Abstract
The major causative agent of cervical cancer is human papilloma virus (HPV); the viral proteins E6 and E7 induce carcinogenesis through the inactivation of the host tumor-suppressor gene. Therefore, the stable expression of specific inhibitors of E6 and E7 in cancer cells is expected to provide effective treatment for cervical cancer without affecting normal tissue. In this study, we propose a novel therapeutic approach using an adeno-associated virus (AAV) vector encoding short hairpin RNA (shRNA) against the onco-proteins E6 and E7 (shE6E7) of HPV type 16 (HPV-16), termed AAV-shE6E7. Three different HPV-16-positive cervical cancer cell lines (BOKU, SiHa and SKG-IIIa cells) were tested for gene transfer efficiency using serotypes of AAV vectors. For in vitro analysis, the cells were transduced AAV-shE6E7; alternatively, in vivo studies were performed via the administration of a direct injection of AAV-shE6E7 into cervical cancer cell-derived tumors in mice. The high gene transfer efficiency was observed using AAV2 in all three cervical cancer cell lines. Following transduction, we observed apoptosis, G1 phase arrest and cell growth inhibition. Additionally, in the transduced cells, the E6, E7 and p16 expression levels decreased, whereas the expression levels of p53, p21 and pRb levels were enhanced. The growth of subcutaneously transplanted tumors was markedly inhibited by the single administration of AAV2-shE6E7, and the tumors were almost completely eradicated without any adverse effects. These results provided evidence of the utility of AAV2-shE6E7 as a novel treatment approach for cervical cancer.
Keywords: cervical cancer, adeno-associated virus vector, short hairpin RNA, human papillomavirus, E6, E7, p53, Rb, apoptosis, gene transfer efficiency
Introduction
Cervical cancer is the third most frequent cause of cancer-related mortality among women worldwide (1). Over 500,000 cases of cervical cancer were reported in 2015, with approximately one third of those cases resulted in death (1). Advanced cervical cancer is treated with surgery, radiotherapy, chemotherapy, or a combination of these modalities; however, the prognosis of patients with cervical cancer remains poor, and major improvements in outcomes have not been reported over the past 30 years (2).
Cervical cancer generally develops from a pre-cancerous cervical intraepithelial neoplasm (CIN) (3). At present, early-stage cervical cancers, including high-grade CIN and early infiltrating cancer, are only surgically treated using cervical conization and total hysterectomy (4,5). Nonetheless, the incidence of complications, such as infertility, miscarriage and premature delivery, is high in patients treated with conization deu to the shortening of the uterine cervix and sensitivity to intra-uterine infection. Therefore, alternative modes of therapy are required in order to avoid these complications (6).
Carcinogenesis of the cervix is triggered by infection with high-risk human papillomavirus (HPV) (3,7,8). High-risk HPV is detected in >90% of cervical cancer cases, and HPV type 16 (HPV-16) accounts for approximately half of these (9). When high-risk HPV continuously infects basal cells of the cervical epithelium and is integrated into the host chromosome, HPV oncoproteins E6 and E7 are overexpressed and inactivate the tumor suppressor genes p53 and Rb, thereby inducing cancer (10–12). Therefore, cervical cancer may be resolved by inhibiting the expression of E6 and E7. The expression of E6 and E7 is localized in lesions, and these proteins are not expressed in normal tissues, including the cervix (13), thereby limiting the effects of specific inhibition of E6/E7 to cancerous lesions.
RNA interference (RNAi) is a superior method of inhibiting gene expression following transcription (14). Among various RNAi techniques, the specificity of RNAi using short small interfering RNA (siRNA) for the target gene is very high, and the inhibitory action of siRNA is potent, but transient (15,16), limiting its clinical application in the treatment of diseases (17). Polymerase III promoter-driven short hairpin RNA (shRNA) has recently been developed as an alternative strategy capable of stably inhibiting target gene expression for longer periods of time, and its effect has been widely recognized (18). Therefore, E6 and E7 of high-risk HPV may be strongly and persistently inhibited using shRNA. However, the application of this strategy in the treatment of cervical cancer would require a vector that efficiently transfers the genes into target cervical cancer cells.
Adeno-associated virus (AAV) vectors are derived from non-pathogenic viruses and can transfer genes into non-dividing cells (19). Many serotypes have been developed as vectors, and specific tissue and organ tropisms have been reported (20). Moreover, gene transfer into a broad range of cells by direct administration and long-term protein expression have been observed in humans; indeed, AAV vectors are one of the most prominent viral vectors used to facilitate gene therapy (21). Therefore, in the present study, we aimed to develop a highly specific, novel treatment for cervical cancer using an AAV vector containing hpV-16 E6/E7-targeting shRNA (AAV-shE6E7).
Materials and methods
Cell culture
The HPV-16-positive human cervical cancer cell lines, BOKU and SKG-IIIa, and the human immortalized cell line, 293, were purchased from the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan). The HPV-16-positive human cervical cancer cell line, SiHa, was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). All cell lines were authenticated by the above-mentioned cell banks by means of short-tandem repeat-polymerase chain reaction (pCR) profiling. The cells were cultured in Dulbecco’s modified Eagle’s medium/F12 (life Technologies, Carlsbad, CA, USA) containing 10% inactivated fetal bovine serum (Sigma-Aldrich, St. Louis, MO, USA) and 1% penicillin/streptomycin (life Technologies) in 5% carbon dioxide at 37°C.
Efficiency of gene transfer into cervical cancer cells using different serotypes of AAV vectors
The BOKU, SiHa and SKG-IIIa cells were seeded in 96-well plates at a density of 1×103 cells/well. After 12 h, the tumor cells were transduced with AAV vectors containing the LacZ gene encoding β-galactosidase (AAV-CMV-LacZ) types 1–9 (22–25) at 3×105 viral genomes (vg)/cell. After 48 h, β-galactosidase expression was measured using a β-galactosidase staining kit (Cell Signaling Technology, Danvers, MA, USA) according to the manufacturer’s instructions.
Construction of a plasmid vector containing shE6E7
The U6 promoter and siRNA sequences were cut out from the PvuII site of the pSilencer U6 vector (Thermo Fisher Scientific, Waltham, MA, USA) and inserted into the blunted MluI site of pAAV-hrGFP (Agilent Technologies, Santa Clara, CA, USA) to prepare pu6si-CmV-green fluorescent protein (gFp). The target site for the shRNA was the following 21-base sequence (564–582) in HPV-16 E6/E7 mRNA, taken from a previously published study (26): sense, 5′-GATCCGCATGGAGATACACCTACAttcaagagaTGTAGGTGTATCTCCATGCTTTTTTG-3′ and antisense, 5′-AATTCAAAAAAGCATGGAGATACACCTACAtctcttgaaTGTAGGTGTATCTCCATGCG-3′. The two oligomers were heated at 90°C for 3 min, annealed by slow cooling to 37°C over a 30-min period, and inserted into the multi-cloning sites of pU6-MCS-CMV-GFP and the BamHI and HindIII sites to prepare an shE6E7-targeting shRNA expression plasmid vector (pU6-shE6E7-CMV-GFP). A control vector (pU6-shNC-CMV-GFP) was prepared by inserting a 21-base sequence (Thermo Fisher scientific) known to have no influence on gene expression into pU6-shNC-CMV-GFP at the same sites.
Preparation of an AAV vector containing shE6E7
shE6E7-containing and control AAV vectors were prepared by transfecting the 293 cells with 3 plasmids, including pU6-shE6E7-CMV-GFP or pU6-shNC-CMV-GFP, the helper plasmid for adenovirus genes, and the helper plasmid for AAV (27,28). Recombinant AAV vectors (AAV-shE6E7 and AAV-shNC) were collected by repeated freezing and thawing of the cells 3 times. The vector solutions were purified by density gradient ultracentrifugation, and the vector titers were measured using real-time PCR, as previously described (29).
