Abstract
Introduction:
Asthma is associated with activation of interleukin-4 (IL-4)/interleukin-13 (IL-13)/signal transducer and activator of transcription factor-6(STAT6) inflammatory response via overexpression of all pathway components: IL-4, IL-13, and STAT6.
Objectives:
To evaluate the association of IL-4, IL-13, and STAT6 expression and immunoexpression with atopic asthma development.
Patients and methods:
Fifty patients with atopic asthma and 20 healthy controls were enrolled into the study. Relative gene expression was analyzed by qPCR method. Immunoexpression was assessed by ELISA method.
Results:
The expression levels of IL-4, IL-13, and STAT6 were higher in patients compared to the controls, but a statistically significant difference was observed only for IL-13 (P = 0.03). In immunoexpression analysis, a statistically significant difference between patients and controls was found for IgE (P = 0.03). Significant positive correlations in the patient group were found between IL-13 gene expression and total level of serum IgE (rho = 0.230, P = 0.033), STAT6 gene/STAT6 protein and total level of serum IgE (STAT6: rho = 0.077, P = 0.038; STAT6: rho = 0.049, P = 0.042), IL-4, and STAT6 expression (rho = 0.098, P = 0.048). Any significant correlations were found between expression/immunoexpression levels of the studied genes and clinical classification, clinical features, or lung function parameters.
Conclusions:
Our data support the role of Th2 cytokines (IL-4, IL-13) and STAT6 in Th1/Th2 imbalance and highlight the etiological relationship between IL-4/IL-13/STAT6 signaling and atopy and asthma.
Keywords: atopic asthma, IL-4, Il-13, STAT6
Introduction
Asthma, a phenotypically varied inflammatory disease, is influenced by genetic and environmental factors.1,2 The disease is defined by the presence of airway hyperreactivity (AHR), mucus overproduction, and chronic eosinophilic inflammation. Asthma is also often characterized by enhanced total serum IgE level upon the exposure to allergens, which is known as an atopy. The involvement of genetic predisposition in the development of atopy in asthmatic patients was confirmed on many family studies, through genome-wide linkage studies.3–8 The elevated IgE production in asthmatic patients results in promotion of acute hypersensitivity responses, chronic eosinophil-predominant allergic inflammation with Th2 cells cytokine production. Among Th2 cytokines (such asIL-4, IL-5, IL-13), which are responsible for the allergic response and IgE production, IL-4/IL-13/STAT6 signaling pathway seems to be the most essential.6–13 Interleukin 5 (IL-5) has been recognized as the most important cytokine in the eosinophil lineage and it has been identified as the key common molecule in inflammatory pathways in asthma. IL-5 plays a key role in eosinophil proliferation, differentiation, migration to tissue sites and survival, as well as in prevention of eosinophil apoptosis.14–16
So far, several studies have confirmed the genetic linkage and association between 5q21-5q33 chromosomal region (involving, among others, loci for IL-3, IL-4, and IL-13) and atopic phenotype with predominance of Th2 balance.17–19 It is claimed that IL-4 and IL-13 in particular and a common subunit of their corresponding receptor complexes (IL-4Rα) regulate allergic inflammation.8,20 Moreover, it has been documented that IL-4 and IL-13 are exclusive cytokines, regarding the stimulation of IgE production in B cells and Th2 type differentiation in T cells.21 Also STAT6, the signaling molecule from JAK/STAT pathway, activated by IL-4 and IL-13 cytokines, plays an important role in IgE production and allergic airway inflammation. This mechanism was precisely documented in STAT6 gene-knockout animal studies, where IL-13 was suggested as a crucial cytokine in inducing asthma in animal model.22–25 IL-4, IL-13, or STAT6 deficiency in mice may influence the IgE synthesis and Th-2 type reactions. Based on animal studies, it can be assumed that molecular changes in transcriptional and/or translation level of IL-4/IL-13/STAT6 signaling pathway may be important in the development of atopic asthma in human. Additionally, so far little is known about the contribution of IL-4/IL-13/STAT6 in atopy development and IgE production in patients with allergic asthma.26,27 The study of interactions between three major molecules in IL-4/IL-13/STAT6 signaling pathway might be essential in understanding the gene/protein expression effect of these molecules on the development and course of atopic asthma. Therefore, the aim of our study was the evaluation of IL-4, IL-13, STAT6 gene expression and protein immunoexpression in the context of their relationship with serum total IgE level and lung function in patients with atopic asthma.
Materials
Patients and control group
The study has been approved by the Ethical Committee of the Medical University of Lodz, Poland, no. RNN/94/11/KE. Written informed consent was obtained from each patient.
A total of fifty (n = 50) patients with atopic asthma, 13 men and 37 women (mean age, 48±6 years) were recruited in the study. Patients were admitted to the Department of Pneumonology and Allergy of N. Barlicki Memorial University Teaching Hospital No 1, Poland, during the years 2012–2014.
