Abstract
Background and aims
The activity of CYP2A6, the major nicotine-inactivating enzyme, is measurable in smokers using the nicotine metabolite ratio (NMR; 3’hydroxycotinine/cotinine). Due to its role in nicotine clearance, the NMR is associated with smoking behaviours and response to pharmacotherapies. The NMR is highly heritable (~80%), and on average lower in African Americans (AA) versus Whites. We previously identified several reduce and loss-of-function CYP2A6 variants common in individuals of African descent. Our current aim was to identify novel genetic influences on the NMR in AA smokers using genome-wide approaches.
Design
Genome-wide association study (GWAS).
Setting
Multiple sites within Canada and the United States.
Participants
AA smokers from two clinical trials: Pharmacogenetics of Nicotine Addiction Treatment (PNAT)-2 (NCT01314001; n=504) and Kick-it-at-Swope (KIS)-3 (NCT00666978; n=450).
Measurements
Genome-wide SNP genotyping, the NMR (phenotype), and population substructure and NMR covariates.
Findings
Meta-analysis revealed three independent chromosome 19 signals (rs12459249, rs111645190, and rs185430475) associated with the NMR. The top overall hit, rs12459249 (P=1.47e-39; beta=0.59 per C (versus T) allele, SE=0.045), located ~9.5kb 3’ of CYP2A6, remained genome-wide significant after controlling for the common (~10% in AA) non-functional CYP2A6*17 allele. In contrast, rs111645190 and rs185430475 were not genome-wide significant when controlling for CYP2A6*17. In total, 96 signals associated with the NMR were identified; many were not found in prior NMR GWASs in European descent individuals. The top hits were also associated with the NMR in a third cohort of AA (KIS2; n=480). None of the hits were in UGT or OCT2 genes.
Conclusions
Three independent chromosome 19 signals account for ~20% of the variability in the nicotine metabolite ratio in African-American smokers. The hits identified may contribute to inter-ethnic variability in nicotine metabolism, smoking behaviours, and tobacco-related disease risk.
Keywords: CYP2A6, genome-wide association study, nicotine metabolism biomarker, cigarette smoking, African Americans, treatment-seeking smokers
Introduction
The prevalence of cigarette smoking remains high despite widespread tobacco control efforts; recent estimates suggest 15% of Americans are current smokers (1). A growing segment of American smokers are light smokers (smoke ≤10 cigarettes/day) who, like heavy smokers, experience elevated risks of disease and mortality compared to never smokers (2). Smoking behaviour and smoking-related morbidity differ by ethnicity. For instance, although African American (AA) smokers on average smoke fewer cigarettes/day than European American (EA) smokers, the level of total nicotine equivalents, a biomarker of total nicotine intake, is similar in AA and EA smokers, suggesting more intensive smoking (e.g., greater puff volume) among AAs (3). At an equivalent number of cigarettes per day, the risk for lung cancer is higher in AA compared to EA smokers, perhaps due in part to more intensive smoking and, therefore, greater exposure to tobacco-specific nitrosamines and other harmful chemicals (4). Of note, AA (vs. EA) smokers are also more likely to make quit attempts and are less likely to achieve cessation (5). Understanding the factors that contribute to this increased risk for lung cancer and reduced likelihood of cessation among AA smokers will help guide treatment interventions for this population.
Nicotine is the predominant psychoactive compound in cigarettes (6). Nicotine undergoes metabolic inactivation by the hepatic CYP2A6 enzyme to cotinine, which is further metabolized to 3’hydroxycotinine exclusively by CYP2A6 (7, 8). The CYP2A6 gene, located on chromosome 19q13.2, contains several functional polymorphisms, leading to inter-individual variation in the rate of nicotine clearance (9). The nicotine metabolite ratio (NMR; 3’hydroxycotinine/cotinine) is an established and validated phenotypic marker of CYP2A6 activity in smokers; it is associated with CYP2A6 genotype and correlates with nicotine clearance (10-15). The NMR is 80% heritable (estimated in Finnish European twins) (16); in addition to genetic influences, the NMR captures relatively minor environmental (e.g., mentholated cigarette use and BMI) (17, 18) sources of variation in CYP2A6 activity.
The NMR has been evaluated as a clinical marker for personalizing smoking cessation treatment. Compared to higher NMR, lower NMR (i.e., slower nicotine metabolism) is associated with higher cessation rates with behavioural counseling (19) and among nicotine patch treated smokers (20-22). In a placebo-controlled bupropion trial, bupropion increased quit rates over placebo in those with higher but not lower NMR (19). In smokers prospectively randomized to treatment based on the NMR (PNAT2 Trial; NCT01314001), those with higher, but not lower, NMR had higher quit rates on varenicline (versus nicotine patch) (23). Number-needed-to-treat analyses in smokers with higher NMR indicated 5 and 26 smokers would need to be treated with varenicline (versus placebo) and nicotine patch (versus placebo), respectively, for one smoker to quit, again indicating the superiority of varenicline for those with higher NMR (23); in those with lower NMR these values were 8 and 10, respectively. In addition, those with lower (versus higher) NMR experienced greater negative side effects on varenicline (versus placebo) (23). Thus, the evidence indicates that smokers with higher NMR show greater benefit from varenicline or bupropion compared to behavioural counseling or nicotine patch, while smokers with lower NMR are treated more effectively and safely with nicotine patch and/or behavioural counseling.
Variability in CYP2A6 genetics and/or the NMR also influences the level of tobacco consumption (from cigarettes and smokeless tobacco), dependence, and risk for tobacco-related disease; smokers with slower metabolism (i.e., slow CYP2A6 metabolism groups or lower NMR values) generally show lower consumption, dependence, and disease risk (17, 24-26).
The NMR varies by ethnicity, with AA smokers having on average lower NMR (and nicotine clearance) versus smokers of EA descent (17, 27), due in part to the higher frequency of known CYP2A6 reduced or loss-of-function variants in AAs (28); many of these variants, including *23 , *24, *25, *28, *35, and *39-*45 (29-32), were identified and functionally characterized by our group using in vitro (e.g., CYP2A6 cDNA expression system), ex vivo (e.g., human liver bank), and in vivo (e.g., human smokers) nicotine metabolism rate assessments. These variants are common (>1% frequency) in AA, but exceedingly rare in EA populations. CYP2A6*17, with an allele frequency of 10% in AA (33), explains ~8% of the variability in the NMR (unpublished observations in the KIS3 trial (34)). Although >40 CYP2A6 variants have been identified and functionally characterized, estimates in Finnish Europeans indicate only ~30% of NMR variation is currently explained by detected CYP2A6 variants (16). To date, three NMR GWASs have been performed, predominantly or exclusively in Whites who were non-treatment-seeking smokers (only 413 AA total among >4000 total participants from three studies) (16, 35, 36); no GWAS has examined the NMR in smokers seeking treatment, for whom personalized medicine approaches based on CYP2A6/the NMR would be targeted.
Here we performed a GWAS of the NMR, assessed at baseline when participants were smoking ad libitum (i.e., when NMR is stable (13)), in AA smokers from two smoking cessation clinical trials and genotyped top hits in a third trial to confirm associations with the NMR. The first involved heavy smokers (≥10 cigarettes/day) screened for the PNAT2 trial (NCT01314001), where smokers were randomized to placebo, nicotine patch, or varenicline (23). The second involved light-smokers (≤10 cigarettes/day) that participated in a placebo-controlled bupropion trial for smoking cessation (KIS3 trial; NCT00666978) (34, 37). To further investigate associations between selected GWAS hits and the NMR, we utilized a third sample of AA light-smokers from a placebo-controlled nicotine gum trial (KIS2 trial) (38). Our goals were to better understand the genetic underpinnings of the NMR in AA smokers, and to compare genetic signals with those previously found in European populations to identify potential common and unique genetic influences on the NMR in AA smokers.
Methods
The original trial protocols were approved by institutional review boards at all participating sites and at the University of Toronto. Individuals providing written informed consent for DNA sample collection and release of de-identified information to investigators underwent genotyping.
PNAT2 Clinical Trial (NCT01314001) (23)
Participant characteristics and trial procedures are described in detail elsewhere (17, 23). Briefly, eligible adult (aged 18-65 years) smokers (≥10 cigarettes/day) from four clinical sites (University of Pennsylvania, University of Toronto/Centre for Addiction and Mental Health, MD Anderson, and the State University of New York at Buffalo) were randomized prospectively based on their pre-treatment NMR to receive placebo, nicotine patch, or varenicline treatment for smoking cessation.
KIS3 Clinical Trial (NCT00666978) (34)
Participant characteristics and clinical trial procedures are described in detail elsewhere (34, 37). Briefly, eligible adult (aged ≥18 years) light-smokers (≤10 cigarettes/day) from Kansas City, Missouri, were randomized to bupropion plus health education or placebo plus health education for smoking cessation.
KIS2 Clinical Trial (38)
Participant characteristics and clinical trial procedures are described in detail elsewhere (38). Briefly, eligible adult (aged ≥18 years) light-smokers (≤10 cigarettes/day) from Kansas City, Missouri, were randomized to nicotine gum or placebo and health education or motivational interviewing for smoking cessation.
Genome-Wide SNP Genotyping
Genome-wide SNP genotyping (PNAT2 and KIS3) was conducted using the Illumina HumanOmniExpressExome-8 v1.2 array (Illumina, San Diego, CA, USA) at the Centre for Applied Genomics at the Hospital for Sick Children (Toronto, ON, Canada). A custom iSelect® add-on comprising 2,688 variants (Table S1) was included based on previous associations with nicotine metabolism and/or smoking behaviours including cessation; these variants cover the CYP2ABFGST cluster (chromosome 19), the CHRNA5-A3-B4 nicotinic receptor cluster (chromosome 15), OCT2 (chromosome 6), and the UGT2B cluster (chromosome 4).
