Abstract
Circular RNAs (circRNAs) are key regulators in the development and progression of human cancers; however their roles in breast tumorigenesis are not yet well understood. Thus, the present study aimed to investigate the expression profiles and potential modulatory effects of circRNAs on breast carcinogenesis. A human circRNA microarray analysis was performed to screen for abnormally expressed circRNAs in breast cancer tissue and circRNA-000911 was identified as a circRNA which was significantly downregulated in breast cancer cells. Mechanistic investigations suggested that the enhanced expression of circRNA-000911 suppressed cell proliferation, migration and invasion, and promoted the apoptosis of breast cancer cells. By using a biotin-labeled circRNA-000911 probe to perform RNA precipitation in breast cancer cells, we identified miR-449a as the circRNA-000911-associated microRNA. Gain- and loss-of-function assays indicated that miR-449a antagonized circRNA-000911 to regulate breast cancer progression. Subsequently, Notch1 was identified as the functional target of miR-449a, and the overexpression of circRNA-000911 in breast cancer elevated Notch1 expression. Furthermore, Cignal Signal Transduction Reporter Array and western blot analysis identified nuclear factor-κB (NF-κB) signaling as a functional target of the circRNA-000911/miR-449a pathway. On the whole, our findings indicate that circRNA-000911 plays an anti-oncogenic role in breast cancer and may thus serve as a promising therapeutic target for patients with breast cancer. Therefore, the overexpression of circRNA-000911 may provide a future direction which may aid in the development of a novel treatment strategy for breast cancer.
Keywords: breast cancer, circRNA-000911, miR-449a, Notch1, nuclear factor-κB
Introduction
Breast cancer is one of the leading causes of cancer-related mortality worldwide and the most common cancer among women (1). The majority of patients with this type of cancer are diagnosed at the advanced stages of the disease, and the prognosis of these patients remains poor (2). Currently, adjuvant chemotherapy and radiotherapy following surgical resection are the most commonly used treatment strategies. However, the prognosis of patients with breast cancer remains poor, and the survival of patients with breast cancer has reached a plateau (3,4). Therefore, the identification of promising therapeutic and prognostic targets are essential for the development of effective therapies for patients with breast cancer.
Over the past few years, studies have indicated that non-coding RNAs are involved in all aspects of the process of carcinogenesis and can be used as biomarkers for early-risk stratification and long-term survival prediction (5,6). High-throughput RNA sequencing (RNA-Seq), an emerging method used for the study of RNA regulatory mechanisms in the whole genome, has been used to detect small non-coding RNAs, such as circular RNAs (circRNAs) (7). circRNAs are a class of non-coding RNAs characterized by covalently closed continuous loops with neither 5′ to 3′ polarity nor a polyadenylated tail (8). They have been found to play an important role in the regulation of multiple diseases, including cancer (9,10). Recently, certain types of circRNAs have been found to be significantly deregulated in gastric cancer, esophageal squamous carcinoma and breast cancer, and these deregulated circRNAs have been suggested to participate in cancer development (11).
MicroRNAs (miRNAs or miRs) are a novel class of endogenous, small, non-coding RNAs that exert their post-transcriptional regulatory effects by targeting the 3′ untranslated region (3′UTR) of corresponding mRNAs (12). Emerging evidence indicates that some circRNAs act as miRNA sponges to modulate gene transcription and interact with RNA-binding proteins (RBPs) involved in tumorigenesis (13,14). For example, the circRNA CDR1 as, also known as ciRS-7, has been identified as a miR-7 sponge, leading to the inhibition of miR-7 activity and activating targeted gene expression (15); the circRNA MTO1 also acts as a sponge of miR-9 to suppress hepatocellular carcinoma progression (16) and the circRNA MYLK acts as a competing endogenous RNA to promote bladder cancer progression by modulating the vascular endothelial growth factor (VEGF)A/VEGFR2 signaling pathway (17), providing novel promising markers for cancer diagnosis and therapy.
In the present study, we utilized circRNA microarray analysis to screen the expression profiles of circRNAs in breast cancer specimens, and identified that circRNA-0000911 was significantly downregulated and was a predictor of a poor prognosis of patients with breast cancer. We revealed that circRNA-0000911 plays an anti-oncogenic role in breast cancer by acting as a sponge of miR-449a to promote Notch1 expression, consequently suppressing breast cancer progression. Therefore, circRNA-0000911 may serve as a prognostic biomarker and may thus also serve as a potential therapeutic target for patients with breast cancer.
Materials and methods
Clinical samples
Thirty-five cancer tissues and paired adjacent non-cancerous tissues from primary breast cancer patients were collected at The First Hospital of Lanzhou University Hospital in 2016. Five pairs of breast samples (tumor tissues and matched non-tumor tissues) were used for circRNA microarray, and 30 pairs were used for reverse transcription-quantitative polymerase chain reaction (RT-qPCR) validation. Tissues specimens were obtained during surgery and immediately frozen at −80°C until further use. Written informed consent was obtained from each participant prior to tissue collection. The study protocol was approved by the Clinical Research Ethics Committee of the First Hospital of Lanzhou University.
Cell culture
The human breast cancer cell lines, MCF-7, MDA-MB-231, MDA-MB-468, MDA-MB-453, SKBR-3 and T47D, and one normal breast epithelial cell line (MCF-10A) were obtained from the Chinese Academy of Sciences (Shanghai, China). All breast cancer cell lines were maintained in DMEM (Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, USA), 100 g/ml streptomycin and 100 U/ml penicillin (Life Technologies, Grand Island, NY, USA) at 37°C in 5% CO2 and 95% air. Normal breast epithelial cells were grown in DMEM/F12 1:1 medium with 10% FBS, 2.5 mM L-glutamine and 0.3 mg/ml G418 at 37°C in 5% CO2 and 95% air. The cell lines passed the DNA profiling test [short tandem repeat (STR)].
Expression profile analysis of circRNAs
Five cancer tissues and paired adjacent noncancerous tissues from primary breast cancer patients were collected at the First Hospital of Lanzhou University Hospital in 2016. Tissues specimens were obtained during operation and immediately frozen at −80°C until further use. The circRNAs chip (ArrayStar Human circRNAs chip; ArrayStar, Rockville, MD, USA) containing 5,639 probes specific for human circular RNAs splicing sites was used. Following hybridization and washing of the samples, 5 pairs of breast samples (tumor tissues and matched non-tumor tissues) were analyzed on the circRNAs chips. Exogenous RNAs developed by the External RNA Controls Consortium (ERCC) were used as controls. circRNAs were enriched by digesting linear RNA with RNase R (Epicentre, Madison, WI, USA). Labeled RNAs were scanned using a Agilent Scanner G2505C (Agilent Technologies, Santa Clara, CA, USA). The circRNA microarray process was performed by KangChen Biotech, Inc. (Shanghai, China). The GEO Accession number is GSE97332.
