Abstract
Childhood neuroblastoma is one of the most common types of extra-cranial cancer affecting children with a clinical spectrum ranging from spontaneous regression to malignant and fatal progression. In order to improve the clinical outcomes of children with high-risk neuroblastoma, it is crucial to understand the tumorigenic mechanisms that govern its malignant behaviors. MYCN proto-oncogene, bHLH transcription factor (MYCN) amplification has been implicated in the malignant, treatment-evasive nature of aggressive, high-risk neuroblastoma. In this study, we used a SILAC approach to compare the proteomic signatures of MYCN-amplified IMR-32 and non-MYCN-amplified SK-N-SH human neuroblastoma cells. Tumorigenic proteins, including fatty-acid binding protein 5 (FABP5), L1-cell adhesion molecule (L1-CAM), baculoviral IAP repeat containing 5 [BIRC5 (survivin)] and high mobility group protein A1 (HMGA1) were found to be significantly upregulated in the IMR-32 compared to the SK-N-SH cells and mapped to highly tumorigenic pathways including, MYC, MYCN, microtubule associated protein Tau (MAPT), E2F transcription factor 1 (E2F1), sterol regulatory element binding transcription factor 1 or 2 (SREBF1/2), hypoxia-inducible factor 1α (HIF-1α), Sp1 transcription factor (SP1) and amyloid precursor protein (APP). The transcriptional knockdown (KD) of MYCN, HMGA1, FABP5 and L1-CAM significantly abrogated the proliferation of the IMR-32 cells at 48 h post transfection. The early apoptotic rates were significantly higher in the IMR-32 cells in which FABP5 and MYCN were knocked down, whereas cellular migration was significantly abrogated with FABP5 and HMGA1 KD compared to the controls. Of note, L1-CAM, HMGA1 and FABP5 KD concomitantly downregulated MYCN protein expression and MYCN KD concomitantly downregulated L1-CAM, HMGA1 and FABP5 protein expression, while survivin protein expression was significantly downregulated by MYCN, HMGA1 and FABP5 KD. In addition, combined L1-CAM and FABP5 KD led to the concomitant downregulation of HMGA1 protein expression. On the whole, our data indicate that this inter-play between MYCN and the highly tumorigenic proteins which are upregulated in the malignant IMR-32 cells may be fueling their aggressive behavior, thereby signifying the importance of combination, multi-modality targeted therapy to eradicate this deadly childhood cancer.
Keywords: neuroblastoma, high mobility group protein A1, L1-cell adhesion molecule, fatty-acid binding protein 5, survivin
Introduction
Neuroblastoma is a malignancy caused by the hyperplasia of naive neural crest cells (1). It is the most common solid tumor affecting children <5 years of age, and the third most common cause of pediatric cancer-related mortality (2), accounting for 7% of childhood malignancies and 15% of childhood cancer-related mortalities (3). The disease is known for its broad range of clinical behavior and variable responses to treatment (4). This clinical heterogeneity is elucidated by the fact that some neuroblastomas can spontaneously regress or differentiate into a benign form, ganglioneuroma, while others are not responsive to current therapies (5). Even though the prognosis for children with neuroblastoma is generally good, the survival rates for children with high-risk neuroblastomas are still poor, despite advanced therapeutic strategies (6).
Several transcription factors have been shown to be involved in the pathogenesis of neuroblastoma by enhancing cancer cell proliferation (7). MYCN proto-oncogene, bHLH transcription factor (MYCN), one of these transcription factors, is a phosphoprotein in the MYC family of transcription factors, encoded by the MYCN oncogene (7). The amplification of MYCN is one of the first most important genetic signatures of neuroblastoma (8). Patients with neuroblastoma carrying a MYCN amplification are classified in the high-risk group, and their 5-year overall survival rate following diagnosis does not exceed 50% (9). An amplification leading to the aberrant expression of MYCN has been associated with tumor aggressiveness, resistance to chemotherapy and the inability to differentiate (10). In fact, MYCN amplification confers cell resistance to apoptosis induced by the tumor necrosis factor-related apoptosis-inducing ligand system (11), whereas MYCN silencing promotes proliferation arrest, differentiation and the apoptosis of human neuroblastoma cells (12).
In the present study, we conducted a pilot proteomics analysis to compare the proteomic signature of the MYCN-amplified IMR-32 cells to that of the non-MYCN-amplified SK-N-SH human neuroblastoma cells using stable isotope labeling by amino acids in cell culture (SILAC) strategy (13). Of the differentially expressed proteins (based on expression value and fold change) between the IMR-32 and SK-N-SH cells, we were particularly interested in proteins upregulated in the IMR-32 compared to the SK-N-SH cells that mapped to upstream regulators [MYCN, MYC, hypoxia-inducible factor-1α (HIF-1α), E2F transcription factor 1 (E2F1), Sp1 transcription factor (SP1), KIT ligand (KITLG), interleukin 3 (IL3), amyloid precursor protein (APP), insulin receptor (INSR) and sterol regulatory element binding transcription factor 1 or 2 (SREBF1/2)] predicted by Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Inc., Redwood City, CA, USA; 2000–2014) to be activated in the IMR-32 cells. These included: L1 cell adhesion molecule (L1-CAM); high mobility group protein A1 (HMGA1); fatty acid-binding protein 5 (FABP5); and baculoviral IAP repeat containing 5 [BIRC5 (survivin)].
In the present study, we aimed to determine the interaction between the above-mentioned molecules and MYCN in the IMR-32 cells and the effect of transcriptional knockdown (KD) of these targets on cellular proliferation, migration and apoptosis. We also wished to determine the cellular bio-function after single-target versus double-target transcriptional KD of the said proteins and whether an added effect would be observed. In addition, we were interested in examining whether a crosstalk exists between these proteins as determined by differential protein expression levels of one target after transcriptional KD of each of the other targets.
Materials and methods
Reagents and human cell lines
The IMR-32 (MYCN-amplified) and SK-N-SH (non-MYCN-amplified) cells are human neuroblastoma/neuroepthelioma cell lines purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) where routine STR testing was conducted and the cells were confirmed to derive from human species. In addition, the cells were routinely tested for mycoplasma, aerobic and anaerobic bacteria, and human pathogenic viruses including human immunodeficiency virus (HIV), hepatitis B (HepB), human papilloma virus (HPV), Epstein-Barr virus (EBV) and cytomegalovirus (CMV), all of which our cells tested negative for and were used within 6 months of purchase from ATCC. The cells were cultured in minimal essential Eagle's medium (EMEM; cat. no. M2279) supplemented with 2 mM L-glutamine (cat. no. 7513; Sigma, St. Louis, MO, USA), 2% penicillin streptomycin (cat. no. P4333SIGMA), 1 mM sodium pyruvate (cat. no. S8636), 2% non-essential amino acids (cat. no. M7145) and 10% fetal bovine serum (cat. no. F9665) (all from Sigma). The cells were cultured to 80% confluence in T25 flasks at 5% CO2 and 37°C. The medium was replenished every 48 h. After 8 days, the cells covering 80% of the flask were collected and transferred into a 15 ml falcon tube to be centrifuged at 200 × g for 10 min at 4°C. The old medium was discarded and the cells were re-suspended in 10 ml of fresh medium and transferred into a T75 flask. The cells harvested from T75 flasks were frozen in Corning® Cryotubes (Corning Inc., New York, NY, USA) using 50% fetal bovine serum (FBS), 40% EMEM and 10% DMSO (D2650; Sigma) to a final volume of 1 ml. The cryotubes were frozen first at −80°C for 24 h in isopropanol to provide a gradual decrease in temperature. They were then transferred to liquid nitrogen for long term storage.
SILAC proteomics sample preparation
The IMR-32 and SK-N-SH neuroblastoma cells were grown in the standard culture conditions until they reached confluency after which they were washed 6 times with serum-free medium and then cultured in serum-free medium supplemented with 'heavy' isotopes where arginine and lysine were replaced with 13C6-Arg and 13C6, 15N2-Lys (Cambridge Isotopes, Andover, MA, USA) for 6 doublings to ensure incorporation of the isotopes into the entire cellular population. After 6 doublings, the cells were collected, lysed, quantified and mixed together into one SuperSILAC mastermix cocktail and saved at −80°C. The cells were then cultured in serum-free growth medium supplemented with 'light' isotopes where arginine and lysine were replaced with 12C6 -Arg and 12C6, 14N2-Lys until they reached ~80% confluency. The cells were lysed at that point and quantified, and a 'spike-in' strategy used to spike-in the 'heavy' labeled cellular lysates into the 'light' labeled cellular lysates at a 1:1 ratio. Approximately 100 μg of total protein mixture (50:50 μg of cell lysate:SuperSILAC cocktail) was loaded onto a 4–12% polyacrylamide gel and electrophoresed until the proteins were well-separated. The gel was fixed and stained with Commassie Blue (Bio-Rad, Hercules, CA, USA) then de-stained overnight in H2O at 4°C. The gels were then sliced into ~35–40 bands which underwent in-gel digestion with trypsin (Promega, Madison, WI, USA) using a previously documented protocol (14) and the resultant peptides derived from each band were dried by vacuum centrifugation (240 × g) at room temperature, and re-suspended in 6 μl of 0.1% TFA for analysis by mass spectrometry and proteomics analysis as previously described (15).
Mass spectrometry analysis and protein identification and quantification
The methods used to conduct the mass spectrometry and protein identification and quantification were exactly as described in the study by Formolo et al (15).
IPA, STRING protein networks and Enrichr analysis tool
Ingenuity Systems, Inc. 2000–2014 (content version, 18030641; release date, December 6, 2013) was used to derive the target molecules within the data set, upstream regulators, molecule type, predicted activation status with activation z-score and P-values of the proteins differentially expressed between the IMR-32 and SK-N-SH cells based on expression of and number of peptides detected per protein, where anything >2-fold is considered upregulated and anything below 0.5-fold is considered downregulated. The Enrichr (16,17) tool was used to annotate the differentially expressed proteins between the IMR-32 and SK-N-SH cells. Proteins in the network that were differentially expressed in the IMR-32 cells were mapped to the top diseases and bio-functions. Differential expression was determined as either upregulated (>2-fold) or downregulated (<0.5-fold) in the IMR-32 compared to the SK-N-SH cells. The STRING protein network tool was used to determine any interactions (experimentally determined, from curated databases and text-mining) between our proteins of interest.
Protein extraction for validation by western blot analysis
The IMR32 and SK-N-SH cells were collected into 15-ml falcon tubes and centrifuged at 200 × g for 10 min at 4°C. The medium was discarded and the pellet was washed twice with Dulbecco's phosphate-buffered saline (PBS; D8537; Sigma). PBS was then discarded and the cells were incubated on ice for 40 min with 120 μl of lysis buffer (for a confluent T75 flaks) composed of 50 Mm tris-HCl (pH 7.5), 1 mM EDTA, 250 mM NaCl, 50 mM NaF, 0.1 mM Na3VO4, 0.5% Titron X-100 and 1X protease inhibitor cocktail (cat no. 11697498001; Roche, Indianapolis, IN, USA). The lysates were transferred to Eppendorf tubes where they were centrifuged at 4°C for 20 min at 8,000 × g to eliminate cellular debris. The supernatants were then transferred to new Eppendorf tubes where they were mixed with 60 μl of the 3X sample buffer (0.5 M tris-HCl, glycerol, 10% SDS, β-mercaptoethanol and 0.05% bromophenol blue) in preparation for western blot analysis.
Western blot analysis
Protein expression was confirmed by western blot analysis as previously described (18). Briefly, For L1-CAM, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and HMGA1 detection, a 10% polyacrylamide (cat. no. 161-0156; Bio-Rad) gel was used; however, for FABP5 and MYCN detection a 12% polyacrylamide gel was used. A 5% stacking gel was used to form the wells. The extracted proteins were separated on gels using a current of 150 mV. Proteins were then transferred to a PVDF membrane (cat. no. 1620177; Bio-Rad) for 90 min at 100 mV which was then blocked using 5% BSA (cat. no. A2153-100G; Sigma) diluted in 1X TBS with 1% Tween-20 (cat. no. 9005-64-5).
