Abstract
Studies of mitochondrial DNA (mtDNA) as well as the non-recombining part of the Y chromosome help to understand the origin and distribution of maternal and paternal lineages. The Kabardian horse from Northern Caucasia which is well-known for strength, stamina and endurance in distance riding has a large gap in its breeding documentation especially in the recent past. A 309 bp fragment of the mitochondrial D-loop (156 Kabardian horses) and six mutations in Y chromosome (49 Kabardian stallions), respectively, were analyzed to get a better insight into breeding history, phylogenetic relationship to related breeds, maternal and paternal diversity and genetic structure. We found a high mitochondrial diversity represented by 64 D-loop haplotypes out of 14 haplogroups. The most frequent haplogroups were G (19.5%), L (12.3%), Q (11.7%), and B (11.0%). Although these four haplogroups are also frequently found in Asian riding horses (e.g. Buryat, Kirghiz, Mongolian, Transbaikalian, Tuvinian) the percentage of the particular haplogroups varies sometimes remarkable. In contrast, the obtained haplogroup pattern from Kabardian horse was more similar to that of breeds reared in the Middle East. No specific haplotype cluster was observed in the phylogenetic tree for Kabardian horses. On Kabardian Y chromosome, two mutations were found leading to three haplotypes with a percentage of 36.7% (haplotype HT1), 38.8% (haplotype HT2) and 24.5% (haplotype HT3), respectively. The high mitochondrial and also remarkable paternal diversity of the Kabardian horse is caused by its long history with a widely spread maternal origin and the introduction of Arabian as well as Thoroughbred influenced stallions for improvement. This high genetic diversity provides a good situation for the ongoing breed development and performance selection as well as avoiding inbreeding.
Keywords: Genetic diversity, Mitochondrial DNA, Phylogenetic analysis, Y chromosome
Introduction
Molecular markers are powerful tools for detecting genetic variation and uniqueness of populations. Such knowledge helps to protect diversity, prevent inbreeding and improve the breeding process. Sequence analyses of the mitochondrial DNA (mtDNA) and the Y chromosome are often used for discovery of maternal and paternal lineages, for analysis of phylogenetic development and breed origin as well as for detection of population admixture (Di Lorenzo et al., 2016). The specific value of mtDNA and non-recombining Y chromosomal sequences in phylogenetic investigations is caused by its clonal mode of inheritance. A study of hypervariable displacement loop (D-loop) region allows a determination of the diversity of maternal ancestry within a population and its phylogenetic relationship. Therefore, mtDNA is widely used as an instrument for uncovering the evolutional and phylogenetic history of populations like cattle (Ludwig et al., 2013), pig (Alves et al., 2003), chicken (Di Lorenzo et al., 2015) as well as horse (Forster et al., 2012; Yang et al., 2017). D-loop studies were also used for description of intrabreed and interbreed differentiation (Bowling et al., 2000; Chauhan et al., 2011). In horses, a huge amount of mitochondrial haplotypes exists and several authors developed classifications into haplogroups (Jansen et al., 2002; Cieslak et al., 2010; Achilli et al., 2012).
On other hand, mutations in the paternally transmitted portion of the Y chromosome can help to investigate paternal lineages. In contrast to the plenty of mutations in the mitochondrial genome and on the Y chromosome variation of pre-domestic horses (Lippold et al., 2011), no diversity was detected in the Y chromosome of domestic horses awhile (Brandariz-Fontes et al., 2013; Lindgren et al., 2004). In the last years, a few polymorphic sites were found in modern horses leading to a small number of haplotypes in contemporary domestic stallions (Ling et al., 2010; Wallner et al., 2013, 2017; Kreutzmann et al., 2014).
This study addresses the maternal and paternal genetic variation of Kabardian horses – a breed with excellent endurance for mountain rides. The Kabardian represents a relative self-contained population from Caucasus; unfortunately without clear knowledge about its origin and reliable centralized studbook registry from the end of the 1990th years till 2007. For further breeding work, its genetic relationship to other breeds from this region as well as to breeds with good stamina performance like Arabians is of interest. Additional, the investigation of the maternal and paternal lines shall help to improve the line classification in the new studbook.