TaqMan SNP Genotyping
Candidate chromosome 19 polymorphisms (rs12459249, rs111645190, rs2644890, and rs111825958) were genotyped in KIS2 using an ABI ViiATM 7 Real-Time PCR System and TaqMan® SNP genotyping assays (Thermo Fisher Scientific, Waltham, Massachusetts, USA) according to the manufacturer’s protocol. The resulting genotype frequencies for all four SNPs were in Hardy-Weinberg Equilibrium (each P>0.05).
Quality Control (QC) Procedures for Genome-Wide SNP Genotyping and Imputation
QC procedures for sample and variant were carried out, and genotypes were imputed, prior to analysis as outlined in Figures S1 and S2. After identifying and removing samples with discordant sex information and excessive missingness of genetic data, the PNAT2 and KIS3 samples were combined to assess relatedness and ancestry to ensure a) an appropriate level of independence of individuals, and b) use of an equivalent threshold for determining ancestry. Individuals of AA ancestry, determined using principal components analysis in combination with data from HapMap 3 (Figure S2), were selected for further analyses. In PNAT2 and KIS3, 98.5% and 96.6% of African descent smokers, respectively, had genetic ancestries concordant with self-reported ancestry. Following QC, the final number of individuals and markers available was: n=506 PNAT2 AAs (251 males, 255 females), 733,629 variants; and n=458 KIS3 AAs (154 males, 304 females), 742,493 variants (Figure S1). QC procedures were performed using PLINK (version 1.07) (39) and R software.
The PNAT2 AA and KIS3 AA genotypes were then phased using SHAPEIT (40) and imputed using IMPUTE2 (41). The genomic data were divided into individual chromosomes (chromosomes 1-22) and aligned against the reference panel (Phase I release of 1000 Genomes). Following the elimination of duplicated SNPs, a second alignment step was performed, followed by pre-phasing and imputation, according to previously established protocols (41-44). Variants with INFO (i.e., quality) scores > 0.4 (threshold of 0.3 or higher is recommended (45)) and a minor allele frequency > 1% were selected for further analyses. Overall, 17,970,591 and 17,919,969 variants in PNAT2 and KIS3, respectively, were available for analysis.
Assessment of Imputation Quality for CYP2A6 Relative to Other Chromosome 19 Genes
CYP2A6 shares high homology with CYP2A7 and CYP2A13, which can confound the accuracy of CYP2A6 calls (46). IMPUTE2 info (i.e., imputation quality) scores for CYP2A6 were compared to those of EIF3K and TGFβ1, located outside of this region of high homology (~2,222kb 5’ and ~480kb 3’ of CYP2A6, respectively).
Phenotype: Nicotine Metabolite Ratio
The levels of cotinine and 3’hydroxycotinine were determined from blood samples collected at intake when participants were smoking ad libitum using identical liquid chromatography-tandem mass spectrometry according to previously established protocols (10, 11, 47). The NMR (3’hydroxycotinine/cotinine) was square-root-transformed to correct for positive skew (Figure S3). Individuals with cotinine values below 10 ng/ml, suggestive of non-daily smoking (48), were excluded from analyses.
Covariates
Analyses in PNAT2 and KIS3 included principal components 1 and 2 as covariates to control for possible effects of population stratification (49). To identify additional covariates, we performed separate linear regression analyses to identify whether factors previously significantly associated with the NMR in the whole PNAT2 sample (sex, age, estrogen-containing therapy use, BMI, alcohol use (17)) were associated with square-root NMR (with P<0.10) in PNAT2 AA and KIS3 AA. In PNAT2, the following were included as covariates: sex (P=0.038), age (P=0.006), BMI (P=0.038), and use of menthol cigarettes (P=0.063). In KIS3, the following were included as covariates: sex (P=0.056), age (P<0.001), and BMI (P<0.001), but not mentholated cigarette use (P=0.60).
Statistical Analyses
GWAS of the NMR
SNPTEST (version 2.5.2) was used to identify genetic associations with the NMR separately in PNAT2 and KIS3; chromosomes 1-22 were analyzed separately. Frequentist additive models were specified, and genotype uncertainty was controlled for by using the “-method expected” option (uses expected genotype counts or genotype dosages). We also performed a separate set of analyses specifying frequentist dominant models, and the “-method score” option, and acquired similar results. Variants with P<5e-8 were considered to be significant at the genome-wide level (50).
A meta-analysis of chromosome 19 results, adjusting for population sub-structure and NMR covariates, was then performed using META (version 1.7) (51). The genomic control inflation factor (λ) (calculated using PLINK) for the full GWAS analysis (chromosomes 1-22) was 1 and the QQ plots showed no deviation from the null (Figure S4). Because the same phenotype (square-root NMR; Figure S3) measured on the same scale was specified in both cohorts, the inverse-variance method based on a fixed-effects model was implemented (16). Variants with INFO scores ≥0.50 were included in the meta-analysis; a total of 367,834 markers were in the union list.
Conditional Analysis of Chromosome 19 NMR Results in PNAT2 and KIS3
To identify putatively independent chromosome 19 signals associated with the NMR, conditional analyses were performed (16); the variant with the smallest P-value in the meta-analysis (i.e., rs12459249) was considered the first independent signal, and then ‘conditioned on’ (i.e., entered as a covariate) in subsequent frequentist additive models performed separately in PNAT2 and KIS3. These results were meta-analyzed, with the variant with the smallest P-value (i.e., the second independent signal) entered as a covariate along with the first independent signal in the second round of conditional analyses. The procedure was repeated until no additional significant (i.e., P<5e-8) signals emerged.
Proportion of Variation in the NMR Accounted for by rs12459249, rs111645190, rs2644890, and rs11879604
Separate linear regression models were used to determine the proportion of NMR variability attributable to selected variants (three-genotype coding), using SPSS version 23 (IBM, Armonk, New York, USA). The outcome measure was square-root NMR. Models in PNAT2 controlled for sex, age, BMI, and the use of mentholated cigarettes, while models in KIS3 and KIS2 controlled for sex, age, and BMI. The proportion of NMR variability accounted for by each variant was calculated by squaring the variant’s part correlation coefficient and multiplying by 100.
Final Sample Sizes
Two of the n=506 PNAT2 AA participants were excluded from further analyses due to missing and outlying (>4 SD from the mean) square-root NMR values (Figure S1). After additionally excluding individuals with missing menthol covariate data (n=98), n=406 PNAT2 participants were available for GWAS analysis. Eight KIS3 AA participants were excluded due to having cotinine levels <10 ng/ml (Figure S1) which suggests non-daily smoking, and one participant was missing BMI data. Thus, n=449 KIS3 participants were available for GWAS analysis. Of the n=495 KIS2 individuals with pre-treatment NMR that provided a blood sample and consented to genetic testing, n=15 were excluded from further analyses due to insufficient quantity of blood remaining (n=7), cotinine level <10 ng/ml (n=6), and outlying (>4 SD from the mean) square-root NMR values (n=2). Thus, the final KIS2 AA sample comprised n=480 individuals.
Results
Characteristics of the final analyzed sample are provided in Table 1. These values are similar to those reported in the full trial samples (17, 34, 38). In PNAT2 and KIS3, the median info score (out of 1, with higher scores indicating higher imputation quality) for CYP2A6 was 0.9, compared to 0.6 and 0.9 for EIF3K and TGFβ1, respectively, suggesting adequate imputation quality for CYP2A6. In each of PNAT2 and KIS3, 98% of the variants significantly associated with the NMR at the genome-wide level (P<5e-8) were located on chromosome 19, within or near to (several kilobases) the CYP2A6 gene. Of note, no genetic variants in UGT enzymes (involved in the glucuronidation of nicotine, cotinine, and 3’hydroxycotinine (52)) or the OCT2 transporter (involved in nicotine transport (52)) reached genome-wide significance in either PNAT2 or KIS3.
Table 1.
Characteristics of African American smokers from the three clinical trial samples
| Characteristic | PNAT2 (N=504) | KIS3 (N=450) | KIS2 (N=480) |
|---|---|---|---|
| % Female (N) | 50.4 (254) | 66.4 (299) | 69.6 (334) |
| Age, mean (SD); range | 47.3 (9.8); 20-65 | 46.8 (11.6); 19-80 | 45.0 (11.2); 19-81 |
| BMI, mean (SD); range | 30.5 (7.1); 17.6-58.3 | 31.2 (7.8); 14.8-68.4 | 30.5 (8.0); 14.0-73.5 |
| Cigarettes/day, mean (SD); range | 16.3 (6.3); 5-40b | 7.8 (2.6); 1-17 | 7.6 (3.3); 0-30 |
| Cotinine in ng/ml, mean (SD), median; range | 274.2 (130.4), 252.7; 32.2-837.3 | 243.7 (122.4), 233.2; 13.7-680.7 | 248.9 (144.9), 235.9; 10.1-927.3 |
| NMR, mean (SD), median; range | 0.33 (0.20), 0.28; 0.0090-1.17 | 0.38 (0.26), 0.33; 0.02-1.79 | 0.33 (0.23), 0.27; 0.02-1.70 |
Abbreviations: PNAT, Pharmacogenetics of Nicotine Addiction Treatment; KIS, Kick-It-At-Swope; SD, standard deviation; BMI, body mass index; NMR, nicotine metabolite ratio
Meta-analysis of Chromosome 19 NMR Results in PNAT2 and KIS3 AA Smokers
Ninety-six genome-wide significant chromosome 19 variants were identified after adjusting for cohort-specific principal components 1 and 2 and NMR covariates (top 10 variants in Table 2, full list in Table S2; all top variants had info (quality) scores >0.9). Of note, the top 10 variants did not differ (similar betas and P-values) when four principal components were adjusted for. The top (smallest P-value) overall variant in the meta-analysis was rs12459249 (I2=0; Heterogeneity P=0.80), with a combined P-value of 1.47e-39 (Table 2); this was the top variant in PNAT2 (Figure 1A), and the second-most significant variant in KIS3 (Figure 1B). Overall, 58 (60.4%) of the 96 significant hits were not genome-wide significant in the GWAS of the NMR performed in ~1,500 Finnish European smokers (16); the most significant of these 58 AA hits in the meta-analysis was rs111825958 (I2=0.66; Heterogeneity P=0.32), with a combined P-value of 5.93e-26 (Table 2). Effect sizes and P-values for the top variants in each population (PNAT2 and KIS3) are also provided in Table 2. In a separate meta-analysis that additionally controlled for cigarettes/day and menthol use in KIS3, the top hit was rs11878604 with a beta of -0.68 (SE=0.069; P=5.65e-23 per C vs. T allele), while rs12459249 was the second top hit with a beta of 0.59 (SE=0.063; P=5.73e-21 per C vs. T allele); these effect sizes did not substantially differ from those in the primary analysis (Table 2).
