Abstract
The homeobox gene BP1 is expressed in over 80% of breast cancers and is associated with tumor progression and invasion. However, the mechanism of BP1 activation in these tumors remains unknown. Therefore our aim in this study is to assess the amplification status of the BP1 gene in breast cancer and to determine whether BP1 protein expression is caused by gene amplification in these tumors. BP1 amplification and expression were assessed in 36 samples. Twenty primary breast tumors (PBT) and 14 sentinel lymph node (SLN) metastases were analyzed using fluorescence in situ hybridization (FISH) and immunohistochemistry (IHC), respectively. Because of the close proximity of BP1 and HER2/NEU genes on 17q, correlation between their amplification/expression was also investigated. Increased BP1 copy number was observed in 33% of the cases, with a frequency of 36% and 29% in the PBT and SLN metastasis, respectively. BP1 protein was expressed in 91% of the samples: in all of the PBT with increased BP1 copy number and 65% of PBT with normal copy number. HER2/NEU amplification was detected in 22% of the cases. Concordance between BP1 and HER2/NEU copy numbers was found in 68% of the PBT and 90% of the SLN metastasis. In conclusion, we demonstrated that the BP1 homeobox gene is amplified in breast cancer, both in PBT and SLN metastasis, with a significant correlation with HER2/NEU amplification. Considering that BP1 expression was observed in cases with both increased and normal BP1 copy number, we conclude that other mechanisms in addition to gene amplification play a role in BP1 protein expression.
1. Introduction
The BP1 gene is a member of the distal-less (DLX) family of homeobox genes and is normally expressed during early hematopoiesis [1]. Several key observations support the hypothesis that BP1 is an oncogene with a major role in breast cancer, and that its expression correlates with breast cancer progression and invasion. BP1 was detectable in 80% of infiltrating ductal carcinomas, but undetectable in matched normal controls, being the first DLX gene to be strongly implicated in breast cancer [2]. BP1 expression was also associated with ER negative tumors [2] and was shown to increase with breast tumor progression [3]. These findings strongly support the involvement of BP1 expression in breast tumor development and aggressiveness. A central question, however, is the mechanism of activation of BP1 in these tumors. The BP1 gene is located in an amplicon at the 17q21 region [4] closed to where several known oncogenes have been shown to be amplified in breast cancer [5]. However, to our knowledge, the amplification status of this gene has not been previously investigated. Therefore, the main objective of this study was to determine the amplification status of the BP1 gene in breast tumors and the possibility that its amplification could be one of the mechanisms leading to the expression of the BP1 protein.
We assessed the BP1 copy number in archived samples of primary breast tumors (PBT) and sentinel lymph node (SLN) metastasis by fluorescence in situ hybridization (FISH) analysis using a BP1 specific probe. BP1 protein expression was also investigated in the same set using immunohistochemistry (IHC) so that a direct correlation of BP1 amplification and protein expression could be made. Because of the close proximity on 17q and the biologic importance of the HER2/NEU [6,7] this gene was also evaluated for copy number and protein expression in the same tissue sections.
2. Material and methods
2.1. Sample collection
Formalin-fixed paraffin-embedded (FFPE) sections of 36 untreated breast cancer lesions, including 22 samples of PBT and 14 samples of SLN metastasis were evaluated. In 12 cases, both the PBT and the corresponding SLN lesion from the same patient were available.
The samples were obtained from the tumor banks of the Federal University of Paraná, Curitiba, PR, Brazil (n = 27) and the Armed Forces Institute of Pathology (AFIP), Washington DC, USA (n = 9), under informed consent and IRB approval (Brazilian Ethics Committee and AFIP-UBNI and UBIF). The tumors were of invasive ductal carcinoma type, grade II and with PBT size ranging from 0.8 cm to 8.5 cm.
The mean age of the patients was 49 years, and the majority of patients were of Caucasian origin. For each case, an H&E section from each tissue block used for the FISH and IHC studies was evaluated by a pathologist who confirmed the presence of invasive cancer and delineated the tissues. Consecutive 5 μm sections were then cut for the FISH and IHC studies.
