Abstract
Significant selection pressure has been exerted on the genomes of human populations exposed to Plasmodium falciparum infection, resulting in the acquisition of mechanisms of resistance against severe malarial disease. Many host genetic factors, including sickle cell trait, have been associated with reduced risk of developing severe malaria, but do not account for all of the observed phenotypic variation. Identification of novel inherited risk factors relies upon high-resolution genome-wide association studies (GWAS). We present findings of a GWAS of severe malaria performed in a Tanzanian population (n = 914, 15.2 million SNPs). Beyond the expected association with the sickle cell HbS variant, we identify protective associations within two interleukin receptors (IL-23R and IL-12RBR2) and the kelch-like protein KLHL3 (all P<10−6), as well as near significant effects for Major Histocompatibility Complex (MHC) haplotypes. Complementary analyses, based on detecting extended haplotype homozygosity, identified SYNJ2BP, GCLC and MHC as potential loci under recent positive selection. Through whole genome sequencing of an independent Tanzanian cohort (parent-child trios n = 247), we confirm the allele frequencies of common polymorphisms underlying associations and selection, as well as the presence of multiple structural variants that could be in linkage with these SNPs. Imputation of structural variants in a region encompassing the glycophorin genes on chromosome 4, led to the characterisation of more than 50 rare variants, and individually no strong evidence of associations with severe malaria in our primary dataset (P>0.3). Our approach demonstrates the potential of a joint genotyping-sequencing strategy to identify as-yet unknown susceptibility loci in an African population with well-characterised malaria phenotypes. The regions encompassing these loci are potential targets for the design of much needed interventions for preventing or treating malarial disease.
Author summary
Malaria, caused by Plasmodium falciparum parasites, is a major cause of mortality and morbidity in endemic countries of sub-Saharan Africa, including Tanzania. Some gene mutations in the human genome, including sickle cell trait, have been associated with reduced risk of developing severe malaria, and have increased in frequency through natural selection over generations. However, new genetic mutations remain to be discovered, and recent advances in human genome research technologies such as genome-wide association studies (GWAS) and fine-scale molecular genotyping tools, are facilitating their identification. Here, we present findings of a GWAS of severe malaria performed in a well characterised Tanzanian population (n = 914). We confirm the expected association with the sickle cell trait, but also identify new gene targets in immunological pathways, some under natural selection. Our approach demonstrates the potential of using GWAS to identify as-yet unknown susceptibility genes in endemic populations with well-characterised malaria phenotypes. The genetic mutations are likely to form potential targets for the design of much needed interventions for preventing or treating malarial disease.
Introduction
Sub-Saharan Africa bears a disproportionately high share of the global Plasmodium falciparum malaria burden, with 90% of the estimated 212 million annual cases and 92% of 429,000 annual deaths, mostly in children under five years of age [1]. Whilst the majority of cases of Plasmodium falciparum infection are asymptomatic or cause only mild to moderate clinical symptoms, a subset of affected individuals present with severe manifestations such as severe malarial anaemia and cerebral malaria. Risk factors for severe malaria and its various clinical subtypes are poorly understood, although host and parasite genotype, age and immune status have all been established as playing a significant role in individual host susceptibility [2]. Plasmodium falciparum has also exerted significant selection pressure upon the human genome, as evidenced by the geographical concurrence of malaria parasite prevalence with sickle cell trait (HbAS) and other haemoglobinopathies, such as the thalassemias and glucose-6-phosphate dehydrogenase (G6PD) deficiency.
Recent studies, set in a region of high malaria transmission in north-eastern Tanzania, estimated that host genetic factors account for approximately 22% of the total variation in severe malaria risk [3], consistent with previous findings in a Kenyan family-based study [2]. Less than half of this variation can be explained by erythrocyte-associated polymorphisms [4], including HbS (sickle cell trait), alpha-thalassaemia, ABO blood group [5] and G6PD deficiency [4]. Novel polymorphisms in or around USP38, FREM3 [3], glycophorins gypA/B/E [6, 7], DDC [8], MARVELD3 and ATP2B4 [9] account for additional variation but, in sum, are less protective than heterozygous carriage of HbS [3]. Moreover, the effects of some of these loci are subtype-, location-, or population-specific [3, 6, 7, 9], reinforcing the need for targeted genome-wide association studies (GWAS) in different African populations. Utilising such an approach with robust malaria phenotypes in parallel with whole genome sequencing of study populations is crucial to unravelling host genetic factors that could lead to a greater understanding of protective immunity and development of new tools for disease prevention.
To identify novel loci associated with severe malaria in north-eastern Tanzania, we applied genome-wide association and haplotype-based selection methods to a case-control dataset with extensive phenotypic data for 914 participants and 15.2 million SNPs. In addition to the expected HbS (sickle cell) association, our analyses reveal multiple novel loci under association or selection. Association analysis highlighted significant SNPs clusters within IL-23R, IL-12RB2, LINC00944, and KLHL3 whilst lone SNP associations were also present within TREML4 and ZNF536. Further, we reveal loci under recent positive selection including GCLC and loci within the Major Histocompatibility Complex (MHC). These analyses were supported by whole genome sequencing of an independent dataset consisting of 247 Tanzanian individuals within parent-child trios, which was used to confirm the allele frequencies of putative associations and determine if there are any linked common structural variants in chromosome regions encoding important polymorphisms.
Results
Phenotypic and genotypic data
All severe malaria cases (n = 449) and controls (n = 465) came from the Tanga region of North-Eastern Tanzania. Severe malaria cases presented with varying combinations of hyperlactataemia (57.0%), severe malarial anaemia (49.2%), respiratory distress (27.6%) and cerebral malaria (26.7%) (Table 1). Compared to controls, malaria cases were younger (t test P<2.2x10-16) and marginally more likely to be male (Chi squared P = 0.012) (Table 1). DNA from all samples (n = 914) was genotyped on the Illumina Omni 2.5 million SNP chip, and imputed against the 1000 Genomes reference panel (Phase 3) [10] and a Tanzanian parent-child trio panel (see below), using Beagle 4.1 [11], leading to 15.2 high quality SNPs. These markers were complemented by 180 SNPs within malaria candidate genes, including HBB (encoding HbS) [3, 4, 5] on the same cases and controls. DNA from a validation cohort of 78 healthy parent and child trios and 13 independent individuals (“Trios dataset”, n = 247) were whole genome sequenced using Illumina HiSeq2500 technology. For the GWAS samples, a principal component analysis (PCA) using all genome-wide SNPs revealed a low degree of stratification by ethnicity and case-control status (S1 Fig) and potential cryptic relatedness due to familial clustering. A similar analysis revealed that GWAS and Trio sample clusters overlap, and there is some separation from the other 1000 Genome African populations, including Yoruba (Nigeria) and Luhya (Kenya) (S1 Fig).
