Abstract
Much of the current research on longevity focuses on the aging process within a single species. Several molecular players (e.g. IGF1 and MTOR), pharmacological compounds (e.g. rapamycin and metformin), and dietary approaches (e.g. calorie restriction and methionine restriction) have been shown to be important in regulating and modestly extending lifespan in model organisms. On the other hand, natural lifespan varies much more significantly across species. Within mammals alone, maximum lifespan differs more than 100 fold, but the underlying regulatory mechanisms remain poorly understood. Recent comparative studies are beginning to shed light on the molecular signatures associated with exceptional longevity. These include genome sequencing of microbats, naked mole rat, blind mole rat, bowhead whale and African turquoise killifish, and comparative analyses of gene expression, metabolites, lipids and ions across multiple mammalian species. Together, they point towards several putative strategies for lifespan regulation and cancer resistance, as well as the pathways and metabolites associated with longevity variation. In particular, longevity may be achieved by both lineage-specific adaptations and common mechanisms that apply across the species. Comparing the resulting cross-species molecular signatures with the within-species lifespan extension strategies will improve our understanding of mechanisms of longevity control and provide a starting point for novel and effective interventions.
Introduction
Although the words “aging” and “longevity” are often used interchangeably, they ought to be viewed as two related but distinct processes (Gladyshev, 2016). The first is the aging process within an organism. Here, aging populations (e.g. humans) can be compared with non-aging organisms (e.g. hydra). The second is the adjustment in the time an organism lives. Here, different species or organisms within a population may be compared with regard to differences in lifespan.
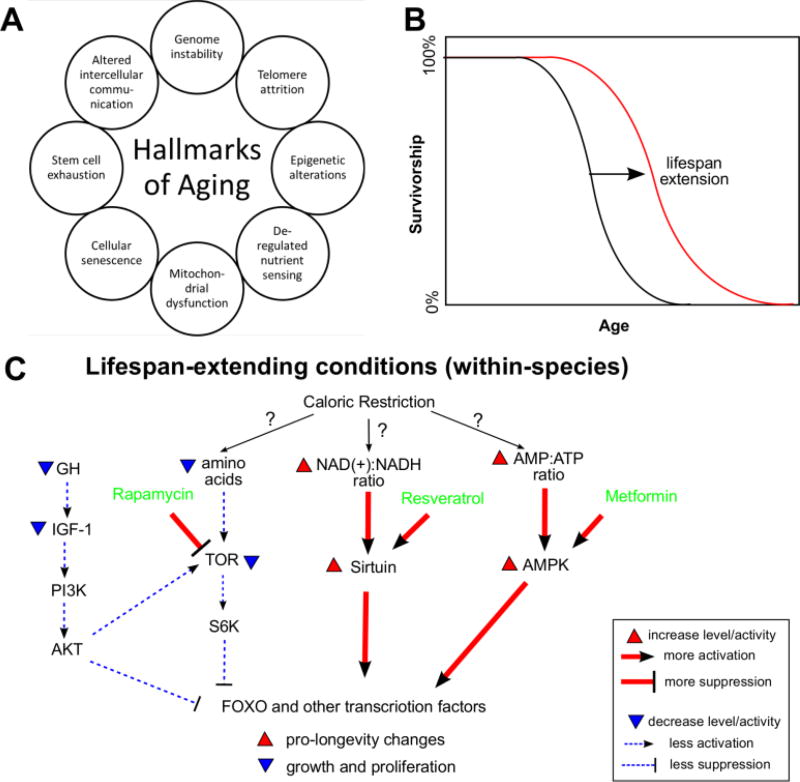
As an individual grows old, many changes occur on the physiological and molecular levels (Figure 1A). Some of the most visible age-related molecular changes can be summarized as 9 hallmarks of aging: increased genome instability, telomere attrition, changes in epigenetic markers, loss of proteostasis, deregulated nutrient sensing, mitochondrial dysfunction, induction of cellular senescence, exhaustion of stem cell population, and altered intercellular communication (Lopez-Otin et al., 2013). However, other molecules, molecular processes, cells and systems within the organism change with age too, so age-related changes are easier to conceptualize as age-related global remodeling. Even if a particular molecule, process, or cell does not change in abundance with age, its interaction with the processes that do change may make it age-related. Absent of extrinsic causes of death, the mortality rate (i.e. the number of deaths per unit population per unit time) of most species increases with age (Jones et al., 2014), producing a concave, downward-sloping survival curve (Figure 1B).
Figure 1. Aging process within a single species.
(A) Hallmarks of aging. The hallmarks are as in (Lopez-Otin et al., 2013).
(B) A typical survival curve. A successful lifespan extension strategy shifts or scales the black curve to the red.
(C) Major pathways implicated in lifespan extension. Only selected components of the pathways are shown. Cross-talks among the pathways are omitted. Caloric restriction is used as an example for illustrative purposes.
Much effort has been directed towards identifying genes, pathways, and treatments that can delay these age-related changes or increase species lifespan. Research on model organisms revealed several key molecular players (Figure 1C), such as the GH/IGF1/FOXO axis (GH: growth hormone; IGF1: insulin-like growth factor 1; FOXO: forkhead box O), TOR/S6K (TOR: target of rapamycin; S6K: ribosomal S6 kinase), sirtuins, and AMPK (AMP-activated protein kinase) (Fontana et al., 2010; Haigis and Sinclair, 2010; Kenyon, 2010). Pharmacological compounds that act on these pathways, such as rapamycin (TOR inhibitor), resveratrol (sirtuin activator), and metformin (AMPK activator), have been reported, at least under some conditions, to extend lifespan in laboratory animals (Harrison et al., 2009; Onken and Driscoll, 2010; Wood et al., 2004). Dietary restriction, in which an animal’s dietary calorie intake is reduced while maintaining normal balance of nutrients, was the first method proven to be effective in extending mammalian longevity (Fontana et al., 2010; McCay et al., 1935). Animals under dietary restriction experience substantial metabolic remodeling and significant changes in endocrine levels, fat oxidation, reactive oxygen species production and protein turnover (Sinclair, 2005). However, the exact mechanisms are not fully clear and its effectiveness in primates is still debated (Colman et al., 2014; Mattison et al., 2012).
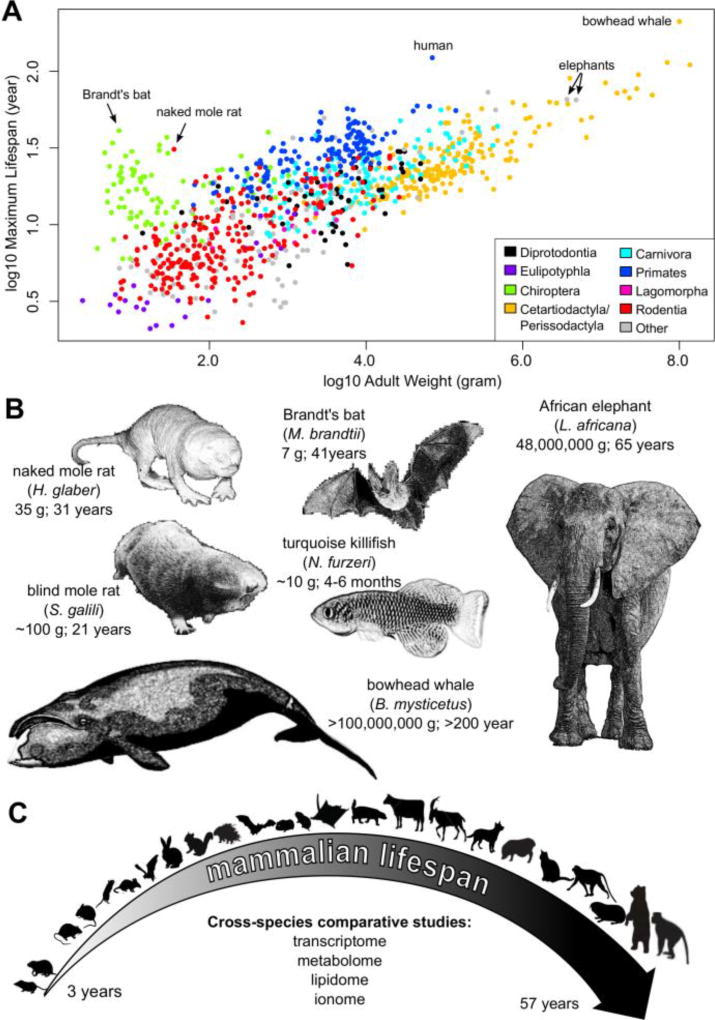
The other area of longevity research looks at lifespan differences across species, which is the focus of this review. The longevity differences in mammals alone are already remarkable (Figure 2A). All extant mammals descend from a common ancestor that lived ~210 million years ago, yet they exhibit > 100-fold difference in lifespan and 50 million-fold variation in body weight (Tacutu et al., 2013). On the one extreme are small and short-lived species: Etruscan shrews (Suncus etruscus) weigh ~ 2 g and live up to 3.2 years. The other extreme are the large and long-lived beasts: African elephant (Loxodonta africana) is the largest land mammal, weighing up to 6 tons and living to 70 years; in the ocean, bowhead whale (Balaena mysticetus) can weigh > 100 tons and are estimated to live > 200 years (Tacutu et al., 2013). There are also lineages that defy the trend of the large long-lived and show exceptional longevity based on deviation from it. For example, despite being smallest mammals (~5–20 g), microbats can live for more than 40 years (compared to < 4 years in shrews and small rodents) (Seim et al., 2013). The naked mole rat (Heterocephalus glaber) lives 10 times longer than other rodents of comparable size (Buffenstein, 2008; Fang et al., 2014b; Kim et al., 2011). Humans are also considered long-lived: Jeanne Calment of France holds the longevity record of 122 years and 164 days (Whitney, 1997), whereas many other similarly sized mammals live only about 30 years. Among vertebrates, the longest-lived animal is perhaps Greenland shark (Somniosus microcephalus), with the age of one individual recently estimated to be 392 ± 120 years (Nielsen et al., 2016).
Figure 2. Lifespan variation across different species.
(A) Maximum lifespan correlates positively with adult weight. Individual points correspond to individual species and are colored by taxonomic orders. Selected species were labeled. Adult body mass and maximum lifespan records of 995 mammalian species used to make the figure are from AnAge Database (Tacutu et al., 2013).
(B) Some recently sequenced species with remarkable longevity. Animal sketches are not drawn to scale.