2.2. FISH analysis of the BP1 and HER2/NEU genes
FISH analysis and scoring were performed using a standard protocol that we have previously described [8,9]. For the BP1 gene, we developed a FISH probe consisting of a contig of 5 overlapping bacterial artificial chromosome (BAC) clones (RP11-723D2, RP11-347L6, RP11-479I21, RP11-298H16, RP11-267M22) containing sequences of the BP1 gene (BACPAC Resources, Oakland, CA). For the HER2 probe, we used the BAC clone RP11-62N23 (BACPAC Resources, Oakland, CA). BAC clone DNA was prepared and labeled using nick translation, as previously described [10].
Briefly, the BP1 and HER2 probes were labeled with biotin-16-dUTP (Roche Applied Sciences, Indianapolis, IN) and digoxigenin-11-dUTP (Roche) respectively, using nick translation. The biotin-labeled probe was visualized with avidin conjugated to fluorescein isothiocyanate (FITC) (Vector Laboratories, Burlingame, CA) and the digoxigenin-labeled probe with a mouse anti-digoxin antibody (Sigma, Saint Louis, MO), followed by detection with a goat anti-mouse antibody conjugated to tetramethylrhodamine isothiocyanate (TRITC) (Sigma, Saint Louis, MO). The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and embedded in antifade to reduce photobleaching. Correct chromosomal localization of the BAC clones to 17q21 was confirmed using standard FISH mapping to normal human chromosomes [10].
The two probe sets showed high quality hybridization signals on both metaphase chromosomes and interphase nuclei. For each clone, two distinct signals were detected in the majority of the metaphase and interphase cells analyzed. No additional non-specific signals were detected. The efficiency of the probe for use with FFPE material was further confirmed by analyzing areas with normal breast morphology, or normal lymph node sections, from each case analyzed. In these normal tissues, the cells displayed two copies for each of the two probes, thus demonstrating their specificity.
Digital image acquisition was performed using a 63X objective mounted on a Leica DMRBE microscope (Leica, Wetzlar, Germany) equipped with optical filters for DAPI Fluorescein and TRITC (Chroma Technologies, Brattleboro, VT) and a cooled charge coupled device (CCD) camera (Photometrics, Tucson, AZ). The IPLab software package (Scanalytics Inc, Fairfax, VA) was used for the image acquisition and processing. Slides were scored by two independent observers. A minimum of 100 nuclei were evaluated in each case. Only intact non-overlapping nuclei were scored. Detection of 2 FISH signals was considered as normal copy number, 3 signals as gain, and 6 signals as high-level gain or amplification. To characterize a tumor as having gain or amplification, at least 30% of the nuclei counted must fall into one of the above categories.
2.3. Immunohistochemistry (IHC) analysis of BP1 and HER2/NEU proteins
IHC was performed as we have previously described, using a BP1 specific antibody (Rabbit Anti-BP1 Polyclonal Antibody, Novus Biologicals, Inc., Littleton, CO) that we have shown to be optimal for BP1 analysis by IHC [3]. An anti-human monoclonal antibody was used against HER2/NEU (Vector). We used a standard protocol for IHC assays (the ABC method) for both the BP1 and the HER2/NEU proteins. A given cell was considered positive for BP1 expression if distinct chromogen coloration was consistently seen in its cytoplasm or nucleus in at least two duplicates of the same IHC analysis. A case was considered positive if more than 5% of its entire cell population showed distinct BP1 immunoreactivity [3]. For HER2/NEU analysis, specimens were considered negative for protein expression when presenting scores of 0 or 1+ and positive when presenting 3+. Score 2+ was considered equivocal and required FISH analysis for clarification [11].
2.4. Statistics analysis
Contingency tables (2x2) were created for FISH (amplified/gain vs. normal for both BP1 and HER2/NEU genes) and IHC (positive vs. negative protein expression) across the 2 sets of breast lesions analyzed (PBT and SLN). Fisher’s exact tests were used to compare the distributions of the FISH (or IHC) results between the PBT and SLN groups. The estimated Spearman’s rank-order correlation coefficient (r) between the BP1 and HER2/NEU were calculated for FISH and IHC, and tests of H0: r = 0 were performed.
