Abstract
Several novel mechanistic findings regarding to arsenic’s pathogenesis has been reported and some of them suggest that the etiology of some arsenic induced diseases are due in part to heritable changes to the genome via epigenetic processes such as DNA methylation, histone maintenance, and mRNA expression. Recently, we reported that arsenic exposure during in utero and early life was associated with impairment in the lung function and abnormal receptor for advanced glycation endproducts (RAGE), matrix metalloproteinase-9 (MMP-9) and tissue inhibitor of matrix metalloproteinase-1 (TIMP-1) sputum levels. Based on our results and the reported arsenic impacts on DNA methylation, we designed this study in our cohort of children exposed in utero and early childhood to arsenic with the aim to associate DNA methylation of MMP9, TIMP1 and RAGE genes with its protein sputum levels and with urinary and toenail arsenic levels. The results disclosed hypermethylation in MMP9 promotor region in the most exposed children; and an increase in the RAGE sputum levels among children with the mid methylation grade; there were also positive associations between MMP9 DNA methylation with arsenic toenail concentrations; RAGE DNA methylation with iAs, and %DMA; and finally between TIMP1 DNA methylation with the first arsenic methylation. A negative correlation between MMP9 sputum levels with its DNA methylation was registered. In conclusion, arsenic exposure during in utero and early life modifies DNA methylation in the evaluated genes. This may contribute to abnormal extracellular matrix remodeling genes expression altering protein levels with a subsequent lung function impairment.
Keywords: Arsenic, children, DNA methylation, MMP9, TIMP1, RAGE
INTRODUCTION
Arsenic contamination is a serious and widespread global public health problem with a significant known groundwater burden in countries such as Argentina, Bangladesh, Chile, China, Ghana, India, Mexico, Taiwan, the United States, and Vietnam. It is estimated that more than 200 million people are at risk of toxic arsenic exposure (Hunt et al., 2014). Chronic arsenic exposure damages a broad range of organs systems in a time-and dose-dependent manner and the severity of its effects have been related with the different stages of the human development. Exposure to arsenic during pregnancy affects human developmental processes resulting in lower birth weight, increased infant mortality, and neurological and intellectual impairments in children (Naujokas et al., 2013). As well, it has been reported that subjects suffering from arsenic toxicity are more susceptible to pulmonary diseases, conditions that also carry higher mortality rates in exposed populations (Naujokas et al., 2013, Parvez et al, 2013). Several authors have suggested that the mechanism of action of As in the lungs is that it can enhance tissue inflammation (De et al., 2004; Nemery B., 1990), inducing respiratory function impairment by oxidative stress (Lantz and Hays, 2006) or by producing or increasing pulmonary fibrosis (Nemery B., 1990; von Ehrenstein et al., 2005).
Some studies have mentioned that a significant proportion of lung disease would originate in utero or during the early infancy (Merkus et al., 2003; Smith et al., 2006; Waalkes et al., 2008; Yuan et al., 2010) suggesting that the etiology of these disease are due in part to heritable changes to the genome (Smeester et al., 2011), which can be induced by the exposure to toxicants during sensitive times of development. In infants born to arsenic exposed mothers an increased inflammatory responses have been reported (Fry et al., 2007). Arsenic exposure alters markers of inflammation such as soluble receptor for advanced glycation endproducts (RAGE), matrix metalloproteinase-9 (MMP-9) and tissue inhibitor of matrix metalloproteinase-1 (TIMP-1)], which are related to remodeling of the extracellular matrix including lung tissue. Recently our research group reported that the arsenic exposure during in utero and early life was associated with impairment in the lung function (Recio-Vega et al., 2015) and with abnormal sRAGE (soluble RAGE), MMP-9 and MMP-9/TIMP-1 levels (Olivas-Calderon et al., 2016). Lantz et al. (2007), found a significant negative correlation between RAGE sputum levels and total urinary inorganic As.
Several novel mechanistic findings regarding arsenic’s pathogenesis has been reported including As induced changes to the genome via epigenetic processes such as DNA methylation (Smeester et al., 2011), histone maintenance, and mRNA expression (Niezwiecki et al., 2013). Alterations in DNA methylation associated with arsenic exposure have been evaluated in cells, animal models and in humans (Jensen et al., 2010; Reichard et al., 2007; Xie et al., 2007; Zhao et al., 1997; Zhong and Mass, 2001). Exposure to As has an impact on both hypomethylation and hypermethylation of repetitive genomic elements and in individual genes (Bailey KA and Fry RC, 2014), with hypermethylation more prevalent in some studies (Jensen et al., 2009). In the traditional model of the effects of DNA methylation, hypermethylation of cytosine-rich gene regions (CpG islands) in the promoters of target genes inhibits the initiation of transcription (Holliday and Pugh, 1975; Riggs, 1975), which in turn, modifies protein expression and/or protein function. Chicoine et al. (2002), found an inverse correlation between levels of methylation of the MMP-9 promoter and the level of MMP-9 expression.
Based on our previous results (Recio-Vega et al., 2015; Olivas-Calderon et al., 2015) and in the reported impacts of arsenic on DNA methylation in individuals genes and proteins important for remodeling of the extracellular matrix, the present work was designed with the aim of determining the association between DNA methylation on promoters of extracellular matrix remodeling proteins (MMP9, TIMP1 and RAGE genes) with the sputum protein levels of these markers and with urinary As levels in our cohort of children exposed in utero and early life to arsenic through drinking water.
MATERIALS AND METHODS
Study population
The subjects included in this report are a subset of those reported in an earlier study (Recio-Vega et al., 2015). More than 500 children were evaluated; however, only 50 children were included in this study. We exclusively included only healthy volunteers who were conceived in the studied rural communities, whose mothers remained throughout their entire pregnancies in these communities, and those volunteers who remained as permanent residents of the same communities. The participants were girls and boys aged 6-12 years residing in four rural communities where the highest arsenic tap water levels detected in the last 20 years ranged from 104-360 ppb. These communities received groundwater through the local water supply and the high As levels in the water were due, in part, to an over-extraction of water from the ground for crops. These communities form part of the geographic area knowns as Comarca Lagunera, which is located in the north-central part of Mexico and known to be an area where increased arsenic toxicity has been reported (Sampayo-Reyes et al., 2010; Recio-Vega et al., 2015).
Written informed consent was obtained from each participant and from their parents to obtain biological samples which were obtained at the time of interview. The study protocol was approved by the Ethics Committee of the School of Medicine at Torreon, University of Coahuila, Mexico.
Questionnaire application
Information was collected through in-person interviews and included socio-demographic variables (education, socioeconomic status, type of kitchen, type of fuel used for cooking), lifetime residential history, lifestyle factors (secondhand smoke defined as someone smoking regularly in the same room at home, and exercise), parent’s occupational history, water source types (municipal tap water, bottled, other), current medications, medical history, and diet. Water consumption habits were ascertained through a standardized questionnaire. Asthmatic volunteers confirmed by a doctor were not included in the study.
