Abstract
Endothelins are expressed in a variety of human tissue and are involved in the processes as proliferation, migration and differentiation. The signal transduction pathway is a result of the endothelin-1-3 (ET1-3) binding to their receptors (ETAR, ETBR). ET-3 is a new candidate tumour suppressor gene, which is often downregulated or silenced in human cancer.
The aim of the study was to examine DNA methylation of ET-3 genes in colorectal cancer (CRC) tissue samples in relation to the clinical stage (CS) of cancer. The paper is a continuation of our previously published results, which showed a four-fold transcriptional silencing of the ET-3 gene in the samples of colorectal cancer in comparison to normal tissues.
A total of 66 paired CRC and normal (surgical margin) tissue samples were used in the study. The tumour tissues were collected from CRC patients in CS I–IV according the 7th edition of UICC TNM Classification of Malignant Tumours (CS I, n = 8; CS II, n = 20; CS III, n = 27; CS IV, n = 11). Assessment of epigenetic silencing of the ET-3 encoding gene was performed in three steps. The silencing of the ET-3 encoding gene was a result from methylation of the promoter sequence using methylation-specific PCR (MS-PCR). Analyses were performed using primers complementary for a CpG island in the first exon of the gene encoding ET-3. An epigenetic silence through methylation of 7.5% (5/66) in comparison to control was observed, including 10% of CS II (2/20), 7% of CS III (2/27) and 9% of CS IV (1/11). The controls and the samples of tumour in CS I showed no epigenetic silencing via methylation. In conclusion, epigenetic silencing of ET-3 in CRC could play a role in the progression than in the induction process. EDN3 would be a future target for epigenetic therapy in colorectal cancer, but further clinical studies are needed.
Keywords: colorectal cancer, endothelin-3, epigenetic silencing, tissue samples
Introduction
The family of endothelins consists of four peptides with different sequences: ET-1, ET-2, ET-3 and ET-4 (also known as vasoactive intestine contractor [VIC]),1,2 encoded by different genes with conservative nucleotide sequences.3,4 Their structure is characteristic: the biologically active N-end (hydrophilic, a single alpha helix determining the affinity for the receptor and with two bisulfide bonds) and the short hydrophobic C-end with a beta-sheet structure responsible for receptor binding. Two types of metabotropic receptors for endothelins as ETAR and ETBR, coupled with a G protein were identified in humans.5–7 They were found in cells from many tissues: endothelium, heart, kidneys, intestines, lungs, brain, spleen and skin.5 Confirmed presence of endothelins’ receptors in the intestine wall and the ability of their agonists to change muscular tension indicates for the role of these proteins in regulation of biological and molecular functions of the gastrointestinal tract.8
The endothelins were shown to play a crucial role in various cancer cells including cell proliferation, escape from apoptosis, angiogenesis, cell survival, invasion and metastatic dissemination, aberrant osteogenesis and modification of nociceptive stimuli.9
Endothelin-3 (ET-3) is excreted by endothelial cells, neuronal brain cells, renal epithelial cells and intestinal cells,10 as well as pancreatic, splenic and prostate cells.11 Its involvement was confirmed in the development of choriocarcinoma12 and malignant melanoma.13 The active form of ET-3 is a 21 amino acid peptide. The active peptide is a ligand for ETBR. ET-3 growth factor protein levels were decreased in cervical cancer compared with normal cervical epithelial cells.14 In our previous paper,15 analysis of gene expression profiles for endothelins and their receptors in the samples of normal colon and adenocarcinoma showed a three-fold increase in the expression of gene encoding endothelin A receptor (ETAR), and an approximately four-fold silencing of ET-3 transcriptional activity. In the study of Wiesmann et al. ET-3 mRNA expression and protein levels were attenuated in human breast cancer tissues compared to normal tissue. According to the authors, this effect was caused by hypermethylation of the ET-3 promoter and subsequent gene silencing.11 The study of Wang et al.16 showed an epigenetic inactivation of ET-2 and ET-3 (as a hypermethylation of EDN2 and EDN3 genes) in rat and human primary colon cancers. Silencing of the gene expression is due to chemical modifications of histon proteins and DNA itself.17
Advances in genomics, molecular pathology and metabolism have generated many risk factors of colorectal cancer (CRC), the third most common malignant neoplasm worldwide.18 Changes in the epigenetic pattern by DNA methylation or histone modifications can lead to the silencing of those genes, which are involved in the regulation of many processes such as cell cycle, apoptosis or DNA repair.19 Practical usage of epigenetics in diagnostics and treatment of cancer is particularly useful for single genes (in particular, tumour suppressors), which can become deregulated or silenced due to a disturbed epigenetic pattern if left unmutated.20,21
ET-3 is a new candidate tumour suppressor gene, which is often downregulated or even silenced by promoter hypermethylation in human cancers. But there are not many reports which have focused on ET-3 gene expression and its implication in human CRC.
