRM analysis with the ITS2 marker. a Normalized melting curves. b Difference curves, with the mean Paris polyphylla var. chinensis curve used as a reference genotype. A color-code table listing the species was used
Table 4.
The list of ten commercial ‘‘Chonglou’’ products that were identified using ITS2 sequencing coupled with high resolution melting temperature values
| Code | Asserted species | Tm (°C) | HRM |
|---|---|---|---|
| Com-1 | Chong-Lou (not define) | 87.30 | Paris polyphylla var. yunnanensis |
| Com-2 | Chong-Lou (not define) | 87.29 | Paris polyphylla var. yunnanensis |
| Com-3 | Chong-Lou (not define) | 87.32 | Paris polyphylla var. yunnanensis |
| Com-4 | Chong-Lou (not define) | 87.57 | Paris tengchongensis |
| Com-5 | Chong-Lou (not define) | 87.55 | Paris tengchongensis |
| Com-6 | Chong-Lou (not define) | 87.52 | Paris tengchongensis |
| Com-7 | Chong-Lou (not define) | 88.40 | Trillium tschonoskii Maxim. |
| Com-8 | Chong-Lou (not define) | 87.31 | Paris polyphylla var. yunnanensis |
| Com-9 | Chong-Lou (not define) | 87.39 | Paris mairei |
| Com-10 | Chong-Lou (not define) | 87.33 | Paris polyphylla var. yunnanensis |
Com Commercial
Table 5.
The results of genotype confidence percentages from the HRM analysis of ten commercial “Chonglou” products
| Taxon | COM-1 | COM-2 | COM-3 | COM-4 | COM-5 | COM-6 | COM-7 | COM-8 | COM-9 | COM-10 |
|---|---|---|---|---|---|---|---|---|---|---|
| P. forrestii | 0.95 | 2.38 | 1.62 | 0.19 | 0.1 | 0.22 | 0 | 1.05 | 21 | 1.84 |
| P. tengchongensis | 75.04 | 61.77 | 68.07 | 97.66* | 99.39* | 98.64* | 0.09 | 76.7 | 14.91 | 61.04 |
| P. mairei | 39.09 | 57.09 | 49.88 | 19.82 | 14.73 | 21.98 | 0 | 41.96 | 99.73* | 49.81 |
| P. polyphylla var. chinensis | 15.75 | 27.23 | 22.04 | 5.84 | 3.87 | 6.49 | 0 | 16.9 | 83.79 | 23.51 |
| P. polyphylla var. yunnanensis | 99.47* | 95.99* | 99.22* | 79.11 | 69.88 | 78.62 | 0.02 | 99.21* | 40.64 | 97.72* |
| P. thibetica | 14.48 | 20.85 | 18.49 | 4.93 | 3.02 | 4.81 | 0 | 13.77 | 48.15 | 23.46 |
| Trillium tschonoskii | 0.01 | 0.01 | 0.01 | 0.04 | 0.06 | 0.04 | 98.93 | 0.02 | 0 | 0.01 |
| Polygonum paleaceum | 0 | 0 | 0 | 0 | 0 | 0 | 0.03 | 0 | 0 | 0 |
| Tupistra spp. | 0.02 | 0.03 | 0.02 | 0.02 | 0.02 | 0.02 | 1.12 | 0.02 | 0.02 | 0.01 |
*Over the 90% confidence percentage threshold are presented in italics
Discussion
Species identification is critical to ensuring the quality of herbal medicine. HRM analysis is a method that measures the dissociation rates of double-stranded DNA into single-stranded DNA in increasing temperatures [26]. Minute difference in amplicon size or composition, even a simple nucleotide, can be detected. In general, different genotypes have their own unique transitions, including changes in curve shape and in the plots of their melting curves, which can be shown in their HRM profiles [24].
