Abstract
Across the expanse of vertebrate evolution, each species produces multiple forms of hemoglobin in erythroid cells at appropriate times and in the proper amounts. The multiple hemoglobins are encoded in two globin gene clusters in almost all species. One globin gene cluster, linked to the gene NPRL3, is preserved in all vertebrates, including a gene cluster encoding the highly divergent globins from jawless vertebrates. This preservation of synteny may reflect the presence of a powerful enhancer of globin gene expression in the NPRL3 gene. Despite substantial divergence in noncoding DNA sequences among mammals, several epigenetic features of the globin gene regulatory regions are preserved across vertebrates. The preserved features include multiple DNase hypersensitive sites, at least one of which is an enhancer, and binding by key lineage-restricted transcription factors such as GATA1 and TAL1, which in turn recruit coactivators such as P300 that catalyze acetylation of histones. The maps of epigenetic features are strongly correlated with activity in gene regulation, and resources for accessing and visualizing such maps are readily available to the community of researchers and students.
Keywords: transcriptional regulation, globin genes, locus control regions, super-enhancers, evolution
Globin genes: a model system for developmental regulation of high-level, tissue-specific gene expression
Hemoglobins play a central role in the physiology of species with multiple organs by carrying oxygen from a source, such as lungs or gills, to peripheral organs, such as muscles, that use the oxygen for aerobic metabolism. Hemoglobins also help carry the product of aerobic metabolism, carbon dioxide, back to the organ from which the carbon dioxide is expired, and they can modulate the effects of nitrogen oxides. The hemoglobins transport these gases within cells called erythrocytes (or red blood cells); indeed the hemoglobins are highly abundant in these cells – and only these cells.
These critical functions of hemoglobins can be understood as an adaptation of multi-organ species to the opportunities of an oxygen-rich environment. Globins and the genes encoding them are ancient, being found in all three major kingdoms of life [1]. The ancestral heme-globin complex likely had catalytic oxidation-reduction activity in nitrogen oxide metabolism [2–4]. However, this catalytic activity is suppressed in some hemoglobins, thereby allowing the hemoglobins to function in gas transport without catalyzing chemical reactions. This is the case for vertebrate and invertebrate hemoglobins. The familiar α2β2 tetrameric structure predominates among vertebrate hemoglobins, but a variety of hemoglobin tertiary structures have been described in invertebrates [5]. Furthermore, several globins in addition to the classic tetrameric erythroid hemoglobins have been discovered in vertebrates. These include myoglobins, cytoglobins, and neuroglobins [6]. Thus the globin superfamily is large and pervasive across the biosphere, and members of the superfamily are responsible for a wide range of activities [4]. In this review, we will focus on the vertebrate hemoglobins (and their genes) responsible for gas transport in the blood.
A remarkable feature of vertebrate hemoglobins is that multiple forms of this protein are used for oxygen transport at different stages of development. In placental mammals (eutherians), one form of hemoglobin is dominant in erythrocytes circulating in embryos (primitive erythrocytes) while a different form is used in adult erythrocytes, and in some cases a distinct fetal form is also produced. The different hemoglobins may be adaptive for the differences in oxygen tension at the source organs, e.g. needing a higher oxygen affinity hemoglobin at the fetal placenta than at the adult lung. However, this production of different hemoglobins at progressive developmental stages is not limited to eutherians. To our knowledge, every vertebrate organism examined makes different forms of hemoglobin, and when they have been studied in a developmental context, distinct forms are made at different stages of development. While the full physiological significance of the developmental diversity of hemoglobins is not yet understood, it is clear that the multiplicity of hemoglobins produced in a developmentally controlled manner is a strongly conserved feature across vertebrates, including the jawless vertebrates (agnathans), which are the most distantly related extant vertebrate relatives to humans.
The production of different hemoglobins at progressive stages of development has particular importance for human health. Hemoglobinopathies such as sickle cell disease and thalassemias are the most common forms of inherited disease world-wide [7]. The pathophysiology of each of these diseases almost always involves the hemoglobins produced during adult life. Thus an enduring hope for potential therapies has been the strategy of reactivating the production of hemoglobins that were previously made in fetal life. Recent progress in this strategy is based on our understanding of the mechanisms of gene regulation in the families of genes encoding the globins.
This review will cover the general themes emerging about regulation of globin gene families from an evolutionary and mechanistic perspective. The evolutionary studies are revealing common features of hemoglobin gene regulation, which can be understood best by combining the DNA sequence comparisons of evolutionary approaches with comparisons of additional biochemical features, such as chromatin accessibility, histone modifications, and transcription factor (TF) occupancy. These latter features are referred to as epigenomic, meaning that they are proteins (e.g. TFs) or biochemical modifications (e.g. DNA methylation or histone acetylation) that lie on top of (epi-) the genetic material (DNA), but do not alter the DNA sequence as such. This review will illustrate how comparisons of genomes and epigenomes lead to insights about regulation and human disease. We also will point readers to resources for examination of epigenomic data for any gene in human or mouse erythroid or related cell types, so that the approaches discussed here can be applied to other genes and gene families.
Globin genes are located in multi-gene loci containing embryonic/fetal and adult genes
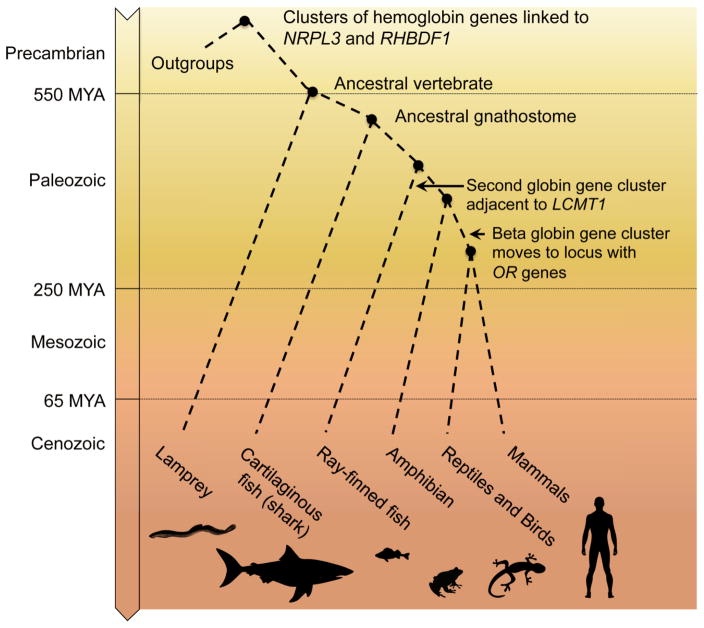
Vertebrates have diversified remarkably since they arose about 550 million years ago (MYA) in the early Paleozoic era (Figure 1). An early divergence separated the jawless vertebrates (agnathans such as lamprey and hagfish) from those with jaws (gnathostomes). The latter large group contains many of the animals familiar to us, including fish (cartilaginous and ray-finned), amphibians, reptiles and birds, and mammals. Each major group had separated from the others by around 250 MYA, in the early Mesozoic era. Diversification of the eutherian mammals is thought to have occurred primarily in the past 65 MY.
Figure 1. Major events in globin gene clusters during vertebrate evolution.
The branching pattern for major vertebrate groups is shown along with a time scale for divergences (millions of years ago, MYA). The labels indicate the inferred times and phylogenetic spans of the presence or movement of specific globin gene clusters.
Despite the long evolutionary time and striking differences between species, some aspects of the arrangements of globin genes have been preserved. Such strong conservation of gene arrangement is indicative of a function, such as regulation, that requires the observed arrangement. When we observe less change than expected, either in DNA sequence or gene arrangement, we infer that changes are disadvantageous and therefore removed from the relevant populations; i.e. we infer that the feature was under purifying or negative selection. Thus mapping the arrangements of globin genes and their neighbors has been of considerable interest for decades. As we will see, the inference that gene arrangement is important has been borne out by the discovery that major regulatory elements are located in genes or intergenic regions distal to the globin genes.
The first preserved feature of vertebrate hemoglobin genes is their presence in multi-gene clusters. The genes encoding hemoglobins have now been mapped across the wide diversity of vertebrates [8–19]. In all cases, multiple hemoglobin genes are found together (Figure 2). For reptiles, birds, and mammals, the genes encoding α-like globins are clustered together on one chromosome, while the genes encoding the β-like globins are clustered on a different chromosome. We refer to the former locus as the α-like globin gene cluster or HBA cluster, and the latter as the β-like globin gene cluster or HBB cluster. In all cases that have been investigated thoroughly, genes at the left side of HBA and HBB clusters (as diagrammed in Figure 2) are expressed in embryonic erythrocytes, while genes on the right side of the clusters display a broader developmental expression pattern and include the globins expressed in adult erythrocytes. While many of the globin genes encode polypeptide components of hemoglobin, no protein product has been discovered from the genes labeled μ and θ in Figure 2, despite the presence of orthologous genes in multiple species [6]. Thus a role for these genes, if any, remains to be determined.
Figure 2. Globin gene clusters across a wide span of vertebrate species.