In this study, we examined DNA methylation of ET-3 gene in CRC tissue samples. This paper is a continuation of our previously published results,15 which showed a four-fold transcriptional silencing of ET-3 gene in the samples of CRC in comparison to normal tissues.
Materials and methods
Patients and tissue sample collection
Cryo-conserved clinical samples – tumour and normal tissues – were obtained from CRC patients treated by primary surgery at the Surgery Department. Patients who received neo-adjuvant chemotherapy or radiotherapy or both, and patients with recurrent CRC were excluded. A total of 66 paired CRC and normal (surgical margin >5 cm) tissue samples were used. The tumours samples were collected from CRC patients in clinical stages (CS) I–IV according to the 7th edition of UICC TNM Classification of Malignant Tumours.22 The patients (26 women, 40 men; age range, 31–86 years) were divided into four groups depending on CS (CS I, n = 8; CS II, n = 20; CS III, n = 27; CS IV, n = 11) (Table 1). The pathological features such as tumour location, type and grade are presented in Table 2.
Table 1.
Demographics and clinic characteristics.
| Age (years) | Gender |
UICC TNM 7th edition |
||||
|---|---|---|---|---|---|---|
| W | M | I | II | III | IV | |
| 65.0 ± 10.6 | 26 | 40 | 8 | 20 | 27 | 11 |
| 31–86 | 40.0% | 60.0% | 12.1% | 30.3% | 40.9% | 16.7% |
Table 2.
Pathological features of the tumour samples.
| Patients (n = 66) |
||||
|---|---|---|---|---|
| n | % | |||
| Tumour location | Colon | 32 | 48.5% | |
| Rectum | 24 | 36.4% | ||
| Type | Adenocarcinoma | 37 | 56.1% | |
| Adenocarcinoma mucosum | 6 | 9.1% | ||
| Adenocarcinoma necroticans | 5 | 7.6% | ||
| Adenocarcinoma exculceratum | 18 | 27.2% | ||
| Grade | G1 | 10 | 15.2% | |
| G2 | 38 | 57.6% | ||
| G3 | 2 | 3% | ||
| Gx | 16 | 24.2% | ||
The study was approved by the Bioethics Committee of the Silesian Medical University, currently the Silesian Medical University in Katowice (NN-6501-79/06).
Methods
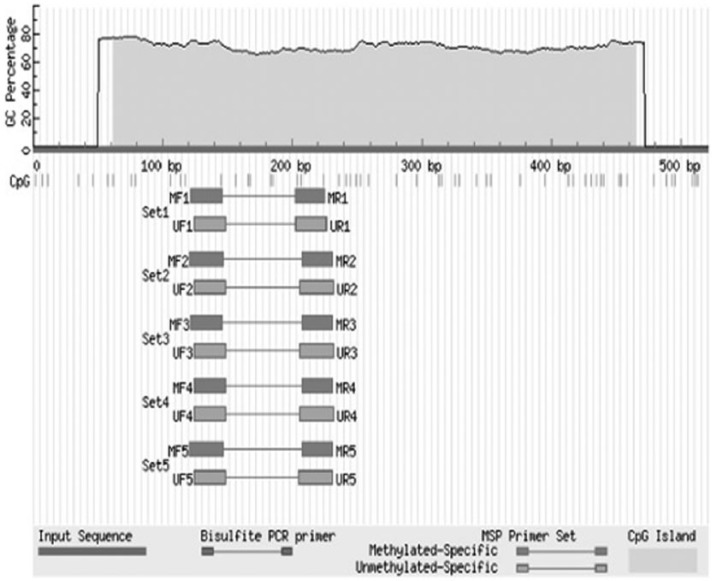
Assessment of epigenetic silencing of the ET-3 encoding gene was performed in three steps. First, DNA was extracted from colorectal using QIAmp DNA Mini kit (Qiagen). A colon/rectal fragment of about 25 mg was digested with proteinase K. Then, DNA from homogenate was isolated according to the instructions of the manufacturer. Next, extracted DNA was treated with sodium bisulfite to convert unmethylated cytosine into uracil using EpiTect Bisulfite kit (QIAGEN). Methylated cytosines do not undergo this reaction. After converting cytosine into uracil, extracted DNA was purified using EpiTect Bisulfite kit (QIAGEN) and used for methyl-specific PCR (MS-PCR) with two pairs of starters. The first pair of starters