Materials and Methods
Material and DNA preparation
Blood from V. jugularis or mane hair samples were taken from 156 Kabardian horses (64 males and 92 females) kept in studs of the Kabardino-Balkarian Republic (16 studs with 24 males and 46 females), Krasnodarsky Krai (2 studs with 3 males and 14 females), Adygea (3 studs with 3 males and 9 females), Karachaevo-Cherkesskaia Republic (1 stud with 5 females) in Russia as well as in Germany (imported horses from Caucasus distributed to 9 studs with 34 males and 18 females). The horses were selected in agreement with the Kabardian horse breeding association to get a representative sample and to avoid the use of closely related horses. All paragraphs of the Federal Law on Protection of Animals against Cruel Treatment were strictly followed (Russia: No. 1571, Germany: 55.2-1-54-2532-128-2014). The genomic DNA was extracted by incubating the roots or the white cell pellets in 180 µL T1 buffer and 20 µL proteinase K (Macherey-Nagel, Berlin, Germany) followed by the salting out procedure (Miller et al., 1988). Addressing the Y chromosome, 49 Kabardian breeding stallions from the 64 male horses used for mtDNA analysis and four stallions of the Przewalski’s horse were investigated.
PCR amplification and sequencing
The amplification of the upstream D-loop region was carried out in a volume of 25 mL containing 50 ng DNA, 0.2 μM of each primer, 0.2 mM dNTP, 2.5 mM MgCl2 and 0.5 U GoTaqflexi polymerase (Promega, Wisconsin, USA). The procedure involved denaturation at 95°C (5 min) followed by 35 cycles of denaturation at 94°C (30 s), annealing at 55°C (30 s) and elongation at 72°C (45 s). The final elongation was carried out at 72°C for 5 min. The separation of the PCR fragment from 2% agarose gel was performed with GeneJet Gel Extraction Kit (Fermentas, Thermo Fisher Scientific, Massachusetts, USA). The sequencing was carried out as Sanger reaction with BigDye Terminator v1.1 Ready Reaction Cycle Sequencing Kit in an ABI PRISM 310 Genetic Analyzer (Life technologies, Thermo Fisher Scientific, Massachusetts, USA) using manufacturer’s protocol.
On Y chromosome, two polymorphic sites found on position 0147 in a prehistoric wild Siberian horse and on position 0201 in a Przewalski’s horse (Lippold et al., 2011) were genotyped using pyrosequencing with 57°C annealing temperature in PCR reaction following manufacturer’s protocol (Qiagen, Hilden, Germany). Four additional polymorphisms known from recent horses (Wallner et al., 2013) were analyzed with KBiosciences Allele Specific PCR (KASP) technology using standard procedure with 57°C touch down PCR (LGC Genomics, Berlin, Germany) for positions 1277 and 25345 as well as with sequencing (like described above) for positions 10594 and 11007-11315. All primer sequences are shown in Table 1.
Table 1.
Primer sequences for polymorphism detection in D-loop region and on Y chromosome.
| Primer name | Nucleotide sequence 5’ – 3’ | Method | Length |
|---|---|---|---|
| H-mtDNA F1) | AGC TCC ACC ATC AAC ACC CAA A | Sequencing | 690 bp |
| H-mtDNA R1) | CCA TGG ACT GAA TAA CAC CTT ATG GTT G | ||
| H-Y0147 F | Biotin-ATG TCA GGA TGC CAA CTG GTC | Pyroseq | 74 bp |
| H-Y0147 R | GTG CCC ACA AGA CAA TGT C | ||
| H-Y0147 S | CCA CTA GGG CCC TGT TTC | ||
| H-Y0201 F | Biotin-AGT GGC CCC CTG AAG ACA TT | Pyroseq | 76 bp |
| H-Y0201 R | CGA CCG GAA AAG GGA CAG TG | ||
| H-Y0201 S | GGC CAG AGC CAC TAC T | ||
| H-Y1277 A1 | TTC TAA ACA GTA ATC TCA AAC TGT GAT | KASP | 57 bp |
| H-Y1277 A2 | CTT TCT AAA CAG TAA TCT CAA ACT GTG AA | ||
| H-Y1277 C | GGC TTC CAT GAA TGA CTC TCG AGT T | ||
| H-Y25345 A1 | AAG TTT TAC GAA AGA CAT AAA CTA CGT TAA AT | KASP | 62 bp |
| H-Y25345 A2 | GTT TTA CGA AAG ACA TAA ACT ACG TTA AAC | ||
| H-Y25345 C | GAG GAT AGA TGG GGA AAA GGT TGA AAA TA | ||
| H-Y10594 F2) | CCC TCT GCT GAG CAT CTA GG | Sequencing | 297 bp |
| H-Y10594 R2) | TTG GAT GAA AGG GAC AGT GA | ||
| H-Y11007 F2) | CCA ACA CAC GTC AAC AGC TC | Sequencing | 444 bp |
| H-Y11007 R2) | GGC TTA GGC CAC TGA TGG TA |
Primer sequences after:
Khanshour and Cothran (2013) and
Wallner et al. (2013).
