Abstract
The single nucleotide polymorphism located within the IFNL3 (also known as IL28B) promoter is one of the host factors associated with hepatitis C virus (HCV) clearance by interferon (IFN)-α therapy; however the mechanism remains unknown. We investigated how IL28B gene polymorphism influences HCV clearance with infected primary human hepatocytes, liver biopsies, and hepatoma cell lines. Our study confirms that the rs12979860-T/T genotype has a strong correlation with ss469415590-ΔG/ΔG single nucleotide polymorphism that produces IFN-λ4 protein. We found that IFN-α and IFN-λ1 antiviral activity against HCV was impaired in IL28B T/T infected hepatocytes compared with C/C genotype. Western blot analysis showed that IL28B TT genotype hepatocytes expressed higher levels of IFN-λ proteins (IL28B, IL-29), preactivated IFN-stimulated gene (ISG) expression, and impaired Stat phosphorylation when stimulated with either IFN-α or IFN-λ1. Furthermore, we showed that silencing IFN-λ1 in T/T cell line reduced basal ISG expression and improved antiviral activity. Likewise, overexpression of IFN-λ (1 to 4) in C/C cells induced basal ISG expression and prevented IFN-α antiviral activity. We showed that IFN-λ4, produced at low level only in T/T cells induced expression of IL28B and IL-29 and prevented IFN-α antiviral activity in HCV cell culture. Our results suggest that IFN-λ4 protein expression associated with the IL28B-T/T variant preactivates the Janus kinase-Stat signaling, leading to impaired HCV clearance by both IFN-α and IFN-λ.
Hepatitis C is a blood-borne viral infection that is prevalent worldwide.1 Hepatitis C virus (HCV) infection can be asymptomatic for many years, and most individuals are diagnosed only when they develop chronic liver disease. Approximately 30% of individuals infected with HCV develop an acute infection and clear the virus naturally, whereas in most cases the infection becomes chronic.2, 3 Chronic HCV infection is the leading cause of end-stage liver diseases, such as liver cirrhosis and hepatocellular carcinoma, in the United States.4 The mechanism by which some individuals clear infection naturally and others develop chronic liver disease is unknown. The role of IFNL3 [also known as IL28B; type III interferon (IFN)] genetic variability is linked with spontaneous and treatment-induced clearance of HCV infection, but mechanisms for this association are not clear.3, 5 IFN plays an important role in viral clearance in the liver through binding to cell surface receptors and activation of Janus kinase (Jak)-Stat signaling, leading to the antiviral gene transcription.6 All three types of IFNs (type I, type II, and type III) have been shown to inhibit HCV replication in cell culture. Therefore, they may contribute to the mechanisms of HCV persistence and viral clearance by antiviral therapy.7 The endogenously produced IFN, during the host's innate immune response, inhibits viral replication by autocrine or paracrine mechanisms, and therefore also contributes to the success of antiviral therapy. The coordinate role of the IFN system in resolving chronic HCV infection of hepatocytes during antiviral therapy is not well understood.