The arrangement of globin genes and their flanking genes are shown for contemporary species ranging from the jawless vertebrate lamprey to humans. Each gene is shown as a rectangle; those positioned above the lines are transcribed from left to right, those positioned below the lines are transcribed from right to left. Orthologous genes are indicated by rectangles of the same color; boxes for α-like globin genes are yellow and those for β-like globin genes are red. Genes flanking the globin genes are named at their first appearance from the top of the diagram, and names of genes diagnostic for the types of clusters are repeated at the bottom. Almost all species have two of three major clusters. The cluster containing NPRL3 is found in all species, and is indicated by flanking genes in shades of purple and orange. The cluster containing LCMT1 is found in ray-finned fish, and is indicated by flanking genes in shades of green. The cluster containing DCHS1 and RRM1 is found in reptiles, birds, and mammals, and it is indicated by flanking genes in shades of blue. The latter clusters contain what can be a large number of olfactory receptor (OR) genes, and thus only representative OR genes are shown in the figure. An orange dot indicates the major distal enhancer for globin gene clusters in species for which experimental evidence has been obtained for such activity. The figure summarizes maps presented in publications (see references in text) or gleaned from annotations of genome sequences.
In fish and amphibians, the genes encoding α-like globins are clustered together with the β-like globin genes, frequently appearing as pairs of an α-like and a β-like globin gene. In zebrafish, the set of α-βgene pairs expressed in early developmental stages is at one end of the gene cluster, while those expressed at later stages are at the other end [10]. A similar pattern is observed in frogs; the globin genes expressed in tadpoles are separated from those expressed in adult frogs [12, 14, 20]. The genome assembly of the frog Xenopus tropicalis is not sufficiently complete to address the gene arrangements unambiguously. One possibility inferred from the current assembly is shown in the model in Figure 2. The current genome assembly of the genome of the elephant shark, representing cartilaginous fish, shows at least one cluster with genes encoding α-globin, β-globin, and cytoglobin.
The most distant vertebrate with a genome sequence assembly is the lamprey, an agnathan. The globins of lampreys are monomeric and appear to be more closely related to vertebrate cytoglobins than to vertebrate hemoglobins, leading to the inference that the gas-transporting activity of erythrocyte hemoglobins has arisen twice by convergent evolution [11]. Notably, the agnathan hemoglobin genes are arranged as clusters in two different loci (Figure 2, [18]).
The second preserved feature of vertebrate globin gene clusters is the presence of more than one multi-gene cluster. In almost all species, two multi-gene clusters have been identified. For shark and frog, no clear evidence is available for a second multi-gene cluster, but this could reflect the incompleteness of the genome assemblies and correlated work.
The genes flanking the globin multi-gene clusters are conserved across vertebrates
Examination of the genes flanking the globin gene clusters reveals three distinct loci, two of which are used in each species. In almost all mammals and birds, the HBA cluster is located between the NPRL3 and LUC7L genes. The mouse HBA cluster no longer has LUC7L downstream of the globin genes (to the right in Figure 2) because of a chromosomal rearrangement, but NPRL3 has been retained upstream [8, 13]. The HBB cluster in birds and mammals is embedded in a large cluster of OR genes encoding olfactory receptors. Single-copy genes can be far away from the HBB cluster, but in all cases with sufficient contiguity to the genome assembly, the DCHS1 gene is located upstream and the RRM1 and STIM1 genes are located downstream of the HBB cluster in birds and mammals. A similar arrangement appears to be present for the HBA and HBB clusters in turtle, except for an inversion downstream of the HBA cluster that may obscure the presence of LUC7L.
The linkage to NPRL3 is also observed for an amphibian (frog) and every species of ray-finned fish examined (three are shown in Figure 2). These cases can be viewed as a combined cluster of HBA and HBB genes adjacent to NPRL3. A second globin gene cluster is found on a different chromosome in the ray-finned fish. This cluster is smaller, sometimes with three genes, but both α-like and β-like globin genes are found in the smaller cluster [10, 21]. This second cluster is adjacent to the genes ARHGAP17 and LCMT1 on one side, and frequently the gene RHBDF1 is on the other side. A paralogous copy of RHBDF1 (i.e. a related gene generated by duplication) is also frequently close to the first gene cluster containing NPRL3. This second locus is clearly distinct from the OR cluster harboring the HBB complex in mammals. The annotation and assembly of the shark genome is less complete than for the genomes of ray-finned fish, and while NPRL3 has not been mapped unequivocally to the globin gene cluster, the genes RHBDF1 and LUC7L flank the cluster. Thus it is possible that the shark globin gene cluster has an arrangement of flanking genes similar to that in other vertebrates.
Remarkably, one cluster of agnathan globin genes is also flanked by NPRL3, and the other is flanked by RHBDF1 [18]. Therefore even in these distantly related species, two different loci harboring globin genes are present, and those loci show similar flanking genes to those in the jawed vertebrates.
Examination of globin gene clusters across the full span of vertebrates consistently shows that the NPRL3 gene is adjacent to one of the hemoglobin multi-gene clusters. In almost every species examined, NPRL3 is located adjacent to the HBA complex or to a combined HBA-HBB complex. Current apparent exceptions such as the shark may simply reflect incomplete assembly and annotation. Furthermore, NPRL3 is adjacent to a globin gene cluster in agnathans [18], despite the substantial divergence of these monomeric hemoglobins from the tetrameric hemoglobins in gnathostomes [11].
In summary, a striking picture is emerging of conservation of synteny, clustering, and gene order around globin gene loci. Two or more genes present on the same chromosome are syntenic, and when such genes are retained in the same order (and often the same orientation) in different species, we can be confident in concluding that an ancestral arrangement of genes has been preserved over evolution. However, over a large enough span of evolutionary distance, chromosomal rearrangements will break synteny. In fact, for comparisons across major groups of vertebrates (e.g. mammals and birds), the conservation of synteny does not extend much further than the regions shown in Figure 2 [8, 13]. Importantly, the phylogenetic span over which synteny and gene order surrounding globin gene clusters is conserved appears to be greater than for many other loci. We infer from this strong conservation that the surrounding genes are important, and as we will examine later, one important function attributed to this arrangement is regulation of globin gene expression.
The HBB cluster in birds and mammals arose by a transposition
The linkage of a globin gene cluster to NPRL3 is found in all vertebrates, and thus we infer that this is a characteristic derived from the same arrangement in the ancestral vertebrate. The evolutionary history of the other globin gene clusters is more complex. The HBB cluster in birds and mammals is in a different locus from the non-NPRL3-linked cluster in fish, and thus they do not share a common ancestral arrangement. It is therefore likely that the HBB cluster transposed into the DCHS1-OR-RRM1-STIM1 locus in the last common ancestor to reptiles, birds, and mammals (Figures 1 and 2, [6, 17]). The source of those globin genes is unclear. It could be the genes that were linked to LCMT1. In mammals, no globin genes are present around ARHGAP17-LCMT1, and thus one could propose movement from that locus to the OR locus. However, it is also possible that one or more HBB genes from the HBA-HBB combined locus, linked to NPRL3, was the source. In either scenario, genes were lost from one or more of the globin gene loci now seen in contemporary fish, and the transposed HBB genes underwent a series of duplications and divergences to form the contemporary HBB gene cluster with developmentally regulated genes.
The evolutionary rationale for having separate HBA and HBB gene clusters is thought to be that this precludes gene conversion events between HBA and HBB genes, thus promoting fine-tuning of the developmental expression patterns and protein sequences of the α-like and βlike globins. An instructive example is provided by the two γ-globin genes, encoding fetal β-like globins, which have been recently acquired through duplication events in Old World monkeys including humans.
Proximal and distal regulation in the multi-gene clusters
The globin genes are expressed exclusively in erythroid cells, they are expressed at extremely high abundance when activated, and different genes are expressed at different developmental stages. All three aspects of regulation, viz. tissue-specificity, high abundance, and developmental control, have been studied intensively, often using groundbreaking biochemical, genetic, and genomic approaches. For this review, we will discuss some aspects of globin gene regulation that correlate with the evolutionary analyses.
Globin genes have a canonical promoter structure that directs transcription to start at the appropriate location. The sequences conferring this promoter-proximal regulation are found in common for many globin genes, and include a TATAA box at −30bp from the transcription initiation site [22], a CCAAT box at −50bp [23], and a CACCC box at −80bp [24]. The TATAA box is a landing platform for the general transcription factor complex TFIID, and is considered a hallmark of strong tissue-specific promoters. The CCAAT box is a potential binding site for an array of transcription factors, such as the ubiquitously expressed hetero-trimeric NF-Y transcription factor [25] and α-CP1 [26]. The CACCC box element is recognized by members of the specificity protein/Krüppel-like factor (SP/KLF) transcription factor family [27]. This element confers tissue-specificity as it is bound by KLF1, the only erythroid-specific member of the 26-strong mammalian SP/KLF family [28, 29]. The importance of these motifs for high-level globin expression is illustrated by promoter variants leading to thalassemic phenotypes in patients [24, 30, 31], and by systematic analysis of transgene expression in cultured cells and transgenic mice [9].
By themselves the promoters are insufficient to drive high-level transcription of the globin genes in all erythroid cells [32–34]. To achieve this, a series of erythroid-specific enhancer elements is typically required. These enhancer elements are also clustered and located distally from the globin genes. The globin gene loci provided the earliest evidence for long-distance regulation in addition to proximal control [35–37]. A series of DNaseI hypersensitive sites (DHS) upstream of the human embryonic ε-globin gene (HBE, Figure 3) [37] was shown to confer position-independent, high-level expression to a linked globin gene in transgenic mice [35]. This region was termed the Locus Control Region (LCR) and is a defining feature of all mammalian HBB clusters studied to date (orange dots in Figure 2). For the human HBA locus, a major regulatory element was identified located in intron 5 of the NPRL3 gene [36], which is now referred to as Multispecies Conserved Sequence R2 (MCS-R2) [13, 38]. Similar to the LCR, the MCS-R2 is also part of a series of erythroid-specific DHS (Figure 3) [8, 39]. Genetic dissection of the distal regulatory elements has shown that both MCS-R1 and MCS-R2 are major regulatory elements [39, 40]. This arrangement of multiple DHS is typical for mammalian HBA clusters (Figure 3).
