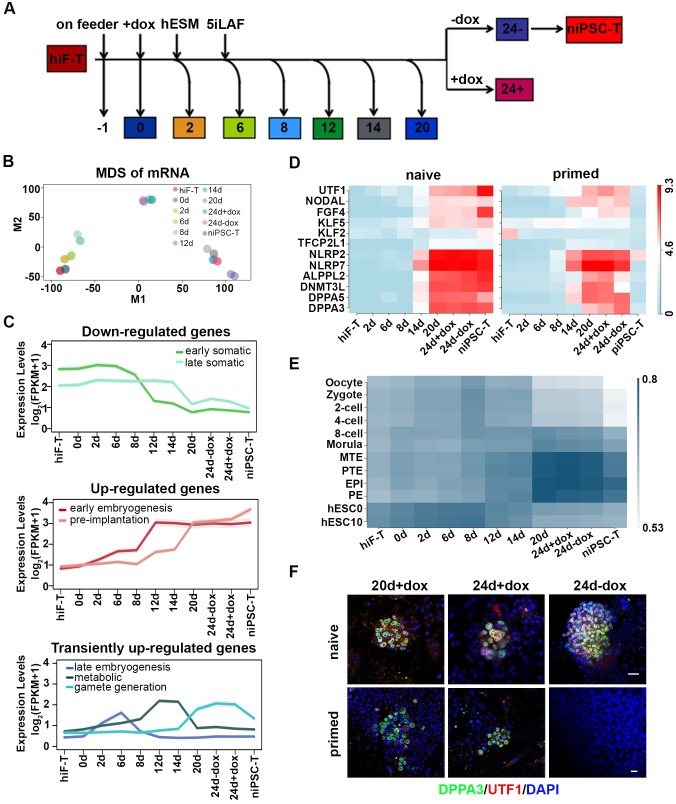
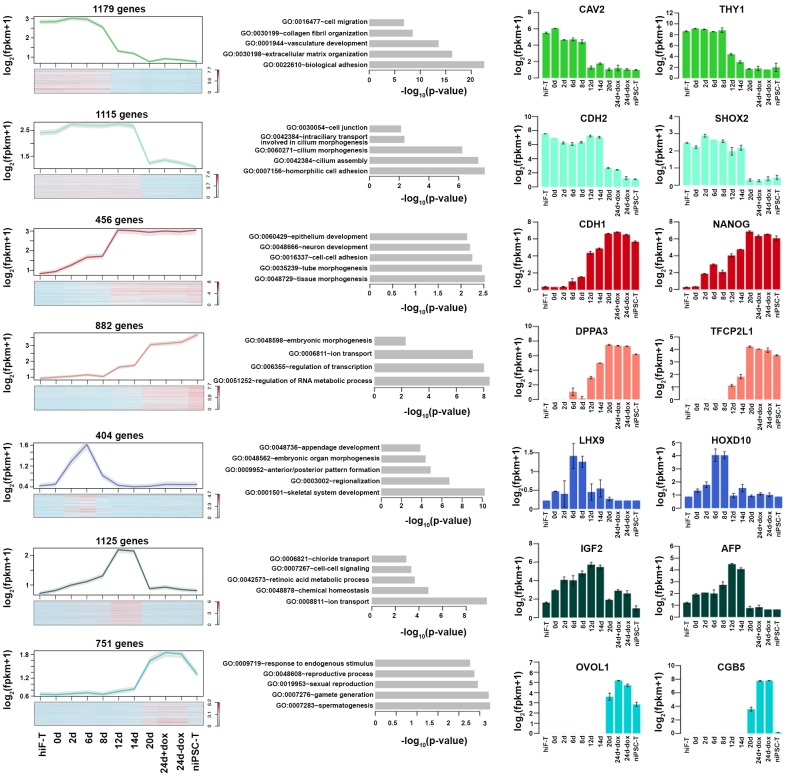
Figure 2. Transcriptional profiling of cells during naïve reprogramming.
(A) Schematic representation of reprogramming intermediate collection at different time points, as indicated. hiF-T cells were first cultured in conventional hESM with dox for 6 days and then switched to 5iLAF culture medium supplemented with dox until day 20. Cells with or without dox treatment for four additional days were collected. (B) MDS analysis of RNA-seq data during the naïve reprogramming process. (C) Line plots showing transcriptional dynamics of differentially expressed genes during the naïve reprograming process. Genes were grouped by k-means clustering. Gray shades represent a 95% bootstrap confidence interval around the mean value. (D) Heatmaps showing the expression patterns of genes with pre-implantation signatures in both the naïve and primed reprogramming process. (E) Correlation analysis of transcriptional profiles between naïve reprogramming and the embryonic development process, with the Pearson correlation coefficient of each pair shown on each cell of the heatmap. (F) Immunostaining images of pluripotency-related marker expression in the reprogramming cells at indicated time points during naïve and primed reprogramming. Scale bar, 50 μm.