Phylogenetic analysis
All sequences were prepared with SnapGene Viewer 2.5.0/2.6.0. The phylogenetic tree was built within MEGA 7.0.18 (Kumar et al., 2016) using a 309 bp fragment (position 15469-15777 after NC001640 from GenBank). For evaluation of haplogroup distribution and frequency in the Kabardian horse, D-loop sequences from 71 Mongolian, 22 Buryat, 24 Transbaikalian, 25 Tuvinian (GenBank JQ936335-JQ936476) and 9 Kirghiz horses (own material) were used as possible closely related breeds from North Asia. For correct classification into haplogroups, 40 sequences selected for mutations in the investigated D-loop region out of 83 (GenBank JN398377-JN398457, EF597513, EF597514) were used and all haplogroups were named or renamed after Achilli et al. (2012). DnaSP software Version 6.10.03 (Rozas et al., 2017) was used for haplotype diversity estimation.
Results and Discussion
In this study, 64 different haplotypes were discovered in 156 Kabardian horses (Table 2). The Asian breeds used for comparison show also a huge number of haplotypes but with regard to the haplogroups, Kabardian horses present the highest diversity. The ratio between number of horses and number of haplotypes illustrates that on average only one to two horses within a breed share the same haplotype and underlines the high variation. With a frequency over 10%, the most haplotypes of Kabardian horses are found in the haplogroups G, L, Q, and B while Kabardian haplotypes clustered rarely in haplogroups N and R. No Kabardian haplotype is found in the haplogroups F, H and K, respectively (Table 3).
Table 2.
Number of horses, haplotypes and haplogroups in different breeds, haplotype diversity as well as the ratio between the number of genotyped horses and number of haplotypes.
| Breed | Number of horses (n) | Number of haplotypes (HT) | Number of haplogroups (HG) | Haplotype diversity (Hd±SD) | Ratio between horses and haplotypes |
|---|---|---|---|---|---|
| Kabardian | 156 | 64 | 14 | 0.980±0.003 | 2.4 |
| Buryat | 22 | 20 | 10 | 0.987±0.020 | 1.1 |
| Kirghiz | 9 | 9 | 7 | 1.000±0.052 | 1.0 |
| Mongolian | 71 | 50 | 12 | 0.987±0.005 | 1.4 |
| Transbaikalian | 24 | 13 | 8 | 0.909±0.039 | 1.8 |
| Tuvinian | 25 | 22 | 10 | 0.993±0.014 | 1.1 |
Table 3.
Haplogroup percentage (in %) of six analyzed breeds (haplogroups O and P were not subdivided, 12 horses don’t share a haplogroup, classification after Achilli et al. (2012).
| Haplogroup | Kabardian | All Asian | Buryat | Kirghiz | Mongolian | Transbaikalian | Tuvinian |
|---|---|---|---|---|---|---|---|
| (n = 154) | (n = 141) | (n = 21) | (n = 9) | (n = 64) | (n = 24) | (n = 23) | |
| A | 6.5 | 4.9 | 0.0 | 0.0 | 6.2 | 12.5 | 0.0 |
| B | 11.0 | 14.2 | 23.8 | 11.2 | 11.0 | 8.3 | 21.8 |
| C | 5.2 | 4.3 | 0.0 | 0.0 | 6.2 | 4.2 | 4.3 |
| D | 2.6 | 4.3 | 4.8 | 0.0 | 3.1 | 8.3 | 4.3 |
| E | 6.5 | 5.7 | 4.8 | 11.1 | 6.2 | 0.0 | 8.7 |
| F | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| G | 19.5 | 11.3 | 4.8 | 11.1 | 3.1 | 45.9 | 4.3 |