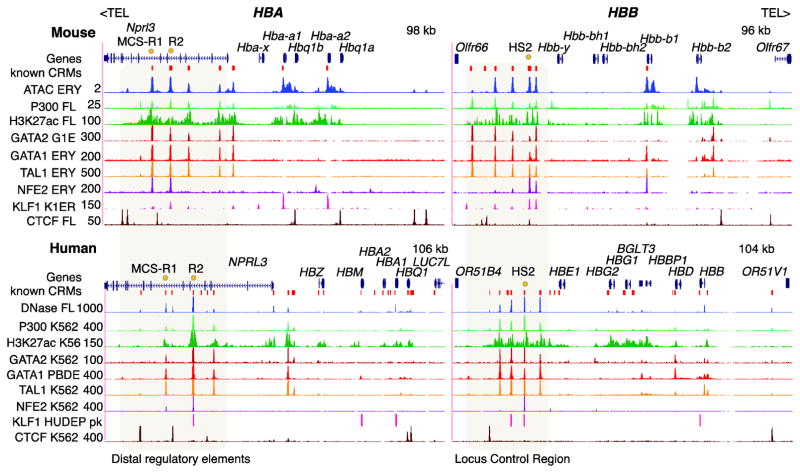
Figure 3. Epigenetic features in HBA and HBB clusters in mouse and human.
For each gene cluster, the first row shows the positions of genes, followed by tracks for known cis-regulatory modules (CRMs, red boxes), accessible chromatin measured by ATAC-seq or DNase-seq, occupancy by the co-activator P300, modification of chromatin at histone H3 acetylated on lysine 27 (H3K27ac), and occupancy by GATA2, GATA1, TAL1, NFE2, KLF1, and CTCF. Numbers to the left of each signal track give the maximum value for the signal shown; only peak positions are shown for KLF1 binding in human HUDEP-2 cells [102]. The values were obtained from different programs and thus are not comparable between tracks, but the signal values along each track are meaningful. The orange dots indicate major distal enhancers. The transcriptional orientation of globin genes is from left to right. Maintaining consistency in orientation of the globin genes required a reversal of orientation from the reference genome sequence for the HBB clusters; this is indicated by showing the direction to the nearest telomere as TEL>. The epigenetic features were determined either in primary erythroid cells and tissues or in cell lines with erythroid character. Abbreviations for cell types are: ERY=erythroblast from mouse bone marrow, FL=fetal liver (mouse or human), G1E=immortalized erythroid-differentiated mouse ES cells with a knockout of the Gata1 gene, K1ER=immortalized erythroid-differentiated mouse ES cells with a knockout of the Klf1 gene that have KLF1 restored as a fusion with an estrogen receptor domain, K562 or K56=transformed human cell line that has some erythroid properties, HUDEP-2=immortalized human erythroid progenitor cell line, and PBDE=peripheral blood derived erythroblasts from humans. The data are from many sources (see references in text), and can be viewed and downloaded from resources of the VISION project (URL is in Table 1).
Interestingly, it was found that the importance of the NPRL3 intron 5 MCS-R2 element for high-level activation of the globin genes differs between species. Deletion of the homologous element in mice resulted in only a modest reduction in α-globin expression [40]. We now know that the clustered DHS work together in an additive fashion [39, 41, 42]; the NPRL3 intron 5 MCS-R2 element appears to have a more dominant role in α-globin gene activation in humans than it does in mice.
The origin of the HBB LCR remains obscure. It could have been derived from the HBA MCS-R2, if that part of NPRL3 were included in the transposition. Regulation of the fish globin clusters flanked by the ARHGAP and RHBDF genes has not been studied in detail yet [10, 21]. One or more DHS have been mapped in this globin gene locus in both Fugu and in zebrafish [10, 21], and it is possible that this could be an important regulatory element (orange dots in Figure 2). Thus the LCR could also have been be derived from this element. Alternatively, the HBB LCR could have arisen de novo, in which case it acquired binding sites for an array of transcription factors very similar to those seen in the globin loci linked to NPRL3 (Figure 3).
Epigenomic features across mammalian HBA and HBB loci
Despite the different evolutionary paths of the HBA and HBB loci, many aspects of their regulation are conserved. Indeed, not only are the genes in both clusters subject to control of tissue-specificity, high abundance, and developmental switches, but the production of proteins from each locus must be balanced to make the globin polypeptides for the α2β2 hemoglobin tetramer. Maps of epigenetic features associated with gene regulation have been produced across the genomes of erythroid-related cell types in mouse and human, both from individual labs and from large consortia. Examination of these maps reveals substantial similarities between the HBB and HBA loci, and strong conservation between mouse and human (Figure 3). These similarities in maps suggest similarities in regulatory mechanisms.
A cluster of regulatory elements, marked by DHS, is distal to the globin genes in both loci (Figure 3). This cluster is referred to as the locus control region, or LCR, for the HBB locus. At least one of the DHS in each distal regulatory region is a strong enhancer as assayed by gain-of-function reporter gene assays or by deletional analysis. In humans, such strong enhancer activity is associated with MCS-R1 and MCS-R2 in the HBA locus and 5′HS2 in the HBB LCR. Activated globin genes are marked by a DHS at the promoter and often broader, proximal nuclease cleavage sensitivity.
The regulatory elements marked by distal and proximal DHS are occupied by the co-activator P300 (Figure 3). This enzyme catalyzes the acetylation of lysine 27 of histone H3, leading to a strong signal for H3K27ac, spreading from the positions occupied by P300. Co-activators are recruited by transcription factors bound to specific sequences. The maps show binding by key transcription factors such as GATA2 and GATA1 (at different stages of erythroid differentiation) and TAL1 to many of the regulatory elements. Indeed, co-binding by GATA factors and TAL1 is strongly predictive of induced expression of target genes [43]. Furthermore, more selective binding of NFE2 is observed for strong enhancers, in keeping with previous observations that the DNA binding motifs for NFE2 were critical for erythroid-specific transcriptional enhancement [8, 36, 44]. Selective binding by the erythroid transcription factor KLF1 is also observed at regulatory elements [45–47]. The protein CTCF is bound at the extremities of the HBA and HBB loci. Some CTCF-bound sites are also bound by components of cohesin, suggesting that they are involved in forming distinct structures within the chromatin, which may in turn play roles in demarcating domains of regulation [48–50]. Overall, the epigenomic maps at the HBA and HBB loci are strikingly similar.
Maps such as those shown in Figure 3 are available genome-wide for a large number features, including chromatin accessibility, multiple histone modifications, and many transcription factors [51–62]. Expression data for protein-coding and noncoding genes, largely from RNA-seq approaches, are also available from a large number of cell types. These data can be powerful resources to generate hypotheses about regulation that can be tested experimentally by individual investigators. Thus it is important to provide easy access to the data. A list of some of these resources, along with URLs, is provided in Table 1. A multi-investigator project, called VISION (for ValIdated Systematic IntegratiON of epigenomic data in hematopoiesis) is an ongoing effort to compile, integrate and model the effects of candidate regulatory elements on expression, to validate those models experimentally, and provide the results freely to the community. Figure 3 was generated by using a subset of the data compiled and displayed by VISION.
Table 1.
Resources for obtaining and visualizing epigenetic data
| Project | Description or goal | URL |
|---|---|---|
| VISION | Generate and compile epigenomic data: integrate, model, and validate. Focus on erythro-myeloid lineages | http://www.bx.psu.edu/~giardine/vision/ |
| CODEX | Curated collection of epigenomic data in hematopoietic cells and stem cells | http://codex.stemcells.cam.ac.uk |
| ENCODE | Generate and integrate epigenomic data across a wide variety of cell types | https://www.encodeproject.org |
| BLUEPRINT | Generate epigenomic in human hematopoietic cells | http://www.blueprint-epigenome.eu/ |
Common epigenomic features across fish and mammalian HBA and HBB loci
Similar regulatory landscapes for globin loci are observed across large phylogenetic distances. In the time since mammals and ray-finned fish diverged, the genome sequences have become quite different. Only a very small subset of the human genome aligns to any fish genome, and the alignments are largely confined to protein-coding exons. The exceptions of noncoding regions conserved between mammals and fish have proven to be dramatic examples of conserved regulatory regions, but these are rare. Not even all protein-coding exons are conserved between human and fish.