(C) Comparative omics studies across mammalian species. Animal sketches are not drawn to scale.
The human life expectancy at birth in developed countries has increased by 27 years over the last century largely due to improvements in public health care (Hayflick, 2007), but heath care alone cannot change the biological limit of species longevity. Even with best provision of nutrients and optional environmental conditions in a laboratory, wildtype mice do not survive for 5 years. When discussing species longevity here, we refer to its maximum lifespan, i.e. the longest time a member of the species has been observed to live. For most animals this refers to their lifespans in captivity (Holliday, 2006), but for the exceptionally long-lived species, one has to rely on unconventional methods. For example, a male Brandt’s bat (Myotis brandtii) was recaptured in the wild in Siberia, Russia, 41 years after it was originally banded (Podlutsky et al., 2005). Harpoon points made of ivory and stone were found in three bowhead whales captured in 2007, and such weapons were last manufactured in New England in 1880s (George and Bockstoce, 2008). The age dating of Greenland sharks relies on the radiocarbon levels in crystalline proteins of eye lens nuclei, and the fallouts from atmospheric tests of nuclear weapons in 1960s help differentiate those individuals born before and born after this period (Nielsen et al., 2016). Since maximum lifespan may suffer from reporting bias, as an alternative one can track female time to maturity (i.e. the time to reach puberty), which may be measured more easily and show strong correlation with maximum lifespan (Fushan et al., 2015).
In general, longer-lived species tend to be larger in size, produce fewer offspring, grow more slowly, and have lower mass-specific metabolic rates (Peters, 1986; Sacher, 1959; Western, 1979). These coordinated changes likely require rewiring of multiple biological pathways. Indeed, natural selection acts not on longevity per se, but on the overall reproductive success and survival. There is little advantage for a mouse to have a lifespan potential of 80 years if it is likely to be eaten by a predator or die of cold or hunger within one year of birth. Therefore, to achieve long lifespan a species must also have the corresponding changes in body size, reproduction, growth rate, development rate and metabolism. Evolution has resulted in many different species with varying life history traits, and the general pattern of lifespan modulation will emerge if we look across the entire spectrum. However, to date many questions remain on the mechanism of longevity control and regulation. For example, how do the long-lived species differ from the short-lived species in terms of genome, transcriptome, proteome and metabolome? Are these patterns unique to certain exceptionally long-lived species, or can they also account for the general lifespan variation (Partridge and Gems, 2002)? Do the naturally long-lived species have transcriptomic and metabolic states similar to those laboratory animal models subjected to lifespan extension treatments?
The past few years have seen the publication of the genomes of several species known for their remarkable lifespan or cancer resistance (Gorbunova et al., 2014) (Figure 2B): naked mole rat (Kim et al., 2011), blind mole rat (Spalax galili) (Fang et al., 2014a), African elephant (Abegglen et al., 2015), Brandt’s bat (Seim et al., 2013), black flying fox (Pteropus alecto) and David’s Myotis (Myotis davidii) (Zhang et al., 2013), bowhead whale (Keane et al., 2015; Seim et al., 2014) and Minke whale (Balaenoptera acutorostrata) (Yim et al., 2014). On the other end of the longevity spectrum, the genome of the African turquoise killifish (Nothobranchius furzeri), which is a particularly short-lived species (4–6 months in captivity), was also published recently by two groups (Reichwald et al., 2015; Valenzano et al., 2015). Many of these studies look at the physiological and biological adaptations of these species and shed some light on the molecular basis of their exceptional longevity (Table 1). For example, a recently published study on hypoxia resistance in naked mole rat revealed the animal’s remarkable ability in utilizing fructose for glycolysis to avoid the lethal effects of oxygen deprivation (Park et al., 2017). In parallel, there are also a number of recent comparative studies on gene expression (Brawand et al., 2011; Fushan et al., 2015), splicing (Merkin et al., 2012), metabolites (Ma et al., 2015b), lipids (Bozek et al., 2017), and element levels (Ma et al., 2015a) across multiple mammalian species (Figure 2C), which start to reveal the molecular signatures associated with longevity (Table 2). Together, these studies provide the first insights into the general principles of lifespan control.
Table 1.
Putative lifespan-related traits and other features associated with longevity in exceptionally long- and short-lived species.
| Animal | Notable features / putative lifespan-related traits |
|---|---|
| Naked mole rat |
|
| Blind mole rat |
|
| African elephant |
|
| Microbats |
|
| Bowhead whale |
|
| African turquois killifish |
|
Table 2.
Summary of key findings from recent cross-species comparative studies.
| Cross-species studies | Key findings |
|---|---|
| Transcripts (33 mammalian species, 3 organs) |
|
| Transcripts (15 mammalian species, fibroblasts) |
|
| Metabolites (26 mammalian species, 4 organs, 262 metabolites) |
|
| Elements (26 mammalian species, 4 organs, 18 elements) |
|
| Lipids (35 mammalian species, 6 organs, 13,000–21,000 compounds) |
|
Naked mole rat
The naked mole rat is a subterranean, exceptionally long-lived eusocial mammal. Even though it is of similar size as mouse, it lives for > 30 years and is the longest-lived rodent. Naked mole rats are also remarkably resistant to cancer, based on multi-year observations of this species’ large colonies (Buffenstein, 2008) and transfection of naked mole rat fibroblasts with activated Ras and SV40 LT to induce oncogenic transformation (Seluanov et al., 2009). Nevertheless, cancer is possible in this species, as recently shown (Delaney et al., 2016; Taylor et al., 2017). Sequencing of the genomes of the naked mole rat (Kim et al., 2011) and a closely related species, the Damaraland mole rat (Fang et al., 2014b), as well as a number of cellular and biochemical studies begin to shed light on the mechanisms for their cancer resistance and long lifespan (Azpurua et al., 2013; Seluanov et al., 2009; Tian et al., 2013). Compared to mouse fibroblasts, naked mole rat fibroblasts was suggested to undergo cell division arrest at a much lower density in cell culture, in a process called “early contact inhibition” associated with p16Ink4a induction (Seluanov et al., 2009). It was reported that early contact inhibition is mediated by high-molecular-mass hyaluronan (HA), an unbranched disaccharide glucuronic acid/N-acetylglucosamine polymer found in extracellular matrix (Tian et al., 2013). HA secreted by naked mole rat fibroblasts is of higher molecular mass and in larger quantity than that secreted by human and mouse fibroblasts, possibly due to a unique sequence change in naked mole rat hyaluronan synthase 2 (HAS2) and reduced activity of HA-degrading enzymes. When the high-molecular-mass HA is removed by either HAS2 knockdown or HA-degrading enzyme overexpression, the naked mole rat cells become susceptible to malignant transformation (Tian et al., 2013), suggesting the high-molecular-mass HA is a part of their cancer resistance strategies.
Other mechanisms have also been proposed to account for their exceptional lifespan. Naked mole rat 28S ribosomal RNA has a cryptic intron near the center of the sequence and is processed into 2 fragments (Fang et al., 2014b). The resulting naked mole rat ribosome has a higher translational fidelity than mouse ribosome and up to 10-fold lower amino acid mis-incorporation rate at 32 °C (Azpurua et al., 2013). The whole genome sequencing (Kim et al., 2011) revealed unique amino acid changes in several proteins involved in cell cycle (e.g. cyclin E1 (CCNE1), uncoupling protein 1 (UCP1), γ-crystallin (CRYGS)) and DNA integrity (e.g. DNA repair enzyme (APEX1), replication factor C (RFC1), and DNA topoisomerase (TOP2A)). There are two early stop codons in the p16Ink4A transcript and four stop codons in the p19Arf transcript, and molecular cloning confirmed the proteins were shorter than those in mouse (comparable to guinea pig) but remained functional (Miyawaki et al., 2015). Interestingly, the naked mole rat INK4a/b locus also encodes a hybrid protein isoform called pALTInk4a/b, which consists of p15Ink4b exon 1 joined to p16Ink4a exons 2 and 3. pALTInk4a/b is activated during early contact inhibition and by a variety of stresses but has higher capacity to induce cell cycle arrest than either p15Ink4b or p16Ink4a, and this product is unique to the naked mole rat and is not observed in human or mouse (Tian et al., 2015). In addition, naked mole rat p19Arf lacks the apoptotic domain of mouse p19Arf. When over-expressed in cell lines, the naked mole rat protein could induce cell cycle arrest but produce less cell death than the mouse protein (Miyawaki et al., 2015).
Transcriptome analyses also revealed lower expression of genes involved in insulin/IGF1 signaling in naked mole rat liver than in mouse liver (Kim et al., 2011). Both naked mole rat and Damaraland mole rat express IGF2 and its binding protein IGF2BP2 in liver, whereas IGF2 expression is significantly downregulated after birth in most other mammals (Fang et al., 2014b). Comparative genomic analyses suggest that the naked mole rat, Damaraland mole rat and other hystricognath rodents harbor a sequence change in insulin β-chain associated with insulin misfolding and diabetes, even though they are able to handle glucose in the absence of conventional insulin (Fang et al., 2014b). Furthermore, liver gene expression profiles revealed that the naked mole rat expressed higher levels of genes associated with oxidation-reduction and mitochondria than mouse (Yu et al., 2011). Comparison of naked mole rats and nine other rodent species also showed positive correlation between species maximum lifespan potential and the activity of nuclear factor erythroid 2-related factor (Nrf2) (a transcription regulator of many cytoprotective molecules), with the longer-lived species suppressing the degradation of Nrf2 by down-regulating Kelch-like ECH-Associated Protein 1 (Keap1) and β-transducin repeat-containing protein (βTrCP) (Lewis et al., 2015). In addition, when exposed to low O2 conditions, naked mole rat liver also showed down regulation of genes associated with energy metabolism (especially homeostasis of triglycerides and lipids) and upregulation of genes involved in immune signaling (including chemokine and IFN-γ pathways) (Kim et al., 2011). These data suggest that a less bioactive insulin, alterations in mitochondria and oxidation-reduction pathways, and augmented Nrf2 activities may have contributed to the enhanced longevity of naked mole rats, whereas they alter their energy metabolism and immune signaling in response to the hypoxic environment.
