Abstract
Resistance to anti-tumor therapeutics is an important clinical problem. Tumor-targeted therapies currently used in the clinic are derived from antibodies or small molecules that mitigate growth factor activity. These have improved therapeutic efficacy and safety compared to traditional treatment modalities but resistance arises in the majority of clinical cases. Targeting such resistance could improve tumor abatement and patient survival. A growing number of such tumors are characterized by prominent expression of the human epidermal growth factor receptor 3 (HER3) on the cell surface. This study presents a “Trojan-Horse” approach to combating these tumors by using a receptor-targeted biocarrier that exploits the HER3 cell surface protein as a portal to sneak therapeutics into tumor cells by mimicking an essential ligand. The biocarrier used here combines several functions within a single fusion protein for mediating targeted cell penetration and non-covalent self-assembly with therapeutic cargo, forming HER3-homing nanobiologics. Importantly, we demonstrate here that these nanobiologics are therapeutically effective in several scenarios of resistance to clinically approved targeted inhibitors of the human EGF receptor family. We also show that such inhibitors heighten efficacy of our nanobiologics on naïve tumors by augmenting HER3 expression. This approach takes advantage of a current clinical problem (i.e. resistance to growth factor inhibition) and uses it to make tumors more susceptible to HER3 nanobiologic treatment. Moreover, we demonstrate a novel approach in addressing drug resistance by taking inhibitors against which resistance arises and re-introducing these as adjuvants, sensitizing tumors to the HER3 nanobiologics described here.
Keywords: HER3, Neuregulin, Target, Tumor, Therapeutic, Nanobiologic, Resistance
1. Introduction
Expression of human EGF receptor 3 (HER3, ERBB3) is increased on many tumor types, including breast, prostate, gastric, colon, lung, pancreatic, head and neck, ovarian, cervical, glioblastoma and skin cancers [1–13] in experimental models, and patient specimens [14]. Increased cell surface HER3 also associates with resistance to a number of targeted tumor treatments, including inhibitors of EGF-R (lapatinib), HER2 (lapatinib, trastuzumab), HER2–3 dimerization (pertuzumab), PI3K, and antibody-drug conjugates such as T-DM1 [1,2,7,15–19]. HER3 is the preferred heterodimerization partner of the HER2 receptor tyrosine kinase on HER2+ tumors, and its phosphorylation activates the PI3K-Akt pathway supporting tumor progression and survival [20–22]. HER3 increase sustains this activation in the face of targeted therapies [1,2,7,15–19]. Patients with such refractory tumors currently have limited treatment options and poor prognoses. There are no currently available HER3-targeted therapies used in the clinic, as the HER3 kinase domain is relatively inactive [23,24]. Moreover, as up to 70% of cases resist or acquire resistance to receptor blockade therapies [2,15,25], an alternative approach addressing this problem has the potential for significant clinical impact.
Here we examine whether nanobiologics that exploit the receptor binding activity of the neuregulin family of ligands show specificity to HER3 and possible therapeutic efficacy on tumors resisting clinically approved ErbB receptor inhibitors. Neuregulins represent the products of several genes (and their splice variants) recognizing different ErbB receptor family members, and can be synthesized as membrane-bound proteins that are cleaved from the cell surface [26]. The recombinant multidomain polypeptide, HerPBK10 (designated HPK in the present study), incorporates a minimal receptor-binding domain from neuregulin-1α1, produced as a recombinant fusion to exogenous protein domains for self-assembly with therapeutic cargo and targeted penetration into tumor cells [27]. Although we have previously evaluated this protein for delivery of various payloads to HER2+ tumors [27–29], its exact receptor family member specificity has been unclear and mostly inferred from known activities of the related ligand, neuregulin-1β1. HER2 lacks its own ligand-binding domain while heterodimerizing with HER1, HER3, and HER4 receptor family members [20–22], which can be present at varying levels on different tumor types and cell lines. The findings from ligand interaction studies conducted mostly on the beta-isoform of neuregulin-1 [30] are not necessarily applicable to the alpha-1 isoform, much less the minimal binding domain used in the present study. However, if the polypeptide used here is indeed specific to HER3, this predicts that nanobiologics derived from this polypeptide may show particular affinity for tumors resisting clinically approved inhibitors due to tumor upregulation of HER3.
Therefore, in the present study we have examined the HER3 specificity of HPK-derived particles and their capacity to direct therapeutic efficacy to HER3-expressing tumor cells, especially those associated with resistance to ErbB receptor inhibitors. These studies provide new insight on the receptor dynamics of the targeting protein, as well as its structure and particle assembly. Importantly, we have investigated whether these nanobiological particles have augmented therapeutic efficacy on resistant over parental tumor cells; and finally, determine whether signal-inhibitors currently used in the clinic prime both naïve and inherently-resistant breast tumor cells for nanobiologic attack through induced elevation of HER3.
2. Material and methods
2.1. Materials
HPK protein, and associated particles (H3-D and H3-G) were produced as described in Supplemental Materials. HER3 and HER4 peptides were obtained from Prospec (Ness-Ziona, Israel) and Abnova (Taiwan), respectively. Betacellulin was purchased from Peprotech (NJ, USA). Tz and lapatinib (Lp) were purchased from the Cedars-Sinai Pharmacy. Pertuzumab (Pz; Clone 2C4) was acquired from Genentech. HER1, HER2, and HER4 antibodies were purchased from Abgent (CA, USA; Anti-ErbB1/Her1, rabbit polyclonal, Cat# AP7628a; Anti-ErbB2/HER2, rabbit polyclonal, Cat# AP7629a; Anti-HER4, rabbit polyclonal, Cat# AP7631a. Anti-HER3 [H3.105.5 (Ab105), Cat# MA5-13008] was purchased from Pierce-ThermoFisher (MA, USA). Anti-RGS-His tag (#34650) was purchased from Qiagen (CA, USA). Note: The HER2 antibody used here recognizes aa 21–52 of the HER2 extracellular domain, and does not overlap with the trastuzumab binding domain [residues 557–561 (loop 1), 570–573 (loop 2) and 593–603 (loop 3)] [31].
BT-474 and SKBR3 breast cancer cell lines obtained from and authenticated by ATCC were maintained at 37 °C/5% CO2 under mycoplasma-free conditions in complete media comprised of DMEM (Dulbecco’s modified Eagle’s medium), 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin. Two SKBR3 Tz-resistant lines were generated by chronic exposure (> 216 h) to Tz (28 μM). Tz-resistant BT-474 lines were generated by long-term culture with increasing concentrations of Tz. JIMT1 cells were a kind gift from Dr. Julia Ljubomova (Cedars-Sinai Medical Center). All cells except JIMT1 were maintained at 37 °C in complete DMEM at 5% CO2. JIMT1 cells were maintained in RPMI (Roswell Park Memorial Institute Media), 10% Fetal Bovine Serum, 100 U/mL penicillin, 100 μg/mL streptomycin and 1 mM sodium pyruvate at 5% CO2. Resistant lines were maintained with low levels of Tz in the media (10 μg/mL). Matrigel was purchased from BD Biosciences, NJ, USA. PBS+: 1% MgCl2, 1% CaCl2 in PBS.
2.2. Cell surface ELISA
JIMT1 and SKBR3 cells plated at 1e4/well and BT474 cells plated at 1.5e4/well were maintained for 24 h, then washed with PBS+ followed by fixation without permeabilization (to detect cell surface proteins only) and ELISA processing as described [28]. The indicated primary and secondary antibodies were used at 1:500 and 1:1000 dilutions, respectively. After ELISA development, the plates were processed for crystal violet staining to normalize for cell number as described previously [32]. Where indicated, cells were treated with 0.1 mg/mL of Tz for 24 h before washing and fixation. Cell surface receptor levels were compared to mock (PBS)-treated cells.
2.3. Receptor-binding
Cells growing in 96-well plates were exposed to indicated proteins, peptides, or reagents on ice for 30 min to promote receptor binding but not internalization, followed by processing for cell surface ELISA as described earlier. HPK was detected using an anti-RGS-His tag antibody (1:1000; Qiagen, MD, USA) and anti-mouse secondary antibody (1:2000).