The blots were incubated with rabbit monoclonal L1-CAM (cat. no. ab20148), HMGA1 (cat. no. ab129153), FABP5 (cat. no. ab84028), survivin (cat. no. ab469) (all from Abcam, Cambridge MA, USA) and MYCN antibodies (cat. no. 9405; Cell Signaling Technology, Danvers, MA, USA) and with mouse monoclonal GAPDH antibody (cat. no. ab9484) (Abcam) overnight (all diluted 1:1,000 in TBS-T +5% BSA blocking solution). The blots were washed 5 times for 5 min with TBST and then incubated with secondary antibodies [goat anti-rabbit (cat. no. 170-5046) or goat anti-mouse (cat. no. 170-5047) HRP-conjugated secondary antibodies from Bio-Rad] for 1 h. After washing, the blots were incubated with Clarity Western ECL substrate (cat. no. 1705060; Bio-Rad) for 3 min and imaged using the Bio-Rad© ChemiDoc system and analyzed using ImageLab® software. The bands obtained were normalized to GAPDH.
Transcriptional KD of target proteins
The 4 siRNA oligonucleotides for L1-CAM (Hs L1-CAM_4 SI00009296, Hs L1-CAM_3 SI00009289, Hs L1-CAM_2 SI00009282 and Hs L1-CAM_1 SI00009275; Qiagen, Valencia, CA, USA), HMGA1 (D-004597-01 HMGA1, D-004597-02 HMGA1, D-004597-03 HMGA1 and D-004597-18 HMGA1; Dharmacon, Lafayette, CO, USA), MYCN (Hs_MYCN_7 SI03113670, Hs_MYCN_6 SI03087518, Hs_MYCN_5 SI03078222 and Hs_MYCN_3 SI00076300; Qiagen) and FABP5 (Hs_FABP5_10 SI04277553, Hs_FABP5_9 SI04210948, Hs_FABP5_8 SI04210941 and Hs_FABP5_5 SI03145835; Qiagen) were reconstituted in RNAase-free water to a final concentration of 10 μM. A total of 5×105 cells were seeded in each well of a 6-well plate in a final volume of 2 ml of antibiotics-free medium. The cells were transfected with the siRNA using the fast-forward Hiperfect transfection protocol following the manufacturer's instructions (cat. no. 301705; Qiagen). A final concentration of 100 nM of each siRNA was achieved by mixing 10 μl of the 10 μM siRNA with 10 μl of Hiperfect (cat. no. 301705; Qiagen) and 370 μl of OptiMEM (cat. no. 31985-062; Life Technologies, Carlsbad, CA, USA). The mixture was incubated at room temperature for 20 min, and added in a drop-wise manner to the corresponding wells. The negative control was prepared by treating the cells with 90 μl of OptiMEM and 10 μl of Hiperfect. The cells were then incubated with the siRNAs for 48 h, lysed and processed for western blot analysis.
WST-1 cell proliferation assay
In order to examine the effects of KD on cellular proliferation, a WST-1 cell proliferation assay was conducted. A total of 5×104 cells were cultured in each well of a 96-well plate in a final volume of 100 μl. Duplicates of each condition were prepared. The cells were transfected with L1-CAM, HMGA1, MYCN, FABP5 or scrambled siRNAs in a final concentration of 100 nM. The cells were incubated in 100 μl growth medium for 24, 48 and 72 h and treated with 10 μl of WST-1 reagent (cat. no. ab155902; Abcam) per well prior to reading. The absorbance was detected after 3 h using an Epoch™ Microplate Spectrophotometer at 450 nm (BioTek, Winooski, VT, USA).
Annexin V-FITC/propidium iodide (PI) apoptosis detection assay
The IMR32 cells were cultured in a 6-well plate (5×105 cells/well), and transfected with the various siRNA constructs as described above. Negative control cells were either unlabeled/untreated or Annexin V-FITC- and PI-labeled and untreated. Positive control cells were transfected with 100 nM siRNA of our target constructs and labeled with Annexin V-FITC and PI or Annexin V-FITC alone. The cells were then collected at 48 h using accutase (cat. no. A11105-01; Thermo Fisher Scientific, Waltham, MA, USA) and centrifuged at 200 × g for 5 min at 4°C. They were then re-suspended in 500 μl of 1X binding buffer, treated with 5 μl of Annexin V-FITC or PI from Abcam (cat. no. ab14085) and incubated in the dark for 5 min prior to flow cytometric analysis using FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). Data were analyzed using IQuest Pro software (version 5.1) (BD Biosciences). IMR32 cells were identified by their forward-scatter (FSC) and side-scatter (SSC) characteristics. Viable, early apoptotic, late apoptotic and dead cell populations were identified as Annexin V−/PI−, Annexin V+/PI−, Annexin V+/PI+ and Annexin V−/PI+, respectively. Cells stained with Annexin V-FITC or PI alone were used to adjust color compensation settings on flow cytometer. A minimum of 20,000 cell events were recorded for each sample.
Wound healing migration assay
The IMR-32 cells were grown to ~70% confluency in triplicates of 12-well plates in standard culture medium and conditions. FABP5 or HMGA1 siRNA transfection was conducted as described above and after 24 h, the cells were scraped down the midline of the plate to resemble a 'wound'. The cells which were scraped off were washed 2X with serum-free medium and fresh medium replenished and the cells were then imaged (0 h time-point) and cultured under standard growth culture conditions for 24 h. The cells were then imaged at several random fields down the scratched 'wound' area and the area (μM2) of 'wound-closure' after 24 h in siRNA KD-compared to control siRNA-transfected cells was measured using AxioVision Systems software (Zeiss, Oberkochen, Germany). The average of the areas of multiple random fields was represented as a fold change compared to untransfected controls and graphed using Microsoft Excel software.
Statistical analysis
Experiments were conducted in triplicate, repeated 3 independent times and the means ± the standard error of the means (SEM) of all 3 experiments was calculated and plotted. A two-sided Student's t-test was used to determine statistically significant differences between groups. A one-way analysis of variance (ANOVA) test was used followed by a post hoc Fisher's least significant difference (LSD) test to determine the significance among the means of multiple groups obtained from ≥3 independent experiments. The means ± SEM of 3 or more experiments was derived and graphed using Microsoft Excel software. Statistical significance was set at a P-value <0.05.
Results
Top networks, diseases and bio-functions to which overexpressed proteins in IMR-32 cells are mapped
We conducted preliminary proteomics analysis of the MYCN-amplified IMR-32 compared to the non-MYCN-amplified SK-N-SH human neuroblastoma cell lines using a highly quantitative SILAC methodology. The significant, differentially expressed proteins were then annotated using IPA in order to predict the upstream regulators of highly tumorigenic proteins in the IMR-32 compared to the SK-N-SH cells. The total numbers of differentially expressed proteins were found to be 875 out of 4,960 proteins (either >2-fold upregulated or <0.5-fold down-regulated) in the IMR-32 compared to the SK-N-SH cells. These proteins were affiliated with upstream regulators that IPA predicted to be activated in the IMR-32 compared to the SK-N-SH cells based on the molecules identified in our dataset (data not shown).
Table I lists the top diseases and disorders, molecular and cellular bio-functions, as well as physiological system development and function categories that proteins significantly overexpressed in the IMR-32 cells are mapped to; the fold change cut-off was set at ≥2-fold. Renal and urological, neurological, hereditary and infectious diseases, as well as organismal injury and abnormalities, were among the top disease categories that the proteins were mapped to Table IA. RNA post-transcriptional modification, molecular transport, RNA trafficking, cellular compromise and gene expression were among the top molecular and cellular functions that the overexpressed proteins in the IMR-32 cells were mapped to Table IB. The physiological system development and function categories that proteins belonged to included tissue development, nervous system development and function, connective tissue development and function and embryonic development (Table IC).
Table I.
Functional annotation of proteins upregulated in the IMR-32 compared to the SK-N-SH human neuroblastoma cell lines.
| A, Diseases and disorders | ||
|---|---|---|
|
| ||
| Name | P-value | No. of molecules |
| Infectious disease | 2.02E-09-1.69E-02 | 135 |
| Organismal injury and abnormalities | 2.02E-09-1.89E-02 | 164 |
| Renal and urological disease | 4.93E-09-1.69E-02 | 70 |
| Hereditary disorder | 8.90e-07-1.89e-02 | 41 |
| Neurological disease | 8.90E-07-1.75E-02 | 98 |
| B, Molecular and cellular functions | ||
|---|---|---|
|
| ||
| Name | P-value | No. of molecules |
| RNA post-transcriptional modification | 2.54E-33-1.73E-02 | 75 |
| Molecular transport | 4.20E-11-1.73E-02 | 75 |
| RNA trafficking | 4.20E-11-1.97E-02 | 23 |
| Cellular compromise | 1.45E-10-1.90E-02 | 38 |
| Gene expression | 1.19E-07-1.07E-02 | 180 |
|
| ||
| Associated network function | Score | |
|
| ||
| RNA post-transcriptional modification, molecular transport, RNA trafficking | 45 | |
| Gene expression, RNA damage and repair, RNA post-translational modification | 45 | |
| Cellular assembly and organization, cellular function and maintenance, tissue development | 40 | |
| Cellular movement, developmental disorder, hereditary disorder | 38 | |
| Cellular compromise, cell cycle, cellular assembly and organization | 38 | |
| C, Physiological system development and function | ||
|---|---|---|
|
| ||
| Name | P-value | No. of molecules |
| Tissue development | 8.61E-06-1.57E-02 | 70 |
| Nervous system development and function | 6.52E-05-1.73E-02 | 75 |
| Connective tissue development and function | 7.68E-04-1.89E-02 | 55 |
| Embryonic development | 7.68E-04-1.73E-02 | 66 |
Fold change and predicted activated up-stream regulators of proteins overexpressed in IMR-32 compared to SK-N-SH cells
IPA was used to predict the activation status of the upstream regulators of the differentially expressed proteins between the IMR-32 and SK-N-SH cells. Table II lists the top molecules and the fold change of upregulated proteins which included, CaM kinase like vesicle associated (CAMKV), tumor protein D52 (TPD52), argininosuccinate synthetase 1 (ASS1), FABP6, L1-CAM, FABP5, DNA polymerase γ, catalytic subunit (POLG), collapsin response mediator protein 1 (CRMP1), ribonuclease H2 subunit B (RNASEH2B) and G protein subunit αO1 (GNAO1). MYCN, hepatocyte nuclear factor 4α (HNF4A), microtubule associated protein Tau (MAPT), APP and E2F were predicted to be activated upstream regulators of the proteins in this dataset, as determined by IPA (based on the target molecules that were found to be >2-fold upregulated in the IMR-32 compared to the SK-N-SH cells). In addition to the top upstream regulators, Table III lists additional upstream regulators predicted to be activated or inhibited in the IMR-32 compared to the SK-N-SH cells. Of note, is the activation of cancer-promoting, angiogenic and cancer stemness players, such as MYC, HIF-1α, eukaryotic translation initiation factor 4γ1 (EIF4G1), IL3, angiopoietin 2 (ANGPT2) and SREBF in the IMR-32 cells. Molecules predicted to be inhibited in the IMR-32 compared to the SK-N-SH cells included Aly/REF export factor (ALYREF), Ankyrin (ANK)2, autophagy related 7 (ATG7), coiled-coil domain containing 88A (CCDC88A), chromatin target of PRMT1 (CHTOP), cleavage and polyadenylation specific factor 1 (CPSF1), CPSF2 and CPSF3 among others, primarily involved in RNA post-transcriptional modification, molecular transport and RNA trafficking.