Flow cytometry
Flow cytometry was used to measure the gene transfer efficiency of the type 2 AAV vector (AAV2) according to GFP expression, and cell cycle analysis was performed using Annexin V (Annexin V-FITC apoptosis detection kit; BD Biosciences, San Jose, CA, USA) and propidium iodide (PI) staining (BD Cycletest Plus DNA Reagent kit; BD Biosciences) to determine the rate of cellular apoptosis after AAV2-shE6E7 transduction. Briefly, the cells were seeded in 12-well plates at 1×104 cells per well. After 12 h, the cells were transduced with 1×105 vg/cell of AAV2-shE6E7 or AAV2-shNC. The cells were then collected by trypsin treatment 48 h after transduction, washed twice with PBS, and stained on ice with Annexin V-FITC for 15 min followed by BD Cycletest for 10 min. The stained cells were measured using fluorescence-activated cell sorting (FACs; BD Biosciences) with a CellQuest software system (BD Biosciences).
Reverse transcription-quantitative PCR (RT-qPCR)
Cellular mRNA was extracted using an RNeasy Mini kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions and was used as a template for cDNA synthesis by reverse transcription (ReverTra Ace; Toyobo, Tokyo, Japan). RT-qPCR was performed using a LightCycler System (Roche Diagnostics, Mannheim, Germany) following the manufacturer’s instructions. pCR was carried out using 40 cycles of heating at 95°C for 15 sec, 58°C for 15 sec and 72°C for 20 sec. The mRNA levels of the target genes were determined relative to the fluorescence signal level of β-actin. Primer sequences are as follows: human β-actin forward, 5′-AGCCATGTACGTTGCTATCC-3′ and reverse, 5′-TTGGCGTACAGGTCTTTGC-3′; HPV16E6: forward, 5′-TTACCACAGTTATGCACAGA-3′ and reverse, 5′-ACAGTGGCTTTTGACAGTTA-3′; and HPV16E7 forward, 5′-AGAAACCCAGCTGTAATCAT-3′ and reverse, 5′-TTATGGTTTCTGAGAACAGA-3′.
Western blot analysis
The cells were seeded in 6-well plates at 1×106 cells/well. After 12 h, the cells were transduced with AAV2-shE6E7 or AAV2-shNC at 1×105 vg/cell. After 48 h, the cells were lysed using lysis buffer (1% NP-40, 150 mM NaCl, 50 mM Tris-HCl, pH 8.0), and protein was extracted. Protein samples were mixed with 1% SDS sample buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% SDS, EDTA-free protease inhibitor cocktail (Roche Diagnostics), separated by 10% polyacrylamide gel electrophoresis, and transferred onto polyvinylidene difluoride (pVDF) membranes (Merck Millipore, Billerica, MA, USA). Membranes were placed in PVDF Blocking Reagent for Can Get Signal (Toyobo) at room temperature for 1 h and then reacted with anti-hpV-16/18 E6 (sc-460; Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-HPV-16 E7 (Cat. no. sc-6981; Santa Cruz Biotechnology), anti-p53 (Cat. no. sc-126; Santa Cruz Biotechnology), anti-p21 (Cat. no. 2947S; Cell Signaling Technology), anti-p16 (Cat. no. 4824; Cell Signaling Technology), anti-Rb (Cat. no. 9313; Cell Signaling Technology), anti-phosphorylated-Rb (p-Rb; Cat. no. 2181; Cell Signaling Technology), or anti-β-actin monoclonal antibodies (sc-1616-R; Santa Cruz Biotechnology) at 4°C overnight using Can get Signal Immunoreaction Enhance Solution 1 (Toyobo). After washing, the membranes were reacted with peroxidase-labeled anti-mouse antibodies (GE Healthcare Japan, Tokyo, Japan) using Can Get Signal Immunoreaction Enhance Solution 2 (Toyobo) at room temperature for 1 h. Protein bands were detected using ECL Western Blotting Detection Reagents (GE Healthcare Japan) and a cold CCD system (LAS-4000mini; GE Healthcare Japan).
In vitro cell growth
The cells were seeded in 96-well plates at 1×103 cells per well. After 12 h, the cells were transduced with AAV2-shE6E7 at 0–1×107 vg/cell or AAV2-shNC at 1×105 vg/cell. After 72 h, 10 μl premix WsT-1 reagent (Takara Bio, Otsu, Japan) was added, and the absorbance at 450 nm was measured using a SpectraMax 190 microplate reader (Molecular Devices, Sunnyvale, CA, USA) after 1 h.
Animal experiment
Five-week-old severe combined immunodeficiency (sCID) nude mice (n=39) (Charles River Japan, Yokohama, Japan), weighing 15–20 g, were inoculated with BOKu or siha cells, and 5-week-old BAlB/c nude mice (n=15) (Clea Japan, Tokyo, Japan), weighing 15–20 g, were inoculated with SKG-IIIa cells. The animals were maintained under specific pathogen-free conditions. The animal experiments were performed according to the animal experimental guidelines and following the approval of the Ethics Committee of Jichi Medical University (Tochigi, Japan).
Subcutaneous tumors with 5- and 8-mm major axes were investigated as early-stage and infiltrating cancer models, respectively (30). Briefly, nude mice were subcutaneously inoculated with 1×107 tumor cells in the dorsal region to form a subcutaneous tumor, and 2.5×1011 vg/mouse AAV2-shE6E7 or AAV2-shNC was directly injected once into the tumors after the tumors had reached the target size for each purpose. The tumor size was measured twice weekly using calipers, and the tumor volume was calculated as the major axis x the minor axis2 × 1/2. The mice were sacrificed when the tumor volume exceeded 500 mm3. Multiple tumors were not observed in our study.
Immunohistochemical staining
The cancer-bearing mice were sacrificed by decapitation, and the tumors were excised and embedded in paraffin. TuNEl staining was performed using an in situ apoptosis detection kit (Takara Bio) following the manufacturer’s instructions. Briefly, paraffin-embedded tissue sections were deparaffinized and treated with proteinase K (10–20 μg/ml, 15 min; Takara Bio). Endogenous peroxidase was blocked with 3% H2O2 aqueous solution for 5 min. After washing, 50 μl ice-cold labeling reaction solution was added to the sections, and the samples were incubated in a moist container at 37°C for 60 min. After washing, the sections were reacted with 70 μl anti-FITC hRp conjugate (Takara Bio) at 37°C for 30 min, followed by color development with DAB/h2O2 reaction solution (Takara Bio) at room temperature for 10 min. The sections were then stained with hematoxylin (Takara Bio), dehydrated, permeabilized, sealed and observed under a light microscope (IX73, Olympus, Tokyo, Japan). TUNEL-positive cells were counted in 5 visual fields at x40 magnification.
Statistical analysis
Student’s t-tests were used for between-group comparisons, and differences with P-values <0.05 were considered to indicate a statistically significant difference. Tukey’s tests were used for multiple group comparisons, and differences with P-values <0.05 were considered to indicate a statistically significant difference.