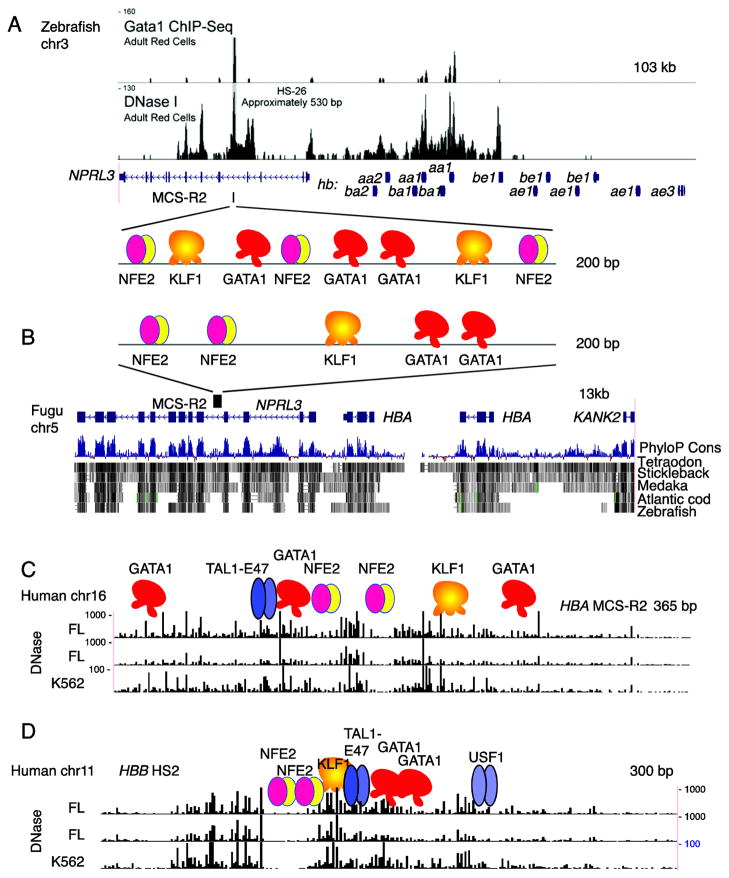
This sparse conservation makes the conserved synteny of HBA loci in mammals with the NPRL3-linked globin genes in fish (Figure 2) even more striking. However, when the intron in Fugu or zebrafish NPRL3 that should be orthologous to the mammalian intron harboring MCS-R2 was searched for alignments with human NPRL3, no meaningful matches were detected above the background of random matches [8]. Reasoning that short matches such as the binding site motifs for transcription factors may not be detected by large-scale alignments, the relevant introns from Fugu and zebrafish NPRL3 were tested experimentally and shown to be active erythroid enhancers [8, 10] (Figure 4). Furthermore, these introns have matches to binding sites for the battery of erythroid transcription factors found in mammalian globin gene regulatory elements, and ChIP-seq experiments confirm binding. This inability to find regulatory elements by interspecies sequence alignments even applies within the clade of ray-finned fish. The Fugu Nprl3 intron 5 harboring MCS-R2 aligns with the orthologous sequence from several fish, but not zebrafish. Despite the lack of alignment, both the Fugu and zebrafish introns have enhancer activity and appear to bind a similar set of transcription factors [8, 10, 21]. These examples illustrate the power of epigenomic analysis for deep interrogation of regulatory mechanisms. Indeed, conservation of epigenomic features may be a particularly effective means of finding active regulatory elements.
Figure 4. Conservation of epigenetic signals versus genome sequences in distal regulatory regions of globin gene clusters.
(A) Binding of GATA1 and DNase accessibility at the major regulatory element (MCS-R2) for the globin gene cluster on chromosome 3 of zebrafish. The signal tracks are from Figure 4 of Ganis et al. [10]; they are aligned with a gene map from the genome assembly. The proteins inferred to be bound at MCS-R2 are shown as colored icons in the zoomed-in view. (B) Genome sequence conservation and divergence at MCS-R2 in other fish. The HBA cluster from Fugu on chromosome 5 is shown, highlighting the MCS-R2 by showing the inferred proteins bound. Underneath the gene map is a track showing the likelihood that DNA segments are under purifying selection. That PhyloP Cons score is estimated from sequence alignments of multiple species, which are shown as dark rectangles indicating aligned sequences. Note that the intron containing the Fugu MCS-R2 aligns with sequences from three other fish, but not to zebrafish. (C) DNase footprints for the HBA MCS-R2 in humans. The high density DNase-seq analysis [63, 64] was done on highly erythroid human fetal liver (FL) tissues and in K562 cells. Regions of frequent cleavage (greater accessibility in chromatin) have a high signal on the tracks. Positions of bound transcription factors, determined from other studies, are shown as colored icons. (D) DNase footprints for the HBB LCR 5′HS2 in humans.
The collection of transcription factors binding to MCS-R2 in ray-finned fish is strikingly similar to those observed at distal regulatory elements in mammals (Figure 4, C and D). High resolution DNase sensitivity maps show footprints, i.e. regions of protection (presumably from transcription factor binding) separated by regions of higher cutting [63, 64]. Those footprints correspond well to the binding site motifs for transcription factors, each of which has been shown by ChIP-seq or other assays to be bound. While the exact number and pattern of binding sites differs among the regulatory elements, the transcription factors bound at the active elements tend to be the same.
Distal regulatory elements are required for high-level expression of the globin genes
The mechanism by which the promoters of the globin genes are activated by the distal regulatory elements was the subject of fierce scientific debates during the 90s of the previous century. A fairly bewildering variety of models were proposed, and while some models were more credible than others, most were not mutually exclusive. Even to date, a detailed time-resolved description of the molecular mechanism is lacking; this will require further development of advanced high-resolution microscopy to follow the dynamic changes in three-dimensional organization of the globin loci in living cells. One model proposed release of RNA polymerase II from the LCR, which would then track along the DNA and start transcription at the first available promoter it encountered [65]. For the chicken HBA locus, a full locus transcript including all the globin genes was reported [66]. Linking of the LCR to the globin promoters via extended protein bridges was another model for transcriptional activation [67]; local repression of embryonic/fetal promoters would prevent activation of these genes in adult erythroid cells. An alternative model, not necessarily excluding a role for most of the other proposed mechanisms, was derived from observations made in bacterial systems in which the formation of DNA loops was observed to accommodate protein-protein interactions between DNA-bound transcriptional regulators [68]. Indirect support for the looping model was obtained from transgenesis experiments in which the position and/or order of genes and regulatory elements was changed [69–72]. The first direct support was obtained using RNA-fishtrap, a method that tags and recovers chromatin in the immediate vicinity of an actively transcribed gene. This revealed that 5′HS2 of the LCR is in close physical proximity to the actively transcribed β-globin gene [73]. Development of Chromosome Conformation Capture (3C) enabled investigation of the three-dimensional structure of loci inside the nucleus independent of transcriptional status [74]. Initially developed in vitro [75], then in yeast [74], adaptation of 3C to analysis of mammalian cells showed that, in adult erythroid cells, the DHS of the LCR come in close spatial proximity with the promoter of the β-globin gene, with the intervening DNA looping out [76]. No such interactions were observed in non-erythroid cells. The LCR-promoter interactions were developmentally regulated, leading to the proposal that the DHS form a holo-complex which facilitates activating interactions preferentially with the nearest accessible globin promoter [77]. Binding of repressors to embryonic/fetal globin promoters in adult erythroid cells would exclude participation in this structure, which was termed the active chromatin hub (ACH). Key erythroid transcription factors KLF1 [78] and GATA1 [79] were found to be required for ACH formation in adult erythroid cells. Remarkably, tethering the self-association domain of the GATA1 cofactor LDB1 to the γ-globin promoter forced LCR looping to the γ-globin promoter and resulted in significant reactivation of the fetal gene in adult erythroid cells [80]. For the HBA locus, similar mechanisms are operational [39]. Of note, a single nucleotide polymorphism (SNP), located between the upstream DHS and the α-globin genes, was shown to create a decoy promoter interfering with normal activation of the downstream α-globin genes. This single SNP is the cause of α-thalassemia in individuals from Melanesia, illustrating that altered chromatin loop formation can be the underlying cause of human disease [81].
How loop formation is achieved remains to be elucidated. Looping requires that regulatory elements sample the nuclear space in order to come in close proximity to each other. We know that the likelihood of in cis enhancer – promoter interactions decreases with distance [69, 70, 82]. Recent investigations of the three-dimensional organization of the genome have revealed that chromatin is compartmentalized by several mechanisms, such as association with the nuclear lamina [83, 84] and division in topologically associated domains [85]. In addition, the high local densities of proteins and nucleic acids at enhancers and promoters may result in the formation of membraneless organelles, called cellular bodies, which are formed by a process termed phase separation. It has been proposed that the formation of such phase-separated multi-molecular assemblies are an essential feature for the function of super-enhancers [86], clusters of DHS such as those found in the HBA and HBB loci.
Super-enhancers for robust regulation
Recently, clusters of hypersensitive sites such as those found in the HBA and HBB loci have been re-branded as super-enhancers [87]. The multiple DHS and extensive histone modifications in the super-enhancers have been interpreted as indicating a large, interacting complex of regulatory elements that together produce a stronger regulatory effect than the individual elements acting separately. Such a model predicts that the regulatory elements in a super-enhancer would act synergistically. However, a recent study examining the effects of deleting each of the five DHS of the HBA locus super-enhancer, singly and in combination, demonstrated that individual DHS act independently of the other four elements. The DHS operated in an additive fashion with respect to hematological phenotype, gene expression, chromatin structure, and chromosome conformation [39]. These results are entirely consistent with earlier studies on the HBB LCR [41, 42, 88–94]. The magnitude of the effects of the deletions of individual elements differed widely. In the HBA complex, deletion of each of two of the candidate regulatory elements with all the hallmarks of enhancers (DNase hypersensitivity, histone modifications indicative of active chromatin, binding by key transcription factors, enhancer RNAs, interactions with promoters, interspecies sequence conservation) had almost no impact on expression [39]. This result shows that these candidate regulatory elements are dispensable for globin gene expression in a laboratory setting, but it does not preclude a role under other conditions.
We suggest that, rather than facilitating synergistic interactions or higher-order effects on the 3D structure of the hemoglobin loci, the super-enhancer architecture in the HBA and HBB loci provides robustness to the system. Such robustness may be the main force driving evolutionary selection on the complex enhancers of the globin loci. Multiple regulatory elements acting independently ensure that expression of the globin genes is fully activated in the vast majority of red cells being produced [95]. Given that an adult human needs to produce over 2 million new erythrocytes every second to replenish worn-out erythrocytes, this is not a trivial consideration.
