James and Zagotta discuss how recent cryoEM structures inform our understanding of cyclic nucleotide–binding domain channels.
Abstract
Cyclic nucleotide-binding domain (CNBD) channels are a family of ion channels in the voltage-gated K+ channel superfamily that play crucial roles in many physiological processes. CNBD channels are structurally similar but functionally very diverse. This family includes three subfamilies: (1) the cyclic nucleotide-gated (CNG) channels, which are cation-nonselective, voltage-independent, and cyclic nucleotide-gated; (2) the hyperpolarization-activated cyclic nucleotide-gated (HCN) channels, which are weakly K+ selective, hyperpolarization-activated, and cyclic nucleotide-gated; and (3) the ether-à-go-go-type (KCNH) channels, which are strongly K+ selective, depolarization-activated, and cyclic nucleotide-independent. Recently, several high-resolution structures have been reported for intact CNBD channels, providing a structural framework to better understand their diverse function. In this review, we compare and contrast the recent structures and discuss how they inform our understanding of ion selectivity, voltage-dependent gating, and cyclic nucleotide–dependent gating within this channel family.
Introduction
Ion channels are membrane-embedded, ion-permeable proteins that are responsible for the electrical activity of excitable cells. Ion conduction through these channels, driven by electrochemical gradients across the membrane, produces an electrical current that cells use to interact with the outside world and regulate intracellular processes (Hille, 2001; Zheng and Trudeau, 2015). The central feature of all ion channels is the pore, which provides a pathway for ions to cross the membrane. Depending on the pore architecture, certain ions may permeate more readily, imparting ion selectivity to the channel. Ion permeation through the pore is often gated, with opening of the pore occurring when specific stimuli are applied to the channel. Different modules coupled to the pore, such as voltage-sensor domains or cyclic nucleotide–binding domains (CNBDs), detect stimuli and transduce them into pore opening (i.e., channel activation). The structural basis for ion selectivity and channel gating, particularly the coupling between stimuli-sensing modules and the pore, has been the subject of intense study. To this end, atomic (or near atomic)-resolution structural information from channel fragments, and more recently full-length channels, has proven valuable for determining the mechanisms for ion channel function.
A wealth of high-resolution structural information has recently become available for the CNBD family of cation channels (Whicher and MacKinnon, 2016; James et al., 2017; Lee and MacKinnon, 2017; Li et al., 2017; Wang and MacKinnon, 2017). This family is part of the voltage-gated K+ (KV) channel superfamily and consists of three subfamilies: the cyclic nucleotide–gated (CNG) channels, the hyperpolarization-activated cyclic nucleotide–gated (HCN) channels, and the ether-à-go-go-type (KCNH) channels (Fig. 1). Members of this family share a common core architecture, including a C-terminal CNBD and a C-linker domain that connects the CNBD to the pore. Although they are structurally related, these channels play distinct physiological roles. CNG channels are expressed in photoreceptor cells and olfactory sensory neurons, where they participate in the phototransduction and olfactory transduction pathways, respectively (Fesenko et al., 1985; Nakamura and Gold, 1987; Kaupp and Seifert, 2002; Craven and Zagotta, 2006; Dai and Varnum, 2015). Upon activation (i.e., photon absorption or odorant binding), both pathways produce a transient change in the cytoplasmic cyclic nucleotide concentration. CNG channels, being directly gated by cyclic nucleotides, detect this change and convert it into an electrical signal that regulates neurotransmitter release from the cell. HCN channels, in comparison, are notable for contributing to the spontaneous and rhythmic firing activity of several cell types, including sinoatrial node cells in the heart and thalamocortical relay neurons in the central nervous system (Craven and Zagotta, 2006; Biel et al., 2009; Wahl-Schott and Biel, 2009; Benarroch, 2013). In these cells, HCN channels open during the hyperpolarizing phase of an action potential and conduct an inward current—the “funny” (If) or “hyperpolarization-activated” (Ih) current—that depolarizes the membrane sufficiently to initiate another action potential. Finally, KCNH channels help regulate excitability in a variety of cardiac and neuronal cells (Ganetzky et al., 1999; Morais-Cabral and Robertson, 2015). In neurons, KCNH channels fulfill this role by producing an outward, hyperpolarizing K+ current that excitatory (i.e., membrane-depolarizing) stimuli must overcome to elicit an action potential. The KCNH subfamily consists of the ether-à-go-go (EAG), EAG-like (ELK), and EAG-related (ERG) subgroups (Warmke et al., 1991; Warmke and Ganetzky, 1994; Fig. 1), the latter subgroup being responsible for the fast delayed-rectifier current (IKr) that helps terminate cardiac action potentials (Sanguinetti et al., 1995; Trudeau et al., 1995).
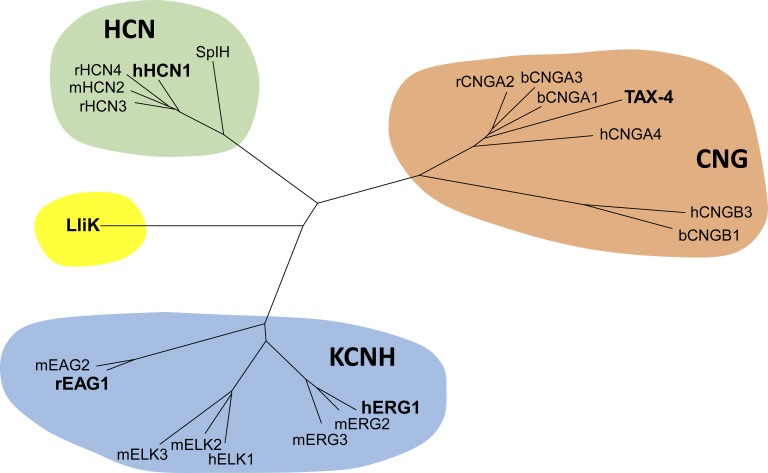
Figure 1.
Dendrogram of the CNBD cation channel family. This family contains three subfamilies: CNG channels, HCN channels, and KCNH channels. The five channels whose structures are discussed in this review (rEAG1, hERG1, hHCN1, TAX-4, and LliK) are highlighted in bold.
This review focuses on five recent CNBD channel structures resolved by cryo-electron microscopy (cryoEM), including members from all three subfamilies. Through collective analysis of these structures, it highlights how the three CNBD channel subfamilies may share common elements in voltage- and ligand-dependent gating, despite being functionally divergent.
Diverse functional properties
The CNBD channel subfamilies are functionally distinct, differing in their cation selectivity, voltage dependence, and cyclic nucleotide dependence. Within each subfamily, functional differences also arise between channel isoforms (e.g., HCN1 or HCN2) or subgroups (e.g., EAG or ERG), as summarized below.
Ion selectivity
CNBD channels vary widely in their selectivity for different cations. CNG channels are essentially nonselective for monovalent cations and conduct K+, Na+, Li+, and Rb+ almost equally well (Kaupp and Seifert, 2002). CNG channels also conduct Ca2+ (Frings et al., 1995; Dzeja et al., 1999), which serves as a second messenger in both the phototransduction and olfactory transduction pathways (Burns et al., 2002; Matthews and Reisert, 2003). HCN channels are weakly selective for K+ over Na+ ions, possessing a permeability ratio (PK/PNa) of 3–5, and are also considerably less permeable to Ca2+ (Gauss et al., 1998; Ludwig et al., 1998; Santoro et al., 1998). Weak selectivity between K+ and Na+ is an important feature of HCN channels, allowing them to conduct an inward, depolarizing Na+ current at the hyperpolarized potentials where they are active. KCNH channels exhibit the greatest selectivity among CNBD channels, and are highly selective for K+ over Na+ (PK/PNa >100; Ludwig et al., 1994). As with CNG and HCN channels, the ion selectivity of KCNH channels is matched to their physiological roles. The high K+ selectivity of ERG channels, for example, allows them to conduct a hyperpolarizing outward K+ current that helps repolarize the cardiac action potential (Sanguinetti and Tristani-Firouzi, 2006).
Voltage dependence
Each CNBD channel subfamily exhibits a distinct response to changes in the membrane electrical potential. CNG channels are largely insensitive to membrane voltage, showing only a slight increase in conductance at depolarized potentials (Karpen et al., 1988; Kaupp et al., 1989; Dhallan et al., 1990; Goulding et al., 1992; Fig. 2 A). In comparison, HCN channels activate with membrane hyperpolarization (Gauss et al., 1998; Ludwig et al., 1998; Santoro et al., 1998; Fig. 2 B) and also exhibit “voltage-dependent potentiation” (also called “mode-shift” or “hysteresis”), where prior exposure to hyperpolarizing (i.e., activating) potentials shifts the voltage dependence of activation to more depolarized potentials (Männikkö et al., 2005; Elinder et al., 2006). Finally, KCNH channels behave more like canonical shaker-type channels and activate with membrane depolarization (Brüggemann et al., 1993; Trudeau et al., 1995; Ganetzky et al., 1999; Fig. 2 C). The response of KCNH channels to membrane voltage is complex and varies between the EAG, ELK, and ERG subgroups. ERG channels, for example, undergo rapid and voltage-dependent inactivation from the open state (Smith et al., 1996), whereas EAG channels are non-inactivating (Ludwig et al., 1994). In addition, ERG and ELK channels both exhibit voltage-dependent potentiation, where prior exposure to depolarizing potentials shifts the voltage dependence of activation to more hyperpolarized potentials (Tan et al., 2012; Li et al., 2015; Dai and Zagotta, 2017).
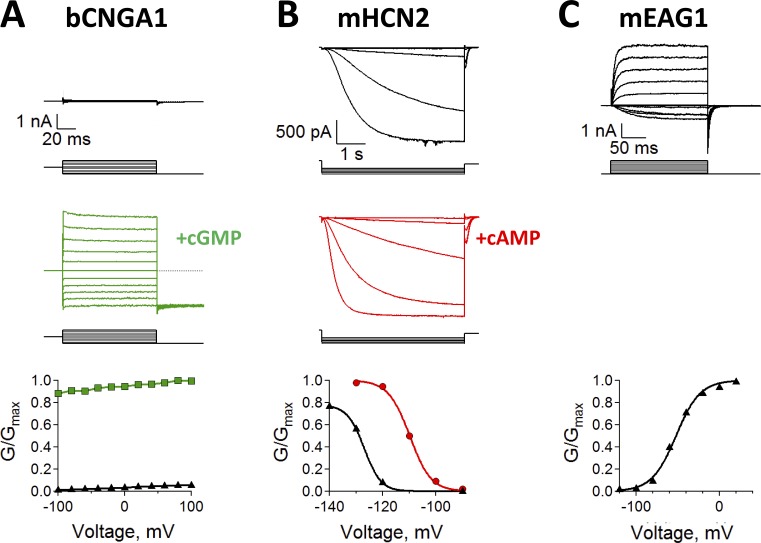
Figure 2.
Diverse functional properties of CNBD channels. (A) Na+ currents recorded from bovine CNGA1 (bCNGA1) channels in the absence (top, black) and presence (middle, green) of saturating cGMP concentrations. Scale bar applies to both top and middle panels. Currents were measured by stepping from 0 mV to test potentials ranging from −100 mV to 100 mV, before stepping to a −100-mV tail potential. The conductance–voltage (G-V) relationship (bottom) was obtained from normalized tail currents. (B) K+ currents recorded from mouse HCN2 (mHCN2) channels in the absence (top, black) and presence (middle, red) of saturating cAMP concentrations. Scale bar applies to both top and middle panels. Currents were measured by stepping from 0 mV to test potentials ranging from −90 mV to −140 mV, before stepping to a 0-mV tail potential. The G-V curve (bottom) was obtained from normalized tail currents. (C) K+ Currents recorded from mouse EAG1 (mEAG1) channels in the absence of cyclic nucleotides (top, black). Currents were measured by stepping from −120 mV to test potentials ranging from −120 mV to 100 mV, before stepping to a −120-mV tail potential. The G-V curve (bottom) was obtained from normalized tail currents.
Cyclic nucleotide regulation
CNG and HCN channels are both regulated by the direct binding of cyclic nucleotides to an intracellular CNBD (Fesenko et al., 1985; Craven and Zagotta, 2006; Fig. 2, A and B), whereas KCNH channels do not bind and are not directly regulated by cyclic nucleotides (Robertson et al., 1996; Brelidze et al., 2009). In CNG channels, cyclic nucleotide binding is sufficient for activation, although the efficacy of activation depends on the specific cyclic nucleotide and varies between CNG channel types. Rod CNG channels activate almost completely in the presence of saturating concentrations of cGMP, whereas cAMP elicits very little current (Ildefonse et al., 1992; Gordon and Zagotta, 1995; Varnum et al., 1995). In contrast, olfactory CNG channels and the TAX-4 CNG channel from Caenorhabditis elegans (whose structure is discussed in this review) are fully activated by both saturating cAMP and cGMP concentrations (Nakamura and Gold, 1987; Dhallan et al., 1990; Paoletti et al., 1999).