Table 2.
Top 10 overall genetic variants significantly associated with the NMR in the meta-analyzed GWAS results from PNAT2 and KIS3 African American smokers
| Variant | Genotyped or Imputed (Imputation Quality Score) | Gene/location | Base-Pair Location (GRCh37) | Ref. Allele | Test Allele | MAF (%) in PNAT2 | MAF (%) in KIS3 | Beta (SE); P-Valuea,b in Meta-Analysis | Beta (SE); P-Valuea in PNAT2 | Beta (SE); P-Valueb in KIS3 |
|---|---|---|---|---|---|---|---|---|---|---|
| rs12459249 | Imputed in PNAT2 (0.99) and KIS3 (0.99) | ~9.5kb 3’ of CYP2A6 | 41339896 | T | C | 31.2 | 35.9 | 0.59 (0.045); 1.47e-39 | 0.61 (0.066); 1.59e-18 | 0.58 (0.062); 3.41e-19 |
| rs10853742 | Imputed in PNAT2 (0.99) and KIS3 (0.99) | ~8.9kb 3’ of CYP2A6 | 41340573 | G | C | 31.0 | 36.0 | 0.59 (0.045); 2.10e-39 | 0.60 (0.066); 1.92e-18 | 0.58 (0.062); 3.79e-19 |
| rs11667314 | Imputed in PNAT2 (0.98) and KIS3 (0.98) | ~8.5kb 3’ of CYP2A6 | 41340983 | T | C | 30.8 | 35.8 | 0.59 (0.045); 5.00e-39 | 0.60 (0.066); 2.90e-18 | 0.58 (0.062); 5.17e-19 |
| rs11878604 | Imputed in PNAT2 (0.97) and KIS3 (0.95) | ~16kb 3’ of CYP2A6 | 41333284 | T | C | 22.7 | 22.8 | -0.65 (0.050); 7.36e-39 | -0.64 (0.074); 9.60e-17 | -0.67 (0.069); 2.19e-20 |
| rs11083569 | Imputed in PNAT2 (0.95) and KIS3 (0.95) | ~9.1kb 3’ of CYP2A6 | 41340321 | C | G | 37.5 | 41.1 | 0.53 (0.045); 3.97e-32 | 0.50 (0.066); 2.09e-13 | 0.56 (0.061); 4.88e-18 |
| rs56267346 | Imputed in PNAT2 (0.97) and KIS3 (0.96) | CYP2A6 (intronic) | 41353338 | A | G | 38.8 | 39.6 | -0.50 (0.047); 5.49e-27 | -0.56 (0.068); 4.02e-15 | -0.46 (0.064); 5.83e-12 |
| rs111825958c | Genotyped in PNAT2 (N/A); imputed in KIS3 (0.92) | ~17kb 3’ of EGLN2 | 41331209 | C | A | 12.1 | 12.4 | -0.71 (0.067); 5.93e-26 | -0.64 (0.099); 4.11e-10 | -0.77 (0.092); 5.28e-16 |
| rs12986371 | Genotyped in PNAT2 (N/A) and KIS3 (N/A) | ~5.7kb 3’ of CYP2A6 | 41343698 | G | A | 21.3 | 26.7 | 0.52 (0.051); 6.79e-24 | 0.53 (0.078); 3.70e-11 | 0.51 (0.069); 5.40e-13 |
| rs111645190 c | Imputed in PNAT2 (1.0) and KIS3 (0.99) | ~5.5kb 5’ of CYP2A6 | 41361808 | G | A | 13.7 | 14.2 | -0.62 (0.062); 1.06e-23 | -0.59 (0.092); 4.10e-10 | -0.65 (0.085); 6.88e-14 |
| rs145638254c | Imputed in PNAT2 (1.0) and KIS3 (0.99) | ~4.7kb 5’ of CYP2A6 | 41361027 | G | A | 13.7 | 14.2 | -0.62 (0.062); 1.07e-23 | -0.59 (0.092); 4.10e-10 | -0.65 (0.085); 6.88e-14 |
MAF, minor allele frequency; N/A, not applicable
In PNAT2, GWAS results were adjusted for principal components 1 and 2, sex, age, BMI, and the use of mentholated cigarettes.
In KIS3, GWAS results were adjusted for principal components 1 and 2, sex, age, and BMI. The genomic inflation factor score (λ) in each population was 1 and therefore not adjusted for in the meta-analysis.
These variants were not genome-wide significant in a study of ~1500 Finnish European smokers (16)
Figure 1. rs12459249 and rs111645190 tagged the top two independent signals significantly associated with the NMR from the meta-analysis of conditional analyses in PNAT2 and KIS3 African American smokers.
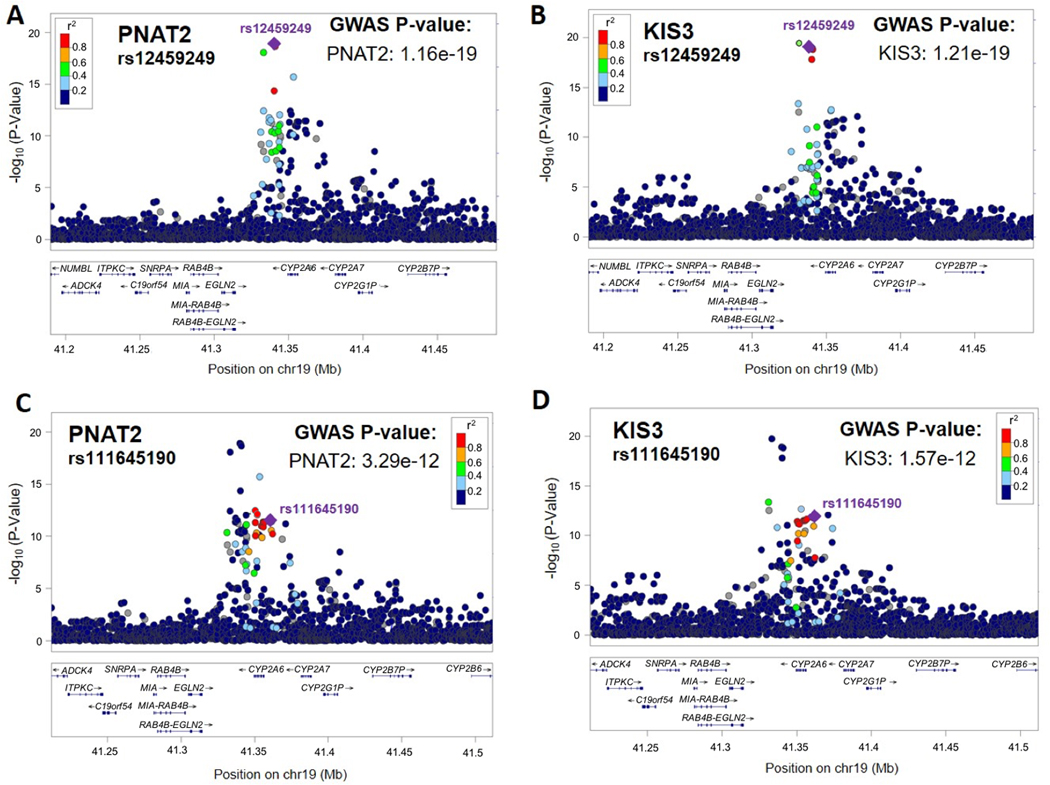
The top (i.e., smallest P-value) overall variant associated with the NMR was rs12459249. LocusZoom plots depicting rs12459249 (indicated with a purple diamond) in PNAT2 and KIS3 are shown in (A) and (B), respectively. P-values are adjusted for principal components 1 and 2. The second independent signal associated with the NMR was tagged by rs111645190. LocusZoom plots depicting rs111645190 (indicated with a purple diamond) in PNAT2 and KIS3 are shown in (C) and (D), respectively. P-values are adjusted for principal components 1 and 2. LD patterns are based upon the hg19/1000 Genomes November 2014 release AFR reference population.
