Abstract
Staphylococcus aureus is a major cause of nosocomial infections and secretes a diverse spectrum of virulence determinants as well as forms biofilm. The emergence of antibiotic-resistant S. aureus highlights the need for alternative forms of therapeutics other than conventional antibiotics. One route to meet this need is screening small molecule derivatives for potential anti-infective activity. Using a previously optimized C. elegans – S. aureus small molecule screen, we identified a benzimidazole derivative, UM-C162, which rescued nematodes from a S. aureus infection. UM-C162 prevented the formation of biofilm in a dose-dependent manner without interfering with bacterial viability. To examine the effect of UM-C162 on the expression of S. aureus virulence genes, a genome-wide transcriptome analysis was performed on UM-C162-treated pathogen. Our data indicated that the genes associated with biofilm formation, particularly those involved in bacterial attachment, were suppressed in UM-C162-treated bacteria. Additionally, a set of genes encoding vital S. aureus virulence factors were also down-regulated in the presence of UM-C162. Further biochemical analysis validated that UM-C162-mediated disruption of S. aureus hemolysins, proteases and clumping factors production. Collectively, our findings propose that UM-C162 is a promising compound that can be further developed as an anti-virulence agent to control S. aureus infections.
Introduction
Staphylococcus aureus is a clinical pathogen that causes both human and animal infections, ranging from mild superficial infections to toxin-associated diseases and severe life-threatening invasive infections1. S. aureus is acknowledged as a key agent implicated in nosocomial infections resulting in significant morbidity and mortality among hospitalized patients. This is in part due to its ability to adhere to the surface of indwelling medical devices and develop biofilm, a multilayered structure comprising of bacterial communities embedded within the extracellular hydrated polymeric matrix2.
To survive the onslaught of the host immune response and establish an infection, S. aureus expresses a comprehensive array of virulence determinants, including extracellular toxins (e.g. hemolysins, leukotoxins and enterotoxins), enzymes (e.g. proteases and coagulases) and surface proteins (e.g. clumping factors, adhesins and S. aureus surface proteins)3. The secretion of these virulence factors and formation of biofilm are controlled by a sophisticated network of virulence regulators such as the agr, sarA, saeR/S and arlRS4. The diversity of S. aureus virulence factors along with the ability to form disease-associated biofilm presents a challenge to the host immune defence to fully eradicate the bacteria whilst rendering the bacteria highly tolerant to antibiotics, leading to persistent chronic infections2.
The growing frequency of antibiotic-resistant S. aureus strains as exemplified by methicillin-resistant S. aureus (MRSA) is a serious threat to the community and highlights a pressing need for alternative therapies that impose minimum selection pressure upon the bacteria. The cumulative understanding of bacterial virulence and pathogenesis opens up new perspectives for developing novel therapies by attenuating bacterial virulence and/or biofilm as alternative treatment options for bacterial infections5–8. Recent studies have focused on development of small molecules and phytochemicals to inhibit the formation of biofilm and/or other virulence factors in S. aureus9–12. As most virulence traits are non-essential for bacterial survival, these strategies hold great promise as effective means to control bacterial infections with less probability for selection of resistant populations13,14.
Benzimidazole is structurally analogous to naturally occurring nucleotides, making it a key intermediate in the synthesis of new chemical entities of biological interest. The benzimidazole class of compounds are known to possess an extensive range of pharmacological activities such as anti-malarial, anti-cancer and antioxidative effects15. In light of the notable therapeutic properties of the benzimidazole scaffold, we performed an in vivo screen on a collection of 35 benzimidazole derivatives for potential anti-infective effects using a previously established C. elegans – S. aureus anti-infective screen16. In an anti-biofilm assay, several compounds reduced the formation of biofilm without inhibiting the growth of S. aureus and the most potent derivative, 2-(4′-((3-acetamidobenzyl)oxy)-[1,1′-biphenyl]-4-yl)-6-methyl-1H-benzimidazole-4-carboxylic acid (referred to as UM-C162 henceforth) was selected for further analysis. Transcriptome profiling was also performed on UM-C162-treated S. aureus to identify the bacterial virulence factors targeted by UM-C162.
Results
Benzimidazole derivatives exhibit anti-infective and anti-biofilm activities
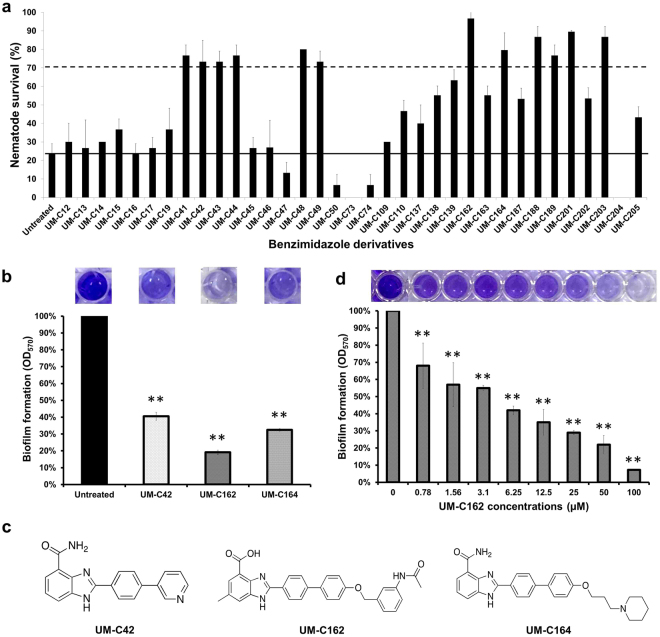
In the current study, we extended the utility of a previously optimized C. elegans – S. aureus anti-infective screen16 to explore the potential anti-infective property of a series of benzimidazole compounds. The premise of the screen was that the addition of individual compounds that are able to rescue the nematodes from a lethal S. aureus infection are deemed to exhibit potential anti-infective effects. We screened 35 benzimidazole derivatives (Table S1) and observed that 12 compounds increased the survival of infected worms to > 70% at a point when ~80% of the untreated worms succumbed to the infection (Fig. 1a). The 12 positive hits are compounds UM-C41, UM-C42, UM-C43, UM-C44, UM-C48, UM-C49, UM-C162, UM-C164, UM-C188, UM-C189, UM-C201 and UM-C203. All positive hits (at 100 µM) enhanced the survival of S. aureus infected-worms by 3 to 4-fold relative to the untreated control after 96 hours post-infection. On the other hand, we also noticed a small number of compounds (namely UM-C50, UM-C73, UM-C74 and UM-C204) that accelerated the killing of S. aureus-infected worms, resulting in a lower survival of nematodes (<10%) as compared to the untreated infected worms (~20%) (Fig. 1a). To address the possible toxic effect of these four compounds on the host, we repeated the assay on worms exposed to heat-killed Escherichia coli OP50 in the presence of these compounds. As expected, the survival of the worms decreased significantly as early as 48 hours post-exposure (Fig. S1), indicating that these compounds are indeed toxic to the host.
Figure 1.
Benzimidazole derivatives rescued C. elegans from a S. aureus infection and altered S. aureus biofilm formation. (a) Survival of S. aureus-infected nematodes upon treatment with individual benzimidazole compounds (100 µM). Results are shown as mean ± SD from a single replicate of two independent screens. The straight line shows the survival of untreated worms while the dashed line demarcates the positive hits. Compounds that promoted the survival of infected worms to > 70% were considered as positive in the screen. (b) Effect of benzimidazole compounds (100 µM) towards S. aureus biofilm formation. (c) Molecular structures of compounds UM-C42, UM-C162 and UM-C164 that demonstrated significant anti-biofilm activity. (d) Dose-dependent anti-biofilm effect of compound UM-C162. The graphs in (b) and (d) depict the percentage of biofilm formation after 24 hours incubation of S. aureus in the presence and absence of compounds. Three independent experiments were conducted. Error bars indicate SEM. Images of representative wells from the crystal-violet biofilm assay are shown. (**) denotes significant difference between untreated and compound-treated bacteria (p < 0.01).
Previously, Begun et al. reported that S. aureus that overexpressed the icaADBC locus and constitutively produced excess biofilm exopolysaccharide, killed worms significantly faster as compared to a strain that produced less biofilm, suggesting that staphylococcal biofilm is a virulence mechanism employed by the bacteria that contributes to death of the infected host17. In another study, a benzimidazole-derived molecule (ABC-1) was identified as a broad-spectrum biofilm inhibitor towards both Gram-positive and Gram-negative bacteria18. ABC-1 suppresses S. aureus protein A (SpA) expression and prevents the accumulation of polysaccharide intercellular adhesin (PIA)19. Therefore, it is possible that some of the hits identified from the C. elegans anti-infective screen may regulate the production of biofilm by S. aureus, thereby extending the survival of infected nematodes. To examine the potential anti-biofilm property of the hits, bacteria were cultured in the presence of the individual compounds and biofilm formation was screened using the 96-well crystal violet biofilm assay20. Following 24 hours incubation under static conditions, three of the 12 hits (UM-C42, UM-C162 and UM-C164) dramatically inhibited S. aureus biofilm development by > 70% (p < 0.01). The most pronounced anti-biofilm activity was observed for UM-C162 (Fig. 1b). UM-C42, UM-C162 and UM-C164 at a concentration of 100 µM were able to inhibit biofilm formation to 41 ± 2.4%, 19 ± 1.4% and 32 ± 0.8% of the untreated bacteria, respectively. The molecular structures of these compounds are presented in Fig. 1c. The ability of UM-C42, UM-C162 and UM-C164 to diminish the biofilm-forming capability of S. aureus suggests that these compounds rescued the nematodes from infection via attenuation of this bacterial virulence factor. As compound UM-C162 was most effective in preventing S. aureus biofilm formation, we further assessed the effect of this compound on S. aureus biofilm production across a series of lower concentrations. The results showed that UM-C162 reduced S. aureus biofilm formation in a concentration-dependent manner after treatment for 24 hours, where supplementation of UM-C162 from 0.78 µM to 100 µM was able to reduce biofilm formation by 8.9% to 68% (Fig. 1d). Based on these data, inhibition of >50% biofilm formation was achieved with 6.25 µM of UM-C162. We thus selected 6.25 µM as the lowest working concentration for the rest of the assays.