3. Results
Results from FISH and IHC analyses for both BP1 and HER2/NEU genes are summarized in Table 1. Increased copy number (gain and/or amplification) of the BP1 gene was found in 12/36 (33%) of the breast lesions analyzed: 8/22 (36.3%) PBT and 4/14 (28.6%) SLN metastases. This difference was not statistically significant (p = 0.73). In 12 cases, it was possible to analyze both the PBT and the corresponding SLN metastasis from the same patient. Among these pairs, 8/12 (66.7%) showed concordant results (same gene copy number status in both PBT and SLN) for BP1 gene copy number. In contrast, IHC analysis showed that the BP1 protein was expressed in most cases (29/32, 91%): in 17/20 of the PBT (85%) and in all the 12 SLN metastasis. There was no statistically significant difference between the IHC results in the 2 groups at 5% level (p = 0.27).
Table 1.
Primary breast tumor (PBT) and sentinel lymph node (SLN) metastasis analyzed by FISH and IHC for BP1 and HER2/NEU gene copy number and protein expression, respectively
| PATIENT# | TISSUE | FISH BP1 | IHC BP1 | FISH HER2/NEU | IHC HER2/NEU |
|---|---|---|---|---|---|
| 1 | PBT | Normal | pos | Gain | pos |
| 2 | PBT | Gain | pos | Amplification | pos |
| 3 | PBT | Amplification | pos | Amplification | pos |
| 4 | PBT | Normal | pos | Normal | neg |
| 5 | PBT | Normal | pos | Amplification | pos |
| 6 | PBT | Normal | neg | Gain | neg |
| 7 | PBT | Normal | pos | Normal | neg |
| 8 | PBT | Normal | neg | Amplification | neg |
| 9 | PBT | Normal | pos | Amplification | neg |
| 10 | PBT | Normal | pos | Normal | pos |
| 11 | SLN | Normal | pos | – | – |
| 12 | SLN | Gain | – | – | – |
| 13 | PBT | Amplification | pos | Gain | neg |
| SLN | Normal | pos | – | – | |
| 14 | PBT | Amplification | pos | Amplification | neg |
| SLN | Gain | pos | Gain | neg | |
| 15 | PBT | Gain | pos | Normal | neg |
| SLN | Normal | pos | Normal | neg | |
| 16 | PBT | Normal | pos | Normal | neg |
| SLN | Normal | pos | – | – | |
| 17 | PBT | Amplification | pos | Amplification | pos |
| SLN | Normal | pos | Normal | neg | |
| 18 | PBT | Normal | neg | Normal | neg |
| SLN | Normal | pos | Normal | neg | |
| 19 | PBT | Normal | pos | Normal | neg |
| SLN | Normal | pos | Normal | neg | |
| 20 | PBT | Gain | – | Gain | pos |
| SLN | Gain | pos | Gain | neg | |
| 21 | PBT | Normal | pos | Normal | neg |
| SLN | Normal | pos | Normal | neg | |
| 22 | PBT | Amplification | – | Normal | neg |
| SLN | Amplification | – | Normal | neg | |
| 23 | PBT | Normal | pos | Normal | neg |
| SLN | Normal | pos | Normal | neg | |
| 24 | PBT | Normal | pos | Normal | neg |
| SLN | Normal | pos | Normal | neg |
Abbreviations: pos, positive; neg, negative; –, result not obtained.
BP1 protein was expressed in all the PBT that showed an increased copy number of the BP1 gene as well as in 11/17 (65%) of the cases that presented normal BP1 copy number (2 copies) by FISH (Fig. 1). No statistically significant association was detected between copy number changes and protein expression for BP1 in the combined samples (r = 0.1284; p = 0.46).
Fig. 1.
