Abstract
As the second largest transcription factor family in plant, the basic helix-loop-helix (bHLH) transcription factor family, characterized by the conserved bHLH domain, plays a central regulatory role in many biological process. However, the bHLH transcription factor family of strawberry has not been systematically identified, especially for the anthocyanin biosynthesis. Here, we identified a total of 113 bHLH transcription factors and described their chromosomal distribution and bioinformatics for the diploid woodland strawberry Fragaria vesca. In addition, transcription profiles of 113 orthologous bHLH genes from various tissues were analyzed for the cultivar ‘Benihoppe’, its white-flesh mutant ‘Xiaobai’, and the ‘Snow Princess’ from their fruit development to the ripening, as well as those under either the ABA or Eth treatment. Both the RT-PCR and qRT-PCR results show that seven selected FabHLH genes (FabHLH17, FabHLH25, FabHLH27, FabHLH29, FabHLH40, FabHLH80, FabHLH98) are responsive to the fruit anthocyanin biosynthesis and hormone signaling according to transcript profiles where three color modes are observed for strawberry’s fruit skin and flesh. Further, prediction for the protein interaction network reveals that four bHLHs (FabHLH25, FabHLH29, FabHLH80, FabHLH98) are involved in the fruit anthocyanin biosynthesis and hormone signaling transduction. These bioinformatics and expression profiles provide a good basis for a further investigation of strawberry bHLH genes.
Introduction
The basic helix-loop-helix (bHLH) proteins, named by their signature conserved domain, form a large superfamily of transcription factor. They are widely distributed from yeast to human1,2 and play a central role in many different functions in the development of animals and plants3,4. Typically, a bHLH domain consists of ~60 amino acids with two functionally distinct regions, and it comprises a stretch of about 13–17 hydrophilic basic amino acids at the N-terminal (basic region), followed by two regions of hydrophobic residue α-helix separated by an intervening loop (HLH region)5. The basic region, which contains six typical residues with a highly conserved HER motif (His-Glu-Arg) and is thus relevant to its binding to DNA sequences, allows HLH proteins to specifically adhere to the E-box (5′-CANNTG-3′) or the variant G-box (5′-CACGTG-3′), where N corresponds to any nucleotide3,6–8. The HLH region functions as a dimerization domain which promotes the formation of homodimers or heterodimers complex, and has been found to be highly conserved in organisms3,5,9,10. Furthermore, the bHLH motifs have been characterized to be able to modulate gene expression by binding to DNA sequences and further participate in plant development3,11.
With the completion of the plant genome sequencing, a large number of bHLH sequences are identified. Recently, more and more bHLH gene families have been located with the investigation of their functions for plants, including Arabidopsis (Arabidopsis thaliana)9,12, tomato (Solanum lycopersicum)3,4, common bean (Phaseolus vulgaris)7, apple (Malus × domestica)8, Chinese cabbage (B. rapa ssp. pekinensis)13, cotton (Gossypium)14. According to their bioinformatics and evolutionary relationships2–4,7,8,13, bHLH genes are classified into 15–26. For Arabidopsis, 162 bHLH genes have been identified from genome sequences and consequently been divided into 26 subfamilies according to the topology of trees, clade support values, branch lengths, and visual inspection2. In addition, for the ‘Golden Delicious’ apple, 188 MdbHLH (Malus × domestica bHLH) transcription factors are sorted out and classified into 18 subfamilies8. Furthermore, transcription factors belonging to the same subfamily show similar structure, motif and protein function in plant1,9.
bHLH transcription factors are important regulators in plant physiology, such as anthocyanin biosynthesis2,10,15, biotic and abiotic stress12,16,17, organ development1,12,18, etc. To date, certain types of plant bHLH genes have been studied in-depth, providing insights into their biochemical functions5 and central roles of transcription factors. For example, genes, located in the IIIf subfamily of bHLH for Arabidopsis, have been proved to be involved in both the flavonoid biosynthesis and trichome formation5. Molecular interaction of TT8 (Transparent testa 8), GL3 (Glabra 3) and EGL3 (Enhancer of Glabra 3) from the IIIf subfamily with TTG1 (Transparent testa glabra1, WD40 protein family) and MYB (myeloblastosis protein, MYB protein family) forms a MBW (MYB-bHLH-WD40) complex, which regulates genes in the anthocyanin biosynthesis for Arabidopsis and tomato6,19–22. Besides, genes from bHLH III(d + e) subfamily have been demonstrated to be able to regulate the JA signal pathway to enhance the plant defense and promote the anthocyanin biosynthesis23–27. The underlying mechanism for the former is that MYC2 (myelocytomatosis 2) is crucial to the plant growth and thus may enhance disease resistance for apple28. For the latter, low temperature facilitates the expression of MdbHLH3, which regulates anthocyanin accumulation and fruit coloration for apple27,29.
Strawberry (Fragaria × ananassa Duch.) is well recognized universally as a delicious and healthy food30. In recent years, white strawberry is more and more favored by consumers, such as ‘Xiaobai’31, ‘Snow Princess’ and ‘Tokun’ varieties. As a result, numerous researchers have been casting their eyes on the fruit ripening, ABA (abscisic acid) signaling pathway32–36 and anthocyanin biosynthesis37,38. Roles of MYB transcription factors have been highlighted in the anthocyanin biosynthesis20,37,38, while very few reports on bHLH transcription factors have been made38–40 and they are mostly limited to the single bHLH. For example, anthocyanin biosynthesis is essentially regulated by the FvDFR (F. vesca DFR, dihydroflavonol 4-reductase) and FvUFGT (F. vesca UFGT, 3-O-glucosyltransferase), which can be activated by FvbHLH33 (F. vesca bHLH33) with the co-expression of FvMYB10 (F. vesca MYB10)39. Moreover, FabHLH3 (F. ananassa bHLH3) and FabHLH3∆ (encode putative negative regulator), by interacting with the four MYBs, are found to be involved in the proanthocyanidins biosynthesis for strawberry38. In order to systematically explore the molecular basis of bHLH from all of FvbHLHs involved in the anthocyanin biosynthesis and hormone response pathway, we will first analyze the bioinformation of 113 bHLH genes for the diploid woodland strawberry, F. vesca, and reveal their structure, evolution and function. Furthermore, we will study the transcript profiles of FabHLH genes from various tissues for the cultivar ‘Benihoppe’, its white flesh mutant ‘Xiaobai’, and the ‘Snow Princess’ from their fruit development to the ripening period, as well as those under either the ABA or Eth (ethephon) treatment. We finally discover that seven FabHLHs are crucial to the anthocyanin biosynthesis and fruit ripening for the strawberry fruit. We hope that this work will serve as a solid foundation for further investigations into functions of bHLH genes for the anthocyanin biosynthesis.
Results
Identification and annotation of bHLH transcription factors in strawberry
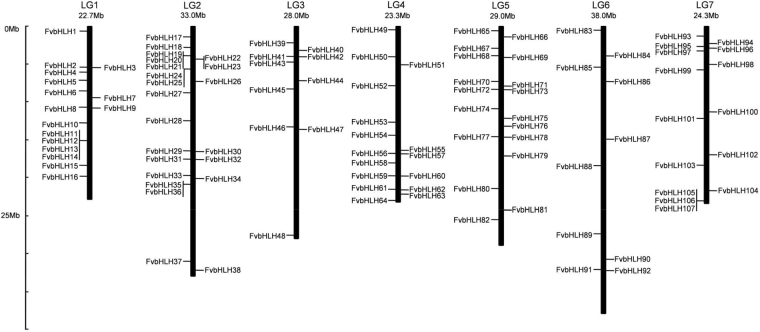
To identify bHLH transcription factors for F. vesca, a total of 166 bHLH members for strawberry via the BLAST-P (Basic Local Alignment Search Tool) search in the database of NCBI (National Center for Biotechnology Information) were obtained by comparing with the 112 strawberry bHLH amino acid sequences from the Plant Transcription Factor Database for the diploid woodland strawberry accession Hawaii-4 genome. Subsequently, to verify the reliability of the selection, a survey was conducted to confirm the presence of the conserved bHLH domain in protein sequences using the online CDD (Conserved Domains Database), SMART (Simple Modular Architecture research tool), and InterProScan database. The unique hits are kept, and duplications and similar DNA or protein sequences (with several bases different) are ruled out with only one of them left10. For example, there are four alternative variants for the sequence of FvbHLH64, only the longest variant is kept for the further analysis. In the end, 113 out of the 166 FvbHLH members are eventually selected (Table 1) out, forming the bHLH family for strawberry. The first 107 genes are renamed from FvbHLH1 to FvbHLH107 according to their distributions on the chromosome 1–7 from NCBI database8,41 (Table 1; Fig. 1). In particular, the left 6 on unknown chromosome are renamed from FvbHLH108 to FvbHLH113 by their position value from the minimum to the maximum(Table 1). The acquired 113 bHLH genes will be further used to study their bioinformation and biofunction, specially for the anthocyanin biosynthesis.
Table 1.
