Abstract
Although next-generation sequencing (NGS) is a robust technology for comprehensive assessment of EGFR-mutant lung adenocarcinomas with acquired resistance to tyrosine kinase inhibitors, it may not provide sufficiently rapid and sensitive detection of the EGFR T790M mutation, the most clinically relevant resistance biomarker. Here, we describe a digital PCR (dPCR) assay for rapid T790M detection on aliquots of NGS libraries prepared for comprehensive profiling, fully maximizing broad genomic analysis on limited samples. Tumor DNAs from patients with EGFR-mutant lung adenocarcinomas and acquired resistance to epidermal growth factor receptor inhibitors were prepared for Memorial Sloan-Kettering-Integrated Mutation Profiling of Actionable Cancer Targets sequencing, a hybrid capture-based assay interrogating 410 cancer-related genes. Precapture library aliquots were used for rapid EGFR T790M testing by dPCR, and results were compared with NGS and locked nucleic acid-PCR Sanger sequencing (reference high sensitivity method). Seventy resistance samples showed 99% concordance with the reference high sensitivity method in accuracy studies. Input as low as 2.5 ng provided a sensitivity of 1% and improved further with increasing DNA input. dPCR on libraries required less DNA and showed better performance than direct genomic DNA. dPCR on NGS libraries is a robust and rapid approach to EGFR T790M testing, allowing most economical utilization of limited material for comprehensive assessment. The same assay can also be performed directly on any limited DNA source and cell-free DNA.
The use of epidermal growth factor receptor (EGFR)-targeted agents has dramatically improved the clinical management of patients with lung adenocarcinomas harboring EGFR sensitizing mutations. However, after initial and often dramatic responses, patients inevitably progress due to the development of resistance mechanisms. The best characterized of these mechanisms is the secondary EGFR T790M mutation that is detected in approximately 60% of patients.1, 2 Rarely, secondary mutations in other genes, such as BRAF, are detected.3 MET or ERBB2 amplification have also been associated with the development of resistance in small subsets of patients,4, 5 but up to one-third of patients exhibit acquired resistance through mechanisms that are yet to be defined.
Identifying the mechanism underlying secondary resistance is critical to the further management of the patient.6 The detection of the T790M mutation is the most crucial in current practice, given the availability of third-generation EGFR inhibitors active in the presence of this mutation, such as osimertinib and rociletinib.7, 8, 9 Further assessment for resistance mechanisms beyond T790M is important in the development of other treatment strategies and to better understand the biology of these tumors. The need to detect both mutations and amplifications in these resistance samples make hybrid capture-based next-generation sequencing (NGS) assays especially attractive.
In the clinical resistance setting, tumor samples are often scant and insufficient for multimarker evaluation based on single gene or low multiplexed assays. Although comprehensive biomarker testing using NGS methods is an ideal approach in such cases, their value in the assessment of T790M status, specifically, may be limited by their often longer turnaround time and analytical sensitivities that may be insufficient in this setting. The T790M mutation may be present at subclonal levels, with allele fractions sometimes <5% of tumor cells.10, 11 Resistance samples may also harbor a higher proportion of nonneoplastic cells than untreated primary tumors, further hampering the detection of an already subclonal event.12
From current guidelines,13 assays for detection of a T790M should achieve an analytical sensitivity sufficient to detect as few as 5% of T790M-positive cells (analytical sensitivity of 2.5% for a heterozygous mutation) and a maximum turnaround time of 10 working days from receipt of the tissue. Rapid single gene assays for detection of EGFR T790M are therefore broadly used in clinical laboratories, but this often leads to exhaustion of already limited material, precluding assessment for other known resistance mechanisms.
To address these issues and to optimize tissue utilization for comprehensive NGS testing, we validated a high sensitivity picodroplet digital PCR (dPCR) assay to rapidly and accurately detect the T790M mutation on the same NGS libraries prepared for comprehensive profiling. This method can also be used on DNA samples from limited material or very low tumor content, which are unsuitable for other testing, and on cell-free plasma DNA.
Materials and Methods
Samples
Seventy-one EGFR-mutant lung adenocarcinoma samples submitted for routine mutation analysis at the time of clinical progression on an EGFR tyrosine kinase inhibitor (TKI) were identified. Samples were received as sections from formalin-fixed, paraffin-embedded (FFPE) tumor tissue (10 to 20 unstained sections, 5 μ each with corresponding hematoxylin and eosin-stained slide). Manual macrodissection to ensure >10% tumor was performed as needed, and DNA was extracted using standard laboratory protocols. Total DNA yield was measured using the Qubit dsDNA HS assay (Life Technologies, Carlsbad, CA). Libraries of DNA samples extracted from non-lung adenocarcinoma cases and normal blood samples were selected as negative controls.