UM-C162 disrupts biofilm formation without affecting S. aureus growth
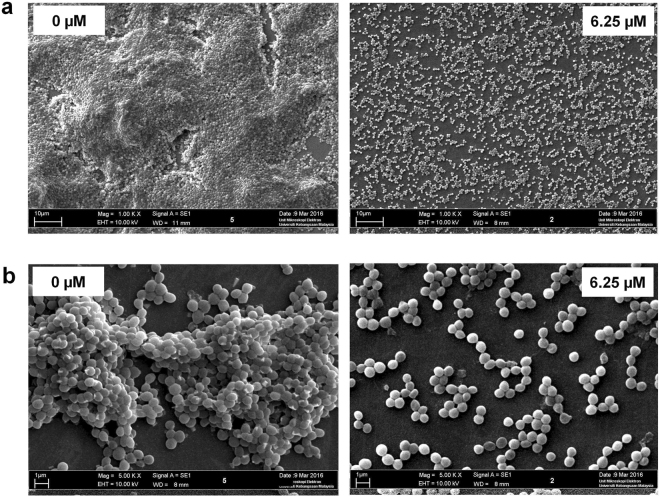
To further characterize the effect of UM-C162 on S. aureus biofilm, S. aureus was supplemented with 6.25 µM, 12.5 µM and 25 µM UM-C162 and biofilm was allowed to develop on the surface of glass slides in a multi-well plate. The morphology of UM-C162-treated and untreated S. aureus biofilm architecture formed on the glass slides following 24 hours incubation was visualized using scanning electron microscopy (SEM). Under 1000x magnification, SEM analysis revealed a thick, dense and fully established biofilm consisting of multi-layered bacterial cells (Fig. 2a). Upon treatment with UM-C162, biofilm production was disrupted and a negligible biofilm mass was detected on the glass slide. The bacteria appeared as a monolayer of dispersed cells scattered on the surface (Fig. 2a). At higher magnification (5000x), untreated S. aureus appeared as large aggregates of cells in overlapped layers (Fig. 2b). In contrast, bacteria treated with 6.25 µM of UM-C162 presented as a uniform layer of cells with negligible clumping (Fig. 2b). A similar observation was noted upon supplementation with higher concentrations of UM-C162 (Fig. S2). These images validate the data from the crystal-violet biofilm assay.
Figure 2.
Scanning electron microscopy micrographs of the S. aureus biofilm structure. (a) Biofilm formation by untreated (0 µM) and UM-C162-treated S. aureus (6.25 µM) following 24 hours incubation (1000x magnification). (b) Bacterial cells cluster in untreated S. aureus (0 µM) and the absence of cell aggregates in UM-C162-treated bacteria (6.25 µM) under 5000x magnification.
We further evaluated the biofilm inhibitory effect and biofilm eradication activity of UM-C162 against two different strains of S. aureus (S. aureus ATCC25923 and a MRSA clinical strain) using an artificial dermis wound model21,22. To assess the biofilm inhibitory activity, the dermis was supplemented with UM-C162 before the formation of biofilm. On the other hand, in the biofilm eradication assay, we exposed the 24-hour old biofilm to UM-C162 for 24 hours before harvesting the biofilm cells to enumerate bacterial colony forming units (CFU). In the inhibition assay, formation of biofilm was noted in the untreated infected wound whilst no visually detectable biofilm was observed in artificial dermis wounds treated with 50, 100 and 200 µM of UM-C162 for both S. aureus (Fig. S3a) and MRSA (Fig. S3b). This concentration range was selected based on a preliminary screen across a series of concentrations using the similar artificial dermis model. For the inhibition assay, in the absence of UM-C162, the bacterial load was 3.2 × 106 CFU for S. aureus and 2.6 × 106 for MRSA. Upon treatment with UM-C162, we observed a significant reduction (p < 0.001) in the number of S. aureus harvested from the artificial dermis i.e. 3.2 × 105 (50 µM), 2.7 × 105 (100 µM) and 1.0 × 105 (200 µM) whilst for MRSA, the number of bacteria was 1.7 × 105 (50 µM), 6.5 × 104 (100 µM) and 4.0 × 104 (200 µM) (Fig. 3a). Similarly, in the eradication assay, UM-C162-treated wounds demonstrated a substantial reduction in bacterial loads for S. aureus and MRSA (p < 0.001) (Fig. 3b). Bacterial loads recovered from the untreated 24-hour old biofilm were 2.7 × 108 CFU for S. aureus and 4.4 × 108 for MRSA. Bacterial CFU were 1.3 × 107, 2.1 × 107, 1.4 × 106 for S. aureus and 1.3 × 107, 8.6 × 106 and 3.2 × 105 for MRSA after exposure to UM-C162 at 50, 100 and 200 µM respectively, suggesting that UM-C162 might be able to eradicate the pre-formed biofilms or prevent further biofilm formation in an infected wound.
Figure 3.
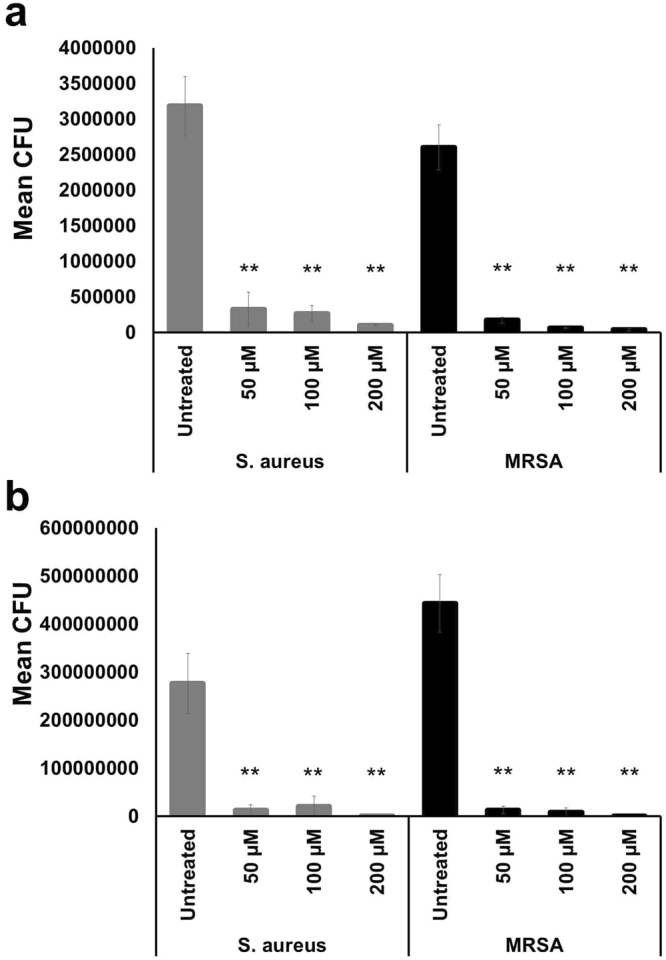
UM-C162 displays anti-biofilm activity in an artificial dermis wound model. Number of S. aureus and MRSA CFU recovered from biofilm formed on artificial dermis after exposure to various concentrations of UM-C162 for 24 hours in (a) biofilm inhibition and (b) biofilm eradication assays. Data represent the mean ± SD of two biological replicates. (**) denotes significant difference between untreated and compound-treated bacteria (p < 0.001).
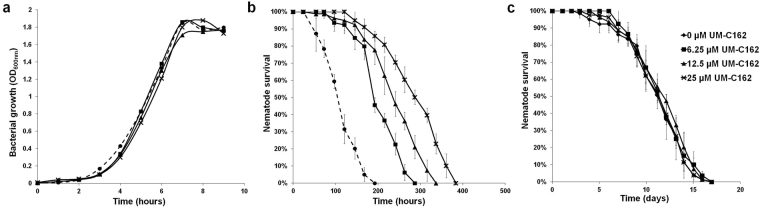
S. aureus susceptibility towards UM-C162 was tested across a range of concentrations using the broth microdilution MIC test. Gentamicin was tested in parallel as the positive control and was antimicrobial towards S. aureus with an MIC of < 30 µg/mL (Fig. S4). On the other hand, S. aureus supplemented with different concentrations of UM-C162 appeared turbid after an overnight incubation, indicating that over a dose of 0.05 µM to 200 µM, UM-C162 did not inhibit S. aureus growth (Fig. S4). To confirm the MIC data, the effect of UM-C162 on S. aureus growth was evaluated by monitoring the absorbance at OD600nm of batch cultures. S. aureus showed a similar growth rate in the presence and absence of UM-C162 (Fig. 4a), confirming that UM-C162 at concentrations ranging from 6.25 µM to 25 µM did not interfere with S. aureus growth. These findings also propose that the attenuation of S. aureus virulence by UM-C162 is mainly due to the compound’s ability to retard biofilm production and not to its antimicrobial activity against planktonic cells.
Figure 4.
UM-C162 improves the lifespan of S. aureus-infected nematodes without affecting bacterial growth. (a) UM-C162 does not inhibit the growth of S. aureus in the presence of 6.25 µM, 12.5 µM and 25 µM UM-C162. (b) Survival curves of S. aureus-infected C. elegans treated with 6.25 µM, 12.5 µM and 25 µM of UM-C162 compared to untreated control (0 µM). UM-C162 at all concentrations tested confers significant enhanced survival in infected nematodes (p < 0.0001). (c) UM-C162 does not alter the basal lifespan of worms fed on heat killed E. coli OP50. Graphs in (b) and (c) show the mean ± SD of six technical replicates (20 worms/replicate) from a representative of two independent experiments.