Details of bHLH gene family for strawberry.
| Name | Accession no. | length(bp) | No. of aa | Mw(Da) | pI | Chr. | Location | Group |
|---|---|---|---|---|---|---|---|---|
| FvbHLH1 | XM_004287073 | 1999 | 459 | 39465.8 | 6.67 | LG1 | 774071..777509 | VII(a + b) |
| FvbHLH2 | XM_011472368 | 1508 | 353 | 39258.2 | 4.83 | LG1 | 5628786..5630945 | III(a + c) |
| FvbHLH3 | XM_011472373 | 999 | 328 | 35846.2 | 5.09 | LG1 | 5633593..5635348 | III(a + c) |
| FvbHLH4 | XM_004288907 | 2261 | 609 | 68051 | 7.62 | LG1 | 6270695..6273919 | IVd |
| FvbHLH5 | XM_011462840 | 1688 | 422 | 46054.2 | 4.71 | LG1 | 7257743..7260881 | XI |
| FvbHLH6 | XM_004287765 | 1722 | 321 | 35480.6 | 7.04 | LG1 | 8558473..8560804 | IVb |
| FvbHLH7 | XM_011464708 | 1979 | 378 | 41465.2 | 9.25 | LG1 | 9553758..9555736 | VIII |
| FvbHLH8 | XM_004287975 | 1584 | 376 | 40793.2 | 5.27 | LG1 | 10770968..10773625 | VII(a + b) |
| FvbHLH9 | XM_004287983 | 1126 | 250 | 27603.6 | 9.81 | LG1 | 10839398..10840523 | VIIIb |
| FvbHLH10 | XM_004288109 | 1417 | 335 | 37562.9 | 6.2 | LG1 | 12901813..12903720 | Ia |
| FvbHLH11 | XM_011468187 | 1629 | 351 | 39316.2 | 6.03 | LG1 | 15128707..15131319 | IVa |
| FvbHLH12 | XM_004288253 | 1878 | 352 | 39142.8 | 6.13 | LG1 | 15140049..15142329 | IVa |
| FvbHLH13 | XM_004288264 | 1519 | 365 | 40767.6 | 4.64 | LG1 | 15309610..15311394 | IIIb |
| FvbHLH14 | XM_004289363 | 1400 | 296 | 33300.1 | 6.19 | LG1 | 15852165..15853564 | VIIIb |
| FvbHLH15 | XM_004288393 | 968 | 240 | 27430.5 | 9.35 | LG1 | 18393887..18395702 | Ib |
| FvbHLH16 | XM_004289479 | 880 | 216 | 23826 | 10.2 | LG1 | 19941466..19942513 | Vb |
| FvbHLH17 | XM_004289620 | 1724 | 422 | 46479.1 | 6.06 | LG2 | 1449671..1452519 | XII |
| FvbHLH18 | XM_004289703 | 1445 | 273 | 29597.9 | 7.62 | LG2 | 2948918..2950935 | X |
| FvbHLH19 | XM_004289751 | 2926 | 454 | 48833.1 | 8.64 | LG2 | 3904357..3908604 | VII(a + b) |
| FvbHLH20 | XM_004292083 | 810 | 269 | 30282.7 | 4.95 | LG2 | 3955128..3956367 | VIIIc |
| FvbHLH21 | XM_011461385 | 1084 | 349 | 38855.1 | 5.38 | LG2 | 3963999..3965470 | VIIIc |
| FvbHLH22 | XM_011459605 | 1565 | 332 | 37043.7 | 8.94 | LG2 | 4390466..4392690 | IVa |
| FvbHLH23 | XM_011459618 | 1462 | 325 | 35939.1 | 5.95 | LG2 | 4612200..4615547 | IVb |
| FvbHLH24 | XM_004289847 | 1561 | 292 | 31061.1 | 5.91 | LG2 | 5528604..5532429 | XI |
| FvbHLH25 | XM_011459681 | 2241 | 504 | 55378.5 | 5.77 | LG2 | 5784856..5787096 | III(d + e) |
| FvbHLH26 | XM_011459763 | 2595 | 518 | 56543.3 | 5.83 | LG2 | 7260709..7265049 | VII(a + b) |
| FvbHLH27 | XM_004292295 | 2144 | 544 | 59176.8 | 6.42 | LG2 | 8870693..8873438 | XII |
| FvbHLH28 | XM_004290363 | 1260 | 244 | 27544 | 6.2 | LG2 | 12507957..12509504 | Ib |
| FvbHLH29 | XM_004290615 | 2955 | 702 | 77736.9 | 5.54 | LG2 | 16556642..16562043 | IIIf |
| FvbHLH30 | XM_004290623 | 1197 | 296 | 33052.3 | 5.3 | LG2 | 16628469..16632121 | VIII |
| FvbHLH31 | XM_011460379 | 2134 | 583 | 66027.7 | 5.22 | LG2 | 17537839..17540937 | IIIb |
| FvbHLH32 | XM_011460381 | 2473 | 674 | 76146.1 | 5.16 | LG2 | 17543996..17547612 | IIIb |
| FvbHLH33 | XM_011461514 | 1208 | 381 | 42148.2 | 5.05 | LG2 | 19839411..19840894 | VIIIc |
| FvbHLH34 | XM_004290949 | 2921 | 430 | 46646.8 | 5.58 | LG2 | 20151639..20155328 | XII |
| FvbHLH35 | XM_011460622 | 1425 | 324 | 35244.1 | 5.88 | LG2 | 20942790..20945565 | VII(a + b) |
| FvbHLH36 | XM_004292764 | 1164 | 244 | 27373 | 7.96 | LG2 | 21026021..21027184 | VIIIb |
| FvbHLH37 | XM_011461186 | 1047 | 262 | 29141.9 | 6.61 | LG2 | 30979646..30981384 | Vb |
| FvbHLH38 | XM_004291800 | 2076 | 468 | 50979.6 | 7.79 | LG2 | 32278737..32282900 | X |
| FvbHLH39 | XM_004293556 | 1718 | 244 | 27140 | 4.9 | LG3 | 2254773..2258017 | XIV |
| FvbHLH40 | XM_011461973 | 1389 | 93 | 10385.6 | 5.07 | LG3 | 3263115..3264716 | XV |
| FvbHLH41 | XM_011462057 | 1986 | 484 | 51788 | 5.46 | LG3 | 4011392..4014350 | X |
| FvbHLH42 | XM_011462059 | 1003 | 216 | 24434 | 5.44 | LG3 | 4026675..4028036 | III(a + c) |
| FvbHLH43 | XM_011462115 | 1885 | 286 | 32432.7 | 5.44 | LG3 | 4881126..4883099 | VIII |
| FvbHLH44 | XM_011462301 | 2375 | 262 | 28591.3 | 7.74 | LG3 | 7317885..7322470 | Vb |
| FvbHLH45 | XM_004293965 | 1075 | 187 | 20972.7 | 7.02 | LG3 | 8396475..8397892 | Ib |
| FvbHLH46 | XM_004294266 | 2082 | 448 | 49286.4 | 6.4 | LG3 | 13388482..13391583 | X |
| FvbHLH47 | XM_004294288 | 1229 | 231 | 25421 | 8.44 | LG3 | 13637964..13641643 | IVc |
| FvbHLH48 | XM_004295162 | 3075 | 431 | 47638.7 | 6.09 | LG3 | 27564756..27568579 | IX |
| FvbHLH49 | XM_004296310 | 1113 | 231 | 25855.3 | 6.02 | LG4 | 53766..56495 | IVc |
| FvbHLH50 | XM_004296450 | 1106 | 244 | 27632.5 | 5.46 | LG4 | 4132264..4135925 | III(a + c) |
| FvbHLH51 | XM_004296502 | 1059 | 94 | 10600 | 7.93 | LG4 | 5165859..5167450 | XV |
| FvbHLH52 | XM_004296662 | 1799 | 350 | 37266.4 | 8.78 | LG4 | 7927705..7935898 | IX |
| FvbHLH53 | XM_004296894 | 1024 | 202 | 22735.5 | 9.27 | LG4 | 12705986..12707777 | Ia |
| FvbHLH54 | XM_004296937 | 2317 | 550 | 59476.9 | 6.3 | LG4 | 14464166..14467207 | XII |
| FvbHLH55 | XM_004297098 | 1619 | 345 | 38323.1 | 6.07 | LG4 | 16335664..16338026 | Vb |
| FvbHLH56 | XM_004297136 | 3010 | 550 | 59018.9 | 5.25 | LG4 | 16797062..16800740 | XII |
| FvbHLH57 | XM_004297147 | 1599 | 331 | 36302.8 | 5.95 | LG4 | 16909459..16912235 | Va |
| FvbHLH58 | XM_004297270 | 1047 | 98 | 11187.5 | 9.03 | LG4 | 18127224..18128727 | XV |
| FvbHLH59 | XM_004297447 | 2201 | 535 | 58210 | 5.81 | LG4 | 19786891..19789971 | IIIb |
| FvbHLH60 | XM_004298488 | 1789 | 339 | 37867.3 | 7.12 | LG4 | 19872469..19875066 | XII |
| FvbHLH61 | XM_011465048 | 1910 | 436 | 48434.6 | 6.18 | LG4 | 21544358..21546437 | VIIIb |
| FvbHLH62 | XM_004298520 | 1812 | 464 | 52734.2 | 6.36 | LG4 | 21671222..21673829 | III(a + c) |
| FvbHLH63 | XM_004297644 | 1599 | 275 | 29997.3 | 6.02 | LG4 | 22264260..22267220 | XII |
| FvbHLH64 | XM_011465190 | 2108 | 533 | 58065.7 | 6.78 | LG4 | 23023913..23027032 | orphans |
| FvbHLH65 | XM_004300691 | 1309 | 262 | 29626.9 | 6.42 | LG5 | 686954..688474 | Ib |
