Abstract
Objective
The Gynecologic Oncology Group (GOG) examined the prognostic relevance of c-MYC amplification and polysomy 8 in epithelial ovarian cancer (EOC).
Methods
Women with suboptimally-resected, advanced stage EOC who participated in GOG-111, a multicenter randomized phase III trial of cyclophosphamide + cisplatin vs. paclitaxel + cisplatin, and who provided a tumor block through GOG-9404 were eligible. Fluorescence in situ hybridization (FISH) with probes for c-MYC and the centromere of chromosome 8 (CEP8) was used to examine c-MYC amplification (≥2 copies c-MYC/CEP8) and polysomy 8 (≥4 CEP8 copies).
Results
c-MYC amplification, defined as ≥2 copies c-MYC/CEP8, was observed in 29% (28/97) of EOCs and levels were ranged from 2.0–3.3 copies of c-MYC/CEP8. c-MYC amplification was not associated with patient age, race, GOG performance status, stage, cell type, grade, measurable disease status following surgery, tumor response or disease status following platinum-based combination chemotherapy. Women with vs. without c-MYC amplification did not have an increased risk of disease progression (hazard ratio [HR] = 1.03; 95% confidence interval [CI] = 0.65–1.64; p = 0.884) or death (HR = 1.08; 95% CI = 0.68–1.72; p = 0.745). c-MYC amplification was not an independent prognostic factor for progression-free survival (HR = 1.03, 95% CI = 0.57–1.85; p = 0.922) or overall survival (HR = 1.01, 95% CI = 0.56–1.80; p = 0.982). Similar insignificant results were obtained for c-MYC amplification categorized as ≥1.5 copies c-MYC/CEP8. Polysomy 8 was observed in 22 patients without c-MYC amplification and 3 with c-MYC amplification, and was associated with age and measurable disease status, but not other clinical covariates or outcomes.
Conclusions
c-MYC amplification and polysomy 8 have limited predictive or prognostic value in suboptimally-resected, advanced stage EOC treated with platinum-based combination chemotherapy.
Keywords: c-MYC, FISH, Ovarian cancer, Polysomy 8, Gene amplification
Introduction
Ovarian cancer is the leading cause of cancer-related death among the gynecologic malignancies [1]. It is estimated that in the United States 21,850 new cases of ovarian cancer will be diagnosed, 68% of whom will present with advanced disease, and 15,520 women would die from their cancer in 2008 [1]. Currently, surgical staging followed by platinum-based chemotherapy is the standard of care for this disease [2,3]. Even though 70% of women with advanced stage disease respond to first-line platinum-based chemotherapy, five-year survival for women in this setting remains around 30% [1–3]. Researchers continue to study molecular and biochemical defects in epithelial ovarian cancer (EOC) with the hope of identifying biomarkers with prognostic and/or predictive value in this patient setting.
c-MYC is a multifunctional proto-oncogene that exhibits diverse and at times, opposing functions [4–9]. c-MYC protein promotes tumorigenesis by inducing proliferation, inhibiting cells from exiting the cell cycle, stimulating blood vessel formation and cell migration, enhancing genomic instability, and helping tumor cells adapt and thrive in hypoxic environments [4,8,10–13]. In contrast, c-MYC inhibits tumorigenesis by sensitizing cells to apoptosis [14]. c-MYC is activated in about 20% of human cancers by several mechanisms including gene amplification, mutation or rearrangement, promoter insertion, transcriptional and post-translational [4–9], and appears to be a feasible target for cancer therapeutics [15–16].
Amplification of c-MYC has been described in a variety of human cancers [17–32] including ovarian cancer [33–51]. Investigators have shown an association between c-MYC amplification and poor outcome in breast cancer [18–23], prostate cancer [24–27] and chondrosarcoma [28], but not in non-small cell lung cancer (NSCLC) [29–30], colorectal cancer [31] or esophageal squamous cell carcinoma (SCC) [32]. In EOC, amplifications of c-MYC have been shown to be associated with stage [43], cell type [37,51] and/or grade [41,43], or conversely to not be associated with stage [46,51], cell type [43,46], grade [36,37,46,51], progression-free survival (PFS) [45,46], overall survival (OS) [45,46,48,49] and/or response to platinum-based chemotherapy [36].
The Gynecologic Oncology Group (GOG) undertook a study of c-MYC amplification in women with suboptimally-resected, advanced stage EOC who participated in a multicenter randomized phase III trial and provided a tumor block for research. Fluorescence in situ hybridization (FISH) with probes for c-MYC and the centromeric region of chromosome 8 (CEP8) was used to examine c-MYC amplification defined as ≥2 copies of c-MYC/CEP8 per cell [23,28,21] or ≥1.5 copies c-MYC/CEP8 [22,24] and polysomy for chromosome 8 defined as ≥4 copies of CEP8 per cell (as recommended by the manufacturer). Our results will be discussed in context with the other studies of c-MYC amplification in invasive EOC and our current understanding of the c-MYC proteins.
Methods
Patients
Women who participated in GOG-111, a phase III treatment protocol, and provided a tumor block through GOG-9404 were eligible for this translational research study. Women on the GOG-111 protocol had to have previously-untreated, histologically-confirmed, surgically-staged EOC with evaluable or measurable stage III disease that was suboptimally-resected (>1 cm residual disease) or stage IV disease, a GOG performance status of 0 to 2, and adequate bone marrow counts, renal function and hepatic function as previously described [52]. Women with a borderline tumor or optimally-resected stage III disease were specifically excluded from GOG-9404. Women were required to provide written informed consent, and participating institutions were required to obtain annual Institutional Review Board approval for GOG-111 and GOG-9404 consistent with federal, state and local requirements.
Platinum-based combination chemotherapy
Women on GOG-111 were randomly allocated to receive either 75 mg/m2 cisplatin intravenously on day 1 and 750 mg/m2 cyclophosphamide intravenously on day 1 every 3 weeks for a total of 6 cycles, or a 24-hour continuous intravenous infusion of 135 mg/m2 paclitaxel on day 1 and 75 mg/m2 cisplatin intravenously on day 2 every 3 weeks for a total of 6 cycles [52]. Treatment at the time of disease progression was left to the discretion of the treating physician and patient.
Clinical end-points
Women on GOG-111 were followed quarterly for 2 years, semiannually for the next 3 years and then annually until death from the time they went off-treatment due to completion of protocol-specified therapy, toxicity, or disease progression [52]. Progression-free survival (PFS) was calculated as the time in months from enrollment on GOG-111 to disease progression or death (failure), or to the date of last contact for women who were still alive with no evidence of disease progression (censored). Overall survival (OS) was calculated as the time in months from enrollment on GOG-111 to death (regardless of cause) or to the date of last contact for women who were alive. Tumor response was evaluated after every 2 cycles of treatment in the women with measurable disease. Complete disappearance of all disease was required for a complete response. A partial response required a ≥50% reduction in tumor (product of perpendicular diameters). Progressive disease required a ≥50% increase in the dimensions of any lesion documented within 6 weeks of study entry or the appearance of new lesions within 8 weeks of study entry. Stable disease was defined as any condition not meeting the above categories. Women on GOG-111 who were clinically-free of disease after primary chemotherapy, or who had CA125<100 U/ml and were entered with non-measurable disease were required to undergo a reassessment laparotomy as specified in the protocol. Disease status was assessed following primary chemotherapy and classified as negative when the reassessment laparotomy showed no evidence of disease, or positive when disease progression was documented during treatment or when microscopic or macroscopic evidence of disease was observed during the reassessment laparotomy.