As measurement in drinking water, urine and toenails
Drinking water samples (well) were collected from each rural community included in the study and analyzed for inorganic arsenic levels. No other contaminants in the drinking water were assessed. Well water samples from each rural community are representative of the water that participants drank and it is provided through the unique local water supply system. Individual exposure was assessed based on the urinary concentration of the total arsenic level. A first morning void urine sample was collected in two sterile 120-mL screw-topped polypropylene containers. The urine samples were obtained during the late autumn and winter seasons to avoid the hottest seasons when there is much higher water consumption and children have increased outdoor activities. Also we cut children’s nails with a new sterilized nail clipper for each child. Parents were instructed to wait two weeks before study and then we clipped nails from all ten of their children’s toes and placed the clippings in small brown manila envelopes. The in utero and childhood As exposure was calculated as the As tap water level mean registered in the studied communities during the period of 2000-2013. Arsenic content were analyzed at Arizona Laboratory for Emerging Contaminants University of Arizona, Tucson, Arizona. Total As concentration in urine and toenails were measured by inductively coupled plasma mass spectrometry (ICP-MS) (7700x ICP-MS, Agilent Technologies, Santa Clara, CA, USA). Briefly, arsenic species in urine [AsV, AsIII, monomethylarsonic acid (MMAV), dimethylarsinic acid, (DMAV) and arsenobetaine] was performed using high performance liquid chromatography (Agilent 1100) with inductively coupled plasma mass spectroscopy (Agilent 7500) (HPLC/ICP-MS) (Agilent 1100 HPLC with Hamilton PRP-X100 column) with a method adapted from Milstein et al. (2003). Additional exposure to DMA or to organic arsenic metabolized in the body to DMA, which usually is attributable to consumption of seafood such as bivalves and seaweeds, was considered minimal because such sea-foods are essentially never eaten in this area. Arsenic metabolism efficiency was calculated using the following formulas proposed by Del Razo et al. (1997), first methylation = MMAV/(AsV+AsIII); second methylation = DMAV/MMAV.
According with their total urinary arsenic level, subjects were divided in three study groups (tertiles) with the aim to search the effects related to exposure gradients (Group 1: <59 ug/L; Group 2: ≥59 <256 ug/L; Group 3: ≥256 <664 ug/L).
Sputum Analysis
Sputum induction was performed as previously described (Burgess et al., 2013). Sputum induction was performed by using a sterile 5% saline aerosol (Baxter, Deerfield, IL, USA) generated by using Comp Air XLT, Compressor Nebulizer Systems (Model NE-C25, Omcron Healthcare, Bannockburn, Illinois, USA) set on maximum output. Subjects were encouraged to cough up every 2 min for a period of 30 min and the sputum was collected. Prior to each cough, subjects were asked to discard any saliva to minimize salivary contamination of the sputum sample. Sputum samples were treated with 10% sputolysin (Calbiochem, San Diego, CA, USA) containing penicillin streptomycin to inhibit bacterial growth. Samples were processed in the Environmental Health Laboratory. The sputum samples were centrifuged to separate cells from the supernatant, which was stored at -70 °C. Sputum supernatant samples were analyzed in duplicate for levels of sRAGE, MMP-9 and TIMP1 using a commercially available enzyme-linked immunoabsorbent assay (ELISA) (Biovendor LLC, Asheville NC 28806 USA). A uniform initial sample dilution was performed on all samples to maximize the number of samples with concentrations within the standard range for the sRAGE, MMP-9, TIMP-1.
DNA methylation analysis by MassARRAY
Sodium bisulfite-treated genomic DNA was prepared according to the manufacturer’s instructions of EZ DNA Methylation-Gold ™ Kit (Zymo Research). Sodium bisulfite-treated DNA (5 ng) was seeded into a region-specific PCR incorporating a T7 RNA polymerase sequence as described by the manufacturer (Sequenom). Resultant PCR product was then subjected to in vitro transcription and RNase A cleavage using the MassCLEAVE T-only kit, spotted onto a Spectro CHIP array, and analyzed using the MassARRAY Compact System matrix-assisted laser desorption/ionization-time-of-flight mass spectrometer (Sequenom). Each sodium bisulfite-treated DNA sample was processed in two independent experiments. Data were analyzed using EpiTyper software (Sequenom) as described previously (Novak et al., 2008; Coolen et al., 2007). Primer sequences were designed using EpiDesigner. Primer sequences are available upon request.
The methyl DNA immunoprecipitation microarrays and data analysis
The methylated fraction of DNA was obtained by immunoprecipitation as described (Novak P et al., 2006). Input and immune-precipitated DNAs were amplified, labeled and analyzed on Human Promoter arrays as described (Jensen TJ et al., 2009). The magnitude of the microarray methylation was directly correlated with the degree of methylation as determined by MassARRAY.
Subjects were divided in three study groups (tertiles) according with their DNA methylation levels with the aim to search the effects related to DNA methylation gradients (MMP9: methylation groups: Low=0.3–0.12; Mid=0.13-0.17; High=0.18-0.33. TIMP1: methylation groups: Low=0.02-0.04; Mid=0.4-0.21; High=0.23-0.32. RAGE: methylation groups: Low=0.2-0.64; Mid=0.65-0.75; High=0.77-0.98).
Statistics
Independent and dependent variables were described according to their frequency and distribution measurements (arithmetic mean and standard deviation). The F test was used when the variable was divided into more than two categories, and the Mann Whitney test was used to compare different frequencies of DNA methylation and levels of arsenic when dichotomous variable categories were analyzed. This method permitted us to establish statistical differences among groups for each dependent variable. Linear regression models were used to assess crude or independent associations between the different DNA methylation levels with arsenic urine and toenail concentrations. In all multivariable models, we included those statistical significant variables (P<0.05) identified in the bivariate model and those possessing biological plausibility (age, gender, second hand smoking). All analyses were performed using the statistical software STATA 11.0 (Stata Corp., College Station, TX).
RESULTS
Socio-demographic, anthropometric characteristics and life style
Most of the socio-demographic, anthropometric characteristics and life style variables of the volunteers were similar between groups of study, except gender, there were more girls in the most exposed groups (2 and 3) than in the less exposed group (1) (p<0.05). On the other hand, the frequency of use of tap water for cooking was slightly higher in participants from Group 3, however, it was not significantly different from the other groups (p=0.30). No differences were found in drinking of tap water at home or at school between the studied groups (P>0.05) (Table 1).
Table 1.
Socio-demographic, anthropometric characteristics and life style of the participants by study groups. Results are shown as arithmetic mean and standard deviation and percentage.
| Group 1 [<59 ug/L] (n=16) |
Group 2 [≥59 <256 ug/L] (n=17) |
Group 3 [≥256 <664 ug/L] (n=17) |
|
|---|---|---|---|
| Age (years) | 8.68 ± 1.74 | 9.29 ± 1.96 | 8.52 ± 1.66 |
| Gender (%) | |||
| Female | 56.25 | 58.82* | 58.82* |
| Male | 56.25 | 41.18 | 41.18 |
| BMI** | 17.98 ± 2.99 | 18.48 ± 3.61 | 16.73 ± 3.95 |
| Type of water used at home (%) | |||
| Tap water | 43.75 | 29.41 | 41.18 |
| Bottled water | 50.00 | 64.71 | 52.94 |
| Both | 6.25 | 5.88 | 5.88 |
| Type of water used at school (%) | |||
| Tap water | 100 | 94.12 | 88.24 |
| Bottled water | 0 | 5.88 | 5.88 |
| Both | 0 | 0 | 5.88 |
| Type of water for cooking (%) | |||
| Tap water | 62.50 | 88.24 | 88.24 |
| Bottled water | 31.25 | 11.74 | 11.76 |
| Both | 6.25 | 0 | 0 |
| Use of folic acid during pregnancy | |||
| Yes | 87.50 | 82.35 | 100 |
| No | 12.50 | 17.65 | 0 |
| Second hand smoking | |||
| Yes | 87.50 | 58.82 | 70.59 |
| No | 12.50 | 41.18 | 29.41 |
| Living near to an industry | |||
| Yes | 31.25 | 41.18 | 29.41 |
| No | 68.75 | 58.82 | 70.59 |
P<0.05: Mann–Whitney test o Chi2 (Groups 2 or 3 vs Group 1)
BMI weight/heigh2
Urinary and toenail arsenic concentrations
Mean total arsenic concentration in the entire population in urine and toenails was 191.5±165.5 (ug/L) and 2.84±1.61 (ug/g), respectively. Total As, AsIII, MMA, DMA and inorganic As (iAs) levels were significantly higher (P<0.05) in subjects from Groups 2 and 3 than in Group 1; meanwhile, AsV was lower in Groups 2 and 3 (Table 2). The percentages of MMA and DMA were similar between groups (P>0.05) (Table 2). The first methylation rate was significantly higher in Group 3; as well as, the second methylation in Group 2 (P<0.05). Total As in toenails was higher in Group 2 and 3 compared with that found in subjects from Group 1 (Table 2).