Results
Optimization of AAV serotypes for gene transfer into cervical cancer cells
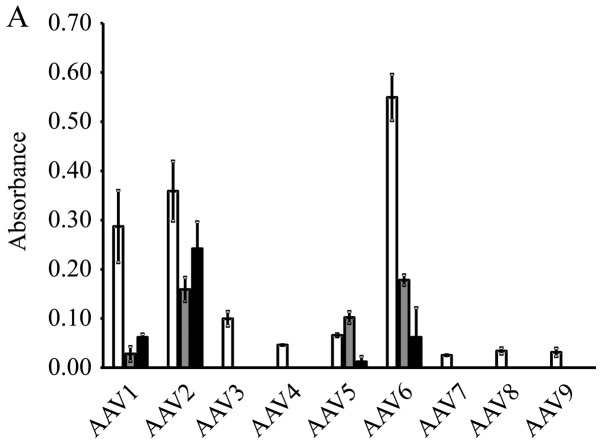
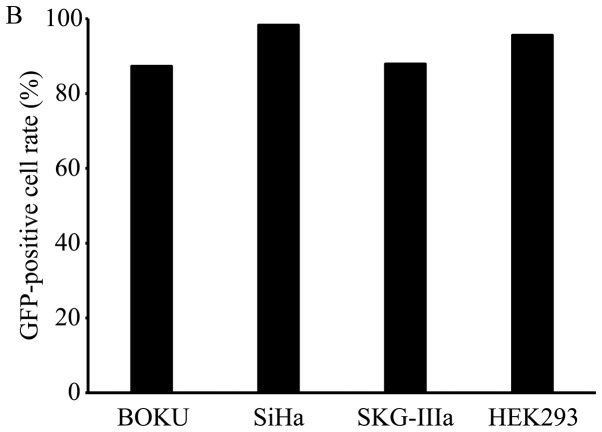
The results of β-galactosidase expression in the cervical cancer cell lines are shown in Fig. 1A. A high expression level was obtained by AAV serotype 2 in all 3 cell lines. Therefore, we selected to perform the subsequent experiments using the AAV2-based vectors. High levels of GFP expression were observed in the cells that were transduced with AAV2-shNC, a GFP-encoding vector (Fig. 1B).
Figure 1.
Gene transfer efficiency of each AAV vector serotype in cervical cancer cells. (A) β-galactosidase expression levels at 48 h following the transduction of BOKU, SiHa and SKG-IIIa cells with the AAV vector (AAV-CMV-LacZ) types 1–9 containing the LacZ gene encoding β-galactosidase (white bars, BOKU cells; gray bars, SiHa cells; black bars, SKG-IIIa cells). The results are presented as the means ± sD. (B) gene transfer efficiency of AAV2 in cervical cancer cell lines. The GFP-positive cell rate 48 h after AAV2-shNC transduction was determined using flow cytometry.
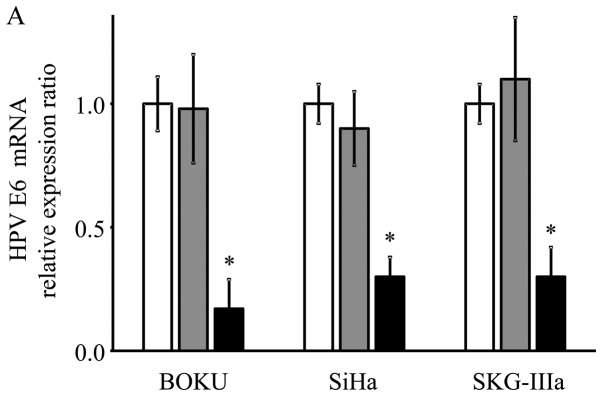
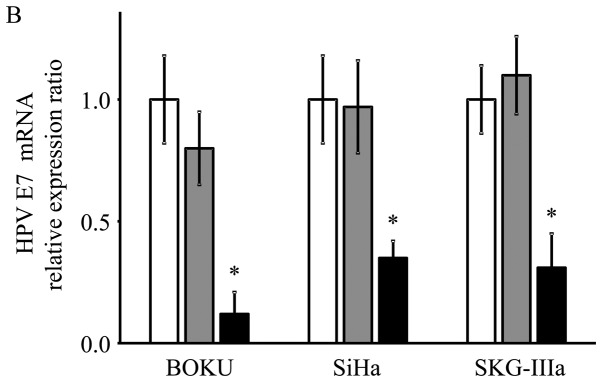
HPV-16 E6/E7 mRNA expression following AAV2-shE6E7 transduction
As shown using RT-qPCR, the mRNA expression levels of E6 and E7 were significantly decreased in the AAV2-shE6E7-transduced cervical cancer cells compared with the control cells (E6: 13, 29 and 21% vs. that of the control for the BOKU, SiHa and SKG-IIIa cells, respectively; and E7: 12, 33 and 34% vs. of the control for the BOKU SiHa SKG-IIIa, respectively; Fig. 2).
Figure 2.
mRNA expression levels of the HPV-16 E6 and E7 transcripts following AAV2-shE6E7 transduction. (A) HPV-16 E6 (B) and E7 mRNA levels in the 3 cervical cancer cell lines 48 h following AAV2-shE6E7 or AAV-shNC transduction, as measured by RT-qPCR (white bars, control cells; gray bars, AAV2-shNC-transduced cells; black bars, AAV2-shE6E7-transduced cells); *P<0.05 as compared to the AAV2-shNC-transduced cells. The results are presented as the means ± SD.
Protein expression following AAV2-shE6E7 transduction
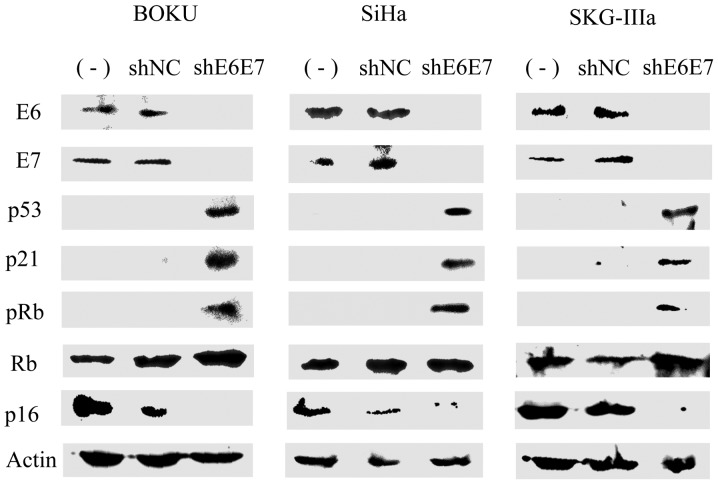
Subsequently, western blot analysis was used to measure protein expression in the cervical cancer cells following transduction with AAV2-shE6E7 or AAV2-shNC. In all cell lines, the expression levels of E6, E7 and p16 were decreased, while the expression levels of p53, p21 and pRb were markedly increased in the AAV2-shE6E7-transduced cells compared with those in the control cells (Fig. 3).
Figure 3.
Protein expression following AAV2-shE6E7 transduction. E6, E7, p53, p21, pRb, Rb and p16 protein levels were measured by western blot analysis in the 3 cervical cancer cell lines 48 h following AAV2-shE6E7 or AAV-shNC transduction.
Induction of apoptosis following AAV2-shE6E7 transduction
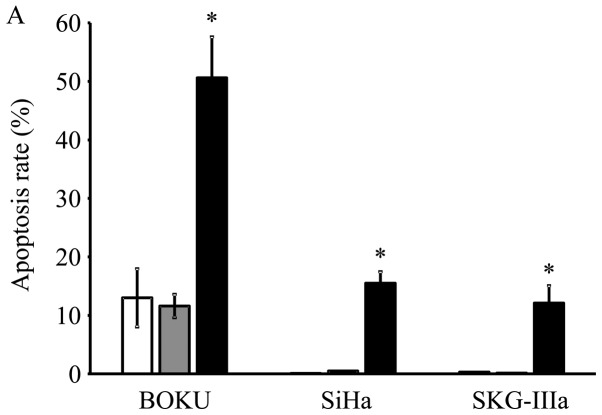
Transduction with AAV2-shE6E7 increased the rates of apoptosis in all 3 cell lines as compared with the control and AAV2-shNC transduction, with much more evident increases observed in the BOKU, SiHa and SKG-IIIa cells (Fig. 4A). Thus, a marked induction of apoptosis was noted in all 3 cervical cancer cell lines transduced with AAV2-shE6E7.
Figure 4.