Conditional Analysis of Chromosome 19 NMR Results in African American Smokers
Conditional analyses of the chromosome 19 NMR results in PNAT2 and KIS3 AA smokers revealed a total of three independent signals associated with the NMR; the first two were tagged by rs12459249 and rs111645190 (Table 3). In PNAT2 and KIS3, rs12459249, located ~9.5kb 3’ of CYP2A6, substantially altered NMR (Figure 2A and 2B), explaining 17.1% and 15.3% of the variability in the NMR, respectively. The association between the rs12459249 variant and the NMR also replicated in KIS2 (P=1.30e-17; Figure 2C). After conditioning on rs12459249, rs111645190 (located ~5.5kb 5’ of CYP2A6) had a P-value of 1.19e-11 (beta=-0.42 per A versus G allele; SE=0.062; Table 3 and Figure 1C and 1D). In PNAT2 and KIS3, the influence of rs111645190 on the NMR was also pronounced (Figure 2D and 2E), explaining an additional 2.9% and 5.2% of the variation in the NMR, respectively, after controlling for rs12459249. Of note, in a separate meta-analysis that additionally controlled for cigarettes/day and menthol use in KIS3, the effect size for rs111645190 was similar to the primary analysis (Table 2) (beta=-0.67, SE=0.085; P=4.10e-15 per A vs. G allele). The association for rs111645190 also replicated in KIS2 (P=1.77e-7; Figure 2F). After conditioning on both rs12459249 and rs111645190, a third independent signal emerged, tagged by rs185430475 (MAF = 2%; located >10MB 3’ of CYP2A6), with a P-value of 1.94e-8 (beta=1.27 per G versus C allele; SE=0.23). Of note, the rs185430475 variant was not significantly associated with the NMR in the meta-analysis (beta = 1.25 per G versus C allele; SE=0.26; P=9.26e-7), nor in PNAT2 (beta = 1.02 per G versus C allele; SE=0.33; P=0.0023) or KIS3 (beta = 1.47 per G versus C allele; SE=0.34; P=2.39e-5).
Table 3.
The variants tagging the top two independent signals from the meta-analysis of conditional analyses in PNAT2 and KIS3 African American smokers
| Variant | Gene/location | Ref. Allele | Test Allele | MAF (%) in PNAT2 | MAF (%) in KIS3 | Beta (SE); P-Value in PNAT2a | Beta (SE); P-Value in KIS3a | Beta (SE); P-Value in Meta-Analysisb |
|---|---|---|---|---|---|---|---|---|
| rs12459249 | ~9.5kb 3’ of CYP2A6 | T | C | 31.2 | 35.9 | 0.60 (0.063); 1.16e-19 | 0.61 (0.064); 1.21e-19 | 0.59 (0.045); 1.47e-39 |
| rs111645190 | ~5.5kb 5’ of CYP2A6 | G | A | 13.7 | 14.2 | -0.63 (0.088); 3.29e-12 | -0.65 (0.089); 1.57e-12 | -0.62 (0.06); 1.06e-23 |
MAF, minor allele frequency
Adjusted for cohort-specific principal components 1 and 2
In PNAT2, GWAS results were adjusted for principal components 1 and 2, sex, age, BMI, and the use of mentholated cigarettes. In KIS3, GWAS results were adjusted for principal components 1 and 2, sex, age, and BMI. The genomic inflation factor score (λ) in each population was 1 and therefore not adjusted for in the meta-analysis.
Figure 2. Influence of the top two independent signals, tagged by rs12459249 and rs111645190, identified in the meta-analysis of conditional analyses on the NMR in PNAT2, KIS3, and KIS2 smokers.
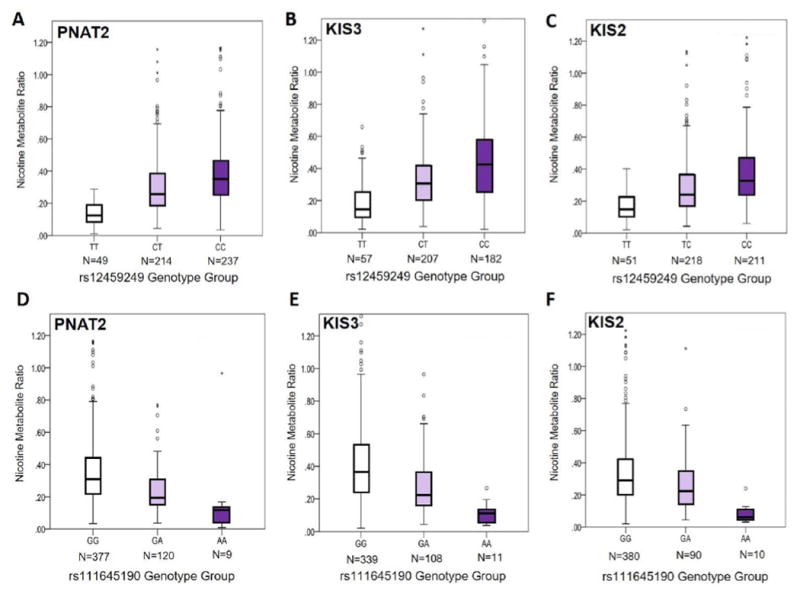
Associations between rs12459249 and the NMR (not transformed) are shown in PNAT2 (P=1.59e-18) (A), KIS3 (P=3.41e-19) (B), and KIS2 (P=1.30e-17) (C) African American smokers using boxplots. Associations between rs111645190 and the NMR (not transformed) are shown in PNAT2 (P=4.10e-10) (D), KIS3 (P=6.88e-14) (E), and KIS2 (P=1.77e-7) (F) African American smokers using boxplots. The box represents the interquartile (IQ) range. The line across the box indicates the median NMR value. Open circles represent NMR values that are between 1.5X and 3X the IQ range, while asterisks represent NMR values that are greater than 3X the IQ range. The P-values for PNAT2 and KIS3 are derived from the square-root NMR GWAS conducted separately in each sample, and are adjusted for cohort-specific principal components 1 and 2 and NMR covariates. The P-values for KIS2 are derived from additive linear regression models of square-root NMR adjusting for sex, age, and BMI. In KIS2, n=5 individuals with NMR values of 1.70, 1.52, 1.45, 1.37, and 1.36 are omitted from the graph but were included in the analysis. In KIS3, n=5 individuals with NMR values of 1.79, 1.78, 1.56, 1.52, and 1.48 are omitted from the graph but were included in the analysis.
Genetic Variants Associated with the NMR in African American Smokers (PNAT2 and KIS3 Analyzed Separately)
In PNAT2, 56 chromosome 19 variants significantly (P<5e-8) associated with the NMR were identified after adjusting for population sub-structure; 53 remained significant after additionally controlling for NMR covariates (Table S3). Controlling for clinical site did not substantially alter the findings (53 hits were still observed; rs12459249 remained the top hit with a beta (SE) per C vs. T allele of 0.61 (0.066); P=1.39e-18). A variant within chromosome 2 was also significantly associated with the NMR (rs16984355; P=2.1e-9). The top overall variant identified in PNAT2 was rs12459249 (Figure 1A and 2A), explaining 17.1% of NMR variation. Thirty-five of the 56 significant hits were not genome-wide significant in the ~1500 Finnish Europeans (16); the most significant of these hits was rs2644890 (Table 2 and Figure 3A and 4A), explaining 2.3% of NMR variation after controlling for rs12459249. Per 1000 Genomes, rs12459249 and rs2644890 are not in appreciable linkage disequilibrium (LD) in individuals of African descent (r2<0.20). The rs2644890 variant was also significantly associated with the NMR in KIS3 (P=1.24e-7; Figure 3B and 4B) and KIS2 (P=5.60e-5; Figure 4C).
Figure 3. rs2644890 and rs111825958 were the top unique variants significantly associated with the NMR in PNAT2 and KIS3 African American smokers, respectively.
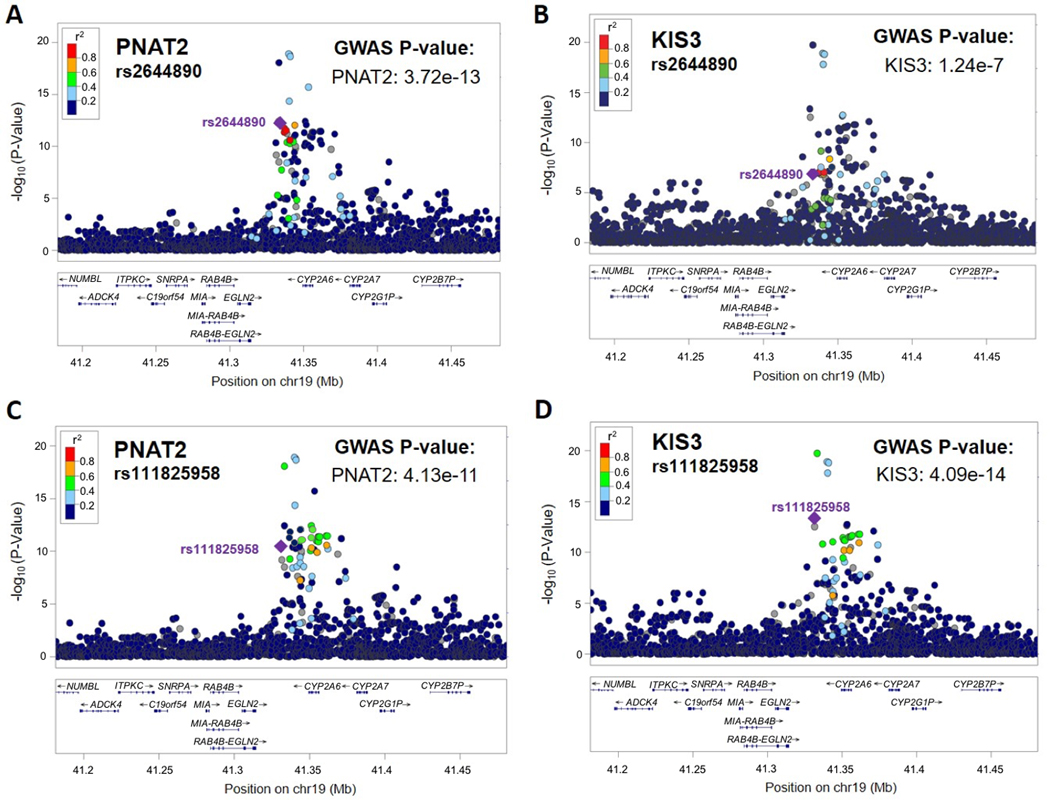
The top (i.e., smallest P-value) unique variant associated with the NMR in PNAT2 was rs2644890. LocusZoom plots depicting rs2644890 (indicated with a purple diamond) in PNAT2 and KIS3 are shown in (A) and (B), respectively. P-values are adjusted for principal components 1 and 2. The top (i.e., smallest P-value) unique variant associated with the NMR in KIS3 was rs111825958. LocusZoom plots depicting rs111825958 (indicated with a purple diamond) in PNAT2 and KIS3 are shown in (C) and (D), respectively. P-values are adjusted for principal components 1 and 2. LD patterns are based upon the hg19/1000 Genomes November 2014 release AFR reference population.