NGS Library Preparation
Memorial Sloan-Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) library construction was performed using 50 to 250 ng of DNA according to previously published protocols.14 Briefly, genomic DNA was sheared, and pre-enrichment sequencing libraries were prepared using the KAPA HTP protocol, including end repair, A-base addition, ligation of Illumina sequence adaptors followed by 12 cycles of PCR amplification, clean-up, and quantification. Aliquots of the barcoded libraries were dedicated for rapid T790M testing. The remaining libraries were pooled, captured, and sequenced on an Illumina HiSeq 2500 system (Illumina, San Diego, CA) using the MSK-IMPACT assay designed for targeted sequencing of all exons and selected introns of 410 genes as previously described.14
dPCR Assay
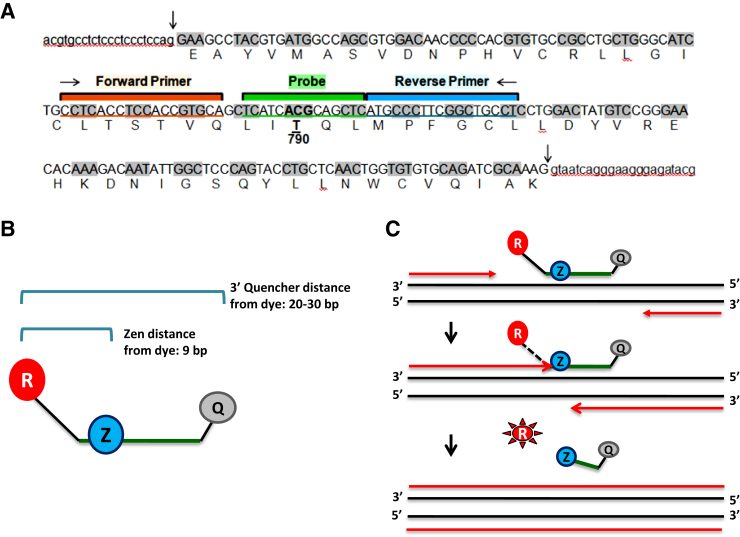
Rapid testing for T790M was performed using the Raindrop dPCR system (RainDance Technologies, Billerica, MA), adapting previously described protocols.15 The design of the assay is summarized in Figure 1. The sequences of the forward and reverse PCR primers were 5′-CCTCACCTCCACCGTGCA-3′ and 5′-AGGCAGCCGAAGGGCA-3′, respectively. Probes were designed with standard 5′ fluorophores, TET or 6FAM, and a 3′ quencher IABkFQ. Each probe contained locked nucleic acid (LNA) nucleotides, denoted with ‘+’ preceding the nucleotide and an internal additional Zen Quencher as follows: 5′-TET/T+CATC+A+C+GC/ZEN/A+GCTC/-3′ IABkFQ for the T790 wild-type probe and 5′-6FAM/T+CATC+A+T+GC/ZEN/A+GC+TC/-3′ IABkFQ for the T790M mutant probe. Small aliquots ranging from 1 to 40 ng of post-PCR cleanup NGS libraries (or 60 ng of DNA) were loaded onto the RainDrop Source instrument (RainDance Technologies) for droplet generation in the presence of TaqMan Genotyping PCR Master Mix, 1× (Applied Biosystems, Foster City, CA), droplet stabilizer 1× (Raindance Technologies), 10 μmol/L Tris-HCl, 0.25 μmol/L PCR primers flanking the EGFR codon 790 (Clinical and Commercial Manufacturing, production from EGFR T790M mutant probes), 0.1 μmol/L TET fluorescent EGFR wild-type and FAM fluorescent EGFR T790M mutant probes in a 50-μL reaction volume. PCR amplification was performed under the following conditions: 10 minutes of 95°C polymerase activation, followed by 50 cycles of 15 seconds of denaturation at 95°C, 15 seconds of annealing at 58°C, and 45 seconds of extension at 60°C, followed by polymerase inactivation at 98°C for 10 minutes. Amplified samples were loaded onto the Raindrop Sense instrument for droplet counting and fluorescence intensity readout. Data from cluster plots were analyzed using the RainDrop Analyst data analysis software version 10.0.7r2 (RainDance Technologies) using a conservative gate setting.15
Figure 1.
Assay design. A: Genomic sequence of the EGFR exon 20 with primer and probe localization. Intron sequence in lower case; exons in capital letters with corresponding amino acid under each codon; the beginning and end of the exon are marked by downward arrows. B: The probes are designed with standard 5′ fluorophores TET or 6FAM and a 3′ Q. The probe sequence contains LNA nucleotides. All probes also contain a second internal Z Q. The incorporation of an internal Z Q decreases the distance between the 5′ dye and Q and, in concert with the 3′ Q, provides greater overall dye quenching, lowering background, and increasing signal detection. C: The 5′ to 3′ exonuclease activity of the polymerase degrades the probe, physically separating the Qs from the fluorophore, which results in measurable fluorescence. LNA, locked nucleic acid; Q, quencher; R, reporter; Z, zen.
High-Sensitivity LNA-PCR Sequencing Assay
A high-sensitivity LNA-PCR sequencing assay for assessment of T790M was used as the reference method following previously described methods.1 The limit of sensitivity for this assay has been previously established at 0.1%.1
Results
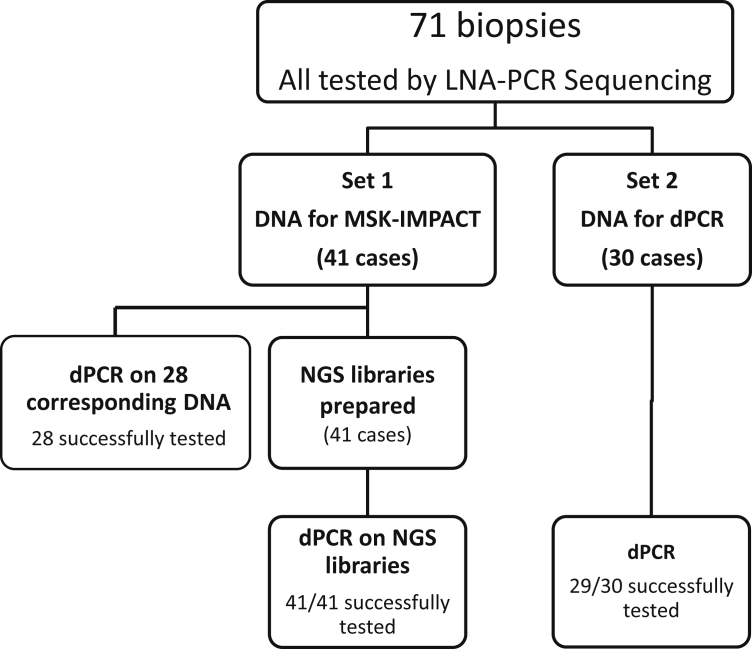
A total of 71 EGFR-mutant resistance samples were tested using the dPCR assay as detailed in Figure 2. All samples were tested upfront using the LNA-PCR sequencing assay as the reference standard to establish the T790M status. MSK-IMPACT was performed on 41 cases, and library aliquots were obtained for rapid T790M assessment by dPCR. A subset of these (n = 28) was also tested by dPCR directly from DNA for direct comparison to assess the effect of library preparation on results. Thirty additional samples, for which MSK-IMPACT testing was not requested, were assessed by dPCR directly on tumor biopsy DNA. Thirty-five non-lung, non-resistance samples (true negatives) were tested as a separate set to establish intrinsic levels of background noise and to assess potential artifacts that would limit interpretation.