Developmental regulation of globin gene expression: hemoglobin switching
The recent insights in molecular control of hemoglobin switching elegantly combine the concept of activation of the individual globin genes by the distal regulatory elements via interactions with the globin promoters. The appearance of specific repressor proteins during development renders the promoters of the embryonic/fetal globin genes inaccessible for activation, shifting the DHS-promoter interactions to the adult globin genes. Notably, in the vast majority of cases the embryonic/fetal globin genes are located closer to the DHS along the genomic DNA, with the adult genes located more distally in the locus. Experimentally, the importance of gene order, direction, and distance to the DHS has also been demonstrated using a variety of transgenic approaches in mice [69–72]. The essential role of repressor proteins in orchestrating the switch from embryonic/fetal to adult globin gene expression has now been firmly established with the identification of a regulatory circuit involving MYB [96], KLF1 [97, 98], BCL11A [99] and LRF (also called Pokemon or ZBTB7A) [100]. In adult erythroid cells, MYB activates expression of KLF1, a major activator of terminal erythroid differentiation [101]. KLF1 is a positive regulator of BCL11A [97, 98] and LRF [102] expression, two transcription factors which act as direct repressors of the embryonic/fetal globin genes. Since KLF1 preferentially activates adult globin genes [103, 104], this MYB-KLF1-BCL11A-LRF regulatory circuit results in high-level expression of adult globin genes and very efficient repression of the embryonic/fetal genes in adult erythroid cells. Clinically, this regulatory circuit provides rational targets for directed genome editing in somatic cells or development of novel drugs aimed at reactivation of the fetal β-like globin genes in patients with β-thalassemia and sickle cell disease. A promising recent study showed that removal of a repressor binding site upstream of the γ-globin genes led to substantial increase in fetal hemoglobin and reduced sickling in cells derived from sickle cell patients [105]. While classical transcription factors lack domains with catalytic activity and are therefore as such not very attractive drug targets, they are known to require a host of co-factors in order to exert their functions. These co-factors include histone- and DNA modification enzymes and chromatin remodelers, for which an arsenal of pharmacologic inhibitors is available. Thus, targeting (a combination of) these co-factors is currently a very active area of research. An early example is provided by treatment of a β-thalassemia patient with the DNA methyltransferase inhibitor 5-azacytidine, which resulted in increased fetal hemoglobin expression [106, 107]. This experimental treatment was stopped because of concerns about toxicity, and it remains controversial whether the effects of 5-azacytidine are directly related to inhibition of DNA methylation or due to other metabolic changes in the erythroid cells [108]. Mixed results have been reported on the fetal hemoglobin inducing activities of inhibitors of histone deacetylases [109, 110] and the histone demethylase LSD1 [111, 112]. It is nevertheless encouraging that the increasingly detailed knowledge of the developmental regulation of globin gene expression provides guidance to the development of desperately needed novel pharmacological regimes for the treatment of β-hemoglobinopathy patients. In addition, successful gene therapy of β-hemoglobinopathy patients has been reported for a small number of cases [113, 114]. The gene therapy vectors are based on what could be viewed as an ultra-condensed version of the HBB locus, depending entirely on the inclusion of core regulatory elements of the LCR and the β-globin gene to drive high-level erythroid-specific expression of the therapeutic globin gene [115].
In conclusion, the study of globin loci across the vertebrate kingdom has yielded a wealth of information about developmental regulation of multigene loci and provided a paradigm for understanding spatio-temporal transcriptional control of more complex gene clusters such as the HOX loci [116]. Furthermore, the detailed studies on evolution of the globin gene clusters have helped to reveal the molecular mechanisms underlying gene regulation in higher eukaryotes. This has profoundly contributed to our understanding of human genetic disease in general, and paved the way for development of novel treatments of the hemoglobinopathies, the most common monogenic disorders in the human population.
Acknowledgments
This work was supported by the Landsteiner Foundation for Blood Transfusion Research (LSBR 1627), Netherlands Organization for Scientific Research (NWO/ZonMw TOP 40-00812-98-12128) and EU fp7 Specific Cooperation Research Project THALAMOSS (306201) to SP, and by grants R24DK106766, R01DK054937, and 1R01CA178393 from the National Institutes of Health to RCH. We thank Dr. Doug Vernimmen for his help in the generation of Figure 1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vinogradov SN, Hoogewijs D, Bailly X, Mizuguchi K, Dewilde S, Moens L, Vanfleteren JR. A model of globin evolution. Gene. 2007;398:132–142. doi: 10.1016/j.gene.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 2.Hardison R. Hemoglobins from bacteria to man: evolution of different patterns of gene expression. J Experimental Biology. 1998;201:1099–1117. doi: 10.1242/jeb.201.8.1099. [DOI] [PubMed] [Google Scholar]
- 3.Poole RK, Hughes MN. New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol Microbiol. 2000;36:775–783. doi: 10.1046/j.1365-2958.2000.01889.x. [DOI] [PubMed] [Google Scholar]
- 4.Gell D. Structure, stability, and function of haemoglobins. Blood Cells, Molecules, and Diseases. 2017 doi: 10.1016/j.bcmd.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Gel D. Structure, stability, and function of haemoglobins. Biomolecules. 2017 [Google Scholar]
- 6.Hardison RC. Evolution of hemoglobin and its genes. Cold Spring Harb Perspect Med. 2012;2:a011627. doi: 10.1101/cshperspect.a011627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams TN, Weatherall DJ. World distribution, population genetics, and health burden of the hemoglobinopathies. Cold Spring Harb Perspect Med. 2012;2:a011692. doi: 10.1101/cshperspect.a011692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flint J, Tufarelli C, Peden J, Clark K, Daniels RJ, Hardison R, Miller W, Philipsen S, Tan-Un KC, McMorrow T, Frampton J, Alter BP, Frischauf AM, Higgs DR. Comparative genome analysis delimits a chromosomal domain and identifies key regulatory elements in the alpha globin cluster. Hum Mol Genet. 2001;10:371–382. doi: 10.1093/hmg/10.4.371. [DOI] [PubMed] [Google Scholar]
- 9.Forget BG, Hardison RC. The normal structure and regulation of human globin gene clusters. In: Steinberg MH, Forget BG, Higgs DR, Weatherall DJ, editors. Disorders of Hemoglobins: Genetics, Pathophysiology and Clinical Management. Cambridge University Press; Cambridge, U.K: 2009. pp. 46–61. [Google Scholar]
- 10.Ganis JJ, Hsia N, Trompouki E, de Jong JL, DiBiase A, Lambert JS, Jia Z, Sabo PJ, Weaver M, Sandstrom R, Stamatoyannopoulos JA, Zhou Y, Zon LI. Zebrafish globin switching occurs in two developmental stages and is controlled by the LCR. Dev Biol. 2012;366:185–194. doi: 10.1016/j.ydbio.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann FG, Opazo JC, Storz JF. Gene cooption and convergent evolution of oxygen transport hemoglobins in jawed and jawless vertebrates. Proc Natl Acad Sci U S A. 2010;107:14274–14279. doi: 10.1073/pnas.1006756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hosbach HA, Wyler T, Weber R. The Xenopus laevis globin gene family: Chromosomal arrangement and gene structure. Cell. 1983;32:45–53. doi: 10.1016/0092-8674(83)90495-6. [DOI] [PubMed] [Google Scholar]
- 13.Hughes JR, Cheng JF, Ventress N, Prabhakar S, Clark K, Anguita E, De Gobbi M, de Jong P, Rubin E, Higgs DR. Annotation of cis-regulatory elements by identification, subclassification, and functional assessment of multispecies conserved sequences. Proc Natl Acad Sci U S A. 2005;102:9830–9835. doi: 10.1073/pnas.0503401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeffreys AJ, Wilson V, Wood D, Simons JP, Kay RM, Williams JG. Linkage of adult alpha- and beta-globin genes in X. laevis and gene duplication by tetraploidization. Cell. 1980;21:555–564. doi: 10.1016/0092-8674(80)90493-6. [DOI] [PubMed] [Google Scholar]