covered the unmodified ET-2 promoter region. The second pair covered a region of CpG islands, which were unmodified due to conversion of unmethylated cytosine into uracil. Assessment whether transcriptional silencing of ET-3 encoding gene was a result from methylation of the promoter sequence using Methylation Specific PCR (MS-PCR).23 MS-PCR method involves amplification of a sodium bisulfite treated template DNA with the use of two primer pairs. One pair amplifies the methylated CpG island, recognising 5-methylocytosine as cytosine. The second pair is complementary to the unmethylated sequence, where cytosine was converted to uracil and it is recognised as uracil/thymidine. Depending on the primer pair, which gives reaction, we obtain information on the presence or lack of methylation in a specific genomic position. Assessment of DNA methylation was started with identification of CpG islands in the EDN3 gene sequence, consisting of 32,549 bp: including 5000 bp domain before first exon mRNA with a promoter. A CpG island of 520 bp was found between this sequence and the first exon of the ET-3 gene. This sequence was used to construct starter pairs for the detection of methylated and unmethylated sequence with PCR (Figure 1).
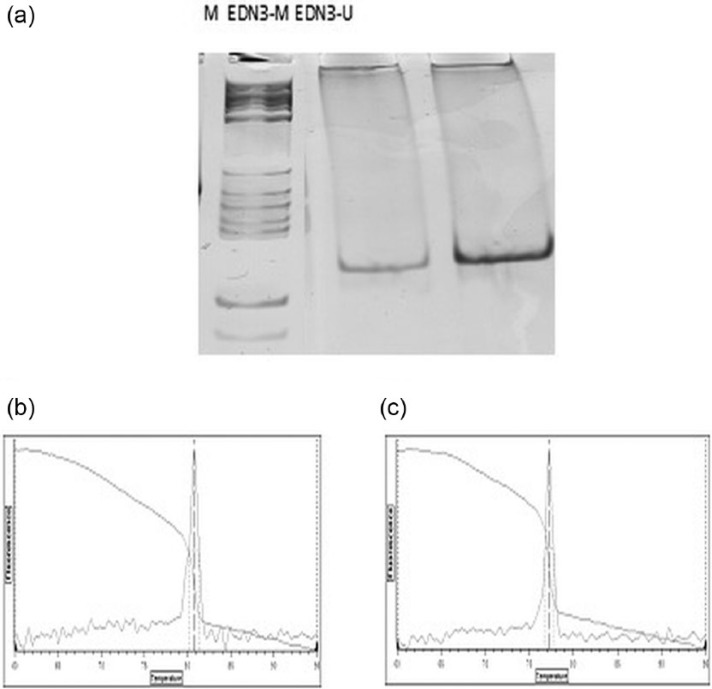
Figure 1.
The assessment of PCR specificity on the basis of electrophoresis in an acrylamide gel and the amplimer dissociation curve with the melting temperature Tm for amplimers in the each analysed sample. (a) The image was showing electrophoresis results of PCR products separation in a polyacrylamide gel stained with silver salts. (b) EDN-3M Tm = 80.8 control for methylated endothelin-3 gene. (c) EDN-3U Tm = 77.2 control for unmethylated ET-3 gene after conversion.
Validation of the matrix experiment was performed using the MS-PCR method and defining the EDN3 DNA methylation profile to confirm epigenetic silencing of EDN3 gene.
Results
Structure of ET-3 encoding sequence
Assessment of DNA methylation started with identification of CpG islands in the EDN3 gene sequence, consisting of 32,549 bp: including 5000 bp domain before first exon mRNA with a promoter. A CpG island of 520 bp was identified between this sequence and the first exon of the ET-1 gene sequence. This sequence was used to construct starters for detection of methylated and unmethylated sequence using PCR. Primers were constructed with MethPrimer software23 according to the following criteria of CG island identification:
- an island must be longer than 100 nucleotides,
- a GC percentage that is greater than 50%,
- an observed-to-expected ratio that is greater than 0.6.24
The ET-3 sequence with an indicated CpG island at 5’ exon 1; accession number: NM000114 (Figure 2).25
Figure 2.