Figure 2.
EGFR T790M assessment by dPCR. A total of 71 cases were tested. Initial assessment was based on a high-sensitivity LNA-PCR Sanger Sequencing assay to establish the T790M status. Cases were divided into two sets. Set 1 encompassed cases submitted for comprehensive testing by NGS. Set 2 encompassed cases in which MSK-IMPACT testing was not ordered. dPCR was performed on both sets either based on MSK-IMPACT libraries or directly from DNA. For a subset of cases in set 1 (28 cases) testing was also performed on direct DNA based on availability for paired direct comparisons. Among cases tested on the libraries, there was 100% success. For cases tested directly from DNA, one case was nonevaluable and two (from the paired set 1) were borderline. dPCR, digital PCR; LNA, locked nucleic acid; MSK-IMPACT, Memorial Sloan-Kettering-Integrated Mutation Profiling of Actionable Cancer Targets; NGS, next-generation sequencing.
Accuracy Study (Comparison with Reference Method of High Sensitivity)
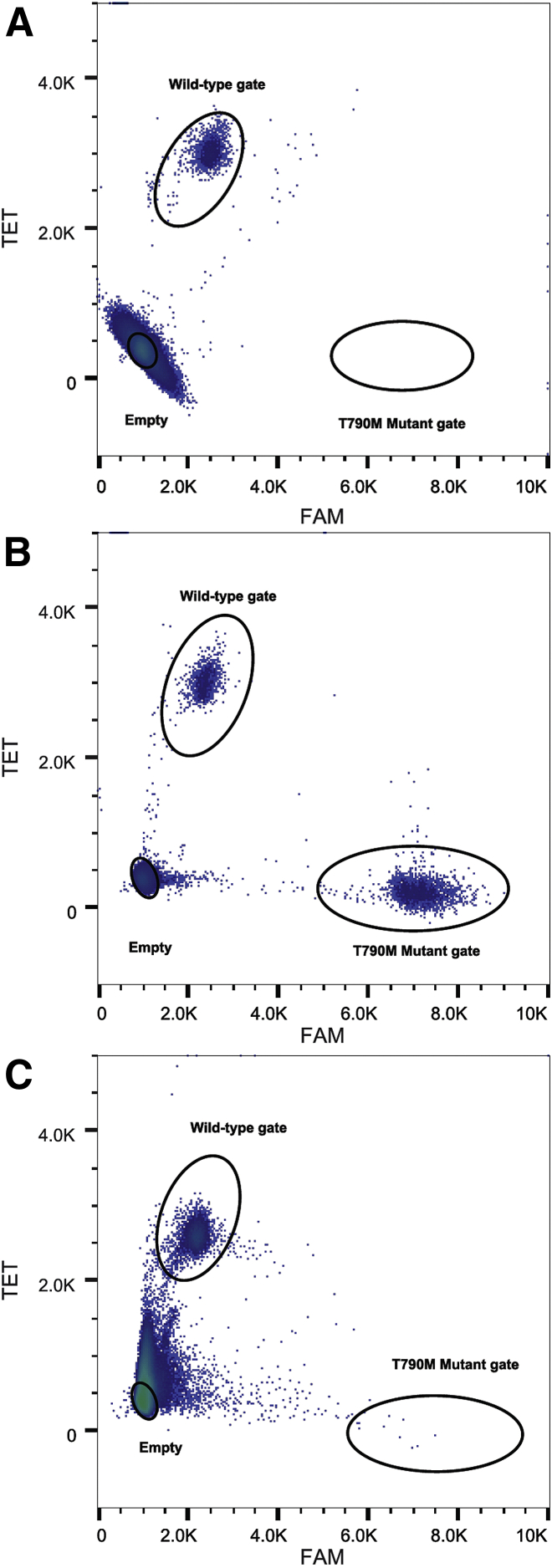
Results of the accuracy study are summarized in Table 1 and detailed in Supplemental Table S1. Significant differences were seen between the numbers of occupied droplets generated from NGS libraries compared directly with genomic DNA: 626 ng versus 166 ng, respectively. One sample tested directly from genomic DNA was not evaluable due to high background. The overall concordance between the dPCR assay and the high-sensitivity LNA-PCR sequencing among the successfully tested samples was 99% (69 of 70). Representative cluster plots of positive, negative, and equivocal/non-evaluable samples are displayed in Figure 3.
Table 1.