| FvbHLH66 | XM_004298754 | 3254 | 636 | 71225.1 | 5.51 | LG5 | 1485513..1490045 | IIIf |
| FvbHLH67 | XM_004298887 | 836 | 191 | 21670.1 | 9.72 | LG5 | 2965347..2966500 | Ib |
| FvbHLH68 | XM_004300913 | 1850 | 412 | 45859.4 | 6.39 | LG5 | 4025201..4029399 | orphans |
| FvbHLH69 | XM_004299007 | 1180 | 276 | 31128.1 | 6.45 | LG5 | 4234865..4236694 | XII |
| FvbHLH70 | XM_004299222 | 2515 | 571 | 62714.7 | 8 | LG5 | 7347973..7353265 | Va |
| FvbHLH71 | XM_004301080 | 1492 | 394 | 44179.6 | 7.83 | LG5 | 7937035..7939413 | VII(a + b) |
| FvbHLH72 | XM_004299332 | 1209 | 246 | 26920.3 | 5.99 | LG5 | 8368043..8370357 | IVb |
| FvbHLH73 | XM_004299336 | 984 | 92 | 10426.7 | 9.17 | LG5 | 8419238..8420695 | XV |
| FvbHLH74 | XM_004299574 | 1299 | 274 | 29662.9 | 5.68 | LG5 | 10962434..10965206 | XII |
| FvbHLH75 | XM_004299639 | 1500 | 340 | 38514.2 | 6.16 | LG5 | 12157286..12159348 | Ia |
| FvbHLH76 | XM_004301340 | 1020 | 339 | 38024.9 | 4.71 | LG5 | 13269137..13271499 | VIIIc |
| FvbHLH77 | XM_011466454 | 1192 | 224 | 24295.2 | 5.29 | LG5 | 14535826..14540081 | IX |
| FvbHLH78 | XM_004299778 | 1442 | 354 | 40009.1 | 4.85 | LG5 | 14623387..14625157 | IIIb |
| FvbHLH79 | XM_011466590 | 1909 | 386 | 41948.8 | 6.7 | LG5 | 17225517..17229489 | X |
| FvbHLH80 | XM_004300191 | 2690 | 682 | 74856.2 | 5.52 | LG5 | 21462279..21464968 | III(d + e) |
| FvbHLH81 | XM_004300360 | 1268 | 321 | 35418.5 | 6.67 | LG5 | 24315240..24317523 | Ia |
| FvbHLH82 | XM_004300469 | 1196 | 295 | 32841.5 | 5.45 | LG5 | 25509922..25511575 | XII |
| FvbHLH83 | XM_004302005 | 2095 | 339 | 36374.6 | 6.07 | LG6 | 538981..543382 | XI |
| FvbHLH84 | XM_004302277 | 2778 | 710 | 76643.7 | 6.08 | LG6 | 3989352..3994265 | VII(a + b) |
| FvbHLH85 | XM_004302404 | 1427 | 342 | 39041.4 | 5.67 | LG6 | 5477305..5478975 | Ia |
| FvbHLH86 | XM_011470140 | 525 | 174 | 19761.3 | 8.71 | LG6 | 7363269..7363793 | VIIIb |
| FvbHLH87 | XM_011468604 | 997 | 230 | 26241.7 | 6.14 | LG6 | 14930398..14931670 | Ib |
| FvbHLH88 | XM_004305478 | 1408 | 298 | 32732.1 | 6.59 | LG6 | 18431373..18433464 | VIIIc |
| FvbHLH89 | XM_004303853 | 2182 | 540 | 59336.8 | 7.66 | LG6 | 27360228..27363650 | VII(a + b) |
| FvbHLH90 | XM_004304146 | 1469 | 322 | 36058.5 | 5.7 | LG6 | 30729849..30733592 | Va |
| FvbHLH91 | XM_004304266 | 1728 | 319 | 35277.2 | 8.32 | LG6 | 32166984..32169818 | IX |
| FvbHLH92 | XM_004304269 | 1670 | 352 | 38982.9 | 5.97 | LG6 | 32238590..32243935 | XII |
| FvbHLH93 | XM_004308165 | 1937 | 429 | 47554.5 | 8.19 | LG7 | 1392304..1395438 | IX |
| FvbHLH94 | XM_011470561 | 1430 | 294 | 33574.5 | 6.15 | LG7 | 2332756..2334886 | VIII |
| FvbHLH95 | XM_004306563 | 2160 | 420 | 46844.5 | 6.1 | LG7 | 2743026..2745885 | Ia |
| FvbHLH96 | XM_004306579 | 1955 | 491 | 54529.8 | 5.03 | LG7 | 2955020..2956974 | III(d + e) |
| FvbHLH97 | XM_004306609 | 3013 | 616 | 67483 | 6.11 | LG7 | 3395170..3398461 | III(d + e) |
| FvbHLH98 | XM_004308329 | 2615 | 643 | 71423.8 | 5.42 | LG7 | 5102380..5106265 | IIIf |
| FvbHLH99 | XM_011470775 | 1286 | 361 | 39894.6 | 6.6 | LG7 | 5799342..5800960 | Ia |
| FvbHLH100 | XM_004308575 | 2010 | 465 | 52012.7 | 5.61 | LG7 | 11372618..11375286 | II |
| FvbHLH101 | XM_011471173 | 1907 | 243 | 27413.7 | 5.92 | LG7 | 12278020..12280123 | Ib |
| FvbHLH102 | XM_004307508 | 1708 | 446 | 50855 | 6.11 | LG7 | 17017617..17022889 | X |
| FvbHLH103 | XM_004307626 | 1299 | 261 | 28984 | 9.04 | LG7 | 18389468..18391089 | Vb |
| FvbHLH104 | XM_004309112 | 1767 | 366 | 40022.1 | 5.39 | LG7 | 21789731..21792322 | Ia |
| FvbHLH105 | XM_004308085 | 1469 | 254 | 28708.4 | 5.51 | LG7 | 23053917..23055551 | Ib |
| FvbHLH106 | XM_011472396 | 792 | 196 | 21478.6 | 9.61 | LG7 | 23057596..23058575 | Ib |
| FvbHLH107 | XM_004308087 | 1609 | 261 | 29145.2 | 9.42 | LG7 | 23060616..23062531 | Ib |
| FvbHLH108 | XM_004309700 | 2236 | 365 | 40673.8 | 7.06 | Un | 17881..23082 | Ib |
| FvbHLH109 | XM_004309608 | 1230 | 245 | 26615.1 | 5.83 | Un | 32335..34765 | IVc |
| FvbHLH110 | XM_004309546 | 1181 | 166 | 18402 | 9.1 | Un | 56243..60570 | X |
| FvbHLH111 | XM_011459272 | 3679 | 780 | 85359.2 | 5.24 | Un | 147469..152520 | XIII |
| FvbHLH112 | XM_011459212 | 949 | 175 | 19447.2 | 5.77 | Un | 1751808..1753619 | XII |
| FvbHLH113 | XM_004310203 | 1028 | 171 | 19329.6 | 11.5 | Un | 1966158..1967185 | XV |
Accession numbers are available in the National center for Biotechnology Information database. Mw, molecular weight; pI, isoelectric point.
Figure 1.
Chromosomal distributions of FvbHLH genes. The name on the side of each chromosome corresponds to the approximate location of each bHLH gene.
Phylogenetic analysis and multiple sequence alignments of the strawberry FvbHLH proteins
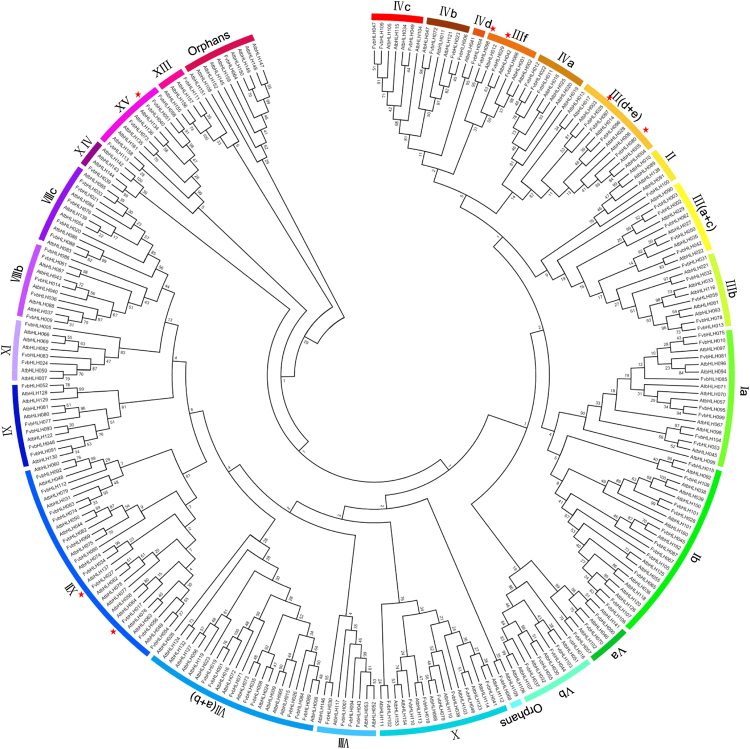
Reflecting on the past researches, the exact number of the classified subfamily for bHLH proteins has barely been reported8. To investigate the classification and evolution as well as to gain insights into the potential function of FvbHLH proteins for strawberry, we constructed a phylogenetic tree (Fig. 2) for the 113 FvbHLHs from F. vesca and 158 AtbHLHs from Arabidopsis. 26 of bHLH subfamilies are further classified according to the nomenclature protocol proposed by Heim et al.5, with some modifications. For example, I(a + b) is divided into Ia and Ib, and IIIa and IIIc are combined into III(a + c); bHLHs that are not located in any of the 24 subfamilies are classified as “orphans” (Fig. 2). We find that FvbHLH protein is persistently present in all subfamilies and the number of it varies hugely from subfamily to subfamily. For instance, each of the smallest group II, IVd, XIII and XIV contains one FvbHLH gene, while the largest clade group XII contains twelve. Consequently, the classification of bHLH genes provides an evidence for relationships among genes during their evolution.
Figure 2.
Phylogenetic tree constructed from the neighbor-joining method using the bHLH transcription factor domain for strawberry. Genes marked by the red asterisk indicates the seven candidates of FvbHLH involved in the anthocyanin biosynthesis.