ET-3 sequence with a marked CpG island (sequence within a rectangle) at 5’ end of exon 1.
Bolded sequence means primers for methylated sequence (forward primer 121-146; reverse primer 207-231), underlined sequence means primers for the unmethylated sequence (forward primer 124-148; reverse primer 205-231). The sequence in the frame indicates a CpG island of 251 base pairs (start 62 – end 312).
The sizes of amplified products were 144 bp for no-methylation specific primers and 140 bp for methylation specific primers, respectively (Figures 3 and 4). Lack of proper product in both PCR reactions was treated as wrong modification or wrong DNA purification and an indication for repeated analysis. Correctness of reactions was checked using a control set – EpiTect Control DNA (Qiagen). Amplification specificity was based on a DNA melting curve and determination of Tm values.
Figure 3.
The result primer design methylated and unmethylated CpG islands sequences.
Figure 4.
The DNA nucleotide sequence for ET-3 in the reaction of converting unmethylated cytosine to thymidine (:– means cytosine, which certainly becomes converted to thymidine, ++ – means CG sites, where cytosine will not necessarily be converted to thymidine).
Methylation profile for ET-3
Analyses were performed using primers complementary for a CpG island in the first exon of the gene encoding ET-3. When assessing methylation in samples of CRC, we observed an epigenetic silencing through methylation of 7.5% (5/66) in comparison to control, including 10% of CS II (2/20), 7% of CS III (2/27), and 9% of CS IV (1/11). The controls and the samples of tumour in CS I showed no epigenetic silencing via methylation.
Discussion
Experimental, genetic, epidemiologic and socioeconomic studies have suggested that the incidence of CRC depends on the interactions between inherited susceptibility (Lynch syndrome I and II, familial polyposis), clinical conditions (ulcerative colitis, Crohn’s disease) and environmental / lifestyle-related risk factors (physical inactivity, smoking, excessive alcohol consumption, high-fat / low-fibre diet, overweight / obesity).26 Kim et al.27 concluded that accumulation of genetic and epigenetic alterations in colonic epithelial cells results in neoplastic transformations. Epigenetic modifications including changes in DNA methylation, histone modifications and non-coding RNAs could play a critical role in carcinogenesis.28 Some studies showed that DNA methylation mediated tumour suppressor gene silencing may contribute to tumour progression.29–32 In humans, DNA methylation occurs at the 5′ position of the pyrimidine ring of the cytosine residues within CpG dinucleotides through the addition of a methyl moiety to form 5-methylcytosines.33 Aberrant DNA methylation of loci have become promising biomarkers in the early diagnosis of diseases and predictors of metastatic or aggressive CRC.34 Epigenetic regulation by DNA methylation is an unknown, dynamic process.35 The endothelins and their receptors play an important role in carcinogenesis, by regulating epithelial-mesenchymal transition (EMT) through different signalling pathways.36 A balance regulation of the endothelin axis (ETs, ETAR, ETBR – ET-axis) is essential for homing processes to tissue destination. The changes in the regulation of ET-axis are important in cancer and progression. The signal transduction pathway is a result of the ET-1 and ET-2 binding to ETAR.37 ETAR exhibits an affinity for ET-3, but that is lower than that for ET-1 and ET-2. The activation of the ETAR pathway by ET-1 (a powerful mitogenic and antiapoptotic peptide) may indirectly lead to the stimulation of the AKT and the mTOR signalling pathway.9 ET-1 and ET-2 are overexpressed in many cancers.12,38,39 Only a few analysis of ET-3 expression in human cancer were published.9,11,15,16 Wiesmann et al.16 emphasised that there is still little knowledge about the role of ET-3 in cancer, but probably EDN3 may exert a functional role divergent to that of EDN1/EDN2. The loss of EDN3 expression by methylation could change the balance of ET-axis. The results of previous studies indicate a statistically significant decrease in ET-3 expression in tumours.9,11,15,16 The remaining question is whether silencing of this gene is associated only with DNA hypermethylation, which can be of potential diagnostic and therapeutic importance. The prerequisite to obtain the presented results was the hypothesis that changes at the level of genes expression profile are related to the endothelin’s biological activity and diversified mRNA concentration profile depends on the pathological changes advancements. It can be the basis to elaborate new diagnostic and therapeutic methods in case of CRC.