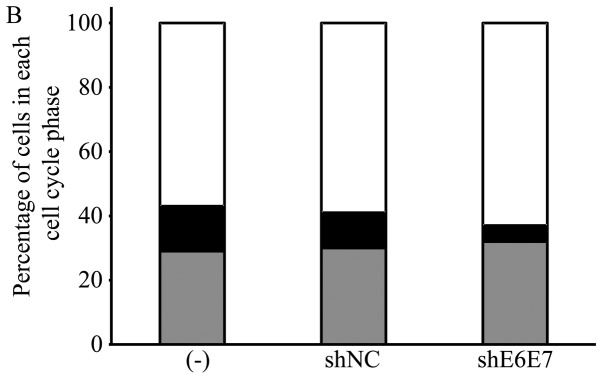
In vitro apoptosis and cell cycle analysis following transduction with AAV2-shE6E7. (A) Apoptosis-positive rates using Annexin V as an index 48 h following AAV2-shE6E7 or AAV-shNC infection by flow cytometry (white bars, uninfected control cells; gray bars, AAV2-shNC-infected group; black bars, AAV2-shE6E7-infected group). The induction of apoptosis was noted in the cells following AAV2-shE6E7 infection in all 3 cell lines. *P<0.05 as compared to the AAV2-shNC-infected group. The results are presented as the means ± SD. (B) Flow cytometric analysis of the cell cycle at 48 h following AAV2-shE6E7 or AAV-shNC infection using PI staining as an index (white, G0/g1 phase; black, S phase; gray, G2M phase). The numbers of BOKU cells in the G0/g1 phase were 57, 58 and 63% in the control, AAV2-shNC-infected and AAV2-shE6E7-infected groups, respectively, and those of the cells in the S phase were 14, 11 and 5%, respectively, showing that cells in the S phase decreased in the AAV2-shE6E7-infected group.
Changes in cell cycle distribution following AAV2-shE6E7 transduction
Subsequently, the BOKU cells were transduced with AAV2-shE6E7 or AAV2-shNC, and changes in the cell cycle distribution were analyzed using PI staining. The results are shown in Fig. 4B. As shown this band graph, the proportions of BOKU cells in the G0/g1 phase in the control group, the AAV2-shNC infected group and the AAV2-shE6E7-infected group were 57, 58 and 63%, respectively, and the proportions in the S phase were 14, 11 and 5%, respectively. The proportion of cells in the G0/g1 phase increased, and the proportion of cells in the S phase decreased in the AAV2-shE6E7-infected group.
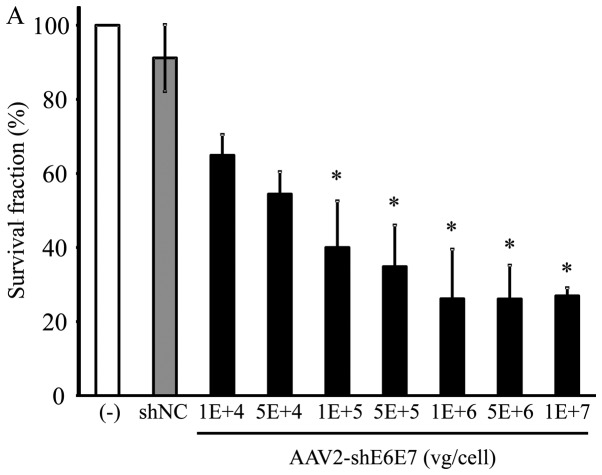
Changes in cell growth following transduction with AAV2-shE6E7
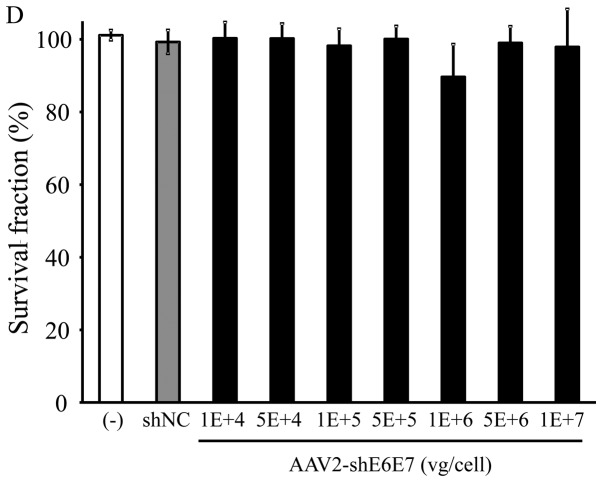
The BOKU, SiHa, SKG-IIIa and 293 cells were transduced with AAV2-shE6E7 or AAV2-shNC, and the viable cells were counted. In all 3 cervical cancer cell lines, the numbers of viable cells were decreased following transduction with AAV2-shE6E7 in a concentration-dependent manner. By contrast, AAV2-shE6E7 had no effect on the growth of 293 cells (Fig. 5).
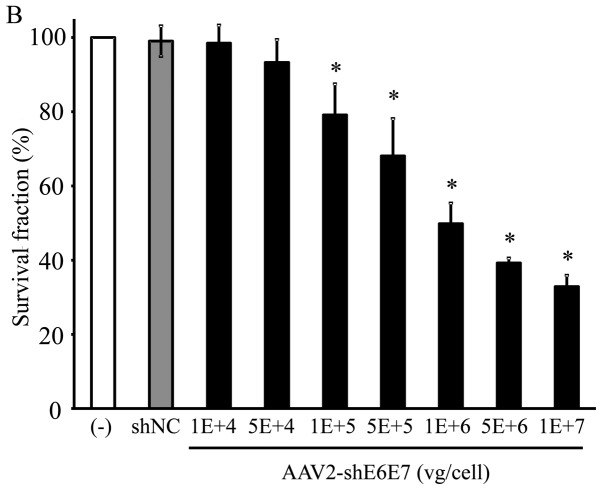
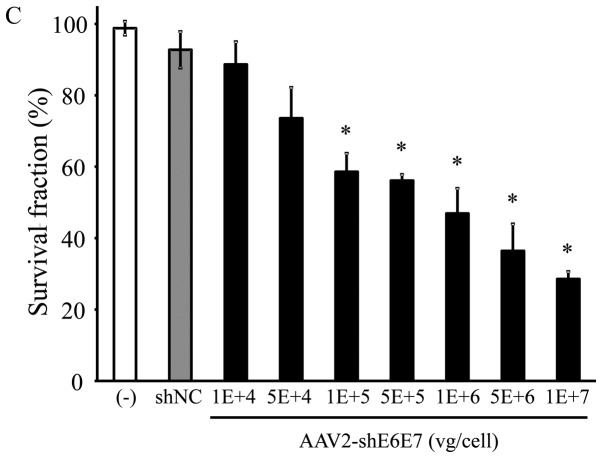
Figure 5.
Cell growth following AAV2-shE6E7 transduction. Viable cells were measured by WST-1 assay at 72 h folllowing AAV2-shE6E7 or AAV-shNC transduction (A) BOKU cells, (B) SiHa cells, (C) SKG-IIIa cells, and (D) 293 cells; *P<0.05 as compared to the AAV-shNC transduction cells. The results are presented as the means ± SD.