In order to confirm the diagnosis of atopic asthma, the patient should fulfill the following criteria: positive spirometric reversibility test or methacholine challenge; disease onset before the age of 40 years; and atopy confirmed by the positive skin-prick tests or IgE in the peripheral blood and positive family history.28 Moreover, in controls IgE evaluation and lung function tests were performed. Current or ex-smokers, children, adolescents, and pregnant women were excluded from the study.
The skin prick tests were performed using commercially available tests (Allergopharma, Joahim Ganzer KG, Germany). The positive result was wheal size over 3 mm.
Measurements of total serum IgE were performed using commercially available, specific enzyme-linked immunosorbent assays (IgE ELISA kit, Allergopharma, Warsaw, Poland). The detection limit for the assay was 1 IU/mL. The concentration over 100 IU/mL was considered as high total serum IgE concentration.
Spirometry was performed according to European Respiratory Society (ERS) standards.29 The reversibility test was done 20–30 min after 400 μg of inhaled salbutamol. Pulmonary function tests were performed: forced vital capacity (FVC), forced expiratory volume in 1 s (FEV1), and peak expiratory flow rate (PEFR) were measured. Results were expressed as percentage of the predicted values (% pred.). The ratio of forced expiratory volume in the first second to forced vital capacity (FEV1/FVC) was calculated (Table 1).
Table 1.
Clinical and biological characteristics of the study patients with diagnosed atopic asthma.
| Patients | Gender | Age (years) | PEFR (l/s/%pred.) | FEV1% | FVC% | FEV1/FVC %pred. | |
|---|---|---|---|---|---|---|---|
| Atopic asthma (n = 50) | F 37 | M 13 | 48 ± 6.09 | 4.13/80– 10.34/141 | 53–143 ± 4.23 | 63–131 ± 4.12 | 63–94 ± 4.33 |
| Controlled (n = 40) | 21 | 19 | 45 ± 8.01 | 2.35/98–11.23/138 | 45–112 ± 3.28 | 58–125 ± 3.56 | 56–78 ± 4.21 |
| Uncontrolled (n = 10) | 6 | 4 | 49 ± 7.45 | 4.74/77– 9.97/146 | 59–152 ± 4.87 | 67–139 ± 4.53 | 69–106 ± 4.41 |
F, female; FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity; M, male; PEFR, peak expiratory flow rate.
Twenty gender- and age-matched normal non-asthmatic volunteers were involved in this study as controls. The small number of controls was related to the technical problems – we did not manage to collect more healthy volunteers.
Methods
Blood sample collection
Blood samples were collected into 2 mL EDTA (anticoagulant) containing tubes. Lymphocytes were isolated using Histopaque-1077 (Sigma-Aldrich, Poznan, Poland), the density gradient cell separation medium, according to the producer’s protocol.
Serum collection
The blood was collected in Eppendorf tubes, and left for approximately 30–45 min at 37°C (until clot formation). Then it was placed in a refrigerator at a temperature of 4°C for several hours (0.5–24 h) up to the total organization of the clot. Next, the tube was centrifuged (1200 × g 10 min, 4°C), and serum was separated from the clot carefully into a new sterile tubes, frozen, and stored at −20°C.
RNA extraction, real-time PCR
RNA isolation from lymphocytes was performed using RNA Isolation Kit (EURx, Poland), according to the manufacturer’s protocol. The quality and quantity assessments of RNA samples were determined by mini-electrophoreses in polyacrylamide gel (Agilent 2100 Bioanalyzer, Agilent, USA), using RNA 6000 Pico/Nano LabChip kit (Agilent Technologies, USA). Complementary DNA (cDNA) was transcribed from 100 ng of total RNA, using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA) in a total volume of 20 µL per reaction. Reverse transcription (RT) master mix contained: 10× RT buffer, 25× dNTP Mix (100 mM), 10× RT Random Primers, MultiScribe™ Reverse Transcriptase, RNase Inhibitor, and nuclease-free water. RT reaction was performed in a Personal Thermocycler (Eppendorf, Germany) in the following conditions: 10 min at 25°C, followed by 120 min at 37°C, then the samples were heated to 85°C for 5 s, and held at 4°C. The relative expression was assessed using TaqMan probes: Hs00932431_m1, Hs00174379_m1, Hs00598625_m1, for the studied genes IL-4, IL-13, and STAT6, respectively, as well as for β-actin (ACTB, Hs99999903_m1), as the reference gene. The qPCR mixture contained: cDNA (1 to 100 ng), 20× TaqMan® Gene Expression Assay, 2× TaqMan® Gene Expression Master Mix, RNase-free water in a total volume of 20 µL. The qPCR reactions were performed in Applied Biosystems 7900HT Fast Real-Time PCR System for 39 cycles, with annealing temperature of 60°C, repeated three times for each sample. The relative expression of the studied samples was assessed using the comparative delta-delta CT method (TaqMan Relative Quantification Assay software) and presented as RQ value, adjusted to β-actin expression level. RNA isolated from lymphocytes of healthy person served as calibrator sample.
Immunoexpression analysis
Immunoexpression analysis was performed using commercial ELISA kits for IL-4, IL-13 (Diaclone, Besancon Cedex, France), and STAT6 (RayBiotech, USA). The intensity of the final colorimetric reaction, in proportion to the amount of protein bound, was measured in a plate reader (ELx800, BioTek) at 450 nm. The obtained results were compared to the standard solution of known concentrations (10–1000 pg/mL).
