Abstract
Cyclic nucleotide phosphodiesterase (PDE) exists as multiple molecular forms. Of the 11 families of PDE identified so far, PDE4, a cAMP-specific PDE, has been identified as the major isoform regulating inflammatory activity. The principle aim of the present study was to determine whether human basophils and human lung mast cells express PDE4. Four sub-classes of PDE4 (A, B, C, and D) have been identified and expression of these was determined by RT-CPR and by western blotting. In basophils, prominent expression of mRNA for PDE4A and PDE4D was observed whereas little if any expression of PDE4B and PDE4C was detected. These findings were paralleled by immunoblotting experiments as human basophils were found to express PDE4A and PDE4D with little evidence for the presence of either PDE4B or PDE4C. By contrast, human lung mast cells expressed very little, if any, mRNA for PDE4 sub-classes although, in some preparations, some modest levels of mRNA for PDE4D were detected. However, there was no evidence, at the protein level, that mast cells express PE4. Overall, these data indicate that basophils express PDE4 (4A and 4D) whereas human lung mast cells do not.
Keywords: allergy, basophil, drug, lung, mast cell, PCR
Introduction
Recent studies indicate that cyclic nucleotide phosphodiesterase (PDE) exists as multiple molecular forms.1–4 At least 11 families of PDE are known to exist and, as products of different genes, these can be disguised structurally.3,4 Moreover, different isoforms can be discriminated in terms of preferential hydrolysis of substrate (cAMP or cGMP) and regulation by both naturally-occurring and pharmacological agents.2 Indeed, the development of isoform-selective drugs has aided significantly in the identification of isoforms in tissues and in determining whether a given PDE isoform is involved in a biological process.2,5 As a consequence of studies of this type, a large body of evidence has emerged demonstrating that inhibitors of the cAMP-specific PDE, PDE4, are effective at stabilizing the activity of a wide variety of inflammatory cells.5 This has led to suggestions that targeting PDE4 could be an effective anti-inflammatory strategy.6
The mast cell and basophil are thought to be central to inflammation that has an allergic basis as allergens activate these cells in an IgE-dependent manner to generate mediators such as histamine, eicosanoids, and cytokines.7–9 It has been known for some time that non-selective inhibitors of PDE, such as IBMX and theophylline, are effective inhibitors of IgE-mediated histamine release.10,11 Later studies have shown that these same compounds can inhibit the stimulated generation of eicoanoids and cytokines from these cells.12–16 These inhibitory effects of IBMX and theophylline are likely to be mediated by cAMP rather than cGMP as non-hydrolysable analogues of cAMP, but not analogues of cGMP, inhibit mast cell and basophil activity.14 Moreover, extracts of mast cells and basophils hydrolyze cAMP but neither cell contains any noteworthy cGMP hydrolytic activity.13,14
The effects of selective inhibitors of PDE1 (8-methoxy-methyl-IBMX), PDE3 (siguazodan), PDE4 (rolipram), and PDE5 (zaprinast) on human basophil and mast cell function demonstrate that, of these selective inhibitors, only inhibitors of PDE4 are effective at preventing the stimulated release of histamine, eicosanoids, and cytokines from basophils. By contrast, none of the isoform-selective inhibitors, including several PDE4-selective inhibitors, has any effect on mediator release from human lung mast cells.14 These data suggest that inhibiting PDE4 stabilizes basophil but not mast cell activity. As a corollary to these functional data, the principal aim of the present study was to determine whether PDE4, which is known to exist as four sub-classes (4A, 4B, 4C, and 4D), is expressed by basophils and mast cells.
Materials and methods
Materials
The following were purchased from the sources indicated; aprotinin, BSA, Collagenase, DNase, Dowex AG-50W, DTT leupeptin, Percoll, PMSF, SBTI, Triton X-100 (all Sigma, Poole, UK); EDTA calcium chloride and magnesium chloride (BDH, Poole, UK); Tris, SDS, (Bio-Rad, Hemel Hempstead, UK); monoclonal (IgG1) mouse anti-human c-kit and monoclonal (IgG2) mouse anti-human IgE ( Immunotech, Marseilles, France); magnetic beads coated with rat anti-mouse IgG1 or IgG2 antibody (Dynal, Wirral, UK); anti-mouse and anti-rabbit HRP-conjugated antibodies (Santa Cruz, CA, USA). The following were gifts; recombinant PDE4A5, PDE4B2, and PDE4D1 and monoclonal antibodies to these were kindly provided by ICOS Corporation (Bothell, WA, USA); recombinant PDE4C and PDE7A2 and rabbit polyclonal antibodies were generously by Celltech Therapeutics, Ltd. (Slough, UK).
Buffers
Tyrode’s buffer contained (mM): NaCl 137; HEPES 1.2; KCL 2.7; NaH2PO4.H2O 0.04; glucose 5.6. Tyrode’s-BSA was Tyrode’s which additionally contained: CaCl2.2H2O 0.5 mM; MgCl2.6H2O 1 mM; BSA 1 mg/mL; DNase 15 μg/mL. The pH of Tyrode’s buffers was titrated to 7.3.
PBS contained (mM): NaCl 137; NaH2PO4.12H2O 8; KCl 2.7; KH2PO4 1.5. The pH of PBS buffer was titrated to 7.5.
Western lysis buffer contained: Tris 50 mM; NaCl 150 mM; PMSF 50 μg/mL; SBTI 50 μg/mL; leupeptin 5 μg/mL; aprotinin 5 μg/mlL Triton X-100 0.5%. The pH was titrated to 8.0.
Lung tissue
Macroscopically normal lung tissue from resections was used in this study. Background, non-lesion tissue from lung resections of patients was obtained following surgery. Most of patients were undergoing surgery for carcinoma. The male:female split was 60:40 and 95% of the patients were white Caucasians. The provision of lung tissue and the use of the tissue in this study were approved by the Local Research Ethics’ Committee.
Isolation of mast cells
Mast cells were isolated from normal human lung tissue by a modification of the method described.18 The tissue was stripped of its pleura and chopped vigorously for 15 min with scissors in a small volume of Tyrode’s buffer. The chopped tissue was washed over a nylon mesh (100 μm pore size; Casdisch and Sons, London, UK) with 0.5–11 of Tyrode’s buffer to remove lung macrophages. The tissue was reconstituted in Tyrode’s BSA 910 mL per g tissue) containing collagenase Ia (350 U per mL of Tyrode’s-BSA) and agitated by using a water-driven magnetic stirrer immersed in a water bath set at 37°C. The supernatant was separated from the tissue by filtration over nylon mesh. The collagenase-treated tissue was then reconstituted in a small volume of Tyrode’s-BSA buffer and disrupted mechanically with a syringe. The disrupted tissue was then washed over nylon gauze with Tyrode’s-BSA (300–600 mL). The pooled filtrates were sedimented (120 × g, room temperature, 8 min), the supernatant discarded, and the pellets reconstituted in Tryode’s-BSA (100 mL). The pellet was washed a further two times. The dispersion procedure generated 0.2–1 × 106 mast cells per g of lung tissue at 5–20% purity as assessed by Alcian blue staining.19 Mast cells were further purified by immunomagnetic bead separations as previously described.14 In brief, the lung cell preparations were incubated (1 h over ice with monoclonal [IgG1] mouse anti-human c-kit and then incubated (30 min) with Dynal magnetic beads coated with a rat anti-mouse IgG1 antibody at a ratio of beads to cells of 4 to 1. The magnetic fraction was harvested, using a Dynal MPC-1 magnet and the magnetically adherent cells counted with Alcian blue to determine mast cell purities (Gilbert and Ornstein, 1975). This fraction typically contained 1–3 × 106 mast cells at purities in the range of 91–99%.