Table 2.
Urinary arsenic and its metabolites concentrations in the studied population. Results are shown as arithmetic mean and standard deviation.
| Group 1 (n=16) |
Group 2 (n=17) |
Group 3 (n=17) |
|
|---|---|---|---|
| Total As | 39.14 ± 15.38 | 147.68 ± 83.62* | 378.57 ± 119.65* |
| AsIII | 3.14 ± 2.05 | 19.00 ± 19.48* | 53.17 ± 22.65* |
| AsV | 16.30 ± 16.09 | 10.67 ± 16.86* | 10.99 ± 8.90* |
| MMAV | 5.37 ± 2.75 | 20.88 ± 15.54* | 52.17 ± 18.34* |
| DMAV | 14.04 ± 15.63 | 95.88 ± 60.83* | 225.16 ± 76.13* |
| iAs | 19.45 ± 16.45 | 29.67 ± 21.44 | 64.16 ± 28.39* |
| First methylation | 0.54 ± 0.49 | 0.84 ± 0.48 | 0.87 ± 0.19* |
| Second methylation | 3.05 ± 3.36 | 5.30 ± 2.92* | 4.52 ± 1.31 |
| %iAs | 51.45 ± 35.63 | 23.28 ± 23.85 | 17.05 ± 6.07 |
| %MMA | 13.32 ± 3.29 | 13.05 ± 3.88 | 14.08 ± 3.73 |
| %DMA | 35.16 ± 35.54 | 62.39 ± 23.85 | 60.48 ± 13.01 |
| Total As in toenails | 2.10 ± 1.67 | 2.83 ± 1.26* | 3.54 ± 1.69* |
P<0.05: Mann–Whitney test (Groups 2 or 3 vs Group 1).
DNA methylation in promoter regions of MMP9, TIMP1 and RAGE genes and arsenic exposure
DNA methylation gradient in promoter region of MMP9 was significantly higher (p=0.03) in the subjects most exposed (Group 3) than in the less exposed (Group 1) (Table 3). A slight increased DNA methylation in the promoter region of TIMP1 was recorded in participants from Groups 3 but this increase was not statistically significant (P>0.05) (Table 3). Likewise, DNA methylation in the promoter region of RAGE was higher in Groups 2 and 3 than in Group 1, but this difference was again not statistically significant (P>0.05) (Table 3).
Table 3.
DNA methylation in the promoter region of MMP9, TIMP1 and RAGE by study groups. Results are shown as arithmetic mean and standard deviation.
| Gene | Group 1 (n=16) |
Group 2 (n=17) |
Group 3 (n=17) |
|---|---|---|---|
| MMP9 | 0.13 ± 0.07 | 0.16 ± 0.07 | 0.17 ± 0.03* |
| TIMP1 | 0.13 ± 0.10 | 0.12 ± 0.11 | 0.14 ± 0.10 |
| RAGE | 0.68 ± 0.16 | 0.73 ± 0.12 | 0.73 ± 0.10 |
P<0.05: Mann–Whitney test (Groups 2 or 3 vs Group 1).
MMP9, TIMP1 and RAGE sputum levels and DNA methylation
A reduction of 47% of MMP9 sputum levels were observed in those subjects with the higher gradient of DNA methylation when compared with that seen in those with the lower methylation gradient; however, that difference was not statistically significant (P>0.05). TIMP1 sputum levels were similar between methylation groups. RAGE sputum levels were significantly higher (62%) (P<0.05) in subjects with the mid methylation gradient when compared with the observed in subjects from the low methylation gradient group (Table 4).
Table 4.
MMP9, TIMP1 and RAGE sputum levels by DNA methylation gradients. Results are shown as arithmetic mean and standard deviation.
| Low methylation& (n=16) |
Mid methylation& (n=17) |
High methylation& (n=17) |
|
|---|---|---|---|
| MMP9 (pg/mL) | 316.26 ± 292.69 | 343.77 ± 332.42 | 168.35 ± 122.39 |
| TIMP1 (pg/mL) | 96.52 ± 67.86 | 96.34 ± 60.84 | 75.06 ± 65.45 |
| RAGE (pg/mL) | 381.62 ± 275.55 | 619.36 ± 276.11* | 473.68 ± 386.53 |
Methylation groups: MMP9: Low=0.3-0.12; Mid=0.13-0.17; High=0.18-0.33; TIMP1: Low=0.02-0.04; Mid=0.4-0.21; High=0.23-0.32 and RAGE: Low=0.2-0.64; Mid=0.65-0.75; High=0.77-0.98.
P<0.05: Mann–Whitney test (Groups 2 or 3 vs Group 1).
Associations between DNA methylation and arsenic exposure
In the crude linear regression model, no associations were found between the As species measured with the grade of DNA methylation in the promoter regions of MMP9 and TIMP1, but a positive association was registered between MMP9 methylation with toenail As levels (Table 5). With regard to the DNA methylation in the promoter of RAGE, three positive associations were observed with iAs, MMAV, and %MMA (P<0.05); meanwhile, a negative association (P<0.05) was found with the second methylation (Figure 1). In the adjusted linear regression model, all associations recorded in the crude model remained statistically significant (Table 5).
Table 5.
Association between DNA methylation in promoter region of MMP9, TIMP1 and RAGE with arsenic levels.
| MMP9 | TIMP1 | RAGE | ||||
|---|---|---|---|---|---|---|
| β | P | β | P | β | P | |
| Total As | 0.0001 | 0.224 | 0.00002 | 0.799 | 0.0001 | 0.412 |
| AsIII | 0.0005 | 0.278 | 0.0001 | 0.801 | 0.001 | 0.237 |
| AsV | 0.0033 | 0.201 | −0.0011 | 0.294 | 0.0014 | 0.430 |
| MMAV | 0.0007 | 0.217 | 0.0002 | 0.671 | 0.0016 | 0.050* |
| DMAV | 0.0002 | 0.117 | 0.0001 | 0.561 | 0.0002 | 0.423 |
| iAs | 0.0005 | 0.231 | −0.0001 | 0.848 | 0.0013 | 0.044* |
| First methylation | 0.0412 | 0.313 | 0.025 | 0.032* | 0.0022 | 0.965 |
| Second methylation | −0.0082 | 0.249 | 0.001 | 0.916 | −0.017 | 0.034* |
| %iAs | −0.0001 | 0.979 | −0.003 | 0.531 | 0.001 | 0.367 |
| %MMA | 0.003 | 0.441 | 0.002 | 0.518 | 0.015 | 0.016* |
| %DMA | −0.0001 | 0.922 | 0.0003 | 0.514 | −0.001 | 0.303 |
| Total As in toenails | 0.017 | 0.036* | 0.004 | 0.568 | 0.0001 | 0.994 |
Adjusted by age, gender, second hand smoking.
Figure 1.