A recently published study on hypoxia resistance in naked mole rat revealed the interesting metabolic rewiring in this animal (Park et al., 2017). Given their subterranean and eusocial environment, naked mole rats routinely live under low-O2 and high-CO2 conditions, with CO2 levels in their burrows reaching up to 10%. Subjecting naked mole rates to 5% O2 for 5 hours produced no apparent side effects, whereas mice died within 15 minutes. Even under 0% O2 condition for 18 minutes, naked mole rates could be revived successfully upon reoxygenation (Park et al., 2017). Gene expression and metabolic analyses revealed that naked mole rats express higher levels of fructose transporter GLUT5 and ketohexokinase KHK-A and KHK-C in multiple organs than mice, and under anoxia condition they are able to utilize fructose for glycolysis (Park et al., 2017). It was suggested that naked mole rats may have evolved the fructose-driven glycolysis in order to bypass the metabolic block at phosphofructosekinase and ensure the continued supply of energy to vital organs such as brain and heart in anaerobic conditions (Park et al., 2017). In mammals, a switch to fructose metabolism is often associated with cancer malignancy of pancreas and small intestines (Liu et al., 2010; Port et al., 2012), but naked mole rats are highly resistant to cancer and can utilize fructose with no apparent physiological drawback (Park et al., 2017). It is worth exploring further the link between the metabolic rewiring and hypoxia resistance in naked mole rats and their exceptional longevity.
Blind mole rat
The blind mole rat is another subterranean rodent characterized by remarkable cancer resistance. In over 50 years of research, no individuals were reported to have spontaneous tumors, and the animals were also very resilient to treatments with potent chemical carcinogens (Manov et al., 2013). As naked mole rat and blind mole rat are evolutionarily distant rodents (the former groups with guinea pigs while the latter with mice and rats), they may have evolved independent mechanisms for cancer resistance (Gorbunova et al., 2014). Blind mole rat fibroblasts cultured in vitro always undergo massive necrotic cell death after 7–20 population doublings (Gorbunova et al., 2012). This “concerted cell death” phenomenon was caused by interferon β (IFN-β) secreted by aging blind mole rat cells into the culture, and the conditioned media could trigger similar massive necrosis in mouse cells (Gorbunova et al., 2012).
The publication of the blind mole rat genome (Fang et al., 2014a) and other molecular studies provided further insights into their unique physiologies and cancer resistance mechanisms. Given the highly hypoxic subterranean environment, the blind mole rat has evolved a number of adaptive strategies, including improved myocardial oxygen delivery and function (Edoute et al., 1988), adaptive heart rate and breathing frequencies (Arieli et al., 1986b), high blood hemoglobin and hematocrit concentrations (Arieli et al., 1986a), and increased tissue mitochondrial densities (Widmer et al., 1997). On the molecular levels, there were also distinct gene expression patterns in blind mole rat compared to rat under hypoxic conditions (Shams et al., 2005), including down-regulation of genes coding for p53 regulator MDM2 (MDM2), caspase 9 (CASP9), cyclin G1 (CCNG1), and cytochrome C (CYTC) (Fang et al., 2014a), and up-regulation of myoglobin in muscle, neuroglobin in brain, and cytoglobin in fibroblast-like cells (Avivi et al., 2010). In particular, both cyclin G1 and MDM2 are transcriptional targets of p53. High levels of cyclin G1 stimulate p53 activities in a positive feedback loop, whereas MDM2 mediates the degradation of p53 and cyclin G1 by the proteasome (Zhao et al., 2003). The coordinated changes in MDM2 and cyclin G1 expression are therefore consistent with altered p53 activities during hypoxia in blind mole rats.
Interestingly, the blind mole rat p53 protein has an amino acid change corresponding to a mutation (R174K in human) frequently found in human tumors, which leads to partial inactivation of p53 functions and favoring the binding to promoters of cell cycle arrest genes rather than apoptotic targets (Ashur-Fabian et al., 2004; Avivi et al., 2005; Kato et al., 2003). The sequences of the activation and regulatory domains of the blind mole rat p53 are more similar to other hypoxia-tolerant species than expected from phylogeny, and 3D modeling showed the interactions with coactivator p300 and DNA repair protein RPA70 were reduced in the blind mole rat p53 (Domankevich et al., 2016). APAF1 promoter, a central player in the mitochondrial apoptotic pathway, is completely non-responsive to the blind mole rat p53 (Avivi et al., 2005). It was hypothesized that such a mutation arose in human tumors due to the selection pressure of hypoxia to favor the cells less prone to hypoxia-induced apoptosis (Achison and Hupp, 2003), and blind mole rats probably utilize the same mechanism as an adaptation to the hypoxic subterranean environment (Ashur-Fabian et al., 2004).
Furthermore, the B1 SINE repeat showed significant hypoxia-induced transcriptional upregulation in the blind mole rat (but not in rat), which is consistent with weak p53 control and activation of interferon response (Fang et al., 2014a). The gene coding for IFN-β1 (IFNB1) underwent a duplication event in the blind mole rat genome, effectively increasing the gene dosage. The MX1 gene from the interferon signaling pathways and several genes involved in cell death and inflammation regulation also underwent expansion or showed evidence of positive selection (Fang et al., 2014a). In addition, heparanase is an endoglycosidase that degrades heparan sulfate in the extracellular matrix and often correlates with tumor aggressiveness, metastasis and angiogenesis. A unique splice variant of this enzyme is expressed in the blind mole rat, which functions as a dominant negative to the wild-type enzyme and can inhibit heparan sulfate degradation, suppress glioma tumor growth and reduce cancer cell colonization of lung in mouse model (Nasser et al., 2009).
Analysis of the stress response Nrf2-Keap1 pathway revealed a key residual change in the blind mole rat Keap1 protein important for binding and degradation of Nrf2 (Schmidt et al., 2016). As noted above, naked mole rats also suppressed the degradation of Nrf2 by down-regulating Keap1 (Lewis et al., 2015), indicating the convergent evolution of the Nrf2-Keap1 pathway. Overall, the data suggest that, in order to adapt to the low-oxygen subterranean environment, blind mole rats have evolved to dampen the apoptotic function of p53. As compensation, they have strengthened the immune-inflammatory response and interferon pathways and undergone additional changes at heparanase and Keap1, which may function as cancer resistance mechanisms.
African elephant
In the 1970s, epidemiologist Richard Peto of the University of Oxford observed that contrary to expectation, there is little relationship between cancer incidence and the number of cells in an organism (Peto et al., 1975). This so-called “Peto’s paradox” suggests that larger animals may have evolved additional mechanisms to reduce cancer rates. A recent study of the African elephant genome suggested the answer may lie in multiple copies of tumor suppressor gene TP53 (Abegglen et al., 2015). Analysis of the genome of African elephant genome revealed one TP53 copy with comparable gene structure to those found in other mammalian species, as well as 19 additional copies that lack true introns, correspond to ORFs with partial sequences or premature stop codons, and may have originated from retro-transposition. Similarly, Asian elephant DNA also contains 15 to 20 copies of these retrogenes (pseudogenes), suggesting it may be a common evolutionary feature among elephants.
When exposed to radiation, African elephant peripheral blood lymphocytes (PBLs) showed measurable TP53 retrogene expression and Mdm2 binding, underwent apoptosis at much higher rate than normal human PBLs, and showed elevated p21 protein expression (Abegglen et al., 2015). Similar responses were also observed in Asian elephant PBLs. In contrast, human patients suffering from Li-Fraumeni syndrome lack one functional allele of TP53 and have > 90% lifetime risk for cancer, and their PBLs showed significantly less apoptotic response to ionizing radiation (Abegglen et al., 2015). It was estimated that elephant cells were twice as sensitive to DNA damage-induced apoptosis as human cells, and a 2-fold decrease in somatic mutation rate can result in 100X increase in cell mass without cancer transformation (Abegglen et al., 2015). Therefore, it was suggested that, through the multiple copies of TP53 retrogenes, elephants may have reduced their cancer incidence by increasing the cellular sensitivity to DNA damages, lowering the threshold of apoptosis, and efficiently removing mutant cells. The nature of these effects and the associated mechanisms, however, remain unclear, since the number of functional TP53 in the elephant has not been altered compared to other mammals. Resistance to cancer should also be examined more thoroughly in elephants, as the initial report did not reveal a clear age-related increase in cancer incidence in this species (Abegglen et al., 2015), which is one of the defining features of cancer in general. It should also be noted that while modulation of p53 activities was involved in both blind mole rats and African elephants, the strategies were somewhat different: blind mole rat p53 (due to the amino acid change) favors cell cycle arrest over apoptosis, whereas African elephant p53 (due to multiple copies of retrogenes) was proposed to sensitize the cells to DNA damage-induced apoptosis. Therefore, the same molecular player may have evolved along different paths to adopt to the long lifespans of these animals.
Microbats
Bats have a number of remarkable features. They are the only mammals that have evolved powered flight, are among the few animals that echolocate, and despite their relatively small sizes, are exceptionally long-lived as a lineage (Seim et al., 2013). Brandt’s bats hold the longevity record of at least 41 years. While other Myotis species and black flying fox are reported to live for about 20 years (Tacutu et al., 2013), accurate records are rarely available, and it is likely that many other microbats are long-lived as well. The genomes of Brandt’s bat, David’s Myotis and black flying fox have offered insights into their remarkable physiology and lifespan (Seim et al., 2013; Zhang et al., 2013). A large number of genes involved in DNA damage checkpoint and DNA repair pathway were found to be under positive selection in the bat ancestor, including ATM, RAD50, KU80 (XRCC5), and MDM2, which may reflect adaptation to minimize or repair the negative effects of reactive oxygen species generated during flight (Zhang et al., 2013). Remarkably, XRCC5 also showed positive selection signals in the short-lived killifish (Reichwald et al., 2015; Valenzano et al., 2015), and its gene expression level correlated positively with maximum lifespan of mammals (Fushan et al., 2015).
The gene families involved in immune response have undergone expansion in bats, including those coding for immunoglobulin subtype and immunoglobulin V-set subgroup (Seim et al., 2013). c-REL, whose gene product belongs to the nuclear factor κB (NF-κB) family of transcription factor and is involved in innate and adaptive immunity, also exhibits a feature of positive selection (Zhang et al., 2013). As bats are known to carry several deadly viruses without being adversely affected, the long-term coexistence of bats and viruses may have led to strong selective pressures on the genes involved in immunity. Further, the link between immune response and longevity was also noted in other studies. As mentioned above, the long-lived naked mole rats upregulated their immune response genes under hypoxic conditions (Kim et al., 2011), whereas in the blind mole rats several genes involved in inflammation regulation (e.g. Ifnb1 and Mx1) undergo duplication or show positive selection signals (Fang et al., 2014a). In a study of gene expression variation across 33 mammalian species (discussed more below), the expression of immune response genes in brain, kidney, and liver all showed positive correlation with species maximum lifespan (Fushan et al., 2015). Animal aging also associates with the upregulation of genes involved in immune response and complement activation, as observed in the aging brain in the short-lived killifish (Baumgart et al., 2014), and in a meta-analysis of 27 databases of age-related gene expression profiles in mice, rats and humans (de Magalhães et al., 2009).