Isolation of basophils
Mononuclear cells were isolated from venous blood of volunteer healthy people by Percoll density centrifugation according to methods described in detail elsewhere.14 Flotation of blood over Percoll density gradients also generated a basophil-enriched (5–15% purity) fraction which was purified further by immunomagnetic bead separation according to the method described.14 In brief, the basophil-enriched fraction was incubated (1 h) over ice with monoclonal (IgG2A) mouse anti-human IgE and then incubated (30 min) with Dynal magnetic beads coated with a rat anti-mouse IgG2A antibody at a ratio of beads to cells of 4 to 1. The magnetic fraction was harvested, using a Dynal MPC-1 magnet, and the magnetically adherent cells counted with Alcian blue to determine basophil purities.19 This fraction typically contained 1–3 × 106 basophils at purities in the range of 91–99%.
RT-PCR
Total RNA was extracted from cells (0.5–3 × 106 cells) using Trizol (1 mL). RNA content was quantified spectrophotometrically. The extracted RNA (1–2 μg) was denatured (65°C, 10 min) and then was reserve transcribed (37°C, 2 h) using the Moloney Murine Leukaemia reverse transcriptase in the presence of RNasin (30 U), dNTPs (1 mM), buffer, and random hexamers (113 μg/mL) in a total reaction volume of 40 μL. Samples were then incubated at 65°C for 10 min in order to inactivate the reverse transcriptase. As a control for genomic DNA, each sample of RNA was processed with and without the reverse transcriptase.
Reverse transcriptase-generated cDNAs were amplified by using specific primers20–22 and conditions designed to identify cDNAs for PDE3A, PDE3B, PDE4A, PDE4B, PDE4C, PDE4D, and PDE7 (Table 1). Also, to confirm the integrity of RNA and to gain an estimate of the level of PDE expression, we performed RT-PCR of the house-keeping gene, GADPH. The PCR reaction was carried out in a reaction volume of 50 μL containing cDNA, dNTPs (200 µM of each), Mg2+ (1–1.8 mM), primers (1 µM), and Taq polymerase (1 µL). Initial denaturation was at 94°C for 1 min with annealing temperatures, as indicated in Table 1, and polymerization was performed at 72°C for 1 min. Amplification was performed for 25 (GADPH) or 35 (PDEs) cycles. The PCR products were size fractionated alongside a 100 bp ladder on 1–1.5% agarose gels in a TBE buffer containing ethidium bromide. The PCR products were visualized and photographed under ultraviolet light. In order to control for potential contamination, water blanks were run alongside cDNA samples and also subjected to the PCR protocol.
Table 1.
PDE primers and conditions used for RT-PCR.
| PDE variant | Accession number | Primer sequences (5’-3’) | Tm of primers (ºC) | Optimal Mg2+ (mM) | Conditions | Amplicon length (base pairs) |
|---|---|---|---|---|---|---|
| PDE3A | M91667 | Sense: GAA CAG GGT GAT GAA GAG GC Antisense : CAC TGG TCT GGC TTTT GGG TTG |
62 68 |
1.8 | Denaturation, 94°C, 30 s Annealing, 60°C, 1 min Polymerization, 72°C, 1 min Elongation, 72°C, 10 min |
506 |
| PDE3B | X95520 | Sense: AAT TCT TCC AAC CAT GGA CC Antisense: GCA TGT AGC ACA TCT GTG GC |
58 62 |
1.8 | Denaturation, 94°C, 30 s Annealing, 64°C, 45 s Polymerization, 68°C, 1 min Elongation, 68°C, 1 min |
676 |
| PDE4A | L20965 | Sense: AAG AGG AGG AAG AAA TAT CAA TGG Antisense: TTA CAG CAA CCA CGA ATT CCT CC |
66 68 |
1.8 | Denaturation, 94°C, 30 s Annealing, 64°C, 30 s Polymerization, 68°C, 1 min Elongation, 68°C, 10 min |
272 |
| PDE4B1A | L20966 | Sense: AGG GCA GCC TGA GGT ATT AAA Antisense: CAC TCC TGG CTT ACA GTT GTA |
62 60 |
1.6 | Denaturation, 94°C, 30 s Annealing, 60°C, 30 s Polymerization, 68°C, 1 min Elongation, 68°C, 10 min |
364 |
| PDE4C | Z46632 | Sense: GCTTTGGGGTCCAGACTGAC Antisense: CTGAATATGATAGCTGTGAGGGG |
53 52 |
1.4 | Denaturation, 94°C, 30 s Annealing, 62°C, 30 s Polymerization, 68°C, 1 min Elongation, 68°C, 10 min |
132 |
| PDE4D1/2 | U50157/ U50158 | Sense: ATA TGA AGG AGC CCT CAT G Antisense: CCA GAC CGA CTC ATT TCA GAG A |
56 66 |
1.5 | Denaturation, 94°C, 30 s Annealing, 62°C, 1 min Polymerization, 72°C, 1 min Elongation, 72°C, 10 min |
D1 221 D2 307 |
| PDE4D3A | L20970 | Sense: GCA AGA TCG AGC ACC TAG CAA Antisense: CGG TTA CCA GAC AAC TCT GCT |
64 64 |
1.6 | Denaturation, 94°C, 30 s Annealing, 62°C, 30 s Polymerization, 68°C, 1 min Elongation, 68°C, 10 min |
516 |
| PDE7 | L12052 | Sense: GGA CGT GGG AAT TAA GCA AGC Antisense: TCC TCA CTG CTC GAC TGT TCT |
64 64 |
1.6 | Denaturation, 94°C, 30 s Annealing, 60°C, 30 s Polymerization, 68°C, 1 min Elongation, 68°C, 10 min |
285 |
| GAPDH | JO4038 | Sense: CCA CCC ATG GCA AAT TCC ATG GCA Antisense: TCT AGA CGG CAG GTC AGG TCC ACC |
74 74 |
1.6 | Denaturation, 95°C, 30 s Annealing, 58°C, 30 s Polymerization, 72°C, 1 min Elongation, 72°C, 10 min |
587 |
Immunoblotting
Cell extracts were prepared for use in immunoblotting according to methods described elsewhere.23 Purified cells were incubated (10 min on ice ) with western lysis buffer (10 μL per 1 × 106 cells) and then the lysates clarified by centrifugation (13,000 × g, 3min ). The lysates were subjected to SDS-PAGE using 10% gels and then the separated proteins transferred (14 h) electrophoretically to nitrocellulose membranes. Membranes were probed with monoclonal (PDE4A, 4B, 4C, and 4D) and polyclonal (PDE7) antibodies\ by testing decreasing different concentrations of recombinant protein. Protein bands of interest were visualized by the addition of enhanced chemiluminescence (ECL) reagents and signals detected on ECL film (Table 2).