To further assess the protective role of UM-C162 towards the host in vivo, the lifespan of infected nematodes in the presence and absence of UM-C162 was monitored in a survival assay with additional time points and a greater number of worms. S. aureus killed the nematodes with a mean-time-to-death (TDmean) of 114.5 ± 3.7 hours (4.7 days) (Fig. 4b). Complete killing of all nematodes was achieved in 7–8 days. Upon treatment with 6.25 µM, 12.5 µM and 25 µM of UM-C162, the TDmean of S. aureus-infected worms was markedly improved to 207.1 ± 5.5, 244.5 ± 7.0 and 290.2 ± 7.8 hours, respectively (p < 0.0001) (Fig. 4b). Hence, the killing kinetics of infected worms was significantly delayed in a concentration-dependent manner for worms treated with UM-C162.
We then asked if UM-C162 is toxic towards a multicellular eukaryotic organism. To test this, we exposed the worms to various concentrations of UM-C162 in the absence of infection and lifespan of the worms was monitored. No significant difference was observed between the lifespan of UM-C162-treated and untreated worms, where both survived for ~17 days at 25 °C (Fig. 4c), ruling out the possibility that the compound could cause adverse effects onto the host. Moreover, the compound did not further extend the basal lifespan of nematodes, implying that the increased survival of nematodes infected by S. aureus is unlikely to be an outcome of any lifespan extending property of UM-C162.
S. aureus global transcriptional profile in response to treatment with UM-C162
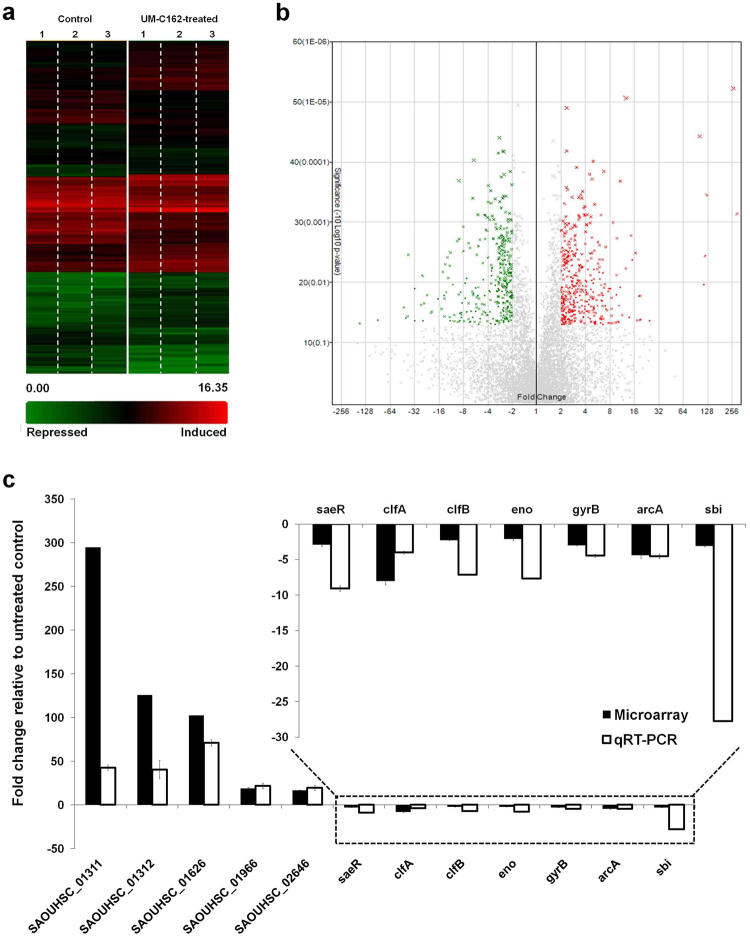
The expression of S. aureus virulence traits and components required for biofilm formation is tightly regulated by a complicated coordination of two-component systems and various transcriptional regulators4. Therefore, we postulated that a compound that can alter the formation of biofilm might also be able to reduce bacterial virulence. To probe the transcriptional changes of S. aureus in response to UM-C162, total RNA was isolated from the bacteria after overnight incubation in the presence of 6.25 µM UM-C162. Three independent microarray experiments were conducted to compare the gene expression profile of UM-C162-treated and untreated S. aureus. GeneChip analysis revealed a substantial number of genes (456) that were significantly modulated by UM-C162 (fold change ≤ −2 and ≥ 2, p < 0.05). Of these, 235 genes were up-regulated whilst 221 genes were down-regulated in UM-C162-treated S. aureus (Fig. 5a,b). A complete list of all genes differentially expressed by UM-C162 is available in Table S2. The microarray data is deposited in the NCBI Gene Expression Omnibus (GEO) (Accession number: GSE84485).
Figure 5.
Transcriptome dynamics of UM-C162-treated and untreated S. aureus. (a) Hierarchically clustered expression profile (rows) for 456 S. aureus genes differentially expressed upon exposure to UM-C162 as compared to the untreated control. Data from three independent experiments (columns) are shown. (b) Volcano plot showing the gene level analysis to select differentially expressed genes after treatment with UM-C162. The data for all genes are plotted as fold change versus −log10 of the adjusted p-value. Green, grey and red correspond to genes with < −2-fold, −2 to 2 fold and > 2-fold differential expression, respectively. (c) Verification of microarray data by qRT-PCR. Transcript levels of twelve differentially expressed S. aureus genes from qRT-PCR and microarray derived data were compared. Results displayed are the average relative fold-change for transcripts from UM-C162-treated bacteria compared to untreated control. The relative quantification of S. aureus RNA was determined by the change in expression of genes of interest normalized to S. aureus 16 s rRNA housekeeping gene.
We validated the microarray derived expression data of a subset of genes by quantitative real-time PCR (qRT-PCR). The relative expression levels of five up-regulated genes and seven down-regulated genes were compared using RNA extracted from three independent bacterial cultures. A strong positive correlation was observed for the qRT-PCR and microarray derived data for all twelve genes tested, albeit with different magnitudes (Fig. 5c).
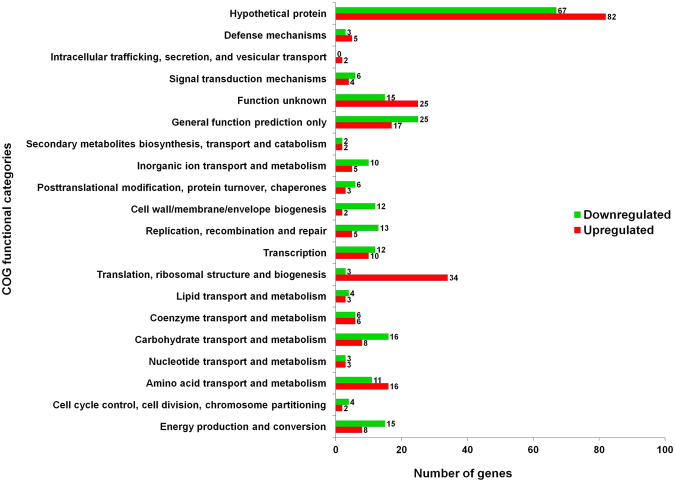
The differentially expressed genes were classified into their respective functional groups using clusters of orthologous groups (COGs) designations. The distribution of UM-C162-modulated genes and their biological roles according to COGs classification is shown in Fig. 6. Of note, a majority of the genes were grouped as hypothetical proteins (32%), general function prediction only (9%) and genes with function unknown (8.7%). By comparing the number of up-regulated and down-regulated genes in each functional group, we noted that the group of translation-, ribosomal structure- and biogenesis-encoding genes contained a higher number of up-regulated genes (34) and only 3 down-regulated genes whilst genes involved in cell wall/membrane/envelope biogenesis and replication, recombination and repair were mainly down-regulated (12 and 13 genes, respectively) and only 2 or 5 up-regulated genes, respectively (Fig. 6). Other UM-C162-regulated genes were generally classified as genes involved in amino acid transport and metabolism, carbohydrate transport and metabolism, energy production and conversion, and transcription.
Figure 6.
Functional classification of S. aureus genes significantly modulated by UM-C162. Genes were divided into their respective functional groups based on clusters of orthologous groups (COGs) designations. The red bars correspond to induced genes while green bars show the repressed genes when S. aureus was cultured in the presence of 6.25 µM UM-C162.The number at the end of each bar represents the total number of genes in each functional class.
Further gene function enrichment analysis on the induced genes based on Gene Ontology (GO) Term identified significant enrichment (p < 0.0001) of genes involved in translation. In addition, KEGG pathway analysis showed enrichment of prokaryotic-type ATP-binding cassette (ABC) transporter/permease proteins (p < 0.001). On the other hand, the genes repressed by UM-C162 were mainly involved in cell wall/external encapsulating structure (p < 0.0001), virulence/pathogenesis (p < 0.0001) and response to stress/DNA repair (p < 0.01) (Table 1).
Table 1.
Functional enrichment of S. aureus genes induced and suppressed by UM-C162.
| Function | No. of genes | Genes | P value |
|---|---|---|---|
| Up-regulated | |||
| Translation | 30 | SAOUHSC_02504, SAOUHSC_01666, rpsC, rpsD, rpsE, rpsH, rpsJ, rpsQ, rpsS, rpsT, rplB, rplC, rplD, rplE, rplF, rplJ, rplN, rplP, rplQ, rplR, rplT, rplV, rplW, rplX, rpmA, rpmC, rpmD, def, pheT, thrS, | p < 0.0001 |
| ABC transporter/permease protein | 8 | SAOUHSC_00074, SAOUHSC_00136, SAOUHSC_00169, SAOUHSC_00693, SAOUHSC_01046, SAOUHSC_01311, SAOUHSC_01312, SAOUHSC_02821 | p < 0.001 |
| Down-regulated | |||
| Cell wall/external encapsulating structure | 23 | SAOUHSC_00114, SAOUHSC_00356, SAOUHSC_00383, SAOUHSC_00386, SAOUHSC_00437, SAOUHSC_01127, SAOUHSC_02549, SAOUHSC_02708, SAOUHSC_02971, capF, capL, capA, clfA, clfB, eno, hlgB, hlgC, sspA, sbi, hlb, sasA, sdrC, atl | p < 0.0001 |
| Pathogenesis/virulence | 19 | SAOUHSC_00383, SAOUHSC_00386, SAOUHSC_01127, SAOUHSC_02708, clfA, clfB, eno, hlgB, hlgC, hlb, sspA, sbi, sasA, sdrC, saeR, yent1, arlS, clpL, clpC | p < 0.0001 |
| Response to stress/DNA repair | 9 | SAOUHSC_00503, SAOUHSC_01363, SAOUHSC_01667, clpC, clpL, recO, recF, recA, recN | p < 0.01 |
Genes encoding the ABC transporters SAOUHSC_01311 and SAOUHSC_01312 were among the most highly induced genes, with an induction magnitude of 294.62 and 125.66 fold, respectively (Table S2). ABC transporters are transmembrane proteins with diverse roles in cellular function, including the translocation of various substances (amino acids, metabolic products, lipids and drug molecules) across membranes23. Hence, the observed induction of ABC transporter genes may be associated with the active transport of UM-C162 molecules in and out of the bacterial cells. Exposure to UM-C162 also resulted in an up-regulation of ribosomal protein subunits encoded by rps, rpl and rpm (2 to 4-fold induction) (Table S2). Genes encoding ABC transporters and ribosomal proteins were also up-regulated in S. aureus exposed to ortho-phenylphenol, a phenolic compound with known antimicrobial properties24.