Table 1. Study participants.
| Controls (n = 465) |
Cases (n = 449) |
Difference P-value |
|||
|---|---|---|---|---|---|
| Age* (median, range) | 2.8 | 0.9–10.9 | 1.7 | 0.2–10.0 | <2.2x10-16 |
| Female | 252 | 54.2% | 205 | 45.7% | 0.012 |
| Ethnicity** | 0.52 | ||||
| Mzigua | 151 | 32.5% | 146 | 32.5% | |
| Wasambaa | 142 | 30.5% | 135 | 30.1% | |
| Wabondei | 83 | 17.8% | 86 | 19.2% | |
| Mmbena | 26 | 5.6% | 23 | 5.1% | |
| Mngoni | 17 | 3.7% | 18 | 4.0% | |
| Pare | 11 | 2.4% | 8 | 1.8% | |
| Mmakonde | 11 | 2.4% | 8 | 1.8% | |
| Mgogo | 7 | 1.5% | 8 | 1.8% | |
| Chagga | 9 | 1.9% | 7 | 1.6% | |
| Other | 8 | 1.7% | 10 | 2.2% | |
| Mixed Ethnicity*** | 150 | 32.3% | 172 | 38.2% | 0.065 |
| Hyperlactatemia/acidosis | - | - | 256 | 57.0% | - |
| Severe Malarial Anaemia | - | - | 221 | 49.2% | - |
| Respiratory Distress | - | - | 124 | 27.6% | - |
| Cerebral Malaria | - | - | 120 | 26.7% | - |
* months
** based on paternal ethnicity
*** if parental ethnicities were different
Association analysis
GWAS analysis was undertaken with EMMAX mixed model regression [12], controlling for age as a fixed effect and relatedness (represented in a kinship matrix) as a random effect to account for the cryptic population clustering. Separate models of association were fitted for each SNP (additive, heterozygous, dominant, recessive), with their respective genomic inflation factors all being close to one (see S1 Fig for the heterozygous results), consistent with reliable adjustment for stratification. A total of 53 SNPs (in 16 genomic regions) were identified with a significance level below our threshold (P<1x10-6) (Fig 1, Table 2, S1 Table). Relaxing the stringency would lead to 258 SNPs with a p-value below 1x10-5 and 2,322 below a threshold of 1x10-4. As expected, the most significant association was with the sickle cell locus, rs334 (P = 8.59x10-13, heterozygous odds ratio = 0.07) (Table 2). Controlling for HbS status through a complementary conditional GWAS demonstrated our top associations as robust against linkage with rs334 (Table 2, S1 Table).
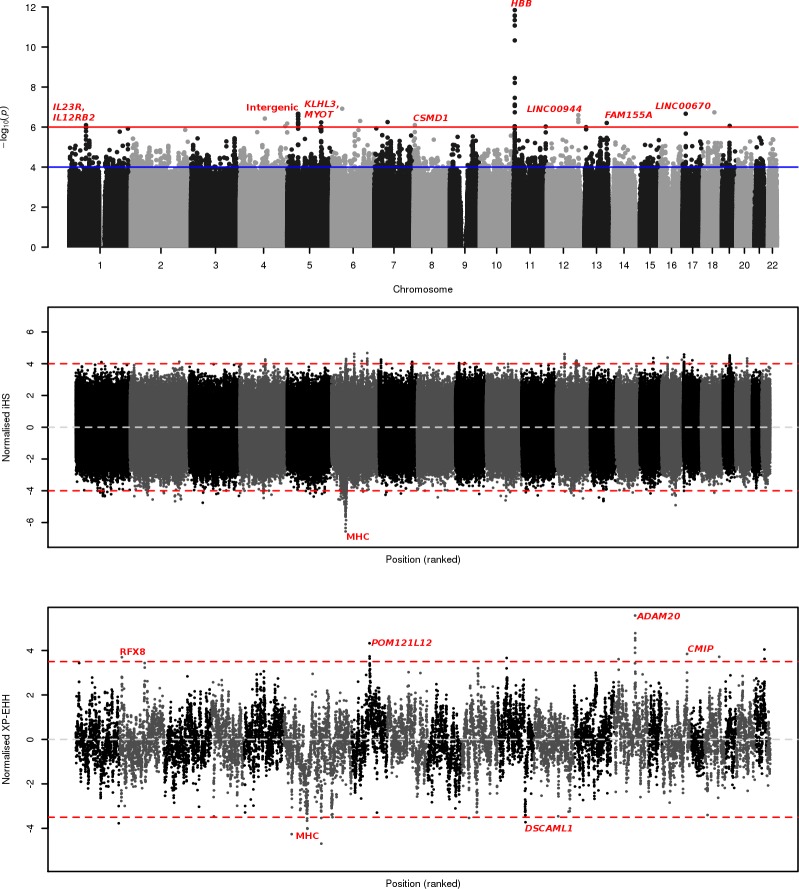
Fig 1. Genome-wide association and selection plots.
a) Case-Control SNP Associations. Red line indicates genome-wide significance at 1x10-6, blue line indicates genome-wide suggestive significant at 1x10-4; b) Combined population iHS selection. Red lines indicate significance for absolute iHS scores of 4 or greater; c) Case-Control XP-EHH selection. Red lines indicate significance for absolute XP-EHH scores of 4 or greater.