Statistical analysis
The Mann–Whitney U test, ANOVA Kruskal–Wallis test, and Spearman’s rank correlation coefficient were performed in order to evaluate the relationship between the expression (RQ values)/immunoexpression levels of the studied genes/proteins (IL-4, IL-13, and STAT6), between RQs/protein concentration and patient characteristics: age, gender and classification of patients according to the degree of asthma control (controlled and uncontrolled asthma), and lung functional parameters (PEFR%, FEV1%, FVD%, FEV/ FVC). Statistica for Windows 10.0 program was applied for calculations.
Results
Relative gene expression analysis in PB lymphocytes of patients vs. controls
In PB lymphocytes all studied genes: IL-4, IL-13, and STAT6 revealed higher relative expression (mean RQ values) in asthmatic patients when compared with controls. Statistically significant differences between these two groups have been observed only for IL-13 gene (P = 0.03, Mann–Whitney U test), with higher gene expression in asthmatic patients. The mean RQ values obtained for the studied genes in PB lymphocytes are shown in Table 2.
Table 2.
Mean RQ values of all studied genes: IL-4, IL-13, STAT6 in PB lymphocytes in patient and control groups.
| Study group | Mean (SD) RQ value |
||
|---|---|---|---|
| IL-4 | IL-13 | STAT6 | |
| Patients with atopic asthma (n = 50) | 1.28 ± 8.25 | 3.53 ± 3.25 | 1.48 ± 6.53 |
| Controls (n = 20) | 0.98 ± 0.81 | 0.53 ± 1.45 | 0.84 ± 0.24 |
| Significance level | P = 0.17 | P = 0.03 | P = 0.23 |
The expression of the studied genes was increased in relation to calibrator (RQ>1) in 32% samples in case of IL-4 gene (P = 0.17; Mann–Whitney U test), in 58% samples for IL-13 (P = 0.03; Mann–Whitney U test) and in 63% samples for STAT6 gene (P = 0.23; Mann–Whitney U test).
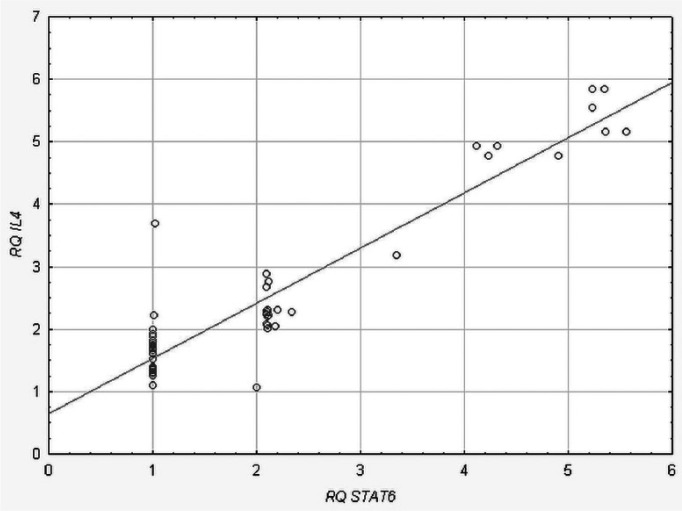
We found a statistically significant positive correlation between IL-4 and STAT6 (rho = 0.098, P = 0.048, Spearman’s rank correlation) in the patient group (Figure 1). There were no statistically significant correlations between expression levels of the studied genes in the control group (data not shown).
Figure 1.
Positive correlation between STAT6 and IL-4 gene expression levels (RQ values) in atopic asthma patients.
Relative expression analysis of the studied genes in PB lymphocytes of patients in relation to the degree of asthma control
In PB lymphocytes higher relative expression (mean RQ value) was observed for IL-13 and lower for IL-4 and STAT6 genes for patients with uncontrolled asthma as compared to controlled asthma group (Mann–Whitney U test), however, without statistically significant differences. Mean RQ values for all studied genes in PB lymphocytes are shown in Table 3.
Table 3.
Mean RQ values of all studied genes: IL-4, IL-13, STAT6 in PB lymphocytes in patients with controlled asthma and with uncontrolled asthma.
| Patient groups | Mean (SD) RQ |
||
|---|---|---|---|
| IL-4 | IL-13 | STAT6 | |
| Controlled asthma (n = 40) | 1.67 ± 3.42 | 3.41 ± 1.25 | 1.33 ± 3.52 |
| Uncontrolled asthma (n = 10) | 1.17 ± 1.87 | 3.66 ± 2.34 | 1.17 ± 2.81 |
| Significance level | P = 0.12 | P = 0.34 | P =0.25 |
Immunoexpression analysis of the studied proteins in serum of patients vs. controls
In blood serum, all studied proteins: IL-4, IL-13, and STAT6 revealed higher immunoexpression levels in patients with allergic asthma as compared to control group, but the differences were not statistically significant (P >0.05, Mann–Whitney U test). Mean immunoexpression levels of the studied proteins in serum are shown in Table 4.