UM-C162 represses the expression of biofilm-associated genes in S. aureus
To decipher the molecular mechanism underlying the ability of UM-C162 to inhibit S. aureus biofilm, we compared our list of differentially expressed genes to reported S. aureus biofilm-related genes25–27. Generally, many biofilm-related genes were indeed suppressed following S. aureus exposure to UM-C162 (Table 2). Specifically, genes encoding proteins that mediate cell adhesion and bacterial attachment (SAOUHSC_02690, clfA, clfB, sdrC, sdrH, eno and arlS) were among the major group of genes suppressed by UM-C162. The clumping factors (Clf) and serine-aspartate repeat-containing proteins (Sdr) are typical S. aureus microbial surface component-recognizing adhesive matrix molecules (MSCRAMMs) that promote adherence of bacteria to a substratum and induce staphylococcal biofilm formation28. Clumping factors enable the binding of bacterial cells to fibrinogen and have been identified as an important virulence determinant in various infection models29,30. Enolase (eno) mediates S. aureus pathogenesis by adhering to laminin-containing extracellular matrix31 and has also been shown to enhance bacterial invasiveness and metastasis to secondary sites of infection in other pathogens32. ArlS is a member of the two-component system ArlS-ArlR involved in the regulation of S. aureus adhesion, autolysis, multidrug resistance and production of virulence factors33. An arlS mutant exhibited increased biofilm production when cultured on a polystyrene surface; however, intercellular adhesion and cell aggregation properties of this mutant were impaired34. The abrogation of arlS promoted peptidoglycan hydrolase activity leading to cell autolysis. Thus, it was proposed that the arl system might be required for cell growth and division as well as bacterial adherence to polymer surfaces34. In general, the development of biofilm involves three key steps which are initial attachment, proliferation and maturation of biofilm, followed by detachment or dispersal of cells and dissemination of the bacteria35. Collectively, our findings propose that UM-C162 most likely blocks the primary attachment/adhesion during biofilm formation, preventing further development of the biofilm.
Table 2.
Suppression of known S. aureus genes associated with biofilm formation.
| Role in biofilm formation | Genes | Fold change (UM-C162-treated vs control) |
|---|---|---|
| Cell adhesion and bacterial attachment | SAOUHSC_02690 | −3.39 |
| clfA | −8.03 | |
| clfB | −2.25 | |
| sdrC | −2.55 | |
| sdrH | −2.56 | |
| eno | −2.07 | |
| arlS | −2.32 | |
| Capsular polysaccharide biosynthesis | SAOUHSC_00114 | −2.91 |
| capF | −5.36 | |
| capL | −3.83 | |
| capA | −2.91 | |
| Intracellular multiplication | clpB | −9.54 |
| clpC | −5.23 | |
| ctsR | −8.61 | |
| Surface protein of Gram positive cocci | SAOUHSC_00437 | −3.69 |
| SAOUHSC_02549 | −2.75 | |
| Teichoic acid biosynthesis protein | SAOUHSC_00974 | −25.4 |
| SAOUHSC_00643 | −2.6 | |
| Peptidoglycan biosynthesis | SAOUHSC_01987 | −2.12 |
In addition to genes encoding cell adhesion and attachment, treatment of S. aureus with UM-C162 also led to a down-regulation of genes encoding for capsular polysaccharide (SAOUHSC_00114, capF, capL and capA), teichoic acid (SAOUHSC_00974 and SAOUHSC_00643) and peptidoglycan (SAOUHSC_01987) biosynthesis as well as Gram positive cocci surface proteins (Table 2). Among them, SAOUHSC_00974 encoding a poly (glycerol-phosphate) alpha-glucosyltransferase for teichoic acid biosynthesis was suppressed up to 25.4-fold. Polysaccharide capsule, cell wall teichoic acid, peptidoglycan and surface proteins are all essential constituents of the biofilm produced by Gram-positive bacteria such as S. aureus and S. epidermidis35. A reduction in the synthesis of these components is indicative of low biofilm extracellular polymeric substance production. From the analysis of the microarray data, we also noted reduced expression of ATP-dependent Clp proteases, clpB (−9.54-fold), clpC (−5.23-fold) (Table 2) and clpL (−2.64-fold) (Table S2). Inactivation of clpC in an S. aureus mutant decreased biofilm formation, proposing that ClpC is required for biofilm formation36. ClpC was also reported to affect S. aureus capsule production37. Furthermore, ClpB and ClpC, and to a minor extent ClpL, are required for bacterial long-term survival and intracellular multiplication in a cell-culture based assay36. However, we also noted that the expression of ctsR, a negative regulator of clp expression38, was repressed (−8.61-fold) in UM-C162-treated samples. Hence, clp expression may be regulated by more than one regulator or pathway.
UM-C162 diminishes the production of key virulence factors in S. aureus
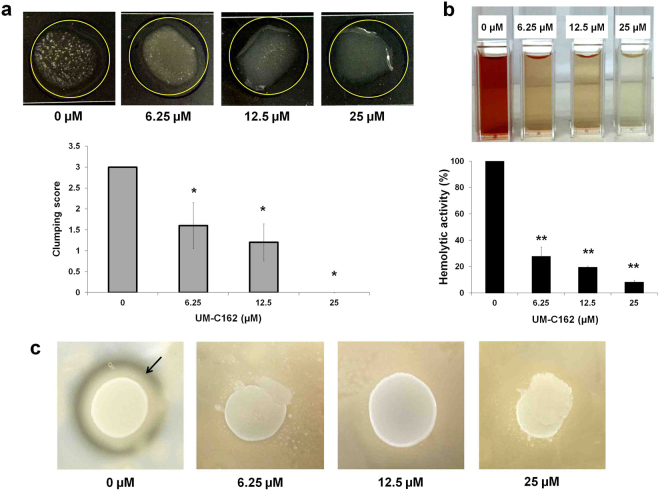
Biofilm has been recognized as a major bacterial virulence factor. The components that contribute to the formation of biofilm, for example, clf, eno, clp and cap are also classified as important virulence factors for S. aureus. ClfA mediates attachment to plasma clots and platelets whilst ClfB enhances clumping of cells in the presence of fibrinogen39. We noted that both clfA and clfB transcript levels in UM-C162 treated S. aureus were reduced relative to the untreated bacteria. To validate the ability of UM-C162 to inhibit S. aureus clumping factors, we assessed bacterial cell clumping activity using the slide coagulation test with rabbit plasma. Scores from 0 to 3 + were assigned based on the ability of S. aureus cells to form macroscopic clumps on the slides (Fig. S5). In the absence of UM-C162, S. aureus consistently agglutinated in rabbit plasma, whereas in bacteria-treated with UM-C162, less clumping activity was observed (p < 0.05) (Fig. 7a). The aggregates or clumps formed on the slides decreased with increasing concentration of UM-C162 and treatment with 25 µM UM-C162 completely abolished the clumping activity of S. aureus (Fig. 7a).
Figure 7.
Effect of UM-C162 on S. aureus virulence factors. (a) Results of S. aureus slide clumping factor assay. Shown are the pictures of S. aureus cell clumping in rabbit plasma in the presence and absence of UM-C162. Cultures supplemented with UM-C162 at 6.25 µM and 12.5 µM demonstrate weak agglutination with turbid background while cells treated with 25 µM show no cell agglutination. Bars represent the mean ± SD of clumping scores (expressed in arbitrary units) corresponding to the formation of bacteria cell clumps (n = 5 for each treatment) according to the criteria presented in Fig. S5. (b) Anti-hemolytic property of UM-C162 against S. aureus. Hemolysis of red blood cells by S. aureus in the presence of UM-C162 at 6.25 µM, 12.5 µM and 25 µM was measured following 4 hours incubation at 37 °C. Treatment with UM-C162 at various concentrations dramatically reduces the ability of S. aureus to lyse red blood cells. Pictures of spectrophotometer cuvettes indicating hemolytic activity of S. aureus are shown. (c) Effect of UM-C162 on the production of S. aureus proteases. Shown are the representative micrographs of halo formation (indicated by black arrow) in untreated S. aureus culture (0 µM) and absence of halo formation in UM-C162-treated bacteria on 3% skim milk agar after 24 hours incubation. (*) marks significant difference between untreated (0 µM) and UM-C162-treated bacteria at p < 0.05, (**) p < 0.01.