2.4. Cell uptake and intracellular trafficking
MDA-MB-435 cells were plated on coverslips in a 12-well plate (100,000 cells/well) and allowed to grow for 36 h. The cells were then pre-chilled by placing plates on ice, and the media replaced with cold Buffer A (20 mM HEPES, pH 7.4; 2 mM MgCl2; and 3% BSA in DMEM) containing HPK or Tz (10 μg, or 0.1 nmol, each). Plates were agitated on ice for 1 h to promote receptor binding but not internalization, followed by aspiration and washing with Buffer A to remove unbound protein. Wells then received pre-warmed complete cell media and plates incubated at 37 °C/5% CO2 to promote receptor-mediated uptake. At the indicated time points, separate coverslips were removed from the plates, fixed and processed for immunocytofluorescence as described previously [33]. Specifically, coverslips were washed with 1% MgCl2/PBS, then fixed in 4% PFA/PBS (15 min), followed by washing in PBS and incubation for 5 min in 50 mM ammonium chloride/PBS to quench endogenous fluorescence. Cells were then washed with PBS and permeabilized in 0.1% Triton X-100/PBS (5 min), washed again, and then incubated in 1% BSA/PBS (> 1 h) to block non-specific sites. For the HPK-treated cells, coverslips were transferred to blocking buffer containing rabbit primary antibody against HPK (1:150 dilution of #Ab6982, which recognizes the penton base domain; Abcam, MA, USA) overnight at 4 °C. After washing to remove nonspecifically bound antibodies, coverslips were incubated in Alexafluor 488-conjugated secondary antibody (1:500; Life Technologies/Thermo Fisher, CA, USA) against either rabbit or human IgG (to identify HPK or Tz, respectively) for 1 h in the dark. Cells were counterstained with rhodamine phalloidin and DAPI to identify actin and nuclei, respectively, followed by washing and mounting. Images were obtained using a Leica SPE laser scanning confocal microscope.
2.5. Co-precipitation with nickel beads
HPK (~200 μg) was bound to pre-equilibrated nickel (Ni-NTA; Qiagen) beads in a 100 μL 50% slurry of incubation buffer (50 mM NaH2PO4, pH 8.0; 0.1 M NaCl; 5 mM imidazole; 10% glycerol) for 1 h on ice with agitation, followed by washing 3× to remove unbound protein. Pre-formed DNA-Dox was incubated with beads (with or without pre-bound protein) for 45 min at RT, followed by pelleting and washing 3×. Beads were then incubated in 100 μL elution buffer (50 mM NaH2PO4, pH 8.0; 0.1 M NaCl; 400 mM imidazole; 10% glycerol), and supernatants isolated from bead pellets.
2.6. In vitro cytotoxicity
Cells were plated at 10,000 cells per well (JIMT1 and SKBR3) or 15,000 cells per well (BT-474) in 96-well plates. At 48 h after plating, the media was replaced with 40 μL of complete media containing the indicated concentrations of H3-D, Tz, Pz, Tz/Pz combination, Lp, or vehicle. Plates were rocked for 4 h at 37 °C and then 60 μL of complete media was added to each well to bring the total volume to 100 μL, followed by continued incubation without rocking for 44 h at 37 °C, 5% CO2. At the conclusion of the incubation, relative cell viability was determined by measuring metabolic activity (MTS assay, Promega) according to manufacturer’s instructions, followed by crystal violet stain to assess relative cell number as described previously [32]. Where indicated, H3-D was incubated with HER3 peptide (Prospec) at 1:1 M (peptide:protein) ratio on ice for 1 h before the particle was added to cells. Particles, Tz or Tz+ Pz were administered at concentration ranges previously established for eliciting therapeutic efficacy [1,2,29,34,35].
2.7. In vivo procedures
Immunodeficient (NU/NU) mice were obtained from Charles River Laboratories, Inc. All procedures involving mice were performed following IACUC-approved protocols #6037 and #5790 in accordance with the institutional and national guide for the care and use of laboratory animals. Specifically, female nu/nu mice (6–8 weeks) receiving bilateral xenograft implants of indicated tumor cells (1e7 cells/implant) in matrigel (1:1 vol:vol) were randomized at tumor establishment (≥100 mm3). Tumor volumes (height× width ×depth) were monitored ~3×/week under single-blinded conditions (treatment groups unknown to the individual acquiring measurements). Mice receiving BT474 cells were implanted with an estradiol pellet (Innovative Research of America) one week before tumor implant. BT474-TR resistance was maintained by weekly IP administration of 2 mg/kg Tz. When tumors reached 200–300 mm3, saline or H3-G (0.004 mg/kg final corrole dose, as determined previously) [28] was i.v. administered daily for one week, then continued weekly for the remainder of the experiment. When JIMT-1 tumors reached 100–200 mm3, mice received daily i.v. injections of H3-D or Dox (both at 0.02 mg/kg doxorubicin, or HPK (at equivalent protein dosage to H3-D) for 1 week, then 2×/week thereafter. Tz treatment for both mouse models entailed 2 mg/kg twice weekly via tail vein injection for the duration of experiments. At the experiment’s termination, all animals were sacrificed and tissues collected.
2.8. Immunohistofluorescence
Tissues harvested from mice were preserved in 4% paraformaldehyde in PBS (phosphate buffered saline) and then transferred to 70% ethanol. Tissues were then paraffin embedded, sectioned, and mounted onto slides. The slides were deparafinized by incubating in a dry oven for 1 h and then washing slides in xylene 5 times for 4 min each, followed by sequential rinses in 100%, 95%, 90%, 80%, and 70% ethanol, 2× each for 3 min. The slides were then submerged in water. Epitope retrieval was performed by incubating the slides for 30 min at 37 °C in 20 μg/mL Proteinase K in 10 mM Tris pH 7.8. A TUNEL assay was then performed on the slides according to the manufacturer’s instructions (Roche). Following treatment, slides were counterstained with DAPI (Thermofisher) and mounted with Prolong Antifade (Thermofisher). Images of the tissues were captured using a Leica SPE laser scanning confocal microscope. Images were analyzed using ImageJ.
2.9. Human subjects
De-identified specimens were obtained by informed consent under IRB-approved protocols #3870 and #29973.
2.10. Processing and plating of patient-derived tissue
Live human breast tumor tissue was dissociated as described in StemCell Technical Bulletin for Mammary and Prostate Tissue Dissociation [36]. Briefly, after removal from the patient, tissue was placed in DMEM/F12 media supplemented with 5% BSA. Tissue was finely minced using a sterile razor blade. Minced tissue was placed into 50 mL of the above media supplemented with 5 mL of 10× Collagenase/Hyaluronidase (StemCell) and 2% BSA in a 250 mL Erlenmeyer flask. The minced tissue and media were incubated for 16 h in a table-top shaker at 37 °C and 210 RPM. Media/cell mixture was spun down in a 50 mL conical tube at 80 ×g for 30 s in a clinical centrifuge. Supernatant was poured into a new conical tube and the pellet (Pellet #1) was saved and processed as described below. The supernatant was spun down for 3 min at 1200 ×g. The supernatant was discarded and the pellet (Pellet #2) was saved. Pellets 1 and 2 were processed by resuspending the cells in 2 mL of pre-warmed Trypsin-EDTA (Corning). The cell mixture was gently pipetted up and down for 3 min. Next, 10 mL of cold HBSS supplemented with 2% FBS was added to the cell/media mixture and spun down for 5 min at 350×g. The supernatant was removed, and 2 mL of pre-warmed 5 mg/mL Dispase (StemCell) and 0.2 mL of 1 mg/mL DNase I were added to the pellet. The sample was pipetted with a P1000 tip for 1 min. The cell suspension was diluted with an additional 10 mL of cold HBSS supplemented with 2% FBS and put through a 40 μm cell strainer into a 50 mL conical tube. The cells were spun down at 350×g for 5 min. The supernatant was discarded, and the cell pellet was resuspended in DMEM/F12 with 10% FBS and plated in a T-75 dish. Once the primary cells were confluent, they were split into 96-well dishes at 7000 cells per well and treated as indicated.
2.11. Statistical methods
Except where indicated, in vitro data are presented as the mean of triplicate samples ± s.e.m. from three independent experiments (hence, n= 9). For normally distributed in vitro data, significances were determined by one-way ANOVA followed by Tukey post hoc analyses unless otherwise indicated. In vivo data are presented as normalized mean ± s.e.m. Significances within in vivo experiments were determined by a Kruskall-Wallis test followed by a Mann-Whitney post hoc analysis.
3. Results
3.1. HER3 specificity and effect of human patient sera
The functional domains of our polypeptide (designated here as HPK) are independently derived from naturally occurring proteins used to promote assembly with therapeutic cargo, tumor targeting, and penetration across the plasma membrane (Fig. 1A). The targeting ligand, drawn from the minimal receptor-binding domain of neuregulin-1α1 [37], exhibits reduced activation of HER3 (present on the tumor cells used in this study; Fig. S1) compared to neuregulin-1β1 (Fig. S2A). Additionally, this ligand and full-length HPK have no growth-inducing effect on HER3-expressing tumor cells (Fig. S2B–C). In the context of HPK, this ligand is produced as a recombinant fusion to a membrane-penetrating moiety extracted from the adenovirus penton base capsid protein (Fig. 1A) [33]. A cationic domain mediates assembly with anionic payloads, including nucleic acids and sulfonated corroles [27,28,38] (Fig. 1A).