Compared with CNG channels, HCN channels are activated by both membrane hyperpolarization and cyclic nucleotide binding to an intracellular CNBD. Cyclic nucleotides have multiple effects on HCN channel function, including (a) an acceleration of channel activation kinetics; (b) a depolarizing shift in the voltage of half-maximal activation (V1/2); and (c) an increase in the maximal current (Wang et al., 2001; Craven and Zagotta, 2004; Zhou et al., 2004; Fig. 2 B). However, these effects vary between different HCN channel isoforms, of which there are four in mammals (HCN1–4; Santoro et al., 1998). In HCN4, the primary isoform expressed in sinoatrial node cells, cAMP binding produces a 10–25-mV depolarizing shift in V1/2 (Ludwig et al., 1998; Altomare et al., 2003). In HCN1, which is expressed in both the heart and the central nervous system, cAMP binding produces only a ∼2-mV depolarizing shift in V1/2. Associated regulatory and trafficking proteins also influence the cyclic nucleotide dependence of HCN channels. For example, binding of the trafficking protein TRIP8b reduces or eliminates cyclic nucleotide–dependent activation in HCN channels (Santoro et al., 2009; Zolles et al., 2009; Bankston et al., 2012; Hu et al., 2013; Saponaro et al., 2014; Han et al., 2017).
Common structural features
CNBD channels share a common core architecture composed of four domains: a transmembrane voltage-sensor domain (VSD), a transmembrane pore domain, an intracellular C-linker domain, and an intracellular CNBD (Fig. 3). This architecture may also be shared by the K+ selective, hyperpolarization-activated KAT- and AKT-type channels found in plants (Anderson et al., 1992; Sentenac et al., 1992; Gaymard et al., 1996), although it is unclear whether these channels are directly regulated by cyclic nucleotides (Hoshi, 1995; Gaymard et al., 1996). In KCNH channels, the CNBD does not bind cyclic nucleotides and is referred to as a cyclic nucleotide–binding homology domain (CNBHD; Brelidze et al., 2012, 2013; Marques-Carvalho et al., 2012).
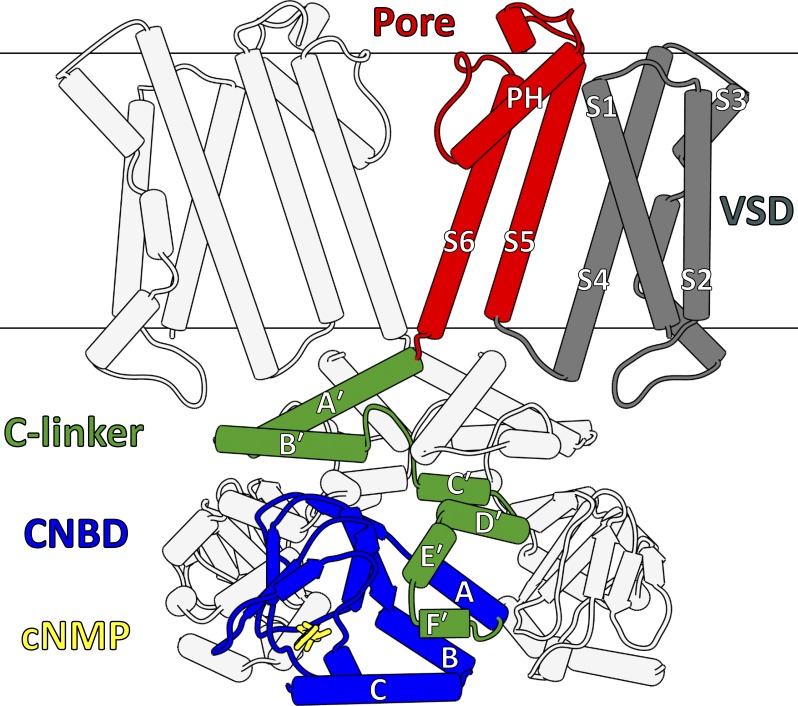
Figure 3.
Core architecture of CNBD channels. For clarity, the diagram omits the transmembrane regions (VSDs, pore domains) of the front and back subunits, along with the CNBD of the back subunit. The core domains of one subunit are highlighted, and include a VSD (gray), a pore domain (red), a C-linker domain (green), and a CNBD (blue). A cyclic nucleotide (cNMP, yellow) is also shown within CNBD binding pocket. All figures depicting protein structure were generated using the UCSF Chimera software package (Pettersen et al., 2004).
Voltage-sensor and pore domains
CNBD channels share structural similarities with the KV1–9, KCa, and transient receptor potential (TRP) channel families (Zheng and Trudeau, 2015). They are tetramers, with each subunit containing six transmembrane helices (S1–S6) divided into a VSD (S1–S4) and a pore domain (S5–S6) connected by an S4–S5 linker. The subunits tetramerize in the membrane through interactions between their pore domains, which associate to create a centrally located pore (i.e., the ion permeation pathway), with the VSDs surrounding and contacting the pore perimeter (Fig. 3). Between S5 and S6 is a short, reentrant helix (the P-helix) followed by residues comprising the ion selectivity filter, which forms the extracellular entrance to the pore. The pore also contains the gate, which stimuli-sensing modules (VSDs, CNBDs) act on to control ion permeation.
The VSD contains two important components for voltage sensing: an S4 helix with positively charged residues (Lys, Arg) distributed along its length, and a “charge-transfer center” composed of an aromatic residue (Phe, Tyr) on S2 and negatively charged residues (Asp, Glu) on S2 and S3. The positively charged residues along S4 are responsible for detecting changes in membrane voltage. The membrane electrical potential exerts force on these residues, drawing them through the charge-transfer center, and the resulting movement of S4 is coupled to the pore gate to control ion permeation (Long et al., 2005a,b, 2007; Tao et al., 2010; Guo et al., 2016; Kintzer and Stroud, 2016).
In KV1–9 channels, the voltage-sensor and pore domains are connected by a long, helical S4–S5 linker (Long et al., 2005a, 2007). This allows the subunits to tetramerize within the membrane in a domain-swapped configuration, in which the pore domain of one subunit interacts with the VSD of an adjacent subunit. Unexpectedly, the S4–S5 linker of CNBD channels instead forms a short loop that permits direct interactions between the VSD and pore domain of the same subunit (Fig. 3). This results in a non–domain-swapped transmembrane architecture, which has important consequences for voltage-dependent gating in these channels. Non–domain-swapped architecture is not unique to CNBD channels and has also been observed recently in the structures of KCa channels Slo1 and Slo2.2 (Hite and MacKinnon, 2017; Tao et al., 2017). Interestingly, the bacterial CNG channel MloK1 (alternatively MlotiK1), which lacks a C-linker domain, appears to adopt a domain-swapped transmembrane architecture like that observed in shaker-type channels (Nimigean et al., 2004; Clayton et al., 2008; Kowal et al., 2014).
C-linker domain
The C-linker resides between the pore domain and the CNBD and consists of two antiparallel helix-turn-helix motifs (A′-B′ and C′-D′), followed by another two helices (E′ and F′) that associate closely with the CNBD (Fig. 3). The C-linker forms intersubunit contacts via “elbow-on-shoulder” interactions, where the A′-B′ (elbow) motif of one subunit rests atop the C′-D′ (shoulder) motif of an adjacent subunit to form a fourfold symmetric ring structure below the pore. The elbow-on-shoulder contacts are maintained by a combination of hydrophobic, hydrogen-bonding, and salt-bridge interactions (Zagotta et al., 2003; Craven and Zagotta, 2004; Craven et al., 2008).
CNBD
The CNBD follows the C-linker and makes close contact with the E′ and F′ helices (Fig. 3). CNBDs are found in many proteins regulated by cyclic nucleotides, including the regulatory subunit of cAMP-dependent protein kinase A (Su et al., 1995) and Escherichia coli catabolite activator protein (McKay et al., 1982). These domains consist of an α-helix (A-helix) followed by an eight-stranded β-roll containing a short α-helix (P-helix) between β6 and β7, followed finally by two additional α-helices (B- and C-helices). The β-roll forms the cyclic nucleotide–binding pocket and contains several residues that form specific electrostatic and hydrogen-bonding interactions with bound cyclic nucleotides, including a highly conserved Arg residue (e.g., Arg549 in human HCN1; Fig. 4) that interacts with the negatively charged cyclic phosphate moiety (Zagotta et al., 2003; Zhou and Siegelbaum, 2007). Upon cyclic nucleotide binding, the B- and C-helices appear to rotate as a single unit (hinged near the B-helix N terminus) toward the β-roll, allowing the C-helix to interact with the base moiety and thereby cap the binding pocket (Varnum et al., 1995; Zagotta et al., 2003; Akimoto et al., 2014; Puljung et al., 2014; Saponaro et al., 2014; Goldschen-Ohm et al., 2016). The β-roll and C-helix play distinct roles in cyclic nucleotide–dependent activation (Tibbs et al., 1998; Matulef et al., 1999), with the β-roll establishing the initial binding affinity of different cyclic nucleotides and the C-helix contributing to the efficacy of channel activation by different cyclic nucleotides (Gordon and Zagotta, 1995; Varnum et al., 1995; Gordon et al., 1996; Flynn et al., 2007; Zhou and Siegelbaum, 2007).
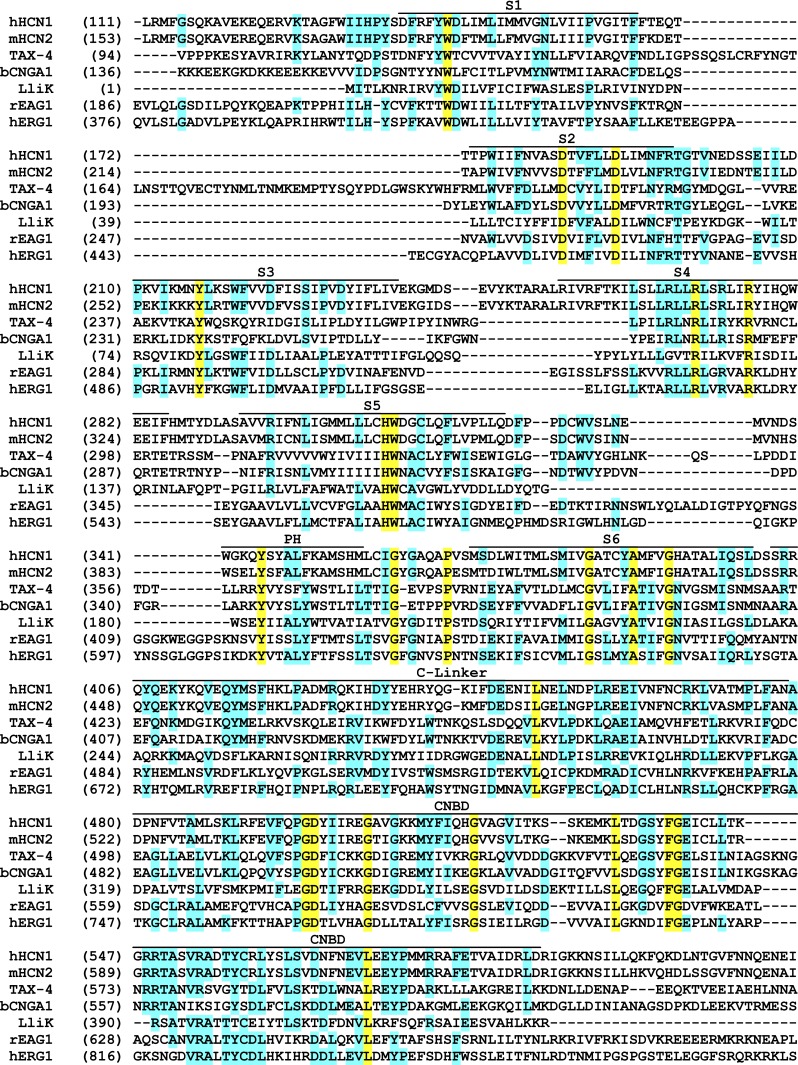
Figure 4.
Protein sequence alignment covering the core architecture of CNBD channels. Alignment includes the five channels discussed in this review (rEAG1, hERG1, hHCN1, TAX-4, and LliK), along with mouse HCN2 (mHCN2) and bovine CNGA1 (bCNGA1). Approximate boundaries of the transmembrane helices (S1–S6), pore helix (PH), and intracellular domains (C-linker, CNBD) are shown above the alignment. Cyan highlighting indicates strong sequence conservation, whereas yellow highlighting indicates complete conservation.
In place of a CNBD, KCNH channels have a structurally similar CNBHD. These domains lack several of the conserved β-roll residues that interact with bound cyclic nucleotides in CNG and HCN channels, most notably the Arg residue (e.g., Ser630 in rat EAG1; Fig. 4) that forms electrostatic interactions with cyclic phosphate moiety. Predictably, KCNH channels do not bind cyclic nucleotides (Brelidze et al., 2009). The β-roll binding pocket in all KCNH channels is instead occupied by a three-residue β-strand motif, Tyr(Phe)-Asn-Leu, immediately following the C-helix (Brelidze et al., 2012, 2013; Marques-Carvalho et al., 2012; Whicher and MacKinnon, 2016; Wang and MacKinnon, 2017). Here, the side-chains of the Tyr(Phe) and Leu residues are oriented into the binding cavity, with the Leu side chain filling the space otherwise occupied by the cyclic phosphate moiety, and the Tyr(Phe) side chain taking the place of the planar base moiety. Thus, the β-strand motif appears to act as an “intrinsic ligand” that mimics the structural features of a bound cyclic nucleotide. Notably, deletion or mutation of the intrinsic ligand alters the functional properties of KCNH channels (Brelidze et al., 2012, 2013; Marques-Carvalho et al., 2012; Zhao et al., 2017).