S. aureus produces a series of hemolysins including γ-hemolysin which lyse rabbit erythrocytes and various leukocytes such as neutrophils, monocytes, granulocytes and macrophages40. γ-hemolysin corresponds to two functional bi-component pore-forming toxins, HlgAB and HlgCB, which share the common HlgB protein. From the transcription profile, there is a significant decrease in the expression of genes encoding the bi-component γ-hemolysins (hlgA (−3.63 fold), hlgB (−4.52 fold) and hlgC (−6.61 fold)) (Table 3) in UM-C162-treated bacteria when compared to untreated bacteria. The hlb gene encoding ß-hemolysin or phospholipase C and SAOUHSC_02708, a putative leukocidin, were also down-regulated in UM-C162-treated S. aureus whilst the α-toxin (hla) was not significantly modulated by UM-C162. To validate the suppression of S. aureus hemolysin production by UM-C162, rabbit erythrocytes were exposed to UM-C162 treated bacteria. We noted that UM-C162 treated S. aureus exhibited weak hemolytic activity (Fig. 7b). Up to 70% inhibition in hemolytic activity was noted upon treatment with UM-C162 from as low as 6.25 µM (p < 0.01) (Fig. 7b), confirming a disruption in hemolysin production by S. aureus. Treatment with UM-C162 at 12.5 µM and 25 µM led to a greater degree of hemolysis inhibition of 80% and 92% inhibition respectively.
Table 3.
Down-regulation of S. aureus genes encoding major virulence factors.
| Virulence factors | Genes | Fold change (UM-C162-treated vs control) |
|---|---|---|
| Hemolysins/leukocidin | hlgA | −3.63 |
| hlgB | −4.52 | |
| hlgC | −6.61 | |
| SAOUHSC_02708 | −3.63 | |
| Phospholipase C | hlb | −2.31 |
| Serine protease (V8 protease) | sspA | −2.07 |
| Staphylococcal superantigen-like (SSL) proteins | SAOUHSC_00383 | −2.05 |
| SAOUHSC_00386 | −2.78 | |
| SAOUHSC_01127 | −2.13 | |
| Immunoglobulin-binding protein | sbi | −3.09 |
| S. aureus surface protein A | sasA | −2.84 |
| S. aureus exoprotein expression protein | saeR | −2.9 |
In addition to ATP-dependent Clp proteases (Table 2), expression of sspA encoding a V8 serine protease, was also reduced in UM-C162-treated samples as compared to untreated bacteria (Table 3). Clp proteases direct cellular functions through proteolysis of proteins such as casein and albumin, in the presence of ATP. In S. aureus, virulence of clp mutants is impaired in animal infection models36, suggesting that Clp proteases are crucial for normal cellular physiology as well as the control of the pathogen virulence machinery. V8 protease is thought to facilitate host tissue invasion as well as nutrient acquisition. The importance of S. aureus V8 protease for bacterial survival and virulence in vivo has been revealed in a number of animal infection models41. To assess the effect of UM-C162 on the production of S. aureus proteases, we performed the skim milk agar-based protease assay for both treated and untreated bacterial cultures. In the absence of UM-C162, S. aureus proteases hydrolyzed the milk casein resulting in the formation of a clear zone around the bacterial colony (Fig. 7c). When the bacteria were grown in the presence of various concentrations of UM-C162, no halo formation was evident (Fig. 7c), proposing that UM-C162 prevented the secretion of proteases by S. aureus.
In addition to hemolysins, proteases, clumping factors and the virulence factors involved in biofilm formation, treatment with UM-C162 also resulted in reduced expression of other virulence-related genes, including staphylococcal superantigen-like (SSL) proteins (SAOUHSC_00383, SAOUHSC_00386 and SAOUHSC_01127), immunoglobulin-binding protein (sbi), S. aureus surface protein A (sasA) and S. aureus exoprotein expression protein (saeR) (Table 3). Taken together, these biochemical tests validate the proposal that UM-C162 is a promising anti-biofilm and anti-virulence agent towards S. aureus.
Discussion
S. aureus is a key etiological agent responsible for infections that impose substantial financial burden on hospitals globally as well as high mortality and morbidity rates. The alarming increase in antibiotic-resistant S. aureus advocates the need to search for novel anti-infective drugs. In this study, we screened a collection of 35 benzimidazole derivatives for potential anti-infective activity towards S. aureus16. From this screen, we identified 12 positive hits of which three are able to alleviate S. aureus biofilm formation. Previously, a similar screen successfully identified a number extracts and compounds with anti-bacterial effects16. In addition, a plant extract that did not affect bacterial growth was able to stimulate the host antimicrobial response to clear the infection42. Hence, it is possible that some of the positive hits may permit increased survival of infected hosts by inhibiting bacterial growth or activating the host immune response to eradicate the bacteria. Benzimidazoles are widely used as anti-helminthics to treat nematode and trematode infections in domestic animals43. In our study, we also noted a small number of benzamidazole derivatives that killed C. elegans even in the absence of infection supporting the advantage of this C. elegans screen in facilitating the elimination of compounds that are toxic to the host.
Rapid developments and advances in structure-based rational design of novel molecular entities that function or target certain biomolecular pathways have led to the discovery of potential therapies for infectious diseases. The collection of benzimidazole derivatives used in this study have previously been tested for their inhibitory effect on poly (ADP-ribose) polymerase-1 (PARP-1) and dihydroorotate dehydrogenase (DHODH) and a number of these compounds showed promising anti-PARP-1 and DHODH effects, suggesting their prospective role in anti-cancer therapy44. Of note, two positive hits in our anti-infective screen, UM-C49 and UM-C201, were also able to inhibit both PARP-1 and DHODH enzymes. UM-C162 demonstrated modest DHODH inhibition and a compound with a similar structure (UM-C187) showed dual potency on both DHODH and PARP-144. Recently, several other compounds that exhibit both anti-cancer and anti-biofilm effects in vitro have been discovered45,46. Nevertheless, the link between anti-cancer and anti-biofilm activities is yet to be clarified.
An anti-infective agent that reduces bacterial virulence without disrupting bacterial viability is thought to avoid the selective pressure that promotes antibiotic resistance5. Benzimidazole derivatives with potent antimicrobial activity against both bacteria and fungus have previously been reported15. Conversely, in our study, both the antimicrobial test (Fig. S4) and growth curve analysis (Fig. 4a) demonstrated that UM-C162 was not antibacterial towards S. aureus, advocating the potential of UM-C162 as an anti-virulence candidate that is not likely to stimulate bacterial resistance. Furthermore, an anti-virulence agent that does not impede bacterial growth is not likely to interfere with the preservation of host endogenous flora.
Previously, a benzimidazole analog, 5-methoxy-2-[(4-methylbenzyl)sulfanyl]-1H-benzimidazole (also known as ABC-1), was identified as a wide-spectrum biofilm inhibitor towards both Gram-positive and Gram-negative bacterial biofilm (including S. aureus, MRSA, Vibrio cholerae, Pseudomonas aeruginosa) without having any impact on bacterial growth18. At 25 µM, ABC-1 inhibited S. aureus biofilm by < 50% whilst in this study, UM-C162 was able to inhibit S. aureus biofilm by > 50% even at 6.25 µM. In addition, the 2-aminobenzimidazole class of compounds also exhibits biofilm inhibitory effects45, proposing that compounds containing a benzimidazole functional group are a promising scaffold for the discovery of novel anti-biofilm candidates.
The diversity and functional redundancy of S. aureus virulence factors is important to the overall fitness and disease causation of S. aureus in the host. As a result, attenuating a single virulence factor does not usually lead to the diminution of S. aureus virulence. Our genome-wide transcriptome profile and SEM analysis indicate that UM-C162 most likely blocks S. aureus adherence or cell aggregation, which is the first step in the process of biofilm production. A cohort of virulence factor-encoding genes was suppressed in UM-C162-treated bacteria but not in the untreated control. Together with the outcomes from the biochemical assays, we provide evidence that compound UM-C162 inhibits the production of several S. aureus virulence factors, namely biofilm, clumping factors, hemolysins and proteases. These surface proteins, toxins and enzymes are vital for the pathogen to attach to host cells, establish an infection, impair the host immune reaction and result in host tissue deterioration. A previous S. aureus - C. elegans host-pathogen interaction study revealed that the virulence determinants, including hemolysins, V8 serine proteases and biofilm, are needed for full pathogenicity of S. aureus in the nematode model of infection17,47. S. aureus mutants for these crucial virulence factors were attenuated in C. elegans killing47. By blocking these virulence traits, the bacteria are rendered less virulent and less able to disseminate within the host. This allows the host immune response to initiate elimination of the less pathogenic bacteria from the host, ensuring the survival of the infected host.
A complex process of multiple systems is required to regulate the expression of virulence-associated genes. Among the well-studied regulators are the two component systems Agr operon, SaeRS, ArlSR and SarA global transcriptional regulators4. We noted a down-regulation of the transcript levels for saeR and arlS while the expression of agr and sarA remained unchanged upon treatment with UM-C162, suggesting that UM-C162 might target the regulator/s upstream of saeRS and arlSR. SaeRS and ArlSR regulate the expression of genes encoding S. aureus virulence factors such as hemolysins, coagulases, lipase and S. aureus surface protein A48,49. Through various biochemical assays, we consistently observed suppression of hemolysins, coagulases and proteases by UM-C162.
S. aureus biofilms are often associated with chronic infections and contaminated implanted medical devices and the presence of biofilm renders the bacteria highly tolerant to antibiotics and able to resist phagocytosis. Additionally, S. aureus expresses an arsenal of virulence determinants that contribute to its versatility in surviving the host defence response. In our search for novel anti-infectives that target bacterial virulence, we identified a benzimidazole-based molecule, UM-C162, as a potential anti-virulence agent that interferes with the production of bacterial virulence determinants (e.g. toxins, enzymes and biofilm). In light of its ability to inhibit a myriad of key S. aureus virulence factors, UM-C162 is a promising anti-virulence agent to be further developed as an anti-infective agent. No detectable toxic effect on adult worms was observed signifying that UM-C162 is non-toxic to the eukaryotic host. Our results strongly propose UM-C162 as a potential anti-virulence candidate with minimal toxicity, to be developed as an alternative therapeutic for S. aureus infections.