Eradication of in vivo xenograft tumors following AAV2-shE6E7 transduction
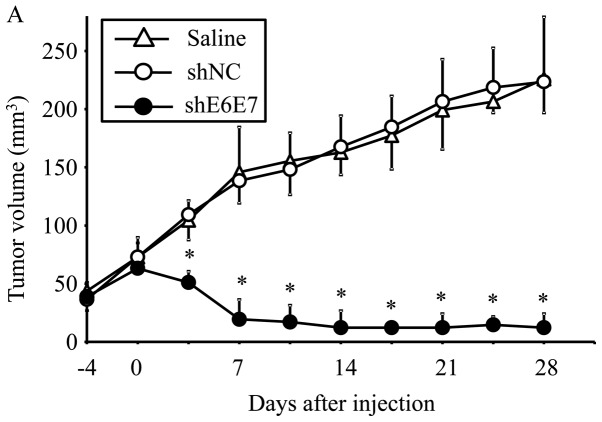
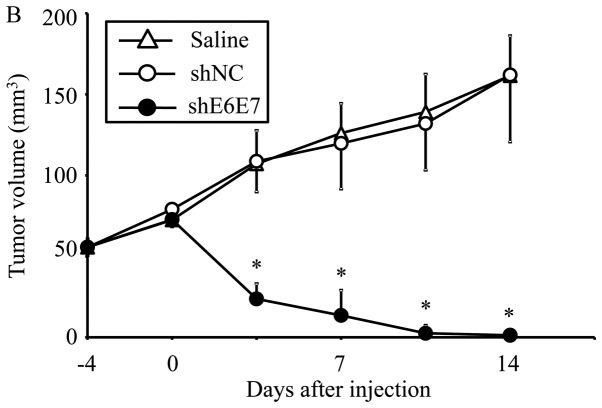
AAV2-shE6E7, AAV2-shNC, or saline was injected in a single dose into tumors formed by the subcutaneous transplantation of BOKU, SiHa, or SKG-IIIa cells into mice, and tumor growth was observed. First, we analyzed the efficacy of E6 and E7 knockdown using smaller tumors (with a 5-mm major axis). The mean volume of BOKU-derived tumors at 28 days after administration (n=4) was significantly smaller than that of tumors that formed in mice treated with saline (n=4) or AAV2-shNC (n=4) (p<0.01; Fig. 6A). Similarly, the mean volume of SiHa-derived tumors at 14 days after administration was significantly lower in the AAV2-shE6E7 group (n=4) than in the saline (n=4) and AAV2-shNC groups (n=4) (p<0.01; Fig. 6B and C).
Figure 6.
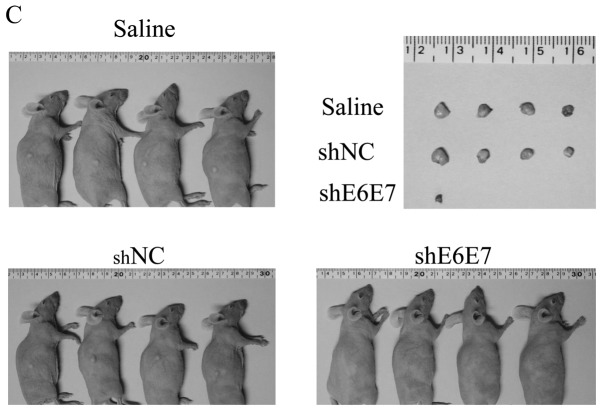
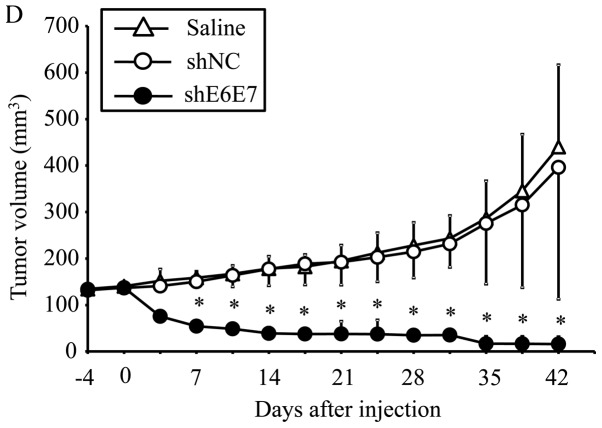
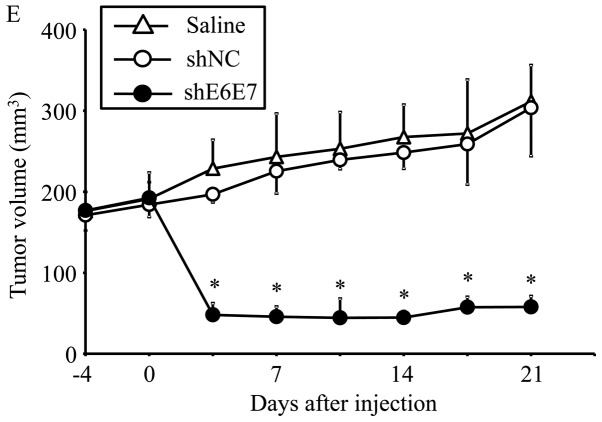
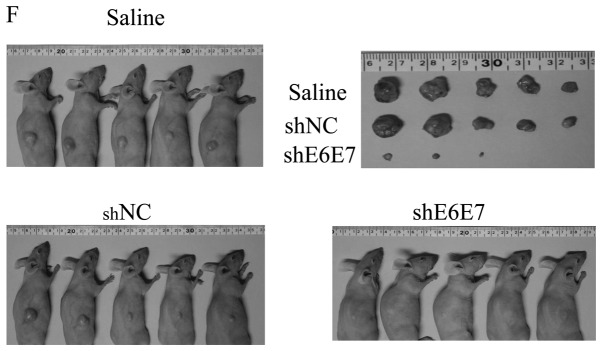
In vivo tumor growth following AAV2-shE6E7 transduction. (A and B) Tumor growth curves after a single injection of AAV2-shE6E7, AAV-shNC, or saline into subcutaneous tumors with a 5-mm major axis (volume, 60 mm3); (A) BOKU cells, (B) SiHa cells; *P<0.05. The results are presented as the means ± SD. (C) SiHa-derived tumors at 14 days after administration. (D and E) Tumor growth curves after a single injection of AAV2-shE6E7, AAV-shNC, or saline into subcutaneous tumors with an 8-mm major axis; (D) SiHa cells, (E) SKG-IIIa cells. (F) SiHa-derived tumors at 42 days after administration.
The efficacy of this treatment was then tested in larger tumors (with an 8-mm major axis). At 42 days after administration, the mean tumor volume was significantly decreased in the AAV2-shE6E7-injected mice (n=5) with siha-derived tumors compared with those in the saline (n=5) and AAV2-shNC groups (n=5) (p<0.01; Fig. 6D and F). Notably, no regrowth was observed in all tumors during the test period of 8 weeks. Similar results were obtained in the mice with SKG-IIIa-derived tumors (P<0.01 for the AAV2-shE6E7 group (n=5) compared with those in the saline (n=5) and the AAV2-shNC group (n=5); Fig. 6E).
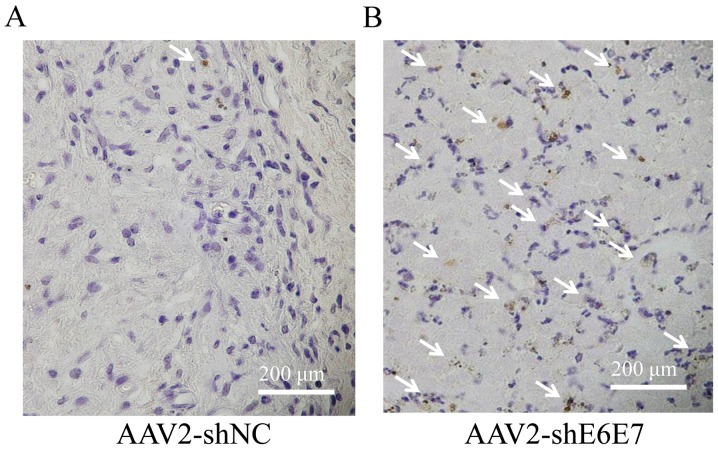
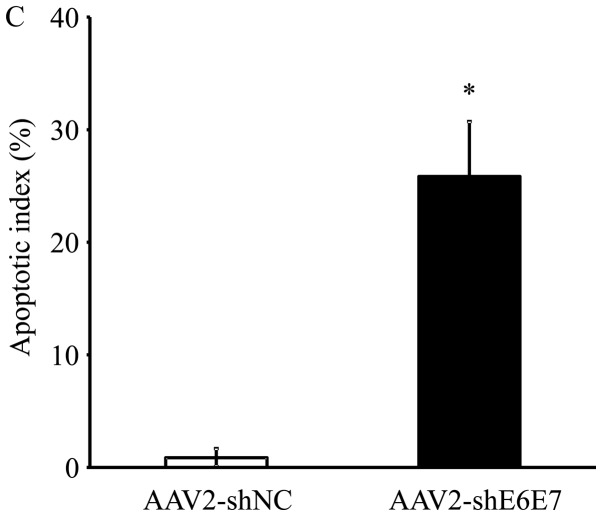
The results of TUNEL staining of the SiHa-derived tumor tissues are shown in Fig. 7A and B. The mean TUNEL-positive cell rate was significantly higher in the AAV2-shE6E7 group than in the AAV2-shNC group (P<0.01; Fig. 7C). No macroscopic changes, including edema or inflammation, were noted at the sites of vector inoculation, nor were there any differences in body weights among the groups (data not shown).