Table II.
Fold change of top 10 proteins upregulated in the IMR-32 versus the SK-N-SH human neuroblastoma cells.
| Molecules | Fold change | |
|---|---|---|
| CAMKV | 103.800 | |
| TPD52 | 39.976 | |
| ASS1 | 24.995 | |
| FABP5 | 18.000 | |
| POLG | 16.726 | |
| L1-CAM | 16.708 | |
| CRMP1 | 15.295 | |
| FABP6 | 15.268 | |
| RNASEH2B | 15.250 | |
| GNAO1 | 14.943 | |
|
| ||
| Upstream regulator | P-value of overlap predicted | Activation state |
|
| ||
| MYC | 1.52E-18 | Activated |
| HNF4A | 2.15E-15 | |
| MAPT | 7.36E-12 | |
| APP | 1.10E-11 | |
| E2F | 1.37E-11 | |
Table III.
Upstream regulators predicted to be either activated (in addition to the above top 5 upstream regulators), or inhibited in the IMR-32 compared to the SK-N-SH cells based on the statistically significant over- or underexpression of molecules within the IPA-derived dataset.
| Predicted to be activated | MYC, ESRRA, MYCN, INSR, HIST1H1T, HIST1H1A, HIF1A, EIF4E, EIF4G1, SREBF2, ANGPT2, FLI1, IL3, SREBF1, SCAP, RUVBL1, SP1, CD40LG, EPAS1, KITLG, ADORA2A, PGR, KAT5, PSEN1 |
| Predicted to be inhibited | ALYREF, ANK2, ATG7, CCDC88A, CHTOP, CPSF1, CPSF2, CPSF3, CSTF1, DDX39A, DDX39B, DENR |
Proteins overexpressed in the IMR-32 cells belong to tumorigenic pathways
The proteins overexpressed in the IMR-32 compared to the SK-N-SH cells that belonged to the upstream regulators predicted to be activated by IPA (SREBF1, SP1, SCAP, SREBF2, IL3, KITLG, FLI1, INSR, HIF-1α, MYC, EIF4E and ANGPT2 among others), mapped to signaling pathways that drive cancer stem cell maintenance and propagation (Notch) (19), invasion and metastasis [Rac1/Pak1/p38/MMP-2 (20) and the leptin signaling pathway (21)], cancer malignancy [SREBP signaling (22) and the AGE/RAGE pathway (23)] and proliferation [AMPK pathways (24)]. Table IV lists the upstream regulators predicted to be activated by IPA, with adjusted P-values for the overexpression level in the IMR-32 cells and the pathways in which these regulators have been demonstrated to play pivotal roles.
Table IV.
Functional annotation of upstream regulators of the overexpressed proteins in the IMR-32 cells and the signaling pathways they are affiliate witha.
| Pathways | Adjusted P-value | Genes overexpressed |
|---|---|---|
| SREBP signaling_WP1982 | 8.04E-05 | SREBF1, SP1, SCAP, SREBF2 |
| Adipogenesis_WP236 | 4.00E-04 | SREBF1, EPAS1, SP1, HIF1A |
| Hematopoietic stem cell differentiation | 4.55E-04 | IL3, KITLG, FLI1 |
| AGE/RAGE pathway_WP2324 | 9.50E-04 | SP1, INSR, HIF1A |
| SREBF and miR-33 in cholesterol and lipid homeostasis_WP2011 | 2.79E-03 | SREBF1, SREBF2 |
| Mitochondrial gene expression_WP391 | 3.11E-03 | ESRRA, SP1 |
| Nuclear receptors_WP170 | 9.47E-03 | ESRRA, PGR |
| Integrated pancreatic cancer pathway_WP2377 | 1.20E-02 | IL3, SP1, MYC |
| Differentiation pathway_WP2848 | 1.20E-02 | IL3, KITLG |
| Translation factors_WP107 | 1.20E-02 | EIF4E, EIF4G1 |
| Notch signaling pathway_WP61 | 1.67E-02 | MYC, HIF1A |
| Rac1/Pak1/p38/MMP-2 pathway_WP3303 | 1.84E-02 | ANPGPT2, MYC |
| AMPK signaling_ WP1403 | 1.85E-02 | SREBF1, INSR |
| Leptin signaling pathway_WP2034 | 2.08E-02 | SP1, EIF4E |
| Androgen receptor signaling pathway_WP138 | 2.78E-02 | KAT5, SP1 |
| DNA damage response (only ATM dependent)_WP710 | 4.08E-02 | MYC, INSR |
| miR-148/miR-31/FIH1/HIF1? - Notch signaling in glioblastoma_WP3593 | 5.00E-02 | HIF1A |
As derived from Enrichr using the IPA-predicted activated up-stream regulators dataset.
IMR-32 cells significantly overexpress the tumorigenic proteins, HMGA1, L1-CAM, FABP5 and BIRC5
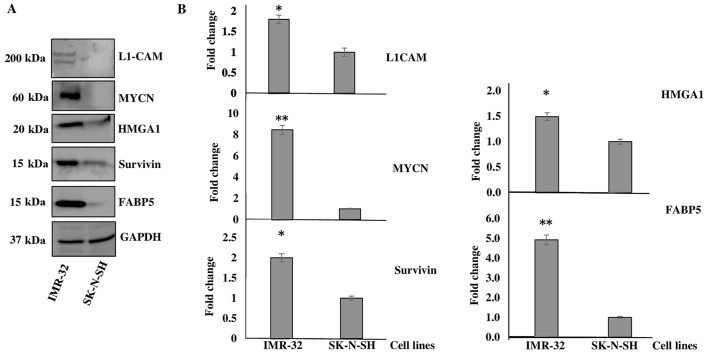
Of the highly tumorigenic proteins identified to be upregulated in the IMR-32 compared to the SK-N-SH cells, we decided to focus our investigation on L1-CAM, HMGA1, FABP5, BIRC5 and MYCN as they mapped to several of the activated upstream regulators, as predicted by IPA and due to their tumorigenic roles played in various types of cancer as elaborated below in the 'Discussion'. Western blot analysis was used to determine the levels of HMGA1, FABP5, BIRC5 and L1-CAM, as well as MYCN protein expression between the IMR-32 and SK-N-SH cells. As expected, we found all these targets to be significantly overexpressed in the IMR-32 compared to the SK-N-SH neuroblastoma cells. Fig. 1A is a representative demonstration of multiple western blotting experiments verifying the upregulation of these targets, as indicated by the SILAC proteomics data. Densitometric analysis revealed a significant upregulation of the protein expression of L1-CAM (~1-fold overexpressed), MYCN (~7-fold overexpressed), HMGA1 (~0.5-fold overexpressed), BIRC5 (~1-fold overexpressed) and FABP5 (~4-fold overexpressed) in the IMR-32 compared to the SK-N-SH cells (Fig. 1B).
Figure 1.
Verification of proteomics data. (A) Representative western blot images of L1-cell adhesion molecule (L1-CAM), MYCN proto-oncogene, bHLH transcription factor (MYCN), high mobility group protein A1 (HMGA1), survivin and fatty-acid binding protein 5 (FABP5) protein expression in the IMR-32 compared to the SK-N-SH cells. (B) Densitometric analysis verified the proteomics data and revealed the significant overexpression of L1-CAM, MYCN, HMGA1, survivin and FABP5 in the IMR-32 compared to the SK-N-SH cells. Experiments were run in triplicate and repeated >3 times. Results represent the means ± SEM; *P<0.05 and **P<0.01.
Upregulated proteins in IMR-32 cells mapped to the highly tumorigenic MYCN pathway
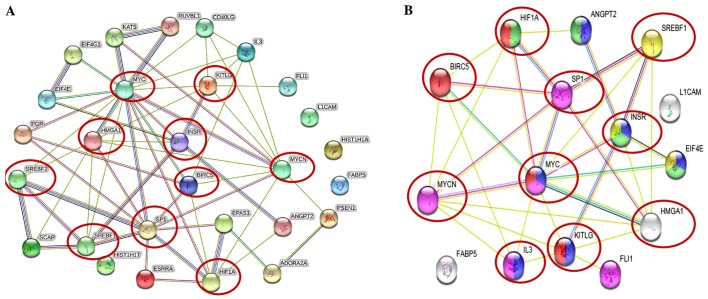
The STRING protein network database was used to analyze the interactions (experimentally determined and from curated databases) between the activated upstream regulators of the overexpressed proteins in the IMR-32 cells (HMGA1, L1-CAM, BIRC5, FABP5, MYC, INSR, SP1, HIF1A and SREBF1/2 among others) and MYCN (Fig. 2A). In addition, we used STRING protein network analysis to focus on the interactions between the activated upstream regulators, which our validated proteins (by western blot analysis; HMGA1, L1-CAM, FABP5 and BIRC5) mapped to Fig. 2B). Of particular interest, are the various hubs (proteins with multiple edges) identified, including INSR, SP1, HIF-1α, MYC, MYCN, HMGA1 and SREBF that show strong interactions (experimentally-determined, from curated databases and text-mining) with many other tumorigenic proteins (Fig. 2A). We further highlight the important interactions (experimentally determined, from curated databases and text-mining) between our validated targets (by western blot analysis; HMGA1, BIRC5 and MYCN) and the activated upstream regulators, SP1, SREBF1, MYC, KITLG, IL3, INSR and HIF-1α (Fig. 2B). KEGG pathway analysis mapped the number of molecules to various pathways, most of which are known tumorigenic drivers, including PI3K/Akt, Ras, Rap1, Wnt, HIF-1α, transforming growth factor-β (TGF-β) and mammalian target of rapamycin (mTOR) and insulin signaling pathways (Table V). This intricate network of interplay between these highly tumorigenic proteins warrants further investigation, particularly in pre-clinical, animal models, whereby multiple targeting of more than one of these players may yield beneficial, antitumor effects and improve therapeutic outcomes.
Figure 2.
Tumorigenic proteins significantly upregulated in the IMR-32 cells mapped to the MYCN proto-oncogene, bHLH transcription factor (MYCN) pathway. (A) STRING pathway analysis revealed interactions between molecules of activated upstream regulators in IMR-32 cells as predicted by IPA. Of importance are the various proteins with hubs (multiple edges, red circles), including Sp1 transcription factor (SP1), MYC, MYCN, hypoxia-inducible factor-1α (HIF-1α), high mobility group protein A1 (HMGA1), INSR, KIT ligand (KITLG), sterol regulatory element binding transcription factor 2 (SREBF2) and SREBF1. (B) STRING pathway analysis also focused on the strong interactions between the activated upstream regulators which HMGA1, MYCN and BIRC5 mapped to. Of particular interest are the hubs, including MYCN, MYC, SP1, HIF-1α, SREBF1, BIRC5, HMGA1 and INSR that affiliated with one or more of our validated targets.
Table V.