| H | 0.0 | 2.8 | 0.0 | 0.0 | 1.6 | 0.0 | 13.1 |
| I | 7.2 | 2.8 | 19.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| J | 3.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| K | 0.0 | 0.7 | 0.0 | 0.0 | 0.0 | 0.0 | 4.3 |
| L | 12.3 | 14.9 | 4.8 | 22.2 | 17.2 | 8.3 | 21.8 |
| M | 5.2 | 7.8 | 9.5 | 11.1 | 11.0 | 4.2 | 0.0 |
| N | 1.9 | 0.7 | 0.0 | 11.1 | 0.0 | 0.0 | 0.0 |
| O’P | 5.8 | 7.1 | 9.5 | 0.0 | 12.5 | 0.0 | 0.0 |
| Q | 11.7 | 14.2 | 9.5 | 22.2 | 18.8 | 8.3 | 8.7 |
| R | 1.3 | 4.3 | 9.5 | 0.0 | 3.1 | 0.0 | 8.7 |
A direct comparison with the Asian riding horse breeds shows that haplogroups L, B and Q are frequently present in these breeds. Though, there are some distinct differences in haplotype frequencies between the breeds. The haplogroups Q and L are very frequent in Mongolian horses while Tuvinian horses cluster mainly in haplogroups B and L. The main haplogroup of Buryat horse is B while the frequent haplogroup L is rare in this breed. In Transbaikalian horses, the haplogroup G being the most frequent haplogroup in Kabardian horses is the clear dominant one. In the other breeds, this haplogroup is rather rare. The small number of Kirghiz horse is present in these haplogroups. In contrast to the Kabardian horses, Buryat horses show high frequencies in haplogroup I, Mongolian horses in haplogroups M and O’P, and Tuvinian horses in haplogroup H, respectively. Only the Transbaikalian horses show a high percentage of haplogroup A which seems to be common in Asian breeds (Achilli et al., 2012).
Altogether, the Kabardian horses distinguish from the Asian breeds in many haplogroup frequencies. Otherwise, the haplogroup structure of West European breeds with the dominant L (Cieslak et al., 2010) and Bulgarian mountain pony breeds with higher percentage of M and J is also quite different (Hristov et al., 2016). The best congruency can be found to horses from the Middle East (Achilli et al., 2012) with exception of East Anatolian horses (Koban et al., 2012) although the group G has a higher percentage in Kabardian horses at expense of group L.
In breeds from Middle East (Ahkal Teke, Caspian), the haplogroup L is dominant followed by a mixture of A/B/G (McGahern et al., 2006a; Cieslak et al., 2010). But overall, the majority of haplotypes and haplogroups are not restricted to a certain geographical area or a specific breed. The phylogenetic tree of D-loop sequences produces no specific cluster (Fig. 1) albeit two or more breeds share the same haplotype only in 15% of all case. Differences only emerge in numbers and frequencies of haplotypes.
Fig. 1.
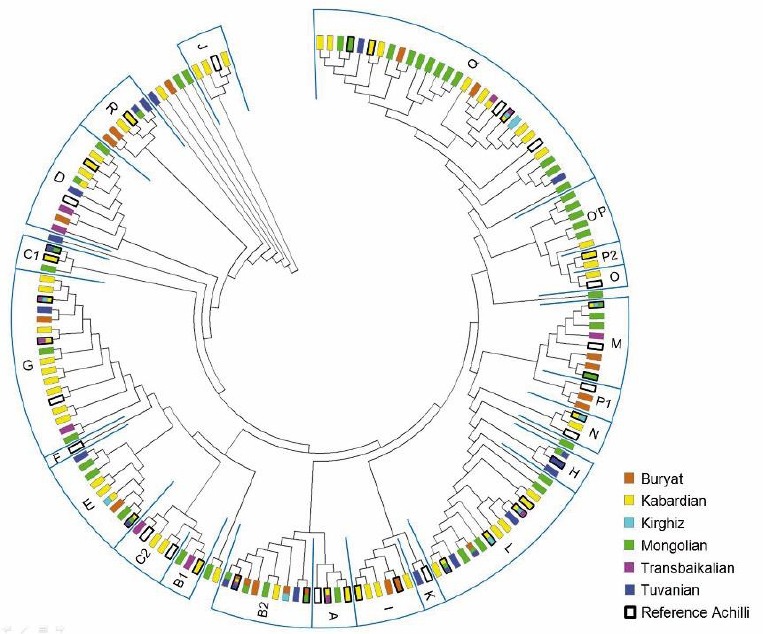
Phylogenetic tree based on mtDNA D-loop sequences (309 bp from 15469-15777 with NC001640 from GenBank as reference) within 161 haplotypes (64 Kabardian, 20 Buryat, 9 Kirghiz, 50 Mongolian, 13 Transbaikalian, 22 Tuvinian). From GenBank, 40 additonal sequences – framed with thick black lines – were used for identification of haplogroup cluster (letters) after Achilli et al. (2012).