Methods
Bacteria and nematode growth conditions
S. aureus strains used were NCTC8325-4, ATCC25923 and a MRSA clinical strain22 (Adelaide Pathology Partners (Mile End, Australia)). S. aureus NCTC8325-4 was used in all experiments unless otherwise specified. S. aureus was routinely grown in Trypticase Soy (TS) media (Oxoid/Pronadisa) and incubated at 37 °C under aerobic conditions. A uracil-requiring strain of Escherichia coli OP50 was propagated on Luria-Bertani (LB) media (Pronadisa) with streptomycin (100 µg/mL) at 37 °C. The wild type C. elegans N2 strain was cultivated on nematode growth medium (NGM) with E. coli OP50 as their food. The nematodes were rendered sterile by RNAi knockdown of the pos-1 gene that caused the gravid worms to lay unhatched eggs50. This eliminates any confusion in the scoring as a result of nematode reproduction. The pos-1 RNAi treated nematodes were allowed to develop into young adult worms to be used in the screen.
Synthesis and preparation of benzimidazole derivatives
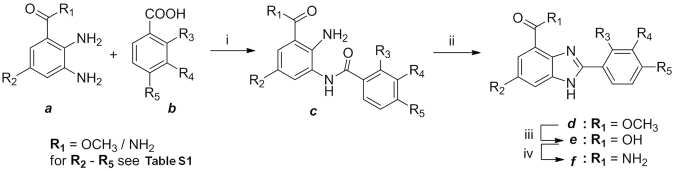
Thirty five compounds with the benzimidazole backbone were synthesized using the previously described procedures44 as outlined in the scheme in Fig. 8. Briefly, various functional groups were placed at the R3, R4 and R5 positions of the corresponding carboxylic acid intermediates by de novo synthesis. Two different routes were employed to prepare the compounds. The first route (for R1 = OCH3) involves coupling of diamine a with carboxylic acid b by using 1-[bis(dimethylamino) methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium-3-oxidhexafluoro-phosphate (HATU). Subsequent thermal cyclization of the resulting amide c in acetic acid gave benzimidazole ester d. Saponification of the ester d with lithium hydroxide resulted in the benzimidazole carboxylic acids e which were then converted to the corresponding benzimidazole carboxamide f with 1-hydroxybentzotriazole (HOBt). The second route (for R1 = NH2) involves direct coupling of diamine a with carboxylic acid b in the presence of 1,1′-carbonyldiimidazole (CDI). Subsequent thermal cyclization of the resulting amide c in one-pot afforded benzimidazole carboxamide f in two-steps. DMSO was used as the solvent and all reconstituted compounds were filter sterilized using a 0.2 µm membrane and stored at −20 °C before use.
Figure 8.
General synthesis of benzimidazole derivatives. Reaction conditions: For R1 = OCH3 (i) HATU, DIPEA, DMF (ii) AcOH, 110 °C (iii) LiOH, MeOH/THF (iv) HOBt, EDCl, NH4Cl, DIPEA, DMF; For R1 = NH2 (i) CDI, pyridine, DMF (ii) AcOH, 110 °C.
C. elegans – S. aureus anti-infective screen
Screening for potential anti-infective agents was performed as previously described16. Briefly, medium containing worm M9 buffer, S. aureus culture and 10 µg/mL cholesterol was prepared and added into individual wells in a 48-well plate. Each benzimidazole derivative was individually added to a final concentration of 100 µM. The controls were prepared by replacing the compound with DMSO (untreated control) or by replacing S. aureus with E. coli OP50 (uninfected control). Ten age-matched pos-1 treated nematodes were manually transferred into the liquid medium and the plate was incubated at 25 °C without agitation. The number of live and dead worms was scored every 24 hours. Positive hits were selected based on ≥70% survival of S. aureus-infected compound-treated worms in at least two replicates at a point when ~20% of the untreated control worms survived.
Antimicrobial tests
To determine the antimicrobial activity of benzimidazole compounds, the broth microdilution test and bacterial growth analysis were performed. S. aureus was cultured in 3 mL of TS broth and incubated overnight (16–18 hours) at 37 °C in a shaking incubator. The initial bacterial inoculum size was standardized by diluting the S. aureus overnight culture to OD600 = 0.3. The bacterial suspension was further diluted 1:100 to obtain 106 cfu/mL. To determine the minimum inhibitory concentration (MIC), a series of different concentrations of compound was prepared by two-fold serial dilution in a 96-well plate, followed by inoculation with 106 cfu/mL S. aureus and overnight incubation at 37 °C. Gentamicin (0.03 mg/mL to 50 mg/mL) and DMSO (0.0005% to 1%) served as the positive and negative controls, respectively. To evaluate the effect of compound UM-C162 on S. aureus growth, a standardized suspension of 108 cfu/mL S. aureus was diluted (1:100) in TS broth supplemented with the compound at different concentrations, followed by incubation at 37 °C with shaking. At selected time intervals, the turbidity of the culture was measured at 600 nm.
Crystal-violet biofilm assay
The biofilm assay was conducted in a 96-well polystyrene flat-bottomed microplate (Greiner Bio-One). Firstly, the standardized S. aureus suspension at 106 cfu/mL was prepared and dispensed into each well in the 96-well plate. To screen for anti-biofilm activity, individual benzimidazole compounds were added to the bacterial suspension to a final concentration of 100 µM and tested in duplicate wells, representing two technical replicates. For the dose-dependent anti-biofilm assay, the selected compound was tested across of series of concentrations, ranging from 100 to 0.78 µM. As a control, the compound was replaced with DMSO. Uninoculated TS broth was also included as the blank control. The plate was incubated at 37 °C for 24 hours under static conditions. After incubation, the wells were washed three times with 1X phosphate buffered saline (PBS) to eliminate non-adherent bacteria and fixed with 99% (v/v) methanol for 15 minutes. The wells were allowed to dry in a laminar flow. The attached biofilm cells were stained using filtered 0.5% crystal violet (Sigma-Aldrich) for 5 minutes at room temperature. The excess stain was removed by rinsing with water and crystal violet bound cells were solubilised with 33% acetic acid. The released stain was measured at 570 nm using a microplate reader (Sunrise, Tecan). Analysis was performed on three independent occasions and two technical replicates for each compound.
Scanning electron microscopy (SEM) analysis of biofilm formation
An overnight culture of S. aureus was adjusted to OD600 = 0.3 and further diluted 1:100 in TS broth to obtain 106 cfu/mL S. aureus. UM-C162 was tested at 6.25 µM, 12.5 µM and 25 µM. For each concentration, 4 mL of bacterial suspension was supplemented with the desired amount of compound and added into individual wells in a 6-well plate. A glass slide (about 10 mm × 10 mm) was positioned in the middle of each well. The plate was incubated at 37 °C for 24 hours to allow the formation of biofilm on the slides. The bacteria cells were processed for SEM as described previously51. Briefly, the samples were fixed overnight at 4 °C in 4% (v/v) glutaraldehyde, rinsed three times with 1X PBS and dehydrated through a graded ethanol series (35%, 50% and 70%), followed by critical-point drying in a drying apparatus with liquid CO2. The samples were gold-coated using a gold sputtering unit and observed using a LEO 1450VP variable pressure scanning electron microscope (Electron Microscopy Unit, Universiti Kebangsaan Malaysia).
Artificial dermis wound biofilm model
An artificial dermis was prepared as previously described22. The dermis consisting of hyaluronic acid and collagen spongy sheets was sterilized by UV light treatment followed by heating at 110 °C for an hour before use. A mixture of lyophilized bovine plasma, Bolton broth, horse blood and heparin was prepared and added to the dermis in a 24-well plate. Overnight cultures of S. aureus ATCC 25923 and a clinical strain of MRSA was adjusted to 1 × 106 cfu/mL and 10 µL of adjusted bacterial inoculum was inoculated onto the surface of each dermis. To assess for a potential biofilm inhibitory effect, UM-C162 was added to each artificial dermis immediately after the dermis was inoculated with bacteria. On the other hand, to assess for the eradication of biofilm, following bacterial inoculation, biofilm was allowed to form on the dermis at 37 °C for 24 hours. After the formation of biofilm on each dermis, sterile filter paper impregnated with UM-C162 was placed in contact with the biofilm, followed by incubation at 37 °C for 24 hours. The dermis was transferred to 10 mL saline and biofilm was extracted by vortexing and sonication. Recovered bacterial cells were serially diluted and plated on TSA supplemented with 7.5% NaCl. Plates were incubated overnight at 37 °C and colonies were counted. All experiments were repeated twice independently with at least two technical replicates.
Nematode survival and lifespan assays
C. elegans survival and lifespan assays were conducted to examine the effect of benzimidazole compounds in a live-animal model as previously described16,42. The survival assay was performed to confirm the anti-infective effect of the selected benzimidazole compound, UM-C162, at different concentrations whilst the lifespan assay was performed to assess potential toxicity on uninfected worms. These assays were performed using a liquid-based medium in the same manner as the anti-infective screen except the number of worms used was increased from ten to twenty worms per well and six technical replicates were analysed. The lifespan assessment experiment was conducted by feeding the worms with heat-killed E. coli OP50 in the presence of UM-C162. Heat-killed (65 °C for 30 minutes) E. coli OP50 were used to avoid any possible effect of UM-C162 on the bacteria which may contribute to a reduction in C. elegans lifespan. The bacteria were added into M9 medium supplemented with different concentrations of compound before transferring age-matched pos-1 RNAi treated nematodes into the medium. The plates were kept at 25 °C. Survival of treated and untreated worms was monitored over time until all worms died. The experiment was performed in duplicate.