RNA oligoribonucleotides and cell transfection
The miR-449a mimics, and specific anti-miR-449a RNAs were synthesized by GenePharma (Shanghai, China). The circular transcript expression vector possesses two elements termed as the front circular and the back circular frame which were specially designed containing inverted repeat sequences flank. The full-length cDNA of circRNA-000911 was amplified in the MCF-7 cells, and was cloned into the specific vector between two frames, while the mock plasmid without the circRNA-000911 cDNA served as the control. The cDNA sequence of circRNA-000911 was synthesized by RiboBio Co., Ltd. (Guangzhou, China) and then cloned into the lentiviral expression vector, pLVXIRES-neo (Clontech Laboratories Inc., San Francisco, CA, USA). Lentiviral production and transduction were conducted by following previously published procedures (18). The breast cancer cells were maintained in a 6-well plate in DMEM supplemented with 10% FBS and cultured until 50–70% confluent. RNA oligoribonucleotides were mixed with Lipofectamine 3000 (Invitrogen) in reduced serum medium (Opti-MEM; Gibco, Carlsbad, CA, USA) according to the manufacturer’s instructions and the final concentration of the RNA oligoribonucleotides was 100 nM. The effects of knockdown or overexpression were examined by RT-qPCR using RNA extracted 48 h after transfection.
RT-qPCR
Total RNA was isolated from breast cancer cell lines using TRIzol reagent (Invitrogen). Subseequently, cDNA was synthesized from 200 ng extracted total RNA using the SuperScript III® (Invitrogen) and amplified by RT-qPCR based on the TaqMan method on an ABI PRISM 7500 Sequence Detection System (Life Technologies, Grand Island, NY, USA) with the housekeeping gene GAPDH or U6 as an internal control. The 2−ΔΔCq method was used to determine the relative quantification of gene expression levels (19). All the primer sequences were synthesized by RiboBio Co., Ltd., and their sequences are shown as follows: circRNA-000911 forward, 5′-AAAAGCAAGCAGTGCCCATA-3′ and reverse, 5′-GCTCGAATCAGGTCCACCA-3′; Notch1 forward, 5′-GCAAGAAGAAGCGGAGAG-3′ and reverse, 5′-AGCTGGCACCCTGATAGATG-3′; and GAPDH forward, 5′-AGATCCACAACGGATACATT-3′ and reverse, 5′-TCCCTCAAGATTGTCAGCAA-3′.
Dual-luciferase reporter assay
The circRNA-000911 sequence in the MCF-7 cells was subcloned into the luciferase reporter psiCHECK2 (Promega, Madison, WI, USA) and designated as psiCHECK2-circRNA-000911-WT. The circRNA-000911 sequence with mutation of miR-449a binding site was synthesized using overlap extension PCR and cloned into psiCHECK2 vector designated as psiCHECK2-circRNA-000911-Mut. The Notch1 3′UTR cDNA in MCF-7 cells was amplified and cloned to psiCHECK2 and termed as psiCHECK2-Notch1-3′-UTR-WT. The mutant vector for the miR-449a binding site was constructed and termed as psiCHECK2-Notch1-3′UTR-Mut. A total of 3×104 breast cancer cells were seeded in 24-well plates in triplicate. At 48 h following transfection with miR-449a mimics, luciferase reporter assays were conducted using the dual-luciferase reporter assay system (Promega) according to the manufacturer’s instructions. Relative luciferase activity was normalized to the Renilla luciferase internal control.
Cell proliferation assay
Cell proliferation was quantified using the cell counting kit-8 (CCK-8; Sigma-Aldrich). Briefly, 100 μl of cells from the different transfection groups were seeded onto a 96-well plate at a concentration of 2,000 cells per well and incubated at 37°C. At 72 h, the optical density was measured at 450 nm using a microtiter plate reader (SpectraMax M serious; Molecular Devices, Sunnyvale, CA, USA), and the rate of cell survival was expressed as the relative absorbance. The results represent the means of 3 replicates under the same conditions.
FACS apoptosis assay
The cells (1×105/well) were collected at 48 h following transfection, and were stained with Annexin V/FITC and propidium iodide (PI) according to the manufacturer’s instructions (BD Biosciences, Erembodegem, Belgium). Apoptosis was assessed by flow cytometry (BD FACSCalibur flow cytometer; BD Biosciences).
Colony formation assays
The MCF-7 and MDA-MB-231 cells were infected with the circRNA-000911 vectors and cultured for 72 h, and they were then re-plated in 6-well plates at a density of 5×102 cells/well and maintained for 10 days. The colonies were fixed and stained with 0.5% crystal violet (Beyotime Institute of Biotechnology, Shanghai, China) for 15 min. After rinsing well, images were captured using an Olympus microscope (Olympus Inc., Tokyo, Japan) visible colonies were counted using ImageJ 1.47 software.
Cell migration and invasion assay
The cell migratory ability was detected by wound healing assay. Briefly, the transfected cells were seeded in 6-well plates and cultured to near confluence. Artificial wounds were created on the cell monolayer using culture inserts for live cell analysis, and the migrated cells and wound healing were visualized. For each group, at least 3 artificial wounds were photographed with an Olympus microscope (Olympus Inc.) immediately and at the time points indicated after the wound formation. After the wounds were created, the cells were incubated in culture medium without FBS and then photographed at 48 h. The percentage of wound closure was calculated using ImageJ 1.47 software.
The cell invasive ability was detected using Transwell permeable supports (Corning, New York, NY, USA) according to manufacturer’s instructions. Briefly, the transfected cells were plated onto a Matrigel-coated membrane in the upper chamber of a 24-well insert containing medium free of serum. The bottom chamber contained DMEM with 10% FBS. The cells were incubated at 37°C with 5% CO2 for 48 h after plating. The bottom of the chamber insert was then fixed with methanol and stained with DAPI for 5 min. The number of cells that invaded through the membrane was determined from digital images captured on an Olympus microscope (Olympus Inc.) and calculated using ImageJ 1.47 software.
circRNA immunoprecipitation (circRIP)
The biotin-labeled circRNA-000911 probe (5′-CATTCGTGAACGAATACAATCTGGAACA-3′-biotin) was synthesized by Sangon Biotech (Shanghai, China) and the circRIP assay was performed as previously described with minor modifications (20). The MCF-7 cells were fixed by 1% formaldehyde for 10 min, lysed and sonicated (Bioruptor Sonicator; Diagenode Corp., Denville, NJ, USA). Following a 12,000 × g centrifugation for 5 min at 4°C, 50 μl of the supernatant was retained as input and the remaining part was incubated with circRNA-000911 specific probes-streptavidin dynabeads (M-280; Invitrogen) mixture overnight at 30°C. The following day, M-280 dynabeads-probes-circRNAs mixture was washed and incubated with 200 μl lysis buffer and proteinase K to reverse the formaldehyde cross-linking. Finally, TRIzol reagent was added to the mixture for RNA extraction and RT-qPCR detection.
The RIP experiment using Notch1 antibody (1:1,000; Santa Cruz Biotechnology, Santa Cruz, CA, USA) to pull down miR-449a was also performed. According to the manufacturer’s instructions, the Magna RIP RNA-Binding Protein Immunoprecipitation kit (Millipore, Billerica, MA, USA) was used. Total RNA and controls were also assayed to demonstrate that the detected signals were from RNAs specifically binding to Notch1. The final analysis was performed by using RT-qPCR and shown as the fold enrichment of miR-449a.