Diversity in N- and distal C-terminal domains
The N- and C-terminal regions falling outside the core CNBD channel architecture also play important roles in channel function. In addition to the four core domains, KCNH channels possess an N-terminal, intracellular eag domain (Fig. 5, A and B), which consists of a Per-ARNT-Sim (PAS) domain preceded by a PAS-cap motif. The eag domains of KCNH channels appear to interact dynamically with the C-terminal CNBHDs (Gustina and Trudeau, 2009, 2011; Haitin et al., 2013; Dai and Zagotta, 2017) to control the activation, deactivation, and inactivation kinetics of these channels (Terlau et al., 1997; Wang et al., 1998; Gustina and Trudeau, 2011; Gianulis et al., 2013). The intracellular N-terminal regions in CNG and HCN channels also have distinct structural and functional features. For example, the N-terminal regions of the rod CNGB1 and olfactory CNGA2 channel subunits contain an inhibitory Ca2+-calmodulin (Ca2+-CaM) binding site (Munger et al., 2001; Trudeau and Zagotta, 2003; Bradley et al., 2004; Dai and Varnum, 2015). In addition, the recent structure of human HCN1 (discussed in this review) revealed a unique N-terminal “HCN domain” immediately preceding the VSD (Fig. 5 C), although its role in channel function remains unclear (Lee and MacKinnon, 2017).
Figure 5.
Overview of the five CNBD channel structures. For each channel, the core domains of one subunit are colored as in Fig. 3 (VSD in gray, pore domain in red, C-linker domain in green, and CNBD in blue). Additional domains and/or associated regulatory proteins are colored separately, whereas bound cyclic nucleotides or intrinsic ligand residues are shown as yellow sticks. (A) The rEAG1 (PDB ID: 5K7L) channel, with the N-terminal eag domain shown in orange and one bound Ca2+-calmodulin shown in cyan. (B) The hERG1 (PDB ID: 5VA1) channel, with the N-terminal eag domain shown in orange. (C) The hHCN1 channel bound by cAMP (PDB ID: 5U6P), with the N-terminal HCN domain shown in light brown. (D) C. elegans CNG channel (TAX-4, PDB ID: 5H3O) bound by cGMP. (E) Leptospira licerasiae bacterial CNG channel (LliK, PDB ID: 5V4S) in the presence of cAMP.
The distal C-terminal region following the CNBD/CNBHD also harbors many important features. A portion of the distal C terminus following the CNBHD in rat EAG1 forms a coiled-coil domain that is necessary for channel tetramerization (Ludwig et al., 1997). Furthermore, both human and rat EAG1 channels are inhibited by Ca2+-CaM binding, which is partially mediated by two contact points in the C-terminal region following the CNBHD (Schönherr et al., 2000; Ziechner et al., 2006; Whicher and MacKinnon, 2016). In HCN channels, the three most C-terminal residues form one of two binding sites for the regulatory and trafficking protein TRIP8b (Lewis et al., 2009; Santoro et al., 2011; Bankston et al., 2012; DeBerg et al., 2015; Han et al., 2017). In native rod CNG channels, which are heterotetramers composed of three CNGA1 subunits and one CNGB1 subunit (Weitz et al., 2002; Zheng et al., 2002; Zhong et al., 2002), part of the distal C terminus forms a coiled-coil domain that is responsible for controlling subunit stoichiometry (Zhong et al., 2002; Shuart et al., 2011).
Overall summary of structures
Recently, five intact CNBD channel structures have been reported, including at least one structure from each subfamily. Although all five channels possess the same core domains (VSD, pore, C-linker, and CNBD/CNBHD), each structure contains distinct features or additional domains that are summarized here.
Rat EAG1 (rEAG1)
The rEAG1 channel is a K+-selective, depolarization-activated KCNH channel (Ludwig et al., 1994) that is inhibited by the binding of Ca2+-CaM (Schönherr et al., 2000; Ziechner et al., 2006). The structure of detergent-solubilized rEAG1 in complex with Ca2+-CaM was resolved by cryoEM to an overall resolution of 3.8 Å, with local resolution ranging from 3.3 to 6 Å (Whicher and MacKinnon, 2016; Fig. 5 A). The construct used for structure determination lacks residues 773–886 from the distal C-terminal region but is otherwise intact. The rEAG1 structure resolves five domains per subunit, including (from N to C terminus) an eag domain, transmembrane voltage-sensor and pore domains, a C-linker domain, and a CNBHD. The transmembrane domains are non–domain-swapped, with the VSDs adopting an apparently depolarized conformation while the pore is closed (presumably because of the bound Ca2+-CaM). The C-linkers and CNBHDs adopt a quaternary architecture like that observed in the structures of HCN channel C-terminal fragments (Zagotta et al., 2003; Xu et al., 2010; Lolicato et al., 2011). The C-linkers form elbow-on-shoulder contacts to create a fourfold symmetric ring below the pore, with the CNBHDs docking to the intracellular face of this ring. The rEAG1 C-terminal region is surrounded by the N-terminal eag domains, which dock to the outer faces of the CNBHDs. Four Ca2+-CaM molecules are also observed in the structure. Here, the N- and C-lobes of each Ca2+-CaM are separated by ∼12 Å and interact with distinct parts of the channel. The N-lobe binds to a single site on the eag domain, and the C-lobe is gripped by two helices located near the rEAG1 C terminus (Schönherr et al., 2000; Ziechner et al., 2006; Fig. 5 A). Each Ca2+-CaM lobe interacts with two channel subunits, which sit crosswise (rather than adjacent) in the tetrameric assembly.
Human ERG1 (hERG1)
The hERG1 channel is a K+-selective, depolarization-activated KCNH channel that undergoes rapid, voltage-dependent inactivation (Trudeau et al., 1995; Schönherr and Heinemann, 1996). The structure of detergent-solubilized hERG1 was resolved by cryoEM to an overall resolution of 3.8 Å, with local resolution ranging from 3.2 to 5.6 Å (Wang and MacKinnon, 2017; Fig. 5 B). The construct used for structure determination contains two internal deletions, one in the linker region connecting the eag domain to the VSD (residues 141–350) and one in the distal C-terminal region following the CNBHD (residues 871–1005). The structure of a slowly inactivating hERG1 mutant (Ser631Ala; Schönherr and Heinemann, 1996) with a slightly larger internal N-terminal deletion (residues 141–380) was also resolved to an overall resolution of 4.0 Å, although only the nonmutant hERG1 structure will be discussed in this review. The overall structure of hERG1 strongly resembles that of rEAG1: the transmembrane region is non–domain-swapped, the voltage sensors are in an apparently depolarized state, and the intracellular region (eag domains, C-linkers, CNBHDs) adopts a similar quaternary architecture; however, the hERG1 pore appears to be open.
Human HCN1 (hHCN1)
The hHCN1 channel is a weakly K+-selective channel activated by membrane hyperpolarization, with cAMP binding producing a small (∼2 mV) depolarizing shift in the voltage dependence of activation (Wainger et al., 2001). The structure of detergent-solubilized hHCN1 was determined by cryoEM in both the unliganded and cAMP-bound states to an overall resolution of 3.5 Å, with local resolution ranging from 3 to 6.6 Å (Lee and MacKinnon, 2017; Fig. 5 C). The construct used for structure determination lacks residues 636–865 from the distal C-terminal region but is otherwise intact. The unliganded and cAMP-bound structures both resolve four core domains per subunit, including a non–domain-swapped VSD and closed pore domain, followed by a C-linker domain and a CNBD. Additionally, both structures resolve an unexpected fifth domain near the N terminus—the HCN domain—which docks into a crevice between the VSD and the C-linker/CNBD. This domain encompasses ∼45 residues preceding S1 and adopts a unique fold consisting of three short α-helices. Coimmunoprecipitation experiments have identified this region as a potential interaction site between HCN channels and the β2 adrenergic receptor (Greene et al., 2012), suggesting that the HCN domain may help mediate protein–protein interactions in vivo.
TAX-4
The TAX-4 channel is a nonselective CNG channel from C. elegans that displays near-complete activation by cGMP (Paoletti et al., 1999). The structure of TAX-4 bound to cGMP was determined by cryoEM to an overall resolution of 3.5 Å, with local resolution ranging from 3 to 5 Å (Li et al., 2017; Fig. 5 D). The full-length channel reconstituted into amphipol A8-35 was used for structure determination. The overall architecture of TAX-4 is similar to the other channel structures, but the pore domain is open. The C-linkers and CNBDs of TAX-4 adopt a structure similar to the C-terminal fragment of cGMP-bound mouse HCN2 (Zagotta et al., 2003).
LliK
The structure of detergent-solubilized bacterial CNG channel LliK in the presence of 5 mM cAMP was determined to an overall resolution of 4.5 Å, with local resolution in the transmembrane region and C-linker domain averaging 4.2 Å (James et al., 2017; Fig. 5 E). From fluorescence-based flux measurements and homology to known bacterial CNG channels (Brams et al., 2014), LliK is likely a K+-selective CNG channel. The transmembrane region of LliK shares many similarities with the CNBD channels discussed previously, particularly rEAG1 and hERG1. However, the C-linker/CNBD of LliK adopts a quaternary architecture that is unique among the structures discussed here. The CNBDs adopt a compact, cyclic nucleotide–bound conformation but are splayed and rotated away from the channel fourfold symmetry axis. Moreover, the C-linker domains adopt a “flattened” conformation, with both the A′- and B′-helices lying nearly parallel to the membrane surface. At the interface between the LliK pore and C-linker domains, the kink between S6 and the A′-helix occurs six residues later than in the other structures, making S6 considerably longer at its intracellular end and the A′-helix proportionately shorter.
Ion selectivity
CNBD channels range from highly K+-selective to essentially nonselective for monovalent cations. The recent structures support established models for high K+ selectivity, while providing a framework for understanding weak selectivity in HCN channels and nonselectivity in eukaryotic CNG channels.
High selectivity
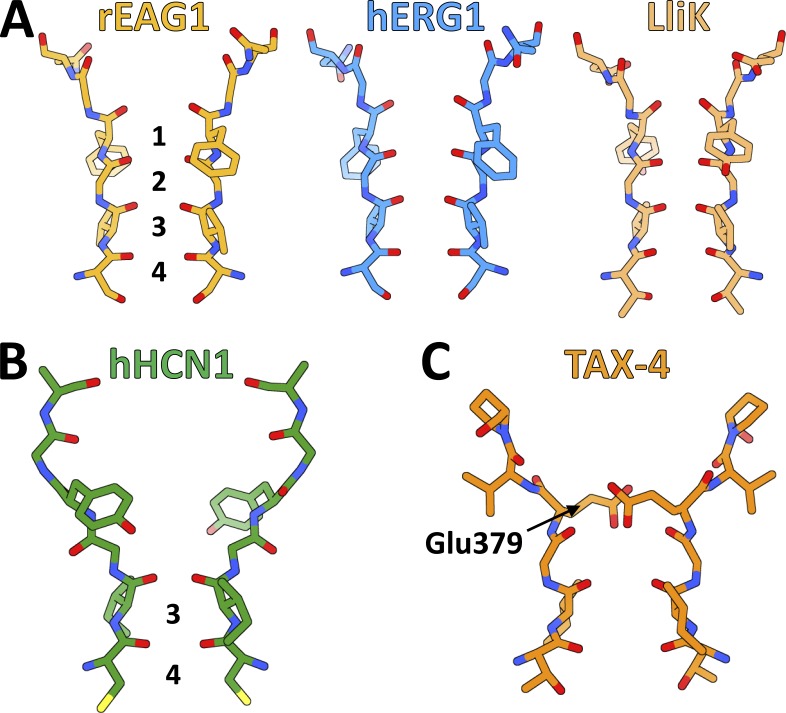
Both rEAG1 and hERG1 are highly selective for K+ over Na+ (Ludwig et al., 1994), whereas LliK seems to exhibit similar selectivity (James et al., 2017). These channels also possess the signature Thr(Ser)-Val-Gly-Tyr(Phe)-Gly motif common to highly K+-selective channels (Fig. 4). Predictably, their selectivity filters all adopt a similar structure (Fig. 6 A) that resembles the selectivity filters found in other K+-selective channels, such as KcsA (Doyle et al., 1998; Zhou et al., 2001) and the KV1.2-2.1 chimera (Long et al., 2007). The rEAG1, hERG1, and LliK selectivity filters are constructed such that the backbone carbonyl groups, along with a Ser(Thr) side-chain hydroxyl group, are oriented toward the permeation pathway. In the KcsA and KV structures, this creates a series of four cation binding sites (1–4) with the proper geometry to “hydrate” de-solvated K+ ions (but not Na+ ions) as they pass through the filter (Zhou et al., 2001).
Figure 6.
Selectivity filters of CNBD channels. (A) Stick representations of selectivity filters from the highly K+-selective rEAG1, hERG1, and LliK channels. Cation-binding sites 1–4, composed of backbone carbonyl groups and Ser(Thr) side chains, are labeled in the rEAG1 selectivity filter. (B) Stick representation of the weakly selective hHCN1 selectivity filter, with retained cation binding sites 3 and 4 labeled. (C) Stick representation of the nonselective TAX-4 selectivity filter, with highly conserved residue Glu379 labeled.