Recently, the sustained virologic response of patients with HCV has improved significantly by the development of direct-acting antivirals (DAAs) compared with the previously used IFN-α and ribavirin (RBV) combination therapy.8 However, understanding the host's genetic factor and the contribution to HCV clearance still remains an important area of HCV research. An important discovery made through the genome-wide association studies in 2009 showed that genetic variation near the IFNL gene (IL28B) predicts HCV clearance with IFN-α/RBV combination therapy.3, 9, 10, 11, 12 IL28B genotype single nucleotide polymorphism (SNP) rs12979860 is located 3 kb upstream of the IFNL3 (IL28B) gene. Individuals who carry the IL28B C/C allele are two to five times more likely to achieve viral clearance by IFN-α plus RBV treatment than subjects with IL28B T/T makeup.9, 10, 11, 12 These studies concluded that the SNPs rs12979860 and rs8099917 are highly correlated with HCV clearance among Asian and European patients, whereas the rs12979860-T/T allele was found more often in African Americans, which should explain why African Americans respond less to IFN-α and RBV treatment.3, 12 Recently, Prokunina-Olsson et al13 discovered a new SNP ss469415590-ΔG/ΔG associated with production of a new protein called IFN-λ4. They found that rs12979860-T/T SNP shows a strong linkage disequilibrium with ss469415590-ΔG/ΔG and is located within the intron of IFNL4 gene. Some recent studies claim that rs12979860 SNP T/T of the IL28B gene is also associated with slower viral clearance than the rs12979860 SNP C/C, using DAA combination therapy.14, 15 The role of endogenous intrahepatic IFN production has been claimed to be associated with HCV clearance by DAA combination therapy.16 From this evidence it is anticipated that IL28B genotype prediction might decrease the duration of DAA combination therapy and reduce the cost of HCV treatment.17 The relation between the IL28B T/T genotype and HCV clearance by antiviral therapy has been confirmed by many clinical studies. The mechanism of treatment induced viral clearance in chronic HCV patients with IL28B C/C genotype has been found to be correlated with higher IL28B levels in serum,18, 19 production of IFN-λ by dendritic cells,20, 21 high IL28B mRNA expression in the lymphocytes,11, 12 and increased stability of IL28B mRNA.22 Although these results provide an explanation for improved treatment response to IFN-α and RBV treatment for favorable C/C genotypes, the mechanism for impaired intrahepatic HCV clearance among unfavorable IL28B T/T genotype patients is unknown.
Investigations in our laboratory have focused on understanding the host cellular factors responsible for HCV resistance to IFN-α with in vitro cell culture. We demonstrated that HCV infection induces endoplasmic reticulum stress and autophagy response that degrades IFN-α receptor 1 (IFNAR1) and RBV transporters, which impaired HCV clearance by IFN-α and RBV treatment.23, 24, 25 We also verified that it impaired expression of IFNAR1 in chronic liver disease and liver cirrhosis, whereas the expression of IFN-λ receptor (IFNLR) was not altered.26 This suggests that the IFN-λ axis could be playing an important role in inducing HCV clearance by antiviral therapy.27 From our understanding of HCV clearance by type I and type III IFN in cell culture models, we propose that IFN-α induces HCV clearance in the hepatocytes of IL28B C/C or T/T make up in relation to IFN-λ signaling. This study was initiated to explore the mechanisms by which IL28B genetic polymorphism contributes to the inhibition of HCV replication and translation by exogenous IFN-α and IFN-λ. Our results show that hepatocytes with the IL28B T/T genotype are associated with increased production of IFN-λ, which preactivates antiviral IFN-stimulated gene (ISG) expression and prevents the antiviral activity of exogenous IFN-α and IFN-λ.
Materials and Methods
Antibodies and Reagents
The following reagents were obtained from commercially available sources: IFN-α (EMD Merck, Billerica, MA) and IFN-λ1 (IL29; ReproTech, Rocky Hill, NJ); siRNA against IL29 was obtained from Qiagen (Germantown, TN); antibodies against Stat1, pStat1, Stat2, pStat2, eIF2a, peIF2a, PKR, pPKR, GFP, and DYDDDDK-flag were obtained from Cell Signaling (Beverly, MA); antibodies for Mxa, OAS1 was purchased from Santa Cruz (Dallas, TX); IL28B was purchased from Abgent (San Diego, CA); IL29 was purchased from Thermo Scientific (Waltham, MA); HCV-NS3 was purchased from Virogen (Watertown, MA); and IFN-λ receptor (IL-10Rβ) antibody was obtained from R&D Systems (Minneapolis, MN).
