Abstract
Chemotherapeutic resistance remains a critical clinical issue is responsible for treatment failure in patients with ovarian cancer. Evidence of the involvement of miRNAs in chemoresistance in ovarian cancer has been recently emerging. However, the underlying molecular links between chemoresistance and miRNAs remain largely unknown. In this study, we report that miR-149-5p expression is markedly elevated in chemoresistant ovarian cancer tissues compared with the chemosensitive ovarian cancer tissues. Furthermore, the silencing of miR-149-5p enhanced the chemosensitivity of ovarian cancer cells to cisplatin in vitro and in vivo. Conversely, the upregulation of miR-149-5p aggravated chemoresistance in ovarian cancer cells. Our results further revealed that miR-149-5p directly targeted the core kinase components of the Hippo signaling pathway, STE20-like kinase (MST)1 and protein salvador homolog 1 (SAV1), resulting in the inactivation of TEA domain (TEAD) transcription. On the whole, our findings reveal a novel mechanism of of action miR-149-5p in inducing chemotherapeutic resistance in ovarian cancer, indicating that miR-149-5p may serve as a chemotherapeutic response indicator and a potential therapeutic target in ovarian cancer.
Keywords: microRNA-149-5p, chemotherapeutic resistance, Hippo signaling pathway and ovarian cancer
Introduction
Ovarian cancer is one of the most common types of gynecological cancers and the 5th leading cause of cancer-related mortality in females (1,2). Despite enormous improvements over the past years, the prognosis of patients with ovarian cancer remains dismal, and this is largely attributed to chemotherapeutic resistance following several cycles of treatment. Hence, a better understanding of the underlying mechanisms responsible for chemotherapeutic resistance in ovarian cancer may aid in the development of novel targets for therapeutic intervention in ovarian cancer.
The Hippo pathway represents a novel tumor suppressor pathway and the dysregulation of Hippo signaling has been demonstrated to be involved in several biological process and tumor development (3–5). From a molecular perspective, 4 core kinase components have been reported to be essential for the tumor suppressive function of Hippo signaling, including mammalian STE20-like kinase (MST)1/2, protein salvador homolog 1 (SAV1), large tumor-suppressor kinase (LATS)1/2 and Mps one binder kinase activator-like 1 (MOB1) (5). Physiologically, the Hippo signaling pathway is active, which tightly controls the activity of 2 critical downstream effectors of the Hippo pathway, YAP and tafazzin (TAZ), both spatially and temporally via the phosphorylation of YAP and TAZ by these 4 core kinase cassettes, leading to the degradation of YAP and TAZ (6,7). Conversely, when the activity of Hippo signaling is inhibited by unknown mechanisms, phosphorylated YAP and TAZ translocate to the nucleus and elicit the transcriptional activity of TEA domain (TEAD) family members, including TEAD1-4 (8–10), which further transcriptionally activates downstream effectors to exert its versatile functions (11–13). Recently, accumulating evidence has indicated that the inactivation of Hippo promotes the development of chemotherapeutic resistance in numerous human cancers. For example, the study by Lai et al reported that upregulation of TAZ induced Taxol resistance in breast cancer cells via transcriptionally activating Cyr61 and CTGF (14); moreover, the hyperactivation of YAP has been reported to be positively associated with a poor chemotherapeutic response to taxanes and a poor prognosis in patients with ovarian cancer (15), indicating that the inactivation of the Hippo pathway is crucial for the development of cancer chemoresistance. Therefore, a better understanding of the mechanisms responsible for the inactivation of the Hippo pathway may provide new insight and may aid in the development of more effective treatment strategies for ovarian cancer.
MicroRNAs (miRNAs or miRs) are small endogenous non-coding RNAs that are able to repress a variety of target genes at the post-transcriptional level by binding with the 3′ untranslated region (3′UTR) of the target gene mRNAs (16). Numerous studies have reported that miRNAs play crucial roles in tumorigenesis, malignant progression and the metastasis of cancers through various mechanisms (17–22). Furthermore, studies have also supported that the up- or downregulation of a certain miRNA can be directly tied to the response to chemotherapeutic agents (23–26). In this study, from a publically available miRNA dataset from The Cancer Genome Atlas (TCGA) and our experiments, we found that miR-149-5p expression was significantly elevated in chemo-resistant ovarian cancer tissues and cell lines compared with chemosensitive ovarian cancer ones. Furthermore, the silencing of miR-149-5p increased the apoptotic ratio and decreased the mitochondrial potential of ovarian cancer cells in response to cisplatin (CDDP) in vitro, and also enhanced the chemotherapeutic sensitivity of ovarian cancer cells to CDDP in vivo. Conversely, the upregulation of miR-149-5p decreased the apoptotic ratio and increased the mitochondrial potential of ovarian cancer cells in response to to CDDP in vivo. Our results further demonstrated that miR-149-5p directly targeted MST1 and SAV1, leading to the inactivation of TEAD transcription. Importantly, the effects of miR-149-5p silencing on the apoptotic ratio, mitochondrial potential and caspase-3 activity in ovarian cancer cells were attenuated by the individual knockdown of MST1 and SAV1. Therefore, our findings reveal a novel mechanism through which miR-149-5p induces chemotherapeutic resistance in ovarian cancer, suggesting that miR-149-5p may be used as a chemotherapeutic response indicator in ovarian cancer.
Materials and methods
Cell culture and agents
The ovarian cancer cell lines, TOV-21G, A2780, OVCAR-3, Caov-3, ES-2, HO-8910 and SK-OV-3, were obtained from the Shanghai Chinese Academy of Sciences cell bank (Shanghai, China), and all human ovarian cancer cell lines were maintained in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS (HyClone, Logan, UT, USA). CDDP (Cat. no. P4394; Sigma-Aldrich, St. Louis, MO, USA), which undergoes relatively slow solvolysis in DMSO, was dissolved in 0.9% NaCl at 4 mM, immediately aliquoted and stored at −70°C. Thawed aliquots were used only once. The stock solutions were further diluted with DMSO and added to the cell cultures with the final DMSO concentrations being <0.05%. The ovarian cancer cells were treated with cisplatin (5 µM, 48 h) before further functional assays.
Patients and tumor tissues
A total of 20 ovarian cancer tissues and 10 benign ovarian lesion tissues, including 4 simple ovarian cyst and 6 simple ovarian serous cystadenoma were obtained during surgery at the Sun Yat-sen University Cancer Center (Guangzhou, China) (Tables I and II). Patients were diagnosed based on clinical and pathological evidence, and the specimens were immediately snap-frozen and stored in liquid nitrogen tanks. For the use of these clinical samples for research purposes, prior patient consents and approval from the Institutional Research Ethics Committee of Sun Yat-sen University Cancer Center were obtained (ethics approval no. 308-2016-03-01).
Table I.
The basic information of 20 patients with ovarian serous cystadenocarcinoma for miR-149-5p expression analysis.
| Characteristic | Cases (n) | Percentage |
|---|---|---|
| Age (years) | ||
| <55 | 6 | 30 |
| ≥55 | 14 | 70 |
| Stage | ||
| Stage I | 1 | 5 |
| Stage II | 1 | 5 |
| Stage III | 14 | 70 |
| Stage IV | 4 | 20 |
| Grade | ||
| G1 | 0 | 0 |
| G2 | 3 | 15 |
| G3 | 17 | 85 |
| Treatment | ||
| Complete response | 10 | 50 |
| Partial response | 1 | 5 |
| Stable disease | 3 | 15 |
| Progressive disease | 6 | 30 |
Table II.