At present, several methods, such as high performance liquid chromatography (HPLC) and near infrared spectroscopy (NIR), have been introduced to discriminate the controversial Paris species [27, 28]. However, the HPLC method is relatively expensive and NIR spectroscopy suffers from low precision and tedious calibration, which is not ideal for the rapid screening of large numbers of samples. For these reasons, molecular biology-based analysis is becoming increasingly popular for the differentiation and identification of species in several scientific fields [29]. Among the various molecular biology techniques, DNA barcoding, ITS2, has been successfully applied to distinguish Paris species [23]. However, DNA sequencing technology is relatively expensive and time-consuming since the sequencing process is about 1–2 days. The use of Bar-HRM can overcome the cost and time issues (the whole process only takes 4 h). Moreover, this new approach has its own advantage compared with previously employed methods. The Bar-HRM analysis is performed in one completely “closed” tube and the results are available for analysis at the end of the run.
In this study, we have successfully applied Bar-HRM analysis for the rapid detection and identification of two major Paris species and their seven adulterants. The results indicated that Bar-HRM is a sensitive, fast, cheap, and reliable method for identifying and tracking herbal medicines. To validate the accuracy of polymorphisms detected by HRM analysis, the sequences of the reference plant specie were analyzed. Each PCR product was amplified and both strands were sequenced in opposite directions. Although the PCR products of Tupistra spp. were clearly visible as a single band on agarose gel, the sequence information of Tupistra spp. was not obtained, and there are no documented reports on its ITS2 sequences information in the GeneBank. The multi-copy of ITS2 region may lead to the failure of sequence. However, the HRM melting curves could be used to distinguish Chonglou from its adulterants clearly, included Tupistra spp. Considering these features, Bar-HRM can be a useful tool for quality control of herbal medicines in the supply chain of the natural health product industry (NHP) and medicine supervisory institutions. In conclusion, this study substantially contributes to the protection of public health worldwide.
Conclusions
The Bar-HRM analysis has been used in this study to differentiate Chonglou, namely P. polyphylla var. yunnanensis and P. polyphylla var. chinensis, from their adulterate species. Species-specific PCR amplification of the ITS2 region was performed to validate the results of HRM analysis. The difference between Chonglou species and their adulterants can be observed from their sequencing information. The proposed method has been proved to be accurate, reliable, rapid, cost-effective and robust for identifying Chonglou from its adulterants. Bar-HRM analysis is also feasible for high-throughput assays, and could be well applied to other herbal species in the future, ensuring the perceived value and quality of herbal medicines.
Additional file
Additional file 1. Minimum Standards of Reporting Checklist.
Authors’ contributions
BZD and HLF carried out the molecular experiments, analyzed the data and wrote the manuscript. YPW wrote the manuscript, CX carried out the data analyses, PW thoroughly revised the manuscript. XWL and SLC conceived of the study, contributed to the design and interpretation of the research. All authors read and approved the final manuscript.
Acknowledgements
We acknowledge the help of Conglong Xia and Zhigang Duan (Dali University) for their help in the collection of plant samples and the identification of species.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The dataset supporting the conclusions of this article is included within the article.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This study was supported by Grants from The National Natural Science Foundation of China (31460084), the National Key Technology Support Program (2015BAI05B02), the Science and Technology Cooperation Project of Hong Kong, Macao and Taiwan (2015DFM30030), the Training Scheme of reserve Talents for Young and Middle-aged Academic and Technical Leaders of the Yunnan Province (2015HB058), and the Doctoral Science Foundation of Dali University (KYBS201737).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- Bar-HRM
barcoding coupled with high resolution melting
- HRM
high resolution melting
- ITS2
internal transcribed spacer 2
- IUCN
International Union for Conservation of Nature
- GCPs
genotype confidence percentages
- PCR
polymerase chain reaction
- Tm
melting temperature
- HPLC
high performance liquid chromatography
- NIR
near infrared spectroscopy
- NHP
natural health product industry
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13020-018-0162-4) contains supplementary material, which is available to authorized users.