A number of unique sequence changes in the bat genomes may help to explain their exceptional longevity. Ablation of signaling along the GH/IGF1 axis is known to be associated with increased resistance to diabetes and cancer in humans and mice (Coschigano et al., 2000; Guevara-Aguirre et al., 2011), and the grown hormone receptor (GHR) of the Brandt’s bat and several other bat species lacks a key leucine residue in the transmembrane domain (Seim et al., 2013). There are also multiple unique amino acid changes in the transmembrane region and the sequences immediately preceding and following it in IGF1 receptor (IGF1R) of several bat species, which may alter its signaling function. Furthermore, the gene expression profile of the Brandt’s bat was very similar to those of long-lived mice models, with 11 out of 15 orthologous genes moving in the expected directions. Small islet of Langerhans and decreased β-cell mass often found in GHR−/− mice were also observed in the pancreas of the Brandt’s bat (Seim et al., 2013). Changes in GHR and IGF1R are associated with dwarfism in rodents and other species, including humans (Laron syndrome) and are consistent with the reduced body size of microbats, suggesting this is one of the features contributing to the exceptional longevity of microbats. Comparison of gene expression in hibernating versus summer active Brandt’s bats revealed downregulation of genes involved in protein synthesis, glycolysis, splicing, and respiration, and upregulation of genes involved in lipid metabolism, coagulation, maintenance of oxidative stress and adaptation to starvation. In addition, unique amino acid changes in the receptor-ligand interface of follicle-stimulating hormone beta-subunit (FSHβ) may explain the delay ovulation in hibernating bats during winter (Seim et al., 2013). Given that hibernation is linked to increased survival and slower life histories in mammals (Turbill et al., 2011), these may represent the evolutionary changes associated with the extended lifespan.
Bowhead whale
Bowhead whales are one of two largest and also longest-lived mammals, with estimated maximum lifespan of over 200 years (George and Bockstoce, 2008). Similar to other long-lived mammals, they evolved to have reduced fecundity and extended developmental time (Tacutu et al., 2013). The publication of the genomes of the bowhead whale (Keane et al., 2015) and the related Minke whale (which is smaller and shorter lived) (Yim et al., 2014) enabled the use of comparative genomics to understand the evolution of bowhead whale longevity. A number of genes related to aging and cancer showed dN/dS values above 1 in whales, including suppressor of cytokine signaling 2 (SOCS2), aprataxin (APTX), noggin (NOG), and leptin (LEP). In particular, the genes FOXO3 (Forkhead box protein), ERCC3 (involved in DNA repair), FGFR1 (fibroblast growth factor receptor) showed high dN/dS values in bowhead whale specifically, whereas amino acid changes unique to bowhead whale were found in ERCC1 (DNA repair), HDAC1 and HDAC2 (histone deacetylases) (Keane et al., 2015). The DNA replication processivity factor PCNA and the amino acid sensor and mTORC1 activator LAMTOR1 also underwent both gene duplication and unique amino acid changes. Transcriptional analysis suggested differential expression of genes involved in DNA replication in the bowhead whale liver compared to other mammals (Seim et al., 2014). Interestingly, the UCP1 gene (coding for uncoupling protein 1), which has several unique amino acid changes in the naked mole rat (Kim et al., 2011), also has a premature stop codon in C-terminal region in the bowhead whale (Keane et al., 2015). This may reflect their need for thermoregulation that are unique from other mammals (naked mole rats are poikilothermic and vary their temperatures with the environment, whereas bowhead whales need to maintain stable body temperatures in the cold ocean water).
African turquoise killifish
Unlike other species mentioned above, the African turquoise killifish (Nothobranchius furzeri) is known for its exceptionally short lifespan. Found in the ephemeral ponds in Zimbabwe and Mozambique that are present only during the wet season, the turquoise killifish has evolved a state of embryonic diapause to survive through the dry season. The strains derived from different killifish population can exhibit different experimental lifespans from 3 to 12 months, depending on the genetic background (Reichwald et al., 2015; Terzibasi et al., 2008; Valenzano et al., 2015). The killifish genome, based on the short-lived and inbred GRZ strain, was recently published by two groups and revealed a number of interesting features related to aging and longevity (Reichwald et al., 2015; Valenzano et al., 2015). Using the available fish genomes as outgroup, Valenzando et al. reported ~500 genes under positive selection, including 22 previously known aging-related genes, such as insulin receptor A (INSRA), IGF1R (IGF1RA), insulin receptor substrate 1 (IRS1), nuclear Lamin-A/C (LMNA3), BCL-2 associated X protein (BAX) and the DNA repair protein XRCC5 (XRCC5) (Valenzano et al., 2015). Taking a complementary approach, Reichwald et al. used the de novo assembled transcriptomes of five additional Nothobranchius species as well as the non-annual killifish Aphyosemion striatum as outgroup, and reported several genes under positive selection, including inhibitor of DNA binding 3 (ID3) and Kappa B Kinase Interacting Protein (IKBIP) (Reichwald et al., 2015). ID3 is a key component of TGF-β signaling and IKBIP is a pro-apoptotic gene; they and several other genes with positive selection signals all showed up- or down-regulation during aging in killifish (Reichwald et al., 2015). Enrichment analysis of the expression changes during killifish aging and diapause also revealed upregulation of translational elongation and ribonucleoprotein complex biogenesis, and downregulation of cell cycle and DNA replication (Reichwald et al., 2015).
A more careful analysis on the positively selected sites reported by Valenzado et al. also revealed the origin of the positive selection signals (Sahm et al., 2016). While some sites were either false positives due to gene model error or also found in the non-annual killifish, there were 4 sites in XRCC5, 4 sites in IGF1RA, and 1 site each in BAX and IRS1 that occurred specifically among the Nothobranchius genus and were therefore associated with diapause and shorter lifespan (Sahm et al., 2016). Furthermore, two sites in XRCC5 were unique to the short-lived N. furzeri, while one of the sites in IGF1RA was located within the tyrosine kinase domain and was predicted to have a strong functional effect (Sahm et al., 2016). Reduction in insulin/IGF1R function is associated with lifespan extension in worms, flies, and mice (Fontana et al., 2010; Kenyon, 2010), and functionally significant mutations in IGF1R have been reported in human centenarians (Suh et al., 2008).
Interestingly, comparison of the positive selected residues in killifish IGF1R and in Brandt’s bat IGF1R, and the longevity mutations in IGF1R of long-lived C. elegans DAF-2 mutant, revealed that these residues all locate in the key functional domains but do not overlap, suggesting that those proteins acting as central nodes in longevity controls may be harnessed for both lifespan extension or lifespan contraction, depending on the residues involved (Valenzano et al., 2015). The same was also observed for several other genes with positive selection signals in killifish (Sahm et al., 2017). POLRMT codes for mitochondrial RNA polymerase and showed positive selection signals in two branches of the short-lived killifish as well as in the long-lived Brandt’s bat. Tenascin (TNC) was positively selected in humans and the last common ancestor of mole rats, while collagen type IV alpha 2 (COL4A2) was positively selected in naked mole rat and the last common ancestor of mole rats, and both genes also showed positive selection signals along the branch leading to the killifish N. furzeri, N. kadleci and N. kuhntae (Sahm et al., 2017). Furthermore, a significant number of genes coding for components of mitochondrial respiratory chain complex I, mitochondrial ribosome proteins, and proteins involved in mitochondrial biogenesis and respiratory chain complex assembly also showed positive selection in killifish (Sahm et al., 2017). In agreement, mitochondrial respiratory chain complex I was previously identified as a hub of gene modules whose expression correlated negatively with lifespan in killifish (Baumgart et al., 2016) and with lifespan of mammalian species (Fushan et al., 2015). Partial pharmacological inhibition of complex I by the small molecule rotenone could result in 15% lifespan extension in killifish (Baumgart et al., 2016). Hence, the regulation of longevity and aging in both long-lived and short-lived species may involve the parallel evolution of the same set of genes and pathways (Sahm et al., 2017; Valenzano et al., 2015).
Challenges in cross-species studies
Besides looking at the exceptionally long-lived or short-lived species, one can approach the question of lifespan regulation by examining molecular patterns across different species along the continuum of longevity. Several early studies quantified a number of biochemical and enzymatic parameters, revealing that the longer-lived species tend to have greater maintenance capacity, suffer less damage, and are more efficient at repair than the shorter-lived species (Holliday, 1997, 2006). However, until recently there had been very few cross-species comparative studies analyzing the entire mammalian transcriptome or metabolome (Bozek et al., 2017; Brawand et al., 2011; Fushan et al., 2015; Ma et al., 2015a; Ma et al., 2015b; Merkin et al., 2012). Besides the difficulty in obtaining reliable biological samples and the costs of large-scale omics studies, there are also some unique challenges in cross-species analyses that require particular attention.
The relationship between two variables can be modeled by regression analysis. In the context of longevity research, regression can be performed between any molecular measurement (e.g. gene expression or metabolite level) and longevity trait (e.g. maximum lifespan) to identify the genes or metabolites showing statistically significant correlation. Linear regression by ordinary least squares is the simplest and commonly used form in biological research, but it requires the input data to meet a number of statistical assumptions. In particular, the variance of residuals is assumed to be equal and uncorrelated (i.e. homoscedasticity and independence) (Logan, 2010). While these assumptions are likely valid for random samples from a single species, the assumption of independence may be violated for samples collected from many different species. For example, for a dataset involving several rodent species and several primate species, the rodents are naturally more closely related to each other and the primates are more closely related to each other, owing to their respective phylogenetic relationship. The data variance is likely to be smaller than if these species are completely independent, and the statistical methods that fail to account for this relatedness will be unreliable. The solution has been provided by several studies in comparative and evolutionary biology (Butler and King, 2004; Felsenstein, 1985; Garland et al., 1993; Grafen, 1989). Known as “phylogenetic regression”, “phylogenetically independent contrast”, or “phylogenetic generalized least squares”, this approach incorporates the phylogenetic relationship of the species and performs regression analysis using generalized least squares approach. The overall effect is to increase the stringency for statistical significance and require that a relationship across the species must persist above and beyond what can be expected based on phylogeny. The original approach was devised for studying a small number of traits, but it has been adapted for cross-species expression analyses (Brawand et al., 2011; Dunn et al., 2013; Fushan et al., 2015).