The basic information of 10 patients with benign ovarian lesions for miR-149-5p expression analysis.
| Characteristic | Cases (n) | Percentage |
|---|---|---|
| Age (years) | ||
| <55 | 4 | 40.0 |
| ≥55 | 6 | 60.0 |
| Type of disease | ||
| Simple cyst | 4 | 40.0 |
| Simple serous cystadenoma | 6 | 60.0 |
RNA extraction, reverse transcription and real-time PCR
Total RNA from the tissues or cells was extracted using TRIzol reagent (Life Technologies/Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions. Messenger RNAs (mRNAs) were polyadenylated using a poly-A polymerase-based First-Strand Synthesis kit (Takara, Dalian, China) and the reverse transcription (RT) from total mRNA was performed using a PrimeScript RT Reagent kit (Takara) according to the manufacturer's instructions. miRNAs were reverse transcribed from total mRNA using the RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific) according to the manufacturer's instructions. Complementary DNA (cDNA) was amplified and quantified on an ABI 7500HT system (Applied Biosystems, Foster City, CA, USA) using SYBR-Green I (Applied Biosystems). The RT-PCR reaction was performed with the following parameter values: 15 min at 37°C, 10 min at 65°C, 5 min at 85°C and −20°C until use. RT-PCR analysis was performed on an iQ5 Real Time PCR Detection System (Bio-Rad, Hercules, CA, USA) with a 20 µl volume reaction containing 2 µl reverse transcription product, 10 µl 2All-in-One™ Q-PCR Mix, 2 µl PCR Forward Primer (2 µM), 2 µl Universal Adaptor PCR Primer (2 µM) and 4 µl ddH2O. The reactions were incubated in 96-well plates at 95°C for 10 min, following by 40 cycles, and then ramped from 66°C to 95°C to obtain the melting curve. Each sample was analyzed in triplicate. The primers sequence and the product size were listed in Table III. Real-time PCR was performed according to a standard method, as previously described (27). Primers for U6 and miR-149-5p (miRQ0000450-1-1) were synthesized and purified by RiboBio (Guangzhou, China). U6 or glycer-aldehyde-3-phosphate dehydrogenase (GAPDH) was used as endogenous controls for miRNA or mRNA respectively. Relative fold expression was calculated with the comparative threshold cycle (2−ΔΔCq) method as previously described (28).
Table III.
List of primers used in the reactions for RT-qPCR.
| Gene | Sequence (5′-3′) | Product size (bp) |
|---|---|---|
| BCL2-up | GGTGGGGTCATGTGTGTGG | 89 |
| BCL2-dn | CGGTTCAGGTACTCAGTCATCC | |
| BCL2L1-up | CTGCTGCATTGTTCCCATAG | 289 |
| BCL2L1-dn | TTCAGTGACCTGACATCCCA | |
| CTGF-up | TGGAGATTTTGGGAGTACGG | 107 |
| CTGF-dn | CAGGCTAGAGAAGCAGAGCC | |
| CYR61-up | GGTCAAAGTTACCGGGCAGT | 115 |
| CYR61-dn | GGAGGCATCGAATCCCAGC | |
| HOXA1-up | TCCTGGAATACCCCATACTTAGC | 95 |
| HOXA1-dn | GCACGACTGGAAAGTTGTAATCC | |
| SOX9-up | AGCGAACGCACATCAAGAC | 85 |
| SOX9-dn | CTGTAGGCGATCTGTTGGGG | |
| GAPDH-up | TCCTCTGACTTCAACAGCGACAC | 126 |
| GAPDH-dn | CACCCTGTTGCTGTAGCCAAATTC |
Plasmids, small interfering RNA and transfection
The miR-149-5p expression plasmid was generated by cloning the genomic pre-miR-149-5p gene into the retroviral transfer plasmid pMSCV-puro (Clontech Laboratories Inc., Tokyo, Japan) to generate plasmid pMSCV-miR-149-5p. Subsequently, pMSCV-miR-149-5p was co-transfected with the pIK packaging plasmid (Cat. no. 75483; ATCC, VA, USA) into 293FT (CRL-1573) cells using the standard calcium phosphate transfection method, as previously described (29). At 36 h after the co-transfection, the supernatants were collected and incubated with the cells to be infected for 24 h in the presence of polybrene (2.5 µg/ml). Following infection, puromycin (1.5 µg/ml) was used to select stably transduced cells over a 10-day period. The luciferase reporter system of pTEAD-luc (Cat. no. 83467; Addgene, Cambridge, MA, USA) was used to examine the transcriptional activity of TEAD. The 3′-untranslated region (3′UTR) regions of human MST1 and SAV1 were PCR-amplified from genomic DNA and cloned into the pmirGLO luciferase reporter vector (Promega, Madison, WI, USA) and the list of primers used in clone reactions are presented in Table IV. The miArrest plasmids for anti-miR-149-5p and negative control plasmids were constructed and cloned into phU6 plasmids by GeneChem (Shanghai, China). The sequence of anti-miR-149-5p was GGGAGTGAAGACACGGAGCCAGA. Small interfering RNA (siRNA) for YAP, TAZ, MST1 and SAV1 knockdown were obtained from Ribobio. The transfection of siRNAs and the plasmids was performed using Lipofectamine 3000 (Life Technologies/Thermo Fisher Scientific) according to the manufacturer's instructions.
Table IV.
List of primers used in the reactions for clone PCR.
| Gene | Sequence (5′-3′) |
|---|---|
| miR-149-up | GGGCACATTGCTCTCCATTCAG |
| miR-149-dn | AGGCCCGAAACACCCGTAAG |
| MST1-3′UTR-34nt-up | CAGGCTTTGGGTGAATTCTGGATG |
| MST1-3′UTR-1577nt-dn | GTATCCTGAGGTGCAACCCAGTC |
| SAV1-3′UTR-74nt-up | CTCTGGTTAATAAATGCTGCTTCCTTTGTG |
| SAV1-3′UTR-1449nt-dn | CTGGGAGTAGTTTGTATTCTGGAAGAC |
Western blot analysis
Nuclear/cytoplasmic fractionation was carried out using the Cell Fractionation kit (Cell Signaling Technology, Danvers, MA, USA) according to the manufacturer's instructions, and the whole cell lysates were extracted using RIPA Buffer (Cell Signaling Technology). Western blot analysis was performed according to a standard method, as previously described (30). Antibodies against YAP (Cat. no. ab81183), TAZ (Cat. no. ab93362), MST1 (Cat. no ab76822), SAV1 (Cat. no. ab105105), Bcl-2 (Cat. no. ab194583) and Bcl-xL (Cat. no. ab32370) were purchased from Abcam (Cambridge, MA, USA), and antibodies against caspase-3 (Cat. no. 9662) and caspase-9 (Cat. no. 9501) were from Cell Signaling Technology. The membranes were stripped and reprobed with an anti-α-tubulin antibody (Sigma-Aldrich) as the loading control.
Flow cytometric analysis
Flow cytometric analysis of apoptosis was carried out using the FITC Annexin V Apoptosis Detection kit I (BD Biosciences, San Jose, CA, USA), and was presented as the protocol described below. Briefly, the cells were dissociated with trypsin and resuspended at 1×106 cells/ml in binding buffer with 50 µl/ml FITC Annexin V and 50 µl/ml PI. The cells were subsequently incubated for 15 min at room temperature, and were then were analyzed using a Gallios flow cytometer (Beckman Coulter, Atlanta, GA, USA). The cell inner mitochondrial membrane potential (Δψm) was detected by flow cytometry using the MitoScreen JC-1 staining kit (BD Biosciences), and was presented as the protocol described below. Briefly, the cells were dissociated with trypsin and resuspended at 1×106 cells/ml in Assay Buffer, and then incubated at 37°C for 15 min with 10 µl/ml JC-1. Before analyzed by flow cytometer, cells were washed twice by Assay Buffer. Flow cytometry data were analyzed using FlowJo 7.6 software (TreeStar Inc., Ashland, OR, USA).
Caspase-9 or caspase-3 activity assays
The activity of caspase-9 or caspase-3 was analyzed by spectrophotometry using the Caspase-9 Colorimetric Assay kit or Caspase-3 Colorimetric Assay kit (KeyGen BioTech, Nanjing, China), and was presented as the protocol described below. Briefly, 5×106 cells or 100 mg fresh tumor tissues were washed with cold PBS and re-suspended in lysis buffer and incubated on ice for 30 min. Subsequently, 50 µl cell suspension was mixed with 50 µl reaction buffer, and 5 µl caspase-3/-9 substrate, followed by incubation at 37°C for 4 h. The absorbance was measured at 405 nm, and BCA protein quantitative analysis was used as the reference to normalize each experimental group.