Figure 7.
Analysis of apoptosis in vivo following AAV2-shE6E7 transduction. TUNEL staining of SiHa-derived subcutaneous tumors in mice at 48 h following (A) AAV2-shNC or (B) AAV-shE6E7 administration (white arrows indicate TuNEl-positive cells). Scale bars, 200 μm. (C) The rate of apoptosis was determined ny TUNEL staining as an index; *P<0.05. The results are presented as the means ± SD.
Discussion
In this study, a basic analysis was performed aiming at developing a novel, highly specific cervical cancer treatment method using AAV-shE6E7. Firstly, the serotype of an AAV vector with the highest transfer efficiency for cervical cancer cells was screened, and the gene transfer efficiency of the AAV2 vector was found to be the highest. AAV vectors are known to be highly organ-specific, and serotypes with high transfer efficiency for specific organs are known. For example, the transfer efficiencies of serotypes 1 and 7 for skeletal muscles, serotypes 2 and 3 for nerves, serotype 5 for the retina, and serotype 8 for the liver are high (20). Our study clarified that type 2 most efficiently transfers genes into cervical cancer cells.
As regards HPV, >120 types have been reported due to differences in the L1 gene base sequence (31). Approximately 20 types are high-risk HPV causing cervical cancer (32), and type 16 is most frequently detected in cervical cancer (32). Thus, in this study, we examined type 16. Since cancer genes of HPV16, E6 and E7, are encoded by a single mRNA (33,34), both E6 and E7 expression levels can be inhibited with one type of shRNA (35). An shRNA sequence that efficiently inhibits HPV16 E6 and E7 has been reported (26), and we prepared an AAV2 vector (AAV2-shE6E7) containing this sequence. The GFP-encoding gene was also contained in AAV2-shE6E7 and AAV2-shNC as a reporter gene driven by the CMV promoter. When the 3 types of cervical cancer cell were infected with AAV2-shNC in vitro, high gene transfer efficiency (approximately 90%) was observed, and this was comparable to that of AAV vectors for 293 cells known to show very high efficiency. The gene transfer efficiency was also almost 100% on macroscopic measurement using a hemocytometer in all cell lines (data not shown). The treatment strategy targeted in this study is local therapy directly transferring a gene into cancer cells, for which high gene transfer efficiency is essential. It was demonstrated that the AAV2 vector transfers genes into cervical cancer at a high efficiency.
To confirm the effect of AAV2-shE6E7, the mRNA expression levels of the direct targets, E6 and E7, were measured by RT-qPCR. In the 3 types of cervical cancer cell infected with AAV2-shE6E7 in vitro, the E6 and E7 mRNA expression levels decreased to 1/3–1/10 of those in the control. Subsequently, the expression levels of E6, E7 and related factors were examined by western blot analysis. In all 3 types of cervical cancer cell cells infected with AAV2-shE6E7, the E6, E7 and p16 expression levels were decreased, whereas those of p53, p21, and pRb were increased. Following the investigation of apoptosis using Annexin V as an index, a marked induction of apoptosis was observed in all 3 types of cervical cancer cell infected with AAV2-shE6E7. Following the analysis of the cell cycle using flow cytometry, the numbers of cells in the g1 phase increased following AAV2-shE6E7 infection, i.e., G1 arrest occurred. In cervical cancer, E6 degrades p53, which results in the inhibition of downstream p21 expression, leading to the inhibition of apoptosis induction (36). E7 inactivates pRb, which causes the dysfunction of the G1 checkpoint and avoids G1 arrest (36). To adjust these abnormalities in the cell cycle, p16 expression is enhanced for emergency avoidance of the condition (36). The inhibitory effect of AAV2-shE6E7 on E6 and E7 expression may have resolved these abnormalities and each factor may have been corrected to function normally, leading to apoptosis and normal functioning of the G1 checkpoint.
Subsequently, we investigated the actual inhibitory effect of AAV2-shE6E7 on cervical cancer cell growth in vitro. AAV2-shE6E7 showed a growth-inhibitory effect on all 3 types of cervical cancer cell in a concentration-dependent manner.
Based on the in vitro experimental results, the effect of AAV2-shE6E7 was investigated in in vivo animal experiments. Mice were subcutaneously inoculated with cervical cancer cells. After macroscopically confirming tumor formation, AAV2-shE6E7 was subcutaneously injected around the tumor, and changes in the tumor were observed. Firstly, the experiment was performed with tumors with a 5-mm major axis (volume, approximately 60 mm3) as an early cancer model. In all tumors formed by the 3 types of cervical cancer cells, a marked tumor size reduction was noted after AAV2-shE6E7 administration, and the tumors were mostly resolved. The experiment was then performed with tumors with an 8-mm major axis (volume, approximately 250 mm3) as an advanced cancer model. Similarly, a marked decrease in tumor size reduction was noted following the injection of AAV2-shE6E7 in all tumors formed by the 3 types of cervical cancer cells, and the tumors were mostly resolved. In addition, no tumor re-growth was noted over long-term observation. Many apoptotic bodies were observed in the tumors injected with pAAV2-shE6E7, clarifying that AAV2-shE6E7 reduced the size of and resolved the tumor by inducing apoptosis.
Several animal experiments with siRNA targeting E6 and E7 in cervical cancer have been reported (37–39). In these studies, experiments were conducted in which siRNA targeting HPV E6E7 was administered under the protection of atelocollagen (37,38). All of these results were limited in tumor growth suppression despite multiple administrations. By contrast, in our study using AAV2-shE6E7, tumor growth was completely inhibited by only a single administration. These differences in the effects may have reflected a difference in the gene transfer efficiency for cancer cells between the siRNA protected by atelocollagen and AAV2-shE6E7. An ex vivo study using an AAV vector encoding E6E7 antisense RNA also reported significant tumor growth inhibitory effect (40).
Recently, attempts to knock out E6 or E7 in cervical cancer using genome editing technology have been reported (41–43). In these studies, the effects in vitro or ex vivo were confirmed. In order to obtain sufficient antitumor effect in vivo, the combination with the AAV vector may be effective as in this study. Recently, a CRIspR/Cas 9 derived from Staphylococcus aureus that can be encoded to an AAV vector was reported (44). We are currently conducting fundamental research on cervical cancer treatment applying this new genome editing technology.
As regards adverse effects, no weight loss or abnormalities at the administration site was noted in mice treated with AAV2-shE6E7, and these may have been due to the following 2 reasons: Firstly, AAV vectors have no toxicity, and secondly, shE6E7 does not affect normal tissue as E6 and E7 are expressed only in cervical cancer cells, not in normal tissue. It was thus suggested that treatment with AAV-shE6E7 is safe.