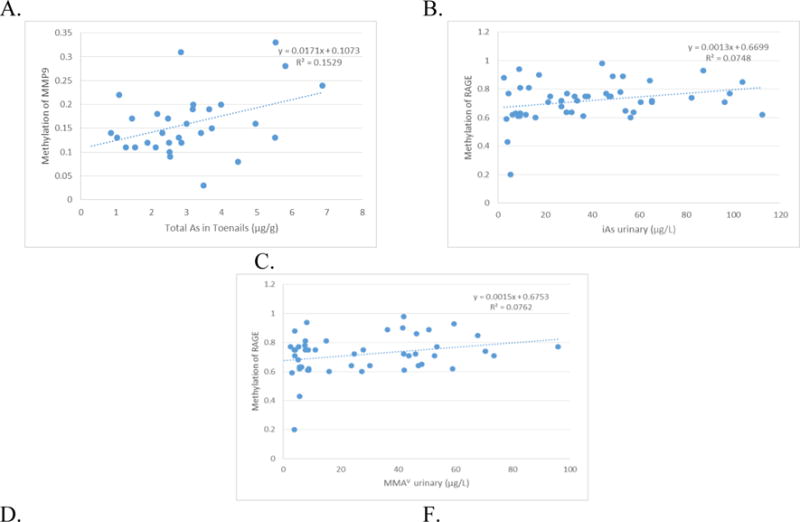
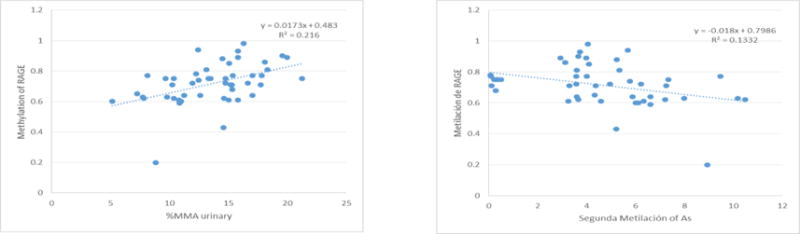
Significant association between DNA methylation in promoter region and arsenic where: A. Methylation of MMP9 and total As in toenails (ug/g). B. Methylation of RAGE and iAs in urine (ug/L). C. Methylation of RAGE and MMAV in urine. D. Methylation of RAGE and %MMA in urine. E. Methylation of RAGE and Second Methylation of As.
Association between DNA methylation with MMP9, TIMP1 and RAGE sputum levels
In the bivariate or multivariate linear regression models, no significant associations between the DNA methylation gradients of promoter regions assessed with the sputum protein concentrations were registered (Table 6). Figure 2 shows the relationship between DNA methylation gradient of the promoter region of MMP9 with MMP9 sputum levels.
Table 6.
Adjusted linear regression between DNA methylation of promoter region of MMP9, TIMP1 and RAGE with their sputum protein levels.
| β | P | IC 95% | |
|---|---|---|---|
| MMP9 | −4.44 | 0.155 | −10.66 – 1.77 |
| TIMP1 | −0.079 | 0.969 | −4.19 – 4.03 |
| RAGE | −0.028 | 0.978 | −2.11 – 2.05 |
Adjusted by age, gender, second hand smoking.
Figure 2.
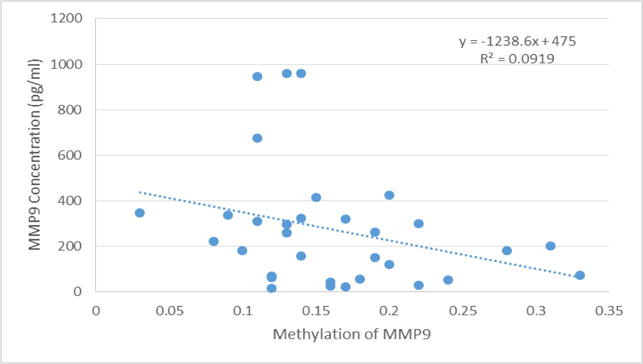
Association between MMP9 sputum levels (pg/mL) with DNA methylation in promoter region of MMP9.
DISCUSION
To our knowledge, this is the first study evaluating the association between the DNA methylation of three genes implicated in remodeling of extracellular matrix (MMP9, TIMP1 and RAGE) with the sputum inflammatory marker levels and with urinary As concentration in a cohort of children exposed in utero and early life to this metalloid.
Several reports have shown that some As’s health risks are associated with As levels above 50 ug/L (ACGIH, 2001; WHO, 2001; Tseng et al., 2005; Valenzuela et al., 2005) and with abnormal As biotransformation (methylation profile). The studied subjects have been chronically exposed to high As levels in utero and during their childhood through drinking water (mean 152.1±49.3 ug/L; range 104-275 ug/L). They are still exposed because their present As urinary levels (mean 191.5±165.5 ug/L) show that more than 75% of them had urinary As levels >50 ug/L. These high levels are impacting the children’s health as reveled in our previous studies in which we found that chronic arsenic exposure modified negatively the sRAGE, MMP-9 and MMP-9/TIMP-1 sputum levels and increased significantly the frequency of abnormal spirometric patterns (Calderon-Olivas et al., 2015). Arsenic mediates its toxicity by generating oxidative stress, causing immune dysfunction, promoting genotoxicity, hampering DNA repair, disrupting signal transduction (Hunt et al., 2014) and inducing changes to the genome via epigenetic processes such as DNA methylation (Argos M, 2015; Boellmann et al., 2010; Green et al., 2016; Rager et al., 2015; Ren et al., 2011). Several studies have evaluated DNA methylation in vitro, in vivo and in human populations exposed to different doses of arsenic (Chanda et al., 2006). All these studies have reported contradictory results in terms of DNA methylation gradient, some of them demonstrated that arsenic exposure is associated with hypomethylation (Pilsner et al., 2009; Wilhelm et al; 2010; Intarasunanont et al., 2012; Tajuddin et al., 2013) and others with hypermethylation (Chen et al., 2007; Pilsner et al., 2007; Majumdar et al., 2010; Hossain et al., 2012; Kile et al., 2012; Pilsner et al., 2012; Lambrou et al., 2012) at various genetic loci. With the aim to try to explain these controversial results, Chanda et al. (2006) suggested that DNA hypermethylation is the main effect after acute As exposure and hypomethylation occur just after a long exposure to high doses. However, in a chronic exposed population to As through drinking water (100 ug/L), Smeester et al. (2011), found 183 genes with altered methylation of which 182 were hypermethylated. Another factor that probably plays an important role in the DNA methylation gradient is the dose at which subjects are exposed. Indeed, Majumdar et al. (2010), showed that exposures to 250–500 ug/L of arsenic in drinking water results in global hypermethylation, but concentrations above 500 ug/L of arsenic results in global hypomethylation. In our studies, subjects have been chronically exposed to As through drinking water (152.1 ug/L) and have high As urinary levels (191.5 ug/L). We found a hypermethylation in the promotor region of MMP9 and RAGE in the two most exposed groups; however, only the increase in DNA methylation of MMP9 was significantly higher (P<0.05) and was associated with the long term biomarker of As exposure (toenail As levels). Liu et al. (2014), evaluated toenail arsenic concentration in relation to white blood cell global DNA methylation and they did not find statistically significant associations. Differences between our results and those obtained by Liu et al. (2014) could be due to the fact that toenail As concentration in Liu’s population was lower (0.144 ug/g) compared to that registered in our population (2.84 ug/g). Our data support the ideas that time of exposure and the magnitude (dose) of exposure play an important role in DNA methylation in the promoter of MMP9. On the other hand, our data suggest too, that genes exhibit different susceptibility for abnormal DNA methylation in terms of exposure time, because the DNA methylation in the promoter of RAGE was positively associated with three acute As exposure measurements and not with the long term As biomarker.
Another major area of interest has been the relationship between iAs biotransformation and the DNA methylome (Baley and Fry, 2014). The efficiency of iAs biotransformation is believed to be an important factor in the development of iAs-associated diseases, because several iAs metabolites are biologically reactive and toxic (Tseng, 2007; Baley and Fry, 2014). In our study, we quantified As metabolites and calculated percentages of As and efficiency of methylation and their association with DNA methylation of promoter regions. We found a positive association between DNA methylation of RAGE with iAs, MMA and %DMA. Similar results were found by Hossain et al. (2012), they found a positive association with %iAs and methylation of p16 and a negative association between %DMA and methylation of p16. Contrary, Bailey et al. (2014) found a negative association between the DNA methylation in promoter regions of almost all evaluated genes with urinary As metabolites concentrations, as well as, Niedzwiecki et al. (2013), did not find any correlation between As metabolites and DNA methylation. The differences between our and Hossain et al. (2012) results with those reported by Bailey et al. (2014), and Niedzwiecki et al. (2013), might by due to the differences in age, race, genetics, environment, style life, health status, kind of cells evaluated, time of exposure, dose of exposure, As biotransformation and genes evaluated.