Tumor specimens and fluorescence in situ hybridization
Previously-untreated, primary tumor was excised during cytoreductive surgery, fixed in formalin and then embedded in paraffin. Unstained tissue sections, 5 µm in thickness, were prepared on charged glass slides. Dual-label FISH was used to quantify the number of copies of c-MYC and chromosome 8 in unstained slides as previously described [24]. Briefly, unstained sections were baked overnight at 60 °C, deparaffinized, dehydrated, treated for 10 min with 4% (w/v) pepsin at 45 °C, denatured for 10 min at 90 °C, hybridized for 12 to 16 h at 37 °C with a probe cocktail consisting of an alpha satellite probe to chromosome 8 (CEP8) directly labeled with Spectrum Green and a c-MYC region probe directly labeled with Spectrum Orange (Vysis Inc., Naperville, IL). The slides were post-washed for 5 min with 2×SSC buffer (0.3 M NaCl and 30 mM sodium citrate tribasic dihyrate, pH 7.0) at 72 °C and counterstained with 4′,6′-diamidino-2-phenyindole (DAPI). Red staining for chromosome 8 and green staining for c-MYC were visualized using a Zeiss Axiophot fluorescence microscope (Carl Zeiss MicroImaging, Inc. Thornwood, NY, USA) with appropriate filters and an Applied Imaging Cytovision system (Pittsburgh, PA, USA), and quantified in at least 50 tumor cells per case by one or two technicians (CC and GV; see Acknowledgements) working under the direct supervision of one of the authors (JKB). Average copies of c-MYC and CEP8 per case were calculated for the 28% of equivocal cases scored by two reviewers. c-MYC amplification is defined as ≥2 copies of c-MYC/CEP8 per cell, and unless specifically stated otherwise, this is the definition used. Supportive analyses were done defining c-MYC amplification as ≥1.5 copies of c-MYC/CEP8 per cell based on studies in breast cancer [22] and prostate cancer [24] showing that low level amplification increased protein expression.
Statistical methods
Biomarker and clinical data were analyzed using SPSS versions 14.0 (SPSS Inc., Chicago, IL) and SAS® version 9.1 software (SAS Institute, Inc. Cary, NC). All tests were two-sided and the level of significance was set at 0.05. Associations between categorical variables were evaluated using Fisher's Exact Test [53,54]. Estimates of survival probabilities were calculated using the Kaplan–Meier method [55]. Logrank tests [56] were used to test the equality of survival distributions between groups. Cox proportional hazards regression [57] was used to model associations of variables with PFS or OS.
Results
GOG-111 was a randomized phase III trial that enrolled 410 women between April 13, 1990 and March 2, 1992 [52]. Ninety-seven women on GOG-111 were enrolled on the companion protocol, GOG-9404 and provided primary tumor tissue for research. The patient characteristics for the 97 women in this cohort are summarized in Table 1 and are representative of that observed in the entire GOG-111 cohort [52]. At the time of the final analysis, five women were alive with no evidence of disease, four women were alive with disease progression and 88 women died due to disease progression. Median follow-up for the nine women who were still alive at the time of the final analysis was 127 (range 15 to 207) months including three women who were lost to follow up after 15, 24 or 87 months of enrollment.
Table 1.
Association between clinical characteristics and c-MYC amplification or polysomy for chromosome 8.
| Total Cases |
c-MYC amplificationa | c-MYC amplificationb | Polysomy for chromosome 8c | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||
| ≥ 2 c-MYC/CEP8 copies | ≥ 1.5 c-MYC/CEP8 copies | ≥ 4 CEP8 copies | ||||||||
|
|
|
|
||||||||
| No | Yes | P* | No | Yes | P* | No | Yes | p* | ||
| Patient age in years | 0.479 | 0.382 | <0.001 | |||||||
| <50 | 30 | 20 (66.7) | 10 (33.3) | 10 (33.3) | 20 (66.7) | 27 (90.0) | 3 (10.0) | |||
| 50–59 | 26 | 17 (65.4) | 9 (34.6) | 10 (38.5) | 16 (61.5) | 19 (73.1) | 7 (26.9) | |||
| 60–69 | 28 | 23 (82.1) | 5 (17.9) | 15 (53.6) | 13 (46.4) | 13 (46.4) | 15 (53.6) | |||
| ≥ 70 | 13 | 9 (69.2) | 4 (30.8) | 4 (30.8) | 9 (69.2) | 13 (100.0) | 0 (0.0) | |||
| Race | 0.430 | 0.489 | 0.860 | |||||||
| Caucasian | 86 | 59 (68.6) | 27 (31.4) | 36 (41.9) | 50 (58.1) | 64 (74.4) | 22 (25.6) | |||
| African American | 6 | 5 (83.3) | 1 (16.7) | 1 (16.7) | 5 (83.3) | 4 (66.7) | 2 (33.3) | |||
| Otherd | 5 | 5 (100.0) | 0 (0.0) | 2 (40.0) | 3 (60.0) | 4 (80.0) | 1 (20.0) | |||
| GOG performance status | 1.000 | 0.321 | 0.775 | |||||||
| Asymptomaticscore 0/1 | 77 | 55 (71.4) | 22 (28.6) | 33 (42.9) | 44 (57.1) | 58 (75.3) | 19 (24.7) | |||