Common tumorigenic pathways that MYCN, BIRC5 and the activated upstream regulators mapped to.
| Pathway ID no. | Pathway description | Observed gene count | False discovery rate | Matching proteins in your network (labels) |
|---|---|---|---|---|
| 4151 | PI3K-Akt signaling pathway | 6 | 1.26E-05 | ANGPT2, EIF4E, IL3, INSR, KITLG, MYC |
| 5202 | Transcriptional misregulation in cancer | 5 | 1.26E-05 | FLI1, IL3, MYC, MYCN, SP1 |
| 4066 | HIF-1 signaling pathway | 4 | 7.58E-05 | ANGPT2, EIF4E, HIF1A, INSR |
| 5200 | Pathways in cancer | 5 | 5.02E-03 | BIRC5, HIF1A, KITLG, MYC, EPAS1 |
| 4910 | Insulin signaling pathway | 3 | 6.48E-03 | EIF4E, INSR, SREBF1 |
| 4015 | Rap1 signaling pathway | 4 | 1.95E-02 | ADORA2A, ANGPT2, INSR, KITLG |
| 4014 | Ras signaling pathway | 3 | 2.05E-02 | ANGPT2, INSR, KITLG |
| 4310 | Wnt signaling pathway | 3 | 1.84E-02 | MYC, PSEN1, RUVBL1 |
| 4150 | mTOR signaling pathway | 2 | 2.67E-02 | EIF4E, HIF1A |
| 5210 | Colorectal cancer | 2 | 2.67E-02 | BIRC5, MYC |
| 4350 | TGF-β signaling pathway | 2 | 4.08E-02 | MYC, SP1 |
| 4640 | Hematopoietic cell lineage | 2 | 4.51E-02 | IL3, KITLG |
When we combined the predicted activated upstream regulators and MYCN in the STRING protein network analysis, the common pathways identified were highly tumorigenic, including PI3K-Akt, transcriptional misregulation in cancer, pathways in cancer, HIF-1, Wnt, Rap1, Ras, mTOR, TGF-β and insulin signaling pathways.
Novel pathways link the MYCN-affected proteins in the IMR-32 cells to cancer malignancy
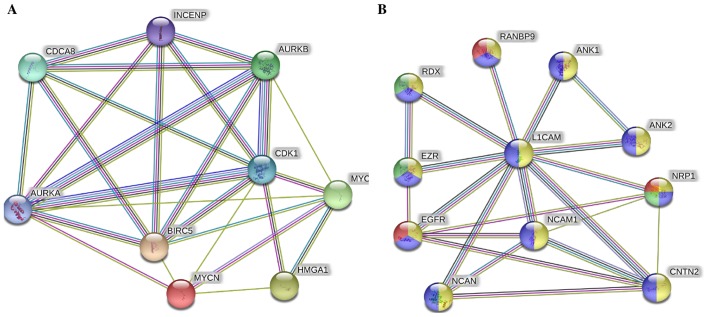
In addition to the above-mentioned pathways, when we combined only MYC, MYCN and our validated targets in the STRING protein network analysis (Fig. 3A), we found very strong interactions between MYC, MYCN, BIRC5 and HMGA1 with Aurora kinase (AURK)A/B, cyclin-dependent kinase 1 (CDK1), cell division cycle associated 8 (CDCA8) and INCENP. These players were mapped to cellular functions highly affiliated with mitosis and cellular division and proliferation (Table VI). Lastly, L1-CAM focused network analysis revealed its strong interaction with neural cell adhesion molecule 1 (NCAM1), neurocan (NCAN), ezrin (EZR), RDX, ANK1, ANK2, contactin 2 (CNTN2), neuropilin 1 (NRP1), epidermal growth factor receptor (EGFR) and RAN binding protein 9 (RANBP9) (Fig. 3B), most of which drive angiogenic and migratory processes to fuel cellular invasion, migration and metastatic spread.
Figure 3.
Novel pathways link the MYCN proto-oncogene, bHLH transcription factor (MYCN)-affected proteins in IMR-32 cells to cancer malignancy. (A) STRING pathway analysis revealed very strong interactions between MYCN, BIRC5, high mobility group protein A1 (HMGA1) and the cellular mitosis and proliferation players AURKA/B, cyclin-dependent kinase 1 (CDK1), CDCA8 and INCENP; (B) whereas L1-cell adhesion molecule (L1-CAM) showed very strong interactions with neural cell adhesion molecule 1 (NCAM1), neuropilin-1 (NRP1), NCAN, contactin 2 (CNTN2), RANBP1 and epidermal growth factor receptor (EGFR) among others most of which are angiogenic markers.
Table VI.
Common tumorigenic pathways that MYC and our validated targets (BIRC5, HMGA1 and MYCN) mapped to.
| Pathway ID no. | Pathway description | Observed gene count | False discovery rate | Matching proteins in your network (labels) |
|---|---|---|---|---|
| GO.0051301 | Cell division | 6 | 0.000118 | AURKA, AURKB, BIRC5, CDCA8, CDK1, INCENP |
| GO.0007067 | Mitotic nuclear division | 5 | 0.000834 | AURKB, BIRC5, CDCA8, CDK1, INCENP |
| GO.0007052 | Mitotic spindle organization | 3 | 0.0017 | AURKA, AURKB, BIRC5 |
| GO.0007059 | Chromosome segregation | 4 | 0.0017 | AURKB, BIRC5, CDCA8, INCENP |
| GO.0043146 | Spindle stabilization | 2 | 0.00178 | AURKA, AURKB |
| GO.0051225 | Spindle assembly | 3 | 0.00178 | AURKA, AURKB, BIRC5 |
| GO.0051302 | Regulation of cell division | 4 | 0.00382 | AURKA, AURKB, BIRC5, MYC |
| GO.0007264 | Small GTPase-mediated signal transduction | 5 | 0.00405 | AURKB, BIRC5, CDCA8, CDK1, INCENP |
| GO.0000910 | Cytokinesis | 3 | 0.00568 | AURKB, BIRC5, INCENP |
| GO.0031145 | Anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | 3 | 0.00568 | AURKA, AURKB, CDK1 |
| GO.0010941 | Regulation of cell death | 6 | 0.00604 | AURKA, AURKB, BIRC5, CDK1, MYC, MYCN |
| GO.0006468 | Protein phosphorylation | 5 | 0.00685 | AURKA, AURKB, BIRC5, CDK1, MYC |
| GO.0018105 | Peptidyl-serine phosphorylation | 3 | 0.00685 | AURKA, AURKB, CDK1 |
| GO.0034501 | Protein localization to kinetochore | 2 | 0.00685 | AURKB, CDK1 |
| GO.0035404 | Histone-serine phosphorylation | 2 | 0.00685 | AURKA, AURKB |
| GO.0043066 | Negative regulation of apoptotic process | 5 | 0.00685 | AURKA, AURKB, BIRC5, CDK1, MYC |
| GO.0051276 | Chromosome organization | 5 | 0.00736 | AURKA, AURKB, CDCA8, HMGA1, MYC |
| GO.0006996 | Organelle organization | 7 | 0.00837 | AURKB, BIRC5, CDCA8, CDK1, HMGA1, INCENP, MYC |
| GO.0051781 | Positive regulation of cell division | 3 | 0.0103 | AURKA, AURKB, BIRC5 |
| GO.0035556 | Intracellular signal transduction | 6 | 0.0104 | AURKB, BIRC5, CDCA8, CDK1, INCENP, MYC |
| GO.0000086 | G2/M transition of mitotic cell cycle | 3 | 0.0122 | AURKA, BIRC5, CDK1 |
| GO.0022402 | Cell cycle process | 5 | 0.0122 | AURKB, BIRC5, CDCA8, INCENP, MYC |
| GO.1902589 | Single-organism organelle organization | 6 | 0.0131 | AURKB, BIRC5, CDCA8, CDK1, HMGA1, INCENP |
| GO.0033043 | Regulation of organelle organization | 5 | 0.0134 | AURKA, AURKB, BIRC5, HMGA1, MYC |
| GO.0090307 | Mitotic spindle assembly | 2 | 0.0136 | AURKB, BIRC5 |
| GO.0016568 | Chromatin modification | 4 | 0.0151 | AURKA, AURKB, HMGA1, MYC |
| GO.0007049 | Cell cycle | 5 | 0.026 | AURKB, BIRC5, CDCA8, INCENP, MYC |
| GO.0000075 | Cell cycle checkpoint | 3 | 0.0275 | AURKB, BIRC5, CDK1 |
| GO.0002053 | Positive regulation of mesenchymal cell proliferation | 2 | 0.0275 | MYC, MYCN |
| GO.0007098 | Centrosome cycle | 2 | 0.0275 | AURKA, CDK1 |
| GO.0090068 | Positive regulation of cell cycle process | 3 | 0.0338 | AURKA, AURKB, BIRC5 |
| GO.0031577 | Spindle checkpoint | 2 | 0.0342 | AURKB, BIRC5 |
| GO.1903047 | Mitotic cell cycle process | 4 | 0.0372 | AURKB, BIRC5, CDCA8, INCENP |
| GO.0030162 | Regulation of proteolysis | 4 | 0.0399 | AURKA, BIRC5, CDK1, MYC |
| GO.0045840 | Positive regulation of mitotic nuclear division | 2 | 0.0412 | AURKA, BIRC5 |
| GO.0051303 | Establishment of chromosome localization | 2 | 0.0434 | BIRC5, CDCA8 |
| GO.0010639 | Negative regulation of organelle organization | 3 | 0.0455 | AURKA, AURKB, HMGA1 |
As indicated, there was a strong interaction between BIRC5, HMGA1, MYC, MYCN and AURKA/B, CDK1 and CDCA8, all of which are highly affiliated with processes that drive cellular mitosis.
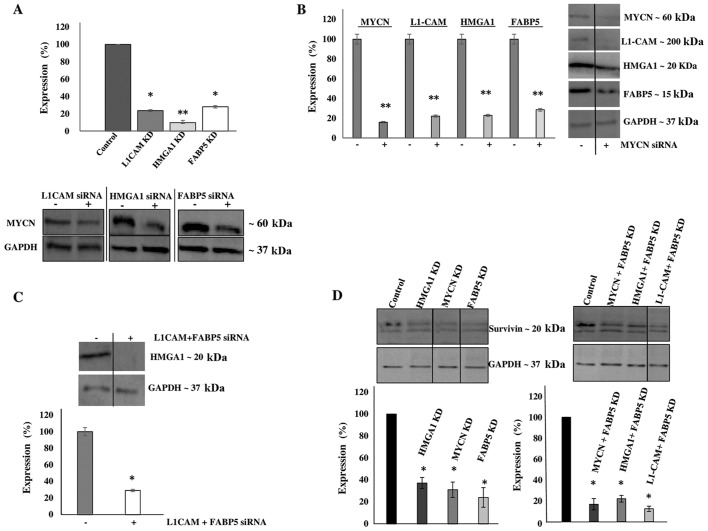
Transcriptional KD experiments reveal interplay between tumorigenic proteins upregulated in IMR-32 cells
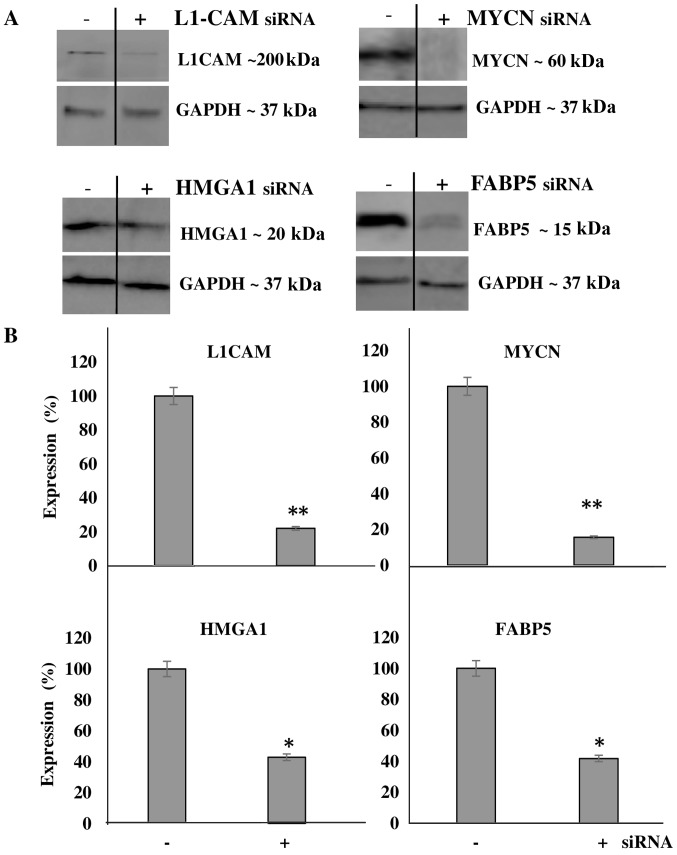
We then aimed to transcriptionally KD the protein expression of these molecules and determine the effects of this KD on cellular bio-function. Additionally, we wished to determine whether the downregulation of one protein would affect the expression level of the other targets, thereby implicating a potential interplay between the proteins. Using transient siRNA transfection, we successfully downregulated the protein expression (Fig. 4A) of MYCN (~85%), HMGA1 (~60%), FABP5 (~60%) and L1-CAM (~80%) as determined by western blot analysis at 48 h after siRNA transfection (Fig. 4B). Of note, L1-CAM, HMGA1 and FABP5 transcriptional KD led to the significant concomitant downregulation of MYCN protein expression by ~80, 90 and 75%, respectively (Fig. 5A). In addition, MYCN transcriptional KD led to the significant concomitant downregulation of L1-CAM, HMGA1 and FABP5 protein expression by ~77, 75 and 70, respectively (Fig. 5B). Moreover, the combined transcriptional KD of L1-CAM and FABP5 led to the significant concomitant downregulation of HMGA1 protein expression by ~70% (Fig. 5C), while survivin protein expression was abrogated following the transcriptional KD of MYCN, HMGA1 and FABP5 (Fig. 5D). This interplay between these tumorigenic proteins is extremely interesting and warrants further investigation into the mechanisms utilized by this network to drive cancerous progression and malignant, treatment-evasive recurrence in high-risk, MYCN-amplified pediatric neuroblastomas.