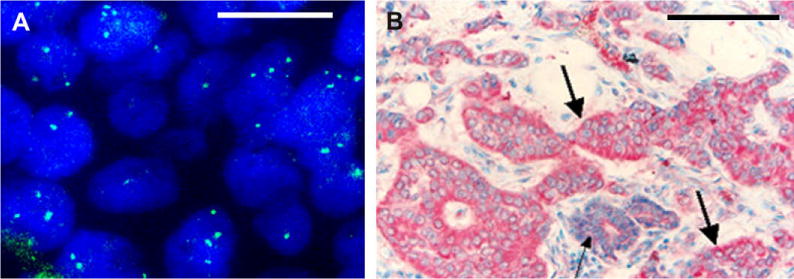
Formalin-fixed, paraffin-embedded (FFPE) section from a representative primary breast tumor sample showing: A: Amplification of BP1 gene (green signals) by FISH (100×; magnification bar is 100 μm); B: Immunostaining for BP1 (red). Thick arrows identify invasive lesions with strong BP1 expression. Thin arrows identify normal area with no or weaker BP1 expression (40×; magnification bar is 2.5 mm).
HER2/NEU FISH analysis was performed in 32 of the samples (22 PBT and 10 SLN). HER2/NEU amplification was observed in 7 samples (32%), and all 7 were primary tumors. In the 22 PBT analyzed for both BP1 and HER2/NEU copy number, 15 (68%) had concordant findings for both genes, whereas in the 10 SLN cases, 9 (90%) had concordant findings (r = 0.4731, p = 0.01) (Figs. 2 and 3). In 25% of the samples, increase copy number was observed for both BP1 and HER2/NEU genes. Of note, three samples (#14 PBT, #21 PBT, and #21 SLN) presented gain/amplification of BP1 but no gain/amplification of HER2/NEU gene).
Fig. 2.
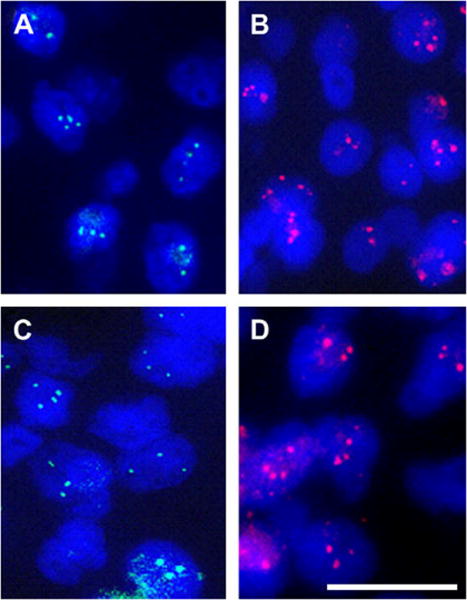
FISH analysis of formalin-fixed, paraffin-embedded (FFPE) sections of a pair of primary breast tumor (A and B) and corresponding sentinel lymph node metastasis (C and D) showing hybridization for the BP1 (green signals) and HER2/NEU (red signals) genes. Note the detection of increased copy number for both genes in both of the lesions (100x; magnification bar is 100 μm).
Fig. 3.
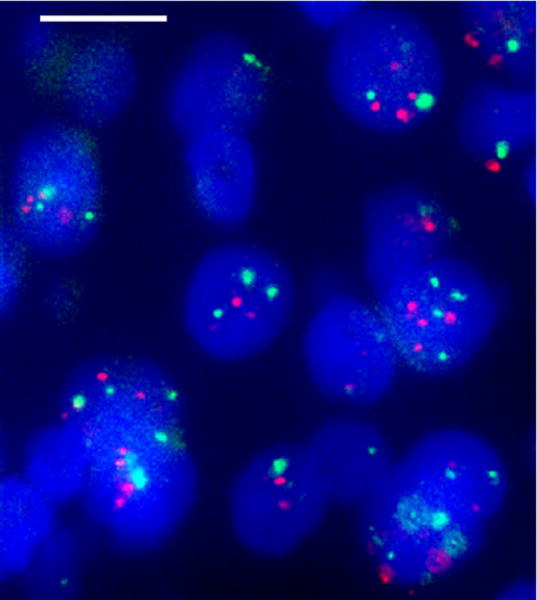
FISH analysis of a formalin-fixed, paraffin-embedded (FFPE) section of a representative primary breast tumor sample showing increased copy number for the BP1 (green signals) and HER2/NEU (red signals) genes (magnification bar is 100 μm).