In addition, discrepancies in the haplogroup percentage result often from the limited number of investigated horses, which is often not representative for the entire breed. In consideration of the high diversity of D-loop region, these results can only show a tendency of haplogroup distribution in regions or breeds. Significant geographical separation or breeding effects on the haplogroup structure can only be detected in exceptional cases like breed isolation, limited number of founders as well as extremely rare haplotypes (Cardinali et al., 2016; McGahern et al., 2006a, b).
The lack of such specific haplotypes and the wide variability in Kabardian horses precludes a classification into a region or a group of breeds. But the very high mitochondrial haplotype diversity in horses is a well-known fact (Cardinali et al., 2016; Gemingguli et al., 2016).
Multiple, widespread and repeated domestication of diverse founder mares as well as mixed origin of most breeds are considered as likely causes for this phenomenon (Cozzi et al., 2004; McGahern et al., 2006a; Lei et al., 2009). In agreement with previous studies, only one to three horses share the same haplotype (Cozzi et al., 2004; Lei et al., 2009; Achilli et al., 2012; Bigi et al., 2014).
The haplotype diversity of Kabardian (0.980) is similar to that of the additional investigated Asian horses or higher. That also applies to comparison with other breeds like Arabian (0.784 and 0.962), Thoroughbred (0.883 and 0.942), Kazakh (0.977), Chinese (0.978 and 0.989) as well as Brazilian (varying from 0.200 to 0.944) horses (Zhang et al., 2012; Ianella et al., 2017; Othman et al., 2017).
However, the Kabardian horse has a huge genetic variability in its maternal lineages representing a diverse origin and a longstanding history, which is in agreement with recently published microsatellite analysis (Duduev et al., 2014).
Only a small diversity was found in the Y chromosome sequences (Table 4). Neither the Kabardian nor the Przewalski’s stallions presented mutations proved in wild Siberian (M0147), Icelandic (M25345) and Fjord (M11007-11315) horses, respectively.
Table 4.
Genotyping results for several mutations on equine Y chromosome (dash: not discovered).
| n | M01471) | M02011) | M253452) | M105942) | M11007-113152) | M12772) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | T | C | G | A | G | T | Del | Normal | Mutated | A | T | ||
| aDNA1) | 4 | 3 | 1 | 0 | 4 | - | - | - | - | - | - | - | - |
| Przewalski’s | 4 | 4 | 0 | 0 | 4 | 0 | 4 | 4 | 0 | 4 | 0 | 0 | 4 |
| Kabardian | 49 | 49 | 0 | 49 | 0 | 0 | 49 | 37 | 12 | 49 | 0 | 31 | 18 |
Mutation position and wild ancient horse (aDNA) results from Siberia after Lippold et al. (2011).
Mutation position after Wallner et al. (2013).
An old mutation (M0201) typical for pre-domestic horses and Przewalski’s stallions (Lippold et al., 2011; Der Sarkissian et al., 2015) was also not found in Kabardian horse. Only two common domestic horse polymorphisms (M10594 and M1277) were present in Kabardian stallions leading to three haplotypes. The haplotype HT1 (T on position M1277 and M10594; after Wallner et al. (2013)) which was present in 36.7% of the Kabardian stallions is the ancestral haplotype of domestic horses and the non-mutated variant T was also found in Przewalski’s horse.
Haplotype HT2 (A on position M1277 and T on M10594) originating from Arabian founders was also present with a high frequency of 38.8%. This haplotype occurs frequently in breeds from Eastern and Central Europe as well as in breeds of the Middle East like Ahkal teke.
Beside these two old and widely spread haplotypes, 24.5% of the Kabardian stallions had haplotype HT3 (A on position M1277 and deletion on M10594). These stallions obtained their Y chromosome from the introgression of Thoroughbred. In the 19th and at the beginning of the 20th century, Thoroughbred stallions were carefully introduced for improving speed in combination with endurance. Around 1960, a second introduction of Thoroughbred created the Anglo Kabardian lineage (Dragilev, 1953) but this population was not investigated in our study.
Compared with the maternal variation, the paternally-inherited non-recombining Y chromosome diversity in horses is limited (Wallner et al., 2003; Lindgren et al., 2004; Ling et al., 2010; Kreutzmann et al., 2014) but with three paternal haplotypes, the Kabardian horse has a remarkable diversity.