Table 4.
Immunoexpression levels (pg/mL) of IL-4, IL-13, and STAT6 proteins in patients and controls.
| Patient groups | Mean (SD) immunoexpression levels |
||
|---|---|---|---|
| IL-4 | IL-13 | STAT6 | |
| Patients with atopic asthma (n = 50) | 16.72 pg/mL ± 3.64 | 81.67 pg/mL ± 2.03 | 465.21 pg/mL ± 0.91 |
| Control group (n = 20) | 12.87 pg/mL ± 12.57 | 51.34 pg/mL ± 8.21 | 234.27 pg/mL ± 5.08 |
| Significance level | P = 0.43 | P = 0.82 | P = 0.67 |
Immunoexpression analysis of the studied proteins in serum of patients in relation to the degree of asthma control
In blood serum all studied proteins: IL-4, IL-13, and STAT6 revealed higher immunoexpression levels in patients with uncontrolled asthma as compared to patients with controlled asthma, however the differences were not statistically significant (P >0.05; Mann–Whitney U test). The immunoexpression for the study proteins IL-4, IL-13, STAT6 in serum of patients in relation to degree of asthma control are shown in Table 5.
Table 5.
Immunoexpression levels (pg/mL) of IL-4, IL-13, and STAT6 proteins in patients in relation to the degree of asthma control.
| Protein | Mean (SD) immunoexpression levels |
||
|---|---|---|---|
| IL-4 | IL-13 | STAT6 | |
| Controlled asthma (n = 40) | 15.42 pg/mL ± 1.63 | 80.28 pg/mL ± 2.84 | 466.82 pg/mL ± 2.02 |
| Uncontrolled asthma (n = 10) | 17.87 pg/mL ± 2.54 | 87.92 pg/mL ± 5.55 | 473.72 pg/mL ± 1.98 |
| Significance level | P = 0.21 | P = 0.36 | P = 0.15 |
Total serum IgE levels in patients vs. controls
Mean values of serum IgE levels in patients and controls were 93.2 IU/mL and 55.1 IU/mL, respectively. The difference was statistically significant (P = 0.03; Mann–Whitney U test). Serum IgE level exceeded 100 IU/mL in 82% of patients as compared to controls.
Gene expression analysis in relation to patient age, gender, smoking history, lung functional parameters (PEFR%, FEV1%, FVD%, FEV/FVC), degree of asthma control, and IgE level
There were no statistically significant differences between the levels of gene expression and patient classification in relation to the degree of asthma control (controlled and uncontrolled asthma), clinical features of patients (age, gender, smoking history), and lung functional parameters (PEFR%, FEV1%, FVD%, FEV/FVC) (P >0.05, Mann–Whitney U test, Kruskal–Wallis test, Neuman–Keuls’ multiple comparison test, and Spearman’s rank correlation).
Positive correlations were found between: the expression level of IL-13 gene and total level of serum IgE (rho = 0.23; P = 0.033; Spearman’s rank correlation rho) and the expression level of STAT6 gene and total level of serum IgE (rho = 0.077, P = 0.038; Spearman’s rank correlation). There was no significant correlation between the level of serum IgE and IL-4 expression level.
Protein expression analysis in relation to patient age, gender, smoking history, lung functional parameters (PEFR%, FEV1%, FVD%, FEV/FVC), degree of asthma control, and IgE level
Statistical analysis showed no significant correlations between the levels of protein immunoexpression and classification of patients according to the degree of asthma control (controlled and uncontrolled asthma), patients’ clinical features (age, gender, smoking history), and lung functional parameters (PEFR%, FEV1%, FVD%, FEV/FVC) (P >0.05, Mann–Whitney U test, Kruskal–Wallis test, Neuman–Keuls’ multiple comparison test, and Spearman’s rank correlation). We found no significant correlations between IL-4, IL-13 immunoexpression levels and IgE levels.
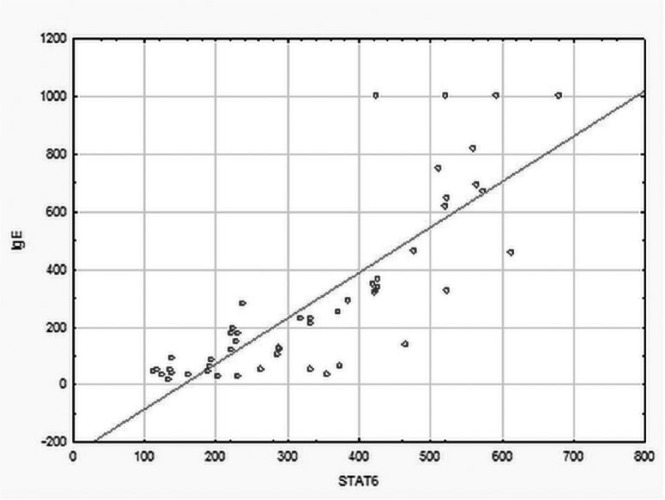
Spearman’s rank correlation coefficient revealed statistically significant positive correlation between STAT6 level and IgE levels (rho = 0.49, P = 0.042; Spearman’s rank correlation) (Figure 2).