Figure 4.
The transcriptional knockdown of tumorigenic proteins significantly decreases protein expression. (A) Transient transfection with siRNA targeting L1-cell adhesion molecule (L1-CAM), high mobility group protein A1 (HMGA1), MYCN proto-oncogene, bHLH transcription factor (MYCN) and fatty-acid binding protein 5 (FABP5) abrogated their protein expression. (B) Densitometric analysis of multiple experimental repeats revealed a significant decrease in the expression of the said proteins compared to control siRNA-transfected cells. Experiments were run in triplicate and repeated >3 times. Results represent the means ± SEM; *P<0.05 and **P<0.001.
Figure 5.
Transcriptional knockdown experiments reveal an interplay between tumorigenic proteins. (A) Western blot analysis revealed a concomitant significant downregulation of MYCN proto-oncogene, bHLH transcription factor (MYCN) protein expression following the knockdown of L1-cell adhesion molecule (L1-CAM), high mobility group protein A1 (HMGA1) and fatty-acid binding protein 5 (FABP5) by siRNA in the IMR-32 cells compared to the controls. (B) Conversely, MYCN knockdown by siRNA led to the concomitant significant downregulation of L1-CAM, HMGA1 and FABP5 protein expression, whereas (C) the combined knockdown of L1-CAM and FABP5 by siRNA led to the concomitant significant downregulation of HMGA1 protein expression. (D) MYCN, HMGA1 and FABP5 knockdown and the combined dual-target knockdown led to the concomitant downregulation of survivin protein expression. Non-adjacent bands of western blot experiments were re-aligned side by side to increase the clarity of the presented data and designated by a straight vertical line. Experiments were run in triplicate and repeated >3 times. Results represent the means ± SEM; *P<0.05 and **P<0.01.
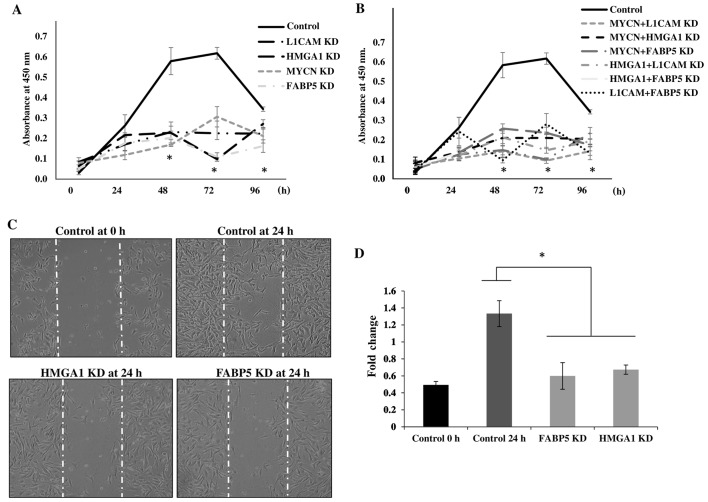
Transcriptional KD of tumorigenic proteins inhibits the proliferation and migration of IMR-32 cells
To determine the cellular bio-functional effects of the transcriptional downregulation of our protein targets, we examined the rate of proliferation of the IMR-32 cells over 5 days following the transcriptional KD of these tumorigenic proteins. We observed no significant differences in the rate of the proliferation of IMR-32 cells at 0 and 24 h following the transcriptional KD of all the above-mentioned siRNA-targeted, tumorigenic proteins compared to the controls. However, there was a statistically significant reduction in the rate of cellular proliferation between the MYCN, L1-CAM, HMGA1 and FABP5 siRNA-transfected cells and the controls at 48, 72 and 96 h post-siRNA transfection (Fig. 6A), implicating the role of these proteins in highly proliferative signaling pathways. The rate of the proliferation of cells targeted with double-target siRNA transfection (MYCN + L1-CAM, MYCN + HMGA1, MYCN + FABP5, HMGA1 + L1-CAM, HMGA1 + FABP5 and FAB5 + L1-CAM) was also significantly decreased from 48 to 96 h post-transfection compared to the controls (Fig. 6B). Double-target siRNA transfection did not exert an additive effect on the rate of cell proliferation compared to the single-target siRNA-transfected cells (data not shown); thus, we speculate on the possible redundancy of the proliferative signaling pathways of these proteins. We then sought to examine the effects of FABP5 and HMGA1 transcriptional KD on the migration of IMR-32 cells, since L1-CAM (25) and MYCN (26,27) have been previously reported to affect neuroblastoma cell migration. Using the 'wound healing' scratch assay (Fig. 6C) we observed a significant decrease in the migratory capacity of the IMR-32 cells subjected to FABP5 or HMGA1 transcriptional KD at 24 h after 'wound induction' (Fig. 6D).
Figure 6.
Transcriptional knockdown of tumorigenic proteins inhibits the proliferation and migration of IMR-32 cells. (A) Transient siRNA transfection and knockdown of MYCN proto-oncogene, bHLH transcription factor (MYCN), L1-cell adhesion molecule (L1-CAM), high mobility group protein A1 (HMGA1) and fatty-acid binding protein 5 (FABP5) protein expression led to a significant decrease in IMR-32 cell proliferation from 48–96 h compared to the controls, as determined using a WST-1 cell proliferation assay. (B) Double-siRNA targeted knockdown (MYCN + L1-CAM, MYCN + HMGA1, MYCN + FABP5, L1-CAM + HMGA1, L1-CAM + FABP5 and HMGA1 + FABP5) also led to significant decrease in the IMR-32 cellular proliferation rate from 48–96 h compared to the controls. (C) Representative images of 'wound closure' illustrate a reduction in the migratory capacity of the IMR-32 cells subjected to FABP5 and HMGA1 siRNA knockdown. (D) The average area of 'wound closure' in multiple random fields, measured using AxioVision Systems software, revealed a statistically significant decrease in IMR-32 cell migration compared to the controls. Experiments were run in triplicate and repeated >3 times. Results represent the means ± SEM; *P<0.05.
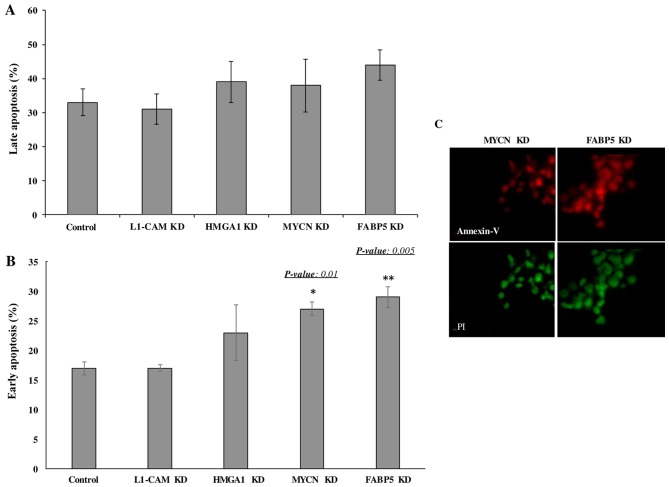
Transcriptional KD of FABP5 and MYCN induces the early apoptosis of IMR-32 cells
Lastly we wished to determine whether the transcriptional KD of the tumorigenic proteins would affect the rate of apoptosis of IMR-32 cells. The late apoptotic rate was not observed to differ significantly between the controls and siRNA-transfected cells of any target at 48 h following transcriptional KD (Fig. 7A). However, early apoptosis was significantly higher in the cells subjected to FABP5 and MYCN transcriptional siRNA KD at 48 h, as determined by Annexin V/PI staining and FACS analysis (Fig. 7B). This was confirmed by immunofluorescence staining of the cells with Annexin V and PI (Fig. 7C). Perhaps a difference in the rate of late apoptosis would be observed had we conducted the apoptosis assay at 72 or 96 h post-transcriptional KD.
Figure 7.
Transcriptional knockdown of MYCN proto-oncogene, bHLH transcription factor (MYCN) and fatty-acid binding protein 5 (FABP5) induces cellular apoptosis of IMR-32 cells. (A) The rate of late cellular apoptosis between MYCN, L1-cell adhesion molecule (L1-CAM), high mobility group protein A1 (HMGA1) and FABP5 siRNA-transfected and control IMR-32 cells did not differ significantly at 48 h post-transfection as determined using an Annexin V/propidium iodide (PI) cell apoptosis assay and flow cytometry analysis. (B) The rate of early apoptosis in MYCN and FABP5 siRNA-transfected IMR-32 cells was significantly higher compared to controls (~27 and ~29% compared to ~17%, respectively) at 48 h post-transfection. (C) Fluorescence microscopy images of MYCN and FABP5 siRNA-transfected cells stained with Annexin V (red) and PI (green) at 48 h post-transfection. Experiments were run in triplicates and repeated >3 times. Results represent the mean ± SEM.
Discussion
Neuroblastoma is a devastating childhood cancer with a dismal prognosis if presented in the high-risk group at diagnosis. IMR-32 is a MYCN-amplified, highly malignant human neuroblastoma cell line. On the contrary, SK-N-SH, while a metastatic human neuroblastoma cell line derived from an epithelial origin (28), is non-MYCN-amplified (29), and hence, is considered less invasive than the IMR-32 cells and a good cell line to use in comparison to IMR-32. In addition to the well-established pathways known to affiliate with MYCN in tumorigenic processes, we sought to discover whether other tumorigenic proteins and/or pathways are also 'accomplices' of the main 'culprit' MYCN, in its pursuit of cancerous-progression, malignancy, anti-apoptosis and resistance to treatment in high-risk neuroblastoma.
Our preliminary analysis of the proteomic signatures in the MYCN-amplified IMR-32 compared to the non-MYCN-amplified SK-N-SH cells revealed some noteworthy, upregulated tumorigenic proteins in the IMR-32 compared to the SK-N-SH cells. Enrichr mapped these proteins to highly cancerous pathways, including leptin signaling, AGE/RAGE signaling, AMPK signaling, Notch, SREBP and Rac1/Pak1/p38/MMP-2 signaling pathways, among others. The leptin signaling pathway is highly affiliated with cancer migration and invasion (30), whereas SREBP, AGE/RAGE, Notch, Rac and AMPK have all been implicated in the tumorigenesis of various nervous system cancers, as well as other solid tumors (19–24). IMR-32 cells represent a highly malignant, invasive, treatment-resistant form of neuroblastoma and the identification of these activated pathways within them has shed some light on the understanding of their malignant mechanisms.