Establishment of tumor xenograft model
For the establishment of a tumor xenograft model, 20 BALB/c-nu mice (6 weeks old, weighing 10–12 g) were purchased from Hunan Silaikejingda Laboratory Animal Technology Co. (Hunan, China) and randomly divided into 4 groups (n=5 per group) and the indicated cells (1×106) were inoculated subcutaneously into the inguinal folds of the nude mice. At 7 days after the cell inoculation, the mice were injected intraperitoneally 15 mg/kg CDDP twice per week for 5 weeks in one group of mice injected with the vector cells and one group of mice injected with the anti-miR-149-5p cells respectively. Tumor volume was determined using an external caliper and calculated using the equation (L × W2)/2. On day 40, the animals were euthanized, and the tumors were excised, weighed and stored in liquid nitrogen tanks. For survival analyses, the mice were monitored daily for signs of discomfort, and were either euthanized all at one time or individually when presenting signs of distress, such as a 10% loss of body weight, or head tilting. Overall survival was assessed by the Kaplan-Meier method, using an exact log-rank test. Ethics approval for the animal experiments was provided by The Institutional Animal Care and Use Committee of Sun Yat-sen University Cancer Center (ethics approval no. 102012016008I).
Luciferase assay
The cells (4×104) were seeded in triplicate in 24-well plates and luciferase assay was carried out as previously described (31). The cells were transfected with 100 ng of HOP-flash reporter luciferase plasmid, HIP-flash (HOP-flash mutant), pmirGLO-MST1-3′UTR or -SAV1-3′UTR, luciferase plasmid, plus 5 ng pRL-TK Renilla plasmid (Promega) using Lipofectamine 3000 (Invitrogen) according to the manufacturer's instructions. Luciferase and Renilla signals were measured at 36 h following transfection using a Dual Luciferase Reporter Assay kit (Promega) according to the manufacturer's instructions.
miRNA immunoprecipitation
The cells were co-transfected with the HA-Ago2 plasmid (10822; Addgene), followed by HA-Ago2 immunoprecipitation using HA-antibody (Cat. no. H3663, Sigma-Aldrich) as previously described (32). Real-time PCR analysis of the IP material was used to test the association of the mRNA of YAP1 and TAZ with the RISC complex.
Gene set enrichment analysis (GSEA)
The miRNA dataset of ovarian cancer from TCGA was downloaded and we then procured the expression value of the corresponding genes from the Level 3 data of each sample (the unit was RNA-Seq by expectation maximization, RKPM). We then analyzed the log2 value of each sample using Excel 2010 and GraphPad 5 software, as well as statistically analyzed the miRNA expression levels of all ovarian cancer tissues using a paired t-test or unpaired t-test. GSEA was performed with RNAseqV2 dataset of ovarian cancer from TCGA as the MSigDB dataset. The high and low expression level of miR-149-5p was stratified by the medium expression level of miR-149-5p in ovarian cancer tissues. Gene set analysis was performed by Molecular Signatures Database version 5.2.
Identification of potential targets of miR-149-5p
As previously described (33,34), the TargetScan (http://www.targetscan.org/vert_71/) and miRanda (http://34.236.212.39/microrna/home.do) databases were used to identify possible targets of miR-149-5p.
Statistical analysis
All values are presented as the means ± standard deviation (SD). Significant differences were determined using GraphPad 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA). One-way ANOVA with Tukey's post hoc test was used to determine statistical differences between multiple groups. An unpaired or paired t-test was used to determine statistical differences between 2 groups and confidence intervals, 95%. A value of p<0.05 was considered to indicate a statistically significant difference. All the experiments were repeated 3 times.
Results
miR-149-5p is upregulated in chemoresistant ovarian cancer tissues
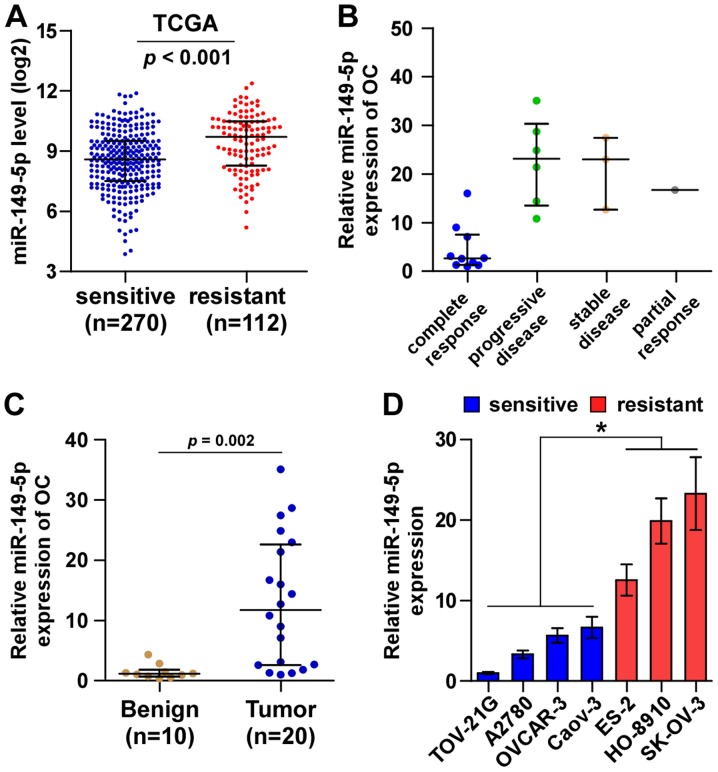
By analyzing TCGA ovarian cancer miRNA sequencing datasets, we found that miR-149-5p expression was elevated in chemoresistant ovarian cancer tissues compared with chemosensitive ovarian cancer tissues (Fig. 1A). We further examined miR-149-5p expression in our own 20 ovarian cancer tissues, including 10 complete response, 1 partial response, 3 stable disease and 6 progressive disease ovarian cancer tissues perspectively. Consistently, we found that miR-149-5p expression in the ovarian cancer tissues from patients with stable and progressive disease was higher than that in ovarian cancer tissues from patients with complete response (Fig. 1B). Furthermore, miR-149-5p expression was markedly upregulated in ovarian cancer tissues compared with benign ovarian disease tissues (Fig. 1C). The expression levels of miR-149-5p in the ovarian cancer cell lines were evaluated and the results revealed that compared with the chemosensitive ovarian cancer cell lines (TOV-21G, A2780, OVCAR-3 and Caov-3), miR-149-5p expression was differentially upregulated in the chemoresistant ovarian cancer cell lines (ES-2, HO-8910, SK-OV-3) (Fig. 1C). Thus, these results suggest that a high expression of miR-149-5p positively correlates with a poor chemotherapeutic response in patients with ovarian cancer.
Figure 1.
miR-149-5p expression is upregulated in chemoresistant ovarian cancer tissues. (A) The expression level of miR-149-5p in chemoresistant and chemosensitive ovarian cancer tissues from the TCGA ovarian cancer miRNA sequencing dataset (chemosensitive tissue, n=270; chemoresistant tissues, n=112). (B) miR-149-5p expression level in 10 complete response, 1 partial response, 3 stable disease and 6 progressive disease ovarian cancer tissues. (C) miR-149-5p expression in 10 benign ovarian lesions, including 4 simple ovarian cyst and 6 simple ovarian serous cystadenoma, and 20 ovarian cancer tissues. Transcript levels were normalized to U6 expression. (D) Real-time PCR analysis of miR-149-5p expression in ovarian cancer cell lines. Transcript levels were normalized to U6 expression. *p<0.05.