In order to know sequence features of strawberry bHLH domains and further to understand FvbHLH gene’s function, we performed multiple sequence alignment of amino acid sequences of the 113 strawberry bHLH (Figs 3; S1). It is revealed that there are four conserved regions for a bHLH domain, including one basic region, two helix regions and one loop region. We find that residues of His-2, Glu-6, Arg-7, Arg-9, Arg-10, Leu-20, Leu-23, Leu-36, Leu-46, etc., in the bHLH domain are conserved, implying that the amino acid residues may play an important role in strawberry’s evolution. In addition, we notice that the basic region of the bHLH domain can bind to DNA and it is critical to the gene biofunction4. It also has been known that both Glu-6 and Arg-9 in basic region of bHLH domain play important roles in the DNA binding4,9,13 and recognition of G-box and E-box (binding mode). As a result, we divided the FvbHLH binding into three modes: G-box (with the presence of His/Lys-2, Glu-6 and Arg-9), E-box (with the presence of Glu-6 and Arg-9) and non-E-box (without the simultaneous presence of Glu-6 and Arg-9) binding42. As is demonstrated in Fig. S1, FvbHLH proteins are divided into three types: 57 for the G-box-binding, 25 for the E-box-binding, 31 for the non-E-box-binding.
Figure 3.
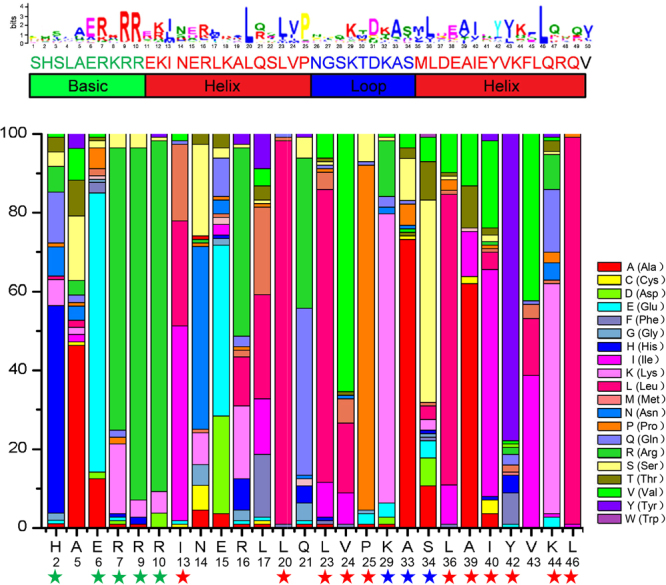
The characterization and distribution of bHLH domains. The top: sequence logo of the FvbHLH domain by MEME. The bottom: distribution of amino acids in the bHLH consensus motif among strawberry. Asterisk symbol corresponds to each column above, which stands for the percentage of presence of amino acids at each site and the color of the asterisk symbol corresponds bHLH regions from the top insert. The analysis of the amino acids composition at each site marked by the asterisk indicates that the conservation of conserved amino acids is over 50%.
Gene structure and conserved motif analysis of FvbHLH genes
Gene structure and conserved motif analysis of Arabidopsis and strawberry bHLH were performed to acquire more information about gene families5. By scanning all aspects of gene structure and conserved motif, genes within each subfamily are discovered to contain a similar number of intron and conserved motif, while the number of them is strikingly different on genes from different subfamily (Figs S2; S3), in consistent with the previous bootstrap analysis43–45. For instance, each gene from III(d + e) subfamily contains one exon except for FvbHLH97 and AtbHLH14 genes. In sharp contrast to this, 77.8% of bHLH genes from Ia subfamily contain three exons and two introns.
It has been pointed out that part of motifs, acting as activation domain, are important for the interaction with other modules of the transcription complex, and are the targets of signal transduction chains5,10. It might be inspiring to see how the motif structure is related to the gene classification. Thus, we searched 24 conserved motifs by MEME (Multiple Expectation Maximization for Motif Elicitation) program to obtain their distributions on bHLH sequences (Figs S2; S3). As is shown in Fig. S2, the bHLH proteins identified from the same subfamily share similar conserved motif. For example, motif 21 is exclusively located in all members from the XIII subfamily, whereas all bHLH sequences from IVc subfamily contain motif 1, motif 2, motif 10 and motif 15 at the C-terminal region. As bHLH is composed of motif 1 and motif 2, both of which are consistently identified in all strawberry and Arabidopsis bHLH proteins (Figs S2; S3). Hence, the classification of 26 subfamilies is thus further supported by the gene structure and motif analysis.
Transcript patterns of FabHLH genes among different tissues in three cultivated strawberry varieties
To reveal FabHLH genes (F. ananassa bHLH)’ role in regulating strawberry’s development, we focus on their temporal and spatial transcript patterns from eight different organs/tissues for three cultivated strawberry varieties (‘Benihoppe’, ‘Xiaobai’, and ‘Snow white’) under standard growth conditions (Figs 4A; 5A). We observe from the Fig. 5A that 78 bHLH genes are highly transcripted in certain tissues and their transcript patterns are similar to each other. For example, the FabHLH31 and FabHLH32 from IIIb subfamily are only transcript in anthotaxy for the two varieties (‘Benihoppe’ and ‘Snowwhite’); the FabHLH6 and FabHLH49 show similar transcript mode in all tissues for the three varieties (‘Benihoppe’, ‘Xiaobai’, and ‘Snowwhite’). However, some bHLH genes show observable different transcript behavior for the three varieties. For instance, FabHLH5 carries on the same transcript pattern with certain degree of expression in tissues from ‘Benihoppe’ and ‘Xiaobai’, while it is barely expressed in ‘Snow Princess’, resulting into a considerable deviation from the transcript pattern for the other two. Specially, expression mode of FabHLH18 differs for all the three varieties: highly transcripted in all tissues from ‘Xiaobai’, highly transcripted in some tissues from ‘Benihoppe’, lowly expressed in all tissues from ‘Snow Princess’.
Figure 4.

Materials of strawberry used in this study. (A) The fruit of ‘Benihoppe’, ‘Xiaobai’, and ‘Snow Princess’. (B) Seven fruit development and ripening stages of ‘Benihoppe’, ‘Xiaobai’, and ‘Snow Princess’. Bar = 1 cm.
Figure 5.
Transcript accumulation profiles of 113 FabHLH genes from different tissues and seven fruit development and ripening stages using semi-quantitative PCR for the three cultivated strawberry varieties. (A) Lanes: R, roots; YL, young leaves; ML, mature leaves; RN, runners; RT, runner tips; RTL, runner with tips and one leaf; A, anthotaxy; F, flowers. (B) Lanes: S1, small green fruit; S2, middle green fruit; S3, large green fruit; S4, white fruit; S5, initial red; S6, partial red; S7, full red. FvActin, FvRib413 and FvGAPDH2 were used as an internal control.
Transcript patterns of FabHLH genes during the fruit development and ripening for the white-flesh mutant strawberry
In order to identify bHLH genes involved in the color formation of strawberry fruit, three cultivated strawberry varieties were used in this study: Benihoppe, Xiaobai and Snow Princess. Colors of both the fruit flesh and skin of ‘Benihoppe’ are red. As the mutant of ‘Benihoppe’, ‘Xiaobai’ carries on the white or yellow color for its flesh with its fruit skin red or pink31. White is found for the color of ‘Snow Princess’ fruit flesh and skin (Fig. 4A). Additionally, strawberry fruit development and ripening are divided into seven stages: S1 small green fruit, S2 middle green fruit, S3 large green fruit, S4 white fruit, S5 initial red, S6 partial red, S7 full red (Fig. 4B). Because of the strong correlation between the gene expression pattern with its function, transcript patterns of 113 FabHLH genes for the color formation during the fruit development and ripening stages for the three varieties are tracked and summarized in Fig. 5B, in which the synthesis of anthocyanin is recorded from the turning stage to the red stage28. To examine the transcript of FvbHLH genes involved in the anthocyanin biosynthesis, both the RT-PCR (semi-quantitative reverse-transcription PCR) and qRT-PCR (quantitative RT-PCR) techniques are adopted to analyze genes’ expression level.