Summary of Samples Tested and Corresponding Results by Each Method Used
| Set | Number of cases (tests performed) | Number of occupied droplets | Result | LNA-PCR Sanger | dPCR | MSK-IMPACT |
|---|---|---|---|---|---|---|
| 1 | 41 (dPCR on libraries) | Average 626/ng (range, 129–4131) | Positive | 33 | 34 | 32 |
| Negative | 8 | 7 | 9 | |||
| Failure | 0 | 0 | 0 | |||
| 2 | 30 (dPCR on DNA) | Average 166/ng (range, 17–694) | Positive | 20 | 20 | NA |
| Negative | 10 | 9 | NA | |||
| Failure | 0 | 1∗ | NA |
dPCR, digital PCR; LNA, locked nucleic acid; MSK-IMPACT, Memorial Sloan-Kettering-Integrated Mutation Profiling of Actionable Cancer Targets; NA, not available.
Not evaluable due to high background.
Figure 3.
Representative dPCR plots. A–C: Negative (A), positive (B), and equivocal (C) representative plots from three FFPE tumor samples using 40-ng library inputs. Ovals designate the EGFR T790 wild-type (upper left), T790M mutant gate (lower right), and empty droplet gate (lower left). Negative case shows no droplets in the T790M gate. In positive samples, clusters are typically tight and exhibit little variability. Depending on the amount of background noise, droplets from outside the gate may extend into the positive T790M gate, but these are dispersed and do not form a tight cluster. These cases are interpreted as equivocal because very low levels of a mutation may be hidden by the background. These cases may be resolved by repeating the assay with higher DNA input. dPCR, digital PCR; FFPE, formalin-fixed, paraffin-embedded.
For set 1 (tested by all methods; n = 41), the variant frequency (VF) of positive samples ranged from 0.86% to 65.1%. The concordance between the three methods is summarized in Table 1 and detailed in Supplemental Table S1. One discrepant result with our high-sensitive platform of reference, LNA-Sanger sequencing, was identified on this set and was associated with a sample with low-level T790M. This case was positive by dPCR (VF of 0.86%) and negative by the LNA-PCR and NGS by initial pipeline calling. Further manual review of the NGS data confirmed the presence of EGFR T790M at a VF of 1.23%.
Comparison of VF obtained by dPCR and the NGS assay showed a high correlation in all cases (Pearson coefficient of 0.995) (Figure 4). Two cases (2 of 41) were not initially called by our pipeline, based on an established cutoff of 2%. Both mutations were detected on manual review at VF of 1.23 (described above) and 1%.
Figure 4.
Correlation between dPCR and MSK-IMPACT variant frequencies. Scatterplot showing variant frequencies for the same samples analyzed with the dPCR platform as a function of the variant frequencies obtained with the MSK-IMPACT platform shows excellent linearity with Pearson's correlation coefficient of r = 0.995. Inset shows the expansion of the area of lowest region for details. dPCR, digital PCR; MSK-IMPACT, Memorial Sloan-Kettering-Integrated Mutation Profiling of Actionable Cancer Targets.
For set 2 (samples tested only from DNA), 29 samples were evaluable (29 of 30; 97%). All 29 cases showed 100% concordance with the LNA-PCR method. Results are summarized in Table 1. One nonevaluable sample (negative by LNA-PCR method) had high background, limiting the analytical sensitivity of the assay for T790M mutation if present at low VF.
Direct Comparison of dPCR on NGS Libraries with Nonlibrary DNA
To demonstrate that the 12 cycles of random oligonucleotide-primed PCR amplification used to prepare the NGS libraries were not introducing significant genome representation artifacts in our samples, we compared the allele frequencies obtained by dPCR on libraries with dPCR on direct genomic DNA. Results are summarized in Figure 5 and Supplemental Table S2. Comparison of the number of occupied droplets generated per nanogram for each sample showed significantly higher numbers for the library (490 droplets/ng) versus direct genomic DNA (241 droplets/ng). Four samples were associated with significantly higher than expected droplet occupancy with 2.1-, 2.6-, 3.2-, and 9.8-fold increases and corresponded to high EGFR amplification. Copy number analysis by MSK-IMPACT showed corresponding increased fold-changes of 2.4, 2.8, 2.8, and 11.2, respectively. Despite differences in total occupied droplets, there was high concordance of variant frequencies.
Figure 5.
Comparison of dPCR performed on NGS libraries versus direct DNA. Twenty-eight FFPE samples were tested concurrently using dPCR on NGS libraries and direct DNA to assess assay performance. Results demonstrate that 12 cycles of random oligonucleotide-primed PCR amplification used to prepare the NGS libraries do not introduce major genome representation artifacts in our samples. A: Scatterplot shows direct comparison of T790M variant frequencies obtained on libraries and DNA and shows good correlation at all frequencies (correlation coefficient R2 of 0.98). B: Comparison of droplet occupancy for testing on DNA and libraries. Results are normalized and expressed as the percentage of expected occupancy based on the DNA input (total occupied droplets generated/expected occupied droplet count for input, ×100). Red box shows the distribution based on the NGS libraries; green displays the distribution based on testing directly from genomic DNA. One hundred percent expected occupancy corresponds to theoretical maximal yield of 308 occupied droplets/ng DNA, assuming two EGFR copies per cell. In most samples analyzed directly from genomic DNA, yield is less than expected. Preparations from libraries have higher occupancy, reflecting higher quality and more consistent amount of amplifiable DNA after barcoding and clean-up steps. Higher-than-expected occupancy is related to copy number gains. The bar at 200% expected occupancy level corresponds to a twofold increase in the number of expected EGFR droplets. Samples yielding droplets above this level (with fold increases of 2.1, 2.6, 3.2, and 9.8) are associated with EGFR amplification. Corresponding MSK-IMPACT copy number analysis for four outlier samples above the red box are associated with equivalent EGFR amplification with fold changes of 2.4, 2.8, 2.8, and 11.2, respectively. dPCR, digital PCR; FFPE, formalin-fixed, paraffin-embedded; MSK-IMPACT, Memorial Sloan-Kettering-Integrated Mutation Profiling of Actionable Cancer Targets; NGS, next-generation sequencing; VF, variant frequency.