Literature has described other variables that may influence the degree of DNA methylation and the most important are age, gender and intake of acid folic as a promoter of methyl group synthesis.
Evidence supports that global DNA methylation in white blood cells changes with age (Terry et al., 2011). Pilsner et al. (2007), found that age was positively associated with [3H]-methyl incorporation. Similarly, Bollati et al. (2009), described low levels of methylation in the Long Interspersed Nuclear Elements-1 (LINE-1) with an increase of age. Although other studies did not find a significant correlation between age and [3H]-methyl incorporation like our results, it is possible that this is attributable to the demographics of the study populations.
Several studies have found that global DNA methylation is higher in women than in men (Wilhelm et al., 2010; Hsiung et al., 2007; Fuke, 2004; Zhu et al., 2010; El-Maarri et al., 2011; Zhang et al., 2011). Fuke C. et al. (2004) reported a higher number of methylated cytosines in men than in women; however, these findings could not be replicated in a study with a greater number of subjects where methylated cytosines were also quantified (Jones PA et al., 2002). In our population we did not find statistical differences between DNA methylation by gender, which may suggest that other gender-specific factors such as hormone levels, socio-economic factors and lifestyles may subsequently influence other stages of life, resulting in differences in DNA methylation.
Epidemiologic studies in adults have reported that associations between As exposure and global DNA methylation were modified by folate status (Niedzwiecki et al. 2013). Folate is required for the synthesis of methionine, a precursor of the universal methyl donor SAM. SAM provides methyl groups in a variety of biochemical reactions, including methylation of DNA. One would expect that a deficiency of methyl groups in the diet might lead to hypomethylation of DNA in several tissues, including blood. However, associations between global DNA methylation in blood and dietary folate or plasma levels of folate have not been generally detected in observational studies (Terry et al., 2011; Niedzwiecki et al., 2013). In another study where supplemental folic acid was administered to the study population, those who have sufficient levels of folate at the beginning of the study did no show any increase in DNA methylation (Basten et al., 2006, Jung et al., 2011). In support of this hypothesis, folic acid supplementation was not associated with higher global DNA methylation in study populations whose participants were folate sufficient at baseline (Basten et al. 2006; Jung et al. 2011). When we compare the degree methylation of DNA between children from mothers who took folic acid during pregnancy (85%) with those who did not, we did not find statistical differences between DNA methylation. Our results support that folate might influence DNA methylation only under conditions in which folate is limiting (Niedzwiecki et al., 2013).
Finally, it is important to point out that the degree of methylation of specific genes may vary according to the type of cell used to assess it. DNA methylation patterns are tissue specific, and one critical limitation for human epigenetic studies is that tissues that are relevant for disease etiology cannot be easily obtained in most cases from patients and study participants (Baccarelli et al., 2012; Terry et al., 2011). Human in vivo studies of DNA methylation have often used blood (Tarantini et al., 2009; Bacarelli et al., 2009; Hou et al., 2010; Wright et al., 2010; Bailey KA et al., 2012; Bollati et al., 2009) or buccal cells as easily obtainable specimens in patients as well as in healthy individuals (Baccarelli et al., 2012). Characterizing methylation signatures in DNA from peripheral blood may have important implications as a biomarker for early diagnosis and prognosis (Qiu, et al., 2012; Terry et al., 2011). In our study, we used peripheral blood white cells, which are not known to be direct target of As-induced carcinogenesis (Niedzwiecki et al., 2013) or diseases. However, As trioxide is a highly effective therapeutic drug for the treatment of acute promyelocytic leukemia (Powell et al. 2010). Therefore, we can assume that As distributes to bone marrow progenitor cells and influences their cellular function including white cells (Petrie et al. 2009; Soignet et al. 1998); nevertheless, we do not know the extent to which methylation of peripheral blood white cell DNA reflects the methylation of other target tissues (Niedzwiecki et al., 2013). Therefore, the previous factors must be considered and analyzed integrally when results from different studies are discrepant.
The mechanism by which As influences DNA methylation after exposure to the metalloid is not clear, however is widely known that arsenic methyltransferase uses the same methyl donor (SAM) as DNA methyltransferase and other methyltransferases. It has been reported that As exposure might cause induction of cytosine methyltransferase (Goeringet al., 1999), which might explain the hypermethylation; however, other studies have suggested, that long-term arsenic exposure may cause depletion of the SAM methyl donor pool due to over consumption of the methyl groups by arsenic methyltransferase which would result in the inability to maintain methylated cytosine in DNA (hypomethylation) (Mass and Wang, 1997). Changes in SAM availability due to As exposure/biotransformation may therefore disturb DNA methylation patterns and subsequently influence disease risk by altering the expression of key genes (Bailey et al., 2013).
DNA methylation is a fundamental determinant of chromatin structure. In general, the extent of methylation at CpG islands, DNA sequence clusters rich in CG dinucleotides near gene promoters, correlates with the long-term silencing of gene transcription (Reichard et al., 2007). Genes with high levels of DNA methylation had, on average, lower expression levels, whereas genes with low expression displayed higher levels of DNA methylation. This finding is consistent with previous observations comparing the methylation levels of genes according to their expression category (Rojas et al., 2015). Chicoine et al. (2002) evaluated the correlation between the level of methylation of the MMP-9 promoter and the level of MMP-9 expression and found an inverse relationship between these variables which indicates that methylation may contribute to the suppression on transcriptional activity of MMP-9 promoter.
We evaluated DNA methylation in promoter regions of MMP9, TIMP1 and RAGE because these are rich regions in CpG islands and can be the most predictive of gene expression levels. Although we did not evaluate gene expression, we quantified in sputum the levels of their respective proteins. We did not find significant associations between protein amounts and DNA methylation; however, there was a decrease in the protein levels with higher DNA methylation in the promoter regions of MMP9 and TIMP1. These results are in agreement with previous studies in which a negative relationship between promoter CpG island/CpG island shore methylation and gene expression was observed (Bell et al., 2011; van Eijk et al., 2012). Alterations in the DNA methylation gradient of genes influences the synthesis of their respective proteins, which in turn, modifies the biological process in which they are involved causing or predisposing to disease.
In many cases, the biological consequences of these changes (e.g., corresponding changes in mRNA or protein levels) or significance of relationship (e.g., causal relationship between iAs exposure, DNA methylation alterations, and malignant transformation) have not been established (Bailey KA, 2014). An increase in global DNA methylation might be associated with an increased spontaneous mutation rate. Methylated cytosines are more prone to deamination than unmethylated cytosines, which increase the likelihood of C→T transitions (Niedzwiecki et al. 2013). In addition, DNA methylation is associated with heterochromatic regions (Beisel and Paro 2011), and somatic mutation density in human cancer genomes have been found to be highest in heterochromatin-like domains (Schuster-Bockler and Lehner 2012).
CONCLUSION
Arsenic exposure during in utero and early life modify DNA methylation in the evaluated genes, which may contribute to abnormal extracellular matrix remodeling genes expression and function, altering protein levels with subsequent lung function impairment.
Acknowledgments
We thank Dr. R. Clark Lantz and Binh Chau (Cellular & Molecular Medicine - University of Arizona) for outstanding financial and technical support. Also thanks Dr. Bernard W Futscher, Dr. Marc M. Oshiro, (The University of Arizona Cancer Center), MB(ASCP)CM Colleen Ramsower and RS Crystal Richt (University of Arizona Genetics Core) for outstanding technical support. Supported in part by grants P42 ES004940 and P30 ES006694.