- 15.Opazo JC, Lee AP, Hoffmann FG, Toloza-Villalobos J, Burmester T, Venkatesh B, Storz JF. Ancient Duplications and Expression Divergence in the Globin Gene Superfamily of Vertebrates: Insights from the Elephant Shark Genome and Transcriptome. Mol Biol Evol. 2015;32:1684–1694. doi: 10.1093/molbev/msv054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel VS, Cooper SJ, Deakin JE, Fulton B, Graves T, Warren WC, Wilson RK, Graves JA. Platypus globin genes and flanking loci suggest a new insertional model for beta-globin evolution in birds and mammals. BMC Biol. 2008;6:34. doi: 10.1186/1741-7007-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel VS, Ezaz T, Deakin JE, Graves JA. Globin gene structure in a reptile supports the transpositional model for amniote alpha- and beta-globin gene evolution. Chromosome Res. 2010;18:897–907. doi: 10.1007/s10577-010-9164-5. [DOI] [PubMed] [Google Scholar]
- 18.Schwarze K, Campbell KL, Hankeln T, Storz JF, Hoffmann FG, Burmester T. The globin gene repertoire of lampreys: convergent evolution of hemoglobin and myoglobin in jawed and jawless vertebrates. Mol Biol Evol. 2014;31:2708–2721. doi: 10.1093/molbev/msu216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwarze K, Singh A, Burmester T. The Full Globin Repertoire of Turtles Provides Insights into Vertebrate Globin Evolution and Functions. Genome Biol Evol. 2015;7:1896–1913. doi: 10.1093/gbe/evv114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patient RK, Elkington JA, Kay RM, Williams JG. Internal organization of the major adult alpha- and beta-globin genes of X. laevis. Cell. 1980;21:565–573. doi: 10.1016/0092-8674(80)90494-8. [DOI] [PubMed] [Google Scholar]
- 21.Gillemans N, McMorrow T, Tewari R, Wai AW, Burgtorf C, Drabek D, Ventress N, Langeveld A, Higgs D, Tan-Un K, Grosveld F, Philipsen S. Functional and comparative analysis of globin loci in pufferfish and humans. Blood. 2003;101:2842–2849. doi: 10.1182/blood-2002-09-2850. [DOI] [PubMed] [Google Scholar]
- 22.Antonarakis SE, Irkin SH, Cheng TC, Scott AF, Sexton JP, Trusko SP, Charache S, Kazazian HH., Jr beta-Thalassemia in American Blacks: novel mutations in the “TATA” box and an acceptor splice site. Proc Natl Acad Sci U S A. 1984;81:1154–1158. doi: 10.1073/pnas.81.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dierks P, van Ooyen A, Cochran MD, Dobkin C, Reiser J, Weissmann C. Three regions upstream from the cap site are required for efficient and accurate transcription of the rabbit beta-globin gene in mouse 3T6 cells. Cell. 1983;32:695–706. doi: 10.1016/0092-8674(83)90055-7. [DOI] [PubMed] [Google Scholar]
- 24.Orkin SH, Antonarakis SE, Kazazian HH., Jr Base substitution at position −88 in a beta-thalassemic globin gene. Further evidence for the role of distal promoter element ACACCC. J Biol Chem. 1984;259:8679–8681. [PubMed] [Google Scholar]
- 25.Ronchi AE, Bottardi S, Mazzucchelli C, Ottolenghi S, Santoro C. Differential binding of the NFE3 and CP1/NFY transcription factors to the human gamma- and epsilon-globin CCAAT boxes. J Biol Chem. 1995;270:21934–21941. doi: 10.1074/jbc.270.37.21934. [DOI] [PubMed] [Google Scholar]
- 26.Kim C, Sheffery M. Physical characterization of the purified CCAAT transcription factor a-CP1. J Biol Chem. 1990;265:13362–13369. [PubMed] [Google Scholar]
- 27.Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics. 2005;85:551–556. doi: 10.1016/j.ygeno.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Feng WC, Southwood CM, Bieker JJ. Analyses of beta-thalassemia mutant DNA interactions with erythroid Kruppel-like factor (EKLF), an erythroid cell-specific transcription factor. J Biol Chem. 1994;269:1493–1500. [PubMed] [Google Scholar]
- 29.Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol Cell Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orkin S. Disorders of hemoglobin synthesis: the thalassemias. In: Stamatoyannopoulos G, Nienhuis A, Leder P, Majerus P, editors. The Molecular Basis of Blood Diseases. W. B. Saunders Co; Philadelphia: 1987. pp. 106–126. [Google Scholar]
- 31.Giardine B, Borg J, Higgs DR, Peterson KR, Philipsen S, Maglott D, Singleton BK, Anstee DJ, Basak AN, Clark B, Costa FC, Faustino P, Fedosyuk H, Felice AE, Francina A, Galanello R, Gallivan MV, Georgitsi M, Gibbons RJ, Giordano PC, Harteveld CL, Hoyer JD, Jarvis M, Joly P, Kanavakis E, Kollia P, Menzel S, Miller W, Moradkhani K, Old J, Papachatzopoulou A, Papadakis MN, Papadopoulos P, Pavlovic S, Perseu L, Radmilovic M, Riemer C, Satta S, Schrijver I, Stojiljkovic M, Thein SL, Traeger-Synodinos J, Tully R, Wada T, Waye JS, Wiemann C, Zukic B, Chui DH, Wajcman H, Hardison RC, Patrinos GP. Systematic documentation and analysis of human genetic variation in hemoglobinopathies using the microattribution approach. Nat Genet. 2011;43:295–301. doi: 10.1038/ng.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chada K, Magram J, Raphael K, Radice G, Lacy E, Costantini F. Specific expression of a foreign beta-globin gene in erythroid cells of transgenic mice. Nature. 1985;314:377–380. doi: 10.1038/314377a0. [DOI] [PubMed] [Google Scholar]
- 33.Kollias G, Wrighton N, Hurst J, Grosveld F. Regulated expression of human A gamma-, beta-, and hybrid gamma beta-globin genes in transgenic mice: manipulation of the developmental expression patterns. Cell. 1986;46:89–94. doi: 10.1016/0092-8674(86)90862-7. [DOI] [PubMed] [Google Scholar]
- 34.Townes TM, Lingrel JB, Chen HY, Brinster RL, Palmiter RD. Erythroid-specific expression of human beta-globin genes in transgenic mice. EMBO J. 1985;4:1715–1723. doi: 10.1002/j.1460-2075.1985.tb03841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grosveld F, van Assendelft GB, Greaves DR, Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 36.Higgs DR, Wood WG, Jarman AP, Sharpe J, Lida J, Pretorius IM, Ayyub H. A major positive regulatory region located far upstream of the human alpha-globin gene locus. Genes Dev. 1990;4:1588–1601. doi: 10.1101/gad.4.9.1588. [DOI] [PubMed] [Google Scholar]
- 37.Tuan D, London IM. Mapping of DNase I-hypersensitive sites in the upstream DNA of human embryonic epsilon-globin gene in K562 leukemia cells. Proc Natl Acad Sci U S A. 1984;81:2718–2722. doi: 10.1073/pnas.81.9.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Higgs DR, Wood WG. Long-range regulation of alpha globin gene expression during erythropoiesis. Curr Opin Hematol. 2008;15:176–183. doi: 10.1097/MOH.0b013e3282f734c4. [DOI] [PubMed] [Google Scholar]
- 39.Hay D, Hughes JR, Babbs C, Davies JO, Graham BJ, Hanssen LL, Kassouf MT, Oudelaar AM, Sharpe JA, Suciu MC, Telenius J, Williams R, Rode C, Li PS, Pennacchio LA, Sloane-Stanley JA, Ayyub H, Butler S, Sauka-Spengler T, Gibbons RJ, Smith AJ, Wood WG, Higgs DR. Genetic dissection of the alpha-globin super-enhancer in vivo. Nat Genet. 2016;48:895–903. doi: 10.1038/ng.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anguita E, Sharpe JA, Sloane-Stanley JA, Tufarelli C, Higgs DR, Wood WG. Deletion of the mouse alpha-globin regulatory element (HS -26) has an unexpectedly mild phenotype. Blood. 2002;100:3450–3456. doi: 10.1182/blood-2002-05-1409. [DOI] [PubMed] [Google Scholar]
- 41.Fiering S, Epner E, Robinson K, Zhuang Y, Telling A, Hu M, Martin DI, Enver T, Ley TJ, Groudine M. Targeted deletion of 5′HS2 of the murine beta-globin LCR reveals that it is not essential for proper regulation of the beta-globin locus. Genes Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- 42.Ley TJ, Hug B, Fiering S, Epner E, Bender MA, Groudine M. Reduced beta-globin gene expression in adult mice containing deletions of locus control region 5′ HS-2 or 5′ HS-3. Ann N Y Acad Sci. 1998;850:45–53. doi: 10.1111/j.1749-6632.1998.tb10461.x. [DOI] [PubMed] [Google Scholar]
- 43.Soler E, Andrieu-Soler C, de Boer E, Bryne JC, Thongjuea S, Stadhouders R, Palstra RJ, Stevens M, Kockx C, van Ijcken W, Hou J, Steinhoff C, Rijkers E, Lenhard B, Grosveld F. The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev. 2010;24:277–289. doi: 10.1101/gad.551810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuan DY, Solomon WB, London IM, Lee DP. An erythroid-specific, developmental-stage-independent enhancer far upstream of the human “beta-like globin” genes. Proc Natl Acad Sci U S A. 1989;86:2554–2558. doi: 10.1073/pnas.86.8.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pilon AM, Ajay SS, Kumar SA, Steiner LA, Cherukuri PF, Wincovitch S, Anderson SM, Center NCS, Mullikin JC, Gallagher PG, Hardison RC, Margulies EH, Bodine DM. Genome-wide ChIP-Seq reveals a dramatic shift in the binding of the transcription factor erythroid Kruppel-like factor during erythrocyte differentiation. Blood. 2011;118:e139–148. doi: 10.1182/blood-2011-05-355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su MY, Steiner LA, Bogardus H, Mishra T, Schulz VP, Hardison RC, Gallagher PG. Identification of biologically relevant enhancers in human erythroid cells. J Biol Chem. 2013;288:8433–8444. doi: 10.1074/jbc.M112.413260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tallack MR, Whitington T, Yuen WS, Wainwright EN, Keys JR, Gardiner BB, Nourbakhsh E, Cloonan N, Grimmond SM, Bailey TL, Perkins AC. A global role for KLF1 in erythropoiesis revealed by ChIP-seq in primary erythroid cells. Genome Res. 2010;20:1052–1063. doi: 10.1101/gr.106575.