Figure 4. Influence of the top unique variants, rs2644890 and rs111825958, on the NMR in PNAT2, KIS3, and KIS2 smokers.
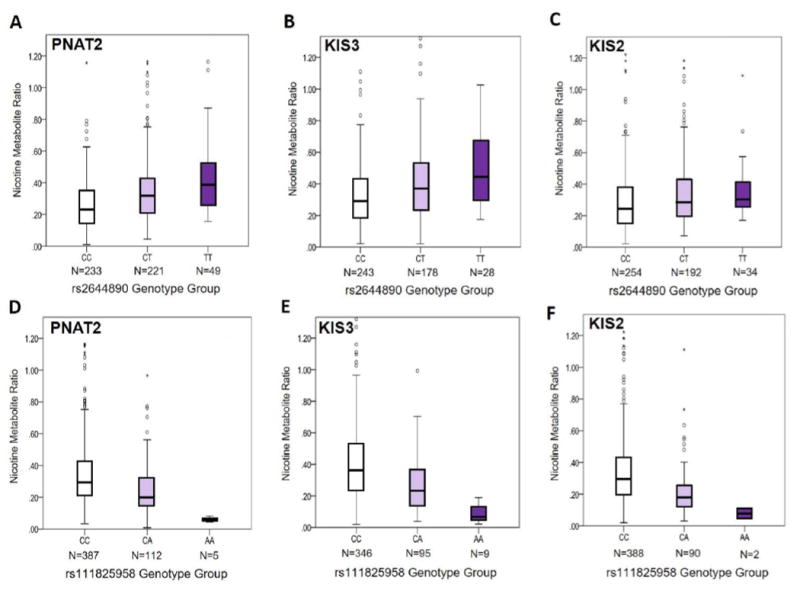
Associations between rs2644890 and the NMR (not transformed) are shown in PNAT2 (P=2.04e-12) (A), KIS3 (P=5.95e-7) (B), and KIS2 (P=5.60e-5) (C) African American smokers using boxplots. Associations between rs111825958 and the NMR (not transformed) are shown in PNAT2 (P=4.11e-10) (D), KIS3 (P=5.28e-16) (E), and KIS2 (P=4.25e-11) (F) African American smokers using boxplots. The box represents the interquartile (IQ) range. The line across the box indicates the median NMR value. Open circles represent NMR values that are between 1.5X and 3X the IQ range, while asterisks represent NMR values that are greater than 3X the IQ range. The P-values for PNAT2 and KIS3 are derived from the square-root NMR GWAS conducted separately in each sample, and are adjusted for cohort-specific principal components 1 and 2 and NMR covariates. The P-values for KIS2 are derived from additive linear regression models of square-root NMR adjusting for sex, age, and BMI. In KIS2, n=5 individuals with NMR values of 1.70, 1.52, 1.45, 1.37, and 1.36 are omitted from the graph but were included in the analysis. In KIS3, n=5 individuals with NMR values of 1.79, 1.78, 1.56, 1.52, and 1.48 are omitted from the graph but were included in the analysis.
In KIS3, 46 chromosome 19 variants significantly (P<5e-8) associated with the NMR were identified after adjusting for population sub-structure; 38 (>80%) of these variants were also genome-wide significant in PNAT2. After additionally controlling for NMR covariates, 44 chromosome 19 variants remained significant (Table S4). A variant within chromosome 2 was also significantly associated with the NMR (rs139278877; P=5.2e-9). The top overall variant in KIS3 was rs11878604 (Table 2), accounting for 17.1% of NMR variation; rs11878604 was also significant in PNAT2 (P=9.60e-17; Table 2). Twenty-eight of the 46 significant hits in KIS3 were not genome-wide significant in the ~1500 Finnish Europeans (16); the most significant of these hits was rs111825958 (Table 2 and Figure 3D and 4E), which explained 0.8% of NMR variation after controlling for rs11878604. Per 1000 Genomes, rs11878604 and rs111825958 are in moderate LD in individuals of African descent (r2=0.39). The rs111825958 variant was also significantly associated with the NMR in PNAT2 (P=4.11e-10; Figure 3C and 4D) and KIS2 (P=4.25e-11; Figure 4F).
Of note, the previously characterized nonsynonymous rs28399454 (C>T) variant in exon 7 of CYP2A6, which defines the non-functional CYP2A6*17 allele present at high frequency in AAs (33), was significantly associated with the NMR in both PNAT2 (P=4.56e-11; beta=-0.68 per T versus C allele; SE=0.10, allele frequency=10.5%) and KIS3 (P=5.90e-11; beta=-0.68 per T versus C allele; SE=0.10 allele frequency=11.0%). In a model that controlled for population sub-structure, cohort-specific NMR covariates, and additionally for rs28399454, the P-values for rs12459249 in PNAT2 and KIS3 increased somewhat (from 1.59e-18 to 1.03e-11, and from 3.41e-19 to 1.13e-11, respectively). However, the P-value for rs111645190 was not genome- wide significant in each of PNAT2 and KIS3 after controlling for rs28399454: P-values increased from 4.10e-10 to 0.023 in PNAT2, and from 6.88e-14 to 0.011 in KIS3.
Discussion
This is the first NMR GWAS conducted exclusively in African Americans. We identified three independent signals tagged by rs12459249, rs111645190, and rs185430475. These three signals were not in LD (r2<0.20) with (in the 1000 Genomes Project AFR population) the four independent signals (rs56113850, rs113288603, esv2663194, and rs12461964) identified in the first NMR GWAS, which we conducted in ~1500 Finnish European smokers (16). Together these findings extend our prior work (e.g., for *23 , *24, *25, *28, *35, and *39-*45 (29-32)) showing the existence of unique genetic influences on CYP2A6 function and the NMR in AA. The top independent signal, rs12459249, located ~9.5kb 3’ of CYP2A6, was also genome-wide significant in the Finnish sample (16), suggesting a common ancestral origin; however, it is possible that rs12459249 tags different functional variants in different populations. After controlling for rs28399454, the defining variant in the CYP2A6*17 allele present at high frequencies in AA (33), the P-values for rs12459249 in PNAT2 and KIS3 increased only somewhat (from ~10-18 to 10-11), suggesting at least a portion of the influence of rs12459249 on the NMR is independent of CYP2A6*17. A recent study examined the NMR following oral or i.v. administration of labeled nicotine and cotinine in n=212, n=51, and n=49 individuals of EA, Asian American, and AA ancestry, respectively, and identified rs12459249 as the top-ranked SNP overall; rs12459249 was also non-significantly associated with the NMR (P=5.76e-6) in the small sample of AA (35).
The second independent signal was tagged by rs111645190, located ~5.5kb 5’ of CYP2A6. The top two independent variants (rs12459249 and rs111645190) explained ~20% of NMR variation, comparable to the amount of variability captured in the ~1500 Finnish European smokers (16), where the independent signals explained ~18-31% of NMR variation. However, the influence of rs111645190 on the NMR was no longer significant in either PNAT2 or KIS3 after controlling for rs28399454 (CYP2A6*17 allele), suggesting this second independent signal is largely driven by rs28399454 (33).
Of note, over half (~60%) of the 96 hits found in the meta-analysis were not genome-wide significant in the ~1500 Finnish Europeans (16), in part reflecting unique population LD structure. The top unique variant in the meta-analysis, rs111825958, was also associated with the NMR in KIS2 (38). After controlling for rs28399454 (CYP2A6*17), the P-values in PNAT2 and KIS3 increased from 4.11e-10 to 0.018, and from 5.28e-16 to 1.18e-5, respectively, suggesting, as for rs111645190, that rs28399454 explains a large portion of the influence of rs111825958 on the NMR.