References
- American Conference of Governmental Industrial Hygienists (ACGIH) TLVs and BEIs. Cincinnati, OH: Signature Publications; 2005. [Google Scholar]
- Argos M. Arsenic Exposure and epigenetic alterations: Recent findings based on the Illumina 450K DNa methylation Array. Curr Environ Health Rep. 2015;2(2):137–44. doi: 10.1007/s40572-015-0052-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Rusconi F, Bollati V, Catelan D, Accetta G, Hou L, Barbone F, Bertazzi PA, Biggeri A. Nasal cell DNA methylation, inflammation, lung function and wheezing in children with asthma. Epigenomics. 2012;4(1):91–100. doi: 10.2217/epi.11.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179(7):572–578. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey KA, Wu MC, Ward WO, Smeester L, Rager JE, García-Vargas G, Del Razo LM, Drobná Z, Stýblo M, Fry RC. Arsenic and the Epigenome: Inter-individual differences in arsenic metabolism related to distinct patterns of DNA methylation. J Biochem Mol Toxicol. 2013;27(2):106–115. doi: 10.1002/jbt.21462. Epub 2013 Jan 11. doi: 10.1002/jbt.21462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey KA, Fry RC. Arsenic-Associated Changes to the Epigenome: What Are the Functional Consequences? Curr Envir Health Rpt. 2014;1:22–34. doi: 10.1007/s40572-013-0002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey KA, Fry RC. Arsenic-induced Changes to the Epigenome, in Toxicology and Epigenetics. Chichester: John Wiley & Sons, Ltd; 2012. [DOI] [Google Scholar]
- Basten GP, Duthie SJ, Pirie L, Vaughan N, Hill MH, Powers HJ. Sensitivity of markers of DNA stability and DNA repair activity to folate supplementation in healthy volunteers. Br J Cancer. 2006;94(12):1942–1947. doi: 10.1038/sj.bjc.6603197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel C, Paro R. Silencing chromatin: comparing modes and mechanisms. Nat Rev Genet. 2011;12(2):123–135. doi: 10.1038/nrg2932. [DOI] [PubMed] [Google Scholar]
- Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, Gilad Y, Pritchard JK. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boellmann F, Zhang L, Clewell HJ, Schroth GP, Kenyon EM, Andersen ME, Thomas RS. Genome-wide Analysis of DNA Methylation and Gene Expression Changes in the Mouse Lung following Subchronic Arsenate Exposure. Toxicologcal Sciences. 2010;117(2):404–417. doi: 10.1093/toxsci/kfq225. [DOI] [PubMed] [Google Scholar]
- Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, Sparrow D, Vokonas P, Baccarelli A. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev. 2009;130:234–9. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess J, Kurzius-Spencer M, Poplin G, Littau S, Kopplin M, Sturup S, Boitano S, Lantz RC. Environmental arsenic exposure, selenium and sputum alpha-1 antitrypsin. J Expo Sci Environ Epidemiol. 2013;24(2):150–5. doi: 10.1038/jes.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda S, Dasgupta UB, Guhamazumder D, Gupta M, Chaudhuri U, Lahiri S, Das S, Ghosh N, Chatterjee D. DNA hypermethylation of promoter of gene p53 and p16 in arsenic-exposed people with and without malignancy. Toxicol Sci. 2006;89:431–437. doi: 10.1093/toxsci/kfj030. [DOI] [PubMed] [Google Scholar]
- Chen WT, Hung WC, Kang WY, Huang YC, Chai CY. Urothelial carcinomas arising in arsenic-contaminated areas are associated with hypermethylation of the gene promoter of the death-associated protein kinase. Histopathology. 2007;51(6):785–92. doi: 10.1111/j.1365-2559.2007.02871.x. [DOI] [PubMed] [Google Scholar]
- Chicoine E, Estève PO, Robledo O, Van Themsche C, Potworowski EF, St-Pierre Y. Evidence for the role of promoter methylation in the regulation of MMP-9 gene expression. Biochem Biophys Res Commun. 2002;297(4):765–72. doi: 10.1016/s0006-291x(02)02283-0. [DOI] [PubMed] [Google Scholar]
- Coolen MW, Statham AL, Gardiner-Garden M, Clark SJ. Genomic profiling of CpG methylation and allelic specificity using quantitative high-throughput mass spectrometry: critical evaluation and improvements. Nucleic Acids Res. 2007;35(18):e119. doi: 10.1093/nar/gkm662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De BK, Majumdar D, Sen S, Guru S, Kundu S. Pulmonary involvement in chronic arsenic poisoning from drinking contaminated ground-water. J Assoc Physicians India. 2004;52:395–400. [PubMed] [Google Scholar]
- Del Razo LM, Hernandez JL, Garcia-Vargas GG, Ostrosky-Wegman P, Cortinas de Nava C, Cebrian ME. Urinary excretion of arsenic species in a human population chronically exposed to arsenic via drinking water: a pilot study. In: Chappell WR, Abernathy CO, Cothern CR, editors. Arsenic Exposure and Health. Northwood, England: Science and Technology Letters; 1994. pp. 91–100. https://hero.epa.gov/hero/index.cfm/reference/details/reference_id/1372999. [Google Scholar]
- El-Maarri O, Walier M, Behne F, van uum J, Singer H, Diaz-Lacava A, Nüsgen N, Niemann B, Watzka M, Reinsberg J, van der Ven H, Wienker T, Stoffel-Wagner B, Schwaab R, Oldenburg J. Methylation at global LINE-1 repeats in human blood are affected by gender but not by age or natural hormone cycles. PLoS One. 2011;6:16252. doi: 10.1371/journal.pone.0016252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry RC, Navasumrit P, Valiathan C, Svensson JP, Hogan BJ, Luo M, Bhattacharya S, Kandjanapa K, Soontararuks S, Nookabkaew S, Mahidol C, Ruchirawat M, Samson LD. Activation of inflammation/NF-kappaB signaling in infants born to arsenic-exposed mothers. PLoS Genet. 2007;(11):e207. doi: 10.1371/journal.pgen.0030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuke C, Shimabukuro M, Petronis A, Sugimoto J, Oda T, Miura K, Miyazaki T, Ogura C, Okazaki Y, Jinno Y. Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: An HPLC-based study. Ann Hum Genet. 2004;68:196–204. doi: 10.1046/j.1529-8817.2004.00081.x. [DOI] [PubMed] [Google Scholar]
- Green BB, Karagas MR, Punshon T, Jackson BP, Robbins DJ, Houseman EA, Marsit CJ. Epigenome-wide assesment of DNA methyltation in the placenta and arsenic exposure in the New Hampshire birth cohort study. Environ Health Perspect. 2016;124(8):1253–60. doi: 10.1289/ehp.1510437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goering PL, Aposhian HV, Mass MJ, Cebrian M, Beck BD, Waalkes MP. The enigma of arsenic carcinogenesis: Role of metabolism. Toxicol Sci. 1999;49(1):5–14. doi: 10.1093/toxsci/49.1.5. [DOI] [PubMed] [Google Scholar]
- Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- Hossain MB, Vahter M, Concha G, Broberg K. Environmental arsenic exposure and DNA methylation of the tumor suppressor gene p16 and the DNA repair gene MLH1: effect of arsenic metabolism and genotype. Metallomics. 2012;4(11):1167–75. doi: 10.1039/c2mt20120h. [DOI] [PubMed] [Google Scholar]