Signal transduction reporter array
Cignal Signal Transduction Reporter Array (Qiagen, Valencia, CA, USA) was used to simultaneously investigate alternations in the activities of 50 canonical signaling pathways in response to UCA1 knockdown. The cells were transfected with antisense oligonucleotides targeting UCA1 for 24 h and were subsequently transfected with a mixture of a transcription factor-responsive firefly luciferase reporter and a constitutively expressing Renilla construct. The relative activity of each pathway was decided by luciferase/Renilla and normalized to the untreated controls. Experiments were performed in triplicate.
Bioinformatics analysis
The online target-predicting database miRBase (http://www.mirbase.org/) was used for the prediction of potential targeted sequences between circRNA-000911 and miR-449a. Another two databases, TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org/microrna/home.do), were used for the prediction of potential targeted sequences between miR-449a and the Notch1 gene.
Western blot analysis and antibodies
The breast cancer cells were lysed with RIPA buffer with protease inhibitors (Sigma-Aldrich). Protein quantification was carried out using a BCA protein assay kit (Promega). The primary antibodies used for western blot analysis were as follows: Rabbit anti-human Notch1 antibody (#sc6014; 1:500; Santa Cruz Biotechnology), anti-p65-nuclear factor-κB (NF-κB) antibody (#8242; 1:1,000), anti-p50-NF-κB antibody (#3035; 1:1,000) and rabbit anti-human β-actin antibody (#4967, 1:1,000) (all from Cell Signaling Technology, Beverly, MA, USA). Horseradish peroxidase-conjugated (HRP) anti-rabbit antibodies (#e62238, 1:5,000; Santa Cruz Biotechnology) were used as the secondary antibodies. A total of 25 μg protein from each sample was separated on 10% Bis-Tris polyacrylamide gel through electrophoresis and then blotted onto polyvinylidene fluoride (PVDF) membranes (GE Healthcare, Piscataway, NJ, USA). The membranes were then blocked with 5% (5 g/100 ml) non-fat dry milk (Bio-Rad, Hercules, CA, USA) in Tris-buffered saline plus Tween (TBS-T) buffer for 2 h. The blots were immunostained with primary antibody at 4°C overnight and with secondary antibody at room temperature for 1 h. Immunoblots were visualized using Immobilon™ Western Chemiluminescent HRP Substrate (Millipore). Protein levels were normalized to β-actin.
Statistical analysis
The Kolmogorov-Smirnov test was used to determine the normality of the distribution of data in each group. For breast cancer vs. normal cell lines, differences are shown as the median expression and were determined using the Mann-Whitney U test or Kruskal-Wallis test. Count data were described as frequency and examined using Fisher’s exact test. The results were considered statistically significant at P<0.05. Error bars in the figures represent the means ± standard deviation (SD). Statistical analyses were performed using GraphPad Prism (version 5.01; GraphPad Software, Inc., La Jolla, CA, USA) software.
Results
Analysis of circRNA expression profiles
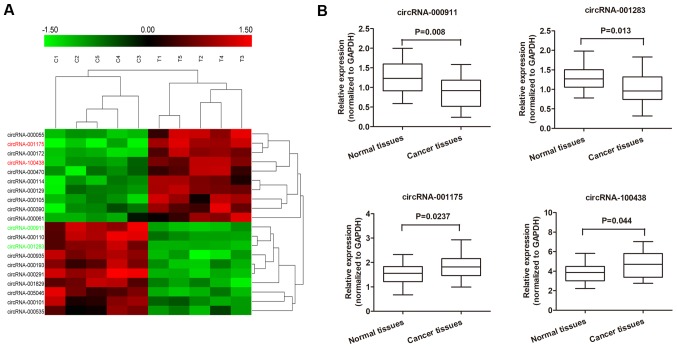
To identify specific circRNAs that are differentially expressed between patients with breast cancer and normal subjects, 5 tissues samples and control non-tumor tissue samples pooled from patients with breast cancer were subjected for circRNA microarray assay. A total of 716 circRNAs that were differently expressed by >1.5-fold between the breast cancer tissues and the normal tissues were identified. Subsequently, we narrowed the scope of the analysis to the 20 most aberrantly expressed circRNAs, including 10 upregulated circRNAs and 10 downregulated circRNAs (shown in the heatmap in Fig. 1A). We then detected the expression of these 20 mostly significantly altered circRNAs in breast cancer and matched non-tumor tissue samples from 30 patients to confirm their expression. As shown in Fig. 1B, 4 circRNAs were found to be significantly deregulated between the breast cancer and paired non-cancerous tissues (P<0.05), including 2 upregulated circRNAs, circRNA- 001175 and circRNA-100438, and 2 downregulated circRNAs, circRNA-000911 and circRNA-001283.
Figure 1.
Deregulated circular RNAs (circRNAs) in breast cancer tissues. (A) The heatmap shows the top 10 most significantly increased and decreased circRNAs in breast cancer tissues as compared to those in the matched non-tumor tissues analyzed by circRNAs ArrayStar Chip. (B) Relative expression of the 4 indicated circRNAs from 30 breast cancer tissues and adjacent non-tumor tissues listed in (A) measured by RT-qPCR.