Establishing Assay Sensitivity and Limit of Detection
The sensitivity and limit of detection were initially established based on maximum library inputs of 40 ng. Nine serial dilutions were prepared using a positive patient sample with T790M at a VF of 45.5%, based on MSK-IMPACT testing. The sample was diluted with a T790M-negative library to obtain dilutions of 50%, 25%, 12.5%, 6.25%, 3%, 1.5%, 1%, 0.1%, and 0.01% mutant sample. The T790M mutation was detected at all dilutions and showed excellent linearity over the entire range of mutation levels (Figure 6).
Figure 6.
Sensitivity study results. A: dPCR plots of sequential dilutions from 50% to 0.01%. B: Linearity at all dilution levels. Y axis was calculated T790M variant frequencies are log10 transformed. X axis is dilution levels on the x axis: 1 is undiluted, 2 (50%), 3 (25%), 4 (12.5%), 5 (6.25%), 6 (3.13%), 7 (1.56%), 8 (1%), 9 (0.1%) to 10 (0.01% dilution). dPCR, digital PCR; VF, variant frequency. Ovals designate the EGFR T790 wild-type (upper left), T790M mutant gate (lower right), and empty droplet gate (lower left).
To establish the minimum input required to maintain an analytical sensitivity of at least 1%, decreasing amounts of a positive control library with known T790M at a VF of 1% were also studied. Results from total inputs of 80, 40, 20, 10, 5, 2.5, 1.25, and 0.625 ng of library DNA showed accurate detection of the mutation with inputs as low as 2.5 ng (Supplemental Figure S1). Below this level, a cluster is difficult to appreciate, and droplets appear at the level of the background.
Establishing Cutoffs for Inherent Background Noise and Low-Frequency Errors
To further assess the signal-to-noise limitations of the assay, we analyzed a wide range of negative control library samples. Twenty-three FFPE tumors (non-lung cancer malignancies) known to be negative for any EGFR mutations and TKI naive were selected. Cases encompassed a wide range of histologies with variable levels of necrosis and differences in fixation protocols in an effort to further assess potential artifacts that could interfere with interpretation. Twelve normal blood samples were also included for comparison. Blood samples showed limited noise with occasional scattered droplets within the T790M-positive gate and corresponding positive frequencies of up to 0.02%. By contrast, paraffin-embedded tissue samples showed a higher noise pattern with up to 17 droplets in the region of the T790M-positive gate with corresponding positive frequencies of up to 0.2%. In all cases, analysis of the plots showed no distinct cluster in the region of the gate but rather dispersed droplets, and, in virtually all, these could be identified as direct extension from areas of high background outside the gate (Supplemental Figure S2).
Repeat testing of cases with high background, demonstrated a change in the pattern from run to run following a dispersed distribution that would not cluster equally in the region of the gate (Supplemental Figure S3). From this set of samples, the inherent background level for FFPE tissue was set at 0.099%. Rules for unequivocal positive calls were established, requiring at least five droplets forming a tight cluster in the T790M-positive gate and accounting for at least 0.1% of the total events.
Interassay and Intra-Assay Reproducibility
Assay precision (intra-assay variability) was evaluated by determining the variability within a single run. Measurements were performed by testing eight independent libraries (four positive and four negative) from FFPE tissue in triplicate. Positive libraries were chosen to represent the high, intermediate, and low end of the assay dynamic range using 40 ng of total library input for each. The same eight samples were used to determine the assay reproducibility (interassay variability). All runs showed excellent interassay and intra-assay reproducibility. Results are summarized in Supplemental Figure S4 and Supplemental Tables S3 and S4.
Discussion
With the development of agents specifically targeting the EGFR T790M mutation, establishing rapid and accurate methods to identify patients eligible for treatment has gained major clinical importance. The existence of alternate resistance mechanisms, including gene amplifications and other mutations in patients treated with EGFR TKIs, highlights the need for more comprehensive testing by hybrid capture-based NGS assays. However, the ability of such assays to establish the EGFR T790M status within current guidelines for TAT and sensitivity may be suboptimal, depending on the specific technology, assay design, and testing workflows within individual clinical laboratories.
Here, we have described our approach and clinical validation of a rapid picodroplet dPCR assay to accurately, reproducibly, and sensitively establish the EGFR T790M status of lung carcinoma patients with acquired resistance to EGFR TKIs. The assay uses small aliquots of precapture NGS libraries from FFPE tumor tissue, which are prepared upfront to assess genomic alterations through our targeted MSK-IMPACT assay, whole genome, or whole exome studies. The same assay can also be used for detection of the mutation on direct genomic DNA extracted from the tumor when libraries are not prepared or on cell-free DNA. The turnaround time is 1 day after library preparation, compared with approximately 10 days for some comprehensive NGS assays.
dPCR technology can be highly sensitive and precise for the detection of mutations in low frequency. Because amplicons are designed to cover short regions of DNA (52 bp for the T790M assay), dPCR can be used to detect mutations in highly fragmented DNA as in the case of cell-free DNA, degraded samples, or already sheared fragments in NGS library preparations. Importantly, dPCR can also provide absolute quantification with highly accurate T790M allelic frequencies, which is emerging as an important factor in predicting and monitoring response to treatment.16
The use of dPCR on NGS libraries, specifically, has been described as a way to increase the accuracy of library quantification and to enable accurate balancing of the libraries.17, 18, 19, 20, 21 Although it can also be used to validate NGS findings in the research setting, to our knowledge, this is the first report of the use of dPCR on NGS libraries as a clinically validated assay for T790M detection.