Table 2.
Presence of PDE isoforms protein in basophils and HLMC.
| PDE4A | PDE4B | PDE4C | PDE4D | PDE3B | PDE7A | |
|---|---|---|---|---|---|---|
| Basophil | ++ | – | – | + | + | – |
| HLMC | – | – | – | + | – | – |
+Expression; – None detected.
Results
The expression of cAMP-specific PDEs in HLMC, basophils, and mononuclear cells (MNC) as a control, was determined by RT-PCR and immunoblotting.
Expression of GAPDH mRNA and actin
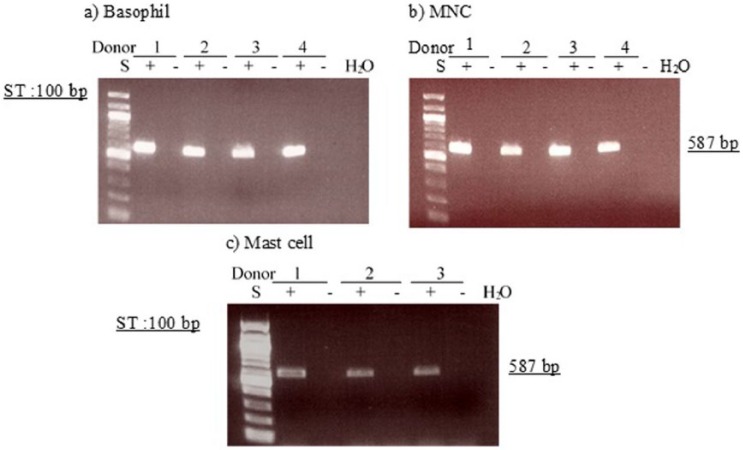
In initial experiments to confirm the integrity of RNA and to gain an estimate of the level of PDE expression, we performed RT-PCR of the house-keeping gene, GAPDH (glyceraldehyde-3-phosphate dehydrogenase), for all samples obtained from basophils, HLMC, and MNC. In further experiments to determine the presence of PDE transcripts, approximately equivalent levels of transcribed material were used. In all RT-PCR experiments, GAPDH bands were detected at the expected size (587 bp). However, GAPDH expression was greater in basophils and MNC than in mast cells. Since actin is an important scaffold protein and a component of all eukaryotic cells, it was used as a semi-quantitative marker and a control for cellular material in blots. A protein band at the expected molecular weight (40 kDa) for actin was detected after immunoblotting of basophils and mast cells (Figures 14 and 15).
Figure 14.

Western blot for PDE7. Mast cells (HLMC, 0.5 × 106 cell equivalents) and basophils (BAS, 0.5 × 106 cell equivalents) and Hut78 cells were solubilized and along with recombinant PDE7A2 (r-PDE7A) subjected to SDS-PAGE. The proteins were then transferred electrophoretically to nitrocellulose membranes and membranes probed with monoclonal antibody to PDE7A2. Molecular weights (kDa) of standards (S) are indicated. The recombinant protein PDE7A2 has a calculated molecular weight of 55 kDa. The blot is representative of a total three of similar design. Mast cell and basophil purities were 99% and 92%, respectively.
Figure 15.
Determination of message for GAPDH. RNA was extracted from four basophils (a), four MNC (b), and four HLMC preparations (c) and reverse transcribed in the absence (-) or presence (+) of MMLV-RT. Amplification of cDNAs was performed using primers for GAPDH and a water blank was also subjected to the amplification step. PCR products and a 100 bp ladder, serving as molecular weight standards (S), were then subjected to electrophoresis and bands visualized with ethidium bromide. The expected molecular weight of the amplified product is 587 bp. Data from four (a, b) and three (c) different donors are shown and are represented of a total of seven different donors. Mast cell and basophil purities were 99% and 92%, respectively.
Expression of PDE4A mRNA and protein
Experiments to determine whether PDE4A mRNA is expressed by basophils or mast cells, in parallel with MNC, were performed by RT-PCR using primers specific for PDE4A. The data demonstrated that basophils express PDE4A, as do MNC. In some mast cell preparations, modest expression of PDE4A was observed but in other mast cells preparations there was no evidence for PDE4A (Figure 1).
Figure 1.
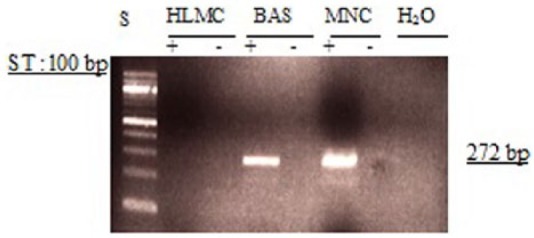
Determination of message for PDE4A. RNA was extracted from mast cells (HLMC), basophils (BAS), and mononuclear cells (MNC) and reverse transcribed in the absence (-) or presence (+) of MMLV-RT. Amplification of cDNAs was performed using primers for PDE4A and a water blank was also subjected to the amplification step. PCR products and a 100 bp ladder, serving as molecular weight standards (S), were then subjected to electrophoresis and bands visualized with ethidium bromide. The expected molecular weight of the amplified product is 272 bp. Results are representative of a total of six experiments. Mast cell and basophil purities were 99% and 92%, respectively.
Further studies to determine whether PDE4A could be detected by immunoblotting, demonstrated that basophils express a protein that is recognized by antibody to PDE4A and that this has the same molecular weight (~130 kDa) as recombinant PDE4A5 (Figure 2). A lower molecular weight moiety (~51 kDa) was also prominently expressed by basophils. No known PDE4A variants run to this molecular weight. By contrast, mast cells do not express PDE4A.
Figure 2.
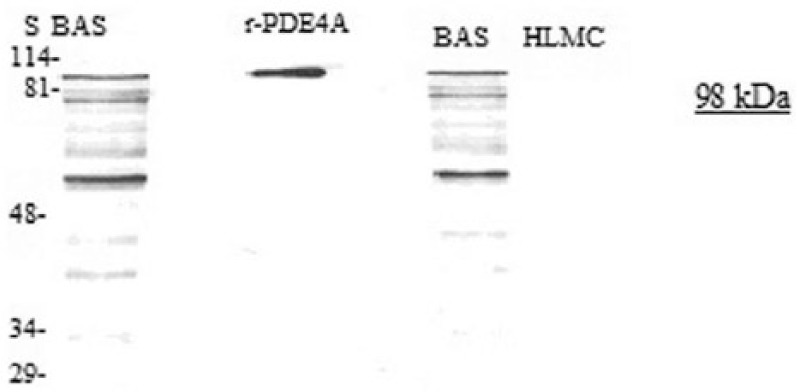
Western blot for PDE4A. Mast cells (HLMC, 0.8 × 106 cell equivalents) and basophils (BAS, 0.5 × 106 cell equivalents) were solubilized and along with recombinant r-PDE4A subjected to SDS-PAGE. The proteins were then transferred electrophoretically to nitrocellulose membranes and membranes probed with monoclonal antibody to PDE4A. Molecular weights (kDa) of standards (S) are indicated. The recombinant protein PDE4A has a calculated molecular weight of 98 kDa. Results are representative of a total of six experiments. Mast cell and basophil purities were 99% and 92%, respectively.