To date, there are only ~100 mammalian species with publicly available complete genomes, posing significant challenges on the read alignment of RNA sequencing data for those species without reference genomes. Aligning reads to the genome of a related species is often far from ideal: for example, only 13% of the reads of African grass rat fibroblasts could be uniquely mapped to the mouse genome (even though both belong to the same Family Muridae), and the alignment rate was even lower for the red squirrel (about 5%) (Ma et al., 2016). The identification of gene orthologs (i.e. genes in different species that evolved from a common ancestral gene) can also be challenging. In order to make meaningful comparison of read counts of a particular gene across different species, one needs to identify the ortholog of this gene in each species. The ortholog sets for the commonly studied species are available from a number of online databases (e.g. HomoloGene of NCBI; BioMart of Ensembl; and Multiz Alignment of UCSC Genome Browser), but there is little information on gene orthology relationship for the less common species. To address these problems, de novo assembly of RNA sequencing transcriptome (e.g. using Trinity (Grabherr et al., 2011)) can be used to recover the coding sequences, whereas gene ortholog sets can be defined as reciprocal best hits in BLAST (Altschul et al., 1997; Tatusov et al., 1997). While these methods are less stringent than those used in phylogeny reconstruction, they can typically identify 5,000~10,000 orthologs (Brawand et al., 2011; Fushan et al., 2015; Ma et al., 2016), which will be sufficient as a starting point for cross-species analyses.
Cross-species transcriptome
One of the recent studies examined longevity associated gene expression patterns in brain, kidney and liver of mammals (Fushan et al., 2015). The dataset covered 33 mammalian species across 10 taxonomic orders, with maximum lifespan ranging from 3.2 years (house shrew) to 100 years (human), representing the largest to date study to examine how gene expression is related to lifespan control across species. Regression with life history traits identified positive correlation with genes involved in DNA repair (including base-excision repair and nonhomologous end-joining pathways, e.g. XRCC5, XRCC6, PRKDC), regulation of immune response, and regulation of defense response across the three organs. In terms of negative correlation, the downregulated pathways in liver of long-lived species included lipid oxidation, fatty acid metabolism, degradation of amino acids and derivatives, tricarboxylic acid cycle, mitochondrial respiratory chains, and ubiquitin complex. In brain of the long-lived species, the downregulated functions included axonogenesis, neuron differentiation, synaptic transmission, inositol and calcium mediated signaling pathways, and transmembrane channel transport (Fushan et al., 2015). Many of these pathways remained significant even after adjustment for body mass differences. Notably, many of these genes and pathways also corresponded to longevity-related changes in the long-lived or short-lived species mentioned above. For example, XRCC5 showed positive selection signals in Brandt’s bat (Seim et al., 2013) and African killifish (Reichwald et al., 2015; Sahm et al., 2016; Valenzano et al., 2015). The central role of immune response was noted in naked mole rats (Kim et al., 2011), blind mole rats (Fang et al., 2014a), microbats (Seim et al., 2013; Zhang et al., 2013), and the brain of aging killifish (Baumgart et al., 2014). In killifish, mitochondrial respiratory chains showed positive selection signals (Sahm et al., 2017) and negative correlation with lifespan (Baumgart et al., 2014; Baumgart et al., 2016), and inhibition of complex I with rotenone could achieve 15% lifespan extension (Baumgart et al., 2016).
In another study, primary skin fibroblasts were obtained from 13 species of rodents, 2 species of bats, and 1 species of shrew and subjected to RNA sequencing and metabolite profiling (Ma et al., 2016). Phylogenetic regression was performed to identify gene expression with significant association to maximum lifespan or female time to maturity. The expression levels of the genes involved in DNA repair were also found in positive correlation with species longevity, whereas those involved in proteolysis were in negative correlation, which were consistent with the trends observed in the mammalian brain and liver (Fushan et al., 2015; MacRae et al., 2015) and previous cellular studies (Cortopassi and Wang, 1996; Hart and Setlow, 1974). When these fibroblasts were treated with stress-inducing agents such as cadmium and paraquat, those from the long-lived species were more resistant (Ma et al., 2016). Other up-regulated pathways in the fibroblasts from long-lived species included nucleotide binding, glucose metabolic process, and chromosome organization, while the down-regulated pathways included protein transport and localization, and regulation of transcription. Interestingly, a number of genes involved in apoptosis regulation, including the tumor suppressor TP53 (encoded by Trp53), BCL-2 associated X protein BAX (encoded by Bax), transcription factor FOXO3 (encoded by Foxo3), as well as mitogen-activated protein (MAP) kinase (encoded by Mapk1) were also down-regulated in the long-lived species fibroblasts (Ma et al., 2016), suggesting the long-lived species may generate less damage and/or have better repair mechanisms, so that the cells rely less on proteolysis, autophagy and apoptosis.
Cross-species metabolome, ionome, and lipidome
Gene expression is informative with regard to molecular features associated with longevity across species, but this is only one of the ways to assess these features. Two studies reported the analyses of metabolite and ion levels in brain, heart, kidney, and liver of 26 mammalian species across 10 taxonomic orders (Ma et al., 2015a; Ma et al., 2015b). Among the 162 water soluble metabolites and 100 lipids, positive correlation with species longevity traits was observed for sphingomyelins (in brain), whereas negative correlation was observed for amino acids (in brain), lysophosphatidyl-cholines (in brain and heart), lysophosphatidyl-ethanolamines (in brain and kidney), and triacylglycerols (in kidney) (Ma et al., 2015b). In particular, only those triacylglycerols with polyunsaturated fatty acid (PUFA) side chains showed significant negative correlation with species lifespan. A recent study on human plasma lipidomes of middle-aged offspring of nonagenarians revealed a signature of 19 lipid species associating with female familial longevity, including high levels of sphingomyelins and low levels of PUFA triacylglycerols (Gonzalez-Covarrubias et al., 2013). Analysis of phospholipids in heart of a number of mammals also revealed a negative correlation between double bond content and maximum lifespan (Pamplona et al., 2000). Naked mole rat tissues contain much lower levels of docosahexaenoic acid-containing (with 6 double bonds) phospholipids compared to mouse (Mitchell et al., 2007). Since PUFA are particularly sensitive to peroxidation damage (especially when present in membrane) (Hulbert, 2008), reduced level of polyunsaturated TAG in long-lived species may reflect their enhanced resistance to oxidative stress. Indeed, the peroxidation index for membrane composition is inversely correlated to longevity in mammals, birds, bivalve mollusks, honeybees and C. elegans (Hulbert et al., 2014).
In addition, high urate:allantoin ratio was observed among the long-lived species, which also expressed lower levels of uricase in liver (the enzyme that converts urate to allantoin). In fact, the uricase expression levels were exceptionally low in the naked mole rat, and this gene is a pseudogene in human and other higher primates (Ma et al., 2015b). The liver concentrations of two tryptophan degradation products, anthranilic acid and kynurenine, were also low among the long-lived species. Knockdown of the enzyme involved in breaking down tryptophan (tryptophan 2,3-dioxygenase, TDO) was shown to increase lifespan in C. elegans and fruit flies (Oxenkrug, 2010; van der Goot et al., 2012), whereas the kynurenine:tryptophan ratio in human increases with aging (Capuron et al., 2011).
In the ionome study, the levels of 18 elements in brain, heart, kidney and liver samples of the mammals were quantified by inductively-coupled plasma mass spectrometry. Each organ showed a distinctive pattern of ion distribution: lithium, sodium, and calcium levels were relatively high in kidney; phosphorus and potassium were high in brain; and 13 out of the 18 elements were high in liver (Ma et al., 2015a). In terms of correlation with longevity, zinc in kidney and liver showed significant positive correlation with species lifespan, although the effect was largely due to body mass. On the other hand, liver selenium levels showed modest negative correlation, and the effect remained significant after adjusting for body mass. This is interesting, because this element is considered an antioxidant, due to its presence in 25 mammalian proteins in the form of the 21st amino acid, selenocysteine (Kryukov et al., 2003). Accordingly, selenium supplements have been advocated that would maximize selenoprotein expression. The finding of decreased utilization of selenium by long-lived mammals suggests that the long-term use of this element is deleterious, likely to its reactivity leading to generation of reactive oxygen species and other toxic molecules. Surprisingly, cadmium levels in kidney and liver showed strong and robust positive correlation with longevity (even after body mass adjustment), although cadmium has no known biological role in mammals and high cadmium is toxic. Since cadmium mostly comes from the soil or food, one possibility is that longer-lived mammals simply consume a greater amount of food over their life time and, as they cannot efficiently excrete this metal, accumulate more cadmium (Ma et al., 2015a).
In a recently published lipidome study, the levels of 13,000 to 21,000 hydrophobic compounds were quantified across 6 organs (brain cortex, cerebellum, heart, kidney, liver, and muscle) in 35 mammalian species belonging to rodents, primates, and bats (Bozek et al., 2017). Using logistic regression with elastic net penalty and adjusting for the confounding effects between lifespan and body mass, the authors constructed predictive models of the species’ maximum lifespans based on the lipid concentrations, with average prediction accuracy above 0.9. The most consistent predictors of long lifespans included increased levels of triacylglycerols and decreased levels of glycerophospholipids and sphingolipids. The numbers of double bonds were also associated with lifespan variations, although the exact effects depended on the lipid classes (Bozek et al., 2017). Furthermore, genome analyses across these species revealed that the enzymes linked to these lipid classes and pathways displayed signatures of greater stabilizing selection among the long-lived species (Bozek et al., 2017), indicative of the concerted changes at both genomic and lipidomic levels in relation to the extended lifespans.
Comparison with long-lived mouse models
As the model for studying aging in mammals, lifespan of mice can be successfully increased using several different strategies, including knockout of IGF1R (Holzenberger et al., 2003) or GHR (Coschigano et al., 2000), restriction of food or methionine (Flurkey et al., 2010; Weindruch et al., 1986), defects in anterior pituitary development (Ames and Snell dwarf mice) (Brown-Borg et al., 1996; Flurkey et al., 2001), administration of rapamycin (Harrison et al., 2009) or acarbose (Harrison et al., 2014), and in the context of high calorie diet, feeding with resveratrol (Baur et al., 2006). While these methods achieve different degrees of lifespan extension, it is still not clear if they all act via a common pathway or fall into different sub-categories. Their effects on gene expression and metabolite levels in different organs as well as their overlaps with cross-species longevity signature have not been extensively explored.