Figure 2.
Positive correlation between IgE and STAT6 levels in atopic asthma patients.
Discussion
Atopic asthma is influenced by genetic and environmental factors, and the level of atopy in particular is an inherited tendency, characterized by high non-specific IgE and/or high specific IgE against common antigens.27 Particularly, non-antigen specific IgE responses (total serum IgE level), involving interactions between mast cells, basophils, and T and B cells, require complex gene–gene and gene–environment interactions. Recently, many genetic and functional studies have highlighted the significance of IL-4/IL-13 signaling in the development of asthma and atopy.1–10,16–18,26,27 Specifically, several reports confirmed the correlations of IL-4Ralpha gene polymorphisms with atopy.6,7,30 Surprisingly, the studies focused on mRNA/protein levels of molecules of IL-4/IL-13/STAT6 signaling pathway and their significance in development and course of atopic asthma are rare.
In our study we confirmed higher mRNA level of the studied genes: IL-4, IL-13, and STAT6 in patients with atopic asthma when compared to controls, and in case of IL-13 this difference was statistically significant. This finding is in accordance with the observations of other authors who reported significantly elevated mRNA IL-13 levels in PB or bronchial mucosal biopsy specimens from asthmatic (atopic and non-atopic) subjects.26,27 Based on these results, we may highlight the hypothesis that IL-13 cytokine, especially on a transcription level, may have a regulatory function in the development of atopic asthma. Our finding may also be reinforced in the light of observations of Humbert et al.,19 who found a correlation between the IL-13 mRNA level and severity of atopic asthma.27 The International Severe Asthma Forum (ISAF)28 proposed the definition of severe asthma which causes much controversy among clinicians. According to the ISAF definition, severe asthma is distinguished on the basis of clinical signs and deterioration of lung function parameters, and also in untreated patients. Recently, according to the GINA guidelines and the World Health Organization (WHO)32,33 new definition of asthma, attention has been drawn to the degree of asthma control, and severe asthma has been well-defined as an uncontrolled severe asthma, with frequent exacerbations in the course of disease. In the light of this definition, we did not observe any significant differences in mRNA expression of IL-13 gene between controlled and uncontrolled asthma, however IL-13 expression was higher in patients with uncontrolled atopic asthma. Interestingly, Truyen et al.34 demonstrated that IL-13 mRNA expression level significantly correlated with the percentage of eosinophils and was higher in subjects with allergic asthma. In our study we did not assess the percentage of eosinophils. However, another study showed that IL-13 expression was increased generally in asthma as compared to non-asthmatic eosinophilic bronchitis.35 It points out a lack of correlation between IL-13 expression and eosinophil levels in the course of asthma.
Nevertheless, the relationship between IL-13 and IgE seems to be interesting because IL-13 and IL-4 were recognized as potent switch factors for IgE synthesis in human B cells in allergic patients.18 Doleck et al.18 confirmed that adding of IL-4, IL-13 to the cultures alone or in combinations significantly induced total IgE production. However, Wills-Karp et al.36 suggested that IL-13 was one of the cytokines that induced asthma via the mechanisms independent of IL-4, IgE, and eosinophils. Also, Afshari et al.27 found that IgE production in the asthmatic patients did not correlate with expression of IL-13 mRNA. Surprisingly, in our study the expression level of IL-13 correlated with IgE immunoexpression level in patients with atopic asthma. Our results are similar to those obtained by Metwally et al.37 who found highly significant positive correlation between serum levels of IgE and the levels of IL-13 mRNA expression in patients with atopic dermatitis. This observation indicated that IL-13 overexpression is related rather to atopy than asthma pathomechanism.38
In our study we also assessed IL-4 on transcription and translation levels. We did not find statistically significant differences between controlled/uncontrolled atopic asthma, however the highest IL-4 expression was recognized in uncontrolled asthma patients and it was also higher in patients as compared to controls. This observation may suggest that IL-4 expression may play a significant role in asthma course but it is not a sufficient inducer for asthma.6,19 Additionally, the expression/immunoexpression level of IL-4 is functionally dependent on genetic variants of IL-4 (SNP).36 In light of the association studies, it can be hypothesized that some IL-4 SNPs may lead to an overexpression of IL-4 gene and thus influence the immunological reaction.39 In our study we did not observe the correlation between IL-4 expression/immunoexpression and IgE level in patients. Our findings are consistent with the results of other authors. In the study performed by Kraan et al.40 the correlation between the production of IgE and IL-4 in atopic patients with asthma was not recognized, and thus emphasized the fact that IgE synthesis is distinguished in different patient groups. On the other hand, the study performed by Humbert et al.19 indicated that expression of IL-4 mRNA positively correlated with total serum IgE level and, similarly like in our study, did not correlate with asthma severity.
Park et al.41 showed the association between single nucleotide polymorphism of IL-13 and lung function in early childhood. In COPD the association was found between IL-13 level and increased lung functional parameters, e.g. % FEV1.42 In our study we did not observe any correlations between IL-13 expression/immunoexpression and lung functional parameters.