Our previous NMR GWAS in ~1500 Finnish European smokers (16) identified >700 hits, all found on chromosome 19q13 in or near to the CYP2A6 locus. The top hit, rs56113850, located in intron 4 of CYP2A6, also replicated in the EA participants from the smaller GWAS of laboratory-based NMR (35). A subsequent GWAS of urinary NMR, conducted in ~2,200 smokers (including n=364 AA) from a prospective multi-ethnic cohort study, identified 248 variants (~99% of which were within or near CYP2A6) significantly associated with the NMR, and replicated this top hit (rs56113850) (36). The rs56113850 variant was also significantly associated with the NMR in PNAT2 (P=1.30e-10, beta = 0.46 per C versus T allele, SE= 0.069) and KIS3 (P=8.02e-12, beta = 0.45 per C versus T allele, SE = 0.064) AA smokers, suggesting, as for rs12459249, a common ancestral origin. The demonstrated influence of rs12459249 and rs56113850 on the NMR in a variety of ethnic groups combined with their high variant allele frequencies (~30-60% in individuals of European and African descent) and only moderate LD (r2=0.46 and <0.20 in European and African descent individuals, respectively; 1000 Genomes data), suggest that these SNPs should be routinely included in genotyping platforms for genomic investigations of nicotine metabolism and smoking cessation. GTEx expression quantitative trait loci analyses suggest rs12459249 is associated with CYP2A6 protein expression in the lung, and possibly liver (effect size=0.12 for C vs. T), while rs56113850 has a greater relative (vs. rs12459249) influence on liver CYP2A6 mRNA expression (effect size=0.26 for C vs. T).
Because the NMR is 80% heritable (estimated in Finnish twins) (16), largely mediated by a single enzyme (i.e., CYP2A6), and not appreciably altered by environmental factors (17), the usefulness of CYP2A6 genetics for personalizing therapy and understanding tobacco-related disease risk shows great promise. However, the NMR can only be reliably used to assess CYP2A6 activity in current, regular (i.e., daily) smokers (12-14), while CYP2A6 genetics could be used to predict activity phenotype in current, former, and non-smokers in epidemiological investigations of cancer risk, for example. Thus, it is likely that a CYP2A6 genetics-based approach could have greater utility and wider applicability compared to the NMR. In addition, because CYP2A6 also metabolizes therapeutic drugs including letrozole (53) and tegafur (54), two chemotherapeutics, as well as other drugs (e.g., efavirenz, metronidazole, artemisinin, valproic acid) (55), the usefulness of CYP2A6 genetics in personalized medicine approaches extends beyond tobacco dependence.
Several limitations of our work warrant mention. By virtue of the genome-wide genotyping chip, we were unable to adequately examine structural variation in CYP2A6. Copy number variation, such as the CYP2A6*1XA duplication and CYP2A6*4 deletion variants (46), is known to alter CYP2A6 activity; it is possible that known and/or novel copy number variants in CYP2A6 are in LD with the variants identified in our study. In addition, the lack of overlap in signals observed between AA and European descent smokers may be due, in part, to differences in the genotyping platforms used, reference panels for imputation, quality control/imputation/MAF filtering pipelines, as well as potential inter-ethnic variation in environmental confounding factors and/or additional potential differences between smokers seeking treatment versus those that are not. However, head-to-head comparisons of NMR GWAS signals in PNAT2 AA and PNAT2 European descent smokers, analyzed using an identical genotyping platform and phase I release of 1000 Genomes, also indicate a substantial lack overlap (unpublished observations). Finally, analyzing treatment-seeking smokers may limit generalizability to general smoking populations, however personalized medicine approaches based on CYP2A6 or the NMR would be targeted to treatment-seeking smokers and future GWAS in treatment-seeking smokers from other ethnic backgrounds should be considered. Future larger studies may identify important signals outside of CYP2A6 that influence the NMR.
In summary, we identified three independent signals in the largest NMR GWAS of AA smokers performed to date, accounting for ~20% of the total variability in the NMR. Over half (~60%) of the 96 total hits were not found in the largest NMR GWAS of European descent smokers (16), and might contribute to unique regulation of CYP2A6 in AA. Further investigation of these hits, including haplotype characterization and functional assessments will help identify which variants are causally influencing the NMR beyond known functional variants (e.g., CYP2A6*17 (33)). There may also be rare CYP2A6 variants (56, 57) with substantial impacts on the NMR; future sequencing-based studies will complement GWAS approaches and may further improve our understanding of the genetic influences on the NMR. Determining whether these genetic variants influence other phenotypes including smoking cessation will set the stage for genomics-based personalization of tobacco dependence treatment. Functional characterization studies may also provide insight into inter-ethnic variability in nicotine metabolism/CYP2A6 activity and resulting smoking behaviours and tobacco-related disease risk.
Supplementary Material
The quality control (QC) steps for both samples and markers (i.e., variants) carried out prior to analysis are depicted in chronological order. The original sample sizes were: n=1,687 PNAT2 smokers (self-reported Caucasian, African or African American, Asian, Native American, Hawaiian/Polynesian, multi-racial, or other), and n=502 KIS3 smokers (self-reported African American). Sample QC on PNAT2 and KIS3 was performed separately for steps 1 and 2. The PNAT2 and KIS3 samples remaining following sample QC step 2 were analyzed together for relatedness (step 3) and ancestry determination (step 4). Remaining sample QC (step 5; heterozygosity determination) and all marker QC (steps 6-8) were performed separately for PNAT2 AA and KIS3 AA. The final numbers of samples and markers remaining after sample and marker QC are indicated in the grey boxes. The final number of PNAT2 AA and KIS3 AA samples remaining after sample, marker, and biomarker QC (step 9) are indicated in the black boxes. There were n=406 PNAT2 AA individuals available for final analyses after excluding those with missing menthol data (n=98), and n=449 KIS3 AA individuals were available for final analyses after excluding one participant with missing BMI data.
Abbreviations: QC, quality control; IBS, identity by state; PCA, principal components analysis; PC, principal component; SD, standard deviation; AA, African American
Principal Components 1 and 2, generated from Sample QC Step 4 (see Figure S1), are plotted for the whole PNAT2 clinical sample (turquoise open circles), KIS3 (black open circles), and HapMap reference populations: CEU (Utah residents with Northern and Western European ancestry; red open circles), ASW (African ancestry in Southwest USA; purple open circles), YRI (Yoruba in Ibadan, Nigeria; green open circles), and CHB (Han Chinese in Beijing, China; blue open circles). African American ancestry for PNAT2 and KIS3 was determined using the following cut-points: Principal Component 1 ≤-0.025, and Principal Component 2 ≤0.01, indicated with black dotted lines. These conservative thresholds were selected to ensure homogeneity of the population and were chosen based on visual inspection of the plot.
The NMR distribution is shown in PNAT2 (A), KIS3 (B), and KIS2 (C) African American smokers. The square-root NMR distribution is shown in PNAT2 (D), KIS3 (E), and KIS2 (F) African American smokers. NMR was square-root transformed to help correct for positive skew.
Abbreviation: SD, standard deviation
QQ plots of the full results of the square-root NMR GWAS are shown in PNAT2 (A) and KIS3 (B), and after excluding genome-wide significant hits in PNAT2 (C) and KIS3 (D). Expected P-values are those expected under the null hypothesis. The shaded area around the red line indicates the 95% confidence interval under the null.
Table S1. 2,688 variants included as a custom iSelect® add-on to the Illumina HumanOmniExpressExome-8 v1.2 array.
Table S2. Complete list of chromosome 19 variants significantly (P<5e-8) associated with the NMR in the meta-analysis of PNAT2 and KIS3 African American smokers.
Table S3. Complete list of chromosome 19 variants significantly (P<5e-8) associated with the NMR in PNAT2 African American smokers.
Table S4. Complete list of chromosome 19 variants significantly (P<5e-8) associated with the NMR in KIS3 African American smokers.
Acknowledgments
Computations were performed on the Centre for Addiction and Mental Health (CAMH) Specialized Computing Cluster (SCC), funded by the Canada Foundation for Innovation Research Hospital Fund. J. J. Ware contributed to this work while supported by a Post-Doctoral Research Fellowship from the Oak Foundation (http://www.oakfnd.org/). J. J. Ware was also a member of the Medical Research Council (MRC) Integrative Epidemiology Unit at this time; support from the MRC is gratefully acknowledged (MC_UU_12013/6). We also acknowledge these sources of funding: Canada Research Chair in Pharmacogenomics (R. F. Tyndale); National Institutes of Health (NIH) grants PGRN DA020830 (R. F. Tyndale and C. Lerman) and CA091912 (L. Sanderson Cox); Canadian Institutes of Health Research (CIHR) grant TMH-109787 (R. F. Tyndale); the Campbell Family Mental Health Research Institute of the Centre for Addiction and Mental Health (CAMH); the CAMH Foundation; the Canada Foundation for Innovation (#20289 and #16014); and the Ontario Ministry of Research and Innovation. Members of the Pharmacogenomics Research Network: Pharmacogenetics of Nicotine Addiction Treatment (PGRN-PNAT) Research Group include Frank Leone, Henry Glick, Angela Pinto, Paul Sanborn, Peter Gariti, Richard Landis (University of Pennsylvania); Maria Novalen, Bin Zhao, Ewa Hoffmann, Qian Zhou, Adel Aziziyeh (CAMH/University of Toronto); Martin Mahoney (Roswell Cancer Center, University of Buffalo); Maher Karam-Hage (The University of Texas M.D. Anderson Cancer Center); David Conti (University of Southern California); Andrew Bergen (SRI International).