The metabolite profiles of brain and liver of 5 long-lived mouse models (calorie restriction, rapamycin treatment, acarbose treatment, GHR knockout, and Snell dwarf), in both males and females, revealed some interesting results (Ma et al., 2015b). Surprisingly, except for the Snell dwarf (which had defective anterior pituitary development), these mouse models showed very little metabolic changes in brain compared to the wild-type controls, suggesting that most of their brain metabolites remained constant despite extended lifespan. On the other hand, liver metabolites showed significant changes in these long-lived models, and some of these patterns (e.g. up-regulation of sphingomyelins and down-regulation of PUFA triacylglycerols) were consistent with the cross-species longevity signature. Furthermore, changes in metabolite abundance in response to calorie restriction and acarbose treatment were very similar, probably because acarbose acts by inhibiting glycoside hydrolases (enzymes that digest complex carbohydrates to absorbable sugars in the gastrointestinal tract) and the inhibition of glucose uptake produces the effects similar to calorie restriction. However, GHR knockout and rapamycin treatment led to rather distinctive metabolite patterns, likely indicative of different lifespan modulation mechanisms.
Conclusions
Over the last few years, the focus of longevity research has started to move beyond the classical model organisms and towards the naturally long-lived or short-lived species and comparative cross-species studies. Genomic and cellular studies have uncovered several interesting (albeit somewhat unconventional) anti-cancer mechanisms, such as the production of high-molecular-mass HA in the naked mole rat and secretion of IFN-β by blind mole rat cells. The genomes of bats, whales, and killifish reveal that the genes involved in DNA repair and insulin/IGF signaling often show positive selection signals or have unique amino acid changes in key functional domains, although the exact longevity effects may depend on the actual amino acid residues (Table 1). Cross-species transcriptomic analyses also showed that up-regulation of DNA repair and down-regulation of proteolysis seem to be the common trend among the long-lived species, whereas the metabolic studies highlighted some of the longevity-associating molecules such as triacylglycerols, glycerophospholipids, and sphinolipids (Table 2).
Looking at all of these studies together, some common molecular players and pathways have emerged repeatedly and may point towards certain central modules of lifespan regulation (Table 3). For example, p53 and the associated genes involved in cell cycle and apoptosis often show positive selection signals, unique amino acid changes, and gene expression changes in the species with exceptional lifespans. However, while naked mole rats and blind mole rats seem to favor cell cycle arrest over apoptosis, African elephants may go in the opposite direction and sensitize the cells to apoptosis, suggesting the same pathway may be modulated in a different manner to achieve longevity. In terms of DNA repair, there are often strong positive selection signals for the genes involved in the pathway and upregulation of expression in long-lived species, whereas for respiratory and mitochondria functions, down regulation is more often associated with long lifespan. Down regulation of the GH/IGF1 axis and upregulation of immune response seem to be the common theme among long-lived species, while both naked mole rats and blind mole rats also show augmentation of Nrf2 activities. The processes of protein synthesis, proteolysis, and lipid metabolisms are also noted in many of these studies, although the exact directions of change and mechanisms will require further clarification.
Table 3.
Some of the common pathways and molecular players identified in cross-species studies.
| Pathways | Evidence |
|---|---|
| p53 / cell cycle / apoptosis |
|
| DNA repair |
|
| Respiration / mitochondria |
|
| GH/IGF1 axis |
|
| Immune response |
|
| Nrf2 |
|
| Protein synthesis / proteolysis |
|
| Lipid metabolism |
|
In addition, it should be noted that the various lifespan extension strategies in laboratory setting may have different effects on different organs: for example, among the long-lived mouse models many changes were observed in liver metabolites, whereas brain metabolites remain generally stable. It is also possible that some of the known lifespan extension strategies (e.g. GH/IGF1 axis ablation, calorie restriction, and rapamycin treatment) may act, at least in part, via different pathways, and a better understanding of their molecular mechanisms will be needed to fully appreciate the regulator modules of aging and longevity. Lastly, while correlation itself does not imply causation, the longevity signatures identified in comparative genomic, transcriptomic, and metabolomic studies can serve as a valuable starting point to test and validate the potential molecular or pharmacological approaches to lifespan extension.
Acknowledgments
Supported by grants from the National Institute on Aging and the Russian Federation grant 14.W03.31.0012.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abegglen LM, Caulin AF, Chan A, Lee K, Robinson R, Campbell MS, Kiso WK, Schmitt DL, Waddell PJ, Bhaskara S, et al. Potential Mechanisms for Cancer Resistance in Elephants and Comparative Cellular Response to DNA Damage in Humans. Jama. 2015;314:1850–1860. doi: 10.1001/jama.2015.13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achison M, Hupp TR. Hypoxia attenuates the p53 response to cellular damage. Oncogene. 2003;22:3431–3440. doi: 10.1038/sj.onc.1206434. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arieli R, Heth G, Nevo E, Hoch D. Hematocrit and hemoglobin concentration in four chromosomal species and some isolated populations of actively speciating subterranean mole rats in Israel. Experientia. 1986a;42:441–443. doi: 10.1007/BF02118650. [DOI] [PubMed] [Google Scholar]
- Arieli R, Heth G, Nevo E, Zamir Y, Neutra O. Adaptive heart and breathing frequencies in 4 ecologically differentiating chromosomal species of mole rats in Israel. Experientia. 1986b;42:131–133. [Google Scholar]
- Ashur-Fabian O, Avivi A, Trakhtenbrot L, Adamsky K, Cohen M, Kajakaro G, Joel A, Amariglio N, Nevo E, Rechavi G. Evolution of p53 in hypoxia-stressed Spalax mimics human tumor mutation. Proc Natl Acad Sci U S A. 2004;101:12236–12241. doi: 10.1073/pnas.0404998101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avivi A, Ashur-Fabian O, Amariglio N, Nevo E, Rechavi G. p53 - a key player in tumoral and evolutionary adaptation: a lesson from the Israeli blind subterranean mole rat. Cell Cycle. 2005:4. doi: 10.4161/cc.4.3.1534. [DOI] [PubMed] [Google Scholar]
- Avivi A, Gerlach F, Joel A, Reuss S, Burmester T, Nevo E, Hankeln T. Neuroglobin, cytoglobin, and myoglobin contribute to hypoxia adaptation of the subterranean mole rat Spalax. Proc Natl Acad Sci U S A. 2010;107:21570–21575. doi: 10.1073/pnas.1015379107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpurua J, Ke Z, Chen IX, Zhang Q, Ermolenko DN, Zhang ZD, Gorbunova V, Seluanov A. Naked mole-rat has increased translational fidelity compared with the mouse, as well as a unique 28S ribosomal RNA cleavage. Proc Natl Acad Sci U S A. 2013;110:17350–17355. doi: 10.1073/pnas.1313473110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart M, Groth M, Priebe S, Savino A, Testa G, Dix A, Ripa R, Spallotta F, Gaetano C, Ori M, et al. RNA-seq of the aging brain in the short-lived fish N. furzeri -conserved pathways and novel genes associated with neurogenesis. Aging Cell. 2014;13:965–974. doi: 10.1111/acel.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart M, Priebe S, Groth M, Hartmann N, Menzel U, Pandolfini L, Koch P, Felder M, Ristow M, Englert C, et al. Longitudinal RNA-Seq Analysis of Vertebrate Aging Identifies Mitochondrial Complex I as a Small-Molecule-Sensitive Modifier of Lifespan. Cell Systems. 2016;2:122–132. doi: 10.1016/j.cels.2016.01.014. [DOI] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozek K, Khrameeva EE, Reznick J, Omerbašić D, Bennett NC, Lewin GR, Azpurua J, Gorbunova V, Seluanov A, Regnard P, et al. Lipidome determinants of maximal lifespan in mammals. Sci Rep. 2017;7:5. doi: 10.1038/s41598-017-00037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, Soumillon M, Necsulea A, Julien P, Csardi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- Brown-Borg HM, Borg KE, Meliska CJ, Bartke A. Dwarf mice and the ageing process. Nature. 1996;384:33. doi: 10.1038/384033a0. [DOI] [PubMed] [Google Scholar]
- Buffenstein R. Negligible senescence in the longest living rodent, the naked mole-rat: insights from a successfully aging species. J Comp Physiol B. 2008;178:439–445. doi: 10.1007/s00360-007-0237-5. [DOI] [PubMed] [Google Scholar]
- Butler MA, King AA. Phylogenetic comparative analysis: A modeling approach for adaptive evolution. Am Nat. 2004;164:683–695. doi: 10.1086/426002. [DOI] [PubMed] [Google Scholar]
- Capuron L, Schroecksnadel S, Feart C, Aubert A, Higueret D, Barberger-Gateau P, Laye S, Fuchs D. Chronic low-grade inflammation in elderly persons is associated with altered tryptophan and tyrosine metabolism: role in neuropsychiatric symptoms. Biol Psychiatry. 2011;70:175–182. doi: 10.1016/j.biopsych.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Colman RJ, Beasley TM, Kemnitz JW, Johnson SC, Weindruch R, Anderson RM. Caloric restriction reduces age-related and all-cause mortality in rhesus monkeys. Nat Commun. 2014;5:3557. doi: 10.1038/ncomms4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortopassi GA, Wang E. There is substantial agreement among interspecies estimates of DNA repair activity. Mech Ageing Dev. 1996;91:211–218. doi: 10.1016/s0047-6374(96)01788-5. [DOI] [PubMed] [Google Scholar]