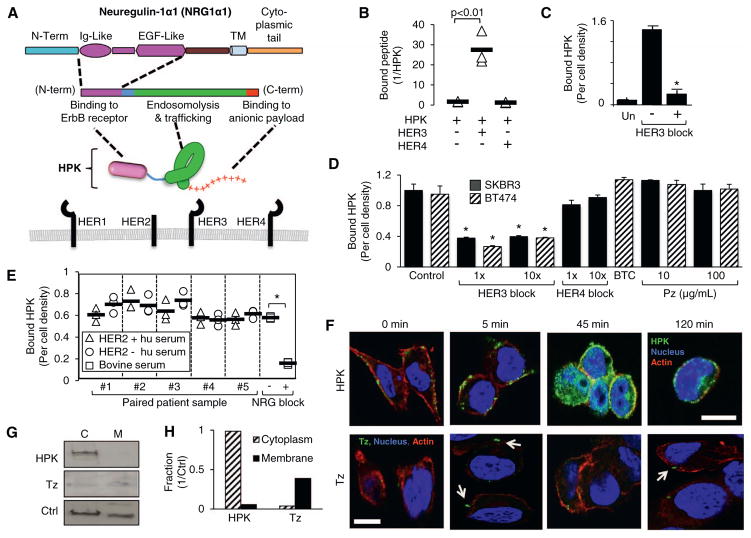
Fig. 1.
Characterization of HPK receptor binding and cell uptake. A, Schematic of HPK, showing Ig- and EGF-like region of heregulin-alpha1 that comprises the ‘Her’ targeting ligand. HER-family receptors represented below. Objects are not drawn to scale. B–C, Evaluation of HPK binding to HER3 or HER4 peptides. In B, immobilized HPK was incubated with respective peptides and assessed for N-terminal blockage by either HER3 or HER4. In C, HPK was incubated with immobilized HER3 −/+ pre-incubation with soluble HER3 peptide as a competitive inhibitor (HER3 block). Un, no HPK. n =9. *, p < 0.05 compared with no HER3 block. D, HPK binding to HER2+/HER3+cells −/+ pre-incubation with: a 1× and 10× molar ratio of soluble HER3 peptide, soluble HER4 peptide, betacellulin (10 μg/mL), or pertuzumab (Pz) as competitive inhibitors. Control, HPK bound to cells with no competitive inhibitor. n =9. *, p < 0.05 compared with corresponding control. E, ELISA of HPK binding to HER2+/HER3+ MDA-MB-435 cells in serum from HER2+ and HER2− patients. Cells were treated with HPK (1.2 μg/well) in media containing 10% human serum (isolated from whole blood) from five HER2+ breast cancer patients or age matched HER2− controls, each obtained pre-chemotherapy treatment. Control samples show binding in 10% bovine serum −/+ 100× molar excess competitive minimal ligand (NRG block). The mean is indicated for each set of measurements (n =3). *, p < 0.05. F, Fluorescence confocal micrographs of MDA-MB-435 cells at different time points of internalization after cell surface binding by HPK (green) or trastuzumab (Tz) (green; arrows highlight Tz signal). Blue, nucleus. Red, actin. Bar, 10 μm. G–H, Western blotting and relative quantification of cytoplasmic (C) and membrane (M) fractions of MDA-MB-435 cells after binding and uptake of HPK or Tz. Relative distributions normalized by marker partitioned in both compartments (Ctrl, control protein recognized by HSP60 antibody). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
HPK binds considerably to a peptide containing the extracellular domain (ECD) of human HER3 (Fig. 1B–C) but not the HER4 ECD (Fig. 1B). This binding was inhibited by pre-adsorption with soluble HER3 peptide in vitro (Fig. 1C). Binding to HER2+ BT-474 and SKBR3 cells (which display HER3 on the cell surface; Fig. S1A–B) was also inhibited by the same HER3 peptide (Fig. 1D). However, cell binding was not inhibited by pre-incubation with a HER4 peptide, or by blocking HER1/EGF-R and HER4 with betacellulin [39] (Fig. 1D), despite the expression of both receptors on these cells (Fig. S1A–B). In further support, cell binding and uptake is significantly reduced on cells deficient in cell surface HER3 (Fig. S2D–E). Whereas HER2-3 heterodimers are prevalent on HER2+ tumor cells [21], pertuzumab (Pz), which inhibits HER2 dimerization with other HER family members including HER3 [40], did not prevent cell binding (Fig. 1D). Finally, whereas the extracellular domains of HER2 and HER4 can be shed in patient sera [41,42], here we show that sera from HER2+ patients and age-matched controls did not prevent HPK binding to HER3-expressing cells in culture (Fig. 1E).
Compared to Tz, which has been evaluated as an antibody-drug conjugate [43], HPK exhibited robust cell surface clustering followed by significant endocytosis within 45 min after cell uptake, whereas Tz remained in sparsely punctate areas on the cells up to 2 h after cell binding (Fig. 1F). Analyses of subcellular compartments show that HPK distributes to the cytoplasm whereas the membrane fraction retains a significant portion of cell-bound Tz (Fig. 1G–H).
3.2. Efficacy on tumors with acquired resistance to trastuzumab
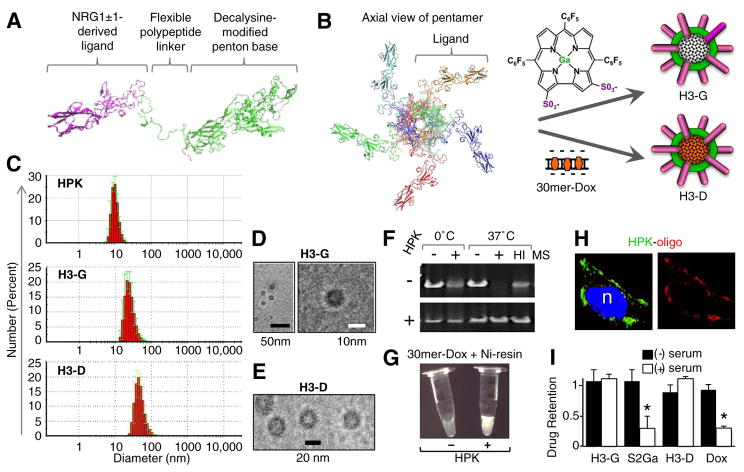
Here we tested the delivery of two therapeutic payloads: the DNA-intercalating agent, doxorubicin (Dox) and a sulfonated corrole. Notably, sulfonated corroles require a cell-penetrating carrier to breach the plasma membrane and are cytotoxic upon entering the cell cytoplasm [28,32,38]. Molecular modeling of HPK predicts that it oligomerizes in solution, and may likely form pentamers driven by the penton base domain (Fig. 2A–B). The hydrodynamic diameter of HPK in solution (~10 nm dia.) is consistent with oligomerization (Fig. 2C). The nanobiologic designated here as H3-G (also known as HerGa) [28] forms from the serum-stable, noncovalent assembly of HPK with sulfonated gallium-metallated corroles (Fig. 2B; described in Supplemental Materials). The alternative nanobiologic, designated H3-D (also known as HerDox) [29], results from non-covalent packaging of DNA-intercalating drugs via a small nucleic acid as a molecular net for drug capture and retention (Fig. 2B). Assembly of HPK with anionic payloads yields a multivalent micellar-like complex in which the protein coats the cargo (Fig. 2B), resulting in ~20–40 nm diameter round particles (Fig. 2C–E). The capacity for HPK to encapsulate and retain payload molecules is supported by its ability to protect small nucleic acids from serum nuclease digestion (Fig. 2F), and to retain such molecules in vitro during precipitation by affinity resin (Fig. 2G), in situ during uptake into cells (Fig. 2H), and in vivo after systemic delivery to mice bearing HER3-expressing tumors (Fig. S2G). Importantly, the latter finding also demonstrates the advantage afforded by HPK, as systemic H3-D exhibits tumor-preferential accumulation and prolonged tumor retention of drug, whereas the free drug exhibits comparatively higher accumulation in non-tumor tissues (Fig. S2G). Finally, each particle (H3-G and H3-D) shows high retention of its respective payload after prolonged exposure (30–60 min) to 100% mouse serum, with no detectable drug loss (Fig. 2I).
Fig. 2.