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Junier I, Dale RK, Hou C, Kepes F, Dean A. CTCF-mediated transcriptional regulation through cell type-specific chromosome organization in the beta-globin locus. Nucleic Acids Res. 2012;40:7718–7727. doi: 10.1093/nar/gks536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saitoh N, Bell AC, Recillas-Targa F, West AG, Simpson M, Pikaart M, Felsenfeld G. Structural and functional conservation at the boundaries of the chicken beta-globin domain. EMBO J. 2000;19:2315–2322. doi: 10.1093/emboj/19.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valdes-Quezada C, Arriaga-Canon C, Fonseca-Guzman Y, Guerrero G, Recillas-Targa F. CTCF demarcates chicken embryonic alpha-globin gene autonomous silencing and contributes to adult stage-specific gene expression. Epigenetics. 2013;8:827–838. doi: 10.4161/epi.25472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson NK, Schoenfelder S, Hannah R, Castillo MS, Schutte J, Ladopoulos V, Mitchelmore J, Goode DK, Calero-Nieto FJ, Moignard V, Wilkenson AC, Jimenez-Madrid I, Kinston S, Spivakov M, Fraser P, Göttgens B. Integrated genome-scale analysis of the transcriptional regulatory landscape in a blood stem/progenitor cell model. Blood. 2016 doi: 10.1182/blood-2015-10-677393. in press. [DOI] [PubMed] [Google Scholar]
- 52.Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, Pimanda JE, de Bruijn MF, Gottgens B. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 53.Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, Shen Y, Pervouchine DD, Djebali S, Thurman RE, Kaul R, Rynes E, Kirilusha A, Marinov GK, Williams BA, Trout D, Amrhein H, Fisher-Aylor K, Antoshechkin I, DeSalvo G, See LH, Fastuca M, Drenkow J, Zaleski C, Dobin A, Prieto P, Lagarde J, Bussotti G, Tanzer A, Denas O, Li K, Bender MA, Zhang M, Byron R, Groudine MT, McCleary D, Pham L, Ye Z, Kuan S, Edsall L, Wu YC, Rasmussen MD, Bansal MS, Kellis M, Keller CA, Morrissey CS, Mishra T, Jain D, Dogan N, Harris RS, Cayting P, Kawli T, Boyle AP, Euskirchen G, Kundaje A, Lin S, Lin Y, Jansen C, Malladi VS, Cline MS, Erickson DT, Kirkup VM, Learned K, Sloan CA, Rosenbloom KR, Lacerda de Sousa B, Beal K, Pignatelli M, Flicek P, Lian J, Kahveci T, Lee D, Kent WJ, Ramalho Santos M, Herrero J, Notredame C, Johnson A, Vong S, Lee K, Bates D, Neri F, Diegel M, Canfield T, Sabo PJ, Wilken MS, Reh TA, Giste E, Shafer A, Kutyavin T, Haugen E, Dunn D, Reynolds AP, Neph S, Humbert R, Hansen RS, De Bruijn M, Selleri L, Rudensky A, Josefowicz S, Samstein R, Eichler EE, Orkin SH, Levasseur D, Papayannopoulou T, Chang KH, Skoultchi A, Gosh S, Disteche C, Treuting P, Wang Y, Weiss MJ, Blobel GA, Cao X, Zhong S, Wang T, Good PJ, Lowdon RF, Adams LB, Zhou XQ, Pazin MJ, Feingold EA, Wold B, Taylor J, Mortazavi A, Weissman SM, Stamatoyannopoulos JA, Snyder MP, Guigo R, Gingeras TR, Gilbert DM, Hardison RC, Beer MA, Ren B, Mouse EC. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, Bernstein BE, Fraenkel E, Cantor AB. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell. 2009;36:682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu W, Cheng Y, Keller CA, Ernst J, Kumar SA, Mishra T, Morrissey C, Dorman CM, Chen KB, Drautz D, Giardine B, Shibata Y, Song L, Pimkin M, Crawford GE, Furey TS, Kellis M, Miller W, Taylor J, Schuster SC, Zhang Y, Chiaromonte F, Blobel GA, Weiss MJ, Hardison RC. Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Res. 2011;21:1659–1671. doi: 10.1101/gr.125088.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, Li P, Durand EM, Mosimann C, Heffner GC, Daley GQ, Paulson RF, Young RA, Zon LI. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.The_ENCODE_Project_Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pimkin M, Kossenkov AV, Mishra T, Morrissey CS, Wu W, Keller CA, Blobel GA, Lee D, Beer MA, Hardison RC, Weiss MJ. Divergent functions of hematopoietic transcription factors in lineage priming and differentiation during erythro-megakaryopoiesis. Genome Res. 2014;24:1932–1944. doi: 10.1101/gr.164178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kowalczyk MS, Hughes JR, Garrick D, Lynch MD, Sharpe JA, Sloane-Stanley JA, McGowan SJ, De Gobbi M, Hosseini M, Vernimmen D, Brown JM, Gray NE, Collavin L, Gibbons RJ, Flint J, Taylor S, Buckle VJ, Milne TA, Wood WG, Higgs DR. Intragenic enhancers act as alternative promoters. Mol Cell. 2012;45:447–458. doi: 10.1016/j.molcel.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 60.Kassouf MT, Hughes JR, Taylor S, McGowan SJ, Soneji S, Green AL, Vyas P, Porcher C. Genome-wide identification of TAL1’s functional targets: insights into its mechanisms of action in primary erythroid cells. Genome Res. 2010;20:1064–1083. doi: 10.1101/gr.104935.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng Y, Wu W, Kumar SA, Yu D, Deng W, Tripic T, King DC, Chen KB, Zhang Y, Drautz D, Giardine B, Schuster SC, Miller W, Chiaromonte F, Zhang Y, Blobel GA, Weiss MJ, Hardison RC. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 2009;19:2172–2184. doi: 10.1101/gr.098921.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng Y, Ma Z, Kim BH, Wu W, Cayting P, Boyle AP, Sundaram V, Xing X, Dogan N, Li J, Euskirchen G, Lin S, Lin Y, Visel A, Kawli T, Yang X, Patacsil D, Keller CA, Giardine B, Mouse EC, Kundaje A, Wang T, Pennacchio LA, Weng Z, Hardison RC, Snyder MP. Principles of regulatory information conservation between mouse and human. Nature. 2014;515:371–375. doi: 10.1038/nature13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, Maurano MT, Humbert R, Rynes E, Wang H, Vong S, Lee K, Bates D, Diegel M, Roach V, Dunn D, Neri J, Schafer A, Hansen RS, Kutyavin T, Giste E, Weaver M, Canfield T, Sabo P, Zhang M, Balasundaram G, Byron R, MacCoss MJ, Akey JM, Bender MA, Groudine M, Kaul R, Stamatoyannopoulos JA. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vierstra J, Reik A, Chang KH, Stehling-Sun S, Zhou Y, Hinkley SJ, Paschon DE, Zhang L, Psatha N, Bendana YR, O’Neil CM, Song AH, Mich AK, Liu PQ, Lee G, Bauer DE, Holmes MC, Orkin SH, Papayannopoulou T, Stamatoyannopoulos G, Rebar EJ, Gregory PD, Urnov FD, Stamatoyannopoulos JA. Functional footprinting of regulatory DNA. Nat Methods. 2015;12:927–930. doi: 10.1038/nmeth.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ling J, Ainol L, Zhang L, Yu X, Pi W, Tuan D. HS2 enhancer function is blocked by a transcriptional terminator inserted between the enhancer and the promoter. J Biol Chem. 2004;279:51704–51713. doi: 10.1074/jbc.M404039200. [DOI] [PubMed] [Google Scholar]
- 66.Broders F, Zahraoui A, Scherrer K. The chicken alpha-globin gene domain is transcribed into a 17-kilobase polycistronic RNA. Proc Natl Acad Sci U S A. 1990;87:503–507. doi: 10.1073/pnas.87.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- 68.Griffith J, Hochschild A, Ptashne M. DNA loops induced by cooperative binding of lambda repressor. Nature. 1986;322:750–752. doi: 10.1038/322750a0. [DOI] [PubMed] [Google Scholar]
- 69.Dillon N, Trimborn T, Strouboulis J, Fraser P, Grosveld F. The effect of distance on long-range chromatin interactions. Mol Cell. 1997;1:131–139. doi: 10.1016/s1097-2765(00)80014-3. [DOI] [PubMed] [Google Scholar]
- 70.Hanscombe O, Whyatt D, Fraser P, Yannoutsos N, Greaves D, Dillon N, Grosveld F. Importance of globin gene order for correct developmental expression. Genes Dev. 1991;5:1387–1394. doi: 10.1101/gad.5.8.1387. [DOI] [PubMed] [Google Scholar]
- 71.Peterson KR, Stamatoyannopoulos G. Role of gene order in developmental control of human gamma- and beta-globin gene expression. Mol Cell Biol. 1993;13:4836–4843. doi: 10.1128/mcb.13.8.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tanimoto K, Liu Q, Bungert J, Engel JD. Effects of altered gene order or orientation of the locus control region on human beta-globin gene expression in mice. Nature. 1999;398:344–348. doi: 10.1038/18698. [DOI] [PubMed] [Google Scholar]
- 73.Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 74.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 75.Cullen KE, Kladde MP, Seyfred MA. Interaction between transcription regulatory regions of prolactin chromatin. Science. 1993;261:203–206. doi: 10.1126/science.8327891. [DOI] [PubMed] [Google Scholar]
- 76.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 77.Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The beta-globin nuclear compartment in development and erythroid differentiation. Nat Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 78.Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 80.Deng W, Rupon JW, Krivega I, Breda L, Motta I, Jahn KS, Reik A, Gregory PD, Rivella S, Dean A, Blobel GA. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014;158:849–860. doi: 10.1016/j.cell.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.De Gobbi M, Viprakasit V, Hughes JR, Fisher C, Buckle VJ, Ayyub H, Gibbons RJ, Vernimmen D, Yoshinaga Y, de Jong P, Cheng JF, Rubin EM, Wood WG, Bowden D, Higgs DR. A regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science. 2006;312:1215–1217. doi: 10.1126/science.1126431. [DOI] [PubMed] [Google Scholar]