- Coschigano KT, Clemmons D, Bellush LL, Kopchick JJ. Assessment of growth parameters and life span of GHR/BP gene-disrupted mice. Endocrinology. 2000;141:2608–2613. doi: 10.1210/endo.141.7.7586. [DOI] [PubMed] [Google Scholar]
- de Magalhães JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25:875–881. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney MA, Ward JM, Walsh TF, Chinnadurai SK, Kerns K, Kinsel MJ, Treuting PM. Initial Case Reports of Cancer in Naked Mole-rats (Heterocephalus glaber) Veterinary Pathology. 2016;53:691–696. doi: 10.1177/0300985816630796. [DOI] [PubMed] [Google Scholar]
- Domankevich V, Opatowsky Y, Malik A, Korol AB, Frenkel Z, Manov I, Avivi A, Shams I. Adaptive patterns in the p53 protein sequence of the hypoxia- and cancer-tolerant blind mole rat Spalax. BMC Evol Biol. 2016;16:177. doi: 10.1186/s12862-016-0743-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CW, Luo X, Wu Z. Phylogenetic Analysis of Gene Expression. Integr Comp Biol. 2013;53:847–856. doi: 10.1093/icb/ict068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edoute Y, Arieli R, Nevo E. Evidence for improved myocardial oxygen delivery and function during hypoxia in the mole rat. J Comp Physiol B. 1988;158:575–582. doi: 10.1007/BF00692566. [DOI] [PubMed] [Google Scholar]
- Fang X, Nevo E, Han L, Levanon EY, Zhao J, Avivi A, Larkin D, Jiang X, Feranchuk S, Zhu Y, et al. Genome-wide adaptive complexes to underground stresses in blind mole rats Spalax. Nat Commun. 2014a;5:3966. doi: 10.1038/ncomms4966. [DOI] [PubMed] [Google Scholar]
- Fang X, Seim I, Huang Z, Gerashchenko MV, Xiong Z, Turanov AA, Zhu Y, Lobanov AV, Fan D, Yim SH, et al. Adaptations to a subterranean environment and longevity revealed by the analysis of mole rat genomes. Cell Rep. 2014b;8:1354–1364. doi: 10.1016/j.celrep.2014.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the Comparative Method. Am Nat. 1985;125:1–15. [Google Scholar]
- Flurkey K, Astle CM, Harrison DE. Life extension by diet restriction and N-acetyl-L-cysteine in genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci. 2010;65:1275–1284. doi: 10.1093/gerona/glq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flurkey K, Papaconstantinou J, Miller RA, Harrison DE. Lifespan extension and delayed immune and collagen aging in mutant mice with defects in growth hormone production. Proc Natl Acad Sci USA. 2001;98:6736–6741. doi: 10.1073/pnas.111158898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushan AA, Turanov AA, Lee SG, Kim EB, Lobanov AV, Yim SH, Buffenstein R, Lee SR, Chang KT, Rhee H, et al. Gene expression defines natural changes in mammalian lifespan. Aging Cell. 2015;14:352–365. doi: 10.1111/acel.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T, Dickerman AW, Janis CM, Jones JA. Phylogenetic Analysis of Covariance by Computer Simulation. Syst Biol. 1993;42:265–292. [Google Scholar]
- George JC, Bockstoce JR. Two historical weapon fragments as an aid to estimating the longevity and movements of bowhead whales. Polar Biology. 2008;31:751–754. [Google Scholar]
- Gladyshev VN. Aging: progressive decline in fitness due to the rising deleteriome adjusted by genetic, environmental, and stochastic processes. Aging Cell. 2016;15:594–602. doi: 10.1111/acel.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Covarrubias V, Beekman M, Uh HW, Dane A, Troost J, Paliukhovich I, van der Kloet FM, Houwing-Duistermaat J, Vreeken RJ, Hankemeier T, et al. Lipidomics of familial longevity. Aging Cell. 2013;12:426–434. doi: 10.1111/acel.12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V, Hine C, Tian X, Ablaeva J, Gudkov AV, Nevo E, Seluanov A. Cancer resistance in the blind mole rat is mediated by concerted necrotic cell death mechanism. Proc Natl Acad Sci U S A. 2012;109:19392–19396. doi: 10.1073/pnas.1217211109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V, Seluanov A, Zhang Z, Gladyshev VN, Vijg J. Comparative genetics of longevity and cancer: insights from long-lived rodents. Nat Rev Genet. 2014;15:531–540. doi: 10.1038/nrg3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafen A. The phylogenetic regression. Philos Trans R Soc Lond B Biol Sci. 1989;326:119–157. doi: 10.1098/rstb.1989.0106. [DOI] [PubMed] [Google Scholar]
- Guevara-Aguirre J, Balasubramanian P, Guevara-Aguirre M, Wei M, Madia F, Cheng CW, Hwang D, Martin-Montalvo A, Saavedra J, Ingles S, et al. Growth hormone receptor deficiency is associated with a major reduction in pro-aging signaling, cancer, and diabetes in humans. Sci Transl Med. 2011;3:70ra13. doi: 10.1126/scitranslmed.3001845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Allison DB, Ames BN, Astle CM, Atamna H, Fernandez E, Flurkey K, Javors MA, Nadon NL, et al. Acarbose, 17-alpha-estradiol, and nordihydroguaiaretic acid extend mouse lifespan preferentially in males. Aging Cell. 2014;13:273–282. doi: 10.1111/acel.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart RW, Setlow RB. Correlation between deoxyribonucleic acid excision-repair and life-span in a number of mammalian species. Proc Natl Acad Sci U S A. 1974;71:2169–2173. doi: 10.1073/pnas.71.6.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayflick L. Biological aging is no longer an unsolved problem. Ann N Y Acad Sci. 2007;1100:1–13. doi: 10.1196/annals.1395.001. [DOI] [PubMed] [Google Scholar]
- Holliday R. Understanding ageing. Philos Trans R Soc Lond B Biol Sci. 1997;352:1793–1797. doi: 10.1098/rstb.1997.0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. Aging is no longer an unsolved problem in biology. Ann N Y Acad Sci. 2006;1067:1–9. doi: 10.1196/annals.1354.002. [DOI] [PubMed] [Google Scholar]
- Holzenberger M, Dupont J, Ducos B, Leneuve P, Geloen A, Even PC, Cervera P, Le Bouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- Hulbert AJ. Explaining longevity of different animals: is membrane fatty acid composition the missing link? Age (Dordr) 2008;30:89–97. doi: 10.1007/s11357-008-9055-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert AJ, Kelly MA, Abbott SK. Polyunsaturated fats, membrane lipids and animal longevity. J Comp Physiol B. 2014;184:149–166. doi: 10.1007/s00360-013-0786-8. [DOI] [PubMed] [Google Scholar]
- Jones OR, Scheuerlein A, Salguero-Gomez R, Camarda CG, Schaible R, Casper BB, Dahlgren JP, Ehrlen J, Garcia MB, Menges ES, et al. Diversity of ageing across the tree of life. Nature. 2014;505:169–173. doi: 10.1038/nature12789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, Ishioka C. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci U S A. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane M, Semeiks J, Webb AE, Li YI, Quesada V, Craig T, Madsen LB, van Dam S, Brawand D, Marques PI, et al. Insights into the evolution of longevity from the bowhead whale genome. Cell Rep. 2015;10:112–122. doi: 10.1016/j.celrep.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kim EB, Fang X, Fushan AA, Huang Z, Lobanov AV, Han L, Marino SM, Sun X, Turanov AA, Yang P, et al. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature. 2011;479:223–227. doi: 10.1038/nature10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- Lewis KN, Wason E, Edrey YH, Kristan DM, Nevo E, Buffenstein R. Regulation of Nrf2 signaling and longevity in naturally long-lived rodents. Proc Natl Acad Sci U S A. 2015;112:3722–3727. doi: 10.1073/pnas.1417566112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Huang D, McArthur DL, Boros LG, Nissen N, Heaney AP. Fructose induces transketolase flux to promote pancreatic cancer growth. Cancer Res. 2010;70:6368–6376. doi: 10.1158/0008-5472.CAN-09-4615. [DOI] [PubMed] [Google Scholar]
- Logan M. Biostatistical design and analysis using R : a practical guide. Chichester, UK; Hoboken, NJ: Wiley-Blackwell; 2010. [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Lee SG, Kim EB, Park TJ, Seluanov A, Gorbunova V, Buffenstein R, Seravalli J, Gladyshev VN. Organization of the Mammalian Ionome According to Organ Origin, Lineage Specialization, and Longevity. Cell Rep. 2015a;13:1319–1326. doi: 10.1016/j.celrep.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Upneja A, Galecki A, Tsai YM, Burant CF, Raskind S, Zhang Q, Zhang ZD, Seluanov A, Gorbunova V, et al. Cell culture-based profiling across mammals reveals DNA repair and metabolism as determinants of species longevity. Elife. 2016:5. doi: 10.7554/eLife.19130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Yim SH, Lee SG, Kim EB, Lee SR, Chang KT, Buffenstein R, Lewis KN, Park TJ, Miller RA, et al. Organization of the Mammalian Metabolome according to Organ Function, Lineage Specialization, and Longevity. Cell Metab. 2015b;22:332–343. doi: 10.1016/j.cmet.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae SL, Croken MM, Calder RB, Aliper A, Milholland B, White RR, Zhavoronkov A, Gladyshev VN, Seluanov A, Gorbunova V, et al. DNA repair in species with extreme lifespan differences. Aging (Albany NY) 2015;7:1171–1184. doi: 10.18632/aging.100866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manov I, Hirsh M, Iancu TC, Malik A, Sotnichenko N, Band M, Avivi A, Shams I. Pronounced cancer resistance in a subterranean rodent, the blind mole-rat, Spalax: in vivo and in vitroevidence. BMC Biology. 2013;11:91. doi: 10.1186/1741-7007-11-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison JA, Roth GS, Beasley TM, Tilmont EM, Handy AM, Herbert RL, Longo DL, Allison DB, Young JE, Bryant M, et al. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature. 2012;489:318–321. doi: 10.1038/nature11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCay CM, Crowell MF, Maynard LA. The Effect of Retarded Growth Upon the Length of Life Span and Upon the Ultimate Body Size: One Figure. J Nutr. 1935;10:63–79. [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell TW, Buffenstein R, Hulbert AJ. Membrane phospholipid composition may contribute to exceptional longevity of the naked mole-rat (Heterocephalus glaber): a comparative study using shotgun lipidomics. Exp Gerontol. 2007;42:1053–1062. doi: 10.1016/j.exger.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Miyawaki S, Kawamura Y, Hachiya T, Shimizu A, Miura K. Molecular cloning and characterization of the INK4a and ARF genes in naked mole-rat. Inflammation and Regeneration. 2015;35:042–050. [Google Scholar]