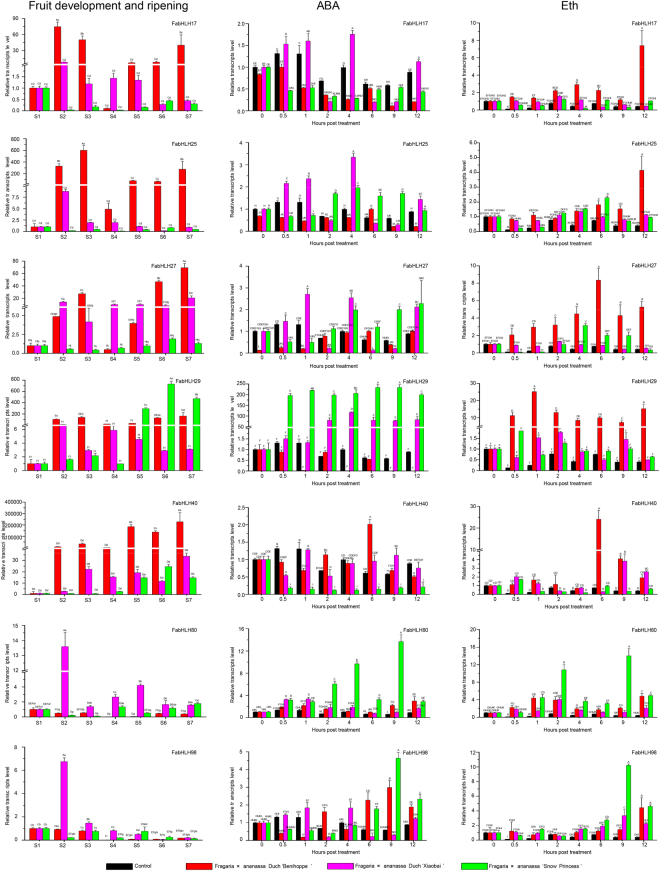
Figure 5B reveals that the number of up-regulated expression of FabHLH genes from ‘Benihoppe’ is 71 during the fruit ripening and this number from the ‘Snow Princess’ and ‘Xiaobai’ continuously falls down to 45 and 24, respectively. Depending on the consistency between the expression level of the up-regulated genes and anthocyanin content (Fig. 4B), 7 FabHLH genes are chosen out of the 113 genes to further investigate the possible expression patterns of bHLHs involved in the anthocyanin biosynthesis (Fig. 5): FabHLH17, FabHLH25, FabHLH27, FabHLH29, FabHLH40, FabHLH80, FabHLH98. In the following will be reported three relevant gene expression patterns: First, we will focus on the FabHLH25. Its expression is significantly up-regulated during all stages for ‘Benihoppe’ fruit, in accordance with its color of fruit skin and flesh, indicating that FabHLH25 promotes the anthocyanin biosynthesis for ‘Benihoppe’; for ‘Xiaobai’ fruit, it is up-regulated at S2 stage and subsequently down-regulated at S5 stage, in discordance with the color of fruit skin while coinciding with the color of fruit flesh, suggesting that FabHLH25 is not relevant to the anthocyanin biosynthesis for ‘Xiaobai’; however, the expression of FabHLH25 is always down-regulated in the whole life for ‘Snow Princess’ fruit, agreeing well with the color of fruit skin and flesh, implying that FabHLH25 is barely related to the anthocyanin biosynthesis for ‘Snow Princess’. As a consequence, expression level of FabHLH25 shows significant difference between ‘Benihoppe’ and ‘Xiaobai’, and no observable difference between ‘Xiaobai’ and ‘Snow Princess’ is found from S4 to S7. This result implies that the FabHLH25 might be involved in the anthocyanin biosynthesis for the fruit flesh. Second, we will turn to FabHLH27 gene. Its expression is up-regulated during the overall stages for both the ‘Benihoppe’ and ‘Xiaobai’ fruits. This mode coincide with the color of fruit skin for ‘Benihoppe’ and ‘Xiaobai’ and the color of fruit flesh for ‘Benihoppe’, and is inconsistent with the color of fruit flesh for ‘Xiaobai’. The consistency here indicates that FabHLH27 promotes the anthocyanin biosynthesis for both the ‘Benihoppe’ and ‘Xiaobai’. Nevertheless, FabHLH27 gene’s expression is always down-regulated for ‘Snow Princess’ fruit, in perfect agreement with the color of fruit skin and flesh for ‘Snow Princess’, implying that FabHLH27 is not in charge of the anthocyanin biosynthesis for ‘Snow Princess’. In brief, expression level of FabHLH27 shows significant difference among ‘Benihoppe’, ‘Xiaobai’ and ‘Snow Princess’ from S4 to S7. This feature signifies that the FabHLH27 could promote the anthocyanin biosynthesis for the fruit skin. Third, we will cast our eyes on the FabHLH80 gene. Its expression is constantly down-regulated for ‘Benihoppe’ fruit, in good accordance with the color of fruit skin and flesh for ‘Benihoppe’, suggesting that FabHLH80 is not involved in the anthocyanin biosynthesis for ‘Benihoppe’. FabHLH80 gene’s expression is up-regulated at S2 stage and subsequently down-regulated at S5 stage for ‘Xiaobai’ fruit, going inversely with the color of fruit skin and flesh for ‘Xiaobai’, indicating that FabHLH80 does not promote the anthocyanin biosynthesis for ‘Xiaobai’; nevertheless, FabHLH80 becomes down-regulated at S2 stage and up-regulated at S4 stage for ‘Snow Princess’ fruit, in good accordance with the color of fruit skin and flesh for ‘Snow Princess’, implying that FabHLH80 does not promote the anthocyanin biosynthesis for ‘Snow Princess’ either. As a short summarize, expression level of FabHLH80 shows significant difference from S4 to S7 for three varieties. Such a mode leads us to the conclusion that the FabHLH80 may inhibit the anthocyanin biosynthesis. Based on those observations and our more extensive data on expression patterns of the 7 previously selected bHLH genes, it is shown that they are indeed related to the anthocyanin biosynthesis.
Transcript patterns of the FabHLHs genes’ response to hormone treatment
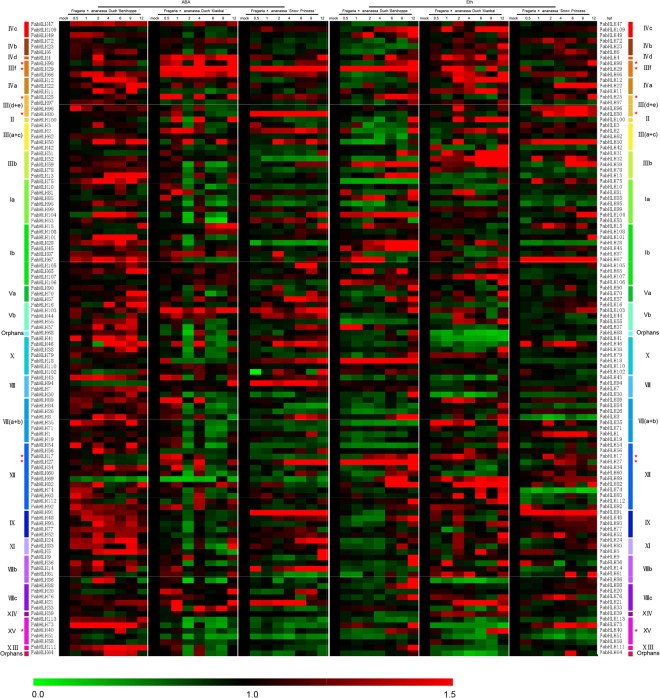
Regarding to the fact that both ABA and Eth are critical plant hormone involved in the plant response to abiotic stress at the fruit ripening9,46,47, we further investigated responding transcript patterns of 113 FabHLH genes for the three varieties under the treatment of either ABA or Eth (Figs 6; 7; S4; S5). With the implement of ABA, numbers of responsive FabHLH genes from ‘Benihoppe’, ‘Xiaobai’ and ‘Snow Princess’ are 62, 47, and 43, respectively, in which 35 shared genes are founded for all the three. In parallel, numbers of responsive FabHLH genes from ‘Benihoppe’, ‘Xiaobai’ and ‘Snow Princess’ under the exposure to Eth are 67, 75 and 57, respectively, with a shared number of 34 for the three. For the two treatments, 25 genes are discovered to be simultaneously responsive for the three varieties. For example, the expression level of FabHLH29 from IIIf subfamily strikingly increases at the initial stage (0.5 hpt (hour post treatment) to 2 hpt) and maintains a high value afterwards in response to ABA treatment for ‘Xiaobai’ and ‘Snow Princess’, while it decreases thoroughly under the ABA treatment for ‘Benihoppe’. When subjected to the Eth, FabHLH29 expresses highly for ‘Benihoppe’ and keeps relatively low yet higher than the control for both ‘Xiaobai’ and ‘Snow Princess’. In addition, expression level of FabHLH98 from IIIf subfamily is invariably high for the three varieties under both treatments compared with the control: the increase of it is significantly induced at early stages (0.5 hpt to 2 hpt), and it reaches the peak at later stages (4 hpt to 9 hpt) in response to the ABA treatment for ‘Benihoppe’ and ‘Xiaobai’. However, it is induced and starts to reach its maximum from 6 hpt to 9 hpt in response to ABA treatment for ‘Snow Princess’; under the treatment of Eth, FabHLH98 ‘s expression is induced and begins to reach the peak at later stages (4 hpt to 12 hpt) for the three varieties. Besides, bHLH genes from III(d + e) subfamily are realized to be responsive to both treatments for the three varieties as well. This finding demonstrates that subfamilies of III(d + e) and IIIf might be involved in the fruit ripening and plant response to abiotic stress.
Figure 6.
Transcript accumulation patterns of 113 bHLH genes for the three strawberry varieties under hormone stress (ABA and Eth). FvActin, FvRib413 and FvGAPDH2 were used as an internal control. The transcript accumulation profiles were generated by semi-quantitative PCR and were visualized as heat maps. The color scale represents the relative transcript level with increased (red) and decreased (green) transcript abundance. The FvbHLH genes marked by red asterisk indicate their candidacy in the anthocyanin biosynthesis.
Figure 7.
qRT-PCR transcription analysis of seven selected FabHLH genes during fruit development and ripening stages, and them under either ABA or Eth treatment for the three strawberry varieties. FvRib413 is used as an internal control. The experiments were repeated three times and gave consistent results. The mean values and SDs were obtained from three biological and technical replicates. Different letters indicate the statistical difference among samples at P ≤ 0.01 and P ≤ 0.05 (fruit development and ripening), and P ≤ 0.01 (ABA and Eth treatments) according to Duncan’s multiple range test.