Assembly and characterization of HPK-particles. A–B, HPK simulated monomer and “predicted” pentamer. A, Computer-generated modeling of the HerPBK10 monomer, showing major protein domains. B, Pentamer formed in computer-simulated “solvent-box” mimicking physiological conditions supporting the natural interactions of protein monomers in solution. Each monomer is designated by a different colour. Computer simulated structures were generated using a high-performance computing cluster suited for molecular dynamics (MD) simulations and molecular visualization of large MD trajectory data sets (Cedars-Sinai High-Performance Computing team directed by Dr. Ravinder Abrol). Schematic includes chemical structure of sulfonated gallium-metallated corrole and DNA-intercalated Dox, and putative assemblies formed when each is combined with HPK. Particle assembly is described in detail in Supplemental Procedures. C, Dynamic light scattering (DLS) measurement of HPK alone and HPK particles. D–E, Electron microscopy of HPK particles after assembly. F, Electrophoresis of oligoduplex before (upper panel) and after (lower panel) binding with HPK. Oligoduplexes (6 pmoles) were incubated for 1 h on ice or at 37 °C with 100% mouse serum (MS) before electrophoresis. Where indicated, oligoduplexes were incubated with HPK (60 pmol) before electrophoresis. HI, heat-inactivated mouse serum. G, UV imaging of microfuge tubes containing nickel bead eluates. DNA-Dox complexes were incubated with nickel beads containing (+) or lacking (−) pre-chelated HPK (detailed procedures in Supplemental Procedures). After incubation, samples were pelleted and washed 3× before imidazole was added to the beads to elute bound molecules. H, Confocal fluorescence microscopy of MDA-MB-435 cells after treatment with HPK bound to a labeled oligonucleotide compared with the same oligonucleotide with Lipofectamine 2000. Red, oligonucleotide; Green, HPK; Blue, nucleus. I, Measurement of payload retention after incubation in mouse serum. Particles (H3-G and H3-D) were incubated for 1 h at 37 °C in 100% active (not heat inactivated) serum drawn from C57BL/6 mice. Each particle was isolated from serum after incubation and the relative quantity of retained payload (gallium corrole or Dox) measured, and compared to that before incubation. Free payloads underwent identical procedures. Detailed procedures are provided in Supplemental procedures. S2Ga, free gallium corrole. *, p < 0.05 compared to sample without serum incubation. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
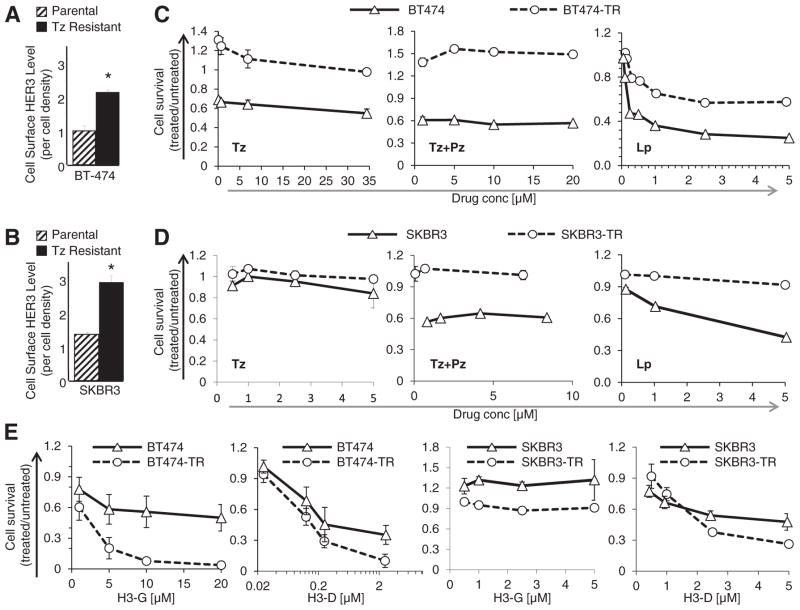
Sublines of HER2+ breast cancer with acquired resistance to Tz (BT474-TR and SKBR3-TR) display increased cell surface HER3 compared to parental counterparts (Fig. 3A–B; Fig. S1A–B) and correspondingly augmented sensitivity to H3-G and H3-D (Fig. 3E). The IC50 of H3-D and H3-G shifted from ~1 to 0.3 μM, and ~10 to 1 μM respectively when comparing activity on parental and Tz-resistant BT474 cells (Fig. 3E; Fig. S1D–E). Likewise, the IC50 of H3-D shifted from ~4 to ~2 μM when comparing activity on parental and Tz-resistant SKBR3 cells, respectively (Fig. 3E). These activities contrast with Tz, which yielded an IC50 of ~35 μM or higher on parental BT474 cells. Whereas SKBR3 generally showed less sensitivity to H3-G compared to H3-D, SKBR3-TR was still more sensitive to H3-G compared to the parental line (Fig. 3E). Both resistant lines showed negligible responses to Tz and combination therapy (Tz + Pz) (Fig. 3C–D). BT474-TR cells partially responded to lapatinib (Lp) but with considerably reduced sensitivity (IC50 > 5 μM) compared to the parental line (IC50 =~0.25 μM) (Fig. 3C), whereas SKBR3-TR cells exhibited Lp-resistance (Fig. 3D). Both H3-G and H3-D yielded similar toxicity as Tz, Tz+ Pz, and Lp on parental BT474 while showing enhanced efficacy on BT474-TR in contrast to the reduced efficacy of all three inhibitors on these cells (Fig. 4A–C). H3-D was more effective than Tz and Tz+ Pz on both parental and resistant SKBR3 (Fig. 4A). Lp efficacy was nearly identical to H3-D on parental SKBR3, but was ineffective on SKBR3-TR in contrast to H3-D (Fig. 4B). Notably, H3-D also improved Dox efficacy on HER3-expressing tumor lines compared to untargeted drug (liposomal doxorubicin), lowering Dox IC50’s up to two orders of magnitude (Fig. S1, D–F and H–I), and elsewhere rendering resistant cells sensitive to Dox (Fig. S1G).
Fig. 3.
In vitro toxicity on tumors with acquired resistance. A–B, Relative surface levels of HER3 on parental and traztuzumab (Tz)-resistant breast cancer cell lines, detected by cell surface ELISA; n = 9; *, p < 0.05 compared with parental. C–D, In vitro tumor cell survival after 48 h exposure to indicated reagents (Tz, trastuzumab; Pz, pertuzumab; Lp, lapatinib). “BT474-TR” and “SKBR3-TR” refer to Tz-resistant lines isolated from corresponding parental BT474 and SKBR3 cell lines. X-axes represent drug concentration. n = 9. E, In vitro tumor cell survival after 48 h exposure to indicated particles (H3-G, HPK particles delivering gallium corrole; H3-D, HPK particles delivering DNA-intercalated Dox).
Fig. 4.
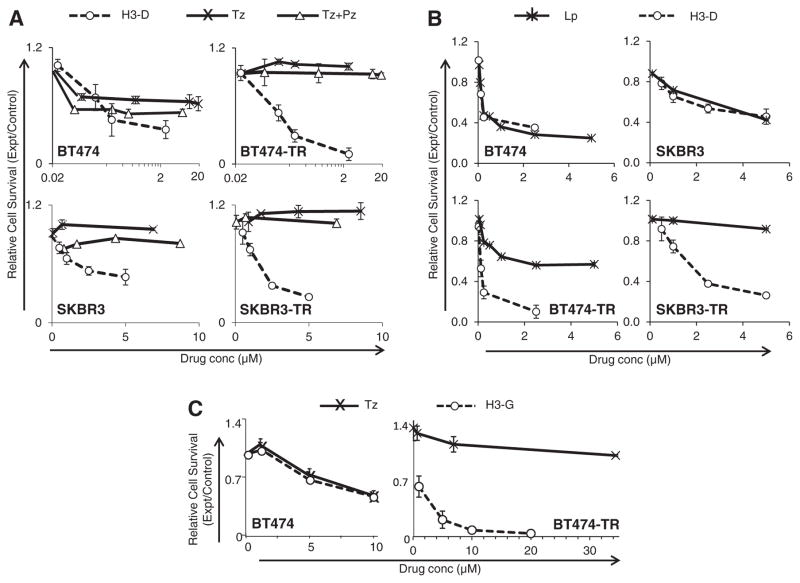
Comparison to ErbB inhibitors. In vitro tumor cell survival after 48 h exposure to (A–B) H3-D or (C) H3-G in comparison with indicated inhibitors (Tz, trastuzumab; Pz, pertuzumab; Lp, lapatinib). “BT474-TR” and “SKBR3-TR” refer to Tz-resistant lines isolated from corresponding parental BT474 and SKBR3 cell lines. X-axes represent drug (Dox, corrole, Tz, Pz, or Lp) concentration. n =9 −/+ s.e.m. Cell survival was measured by in vitro cytotoxicity assay as described in Material and methods. Experimental results have been replicated at least 3×.