Expression of PDE4B mRNA and protein
Further investigations were performed to determine whether basophils and mast cells express PDE4B. These experiments showed little evidence that either basophils or mast cells express this subclass of PDE4 to an appreciable extent at the mRNA level (Figure 3).
Figure 3.
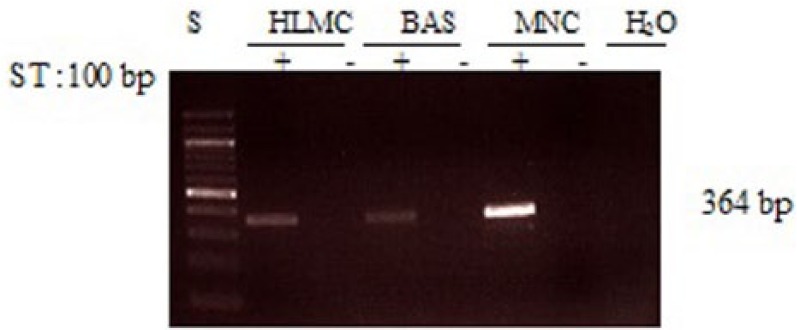
Determination of message for PDE4B. RNA was extracted from mast cells (HLMC), basophils (BAS), and mononuclear cells (MNC) and reverse transcribed in the absence (-) or presence (+) of MMLV-RT. Amplification of cDNAs was performed using primers for PDE4B and a water blank was also subjected to the amplification step. PCR products and a 100 bp ladder, serving as molecular weight standards (S), were then subjected to electrophoresis and bands visualized with ethidium bromide. The expected molecular weight of the amplified product is 364 bp. Results are representative of a total of six experiments. Mast cell and basophil purities were 99% and 92%, respectively.
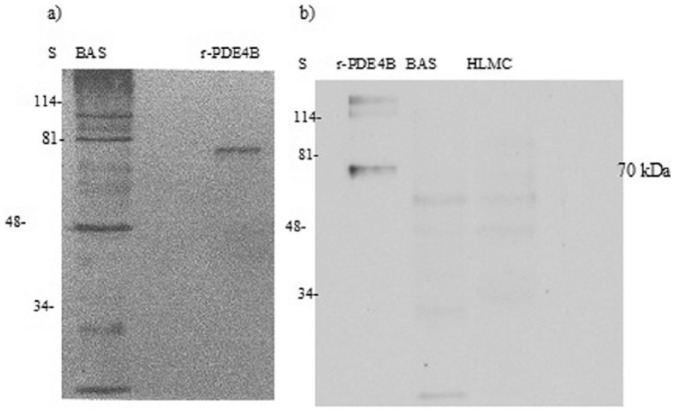
Moreover, in immunoblotting experiments, when the PDE4B-specific antibody was used, no band coincident with the recombinant PDE4B protein (~70 kDa) was detected in either basophils or mast cells (Figure 4). However, other immunoreactive bands were present at molecular weights that were higher or lower than the recombinant protein. No known PDE4B variants run to these molecular weights.
Figure 4.
Western blot for PDE4B. HLMC (0.5 × 106 cell equivalents) and basophils (BAS, 0.5 × 106 cell equivalents) were solubilized and along with recombinant PDE4B (r-PDE4B) subjected to SDS-PAGE. The proteins were then transferred electrophoretically to nitrocellulose membranes and membranes probed with monoclonal antibody to PDE4B. Molecular weights (kDa) of standards (S) are indicated. The recombinant protein has a predicted molecular weight of 70 kDa. Data are representative of a total of four experiments performed for each cell type. Purities of HLMC and basophils were 99% and 92%, respectively.
Expression of PDE4C mRNA and protein
To determine whether basophils and mast cells express PDE4C mRNA, further experiments were performed using RT-PCR. There was no evidence that either cell type expresses message for PDE4C (Figure 5). In addition, MNC did not express message for PDE4C so, as an internal control to ensure that PCR was unaffected, genomic DNA, isolated from lung tissue, was included. Immunoblotting studies further confirmed that neither basophils nor mast cells express PDE4C (Figure 6).
Figure 5.
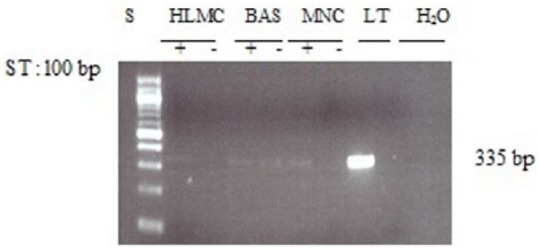
Determination of message for PDE4C. RNA was extracted from mast cells (HLMC), basophils (BAS), and mononuclear cells (MNC) and reverse transcribed in the absence (-) or presence (+) of MMLV-RT. Amplification of cDNAs and genomic DNA isolated from lung tissue (LT) was performed using primers for PDE4C and a water blank was also subjected to the amplification step. PCR products and a 100 bp ladder, serving as molecular weight standards (S), were then subjected to electrophoresis and bands visualized with ethidium bromide. The expected molecular weight of the amplified product is 336 bp. Results are representative of four or five experiments. Mast cell and basophil purities were 99% and 92%, respectively.
Figure 6.
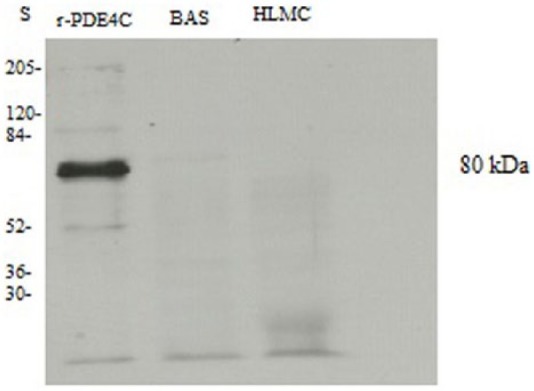
Western blot for PDE4C. Mast cells (HLMC, 1 × 106 cell equivalents) and basophils (BAS, 1.5 × 106 cell equivalents) were solubilized and along with recombinant PDE4C (r-PDE4C) subjected to SDS-PAGE. The proteins were then transferred electrophoretically to nitrocellulose membranes and membranes probed with monoclonal antibody to PDE4C. Molecular weights (kDa) of standards (S) are indicated. The recombinant protein has a predicted molecular weight of 80 kDa. The blot is representative of five different experiments of similar design. Mast cell and basophil purities were 99% and 90%, respectively.
Expression of PDE4D mRNA and protein
Determination of PDE4D mRNA expression is complicated by the existence of multiple N-terminally-spliced variants sometimes referred to as “long” and “short” forms.17 By using a primer pair designed to recognize any of the long forms, we were able to establish that basophils express mRNA for “long” forms of PDE4D (Figure 7). In addition, analysis of the expression of prominent “short” forms of PDE4D18 indicated that basophils express these too (Figures 8 and 9).