This inside into the genetic structure of the Kabardian breed is helpfully for its future improvement. The great mitochondrial diversity without specific haplotype cluster can be considered as result of long-standing genetic exchange and admixture with other domestic horse lineages. The major haplogroup frequency is similar to that of Middle East breeds which argues for a closer relationship. The fact that two-thirds of the stallions go back to performance breeds (Original Arabian and Thoroughbred) emphasizes a continuous selection of the local stock for riding and endurance ability. There is no risk of inbreeding neither from maternal nor from paternal side.
Acknowledgements
This study was funded by the Volkswagen Stiftung, Germany (Az.: 85 858). We thank all Kabardian horse breeders for their support and Frau Schemmel (Humboldt University, Berlin) for helping with laboratory analysis.
Conflict of interest
The authors declare that there is no conflict of interests.
References
- Achilli A, Olivieri A, Soares P, Lancioni H, Hooshiar Kashani B, Perego UA, Nergadze SG, Carossa V, Santagostino M, Capomaccio S, Felicetti M, Al-Achkar W, Penedo MC, Verini-Supplizi A, Houshmand M, Woodward SR, Semino O, Silvestrelli M, Giulotto E, Pereira L, Bandelt HJ, Torroni A. Mitochondrial genomes from modern horses reveal the major haplogroups that underwent domestication. Proc. Natl. Acad. Sci. U S A. 2012;109(7):2449–2454. doi: 10.1073/pnas.1111637109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves E, Ovilo C, Rodriguez MC, Silio L. Mitochondrial DNA sequence variation and phylogenetic relationships among Iberian pigs and other domestic and wild pig populations. Anim. Genet. 2003;34(5):319–324. doi: 10.1046/j.1365-2052.2003.01010.x. [DOI] [PubMed] [Google Scholar]
- Bigi D, Perrotta G, Zambonelli P. Genetic analysis of seven Italian horse breeds based on mitochondrial DNA D-loop variation. Anim. Genet. 2014;45(4):593–595. doi: 10.1111/age.12156. [DOI] [PubMed] [Google Scholar]
- Bowling AT, Del Valle A, Bowling M. A pedigree-based study of mitochondrial D-loop DNA sequence variation among Arabian horses. Anim. Genet. 2000;31(1):1–7. doi: 10.1046/j.1365-2052.2000.00558.x. [DOI] [PubMed] [Google Scholar]
- Brandariz-Fontes C, Leonard JA, Vega-Pla JL, Backstrom N, Lindgren G, Lippold S, Rico C. Y-chromosome analysis in Retuertas horses. PLoS One. 2013;8(5):e64985. doi: 10.1371/journal.pone.0064985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinali I, Lancioni H, Giontella A, Capodiferro MR, Capomaccio S, Buttazzoni L, Biggio GP, Cherchi R, Albertini E, Olivieri A, Cappelli K, Achilli A, Silvestrelli M. An Overview of Ten Italian Horse Breeds through Mitochondrial DNA. PLoS One. 2016;11(4):e0153004. doi: 10.1371/journal.pone.0153004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan M, Gupta AK, Dhillon S. Genetic diversity and population structure of three Indian horse breeds. Mol. Biol. Rep. 2011;38:3505–3511. doi: 10.1007/s11033-010-0461-z. [DOI] [PubMed] [Google Scholar]
- Cieslak M, Pruvost M, Benecke N, Hofreiter M, Morales A, Reissmann M, Ludwig A. Origin and history of mitochondrial DNA lineages in domestic horses. PLoS One. 2010;5(12):e15311. doi: 10.1371/journal.pone.0015311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzi MC, Strillacci MG, Valiati P, Bighignoli B, Cancedda M, Zanotti M. Mitochondrial D-loop sequence variation among Italian horse breeds. Genet. Sel. E. 2004;36:663–672. doi: 10.1186/1297-9686-36-6-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der Sarkissian C, Ermini L, Schubert M, Yang MA, Librado P, Fumagalli M, Jonsson H, Bar-Gal GK, Albrechtsen A, Vieira FG, Petersen B, Ginolhac A, Seguin-Orlando A, Magnussen K, Fages A, Gamba C, Lorente-Galdos B, Polani S, Steiner C, Neuditschko M, Jagannathan V, Feh C, Greenblatt CL, Ludwig A, Abramson NI, Zimmermann W, Schafberg R, Tikhonov A, Sicheritz-Ponten T, Willerslev E, Marques-Bonet T, Ryder OA, McCue M, Rieder S, Leeb T, Slatkin M, Orlando L. Evolutionary Genomics and Conservation of the Endangered Przewalski's Horse. Curr. Biol. 2015;25(19):2577–2583. doi: 10.1016/j.cub.2015.