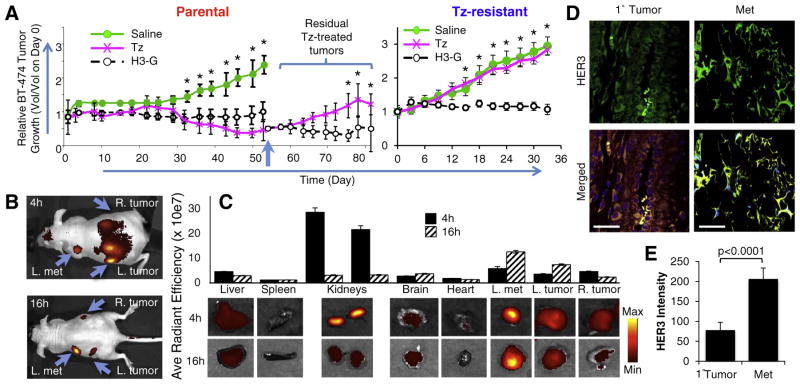
The substantially augmented efficacy of H3-G on resistant compared to parental BT474 cells (Fig. 3E and 4C) provided a basis for evaluating therapeutic efficacy on an in vivo model of acquired resistance. Systemic delivery of H3-G showed similar activity as Tz in mice bearing Tz-sensitive BT474 tumors, whereas Tz-resistant tumors were sensitive to H3-G but non-responsive to Tz (Fig. 5A). Importantly, free gallium corrole (S2Ga) alone had no effect on tumor growth (Fig. S3C). Over time, Tz-sensitive tumors eventually became Tz-resistant (Fig. 5A). Treating a cohort of these mice with systemic H3-G, however, effectively reversed the tumor burden (Fig. 5A). These findings are attributed in part to the tumor-preferential distribution of HPK-particles, accompanied by rapid clearance from non-tumor tissue (Fig. 5B–C). Of note, mice undergoing long-term maintenance of resistance eventually developed secondary tumors (Fig. 5B) exhibiting nearly doubled HER3 compared to primary tumors (Fig. 5D–E), and correspondingly two to three-fold higher particle accumulation (Fig. 5C; Fig. S3A). Importantly, particle accumulation in resistant tumors was at least doubled compared to parental tumors after systemic delivery in mice (Fig. S3B), reflecting the nearly doubled HER3 levels of resistant vs parental tumor cells (Fig. 3A–B) and suggesting that increased uptake by resistant tumors contributes to their augmented particle sensitivity.
Fig. 5.
Targeting acquired resistance in vivo. A, Growth of BT474 parental and resistant tumors during systemic treatment in vivo. Female nude mice bearing the indicated BT-474 tumors received H3-G (0.004 mg/kg/injection), Tz (2 mg/kg), or corresponding volumes of saline i.v. when tumors reached ~100mm3 (daily for 5 days followed by once weekly, except for Tz, which was administered twice weekly only). Day 0 corresponds to the first day of treatment. n =10 tumors per treatment group. *, p≤0.05 comparing H3-G with each control group (saline or Tz where indicated) for both the parental and trastuzumab resistant in vivo tumor models. Short vertical arrow, initiation of H3-G delivery in Tz-treatment cohort. B–C, Biodistribution of labeled particles after systemic delivery in mice bearing BT474-TR tumors. Mice received a single tail vein injection of HPK-particles delivering near-infrared labeled oligonucleotides (described in Supplemental Procedures), followed by fluorescence imaging of (B) whole animals and (C) tissues harvested at indicated timepoints after injection. D, Confocal fluorescence microscopy of primary or metastatic (Met) tumor specimens harvested from mice in B–C. Merged images contain staining of: nuclei (blue), claudin (red), Alexa680 (yellow), and HER3 (green). Bar, ~50 μM. Channel-separated images shown in Fig. S4. E, Quantification of HER3 fluorescence (Ave +/− 1SD) from primary (Tumor) and metastatic (Met) tumor specimens of mice in B–C. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Efficacy on tumors with inherent resistance to trastuzumab
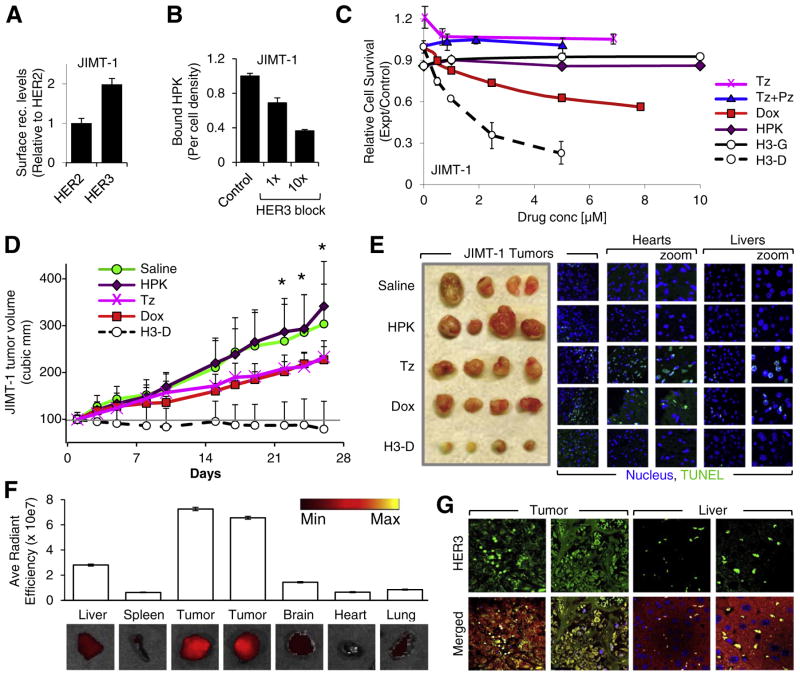
To evaluate therapeutic efficacy on tumors with pre-existing resistance, we assessed the impact of HPK-nanobiologics on a JIMT-1 tumor model. The JIMT-1 cell line is derived from a HER2+ ductal breast cancer that never responded to Tz [44]. These cells sustain insensitivity to HER2-inhibition, thus providing a clinically relevant model for inherent resistance and a challenge for therapeutic testing. The HER3 levels on these cells are ~2× higher than HER2 (Fig. 6A; Fig. S1C). Competitive inhibition by a HER3 peptide confirms that HPK recognizes HER3 on these cells (Fig. 6B). In vitro, these cells remained unresponsive to Tz, Tz+ Pz, and Lp, and only partially responsive to Dox (Fig. 6C) and Lipodox (Fig. S1F). While these cells resisted H3-G, H3-D yielded considerable efficacy (Fig. 6C).
Fig. 6.
Targeting inherent resistance. A, Relative surface levels of HER2 and HER3 on JIMT-1 breast cancer cell lines, detected by cell surface ELISA. n= 9. B, HPK binding to JIMT-1 cells −/+ pre-incubation with a 1× and 10× molar ratio of soluble HER3 peptide. n = 9. C, JIMT-1 cell killing by HerDox in vitro (72 h after treatment) in comparison to traztuzumab (Tz), pertuzumab (Pz), Tz+ Pz, and Dox alone. n = 9. For A–C: Error bars represent s.e.m. D-E, HerDox treatment of mice bearing JIMT-1 tumors. D shows JIMT-1 tumor growth during systemic treatment in vivo. Female nude mice bearing JIMT-1 tumors received either 0.02 mg/kg HerDox (final Dox concentration), equivalent amounts of HPK protein as given in the HerDox treated mice, 0.02 mg/kg Dox, or 2 mg/kg Tz i.v. when tumors reached ~100 mm3. Day 0 corresponds to first day of treatment. n = 10 tumors per treatment group. *, p≤0.05 comparing HerDox to each control group (Saline, HPK, Tz, and Dox). E, Representative tumor, heart, and liver tissue harvested from treated mice in D, showing relative tumor sizes (left panel), and fluorescent TUNEL staining of tumor, heart, and liver tissues (right panels). Blue, nucleus. Green, TUNEL stain. F, Biodistribution of labeled particles after systemic delivery in mice bearing JIMT-1 tumors. Mice received a single tail vein injection of HPK-particles delivering near-infrared labeled oligonucleotides (described in Supplemental Procedures), followed by fluorescence imaging of tissues were harvested at 2.5 h after injection. G, Immunohistofluorescence of tumor and liver specimens from tissues acquired in F. Merged images contain staining of: nuclei (blue), claudin (red), Alexa680 (yellow), and HER3 (green). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Systemic delivery of H3-D to mice bearing JIMT-1 tumors mitigated tumor growth in contrast to Tz and Dox alone (Fig. 6D). Heart tissue extracted from these mice revealed TUNEL-positive cells (indicative of apoptosis) associated with Tz (and somewhat with Dox) treatment, in contrast to the control treatments (saline and HPK alone) (Fig. 6E; Fig. S4B). Tz and Dox also elicited moderate to considerable apoptosis in the liver (Fig. 6E; Fig. S4C). Whereas H3-D treatments yielded substantial apoptosis in residual tumor tissue, no apoptosis was detectable in the heart tissue of these mice (Fig. 6E; Fig. S4A–B). H3-D-treated mice also showed no signs of liver cell apoptosis (Fig. 6E; Fig. S4C) or weight loss (Fig. S4D). These findings are attributed in part to the tumor-preferential biodistribution of the nanobiologic after systemic delivery (Fig. 6F), which in turn reflects the comparatively high HER3 expression in tumors in contrast to the liver (Fig. 6G).