As with any other high-sensitivity technique, the clinical validation of the assay had as a main purpose for the establishment of stringent criteria to ensure high accuracy of results. The use of highly sensitive assays for the detection of EGFR T790M has been historically problematic because such assays are more prone to false-positive results. In samples with low DNA template, for example, artifacts of amplification can be easily detected and are hard to distinguish from a true positive. Importantly, for the FFPE tissue, studies have also shown that the process of formalin fixation itself can lead to rare but widespread DNA sequence alterations that can then be detected by highly sensitive techniques as low-level mutations.22, 23, 24 Although dPCR circumvents some of these issues due to partitioning, we find that FFPE tissue samples are more often associated with higher inherent background levels than plasma and other non-FFPE tissue. This consequently limits the sensitivity of the assay and could lead to false-positive calls if stringent interpretation criteria are not applied. The inherent noise level can vary depending on fixation protocols, cycle conditions, and instrumentation and can be improved but probably not entirely abolished. Establishing conservative rules for unequivocal positive calls in these samples is important and must be specific to each laboratory based on a wide selection of the types of samples received for routine testing.
An important advantage of dPCR rests on absolute quantification and the ability to individually calculate the sensitivity for each sample based on the amplifiable copies. Although dilution studies of a representative tumor sample with normal DNA are routinely used across laboratories for assessment of sensitivity, in practice, the level of sensitivity that can be attained for each sample in a run can be highly variable, because it is influenced by several factors, including the integrity of the DNA, level of background noise, and the degree of amplification of EGFR. For very low-quality samples, where fewer occupied droplets are generated, the sensitivity is markedly decreased, raising the risk of false-negative results. This information, which would not be provided by other assays, would prompt further work up of the case such as repeat testing with higher input or additional biopsy material.
Although testing for T790M by dPCR showed high accuracy and reproducibility when applied to both libraries and directly from genomic DNA, testing on libraries demonstrated better performance. As expected, library preparation leads to enrichment for higher quality of DNA through 12 cycles of PCR for barcoding and subsequent clean-up which was reflected in higher overall droplet occupancy per nanogram of DNA input in each sample. We further confirmed that, despite large differences in overall droplet occupancy, the process of library preparation has no effect on EGFR T790M VF estimations. Given the close correlation between the measured DNA concentration and the amount of amplifiable DNA from library preparations, EGFR gene amplification and copy number gains could also be derived even in the absence of an internal control for comparison. In all cases with occupancies above the 200% level, EGFR amplification was confirmed by MSK-IMPACT data and fluorescence in situ hybridization.
In conclusion, our data support the use of dPCR in conjunction with NGS technology as an ideal approach for the rapid assessment of mutations of high clinical impact, such as EGFR T790M, which allows maximal retention, and most economical use, of limited material for further comprehensive NGS-based testing.
Footnotes
Supported by institutional funds, Memorial Sloan-Kettering Cancer Center, Department of Pathology, and the Cancer Center Support grant (CCSG)/Core grant P30 CA008748.
Disclosures: M.E.A. previously received speaker honoraria from Corporation RainDance Technologies.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2016.07.004.
Supplemental Data
Minimum DNA input requirements. Ovals designate the EGFR T790 wild-type (upper left), T790M mutant gate (lower right), and empty droplet gate (lower left). Decreasing DNA inputs of a positive control library with established T790M mutation at a VF of 1% were analyzed using dPCR. Plots of the 20-, 10-, 2.5-, and 1.5-ng inputs are shown. The T790M mutation could be confidently called with inputs as low as 2.5 ng. With lower inputs, a cluster is difficult to appreciate or droplets are present at the level of background. dPCR, digital PCR; Mut, mutant; VF, variant frequency; WT, wild-type.
Establishing cutoffs for inherent background noise and low-frequency errors. Blood and FFPE tissue samples known to be TKI naive and negative for EGFR mutations were analyzed by dPCR to assess background noise. A: Boxplot showing the distribution of droplets occupying the T790M-positive gate in negative samples. In the set of 35 samples studied, blood samples (non-FFPE) show more limited background noise than FFPE tissue (23 samples). Twenty-six percent of FFPE samples show droplets in the T790M gate above the 0.05% level. Although droplets can be easily identified as background noise in most cases by simple inspection of the dPCR plot, this would interfere with assessment of a low-level mutation. B: dPCR plot of a single case from this set showing excess background and droplets directly extending into the T790M gate (0.20% of total droplets). This level of background is more common with older samples or those with high level of necrosis, as well as with cases tested directly from DNA rather than NGS libraries. dPCR, digital PCR; FFPE, formalin-fixed, paraffin-embedded; NGS, next-generation sequencing; TKI, tyrosine kinase inhibitor.
Assessing potential low-level mutations in the setting of background noise. A: Equivocal case with five droplets in the positive gate (40 ng input). Retesting with increasing inputs of 80 and 160 ng confirms the mutation is absent and the sample is negative. Note the pattern of the background noise changes and clears the T790M gate to allow a confident negative call. B: Equivocal case due to high background. Repeat testing with the same amount shows clearance of the background allowing identification of a dense cluster in the positive gate, confirming positivity of the sample.
Scatter plot showing interassay and intra-assay reproducibility. Minimal variability of values obtained for each sample in both interassay and intra-assay runs. dPCR, digital PCR.