- 82.Hughes JR, Roberts N, McGowan S, Hay D, Giannoulatou E, Lynch M, De Gobbi M, Taylor S, Gibbons R, Higgs DR. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat Genet. 2014;46:205–212. doi: 10.1038/ng.2871. [DOI] [PubMed] [Google Scholar]
- 83.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, van Steensel B. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 84.Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- 85.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A Phase Separation Model for Transcriptional Control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bender MA, Mehaffey MG, Telling A, Hug B, Ley TJ, Groudine M, Fiering S. Independent formation of DnaseI hypersensitive sites in the murine beta-globin locus control region. Blood. 2000;95:3600–3604. [PubMed] [Google Scholar]
- 89.Bender MA, Roach JN, Halow J, Close J, Alami R, Bouhassira EE, Groudine M, Fiering SN. Targeted deletion of 5′HS1 and 5′HS4 of the beta-globin locus control region reveals additive activity of the DNaseI hypersensitive sites. Blood. 2001;98:2022–2027. doi: 10.1182/blood.v98.7.2022. [DOI] [PubMed] [Google Scholar]
- 90.Bender MA, Ragoczy T, Lee J, Byron R, Telling A, Dean A, Groudine M. The hypersensitive sites of the murine beta-globin locus control region act independently to affect nuclear localization and transcriptional elongation. Blood. 2012;119:3820–3827. doi: 10.1182/blood-2011-09-380485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ellis J, Tan-Un KC, Harper A, Michalovich D, Yannoutsos N, Philipsen S, Grosveld F. A dominant chromatin-opening activity in 5′ hypersensitive site 3 of the human beta-globin locus control region. EMBO J. 1996;15:562–568. [PMC free article] [PubMed] [Google Scholar]
- 92.Hug BA, Wesselschmidt RL, Fiering S, Bender MA, Epner E, Groudine M, Ley TJ. Analysis of mice containing a targeted deletion of beta-globin locus control region 5′ hypersensitive site 3. Mol Cell Biol. 1996;16:2906–2912. doi: 10.1128/mcb.16.6.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim CG, Epner EM, Forrester WC, Groudine M. Inactivation of the human beta-globin gene by targeted insertion into the beta-globin locus control region. Genes Dev. 1992;6:928–938. doi: 10.1101/gad.6.6.928. [DOI] [PubMed] [Google Scholar]
- 94.Pruzina S, Hanscombe O, Whyatt D, Grosveld F, Philipsen S. Hypersensitive site 4 of the human beta globin locus control region. Nucleic Acids Res. 1991;19:1413–1419. doi: 10.1093/nar/19.7.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.de Krom M, van de Corput M, von Lindern M, Grosveld F, Strouboulis J. Stochastic patterns in globin gene expression are established prior to transcriptional activation and are clonally inherited. Mol Cell. 2002;9:1319–1326. doi: 10.1016/s1097-2765(02)00558-0. [DOI] [PubMed] [Google Scholar]
- 96.Craig JE, Rochette J, Fisher CA, Weatherall DJ, Marc S, Lathrop GM, Demenais F, Thein S. Dissecting the loci controlling fetal haemoglobin production on chromosomes 11p and 6q by the regressive approach. Nat Genet. 1996;12:58–64. doi: 10.1038/ng0196-58. [DOI] [PubMed] [Google Scholar]
- 97.Borg J, Papadopoulos P, Georgitsi M, Gutierrez L, Grech G, Fanis P, Phylactides M, Verkerk AJ, van der Spek PJ, Scerri CA, Cassar W, Galdies R, van Ijcken W, Ozgur Z, Gillemans N, Hou J, Bugeja M, Grosveld FG, von Lindern M, Felice AE, Patrinos GP, Philipsen S. Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of fetal hemoglobin. Nat Genet. 2010;42:801–805. doi: 10.1038/ng.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhou D, Liu K, Sun CW, Pawlik KM, Townes TM. KLF1 regulates BCL11A expression and gamma- to beta-globin gene switching. Nat Genet. 2010;42:742–744. doi: 10.1038/ng.637. [DOI] [PubMed] [Google Scholar]
- 99.Sankaran VG, Menne TF, Xu J, Akie TE, Lettre G, Van Handel B, Mikkola HK, Hirschhorn JN, Cantor AB, Orkin SH. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 100.Masuda T, Wang X, Maeda M, Canver MC, Sher F, Funnell AP, Fisher C, Suciu M, Martyn GE, Norton LJ, Zhu C, Kurita R, Nakamura Y, Xu J, Higgs DR, Crossley M, Bauer DE, Orkin SH, Kharchenko PV, Maeda T. Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science. 2016;351:285–289. doi: 10.1126/science.aad3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bianchi E, Zini R, Salati S, Tenedini E, Norfo R, Tagliafico E, Manfredini R, Ferrari S. c-myb supports erythropoiesis through the transactivation of KLF1 and LMO2 expression. Blood. 2010;116:e99–110. doi: 10.1182/blood-2009-08-238311. [DOI] [PubMed] [Google Scholar]
- 102.Norton LJ, Funnell APW, Burdach J, Wienert B, Kurita R, Nakamura Y, Philipsen S, Pearson RCM, Quinlan KGR, Crossley M. KLF1 directly activates expression of the novel fetal globin repressor, ZBTB7A, in erythroid cells. Blood Advances. 2017 doi: 10.1182/bloodadvances.2016002303. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 104.Perkins AC, Sharpe AH, Orkin SH. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 105.Traxler EA, Yao Y, Wang YD, Woodard KJ, Kurita R, Nakamura Y, Hughes JR, Hardison RC, Blobel GA, Li C, Weiss MJ. A genome-editing strategy to treat beta-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat Med. 2016;22:987–990. doi: 10.1038/nm.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ley TJ, DeSimone J, Anagnou NP, Keller GH, Humphries RK, Turner PH, Young NS, Keller P, Nienhuis AW. 5-azacytidine selectively increases gamma-globin synthesis in a patient with beta+ thalassemia. N Engl J Med. 1982;307:1469–1475. doi: 10.1056/NEJM198212093072401. [DOI] [PubMed] [Google Scholar]
- 107.Ley TJ, DeSimone J, Noguchi CT, Turner PH, Schechter AN, Heller P, Nienhuis AW. 5-Azacytidine increases gamma-globin synthesis and reduces the proportion of dense cells in patients with sickle cell anemia. Blood. 1983;62:370–380. [PubMed] [Google Scholar]
- 108.Mabaera R, Greene MR, Richardson CA, Conine SJ, Kozul CD, Lowrey CH. Neither DNA hypomethylation nor changes in the kinetics of erythroid differentiation explain 5-azacytidine’s ability to induce human fetal hemoglobin. Blood. 2008;111:411–420. doi: 10.1182/blood-2007-06-093948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Inati A, Kahale M, Perrine SP, Chui DH, Taher AT, Koussa S, Abi Nasr T, Abbas HA, Ghalie RG. A phase 2 study of HQK-1001, an oral fetal haemoglobin inducer, in beta-thalassaemia intermedia. Br J Haematol. 2014;164:456–458. doi: 10.1111/bjh.12635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Patthamalai P, Fuchareon S, Chaneiam N, Ghalie RG, Chui DH, Boosalis MS, Perrine SP. A phase 2 trial of HQK-1001 in HbE-beta thalassemia demonstrates HbF induction and reduced anemia. Blood. 2014;123:1956–1957. doi: 10.1182/blood-2013-11-538470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shi L, Cui S, Engel JD, Tanabe O. Lysine-specific demethylase 1 is a therapeutic target for fetal hemoglobin induction. Nat Med. 2013;19:291–294. doi: 10.1038/nm.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu J, Bauer DE, Kerenyi MA, Vo TD, Hou S, Hsu YJ, Yao H, Trowbridge JJ, Mandel G, Orkin SH. Corepressor-dependent silencing of fetal hemoglobin expression by BCL11A. Proc Natl Acad Sci U S A. 2013;110:6518–6523. doi: 10.1073/pnas.1303976110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F, Down J, Denaro M, Brady T, Westerman K, Cavallesco R, Gillet-Legrand B, Caccavelli L, Sgarra R, Maouche-Chretien L, Bernaudin F, Girot R, Dorazio R, Mulder GJ, Polack A, Bank A, Soulier J, Larghero J, Kabbara N, Dalle B, Gourmel B, Socie G, Chretien S, Cartier N, Aubourg P, Fischer A, Cornetta K, Galacteros F, Beuzard Y, Gluckman E, Bushman F, Hacein-Bey-Abina S, Leboulch P. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ribeil JA, Hacein-Bey-Abina S, Payen E, Magnani A, Semeraro M, Magrin E, Caccavelli L, Neven B, Bourget P, El Nemer W, Bartolucci P, Weber L, Puy H, Meritet JF, Grevent D, Beuzard Y, Chretien S, Lefebvre T, Ross RW, Negre O, Veres G, Sandler L, Soni S, de Montalembert M, Blanche S, Leboulch P, Cavazzana M. Gene Therapy in a Patient with Sickle Cell Disease. N Engl J Med. 2017;376:848–855. doi: 10.1056/NEJMoa1609677. [DOI] [PubMed] [Google Scholar]
- 115.de Dreuzy E, Bhukhai K, Leboulch P, Payen E. Current and future alternative therapies for beta-thalassemia major. Biomed J. 2016;39:24–38. doi: 10.1016/j.bj.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Noordermeer D, Duboule D. Chromatin architectures and Hox gene collinearity. Curr Top Dev Biol. 2013;104:113–148. doi: 10.1016/B978-0-12-416027-9.00004-8. [DOI] [PubMed] [Google Scholar]