3.4. Effect of clinically approved inhibitors
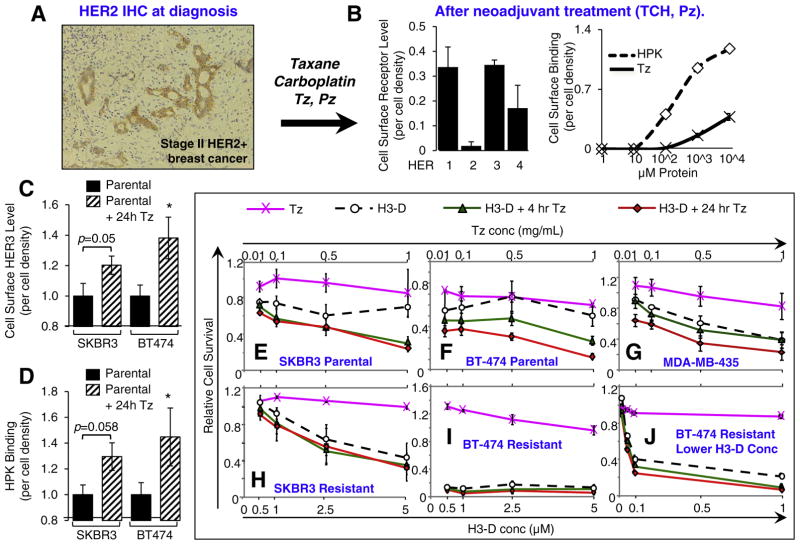
To evaluate whether HPK can recognize HER3 on human primary tissue, we obtained a fresh breast tumor specimen extracted from a HER2+ patient (diagnosed by FISH and IHC; Fig. 7A). Notably, this patient received neoadjuvant Tz and Pz after HER2+ diagnosis and before surgery. HPK and Tz both exhibited concentration-dependent binding to disaggregated cells from this tumor sample, with HPK showing greater binding than Tz (Fig. 7B). This binding curve correlated with the relatively high measured cell surface HER3 and very low HER2 (Fig. 7B). Importantly, Tz and the HER2 antibody used here recognize non-overlapping sites on HER2 (see note in Material and methods).
Fig. 7.
Effect of HER2 neoadjuvant treatment. A, Immunohistology of breast biopsy from a HER2+ patient pre-surgery. Brown, HER2; Blue, nuclei. B, Binding to patient tumor tissue. Disaggregated cells from tissue sample in A were assessed for cell surface receptor levels (left graph) and HPK or traztuzumab (Tz) binding (right graph) by cell surface ELISA. n =3. C–D, Induced elevation of HER3 by Tz and enhanced binding by HPK. Parental cell lines were pre-treated with 0.5 mg/mLTz for 24 h before testing these cells for (C) surface HER3 levels, and (D) HPK binding. n = 9. *, p < 0.05 compared with corresponding parental (non-Tz treated) cells. E–J, Parental or non-resistant cells (E–G) and Tz-resistant cells (H–J) were treated with Tz at indicated concentrations (see upper X-axes) 4 and 24 h before H3-D treatment and assayed for survival at 48–72 h after treatment. n = 9. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Taken together with our in vivo results on parental BT474 tumors (Fig. 5A), these findings raise the possibility that Tz may shift tumor cells to a HER3-elevated phenotype that would facilitate HPK-nanobiologic delivery to resistant cells. To test this, we exposed parental SKBR3 and BT-474 lines to Tz for 24 h and then measured cell surface HER3 levels in comparison with non-treated parental cells. Not only did Tz-treatment yield nearly doubled HER3 levels (Fig. 7C) but HPK binding to these cells also was proportionately increased at least two-fold (Fig. 7D).
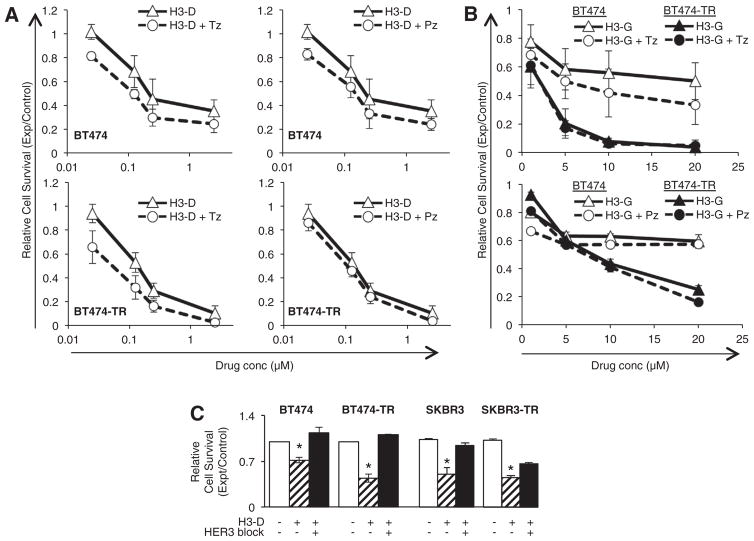
To examine how this phenomenon influences therapeutic efficacy, we treated parental and resistant lines with Tz for 4 h or 24 h before H3-D treatment. H3-D already exhibited improved cytotoxicity compared with Tz on parental SKBR3 cells, while pre-treatment augmented toxicity further (Fig. 7E). H3-D toxicity on parental BT474 cells was modestly improved over Tz, whereas a 4 h and 24 h pre-treatment with Tz yielded correspondingly enhanced H3-D toxicity (Fig. 7F). Even HER3-expressing MDA-MB-435 cells, which are already sensitive to H3-D (Fig. S2F), showed a modest enhancement in toxicity upon 24 h pretreatment with Tz (Fig. 7G). The Tz-resistant lines, on the other hand, already exhibited potent sensitivity to H3-D and thus pre-treatment was for the most part unnecessary (Fig. 7H–J). BT474-TR cells in particular exhibited poor cell viability at all H3-D concentrations tested (Fig. 7I). Five-fold lower H3-D concentrations still potently reduced cell survival, while Tz pre-treatment yielded a slight but significant enhancement in cytotoxicity (Fig. 7J). Importantly, Tz and Pz do not appear to compete with HPK-nanobiologics for cell binding, as co-treatment does not modify the therapeutic efficacy of H3-D or H3-G (Fig. 8A–B), whereas HER3 pre-adsorption significantly blocks H3-D toxicity (Fig. 8C).
Fig. 8.
Tz or Pz do not compete with HPK nanobiologics. Cells received (A) H3-D or (B) H3-G alone or in combination with trastuzumab (Tz) or pertuzumab (Pz) at indicated concentrations, and survival measured by in vitro cytotoxicity assay as described in the Supplemental Procedures. X-axes indicate final concentrations of each drug (Tz, Pz, Dox, or corrole). C, Contribution of HER3 to targeted toxicity. Parental and resistant cell lines were treated with H3-D −/+ HER3 blocking peptide, and tested for cell survival after 48 h. Treatments were compared to mock (saline) –treatment (-HPK, -HER3). n =9. *, p < 0.05 compared with mock.
4. Discussion
These studies show that HPK itself is inert and does not elicit tumor proliferation while triggering rapid and robust endocytosis. Unlike neuregulin, the penton base-derived domain in HPK facilitates endosomal escape and intracellular trafficking [32,33], thus enabling delivery of a variety of therapeutic molecules, especially those that otherwise cannot penetrate the endosomal membrane (i.e., anionic molecules such as nucleic acids and sulfonated corroles) [28,29]. HPK-nanobiologics therefore act as “Trojan horses” by mimicking the natural ligand-receptor interaction on HER3 but resulting in delivery of a tumor-toxic molecule. This effect is augmented on Tz-resistant cells, which increase HER3 expression (1, 15) in response to HER2 blocking, becoming prime targets for HPK-mediated attack. The reduction of HPK binding and particle-mediated cytotoxicity by HER3-blocking peptides affirms that interaction with HER3 contributes to the cytotoxic mechanism of HPK-particles. Several findings in this study suggest that the enhanced sensitivity of resistant cells to HPK-nanobiologics is attributed in part to augmented particle uptake mediated by the heightened HER3 levels on resistant cells. These findings include the observations that: 1. Tz-exposed tumor cells exhibit at least doubled the HER3 levels of naïve tumor cells and bind proportionately (nearly two to three-fold) increased HPK (Fig. 7C–D); 2. accumulation of circulating HPK-particles into Tz-resistant tumors was at least doubled compared to parental tumors in vivo (Fig. S3B), reflecting the nearly doubled HER3 levels on resistant compared to parental tumor cells (Fig. 3A–B); and, 3. metastatic tumors showed at least doubled HER3 receptor levels compared to primary tumors and concomitantly two to three-fold higher particle accumulation (Fig. 5C–E). Importantly, the resistant tumors here show little or no response to Tz, Lp, or Tz +Pz therapy in vitro (though this approach does not assess the impact of antibody-dependent cellular cytotoxicity that contributes to Tz efficacy), but can be effectively mitigated by HPK-therapeutics. This includes the HER2+ JIMT-1 tumor line that, unlike lines with acquired resistance, never responded to HER2-inhibitors [44] and represents one of the most recalcitrant scenarios for therapeutic treatment. Importantly, the resistant tumors here show little or no response to Tz, Lp, or Tz +Pz therapy in vitro (though this approach does not assess the impact of antibody-dependent cellular cytotoxicity that contributes to Tz efficacy), but can be effectively mitigated by HPK-therapeutics. This includes the HER2+ JIMT-1 tumor line that, unlike lines with acquired resistance, never responded to HER2-inhibitors [44] and represents one of the most recalcitrant scenarios for therapeutic treatment.