Network interaction analysis of FabHLHs response to anthocyanin biosynthesis and hormone stress
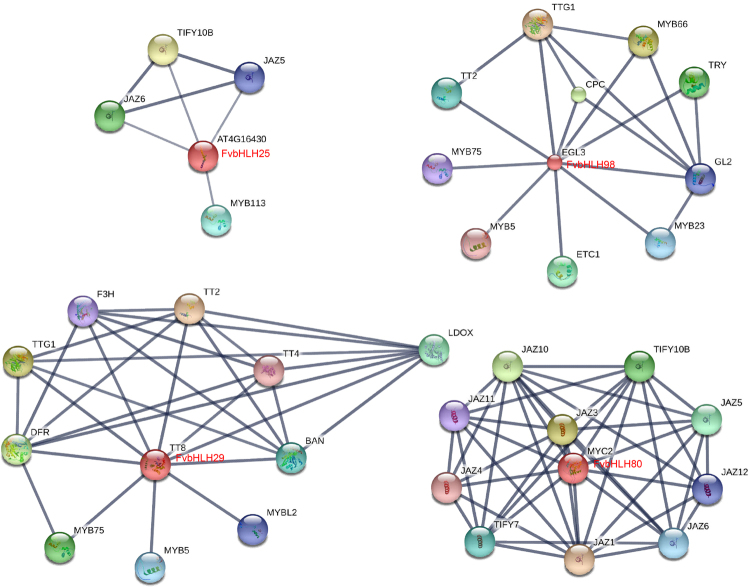
The above results argue that 7 FabHLH genes are highly possible to be involved in the anthocyanin biosynthesis and hormone response pathway for strawberry as a result of the interaction between bHLH and other proteins. Network interaction analysis has been recently demonstrated to be a powerful method to study the gene function. Online software of STRING 10 is used to reconstruct the interaction network of the 7 FvbHLH based on the orthologous gene of Arabidopsis. Only 4 bHLHs (FvbHLH25, FvbHLH29, FvbHLH80, and FvbHLH98) are proved to be able to predict the interacting proteins (Fig. 8; Table S2). According to the database of STRING 10, they are involved in the control of flavonoid pigmentation, epidermal cell fate specification and regulation of ABA-inducible genes under drought stress conditions. As is shown in Fig. 8; Table S2, FvbHLH25 (homologous to AT4G1640 for Arabidopsis) can be associated with MYB113, which could combine with several bHLH proteins in the anthocyanin biosynthesis48. Besides, FvbHLH25 also interacts with JAZ5 (JASMONATE ZIM-Domain 5) and JAZ6, which are the repressor of jasmonate response. FvbHLH29 (homologous to TT8 for Arabidopsis) can interact with MYB75, which promotes the synthesis of anthocyanin biosynthesis by activating the expression of DFR (dihydroflavonol-4-reductase) such that it is eventually involved in the control of flavonoid pigmentation. Moreover, FvbHLH80 (homologous to MYC2 for Arabidopsis) could react with MYB2 in the regulation of ABA-induced genes under drought stress conditions, as well as with MYC3 and MYC4 in the control of subsets of JA-dependent responses. In addition, FvbHLH98 (homologous to EGL3 for Arabidopsis) participates in the anthocyanin accumulation in Arabidopsis1,48,49 and tomato21. These results show that 4 FvbHLHs are involved in the fruit ripening and hormone response pathway25,34,38,47,50.
Figure 8.
Interaction network analysis of bHLH proteins identified for strawberry and related genes for Arabidopsis. Line thickness is related to the combined score (FvbHLH25 score >0.7, the others score >0.9). The homologous genes of strawberry are in red.
Discussion
With the functionality being the transcription, bHLH family are involved in the regulatory process of fruit ripening, hormone signaling and abiotic stress12. In the past few decades, features and functions of the bHLH gene family have been identified and investigated for several plant species3,8,12. Though as one of the most important horticultural crops grown worldwide providing ingredient for processed foods like jams and juices, strawberry has been barely studied for its bHLH family, who participates in the anthocyanin biosynthesis in the fruit ripening. Very few bHLHs have been investigated for the strawberry, such as FabHLH338, FaSPT (spatula)40 and FvbHLH3339. In the present study, we first identified a total of 113 bHLH genes based on the F. vesca genome (Table 1 and Fig. 1), and further implemented their bioinformation analysis (Figs 2; 3; S2) followed by the expression pattern classification during the fruit ripening under hormone treatments for three varieties (Figs 5; 6; 7).
With the rapid development of bioinformation analysis, the information stored in various genomes can be decoded to elucidate mechanisms that regulate fruit ripening and response to abiotic stress4. We firstly identified 113 unique bHLH proteins using the conserved motif of bHLH by filtering candidate genes according to the criteria described by Sun et al.3. Next, based on the phylogenetic analysis of FvbHLH, the selected FvbHLHs were classified into 26 subfamilies (Fig. 3) with the methodology similar to the classification of Arabidopsis (26 subfamilies), tomato (26 subfamilies) and Chinese cabbage (26 subfamilies)2–4,13. Moreover, the analysis of motif and gene structure is performed to gain evidence to support phylogenetic relationship for gene families.
Most bHLH proteins identified so far are mostly functionally characterized for Arabidopsis and tomato, with the revealing of their effects on the regulation of plant development, fruit ripening, anthocyanin biosynthesis and hormone signaling responses6,16. Those results prove that transcript pattern of a gene is closely related to its function, based on which we designed to examine the expression patterns of 113 FvbHLH genes from tissues, at fruit ripening stage, as well as those under the treatment of hormone (Figs 5; 6; 7). We discover that the expression patterns for the 78 out of the 113 genes from various tissues for the three varieties are similar to each other. To comprehensively understand the role of bHLH genes on the anthocyanin biosynthesis, RT-PCR and qRT-PCR analyses for the three varieties with different fruit flesh and skin colors were performed (Figs 4; 5B; 6; 7). 7 FabHLHs are found to be highly responsive for the anthocyanin biosynthesis depending on their different expression levels: FabHLH17, FabHLH25, FabHLH27, FabHLH29, FabHLH40, FabHLH80, FabHLH98. For example, the expression level of FabHLH27 is high for both ‘Benihoppe’ and ‘Xiaobai’ (red or pink skin) at the later stages (S5 → S7), while it stays low for ‘Snow Princess’ (white skin) at the similar stage S5. This implies that this gene is involved in the anthocyanin biosynthesis of fruit skin.
It has been reported that IIIf subfamily matters for the fruit color formation. Hereby, we focus on the 2 out of the 7 candidate FabHLHs that fall into the IIIf subfamily: FabHLH29 and FabHLH98. We found that FabHLH29 is relevant to the anthocyanin biosynthesis according to its expression pattern during the fruit ripening for the three varieties. Besides, gene sequence of FabHLH29 is highly similar to that of AtTT8 (AtbHLH42), which has been reported to be involved in anthocyanin biosynthesis1,6,15. Moreover, the FabHLH29 also is responsive to both the ABA and Eth treatments, thought with certain difference (down-regulated for ‘Benihoppe’ under ABA treatment, up-regulated for rest cases), for the three varieties. More evidence for the involvement FabHLH29 in the anthocyanin biosynthesis comes from the interaction network. Proteins (F3H (Flavanone 3-hydyroxylase), DFR, TTG1 and MYB), located in the pathway of anthocyanin biosynthesis, are predicated to interact with FabHLH29 (AtTT8) (Fig. 8). Researchers have realized that the TT8 from subfamily IIIf is active in regulating the synthesis of anthocyanin and proanthocyanidin for Arabidopsis1,6,50,51 by forming a stabilized MBW complex with TT2 and TTG1, and it is involved in the anthocyanin biosynthesis for rice as well22. We also find that the expression pattern of FabHLH98 (homologous to EGL3) shows no significant difference during the fruit ripening for ‘Benihoppe’, ‘Xiaobai’, and ‘Snow Princess’, which denies the participation of FabHLH98 in the anthocyanin biosynthesis. However, FabHLH98 is responsive to the abiotic stress with the implement of ABA and Eth, which seems to suggest its involvement in the fruit ripening. What’s more, analysis of interaction network of FabHLH98 demonstrates that it also plays a role in the activation of anthocyanin biosynthesis, possibly with MYB75/PAP1, inconsistent with previous results from the analysis of expression pattern during the fruit ripening in this study, yet in good agreement with the precursor reports1,6,27. In brief, expression pattern analysis under hormone treatments fits well with results from the interaction network investigation for the three varieties. However, both are inconsistent with expression pattern results during the fruit ripening. Consequently, FabHLH98 is selected as the candidate gene for the study of anthocyanin biosynthesis and a further study on its precise role is still in demand.
Previous papers inform that genes from bHLH subfamily III(d + e) take part in JA signal pathway, resulting into the regulation of plant defense during developmental process for Arabidopsis23,25,26 and the promotion of anthocyanin biosynthesis24,27 for apple. Moreover, the function of bHLH subfamily IIId, including bHLH3, can negatively regulate JA-mediated plant defence and development13, while the function of bHLH subfamily IIIe can activate JA-induced leaf senescence25. In addition, as a repressor in the JA signaling pathway, MdJAZ can be phosphorylated by MdSnRK1.1 (Snf1-Related protein Kinases) to facilitate its 26S proteasome-mediated degradation, releasing MdbHLH3 which will bind to promoters of the anthocyanin biosynthesis genes MdDFR and MdUFGT, thus finally promotes the biosynthesis of anthocyanin and proanthocyanidin24,27. In our experiments, we find that FabHLH25 from III(d + e) subfamily might be correlated with the anthocyanin biosynthesis of fruit flesh (Figs 5B, 7) from the analysis of the expression pattern for the three varieties during their ripening. Moreover, the FabHLH25 (homologous to AT4G16430, FabHLH3 and MdbHLH3) protein strongly interact with MYB113, JAZ5 and JAZ6 proteins (Fig. 8) according to results from interaction network analysis, in consistent with the known knowledge that FabHLH25 is able to interact with MYB and form the MBW complex to regulate the expression of genes involved in the proanthocyanidin biosynthesis38. What’s more, it has been mentioned that MdMYC2 positively regulates anthocyanin biosynthesis by modulating the expression of positive regulators in JA signaling (MdMYB1, MdbHLH3, MdbHLH33) for the apple52. From our observation, the transcript pattern and interaction network analysis evidence that the FabHLH80 (homologous to MYC2) from III(d + e) subfamily might also be present in the anthocyanin biosynthesis. Therefore, our research hereby paves the way for further studies and understandings of bHLH genes function in the fruit ripening and anthocyanin biosynthesis for strawberry.