circRNA-000911 plays an anti-oncogenic role in breast cancer cells
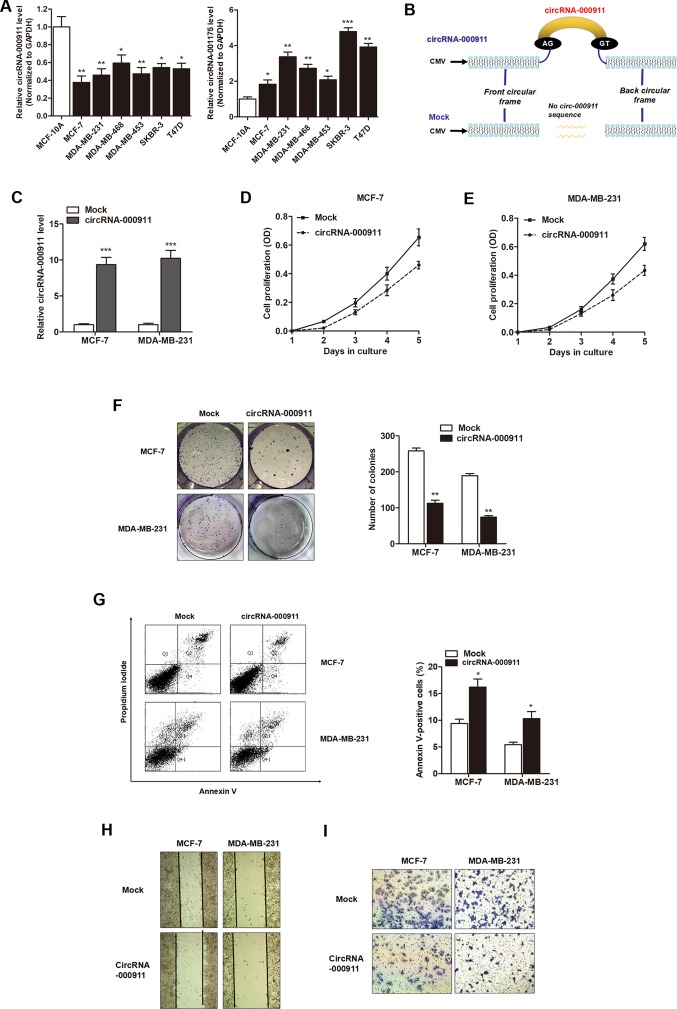
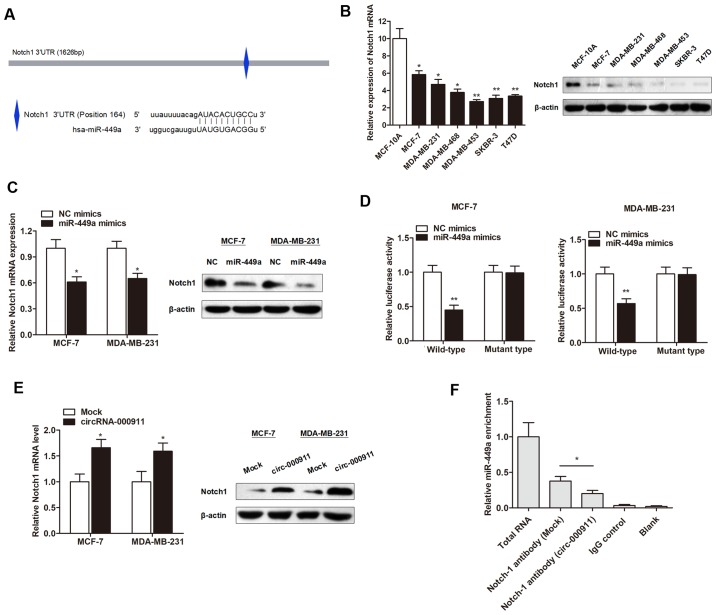
The expression of the 4 circRNAs were then validated in 6 human breast cancer cell lines and one normal human breast epithelial cells to validate their expression. RT-qPCR analysis revealed that only circRNA-000911 and cirRNA-001175 maintained their statistical significance between the breast cancer cell lines and normal breast epithelial cells (Fig. 2A). However, our preliminary results showed that the silencing of cirRNA-001175 had no significant effect on breast cancer cell viability and migration, and the aberrant expression of circRNA-000911 had a significant suppressive effect on cell viability (data not shown). This inspired us to focus on the biological significance of circRNA-000911 in breast cancer progression.
Figure 2.
Circular RNA (circRNA)-000911 plays an anti-oncogenic role in breast cancer cells. (A) The expression levels of circRNA-000911 and cir-cRNA-001175 in 6 breast cancer cell lines and one normal breast epithelial cell line, MCF-10A. (B) A sketch of the structures of circRNA-000911 and mock vector is presented shown. (C) circRNA-000911 was upregulated by transfection with the circRNA vector. (D and E) circRNA-000911 significantly suppressed the proliferation rate of the (D) MCF-7 and (E) MDA-MB-231 cells. (F) The overexpression of circRNA-000911 significantly suppressed the colony formation capacity of both breast cancer cell lines. (G) FACS apoptosis assay indicated that circRNA-000911 increased the proportion of apoptotic cells in both breast cancer cell lines. (H) Wound healing and (I) Matrigel invasion assay suggested that the enhanced expression of circRNA-000911 suppressed the migratory and invasive abilities of the breast cancer cells. *P<0.05, **P<0.01 and ***P<0.001 vs. mock group.
The effect of circRNA-000911 on breast cancer progression was then investigated. The MCF-7 and MDA-MB-231 cells were used for further gain- and loss-of-function assays as the expression of circRNA-000911 was relatively lower in these two cell lines in contrast to the other breast cell lines. The full-length cDNA of circRNA-000911 from the MCF-7 cells was amplified and cloned into the specific vector (Fig. 2B). The results of RT-qPCR revealed that the circRNA-000911 vector significantly increased the expression level of circRNA-000911 in the MCF-7 and MDA-MB-231 cells (Fig. 2C). Subsequently, the effects of circRNA-000911 on breast cancer cell proliferation, apoptosis, invasion and migration were examined. CCK-8 assay revealed that the overexpression of circRNA-000911 significantly suppressed cell proliferation when compared with the control vector in both cell lines (Fig. 2D and E). Similarly, circRNA-000911 inhibited the colony formation ability, and increased the proportion of apoptotic cells (Fig. 2F and G). In addition, circRNA-000911 significantly inhibited the wound healing ability of the breast cancer cells (Fig. 2H). Matrigel invasion assay also revealed that the overexpression of circRNA-000911 noticeably suppressed the invasive capabilities of both cell lines (Fig. 2I).
circRNA-000911 functions as a miRNA sponge for miR-449a in breast cancer cells
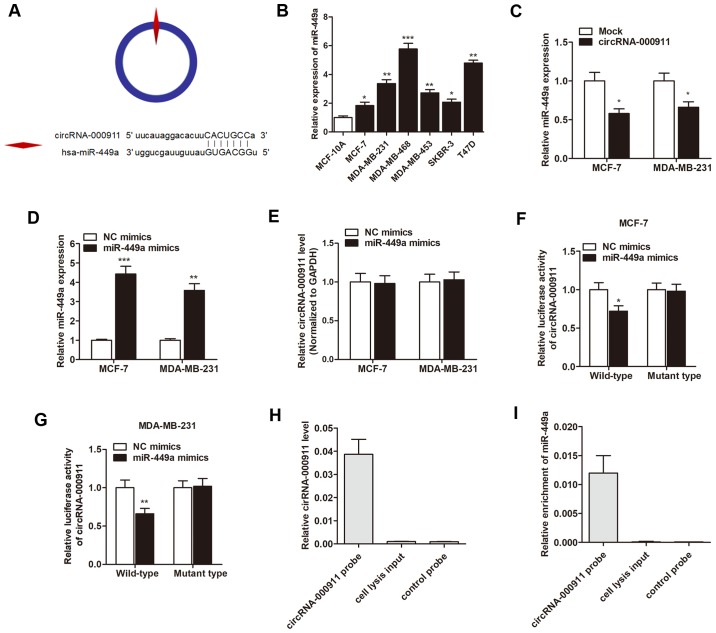
As circRNAs function mainly as miRNA sponges to bind to functional miRNAs and then regulate gene expression, we then examined the potential miRNAs associated with circRNA-000911. According to miRBase prediction, circRNA-000911 possessed a complementary sequence to the miR-449a seed region (Fig. 3A). The results of RT-qPCR also revealed that miR-449a was significantly upregulated in the breast cancer cell lines (Fig. 3B). Moreover, the enhanced expression of circRNA-000911 significantly suppressed miR-449a expression in breast cancer cells (Fig. 3C); however, the overexpression of miR-449a with miR-449a mimics (Fig. 3D) failed to affect the cirRNA-000911 levels (Fig. 3E). Subsequently, luciferase reporter assay was performed to verify that miR-449a directly interacted with circRNA-000911. As shown in Fig. 3F and G, miR-449a significantly inhibited the luciferase activity of the wild-type reporter of cirRNA-000911; however, miR-449a did not inhibit the luciferase activity of the reporter vector containing the mutant binding sites of cirRNA-000911. Finally, we used a cirRNA-000911 specific probe to perform RNA precipitation (RIP). The cirRNA-000911-associated RNAs were purified by the cirRNA-000911 specific probe, and the screened miRNAs were then pulled down by cirRNA-000911 using RT-qPCR assay. As expected, we found a specific enrichment of cirRNA-000911 and miR-449a as compared to the controls (Fig. 3H and I), indicating that cirRNA-000911 specifically interacts with miR-449a in breast cancer cells.