In our study, we found that only a small number of cases showed epigenetic silencing of EDN3. It was observed in 7.5% of tumour tissue (5/66). The silencing via methylation of EDN3 was not found in control and CS I tissue samples. In the study, EDN3 expression was not analysed according to grading, because of a small number of tissue samples in examined groups depending on clinical stages. The study will be continued.
In conclusion
Epigenetic silencing of EDN3 in CRC could play a role in the progression than in the induction process.
EDN3 would be a future target for epigenetic therapy in CRC, but further clinical studies are needed.
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
References
- 1. Dobrek Ł, Thor P. (2010) [Endothelin in cardiovascular diseases pathophysiology]. Polski Merkuriusz Lekarski 166: 289–293. [PubMed] [Google Scholar]
- 2. Kleniewska P, Michalski Ł, Skibska B, et al. (2012) The influence of endothelin-1 and endothelin receptor blocker on the content of white blood cells in the peripheral blood of rat. Polski Merkuriusz Lekarski 32(190): 225–227. [PubMed] [Google Scholar]
- 3. Grimshaw MJ. (2007) Endothelins and hypoxia-inducible factor in cancer. Endocrine-Related Cancer 14(2): 233–234. [DOI] [PubMed] [Google Scholar]
- 4. Paasche JD, Attramadal T, Sandberg C, et al. (2001) Mechanisms of endothelin receptor subtype-specific targeting to distinct intracellular trafficking pathways. Journal of Biological Chemistry 276(36): 34041–34050. [DOI] [PubMed] [Google Scholar]
- 5. Davenport AP. (2002) International Union of Pharmacology. XXIX. Update on endothelin receptor nomenclature. Pharmacological Reviews 54(2): 219–226. [DOI] [PubMed] [Google Scholar]
- 6. Rozengurt E. (2007) Mitogenic signaling pathways induced by G protein-coupled receptors. Journal of Cellular Physiology 213(3): 589–602. [DOI] [PubMed] [Google Scholar]
- 7. Bagnato A, Natali PG. (2004) Endothelin receptors as novel targets in tumor therapy. Journal of Translational Medicine 2(1): 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wanecek M, Weitzberg E, Rudehill A, et al. (2000) The endothelin system in septic and endotoxin shock. European Journal of Pharmacology 407: 1–15. [DOI] [PubMed] [Google Scholar]
- 9. Bagnato A, Loizidou M, Pflug BR, et al. (2011) Role of the endothelin axis and its antagonists in the treatment of cancer. British Journal of Pharmacology 163(2): 220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kedzierski RM, Yanagisawa M. (2001) Endothelin system: The double-edged sword in health and disease. Annual Review Pharmacology and Toxicology 41: 851–876. [DOI] [PubMed] [Google Scholar]
- 11. Wiesmann F, Veeck J, Galm O. (2009) Frequent loss of endothelin-3 (EDN3) expression due to epigenetic inactivation in human breast cancer. Breast Cancer Research 11(3): R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rauh A, Windischhofer W, Kovacevic A, et al. (2008) Endothelin (ET)-1 and ET-3 promote expression of c-fos and c-jun in human choriocarcinoma via ET(B) receptor-mediated G(i)-and G(q)-pathways and MAP kinase activation. British Journal of Pharmacology 154(1): 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Spinella F, Rosanò L, Di Castro V, et al. (2007) Endothelin-1 and endothelin-3 promote invasive behaviour via hypoxia-inducible factor-1alpha in human melanoma cells. Cancer Research 67(4): 1725–1734. [DOI] [PubMed] [Google Scholar]
- 14. Sun de J, Liu Y, Lu DC, et al. (2007) Endothelin-3 growth factor levels decreased in cervical cancer compared with normal cervical epithelial cells. Human Pathology 38: 1047–1056. [DOI] [PubMed] [Google Scholar]
- 15. Olender J, Stachowicz M, Szota J, et al. (2008) Endothelin as a therapeutic target in colorectal cancer. Sc Rev Pharm 9–10:6–9. [Google Scholar]
- 16. Wang R, Löhr Ch, Fischer K, et al. (2013) Epigenetic inactivation of endothelin-2 (ET-2) and ET-3 in colon cancer. International Journal of Cancer 132(5): 1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mueller WC, van Deimling A. (2009) Gene regulation by methylation. Recent Results in Cancer Research 117: 217–239. [DOI] [PubMed] [Google Scholar]