Weak selectivity
HCN channels also possess the Gly-Tyr(Phe)-Gly motif (Fig. 4) but are only weakly selective for K+ over Na+ ions. The hHCN1 structure provides insight into this unexpected functional property. Compared with the selectivity filters in rEAG1, hERG1, and LliK (Fig. 6 A), the extracellular half of the hHCN1 selectivity filter is distorted (Fig. 6 B), with the conserved Tyr side chain (Tyr361 in hHCN1) rotated nearly 180° relative to its counterparts in the highly K+-selective channels (Fig. 6 A). As a consequence, the backbone carbonyl groups flanking the Tyr side-chain are directed away from the permeation pathway. In addition, the hHCN1 selectivity filter is considerably wider at its extracellular end, completely disrupting the first two cation binding sites (1 and 2). Mutational and structural studies of the nonselective bacterial channel NaK indicate that at least four sequential cation-binding sites are necessary for high K+ selectivity (Derebe et al., 2011a,b), suggesting that weak selectivity in hHCN1 results from disruption of the first and second cation-binding sites.
Nonselective
Eukaryotic CNG channels are nonselective for monovalent cations and also allow divalent cations such as Ca2+ to permeate, although Ca2+ (and Mg2+) can block these channels at high concentrations (Dzeja et al., 1999). Unlike the other CNBD channel filters, the TAX-4 selectivity filter is relatively wide throughout (Fig. 6 C), possibly allowing its binding sites to accommodate a variety of cations in different hydration states. The extracellular entrance of the TAX-4 selectivity filter is also constricted by a highly conserved Glu residue (Glu379; Fig. 4) that has been found to contribute to divalent cation and proton block in other eukaryotic CNG channels (Root and MacKinnon, 1993, 1994), and also likely contributes to their poor cation selectivity (Napolitano et al., 2015). In the TAX-4 structure, this residue is well situated to impede ion permeation after protonation or divalent cation binding.
Channel gating mechanisms
Stimuli-sensing domains (e.g., VSDs, CNBDs) control ion permeation through CNBD channels by acting on a gate, located in the pore domain. The recent structures provide insight into the conformational changes each domain undergoes in the process of gating (i.e., pore opening) or stimuli detection (e.g., cyclic nucleotide binding) and how these processes are coupled to one another.
Modular scheme for gating
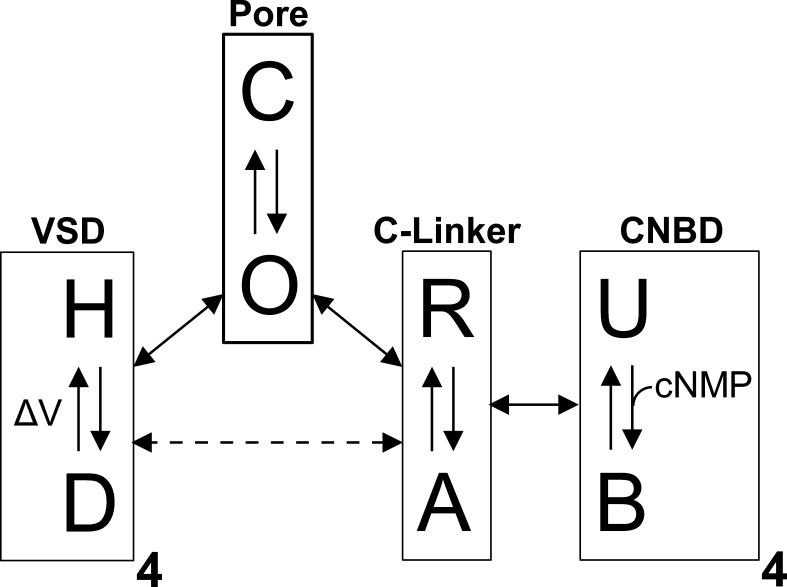
Channels may be conceptualized as a series of energetically coupled “modules,” each capable of transitioning between different conformations (Horrigan and Aldrich, 2002; Craven and Zagotta, 2004; DeBerg et al., 2016). The conformation adopted by one module influences the favorability of an associated module adopting a particular conformation (Fig. 7). For example, the pore functions as a module that transitions between closed (impermeant) and open (permeant) states. The VSDs, meanwhile, act as modules that transition between hyperpolarized and depolarized states in response to changes in membrane voltage. When occupying the depolarized state, the VSDs of depolarization-activated channels increase the favorability of the pore adopting the open state. This coupling is apparently inverted in HCN channels, where the VSD depolarized state promotes channel closure. Similarly, the CNBDs may be treated as ligand-sensing modules that transition between unliganded and ligand-bound states. The C-linker domains, which connect the CNBDs to the pore, function as “transduction modules,” with resting and active states that are influenced by the conformation of the CNBDs. The C-linkers, in turn, influence the state of the pore, and thus serve to energetically couple the CNBDs to the pore. The C-linkers may also directly influence the state of the VSDs, perhaps because of interactions between the C-linker and the S4–S5 linker (Lee and MacKinnon, 2017). Stimuli-sensing modules can be either activating or inhibitory with respect to the pore. For example, the C-linker/CNBD of HCN channels appears to inhibit pore opening when in the resting, unliganded state (Wainger et al., 2001). In truncation mutants whose C-terminal region is deleted, HCN channels exhibit a depolarizing shift in their voltage dependence of activation, similar to the effect of cyclic nucleotide binding to the intact channel.
Figure 7.
Modular gating scheme for ion channel function. CNBD channels may be treated as a collection of energetically coupled modules, each capable of independently transitioning between different states. The VSDs can adopt hyperpolarized (H) or depolarized (D) states; the CNBDs can adopt unbound (U) or bound (B) states; the C-linker can adopt a resting (R) or active (A) state; and the pore can adopt a closed (C) or open (O) state. The conformation adopted by a given module influences the favorability of connected modules occupying a particular state. Energetic coupling between modules is indicated by the double-headed arrows, and equilibration of a module between different conformational states is indicated by the single-headed arrows.
Gating of the pore
Mutagenesis and accessibility studies have established that the ion permeation gate in HCN and KCNH channels resides near the intracellular end of the pore and is composed of S6 residues (Shin et al., 2001; Rothberg et al., 2002, 2003; Thouta et al., 2014; Fig. 8, A and B). Permeation is controlled through dilation of the gate, which must open sufficiently to accommodate hydrated cations. In HCN channels, which undergo both voltage-dependent and cyclic nucleotide–dependent activation, these processes appear to act on the same gate, rather than separate gates (Shin et al., 2004). CNG channels are distinct in that they lack an intracellular ion permeation gate, despite this part of their pore undergoing dilation during channel activation (Flynn and Zagotta, 2001, 2003; Contreras and Holmgren, 2006; Contreras et al., 2008). What structural changes occur in the pore to gate ion permeation, and how do these changes compare between the different subfamilies? Comparison of the closed- and open-state CNBD channel structures helps address these questions.
Figure 8.
Comparison of pore domains in CNBD channels. (A) Structural alignment of rEAG1 and hERG1 pore domains, with intracellular gating residues (Gln476 in rEAG1; Gln664 in hERG1) labeled and shown as sticks. VSDs from one subunit are also shown. Arrows highlight the relative displacement of the hERG1 S5 and S6 helices toward the VSD to permit dilation of the intracellular gate. (B) Structural alignment of the rEAG1 and hHCN1 pore domains, with gating residues (Gln476 in rEAG1; Val390, Thr394, and Gln398 in hHCN1) shown as sticks and overlapping gating residues (Gln476 in rEAG1; Gln398 in hHCN1) highlighted by a dashed circle. (C) Structural alignment of the hHCN1 and LliK pore domains, with gating residues (Tyr224 and Ile228 in LliK; Val390, Thr394, and Gln398 in hHCN1) shown as sticks, and overlapping gating residues (Val390 in hHCN1, Ile228 in LliK) highlighted by a dashed circle. (D) Structural alignment of hERG1 and TAX-4 pore domains, with the hERG1 gating residue Gln664 shown as sticks.
Comparison of open and closed channel structures
Of the five CNBD channel structures, three (rEAG1, hHCN1, LliK) have permeation pathways that are tightly constricted by one or more S6 residues (Fig. 8, A–C), consistent with a closed state. In rEAG1, which is in complex with inhibitory Ca2+-CaM, an intracellular gate is clearly formed by S6 residue Gln476 (Thouta et al., 2014; Fig. 8, A and B), which constricts the pore diameter to <1 Å. In hHCN1, which is closed in the absence of a hyperpolarizing potential, the pore is constricted to 1–2-Å diameter by three residues (Val390, Thr394, and Gln398) lining one face of S6 (Fig. 8, B and C). As expected, these constriction points lie along the intracellular half of S6, with the most C-terminal point (Gln398) aligning with the Gln476 gating residue in rEAG1 (Fig. 8 B). The LliK pore is unusual in that its two constricting S6 residues (Tyr224 and Ile228) reside near the membrane center, with the more C-terminal residue (Ile228) aligning with the most N-terminal constriction point in hHCN1 (Val390; Fig. 8 C). The closed permeation pathway in LliK is also unexpected, as this structure was determined in the presence of saturating cAMP concentrations.
The two remaining CNBD channel structures (hERG1 and TAX-4) are dilated at the intracellular end of their pore (Fig. 8, A and D), consistent with an open state. Dilation is made possible by a kink in S6, which occurs at a conserved Gly “hinge” residue (Gly648 in hERG1, Gly399 in TAX-4) located just below the selectivity filter. Relative to rEAG1, the entire permeation pathway in hERG1 below the selectivity filter is substantially wider (Wang and MacKinnon, 2017), particularly at the conserved Gln residue (Gln664 in hERG1), whose counterpart in rEAG1 serves as the gate (Fig. 8 A). Comparison of hERG1 and rEAG1 offers a simple model for what structural changes occur in the pore during channel activation. In the closed (rEAG1) state, the S6 helices are relatively straight, bringing the Gln gating residues into close proximity (<1-Å pore diameter). In transitioning to the open (hERG1) state, the S6 helices develop a kink at the conserved Gly hinge residue, allowing them (along with the intracellular half of S5) to bend away from the permeation pathway and toward the VSDs. Consequently, the gating residues are moved apart, dilating the pore in this region to ∼11-Å diameter, more than sufficient to accommodate hydrated cations. The pore domains of hHCN1 and LliK also possess the conserved Gly hinge residue (Gly382 in hHCN1, Gly220 in LliK) and may undergo a similar structural change to control ion permeation.
Gating at the selectivity filter
Although dilation at the intracellular end of the pore is associated with activation in eukaryotic CNG channels, it does not directly control ion permeation (Flynn and Zagotta, 2001, 2003; Contreras and Holmgren, 2006). Instead, the selectivity filter itself appears to act as the gate in these channels (Contreras et al., 2008). In TAX-4, the intracellular ends of the S6 helices are bent away from the symmetry axis, similar to hERG1 (Fig. 8 D), suggesting that the TAX-4 structure indeed represents an open state. However, in the absence of an unliganded, closed-state structure, it remains unclear what changes the selectivity filter undergoes to control ion permeation or how cyclic nucleotide binding induces these changes. Scanning cysteine accessibility measurements in the bovine rod CNG channel suggest that the pore helix rotates along its axis during activation (Liu and Siegelbaum, 2000), although it is unknown how this motion would influence the structure of the selectivity filter.
Voltage-dependent gating
In KV1–9 channels, where the transmembrane region is domain-swapped, voltage-dependent gating is likely mediated by the helical S4–S5 linker, which runs parallel to the membrane surface and contacts the pore-lining S6 helix (Long et al., 2007). Upon membrane depolarization, S4 gating residues pass through the charge-transfer center as S4 moves up toward the membrane extracellular surface (Guo et al., 2016; Kintzer and Stroud, 2016). This motion is thought to be coupled to movement of the S4–S5 linker and consequently S6, forcing (or permitting) S6 to bend at a conserved Pro-Val-Pro “hinge” motif to open the intracellular gate (Long et al., 2005b). The transmembrane region of CNBD channels differs from KV1–9 channels in several respects: (a) the transmembrane region is non-domain-swapped; (b) the S4–S5 linker is a short loop that does not interact extensively with S6; and (c) the S6 helix lacks the Pro-Val-Pro hinge motif. Together, these differences indicate that CNBD channels use a different mechanism to couple voltage sensing to pore gating.
Depolarization activation
The VSDs of rEAG1 are structurally similar to their counterparts in KV1–9 channels. In rEAG1, S4 consists of a short 310 helix flanked by α-helical components at either end (Fig. 9 A) and contains six basic residues: Lys327 (K1), Arg330 (R2), Arg333 (R3), Arg336 (R4), Arg339 (R5), and Lys340. A charge-transfer center is also observed within the rEAG1 VSD, composed of residues Phe261 and Asp264 on S2 and Asp299 on S3 (Fig. 9 A). R4 resides within the charge-transfer center, whereas K1–R3 lie above, near the extracellular face of the channel. Mutational studies of closely related hERG1 have demonstrated that only K1–R3 are required for voltage-dependent gating (Zhang et al., 2004), and because these residues sit above the charge-transfer center in rEAG1, its VSDs likely adopt a depolarized conformation. However, because of the inhibitory effects of Ca2+-CaM, the pore remains closed. The VSDs of hERG1 are virtually identical to those of rEAG1, including the presence of a charge-transfer center composed of S2 and S3 residues, along with position and orientation of the S4 gating residues (Fig. 9 A). As such, its VSDs are also likely in a depolarized conformation, and the hERG1 intracellular gate is open.
Figure 9.