Upregulation of miR-149-5p enhances the chemoresistance of ovarian cancer cells to CDDP
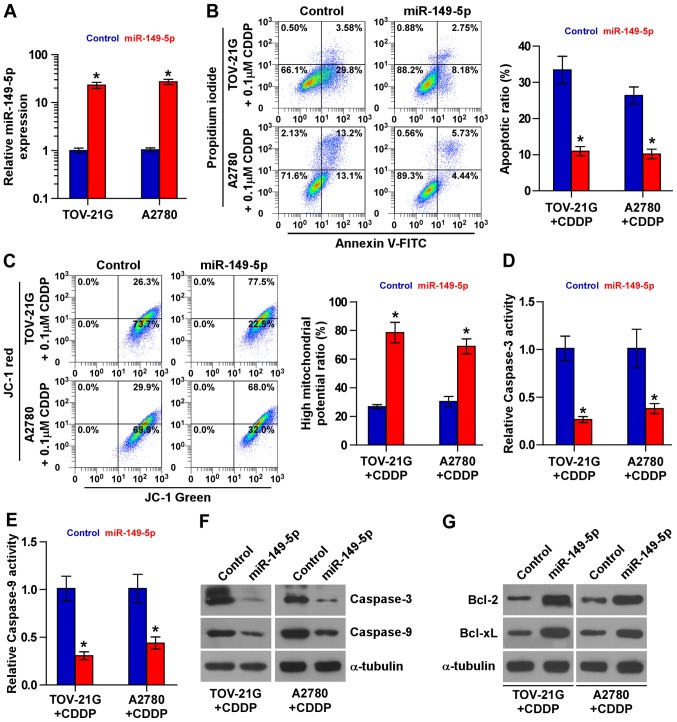
The role of miR-149-5p in the chemoresistance of ovarian cancer was further examined. Based on the results of the expression level of miR-149-5p shown in Fig. 1C, we decided to exogenously overexpress miR-149-5p via viral transduction in the chemosensitive TOV-21G and A2780 cells (Fig. 2A). As shown in Fig. 2B, miR-149-5p overexpression decreased the apoptotic rates of the TOV-21G and A2780 cells treated with CDDP. The effects of miR-149-5p overexpression on mitochondrial potential were further investigated and the results revealed that miR-149-5p overexpression increased the mitochondrial potential of the TOV-21G and A2780 cells treated with CDDP (Fig. 2C). The effects of the upregulation of miR-149-5p on apoptosis and caspase protein expression were further confirmed by examining the activity of caspase-9 and caspase-3, as well as the expression levels of the anti-apoptotic proteins, Bcl-2 and Bcl-xL. The results revealed that the overexpression of miR-149-5p reduced the activity and expression of caspase-3 and caspase-9, and increased the expression Bcl-2 and Bcl-xL (Fig. 2D–G). Therefore, these results demonstrate that miR-149-5p promotes the resistance of ovarian cancer cells to CDDP in vivo.
Figure 2.
Upregulation miR-149-5p promotes chemoresistance in chemosensitive ovarian cancer cells. (A) Real-time PCR analysis of miR-149-5p expression in the indicated ovarian cancer cells. Transcript levels were normalized by U6 expression. Error bars represent the means ± SD of 3 independent experiments; *p<0.05. (B) Annexin V-FITC/PI staining of the indicated cells treated with cisplatin (0.1 µM, 48 h) or DMSO. Error bars represent the means ± SD of 3 independent experiments; *p<0.05. (C) JC-1 staining revealed that the upregulation of miR-149-5p increased the mitochondrial potential in the indicated cells treated with cisplatin (0.1 µM, 48 h) or DMSO. Error bars represent the means ± SD of 3 independent experiments; *p<0.05. (D and E) Analysis of the activities of (D) caspase-3 and (E) caspase-9 were detected by the cleaved forms of these two proteins under treatment of cisplatin (0.1 µM, 48 h) or DMSO. Error bars represent the means ± SD of 3 independent experiments; *p<0.05. (F) Western blot analysis of caspase-3 and caspase-9 expression in the indicated ovarian cancer cells treated with cisplatin (0.1 µM, 48 h) or DMSO. α-tubulin served as the loading control. (G) Western blot analysis of Bcl-2 and Bcl-xL in the indicated cells under treatment of cisplatin (0.1 µM, 48 h) or DMSO. α-tubulin served as the loading control.
Silencing of miR-149-5p sensitizes ovarian cancer cells to CDDP
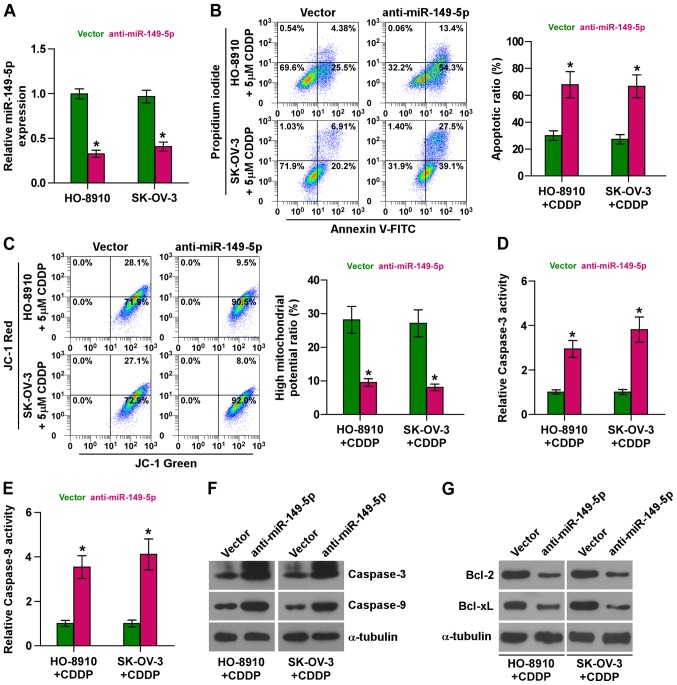
We further endogenously silenced miR-149-5p by transfecting anti-miR-149-5p into the chemoresistant HO-8910 and SK-OV-3 cells (Fig. 3A) due to the higher expression levels of miR-149-5p in the HO-8910 and SK-OV-3 cells (as shown in Fig. 1D). As shown in Fig. 3B, the silencing of miR-149-5p markedly increased the apoptotic rate of the HO-8910 and SK-OV-3 cells treated with CDDP. Furthermore, the silencing of miR-149-5p decreased the mitochondrial potential of the HO-8910 and SK-OV-3 cells treated with CDDP (Fig. 3C). In addition, the silencing of miR-149-5p enhanced the activity and expression of caspase-3 and caspase-9, and decreased the expression levels of Bcl-2 and Bcl-xL (Fig. 3D–G). Collectively, these results demonstrate that the silencing of miR-149-5p promotes the resistance of ovarian cancer cells to CDDP.
Figure 3.
Downregulation miR-149-5p promotes chemosensitivity in chemoresistant ovarian cancer cells. (A) Real-time PCR analysis of miR-149-5p expression in the indicated ovarian cancer cells. Transcript levels were normalized to U6 expression. Error bars represent the means ± SD of 3 independent experiments; *p<0.05. (B) Annexin V-FITC/PI staining of the indicated cells treated with cisplatin (5 µM, 48 h) or DMSO. Error bars represent the means ± SD of 3 independent experiments; *p<0.05. (C) JC-1 staining revealed that the silencing of miR-149-5p decreased the mitochondrial potential in the indicated cells treated with cisplatin (5 µM, 48 h) or DMSO. Error bars represent the means ± SD of 3 independent experiments; *p<0.05. (D and E) Analysis of the activities of (D) caspase-3 and (E) caspase-9 were detected by the cleaved forms of these two proteins under treatment of cisplatin (5 µM, 48 h) or DMSO. Error bars represent the means ± SD of 3 independent experiments; *p<0.05. (F) Western blot analysis of caspase-3 and caspase-9 expression in the indicated ovarian cancer cells treated with cisplatin (5 µM, 48 h) or DMSO. α-tubulin served as the loading control. (G) Western blot analysis of Bcl-2 and Bcl-xL in the indicated cells treated with cisplatin (5 µM, 48 h) or DMSO. α-tubulin served as the loading control.