Total RNA extraction
Total bacterial RNA was extracted using MasterPureTM Complete DNA and RNA Purification Kit (Epicentre) with minor modifications. In brief, S. aureus was cultured for 16–18 hours in the presence and absence of UM-C162 (6.25 µM) at 37 °C with agitation. The bacterial culture was centrifuged at 8000 × g for 1 minute to pellet the cells which were then resuspended in 150 µL of TE buffer (Epicentre MasterPureTM Complete DNA and RNA Purification Kit). Lysozyme (15 µL) (Sigma-Aldrich) was added to a final concentration of 2.5 mg/mL and the mixture was incubated at 37 °C for 30 minutes. Ten µL of Proteinase K (Qiagen) was diluted in 150 µL of Tissue & Cell Lysis Solution and added to the samples, followed by incubation at 65 °C for 15 minutes in a water bath. After incubation, the samples were left to equilibrate to room temperature and placed on ice for 3 minutes. Removal of contaminating DNA was done according to Epicentre protocols. Total RNA was further purified with the Qiagen RNeasy Mini Kit and residual DNA was completely removed using the Qiagen RNase-Free DNase Set according to the manufacturer’s instructions. The presence of DNA contamination in the RNA samples was assessed by PCR targeting the nuc gene using primers and PCR protocol as previously described52. The quantity and quality of RNA was measured with the NanoDrop 2000C spectrophotometer and RIN number was analyszed using the Agilent 2100 Bioanalyser.
Microarray gene expression profiling and data analysis
RNA samples were processed according to the Affymetrix SensationPlus FFPE Amplification and 3′IVT Labeling Kit protocol. Briefly, 50 ng of total RNA was reversed transcribed to produce cDNA/mRNA hybrid molecules, which were subsequently used as template to produce sense RNA via in vitro transcription (IVT). IVT-generated sense RNA was purified, subjected to single-stranded cDNA synthesis following which the cDNA was fragmented, end-labelled and hybridized to the Affymetrix GeneChip S. aureus Genome Array (Affymetrix, Cat. No. 900514) for 16 hours at 45 °C with rotation at 60 rpm. Following hybridization, arrays were washed and stained using the FS450_0005 fluidics protocol and scanned using an Affymetrix 3000 7 G scanner. The scanned images were inspected for hybridization efficiency. To perform data quality control (QC), feature intensity (CEL) files generated from GeneChip® Command Console® Software (AGCC) were imported into the Affymetrix Expression Console (EC) 1.4 software. MAS5 algorithm was used for normalizing probe-level intensity. QC metrics were generated and extracted from the Affymetrix EC report. Raw data were submitted to the Affymetrix EC software to generate probe set summarization (CHP) files from CEL files using the PLIER algorithm. Gene expression analysis was performed using three independent mRNA samples for each group. To perform statistical analysis and identify the differentially expressed genes, the resultant data (CHP files) were submitted to the Affymetrix® Transcriptome Analysis Console (TAC) software for Gene Level Differential Expression Analysis. Genes with a p value < 0.05 and threshold values of ≥ 2 and ≤ – 2-fold change between the UM-C162-treated samples and the untreated controls were defined as significantly differentially expressed. The complete microarray dataset has been deposited in NCBI’s GEO database and can be accessed through GEO accession number GSE84485 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE84485). Functional classifications were carried out based on clusters of orthologous groups (COGs) designations for S. aureus NCTC8325 (http://www.ncbi.nlm.nih.gov/COG/). Gene function enrichment analysis was performed against the Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources version 6.7 (https://david.ncifcrf.gov/) using Fisher Exact test with Benjamini-Hochberg multiple testing correction (p < 0.05).
Quantitative RT-PCR (qRT-PCR) analysis
qRT-PCRs on total DNA-free RNA using the iTaqTM Universal One-Step RT-qPCR kits with SYBR Green (BioRad Laboratories, USA) were performed according to the manufacturer’s recommendations on the Bio-Rad CFX96 Touch Real-Time PCR Detection System. The parameters for the RT-PCR cycle used were 50 °C for 30 minutes, 95 °C for 5 minutes, 95 °C for 30 seconds, 60 °C for 30 seconds, 76 °C for 30 seconds and 60 °C for 7 minutes. Amplification specificity was confirmed by melt curve analysis where the reaction temperature was increased from 55 °C to 95 °C over 10 seconds (0.5 °C increment for 80 cycles) after amplification. The forward and reverse primers used are listed in Table S3. The average cycle threshold (Ct) values were normalized against the S. aureus 16 s rRNA53 that was found to not vary upon treatment. The mRNA expression levels of genes of interest was computed using the CFX ManagerTM software (Bio-Rad Laboratories, USA) on the average normalized Ct value.
Analysis of virulence factors production
Plasma clumping factor test
S. aureus was cultured overnight at 37 °C in the presence and absence of UM-C162. Bacterial cells harvested from the overnight culture were prepared in 1X PBS. Rabbit blood plasma was separated from red blood cells by centrifugation at 900 × g for 5 minutes. Twenty µL of rabbit plasma was placed on the surface of a glass slide, followed by inoculation of 10 µL of S. aureus suspension. The mixture was mixed evenly for 10 seconds and macroscopic clumping/agglutination of bacterial cells was scored from 0 to 3+ (Fig. S5).
Haemolysin test
Lysis of red blood cells was assessed as previously described9 with minor modifications. Briefly, different concentrations of UM-C162 were added to a S. aureus culture with standardized inoculum size, followed by incubation at 37 °C with agitation for 16–18 hours. Rabbit blood was obtained from the Animal House Facility, Universiti Kebangsaan Malaysia. Red blood cells were separated from plasma by centrifugation at 900 × g for 5 minutes at 4 °C, washed 3 times with 1XPBS and diluted in PBS (330 µL of red blood cells in 10 mL of 1X PBS). To measure the haemolytic activity, 200 µL of S. aureus culture (OD600 = 0.3) grown in the presence of compound was added into 10 mL of diluted red blood cells. The negative control (S. aureus supplemented with DMSO only) was run in parallel. The mixture was incubated shaking for 4 hours. Supernatants were collected by centrifugation at 16,600 × g for 10 minutes and the OD readings were measured at 543 nm.
Protease test
The ability of UM-C162-treated and untreated S. aureus to produce protease was determined by spotting the bacterial culture on 3% skim milk agar (Oxoid, UK). S. aureus was grown overnight at 37 °C in the presence and absence of compound and adjusted to the standard inoculum size. Ten µL of standardized S. aureus inoculum was spotted onto skim milk agar, allowed to dry under sterile conditions and incubated at 37 °C for 24 hours. After 24 hours incubation, plates were observed for the formation of a clear zone/halo around the bacteria.
Statistical analysis
At least three replicates were performed for the purpose of statistical analysis unless otherwise specified. For assays involving enumeration of dead and live worms, the worms were defined as dead when no response was detected upon picker touch and no pumping was observed at the pharynx. Dead worms with protruding vulvas were excluded from the analysis. Data from survival assays were analysed using the Kaplan-Meier nonparametric analysis in the StatView® 5.0.1 software (SAS Institute, Inc.). The pair wise comparison was analysed using the Log-rank (Mantel-Cox) significance test. For selection of significantly differentially expressed genes in the microarray analysis, a One-Way Between-Subject Analysis of Variance (ANOVA) (unpaired) was performed for treated and untreated groups. The data from other experiments were expressed as mean ± standard error of mean from at least two independent assays or mean ± standard deviation from one representative replicate. Statistical analyses for the biofilm assays and hemolysin test were performed using Student’s t-test. The ordinal data from the clumping factor assay were analyzed with the Mann-Whitney U-test.
Electronic supplementary material
Acknowledgements
This study was supported by funding from the Ministry of Science, Technology and Innovation, Malaysia (Grant No. 02-05-20-SF11117). We would like to acknowledge the contribution of Iskandar Abdullah and Yean-Kee Lee for the provision of some of the benzimidazole derivatives. The artificial dermis biofilm experiments were supported by a grant from The National Health and Medical Research Council (NHMRC: GNT1090898). The contents of the published material are solely the responsibility of the administering institution, a participating institution or individual authors and do not reflect the views of NHMRC. C.K. was supported by an Australia – APEC Women in Research Fellowship funded by the Department of Education and Training, Australian Government.