References
- 1.Arcila M.E., Oxnard G.R., Nafa K., Riely G.J., Solomon S.B., Zakowski M.F., Kris M.G., Pao W., Miller V.A., Ladanyi M. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res. 2011;17:1169–1180. doi: 10.1158/1078-0432.CCR-10-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu H.A., Arcila M.E., Rekhtman N., Sima C.S., Zakowski M.F., Pao W., Kris M.G., Miller V.A., Ladanyi M., Riely G.J. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19:2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohashi K., Sequist L.V., Arcila M.E., Moran T., Chmielecki J., Lin Y.L., Pan Y., Wang L., de Stanchina E., Shien K., Aoe K., Toyooka S., Kiura K., Fernandez-Cuesta L., Fidias P., Yang J.C., Miller V.A., Riely G.J., Kris M.G., Engelman J.A., Vnencak-Jones C.L., Dias-Santagata D., Ladanyi M., Pao W. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci U S A. 2012;109:E2127–E2133. doi: 10.1073/pnas.1203530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bean J., Brennan C., Shih J.Y., Riely G., Viale A., Wang L., Chitale D., Motoi N., Szoke J., Broderick S., Balak M., Chang W.C., Yu C.J., Gazdar A., Pass H., Rusch V., Gerald W., Huang S.F., Yang P.C., Miller V., Ladanyi M., Yang C.H., Pao W. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takezawa K., Pirazzoli V., Arcila M.E., Nebhan C.A., Song X., de Stanchina E., Ohashi K., Janjigian Y.Y., Spitzler P.J., Melnick M.A., Riely G.J., Kris M.G., Miller V.A., Ladanyi M., Politi K., Pao W. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov. 2012;2:922–933. doi: 10.1158/2159-8290.CD-12-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sequist L.V., Piotrowska Z., Niederst M.J., Heist R.S., Digumarthy S., Shaw A.T., Engelman J.A. Osimertinib responses after disease progression in patients who had been receiving rociletinib. JAMA Oncol. 2016;2:541–543. doi: 10.1001/jamaoncol.2015.5009. [DOI] [PubMed] [Google Scholar]
- 7.Tan C.S., Cho B.C., Soo R.A. Next-generation epidermal growth factor receptor tyrosine kinase inhibitors in epidermal growth factor receptor -mutant non-small cell lung cancer. Lung Cancer. 2016;93:59–68. doi: 10.1016/j.lungcan.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Park K., Lee J.-S., Lee K.H., Kim J.-H., Min Y.J., Cho J.Y., Han J.-Y., Kim B.-S., Kim J.-S., Lee D.H., Kang J.H., Cho E.K., Jang I.-J., Jung J., Kim H.-Y., Sin H.J., Son J., Woo J.S., Kim D.-W. Updated safety and efficacy results from phase I/II study of HM61713 in patients (pts) with EGFR mutation positive non-small cell lung cancer (NSCLC) who failed previous EGFR-tyrosine kinase inhibitor (TKI) J Clin Oncol. 2015;33:8084. [Google Scholar]
- 9.Sequist L.V., Soria J.C., Goldman J.W., Wakelee H.A., Gadgeel S.M., Varga A., Papadimitrakopoulou V., Solomon B.J., Oxnard G.R., Dziadziuszko R., Aisner D.L., Doebele R.C., Galasso C., Garon E.B., Heist R.S., Logan J., Neal J.W., Mendenhall M.A., Nichols S., Piotrowska Z., Wozniak A.J., Raponi M., Karlovich C.A., Jaw-Tsai S., Isaacson J., Despain D., Matheny S.L., Rolfe L., Allen A.R., Camidge D.R. Rociletinib in EGFR-mutated non-small-cell lung cancer. N Engl J Med. 2015;372:1700–1709. doi: 10.1056/NEJMoa1413654. [DOI] [PubMed] [Google Scholar]
- 10.Kaye F.J. A curious link between epidermal growth factor receptor amplification and survival: effect of “allele dilution” on gefitinib sensitivity? J Natl Cancer Inst. 2005;97:621–623. doi: 10.1093/jnci/dji127. [DOI] [PubMed] [Google Scholar]
- 11.Engelman J.A., Mukohara T., Zejnullahu K., Lifshits E., Borras A.M., Gale C.M., Naumov G.N., Yeap B.Y., Jarrell E., Sun J., Tracy S., Zhao X., Heymach J.V., Johnson B.E., Cantley L.C., Janne P.A. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116:2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inukai M., Toyooka S., Ito S., Asano H., Ichihara S., Soh J., Suehisa H., Ouchida M., Aoe K., Aoe M., Kiura K., Shimizu N., Date H. Presence of epidermal growth factor receptor gene T790M mutation as a minor clone in non-small cell lung cancer. Cancer Res. 2006;66:7854–7858. doi: 10.1158/0008-5472.CAN-06-1951. [DOI] [PubMed] [Google Scholar]
- 13.Lindeman N.I., Cagle P.T., Beasley M.B., Chitale D.A., Dacic S., Giaccone G., Jenkins R.B., Kwiatkowski D.J., Saldivar J.S., Squire J., Thunnissen E., Ladanyi M., College of American Pathologists International Association for the Study of Lung Cancer and Association for Molecular Pathology Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Mol Diagn. 2013;15:415–453. doi: 10.1016/j.jmoldx.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Cheng D.T., Mitchell T.N., Zehir A., Shah R.H., Benayed R., Syed A., Chandramohan R., Liu Z.Y., Won H.H., Scott S.N., Brannon A.R., O'Reilly C., Sadowska J., Casanova J., Yannes A., Hechtman J.F., Yao J., Song W., Ross D.S., Oultache A., Dogan S., Borsu L., Hameed M., Nafa K., Arcila M.E., Ladanyi M., Berger M.F. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milbury C.A., Zhong Q., Lin J., Williams M., Olson J., Link D.R., Hutchison B. Determining lower limits of detection of digital PCR assays for cancer-related gene mutations. Biomol Detect Quantif. 2014;1:8–22. doi: 10.1016/j.bdq.