Voltage-sensor domains of CNBD channels. In top panels, the C-terminal portion of S3 is omitted for clarity, and S2 residues (Phe, Asp) contributing to the charge-transfer center and basic residues along S4 (Arg, Lys) are shown as sticks. In bottom panels, the VSDs are rotated by 45° relative to the top panels, with the S4 helices individually colored. (A) Structurally aligned VSDs of rEAG1 and hERG1, with rEAG1 charge-transfer center residues Phe261 and Asp264, along with residue Arg336 (R4) occupying the charge-transfer center, labeled in the top panel. (B) VSD of hHCN1, with residue Arg273 (R5) occupying the charge-transfer center labeled in the top panel. (C) VSD of TAX-4, with the three regions of S4 (S4a, S4b, and S4c) labeled in the bottom panel. (D) VSD of LliK.
Comparison of rEAG1 and hERG1 suggests a possible mechanism for depolarization activation in CNBD channels (Wang and MacKinnon, 2017). In both channels, the S5 and S6 helices form extensive contacts along their entire lengths, whereas S4 and S5 interact at their intracellular ends (Fig. 8 A). During voltage-dependent activation, S4 translates vertically through the membrane (Long et al., 2005b, 2007; Guo et al., 2016), and given the S4–S5 and S5–S6 interactions, S4 movement could be communicated indirectly via S5 to the intracellular end of S6 (near the permeation gate). Within this framework, two possibilities arise. The VSD could function as an inhibitory module where, in the hyperpolarized state, S4 exerts force on S5 (and consequently S6) to hold the permeation gate closed. Alternatively, the VSD could behave as an activating module where, in the depolarized state, it exerts force on S5–S6 to pull the gate open. In both cases, no covalent connection between S4 and S5 is required, as it is the interaction between S4 and S5 (rather than their covalent linkage) that communicates force between the VSD and the pore. This could explain the ability of “split” KCNH channels, where the S4–S5 linker is severed, to assemble into functional, voltage-gated channels (Lörinczi et al., 2015; Tomczak et al., 2017). Notably, splitting EAG1 near the C-terminal end of S4 produces a constitutively open channel, arguing in favor of the VSD acting as an inhibitory module (Tomczak et al., 2017).
Hyperpolarization activation
The VSDs of HCN channels behave like their counterparts in depolarization-activated channels, with S4 moving up toward the membrane extracellular surface upon depolarization and down toward the intracellular surface upon hyperpolarization (Männikkö et al., 2002; Vemana et al., 2004). The VSDs of hHCN1 are also structurally similar to the VSDs of depolarization-activated rEAG1 and hERG1, with the exception that the S4 helix in hHCN1 is considerably longer, extended on its intracellular end by several α-helical turns (Fig. 9 B). The charge-transfer center of hHCN1 is also occupied by voltage-sensing residue R5 (Arg273), rather than R4 as in the rEAG1 and hERG1 structures (Fig. 9, A and B). As in rEAG1 and hERG1, the VSDs in hHCN1 likely adopt a depolarized conformation, probably because of the lack of an electrical potential, which for HCN channels is associated with channel closure. Given these similarities, what is the structural basis for inverse gating in HCN channels? Why is the depolarized VSD conformation in hHCN1 coupled to pore closure, when a similar conformation is coupled to pore opening in rEAG1 and hERG1? One possibility is that inverse gating involves interactions between S4 (or the S4–S5 linker) and the C-linker elbow motif below it (Prole and Yellen, 2006; Kwan et al., 2012; Lee and MacKinnon, 2017). However, HCN channels lacking the C-linker and CNBD still exhibit hyperpolarization activation (Wainger et al., 2001). Without a hyperpolarized-state HCN channel structure, it remains unclear how S4 translation toward the intracellular membrane surface would result in opening of the pore.
Voltage insensitivity
TAX-4 is largely insensitive to the membrane potential (Fig. 2 A; Paoletti et al., 1999), despite its VSDs sharing many structural features with those found in voltage-gated CNBD channels (Fig. 9 C). In particular, the TAX-4 VSDs contain a charge-transfer center composed of residues Tyr215 (S2), Asp218 (S2), and Asp253 (S3), which is occupied by an S4 basic residue (Arg286). Other basic residues are found along S4 both above (Arg280, Arg283) and below (Lys288, Arg289, Arg291, Arg296) the charge-transfer center. However, the S4 helix of TAX-4 is uniquely structured (Fig. 9 C) and can be divided into three distinct regions: an N-terminal loop (S4a), a central 310 helix (S4b), and a C-terminal α-helix (S4c). Between S4b and S4c, the helix kinks sharply, allowing S4c to lie nearly parallel to the membrane surface. Voltage insensitivity could be a consequence of the unusual, segmented nature of S4, which might prevent it from translating vertically through membrane in response to changes in the membrane potential. However, substitution of the Drosophila EAG channel S4 helix with its counterpart from the rat olfactory CNG channel produced a functioning, voltage-dependent channel (Tang and Papazian, 1997), suggesting that VSD elements besides S4 contribute to voltage insensitivity in CNG channels.
The VSDs of LliK share a common architecture with other CNBD channel VSDs (Fig. 9 D). The LliK VSDs appear to adopt a depolarized conformation, with S4 containing four conserved basic residues (Arg122, Lys125, Arg128, and Arg135). Structurally, the S4 helices of LliK appear most similar to those of rEAG1, and contain a central 310-helical segment flanked by α-helical segments at either end. The LliK S4 helix also kinks slightly near its C terminus, although not to the same extent as in TAX-4. LliK is putatively voltage insensitive, based on its homology to other bacterial CNG channels (Brams et al., 2014). However, the electrophysiological properties of LliK have not yet been characterized, which may reveal unexpected voltage dependence.
Cyclic nucleotide–dependent gating
Before the determination of intact CNG and HCN channel structures, our understanding of cyclic nucleotide–dependent activation was based primarily on HCN channel C-terminal fragments (Zagotta et al., 2003; Xu et al., 2010; Akimoto et al., 2014; Puljung et al., 2014; Saponaro et al., 2014; Goldschen-Ohm et al., 2016). These structures revealed that upon cyclic nucleotide binding to the β-roll, the B- and C-helices rotate as a single unit toward the binding pocket, allowing the C-helix to interact with the cyclic nucleotide base moiety. However, the fragment structures fail to reveal any large-scale changes in the C-linker domain between the apo and ligand-bound states, making it unclear how cyclic nucleotide binding is communicated to the pore domain to control ion permeation. The recent intact CNBD channel structures support a mechanism wherein the entire C-terminal region rotates relative to the pore to control opening of the permeation gate.
Comparison of intact channels to fragment structures
The structure of hHCN1 was determined in both the unliganded and cAMP-bound states (Lee and MacKinnon, 2017), and most of the structural differences between these two states are localized to the CNBDs. Here, the B- and C-helices are rotated away from β-roll in the unliganded state (Fig. 10 A), similar to the apo structures of isolated CNBDs from human HCN2 (Saponaro et al., 2014), mouse HCN2 (Goldschen-Ohm et al., 2016), and human HCN4 (Akimoto et al., 2014). In the cAMP-bound structure, which is nearly identical to the fragment structure of cAMP-bound mouse HCN2 (Fig. 10 B; Zagotta et al., 2003), these helices are moved toward the β-roll cavity to permit interaction between the C-helix and cAMP, which is docked into the β-roll in an anti configuration (Fig. 10 A). In the cAMP-bound hHCN1 structure, two additional helices (D- and E-helix) were also observed following the C-helix (not depicted), which interact with both the C-helix and the β-roll. The C-helix and nearby sites on the β-roll, along with residues at the very C terminus of HCN, form interactions with the trafficking protein TRIP8b (Santoro et al., 2011; Saponaro et al., 2014; DeBerg et al., 2015; Han et al., 2017), although it is unclear what role the D- and E-helices play in TRIP8b binding.
Figure 10.
Comparison of intracellular domains. (A) Structural alignment of unliganded (PDB ID: 5U6O) and cAMP-bound hHCN1 CNBDs, with cAMP shown in yellow sticks. Arrows highlight the rotation of the C-helix toward the β-roll after cyclic nucleotide binding, along with upward movement of the C-linker E′- and F′-helices. (B) Structural alignment of the cAMP-bound CNBDs from hHCN1 and mouse HCN2 (PDB ID: 1Q5O), which adopt essentially identical conformations. Bound cAMP shown in yellow sticks. (C) Structural alignment of the cAMP-bound hHCN1 CNBD and the rEAG1 CNBHD. Bound cAMP is shown in yellow sticks, and the rEAG1 intrinsic ligand residues (Tyr672-Asn673-Leu674) are shown as sticks colored like the rEAG1 ribbon structure. (D) Structural alignment of the rEAG1 eag domain (orange) + CNBHD (blue, from an adjacent subunit) with the cocrystal structure of the mEAG1 eag domain + CNBHD (gray, PDB ID: 4LLO). Intrinsic ligand residues (yellow for rEAG1, gray for mEAG1) are shown as sticks.
As with HCN channels, the structures of KCNH channel intracellular fragments (i.e., eag domain, C-linker/CNBHD) correctly predicted many of the features observed in the rEAG1 and hERG1 structures. For example, the fragment structures correctly predicted the presence of an intrinsic ligand within the β-roll binding pocket that mimics a bound cyclic nucleotide (Brelidze et al., 2012, 2013; Marques-Carvalho et al., 2012; Fig. 10, C and D), as well as the interaction between the N-terminal eag domain and the C-terminal CNBHD (Haitin et al., 2013; Whicher and MacKinnon, 2016; Wang and MacKinnon, 2017; Fig. 10 D).
Cyclic nucleotide–dependent activation
Both the fragment and full-length HCN channel structures show that cyclic nucleotide binding to the β-roll causes the B- and C-helices to rotate as a rigid body, bringing the C-helix into contact with the base moiety (Fig. 10 A). How is this structural change within the CNBD communicated to the C-linker? The recent hHCN1 channel structures provide insight into this mechanism. As in the HCN channel fragment structures (Zagotta et al., 2003; Xu et al., 2010; Lolicato et al., 2011), the C-linker E′- and F′-helices are closely associated with the CNBDs in hHCN1, stacking atop the B- and C-helices. Rotation of the B- and C-helices toward the β-roll may exert an upward force on the E′- and F′-helices, moving them closer to the elbow-on-shoulder gating ring beneath the pore (Fig. 10 A). The C-linker/CNBD of HCN channels acts as an inhibitory domain that maintains the pore in a closed state (Wainger et al., 2001), and force exerted by the E′- and F′-helices may alter the structure of the gating ring to relieve this inhibition.
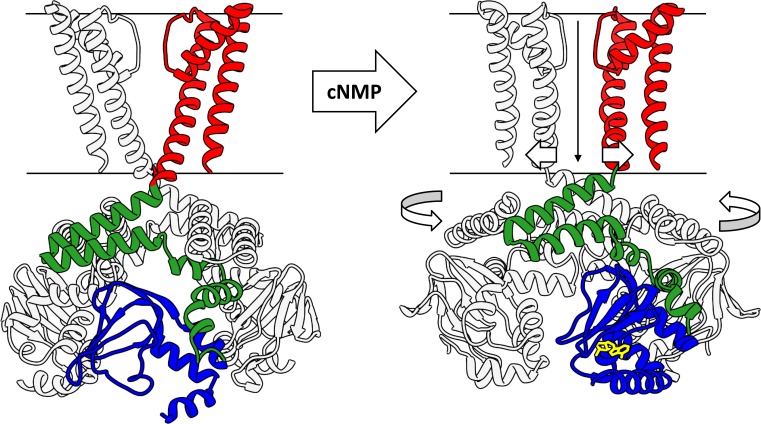
What structural changes does the C-linker gating ring undergo to influence the pore? Comparison of the unliganded and cAMP-bound hHCN1 structures reveals a subtle, counterclockwise rotation (viewed top-down) of the intracellular domains relative to the transmembrane region (Fig. 11 A). This rotation is in opposition to the right-handed twist of the S6 helical bundle, suggesting that further rotation could dilate the bundle near the intracellular gate to allow ion permeation. Comparison of the rEAG1 and hERG1 structures (which possess closed and open intracellular gates, respectively) reveals a more dramatic counterclockwise rotation of the intracellular domains (Fig. 11 B), consistent with the structural change suggested by the unliganded and cAMP-bound hHCN1 structures (Fig. 11 A). Together, these results support a “twist-open” model of channel regulation (Fig. 12), where binding of cyclic nucleotides to the CNBDs induces a large-scale rotation of the intracellular region that unwinds the right-handed S6 helical bundle to dilate the intracellular gate. In the hHCN1 structures, where the cAMP-bound intracellular domain is rotated only slightly relative to its unliganded counterpart, the pore is presumably held closed by the depolarized-state VSDs, which could, in turn, prevent the cAMP-bound intracellular domains from rotating further. Rotation of the intracellular domains has also been proposed as a mechanism for coupling cyclic nucleotide binding to pore gating in CNG channels (Wainger et al., 2001), suggesting that all three CNBD subfamilies use this mechanism for coupling their C-terminal region to the pore.
Figure 11.
Rotation of the C-terminal domains associated with CNBD channel gating. (A) Top-down view of the unliganded and cAMP-bound hHCN1 structures aligned by their pore domains, with the view clipped to highlight the C-linker domains. Alignment reveals a subtle counterclockwise rotation of the C-linker/CNBD in cAMP-bound hHCN1, relative to the apo structure. (B) Top-down view of the rEAG1 (closed gate) and hERG1 (open gate) structures aligned by their pore domains, with the view clipped as in A. Alignment reveals a dramatic counterclockwise rotation of the hERG1 C-linker/CNBHD relative to rEAG1.
Figure 12.