Figure 7.
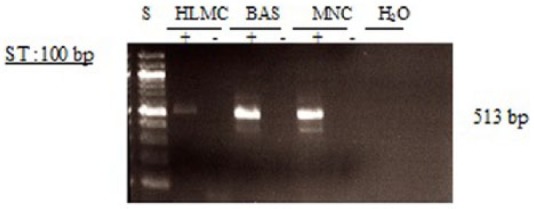
Determination of message for PDE4D3. RNA was extracted from mast cells (HLMC), basophils (BAS), and mononuclear cells (MNC) and reverse transcribed in the absence (-) or presence (+) of MMLV-RT. Amplification of cDNAs was performed using primers for PDE4D3 and a water blank was also subjected to the amplification step. PCR products and a 100 bp ladder, serving as molecular weight standards (S), were then subjected to electrophoresis and bands visualized with ethidium bromide. The expected molecular weight of the amplified product is 516 bp. Results are representative of a total of five or six experiments. Mast cells and basophils purities were 99% and 92%, respectively.
Figure 8.
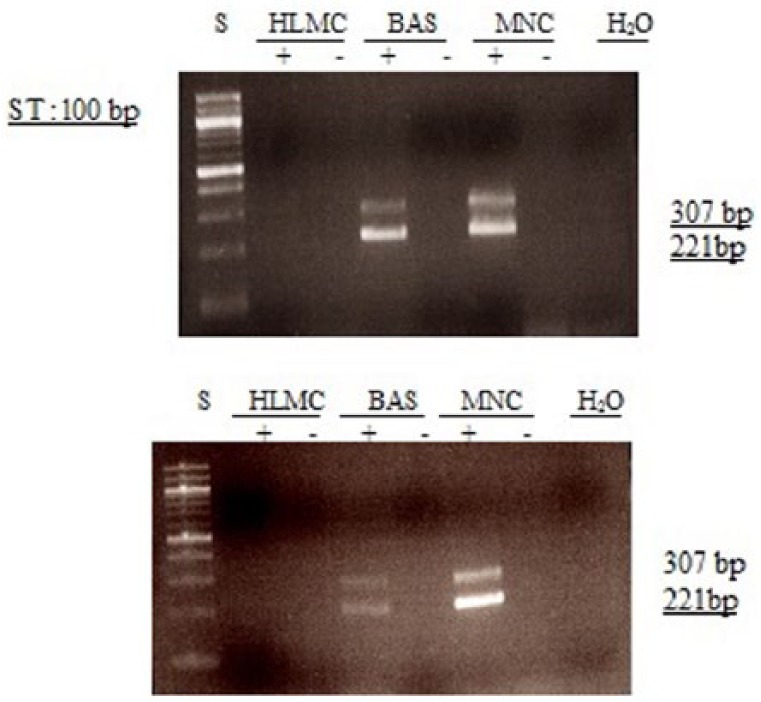
Determination of message for PDE4D1/2. RNA was extracted from mast cells (HLMC), basophils (BAS), and mononuclear cells (MNC) and reverse transcribed in the absence (-) or presence (+) of MMLV-RT. Amplification of cDNAs was performed using primers for PDE4D1/D2 and a water blank was also subjected to the amplification step. PCR products and a 100 bp ladder, serving as molecular weight standards (S), were then subjected to electrophoresis and bands visualized with ethidium bromide. The expected molecular weight of the amplified product is 221 and 307 bp for PDE4D1 and PDE4D2, respectively. Results are representative of a total of six experiments. Mast cell and basophil purities were 99% and 92%, respectively.
Figure 9.
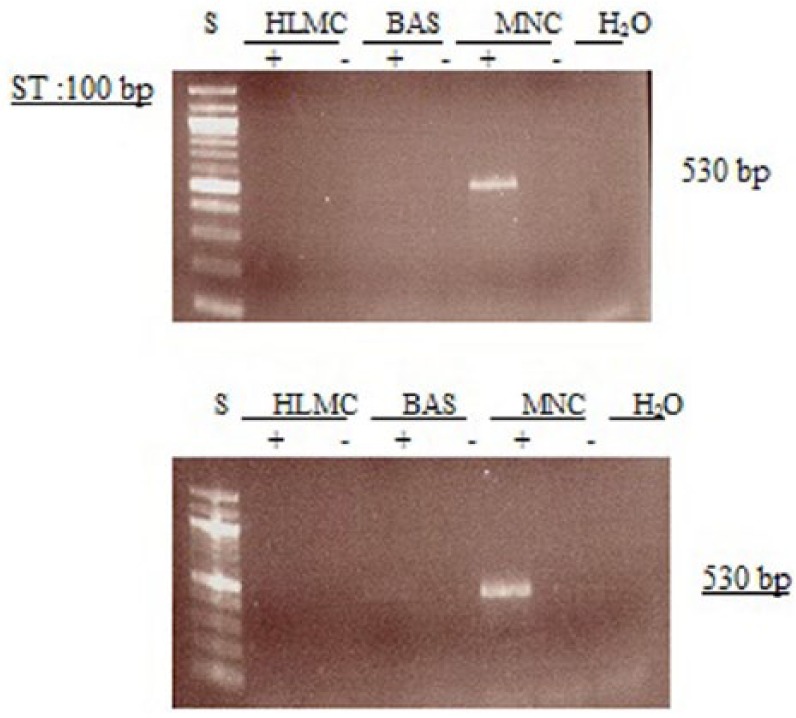
Determination of message for PDE4D1. RNA was extracted from mast cells (HLMC), basophils (BAS), and mononuclear cells (MNC) and reverse transcribed in the absence (-) or presence (+) of MMLV-RT. Amplification of cDNAs was performed using primers for PDE4D1 and a water blank was also subjected to the amplification step. PCR products and a 100 bp ladder, serving as molecular weight standards (S), were then subjected to electrophoresis and bands visualized with ethidium bromide. The expected molecular weight of the amplified product is 530 bp. Results are representative of a total of six experiments. Mast cell and basophil purities were 99% and 92%, respectively.