Figure 3.
Circular RNA (circRNA)-000911 acts as a miRNA sponge for miR-449a in breast cancer cells. (A) The putative sequences of miR-449a and cir-cRNA-000911 with one binding site. (B) miR-449a was upregulated in the 6 breast cancer cell lines when compared with the normal breast epithelial cell line, MCF-10A. (C) miR-449a was downregulated in the breast cancer cells that were transfected with the circRNA-000911 vector. (D) miR-449a was markedly upregulated by transfection with specific miR-449a mimics. (E) miR-449a had no effect on the expression level of circRNA-000911. (F and G) miR-449a significantly inhibited the luciferase activity of the wild-type reporter of cirRNA-000911; however, miR-449a did not inhibit the luciferase activity of the reporter vector containing the mutant binding sites of cirRNA-000911 in both cell lines. (H) circRNA-000911 in MCF-7 cell lysis was pulled down and enriched with a circRNA-000911 specific probe and then detected by RT-qPCR. (I) miR-449a was pulled down and enriched with a circRNA-000911 specific probe and then detected by RT-qPCR. *P<0.05, **P<0.01 and ***P<0.001 vs. the respective control.
miR-449a antagonizes circRNA-000911 to regulate breast cancer progression
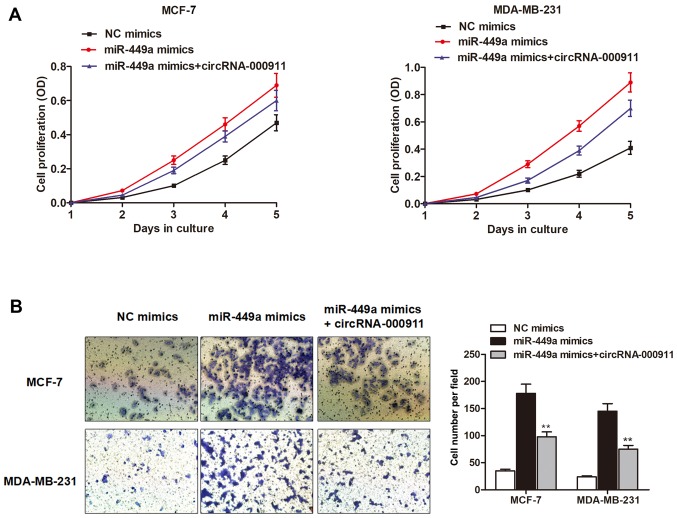
Given that circRNA-000911 serves as a sponge of miR-449a and that circRNA-000911 plays an anti-oncogenic role in breast cancer, we sought to determine whether miR-449a counteracted circRNA-000911 to promote cell progression. We first investigated the functional role of miR-449a in breast cancer cell viability. The results of CCK-8 assay indicated that miR-449a markedly promoted the growth of both the MCF-7 and MDA-MB-231 cells (Fig. 4A). To determine whether miR-449a counteracts the function of circRNA-000911, we co-transfected a circRNA-000911 overexpression vector with miR-449 mimics into the cells. As shown in Fig. 4A, the enhanced cell proliferation rate was significantly abrogated by the co-expression of circRNA-000911. In addition, Matrigel invasion assay indicated that miR-449a promoted the invasive capacity of the breast cancer cells; however, circRNA-000911 suppressed the miR-449a-induced increase in the invasive capacity of the breast cancer cells (Fig. 4B).
Figure 4.
miR-449a antagonizes circular RNA (circRNA)-000911 to regulate breast cancer progression. (A) Cell counting kit-8 (CCK-8) assay indicated that miR-449a markedly promoted the proliferation of both MCF-7 and MDA-MB-231 cells; however, the enhanced cell proliferation rate was significantly abrogated by the co-expression of circRNA-000911. (B) Matrigel invasion assay indicated that miR-449a promoted the invasive capacity of the breast cancer cells; however, circRNA-000911 suppressed the miR-449a-induced increased invasive capacity of the breast cancer cells. **P<0.01 vs. mimics group.
Notch1 is a direct target of miR-449a and positively regulated by circRNA-000911
It is well accepted that miRNAs perform their functions mainly through the direct targeting of mRNAs, and we then aimed to identify the downstream target of miR-449a. On the basis of two major prediction software databases, TargetScan and miRanda, the putative binding sites of miR-449a in the 3’UTR of Notch1 were predicted (Fig. 5A). The results of RT-qPCR and western blot analyses suggested that the Notch1 gene was downregulated in the breast cancer cells compared to the normal breast epithelial cells at both the mRNA transcript and protein levels (Fig. 5B). Moreover, miR-449a markedly inhibited the mRNA and protein expression level of Notch1 in both cell lines (Fig. 5C). To further identify whether Notch1 in breast cancer cells responded to miR-449a through direct interactions with its 3′UTR, we cloned the wild-type or mutant type 3′UTR of the putative miR-449a target into the reporter plasmid containing the luciferase gene. The dual luciferase reporter assay revealed that miR-449a attenuated the fluorescence driven by the wild-type 3′UTR by >1.5-fold compared with the negative control, whereas the mutant 3′UTR was not affected by miR-449a (Fig. 5D). This clearly confirmed that miR-449a targeted the Notch1 gene via a direct interaction with the 3′UTR region. Given that miR-449a was sponged by circRNA-000911, we then examined the association between circRNA-000911 and the Notch1 gene. As expected, the overexpression of circRNA-000911 significantly increased the Notch1 expression level in the breast cancer cells (Fig. 5E). Furthermore, RIP revealed that the enrichment of Notch1 and miR-449a was significantly decreased in the MCF-7 cells transfected with circRNA-000911 (Fig. 5F), suggesting that circRNA-000911 positively regulates Notch1 expression by sponging miR-449a.
Figure 5.
Notch1 is a direct target of miR-449a and positively regulated by circRNA-000911. (A) The putative sequences of miR-449a and Notch1 with one binding site. (B Notch1 was downregulated in breast cell lines at both the mRNA transcript and protein level. (C) Notch1 mRNA and protein levels were downregulated by miR-449a. (D) miR-449a significantly inhibited the luciferase activity of the wild-type reporter for Notch1; however, miR-449a did not inhibit the luciferase activity of the reporter vector containing the mutant binding sites of Notch1 in both cell lines. (E) Notch1 expression was upregulated by transfection with circRNA-000911 expression vector. (F) RIP assays were performed using the Notch1 antibody to immunoprecipitate RNA and a primer to detect miR-449a, and a significantly decreased enrichment of miR-449a was identified in cells transfected with the circRNA-000911 vector. *P<0.05 and **P<0.01 vs. respective control.