| Symptomaticscore 2 | 20 | 14 (70.0) | 6 (30.0) | 6 (30.0) | 14 (70.0) | 14 (70.0) | 6 (30.0) | |||
| Tumor stage | 0.646 | 0.287 | 0.633 | |||||||
| Stage III | 60 | 44 (73.3) | 16 (26.7) | 27 (45.0) | 33 (55.0) | 43 (71.7) | 17 (28.3) | |||
| Stage IV | 37 | 25 (67.6) | 12 (32.4) | 12 (32.4) | 25 (67.6) | 29 (78.4) | 8 (21.6) | |||
| Histologic cell type | 0.179 | 0.051 | 0.234 | |||||||
| Serous | 72 | 53 (73.6) | 19 (26.4) | 29 (40.3) | 43 (59.7) | 51 (70.8) | 21 (29.2) | |||
| Endometrioid | 9 | 8 (88.9) | 1 (11.1) | 7 (77.8) | 2 (22.2) | 9 (100.0) | 0 (0.0) | |||
| Clear cell | 2 | 1 (50.0) | 1 (50.0) | 0 (0.0) | 2 (100.0) | 2 (100.0) | 0 (0.0) | |||
| Mucinous | 3 | 1 (33.3) | 2 (66.7) | 1 (33.3) | 2 (66.7) | 3 (100.0) | 0 (0.0) | |||
| Other cell types | 11 | 6 (54.5) | 5 (45.5) | 2 (18.2) | 9 (81.8) | 7 (63.6) | 4 (36.4) | |||
| Tumor grade | 0.453 | 0.386 | 0.652 | |||||||
| 1well differentiated | 6 | 3 (50.0) | 3 (50.0) | 2 (33.3) | 4 (66.7) | 5 (83.3) | 1 (16.7) | |||
| 2moderately differentiated | 44 | 31 (70.5) | 13 (29.5) | 21 (47.7) | 23 (52.3) | 34 (77.3) | 10 (22.7) | |||
| 3poorly differentiated/not graded | 47 | 35 (74.5) | 12 (25.5) | 16 (34.0) | 31 (66.0) | 33 (70.2) | 14 (29.8) | |||
| Measurable disease | 1.000 | 0.837 | 0.019 | |||||||
| Non-measurable | 44 | 31 (70.5) | 13 (29.5) | 17 (38.6) | 27 (61.4) | 38 (86.4) | 6 (13.6) | |||
| Measurable | 53 | 38 (71.7) | 15 (28.3) | 22 (41.5) | 31 (58.5) | 34 (64.2) | 19 (35.8) | |||
| Treatment | 1.000 | 1.000 | 1.000 | |||||||
| Cyclophos + cisplatin | 52 | 37 (71.2) | 15 (28.8) | 21 (40.4) | 31 (59.6) | 39 (75.0) | 13 (25.0) | |||
| Paclitaxel + cisplatin | 45 | 32 (71.1) | 13 (28.9) | 18 (40.0) | 27 (60.0) | 33 (73.3) | 12 (26.7) | |||
| Total | 97 | 69 (71.1) | 28 (28.9) | 39 (40.2) | 58 (59.8) | 72 (74.2) | 25 (25.8) | |||
c-MYC amplification was categorized as ‘No' when tumors had <2 copies of c-MYC/CEP8 per cell or ‘Yes' when tumors had ≥ 2 copies of c-MYC/CEP8 per cell. Results presented as cases with row percentages in parentheses.
c-MYC amplification was categorized as ‘No' when tumors had < 1.5 copies of c-MYC/CEP8 per cell or ‘Yes’ when tumors had ≥ 1.5 copies of c-MYC/CEP8 per cell. Results presented as cases with row percentages in parentheses.
Polysomy for chromosome 8 was categorized as ‘No' when tumors had < 4 copies of CEP8 per cell or ‘Yes' when tumors had ≥ 4 copies of CEP8 per cell. Results presented as cases with row percentages in parentheses.
Other includes two Asians, one American Indian, and two with an unspecified race.
p-value based on Fisher's Exact Test.
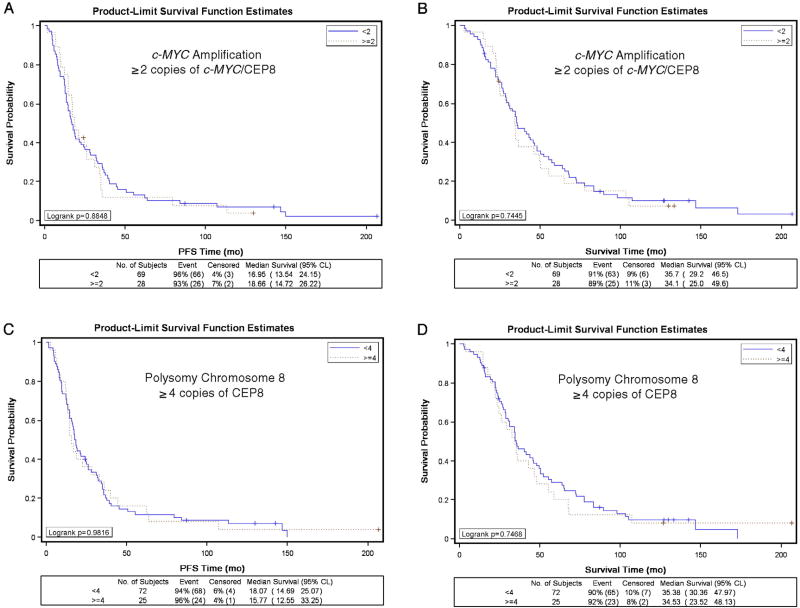
Twenty-eight (29%) of women had tumors that exhibited c-MYC amplification with levels ranging from the lower limit of 2.0 up to 3.3 copies of c-MYC/CEP8 per cell. Of the 69 (71%) of women with tumors without c-MYC amplification, the ratio of c-MYC/CEP8 ranged from 0.42 to 1.98 copies per cell. c-MYC amplification was not associated with patient age, race, GOG performance, stage, cell type, grade or measurable disease status following primary surgery (Table 1). There was no difference in the PFS distributions (Fig. 1A, p = 0.885) or OS distributions (Fig. 1B, p = 0.745) for women with or without c-MYC amplification. Unadjusted Cox modeling demonstrated that women with c-MYC amplification did not have an increased risk of disease progression (hazard ratio [HR] = 1.03; 95% confidence interval [CI] = 0.65–1.64; p = 0.884) or death (HR = 1.08; 95% CI = 0.68–1.72; p = 0.745) compared with women without c-MYC amplification (Table 2). After adjusting for patient age and stratifying by tumor stage, histologic cell type, tumor grade, measurable disease status, and treatment regimen, c-MYC amplification was not an independent prognostic factor for PFS (HR = 1.03, 95% CI = 0.57–1.85; p = 0.922) or OS (HR = 1.01, 95% CI = 0.56–1.80; p = 0.982) in women with suboptimally-resected, advanced stage EOC (Table 2). c-MYC amplification was not associated with tumor response following platinum-based combination chemotherapy or disease status following platinum-based combination chemotherapy (Table 3). c-MYC amplification using the ≥1.5 copies of c-MYC/CEP8 per cell yielded similar insignificant associations with clinical characteristics, PFS, survival, response, and disease status as reported using the ≥2.0 cut point (Tables 1–3).
Fig. 1.
Kaplan–Meier estimate of progression-free survival (A, C) and overall survival (B, D) for women without c-MYC amplification (A, B: <2 copies of c-MYC/CEP8 per cell), with c-MYC amplification (A, B: ≥2 copies of c-MYC/CEP8 per cell), without polysomy 8 (C, D: <4 copies of CEP8 per cell) or with polysomy 8 (C, D: ≥4 copies of CEP per cell).
Table 2.