Contributor Information
Bao-Zhong Duan, Email: bzduan@126.com.
Ya-Ping Wang, Email: 16483820@life.hkbu.edu.hk.
Hai-Lan Fang, Email: hlfang@yntcm.ac.cn.
Chao Xiong, Email: xiongchao080190@126.com.
Xi-Wen Li, Phone: +86 10 6405 8460, Email: xwli@icmm.ac.cn.
Ping Wang, Email: pwang54@aliyun.com.
Shi-Lin Chen, Phone: +86 10 6405 8460, Email: slchen@icmm.ac.cn.
References
- 1.Commission CP . Pharmacopoeia of the People’s Republic of China. Peking: Chin Med Science Press; 2015. [Google Scholar]
- 2.Wu S, Gao W, Duan H, Jia W. Advances in studies on chemical constituents and pharmacological activities of Rhizoma Paridis. Chin Tradit Herb Drug. 2004;35(3):344–346. [Google Scholar]
- 3.Li H. The genus Paris (Trilliaceae) Beijing: Science Press; 1998. [Google Scholar]
- 4.Newmaster SG, Grguric M, Shanmughanandhan D, Ramalingam S, Ragupathy S. DNA barcoding detects contamination and substitution in North American herbal products. BMC Med. 2013;11(1):1–13. doi: 10.1186/1741-7015-11-222. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Ekor M. The growing use of herbal medicines: issues relating to adverse reactions and challenges in monitoring safety. Front Pharmacol. 2013;4(4):177. doi: 10.3389/fphar.2013.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia LY. Handbook of toxicity of traditional Chinese medicine. Hohhot: Inner Mongolia Science Technology Press; 2006. [Google Scholar]
- 7.Chen SL, Song JY, Sun C, Xu J, Zhu YJ, Verpoorte R, Fan TP. Herbal genomics: examining the biology of traditional medicines. Science. 2015;347(6219):S27–S29. doi: 10.1126/science.1259172. [DOI] [Google Scholar]
- 8.Chen SL, Song JY. Herbgenomics. China J Chin Mater Med. 2016;41(21):3881–3889. doi: 10.4268/cjcmm20162101. [DOI] [PubMed] [Google Scholar]
- 9.Cabral P, Soares T, Lima A, De Miranda FD, Souza FB, Gonçalves L. Genetic diversity in local and commercial dry bean (Phaseolus vulgaris) accessions based on microsatellite markers. Gen Mol Res. 2011;10(1):140–149. doi: 10.4238/vol10-1gmr993. [DOI] [PubMed] [Google Scholar]
- 10.Chen SL, Pang XH, Song JY, Shi LC, Yao H, Han JP, Leon C. A renaissance in herbal medicine identification: from morphology to DNA. Biotechnol Adv. 2014;32(7):1237–1244. doi: 10.1016/j.biotechadv.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Gad HA, El-Ahmady SH, Abou-Shoer MI, Al-Azizi MM. Application of chemometrics in authentication of herbal medicines: a review. Phytochem Anal. 2013;24(1):1–24. doi: 10.1002/pca.2378. [DOI] [PubMed] [Google Scholar]
- 12.Al SSRE. Authentication methods for drugs used in Ayurveda, Siddha and Unani Systems of medicine: an overview. Int J Pharm Sci Res. 2012;3(8):2352. [Google Scholar]
- 13.Fu JG, Dai L, Lin Z, Lu HM. Houttuynia cordata Thunb: a review of phytochemistry and pharmacology and quality control. Chin Med. 2013;4(3):101–123. doi: 10.4236/cm.2013.43015. [DOI] [Google Scholar]
- 14.Chen SL, Yao H, Han JP, Liu C, Song JY, Shi LC, Zhu YJ, Ma XY, Gao T, Pang XH. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE. 2010;5(1):e8613. doi: 10.1371/journal.pone.0008613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Techen N, Parveen I, Pan Z, Khan IA. DNA barcoding of medicinal plant material for identification. Curr Opin Biotechnol. 2014;25(2):103–110. doi: 10.1016/j.copbio.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 16.Chen JJ, Zhao QS, Liu YL, Zha SH, Bing Z. Identification of maca (Lepidium meyenii Walp.) and its adulterants by a DNA-barcoding approach based on the ITS sequence. Chin J Nat Med. 2015;13(9):653–659. doi: 10.1016/S1875-5364(15)30062-5. [DOI] [PubMed] [Google Scholar]