- Hou L, Wang H, Sartori S, Gawron A, Lissowska J, Bollati V, Tarantini L, Zhang FF, Zatonski W, Chow WH, Baccarelli A. Blood leukocyte DNA hypomethylation and gastric cancer risk in a high-risk polish population. Int J Cancer. 2010;127(8):1866–1874. doi: 10.1002/ijc.25190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiung D, Marsit C, Houseman E, Eddy K, Furniss C, McClean M, Kelsey KT. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:108–14. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- Hunt KM, Srivastava RK, Elmets CA, Athar M. The mechanistic basis of arsenicosis: pathogenesis of skin cancer. Cancer Lett. 2014;354(2):211–9. doi: 10.1016/j.canlet.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intarasunanont P, Navasumrit P, Waraprasit S, Chaisatra K, Suk WA, Mahidol C, Ruchirawat M. Effects of arsenic exposure on DNA methylation in cord blood samples from newborn babies and in a human lymphoblast cell line. Environ Heal. 2012;11:31. doi: 10.1186/1476-069X-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TJ, Novak P, Wnek SM, Gandolfi AJ, Futscher BW. Arsenicals produce stable progressive changes in DNA methylation patterns that are linked to malignant transformation of immortalized urothelial cells. Toxicol Appl Pharmacol. 2009;241:221–229. doi: 10.1016/j.taap.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Jung AY, Smulders Y, Verhoef P, Kok FJ, Blom H, Kok RM, Kampman E, Durga J. No effect of folic acid supplementation on global DNA methylation in men and women with moderately elevated homocysteine. PLoS One. 2011;6(9):e24976. doi: 10.1371/journal.pone.0024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile ML, Baccarelli A, Hoffman E, Tarantini L, Quamruzzaman Q, Rahman M, Mahiuddin G, Mostofa G, Hsueh YM, Wright RO, Christiani DC. Prenatal arsenic exposure and DNA methylation in maternal and umbilical cord blood leukocytes. Environ Health Perspect. 2012;120(7):1061–6. doi: 10.1289/ehp.1104173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrou A, Baccarelli A, Wright RO, Weisskopf M, Bollati V, Amarasiriwardena C, Vokonas P, Schwartz J. Arsenic exposure and DNA methylation among elderly men. Epidemiology. 2012;23(5):668–76. doi: 10.1097/EDE.0b013e31825afb0b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz RC, Hays AM. Role of oxidative stress in arsenic-induced toxicity. Drug Metab Rev. 2006;38:791–804. doi: 10.1080/03602530600980108. [DOI] [PubMed] [Google Scholar]
- Lantz RC, Lynch BJ, Boitano S, Poplin GS, Littau S, Tsaprailis G, Burgess JL. Pulmonary biomarkers based on alterations in protein expression after exposure to arsenic. Environ Health Perspect. 2007 Apr;115(4):586–91. doi: 10.1289/ehp.9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zheng Y, Zhang W, Zhang X, Lioyd-Jones DM, Baccarelli AA, Ning H, Fornage M, He K, Liu K, Hou L. Blood methylomics in response to arsenic exposure in a low-exposed US population. J Expo Sci Environ Epidemiol. 2014 Mar-Apr;24(2):145–9. doi: 10.1038/jes.2013.89. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar S, Chanda S, Ganguli B, Mazumder DN, Lahiri S, Dasgupta UB. Arsenic exposure induces genomic hypermethylation. Environ Toxicol. 2010;25(3):315–8. doi: 10.1002/tox.20497. [DOI] [PubMed] [Google Scholar]
- Mass MJ, Wang L. Arsenic alters cytosine methylation patterns of the promoter of the tumor suppressor gene p53 in human lung cells: a model for a mechanism of carcinogenesis. Mutat Res. 1997;386(3):263–77. doi: 10.1016/s1383-5742(97)00008-2. [DOI] [PubMed] [Google Scholar]
- Merkus PJ. Effects of childhood respiratory diseases on the anatomical and functional development of the respiratory system. Paediatr Respir Rev. 2003;(1):28–39. doi: 10.1016/s1526-0542(02)00311-1. [DOI] [PubMed] [Google Scholar]
- Milstein LS, Essader A, Pellizzari ED, Fernando RA, Raymer JH, Levine KE, Akinbo O. Development and application of a robust speciation method for determination of six arsenic compounds present in human urine. Environ Health Perspect. 2003;111:293–296. doi: 10.1289/ehp.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naujokas MF, Anderson B, Ahsan H, Aposhian HV, Graziano J, Thompson C, Suk WA. 368 The broad scope of health effects from chronic arsenic exposure: update on a 369 worldwide public health problem. Environ Health Perspect. 2013;121(3):295–302. doi: 10.1289/ehp.1205875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemery B. Metal toxicity and the respiratory tract. Eur Respir J. 1990;3:202–219. [PubMed] [Google Scholar]
- Niedzwiecki MM, Hall MN, Liu X, Oka J, Harper KN, Slavkovich V, Ilievski V, Levy D, van Geen A, Mey JL, Alam S, Siddique AB, Parvez F, Graziano JH, Gamble MV. Dose–Response Study of Arsenic Exposure and Global Methylation of Peripheral Blood Mononuclear Cell DNA in Bangladeshi Adults. 2013;121(11-12):1306–1312. doi: 10.1289/ehp.1206421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak P, Jensen T, Oshiro MM, Wozniak RJ, Nouzova M, Watts GS, Klimecki WT, Kim C, Futscher BW. Epigenetic inactivation of the HOXA gene cluster in breast cancer. Cancer Res. 2006;66:10664–70. doi: 10.1158/0008-5472.CAN-06-2761. [DOI] [PubMed] [Google Scholar]
- Novak P, Jensen T, Oshiro MM, Watts GS, Kim CJ, Futscher BW. Agglomerative epigenetic aberrations are a common event in human breast cancer. Cancer Res. 2008;68(20):8616–8625. doi: 10.1158/0008-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivas-Calderón E, Recio-Vega R, Gandolfi AJ, Lantz RC, González-Cortes T, Gonzalez-De Alba C, Froines JR, Espinosa-Fematt JA. Lung inflammation biomarkers and lung function in children chronically exposed to arsenic. Toxicol Appl Pharmacol. 2015;287(2):161–167. doi: 10.1016/j.taap.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvez F, Chen Y, Brandt-Rauf PW, Bernard A, Dumont X, Slavkovich V, Argos M, D’Armiento J, Foronjy R, Hasan MR, Eunus HE, Graziano JH, Ahsan H. Nonmalignant respiratory effects of chronic arsenic exposure from drinking water among never-smokers in Bangladesh. Environ Health Perspect. 2008;116(2):190–5. doi: 10.1289/ehp.9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie K, Zelent A, Waxman S. Differentiation therapy of acute myeloid leukemia: past, present and future. Curr Opin Hematol. 2009;16(2):84–91. doi: 10.1097/MOH.0b013e3283257aee. [DOI] [PubMed] [Google Scholar]
- Pilsner JR, Hall MN, Liu X, Ilievski V, Slavkovich V, Levy D, Factor-Litvak P, Yunus M, Rahman M, Graziano JH, Gamble MV. Influence of prenatal arsenic exposure and newborn sex on global methylation of cord blood DNA. PLoS One. 2012;7(5):e37147. doi: 10.1371/journal.pone.0037147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsner JR, Liu X, Ahsan H, Ilievski V, Slavkovich V, Levy D, Factor-Litvak P, Graziano JH, Gamble MV. Genomic methylation of peripheral blood leukocyte DNA: influences of arsenic and folate in Bangladeshi adults. Am J Clin Nutr. 2007;86(4):1179–86. doi: 10.1093/ajcn/86.4.1179. [DOI] [PubMed] [Google Scholar]