Associations between c-MYC amplification or polysomy for chromosome 8 and progression-free survival or overall survival.
| Progression-free survivala | Overall survivala | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| Unadjusted model | Adjusted modelb | Unadjusted model | Adjusted modelb | |||||||||
|
|
|
|
|
|||||||||
| HR | 95% CI | p-value | HR | 95% CI | p-value | HR | 95% CI | p-value | HR | 95% CI | p-value | |
| c-MYC amplificationc | ||||||||||||
| ≥ 2 c-MYC/CEP8 copies | ||||||||||||
| No | 1.00 | 1.00 | 1.00 | 1.00 | ||||||||
| Yes | 1.03 | 0.65–1.64 | 0.884 | 1.03 | 0.57–1.85 | 0.922 | 1.08 | 0.68–1.72 | 0.745 | 1.01 | 0.56–1.80 | 0.982 |
| c-MYC amplificationd | ||||||||||||
| ≥ 1.5 c-MYC/CEP8 copies | ||||||||||||
| No | 1.00 | 1.00 | 1.00 | 1.00 | ||||||||
| Yes | 1.37 | 0.89–2.10 | 0.151 | 1.06 | 0.63–1.77 | 0.837 | 1.14 | 0.74–1.76 | 0.555 | 1.03 | 0.60–1.75 | 0.920 |
| Polysomy for chromosome 8e | ||||||||||||
| ≥ 4 CEP8 copies | ||||||||||||
| No | 1.00 | 1.00 | 1.00 | 1.00 | ||||||||
| Yes | 0.99 | 0.62–1.59 | 0.982 | 1.07 | 0.57–2.02 | 0.823 | 1.08 | 0.67–1.74 | 0.747 | 1.04 | 0.54–1.99 | 0.916 |
Estimated hazard ratio (HR), 95% confidence interval (95% CI).
Cox regression analysis for modeling the relative risk of disease progression or death; p-values are based on the Wald test.
Models were adjusted for patient age at enrollment in years, and stratified by tumor stage (III vs. IV), histologic subtype (clear cell or mucinous vs. other histologic subtypes), tumor grade (well differentiated vs. moderately differentiated vs. poorly differentiated), measurable disease status (non-measurable vs. measurable), and treatment regimen (cyclophosphamide + cisplatin vs. paclitaxel + cisplatin).
c-MYC amplification was categorized as ‘No’ when tumors had <2 copies of c-MYC/CEP8 per cell or ‘Yes’ when tumors had ≥2 copies of c-MYC/CEP8 per cell.
c-MYC amplification was categorized as ‘No’ when tumors had <1.5 copies of c-MYC/CEP8 per cell or ‘Yes’ when tumors had ≥1.5 copies of c-MYC/CEP8 per cell.
Polysomy for chromosome 8 was categorized as ‘No’ when tumors had <4 copies of CEP8 per cell or ‘Yes’ when tumors had ≥4 copies of CEP8 per cell.
Table 3.
Association between c-MYC amplification or polysomy for chromosome 8 and clinical response or disease status after completion of first-line treatment.
| Clinical response | Disease status | |||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| No response | Responsea | p-value* | No evidence of disease | Positive for diseaseb | p-value* | |||
| c-MYC amplificationc | ||||||||
| ≥2 c-MYC/CEP8 copies | ||||||||
| No | 7 (18.4%) | 31 (81.6%) | 22 CR | 1.000 | 14 (22.2%) | 49 (77.8%) | 34 + RL | 1.000 |
| 9 PR | 15 + DP | |||||||
| Yes | 3 (20.0%) | 12 (80.0%) | 5 CR | 6 (24.0%) | 19 (76.0%) | 14 + RL | ||
| 7 PR | 5 + DP | |||||||
| c-MYC amplificationd | ||||||||
| ≥ 1.5 c-MYC/CEP8 copies | ||||||||
| No | 3 (13.6%) | 19 (86.4%) | 15 CR | 0.494 | 10 (28.6%) | 25 (71.4%) | 18 + RL | 0.310 |
| 4 PR | 7 + DP | |||||||
| Yes | 7 (22.6%) | 24 (77.4%) | 12 CR | 10 (18.9%) | 43 (81.1%) | 30 + RL | ||
| 12 PR | 13 + DP | |||||||
| Polysomy for chromosome 8e4 CEP8 copies | ||||||||
| ≥ 4 CEP8 copies | ||||||||
| No | 8 (23.5%) | 26 (76.5%) | 17 CR | 0.299 | 15 (22.7%) | 51 (77.3%) | 37 + RL | 1.000 |
| 9 PR | 14 + DP | |||||||
| Yes | 2 (10.5%) | 17 (89.5%) | 10 CR | 5 (22.7%) | 17 (77.3%) | 11 + RL | ||
| 7 PR | 6 + DP | |||||||
Response includes a complete response (CR) or a partial response (PR).
Positive for disease includes those with microscopic or gross disease at reassessment laparotomy (+ RL) or with clinical evidence of disease progression documented during first-line treatment (+ DP).
c-MYC amplification was categorized as ‘No’ when tumors had <2 copies of c-MYC/CEP8 per cell or ‘Yes’ when tumors had ≥2 copies of c-MYC/CEP8 per cell.
c-MYC amplification was categorized as ‘No’ when tumors had <1.5 copies of c-MYC/CEP8 per cell or ‘Yes’ when tumors had ≥1.5 copies of c-MYC/CEP8 per cell.
Polysomy for chromosome 8 was categorized as ‘No’ when tumors had <4 copies of CEP8 per cell or ‘Yes’ when tumors had ≥4 copies of CEP8 per cell.
Fisher's Exact Test.
Polysomy for chromosome 8, defined as a tumor with ≥4 copies of CEP8 per cell, was observed in 22 women without c-MYC amplification and 3 patients with c-MYC amplification. Polysomy 8 was not associated with any of the clinical covariates tested except for patient age at enrollment and measurable disease status (Table 1). Although statistically significant, the relationship between polysomy 8 and age category was not consistent: the proportion of women with polysomy 8 increased incrementally by age for women up to 69 years but none of the women who were ≥70 years old had polysomy 8 (Table 1). The percentage of women with polysomy was statistically significantly higher in women with measurable disease (36%) than in those with non-measurable disease (14%). PFS distributions (Fig. 1C, p = 0.982) and OS distributions (Fig. 1D, p = 0.747) were very similar for women with vs. without polysomy 8. Unadjusted Cox modeling demonstrated that women with polysomy 8 did not have an increased risk of disease progression (HR = 0.99; 95% CI = 0.62–1.59; p = 0.982) or death (HR = 1.08; 95% CI = 0.67–1.74; p = 0.747) compared with women without polysomy 8 (Table 2). After adjusting for patient age and stratifying by tumor stage, histologic cell type, tumor grade, measurable disease status, and treatment regimen, polysomy 8 was not an independent prognostic factor for PFS (HR = 1.07, 95% CI = 0.57–2.02; p = 0.823) or OS (HR = 1.04, 95% CI = 0.54–1.99; p = 0.916) in women with suboptimally-resected, advanced stage EOC (Table 2). Finally, polysomy 8 was not associated with tumor response or disease status following platinum-based combination chemotherapy (Table 3).