Silencing of miR-149-5p enhances the chemosensitivity of ovarian cancer cells to CDDP in vivo
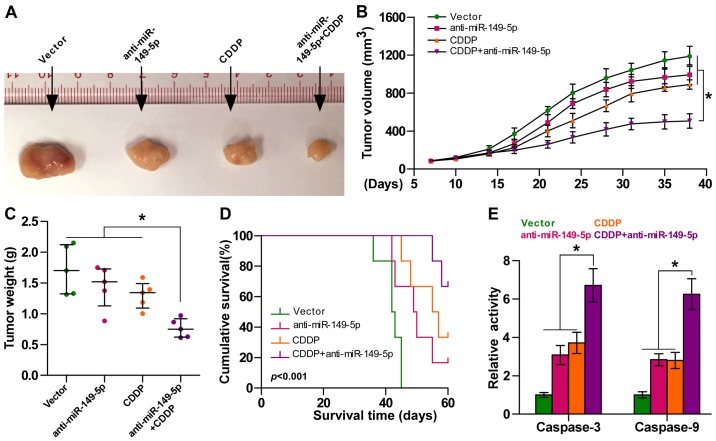
We further examined the effects of miR-149-5p on the chemoresistance of ovarian cancer cells in vivo. Mice were randomly divided into 4 groups (n=5/group). The vector cells were inoculated subcutaneously into 2 groups of mice in the left dorsal flank, and the other 2 groups of mice were inoculated with anti-miR-149-5p cells (1×106 SK-OV-3 cells per mouse). After 7 days, one group of mice injected with the vector cells and one group of mice injected with the anti-miR-149-5p cells were intraperitoneally injected with CDDP twice per week for 5 weeks respectively [15 mg/kg (Fig. 4A)]. As shown in Fig. 4B and C, the tumor volume and tumor weight were differentially decreased in the anti-miR-149-5p group, CDDP treatment alone group and anti-miR-149-5p plus CDDP group compared to the controls, particularly in the anti-miR-149-5p plus CDDP group. Moreover, the mice injected with anti-miR-149-5p-transfected cells in combination with CDDP displayed a longer survival time (Fig. 4D). Caspase-3 and caspase-9 activities were markedly increased in the mice injected with anti-miR-149-5p-transfected cells plus CDDP (Fig. 4E). Taken together, these findings demonstrated that the silencing of miR-149-5p sensitized ovarian cancer cells to CDDP in vivo.
Figure 4.
Silencing of miR-149-5p enhances the sensitivity of ovarian cancer cells to cisplatin in vivo. (A) Representative images of the tumors are shown in the xenograft model of nude mice. (B) At 7 days after the inoculation of the SK-OV-3 cells, the mice were intraperitoneal injected with 15 mg/kg cisplatin (CDDP) twice per week for 5 weeks. Tumor volumes in the indicated groups were measured from the 7th day at 3-day intervals. Data are presented as the means ± SD. (C) Tumor weights of each group. (D) The overall survival of mice in the indicated group. (E) The activities of caspase-3 and caspase-9 in the indicated tumors; *p<0.05.
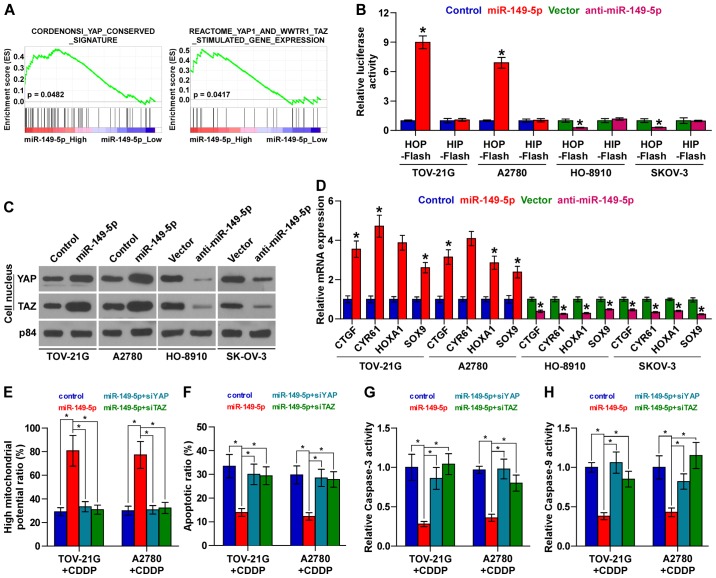
miR-149-5p inactivates the Hippo signaling pathway
To investigate the mechanisms underlying the promoting effect of miR-149-5p on chemoresistance in ovarian cancer, we performed a Gene set enrichment analysis (GSEA) of miR-149-5p expression against the oncogenic signatures collection of the MSigDB and found that miR-149-5p overexpression positively correlated with the YAP and TAZ stimulated signatures (35) (Fig. 5A), suggesting that miR-149-5p regulates the Hippo signaling pathway, which have been reported to play important roles in the development of cancer chemoresistance (14,15). As shown in Fig. 5B, we found that the upregulation of miR-149-5p increased, while the silencing of miR-149-5p decreased TEAD-dependent luciferase activity. Moreover, the results of cellular fractionation and western blot analysis revealed that the overexpression of miR-149-5p enhanced the nuclear accumulation of YAP and TAZ, while the silencing of miR-149-5p suppressed their nuclear expression (Fig. 5C). Real-time PCR analysis also revealed that miR-149-5p overexpression significantly increased, while the silencing of miR-149-5p inhibited the expression levels of multiple downstream genes, including connective tissue growth factor (CTGF), cysteine rich angiogenic inducer 61 (CYR61), homeobox A1 (HOXA1) and SRY-box 9 (SOX9) in ovarian cancer cells (Fig. 5D). Thus, these results demonstrate that the downregulation of miR-149-5p inactivates the Hippo signaling pathway in ovarian cancer cells.
Figure 5.
Upregulation of miR-149-5p inactivates the Hippo signaling pathway in ovarian cancer cells. (A) Gene set enrichment analysis (GSEA) revealed that miR-149-5p expression positively correlated with the YAP and TAZ regulated signatures. (B) TEAD transcriptional activity was assessed by luciferase reporter constructs in the indicated cells. Error bars represent the means ± SD of 3 independent experiments; *p<0.05. (C) Western blot analysis of nuclear YAP and TAZ expression in the indicated cells. The nuclear protein p84 was used as the nuclear protein marker. (D) Real-time PCR analysis of CTGF, CYR61, HOXA1 and SOX9 in the indicated cells. Error bars represent the means ± SD of 3 independent experiments; *p<0.05. (E) Individual silencing of YAP and TAZ attenuated the mitochondrial potential of ovarian cancer cells increased by miR-149-5p overexpression. Error bars represent the means ± SD of 3 independent experiments; *p<0.05. (F) Individual silencing of YAP and TAZ increased the apoptotic rate of ovarian cancer cells which was decreased by miR-149-5p overexpression. Error bars represent the means ± SD of 3 independent experiments; *p<0.05. (G and H) Individual silencing of YAP and TAZ increased caspase-3 or caspase-9 activity in ovarian cancer cells which had been decreased by miR-149-5p overexpression. Error bars represent the means ± SD of 3 independent experiments; *p<0.05.
YAP and TZA are essential for miR-149-5p upregulation-induced chemoresistance in ovarian cancer cells
We then explored the functional significance of YAP and TAZ in the chemoresistance of ovarian cancer cells. As shown in Fig. 5E, the individual inhibition of YAP or TAZ in miR-149-5p-overex-pressing cells abrogated the stimulatory effects of miR-149-5p overexpression on mitochondrial potential. Conversely, the inhibitory effects of miR-149-5p upregulation on the apoptotic ratio, and caspase-3 and caspase-9 activities were reversed by the individual silencing of YAP and TAZ in ovarian cancer cells (Fig. 5F–H). Taken together, our results indicate that YAP and TAZ are functionally relevant effectors of miR-149-5p in the chemotherapeutic response of ovarian cancer cells.