Of the SILAC identified upregulated proteins, we selected to verify with western blot analysis, the protein expression of several that mapped to more than one activated tumorigenic pathways including, L1-CAM (mapped to upstream regulators APP and HIF-1α), HMGA1 (mapped to activated upstream regulators MYC, APP, E2F1, MYCN and SP1), FABP5 (mapped to activated upstream regulators MYC, SREBF2, SREBF1 and KITLG) and survivin or BIRC5 (mapped to activated upstream regulators IL3, MYCN and MYC) to determine their interaction with MYCN in the IMR-32 cells. As shown in Table V, there is intriguing overlap between these upstream regulators in highly tumorigenic pathways including, pathways in cancer, transcriptional misregulation in cancer, PI3K/AKT, Ras, Rap1, TGF-β, HIF-1α, Wnt, insulin and mTOR signaling pathways. The mTOR pathway, one of the most potent signaling pathways involved in cellular proliferation and protein synthesis, is highly activated in MYCN-amplified cell lines (31). Moreover, when MYCN-amplified IMR-32 cells are treated with SU11657 (SUGEN), a selective multi-targeted tyrosine kinase inhibitor with antitumor and anti-angiogenic activity, their growth is significantly inhibited (32).
L1-CAM is a neural cell adhesion molecule L1 found to be expressed in numerous tumors, such as neuroblastomas (25), glioblastomas, melanomas, lymphomas, as well as breast, colon, small cell lung carcinomas and gastrointestinal stromal tumors (33,34). L1-CAM plays a significant role in tumor progression and metastatic behavior (35), and the expression profile analysis in multiple human tumors has identified L1-CAM as a molecular marker for differential diagnosis and targeted therapy (36). Of note, various studies have recently implicated L1-CAM in cancer stem-cell maintenance and propagation (37), the activation of DNA damage checkpoint response that confers resistant to radiation therapy (38) and cancer cell migration and malignancy (39) in glioblastoma and neuroblastoma (40). Moreover, the strong interactions between L1-CAM and EGFR, RANBP9 and NCAM1 (which affiliates with TGF-β) (41) and NRP1 (Fig. 3), may enhance IMR-32 cell invasion and metastasis via an angiogenesis-driven, migratory mechanism.
HMGA1 belongs to a large family of non-histone DNA-binding factors that play important architectural functions in the organization of active chromatin (42). In addition to its role in physiologic processes, a deregulated HMGA1 expression is described in most tumors of epithelial and mesenchymal origin, and is considered a hallmark of cancer (43). Particularly, higher levels of HMGA1 expression are associated with more malignant and metastatic phenotypes in epithelial cancers (44,45). Furthermore, HMGA1 regulation may be controlled by MYCN, which upregulates HMGA1 expression in neuroblastoma cells and in neuroblastoma-like tumors arising in MYCN transgenic mice. Moreover, HMGA1 has been shown to be a direct MYCN transcriptional target, suggesting that HMGA1 is a biologically relevant MYCN target gene (46).
STRING protein network analysis mapped HMGA1 to MYCN, SP1, E2F1 and SREBP1 [a transcription factor with an important role in lipid metabolism, previously found to be highly activated in malignancies (47)], SREBP-cleavage-activating protein [SCAP which plays a role in trafficking SREBP1 from the ER to the Golgi apparatus and recently found to play a role in tumorigenesis (48)], insulin-like growth factor receptor 1 [IGF1R or INSR, a known driver of HIF-2α transcription via PI3K signaling (49)] and the gene KITLG [which codes for the ligand of the tyrosine-kinase receptor c-KIT, STAT1 and STAT4 and implicated in the susceptibility of germ cell tumors such as testicular cancer (50,51)]. Furthermore, when we examined the networks linking only HMGA1, BIRC5 and MYCN, we identified very strong interactions with the highly tumorigenic AURKA/B, CDK1, CDCA8 (52) and INCENP (53). As strong cell proliferation players, we speculate that the IMR-32 cells overexpressing HMGA1 and MYCN have an enhanced mitotic and cell division capacity that favors tumor cell proliferation and the evasion of apoptosis. With HMGA1 being a common player in the above-mentioned tumorigenic pathways, including HIF-1α, SREBP1, KITLG, AURKA/B, CDK1 and CDCA8, it is no surprise that MYCN-amplified IMR-32 cells or high-risk, MYCN-amplified neuroblastomas are highly malignant cancers.
FABP5 binds with a strong affinity to medium and long chain fatty acids, and translocates them into the nucleus where they stimulate their nuclear receptor PPARγ, which in turn stimulates other targets that lead to cancer expansion and progression by enhanced angiogenesis and reduced apoptosis (54). Based on previous studies that have shown FABP5 upregulation in various tumors, its association with cancer malignancy and invasiveness (55) and the demonstrated antitumor activity of the FABP5 chemical inhibitor SBFI26 (56), we decided to evaluate its role in the IMR-32 neuroblastoma cells. We were particularly interested in FABP5 as there are limited studies exploring its role in the aggressive, MYCN-amplified neuroblastoma cells. We were very intrigued by the observation that FABP5 mapped to the highly tumorigenic MYC, SREBF1, SREBF2 and KITLG pathways further explaining the malignant nature of the IMR-32 cells.
Survivin or BIRC5 expression is strongly upregulated in aggressive, high-risk neuroblastoma compared to normal tissues, adult malignancies and non-malignant fetal adrenal neuroblasts (57). This upregulation correlates with an unfavorable prognosis in patients with neuroblastoma (58,59). BIRC5 is a key regulator of mitosis and programmed cell death or apoptosis (60) and the smallest member of the family of inhibitor-of-apoptosis proteins (IAPs) (61,62). Thus, it is an ideal target for killing tumor cells specifically, while sparing healthy normal cells. We found BIRC5 to be highly affiliated with HMGA1, MYC and MYCN in the AURKA/B pathways, which may be critical interactions that fuel IMR-32 cell proliferation and malignancy. In addition, STRING network analysis identified interactions between BIRC5 and IL3 [a stimulator of STATs and AKT pathways (63)] and HIF-1α [an angiogenic driver in oxygen-deprived solid tumors (64) previously reported to protect neuroblastoma cells from hypoxia-induced apoptosis (63)]. The common denominator here is MYC, our validated targets affiliated either directly or indirectly (for example via MYCN) with MYC. Moreover, the intricate network between MYC, MYCN, AURKA/B and NCAM1, and our targets L1-CAM, HMGA1, BIRC5 and FABP5 links them to highly proliferative, angiogenic and metastatic tumorigenic pathways.
The transcriptional KD of all our proteins significantly inhibited the proliferation of IMR-32 cells and FABP5 and MYCN KD inhibited early apoptosis, while FABP5 or HMGA1 KD inhibited cellular migration. The double-target transcriptional KD did not yield an additive effect on cell proliferation compared to single-target KD, implying that these proteins may signal through redundant proliferative pathways. In fact, HMGA1 and MYCN have been found to co-interact and HMGA1 was identified as a direct MYCN transcriptional regulator (46), which would explain the lack of an additive effect with combined, siRNA transcriptional KD of both targets, compared to single-target KD in our cells. Others have demonstrated the interplay between L1-CAM and EGFR in promoting cancer proliferation via ERK activation (65). ERK is a protein downstream of EGFR, platelet-derived growth factor receptor (PDGFR) and vascular endothelial growth factor (VEGFR) signaling, which are receptor tyrosine kinases that our validated proteins may signal through. For example, the FABP5-PPARγ-VEGF signaling transduction axis has been reported to be the pivotal tumorigenic pathway in prostate cancer progression and malignancy (54). Furthermore, L1-CAM (66), HMGA1 (67) and MYCN (68), have all been reported to inhibit p53, thereby affecting cellular proliferation; in addition, L1-CAM (69), FABP5 (54) and MYCN all signal through the mTOR pathway via PI3K/AKT activation, and this may therefor explain the lack of an additive effect between single-target versus double-target KD on cell proliferation.
The fact that FABP5 and MYCN KD were able to induce early apoptosis of our IMR-32 cells at 48 h post-transfection implies that these proteins are crucial for cellular survival and thus early apoptotic rates were observed 48 h following their transcriptional KD. In support of our findings, Kawaguchi et al demonstrated the interaction of FABP5 with SP-1 and c-MYC (both determined by IPA to be upregulated in the IMR-32 compared to SK-N-SH cells) to enhance cellular proliferation in cancer cells and the anti-proliferative effects of FABP5, SP1 or c-MYC silencing in prostate cancer cells (70). We have previously demonstrated the interplay between MYCN and L1-CAM in conferring the radioresistance of IMR-32 cells (25) and L1-CAM has been previously reported to sustain ERK, FAK and PAK phosphorylation in apoptosis-resistant ovarian cancers, whereas its transcriptional KD sensitized ovarian carcinoma cells to apoptotic stimuli (71). We speculate that the interplay between L1-CAM, FABP5 and MYCN in this aggressive neuroblastoma cell line may be driving its malignant, apoptosis-resistant behavior. Of note, a recent study demonstrated the efficacy and safety of using the CE7 monoclonal antibody to target L1-CAM in a (CAR)-redirected T-cell mediated immuno-therapy for children with neuroblastoma (72), which has shown promise in providing both in vitro and in vivo antitumor activity in primary, metastatic and recurrent neuroblastoma.
Our proteins validated by western blot analysis mapped to highly tumorigenic, activated upstream regulators which may help explain the aggressive nature of these malignant MYCN-amplified cells. Seeing all these tumorigenic players highly expressed in the IMR-32 neuroblastoma cells and of particular interest, their mapping to the activated upstream regulator, MYCN is quite intriguing and highlights the possible mechanisms responsible for the aggressive, malignant nature of these cells. For instance, ISNR, SP1, SREBF1, IL3 and HIF-1α, which enhance tumor cell survival under stress-hypoxic conditions that favor proliferative processes and angiogenesis, may function as strong 'accomplices' of the main 'culprit', MYCN, in driving the malignant, tumorigenic behaviors of the IMR-32 cell line. In addition, AURKA/B with its strong interaction with HMGA1, BIRC5, MYC and MYCN, as well as the strong interaction between L1-CAM and the migratory and angiogenic players, including EGFR, RANBP1, NCAM1 and NRP1, may further be fueling the malignant propensity of the MYCN-amplified IMR-32 cells and the high-risk childhood neuroblastomas.
In conclusion, neuroblastoma that presents at diagnosis with high-risk disease is extremely difficult to cure and long-term survival is rarely documented. It is of utmost importance to target the highly malignant, treatment evasive form of this disease with multi-modality therapeutic approaches. As demonstrated in the present study, the MYCN-amplified IMR-32 cells express highly tumorigenic proteins that belong to pathways that govern metastasis, invasion and migration of cancer cells; cancer stem cell propagation and maintenance; cancer angiogenesis, proliferation and evasion of apoptosis. In addition, our observation that the transcriptional KD of one target simultaneously downregulated another tumorigenic target indicates that an important network of interplay exists between highly tumorigenic proteins in these malignant cancer cells.
To the best of our knowledge, we are the first to illustrate the molecular crosstalk between these tumorigenic proteins and their affiliation with highly tumorigenic pathways in the MYCN-amplified IMR-32 neuroblastoma cells. Future studies are warranted to explore these interactions and to elucidate the exact mechanisms of this crosstalk, both at the cellular level and in pre-clinical models. Multi-modality targeted therapy against several non-redundant tumorigenic pathways may cripple this devastating disease and hinder malignant recurrence that almost always resists therapy, leading to mortality in affected children.