Discussion
Our study is unique because it is a multicenter study of c-MYC amplification, detected by FISH using probes for c-MYC and CEP8, in FFPE primary tumor specimens from women with suboptimally-resected, advanced stage EOC treated with platinum-based combination chemotherapy. The availability of detailed clinical, treatment and follow-up data was a major strength of this study. We not only examined the relationship between c-MYC amplification and a full spectrum of clinical covariates but also evaluated the association between c-MYC amplification and multiple measures of clinical outcome. Limited FISH data for c-MYC amplification is currently available in EOC. Our study examined FFPE primary tumor from women with advanced stage EOC (N = 97), adjusted for copy number alterations in chromosome 8, used two different cut points for c-MYC amplification (≥2 and ≥1.5 copies of c-MYC/CEP8 copies) and demonstrated that c-MYC amplification was not associated with tumor stage, cell type, grade, PFS, OS, tumor response or disease status following platinum-based combination chemotherapy. Wang et al. studied mechanically- and enzymatically-dispersed, frozen, early and advanced stage, epithelial and non-epithelial, ovarian cancers, adjusted for copy number alterations in chromosome 8, used ≥1.5 c-MYC/CEP8 copies cut point for c-MYC amplification, and showed that c-MYC amplification was not associated with OS [48]. Dimova et al. studied FFPE tumor from women with invasive, early and advanced stage, epithelial and non-epithelial, ovarian cancers (N = 280), did not adjust for copy number alterations in chromosome 8 or have access to any measures of clinical outcome, used >2 copies c-MYC cut point for c-MYC gain/amplification, and reported that copy number increases in c-MYC were associated with histologic subtype but not with tumor grade or stage [51]. The disparity between our study and Dimova et al. with respect to the relationship of c-MYC amplification and tumor stage may be attributable at least in part to differences in tumor stages, histologic cell type and the adjustment for chromosome 8 copy number alterations.
Our study demonstrated that c-MYC amplification was observed in 29% (28/97) of the women with suboptimally-resected advanced stage EOC. This result is similar to the 25 to 34% levels described in some ovarian cancer studies using Southern/dot/slot blotting [33,34,36,39,45], polymerase chain reaction (PCR) [46] or comparative genome hybridization (CGH) [44,50], but is higher than the 0 to 17% levels reported in some other studies that also used Southern/dot/slot blotting [35,40,42], PCR [38] or CGH [47], and is lower than the 38 to 55% levels provided in still other studies using Southern/dot/slot blotting [37,41,43], PCR [49], CGH [49], or FISH [48,51]. Differences in the source, type and stage of the tumor specimens, method for detecting gene amplification and the definition for c-MYC amplification may offer some explanation for these disparities.
In the 97 EOC studied herein, c-MYC amplification was not associated with tumor stage, cell type, grade, PFS, OS, tumor response or disease status after platinum-based combination chemotherapy. These findings are consistent with studies demonstrating that c-MYC amplification was not associated with tumor stage [46,51], histologic cell type [43,46], tumor grade [36,37,46,51], progression-free survival (PFS) [45,46], overall survival (OS) [45,46,48,49], and/or response to platinum-based chemotherapy [36] but contradict studies showing that c-MYC amplification was associated with tumor stage [43], histologic cell type [37,51] and/or tumor grade [41,43]. The disparities between studies with respect to the relationship of c-MYC amplification and tumor characteristics or outcome may be explained at least in part by differences in sample size, surgery, type of chemotherapy, tumor stage, clinical end-points, follow-up, type of tumor specimen, method for detecting c-MYC amplification, definition for c-MYC amplification and/or adjustment for copy number alterations in chromosome 8 in these studies. The lack of an association between c-MYC amplification and OS was also observed in other diseases including NSCLC [29,30], colorectal cancer [31] and esophageal SCC [32]. In contrast, studies in breast cancer [18–23], prostate cancer [24–27] and chondrosarcoma [28] demonstrated a strong association between c-MYC amplification and poor outcome.
Wang et al. undertook a chromosome 8 centromere study of ovarian cancer using interphase FISH in mechanically- and enzymatically-dispersed, frozen, invasive ovarian cancers and demonstrated that ≥50% of both c-MYC amplified and non-amplified tumors exhibited polysomy 8 [48]. Despite the prevalence of alterations in the centromeric region of chromosome 8 in these 40 ovarian cancers, the presence of polysomy in chromosome 8 did not appear to be correlated with clinical presentation or disease progression [48]. Herein, we utilized a dual color FISH in 97 FFPE EOC and showed that 11% (3/28) of women with c-MYC amplified tumors and 32% (22/69) of women with non-amplified tumors exhibited polysomy 8. Despite the differences in the prevalence of polysomy 8 in women with amplified and non-amplified c-MYC, we confirmed that polysomy 8 was not associated with OS [48] and went on to demonstrate that polysomy 8 was not associated with PFS, tumor response and disease status following platinum-based combination chemotherapy. Unlike studies in prostate cancer [25,27], hematologic malignancies [58], NSCLC [29–30] or chondrosarcoma [28], polysomy 8 does not appear to have prognostic value in ovarian cancer patients.
c-MYC protein is a well recognized transcription factor with a basic helix–loop–helix leucine zipper motif responsible for sequence-specific DNA binding and protein–protein interactions [59–62] that binds to an estimated 25,000 sites in the human genome and regulates as many as 15% of human genes [63]. c-MYC not only upregulates genes involved in cell cycle regulation, metabolism, ribosome biogenesis, protein synthesis, mitochondrial function and apoptosis and represses genes involved in cell growth arrest and adhesion [64–67], but influences DNA replication, translation and chromatin structure [68–71]. In humans, the c-MYC gene encodes three isoforms: the transcription factors c-MYC-1 and c-MYC-2 as well as c-MYC-S, a dominant-negative inhibitor of c-MYC-1 and c-MYC-2 [72–77]. The level and function of the c-MYC isoforms can be influenced by binding other proteins [78,79] and by phosphorylation, ubiquitinylation and acetylation [80–83].
In conclusion, c-MYC amplification and polysomy 8 were not associated with PFS, OS, tumor response or disease status following platinum-based combination chemotherapy and have limited prognostic value in suboptimally-resected, advanced stage EOC. These results, however, do not rule out the potential that c-MYC has prognostic value when evaluated in a panel of biomarkers that exert epistatic interactions or in women with a borderline tumor or optimally-resected stage III EOC. In addition, the c-MYC isoforms which exhibit diverse and at times opposing functions, and the factors that affect their level and function have yet to be fully evaluated in EOC and may have prognostic value in this disease setting.
Acknowledgments
The authors thank Anne Reardon for her assistance in formatting and editing this manuscript, Suzanne Baskerville for coordinating the clinical data for this study, Dr. Mark Brady for his expert work as the statistician for GOG-111, and Christian Cao and Gary Veytsman for scoring the FISH staining for c-MYC and CEP8. Finally, we would like to thank Dr. Heather Lankes and the GOG Publications Subcommittee for their critical review of and thoughtful suggestions for the manuscript.