- Powell BL, Moser B, Stock W, Gallagher RE, Willman CL, Stone RM, et al. Arsenic trioxide improves event-free and overall survival for adults with acute promyelocytic leukemia: North American Leukemia Intergroup Study C9710. Blood. 2010;116(19):3751–3757. doi: 10.1182/blood-2010-02-269621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W, Baccarelli A, Carey VJ, Boutaoui N, Bacherman H, Klanderman B, Rennard S, Agusti A, Anderson W, Lomas DA, DeMeo DL. Variable DNA Methylation Is Associated with Chronic Obstructive Pulmonary Disease and Lung Function. American Journal of Respiratory and Critical care Medicine. 2012;185(4):373–381. doi: 10.1164/rccm.201108-1382OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rager JE, Bailey KA, Smeester L, Miller SK, Parker JS, Laine JE, Drobna Z, Currier J, Douillet C, Olshan AF, Rubio-Andrade M, Stýblo M, García-Vargas G, Fry RC. Prenatal Arsenic Exposure and the epigenome: Altered microRNAs associated with innate and adaptive immune signaling in newborn cord blood. Environmental and Molecular Mutagenesis. 2014;55(3):196–208. doi: 10.1002/em.21842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recio-Vega R, Gonzalez-Cortes T, Olivas-Calderon E, Lantz RC, Gandolfi AJ, Gonzalez-De Alba C. In utero and early childhood exposure to arsenic decreases lung function in children. J Appl Toxicol. 2015;35(4):358–66. doi: 10.1002/jat.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichard JF, Puga A. Effects of arsenic exposure on DNA methylation and epigenetic gene regulation. Epigenomics. 2010;2(1):87–104. doi: 10.2217/epi.09.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichard JF, Schnekenburger M, Puga A. Long term low-dose arsenic exposure induces loss of DNA methylation. Biochem Biophys Res Commun. 2007;352:188–192. doi: 10.1016/j.bbrc.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, McHale CM, Skibola CF, Smith AH, Smith MT, Zhang L. An Emerging Role for Epigenetic Dysregulation in Arsenic Toxicity and Carcinogenesis. Environmental Health Perspectives. 2011;119(1):11–9. doi: 10.1289/ehp.1002114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet. Cell Genet. 1975;14:9–25. doi: 10.1289/ehp.1002114. [DOI] [PubMed] [Google Scholar]
- Rojas D, Rager JE, Smeester L, Bailey KA, Zuzana Drobná Z, Rubio-Andrade M, Stýblo M, García-Vargas G, Fry RC. Prenatal arsenic exposure and the epigenome: Identifying sites of 5-methylcytosine alterations that predict functional changes in gene expression in newborn cord blood and subsequent birth outcomes. Toxicological Sciences. 2015;143(1):97–106. doi: 10.1093/toxsci/kfu210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488(7412):504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- Smeester L, Rager JE, Bailey KA, Guan X, Smith N, García-Vargas G, Del Razo LM, Drobná Z, Kelkar H, Stýblo M, Fry RC. Epigenetic changes in individuals with arsenicosis. Chem Res Toxicol. 2011;24(2):165–167. doi: 10.1021/tx1004419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AH, Marshall G, Yuan Y, Ferreccio C, Liaw J, von Ehrenstein O, Steinmaus C, Bates MN, Selvin S. Increased mortality from lung cancer and bronchiectasis in young adults after exposure to arsenic in utero and in early childhood. Environ Health Perspect. 2006;114:1293–1296. doi: 10.1289/ehp.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soignet SL, Maslak P, Wang ZG, Jhanwar S, Calleja E, Dardashti LJ, Corso D, DeBlasio A, Gabrilove J, Scheinberg DA, Pandolfi PP, Warrell RP. Complete remission after treatment of acute promyelocytic leukemia with arsenic trioxide. N Engl J Med. 1998;339(19):1341–1348. doi: 10.1056/NEJM199811053391901. [DOI] [PubMed] [Google Scholar]
- Tajuddin SM, Amaral AF, Fernández AF, Rodríguez-Rodero S, Rodríguez RM, Moore LE, Tardón A, Carrato A, García-Closas M, Silverman DT, Jackson BP, García-Closas R, Cook AL, Cantor KP, Chanock S, Kogevinas M, Rothman N, Real FX, Fraga MF, Malats N. Genetic and nongenetic predictors of LINE-1 methylation in leukocyte DNA. Environ Health Perspect. 2013;121(6):650–656. doi: 10.1289/ehp.1206068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarantini L, Bonzini M, Apostoli P, Pegoraro V, Bollati V, Marinelli B, Cantone L, Rizzo G, Hou L, Schwartz J, Bertazzi PA, Baccarelli A. Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Environ Health Perspect. 2009;117:217–222. doi: 10.1289/ehp.11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM. DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics. 2011;6(7):828–37. doi: 10.4161/epi.6.7.16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CH. Arsenic methylation, urinary arsenic metabolites and human diseases: current perspective. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2007;25(1):1–22. doi: 10.1080/10590500701201695. [DOI] [PubMed] [Google Scholar]
- Valenzuela OL, Borja-Aburto VH, Garcia-Vargas GG, Cruz-Gonzalez MBB, Garcia-Montalvo EA, Calderon-Aranda ES, Del Razo LM. Urinary trivalent methylated arsenic species in a population chronically exposed to inorganic arsenic. Environ Health Perspect. 2005;113(3):250–254. doi: 10.1289/ehp.7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eijk KR, de Jong S, Boks MP, Langeveld T, Colas F, Veldink JH, de Kovel CG, Janson E, Strengman E, Langfelder P, Kahn RS, van den Berg LH, Horvath S, Ophoff RA. Genetic analysis of DNA methylation and gene expression levels in whole blood of healthy human subjects. BMC Genomics. 2012;13:636. doi: 10.1186/1471-2164-13-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Ehrenstein, Mazumder D, Yuan Y, Samanta S, Balmes J, Sil A, Ghosh N, Hira-Smith Decrements in Lung Function Related to Arsenic in Drinking Water in West Bengal, India. Am J Epidemiol. 2005;162(6):533–41. doi: 10.1093/aje/kwi236. [DOI] [PubMed] [Google Scholar]
- Waalkes MP, Liu J. Early-life arsenic exposure: methylation capacity and beyond. Environ Health Perspect. 2008;116(3):A104. doi: 10.1289/ehp.11276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm CS, Kelsey KT, Butler R, Plaza S, Gagne L, Zens MS, Andrew AS, Morris S, Nelson HH, Schned AR, Karagas MR, Marsit CJ. Implications of LINE1 methylation for bladder cancer risk in women. Clin Cancer Res. 2010;16(5):1682–9. doi: 10.1158/1078-0432.CCR-09-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. IPCS environmental health criteria 224 arsenic and arsenic compounds. Geneva: International Program on Chemical Safety, World Health Organization; 2001. http://www.who.int/ipcs/publications/ehc/ehc_224/en/ [Google Scholar]
- Wright RO, Schwartz J, Wright RJ, Bollati V, Tarantini L, Park SK, Hu H, Sparrow D, Vokonas P, Baccarelli A. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ Health Perspect. 2010;118(6):790–795. doi: 10.1289/ehp.0901429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Liu J, Benbrahim-Tallaa L, Ward JM, Logsdon D, Diwan BA, Waalkes MP. Aberrant DNA methylation and gene expression in livers of newborn mice transplacentally exposed to a hepatocarcinogenic dose of inorganic arsenic. Toxicology. 2007;236:7–15. doi: 10.1016/j.tox.2007.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Marshall G, Ferreccio C, Steinmaus C, Liaw J, Bates M, Smith AH. Kidney cancer mortality: fifty-year latency patterns related to arsenic exposure. Epidemiology. 2010;21:103–108. doi: 10.1097/EDE.0b013e3181c21e46. [DOI] [PubMed] [Google Scholar]
- Zhang FF, Cardarelli R, Carroll J, Fulda KG, Kaur M, Gonzalez K, et al. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics. 2011;6 doi: 10.4161/epi.6.5.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao CQ, Young MR, Diwan BA, Coogan TP, Waalkes MP. Association of arsenic-induced malignant transformation with DNA hypomethylation and aberrant gene expression. Proc Natl Acad Sci USA. 1997;94:10907–10912. doi: 10.1073/pnas.94.20.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong CX, Mass MJ. Both hypomethylation and hypermethylation of DNA associated with arsenite exposure in cultures of human cells identified by methylation-sensitive arbitrarily-primed PCR. Toxicol Lett. 2001;122:223–234. doi: 10.1016/s0378-4274(01)00365-4. [DOI] [PubMed] [Google Scholar]
- Zhu ZZ, Hou L, Bollati V, Tarantini L, Marinelli B, Cantone L, Yang AS, Vokonas P, Lissowska J, Fustinoni S, Pesatori AC, Bonzini M, Apostoli P, Costa G, Bertazzi PA, Chow WH, Schwartz J, Baccarelli Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int J Epidemiol. 2010:1–10. doi: 10.1093/ije/dyq154. [DOI] [PMC free article] [PubMed] [Google Scholar]