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lorenzo P, Ceccobelli S, Panella F, Attard G, Lasagna E. The role of mitochondrial DNA to determine the origin of domestic chicken. World's Poultry. Sci. J. 2015;71(2):311–318. [Google Scholar]
- Di Lorenzo P, Lancioni H, Ceccobelli S, Curcio L, Panella F, Lasagna E. Uniparental genetic systems: a male and a female perspective in the domestic cattle origin and evolution. Electronic J. Biotechnol. 2016;23:69–78. [Google Scholar]
- Dragilev PI. [Working with horses of a new type - in Russian] In Purebred Kabardin Stud Book - Volume III. Ed. Ministry of Agriculture Kabardian ASSR, Nalchik. 1953 [Google Scholar]
- Duduev AS, Khaudov AD, Kokov ZA, Amshokov Kh.K, Zhekamukhov M.Kh, Zaitsev AM, Zaitseva MA, Gavrilicheva IS, Kalinkova LV, Reissmann M. [Genetic structure of the Kabardian horse breed on DNA micosatellite loci and the possiblity of identification on population level by this method - in Russian] Konevodstvo i konnyi sport. 2014;6:18–19. [Google Scholar]
- Forster P, Hurles ME, Jansen T, Levine M, Renfrew C. Origins of the domestic horse. Proc. Natl. Acad. Sci. U S A. 2012;109(46):E3148. doi: 10.1073/pnas.1210326109. author reply E3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemingguli M, Iskhan KR, Li Y, Qi A, Wunirifu W, Ding LY, Wumaierjiang A. Genetic diversity and population structure of Kazakh horses (Equus caballus) inferred from mtDNA sequences. Genet. Mol. Res. 2016;15(4) doi: 10.4238/gmr.15048618. doi:10.4238/gmr.15048618. [DOI] [PubMed] [Google Scholar]
- Hristov P, Yordanov G, Ivanova A, Mitkov I, Sirakova D, Mehandzyiski I, Radoslavov G. Mitochondrial diversity in mountain horse population from the South-Eastern Europe. Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016;28(6):787–792. doi: 10.1080/24701394.2016.1186667. [DOI] [PubMed] [Google Scholar]
- Ianella P, Albuquerque M.S.M, Paiva SR, Egito AA, Almeida LD, Sereno F.T.P.S, Carvalho L.F.R, Mariante AS, McManus CM. D-loop haplotype diversity in Brazilian horse breeds. Genet. Mol. Biol. 2017;40(3):604–609. doi: 10.1590/1678-4685-GMB-2016-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen T, Forster P, Levine MA, Oelke H, Hurles M, Renfrew C, Weber J, Olek K. Mitochondrial DNA and the origins of the domestic horse. Proc. Natl. Acad. Sci. U S A. 2002;99(16):10905–10910. doi: 10.1073/pnas.152330099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanshour AM, Cothran EG. Maternal phylogenetic relationships and genetic variation among Arabian horse populations using whole mitochondrial DNA D-loop sequencing. BMC Genet. 2013;14:83. doi: 10.1186/1471-2156-14-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koban E, Denizci M, Aslan O, Aktoprakligil D, Aksu S, Bower M, Balcioglu BK, Ozdemir Bahadir A, Bilgin R, Erdag B, Bagis H, Arat S. High microsatellite and mitochondrial diversity in Anatolian native horse breeds shows Anatolia as a genetic conduit between Europe and Asia. Anim. Genet. 2012;43(4):401–409. doi: 10.1111/j.1365-2052.2011.02285.x. [DOI] [PubMed] [Google Scholar]
- Kreutzmann N, Brem G, Wallner B. The domestic horse harbours Y-chromosomal microsatellite polymorphism only on two widely distributed male lineages. Anim. Genet. 2014;45(3):460. doi: 10.1111/age.12149. doi:10.1111/age.12149. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. E. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei CZ, Su R, Bower MA, Edwards CJ, Wang XB, Weining S, Liu L, Xie WM, Li F, Liu RY, Zhang YS, Zhang CM, Chen H. Multiple maternal origins of native modern and ancient horse populations in China. Anim. Genet. 2009;40(6):933–944. doi: 10.1111/j.1365-2052.2009.01950.x. [DOI] [PubMed] [Google Scholar]