IHC for the HER2/NEU protein was performed on all the samples tested for HER2/NEU amplification by FISH. HER2/NEU protein was expressed in 22% (7/32) of the samples, and there was concordance between gene and protein status in 75% (24/32) of the lesions (r = 0.4101; p = 0.026). IHC expression between the BP1 and HER2/NEU proteins however, did not show significant correlation (r = 0.1735; p = 0.58).
4. Discussion
This is the first report showing that the BP1 homeobox gene is amplified in breast cancer: one third of the tumors analyzed had an increased copy number of the BP1 gene. BP1 protein expression, measured by IHC analysis, was observed in all the cases that showed increased copy number (gain or amplification) of the BP1 gene, however this correlation was not statistically significant. BP1 expression was also detected in cases with no BP1 copy number alteration. These findings suggest that additional mechanisms other than gene amplification may be responsible for BP1 protein expression.
Gene amplification is a common mechanism of activation, and can result in overexpression of oncogenes in various malignancies, including breast cancer [12,13]. In the 17q21 chromosomal region, where the BP1 gene maps, there is a known amplicon in breast cancer [4,5], in close proximity where numerous genes are highly overexpressed, including members of other homeobox gene families, such as the HOX gene family. The HOXB7 gene, found to be overexpressed in breast cancer cell lines [14–17], was amplified in 10% of PBT in a large series of cases studied by Hyman et al. [17]. These authors suggested that DNA amplification could be a mechanism for overexpressing HOXB7. Other HOX genes, such as HOXA5, HOXA7, and HOXC10 have also been shown to be amplified in breast cancer [18]. Although the effect of gene copy number changes on protein expression levels is largely unknown, several studies have explored gene expression changes in specific amplicons [12,13,19].
HER2/NEU amplification in breast tumors is tightly associated with HER2/NEU protein overexpression. In our study, we observed a significant correlation between HER2/NEU amplification and protein overexpression in 75% of the cases. In contrast, some authors have found that amplification of other genes in breast cancer, such as TOP2A or EGFR, are not necessarily associated with high levels of its corresponding protein expression [20,21].
We observed that BP1 was expressed in all PBT that showed increased copy number of the gene. Some of the genes mapped to 17q21 are up regulated along with the HER2/NEU. TOP2A amplification, for instance, though considered to be an independent event from HER2/NEU amplification, is more frequently seen in tumors with HER2/NEU amplification [13,22,23].
In this study, 25% of the breast lesions analyzed presented increased copy number for both BP1 and HER2/NEU genes. In 2 PBT samples and one SLN sample, however, an increase in BP1 copy number was observed with no corresponding alteration in the HER2/NEU gene copy number. Additional samples need to be investigated to determine if this was a non-random event and if these genes are amplified in an independent manner.
BP1 positive expression was previously found in breast cancer cases that were HER2/NEU positive by IHC [3]. Of the 7 cases that showed HER2/NEU positive expression, BP1 expression was studied in 6 and all 6 were positive. Recently, HER2/NEU protein overexpression was found to be predictive of SLN metastasis in clinically negative nodes [24]. Patients whose tumors were HER2/NEU positive had approximately a 50% higher risk of developing SLN metastasis than those whose tumors did not expressed this marker.
We have recently shown that there was an association between BP1 positivity and positive lymph node status (in preparation). Future studies should include analysis of both BP1 and HER2/NEU protein expression in breast cancer patients with known lymph node status, to determine the prognostic ability of BP1 alone or in combination with HER2/NEU.
In summary, we have demonstrated that the BP1 homeobox gene is amplified in breast cancer (one third of the cases in our study), with significant correlation with HER2/NEU amplification. BP1 expression was observed in all PBT with increased copy number of the BP1 gene, as well as in 65% of the PBTwith normal BP1 copy numbers, indicating that other mechanisms in addition to gene amplification play a role in BP1 protein expression. Further studies investigating the role of amplification of BP1 in breast cancer will be important to determine if BP1 can be considered a new and independent prognostic marker in the 17q region.