Table 2. More significant SNP associations per locus (P<1x10-6).
| SNP | Gene | n SNPs | Location | Minimum P | Conditional P | Subtype P |
|---|---|---|---|---|---|---|
| rs334 | HbS (in HBB) | 40 | 11:5248232 | HET: 8.59E-13 | - | HL: 1.81e-09 |
| rs9296359 | TREML4 | 1 | 6:41205690 | HET: 1.21E-07 | HET: 4.42e-07 | SMA: 3.29e-07 |
| rs149085856 | Intergenic (LINC00670) | 1 | 17:12399526 | ADD: 2.15E-07 | ADD: 1.06e-06 | HL: 2.81e-07 |
| rs113449872 | Intergenic | 20 | 5:43909343 | HET: 2.17E-07 | HET: 2.93e-07 | SMA: 2.92e-05 |
| rs11335470 | LINC00944 | 3 | 12:127237620 | HET: 2.52E-07 | HET: 1.86e-06 | HL: 9.04e-05 |
| rs73832816 | C4orf17 | 1 | 4:100429757 | REC: 3.75E-07 | REC: 9.48e-07 | CM: 1.02e-06 |
| rs17624383 | Intergenic | 3 | 7:53676837 | ADD: 5.62E-07 | ADD: 3.28e-06 | RD: 4.61e-07 |
| rs2967790 | KLHL3, MYOT | 13 | 5:137011761 | ADD: 5.85E-07 | ADD: 2.46e-06 | HL: 8.65e-06 |
| rs144312179 | FAM155A | 6 | 13:108228013 | ADD: 6.24E-07 | ADD: 2.92e-06 | HL: 1.35e-06 |
| rs114169033 | AF146191.4–004 (lincRNA) | 3 | 4:190717704 | ADD: 6.67E-07 | ADD: 1.30e-06 | RD: 5.62e-07 |
| rs6682413 | IL23R, IL12RB2 | 7 | 1:67731614 | REC: 7.98E-07 | REC: 1.03e-06 | SMA: 1.23e-04 |
| rs73505850 | CSMD1 | 5 | 8:4754838 | ADD: 7.98E-07 | ADD: 1.42e-06 | SMA: 1.20e-05 |
| rs8109875 | ZNF536 | 1 | 19:31069639 | REC: 8.69E-07 | REC: 3.57e-06 | SMA: 2.80e-05 |
| rs1878468 | AC108142.1 (antisense) | 1 | 4:182822332 | HET: 8.98E-07 | HET: 1.19e-06 | HL: 8.10e-07 |
| rs3133394 | Intergenic | 4 | 11:130417522 | ADD: 9.41E-07 | ADD: 1.08e-06 | CM: 9.49e-06 |
Allele models: ADD Additive, HET Heterozygous, DOM Dominant, REC Recessive. Subtype significances: HL Hyperlactatemia; SMA Severe Malarial Anaemia; RD Respiratory Distress; CM Cerebral Malaria. Locations correspond to the GRCh37 reference genome. Minimum P indicates the most significant P for SNPs in the locus within the case-control GWAS, whilst Conditional and Subtype Ps indicate the most significant P value for those SNPs when controlling for rs334 status, or considering the severe malarial subtypes.
Novel associations of note also include SNPs within the KLHL3-MYOT region (13 SNPs, Min P = 5.85x10-7, Additive OR = 0.590), the IL23R-IL12RB2 region (7 SNPs, Min P = 7.98x10-7, Recessive OR = 0.479), FAM155A (6 SNPs, Min P = 6.24x10-7, Additive OR = 0.207), and CSMD1 (5 SNPs, Min P = 7.98x10-7, Additive OR = 4.795). (Fig 2). Three significant SNPs are also found within both LINC00943/4 and lincRNA AF146191.4–004.
Fig 2. Region plots for most significant SNP associations.
Green regions highlight genes of interest, as indicated by sub-plot headers. The red line indicates genome-wide significance at 1x10-6, whilst the blue line indicates genome-wide suggestive significant at 1x10-4.
Lone SNP associations are present within proximity of TREML4 (P = 1.21x10-7, Heterozygous OR = 4.087), zinc finger-containing ZNF536 (P = 8.69x10-7, Recessive OR = 0.507), C4orf17 (P = 3.75x10-7, Recessive OR = 0.289), and near LINC00670 (P = 2.15x10-7, Additive OR = 3.867). And finally, three intergenic regions display clusters of significance, most notably a region within chromosome 5 (43,892,232–43,964,366bp; Min P = 2.17x10-7, Heterozygous OR = 0.354), as well as regions within chromosome 7 and 11.
As expected, allele frequencies of the putative polymorphisms within the Trios dataset are generally equivalent to frequencies in our case and control groups, whilst there were some differences from the 1000 Genomes populations, including within the HBB locus (Table 2). Using the Trios dataset, we sought to identify structural variants that could confound the association analysis or be putative hits. We identified no structural variants within HBB, IL-12RBR2 or LINC00943/4, one deletion (2,904bp) within IL23R, and 152 deletions within KLHL3 (63 distinct variants, all singletons except for one 1,325bp deletion in 91 individuals) (S2 Table). None of the common variants are in linkage disequilibrium with the putative GWAS hits, and eight putative regions had structural variants in the Tanzanian trios, but were absent in the 1000 Genomes populations (S2 Table).
Subtype specific association analyses were undertaken for those SNPs found to be significantly associated with severe malaria in the primary GWAS (Table 2). The majority of significant associations are with the hyperlactataemia subtype, a phenotype that includes 57.0% of cases, with variants within FAM155A, and the HBB and KLHL3/MYOT regions exhibiting associations exceeding our 1x10-6 significance threshold. In contrast, variants within IL-23R, IL-12RB2, CSMD1, ZNF536 and TREML4 appear to be most significantly associated with severe malarial anaemia, who comprised 49.2% of cases.
Candidate associations
Candidate SNPs identified in previous studies, with the same individuals, were included to provide appropriate context for novel findings. ABO blood group, USP38, FREM3 and alpha-thalassemia have previously been putatively associated with severe malaria in a Tanzanian population [3, 5], but these associations are no longer statistically significant (P>10−4) at a more stringent GWAS significance threshold (S3 Table). We also performed targeted imputation of HLA haplotypes within the MHC, finding the most significant SNP to be rs1264362, which demonstrated a marginal association with hyperlactatemia (additive model P = 2.33x10-5).