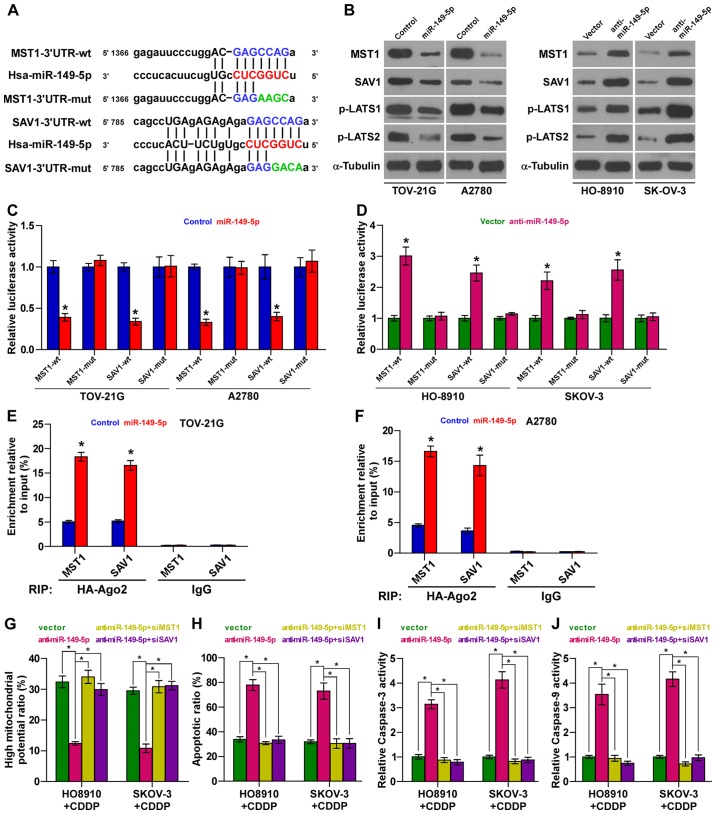
MST1 and SAV1 are authentic targets of miR-149-5p in ovarian cancer cells
Using the publically available algorithms, TargetScan and miRanda, we found that kinase components of Hippo signaling, including MST1 and SAV1, may be potential targets of miR-149-5p (Fig. 6A). Western blot analysis revealed that miR-149-5p overexpression decreased the expression levels of MST1 and SAV1; conversely, the silencing of miR-149-5p increased their expression levels in the ovarian cancer cells (Fig. 6B). Furthermore, luciferase assay revealed that the upregulation of miR-149-5p suppressed, while the silencing of miR-149-5p enhanced the reporter activity driven by the 3′UTRs of the MST1 and SAV1 transcripts, but not by the mutant 3′UTRs of these transcripts within miR-149-5p-binding seed regions in ovarian cancer cells (Fig. 6C and D). Moreover, microribonucleoprotein (miRNP) immunoprecipitation (IP) assay revealed an association of miR-149-5p with MST1 and SAV1 transcripts (Fig. 6E and F), further indicating the direct repressive effects of miR-149-5p on MST1 and SAV1. Importantly, the stimulatory effects of anti-miR-149-5p on the apoptotic ratio, and caspase-3 and caspase-9 activities were abrogated by the individual silencing of MST1 and SAV1; however, the inhibitory effects of miR-149-5p silencing were reversed by the individual silencing of MST1 and SAV1 in ovarian cancer cells (Fig. 6G–J). Collectively, our results indicate that miR-149-5p promotes the chemoresistance of ovarian cancer cells by directly targeting MST1 and SAV1, leading to the inactivation of Hippo signaling.
Figure 6.
miR-149-5p targets MST1 and SAV1 in ovarian cancer cells. (A) Predicted miR-149-5p target sequence in 3′UTRs of MST1 and SAV1. (B) Western blot analysis of MST1 and SAV1 expression in the indicated cells. α-tubulin served as the loading control. (C and D) Luciferase assay of the cells transfected with pmirGLO-3UTR reporter in miR-149-5p overexpressing or silencing cells. Error bars represent the means ± SD of 3 independent experiments; *p<0.05. (E and F) miRNP IP assay showing the association between miR-149-5p and MST1 and SAV1 transcripts in the indicated cells. Pulldown of IgG antibody served as the negative control. Error bars represent the means ± SD of 3 independent experiments; *p<0.05. (G) Individual silencing of MST1 and SAV1 increased the mitochondrial potential of ovarian cancer cells which had been decreased by anti-miR-149-5p. Error bars represent the means ± SD of 3 independent experiments; *p<0.05. (H) Individual silencing of MST1 and SAV1 decreased the apoptotic rate of ovarian cancer cells which had been increased by anti-miR-149-5p. Error bars represent the means ± SD of 3 independent experiments; *p<0.05. (I and J) Individual silencing of MST1 and SAV1 decreased caspase-3 or caspase-9 activity in ovarian cancer cells which had been increased by anti-miR-149-5p. Error bars represent the means ± SD of 3 independent experiments; *p<0.05.
Discussion
In the present study, our results revealed that miR-149-5p expression was upregulated in chemoresistant ovarian cancer tissues and cell lines compared with chemosensitive ovarian cancer tissues and cell lines. The upregulation of miR-149-5p enhanced, while the silencing of miR-149-5p inhibited the chemoresistance of ovarian cancer cells to CDDP in vivo and in vivo. Our results further revealed that miR-149-5p directly suppressed MST1 and SAV1 expression, leading to the inactivation of TEAD transcription and the downregulation of Hippo downstream target genes. Therefore, our findings reveal a novel biological role of miR-149-5p in the chemotherapeutic resistance of ovarian cancer to CDDP.
The dysregulation of the Hippo signaling pathway has been demonstrated to play an important role in the chemotherapeutic resistance of cancer. YAP and TAZ are important downstream effectors of the Hippo pathway and are suppressed by active Hippo signaling (36,37). Studies have shown that the upregulation of YAP and TAZ positively correlates with the chemoresistance of several human cancer types (38,39). Furthermore, the downregulation of the Hippo pathway core kinase components has been reported to be implicated in chemotherapeutic resistance of cancer cells to diverse chemotherapeutic drugs. For example, the downregulation of MST1, which is mediated by heat shock protein 70 (Hsp70) in a proteasomal-dependent manner, promotes the CDDP resistance of prostate cancer cells (40). Additionally, the overexpression of miR-181c contributes to chemoresistance in pancreatic cancer by directly targeting MST1 and SAV1 (41). These studies indicate that the inactivation of the Hippo signaling pathway, either the upregulation of the downstream effectors, YAP and TAZ, or the downregulation of core components of the Hippo signaling pathway, confers chemotherapeutic resistance in human cancers. In this study, our results demonstrated that a high expression of miR-149-5p inactivated Hippo signaling by directly suppressing MST1 and SAV1, which further promoted the chemotherapeutic resistance of ovarian cancer cells to CDDP. Importantly, the inhibitory effects of anti-miR-149-5p on the chemoresistance of ovarian cancer cells to CDDP were attenuated by the individual knockdown of MST1 and SAV1. There finding indicate that miR-149-5p renders CDDP chemoresistance in ovarian cancer cells by targeting MST1 and SAV1, leading to the inactivation of Hippo signaling pathway.
The inactivation of Hippo signaling has been extensively reported to be implicated in the initiation and progression of ovarian cancer through various mechanisms. Fan et al have reported that the disruption of Hippo signaling by Sphingosine 1-phosphate (S1P) contributed to the development of ovarian cancer by promoting unlimited cell proliferation (42). YAP, a major effector of the Hippo signaling pathway, promotes the initiation and progression of ovarian cancer by forming autocrine loops with ERBB signaling (43). Furthermore, YAP confers the resistance of ovarian cancer cells to chemotherapeutic agents, and a high nuclear YAP expression strongly correlates with a poor prognosis in patients with ovarian cancer (44,45). These findings indicate that the disruption of Hippo signaling plays an oncogenic role in ovarian cancer. In this study, our results revealed that miR-149-5p inactivated Hippo signaling by targeting MST1 and SAV1, which further rendered chemotherapeutic resistance in ovarian cancer. Thus, our results present a novel role of miR-149-5p in promoting the chemoresistance of ovarian cancer cells to CDDP via the direct suppression of MST1 and SAV1, leading to the inactivation of Hippo signaling.