NF-κB signaling is identified as a functional target of circRNA-000911/miR-449a pathway
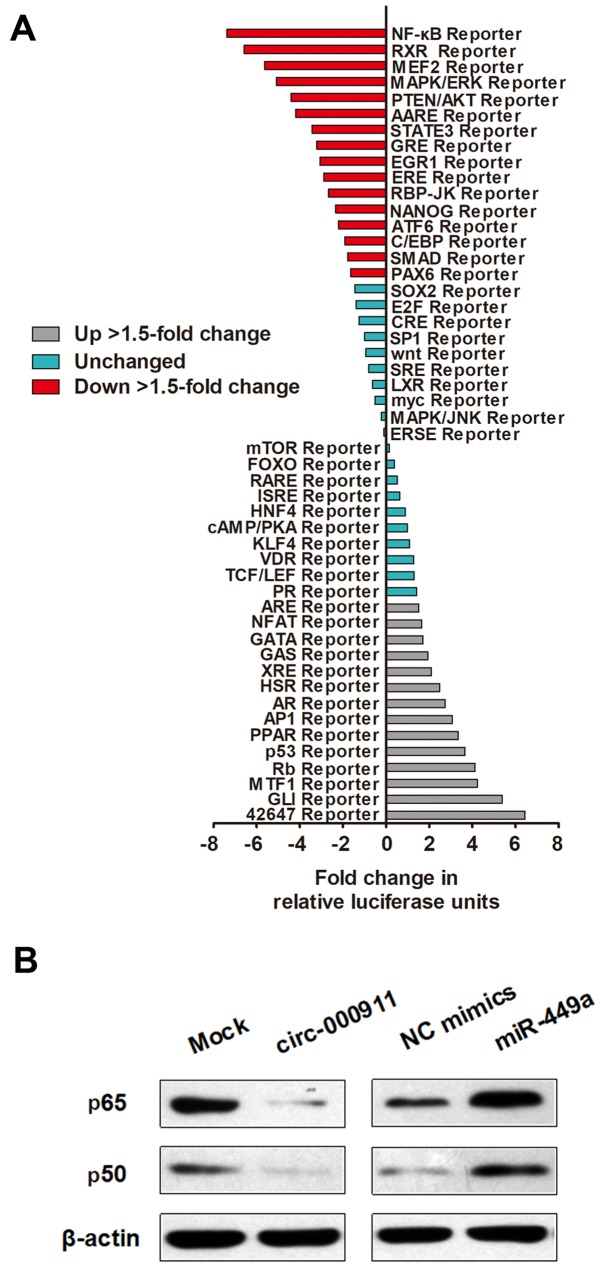
To identify the downstream signaling pathway that may account for the above-mentioned observations, we used Cignal Signal Transduction Reporter Array to simultaneously investigate the activities of 50 canonical signaling pathways upon circRNA-000911 overexpression in MCF-7 cells. Of note, we identified NF-κB signaling as one of the most significantly repressed pathways upon circRNA-000911 overexpression (Fig. 6A). More importantly, the NF-κB signaling pathway participates in the regulation of proliferation, adhesion and invasion in breast cancer, and it has been well accepted that there are functional interactions between Notch1 and NF-κB signaling pathway (21,22). Herein, we sought to determine whether the NF-κB pathway is responsible for the circRNA-000911/miR-449a induced anti-oncogenic role. As expected, the results of western blot analysis revealed that NF-κB signaling was silenced by the overexpression of circRNA-000911 and was promoted by transfection with miR-449a mimics (Fig. 6B).
Figure 6.
Nuclear factor-κB (NF-κB) signaling is identified as a functional target of the circRNA-000911/miR-449a pathway. (A) Histogram shows the fold changes in the activities of different signaling pathways, as indicated by reporter activity. (B) Western blot analysis revealed that the NF-κB signaling pathway (p65 and p50) was silenced by the overexpression of cir-cRNA-000911 and promoted by miR-449a mimics.
Discussion
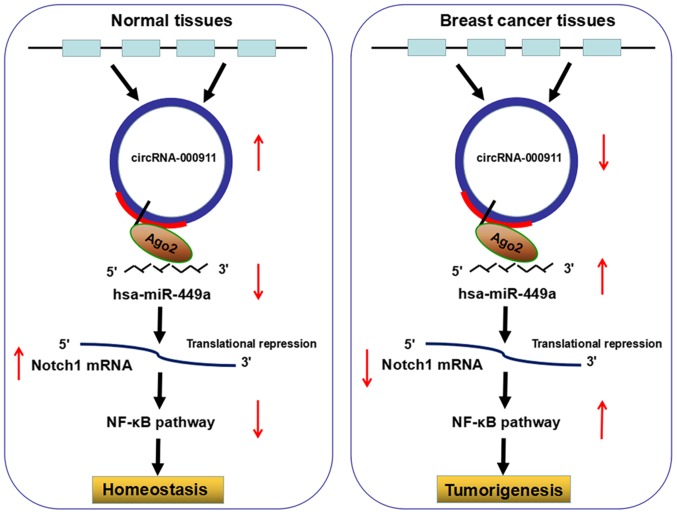
Breast cancer is a highly heterogeneous disease whose underlying pathogenic mechanisms are being increasingly elucidated. Early diagnosis, radical surgery and adjuvant therapy have improved the survival rates and prognosis of patients with breast cancer; however, the mortality rates remain high (23). In this study, we focused on a novel gene regulator, circRNA, and identified the downregulation of circRNA-000911 in breast cancer cells. Mechanistic analysis indicated that circRNA-000911 suppressed the proliferative, migratory and invasive capacity of the breast cancer cells by specifically sponging miR-449a and releasing Notch1. This process commonly accompanied the participation of Ago2, a protein essential for sponge activity (24). Eventually, downstream NF-κB signaling was also identified as the direct functional pathway that may account for the regulatory role of circRNA-000911/miR-449a line in breast cancer progression (Fig. 7).
Figure 7.
Schematic representation of the proposed mechanism of circRNA-000911 in breast cancer cells. Circular RNA (circRNA)-000911 acted as a miR-449a sponge to regulate the miR-449a/Notch1/nuclear factor-κB (NF-κB) pathway in breast cancer cells. Decreased circRNA-000911 expression in breast cancer increased the activity of miR-449a, which downregulated the expression of Notch1 and upregulated the downstream effectors, thereby promoting breast tumorigenesis and progression.