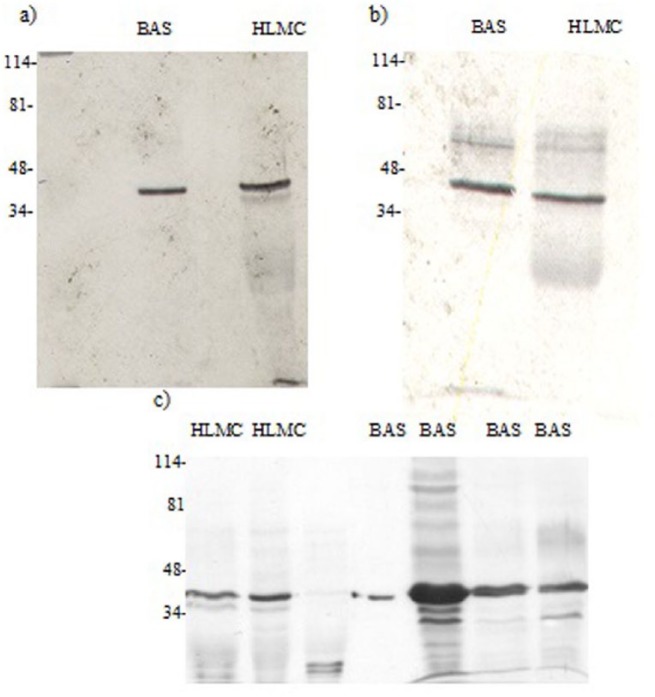
At the protein level, immunoblotting detected a higher molecular weight species (~98 kDa) in basophils consistent with expression of a “long” form of PDE4D (Figure 10). However, there was no evidence for the expression of “short” forms because no bands were detected at around the molecular weight (~72 kDa) of the recombinant protein (PDE4D1, a “short” form) that was used as a control in these experiments (Figures 9 and 10). A lower molecular weight moiety (~55 kDa) was also variably expressed by basophils. No known PDE4D variants run to this molecular weight such that the nature of this protein is uncertain. In some mast cell preparations there was an occasional suggestion that mRNA for PDE4D was present (Figure 7) but this was not accompanied by the presence of protein (Figure 10).
Figure 10.
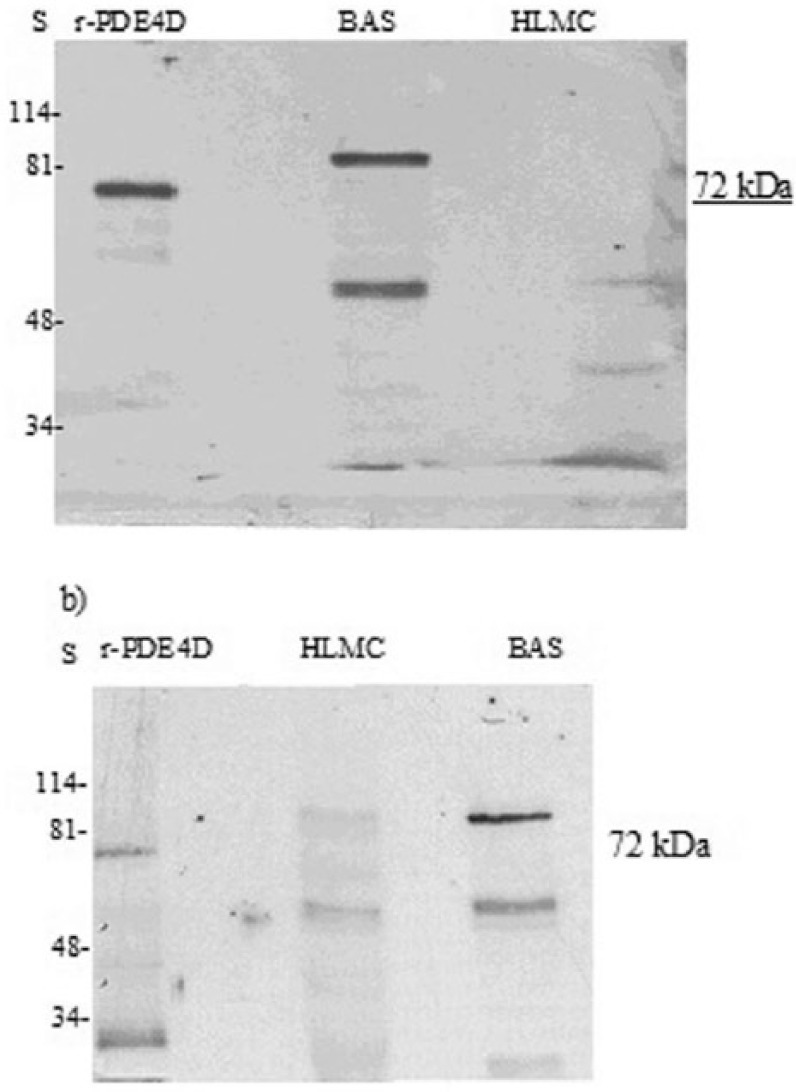
Western blot for PDE4D. Mast cells (HLMC, 0.8 × 106 cell equivalents) and basophils (BAS, 0.8–1 × 106 cell equivalents) were solubilized and along with recombinant PDED1 (r-PDE4D) subjected to SDS-PAGE. The proteins were then transferred electrophoretically to nitrocellulose membranes and membranes probed with monoclonal antibody to PDE4D. Molecular weights (kDa) of standards (S) are indicated. The recombinant protein has a predicted molecular weight of 67 kDa but runs at 72 kDa. Results are representative of a total of six experiments. Mast cell and basophil purities were 99% and 92%, respectively.
Expression of PDE3 and PDE7
The failure to detect PDE4 in mast cells was a probable result given that PDE4-selective inhibitors fail to stabilize mast cell responses.14 However, mast cells are sensitive to the non-selective PDE inhibitors, IBMX and theophylline, and also express a cAMP hydrolytic activity (but not a cGMP hydrolytic activity) that is sensitive to IBMX and theophylline.14 These findings suggest that human lung mast cells express a PDE that hydrolyzes cAMP. In an attempt to determine what this might be, we investigated whether mast cells (and basophils) express PDE3 or PDE7 as these isoforms, in addition to PDE4, are known to hydrolyze cAMP.4,19
Studies to determine whether mast cells and basophils express mRNA for PDE3A indicated that neither cells express this isoform (Figure 11). In addition, MNC did not express message for PDE3A so, as a control to certify that PCR was unaffected, lung cell mRNA was isolated and was also subjected to RT-PCR. The occasional suggestion existed that PDE3B is expressed weakly by basophils but not by mast cells, while in MNC this was prominently expressed (Figure 12).
Figure 11.
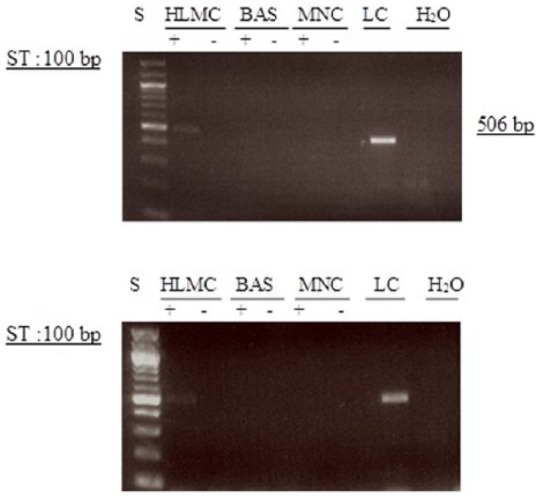
Determination of message for PDE3A. RNA was extracted from mast cells (HLMC), basophils (BAS), mononuclear cells (MNC), and lung cells (LC) and reverse transcribed in the absence (-) or presence (+) of MMLV-RT. In addition, MNC did not express message for PDE3A so, as a control to certify that PCR was unaffected, lung cell mRNA was isolated and was also subjected to RT-PCR. Amplification of cDNAs was performed using primers for PDE3A and a water blank was also subjected to the amplification step. PCR products and a 100 bp ladder, serving as molecular weight standards (S), were then subjected to electrophoresis and bands visualized with ethidium bromide. The expected molecular weight of the amplified product is 506 bp. Results are representative of a total of six experiments. Cells purities were 99% and 92%, respectively.