- Nasser NJ, Avivi A, Shafat I, Edovitsky E, Zcharia E, Ilan N, Vlodavsky I, Nevo E. Alternatively spliced Spalax heparanase inhibits extracellular matrix degradation, tumor growth, and metastasis. Proc Natl Acad Sci U S A. 2009;106:2253–2258. doi: 10.1073/pnas.0812846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J, Hedeholm RB, Heinemeier J, Bushnell PG, Christiansen JS, Olsen J, Ramsey CB, Brill RW, Simon M, Steffensen KF, et al. Eye lens radiocarbon reveals centuries of longevity in the Greenland shark (Somniosus microcephalus) Science. 2016;353:702–704. doi: 10.1126/science.aaf1703. [DOI] [PubMed] [Google Scholar]
- Onken B, Driscoll M. Metformin induces a dietary restriction-like state and the oxidative stress response to extend C. elegans Healthspan via AMPK, LKB1, and SKN-1. PLoS One. 2010;5:e8758. doi: 10.1371/journal.pone.0008758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxenkrug GF. The extended life span of Drosophila melanogaster eye-color (white and vermilion) mutants with impaired formation of kynurenine. J Neural Transm. 2010;117:23–26. doi: 10.1007/s00702-009-0341-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamplona R, Portero-Otin M, Riba D, Requena JR, Thorpe SR, Lopez-Torres M, Barja G. Low fatty acid unsaturation: a mechanism for lowered lipoperoxidative modification of tissue proteins in mammalian species with long life spans. J Gerontol A Biol Sci Med Sci. 2000;55:B286–291. doi: 10.1093/gerona/55.6.b286. [DOI] [PubMed] [Google Scholar]
- Park TJ, Reznick J, Peterson BL, Blass G, Omerbasic D, Bennett NC, Kuich P, Zasada C, Browe BM, Hamann W, et al. Fructose-driven glycolysis supports anoxia resistance in the naked mole-rat. Science. 2017;356:307–311. doi: 10.1126/science.aab3896. [DOI] [PubMed] [Google Scholar]
- Partridge L, Gems D. Mechanisms of ageing: public or private? Nat Rev Genet. 2002;3:165–175. doi: 10.1038/nrg753. [DOI] [PubMed] [Google Scholar]
- Peters RH. The ecological implications of body size. Cambridge University Press; 1986. [Google Scholar]
- Peto R, Roe FJ, Lee PN, Levy L, Clack J. Cancer and ageing in mice and men. Br J Cancer. 1975;32:411–426. doi: 10.1038/bjc.1975.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlutsky AJ, Khritankov AM, Ovodov ND, Austad SN. A new field record for bat longevity. J Gerontol A Biol Sci Med Sci. 2005;60:1366–1368. doi: 10.1093/gerona/60.11.1366. [DOI] [PubMed] [Google Scholar]
- Port AM, Ruth MR, Istfan NW. Fructose consumption and cancer: is there a connection? Curr Opin Endocrinol Diabetes Obes. 2012;19:367–374. doi: 10.1097/MED.0b013e328357f0cb. [DOI] [PubMed] [Google Scholar]
- Reichwald K, Petzold A, Koch P, Downie BR, Hartmann N, Pietsch S, Baumgart M, Chalopin D, Felder M, Bens M, et al. Insights into Sex Chromosome Evolution and Aging from the Genome of a Short-Lived Fish. Cell. 2015;163:1527–1538. doi: 10.1016/j.cell.2015.10.071. [DOI] [PubMed] [Google Scholar]
- Sacher GA. Relation of Lifespan to Brain Weight and Body Weight in Mammals. In: Wolstenholme GEW, O’Conner M, editors. Ciba Foundation Symposium - The Lifespan of Animals (Colloquia on Ageing) Chichester: John Wiley & Sons, Ltd; 1959. pp. 115–141. [Google Scholar]
- Sahm A, Bens M, Platzer M, Cellerino A. Parallel evolution of genes controlling mitonuclear balance in short-lived annual fishes. Aging Cell. 2017;16:488–496. doi: 10.1111/acel.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahm A, Platzer M, Cellerino A. Outgroups and Positive Selection: The Nothobranchius furzeri Case. Trends Genet. 2016;32:523–525. doi: 10.1016/j.tig.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Hangmann J, Shams I, Avivi A, Hankeln T. Molecular evolution of antioxidant and hypoxia response in long-lived, cancer-resistant blind mole rats: The Nrf2-Keap1 pathway. Gene. 2016;577:293–298. doi: 10.1016/j.gene.2015.11.038. [DOI] [PubMed] [Google Scholar]
- Seim I, Fang X, Xiong Z, Lobanov AV, Huang Z, Ma S, Feng Y, Turanov AA, Zhu Y, Lenz TL, et al. Genome analysis reveals insights into physiology and longevity of the Brandt’s bat Myotis brandtii. Nat Commun. 2013;4:2212. doi: 10.1038/ncomms3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seim I, Ma S, Zhou X, Gerashchenko MV, Lee SG, Suydam R, George JC, Bickham JW, Gladyshev VN. The transcriptome of the bowhead whale Balaena mysticetus reveals adaptations of the longest-lived mammal. Aging (Albany NY) 2014;6:879–899. doi: 10.18632/aging.100699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluanov A, Hine C, Azpurua J, Feigenson M, Bozzella M, Mao Z, Catania KC, Gorbunova V. Hypersensitivity to contact inhibition provides a clue to cancer resistance of naked mole-rat. Proc Natl Acad Sci U S A. 2009;106:19352–19357. doi: 10.1073/pnas.0905252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shams I, Avivi A, Nevo E. Oxygen and carbon dioxide fluctuations in burrows of subterranean blind mole rats indicate tolerance to hypoxic-hypercapnic stresses. Comp Biochem Physiol A Mol Integr Physiol. 2005;142:376–382. doi: 10.1016/j.cbpa.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Sinclair DA. Toward a unified theory of caloric restriction and longevity regulation. Mech Ageing Dev. 2005;126:987–1002. doi: 10.1016/j.mad.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Suh Y, Atzmon G, Cho MO, Hwang D, Liu B, Leahy DJ, Barzilai N, Cohen P. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc Natl Acad Sci U S A. 2008;105:3438–3442. doi: 10.1073/pnas.0705467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacutu R, Craig T, Budovsky A, Wuttke D, Lehmann G, Taranukha D, Costa J, Fraifeld VE, de Magalhaes JP. Human Ageing Genomic Resources: integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res. 2013;41:D1027–1033. doi: 10.1093/nar/gks1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Taylor KR, Milone NA, Rodriguez CE. Four Cases of Spontaneous Neoplasia in the Naked Mole-Rat (Heterocephalus glaber), A Putative Cancer-Resistant Species. J Gerontol A Biol Sci Med Sci. 2017;72:38–43. doi: 10.1093/gerona/glw047. [DOI] [PubMed] [Google Scholar]
- Terzibasi E, Valenzano DR, Benedetti M, Roncaglia P, Cattaneo A, Domenici L, Cellerino A. Large differences in aging phenotype between strains of the short-lived annual fish Nothobranchius furzeri. PLoS One. 2008;3:e3866. doi: 10.1371/journal.pone.0003866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Azpurua J, Hine C, Vaidya A, Myakishev-Rempel M, Ablaeva J, Mao Z, Nevo E, Gorbunova V, Seluanov A. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature. 2013;499:346–349. doi: 10.1038/nature12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Azpurua J, Ke Z, Augereau A, Zhang ZD, Vijg J, Gladyshev VN, Gorbunova V, Seluanov A. INK4 locus of the tumor-resistant rodent, the naked mole rat, expresses a functional p15/p16 hybrid isoform. Proc Natl Acad Sci U S A. 2015;112:1053–1058. doi: 10.1073/pnas.1418203112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turbill C, Bieber C, Ruf T. Hibernation is associated with increased survival and the evolution of slow life histories among mammals. Proc Biol Sci. 2011;278:3355–3363. doi: 10.1098/rspb.2011.0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzano DR, Benayoun BA, Singh PP, Zhang E, Etter PD, Hu CK, Clement-Ziza M, Willemsen D, Cui R, Harel I, et al. The African Turquoise Killifish Genome Provides Insights into Evolution and Genetic Architecture of Lifespan. Cell. 2015;163:1539–1554. doi: 10.1016/j.cell.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Goot AT, Zhu W, Vazquez-Manrique RP, Seinstra RI, Dettmer K, Michels H, Farina F, Krijnen J, Melki R, Buijsman RC, et al. Delaying aging and the aging-associated decline in protein homeostasis by inhibition of tryptophan degradation. Proc Natl Acad Sci USA. 2012;109:14912–14917. doi: 10.1073/pnas.1203083109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weindruch R, Walford RL, Fligiel S, Guthrie D. The retardation of aging in mice by dietary restriction: longevity, cancer, immunity and lifetime energy intake. J Nutr. 1986;116:641–654. doi: 10.1093/jn/116.4.641. [DOI] [PubMed] [Google Scholar]
- Western D. Size, life history and ecology in mammals. Afr J Ecol. 1979;17:185–204. [Google Scholar]
- Whitney CR. Jeanne Calment, World’s Elder, Dies at 122. The New York Times 1997 [Google Scholar]
- Widmer HR, Hoppeler H, Nevo E, Taylor CR, Weibel ER. Working underground: respiratory adaptations in the blind mole rat. Proc Natl Acad Sci U S A. 1997;94:2062–2067. doi: 10.1073/pnas.94.5.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- Yim HS, Cho YS, Guang X, Kang SG, Jeong JY, Cha SS, Oh HM, Lee JH, Yang EC, Kwon KK, et al. Minke whale genome and aquatic adaptation in cetaceans. Nat Genet. 2014;46:88–92. doi: 10.1038/ng.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Li Y, Holmes A, Szafranski K, Faulkes CG, Coen CW, Buffenstein R, Platzer M, de Magalhaes JP, Church GM. RNA sequencing reveals differential expression of mitochondrial and oxidation reduction genes in the long-lived naked mole-rat when compared to mice. PLoS One. 2011;6:e26729. doi: 10.1371/journal.pone.0026729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Cowled C, Shi Z, Huang Z, Bishop-Lilly KA, Fang X, Wynne JW, Xiong Z, Baker ML, Zhao W, et al. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339:456–460. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Samuels T, Winckler S, Korgaonkar C, Tompkins V, Horne MC, Quelle DE. Cyclin G1 has growth inhibitory activity linked to the ARF-Mdm2-p53 and pRb tumor suppressor pathways. Mol Cancer Res. 2003;1:195–206. [PubMed] [Google Scholar]