Acknowledgments
The authors wish to acknowledge the significant scientific input of the late Dr. Robert B. Dickson during the planning of this project and his support to Dr. Cavalli through out her scientific career. This research was supported by funds from the Susan G. Komen Foundation (P.E.B.) and by the American Cancer Society – Intramural Research Grant Program (L.R.C).
References
- 1.Chase MB, Fu S, Haga SB, Davenport G, Stevenson H, Do K, Morgan D, Mah AL, Berg P. BP1, a homeodomain-containing isoform of DLX4, represses the -globin gene. Mol Cell Biol. 2002;22:2505–14. doi: 10.1128/MCB.22.8.2505-2514.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu SW, Schwartz A, Stevenson H, Pinzone JJ, Davenport GJ, Orenstein JM, Gutierrez P, Simmens SJ, Abraham J, Poola I, Stephan DA, Berg PE. Correlation of expression of BP1, a homeobox gene, with estrogen receptor status in breast cancer. Breast Cancer Res. 2003;5:82–7. doi: 10.1186/bcr602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Man Y-G, Fu SW, Schwartz A, Pinzone JJ, Simmens SJ, Berg PE. Expression of BP1, a novel homeobox gene, correlates with breast cancer progression and invasion. Breast Cancer Res Treat. 2005;90:241–7. doi: 10.1007/s10549-004-4492-9. [DOI] [PubMed] [Google Scholar]
- 4.Fu S, Stevenson H, Strovel JW, Haga SB, Stamberg J, Do K, Berg PE. Distinct functions of two isoforms of a homeobox gene, BP1 and DLX7, in the regulation of the beta-globin gene. Gene. 2001;278:131–9. doi: 10.1016/s0378-1119(01)00716-8. [DOI] [PubMed] [Google Scholar]
- 5.Mano MS, Rosa DD, De Azambuja E, Ismael GF, Durbecq V. The 17q12-q21 amplicon: Her2 and topoisomerase-II alpha and their importance to the biology of solid tumours. Cancer Treat Rev. 2007;33:64–77. doi: 10.1016/j.ctrv.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Dalifard I, Daver A, Goussard J, Lorimier G, Gosse-Brun S, Lorthlary A, Larra F. p185 overexpression in 220 samples of breast cancer undergoing primary surgery: comparison with c-erbB-2 gene amplification. Int J Mol Med. 1998;1:855–61. doi: 10.3892/ijmm.1.5.855. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell MS, Press MF. The role of immunohistochemistry and fluorescence in situ hybridization for HER2/NEU in assessing the prognosis of breast cancer. Semin Oncol. 1999;26:108–11. [PubMed] [Google Scholar]
- 8.Henke RT, Haddad BR, Kim SE, Rone JD, Mani A, Jessup JM, Wellstein A, Anirban Maitra A, Riegel AT. Overexpression of the nuclear receptor coactivator AIB1 (SRC-3) during progression of pancreatic adenocarcinoma. Clinical Cancer Res. 2004;10:6134–42. doi: 10.1158/1078-0432.CCR-04-0561. [DOI] [PubMed] [Google Scholar]
- 9.Cullen KJ, Newkirk KA, Schumaker LM, Aldosari N, Rone JD, Haddad BR. Glutathione S-transferase pi amplification is associated with cisplatin resistance in head and neck squamous cell carcinoma cell lines and primary tumors. Cancer Res. 2003;63:8097–102. [PubMed] [Google Scholar]
- 10.Haddad B, Pabon-Pena C, Young H, Sun WH. Fine mapping of STAT1 gene by FISH and radiation hybrids. Cytogenet Cell Genet. 1998;83:58–9. doi: 10.1159/000015126. [DOI] [PubMed] [Google Scholar]