- Lindgren G, Backstrom N, Swinburne J, Hellborg L, Einarsson A, Sandberg K, Cothran G, Vila C, Binns M, Ellegren H. Limited number of patrilines in horse domestication. Nat. Genet. 2004;36(4):335–336. doi: 10.1038/ng1326. [DOI] [PubMed] [Google Scholar]
- Ling Y, Ma Y, Guan W, Cheng Y, Wang Y, Han J, Jin D, Mang L, Mahmut H. Identification of Y chromosome genetic variations in Chinese indigenous horse breeds. J. Hered. 2010;101(5):639–643. doi: 10.1093/jhered/esq047. [DOI] [PubMed] [Google Scholar]
- Lippold S, Knapp M, Kuznetsova T, Leonard JA, Benecke N, Ludwig A, Rasmussen M, Cooper A, Weinstock J, Willerslev E, Shapiro B, Hofreiter M. Discovery of lost diversity of paternal horse lineages using ancient DNA. Nat. Commun. 2011;2:450. doi: 10.1038/ncomms1447. [DOI] [PubMed] [Google Scholar]
- Ludwig A, Alderson L, Fandrey E, Lieckfeldt D, Soederlund TK, Froelich K. Tracing the genetic roots of the indigenous White Park Cattle. Anim. Genet. 2013;44(4):383–386. doi: 10.1111/age.12026. [DOI] [PubMed] [Google Scholar]
- McGahern A, Bower MA, Edwards CJ, Brophy PO, Sulimova G, Zakharov I, Vizuete-Forster M, Levine M, Li S, MacHugh DE, Hill EW. Evidence for biogeographic patterning of mitochondrial DNA sequences in Eastern horse populations. Anim. Genet. 2006a;37(5):494–497. doi: 10.1111/j.1365-2052.2006.01495.x. [DOI] [PubMed] [Google Scholar]
- McGahern AM, Edwards CJ, Bower MA, Heffernan A, Park SD, Brophy PO, Bradley DG, MacHugh DE, Hill EW. Mitochondrial DNA sequence diversity in extant Irish horse populations and in ancient horses. Anim. Genet. 2006b;37(5):498–502. doi: 10.1111/j.1365-2052.2006.01506.x. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othman OE, Mahrous KF, Shafey HI. Mitochondrial DNA genetic variations among four horse populations in Egypt. J. Genet. Engineer. Biotechn. 2017;15(2):469–474. doi: 10.1016/j.jgeb.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A. DnaSP v6: DNA Sequence Polymorphism analysis of large Datasets. Mol. Biol. E. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- Wallner B, Brem G, Muller M, Achmann R. Fixed nucleotide differences on the Y chromosome indicate clear divergence between Equus przewalskii and Equus caballus. Anim. Genet. 2003;34(6):453–456. doi: 10.1046/j.0268-9146.2003.01044.x. [DOI] [PubMed] [Google Scholar]
- Wallner B, Palmieri N, Vogl C, Rigler D, Bozlak E, Druml T, Jagannathan V, Leeb T, Fries R, Tetens J, Thaller G, Metzger J, Distl O, Lindgren G, Rubin C.J, Andersson L, Schaefer R, McCue M, Neuditschko M, Rieder S, Schlötterer C, Brem G. Y Chromosome Uncovers the Recent Oriental Origin of Modern Stallions. Curr. Biol. 2017;27(13):2029–2035.e5. doi: 10.1016/j.cub.2017.05.086. doi:10.1016/j.cub.2017.05.086. [DOI] [PubMed] [Google Scholar]
- Wallner B, Vogl C, Shukla P, Burgstaller JP, Druml T, Brem G. Identification of genetic variation on the horse y chromosome and the tracing of male founder lineages in modern breeds. PLoS One. 2013;8(4):e60015. doi: 10.1371/journal.pone.0060015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhu Q, Liu S, Zhao C, Wu C. The origin of Chinese domestic horses revealed with novel mtDNA variants. Anim. Sci. J. 2017;88(1):19–26. doi: 10.1111/asj.12583. [DOI] [PubMed] [Google Scholar]
- Zhang T, Lu H, Chen C, Jiang H, Wu S. Genetic Diversity of mtDNA D-loop and Maternal Origin of Three Chinese Native Horse Breeds. Asian-Australas J. Anim. Sci. 2012;25(7):921–926. doi: 10.5713/ajas.2011.11483. [DOI] [PMC free article] [PubMed] [Google Scholar]