Author Contributions
C.K. conceived and designed the experiments, performed all the experiments, analyzed the data and prepared the manuscript including revisions. K.R. and N.T. were involved in the experimental design of the artificial dermis biofilm model and the relevant data analysis. C.F.C. and N.A.R. synthesized the chemical compounds and were involved in the manuscript preparatio n. S.N. participated in the experimental design, data analysis and manuscript preparation. All authors have read and approved the final version of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21141-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Reddy PN, Srirama K, Dirisala VR. An update on clinical burden, diagnostic tools, and therapeutic options of Staphylococcus aureus. Infect Dis (Auckl) 2017;10:1179916117703999. doi: 10.1177/1179916117703999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paharik, A. E. & Horswill, A. R. The Staphylococcal biofilm: adhesins, regulation, and host response. Microbiol Spectr4, 10.1128/microbiolspec.VMBF-0022-2015 (2016). [DOI] [PMC free article] [PubMed]
- 3.Zecconi A, Scali F. Staphylococcus aureus virulence factors in evasion from innate immune defenses in human and animal diseases. Immunol Lett. 2013;150:12–22. doi: 10.1016/j.imlet.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Arya R, Princy SA. Exploration of modulated genetic circuits governing virulence determinants in Staphylococcus aureus. Indian J Microbiol. 2016;56:19–27. doi: 10.1007/s12088-015-0555-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muhlen S, Dersch P. Anti-virulence strategies to target bacterial infections. Curr Top Microbiol Immunol. 2016;398:147–183. doi: 10.1007/82_2015_490. [DOI] [PubMed] [Google Scholar]
- 6.Heras B, Scanlon MJ, Martin JL. Targeting virulence not viability in the search for future antibacterials. Br J Clin Pharmacol. 2015;79:208–215. doi: 10.1111/bcp.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickey SW, Cheung GYC, Otto M. Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat Rev Drug Discov. 2017;16:457–471. doi: 10.1038/nrd.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richter K, Van den Driessche F, Coenye T. Innovative approaches to treat Staphylococcus aureus biofilm-related infections. Essays Biochem. 2017;61:61–70. doi: 10.1042/EBC20160056. [DOI] [PubMed] [Google Scholar]
- 9.Lee JH, Kim YG, Yong Ryu S, Lee J. Calcium-chelating alizarin and other anthraquinones inhibit biofilm formation and the hemolytic activity of Staphylococcus aureus. Sci Rep. 2016;6:19267. doi: 10.1038/srep19267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma Y, et al. Novel inhibitors of Staphylococcus aureus virulence gene expression and biofilm formation. PLoS One. 2012;7:e47255. doi: 10.1371/journal.pone.0047255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen F, et al. Small-molecule targeting of a diapophytoene desaturase inhibits S. aureus virulence. Nat Chem Biol. 2016;12:174–179. doi: 10.1038/nchembio.2003. [DOI] [PubMed] [Google Scholar]
- 12.Qin N, et al. RNA-Seq-based transcriptome analysis of methicillin-resistant Staphylococcus aureus biofilm inhibition by ursolic acid and resveratrol. Sci Rep. 2014;4:5467. doi: 10.1038/srep05467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mellbye, B. & Schuster, M. The sociomicrobiology of antivirulence drug resistance: a proof of concept. MBio2, 10.1128/mBio.00131-11 (2011). [DOI] [PMC free article] [PubMed]
- 14.Allen RC, Popat R, Diggle SP, Brown SP. Targeting virulence: can we make evolution-proof drugs? Nat Rev Microbiol. 2014;12:300–308. doi: 10.1038/nrmicro3232. [DOI] [PubMed] [Google Scholar]
- 15.Keri RS, Hiremathad A, Budagumpi S, Nagaraja BM. Comprehensive review in current developments of benzimidazole-based medicinal chemistry. Chem Biol Drug Des. 2015;86:19–65. doi: 10.1111/cbdd.12462. [DOI] [PubMed] [Google Scholar]
- 16.Kong C, Yehye WA, Abd Rahman N, Tan MW, Nathan S. Discovery of potential anti-infectives against Staphylococcus aureus using a Caenorhabditis elegans infection model. BMC Complement Altern Med. 2014;14:4. doi: 10.1186/1472-6882-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Begun J, et al. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog. 2007;3:e57. doi: 10.1371/journal.ppat.0030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambanthamoorthy K, et al. Identification of a novel benzimidazole that inhibits bacterial biofilm formation in a broad-spectrum manner. Antimicrob Agents Chemother. 2011;55:4369–4378. doi: 10.1128/AAC.00583-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shrestha L, et al. Inhibitory effects of antibiofilm compound 1 against Staphylococcus aureus biofilms. Microbiol Immunol. 2016;60:148–159. doi: 10.1111/1348-0421.12359. [DOI] [PubMed] [Google Scholar]
- 20.Merritt, J. H., Kadouri, D. E. & O’Toole, G. A. Growing and analyzing static biofilms. Curr Protoc MicrobiolChapter 1, Unit1B 1, 10.1002/9780471729259.mc01b01s00 (2005). [DOI] [PMC free article] [PubMed]
- 21.Brackman G, et al. Dressings loaded with cyclodextrin-hamamelitannin complexes increase Staphylococcus aureus susceptibility toward antibiotics both in single as well as in mixed biofilm communities. Macromol Biosci. 2016;16:859–869. doi: 10.1002/mabi.201500437. [DOI] [PubMed] [Google Scholar]
- 22.Richter, K. et al. A topical hydrogel with deferiprone and gallium-protoporphyrin targets bacterial iron metabolism and has antibiofilm activity. Antimicrob Agents Chemother61, 10.1128/AAC.00481-17 (2017). [DOI] [PMC free article] [PubMed]
- 23.Garmory HS, Titball RW. ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect Immun. 2004;72:6757–6763. doi: 10.1128/IAI.72.12.6757-6763.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jang HJ, Nde C, Toghrol F, Bentley WE. Microarray analysis of toxicogenomic effects of ortho-phenylphenol in Staphylococcus aureus. BMC Genomics. 2008;9:411. doi: 10.1186/1471-2164-9-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beenken KE, et al. Global gene expression in Staphylococcus aureus biofilms. J Bacteriol. 2004;186:4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagarajan V, Smeltzer MS, Elasri MO. Genome-scale transcriptional profiling in Staphylococcus aureus: bringing order out of chaos. FEMS Microbiol Lett. 2009;295:204–210. doi: 10.1111/j.1574-6968.2009.01595.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Resch A, Rosenstein R, Nerz C, Gotz F. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl Environ Microbiol. 2005;71:2663–2676. doi: 10.1128/AEM.71.5.2663-2676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foster TJ, Geoghegan JA, Ganesh VK, Hook M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. 2014;12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mulcahy ME, et al. Nasal colonisation by Staphylococcus aureus depends upon clumping factor B binding to the squamous epithelial cell envelope protein loricrin. PLoS Pathog. 2012;8:e1003092. doi: 10.1371/journal.ppat.1003092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malachowa N, et al. Contribution of Staphylococcus aureus coagulases and clumping factor A to abscess formation in a rabbit model of skin and soft tissue infection. PLoS One. 2016;11:e0158293. doi: 10.1371/journal.pone.0158293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carneiro CR, Postol E, Nomizo R, Reis LF, Brentani RR. Identification of enolase as a laminin-binding protein on the surface of Staphylococcus aureus. Microbes Infect. 2004;6:604–608. doi: 10.1016/j.micinf.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Lahteenmaki K, Edelman S, Korhonen TK. Bacterial metastasis: the host plasminogen system in bacterial invasion. Trends Microbiol. 2005;13:79–85. doi: 10.1016/j.tim.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 33.Yarwood JM, Schlievert PM. Quorum sensing in Staphylococcus infections. J Clin Invest. 2003;112:1620–1625. doi: 10.1172/JCI200320442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fournier B, Hooper DC. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J Bacteriol. 2000;182:3955–3964. doi: 10.1128/JB.182.14.3955-3964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otto M. Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu Rev Med. 2013;64:175–188. doi: 10.1146/annurev-med-042711-140023. [DOI] [PubMed] [Google Scholar]
- 36.Frees D, et al. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol Microbiol. 2004;54:1445–1462. doi: 10.1111/j.1365-2958.2004.04368.x. [DOI] [PubMed] [Google Scholar]
- 37.Luong TT, et al. Staphylococcus aureus ClpC divergently regulates capsule via sae and codY in strain newman but activates capsule via codY in strain UAMS-1 and in strain Newman with repaired saeS. J Bacteriol. 2011;193:686–694. doi: 10.1128/JB.00987-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derre I, Rapoport G, Msadek T. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol Microbiol. 1999;31:117–131. doi: 10.1046/j.1365-2958.1999.01152.x. [DOI] [PubMed] [Google Scholar]
- 39.Foster TJ, Hook M. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 1998;6:484–488. doi: 10.1016/S0966-842X(98)01400-0. [DOI] [PubMed] [Google Scholar]
- 40.Vandenesch F, Lina G, Henry T. Staphylococcus aureus hemolysins, bi-component leukocidins, and cytolytic peptides: a redundant arsenal of membrane-damaging virulence factors? Front Cell Infect Microbiol. 2012;2:12. doi: 10.3389/fcimb.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coulter SN, et al. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol Microbiol. 1998;30:393–404. doi: 10.1046/j.1365-2958.1998.01075.x. [DOI] [PubMed] [Google Scholar]
- 42.Kong C, Tan MW, Nathan S. Orthosiphon stamineus protects Caenorhabditis elegans against Staphylococcus aureus infection through immunomodulation. Biol Open. 2014;3:644–655. doi: 10.1242/bio.20148334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krizova-Forstova V, Lamka J, Cvilink V, Hanusova V, Skalova L. Factors affecting pharmacokinetics of benzimidazole anthelmintics in food-producing animals: the consequences and potential risks. Res Vet Sci. 2011;91:333–341. doi: 10.1016/j.rvsc.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 44.Abdullah I, et al. Benzimidazole derivatives as potential dual inhibitors for PARP-1 and DHODH. Bioorg Med Chem. 2015;23:4669–4680. doi: 10.1016/j.bmc.2015.05.051. [DOI] [PubMed] [Google Scholar]
- 45.Lindsey EA, Brackett CM, Mullikin T, Alcaraz C, Melander C. The discovery of N-1 substituted 2-aminobenzimidazoles as zinc-dependent S. aureus biofilm inhibitors. Medchemcomm. 2012;3:1462–1465. doi: 10.1039/c2md20244a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rashid M, Husain A, Shaharyar M, Sarafroz M. Anticancer activity of new compounds using benzimidazole as a scaffold. Anticancer Agents Med Chem. 2014;14:1003–1018. doi: 10.2174/1871520614666140509153021. [DOI] [PubMed] [Google Scholar]
- 47.Sifri CD, Begun J, Ausubel FM, Calderwood SB. Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect Immun. 2003;71:2208–2217. doi: 10.1128/IAI.71.4.2208-2217.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu, Q., Yeo, W. S. & Bae, T. The SaeRS two-component system of Staphylococcus aureus. Genes (Basel) 7, 10.3390/genes7100081 (2016).
- 49.Fournier B, Klier A, Rapoport G. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol Microbiol. 2001;41:247–261. doi: 10.1046/j.1365-2958.2001.02515.x. [DOI] [PubMed] [Google Scholar]
- 50.Tabara H, Hill RJ, Mello CC, Priess JR, Kohara Y. pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development. 1999;126:1–11. doi: 10.1242/dev.126.1.1. [DOI] [PubMed] [Google Scholar]
- 51.Chin CY, et al. Global transcriptional analysis of Burkholderia pseudomallei high and low biofilm producers reveals insights into biofilm production and virulence. BMC Genomics. 2015;16:471. doi: 10.1186/s12864-015-1692-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ali R, Al-Achkar K, Al-Mariri A, Safi M. Role of Polymerase Chain Reaction (PCR) in the detection of antibiotic-resistant Staphylococcus aureus. Egyptian Journal of Medical Human Genetics. 2014;15:293–298. doi: 10.1016/j.ejmhg.2014.05.003. [DOI] [Google Scholar]
- 53.Atshan SS, et al. Quantitative PCR analysis of genes expressed during biofilm development of methicillin resistant Staphylococcus aureus (MRSA) Infect Genet Evol. 2013;18:106–112. doi: 10.1016/j.meegid.2013.05.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.