Footnotes
This study was supported by the National Cancer Institute grants of the Gynecologic Oncology Group Administrative Office (CA 27469) and the Gynecologic Oncology Group Statistical and Data Center (CA 37517), and the Intramural Research Program of the National Cancer Institute of the National Institute of Health. The following Gynecologic Oncology Group (GOG) institutions participated in this study: University of Alabama at Birmingham, Abington Memorial Hospital, University of Rochester Medical Center, Walter Reed Army Medical Center, Wayne State University, Colorado Gynecologic Oncology Group P.C., University of California at Los Angeles, University of Pennsylvania Cancer Center, Milton S. Hershey Medical Center, Georgetown University Medical Center, Wake Forest University School of Medicine, University of California Medical Center at Irvine, University of Kentucky, The Cleveland Clinic Foundation, Johns Hopkins Oncology Center, Eastern Pennsylvania Gyn/Onc Center, P.C., Cooper Hospital/University Medical Center, Columbus Cancer Council, University of Massachusetts Medical Center, and University of Oklahoma.
Conflict of interest statement
The authors declare that there is no conflict of interest with the exception of William Brady who received payment from Merck and Co, Inc. for working as an independent contractor in the past 2 years.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA- A. Cancer J Clin. 2008;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Thigpen T. First-line therapy for ovarian carcinoma: what's next? Cancer Invest. 2004;22(suppl. 2):21–8. doi: 10.1081/cnv-200030115. [DOI] [PubMed] [Google Scholar]
- 3.Ozols RF. Systemic therapy for ovarian cancer: current status and new treatments. Semin Oncol. 2006;22(suppl. 6):S3–S11. doi: 10.1053/j.seminoncol.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 4.Garte SJ. The c-myc oncogene in tumor progression. Crit Rev Oncogene. 1993;4(4):435–49. [PubMed] [Google Scholar]
- 5.Koskinen PJ, Alitalo K. Role of myc amplification and overexpression in cell growth, differentiation and death. Sem Cancer Biol. 1993;4:3–12. [PubMed] [Google Scholar]
- 6.Boxer LM, Dang CV. Translocations involving c-myc and c-myc function. Oncogene. 2001;20:5595–610. doi: 10.1038/sj.onc.1204595. [DOI] [PubMed] [Google Scholar]
- 7.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2(10):764–76. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 8.Pelengaris S, Khan M. The many faces of c-MYC. Arch Biochem Biophys. 2003;416(2):129–36. doi: 10.1016/s0003-9861(03)00294-7. [DOI] [PubMed] [Google Scholar]
- 9.Adhikary S, Eilers M. Transcriptional regulation and transformation by myc proteins. Nat Rev Mol Cell Biol. 2005;6(8):635–45. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 10.Arvantis C, Felsher DW. Conditional transgenic models define how MYC initiates and maintains tumorigenesis. Semin Cancer Biol. 2006;16(4):313–7. doi: 10.1016/j.semcancer.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 11.Yin XY, Grove L, Datta NA, Long MW, Prochownik EV. C-myc overexpression and p53 loss cooperate to promote genomic instability. Oncogene. 1999;18:1177–84. doi: 10.1038/sj.onc.1202410. [DOI] [PubMed] [Google Scholar]
- 12.Dang CV, Li F, Lee LA. Could MYC induction of mitochondrial biogenesis be linked to ROS production and genomic instability? Cell Cycle. 2005;4(11):1465–6. doi: 10.4161/cc.4.11.2121. [DOI] [PubMed] [Google Scholar]
- 13.Gordon JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12(2):108–13. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer N, Kim SS, Penn LZ. The Oscar-worthy role of Myc in apoptosis. Semin Cancer Biol. 2006;16(4):275–87. doi: 10.1016/j.semcancer.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 15.Ponzielli R, Katz S, Barsyte-Lovejoy D, Penn LZ. Cancer therapeutics: targeting the dark side of Myc. Eur J Cancer. 2005;41:2485–501. doi: 10.1016/j.ejca.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 16.Vita M, Henricksson M. The Myc oncoprotein as a therapeutic target for human cancer. Sem Cancer Biol. 2006;16(4):318–30. doi: 10.1016/j.semcancer.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 17.Brison O. Gene amplification and tumor progression. Biochim Biophys Acta. 1993;1155(1):25–41. doi: 10.1016/0304-419x(93)90020-d. [DOI] [PubMed] [Google Scholar]
- 18.Borg A, Baldetorp B, Ferno M, Olsson H, Sigurdsson H. c-myc amplification is an independent prognostic factor in postmenopausal breast cancer. Int J Cancer. 1992;51(5):687–91. doi: 10.1002/ijc.2910510504. [DOI] [PubMed] [Google Scholar]
- 19.Berns EM, Klijn JG, van Putten WL, van Staveren IL, Portengen H, Foekens JA. c-myc amplification is a better prognostic factor than HER2/neu amplification in primary breast cancer. Cancer Res. 1992;52(5):1107–13. [PubMed] [Google Scholar]
- 20.Deming SL, Nass SJ, Dickson RB, Trock BJ. C-myc amplification in breast cancer: a meta-analysis of its occurrence and prognostic relevance. Br J Cancer. 2000;83(12):1688–95. doi: 10.1054/bjoc.2000.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlotter CM, Vogt U, Bosse U, Mersche B, Wassmann K. C-myc, not HER-2/neu, can predict recurrence and mortality of patients with node-negative breast cancer. Breast Cancer Res. 2003;5(2):R30–6. doi: 10.1186/bcr568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blancato J, Singh BM, Liu AM, Liao DJ, Dickson RB. Correlation of amplification and overexpression of the c-myc oncogene in high-grade breast cancer: FISH, in situ hybridization and immunohistochemical analyses. Br J Cancer. 2004;90:1612–9. doi: 10.1038/sj.bjc.6601703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriquez-Pinilla SM, Jones RL, Lambros MB, Arriola E, Savage K, James M, et al. MYC amplification in breast cancer: a chromogenic in situ hybridization study. J Clin Pathol. 2007;60(9):1017–23. doi: 10.1136/jcp.2006.043869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenkins R, Qian J, Lieber M, Bostwick D. hybridization. Cancer Res. 1997;57:524–31. [PubMed] [Google Scholar]
- 25.Sato K, Qian J, Slezak JM, Lieber MM, Bostwick DG, Bergstralh EJ, et al. Clinical significance of alterations of chromosome 8 in high-grade, advanced, nonmetastatic prostate carcinoma. J Natl Cancer Inst. 1999;91(18):1574–80. doi: 10.1093/jnci/91.18.1574. [DOI] [PubMed] [Google Scholar]