AAV Rep 78 protein is known to inhibit the promoter site of several oncogenes and viral genes. It has been reported that E6 of cervical cancer can be suppressed using Rep 78 protein (45). However, the AAV vector used in our study does not express Rep 78 protein. Therefore, the effect observed in our study is not the same as that of the Rep 78 protein.
HPV vaccine development using HPV L1 antigen has become practically applied, and clinical administration is now widespread (46,47). The reduction of the incidence of cervical cancer by lowering the prevalence of HPV infection is expected. On the other hand, no curative treatment method targeting HPV has been established for tens of millions of individuals already infected with high-risk HPV worldwide and patients with CIN and cervical cancer. Treatment with AAV2-shE6E7 developed by us efficiently inhibited E6 and E7 expression of high-risk HPV and reduced the size of the lesion within a short time, for which a long-term effect can be expected. In clinical application, high-grade CIN and early infiltrating cancer may be firstly treated. This treatment strategy is very likely to be the most effective for these because lesions are localized. High-level safety is essential as the incidence of these diseases is high in young persons and those wishing to become pregnant. No adverse effect of AAV2-shE6E7 was observed; however, further investigations are required aiming at clinical application in the future.
For advanced cervical cancer, the effect of this treatment strategy for broad infiltration and metastasis is limited, and curative treatment cannot be expected. However, it was effective for tumors of a certain size, suggesting that the treatment may be applicable to reduce a bulky tumor size by increasing the dose. It may be a substitute for neoadjuvant chemotherapy (48,49) performed to increase the curability of surgery through size reduction of bulky tumors and increase the effect of radiotherapy. It may also be applicable for some local recurrent carcinomas.
We performed this study using HPV type 16, but cervical cancer caused by type 16 accounts for 50% of all cases (32). To expand the indications of this treatment strategy, it is necessary to develop an shRNA-containing AAV vector effective for high-risk HPV other than type 16. Since the base sequence of E6E7 is not homologous among high-risk HPV types, to treat infection with several high-risk HPV types, it is necessary to design shRNA for individual types. However, the individual preparation of an AAV vector is unnecessary. Since the U6 promoter and shRNA base sequences are comprised of approximately 400 bases in total, theoretically, approximately 10 pairs can be inserted into one AAV vector. If an AAV vector containing shRNAs corresponding to E6E7 of 7 types: HPV type 16, 18, 33, 45, 31, 58 and 52, can be developed, it may be effective for approximately 90% of cervical cancer cases. On the whole, as our data indicated, AAV2-shE6E7 exerted a marked inhibitory effect on cervical cancer for a prolonged period of time. A novel treatment strategy for cervical cancer using AAV2-shE6E7 was thus suggested.
Acknowledgments
The authors would like to thank Mrs. Miyoko Mitsu, Mrs. Satomi Fujiwara and Mrs. Michiko Ohashi for providing excellent technical assistance. This study was partly supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT)-Supported Program for the Strategic Research Foundation at Private Universities, 2013–2017.
Footnotes
Competing interests
The authors declare that they have no competing interests.
References
- 1.Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, Dandona L, et al. Global Burden of Disease Cancer Collaboration Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: A systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3:524–548. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Woodman CB, Collins SI, Young LS. The natural history of cervical HPV infection: Unresolved issues. Nat Rev Cancer. 2007;7:11–22. doi: 10.1038/nrc2050. [DOI] [PubMed] [Google Scholar]
- 4.Sevin BU, Nadji M, Averette HE, Hilsenbeck S, Smith D, Lampe B. Microinvasive carcinoma of the cervix. Cancer. 1992;70:2121–2128. doi: 10.1002/1097-0142(19921015)70:8<2121::AID-CNCR2820700819>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 5.Massad LS, Einstein MH, Huh WK, Katki HA, Kinney WK, Schiffman M, Solomon D, Wentzensen N, Lawson HW, 2012 ASCCP Consensus Guidelines Conference 2012 updated consensus guidelines for the management of abnormal cervical cancer screening tests and cancer precursors. Obstet Gynecol. 2013;121:829–846. doi: 10.1097/AOG.0b013e3182883a34. [DOI] [PubMed] [Google Scholar]
- 6.Bevis KS, Biggio JR. Cervical conization and the risk of preterm delivery. Am J Obstet Gynecol. 2011;205:19–27. doi: 10.1016/j.ajog.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Boshart M, Gissmann L, Ikenberg H, Kleinheinz A, Scheurlen W, zur Hausen H. A new type of papillomavirus DNA, its presence in genital cancer biopsies and in cell lines derived from cervical cancer. EMBO J. 1984;3:1151–1157. doi: 10.1002/j.1460-2075.1984.tb01944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dürst M, Gissmann L, Ikenberg H, zur Hausen H. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc Natl Acad Sci USA. 1983;80:3812–3815. doi: 10.1073/pnas.80.12.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muñoz N, Bosch FX, de Sanjosé S, Herrero R, Castellsagué X, Shah KV, Snijders PJ, Meijer CJ, International Agency for Research on Cancer Multicenter Cervical Cancer Study Group Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 10.Dyson N, Howley PM, Münger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 11.Moody CA, Laimins LA. Human papillomavirus oncoproteins: Pathways to transformation. Nat Rev Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 12.Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 13.Da Silva DM, Eiben GL, Fausch SC, Wakabayashi MT, Rudolf MP, Velders MP, Kast WM. Cervical cancer vaccines: Emerging concepts and developments. J Cell Physiol. 2001;186:169–182. doi: 10.1002/1097-4652(200102)186:2<169::AID-JCP1023>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 14.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. potent and specific genetic interference by double- stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 15.Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc Natl Acad Sci USA. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 17.Scherr M, Eder M. Gene silencing by small regulatory RNAs in mammalian cells. Cell Cycle. 2007;6:444–449. doi: 10.4161/cc.6.4.3807. [DOI] [PubMed] [Google Scholar]
- 18.Paddison PJ, Hannon GJ. RNA interference: The new somatic cell genetics? Cancer Cell. 2002;2:17–23. doi: 10.1016/S1535-6108(02)00092-2. [DOI] [PubMed] [Google Scholar]
- 19.Mueller C, Flotte TR. Clinical gene therapy using recombinant adeno-associated virus vectors. Gene Ther. 2008;15:858–863. doi: 10.1038/gt.2008.68. [DOI] [PubMed] [Google Scholar]
- 20.Wu Z, Asokan A, Samulski RJ. Adeno-associated virus serotypes: Vector toolkit for human gene therapy. Mol Ther. 2006;14:316–327. doi: 10.1016/j.ymthe.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Grieger JC, Samulski RJ. Adeno-associated virus vectorology, manufacturing, and clinical applications. Methods Enzymol. 2012;507:229–254. doi: 10.1016/B978-0-12-386509-0.00012-0. [DOI] [PubMed] [Google Scholar]