Figure 12.
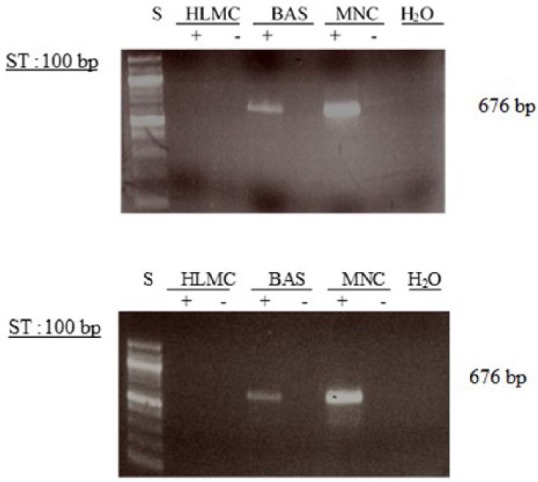
Determination of message for PDE3B. RNA was extracted from mast cells (HLMC), basophils (BAS), and mononuclear cells (MNC) and reverse transcribed in the absence (-) or presence (+) of MMLV-RT. Amplification of cDNAs was performed using primers for PDE3B and a water blank was also subjected to the amplification step. PCR products and a 100 bp ladder, serving as molecular weight standards (S), were then subjected to electrophoresis and bands visualized with ethidium bromide. The expected molecular weight of the amplified product is 676 bp. Results are representative of five or more experiments. Mast cell and basophil purities were 99% and 92%, respectively.
In similar experiments designed to detect PDE7, mast cells, basophils, and MNC all appeared consistently to express substantial levels of mRNA for PDE7 (Figure 13). However, immunoblotting experiments using PDE7A-specific antibody indicated that neither mast cells nor basophils express PDE7A at the protein level (Figure 14). In these same immunoblotting experiments, PDE7A was detected in Hut78 cells, a T cell line, that previously has been shown to express PDE7A1.20 The recombinant protein, PDE7A2, which was used as a positive control in these experiments, has been reported to have a slightly lower molecular weight than the splice variant, PDE7A1.21
Figure 13.
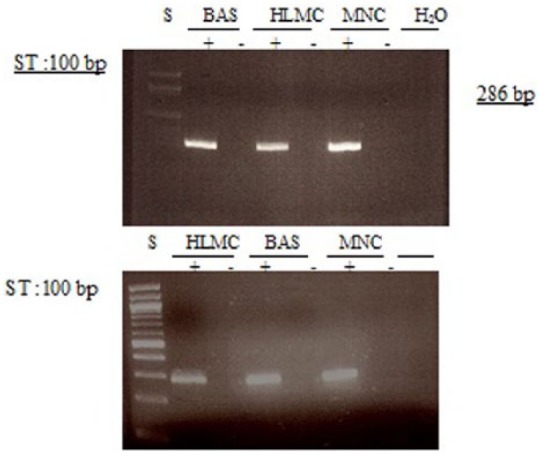
Determination of message for PDE7. RNA was extracted from mast cells (HLMC), basophils (BAS), and mononuclear cells (MNC) and reverse transcribed in the absence (-) or presence (+) of MMLV-RT. Amplification of cDNAs was performed using primers for PDE7A and a water blank was also subjected to the amplification step. PCR products and a 100 bp ladder, serving as molecular weight standards (S), were then subjected to electrophoresis and bands visualized with ethidium bromide. The expected molecular weight of the amplified product is 286 bp. Results are representative of a total of four or five experiments. Mast cell and basophil purities were 99% and 92%, respectively.
Figure 16.
Determination of actin. HLMC (0.5–1 × 106 cell equivalents) and basophils (BAS, 0.5–1 × 106 cell equivalents) were sobuilized and subjected to SDS-PAGE. The proteins were transferred electrophometrically to nitrocellulose membranes and membranes probed with polyclonal antibody to actin. The calculated molecular weight for actin was 40 kDa. Molecular weights (kDa) of standards (S) are indicated. Mast cell and basophil purities were 99% and 92%, respectively. Each of the three panels (a, b, c) represent six different experiments of different preparations of similar design.
Discussion
PDE is known to exist as multiple molecular forms.1 The distribution of these isoforms can vary among tissues and this has led to an appreciation that tissue-selective effects may be obtained by targeting the prominent PDE isoform in a given tissue.2 The therapeutic benefit of such an approach is being realized with the emergence of drugs such as sildenafil, a PDE5 inhibitor, which is used to treat erectile dysfunction.22 Progress in the context of inflammatory disease has also been made with the realization that the main isoform of PDE regulating inflammatory cell activity is PDE4.5 As a consequence, cilomilast and roflumilast, selective PDE4 inhibitors, have been developed and have displayed some promise in targeting respiratory diseases.23
As mast cells and basophils are inflammatory cells, a prominent role for PDE4 in regulating the activity of these cells might be expected. As far as the basophil is concerned, this is indeed the case as PDE4-selective inhibitors have been shown to inhibit the stimulated generation of histamine, leukotrienes, and cytokines from these cells. The present study supports these findings since the basophil expresses both PDE4A and PDE4D although it should be noted that the specific splice variants expressed by basophils have not been delineated. By contrast, previous studies in HLMC have shown that PDE4-selective agents are ineffective at inhibiting stimulated mediator release.14 In accord with these functional studies, the present work has shown that no evidence exists that mast cells express any of the recognized sub-classes of PDE4. Thus, HLMC appear to differ from not only basophils but a whole host of alternative inflammatory cells such as monocytes, macrophages, eosinophils, and lymphocytes all of which have been shown to be sensitive to PDE4-selective inhibitors.14
That mast cells express a PDE seems highly likely since the functional responses of mast cells are sensitive to non-selective PDE inhibitors such as IBMX and theophylline.12–14 Moreover, both IBMX and theophylline are known to increase cAMP in mast cells a finding consistent with effects of these drugs at a PDE.12,13 In addition, extracts of mast cells hydrolyze cAMP (but not cGMP) and this activity is inhibited by IBMX.14 All this evidence points to the existence of a cAMP-specific PDE in mast cells. The most obvious candidate would be PDE4 but in the absence of both functional and biochemical data to support the presence of this isoform in mast cells, alternative candidates include PDE3 and PDE7 both of which are known to hydrolyze cAMP effectively.1–3
In the present study, there was no evidence for the presence of either PDE3A or PDE3B in mast cells and although prominent mRNA expression for PDE7A were detected, no PDE7A was detected by immunoblotting. By contrast, PDE7A was very ably detected by immunoblotting in Hut 78 cells (T cell line) suggesting that if PDE7A is present in mast cells, it is present at imperceptibly low levels questioning the importance of this isoform in the regulation of mast cell activity. The finding that the expression of mRNA for PDE isoforms does not tally with protein expression has also been reported for alternative systems.17,24 However, the suggestion has been made that immunoblotting may be an insufficiently sensitive method to detect low, but functionally important, levels of protein expression.24 By analogy, the possibility that mast cells express very low levels of PDE7A, within discrete cell compartments, and that these low levels may influence mast cell activity, cannot be excluded.