Acknowledgments
This study was supported in part by the Sheik Zayed Institute for Pediatric Surgical Innovation and the Michael Sandler Cancer Research Fund and by grants from the National Center for Scientific Research in Lebanon (grant no. CNRS632). In addition, partial funding in support of this study was provided by the Lebanese American University School of Pharmacy Faculty Research and Development Award, as well as the University of Balamand, Department of Biology, Faculty Research funds. The authors would like to thank Dr Kristy Brown, former Assistant Professor at the Children's national Medical Center, Center for Genetic Medicine Research (Washington, DC, USA) and currently Principle Scientist at Solid Biosciences (Cambridge, MA, USA) for her assistance with the SILAC proteomics analysis. The authors would also like to thank Mr. Samer Bazzi, Research Assistant at the Faculty of Medicine, University of Balamand, for conducting the apoptosis assays and analysis using flow cytometry.
Abbreviations
- APP
amyloid precursor protein
- ANK1 and ANK2
Ankyrin 1 and 2
- AURKA/B
Aurora kinase A and B
- BIRC
baculoviral IAP repeat containing 5 (survivin)
- CDK1
cyclin-dependent kinase 1
- CDCA8
cell division cycle associated 8
- CNTN2
contactin 2
- EGFR
epidermal growth factor receptor
- E2F1
E2F transcription factor 1
- EZR
Ezrin
- FABP5
fatty-acid binding protein 5
- HIF-1α
hypoxiainducible factor-1α
- HMGA1
high mobility group protein A1
- IL3
interleukin 3
- IPA
Ingenuity Pathway Analysis
- KITLG
KIT ligand
- L1-CAM
L1-cell adhesion molecule
- MYCN
MYCN proto-oncogene, bHLH transcription factor
- NCAM1
neural cell adhesion molecule 1
- NCAN
neurocan
- NRP1
neuropilin-1
- RANBP9
RAN binding protein 9
- RDX
radixin
- SP1
Sp1 transcription factor
- SREBF1 or 2
sterol regulatory element binding transcription factor 1 or 2
Footnotes
Competing interests
The authors declare that they have no competing interests.
References
- 1.Louis CU, Shohet JM. Neuroblastoma: Molecular pathogenesis and therapy. Annu Rev Med. 2015;66:49–63. doi: 10.1146/annurev-med-011514-023121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith MA, Seibel L, Altekruse SF, Ries LA, Melbert DL, O'Leary M, Smith FO, Reaman GH. Outcomes for children and adolescents with cancer: Challenges for the twenty-first century. J Clin Oncol. 2010;28:2625–2634. doi: 10.1200/JCO.2009.27.0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 4.Pinto NR, Applebaum MA, Volchenboum SL, Matthay KK, London WB, Ambros PF, Nakagawara A, Berthold F, Schleiermacher G, Park JR, et al. Advances in risk classification and treatment strategies for neuroblastoma. J Clin Oncol. 2015;33:3008–3017. doi: 10.1200/JCO.2014.59.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chevrier L, Meunier AC, Cochaud S, Muller JM, Chadéneau C. Vasoactive intestinal peptide decreases MYCN expression and synergizes with retinoic acid in a human MYCN-amplified neuroblastoma cell line. Int J Oncol. 2008;33:1081–1089. [PubMed] [Google Scholar]
- 6.Matthay KK, Villablanca JG, Seeger RC, Stram DO, Harris RE, Ramsay NK, Swift P, Shimada H, Black CT, Brodeur GM, et al. Children's Cancer Group: Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. N Engl J Med. 1999;341:1165–1173. doi: 10.1056/NEJM199910143411601. [DOI] [PubMed] [Google Scholar]
- 7.Stafman LL, Beierle EA. Cell proliferation in neuroblastoma. Cancers (Basel) 2016;8:E13. doi: 10.3390/cancers8010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicolai S, Pieraccioli M, Peschiaroli A, Melino G, Raschellà G. Neuroblastoma: Oncogenic mechanisms and therapeutic exploitation of necroptosis. Cell Death Dis. 2015;6:e2010. doi: 10.1038/cddis.2015.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suita S, Tajiri T, Kaneko M, Hirai M, Mugishima H, Sugimoto T, Tsuchida Y. Implications of MYCN amplification in patients with stage 4 neuroblastoma who undergo intensive chemotherapy. J Pediatr Surg. 2007;42:489–493. doi: 10.1016/j.jpedsurg.2006.10.056. [DOI] [PubMed] [Google Scholar]
- 10.Brodeur GM. Brodeur. Nat Rev Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 11.Goldsmith KC, Hogarty MD. Targeting programmed cell death pathways with experimental therapeutics: Opportunities in high-risk neuroblastoma. Cancer Lett. 2005;228:133–141. doi: 10.1016/j.canlet.2005.01.048. [DOI] [PubMed] [Google Scholar]
- 12.Kang JH, Rychahou PG, Ishola TA, Qiao J, Evers BM, Chung DH. MYCN silencing induces differentiation and apoptosis in human neuroblastoma cells. Biochem Biophys Res Commun. 2006;351:192–197. doi: 10.1016/j.bbrc.2006.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.M200025-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Jensen ON, Wilm M, Shevchenko A, Mann M. Sample preparation methods for mass spectrometric peptide mapping directly from 2-DE gels. Methods Mol Biol. 1999;112:513–530. doi: 10.1385/1-59259-584-7:513. [DOI] [PubMed] [Google Scholar]
- 15.Formolo CA, Williams R, Gordish-Dressman H, MacDonald TJ, Lee NH, Hathout Y. Secretome signature of invasive glioblastoma multiforme. J Proteome Res. 2011;10:3149–3159. doi: 10.1021/pr200210w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–7. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abouantoun TJ, Castellino RC, MacDonald TJ. Sunitinib induces PTEN expression and inhibits PDGFR signaling and migration of medulloblastoma cells. J Neurooncol. 2011;101:215–226. doi: 10.1007/s11060-010-0259-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bayin NS, Frenster JD, Sen R, Si S, Modrek AS, Galifianakis N, Dolgalev I, Ortenzi V, Illa-Bochaca I, Khahera A, et al. Notch signaling regulates metabolic heterogeneity in glioblastoma stem cells. Oncotarget. 2017;8:64932–64953. doi: 10.18632/oncotarget.18117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez-Villasana V, Fuentes-Mattei E, Ivan C, Dalton HJ, Rodriguez-Aguayo C, Fernandez-de Thomas RJ, Aslan B, Del C, Monroig P, Velazquez-Torres G, Previs RA, et al. Rac1/Pak1/38/MMP-2 axis regulates angiogenesis in ovarian cancer. Clin Cancer Res. 2015;21:2127–2137. doi: 10.1158/1078-0432.CCR-14-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan Y, Gan Y, Shen Y, Cai X, Song Y, Zhao F, Yao M, Gu J, Tu H. Leptin signaling enhances cell invasion and promotes the metastasis of human pancreatic cancer via increasing MMP-P13 production. Oncotarget. 2015;6:16120–16134. doi: 10.18632/oncotarget.3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Zhang J, He J, Zhou W, Xiang G, Xu R. MicroRNA-132 cause apoptosis of glioma cells through blockade of the SREBP-1c metabolic pathway related to SIRT1. Biomed Pharmacother. 2016;78:177–184. doi: 10.1016/j.biopha.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 23.Bao JM, He MY, Liu YW, Lu YJ, Hong YQ, Luo HH, Ren ZL, Zhao SC, Jiang Y. AGE/RAGE/Akt pathway contributes to prostate cancer cell proliferation by promoting Rb phosphorylation and degradation. Am J Cancer Res. 2015;5:1741–1750. [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Qi Q, Feng Z, Zhang X, Huang B, Chen A, Prestegarden L, Li X, Wang J. Berberine induces autophagy in glioblastoma by targeting the AMPK/mTOR/ULK1-pathway. Oncotarget. 2016;7:66944–66958. doi: 10.18632/oncotarget.11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rached J, Nasr Z, Abdallah J, Abou-Antoun T. L1-CAM knock-down radiosensitizes neuroblastoma IMR-32 cells by simultaneously decreasing MycN, but increasing PTEN protein expression. Int J Oncol. 2016;49:1722–1730. doi: 10.3892/ijo.2016.3625. [DOI] [PubMed] [Google Scholar]
- 26.Fabian J, Opitz D, Althoff K, Lodrini M, Hero B, Volland R, Beckers A, de Preter K, Decock A, Patil N, et al. MYCN and HDAC5 transcriptionally repress CD9 to trigger invasion and metastasis in neuroblastoma. Oncotarget. 2016;7:66344–66359. doi: 10.18632/oncotarget.11662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gangoda L, Keerthikumar S, Fonseka P, Edgington LE, Ang CS, Ozcitti C, Bogyo M, Parker BS, Mathivanan S. Inhibition of cathepsin proteases attenuates migration and sensitizes aggressive N-Myc amplified human neuroblastoma cells to doxorubicin. Oncotarget. 2015;6:11175–11190. doi: 10.18632/oncotarget.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preis PN, Saya H, Nádasdi L, Hochhaus G, Levin V, Sadée W. Neuronal cell differentiation of human neuroblastoma cells by retinoic acid plus herbimycin A. Cancer Res. 1988;48:6530–6534. [PubMed] [Google Scholar]
- 29.Schwab M, Alitalo K, Klempnauer KH, Varmus HE, Bishop JM, Gilbert F, Brodeur G, Goldstein M, Trent J. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumor. Nature. 1983;305:245–248. doi: 10.1038/305245a0. [DOI] [PubMed] [Google Scholar]
- 30.Wang T, Zhang Z, Wang K, Wang J, Jiang Y, Xia J, Gou L, Liu M, Zhou L, He T, et al. Inhibitory effects of BMP9 on breast cancer cells by regulating their interaction with pre-adipocytes/adipocytes. Oncotarget. 2017;8:35890–35901. doi: 10.18632/oncotarget.16271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Segerström L, Baryawno N, Sveinbjörnsson B, Wickström M, Elfman L, Kogner P, Johnsen JI. Effects of small molecule inhibitors of PI3K/Akt/mTOR signaling on neuroblastoma growth in vitro and in vivo. Int J Cancer. 2011;129:2958–2965. doi: 10.1002/ijc.26268. [DOI] [PubMed] [Google Scholar]
- 32.Bäckman U, Christofferson R. The selective class III/V receptor tyrosine kinase inhibitor SU11657 inhibits tumor growth and angiogenesis in experimental neuroblastomas grown in mice. Pediatr Res. 2005;57:690–695. doi: 10.1203/01.PDR.0000156508.68065.AA. [DOI] [PubMed] [Google Scholar]
- 33.Rawnaq T, Quaas A, Zander H, Gros SJ, Reichelt U, Blessmann M, Wilzcak W, Schachner M, Sauter G, Izbicki JR, et al. L1 is highly expressed in tumors of the nervous system: A study of over 8000 human tissues. J Surg Res. 2012;173:314–319. doi: 10.1016/j.jss.2010.10.029. [DOI] [PubMed] [Google Scholar]
- 34.Gavert N, Ben-Shmuel A, Raveh S, Ben-Ze'ev A. L1-CAM in cancerous tissues. Expert Opin Biol Ther. 2008;8:1749–1757. doi: 10.1517/14712598.8.11.1749. [DOI] [PubMed] [Google Scholar]
- 35.Raveh S, Gavert N, Ben-Ze'ev A. L1 cell adhesion molecule (L1CAM) in invasive tumors. Cancer Lett. 2009;282:137–145. doi: 10.1016/j.canlet.2008.12.021. [DOI] [PubMed] [Google Scholar]