Proposed mechanism for cyclic nucleotide–dependent activation of CNBD channels. In the absence of cyclic nucleotides (left), the C-linker/CNBD adopts a resting state that exerts a tonic inhibitory force on the pore domain, maintaining it in the closed state. Cyclic nucleotide binding to the CNBDs promotes the transition of the C-linker to its active state, relieving this inhibition. The dis-inhibited pore domain can now dilate at the intracellular gate, inducing a counterclockwise rotation of the C-terminal domains.
Open questions
The recent structures of CNBD channels provide insight into mechanisms of ion selectivity, voltage-dependent gating, and ligand-dependent gating within this channel family. In particular, the structures suggest a mechanistic framework for depolarization activation in rEAG1 and hERG1 that involves direct interactions between the S4, S5, and S6 helices, made possible by the non–domain-swapped transmembrane architecture adopted by these channels. For ligand-dependent activation, comparison of the apo and cAMP-bound hHCN1 structures, together with the rEAG1 and hERG1 structures, suggest a twist-open mechanism wherein cyclic nucleotide binding is communicated via the C-linker E′- and F′-helices to the elbow-on-shoulder gating ring to relieve the tonic, inhibitory force it exerts on the intracellular gate. However, the following questions remain unanswered.
CNG channel gating at the selectivity filter
Mammalian CNG channels do not use an intracellular gate to control ion permeation, and instead the selectivity filter appears to be responsible for obstructing current flow. If CNG channels use the twist-open mechanism, it is unclear how rotation of the intracellular domains is propagated up to the selectivity filter to gate the channel.
HCN channel hyperpolarization activation
It remains unclear how downward movement of S4 upon membrane hyperpolarization would influence the S5 and S6 helices to activate the channel, and whether interactions between the VSD and the C-linker (as observed in the hHCN1 structure) play a role in hyperpolarization activation.
KNCH channel regulation by the PAS domain
The N-terminal eag domain of KCNH channels has profound effects on the activation and deactivation properties of these channels. The rEAG1 and hERG1 channel structures have confirmed that the eag domains interact directly with the C-terminal CNBHDs, although it is not clear how this interaction influences channel gating.
Conclusions
Although high-resolution structures offer valuable insight into channel architecture, allowing for the rationalization of other biophysical data (e.g., mutagenesis and accessibility data) and the creation of detailed mechanistic hypotheses, they are effectively static channel “snapshots” that provide little energetic information. Understanding the coupling between channel modules, along with the energetics of conformational changes within each module, is crucial to understanding the mechanism of CNBD channel function. To this end, electrophysiological measurements provide a wealth of functional and energetic information, although it is often difficult to interpret these results in terms of channel structure. Biophysical approaches that yield both structural and energetic information, particularly spectroscopic techniques such as fluorescence-resonance energy transfer and double electron-electron resonance (DEER) spectroscopy, will prove valuable for developing truly comprehensive models for channel function.
Acknowledgments
We gratefully acknowledge Dr. Galen Flynn for providing the electrophysiological data shown in Fig. 2 and for critically reading the manuscript. We also acknowledge Drs. Jacob Morgan and Gucan Dai for critically reading the manuscript.
This publication was supported by National Eye Institute grant R01EY010329 (to W.N. Zagotta) and National Institute of Mental Health grant R01MH102378 (to W.N. Zagotta). Molecular graphics were generated with the UCSF Chimera software package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by the National Institute of General Medical Sciences grant P41GM103311).
The authors declare no competing financial interests.
Lesley C. Anson served as editor.
References
- Akimoto M., Zhang Z., Boulton S., Selvaratnam R., VanSchouwen B., Gloyd M., Accili E.A., Lange O.F., and Melacini G.. 2014. A mechanism for the auto-inhibition of hyperpolarization-activated cyclic nucleotide-gated (HCN) channel opening and its relief by cAMP. J. Biol. Chem. 289:22205–22220. 10.1074/jbc.M114.572164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altomare C., Terragni B., Brioschi C., Milanesi R., Pagliuca C., Viscomi C., Moroni A., Baruscotti M., and DiFrancesco D.. 2003. Heteromeric HCN1-HCN4 channels: a comparison with native pacemaker channels from the rabbit sinoatrial node. J. Physiol. 549:347–359. 10.1113/jphysiol.2002.027698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.A., Huprikar S.S., Kochian L.V., Lucas W.J., and Gaber R.F.. 1992. Functional expression of a probable Arabidopsis thaliana potassium channel in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 89:3736–3740. 10.1073/pnas.89.9.3736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankston J.R., Camp S.S., DiMaio F., Lewis A.S., Chetkovich D.M., and Zagotta W.N.. 2012. Structure and stoichiometry of an accessory subunit TRIP8b interaction with hyperpolarization-activated cyclic nucleotide-gated channels. Proc. Natl. Acad. Sci. USA. 109:7899–7904. 10.1073/pnas.1201997109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benarroch E.E. 2013. HCN channels: Function and clinical implications. Neurology. 80:304–310. 10.1212/WNL.0b013e31827dec42 [DOI] [PubMed] [Google Scholar]
- Biel M., Wahl-Schott C., Michalakis S., and Zong X.. 2009. Hyperpolarization-activated cation channels: From genes to function. Physiol. Rev. 89:847–885. 10.1152/physrev.00029.2008 [DOI] [PubMed] [Google Scholar]
- Bradley J., Bönigk W., Yau K.-W., and Frings S.. 2004. Calmodulin permanently associates with rat olfactory CNG channels under native conditions. Nat. Neurosci. 7:705–710. 10.1038/nn1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brams M., Kusch J., Spurny R., Benndorf K., and Ulens C.. 2014. Family of prokaryote cyclic nucleotide-modulated ion channels. Proc. Natl. Acad. Sci. USA. 111:7855–7860. 10.1073/pnas.1401917111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelidze T.I., Carlson A.E., and Zagotta W.N.. 2009. Absence of direct cyclic nucleotide modulation of mEAG1 and hERG1 channels revealed with fluorescence and electrophysiological methods. J. Biol. Chem. 284:27989–27997. 10.1074/jbc.M109.016337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelidze T.I., Carlson A.E., Sankaran B., and Zagotta W.N.. 2012. Structure of the carboxy-terminal region of a KCNH channel. Nature. 481:530–533. 10.1038/nature10735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelidze T.I., Gianulis E.C., DiMaio F., Trudeau M.C., and Zagotta W.N.. 2013. Structure of the C-terminal region of an ERG channel and functional implications. Proc. Natl. Acad. Sci. USA. 110:11648–11653. 10.1073/pnas.1306887110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüggemann A., Pardo L.A., Stühmer W., and Pongs O.. 1993. Ether-à-go-go encodes a voltage-gated channel permeable to K+ and Ca2+ and modulated by cAMP. Nature. 365:445–448. 10.1038/365445a0 [DOI] [PubMed] [Google Scholar]
- Burns M.E., Mendez A., Chen J., and Baylor D.A.. 2002. Dynamics of cyclic GMP synthesis in retinal rods. Neuron. 36:81–91. 10.1016/S0896-6273(02)00911-X [DOI] [PubMed] [Google Scholar]
- Clayton G.M., Altieri S., Heginbotham L., Unger V.M., and Morais-Cabral J.H.. 2008. Structure of the transmembrane regions of a bacterial cyclic nucleotide-regulated channel. Proc. Natl. Acad. Sci. USA. 105:1511–1515. 10.1073/pnas.0711533105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras J.E., and Holmgren M.. 2006. Access of quaternary ammonium blockers to the internal pore of cyclic nucleotide-gated channels: Implications for the location of the gate. J. Gen. Physiol. 127:481–494. 10.1085/jgp.200509440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras J.E., Srikumar D., and Holmgren M.. 2008. Gating at the selectivity filter in cyclic nucleotide-gated channels. Proc. Natl. Acad. Sci. USA. 105:3310–3314. 10.1073/pnas.0709809105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven K.B., and Zagotta W.N.. 2004. Salt bridges and gating in the COOH-terminal region of HCN2 and CNGA1 channels. J. Gen. Physiol. 124:663–677. 10.1085/jgp.200409178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven K.B., and Zagotta W.N.. 2006. CNG and HCN channels: Two peas, one pod. Annu. Rev. Physiol. 68:375–401. 10.1146/annurev.physiol.68.040104.134728 [DOI] [PubMed] [Google Scholar]
- Craven K.B., Olivier N.B., and Zagotta W.N.. 2008. C-terminal movement during gating in cyclic nucleotide-modulated channels. J. Biol. Chem. 283:14728–14738. 10.1074/jbc.M710463200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai G., and Varnum M.D.. 2015. Cyclic Nucleotide-Gated Channels. In The Handbook of Ion Channels. Zheng J. and Trudeau M.C., editors. CRC Press, Boca Raton, FL: 361–382. [Google Scholar]
- Dai G., and Zagotta W.N.. 2017. Molecular mechanism of voltage-dependent potentiation of KCNH potassium channels. eLife. 6:e26355 10.7554/eLife.26355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerg H.A., Bankston J.R., Rosenbaum J.C., Brzovic P.S., Zagotta W.N., and Stoll S.. 2015. Structural mechanism for the regulation of HCN ion channels by the accessory protein TRIP8b. Structure. 23:734–744. 10.1016/j.str.2015.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerg H.A., Brzovic P.S., Flynn G.E., Zagotta W.N., and Stoll S.. 2016. Structure and energetics of allosteric regulation of HCN2 ion channels by cyclic nucleotides. J. Biol. Chem. 291:371–381. 10.1074/jbc.M115.696450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derebe M.G., Sauer D.B., Zeng W., Alam A., Shi N., and Jiang Y.. 2011b Tuning the ion selectivity of tetrameric cation channels by changing the number of ion binding sites. Proc. Natl. Acad. Sci. USA. 108:598–602. 10.1073/pnas.1013636108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derebe M.G., Zeng W., Li Y., Alam A., and Jiang Y.. 2011a Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore. Proc. Natl. Acad. Sci. USA. 108:592–597. 10.1073/pnas.1013643108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhallan R.S., Yau K.-W., Schrader K.A., and Reed R.R.. 1990. Primary structure and functional expression of a cyclic nucleotide-activated channel from olfactory neurons. Nature. 347:184–187. 10.1038/347184a0 [DOI] [PubMed] [Google Scholar]
- Doyle D.A., Morais Cabral J., Pfuetzner R.A., Kuo A., Gulbis J.M., Cohen S.L., Chait B.T., and MacKinnon R.. 1998. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 280:69–77. 10.1126/science.280.5360.69 [DOI] [PubMed] [Google Scholar]
- Dzeja C., Hagen V., Kaupp U.B., and Frings S.. 1999. Ca2+ permeation in cyclic nucleotide-gated channels. EMBO J. 18:131–144. 10.1093/emboj/18.1.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinder F., Männikkö R., Pandey S., and Larsson H.P.. 2006. Mode shifts in the voltage gating of the mouse and human HCN2 and HCN4 channels. J. Physiol. 575:417–431. 10.1113/jphysiol.2006.110437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesenko E.E., Kolesnikov S.S., and Lyubarsky A.L.. 1985. Induction by cyclic GMP of cationic conductance in plasma membrane of retinal rod outer segment. Nature. 313:310–313. 10.1038/313310a0 [DOI] [PubMed] [Google Scholar]
- Flynn G.E., and Zagotta W.N.. 2001. Conformational changes in S6 coupled to the opening of cyclic nucleotide-gated channels. Neuron. 30:689–698. 10.1016/S0896-6273(01)00324-5 [DOI] [PubMed] [Google Scholar]
- Flynn G.E., and Zagotta W.N.. 2003. A cysteine scan of the inner vestibule of cyclic nucleotide-gated channels reveals architecture and rearrangement of the pore. J. Gen. Physiol. 121:563–582. 10.1085/jgp.200308819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn G.E., Black K.D., Islas L.D., Sankaran B., and Zagotta W.N.. 2007. Structure and rearrangements in the carboxy-terminal region of SpIH channels. Structure. 15:671–682. 10.1016/j.str.2007.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frings S., Seifert R., Godde M., and Kaupp U.B.. 1995. Profoundly different calcium permeation and blockage determine the specific function of distinct cyclic nucleotide-gated channels. Neuron. 15:169–179. 10.1016/0896-6273(95)90074-8 [DOI] [PubMed] [Google Scholar]
- Ganetzky B., Robertson G.A., Wilson G.F., Trudeau M.C., and Titus S.A.. 1999. The eag family of K+ channels in Drosophila and mammals. Ann. N. Y. Acad. Sci. 868(1):356–369. 10.1111/j.1749-6632.1999.tb11297.x [DOI] [PubMed] [Google Scholar]
- Gauss R., Seifert R., and Kaupp U.B.. 1998. Molecular identification of a hyperpolarization-activated channel in sea urchin sperm. Nature. 393:583–587. 10.1038/31248 [DOI] [PubMed] [Google Scholar]