Overall, findings from the present study indicate that basophils express PDE4 (4A and 4D) whereas mast cells do not. These findings indicate that PDE4-directed anti-inflammatory strategies are likely to stabilize basophil but not mast cell activity. In addition, neither basophils nor mast cells express PDE3 or PDE7. The cAMP hydrolytic activity present in mast cells awaits clarification. Clearly, future work will center on the characterization of PDE3 in mast cells both at the functional and molecular levels. And should PDE3 be found to regulate mast cell responses then this will lend weight to recent suggestions that dual-specificity inhibitors, targeting both PDE3 and PDE4, may be a more successful strategy than single-specificity inhibitors in diseases that feature mast cells, such as asthma.
Footnotes
Declaration of conflicting interests: The author(s) declared on potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
References
- 1. Beavo JA, Conti M, Heaslip RJ. (1994) Multiple cyclic nucleotide phosphodiesterases. Molecular Pharmacology 46: 399–405. [PubMed] [Google Scholar]
- 2. Beavo JA, Reifsnyder DH. (1990) Primary sequence of cyclic nucleotide phosphodiesterase isozymes and the design of selective inhibitors. Trends in Pharmacological Sciences 11: 150–155. [DOI] [PubMed] [Google Scholar]
- 3. Conti M, Richter W, Mehats C, et al. (2003) Cyclic AMP-specific PDE4 phosphodiesterases as critical components of cyclic AMP signaling. Journal of Biological Chemistry 278: 5493–5496. [DOI] [PubMed] [Google Scholar]
- 4. Soderling SH, Beavo JA. (2000) Regulation of cAMP and cGMP signaling: New phosphodiesterases and new functions. Current Opinion in Cell Biology 12: 174–179. [DOI] [PubMed] [Google Scholar]
- 5. Souness JE, Aldous D, Sargent C. (2000) Immunosuppressive and anti-inflammatory effects of cyclic AMP phosphodiesterase (PDE) type 4 inhibitors. Immunopharmacology 47: 127–162. [DOI] [PubMed] [Google Scholar]
- 6. Teixeira MM, Gristwood RW, Cooper N, et al. (1997) Phosphodiesterase (PDE)4 inhibitors: Anti-inflammatory drugs of the future? Trends in Pharmacological Sciences 18: 164–171. [DOI] [PubMed] [Google Scholar]
- 7. Falcone FH, Haas H, Gibbs BF. (2000) The human basophil: A new appreciation of its role in immune responses. Blood 96:4028–4038. [PubMed] [Google Scholar]
- 8. Schroeder J.T., Kagey-Sobotka A, Lichtenstein L.M., The role of the basophil in allergic inflammation. Allergy, 1995. 50(6): p. 463–72. [DOI] [PubMed] [Google Scholar]
- 9. Williams CM, Galli SJ. (2000) The diverse potential effector and immunoregulatory roles of mast cells in allergic disease. Journal of Allergy and Clinical Immunology 105:847–859. [DOI] [PubMed] [Google Scholar]
- 10. Bourne HR, Lichtenstein LM, Melmon KL, et al. (1974) Modulation of inflammation and immunity by cyclic AMP. Science 184:19–28. [DOI] [PubMed] [Google Scholar]
- 11. Lichtenstein LM, Margolis S. (1968) Histamine release in vitro: Inhibition by catecholamines and methylxanthines. Science 161: 902–903. [DOI] [PubMed] [Google Scholar]
- 12. Peachell PT, MacGlashan DW, Jr, Lichtenstein LM, et al. (1988) Regulation of human basophil and lung mast cell function by cyclic adenosine monophosphate. Journal of Immunology 140: 571–579. [PubMed] [Google Scholar]
- 13. Peachell PT, Undem BJ, Schleimer RP, et al. (1992) Preliminary identification and role of phosphodiesterase isozymes in human basophils. Journal of Immunology 148: 2503–2510. [PubMed] [Google Scholar]
- 14. Weston MC, Anderson N, Peachell PT. (1997) Effects of phosphodiesterase inhibitors on human lung mast cell and basophil function. British Journal of Pharmacology 121: 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shichijo M, Shimizu Y, Hiramatsu K, et al. (1997) Cyclic AMP-elevating agents inhibit mite-antigen-induced IL-4 and IL-13 release from basophil-enriched leukocyte preparation. International Archives of Allergy and Immunology 114: 348–353. [DOI] [PubMed] [Google Scholar]
- 16. Gibbs BF, Vollrath IB, Albrecht C, et al. (1998) Inhibition of interleukin-4 and interleukin-13 release from immunologically activated human basophils due to the actions of anti-allergic drugs. Naunyn-Schmiedeberg’s Archives of Pharmacology 357: 573–578. [DOI] [PubMed] [Google Scholar]
- 17. Houslay MD, Sullivan M, Bolger GB. (1998) The multienzyme PDE4 cyclic adenosine monophosphate-specific phosphodiesterase family: Intracellular targeting, regulation, and selective inhibition by compounds exerting anti-inflammatory and antidepressant actions. Advances in Pharmacology 44: 225–342. [DOI] [PubMed] [Google Scholar]
- 18. Barber R, Baillie GS, Bergmann R, et al. (2004) Differential expression of PDE4 cAMP phosphodiesterase isoforms in inflammatory cells of smokers with COPD, smokers without COPD, and nonsmokers. American Journal of Physiology Lung Cellular and Molecular Physiology 287: L332–343. [DOI] [PubMed] [Google Scholar]
- 19. Torphy TJ. (1998) Phosphodiesterase isozymes: Molecular targets for novel antiasthma agents. American Journal of Respiratory and Critical Care Medicine 157: 351–370. [DOI] [PubMed] [Google Scholar]
- 20. Bloom TJ, Beavo JA. (1996) Identification and tissue-specific expression of PDE7 phosphodiesterase splice variants. Proceedings of the National Academy of Sciences of the United States of America 93: 14188–14192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Han P, Zhu X, Michaeli T. (1997) Alternative splicing of the high affinity cAMP-specific phosphodiesterase (PDE7A) mRNA in human skeletal muscle and heart. Journal of Biological Chemistry 272: 16152–16157. [DOI] [PubMed] [Google Scholar]
- 22. Goldstein I, Lue TF, Padma-Nathan H, et al. (1998) Oral sildenafil in the treatment of erectile dysfunction. Sildenafil Study Group. New England Journal of Medicine 338: 1397–1404. [DOI] [PubMed] [Google Scholar]
- 23. Lipworth BJ. (2005) Phosphodiesterase-4 inhibitors for asthma and chronic obstructive pulmonary disease. Lancet 365: 167–175. [DOI] [PubMed] [Google Scholar]
- 24. Smith SJ, Brookes-Fazakerley S, Donnelly LE, et al. (2003) Ubiquitous expression of phosphodiesterase 7A in human proinflammatory and immune cells. American Journal of Physiology Lung Cellular and Molecular Physiology 284: L279–289. [DOI] [PubMed] [Google Scholar]