For the analysis of structural variation within candidate regions in the Trios dataset, we identified 28 distinct deletions within FREM3, of which all but one are present in only one individual, and six distinct deletions in GYPB, for which copy number variation has previously been identified [6]. Nine distinct variants were identified in ABO, including six duplications, one deletion, one insertion and one inversion. All such ABO variants are present in single individuals, though 18 individuals have a 23bp insertion. In contrast to a diversity of structural variation present within HLA and the wider MHC region, minor frequency variants were identified in ATP2B4 (25 deletions across 25 samples), MARVELD3 (five deletions across five samples), HBA2 (3 deletions across three samples), and HBA1 (one sample with one deletion). No structural variants were found in HBB or USP38 (S2 Table).
We imputed structural variants within the wider region of human glycophorin genes (gypA, gypB, gypE) on chromosome four, using 55 distinct large polymorphisms identified in 59 individuals within our Trios dataset (S2 Table). The glycophorin region is structurally highly diverse, and specific individual variants are of low frequency (mean frequency: Case Control dataset = 0.098, Trios dataset = 0.022), consistent with observations in other African populations [7]. Whilst these large variants could be potentially protective against severe malaria, we identified no significant associations looking at each individually (P ≥ 0.301). Grouping these variants into forms based on genomic location and function may enhance signals within this region, but could also introduce experimenter bias. Further, there exists a multitude of potential variant combinations analysis of which, without specific hypotheses, could risk so-called ‘P hacking’. A full and in-depth analysis of this region is required but beyond the scope of this study.
Evidence of positive selection
Two approaches were applied to identify regions under recent positive selection within the Tanzanian GWAS population as a whole (Integrated Haplotype Score, iHS) [13], or between the cases and controls (Cross-Population Extended Haplotype Homozygosity, XP-EHH) [14]. A common genome-wide absolute score threshold of 4 (equivalent to P = 6.3x10-5) was established for both approaches. At this threshold iHS identified 244 loci, 116 (47.5%) within chromosome 6. Ninety-four of these significant signals are within the MHC, with three loci within HLA-DOA having an absolute score greater than 6. Other MHC genes with significant signals include the immunophilin FKBP5 (35732-37931bp, 9 SNPs, iHS: 4.00–4.84), SAMD3 (12490-13053bp, 6 SNPs, iHS: 4.00–4.68) and the exocytosis regulator RIMS1 (72805811-72828559bp, 3 SNPs, iHS: 4.17–4.62) (S4 Table). Most notably, two regions within chromosome 17 (3496105-3689132bp, 6 SNPs, 8 genes including integrin ITGAE) and chromosome 19 (38743962-38900106bp, 14 SNPs, 10 genes including two transmembrane channels) represent regions with a high density of selection signals, akin to those within the MHC. Further signals of note include the transcription factor ZFHX3 (ATBF1) (chr. 16, 16326-73133bp, 3 SNPs, iHS: 4.27–4.91), ABHD5 (chr. 3, 43794949bp, 1 SNP, iHS: 4.75), DUSP19 & NUP35 (chr. 2, 99180–18528, 3 SNPs, iHS: 4.30–4.66), surface tyrosine-kinase receptor ERBB4 (chr. 2: iHS: 4.31–4.54), transcription-associated RORC (chr. 1, 151792842-151817543bp, 2 SNPs, iHS: 4.31–4.66).
No structural variation was identified in ABHD5 or DUSP19, whilst variants were present but rare for the remaining iHS hits (S2 Table). In total, two deletions were identified in RORC, three in ZFHX3, NUP35, and ITGAE, one of which is an 86bp deletion found in seven individuals, and 14 deletions and a 31bp insertion in RIMS1. Particularly variable are ERBB4 and FKBP5 for which we identify 49 and 75 distinct variants respectively. ERBB4 consists of 44 deletions, four insertions, and one inversion, whilst FKBP5 consists of 70 deletions, three duplications, one insertion, and one inversion.
The between case-control XP-EHH approach identifies 10 significant SNPs across six genetic regions (S5 Table). Relative selection for the control population lies within three regions, including POM121L12 (XP-EHH: 4.33), SYNJ2BP, ADAM21, ADAM20 (XP-EHH: 4.12 to 5.57) and ERG (XP-EHH: 4.04), whilst three regions are under relative selection in the case population, including MCUR1 (XP-EHH: -4.26), GCLC (XP-EHH: -4.69) and the MHC (XP-EHH: -4.02) (S5 Table). We identify no structural variants within POM121L12, ADAM21, or ADAM20, but a singleton 75bp deletion in SYNJ2BP, two deletions within MCUR1, three deletions in GCLC, and 20 distinct deletions within ERG, of which 8 individuals share a 106bp deletion and 6 share a 325bp deletion (S2 Table).
Discussion
As expected, the most significant SNP association is the heterozygous protective rs334 effect (P = 2.61x10-13), with thirty-nine further SNPs within HBB also being significantly associated with resistance to severe disease. SNPs in other candidate genes, including FREM3, GYPA, GYPB and USP38 [3], did not exceed a significance threshold of 1x10-6, and their p-values were different (greater) to those previously published because of the use of the more conservative EMMAX mixed model regression [12]. Marginal evidence for a role of HLA association with severe malaria was also identified, and is broadly consistent with previous work in a West African population that demonstrated that carriers of HLA Class I Bw53 and HLA class II DRB1*1302-DQB1*0501 were protected against severe malaria [15]. Note that our targeted imputation of HLA utilised a Caucasian reference panel and may therefore overlook further true associations within the HLA locus. Further, we identified signals of positive selection within the MHC region, this being consistent with malaria as a driver of MHC polymorphism in the human population [16, 17].