- 26.Sato H, Minei S, Hachiva T, Yoshida T, Takimoto Y. Fluorescence in situ hybridization analysis of c-myc amplification in stage TNM prostate cancer in Japanese patients. Int J Urol. 2006;13(6):761–6. doi: 10.1111/j.1442-2042.2006.01399.x. [DOI] [PubMed] [Google Scholar]
- 27.Dvorackova J, Uvirova M. A molecularly genetic determination of prognostic factors of the prostate cancer and their relationships to expression of protein p27kip1. Neoplasma. 2007;54(2):149–54. [PubMed] [Google Scholar]
- 28.Morrison C, Radmacher M, Mohammed N, Suster D, Auer H, Jones S, et al. Mayerson MYC Amplification and polysomy 8 in chrondrosarcoma: array comparative genome hybridization, fluorescent in situ hybridization, and association with outcome. J Clin Oncol. 2005;23(26):936–76. doi: 10.1200/JCO.2005.03.7127. [DOI] [PubMed] [Google Scholar]
- 29.Kubokura H, Koizumi K, Yamamoto M, Tanaka S. Chromosome 8 copy numbers and the c-myc gene amplification in non-small cell lung cancer Analysis by interphase cytogenetics. Nippon Ika Daigaku Zasshi. 1999;66(2):107–12. doi: 10.1272/jnms.66.107. [DOI] [PubMed] [Google Scholar]
- 30.Kubokura H, Tenjin T, Akiyama H, Koizumi K, Nishirmura H, Yamamoto M, et al. Relations of the c-myc gene and chromosome 8 in non-small cell lung cancer: analysis by fluorescence in situ hybridization. Ann Thorac Cardiovasc Surg. 2001;7(4):197–203. [PubMed] [Google Scholar]
- 31.Al-Kuraya K, Novotny H, Bavi P, Siraj AK, Uddin S, Ezzat A, et al. HER2, TOP2A, CCND1, EGFR and C-MYC oncogene amplification in colorectal cancer. J Clin Pathol. 2007;60(7):768–72. doi: 10.1136/jcp.2006.038281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bitzer M, Stahl M, Arjumand J, Rees M, Klump B, Heep H, et al. C-myc gene amplification in different stages of oesophageal squamous cell carcinoma: prognostic value in relation to treatment modality. Anticancer Res. 2003;23(2B):1489–93. [PubMed] [Google Scholar]
- 33.Serova OM. Amplification of c-myc proto-oncogene in primary tumors, metastases and blood leukocytes of patients with ovarian cancer. Eksp Onkol. 1987;9(1):25–7. [PubMed] [Google Scholar]
- 34.Zhou DJ, Gonzalez-Cadavid N, Ahuja H, Battifora H, Moore GE, Cline MJ. A unique pattern of proto-oncogene abnormalities in ovarian adenocarcinomas. Cancer. 1988;62:1573–6. doi: 10.1002/1097-0142(19881015)62:8<1573::aid-cncr2820620819>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 35.Smith DM, Groff DE, Pokul RK, Bear JL, Delgado G. Determination of cellular oncogene rearrangement or amplification in ovarian adenocarcinomas. Am J Obstet Gynecol. 1989;161:911–5. doi: 10.1016/0002-9378(89)90750-3. [DOI] [PubMed] [Google Scholar]
- 36.Baker VV, Borst MP, Dixon D, Hatch KD, Singleton MH, Miller D. c-myc amplification in ovarian cancer. Gynecol Oncol. 1990;38:340–2. doi: 10.1016/0090-8258(90)90069-w. [DOI] [PubMed] [Google Scholar]
- 37.Sasano H, Garrett CT, Wilkson DS, Silverberg S, Camerford J, Hyde J. Protooncogene amplification and tumor ploidy in human ovarian neoplasms. Hum Pathol. 1990;21:382–91. doi: 10.1016/0046-8177(90)90199-f. [DOI] [PubMed] [Google Scholar]
- 38.Schreiber G, Dubeau L. C-myc proto-oncogene amplification detected by polymerase chain reaction in archival ovarian carcinomas. Am J Pathol. 1990;137:653–8. [PMC free article] [PubMed] [Google Scholar]
- 39.Berns EMJJ, Klijn JGM, Henzen_Logmans SC, Rodenburg CJ, va der Burg MEL, Foekens JA. Receptors for hormones and growth factors and (onco)-gene amplification in human ovarian cancer. Int J Cancer. 1992;52:218–24. doi: 10.1002/ijc.2910520211. [DOI] [PubMed] [Google Scholar]
- 40.Csokay B, Papp J, Besznyak I, Bosze P, Sarosi Z, Toth J, et al. Oncogene patterns in breast and ovarian carcinomas. Eur J Surg Oncol. 1993;19(suppl 1):593–9. [PubMed] [Google Scholar]
- 41.Xin XY. The amplification of c-myc, N-ras, c-erb B oncogenes in ovarian malignancies. Zhonghua Fu Chan Ke Za Zhi. 1993;28(7):405–7. [PubMed] [Google Scholar]
- 42.Bellacosa A, de Feo D, Godwin AK, Bell DW, Cheng JQ, Altomare DA, et al. oncogene in ovarian and breast carcinomas. Int J Cancer. 1995;64:280–5. doi: 10.1002/ijc.2910640412. [DOI] [PubMed] [Google Scholar]
- 43.Meilu B, Fan Q, Huang S, Ma J, Lang J. Amplifications of proto-oncogenes in ovarian carcinoma. Chin Med J (Engl) 1995;108(11):844–8. [PubMed] [Google Scholar]
- 44.Iwabuchi H, Sakamoto M, Sakunaga H, Ma YY, Carcangiu ML, Pinkel D, et al. Genetic analysis of benign, low-grade and high-grade ovarian tumors. Cancer Res. 1995;55:6172–80. [PubMed] [Google Scholar]
- 45.Katsaros D, Theillet C, Zola P, Louason G, Sanfilippo B, Isaia E, et al. genes is associated with poor outcome of ovarian cancer patients. Anticancer Res. 1995;15:1501–10. [PubMed] [Google Scholar]
- 46.Diebold J, Suchy B, Baretton GB, Blasenbreu S, Meier W, Schmidt M, et al. ploidy and MYC DNA amplification in ovarian carcinomas Correlations with p53 and bcl-2 expression, proliferative activity and prognosis. Virchows Arch. 1996;429:221–7. doi: 10.1007/BF00198337. [DOI] [PubMed] [Google Scholar]
- 47.Sonoda G, Palazzo J, du Manoir S, Godwin AK, Feder M, Yakushiji, Testa JR. Comparative genomic hybridization detects frequent overrepresentation of chromosomal material from 3q26, 8q24, and 20q13 in human ovarian carcinomas. Genes Chromosomes Cancer. 1997;20:320–8. [PubMed] [Google Scholar]
- 48.Wang ZR, Liu WH, Smith ST, Parrish RS, Young SR. c-myc and chromosome 8 centromere studies of ovarian cancer by interphase FISH. Exp Mol Pathol. 1999;66:140–8. doi: 10.1006/exmp.1999.2259. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki S, Moore DH, Ginzinger DG, Godfrey TE, Barclay J, Powell B, et al. An approach to analysis of large scale correlations between genome changes and clinical end points in ovarian cancer. Cancer Res. 2000;60:5382–5. [PubMed] [Google Scholar]
- 50.Shridhar V, Lee J, Pandita A, et al. Genetic analysis of early- versus late-stage ovarian tumors. Cancer Res. 2001;61:5895–904. [PubMed] [Google Scholar]