- Gaymard F., Cerutti M., Horeau C., Lemaillet G., Urbach S., Ravallec M., Devauchelle G., Sentenac H., and Thibaud J.-B.. 1996. The baculovirus/insect cell system as an alternative to Xenopus oocytes. First characterization of the AKT1 K+ channel from Arabidopsis thaliana. J. Biol. Chem. 271:22863–22870. 10.1074/jbc.271.37.22863 [DOI] [PubMed] [Google Scholar]
- Gianulis E.C., Liu Q., and Trudeau M.C.. 2013. Direct interaction of eag domains and cyclic nucleotide–binding homology domains regulate deactivation gating in hERG channels. J. Gen. Physiol. 142:351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschen-Ohm M.P., Klenchin V.A., White D.S., Cowgill J.B., Cui Q., Goldsmith R.H., and Chanda B.. 2016. Structure and dynamics underlying elementary ligand binding events in human pacemaking channels. eLife. 5:e20797 10.7554/eLife.20797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S.E., and Zagotta W.N.. 1995. Localization of regions affecting an allosteric transition in cyclic nucleotide-activated channels. Neuron. 14:857–864. 10.1016/0896-6273(95)90229-5 [DOI] [PubMed] [Google Scholar]
- Gordon S.E., Oakley J.C., Varnum M.D., and Zagotta W.N.. 1996. Altered ligand specificity by protonation in the ligand binding domain of cyclic nucleotide-gated channels. Biochemistry. 35:3994–4001. 10.1021/bi952607b [DOI] [PubMed] [Google Scholar]
- Goulding E.H., Ngai J., Kramer R.H., Colicos S., Axel R., Siegelbaum S.A., and Chess A.. 1992. Molecular cloning and single-channel properties of the cyclic nucleotide-gated channel from catfish olfactory neurons. Neuron. 8:45–58. 10.1016/0896-6273(92)90107-O [DOI] [PubMed] [Google Scholar]
- Greene D., Kang S., Kosenko A., and Hoshi N.. 2012. Adrenergic regulation of HCN4 channel requires protein association with β2-adrenergic receptor. J. Biol. Chem. 287:23690–23697. 10.1074/jbc.M112.366955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Zeng W., Chen Q., Lee C., Chen L., Yang Y., Cang C., Ren D., and Jiang Y.. 2016. Structure of the voltage-gated two-pore channel TPC1 from Arabidopsis thaliana. Nature. 531:196–201. 10.1038/nature16446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustina A.S., and Trudeau M.C.. 2009. A recombinant N-terminal domain fully restores deactivation gating in N-truncated and long QT syndrome mutant hERG potassium channels. Proc. Natl. Acad. Sci. USA. 106:13082–13087. 10.1073/pnas.0900180106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustina A.S., and Trudeau M.C.. 2011. hERG potassium channel gating is mediated by N- and C-terminal region interactions. J. Gen. Physiol. 137:315–325. 10.1085/jgp.201010582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haitin Y., Carlson A.E., and Zagotta W.N.. 2013. The structural mechanism of KCNH-channel regulation by the eag domain. Nature. 501:444–448. 10.1038/nature12487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Heuermann R.J., Lyman K.A., Fisher D., Ismail Q.-A., and Chetkovich D.M.. 2017. HCN-channel dendritic targeting requires bipartite interaction with TRIP8b and regulates antidepressant-like behavioral effects. Mol. Psychiatry. 22:458–465. 10.1038/mp.2016.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B. 2001. Ionic channels of excitable membranes. Sinauer Associates, Sunderland, Mass. [Google Scholar]
- Hite R.K., and MacKinnon R.. 2017. Structural titration of Slo2.2, a Na+-dependent K+ channel. Cell. 168:390–399.e11. 10.1016/j.cell.2016.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrigan F.T., and Aldrich R.W.. 2002. Coupling between voltage sensor activation, Ca2+ binding and channel opening in large conductance (BK) potassium channels. J. Gen. Physiol. 120:267–305. 10.1085/jgp.20028605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi T. 1995. Regulation of voltage dependence of the KAT1 channel by intracellular factors. J. Gen. Physiol. 105:309–328. 10.1085/jgp.105.3.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., Santoro B., Saponaro A., Liu H., Moroni A., and Siegelbaum S.. 2013. Binding of the auxiliary subunit TRIP8b to HCN channels shifts the mode of action of cAMP. J. Gen. Physiol. 142:599–612. 10.1085/jgp.201311013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ildefonse M., Crouzy S., and Bennett N.. 1992. Gating of retinal rod cation channel by different nucleotides: Comparative study of unitary currents. J. Membr. Biol. 130:91–104. 10.1007/BF00233741 [DOI] [PubMed] [Google Scholar]
- James Z.M., Borst A.J., Haitin Y., Frenz B., DiMaio F., Zagotta W.N., and Veesler D.. 2017. CryoEM structure of a prokaryotic cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. USA. 114:4430–4435. 10.1073/pnas.1700248114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen J.W., Zimmerman A.L., Stryer L., and Baylor D.A.. 1988. Gating kinetics of the cyclic-GMP-activated channel of retinal rods: Flash photolysis and voltage-jump studies. Proc. Natl. Acad. Sci. USA. 85:1287–1291. 10.1073/pnas.85.4.1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaupp U.B., and Seifert R.. 2002. Cyclic nucleotide-gated ion channels. Physiol. Rev. 82:769–824. 10.1152/physrev.00008.2002 [DOI] [PubMed] [Google Scholar]
- Kaupp U.B., Niidome T., Tanabe T., Terada S., Bönigk W., Stühmer W., Cook N.J., Kangawa K., Matsuo H., Hirose T., et al. . 1989. Primary structure and functional expression from complementary DNA of the rod photoreceptor cyclic GMP-gated channel. Nature. 342:762–766. 10.1038/342762a0 [DOI] [PubMed] [Google Scholar]
- Kintzer A.F., and Stroud R.M.. 2016. Structure, inhibition and regulation of two-pore channel TPC1 from Arabidopsis thaliana. Nature. 531:258–262. 10.1038/nature17194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal J., Chami M., Baumgartner P., Arheit M., Chiu P.-L., Rangl M., Scheuring S., Schröder G.F., Nimigean C.M., and Stahlberg H.. 2014. Ligand-induced structural changes in the cyclic nucleotide-modulated potassium channel MloK1. Nat. Commun. 5:3106 10.1038/ncomms4106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan D.C.H., Prole D.L., and Yellen G.. 2012. Structural changes during HCN channel gating defined by high affinity metal bridges. J. Gen. Physiol. 140:279–291. 10.1085/jgp.201210838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-H., and MacKinnon R.. 2017. Structures of the human HCN1 hyperpolarization-activated channel. Cell. 168:111–120.e11. 10.1016/j.cell.2016.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A.S., Schwartz E., Chan C.S., Noam Y., Shin M., Wadman W.J., Surmeier D.J., Baram T.Z., Macdonald R.L., and Chetkovich D.M.. 2009. Alternatively spliced isoforms of TRIP8b differentially control h channel trafficking and function. J. Neurosci. 29:6250–6265. 10.1523/JNEUROSCI.0856-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Zhou X., Wang S., Michailidis I., Gong Y., Su D., Li H., Li X., and Yang J.. 2017. Structure of a eukaryotic cyclic-nucleotide-gated channel. Nature. 542:60–65. 10.1038/nature20819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Anishkin A., Liu H., van Rossum D.B., Chintapalli S.V., Sassic J.K., Gallegos D., Pivaroff-Ward K., and Jegla T.. 2015. Bimodal regulation of an Elk subfamily K+ channel by phosphatidylinositol 4,5-bisphosphate. J. Gen. Physiol. 146:357–374. 10.1085/jgp.201511491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., and Siegelbaum S.A.. 2000. Change of pore helix conformational state upon opening of cyclic nucleotide-gated channels. Neuron. 28:899–909. 10.1016/S0896-6273(00)00162-8 [DOI] [PubMed] [Google Scholar]
- Lolicato M., Nardini M., Gazzarrini S., Möller S., Bertinetti D., Herberg F.W., Bolognesi M., Martin H., Fasolini M., Bertrand J.A., et al. . 2011. Tetramerization dynamics of C-terminal domain underlies isoform-specific cAMP gating in hyperpolarization-activated cyclic nucleotide-gated channels. J. Biol. Chem. 286:44811–44820. 10.1074/jbc.M111.297606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S.B., Campbell E.B., and Mackinnon R.. 2005a Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 309:897–903. 10.1126/science.1116269 [DOI] [PubMed] [Google Scholar]
- Long S.B., Campbell E.B., and Mackinnon R.. 2005b Voltage sensor of Kv1.2: Structural basis of electromechanical coupling. Science. 309:903–908. 10.1126/science.1116270 [DOI] [PubMed] [Google Scholar]
- Long S.B., Tao X., Campbell E.B., and MacKinnon R.. 2007. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 450:376–382. 10.1038/nature06265 [DOI] [PubMed] [Google Scholar]
- Lörinczi É., Gómez-Posada J.C., de la Peña P., Tomczak A.P., Fernández-Trillo J., Leipscher U., Stühmer W., Barros F., and Pardo L.A.. 2015. Voltage-dependent gating of KCNH potassium channels lacking a covalent link between voltage-sensing and pore domains. Nat. Commun. 6:6672 10.1038/ncomms7672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A., Zong X., Jeglitsch M., Hofmann F., and Biel M.. 1998. A family of hyperpolarization-activated mammalian cation channels. Nature. 393:587–591. 10.1038/31255 [DOI] [PubMed] [Google Scholar]
- Ludwig J., Terlau H., Wunder F., Brüggemann A., Pardo L.A., Marquardt A., Stühmer W., and Pongs O.. 1994. Functional expression of a rat homologue of the voltage gated either á go-go potassium channel reveals differences in selectivity and activation kinetics between the Drosophila channel and its mammalian counterpart. EMBO J. 13:4451–4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig J., Owen D., and Pongs O.. 1997. Carboxy-terminal domain mediates assembly of the voltage-gated rat ether-à-go-go potassium channel. EMBO J. 16:6337–6345. 10.1093/emboj/16.21.6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Männikkö R., Elinder F., and Larsson H.P.. 2002. Voltage-sensing mechanism is conserved among ion channels gated by opposite voltages. Nature. 419:837–841. 10.1038/nature01038 [DOI] [PubMed] [Google Scholar]
- Männikkö R., Pandey S., Larsson H.P., and Elinder F.. 2005. Hysteresis in the voltage dependence of HCN channels: Conversion between two modes affects pacemaker properties. J. Gen. Physiol. 125:305–326. 10.1085/jgp.200409130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Carvalho M.J., Sahoo N., Muskett F.W., Vieira-Pires R.S., Gabant G., Cadene M., Schönherr R., and Morais-Cabral J.H.. 2012. Structural, biochemical, and functional characterization of the cyclic nucleotide binding homology domain from the mouse EAG1 potassium channel. J. Mol. Biol. 423:34–46. 10.1016/j.jmb.2012.06.025 [DOI] [PubMed] [Google Scholar]
- Matthews H.R., and Reisert J.. 2003. Calcium, the two-faced messenger of olfactory transduction and adaptation. Curr. Opin. Neurobiol. 13:469–475. 10.1016/S0959-4388(03)00097-7 [DOI] [PubMed] [Google Scholar]
- Matulef K., Flynn G.E., and Zagotta W.N.. 1999. Molecular rearrangements in the ligand-binding domain of cyclic nucleotide-gated channels. Neuron. 24:443–452. 10.1016/S0896-6273(00)80857-0 [DOI] [PubMed] [Google Scholar]
- McKay D.B., Weber I.T., and Steitz T.A.. 1982. Structure of catabolite gene activator protein at 2.9-A resolution. Incorporation of amino acid sequence and interactions with cyclic AMP. J. Biol. Chem. 257:9518–9524. [PubMed] [Google Scholar]
- Morais-Cabral J.H., and Robertson G.A.. 2015. The enigmatic cytoplasmic regions of KCNH channels. J. Mol. Biol. 427:67–76. 10.1016/j.jmb.2014.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger S.D., Lane A.P., Zhong H., Leinders-Zufall T., Yau K.-W., Zufall F., and Reed R.R.. 2001. Central role of the CNGA4 channel subunit in Ca2+-calmodulin-dependent odor adaptation. Science. 294:2172–2175. 10.1126/science.1063224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., and Gold G.H.. 1987. A cyclic nucleotide-gated conductance in olfactory receptor cilia. Nature. 325:442–444. 10.1038/325442a0 [DOI] [PubMed] [Google Scholar]
- Napolitano L.M.R., Bisha I., De March M., Marchesi A., Arcangeletti M., Demitri N., Mazzolini M., Rodriguez A., Magistrato A., Onesti S., et al. . 2015. A structural, functional, and computational analysis suggests pore flexibility as the base for the poor selectivity of CNG channels. Proc. Natl. Acad. Sci. USA. 112:E3619–E3628. 10.1073/pnas.1503334112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimigean C.M., Shane T., and Miller C.. 2004. A cyclic nucleotide modulated prokaryotic K+ channel. J. Gen. Physiol. 124:203–210. 10.1085/jgp.200409133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti P., Young E.C., and Siegelbaum S.A.. 1999. C-Linker of cyclic nucleotide-gated channels controls coupling of ligand binding to channel gating. J. Gen. Physiol. 113:17–34. 10.1085/jgp.113.1.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., and Ferrin T.E.. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Prole D.L., and Yellen G.. 2006. Reversal of HCN channel voltage dependence via bridging of the S4-S5 linker and Post-S6. J. Gen. Physiol. 128:273–282. 10.1085/jgp.200609590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puljung M.C., DeBerg H.A., Zagotta W.N., and Stoll S.. 2014. Double electron-electron resonance reveals cAMP-induced conformational change in HCN channels. Proc. Natl. Acad. Sci. USA. 111:9816–9821. 10.1073/pnas.1405371111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G.A., Warmke J.M., and Ganetzky B.. 1996. Potassium currents expressed from Drosophila and mouse eag cDNAs in Xenopus oocytes. Neuropharmacology. 35:841–850. 10.1016/0028-3908(96)00113-X [DOI] [PubMed] [Google Scholar]