- 36.Ben-Arie A, Huszar M, Ben-Zvi N, Smirnov A, Altevogt P, Fogel M. The role of L1-CAM immunohistochemial staining in the diagnosis of abdominal-pelvic cancer of uncertain primary site in women. Eur J Surg Oncol. 2008;34:795–799. doi: 10.1016/j.ejso.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 37.Bao S, Wu Q, Li Z, Sathornsumetee S, Wang H, McLendon RE, Hjelmeland AB, Rich JN. Targeting cancer stem cells through L1CAM suppresses glioma growth. Cancer Res. 2008;68:6043–6048. doi: 10.1158/0008-5472.CAN-08-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng L, Wu Q, Huang Z, Guryanova OA, Huang Q, Shou W, Rich JN, Bao S. L1CAM regulates DNA damage checkpoint response of glioblastoma stem cells through NBS1. EMBO J. 2011;30:800–813. doi: 10.1038/emboj.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao WJ, Schachner M. Neuregulin 1 enhances cell adhesion molecule L1 expression in human glioma cells and promotes their migration as a function of malignancy. J Neuropathol Exp Neurol. 2013;72:244–255. doi: 10.1097/NEN.0b013e3182863dc5. [DOI] [PubMed] [Google Scholar]
- 40.Zhao W. Zhao. Oncol Lett. 2012;4:812–816. doi: 10.3892/ol.2012.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ackermann MA, Petrosino JM, Manring HR, Wright P, Shettigar V, Kilic A, Janssen PML, Ziolo MT, Accornero F. TGF-β1 affects cell-cell adhesion in the heart in an NCAM1-dependent mechanism. J Mol Cell Cardiol. 2017;112:49–57. doi: 10.1016/j.yjmcc.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grosschedl R, Giese K, Pagel J. HMG domain proteins: Architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 43.Tallini G, Dal Cin P. HMGI(Y) and HMGI-C dysregulation: A common occurrence in human tumors. Adv Anat Pathol. 1999;6:237–246. doi: 10.1097/00125480-199909000-00001. [DOI] [PubMed] [Google Scholar]
- 44.Chiappetta G, Tallini G, De Biasio MC, Manfioletti G, Martinez-Tello FJ, Pentimalli F, de Nigris F, Mastro A, Botti G, Fedele M, et al. Detection of high mobility group I HMGI(Y) protein in the diagnosis of thyroid tumors: HMGI(Y) expression represents a potential diagnostic indicator of carcinoma. Cancer Res. 1998;58:4193–4198. [PubMed] [Google Scholar]
- 45.Abe N, Watanabe T, Sugiyama M, Uchimura H, Chiappetta G, Fusco A, Atomi Y. Determination of high mobility group I(Y) expression level in colorectal neoplasias: A potential diagnostic marker. Cancer Res. 1999;59:1169–1174. [PubMed] [Google Scholar]
- 46.Giannini G, Cerignoli F, Mellone M, Massimi I, Ambrosi C, Rinaldi C, Dominici C, Frati L, Screpanti I, Gulino A. High mobility group A1 is a molecular target for MYCN in human neuroblastoma. Cancer Res. 2005;65:8308–8316. doi: 10.1158/0008-5472.CAN-05-0607. [DOI] [PubMed] [Google Scholar]
- 47.Ru P, Hu P, Geng F, Mo X, Cheng C, Yoo JY, Cheng X, Wu X, Guo JY, et al. Feedback loop regulation of SCAP/SREBP-1 by miR-29 modulates EGFR signaling-driven glioblastoma growth. Cell Reports. 2016;16:1527–1535. doi: 10.1016/j.celrep.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo D. SCAP links glucose to lipid metabolism in cancer cells. Mol Cell Oncol. 2016;3:e1132120. doi: 10.1080/23723556.2015.1132120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mohlin S, Hamidian A, von Stedingk K, Bridges E, Wigerup C, Bexell D, Påhlman S. PI3K-mTORC2 but not PI3K-mTORC1 regulates transcription of HIF2A/EPAS1 and vascularization in neuroblastoma. Cancer Res. 2015;75:4617–4628. doi: 10.1158/0008-5472.CAN-15-0708. [DOI] [PubMed] [Google Scholar]
- 50.Hanes R, Grad I, Lorenz S, Stratford EW, Munthe E, Reddy CC, Meza-Zepeda LA, Myklebost O. Preclinical evaluation of potential therapeutic targets in dedifferentiated liposarcoma. Oncotarget. 2016;7:54583–54595. doi: 10.18632/oncotarget.10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poynter JN, Hooten AJ, Frazier AL, Ross JA. Associations between variants in KITLG, SPRY4, BAK1, and DMRT1 and pediatric germ cell tumors. Genes Chromosomes Cancer. 2012;51:266–271. doi: 10.1002/gcc.20951. [DOI] [PubMed] [Google Scholar]
- 52.Jiao DC, Lu ZD, Qiao JH, Yan M, Cui SD, Liu ZZ. Expression of CDCA8 correlates closely with FOXM1 in breast cancer: public microarray data analysis and immunohistochemical study. Neoplasma. 2015;62:464–469. doi: 10.4149/neo_2015_055. [DOI] [PubMed] [Google Scholar]
- 53.Gohard FH, St-Cyr DJ, Tyers M, Earnshaw WC. Targeting the INCENP IN-box-Aurora B interaction to inhibit CPC activity in vivo. Open Biol. 2014;4:140163. doi: 10.1098/rsob.140163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chamcheu JC, Chaves-Rodriquez MI, Adhami VM, Siddiqui IA, Wood GS, Longley BJ, Mukhtar H. Upregulation of PI3K/AKT/mTOR, FABP5 and PPARβ/δ in human psoriasis and imiquimod-induced murine psoriasiform dermatitis model. Acta Derm Venereol. 2016;96:854–856. doi: 10.2340/00015555-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Connolly RM, Nguyen NK, Sukumar S. Molecular pathways: Current role and future directions of the retinoic acid pathway in cancer prevention and treatment. Clin Cancer Res. 2013;19:1651–1659. doi: 10.1158/1078-0432.CCR-12-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Al-Jameel W, Gou X, Forootan SS, Al Fayi MS, Rudland PS, Forootan FS, Zhang J, Cornford PA, Hussain SA, Ke Y. Inhibitor SBFI26 suppresses the malignant progression of castration-resistant PC3-M cells by competitively binding to oncogenic FABP5. Oncotarget. 2017;8:31041–31056. doi: 10.18632/oncotarget.16055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Althoff K, Lindner S, Odersky A, Mestdagh P, Beckers A, Karczewski S, Molenaar JJ, Bohrer A, Knauer S, Speleman F, et al. miR-542 3p exerts tumor suppressive functions in neuroblastoma by downregulating Survivin. Int J Cancer. 2015;136:1308–1320. doi: 10.1002/ijc.29091. [DOI] [PubMed] [Google Scholar]
- 58.Islam A, Kageyama H, Takada N, Kawamoto T, Takayasu H, Isogai E, Ohira M, Hashizume K, Kobayashi H, Kaneko Y, et al. High expression of Survivin, mapped to 17q25, is significantly associated with poor prognostic factors and promotes cell survival in human neuroblastoma. Oncogene. 2000;19:617–623. doi: 10.1038/sj.onc.1203358. [DOI] [PubMed] [Google Scholar]
- 59.Azuhata T, Scott D, Griffith TS, Miller M, Sandler AD. Survivin inhibits apoptosis induced by TRAIL, and the ratio between survivin and TRAIL receptors is predictive of recurrent disease in neuroblastoma. J Pediatr Surg. 2006;41:1431–1440. doi: 10.1016/j.jpedsurg.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 60.Mita AC, Mita MM, Nawrocki ST, Giles FJ. Survivin: Key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin Cancer Res. 2008;14:5000–5005. doi: 10.1158/1078-0432.CCR-08-0746. [DOI] [PubMed] [Google Scholar]
- 61.Adida C, Berrebi D, Peuchmaur M, Reyes-Mugica M, Altieri DC. Anti-apoptosis gene, survivin, and prognosis of neuroblastoma. Lancet. 1998;351:882–883. doi: 10.1016/S0140-6736(05)70294-4. [DOI] [PubMed] [Google Scholar]
- 62.Hossain MM, Banik L, Ray SK. Survivin knockdown increased anti-cancer effects of (-)-epigallocatechin-3-gallate in human malignant neuroblastoma SK-N-BE2 and SH-SY5Y cells. Exp Cell Res. 2012;318:1597–1610. doi: 10.1016/j.yexcr.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han L, Jorgensen JL, Brooks C, Shi C, Zhang Q, Nogueras González GM, Cavazos A, Pan R, Mu H, Wang SA, et al. Antileukemia efficacy and mechanisms of action of SL-101, a novel anti-CD123 antibody conjugate, in acute myeloid leukemia. Clin Cancer Res. 2017;23:3385–3395. doi: 10.1158/1078-0432.CCR-16-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yin CP, Guan SH, Zhang B, Wang XX, Yue SW. Upregulation of HIF-1α protects neuroblastoma cells from hypoxia-induced apoptosis in a RhoA-dependent manner. Mol Med Rep. 2015;12:7123–7131. doi: 10.3892/mmr.2015.4267. [DOI] [PubMed] [Google Scholar]
- 65.Yoon H, Min JK, Lee DG, Kim DG, Koh SS, Hong HJ. L1 cell adhesion molecule and epidermal growth factor receptor activation confer cisplatin resistance in intrahepatic cholangiocarcinoma cells. Cancer Lett. 2012;316:70–76. doi: 10.1016/j.canlet.2011.10.024. [DOI] [PubMed] [Google Scholar]
- 66.Wang Y, Schachner M. The intracellular domain of L1CAM binds to casein kinase 2α and is neuroprotective via inhibition of the tumor suppressors PTEN and p53. J Neurochem. 2015;133:828–843. doi: 10.1111/jnc.13083. [DOI] [PubMed] [Google Scholar]
- 67.Puca F, Colamaio M, Federico A, Gemei M, Tosti N, Bastos AU, Del Vecchio L, Pece S, Battista S, Fusco A. HMGA1 silencing restores normal stem cell characteristics in colon cancer stem cells by increasing p53 levels. Oncotarget. 2014;5:3234–3245. doi: 10.18632/oncotarget.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mazar J, Rosado A, Shelley J, Marchica J, Westmoreland TJ. The long non-coding RNA GAS5 differentially regulates cell cycle arrest and apoptosis through activation of BRCA1 and p53 in human neuroblastoma. Oncotarget. 2017;8:6589–6607. doi: 10.18632/oncotarget.14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Doberstein K, Pfeilschifter J, Gutwein P. The transcription factor AX2 regulates ADAM10 expression in renal cell carcinoma. Carcinogenesis. 2011;32:1713–1723. doi: 10.1093/carcin/bgr195. [DOI] [PubMed] [Google Scholar]
- 70.Kawaguchi K, Kinameri A, Suzuki S, Senga S, Ke Y, Fujii H. The cancer-promoting gene fatty acid-binding protein 5 (FABP5) is epigenetically regulated during human prostate carcinogenesis. Biochem J. 2016;473:449–461. doi: 10.1042/BJ20150926. [DOI] [PubMed] [Google Scholar]
- 71.Stoeck A, Gast D, Sanderson MP, Issa Y, Gutwein P, Altevogt P. L1-CAM in a membrane-bound or soluble form augments protection from apoptosis in ovarian carcinoma cells. Gynecol Oncol. 2007;104:461–469. doi: 10.1016/j.ygyno.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 72.Künkele A, Taraseviciute A, Finn LS, Johnson AJ, Berger C, Finney O, Chang CA, Rolczynski LS, Brown C, Mgebroff S, et al. Preclinical assessment of CD171-directed CAR T-cell adoptive therapy for childhood neuroblastoma: CE7 epitope target safety and product manufacturing feasibility. Clin Cancer Res. 2017;23:466–477. doi: 10.1158/1078-0432.CCR-16-0354. [DOI] [PubMed] [Google Scholar]