- 18. Siegel R, Naishadham D, Jemal A. (2013) Cancer statistics, 2013. CA: A Cancer Journal for Clinicians 63: 11–12. [DOI] [PubMed] [Google Scholar]
- 19. Flis S, Flis K, Spławiński J. (2007) Epigenetic modification and cancer. Journal of Oncology 4: 427–434. [Google Scholar]
- 20. Mazurek U, Owczarek A, Nowakowska-Zajdel E, et al. (2011) Statistical analysis of differentia genes expression in colorectal cancer using clear-test. Journal of Biological Regulators and Homeostatic Agents 25: 279–283. [PubMed] [Google Scholar]
- 21. Muc-Wierzgoń M, Nowakowska-Zajdel E, Kokot T, et al. (2006) Genetic disregulation of TNF alpha and TNF type II receptors in colon cancer at the II and III stages of diseases. Journal of Biological Regulators and Homeostatic Agents 20: 10–14. [PubMed] [Google Scholar]
- 22. Sobin LH, Gospodarowicz MK, Wittekind Ch. (2009) TNM Classification of Malignant Tumours. Seventh edn Chichester: Wiley-Blackwell. [Google Scholar]
- 23. Li LC, Dahiya R. (2002) MethPrimer: Designing primers for methylation PCRs. Bioinformatics 18(11): 1427–1431. [DOI] [PubMed] [Google Scholar]
- 24. Gardiner-Garden M, Frommer M. (1987) CpG islands in vertebrate genomes. Journal of Molecular Biology 96(2): 261–282. [DOI] [PubMed] [Google Scholar]
- 25. Rao V, Loffler C, Hansmann I. (1991) The gene for the novel vasoactive peptide endothelin 3 (EDN3) is localised to human chromosome 20q13.2-qter. Genomics 10(3): 840–841. [DOI] [PubMed] [Google Scholar]
- 26. Muc-Wierzgoń M, Nowakowska-Zajdel E, Dzięgielewska-Gęsiak S, et al. (2014) Specific metabolic biomarkers as risk and prognostic factors in colorectal cancer. World Journal of Gastroenterology 20: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim MS, Lee J, Sidrnasky D. (2010) DNA methylation markers in colorectal cancer. Cancer Metastasis Reviews 29: 181–206. [DOI] [PubMed] [Google Scholar]
- 28. Schnekenburger M, Dicato M, Diederich M. (2014) Epigenetic modulators from “The Big Blue”: A treasure to fight against cancer. Cancer Letters 351: 182–197. [DOI] [PubMed] [Google Scholar]
- 29. Kulis M, Esteller M. (2010) DNA methylation and cancer. Advances in Genetics 70: 27–56. [DOI] [PubMed] [Google Scholar]
- 30. Baylin SB. (2005) DNA methylation and gene silencing in cancer. Natural Clinical Practice Oncology 2: S4–S11. [DOI] [PubMed] [Google Scholar]
- 31. Szyf M. (2012) DNA methylation signatures for breast cancer classification and prognosis. Genome Medicine 4: 26–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baylin SB, Esteller M, Rountree MR, et al. (2001) Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Human Molecular Genetics 10: 687–692. [DOI] [PubMed] [Google Scholar]
- 33. Hansen KD, Timp W, Bravo HC, et al. (2011) Increased methylation variation in epigenetic domains across cancer types. Nature Genetics 43: 768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Song X, Che H. (2012) Balance of DNA methylation and demethylation in cancer development. Genome Biology 13: 173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bhutani N, Burns DM, Blau HM. (2011) DNA demethylation dynamics. Cell 146: 866–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thiery JP. (2002) Epithelial-mesenchymal transitions in tumour progression. Nature Reviews Cancer 2: 442–454. [DOI] [PubMed] [Google Scholar]
- 37. Masaki T. (2004) Historical review: Endothelin. Trends in Pharmacological Sciences 35: 219–224. [DOI] [PubMed] [Google Scholar]
- 38. Bagnato A, Spinella F, Rosano L. (2005) Emerging role of the endothelin axis in ovarian tumour progression. Endocrine-Related Cancer 12: 761–772. [DOI] [PubMed] [Google Scholar]
- 39. Kim TH, Xiong H, Zhang Z, et al. (2005) β-Catenin activates the growth factor endothelin-1 in colon cancer cells. Oncogene 24: 597–604. [DOI] [PubMed] [Google Scholar]