In atopic asthma it was found that IL-4 expression was stimulated via the activation of the STAT6 pathway thus showing that STAT6 plays a positive role, affecting the expression of IL-4.43 In our study we observed higher expression/immunoexpression of STAT6 in asthmatic patients as compared to controls, and the expression levels of IL-4 and STAT6 showed significant positive correlation, also higher in uncontrolled asthma. So far, it has been documented that SNP variants influence IL-4Rα and STAT6 expression44 and these are essential for the action of both cytokines, and for the development of atopy and asthma. However, no common variants of the genes were confirmed to be associated with phenotypes of asthma across different ethnic groups.44
Interestingly, in our study we observed a positive correlation between IgE serum level and expression/immunoexpression of STAT6. So far it has been claimed that STAT6, the intracellular transcription factor, influences IL-4/IL-13 pathway activation and may be involved in the regulation of Th2 immune response, activating several Th2 specific gene promoters.39 Shikarawa et al.44 showed that mice deficient in IL-4, IL-13, or STAT6 were characterized by an absence of IgE synthesis and Th2-type reactions. It is pointed out that genetic variants (SNP) of molecules of the STAT6/IL-4/IL-13 signaling pathway might be crucial for the development of atopic disorders.44 On the other hand, other data suggested no significant association between STAT6 or STAT4 mRNA and asthma or serum total IgE levels,45 although it was confirmed that it might promote humoral immunity, food allergies, and/or atopic dermatitis.46 Moreover, STAT6 polymorphisms have an important influence on IgE level and development of asthma.47 Lau et al.48 has suggested that loss of STAT6 promotes autoimmune disease and atopy and STAT6 correlates with IgE level.
Studies focused on associations between STAT6 and lung functional parameters are rare. Leung et al.49 suggested that STAT6 might influence lung function growth in asthmatic children. In our study we did not observe any correlations between STAT6 expression/immunoexpression and lung functional parameters.
Reassuringly, we found statistically significant positive correlations between the expression levels of IL-13 gene, STAT6 gene/STAT6, and total level of serum IgE. It could indicate the role of these genes and proteins in asthma progression. In the absence of other reports on these gene expression/imunnoexpression in asthma, the results require confirmation in bigger group of asthma patients.
Additionally, we also found positive correlation between IL-4 and STAT6 expression levels, thus suggesting that IL-4 expression activated the STAT6 pathway.
No correlation was found between expression/immunoexpression level of the studied genes and clinical asthma classification (controlled and uncontrolled atopic asthma), clinical patients’ features (gender, age, smoking behavior) or lung function parameters (PEF%, FEV1%, FVD%, FEV/FVC). In conclusions, we claimed that our data support the role of Th2 cytokines (IL-4, IL-13) and STAT6 in Th1/Th2 imbalance in atopic asthma and highlight the etiological relationship between the molecules of IL-4/IL-13/STAT6 signaling pathway in atopic asthma. The overexpression of STAT6 and IL-13 expression can be useful in the future in biological treatment of patients with atopic asthma by gene expression inhibition.
Such knowledge is extremely important in the context of inhibition of STAT6 in the potential treatment strategy of asthma. Currently, a number of biological molecules are being developed for the treatment of asthma. Moreover, pre-clinical and emerging proof-of-concept data suggest that IL-4/IL-13/STAT-6 pathway plays important role in the pathogenesis of asthma (by promoting airway hyperresponsiveness and inflammation). Additionally, the targeted therapy toward STAT activity might be based on the findings regarding IL-4 and/or IL-13 expression in asthma. Data suggested that the biological compounds targeting these molecules may provide a new therapeutic modality for patients with uncontrolled severe asthma.43
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: This study was financially supported by grants from the Department of Molecular Bases of Medicine, I Chair of Internal Diseases, Medical University of Lodz (no. 502-03/1-151-04/502-14-069) from the Medical University of Lodz, Poland, for completing the research within the financial framework for the development of young scientists and PhD students.