- 11.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, van de Vijver M, Wheeler TM, Hayes DF. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–45. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 12.Monni O, Bärlund M, Mousses S, Kononen J, Sauter G, Heiskanen M, Paavola P, Avela K, Chen Y, Bittner ML, Kallioniemi A. Comprehensive copy number and gene expression profiling of the 17q23 amplicon in human breast cancer. Proc Natl Acad Sci USA. 2001;98:5711–6. doi: 10.1073/pnas.091582298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kauraniemi P, Barlund M, Monni O, Kallioniemi A. New amplified and highly expressed genes discovered in the ERBB2 amplicon in breast cancer by cDNA microarrays. Cancer Res. 2001;61:8235–40. [PubMed] [Google Scholar]
- 14.Chen H, Sukumar S. Role of homeobox genes in normal mammary gland development and breast tumorigenesis. J Mammary Gland Biol Neoplasia. 2003;8:159–75. doi: 10.1023/a:1025996707117. [DOI] [PubMed] [Google Scholar]
- 15.Makiyama K, Hamada J, Takada M, Murakawa K, Takahashi Y, Tada M, Tamoto E, Shindo G, Matsunaga A, Teramoto K, Komuro K, Kondo S, Katoh H, Koike T, Moriuchi T. Aberrant expression of HOX genes in human invasive breast carcinoma. Oncol Rep. 2005;13:673–9. [PubMed] [Google Scholar]
- 16.Wu X, Chen H, Parker B, Rubin E, Zhu T, Lee JS, Argani P, Sukumar S. HOXB7, a homeodomain protein, is overexpressed in breast cancer and confers epithelial-mesenchymal transition. Cancer Res. 2006;66:9527–34. doi: 10.1158/0008-5472.CAN-05-4470. [DOI] [PubMed] [Google Scholar]
- 17.Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, Ringner M, Sauter G, Monni O, Elkahloun A, Kallioniemi OP, Kallioniemi A. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62:6240–5. [PubMed] [Google Scholar]
- 18.Lewis MT. Homeobox genes in mammary gland development and neoplasia. Breast Cancer Res. 2000;2:158–69. doi: 10.1186/bcr49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark J, Edwards S, John M, Flohr P, Gordon T, Maillard K, Giddings I, Brown C, Bagherzadeh A, Campbell C, Shipley J, Wooster R, Cooper CS. Identification of amplified and expressed genes in breast cancer by comparative hybridization onto microarrays of randomly selected cDNA clones. Genes Chromosomes Cancer. 2002;34:104–14. doi: 10.1002/gcc.10039. [DOI] [PubMed] [Google Scholar]
- 20.Mueller RE, Parkes RK, Andrulis I, O’Malley FP. Amplification of the TOP2A gene does not predict high levels of topoisomerase II alpha protein in human breast tumor samples. Genes Chromosomes Cancer. 2004;39:288–97. doi: 10.1002/gcc.20008. [DOI] [PubMed] [Google Scholar]
- 21.Ememura S, Takekoshi S, Suzuki Y, Saitoh Y, Tokuda Y, Osamura RY. Estrogen receptor-negative and human epidermal growth factor receptor 2-negative breast cancer tissue have the highest Ki-67 labeling index and EGFR expression: gene amplification does not contribute to EGFR expression. Oncol Rep. 2005;14:337–43. [PubMed] [Google Scholar]
- 22.Bouchalova K, Trojanec R, Kolar Z, Cwiertka K, Cernakova I, Mihal V, Hajduch M. Analysis of ERBB2 and TOP2A gene status using fluorescence in situ hybridization versus immunohistochemistry in localized breast cancer. Neoplasma. 2006;53:393–401. [PubMed] [Google Scholar]
- 23.Arriola E, Rodriguez-Pinilla SM, Lambros MB, Jones RL, James M, Savage K, Smith IE, Dowsett M, Reis-Filho JS. Topoisomerase II alpha amplification may predict benefit from adjuvant anthracyclines in HER2 positive early breast cancer. Breast Cancer Res Treat. 2007;106:181–9. doi: 10.1007/s10549-006-9492-5. [DOI] [PubMed] [Google Scholar]
- 24.Nathanson SD, Slater R, Debruyn D, Kapke A, Linden M. Her-2/neu expression in primary breast cancer with sentinel lymph node metastasis. Ann Surg Oncol. 2006;13:205–13. doi: 10.1245/ASO.2006.03.032. [DOI] [PubMed] [Google Scholar]