- 17.Han JP, Pang XH, Liao BS, Hui Y, Song JY, Chen SL. An authenticity survey of herbal medicines from markets in China using DNA barcoding. Sci Rep. 2016;6:1–9. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osathanunkul M, Madesis P, de Boer H. Bar-HRM for authentication of plant-based medicines: evaluation of three medicinal products derived from Acanthaceae species. PLoS ONE. 2015;10(5):e128476. doi: 10.1371/journal.pone.0128476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu S, Wirthensohn MG, Hunt P, Gibson JP, Sedgley M. High resolution melting analysis of almond SNPs derived from ESTs. Theor Appl Genet. 2008;118(1):1–14. doi: 10.1007/s00122-008-0870-8. [DOI] [PubMed] [Google Scholar]
- 20.Ganopoulos I, Argiriou A, Tsaftaris A. Adulterations in Basmati rice detected quantitatively by combined use of microsatellite and fragrance typing with high resolution melting (HRM) analysis. Food Chem. 2011;129(2):652–659. doi: 10.1016/j.foodchem.2011.04.109. [DOI] [PubMed] [Google Scholar]
- 21.Osathanunkul M, Suwannapoom C, Osathanunkul K, Madesis P, de Boer H. Evaluation of DNA barcoding coupled high resolution melting for discrimination of closely related species in phytopharmaceuticals. Phytomedicine. 2016;23(2):156–165. doi: 10.1016/j.phymed.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 22.Li JJ, Song M, Xiong C, Zhao B, Sun W. Application of barcode high-resolution melting for rapid authentication of the medicinal plant Psammosilene tunicoides. Biotechnol Biotechnol Equip. 2016;30(4):790–796. doi: 10.1080/13102818.2016.1181988. [DOI] [Google Scholar]
- 23.Zhu YJ, Chen SL, Yao H, Tan R, Song JY, Luo K, Lu J. DNA barcoding the medicinal plants of the genus Paris. Acta Pharm Sin. 2010;45(3):376–382. [PubMed] [Google Scholar]
- 24.Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem. 2003;49(6):853–860. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reed GH, Wittwer CT. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem. 2004;50(10):1748–1754. doi: 10.1373/clinchem.2003.029751. [DOI] [PubMed] [Google Scholar]
- 27.Zhang T, Liu H, Liu XT, Xu DR, Chen XQ, Wang Q. Qualitative and quantitative analysis of steroidal saponins in crude extracts from Paris polyphylla var. yunnanensis and P. polyphylla var. chinensis by high performance liquid chromatography coupled with mass spectrometry. J Pharm Biomed Anal. 2010;51(1):114–124. doi: 10.1016/j.jpba.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 28.Zhao YL, Zhang J, Yuan TJ, Shen T, Li W, Yang SH, Hou Y, Wang YZ, Jin H. Discrimination of wild Paris based on near infrared spectroscopy and high performance liquid chromatography combined with multivariate analysis. PLoS ONE. 2014;9(2):e89100. doi: 10.1371/journal.pone.0089100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li XW, Yang Y, Henry RJ, Rossetto M, Wang YT, Chen SL. Plant DNA barcoding: from gene to genome. Biol Rev. 2015;90(1):157–166. doi: 10.1111/brv.12104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Minimum Standards of Reporting Checklist.
Data Availability Statement
The dataset supporting the conclusions of this article is included within the article.