The advantage of using HPK to deliver therapeutic cargo is demonstrated by comparing the biodistributions of H3-D with free Dox after systemic delivery in mice bearing HER3-expressing tumors. H3-D exhibits tumor-preferential accumulation and prolonged tumor retention whereas Dox exhibits comparatively higher accumulation in non-tumor tissues (Fig. S2G). This is also demonstrated therapeutically, as H3-D shows improved therapeutic efficacy compared to untargeted drug both in vitro (Fig. 6C and Fig. S1D–I) and in vivo (Fig. 6D), as well as improved safety over free Dox in vivo (Fig. 6E–G). Likewise, free gallium corrole has no detectable effect on tumor growth whereas H3-G exhibited effective mitigation of both parental and resistant tumors in vivo (Fig. S3C).
Pre-treating naïve tumors with Tz or Tz+ Pz yielded a HER3-elevated phenotype accompanied by enhanced HPK-binding and augmented sensitivity to HPK particles. Two possible mechanisms, or a combination of the two, may contribute to the increased HER3 observed here. First, in a heterogeneous tumor environment, neoadjuvant treatment of primary tumors can impart selective pressure that enable a subset of resistant cells to expand [45]. Likewise, in these studies, the removal of HER2-sensitive cells by Tz or Tz +Pz pre-treatment may leave a population of pre-existing cells expressing high HER3 to expand. Under such circumstances, HPK particles can enhance the efficacy of current targeted therapeutics by attacking those cells that survive HER2-blocking treatment. On the other hand, it is possible that HER3 is induced as an acute response to HER2 inhibition. HER3 up-regulation can occur within hours of EGFR or HER2 inhibition on HER2+ tumor cell lines [1], leading to acquired resistance. In agreement, we found that Tz or Tz +Pz pre-treatment for as little as 4 h markedly enhanced H3-D efficacy on parental tumor lines. Importantly, co-treatment of the antibodies with the nanobiologic did not necessarily produce an amplified effect nor did they block nanobiologic potency. Altogether, these findings suggest that existing targeted therapies currently used in the clinic can act as adjuvants to prime and sensitize newly diagnosed tumors for HPK-mediated therapy. Thus it is possible to exploit either phenomenon by using HER2 inhibition to set up primary tumors for HPK-mediated attack. Importantly, resistance is not a pre-requisite for our particle to be effective, as HPK-particles exhibit efficacy on both sensitive and resistant tumor cells, but have obvious advantages on resistant tumors compared with existing targeted therapies.
5. Conclusions
The studies here provide support for the association of the HER3 cell surface protein with resistance to clinically approved blockers of the ErbB receptor family, and show that use of HER3-targeted nanobiologics are therapeutically effective on these resistant tumors (which include models of both acquired and inherent resistance). These findings show that nanobiologic targeting and therapeutic efficacy corresponds with HER3 level, and accordingly, potency is heightened on resistant tumor cells. Importantly, nanobiologic potency is enhanced on naïve or parental tumors when applied within hours after trastuzumab treatment due in part to the increased HER3 occurring after exposure to the HER2 blockade. The identification of HER3 upregulation in diverse tumor cell types in response to a growing list of clinically-approved inhibitors suggests that HPK-nanobiologics may be applicable to a broad array of resistant tumors.
Supplementary Material
Acknowledgments
Funding
This work was supported in part by grants to LKMK from the NIH/NCI (R01 CA129822, R01 CA140995), the DoD (W81XWH-15-1-0604), the Avon Foundation (02–2015-060), Margie and Robert E. Petersen Foundation, and the Clinical and Translational Science Institute (CTSI V087). The studies shown in Fig. S1, D-I were supported by Eos Biosciences through a contractual research agreement.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jconrel.2017.12.024.
Competing interests
Dr. Medina-Kauwe and Cedars-Sinai Medical Center hold significant financial interest in Eos Biosciences, Inc., of which Dr. Medina-Kauwe is co-founder and scientific advisor. A patent describing the H3-D (HerDox) nanobiotherapeutic (US Patent No. 9,078,927) has been awarded, and another describing the application of this particle for treating resistant tumors (No. 62/342,829) has recently been filed.
Contributions
The authors thank Josie Bergeron, Catherine Bresee, and Xiao Zhang for assistance with biostatistics; and Accixx Biomedical Consulting (www.Accixx.com) for editorial assistance. LKMK thanks C Rey, A and M Medina-Kauwe, and D Revetto for ongoing support.
References
- 1.Garrett JT, Olivares MG, Rinehart C, Granja-Ingram ND, Sanchez V, Chakrabarty A, et al. Transcriptional and posttranslational up-regulation of HER3 (ErbB3) compensates for inhibition of the HER2 tyrosine kinase. Proc Natl Acad Sci. 2011;108(12):5021–5026. doi: 10.1073/pnas.1016140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garrett JT, Sutton CR, Kuba MG, Cook RS, Arteaga CL. Dual blockade of HER2 in HER2-overexpressing tumor cells does not completely eliminate HER3 function. Clin Cancer Res. 2013;19(3):610–619. doi: 10.1158/1078-0432.CCR-12-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koumakpayi IH, Diallo JS, Le Page C, Lessard L, Gleave M, Begin LR, et al. Expression and nuclear localization of ErbB3 in prostate cancer. Clin Cancer Res. 2006;12(9):2730–2737. doi: 10.1158/1078-0432.ccr-05-2242. Epub 2006/05/06 . [DOI] [PubMed] [Google Scholar]
- 4.Jathal MK, Chen L, Mudryj M, Ghosh PM. Targeting ErbB3: the New RTK(id) on the Prostate Cancer Block. Immunol Endoc Metab Agents Med Chem. 2011;11(2):131–149. doi: 10.2174/187152211795495643. Epub 2011/05/24 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang XL, Yang YS, Xu DP, Qu JH, Guo MZ, Gong Y, et al. Comparative study on overexpression of HER2/neu and HER3 in gastric cancer. World J Surg. 2009;33(10):2112–2118. doi: 10.1007/s00268-009-0142-z. Epub 2009/07/29 . [DOI] [PubMed] [Google Scholar]
- 6.Gespach C. Increasing potential of HER3 signaling in colon cancer progression and therapy. Clin Cancer Res. 2012;18(4):917–919. doi: 10.1158/1078-0432.ccr-11-3143. Epub 2011/12/30 . [DOI] [PubMed] [Google Scholar]
- 7.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–1043. doi: 10.1126/science.1141478. Epub 2007/04/28 . [DOI] [PubMed] [Google Scholar]
- 8.Thomas G, Chardes T, Gaborit N, Mollevi C, Leconet W, Robert B, et al. HER3 as biomarker and therapeutic target in pancreatic cancer: new insights in pertuzumab therapy in preclinical models. Oncotarget. 2014;5(16):7138–7148. doi: 10.18632/oncotarget.2231. Epub 2014/09/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humtsoe JO, Pham E, Louie RJ, Chan DA, Kramer RH. ErbB3 upregulation by the HNSCC 3D microenvironment modulates cell survival and growth. Oncogene. 2016;35(12):1554–1564. doi: 10.1038/onc.2015.220. Epub 2015/06/16 . [DOI] [PubMed] [Google Scholar]
- 10.Jia Y, Zhang Y, Qiao C, Liu G, Zhao Q, Zhou T, et al. IGF-1R and ErbB3/HER3 contribute to enhanced proliferation and carcinogenesis in trastuzumab-resistant ovarian cancer model. Biochem Biophys Res Commun. 2013;436(4):740–745. doi: 10.1016/j.bbrc.2013.06.030. Epub 2013/06/25 . [DOI] [PubMed] [Google Scholar]
- 11.Ocana A, Vera-Badillo F, Seruga B, Templeton A, Pandiella A, Amir E. HER3 overexpression and survival in solid tumors: a meta-analysis. J Natl Cancer Inst. 2013;105(4):266–273. doi: 10.1093/jnci/djs501. Epub 2012/12/12 . [DOI] [PubMed] [Google Scholar]
- 12.Clark PA, Iida M, Treisman DM, Kalluri H, Ezhilan S, Zorniak M, et al. Activation of multiple erbb family receptors mediates glioblastoma cancer stem-like cell resistance to EGFR-targeted inhibition. Neoplasia (New York, NY) 2012;14(5):420–428. doi: 10.1596/neo.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tiwary S, Preziosi M, Rothberg PG, Zeitouni N, Corson N, Xu L. ERBB3 is required for metastasis formation of melanoma cells. Oncogene. 2014;3:e110. doi: 10.1038/oncsis.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. Epub 2015/01/24 . [DOI] [PubMed] [Google Scholar]