Of the novel SNP associations identified here, two of the top candidates are located between the interleukin receptors IL-23R and IL-12RB2, a region that has been identified in GWAS of other inflammatory and immune-linked diseases [18]. IL-12 and IL-23 are related pro-inflammatory cytokines that share both the p40 subunit and the IL-12Rβ1 receptor subunit. IL-12 signals through a receptor comprising IL-12Rβ1 and IL-12Rβ2 and is a potent inducer of IFN-γ which mediates both clearance of infection and immunopathology in infections with Plasmodium parasites. IL-23 signalling (through its receptor, comprising IL-12Rβ1 and IL-23R) promotes transcription of RORC which encodes RORγ, a transcription factor involved in generation of IL-17. RORC was found to be under recent positive selection in our analysis, further supporting the importance of the pathway. Decreased IL-12 levels have been associated with progression from uncomplicated malaria to severe disease, specifically an increased risk of severe malarial anaemia in children [19, 20]. Variants in IL-12B have been linked to P. falciparum parasite density and associated with protection against cerebral malaria in children whilst, variants in the related IL-12A and IL-12RB1 loci have been associated with protection against severe malarial anaemia among children in western Kenya [19]. Conversely, the IL-23/IL-17 immune pathway has been implicated in the development of inflammatory reactions in children that develop severe malarial anaemia [21], in multi-organ dysfunction and acute renal failure in adult P. falciparum cases from India [22] and with the risk of cerebral malaria in Africa [23]. IL-23R haplotypes have also been associated with increased susceptibility to severe malarial anaemia in Kenya [24].
Three significantly associated SNPs are present within LINC00944, with one being 80bp from a known CTCF binding site [25]. Structurally, although the LINC00943/4 region is a known deletion site [25], we identified no such deletions within the region in our ‘Trios’ dataset. Broader functionality of this long intergenic non-coding RNA is unclear, given limited experimental characterisation, making it difficult to determine a role for these SNP variants.
A strong association peak was also identified within KLHL3, kelch-like protein 3, being a region known to contain an enhancer and various deletions [25]. Correspondingly, we identify 152 such deletions within our Trios reference panel, of which 62 distinct variants are present in only one individual and one 1,325bp deletion is present in 91 individuals. This frequent deletion is located within an open chromatin-containing region between 137,022,562 and 137,023,887bp. Mutations of KLHL3 have previously been linked with hypertension and metabolic acidosis [26] suggesting that these novel SNP associations and deletions may prime individuals to have a greater risk for severe malarial acidosis (hyperlactataemia).
A number of the most significantly associated SNPs are present as lone, or paired, associations rather than “stacks”. This includes SNPs within or very near to TREML4 and ZNF536. Whilst this may demonstrate false positive outliers, the existence of these SNPs and their minor frequencies are confirmed in our Trios reference panel.
The broad picture of whole population iHS selection is unsurprising, with the MHC region demonstrating the most striking evidence for recent selective sweeps. Our results are also consistent with a number of previously identified iHS signals, such as those for loci containing the alcohol dehydrogenase ADH7, cadherin PCDH15, synaptotagmin SYT1, the nociception receptor TRPV1, and the transmembrane protein SPINT2 [27]. It should also be emphasised that our iHS signals reflect selection within our case-control dataset and therefore oversample, relative to a general Tanzanian population, for those signals associated with susceptibility to severe malaria.
Recent differential selection between the case and control groups, as determined by XP-EHH, identified very few significant signals. There is likely to be limited differential selection between subsets of a closely related population, despite malaria infection being a strong selector. We identified the MHC, GCLC, MCUR1, POM121L12 and the SYNJ2BP-ADAM21-ADAM20 region. The strongest of these signals covers ADAM20 and ADAM21, both members of a larger family of disintegrins and metalloproteinases that are believed to be exclusively expressed in the testis [28]; this association might simply reflect differences in the gender ratio between the cases and controls, for which XP-EHH does not control. Selection for this region is more likely driven by a variant of SYNJ2BP, a Synaptojanin-2 binding protein with potential roles in membrane trafficking and signalling [29].
Our previous work has demonstrated that novel associations with potentially significant roles in malaria susceptibility remain to be uncovered [3], and here we show that an integrated approach that identifies signals of association, selection and structural variation can empower such studies. However, with only 914 individuals in this study, sample size is a notable limitation for interpretation. Initial approaches to account for this were pursued through robust contextualisation of novel variants within the secondary ‘Trios’ dataset, and the wider 1000 Genomes project. More generally, it remains vital that further validation, through larger scale studies, be undertaken to better characterise the SNP and structural variants uncovered. This is particularly true for structural variation such as within KLHL3, which may impact gene expression and would therefore benefit from incorporation of transcriptomic data.
Distributions of human genetic variants with putative roles in P. falciparum malaria susceptibility are diverse. The HbS sickle cell polymorphism is present across most regions of sub-Saharan Africa but is known to have arisen multiple times leading to a number of distinct haplotypic backgrounds [30]. Similarly, other variants, such as G6PD polymorphism and glycophorin structural variants vary both in frequency across populations and in their direction of association, leading in some cases to allelic heterogeneity that may be subtype specific. Many protective variants identified within our study, such as IL-23R and KLHL3, were found at similar frequencies within the ‘Trios’ dataset but differed from the global 1000 Genomes panel, and may therefore represent examples of Tanzanian- or regional-specific associations. Such variants are informative to our understanding of human-parasite interactions, yet risk being overlooked in inadequately designed studies. Ultimately, human GWAS in parallel with whole genome sequencing of host and parasites in large study populations across Africa will be crucial to unravelling host genetic and parasite interactions that could lead to novel malaria control measures such as vaccines.
Methods
Ethics statement
All DNA samples were collected and genotyped following signed and informed written consent from a parent or guardian. Ethics approval for all procedures was obtained from both LSHTM (#2087) and the Tanzanian National Institute of Medical Research (NIMR/HQ/R.8a/Vol.IX/392).
Study participants and phenotypes
All participants were from the Tanga region of North-Eastern Tanzania, as described previously [3]. Briefly, severe malaria cases (n = 449) were recruited in the Teule district hospital and surrounding villages in Muheza district, Tanga region, Tanzania between June 2006 and May 2007. The controls (n = 465) were recruited, matched on ward of residence, ethnicity and age (given in months), during August 2008 from individuals without a recorded history of severe malaria [3]. Four severe malaria subtypes were identified within case individuals including hyperlactatemia (Blood lactate > 5 mmol/L, n = 256), severe malarial anaemia (Hemocue Hb < 5g/dL, n = 221), respiratory distress (n = 124) and cerebral malaria (Blantyre coma score <5, n = 120) (Table 1). Parasite infection was initially assessed by rapid diagnostic test (HRP-2 –Parascreen Pan/Pf) and confirmed by double read Geimsa-stained thick blood films.