References
- 1. Los H, Koppelman GH, Postma DS. (1999) The importance of genetic influences in asthma. European Respiratory Journal 14: 1210–1227. [DOI] [PubMed] [Google Scholar]
- 2. Custovic A, Marinho S, Simpson A. (2012) Gene-environment interactions in the development of asthma and atopy. Expert Review of Respiratory Medicine 6: 301–308. [DOI] [PubMed] [Google Scholar]
- 3. Ober C, Cox NJ, Abney M, et al. (1998) Genome-wide search for asthma susceptibility loci in a founder population. The Collaborative Study on the Genetics of Asthma. Human Molecular Genetics 7: 1393–1398. [DOI] [PubMed] [Google Scholar]
- 4. Blumenthal MN. (2005) The role of genetics in the development of asthma and atopy. Current Opinion in Allergy and Clinical Immunology 5: 141–145. [DOI] [PubMed] [Google Scholar]
- 5. Blumenthal MN, Langefeld CD, Beaty TH, et al. (2004) A genome-wide search for allergic response (atopy) genes in three ethnic groups: Collaborative Study on the Genetics of Asthma. Human Genetics 114: 157–164. [DOI] [PubMed] [Google Scholar]
- 6. Heinzmann A, Mao XQ, Akaiwa M, et al. (2000) Genetic variants of IL-13 signalling and human asthma and atopy. Human Molecular Genetics 9: 549–559. [DOI] [PubMed] [Google Scholar]
- 7. Shirakawa T, Deichmann KA, Izuhara K, et al. (2000) Atopy and asthma: Genetic variants of IL-4 and IL-13 signalling. Immunology Today 21: 60–64. [DOI] [PubMed] [Google Scholar]
- 8. Hoffjan S, Ober C. (2002) Present status on the genetic studies of asthma. Current Opinion in Immunology 14: 709–717. [DOI] [PubMed] [Google Scholar]
- 9. Venkayya R, Lam M, Willkom M, et al. (2002) The Th2 lymphocyte products IL-4 and IL-13 rapidly induce airway hyperresponsiveness through direct effects on resident airway cells. American Journal of Respiratory Cell and Molecular Biology 26: 202–208. [DOI] [PubMed] [Google Scholar]
- 10. Elias JA, Lee CG, Zheng T, et al. (2003) New insights into the pathogenesis of asthma. Journal of Clinical Investigation 111: 291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kuperman DA, Huang X, Koth LL, et al. (2002) Direct effects of interleukin-13 on epithelial cells cause airway hyperreactivity and mucus overproduction in asthma. Nature Medicine 8: 885–889. [DOI] [PubMed] [Google Scholar]
- 12. Christodoulopoulos P, Cameron L, Nakamura Y, et al. (2001) TH2 cytokine-associated transcription factors in atopic and nonatopic asthma: Evidence for differential signal transducer and activator of transcription 6 expression. Journal of Allergy and Clinical Immunology 107: 586–591. [DOI] [PubMed] [Google Scholar]
- 13. Mullings RE, Wilson SJ, Puddicombe SM, et al. (2001) Signal transducer and activator of transcription 6 (STAT-6) expression and function in asthmatic bronchial epithelium. Journal of Allergy and Clinical Immunology 108: 832–838. [DOI] [PubMed] [Google Scholar]
- 14. Mukherjee M, Sehmi R, Nair P. (2014) Anti-IL5 therapy for asthma and beyond. World Allergy Organization Journal 7: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maeda S, Yanagihara Y. (2001) Inflammatory cytokines (IL-4, IL-5 and IL-13). Nihon Rinsho 59: 1894–1899. [PubMed] [Google Scholar]
- 16. Garcia G, Taillé C, Laveneziana P, et al. (2013) Anti-interleukin-5 therapy in severe asthma. European Respiratory Review 22: 251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adjers K, Pessi T, Karjalainen J, et al. (2004) Epistatic effect of IL1A and IL4RA genes on the risk of atopy. Journal of Allergy and Clinical Immunology 113: 445–447. [DOI] [PubMed] [Google Scholar]
- 18. Dolecek C, Steinberger P, Susani M, et al. (1995) Effects of IL-4 and IL-13 on total and allergen specific IgE production by cultured PBMC from allergic patients determined with recombinant pollen allergens. Clinical and Experimental Allergy 25: 879–889. [DOI] [PubMed] [Google Scholar]
- 19. Humbert M, Corrigan CJ, Kimmitt P, et al. (1997) Relationship between IL-4 and IL-5 mRNA expression and disease severity in atopic asthma. American Journal of Respiratory and Critical Care Medicine 156: 7048. [DOI] [PubMed] [Google Scholar]
- 20. Wynn TA. (2003) IL-13 effector functions. Annual Review of Immunology 21: 425–456. [DOI] [PubMed] [Google Scholar]
- 21. Chatila TA. (2004) Interleukine-4 receptor signaling pathways in asthma pathogenesis. Trends in Molecular Medicine 10: 493–499. [DOI] [PubMed] [Google Scholar]
- 22. Kaplan MH, Schindler U, Smiley ST, et al. (1996) Stat6 is required for mediating responses to IL-4 and for the development of Th2 cells. Immunity 4: 313–319. [DOI] [PubMed] [Google Scholar]
- 23. Shimoda K, van Deursen J, Sangster MY, et al. (1996) Lack of IL-4 induced Th2 response and IgE class switching in mice with disrupted stat6 gene. Nature 380: 630–633. [DOI] [PubMed] [Google Scholar]
- 24. Elias JA, Lee CG, Zheng T, et al. (2003) New insights into the pathogenesis of asthma. Journal of Clinical Investigation 111: 291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grunig G. (1998) Requirement for IL-13 independently of IL-4 in experimental asthma. Science 282: 2261–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Van Der Pouw Krann TC, Van Der Zee JS, Boeije LC, et al. (1998) The role of IL-13 in IgE synthesis by allergic asthma patients. Clinical and Experimental Immunology 111: 129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tavakkol Afshari J, Hosseini FR, Farahabadi HS, et al. (2007) Association of the expression of IL-4 and IL-13 genes, IL-4 and IgE serum levels with allergic asthma. Iranian Journal of Allergy Asthma Immunology 6: 67–72. [PubMed] [Google Scholar]
- 28. Global Strategy for Asthma Management and Prevention, Global Initiative for Asthma (GINA). 2014. Available at: http://www.ginasthma.org/.