In conclusion, the first comprehensive and systematic analysis of strawberry bHLH transcription factors is performed. First, 113 bHLH transcription factors from the entire strawberry genomes are identified as candidate genes responsible for the anthocyanin biosynthesis and further renamed based on their chromosome distribution. Next, the selected genes are divided to 26 subfamilies according to phylogenetic analyses, gene structures and protein motifs. Third, expression patterns of 113 FabHLHs obtained during fruit development and ripening, as well as those under either the ABA or Eth treatment, suggest that seven FabHLHs (FabHLH17, FabHLH25, FabHLH27, FabHLH29, FabHLH40, FabHLH80, FabHLH98) are involved in the anthocyanin biosynthesis of strawberry fruit. Finally, results of interaction network analyses of the four FabHLH genes (FabHLH25, FabHLH29, FabHLH80, FabHLH98) reveal that bHLHs proteins might participate in the anthocyanin biosynthesis during the fruit ripening and in the hormone response pathway. This study will provide an insight into a further understanding of functions of bHLH members in the color formation for fruits.
Materials and Methods
Identification of bHLH transcription factors for strawberry
To identify bHLH transcription factors in the strawberry genome (F. vesca), we performed a search from the NCBI database (F. vesca (taxid:57918)) (https://www.ncbi.nlm.nih.gov/genome/3314). The published Arabidopsis and strawberry bHLH protein sequences were downloaded from the Plant Transcription Factor Database (http://planttfdb.cbi.pku.edu.cn/) and used as queries in BLAST-P searches with default parameters in NCBI database. To further validate all bHLH transcription factors, full-length amino acid sequences of the 166 putative candidates were verified using the CDD (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), the hidden Markov model of SMART (http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1)53,54 and InterProScan program (http://www.ebi.ac.uk/inter-pro/search/sequence-search) to confirm their completeness and the presence of bHLH domain. Details about the bHLH sequences, such as length of amino acid sequences, theoretical molecular weights (Mw) and isoelectric point (pI), were obtained from ExPASy Proteomics server (http://web.expasy.org/compute_pi/).
Bioinformatic analysis of bHLH transcription factors for strawberry
Chromosomal localization data was retrieved from NCBI Map Viewer (https://www.arabidopsis.org/mapview/). Genes were mapped to the chromosomes using MapDraw. These genes were renamed from FvbHLH1 to FvbHLH113 according to their position, from the top to bottom, on the F. vesca chromosome8,41. Multiple domain alignments of strawberry bHLH proteins and domains were performed using ClustalX 2.0.12 with default settings for obtained sequences of the FvbHLH domains, and alignment results were shown and drew by OriginPro 89. To compare the evolutionary relationship between Arabidopsis (AtbHLH) and strawberry (FvbHLH), we obtained the phylogenetic tree for bHLH proteins using MEGA5.1 with the neighbor-joining method and the following parameters: complete deletion, p-distance model and 1000 replicates of bootstrap method4,9. 26 subfamilies were identified according to the clade support values, topology of the trees, branch lengths, visual inspection of the bHLH amino acid sequences and classification of strawberry2,4,10. The online Gene Structure Display Server (GSDS 2.0, http://gsds.cbi.pku.edu.cn/) was used to investigate the exon-intron structure of the FvbHLH transcription factors based on each coding sequence (CDS) and corresponding genomic sequence. Conserved motifs in FvbHLH transcription factors were identified from the online MEME (http://meme-suite.org/tools/meme). The FvbHLH25, FvbHLH29, FvbHLH80 and FvbHLH98 protein sequences were employed as queries for the BLAST-P search in Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org/) to obtain protein sequences of AT4G16430, AtTT8, AtMYC2 and AtEGL3, respectively. Specific interaction network with experimental evidences of AT4G16430, AtTT8, AtMYC2 and AtEGL3 was constructed using online STRING 10 (http://string-db.org/) with option value >0.700 or 0.900.
Plant materials, growth conditions and treatments
Three octoploid cultivated strawberry varieties (F. ananassa Duch. ‘Benihoppe’; F. ananassa Duch. ‘Xiaobai’, the white-flesh mutant of ‘Benihoppe’; F. ananassa ‘Snow Princess’ with white fruit skin and flesh.) were used in this study (Fig. 4A). Plantlets of the three varieties were grown in the strawberry germplasm resource greenhouse of Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, Zhengzhou, Henan, China (Fig. 4A). Strawberry plantlets were transplanted into a plastic pot (diameter: 17 cm, height: 15 cm) containing soil mix (perlite: peat, 1: 4, v/v) and grown in greenhouse with temperatures ranging from 8 °C to 28 °C, relative humidity ranging from 55% to 70%, and without supplemental lighting.
To analyze transcript patterns of bHLH transcription factors, strawberry organs/tissues (roots, young leaves, mature leaves, runners, runner tips, runner with tips and one leaf, anthotaxy, flowers, small green fruit, middle green fruit, large green fruit, white fruit, initial red fruit, partial red fruit, full red fruit) were obtained from different developmental stages. Various vegetative and reproductive tissues were collected and stored at −80 °C for tissue-specific experiments. To analyze the expression level of bHLH transcription factors to different hormones, strawberry plantlets at the stage of the sixth leaf fully expanded were sprayed with ABA at 0.1 mM, Eth at 0.5 g/L, and water, respectively. Leaf samples were collected for RNA extraction at 0, 0.5, 1, 2, 4, 6, 9 and 12 hpt. Leaves with water treatment at 0 hpt were used as control. Each time for each treatment, one leaf from each of the three separate plants, thus three leaves in total, was picked up to conduct one analysis, and all treatments were performed thrice independently.
RNA preparation, semi-quantitative reverse-transcription PCR and quantitative real-time PCR Analysis
Each RNA was extracted from tissue samples using the E.Z.N.A Plant RNA Kit (Omega, China) according to the manufacturers’ instruction. RNA concentration and quality were measured by the NanoDrop 1000 (Thermo, USA). The first-strand cDNA was synthesized using the PrimerScriptTM RT reagent Kit with gDNA Eraser (TaKaRa, China) according to the manufacturers’ instruction. The concentration of cDNA was adjusted based on the strawberry housekeeping genes FvActin, FvRib413 and FvGAPDH244,46. The primers used in this study were designed by the Vector NTI software (Table S1) without any interference with the conserved region, and were amplified the product to a length of of 150 bp to 300 bp. RT-PCR reactions were performed using 2 × Taq Mix (Beijing, China) with the following parameters: annealing temperature between 53 °C and 57 °C with 32–34 cycles. The PCR products were placed on the 1% (w/v) agarose gel with GelStain (10000×) (Tiangen, China) staining and further imaged under the AlphaView SA software. Each reaction was repeated three times. The expression data from the RT-PCR were acquired, analyzed, and visualized using the software AlphaView SA and Mev 4.8.143–45. qRT-PCR was performed according to Wei et al.46. The primers were listed in the Supplemental Table S1.
Statistical analysis
Statistical analysis was performed by the Duncan’s multiple range test module in the SPSS Statistics 17.0 software. Each experiment was independently repeated at least three times. Mean values ± standard deviation of the mean (SD) were presented (Fig. 7), and least significant differences were calculated at the 5% or 1% level of probability.
Electronic supplementary material
Acknowledgements
The authors would also like to thank proofreaders and editors for help on the manuscript. This research is supported by the Central Public-interest Scientific Institution Basal Research Fund (1610192016608, 1612382017204, 1610192017716), the Natural Science Foundation of Henan Province (162300410329) and the Agricultural Science and Technology Innovation Program (CAAS-ASTIP-2017-ZFRI). We thank Menghua Zhao and Aslam Ali for language editing of this manuscript.