- 15.Narayan M, Wilken JA, Harris LN, Baron AT, Kimbler KD, Maihle NJ. Trastuzumab-induced HER reprogramming in “resistant” breast carcinoma cells. Cancer Res. 2009;69(6):2191–2194. doi: 10.1158/0008-5472.CAN-08-1056. [DOI] [PubMed] [Google Scholar]
- 16.Hutcheson I, Barrow D, Hasmann M, Nicholson R. Induction of erbB3/EGFR heterodimers mediates resistance to pertuzumab in a tamoxifen-resistant MCF-7 breast cancer cell line. Mol Cancer Ther. 2007;6:A118. [Google Scholar]
- 17.Phillips GDL, Fields CT, Li G, Dowbenko D, Schaefer G, Miller K, et al. Dual targeting of HER2-positive cancer with Trastuzumab Emtansine and Pertuzumab: critical role for Neuregulin blockade in antitumor response to combination therapy. Clin Cancer Res. 2014;20(2):456–468. doi: 10.1158/1078-0432.ccr-13-0358. [DOI] [PubMed] [Google Scholar]
- 18.Sergina NV, Rausch M, Wang D, Blair J, Hann B, Shokat KM, et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445(7126):437–441. doi: 10.1038/nature05474. Epub 2007/01/09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakrabarty A, Sanchez V, Kuba MG, Rinehart C, Arteaga CL. Feedback upregulation of HER3 (ErbB3) expression and activity attenuates antitumor effect of PI3K inhibitors. Proc Natl Acad Sci U S A. 2012;109(8):2718–2723. doi: 10.1073/pnas.1018001108. Epub 2011/03/04 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hynes NE, Stern DF. The biology of erbB-2/neu/HER-2 and its role in cancer. Biochim Biophys Acta. 1994;1198(2–3):165–184. doi: 10.1016/0304-419x(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 21.Alimandi M, Romano A, Curia MC, Muraro R, Fedi P, Aaronson SA, et al. Cooperative signaling of ErbB3 and ErbB2 in neoplastic transformation and human mammary carcinomas. Oncogene. 1995;10(9):1813–1821. [PubMed] [Google Scholar]
- 22.Olayioye MA, Neve RM, Lane HA, Hynes NE. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 2000;19(13):3159–3167. doi: 10.1093/emboj/19.13.3159. Epub 2000/07/06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Citri A, Skaria KB, Yarden Y. The deaf and the dumb: the biology of ErbB-2 and ErbB-3. Exp Cell Res. 2003;284(1):54–65. doi: 10.1016/s0014-4827(02)00101-5. [DOI] [PubMed] [Google Scholar]
- 24.Boudeau J, Miranda-Saavedra D, Barton GJ, Alessi DR. Emerging roles of pseudokinases. Trends Cell Biol. 2006;16(9):443–452. doi: 10.1016/j.tcb.2006.07.003. Epub 2006/08/02 . [DOI] [PubMed] [Google Scholar]
- 25.Nielsen DL, Kumler I, Palshof JA, Andersson M. Efficacy of HER2-targeted therapy in metastatic breast cancer. Monoclonal antibodies and tyrosine kinase inhibitors. Breast. 2013;22(1):1–12. doi: 10.1016/j.breast.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 26.Montero JC, Rodríguez-Barrueco R, Ocaña A, Díaz-Rodríguez E, Esparís-Ogando A, Pandiella A. Neuregulins and cancer. Clin Cancer Res. 2008;14(11):3237–3241. doi: 10.1158/1078-0432.ccr-07-5133. [DOI] [PubMed] [Google Scholar]
- 27.Medina-Kauwe LK, Maguire M, Kasahara N, Kedes L. Non-viral gene delivery to human breast cancer cells by targeted Ad5 penton proteins. Gene Ther. 2001;8:1753–1761. doi: 10.1038/sj.gt.3301583. [DOI] [PubMed] [Google Scholar]
- 28.Agadjanian H, Ma J, Rentsendorj A, Valluripalli V, Hwang JY, Mahammed A, et al. Tumor detection and elimination by a targeted gallium corrole. Proc Natl Acad Sci U S A. 2009;106(15):6105–6110. doi: 10.1073/pnas.0901531106. (Epub 2009 Apr 2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agadjanian H, Chu D, Hwang JY, Wachsmann-Hogiu S, Rentsendorj A, Song L, et al. Chemotherapy targeting by DNA capture in viral protein particles. Nanomedicine. 2012;7(3):335–352. doi: 10.2217/nnm.11.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breuleux M. Role of heregulin in human cancer. Cell Mol Life Sci. 2007;64(18):2358–2377. doi: 10.1007/s00018-007-7120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho H-S, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, et al. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421(6924):756–60. doi: 10.1038/nature01392. doi: http://www.nature.com/nature/journal/v421/n6924/suppinfo/nature01392_S1.html. [DOI] [PubMed] [Google Scholar]
- 32.Hwang JY, Lubow J, Chu D, Ma J, Agadjanian H, Sims J, et al. A mechanistic study of tumor-targeted corrole toxicity. Mol Pharm. 2011;8(6):2233–2243. doi: 10.1021/mp200094w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rentsendorj A, Xie J, MacVeigh M, Agadjanian H, Bass S, Kim DH, et al. Typical and atypical trafficking pathways of Ad5 penton base recombinant protein: implications for gene transfer. Gene Ther. 2006;13(10):821–836. doi: 10.1038/sj.gt.3302729. [DOI] [PubMed] [Google Scholar]
- 34.Blumenthal GM, Scher NS, Cortazar P, Chattopadhyay S, Tang S, Song P, et al. First FDA approval of dual anti-HER2 regimen: Pertuzumab in combination with Trastuzumab and docetaxel for HER2-positive metastatic breast cancer. Clin Cancer Res. 2013;19(18):4911–4916. doi: 10.1158/1078-0432.ccr-13-1212. Epub 2013/06/27 . [DOI] [PubMed] [Google Scholar]
- 35.Schneeweiss A, Chia S, Hickish T, Harvey V, Eniu A, Hegg R, et al. Pertuzumab plus trastuzumab in combination with standard neoadjuvant anthracycline-containing and anthracycline-free chemotherapy regimens in patients with HER2-positive early breast cancer: a randomized phase II cardiac safety study (TRYPHAENA) Ann Oncol. 2013;24(9):2278–2284. doi: 10.1093/annonc/mdt182. Epub 2013/05/25 . [DOI] [PubMed] [Google Scholar]
- 36.StemCell Technologies. A Guide to Solid Mammary and Prostate Tissue Dissociation. 2009 [Google Scholar]
- 37.Han X, Kasahara N, Kan YW. Ligand-directed retroviral targeting of human breast cancer cells. Proc Natl Acad Sci U S A. 1995;92(21):9747–9751. doi: 10.1073/pnas.92.21.9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agadjanian H, Weaver JJ, Mahammed A, Rentsendorj A, Bass S, Kim J, et al. Specific delivery of corroles to cells via noncovalent conjugates with viral proteins. Pharm Res. 2006;23(2):367–377. doi: 10.1007/s11095-005-9225-1. (Epub 2006 Jan 19) [DOI] [PubMed] [Google Scholar]
- 39.Harskamp LR, Gansevoort RT, van Goor H, Meijer E. The epidermal growth factor receptor pathway in chronic kidney diseases. Nat Rev Nephrol. 2016;12(8):496–506. doi: 10.1038/nrneph.2016.91. [DOI] [PubMed] [Google Scholar]
- 40.Harbeck N, Beckmann MW, Rody A, Schneeweiss A, Muller V, Fehm T, Marschner N, Gluz O, Schrader I, Heinrich G, Untch M, Jackisch C. Breast care. Vol. 8. Basel, Switzerland: 2013. HER2 Dimerization Inhibitor Pertuzumab - Mode of Action and Clinical Data in Breast Cancer; pp. 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shukla S, Singh BK, Pathania OP, Jain M. Evaluation of HER2/neu oncoprotein in serum & tissue samples of women with breast cancer. Indian J Med Res. 2016;143(Suppl 1):S52–S58. doi: 10.4103/0971-5916.191769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carney WP, Neumann R, Lipton A, Leitzel K, Ali S, Price CP. Monitoring the circulating levels of the HER2/neu oncoprotein in breast cancer. Clin Breast Cancer. 2004;5(2):105–116. doi: 10.3816/cbc.2004.n.014. Epub 2004/07/13. [DOI] [PubMed] [Google Scholar]
- 43.Hurvitz SA, Dirix L, Kocsis J, Bianchi GV, Lu J, Vinholes J, et al. Phase II randomized study of trastuzumab emtansine versus trastuzumab plus docetaxel in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer. J Clin Oncol. 2013;31(9):1157–1163. doi: 10.1200/jco.2012.44.9694. Epub 2013/02/06 . [DOI] [PubMed] [Google Scholar]
- 44.Tanner M, Kapanen AI, Junttila T, Raheem O, Grenman S, Elo J, et al. Characterization of a novel cell line established from a patient with Herceptin-resistant breast cancer. Mol Cancer Ther. 2004;3(12):1585–1592. [PubMed] [Google Scholar]
- 45.Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med. 2012;4(127):127ps10. doi: 10.1126/scitranslmed.3003854. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.