A further 247 anonymously sampled individuals, consisting of 78 healthy parent and child trios (156 parents, 78 children, 13 singletons; 80 (32.4%) Chagga, 77 (31.2%) Pare, 90 (36.4%) Wasambaa), were collected between 2007 and 2008. These individuals are those that had no current illness or no history of malaria. The samples were collected from highland, medium and lowland villages near the Kilimanjaro, Pare and West Usambara mountains in the Tanga region of Tanzania. This is a region that experiences low to medium to high levels of malaria transmission. This dataset was used to confirm allele frequencies and identify candidate region structural variation within the general Tanzanian population, as well as to impute variants onto the case-control set.
Sample genotyping, sequencing and imputation
DNA was extracted from processed blood samples, as described previously [3, 5]. The DNA was genotyped on the Illumina Omni 2.5 million SNP chip and SNP genotypes called by the MalariaGEN Resource Centre at the Sanger Institute and the Wellcome Trust Centre for Human Genetics, using previously described methods [6,7]. These data were complemented by Iplex genotyping assays that included 180 single nucleotide polymorphisms (SNP) across 50 loci on the same individuals [3]. 107 additional candidate SNPs, including the HbS SNP rs334, were included from previous candidate genotyping of the same case-control individuals; their collection having been described previously [3]. DNA for the individuals in the Trio dataset (n = 247) was sequenced using Illumina HiSeq2500 technology at the Sanger Institute, and aligned to the GRCh37 build of the human genome [7]. The minimum genome-wide coverage across the samples was 22-fold. SNPs were called from the alignments using the standard samtools-bcftools pipeline [31]. This process led to 2,788,671 high quality SNPs with quality scores of at least 30 (1 error per 1000bp) and perfect trio-consistent genotype calls. Haplotypes were phased from genotypes using SHAPEIT (www.shapeit.fr; default settings). Structural variants, including duplications, deletions, insertions and inversions, were identified within the secondary ‘Trios’ dataset for candidate regions using DELLY version v0.7.3 [32]. This software was applied using default settings, and its use in pipelines has been shown to reliably uncover structural variants from the 1000 Genomes Project, and validation experiments of randomly selected deletion loci show a high specificity [32]. Structural variants greater than 100,000 basepairs in length were removed to conservatively exclude false positives.
To increase genome-wide SNP resolution, our initial case-control dataset was imputed using a combined reference panel of the Phase 3 1000 Genomes project [10] and children within the trio dataset, using Beagle 4.1 [11]. This allowed for the inclusion of 13.5 million additional high quality SNPs, to a total of 15.2 million SNPs. A total of 621,019 SNPs were removed from the pre-imputation dataset due to evidence of: (i) deviations in genotypic frequencies from Hardy-Weinberg equilibrium (HWE) as assessed using a chi-square test (>0.0001); (ii) high genotype call missingness (>10%); or (iii) low minor allele frequency (<0.01). 51 individuals were removed due to: (i) genotypic missingness (>0.1); (ii) abnormal PCA clustering or (iii) missing malaria phenotype data. 849,134 strand flips were identified with snpflip, with these being corrected pre-imputation with Plink v1.07. Raw hybridisation plots were manually verified for all top non-imputed GWAS associations, excluding rs334 for which the data was unavailable. Linkage disequilibrium between SNPs in close genomic distance was calculated using Plink v1.07 [33].
Targeted imputation was performed for HLA haplotypes within the major histocompatibility complex using 9,785 high quality SNPs within the region; for this we utilised SNP2HLA software (version 1.0.3) and the default Caucasian reference panel [34]. Association tests for this targeted analysis were performed through the pipeline described above. Similarly, 1,202 structural variants (698 deletions, 311 duplications, 19 insertions, 174 inversions) within chromosome four were imputed into our primary ‘case-control’ dataset using IMPUTE2 with default parameters, akin to standard SNP imputation. This approach allowed us to perform association analysis on those structural variants using EMMAX mixed model regression [12]. Trio parental SNP data was also used to provide additional context for our case-control SNPs within the wider Tanzanian population, as seen in S1 Table.
Association analysis
Case-Control association analysis of SNPs was undertaken with EMMAX mixed model regression [12], controlling for age as a fixed effect and relatedness (represented by a kinship matrix) as a random effect (to reduce associations relating to familial clustering). Several genotypic models were implemented separately, including additive, heterozygous, dominant and recessive. Minimum P values from each model were utilised for top hit identification. Odds ratios were estimated with Plink v1.07 [33]. Our complementary conditional GWAS shared the pipeline for the main GWAS, but with HbS status added as an additional covariate. To evaluate the statistical potential of our GWAS study, we performed a retrospective power calculation (using http://zzz.bwh.harvard.edu/gpc/cc2.html). A study of 460 cases and 460 controls can detect odds ratios of at least 2 for a high risk allele minor allele frequency of 5% with a statistical power of 85% (and type I error of 10−6). A significance threshold of 10−6 was established using a permutation approach [35]. In particular, both the case-control status of the chromosomes were randomly permuted 10,000 times. From each of the 10,000 random experiments, we determined the maximum chi-square statistics (across the four genotypic tests) over all SNPs genotyped. We ordered these statistics and then calculated the 95 percentile. This was the estimate of the 0.05 significance level for the experiment performed, assuming inference is taken with respect to maximum chi-square statistic observed over all genotyped SNPs, and accounts for the linkage disequilibrium between SNPs and correlation between the results from applying the 4 genotypic tests.
Selection analysis
Whole population Integrated Haplotype Scores (iHS) [13] and case-control Cross-Population Extended Haplotype Homozygosity (XP-EHH) [14] were calculated and normalised over the whole genome using selscan and norm [36]. Core SNPs with a minor allele frequency below 0.01 were excluded from this analysis. In this context, high iHS values indicate a whole population selection signal whilst positive XP-EHH values indicate relative selection within the control population and negative XP-EHH values indicate relative selection within the case population. We looked for structural variants in regions with SNP-based signals of positive selection, as it possible that selection may actually be driven by structural variants (see [37] for an example).