Footnotes
Declaration of Interests
R. F. Tyndale has consulted in the past for Apotex on unrelated topics. N. L. Benowitz has consulted with pharmaceutical companies that market smoking cessation medications and has been a paid expert witness in litigation against tobacco companies. P. M. Cinciripini served on the scientific advisory board of Pfizer, conducted educational talks sponsored by Pfizer on smoking cessation (2006-2008), and has received grant support from Pfizer. R. A. Schnoll has provided consultation to Pfizer and GlaxoSmithKline. Pfizer Inc. provided varenicline and placebo pills at no cost for the PNAT2 clinical trial. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jamal A, King BA, Neff LJ, Whitmill J, Babb SD, Graffunder CM. Current Cigarette Smoking Among Adults - United States, 2005-2015. Mmwr-Morbidity and Mortality Weekly Report. 2016 Nov 11;65(44):1205–11. doi: 10.15585/mmwr.mm6544a2. [DOI] [PubMed] [Google Scholar]
- 2.Inoue-Choi M, Liao LM, Reyes-Guzman C, Hartge P, Caporaso N, Freedman ND. Association of Long-term, Low-Intensity Smoking With All-Cause and Cause-Specific Mortality in the National Institutes of Health-AARP Diet and Health Study. JAMA Intern Med. 2016 Dec 05; doi: 10.1001/jamainternmed.2016.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.St Helen G, Dempsey D, Wilson M, Jacob P, Benowitz NL. Racial differences in the relationship between tobacco dependence and nicotine and carcinogen exposure. Addiction. 2013 Mar;108(3):607–17. doi: 10.1111/j.1360-0443.2012.04077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haiman CA, Stram DO, Wilkens LR, Pike MC, Kolonel LN, Henderson BE, et al. Ethnic and racial differences in the smoking-related risk of lung cancer. N Engl J Med. 2006 Jan 26;354(4):333–42. doi: 10.1056/NEJMoa033250. [DOI] [PubMed] [Google Scholar]
- 5.Goren A, Annunziata K, Schnoll RA, Suaya JA. Smoking Cessation and Attempted Cessation among Adults in the United States. Plos One. 2014 Mar 27;9(3) doi: 10.1371/journal.pone.0093014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benowitz NL. Nicotine addiction. N Engl J Med. 2010 Jun 17;362(24):2295–303. doi: 10.1056/NEJMra0809890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakajima M, Yamamoto T, Nunoya K, Yokoi T, Nagashima K, Inoue K, et al. Role of human cytochrome P4502A6 in C-oxidation of nicotine. Drug Metab Dispos. 1996 Nov;24(11):1212–7. [PubMed] [Google Scholar]
- 8.Nakajima M, Yamamoto T, Nunoya K, Yokoi T, Nagashima K, Inoue K, et al. Characterization of CYP2A6 involved in 3’-hydroxylation of cotinine in human liver microsomes. J Pharmacol Exp Ther. 1996 May;277(2):1010–5. [PubMed] [Google Scholar]
- 9.Benowitz NL, Swan GE, Jacob P, 3rd, Lessov-Schlaggar CN, Tyndale RF. CYP2A6 genotype and the metabolism and disposition kinetics of nicotine. Clinical Pharmacology & Therapeutics. 2006 Nov;80(5):457–67. doi: 10.1016/j.clpt.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 10.Dempsey D, Tutka P, Jacob P, Allen F, Schoedel K, Tyndale RF, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clinical Pharmacology & Therapeutics. 2004 Jul;76(1):64–72. doi: 10.1016/j.clpt.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 11.Tanner JA, Novalen M, Jatlow P, Huestis MA, Murphy SE, Kaprio J, et al. Nicotine metabolite ratio (3-hydroxycotinine/cotinine) in plasma and urine by different analytical methods and laboratories: implications for clinical implementation. Cancer Epidemiol Biomarkers Prev. 2015 Aug;24(8):1239–46. doi: 10.1158/1055-9965.EPI-14-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lea RA, Dickson S, Benowitz NL. Within-subject variation of the salivary 3HC/COT ratio in regular daily smokers: prospects for estimating CYP2A6 enzyme activity in large-scale surveys of nicotine metabolic rate. J Anal Toxicol. 2006 Jul-Aug;30(6):386–9. doi: 10.1093/jat/30.6.386. [DOI] [PubMed] [Google Scholar]
- 13.Mooney ME, Li ZZ, Murphy SE, Pentel PR, Le C, Hatsukami DK. Stability of the nicotine metabolite ratio in ad libitum and reducing smokers. Cancer Epidemiol Biomarkers Prev. 2008 Jun;17(6):1396–400. doi: 10.1158/1055-9965.EPI-08-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.St Helen G, Jacob P, 3rd, Benowitz NL. Stability of the nicotine metabolite ratio in smokers of progressively reduced nicotine content cigarettes. Nicotine Tob Res. 2013 Nov;15(11):1939–42. doi: 10.1093/ntr/ntt065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benowitz NL, St Helen G, Dempsey DA, Jacob P, 3rd, Tyndale RF. Disposition kinetics and metabolism of nicotine and cotinine in African American smokers: impact of CYP2A6 genetic variation and enzymatic activity. Pharmacogenet Genomics. 2016 Jul;26(7):340–50. doi: 10.1097/FPC.0000000000000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loukola A, Buchwald J, Gupta R, Palviainen T, Hallfors J, Tikkanen E, et al. A Genome-Wide Association Study of a Biomarker of Nicotine Metabolism. PLoS Genet. 2015;11(9):e1005498. doi: 10.1371/journal.pgen.1005498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chenoweth MJ, Novalen M, Hawk LW, Jr, Schnoll RA, George TP, Cinciripini PM, et al. Known and novel sources of variability in the nicotine metabolite ratio in a large sample of treatment-seeking smokers. Cancer Epidemiol Biomarkers Prev. 2014 Sep;23(9):1773–82. doi: 10.1158/1055-9965.EPI-14-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swan GE, Lessov-Schlaggar CN, Bergen AW, He Y, Tyndale RF, Benowitz NL. Genetic and environmental influences on the ratio of 3’hydroxycotinine to cotinine in plasma and urine. Pharmacogenet Genomics. 2009 May;19(5):388–98. doi: 10.1097/FPC.0b013e32832a404f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patterson F, Schnoll RA, Wileyto EP, Pinto A, Epstein LH, Shields PG, et al. Toward personalized therapy for smoking cessation: a randomized placebo-controlled trial of bupropion. Clinical Pharmacology & Therapeutics. 2008 Sep;84(3):320–5. doi: 10.1038/clpt.2008.57. [DOI] [PubMed] [Google Scholar]
- 20.Lerman C, Tyndale R, Patterson F, Wileyto EP, Shields PG, Pinto A, et al. Nicotine metabolite ratio predicts efficacy of transdermal nicotine for smoking cessation. Clinical Pharmacology & Therapeutics. 2006 Jun;79(6):600–8. doi: 10.1016/j.clpt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Schnoll RA, Patterson F, Wileyto EP, Tyndale RF, Benowitz N, Lerman C. Nicotine metabolic rate predicts successful smoking cessation with transdermal nicotine: a validation study. Pharmacol Biochem Behav. 2009 Mar;92(1):6–11. doi: 10.1016/j.pbb.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lerman C, Jepson C, Wileyto EP, Patterson F, Schnoll R, Mroziewicz M, et al. Genetic variation in nicotine metabolism predicts the efficacy of extended-duration transdermal nicotine therapy. Clinical Pharmacology & Therapeutics. 2010 May;87(5):553–7. doi: 10.1038/clpt.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lerman C, Schnoll RA, Hawk LW, Cinciripini P, George TP, Wileyto EP, et al. Use of the nicotine metabolite ratio as a genetically informed biomarker of response to nicotine patch or varenicline for smoking cessation: a randomised, double-blind placebo-controlled trial. Lancet Respiratory Medicine. 2015 Feb;3(2):131–8. doi: 10.1016/S2213-2600(14)70294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wassenaar CA, Dong Q, Wei Q, Amos CI, Spitz MR, Tyndale RF. Relationship between CYP2A6 and CHRNA5-CHRNA3-CHRNB4 variation and smoking behaviors and lung cancer risk. J Natl Cancer Inst. 2011 Sep 7;103(17):1342–6. doi: 10.1093/jnci/djr237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sofuoglu M, Herman AI, Nadim H, Jatlow P. Rapid nicotine clearance is associated with greater reward and heart rate increases from intravenous nicotine. Neuropsychopharmacology. 2012 May;37(6):1509–16. doi: 10.1038/npp.2011.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chenoweth MJ, Tyndale RF. Pharmacogenetic Optimization of Smoking Cessation Treatment. Trends Pharmacol Sci. 2016 Oct 3; doi: 10.1016/j.tips.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kandel DB, Hu MC, Schaffran C, Udry JR, Benowitz NL. Urine nicotine metabolites and smoking behavior in a multiracial/multiethnic national sample of young adults. Am J Epidemiol. 2007 Apr 15;165(8):901–10. doi: 10.1093/aje/kwm010. [DOI] [PubMed] [Google Scholar]
- 28.Nakajima M, Fukami T, Yamanaka H, Higashi E, Sakai H, Yoshida R, et al. Comprehensive evaluation of variability in nicotine metabolism and CYP2A6 polymorphic alleles in four ethnic populations. Clinical Pharmacology & Therapeutics. 2006 Sep;80(3):282–97. doi: 10.1016/j.clpt.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 29.Ho MK, Mwenifumbo JC, Zhao B, Gillam EM, Tyndale RF. A novel CYP2A6 allele, CYP2A6*23, impairs enzyme function in vitro and in vivo and decreases smoking in a population of Black-African descent. Pharmacogenet Genomics. 2008 Jan;18(1):67–75. doi: 10.1097/FPC.0b013e3282f3606e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mwenifumbo JC, AlKoudsi N, Ho MK, Zhou Q, Hoffmann EB, Sellers EM, et al. Novel and established CYP2A6 alleles impair in vivo nicotine metabolism in a population of Black African descent. Hum Mutat. 2008 May;29(5):679–88. doi: 10.1002/humu.20698. [DOI] [PubMed] [Google Scholar]