- 29. Miller MR, Hankinson J, Brusasco V, et al. (2005) ATS/ERS Task Force: Standardisation of lung function testing; standardisation of spirometry. European Respiratory Journal 26: 319–338. [DOI] [PubMed] [Google Scholar]
- 30. Heinzmann A, Daser A. (2002) Mouse models for the genetic dissection of atopy. International Archives of Allergy and Immunology 127: 170–180. [DOI] [PubMed] [Google Scholar]
- 31. International Severe Asthma Forum, 11–13 October 2012, Gothenburg, Sweden. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rabe KF, Hurd S, Anzueto A, et al. (2007) Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med 15: 532–555. [DOI] [PubMed] [Google Scholar]
- 33. Łukaszczyk Bush A, Zar HJ. (2011) WHO universal definition of severe asthma. Current Opinion in Allergy and Clinical Immunology 11: 115–121. [DOI] [PubMed] [Google Scholar]
- 34. Truyen E, Coteur L, Dilissen E, et al. (2006) Evaluation of airway inflammation by quantitative Th1/Th2 cytokine mRNA measurement in sputum of asthma patients. Thorax 61: 202–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Siddiqui S, Cruse G, McKenna S, et al. (2009) IL-13 expression by blood T cells and not eosinophils is increased in asthma compared to non-asthmatic eosinophilic bronchitis. BMC Pulmonary Medicine 9: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wills-Karp M, Luyimbazi J, Xu X, et al. (1998) Interleukin-13: central mediator of allergic asthma. Science 282: 2258–2261. [DOI] [PubMed] [Google Scholar]
- 37. Metwally SS, Mosaad YM, Abdel-Samee ER, et al. (2004) IL-13 gene expression in patients with atopic dermatitis: Relation to IgE level and to disease severity. Egyptian Journal of Immunology 11: 171–177. [PubMed] [Google Scholar]
- 38. Fallon PG, Emson CL, Smith P, et al. (2001) IL-13 overexpression predisposes to anaphylaxis following antigen sensitization. Journal of Immunology 166: 2712–2716. [DOI] [PubMed] [Google Scholar]
- 39. Kabesch M, Schedel M, Carr D, et al. (2006) IL-4/IL-13 pathway genetics strongly influence serum IgE levels and childhood asthma. Journal of Allergy and Clinical Immunology 117: 269–274. [DOI] [PubMed] [Google Scholar]
- 40. Van der Pouw Kraan TC, Van der Zee JS, Boeije LC, et al. (1998) The role of IL-13 in IgE synthesis by allergic asthma patients. Clinical and Experimental Immunology 111: 129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Park HW, Lee JE, Kim SH, et al. (2009) Genetic variation of IL13 as a risk factor of reduced lung function in children and adolescents: A cross-sectional population-based study in Korea. Respiratory Medicine 103: 284–288. [DOI] [PubMed] [Google Scholar]
- 42. Lee JS, Rosengart MR, Kondragunta V, et al. (2007) Inverse association of plasma IL-13 and inflammatory chemokines with lung function impairment in stable COPD: A cross-sectional cohort study. Respiratory Research 8: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oh CK, Geba GP, Molfino N. (2010) Investigational therapeutics targeting the IL-4/IL-13/STAT-6 pathway for the treatment of asthma. European Respiratory Review 19: 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shirakawa I, Deichmann KA, Izuhara I, et al. (2000) Atopy and asthma: Genetic variants of IL-4 and IL-13 signalling. Immunology Today 21: 60–64. [DOI] [PubMed] [Google Scholar]
- 45. Pykäläinen M, Kinos R, Valkonen S, et al. (2005) Association analysis of common variants of STAT6, GATA3, and STAT4 to asthma and high serum IgE phenotypes. Journal of Allergy and Clinical Immunology 115: 80–87. [DOI] [PubMed] [Google Scholar]
- 46. Bernstein IL, Li JT, Bernstein DI, et al. (2008) Allergy diagnostic testing: An updated practice parameter. Annals of Allergy Asthma & Immunology 100: 1–148. [DOI] [PubMed] [Google Scholar]
- 47. Godava M, Vrtel R, Vodicka R. (2013) STAT6 - polymorphisms, haplotypes and epistasis in relation to atopy and asthma. Biomedical Papers of the Medical Faculty of the University Palacky, Olomouc, Czechoslovakia 157: 172–180. [DOI] [PubMed] [Google Scholar]
- 48. Lau M, Tsantikos E, Maxwell MJ, et al. (2012) Loss of STAT6 promotes autoimmune disease and atopy on a susceptible genetic background. Journal of Autoimmunity 39: 388–397. [DOI] [PubMed] [Google Scholar]
- 49. Leung TF, Chan IH, Wong GW, et al. (2007) Association between candidate genes and lung function growth in Chinese asthmatic children. Clinical and Experimental Allergy 37: 1480–1486. [DOI] [PubMed] [Google Scholar]