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piotrowska Z., Niederst M.J., Karlovich C.A., Wakelee H.A., Neal J.W., Mino-Kenudson M., Fulton L., Hata A.N., Lockerman E.L., Kalsy A., Digumarthy S., Muzikansky A., Raponi M., Garcia A.R., Mulvey H.E., Parks M.K., DiCecca R.H., Dias-Santagata D., Iafrate A.J., Shaw A.T., Allen A.R., Engelman J.A., Sequist L.V. Heterogeneity underlies the emergence of EGFRT790 wild-type clones following treatment of T790M-positive cancers with a third-generation EGFR inhibitor. Cancer Discov. 2015;5:713–722. doi: 10.1158/2159-8290.CD-15-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boettger L.M., Handsaker R.E., Zody M.C., McCarroll S.A. Structural haplotypes and recent evolution of the human 17q21.31 region. Nat Genet. 2012;44:881–885. doi: 10.1038/ng.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyake K., Yang C., Minakuchi Y., Ohori K., Soutome M., Hirasawa T., Kazuki Y., Adachi N., Suzuki S., Itoh M., Goto Y.I., Andoh T., Kurosawa H., Oshimura M., Sasaki M., Toyoda A., Kubota T. Comparison of genomic and epigenomic expression in monozygotic twins discordant for Rett syndrome. PLoS One. 2013;8:e66729. doi: 10.1371/journal.pone.0066729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor S.D., Ericson N.G., Burton J.N., Prolla T.A., Silber J.R., Shendure J., Bielas J.H. Targeted enrichment and high-resolution digital profiling of mitochondrial DNA deletions in human brain. Aging Cell. 2014;13:29–38. doi: 10.1111/acel.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tewhey R., Warner J.B., Nakano M., Libby B., Medkova M., David P.H., Kotsopoulos S.K., Samuels M.L., Hutchison J.B., Larson J.W., Topol E.J., Weiner M.P., Harismendy O., Olson J., Link D.R., Frazer K.A. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat Biotechnol. 2009;27:1025–1031. doi: 10.1038/nbt.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White R.A., III, Blainey P.C., Fan H.C., Quake S.R. Digital PCR provides sensitive and absolute calibration for high throughput sequencing. BMC Genomics. 2009;10:116. doi: 10.1186/1471-2164-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams C., Ponten F., Moberg C., Soderkvist P., Uhlen M., Ponten J., Sitbon G., Lundeberg J. A high frequency of sequence alterations is due to formalin fixation of archival specimens. Am J Pathol. 1999;155:1467–1471. doi: 10.1016/S0002-9440(10)65461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quach N., Goodman M.F., Shibata D. In vitro mutation artifacts after formalin fixation and error prone translesion synthesis during PCR. BMC Clin Pathol. 2004;4:1. doi: 10.1186/1472-6890-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong C., DiCioccio R.A., Allen H.J., Werness B.A., Piver M.S. Mutations in BRCA1 from fixed, paraffin-embedded tissue can be artifacts of preservation. Cancer Genet Cytogenet. 1998;107:21–27. doi: 10.1016/s0165-4608(98)00079-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Minimum DNA input requirements. Ovals designate the EGFR T790 wild-type (upper left), T790M mutant gate (lower right), and empty droplet gate (lower left). Decreasing DNA inputs of a positive control library with established T790M mutation at a VF of 1% were analyzed using dPCR. Plots of the 20-, 10-, 2.5-, and 1.5-ng inputs are shown. The T790M mutation could be confidently called with inputs as low as 2.5 ng. With lower inputs, a cluster is difficult to appreciate or droplets are present at the level of background. dPCR, digital PCR; Mut, mutant; VF, variant frequency; WT, wild-type.
Establishing cutoffs for inherent background noise and low-frequency errors. Blood and FFPE tissue samples known to be TKI naive and negative for EGFR mutations were analyzed by dPCR to assess background noise. A: Boxplot showing the distribution of droplets occupying the T790M-positive gate in negative samples. In the set of 35 samples studied, blood samples (non-FFPE) show more limited background noise than FFPE tissue (23 samples). Twenty-six percent of FFPE samples show droplets in the T790M gate above the 0.05% level. Although droplets can be easily identified as background noise in most cases by simple inspection of the dPCR plot, this would interfere with assessment of a low-level mutation. B: dPCR plot of a single case from this set showing excess background and droplets directly extending into the T790M gate (0.20% of total droplets). This level of background is more common with older samples or those with high level of necrosis, as well as with cases tested directly from DNA rather than NGS libraries. dPCR, digital PCR; FFPE, formalin-fixed, paraffin-embedded; NGS, next-generation sequencing; TKI, tyrosine kinase inhibitor.
Assessing potential low-level mutations in the setting of background noise. A: Equivocal case with five droplets in the positive gate (40 ng input). Retesting with increasing inputs of 80 and 160 ng confirms the mutation is absent and the sample is negative. Note the pattern of the background noise changes and clears the T790M gate to allow a confident negative call. B: Equivocal case due to high background. Repeat testing with the same amount shows clearance of the background allowing identification of a dense cluster in the positive gate, confirming positivity of the sample.
Scatter plot showing interassay and intra-assay reproducibility. Minimal variability of values obtained for each sample in both interassay and intra-assay runs. dPCR, digital PCR.