- Root M.J., and MacKinnon R.. 1993. Identification of an external divalent cation-binding site in the pore of a cGMP-activated channel. Neuron. 11:459–466. 10.1016/0896-6273(93)90150-P [DOI] [PubMed] [Google Scholar]
- Root M.J., and MacKinnon R.. 1994. Two identical noninteracting sites in an ion channel revealed by proton transfer. Science. 265:1852–1856. 10.1126/science.7522344 [DOI] [PubMed] [Google Scholar]
- Rothberg B.S., Shin K.S., Phale P.S., and Yellen G.. 2002. Voltage-controlled gating at the intracellular entrance to a hyperpolarization-activated cation channel. J. Gen. Physiol. 119:83–91. 10.1085/jgp.119.1.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg B.S., Shin K.S., and Yellen G.. 2003. Movements near the gate of a hyperpolarization-activated cation channel. J. Gen. Physiol. 122:501–510. 10.1085/jgp.200308928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguinetti M.C., and Tristani-Firouzi M.. 2006. hERG potassium channels and cardiac arrhythmia. Nature. 440:463–469. 10.1038/nature04710 [DOI] [PubMed] [Google Scholar]
- Sanguinetti M.C., Jiang C., Curran M.E., and Keating M.T.. 1995. A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell. 81:299–307. 10.1016/0092-8674(95)90340-2 [DOI] [PubMed] [Google Scholar]
- Santoro B., Liu D.T., Yao H., Bartsch D., Kandel E.R., Siegelbaum S.A., and Tibbs G.R.. 1998. Identification of a gene encoding a hyperpolarization-activated pacemaker channel of brain. Cell. 93:717–729. 10.1016/S0092-8674(00)81434-8 [DOI] [PubMed] [Google Scholar]
- Santoro B., Piskorowski R.A., Pian P., Hu L., Liu H., and Siegelbaum S.A.. 2009. TRIP8b splice variants form a family of auxiliary subunits that regulate gating and trafficking of HCN channels in the brain. Neuron. 62:802–813. 10.1016/j.neuron.2009.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro B., Hu L., Liu H., Saponaro A., Pian P., Piskorowski R.A., Moroni A., and Siegelbaum S.A.. 2011. TRIP8b regulates HCN1 channel trafficking and gating through two distinct C-terminal interaction sites. J. Neurosci. 31:4074–4086. 10.1523/JNEUROSCI.5707-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saponaro A., Pauleta S.R., Cantini F., Matzapetakis M., Hammann C., Donadoni C., Hu L., Thiel G., Banci L., Santoro B., and Moroni A.. 2014. Structural basis for the mutual antagonism of cAMP and TRIP8b in regulating HCN channel function. Proc. Natl. Acad. Sci. USA. 111:14577–14582. 10.1073/pnas.1410389111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönherr R., and Heinemann S.H.. 1996. Molecular determinants for activation and inactivation of HERG, a human inward rectifier potassium channel. J. Physiol. 493:635–642. 10.1113/jphysiol.1996.sp021410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönherr R., Löber K., and Heinemann S.H.. 2000. Inhibition of human ether à go-go potassium channels by Ca2+/calmodulin. EMBO J. 19:3263–3271. 10.1093/emboj/19.13.3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentenac H., Bonneaud N., Minet M., Lacroute F., Salmon J.M., Gaymard F., and Grignon C.. 1992. Cloning and expression in yeast of a plant potassium ion transport system. Science. 256:663–665. 10.1126/science.1585180 [DOI] [PubMed] [Google Scholar]
- Shin K.S., Rothberg B.S., and Yellen G.. 2001. Blocker state dependence and trapping in hyperpolarization-activated cation channels: Evidence for an intracellular activation gate. J. Gen. Physiol. 117:91–101. 10.1085/jgp.117.2.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin K.S., Maertens C., Proenza C., Rothberg B.S., and Yellen G.. 2004. Inactivation in HCN channels results from reclosure of the activation gate: Desensitization to voltage. Neuron. 41:737–744. 10.1016/S0896-6273(04)00083-2 [DOI] [PubMed] [Google Scholar]
- Shuart N.G., Haitin Y., Camp S.S., Black K.D., and Zagotta W.N.. 2011. Molecular mechanism for 3:1 subunit stoichiometry of rod cyclic nucleotide-gated ion channels. Nat. Commun. 2:457 10.1038/ncomms1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P.L., Baukrowitz T., and Yellen G.. 1996. The inward rectification mechanism of the HERG cardiac potassium channel. Nature. 379:833–836. 10.1038/379833a0 [DOI] [PubMed] [Google Scholar]
- Su Y., Dostmann W.R.G., Herberg F.W., Durick K., Xuong N.H., Ten Eyck L., Taylor S.S., and Varughese K.I.. 1995. Regulatory subunit of protein kinase A: Structure of deletion mutant with cAMP binding domains. Science. 269:807–813. 10.1126/science.7638597 [DOI] [PubMed] [Google Scholar]
- Tan P.S., Perry M.D., Ng C.A., Vandenberg J.I., and Hill A.P.. 2012. Voltage-sensing domain mode shift is coupled to the activation gate by the N-terminal tail of hERG channels. J. Gen. Physiol. 140:293–306. 10.1085/jgp.201110761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.-Y., and Papazian D.M.. 1997. Transfer of voltage independence from a rat olfactory channel to the Drosophila ether-à-go-go K+ channel. J. Gen. Physiol. 109:301–311. 10.1085/jgp.109.3.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X., Lee A., Limapichat W., Dougherty D.A., and MacKinnon R.. 2010. A gating charge transfer center in voltage sensors. Science. 328:67–73. 10.1126/science.1185954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X., Hite R.K., and MacKinnon R.. 2017. Cryo-EM structure of the open high-conductance Ca2+-activated K+ channel. Nature. 541:46–51. 10.1038/nature20608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terlau H., Heinemann S.H., Stühmer W., Pongs O., and Ludwig J.. 1997. Amino terminal-dependent gating of the potassium channel rat eag is compensated by a mutation in the S4 segment. J. Physiol. 502:537–543. 10.1111/j.1469-7793.1997.537bj.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thouta S., Sokolov S., Abe Y., Clark S.J., Cheng Y.M., and Claydon T.W.. 2014. Proline scan of the HERG channel S6 helix reveals the location of the intracellular pore gate. Biophys. J. 106:1057–1069. 10.1016/j.bpj.2014.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbs G.R., Liu D.T., Leypold B.G., and Siegelbaum S.A.. 1998. A state-independent interaction between ligand and a conserved arginine residue in cyclic nucleotide-gated channels reveals a functional polarity of the cyclic nucleotide binding site. J. Biol. Chem. 273:4497–4505. 10.1074/jbc.273.8.4497 [DOI] [PubMed] [Google Scholar]
- Tomczak A.P., Fernández-Trillo J., Bharill S., Papp F., Panyi G., Stühmer W., Isacoff E.Y., and Pardo L.A.. 2017. A new mechanism of voltage-dependent gating exposed by KV10.1 channels interrupted between voltage sensor and pore. J. Gen. Physiol. 149:577–593. 10.1085/jgp.201611742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudeau M.C., and Zagotta W.N.. 2003. Calcium/calmodulin modulation of olfactory and rod cyclic nucleotide-gated ion channels. J. Biol. Chem. 278:18705–18708. 10.1074/jbc.R300001200 [DOI] [PubMed] [Google Scholar]
- Trudeau M.C., Warmke J.W., Ganetzky B., and Robertson G.A.. 1995. HERG, a human inward rectifier in the voltage-gated potassium channel family. Science. 269:92–95. 10.1126/science.7604285 [DOI] [PubMed] [Google Scholar]
- Varnum M.D., Black K.D., and Zagotta W.N.. 1995. Molecular mechanism for ligand discrimination of cyclic nucleotide-gated channels. Neuron. 15:619–625. 10.1016/0896-6273(95)90150-7 [DOI] [PubMed] [Google Scholar]
- Vemana S., Pandey S., and Larsson H.P.. 2004. S4 movement in a mammalian HCN channel. J. Gen. Physiol. 123:21–32. 10.1085/jgp.200308916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl-Schott C., and Biel M.. 2009. HCN channels: Structure, cellular regulation and physiological function. Cell. Mol. Life Sci. 66:470–494. 10.1007/s00018-008-8525-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainger B.J., DeGennaro M., Santoro B., Siegelbaum S.A., and Tibbs G.R.. 2001. Molecular mechanism of cAMP modulation of HCN pacemaker channels. Nature. 411:805–810. 10.1038/35081088 [DOI] [PubMed] [Google Scholar]
- Wang W., and MacKinnon R.. 2017. Cryo-EM structure of the open human ether-à-go-go-related K+ channel hERG. Cell. 169:422–430.e10. 10.1016/j.cell.2017.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Trudeau M.C., Zappia A.M., and Robertson G.A.. 1998. Regulation of deactivation by an amino terminal domain in human ether-à-go-go-related gene potassium channels. J. Gen. Physiol. 112:637–647. 10.1085/jgp.112.5.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Chen S., and Siegelbaum S.A.. 2001. Regulation of hyperpolarization-activated HCN channel gating and cAMP modulation due to interactions of COOH terminus and core transmembrane regions. J. Gen. Physiol. 118:237–250. 10.1085/jgp.118.3.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmke J.W., and Ganetzky B.. 1994. A family of potassium channel genes related to eag in Drosophila and mammals. Proc. Natl. Acad. Sci. USA. 91:3438–3442. 10.1073/pnas.91.8.3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmke J., Drysdale R., and Ganetzky B.. 1991. A distinct potassium channel polypeptide encoded by the Drosophila eag locus. Science. 252:1560–1562. 10.1126/science.1840699 [DOI] [PubMed] [Google Scholar]
- Weitz D., Ficek N., Kremmer E., Bauer P.J., and Kaupp U.B.. 2002. Subunit stoichiometry of the CNG channel of rod photoreceptors. Neuron. 36:881–889. 10.1016/S0896-6273(02)01098-X [DOI] [PubMed] [Google Scholar]
- Whicher J.R., and MacKinnon R.. 2016. Structure of the voltage-gated K+ channel Eag1 reveals an alternative voltage sensing mechanism. Science. 353:664–669. 10.1126/science.aaf8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Vysotskaya Z.V., Liu Q., and Zhou L.. 2010. Structural basis for the cAMP-dependent gating in the human HCN4 channel. J. Biol. Chem. 285:37082–37091. 10.1074/jbc.M110.152033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta W.N., Olivier N.B., Black K.D., Young E.C., Olson R., and Gouaux E.. 2003. Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature. 425:200–205. 10.1038/nature01922 [DOI] [PubMed] [Google Scholar]
- Zhang M., Liu J., and Tseng G.-N.. 2004. Gating charges in the activation and inactivation processes of the HERG channel. J. Gen. Physiol. 124:703–718. 10.1085/jgp.200409119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Goldschen-Ohm M.P., Morais-Cabral J.H., Chanda B., and Robertson G.A.. 2017. The intrinsically liganded cyclic nucleotide-binding homology domain promotes KCNH channel activation. J. Gen. Physiol. 149:249–260. 10.1085/jgp.201611701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., and Trudeau M.C.. 2015. Handbook of Ion Channels. CRC Press, Boca Raton, FL: 10.1201/b18027 [DOI] [Google Scholar]
- Zheng J., Trudeau M.C., and Zagotta W.N.. 2002. Rod cyclic nucleotide-gated channels have a stoichiometry of three CNGA1 subunits and one CNGB1 subunit. Neuron. 36:891–896. 10.1016/S0896-6273(02)01099-1 [DOI] [PubMed] [Google Scholar]
- Zhong H., Molday L.L., Molday R.S., and Yau K.-W.. 2002. The heteromeric cyclic nucleotide-gated channel adopts a 3A:1B stoichiometry. Nature. 420:193–198. 10.1038/nature01201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., and Siegelbaum S.A.. 2007. Gating of HCN channels by cyclic nucleotides: Residue contacts that underlie ligand binding, selectivity, and efficacy. Structure. 15:655–670. 10.1016/j.str.2007.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Olivier N.B., Yao H., Young E.C., and Siegelbaum S.A.. 2004. A conserved tripeptide in CNG and HCN channels regulates ligand gating by controlling C-terminal oligomerization. Neuron. 44:823–834. 10.1016/j.neuron.2004.11.012 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Morais-Cabral J.H., Kaufman A., and MacKinnon R.. 2001. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 414:43–48. 10.1038/35102009 [DOI] [PubMed] [Google Scholar]
- Ziechner U., Schönherr R., Born A.-K., Gavrilova-Ruch O., Glaser R.W., Malesevic M., Küllertz G., and Heinemann S.H.. 2006. Inhibition of human ether à go-go potassium channels by Ca2+/calmodulin binding to the cytosolic N- and C-termini. FEBS J. 273:1074–1086. 10.1111/j.1742-4658.2006.05134.x [DOI] [PubMed] [Google Scholar]
- Zolles G., Wenzel D., Bildl W., Schulte U., Hofmann A., Müller C.S., Thumfart J.-O., Vlachos A., Deller T., Pfeifer A., et al. . 2009. Association with the auxiliary subunit PEX5R/Trip8b controls responsiveness of HCN channels to cAMP and adrenergic stimulation. Neuron. 62:814–825. 10.1016/j.neuron.2009.05.008 [DOI] [PubMed] [Google Scholar]