- 31.Al Koudsi N, Ahluwalia JS, Lin SK, Sellers EM, Tyndale RF. A novel CYP2A6 allele (CYP2A6*35) resulting in an amino-acid substitution (Asn438Tyr) is associated with lower CYP2A6 activity in vivo. Pharmacogenomics J. 2009 Aug;9(4):274–82. doi: 10.1038/tpj.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piliguian M, Zhu AZ, Zhou Q, Benowitz NL, Ahluwalia JS, Sanderson Cox L, et al. Novel CYP2A6 variants identified in African Americans are associated with slow nicotine metabolism in vitro and in vivo. Pharmacogenet Genomics. 2014 Feb;24(2):118–28. doi: 10.1097/FPC.0000000000000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fukami T, Nakajima M, Yoshida R, Tsuchiya Y, Fujiki Y, Katoh M, et al. A novel polymorphism of human CYP2A6 gene CTP2A6*17 has an amino acid substitution (V365M) that decreases enzymatic activity in vitro and in vivo. Clinical Pharmacology & Therapeutics. 2004 Dec;76(6):519–27. doi: 10.1016/j.clpt.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 34.Cox LS, Nollen NL, Mayo MS, Choi WS, Faseru B, Benowitz NL, et al. Bupropion for smoking cessation in African American light smokers: a randomized controlled trial. J Natl Cancer Inst. 2012 Feb 22;104(4):290–8. doi: 10.1093/jnci/djr513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baurley JW, Edlund CK, Pardamean CI, Conti DV, Krasnow R, Javitz HS, et al. Genome-Wide Association of the Laboratory-Based Nicotine Metabolite Ratio in Three Ancestries. Nicotine Tob Res. 2016 Apr 25; doi: 10.1093/ntr/ntw117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel YM, Park SL, Han Y, Wilkens LR, Bickeboller H, Rosenberger A, et al. Novel Association of Genetic Markers Affecting CYP2A6 Activity and Lung Cancer Risk. Cancer Res. 2016 Oct 1;76(19):5768–76. doi: 10.1158/0008-5472.CAN-16-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cox LS, Faseru B, Mayo MS, Krebill R, Snow TS, Bronars CA, et al. Design, baseline characteristics, and retention of African American light smokers into a randomized trial involving biological data. Trials. 2011 Jan 25;12 doi: 10.1186/1745-6215-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahluwalia JS, Okuyemi K, Nollen N, Choi WS, Kaur H, Pulvers K, et al. The effects of nicotine gum and counseling among African American light smokers: a 2 × 2 factorial design. Addiction. 2006 Jun;101(6):883–91. doi: 10.1111/j.1360-0443.2006.01461.x. [DOI] [PubMed] [Google Scholar]
- 39.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007 Sep;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delaneau O, Marchini J. Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat Commun. 2014;5:3934. doi: 10.1038/ncomms4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009 Jun;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delaneau O, Howie B, Cox AJ, Zagury JF, Marchini J. Haplotype Estimation Using Sequencing Reads. American Journal of Human Genetics. 2013 Oct 3;93(4):687–96. doi: 10.1016/j.ajhg.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howie B, Marchini J, Stephens M. Genotype Imputation with Thousands of Genomes. G3-Genes Genomes Genetics. 2011 Nov 1;1(6):457–69. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Connell J, Gurdasani D, Delaneau O, Pirastu N, Ulivi S, Cocca M, et al. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 2014 Apr;10(4):e1004234. doi: 10.1371/journal.pgen.1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roshyara NR, Kirsten H, Horn K, Ahnert P, Scholz M. Impact of pre-imputation SNP-filtering on genotype imputation results. BMC Genet. 2014 Aug 12;15:88. doi: 10.1186/s12863-014-0088-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wassenaar CA, Zhou Q, Tyndale RF. CYP2A6 genotyping methods and strategies using real-time and end point PCR platforms. Pharmacogenomics. 2016;17(2):147–62. doi: 10.2217/pgs.15.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schnoll RA, George TP, Hawk L, Cinciripini P, Wileyto P, Tyndale RF. The relationship between the nicotine metabolite ratio and three self-report measures of nicotine dependence across sex and race. Psychopharmacology. 2014 Jun;231(12):2515–23. doi: 10.1007/s00213-013-3421-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vartiainen E, Seppala T, Lillsunde P, Puska P. Validation of self reported smoking by serum cotinine measurement in a community-based study. J Epidemiol Community Health. 2002 Mar;56(3):167–70. doi: 10.1136/jech.56.3.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006 Aug;38(8):904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 50.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996 Sep 13;273(5281):1516–7. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 51.Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010 May;42(5):436–40. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benowitz NL, Hukkanen J, Jacob P., 3rd Nicotine chemistry, metabolism, kinetics and biomarkers. Handb Exp Pharmacol. 2009;(192):29–60. doi: 10.1007/978-3-540-69248-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murai K, Yamazaki H, Nakagawa K, Kawai R, Kamataki T. Deactivation of anti-cancer drug letrozole to a carbinol metabolite by polymorphic cytochrome P450 2A6 in human liver microsomes. Xenobiotica. 2009 Nov;39(11):795–802. doi: 10.3109/00498250903171395. [DOI] [PubMed] [Google Scholar]
- 54.Ikeda K, Yoshisue K, Matsushima E, Nagayama S, Kobayashi K, Tyson CA, et al. Bioactivation of tegafur to 5-fluorouracil is catalyzed by cytochrome P-450 2A6 in human liver microsomes in vitro. Clin Cancer Res. 2000 Nov;6(11):4409–15. [PubMed] [Google Scholar]
- 55.McDonagh EM, Wassenaar C, David SP, Tyndale RF, Altman RB, Whirl-Carrillo M, et al. PharmGKB summary: very important pharmacogene information for cytochrome P-450, family 2, subfamily A, polypeptide 6. Pharmacogenet Genomics. 2012 Sep;22(9):695–708. doi: 10.1097/FPC.0b013e3283540217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kozyra M, Ingelman-Sundberg M, Lauschke VM. Rare genetic variants in cellular transporters, metabolic enzymes, and nuclear receptors can be important determinants of interindividual differences in drug response. Genet Med. 2016 Apr 21; doi: 10.1038/gim.2016.33. [DOI] [PubMed] [Google Scholar]
- 57.Fujikura K, Ingelman-Sundberg M, Lauschke VM. Genetic variation in the human cytochrome P450 supergene family. Pharmacogenetics and Genomics. 2015 Dec;25(12):584–94. doi: 10.1097/FPC.0000000000000172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The quality control (QC) steps for both samples and markers (i.e., variants) carried out prior to analysis are depicted in chronological order. The original sample sizes were: n=1,687 PNAT2 smokers (self-reported Caucasian, African or African American, Asian, Native American, Hawaiian/Polynesian, multi-racial, or other), and n=502 KIS3 smokers (self-reported African American). Sample QC on PNAT2 and KIS3 was performed separately for steps 1 and 2. The PNAT2 and KIS3 samples remaining following sample QC step 2 were analyzed together for relatedness (step 3) and ancestry determination (step 4). Remaining sample QC (step 5; heterozygosity determination) and all marker QC (steps 6-8) were performed separately for PNAT2 AA and KIS3 AA. The final numbers of samples and markers remaining after sample and marker QC are indicated in the grey boxes. The final number of PNAT2 AA and KIS3 AA samples remaining after sample, marker, and biomarker QC (step 9) are indicated in the black boxes. There were n=406 PNAT2 AA individuals available for final analyses after excluding those with missing menthol data (n=98), and n=449 KIS3 AA individuals were available for final analyses after excluding one participant with missing BMI data.
Abbreviations: QC, quality control; IBS, identity by state; PCA, principal components analysis; PC, principal component; SD, standard deviation; AA, African American
Principal Components 1 and 2, generated from Sample QC Step 4 (see Figure S1), are plotted for the whole PNAT2 clinical sample (turquoise open circles), KIS3 (black open circles), and HapMap reference populations: CEU (Utah residents with Northern and Western European ancestry; red open circles), ASW (African ancestry in Southwest USA; purple open circles), YRI (Yoruba in Ibadan, Nigeria; green open circles), and CHB (Han Chinese in Beijing, China; blue open circles). African American ancestry for PNAT2 and KIS3 was determined using the following cut-points: Principal Component 1 ≤-0.025, and Principal Component 2 ≤0.01, indicated with black dotted lines. These conservative thresholds were selected to ensure homogeneity of the population and were chosen based on visual inspection of the plot.
The NMR distribution is shown in PNAT2 (A), KIS3 (B), and KIS2 (C) African American smokers. The square-root NMR distribution is shown in PNAT2 (D), KIS3 (E), and KIS2 (F) African American smokers. NMR was square-root transformed to help correct for positive skew.
Abbreviation: SD, standard deviation
QQ plots of the full results of the square-root NMR GWAS are shown in PNAT2 (A) and KIS3 (B), and after excluding genome-wide significant hits in PNAT2 (C) and KIS3 (D). Expected P-values are those expected under the null hypothesis. The shaded area around the red line indicates the 95% confidence interval under the null.
Table S1. 2,688 variants included as a custom iSelect® add-on to the Illumina HumanOmniExpressExome-8 v1.2 array.
Table S2. Complete list of chromosome 19 variants significantly (P<5e-8) associated with the NMR in the meta-analysis of PNAT2 and KIS3 African American smokers.
Table S3. Complete list of chromosome 19 variants significantly (P<5e-8) associated with the NMR in PNAT2 African American smokers.
Table S4. Complete list of chromosome 19 variants significantly (P<5e-8) associated with the NMR in KIS3 African American smokers.