There is evidence to indicate that miRNAs plays an important role in regulating the activation of Hippo signaling via the post-transcriptional suppression of multiple target mRNAs of Hippo signaling components (46,47). For example, miR-129-5p has been shown to inhibit proliferation and survival by targeting the downstream co-activators, YAP and TAZ, of Hippo signaling in ovarian cancer (48); moreover, the upregulation of miR-130b has been shown to promote cancer stem cell characteristics in glioblastoma cells by directly suppressing MST1 and SAV1 (49). These studies demonstrate that the aberrant expression of miRNAs is involved in the development and progression of cancer via the Hippo signaling pathway. In this study, through bioinformatics analysis, we found that the core kinase components of Hippo signaling are potential targets of miR-149-5p. Furthermore, the upregulation of miR-149-5p enhanced the tanscriptional activity of TEAD, nuclear translocation of TAP and TAZ and expression levels of multiple downstream genes of Hippo signaling pathway; conversely, silencing miR-149-5p yielded an opposite effect. Collectively, our results uncover a novel mechanism of the inactivation of the Hippo signaling pathway, which further promotes the chemoresistance of ovarian cells to CDDP.
The upregulation of miR-149-5p has been reported to be implicated in the progression and metastasis of several malignancies, including non-small cell lung cancer, squamous cell carcinomas of the nasopharynx and prostate cancer, and a high expression of miR-149-5p predicts a poor prognosis (50–52). Furthermore, other studies have indicated that miR-149-5p expression is decreased in hepatocellular carcinoma, breast cancer and colorectal cancer (53–55). These studies suggested that the pro- or anticancer roles of miR-149-5p were tumor-type dependent. However, little is known about the biological roles of miR-149-5p in ovarian cancer. In this study, we found that miR-149-5p was markedly upregulated in chemoresistant ovarian cancer tissues and cell lines compared with chemo-sensitive ovarian cancer tissues and cell lines. Moreover, the upregulation of miR-149-5p enhanced, while the silencing of miR-149-5p inhibited the chemoresistance of ovarian cancer cells to CDDP in vivo and in vivo. Our results further revealed that miR-149-5p directly suppressed MST1 and SAV1 expression, leading to the inactivation of TEAD transcription and the downregulation of nuclear YAP and TAZ and Hippo signaling downstream target genes. Therefore, our results demonstrate that miR-149-5p promotes the chemoresistance of ovarian cancer cells to CDDP by inactivating the Hippo signaling pathway.
In conclusion, our study reveals that the upregulation of miR-149-5p contributes to the chemoresistance of ovarian cancer cells to CDDP as a pivotal suppressor of Hippo signaling pathway. Thus, an in-depth understanding of the precise role of miR-149-5p in the chemoresistance of ovarian cancer faciliates to increase our knowledge of the development of chemoresistance in ovarian cancer, which may aid in the development of noveltherapeutic strategies against ovarian cancer.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (no. 81672561).
Footnotes
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Cho KR, Shih IeM. Ovarian cancer. Annu Rev Pathol. 2009;4:287–313. doi: 10.1146/annurev.pathol.4.110807.092246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halder G, Johnson RL. Hippo signaling: Growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 6.Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, Zhao S, Xiong Y, Guan KL. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol. 2008;28:2426–2436. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oka T, Mazack V, Sudol M. Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP) J Biol Chem. 2008;283:27534–27546. doi: 10.1074/jbc.M804380200. [DOI] [PubMed] [Google Scholar]
- 8.Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Liu CY, Zha ZY, Zhao B, Yao J, Zhao S, Xiong Y, Lei QY, Guan KL. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem. 2009;284:13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan SW, Lim CJ, Loo LS, Chong YF, Huang C, Hong W. TEADs mediate nuclear retention of TAZ to promote oncogenic transformation. J Biol Chem. 2009;284:14347–14358. doi: 10.1074/jbc.M901568200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimomura T, Miyamura N, Hata S, Miura R, Hirayama J, Nishina H. The PDZ-binding motif of Yes-associated protein is required for its co-activation of TEAD-mediated CTGF transcription and oncogenic cell transforming activity. Biochem Biophys Res Commun. 2014;443:917–923. doi: 10.1016/j.bbrc.2013.12.100. [DOI] [PubMed] [Google Scholar]
- 12.Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, Wang Y, Halder G, Finegold MJ, Lee JS, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci USA. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imajo M, Miyatake K, Iimura A, Miyamoto A, Nishida E. A molecular mechanism that links Hippo signalling to the inhibition of Wnt/β-catenin signalling. EMBO J. 2012;31:1109–1122. doi: 10.1038/emboj.2011.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai D, Ho KC, Hao Y, Yang X. Taxol resistance in breast cancer cells is mediated by the hippo pathway component TAZ and its downstream transcriptional targets Cyr61 and CTGF. Cancer Res. 2011;71:2728–2738. doi: 10.1158/0008-5472.CAN-10-2711. [DOI] [PubMed] [Google Scholar]
- 15.Jeong W, Kim SB, Sohn BH, Park YY, Park ES, Kim SC, Kim SS, Johnson RL, Birrer M, Bowtell DSL, et al. Activation of YAP1 is associated with poor prognosis and response to taxanes in ovarian cancer. Anticancer Res. 2014;34:811–817. [PMC free article] [PubMed] [Google Scholar]
- 16.Bartel DP. Bartel. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren D, Wang M, Guo W, Huang S, Wang Z, Zhao X, Du H, Song L, Peng X. Double-negative feedback loop between ZEB2 and miR-145 regulates epithelial-mesenchymal transition and stem cell properties in prostate cancer cells. Cell Tissue Res. 2014;358:763–778. doi: 10.1007/s00441-014-2001-y. [DOI] [PubMed] [Google Scholar]
- 18.Ren D, Wang M, Guo W, Zhao X, Tu X, Huang S, Zou X, Peng X. Wild-type p53 suppresses the epithelial-mesenchymal transition and stemness in PC-3 prostate cancer cells by modulating miR-145. Int J Oncol. 2013;42:1473–1481. doi: 10.3892/ijo.2013.1825. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Liu J, Zang D, Wu S, Liu A, Zhu J, Wu G, Li J, Jiang L. Upregulation of miR-572 transcriptionally suppresses SOCS1 and p21 and contributes to human ovarian cancer progression. Oncotarget. 2015;6:15180–15193. doi: 10.18632/oncotarget.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren D, Yang Q, Dai Y, Guo W, Du H, Song L, Peng X. Oncogenic miR-210 3p promotes prostate cancer cell EMT and bone metastasis via NF-κB signaling pathway. Mol Cancer. 2017;16:117. doi: 10.1186/s12943-017-0688-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo W, Ren D, Chen X, Tu X, Huang S, Wang M, Song L, Zou X, Peng X. HEF1 promotes epithelial mesenchymal transition and bone invasion in prostate cancer under the regulation of microRNA-145. J Cell Biochem. 2013;114:1606–1615. doi: 10.1002/jcb.24502. [DOI] [PubMed] [Google Scholar]