Author Contributions
H.Z. conceived the research. F.Z. performed all treatments with the help of P.H., G.L., X.Z., L.L., and W.W. G.L. carried out partly hormone treatment experiments. P.H., X.Z. and L.L. prepared all plant materials. H.Z. and F.Z. analyzed and interpreted the data. F.Z. wrote the manuscript, and H.Z. and J.F. revised it. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21136-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hichri I, et al. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot. 2011;62:2465–2483. doi: 10.1093/jxb/erq442. [DOI] [PubMed] [Google Scholar]
- 2.Pires N, Dolan L. Origin and Diversification of Basic-Helix-Loop-Helix Proteins in Plants. Mol Biol Evol. 2010;27:862–874. doi: 10.1093/molbev/msp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun, H., Fan, H. J. & Ling, H. Q. Genome-wide identification and characterization of the bHLH gene family in tomato. Bmc Genomics16 (2015). [DOI] [PMC free article] [PubMed]
- 4.Wang, J. Y. et al. Genome-wide analysis of bHLH transcription factor and involvement in the infection by yellow leaf curl virus in tomato (Solanum lycopersicum). Bmc Genomics16 (2015). [DOI] [PMC free article] [PubMed]
- 5.Heim MA, et al. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003;20:735–747. doi: 10.1093/molbev/msg088. [DOI] [PubMed] [Google Scholar]
- 6.Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011;66:94–116. doi: 10.1111/j.1365-313X.2010.04459.x. [DOI] [PubMed] [Google Scholar]
- 7.Kavas M, et al. Genome-wide characterization and expression analysis of common bean bHLH transcription factors in response to excess salt concentration. Mol Genet Genomics. 2016;291:129–143. doi: 10.1007/s00438-015-1095-6. [DOI] [PubMed] [Google Scholar]
- 8.Mao, K., Dong, Q. L., Li, C., Liu, C. H. & Ma, F. W. Genome Wide Identification and Characterization of Apple bHLH Transcription Factors and Expression Analysis in Response to Drought and Salt Stress. Front Plant Sci8 (2017). [DOI] [PMC free article] [PubMed]
- 9.Li XX, et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant physiology. 2006;141:1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ledent V, Vervoort M. The basic helix-loop-helix protein family: Comparative genomics and phylogenetic analysis. Genome Res. 2001;11:754–770. doi: 10.1101/gr.177001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carretero-Paulet L, et al. Genome-Wide Classification and Evolutionary Analysis of the bHLH Family of Transcription Factors in Arabidopsis, Poplar, Rice, Moss, and Algae. Plant physiology. 2010;153:1398–1412. doi: 10.1104/pp.110.153593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song XM, et al. Genome-wide analysis of the bHLH transcription factor family in Chinese cabbage (Brassica rapa ssp pekinensis) Mol Genet Genomics. 2014;289:77–91. doi: 10.1007/s00438-013-0791-3. [DOI] [PubMed] [Google Scholar]
- 14.Yan, Q. et al. The Basic/Helix-Loop-Helix Protein Family in Gossypium: Reference Genes and Their Evolution during Tetraploidization. Plos One10 (2015). [DOI] [PMC free article] [PubMed]
- 15.Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53:814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- 16.Goossens J, Mertens J, Goossens A. Role and functioning of bHLH transcription factors in jasmonate signalling. J Exp Bot. 2017;68:1333–1347. doi: 10.1093/jxb/erw440. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi Y, Ebisu Y, Shimazaki K. Reconstitution of Abscisic Acid Signaling from the Receptor to DNA via bHLH Transcription Factors. Plant physiology. 2017;174:815–822. doi: 10.1104/pp.16.01825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sorensen AM, et al. The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 2003;33:413–423. doi: 10.1046/j.1365-313X.2003.01644.x. [DOI] [PubMed] [Google Scholar]
- 19.Nemesio-Gorriz, M. et al. Identification of Norway Spruce MYB-bHLH-WDR Transcription Factor Complex Members Linked to Regulation of the Flavonoid Pathway. Front Plant Sci8 (2017). [DOI] [PMC free article] [PubMed]
- 20.Salvatierra A, Pimentel P, Moya-Leon MA, Herrera R. Increased accumulation of anthocyanins in Fragaria chiloensis fruits by transient suppression of FcMYB1 gene. Phytochemistry. 2013;90:25–36. doi: 10.1016/j.phytochem.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 21.Tominaga-Wada, R. et al. Expression and protein localization analyses of Arabidopsis GLABRA3 (GL3) in tomato (Solanum lycopersicum) root epidermis. Plant Biotechnol-Nar34, 115−+ (2017). [DOI] [PMC free article] [PubMed]
- 22.Zhou LL, Shi MZ, Xie DY. Regulation of anthocyanin biosynthesis by nitrogen in TTG1-GL3/TT8-PAP1-programmed red cells of Arabidopsis thaliana. Planta. 2012;236:825–837. doi: 10.1007/s00425-012-1674-2. [DOI] [PubMed] [Google Scholar]
- 23.Abe H, et al. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu XJ, et al. MdSnRK1.1 interacts with MdJAZ18 to regulate sucrose-induced anthocyanin and proanthocyanidin accumulation in apple. J Exp Bot. 2017;68:2977–2990. doi: 10.1093/jxb/erx150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi TC, et al. Regulation of Jasmonate-Induced Leaf Senescence by Antagonism between bHLH Subgroup IIIe and IIId Factors in Arabidopsis. Plant Cell. 2015;27:1634–1649. doi: 10.1105/tpc.15.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song, S. S. et al. ThebHLH Subgroup IIId Factors Negatively Regulate Jasmonate-Mediated Plant Defense and Development. Plos Genet9 (2013). [DOI] [PMC free article] [PubMed]
- 27.Xie XB, et al. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 2012;35:1884–1897. doi: 10.1111/j.1365-3040.2012.02523.x. [DOI] [PubMed] [Google Scholar]
- 28.Carbone F, et al. Developmental, genetic and environmental factors affect the expression of flavonoid genes, enzymes and metabolites in strawberry fruits*. Plant Cell Environ. 2009;32:1117–1131. doi: 10.1111/j.1365-3040.2009.01994.x. [DOI] [PubMed] [Google Scholar]
- 29.Yang J, et al. Identification and expression analysis of the apple (Malus x domestica) basic helix-loop-helix transcription factor family. Sci Rep. 2017;7:28. doi: 10.1038/s41598-017-00040-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darwish, O. et al. SGR: an online genomic resource for the woodland strawberry. Bmc Plant Biol13 (2013). [DOI] [PMC free article] [PubMed]
- 31.Li CM, Li J. A new variety of strawberry -Xiaobai. Shanxi Fruits. 2014;157:46. [Google Scholar]
- 32.Hawkins C, Caruana J, Schiksnis E, Liu Z. Genome-scale DNA variant analysis and functional validation of a SNP underlying yellow fruit color in wild strawberry. Sci Rep. 2016;6:29017. doi: 10.1038/srep29017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chai YM, Jia HF, Li CL, Dong QH, Shen YY. FaPYR1 is involved in strawberry fruit ripening. J Exp Bot. 2011;62:5079–5089. doi: 10.1093/jxb/err207. [DOI] [PubMed] [Google Scholar]
- 34.Jia HF, et al. Abscisic Acid Plays an Important Role in the Regulation of Strawberry Fruit Ripening. Plant physiology. 2011;157:188–199. doi: 10.1104/pp.111.177311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia HF, et al. Type 2C protein phosphatase ABI1 is a negative regulator of strawberry fruit ripening. J Exp Bot. 2013;64:1677–1687. doi: 10.1093/jxb/ert028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, Y. C. et al. Transcript Quantification by RNA-Seq Reveals Differentially Expressed Genes in the Red and Yellow Fruits of Fragaria vesca. Plos One10 (2015). [DOI] [PMC free article] [PubMed]
- 37.Kadomura-Ishikawa Y, Miyawaki K, Takahashi A, Masuda T, Noji S. Light and abscisic acid independently regulated FaMYB10 in Fragaria x ananassa fruit. Planta. 2015;241:953–965. doi: 10.1007/s00425-014-2228-6. [DOI] [PubMed] [Google Scholar]
- 38.Schaart JG, et al. Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria x ananassa) fruits. New Phytol. 2013;197:454–467. doi: 10.1111/nph.12017. [DOI] [PubMed] [Google Scholar]
- 39.Lin-Wang, K. et al. Engineering the anthocyanin regulatory complex of strawberry (Fragaria vesca). Front Plant Sci5 (2014). [DOI] [PMC free article] [PubMed]
- 40.Tisza V, Kovacs L, Balogh A, Heszky L, Kiss E. Characterization of FaSPT, a SPATULA gene encoding a bHLH transcriptional factor from the non-climacteric strawberry fruit. Plant Physiol Bioch. 2010;48:822–826. doi: 10.1016/j.plaphy.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 41.Ling, J. et al. Genome-wide analysis of WRKY gene family in Cucumis sativus. Bmc Genomics12 (2011). [DOI] [PMC free article] [PubMed]
- 42.Niu X, Guan Y, Chen S, Li H. Genome-wide analysis of basic helix-loop-helix (bHLH) transcription factors in Brachypodium distachyon. Bmc Genomics. 2017;18:619. doi: 10.1186/s12864-017-4044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo C, et al. Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J Exp Bot. 2014;65:1513–1528. doi: 10.1093/jxb/eru007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei, W. et al. Bioinformatics identification and transcript profile analysis of the mitogen-activated protein kinase gene family in the diploid woodland strawberry Fragaria vesca. Plos One12 (2017). [DOI] [PMC free article] [PubMed]
- 45.Wei W, et al. The WRKY transcription factors in the diploid woodland strawberry Fragaria vesca: Identification and expression analysis under biotic and abiotic stresses. Plant Physiol Bioch. 2016;105:129–144. doi: 10.1016/j.plaphy.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 46.Wei, W. et al. Identification and Transcript Analysis of the TCP Transcription Factors in the Diploid Woodland Strawberry Fragaria vesca. Front Plant Sci7 (2016). [DOI] [PMC free article] [PubMed]
- 47.Kumar R, Khurana A, Sharma AK. Role of plant hormones and their interplay in development and ripening of fleshy fruits. J Exp Bot. 2014;65:4561–4575. doi: 10.1093/jxb/eru277. [DOI] [PubMed] [Google Scholar]
- 48.Feyissa DN, Lovdal T, Olsen KM, Slimestad R, Lillo C. The endogenous GL3, but not EGL3, gene is necessary for anthocyanin accumulation as induced by nitrogen depletion in Arabidopsis rosette stage leaves. Planta. 2009;230:747–754. doi: 10.1007/s00425-009-0978-3. [DOI] [PubMed] [Google Scholar]
- 49.Gao CH, et al. Genome-wide identification of GLABRA3 downstream genes for anthocyanin biosynthesis and trichome formation in Arabidopsis. Biochem Bioph Res Co. 2017;485:360–365. doi: 10.1016/j.bbrc.2017.02.074. [DOI] [PubMed] [Google Scholar]
- 50.Baudry A, et al. TT2, TT8 and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- 51.Baudry A, Caboche M, Lepiniec L. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 2006;46:768–779. doi: 10.1111/j.1365-313X.2006.02733.x. [DOI] [PubMed] [Google Scholar]
- 52.An JP, et al. The molecular cloning and functional characterization of MdMYC2, a bHLH transcription factor in apple. Plant Physiol Bioch. 2016;108:24–31. doi: 10.1016/j.plaphy.2016.06.032. [DOI] [PubMed] [Google Scholar]
- 53.Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic acids research. 2015;43:D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of signaling domains. P Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.