The concept of ‘circular RNA’ was first proposed in 1976 by Sanger et al, who found that viroids are single-stranded covalently closed circRNA molecules pathogenic to certain higher plants (25). With the development of gene investigations, it is recognized that circRNAs are widely expressed in human cells, and their expression levels can be ≥10-fold higher compared to their linear isomers (26). The two most important properties of circRNAs are highly conserved sequences and a high degree of stability in mammalian cells (27). Compared with other non-coding RNAs, such as miRNAs and long non-coding RNAs (lncRNAs), these properties provide circRNAs with the potential to become ideal biomarkers in the diagnosis and prognosis of cancers (12,28). To date, only a few circRNAs have been explored. They may serve as oncogenes, tumor suppressor genes, or both, depending on the circumstances. For example, circRNA-MYLK, circRNA-ABCB10 and circRNA-0067934 have been reported to promote cell migration and growth (10,17,29), whereas circRNA-MTO1, circRNA-0003570 and circRNA-LARP4 have been shown to suppress tumor progression via multiple signaling pathways (16,30,31). In this study, we identified one circRNA, circRNA-000911, via microarray sequences. Firstly, circRNA-000911 was significantly downregulated in breast cancer tissues in contrast to paired non-cancerous tissues. The validation array by RT-qPCR further confirmed its expression mode in breast cancer cells.
Taking a step further, we investigated the functional role of circRNA-000911 in breast cancer. The overexpression of circRNA-000911 suppressed cell growth, migration and invasion, while it promoted the apoptosis of breast cancer cells. Han et al found that circRNA-000911 may be downregulated in hepatocellular carcinoma by performing tissue microarray experiments (16). In another study by Liang et al, it was reported that circRNA-000911 was also downregulated in primary breast cancer tissues by tissue microarray; however, the subsequent validation array failed to validate its expression trend in larger clinical and experimental materials (29). To the best of our knowledge, this is the first study to identify the expression and functional role of circRNA-000911 in breast cancer. Our data suggest that circRNA-000911 may serve as a tumor suppressor gene in breast cancer.
We sought to define how circRNA-000911 performs its functions. miRNAs, an abundant class of small non-coding RNAs (~22 nt in length), post-transcriptionally modulate the translation of target mRNAs via corresponding miRNA response elements (MREs) (12). Computational searches for miRNA target sites in circRNAs have identified a portion of circRNA molecules that contain MREs, which may act as miRNA sponges, reducing miRNA binding to target genes, thereby releasing the expression of the miRNA targets indirectly (32–35). Since the first report of circRNAs functioning as miRNA sponges, the potential of circRNAs in regulating cancer-related genes through fine-tuning miRNAs has recently been recognized. For example, the first characterized circRNA to support this model was ciRS-7, which contains >60 miR-7-binding sites, thereby acting as an endogenous miRNA sponge to adsorb and subsequently quench normal miR-7 functions (36,37). More recently, an increasing number of circRNAs have been recognized as miRNA sponges in different types of cancer, such as colorectal cancer, gastric cancer and hepatocellular carcinoma (38). However, reports of this model in breast cancer are very limited. Tang et al revealed that circRNA-001982 promotes breast cancer cell carcinogenesis by decreasing miR-143 expression (39); Liang et al suggested that circRNA-DENND4C promoted the proliferation of breast cancer cells in a hypoxic environment (40). In this study, miR-449a showed a complementary sequence to circRNA-000911 based on bioinformatics analysis, and this miRNA was finally identified as the endogenous competing RNA by luciferase reporter assay and RIP assay. The role of miR-449a in cancer progression is contradictory in different cancer types. Chen et al found that miR-449a suppressed epithelial-mesenchymal transition and the metastasis of hepatocellular carcinoma cells via multiple targets (41); Li et al demonstrated that miR-449a inhibited the malignant progression of nasopharyngeal carcinoma by targeting lactate dehydrogenase A (42). However, the study by You et al indicated that miR-449a suppressed cell invasion by inhibiting MAP2K1 in non-small cell lung cancer (43). As regards the role of miR-449a in breast cancer, studies on this are limited. Shi et al found that miR-449a was implicated functionally in breast cancer pathogenesis, suppressing cysteine-rich protein 2 (CRIP2) and altering cell viability, migration, invasion, in vivo tumor growth and angiogenesis, thereby driving malignant phenotypes (44). In our study, miR-449a promoted cell viability and the invasive ability of breast cancer cells. Moreover, our gain- and loss-of-function assays clearly indicated that miR-449a reversed the circRNA-000911-induced tumor suppressive function, indicating that circRNA-000911 may play an anti-oncogenic role through the sponge activity of miR-449a. Furthermore, the Notch1 gene was then identified as a direct target of miR-449a by performing bioinformatics analysis and subsequent functional validation.
Finally, we sought to define the direct downstream signaling pathways that are regulated by circRNA-000911/miR-449a pathway. We used a Cignal Signal Transduction Reporter Array. This array involved a mixture of a pathway-specific transcription factor-responsive firefly luciferase reporter, which contains a specific transcription factor-responsive element in the promoter, and a constitutively expressed Renilla luciferase reporter, which were co-transfected to monitor alternations in the activity of that signaling pathway. This high-throughput dual-luciferase assay led us to identify the NF-κB pathway as a pathway putatively affected by circRNA-000911. More importantly, there is a tight interaction between Notch1 and NF-κB pathway in a number of diseases, including cancer. The results from western blot analysis also revealed that circRNA-000911 suppressed, whereas miR-449a promoted the expression of related proteins in the NF-κB signaling pathway, which further confirmed the interaction between circRNA-000911/miR-449a and NF-κB pathway.
In conclusion, in this study, and to the best of our knowledge, we report for the first time that circRNA-000911 plays an anti-oncogenic role in breast cancer. Moreover, circRNA-000911 exerts its function by serving as a miRNA sponge for miR-449a and thereby promoting the function of Notch1 and the NF-κB signaling pathway. Therefore, circRNA-000911 may serve as a promising predictive biomarker and therapeutic target for patients with breast cancer.
Acknowledgments
Not applicable.
Footnotes
Funding
This study was supported by the Science Foundation of First Hospital of Lanzhou University.