- 51.Dimova I, Raitcheva S, Dimitrov R, Doganov N, Toncheva D. Correlations between c-myc gene copy-number and clinicopathological parameters of ovarian tumors. Eur J Cancer. 2006;42:674–9. doi: 10.1016/j.ejca.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 52.McGuire W, Hoskins W, Brady M, et al. Cyclophosphamide and cisplatin compared with paclitaxel and cisplatin in patients with stage III and stage IV ovarian cancer. N Engl J Med. 1996;334:1–6. doi: 10.1056/NEJM199601043340101. [DOI] [PubMed] [Google Scholar]
- 53.Fisher RA. from contingency tables, and the calculation of p. J R Stat Soc. 1992;85(1):87–94. [Google Scholar]
- 54.Mehta CR, Patel NR. c contingency tables. J Am Stat Assoc. 1983;78:427–34. [Google Scholar]
- 55.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–81. [Google Scholar]
- 56.Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep. 1966;50:163–70. [PubMed] [Google Scholar]
- 57.Cox DR. Regression model and life tables. J R Stat Soc, B. 1972;34:187–220. [Google Scholar]
- 58.Beyer V, Muhlematter D, Parlier V, Cabrol C, Bougeon-Mamin S, Solenthaler M, et al. Polysomy 8 defines a clinico-cytogenetic entity representing a subset of myeloid hematologic malignancies associated with a poor prognosis: report on a cohort of 12 patients and review of 105 published cases. Cancer Genet Cytogenet. 2005;160(2):97–119. doi: 10.1016/j.cancergencyto.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 59.Amati B, Dalton S, Brooks MW, Littlewood TD, Evan GI, Land H. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature. 1992;359:423–6. doi: 10.1038/359423a0. [DOI] [PubMed] [Google Scholar]
- 60.Amati B, Littlewood TD, Evan GI, Land H. The c-Myc protein induces cell cycle progression and apoptosis through dimerization with Max. EMBO J. 1993;12:5083–7. doi: 10.1002/j.1460-2075.1993.tb06202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Facchini LM, Chen S, Marhin WW, Lear JN, Penn LZ. The Myc negative autoregulation mechanism requires Myc–Max association and involves the c-myc P2 minimal promoter. Mol Cell Biol. 1997;17:100–14. doi: 10.1128/mcb.17.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oster SK, Mao DYL, Kennedy J, Penn LZ. Functional analysis of the N-terminal domain of the Myc oncoprotein. Oncogene. 2003;22:1998–2010. doi: 10.1038/sj.onc.1206228. [DOI] [PubMed] [Google Scholar]
- 63.Hurlin PJ, Huang J. The MAX-interacting transcription factor network. Semin Cancer Biol. 2006;16(4):265–74. doi: 10.1016/j.semcancer.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 64.Cowling VH, Cole MD. Mechanism of transcriptional activation by the Myc oncoproteins. Semin Cancer Biol. 2006;16(4):242–52. doi: 10.1016/j.semcancer.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 65.Dang CV, O'Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16(4):253–64. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 66.Kleine-Kohlbrecher D, Adhikary S, Eilers M. Mechanisms of transcriptional repression by Myc. Curr Top Microbiol Immunol. 2006;302:51–60. doi: 10.1007/3-540-32952-8_3. [DOI] [PubMed] [Google Scholar]
- 67.Lee LA, Dang CV. Myc target transcriptomes. Curr Top Microbiol Immunol. 2006;302:145–67. doi: 10.1007/3-540-32952-8_6. [DOI] [PubMed] [Google Scholar]
- 68.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448(7152):445–51. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 69.Lebofsky R, Waler JC. New Myc-anisms for DNA replication and tumorigenesis? Cancer Cell. 2007;12(2):102–3. doi: 10.1016/j.ccr.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 70.Schmidt EV. The role of c-myc in regulation of translation initiation. Oncogene. 2004;23:3217–21. doi: 10.1038/sj.onc.1207548. [DOI] [PubMed] [Google Scholar]
- 71.Knoepfler PS. Myc goes global: new tricks for an old oncogene. Cancer Res. 2007;67(11):5061–3. doi: 10.1158/0008-5472.CAN-07-0426. [DOI] [PubMed] [Google Scholar]
- 72.Hann SR, King MW, Bentley DL, Anderson CW, Eisenman RNA. non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt's lymphomas. Cell. 1988;52:185–95. doi: 10.1016/0092-8674(88)90507-7. [DOI] [PubMed] [Google Scholar]
- 73.Hann SR. Methionine deprivation regulates the translation of functionally-distinct c-Myc proteins. Adv Exp Med Biol. 1995;375:107–16. doi: 10.1007/978-1-4899-0949-7_10. [DOI] [PubMed] [Google Scholar]
- 74.Spotts GD, Patel SV, Xiao Q, Hann SR. Identification of downstream-initiated c-Myc proteins which are dominant-negative inhibitors of transactivation by full-length c-Myc proteins. Mol Cell Biol. 1997;17:1459–68. doi: 10.1128/mcb.17.3.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xiao Q, Claassen G, Shi J, Adachi S, Sedivy J, Hann SR. Transactivation-defective c-MycS retains the ability to regulate proliferation and apoptosis. Genes Dev. 1998;12(24):3803–8. doi: 10.1101/gad.12.24.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hirst SK, Grandori C. Differential activity of conditional MYC and its variant MYC-S in human mortal fibroblasts. Oncogene. 2000;19(45):5189–97. doi: 10.1038/sj.onc.1203904. [DOI] [PubMed] [Google Scholar]
- 77.Oster SK, Mao DYL, Kennedy J, Penn LZ. Functional analysis of the N-terminal domain of the Myc oncoprotein. Oncogene. 2003;22:1998–2010. doi: 10.1038/sj.onc.1206228. [DOI] [PubMed] [Google Scholar]
- 78.Hurlin PJ, Dezfouli S. Functions of Myc:Max in the control of cell proliferation and tumorigenesis. Int Rev Cytol. 2004;238:183–226. doi: 10.1016/S0074-7696(04)38004-6. [DOI] [PubMed] [Google Scholar]
- 79.Hurlin J, Huang PJ. The MAX-interacting transcription factor network. Semin Cancer Biol. 2006;16(4):265–74. doi: 10.1016/j.semcancer.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 80.Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell. 2003;11(5):1177–88. doi: 10.1016/s1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- 81.Vervoorts J, Luscher-Firzlaff J, Luscher B. The ins and outs of MYC regulation by posttranslational mechanisms. J Biol Chem. 2006;281(46):34725–9. doi: 10.1074/jbc.R600017200. [DOI] [PubMed] [Google Scholar]
- 82.Hann SR. Role of post-translational modifications in regulating c-Myc proteolysis, transcriptional activity and biological function. Semin Cancer Biol. 2006;16(4):288–302. doi: 10.1016/j.semcancer.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 83.Dai MS, Jin Y, Gallegos JR, Lu H. Balance of Yin and Yang: ubiquitylation-mediated regulation of p53 and c-Myc. Neoplasia. 2006;8(8):630–44. doi: 10.1593/neo.06334. [DOI] [PMC free article] [PubMed] [Google Scholar]