- 22.Dai Y, Ren D, Yang Q, Cui Y, Guo W, Lai Y, Du H, Lin C, Li J, Song L, et al. The TGF-β signalling negative regulator PICK1 represses prostate cancer metastasis to bone. Br J Cancer. 2017;117:685–694. doi: 10.1038/bjc.2017.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 24.Bitarte N, Bandres E, Boni V, Zarate R, Rodriguez J, Gonzalez-Huarriz M, Lopez I, Javier Sola J, Alonso MM, Fortes P, et al. MicroRNA-451 is involved in the self-renewal, tumorigenicity, and chemoresistance of colorectal cancer stem cells. Stem Cells. 2011;29:1661–1671. doi: 10.1002/stem.741. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Talmon G, Wang J. MicroRNA-587 antagonizes 5-FU-induced apoptosis and confers drug resistance by regulating PPP2R1B expression in colorectal cancer. Cell Death Dis. 2015;6:e1845. doi: 10.1038/cddis.2015.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren D, Lin B, Zhang X, Peng Y, Ye Z, Ma Y, Liang Y, Cao L, Li X, Li R, et al. Maintenance of cancer stemness by miR-196b-5p contributes to chemoresistance of colorectal cancer cells via activating STAT3 signaling pathway. Oncotarget. 2017;8:49807–49823. doi: 10.18632/oncotarget.17971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang M, Ren D, Guo W, Huang S, Wang Z, Li Q, Du H, Song L, Peng X. N-cadherin promotes epithelial-mesenchymal transition and cancer stem cell-like traits via ErbB signaling in prostate cancer cells. Int J Oncol. 2016;48:595–606. doi: 10.3892/ijo.2015.3270. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Hahn WC, Dessain SK, Brooks MW, King JE, Elenbaas B, Sabatini DM, DeCaprio JA, Weinberg RA. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol. 2002;22:2111–2123. doi: 10.1128/MCB.22.7.2111-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Ren D, Guo L, Wang L, Wu S, Lin C, Ye L, Zhu J, Li J, Song L, et al. Thymosin beta 10 is a key regulator of tumorigenesis and metastasis and a novel serum marker in breast cancer. Breast Cancer Res. 2017;19:15. doi: 10.1186/s13058-016-0785-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Zhang L, Lin B, Chai X, Li R, Liao Y, Deng X, Liu Q, Yang W, Cai Y, et al. Phospholipid Phosphatase 4 promotes proliferation and tumorigenesis, and activates Ca2+-permeable Cationic Channel in lung carcinoma cells. Mol Cancer. 2017;16:147. doi: 10.1186/s12943-017-0717-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Liu F, Lin B, Luo H, Liu M, Wu J, Li C, Li R, Zhang X, Zhou K, et al. miR-150 inhibits proliferation and tumorigenicity via retarding G1/S phase transition in nasopharyngeal carcinoma. Int J Oncol. 2017 Mar 10; doi: 10.3892/ijo.2017.3909. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:4. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. Database. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR, Poletti A, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 36.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 37.Liu CY, Zha ZY, Zhou X, Zhang H, Huang W, Zhao D, Li T, Chan SW, Lim CJ, Hong W, et al. The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCF{beta}-TrCP E3 ligase. J Biol Chem. 2010;285:37159–37169. doi: 10.1074/jbc.M110.152942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huo X, Zhang Q, Liu AM, Tang C, Gong Y, Bian J, Luk JM, Xu Z, Chen J. Overexpression of Yes-associated protein confers doxorubicin resistance in hepatocellullar carcinoma. Oncol Rep. 2013;29:840–846. doi: 10.3892/or.2012.2176. [DOI] [PubMed] [Google Scholar]
- 39.Touil Y, Igoudjil W, Corvaisier M, Dessein AF, Vandomme J, Monté D, Stechly L, Skrypek N, Langlois C, Grard G, et al. Colon cancer cells escape 5FU chemotherapy-induced cell death by entering stemness and quiescence associated with the c-Yes/YAP axis. Clin Cancer Res. 2014;20:837–846. doi: 10.1158/1078-0432.CCR-13-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren A, Yan G, You B, Sun J. Down-regulation of mammalian sterile 20-like kinase 1 by heat shock protein 70 mediates cisplatin resistance in prostate cancer cells. Cancer Res. 2008;68:2266–2274. doi: 10.1158/0008-5472.CAN-07-6248. [DOI] [PubMed] [Google Scholar]
- 41.Chen M, Wang M, Xu S, Guo X, Jiang J. Upregulation of miR-181c contributes to chemoresistance in pancreatic cancer by inactivating the Hippo signaling pathway. Oncotarget. 2015;6:44466–44479. doi: 10.18632/oncotarget.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fan Q, Cheng Y, Chang HM, Deguchi M, Hsueh AJ, Leung PCK. Sphingosine-1-phosphate promotes ovarian cancer cell proliferation by disrupting Hippo signaling. Oncotarget. 2017;8:27166–27176. doi: 10.18632/oncotarget.15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He C, Lv X, Hua G, Lele SM, Remmenga S, Dong J, Davis JS, Wang C. YAP forms autocrine loops with the ERBB pathway to regulate ovarian cancer initiation and progression. Oncogene. 2015;34:6040–6054. doi: 10.1038/onc.2015.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, George J, Deb S, Degoutin JL, Takano EA, Fox SB, Bowtell DD. The Hippo pathway transcriptional co-activator, YAP, is an ovarian cancer oncogene. Oncogene. 2011;30:2810–2822. doi: 10.1038/onc.2011.8. [DOI] [PubMed] [Google Scholar]
- 45.Hall CA, Wang R, Miao J, Oliva E, Shen X, Wheeler T, Hilsenbeck SG, Orsulic S, Goode S. Hippo pathway effector Yap is an ovarian cancer oncogene. Cancer Res. 2010;70:8517–8525. doi: 10.1158/0008-5472.CAN-10-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Lai ZC. Mob as tumor suppressor is regulated by bantam microRNA through a feedback loop for tissue growth control. Biochem Biophys Res Commun. 2013;439:438–442. doi: 10.1016/j.bbrc.2013.08.095. [DOI] [PubMed] [Google Scholar]
- 47.Lin CW, Chang YL, Chang YC, Lin JC, Chen CC, Pan SH, Wu CT, Chen HY, Yang SC, Hong TM, et al. MicroRNA-135b promotes lung cancer metastasis by regulating multiple targets in the Hippo pathway and LZTS1. Nat Commun. 2013;4:1877. doi: 10.1038/ncomms2876. [DOI] [PubMed] [Google Scholar]
- 48.Tan G, Cao X, Dai Q, Zhang B, Huang J, Xiong S, Zhang Y, Chen W, Yang J, Li H. A novel role for microRNA-129-5p in inhibiting ovarian cancer cell proliferation and survival via direct suppression of transcriptional co-activators YAP and TAZ. Oncotarget. 2015;6:8676–8686. doi: 10.18632/oncotarget.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu G, Wang Y, Mijiti M, Wang Z, Wu PF, Jiafu D. Upregulation of miR-130b enhances stem cell-like phenotype in glioblastoma by inactivating the Hippo signaling pathway. Biochem Biophys Res Commun. 2015;465:194–199. doi: 10.1016/j.bbrc.2015.07.149. [DOI] [PubMed] [Google Scholar]
- 50.Lingzi X, Zhihua Y, Xuelian L, Yangwu R, Haibo Z, Yuxia Z, Baosen Z. Genetic variants in microRNAs predict non-small cell lung cancer prognosis in Chinese female population in a prospective cohort study. Oncotarget. 2016;7:83101–83114. doi: 10.18632/oncotarget.13072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang C, Sturgis EM, Chen X, Zheng H, Wei Q, Li G. Pre-miRNA variants as predictors of clinical outcome in patients with squamous cell carcinomas of the nonoropharynx. Oncotarget. 2016;7:26444–26453. doi: 10.18632/oncotarget.8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu J, Wang S, Zhang W, Qiu J, Shan Y, Yang D, Shen B. Screening key microRNAs for castration-resistant prostate cancer based on miRNA/mRNA functional synergistic network. Oncotarget. 2015;6:43819–43830. doi: 10.18632/oncotarget.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luo G, Chao YL, Tang B, Li BS, Xiao YF, Xie R, Wang SM, Wu YY, Dong H, Liu XD, et al. miR-149 represses metastasis of hepatocellular carcinoma by targeting actin-regulatory proteins PPM1F. Oncotarget. 2015;6:37808–37823. doi: 10.18632/oncotarget.5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan SH, Huang WC, Chang JW, Chang KJ, Kuo WH, Wang MY, Lin KY, Uen YH, Hou MF, Lin CM, et al. MicroRNA-149 targets GIT1 to suppress integrin signaling and breast cancer metastasis. Oncogene. 2014;33:4496–4507. doi: 10.1038/onc.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Øster B, Linnet L, Christensen LL, Thorsen K, Ongen H, Dermitzakis ET, Sandoval J, Moran S, Esteller M, Hansen TF, et al. COLOFOL steering group Non-CpG island promoter hypomethylation and miR-149 regulate the expression of SRPX2 in colorectal cancer. Int J Cancer. 2013;132:2303–2315. doi: 10.1002/ijc.27921. [DOI] [PubMed] [Google Scholar]