Supporting information
Visualisation of the first two principal components, by (a) case-control status and (b) father’s ethnicity, highlights the existence of cryptic relatedness; (c) Principal component analysis reveals that the ‘Trios’ and primary ‘Case-Control’ participants overlap and are within the African cluster of the 1000 Genomes dataset; (d) Quantile-quantile plot for the observed and expected P values of the heterozygous model genome-wide association statistic.
(PNG)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank the participants and Tanzanian communities who made this study possible, and the healthcare workers who assisted with this work. We would also like to acknowledge members of the MalariaGEN Resource Centre at the Wellcome Trust Sanger Institute and at the Wellcome Centre for Human Genetics at Oxford University who contributed to this study by carrying out the data production and analysis pipelines for genome-wide SNP genotype calling and sequence alignment: in particular, we thank Quang Si Le, Katja Kivinen, Jim Stalker, Anna Jeffreys, Kate Rowlands, Christina Hubbart, Eleanor Drury, Geraldine Clarke, Chris Spencer, Gavin Band, Dominic Kwiatkowski and Kirk Rockett, as well as members of the Sample Management and DNA pipeline teams at the Wellcome Trust Sanger Institute.
Data Availability
Due to patient confidentiality, data are available upon request from MalariaGEN (https://www.ebi.ac.uk/ega/studies/EGAS00001000638, https://www.ebi.ac.uk/ega/studies/EGAS00001000637, https://www.ebi.ac.uk/ega/studies/EGAS00001000636, and https://www.ebi.ac.uk/ega/studies/EGAS00001001311). Please see https://www.malariagen.net/data/terms-use/human-gwas-data for instructions on how to apply for data access.
Funding Statement
MR is funded by the Biotechnology and Biological Sciences Research Council (grant number BB/J014567/1). The MalariaGEN Project is supported by the Wellcome Trust (WT077383/Z/05/Z) and the Bill and Melinda Gates Foundation through The Foundation for the National Institutes of Health (FNIH, USA) (566) as part of the Grand Challenges in Global Health Initiative. The Resource Centre for Genomic Epidemiology of Malaria is supported by the Wellcome Trust (090770/Z/09/Z). This research was supported by the UK Medical Research Council (G0600718 and G0600230). The Wellcome Trust also provides core awards to the Wellcome Trust Centre for Human Genetics (090532/Z/09/Z) and the Wellcome Trust Sanger Institute (098051/Z/05/Z). TGC is supported by the Medical Research Council UK (Grant no. MR/K000551/1, MR/M01360X/1, MR/N010469/1, MC_PC_15103). SC is funded by the Medical Research Council UK (Grant no. MR/M01360X/1, MC_PC_15103). NS was funded by the Wellcome Trust grant number 091924 and Fundação para a Ciência e Tecnologia through the project UID/MAT/00006/2013. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO. (2015). World Malaria Report 2015. WHO Press. [Google Scholar]
- 2.MacKinnon MJ, Mwangi TW, Snow RW, Marsh K, Williams TN. Heritability of Malaria in Africa. PLoS Medicine 2005; 2(12): e340 doi: 10.1371/journal.pmed.0020340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manjurano A, Sepúlveda N, Nadjm B, Mtove G, Wangai H, Maxwell C, et al. USP38, FREM3, SDC1, DDC, and LOC727982 Gene Polymorphisms and Differential Susceptibility to Severe Malaria in Tanzania. J Infect Dis. 2015; 212:1129–39. doi: 10.1093/infdis/jiv192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manjurano A, Sepulveda N, Nadjm B, Mtove G, Wangai H, Maxwell C, et al. African glucose-6-phosphate dehydrogenase alleles associated with protection from severe malaria in heterozygous females in Tanzania. PLoS Genet. 2015; 11(2):e1004960 doi: 10.1371/journal.pgen.1004960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manjurano A, Clark TG, Nadjm B, Mtove G, Wangai H, Sepulveda N, et al. Candidate human genetic polymorphisms and severe malaria in a Tanzanian population. PLoS One 2012; 7(10):e47463 doi: 10.1371/journal.pone.0047463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malaria Genomic Epidemiology Network. A novel locus of resistance to severe malaria in a region of ancient balancing selection. Nature 2015; 526, 253–257. doi: 10.1038/nature15390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leffler EM, Band G, Busby GBJ, Kivinen K, Le QS, Clarke GM, et al. Resistance to malaria through structural variation of red blood cell invasion receptors. Science 2017; 101126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jallow M, Teo YY, Small KS, Rockett KA, Deloukas P, Clark TG, et al. Genome-wide and fine-resolution association analysis of malaria in West Africa. Nat Genet 2009. 41(6):657–65. doi: 10.1038/ng.388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Timmann C, Thye T, Vens M, Evans J, May J, Ehmen C, et al. Genome-wide association study indicates two novel resistance loci for severe malaria. Nature 2012; 489, 443–446. doi: 10.1038/nature11334 [DOI] [PubMed] [Google Scholar]
- 10.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 2015; 526;68–74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Browning SR and Browning BL. Genotype imputation with millions of reference samples. Am J Hum Genet 2016. 98:116–126. doi: 10.1016/j.ajhg.2015.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang HM, Sui JH, Service SK, Zaitlen NA, Kong S, Freimer NB et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genetics 2010. 42;348–54. doi: 10.1038/ng.548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A Map of Recent Positive Selection in the Human Genome. PloS Biology 2006; (3): e72 doi: 10.1371/journal.pbio.0040072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 449, 913–918. doi: 10.1038/nature06250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill AV, Allsopp CE, Kwiatkowski D, Anstey NM, Twumasi P, Rowe PA et al. Common west African HLA antigens are associated with protection from severe malaria. Nature 1991, 15; 352(6336):595–600. doi: 10.1038/352595a0 [DOI] [PubMed] [Google Scholar]