- 22.Gao G, Vandenberghe LH, Alvira MR, Lu Y, Calcedo R, Zhou X, Wilson JM. Clades of Adeno-associated viruses are widely disseminated in human tissues. J Virol. 2004;78:6381–6388. doi: 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao GP, Alvira MR, Wang L, Calcedo R, Johnston J, Wilson JM. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc Natl Acad Sci USA. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rutledge EA, Halbert CL, Russell DW. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J Virol. 1998;72:309–319. doi: 10.1128/jvi.72.1.309-319.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeda S, Takahashi M, Mizukami H, Kobayashi E, Takeuchi K, Hakamata Y, Kaneko T, Yamamoto H, Ito C, Ozawa K, et al. Successful gene transfer using adeno-associated virus vectors into the kidney: Comparison among adeno-associated virus serotype 1–5 vectors in vitro and in vivo. Exp Nephrol. 2004;96:e119–e126. doi: 10.1159/000077378. [DOI] [PubMed] [Google Scholar]
- 26.Sima N, Wang W, Kong D, Deng D, Xu Q, Zhou J, Xu G, Meng L, Lu Y, Wang S, et al. RNA interference against HPV16 E7 oncogene leads to viral E6 and E7 suppression in cervical cancer cells and apoptosis via upregulation of Rb and p53. Apoptosis. 2008;13:273–281. doi: 10.1007/s10495-007-0163-8. [DOI] [PubMed] [Google Scholar]
- 27.Hata K, Mizukami H, Sadakane O, Watakabe A, Ohtsuka M, Takaji M, Kinoshita M, Isa T, Ozawa K, Yamamori T. DNA methylation and methyl-binding proteins control differential gene expression in distinct cortical areas of macaque monkey. J Neurosci. 2013;33:19704–19714. doi: 10.1523/JNEUROSCI.2355-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsushita T, Elliger S, Elliger C, Podsakoff G, Villarreal L, Kurtzman GJ, Iwaki Y, Colosi P. Adeno-associated virus vectors can be efficiently produced without helper virus. Gene Ther. 1998;5:938–945. doi: 10.1038/sj.gt.3300680. [DOI] [PubMed] [Google Scholar]
- 29.Yagi H, Ogura T, Mizukami H, Urabe M, Hamada H, Yoshikawa H, Ozawa K, Kume A. Complete restoration of phenylalanine oxidation in phenylketonuria mouse by a self-complementary adeno-associated virus vector. J Gene Med. 2011;13:114–122. doi: 10.1002/jgm.1543. [DOI] [PubMed] [Google Scholar]
- 30.Adhim Z, Otsuki N, Kitamoto J, Morishita N, Kawabata M, Shirakawa T, Nibu K. Gene silencing with siRNA targeting E6/E7 as a therapeutic intervention against head and neck cancer-containing HPV16 cell lines. Acta Otolaryngol. 2013;133:761–771. doi: 10.3109/00016489.2013.773405. [DOI] [PubMed] [Google Scholar]
- 31.Fehrmann F, Laimins LA. Human papillomaviruses: Targeting differentiating epithelial cells for malignant transformation. Oncogene. 2003;22:5201–5207. doi: 10.1038/sj.onc.1206554. [DOI] [PubMed] [Google Scholar]
- 32.Clifford GM, Smith JS, Plummer M, Muñoz N, Franceschi S. Human papillomavirus types in invasive cervical cancer worldwide: A meta-analysis. Br J Cancer. 2003;88:63–73. doi: 10.1038/sj.bjc.6600688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherman L, Alloul N. Human papillomavirus type 16 expresses a variety of alternatively spliced mRNAs putatively encoding the E2 protein. Virology. 1992;191:953–959. doi: 10.1016/0042-6822(92)90271-P. [DOI] [PubMed] [Google Scholar]
- 34.Smotkin D, Prokoph H, Wettstein FO. Oncogenic and nononcogenic human genital papillomaviruses generate the E7 mRNA by different mechanisms. J Virol. 1989;63:1441–1447. doi: 10.1128/jvi.63.3.1441-1447.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshinouchi M, Yamada T, Kizaki M, Fen J, Koseki T, Ikeda Y, Nishihara T, Yamato K. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by E6 siRNA. Mol Ther. 2003;8:762–768. doi: 10.1016/j.ymthe.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 37.Fujii T, Saito M, Iwasaki E, Ochiya T, Takei Y, Hayashi S, Ono A, Hirao N, Nakamura M, Kubushiro K, et al. Intratumor injection of small interfering RNA-targeting human papillomavirus 18 E6 and E7 successfully inhibits the growth of cervical cancer. Int J Oncol. 2006;29:541–548. [PubMed] [Google Scholar]
- 38.Yamato K, Yamada T, Kizaki M, Ui-Tei K, Natori Y, Fujino M, Nishihara T, Ikeda Y, Nasu Y, Saigo K, et al. New highly potent and specific E6 and E7 siRNAs for treatment of HP V16 positive cervical cancer. Cancer Gene Ther. 2008;15:140–153. doi: 10.1038/sj.cgt.7701118. [DOI] [PubMed] [Google Scholar]
- 39.Jonson AL, Rogers LM, Ramakrishnan S, Downs LS., Jr Gene silencing with siRNA targeting E6/E7 as a therapeutic intervention in a mouse model of cervical cancer. Gynecol Oncol. 2008;111:356–364. doi: 10.1016/j.ygyno.2008.06.033. [DOI] [PubMed] [Google Scholar]
- 40.Wu S, Meng L, Wang S, Wang W, Xi L, Tian X, Chen G, Wu Y, Zhou J, Xu G, et al. Reversal of the malignant phenotype of cervical cancer CaSki cells through adeno-associated virus-mediated delivery of HPV16 E7 antisense RNA. Clin Cancer Res. 2006;12:2032–2037. doi: 10.1158/1078-0432.CCR-05-2567. [DOI] [PubMed] [Google Scholar]
- 41.Kennedy EM, Kornepati AV, Goldstein M, Bogerd HP, Poling BC, Whisnant AW, Kastan MB, Cullen BR. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRIspR/Cas RNA-guided endonuclease. J Virol. 2014;88:11965–11972. doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhen S, Hua L, Takahashi Y, Narita S, Liu YH, Li Y. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRIspR/Cas9. Biochem Biophys Res Commun. 2014;450:1422–1426. doi: 10.1016/j.bbrc.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 43.Hu Z, Ding W, Zhu D, Yu L, Jiang X, Wang X, Zhang C, Wang L, Ji T, Liu D, et al. TALEN-mediated targeting of HPV oncogenes ameliorates HPV-related cervical malignancy. J Clin Invest. 2015;125:425–436. doi: 10.1172/JCI78206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahn WS, Bae SM, Lee HJ, Kim YW, Lee JM, Namkoong SE, Kim CK, Kim YW, Jin HS. Development of anticancer gene vaccine interact with human papillomavirus oncoprotein inhibition. Int J Gynecol Cancer. 2006;16:270–276. doi: 10.1111/j.1525-1438.2006.00343.x. [DOI] [PubMed] [Google Scholar]
- 46.Paavonen J, Jenkins D, Bosch FX, Naud P, Salmerón J, Wheeler CM, Chow SN, Apter DL, Kitchener HC, Castellsague X, et al. HPV pATRICIA study group Efficacy of a prophylactic adjuvanted bivalent L1 virus-like-particle vaccine against infection with human papillomavirus types 16 and 18 in young women: An interim analysis of a phase III double-blind, randomised controlled trial. Lancet. 2007;369:2161–2170. doi: 10.1016/S0140-6736(07)60946-5. [DOI] [PubMed] [Google Scholar]
- 47.Romanowski B. Long term protection against cervical infection with the human papillomavirus: Review of currently available vaccines. Hum Vaccin. 2011;7:161–169. doi: 10.4161/hv.7.2.13690. [DOI] [PubMed] [Google Scholar]
- 48.Sardi JE, Giaroli A, Sananes C, Ferreira M, Soderini A, Bermudez A, Snaidas L, Vighi S, Gomez Rueda N, di Paola G. long-term follow-up of the first randomized trial using neoadjuvant chemotherapy in stage Ib squamous carcinoma of the cervix: The final results. Gynecol Oncol. 1997;67:61–69. doi: 10.1006/gyno.1997.4812. [DOI] [PubMed] [Google Scholar]
- 49.Benedetti-Panici P, Greggi S, Colombo A, Amoroso M, Smaniotto D, Giannarelli D, Amunni G, Raspagliesi F, Zola P, Mangioni C, et al. Neoadjuvant chemotherapy and radical surgery versus exclusive radiotherapy in locally advanced squamous cell cervical cancer: Results from the Italian multicenter randomized study. J Clin Oncol. 2002;20:179–188. doi: 10.1200/JCO.2002.20.1.179. [DOI] [PubMed] [Google Scholar]