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Authors’ contributions
HW mainly designed and did the research. YX analyzed the data and LW and DM help to collect the data from the patients. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The study protocol was approved by the Clinical Research Ethics Committee of the First Hospital of Lanzhou University. Written informed consent was obtained from each participant prior to tissue collection.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Zhang B, Beeghly-Fadiel A, Long J, Zheng W. Genetic variants associated with breast-cancer risk: Comprehensive research synopsis, meta-analysis, and epidemiological evidence. Lancet Oncol. 2011;12:477–488. doi: 10.1016/S1470-2045(11)70076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodgers RJ, Reid GD, Koch J, Deans R, Ledger WL, Friedlander M, Gilchrist RB, Walters KA, Abbott JA. The safety and efficacy of controlled ovarian hyperstimulation for fertility preservation in women with early breast cancer: A systematic review. Hum Reprod. 2017;32:1033–1045. doi: 10.1093/humrep/dex027. [DOI] [PubMed] [Google Scholar]
- 4.Chen W, Zheng R, Zeng H, Zhang S. The updated incidences and mortalities of major cancers in China, 2011. Chin J Cancer. 2015;34:502–507. doi: 10.1186/s40880-015-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mulrane L, McGee SF, Gallagher WM, O’Connor DP. miRNA dysregulation in breast cancer. Cancer Res. 2013;73:6554–6562. doi: 10.1158/0008-5472.CAN-13-1841. [DOI] [PubMed] [Google Scholar]
- 6.Sidiropoulos KG, Ding Q, Pampalakis G, White NM, Boulos P, Sotiropoulou G, Yousef GM. KLK6-regulated miRNA networks activate oncogenic pathways in breast cancer subtypes. Mol Oncol. 2016;10:993–1007. doi: 10.1016/j.molonc.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorek R, Cossart P. Prokaryotic transcriptomics: A new view on regulation, physiology and pathogenicity. Nat Rev Genet. 2010;11:9–16. doi: 10.1038/nrg2695. [DOI] [PubMed] [Google Scholar]
- 8.Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340:440–441. doi: 10.1126/science.1238522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-β1. Biochem Biophys Res Commun. 2017;487:769–775. doi: 10.1016/j.bbrc.2017.04.044. [DOI] [PubMed] [Google Scholar]
- 10.Xia W, Qiu M, Chen R, Wang S, Leng X, Wang J, Xu Y, Hu J, Dong G, Xu PL, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576. doi: 10.1038/srep35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: A novel type of biomarker for cancer. Breast Cancer. Breast Cancer. 2018;25:1–7. doi: 10.1007/s12282-017-0793-9. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qu S, Zhong Y, Shang R, Zhang X, Song W, Kjems J, Li H. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14:992–999. doi: 10.1080/15476286.2016.1220473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y, Alexandrov PN, Jaber V, Lukiw WJ. Deficiency in the ubiquitin conjugating enzyme UBE2A in Alzheimer’s disease (AD) is linked to deficits in a natural circular miRNA-7 sponge (circRNA; ciRS-7) Genes (Basel) 2016;7:116. doi: 10.3390/genes7120116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 17.Zhong Z, Huang M, Lv M, He Y, Duan C, Zhang L, Chen J. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi: 10.1016/j.canlet.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 18.Zheng J, Deng J, Xiao M, Yang L, Zhang L, You Y, Hu M, Li N, Wu H, Li W, et al. A sequence polymorphism in miR-608 predicts recurrence after radiotherapy for nasopharyngeal carcinoma. Cancer Res. 2013;73:5151–5162. doi: 10.1158/0008-5472.CAN-13-0395. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Su X, Wang H, Ge W, Yang M, Hou J, Chen T, Li N, Cao X. An in vivo method to identify microRNA targets not predicted by computation algorithms: p21 targeting by miR-92a in cancer. Cancer Res. 2015;75:2875–2885. doi: 10.1158/0008-5472.CAN-14-2218. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Zhang J, Xiong N, Li S, Chen Y, Yang H, Wu C, Zeng H, Liu Y. Notch-1 signaling activates NF-κB in human breast carcinoma MDA-MB-231 cells via PP2A-dependent AKT pathway. Med Oncol. 2016;33:33. doi: 10.1007/s12032-016-0747-7. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Su C, Shan Y, Yang S, Ma G. Targeting Notch1 inhibits invasion and angiogenesis of human breast cancer cells via inhibition nuclear factor-κB signaling. Am J Transl Res. 2016;8:2681–2692. [PMC free article] [PubMed] [Google Scholar]
- 23.Krell J, Stebbing J, Frampton AE, Carissimi C, Harding V, De Giorgio A, Fulci V, Macino G, Colombo T, Castellano L. The role of TP53 in miRNA loading onto AGO2 and in remodelling the miRNA-mRNA interaction network. Lancet. 2015;385(Suppl 1):S15. doi: 10.1016/S0140-6736(15)60330-0. [DOI] [PubMed] [Google Scholar]
- 24.Cuzick J, Sestak I, Thorat MA. Impact of preventive therapy on the risk of breast cancer among women with benign breast disease. Breast. 2015;24(Suppl 2):S51–S55. doi: 10.1016/j.breast.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 28.Heneghan HM, Miller N, Kerin MJ. MiRNAs as biomarkers and therapeutic targets in cancer. Curr Opin Pharmacol. 2010;10:543–550. doi: 10.1016/j.coph.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7:1566–1576. [PMC free article] [PubMed] [Google Scholar]
- 30.Fu L, Wu S, Yao T, Chen Q, Xie Y, Ying S, Chen Z, Xiao B, Hu Y. Decreased expression of hsa_circ_0003570 in hepatocellular carcinoma and its clinical significance. J Clin Lab Anal. 2017 May 11; doi: 10.1002/jcla.22239. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Liu H, Hou L, Wang G, Zhang R, Huang Y, Chen X, Zhu J. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16:151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 33.Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14:514–521. doi: 10.1080/15476286.2015.1122162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He J, Xie Q, Xu H, Li J, Li Y. Circular RNAs and cancer. Cancer Lett. 2017;396:138–144. doi: 10.1016/j.canlet.2017.03.027. [DOI] [PubMed] [Google Scholar]
- 37.Peng L, Yuan XQ, Li GC. The emerging landscape of circular RNA ciRS-7 in cancer (Review) Oncol Rep. 2015;33:2669–2674. doi: 10.3892/or.2015.3904. [DOI] [PubMed] [Google Scholar]
- 38.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 39.Tang YY, Zhao P, Zou TN, Duan JJ, Zhi R, Yang SY, Yang DC, Wang XL. Circular RNA hsa_circ_0001982 promotes breast cancer cell carcinogenesis through decreasing miR-143. DNA Cell Biol. 2017;36:901–908. doi: 10.1089/dna.2017.3862. [DOI] [PubMed] [Google Scholar]
- 40.Liang G, Liu Z, Tan L, Su AN, Jiang WG, Gong C. HIF1α-associated circDENND4C promotes proliferation of breast cancer cells in hypoxic environment. Anticancer Res. 2017;37:4337–4343. doi: 10.21873/anticanres.11827. [DOI] [PubMed] [Google Scholar]
- 41.Chen SP, Liu BX, Xu J, Pei XF, Liao YJ, Yuan F, Zheng F. MiR-449a suppresses the epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma by multiple targets. BMC Cancer. 2015;15:706. doi: 10.1186/s12885-015-1738-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Li X, Ge X, Jia L, Zhang Z, Fang R, Yang J, Liu J, Peng S, Zhou M, et al. MiR-34b-3 and miR-449a inhibit malignant progression of nasopharyngeal carcinoma by targeting lactate dehydrogenase A. Oncotarget. 2016;7:54838–54851. doi: 10.18632/oncotarget.10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.You J, Zhang Y, Li Y, Fang N, Liu B, Zu L, Zhou Q. MiR-449a suppresses cell invasion by inhibiting MAP2K1 in non-small cell lung cancer. Am J Cancer Res. 2015;5:2730–2744. [PMC free article] [PubMed] [Google Scholar]
- 44.Shi W, Bruce J, Lee M, Yue S, Rowe M, Pintilie M, Kogo R, Bissey PA, Fyles A, Yip KW, et al. MiR-449a promotes breast cancer progression by targeting CRIP2. Oncotarget. 2016;7:18906–18918. doi: 10.18632/oncotarget.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]