- 16.Garamszegi LZ. Global distribution of malaria-resistant MHC-HLA alleles: the number and frequencies of alleles and malaria risk. Malar J. 2014; 13:349 doi: 10.1186/1475-2875-13-349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leffler E, Gao Z, Pfeifer S, Segurel L, Auton A, Venn O et al. Multiple Instances of Ancient Balancing Selection Shared Between Humans and Chimpanzees. Science 2013. 339:1578–1582. doi: 10.1126/science.1234070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizuki N, Meguro A, Ota M, Ohno S, Shiota T, Kawagoe T, et al. Genome-wide association studies identify IL23R-IL12RB2 and IL10 as Behçet's disease susceptibility loci. Nat Genet. 2010; 42(8):703–6. doi: 10.1038/ng.624 [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Prather D, Vanden Eng J, Crawford S, Kariuki S, ter Kuile F, et al. Polymorphisms in genes of interleukin 12 and its receptors and their association with protection against severe malarial anaemia in children in western Kenya. Malar J. 2010; 9:87 doi: 10.1186/1475-2875-9-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raballah E, Kempaiah P, Karim Z, Orinda GO, Otieno MF, Perkins DJ, Ong'echa JM. CD4 T-cell expression of IFN-γ and IL-17 in pediatric malarial anemia. PLoS One. 2017; 12(4):e0175864 doi: 10.1371/journal.pone.0175864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oyegue-Liabagui SL, Bouopda-Tuedom AG, Kouna LC, Maghendji-Nzondo S, Nzoughe H, Tchitoula-Makaya N, et al. Pro- and anti-inflammatory cytokines in children with malaria in Franceville, Gabon. Am J Clin Exp Immunol. 2017;6(2):9–20. [PMC free article] [PubMed] [Google Scholar]
- 22.Herbert F, Tchitchek N, Bansal D, Jacques J, Pathak S, Bécavin C, et al. Evidence of IL-17, IP-10, and IL-10 involvement in multiple-organ dysfunction and IL-17 pathway in acute renal failure associated to Plasmodium falciparum malaria. Journal of Translational Medicine 2015; 13:369 doi: 10.1186/s12967-015-0731-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marquet S, Conte I, Poudiougou B, Argiro L, Cabantous S, Dessein H, et al. The IL17F and IL17RA Genetic Variants Increase Risk of Cerebral Malaria in Two African Populations. Infect Immun. 2015;84(2):590–7. doi: 10.1128/IAI.00671-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munde EO, Raballah E, Okeyo WA, Ong'echa JM, Perkins DJ, Ouma C. Haplotype of non-synonymous mutations within IL-23R is associated with susceptibility to severe malaria anemia in a P. falciparum holoendemic transmission area of Kenya. BMC Infect Dis. 2017;17(1):291 doi: 10.1186/s12879-017-2404-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 6;489(7414):57–74. doi: 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyden LM, Choi M, Choate KA, Nelson-Williams CJ, Farhi A, Toka HR et al. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature 2012. 482:98–102. doi: 10.1038/nature10814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grossman SR, Andersen KG, Shlyakhter I, Tabrizi S, Winnicki S, Yen A, et al. Identifying recent adaptations in large-scale genomic data. Cell. 2013;152(4):703–13. doi: 10.1016/j.cell.2013.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hooft van Huijsduijnen R. ADAM 20 and 21; two novel human testis-specific membrane metalloproteases with similarity to fertilin-alpha. Gene 1998;206(2):273–82. [DOI] [PubMed] [Google Scholar]
- 29.Nemoto Y, Arribas M, Haffner C and DeCamilli P. Synaptojanin 2, a Novel Synaptojanin Isoform with a Distinct Targeting Domain and Expression Pattern. J Biolog Chem 1997; 272:30817–30821. [DOI] [PubMed] [Google Scholar]
- 30.Serjeant GR. The Natural History of Sickle Cell Disease. Cold Spring Harbor Perspectives in Medicine 2013. doi: 10.1101/cshperspect.a011783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Handsaker B, Wysoker A, Fennell T, Rua J, Homer N, et al. The Sequence Alignment/Map Format and SAMtools. Bioinformatics. 2009; 25(16):2078–9. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rausch T, Zichner T, Schlattl A, Stuetz AM, Benes V, Korbel JO. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 2012; 28: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D et al. PLINK: A toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet 2007; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia X, Han B, Onengut-Gumuscu S, Chen W-M, Concannon PJ, Rich SS, et al. Imputing Amino Acid Polymorphisms in Human Leukocyte Antigenes. PLoS One 2013; 8(6):e64683 doi: 10.1371/journal.pone.0064683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sousa I, Clark TG, Holt R, Pagnamenta AT, Mulder EJ, Minderaa RB, et al. Polymorphisms in leucine-rich repeat genes are associated with autism spectrum disorder susceptibility in populations of European ancestry. Mol Autism. 2010; 1(1):7 doi: 10.1186/2040-2392-1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szpiech ZA, Hernandez RD. selscan: An Efficient Multithreaded Program to Perform EHH-Based Scans for Positive Selection. Molec Biol Evol 2014; 31(10):2824–7. doi: 10.1093/molbev/msu211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ravenhall M, Benavente ED, Mipando M, Jensen AT, Sutherland CJ, Roper C, et al. Characterizing the impact of sustained sulfadoxine/pyrimethamine use upon the Plasmodium falciparum population in Malawi. Malar J. 2016. November 29;15(1):575 doi: 10.1186/s12936-016-1634-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Visualisation of the first two principal components, by (a) case-control status and (b) father’s ethnicity, highlights the existence of cryptic relatedness; (c) Principal component analysis reveals that the ‘Trios’ and primary ‘Case-Control’ participants overlap and are within the African cluster of the 1000 Genomes dataset; (d) Quantile-quantile plot for the observed and expected P values of the heterozygous model genome-wide association statistic.
(PNG)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
Due to patient confidentiality, data are available upon request from MalariaGEN (https://www.ebi.ac.uk/ega/studies/EGAS00001000638, https://www.ebi.ac.uk/ega/studies/EGAS00001000637, https://www.ebi.ac.uk/ega/studies/EGAS00001000636, and https://www.ebi.ac.uk/ega/studies/EGAS00001001311). Please see https://www.malariagen.net/data/terms-use/human-gwas-data for instructions on how to apply for data access.