Plasmid Clones
Plasmid clones (pGL3-IL29, pGL3-IL28B) that contained the promoter region of human IL29 and human IL28B, driving transcription of firefly luciferase, was obtained from our collaborator.28 ISG promoter reporter plasmid (ISRE-Renilla luciferase) was obtained as described previously.28 Mammalian expression plasmid expressing IFN-λ3, IFN-λ1, and IFN-λ4 (pEF2-IFNλ3-FLAG, pEF2-IFNλ1-FLAG, and pEF2-IFNλ4-FLAG, respectively) was obtained from our collaborator.29 Reporter plasmid for studying HCV-internal ribosome entry site (IRES) that mediated translation of Renilla luciferase (pHCV-IRES-RLuc) is described in our earlier publication.7
PHHs
Primary human hepatocytes (PHHs) were obtained from XenoTech LLC (Kansas City, MO) and cultured with hepatocyte culture media supplemented with 10% human serum (Invitrogen, Brown Deer, WI). After 24 hours they were infected with cell culture-grown HCV (JFH-ΔV3-Rluc virus, HCV genotype 2a) with a multiplicity of infection of 0.1 with the use of a standard protocol of our laboratory.23 After 6 days of infection, PHHs were treated with either 100 IU/mL IFN-α or 100 ng/mL IFN-λ for 72 hours. After treatment, all of the uninfected, infected, and IFN-treated hepatocytes were harvested by trypsin-EDTA and subjected to RNA or DNA isolation and Western blot analysis. The success of HCV replication in the infected PHHs was assessed by the detection of positive strand HCV RNA levels by quantitative real-time RT-PCR (RT-qPCR) and Western blot analysis of HCV NS3 protein.
Liver Biopsies
The expression levels of IL28B and IL29 in HCV-infected patients was confirmed by Western blot analysis. The study was conducted after institutional review board approval from the office of the senior vice-president of Tulane University Health Sciences Center (New Orleans, LA). All of the patients were informed about the purpose of this study. Informed written consent was obtained from each patient. A total of six liver biopsy specimens from HCV-infected chronic liver disease patients (three IL28B C/C and three IL28B T/T) collected from Tulane Medical Center (New Orleans, LA) were included in this study. Biopsy samples were weighed and resuspended in RIPA buffer (10 μL for 1 mg of tissue). The liver tissues were homogenized with Bullet Blinder 5 with 3 beads (2 mm zirconium oxide) and centrifuged at speed 8 for 3 minutes (Quasar Instruments, Colorado Springs, CO). The sonicated protein lysates were clarified by centrifugation at 9000 × g for 10 minutes at 4°C. The clear supernatant fluid was then transferred to new tubes, quantified, and stored at −80°C until used.
Cell Lines
Hepatocellular carcinoma cell lines Huh7-CC, Huh7.5.1 (Pawlotsky Laboratory), PLC/PRF (ATCC, Manassas, VA), Huh 7.5 (Rice Laboratory), HLF (Wilkens Laboratory), SK-Hep1, and HepG2 cells (Wu Laboratory) were grown in Dulbecco's Modified Eagle's Medium supplemented with 10% fetal bovine serum, nonessential amino acids, sodium pyruvate, and penicillin, streptomycin, and amphotericin B (Gibco, Carlsbad, CA). The Li-7 cell line was obtained from a Japanese cell bank (Riken BioResource Center, Ibaraki, Japan), cultured in RPMI (Roswell Park Memorial Institute medium) and supplemented with 10% fetal bovine serum, penicillin, streptomycin, and amphotericin B (Gibco). Cells were maintained at 37°C with 5% CO2.
DNA Isolation and rs12979860-C/T and ss469415590-TT/ΔGIFNL4 Genotyping
Genomic DNA was isolated from PHHs and culture cells with a standard protocol described previously.30 Genetic polymorphism located near the IL28B gene (rs12979860-C/T) was determined with allelic discrimination PCR/fluorescence monitoring method, which includes the following primers: forward, 5′-CCTGTCGTGTACTGAACCA-3′; reverse, 5′-TCAGGGTCAATCACAGAAGG-3′; and probes, 5′-/56-FAM/AGGC+G+T+GAACC/3IABkFQ/-3′, 5′-/5-HEX/AGGCG+CGA+ACC/3IABkFQ/-3′ that exactly matched the SNP. The amplification was performed with polymerase mix, Light Cycler 480 from Roche (Indianapolis, IN), according to the PCR program: first cycle at 95°C for 10 minutes, followed by an additional 40 cycles of a denaturation step at 95°C for 10 seconds, then annealing at 61°C for 45 seconds, and extension step at 72°C for 5 seconds. Genetic polymorphism of the IL28B gene (ss469415590) that corresponds to IFNL4 (TT/ΔG) genotyping was performed by direct sequencing of PCR-amplified DNA with the primer forward, 5′-CATTGCCTTCCCTGGGATCCTAAC-3′, and reverse, 5′-GGACCCCTTGGGACAGGAAC-3′, as described previously.30 The amplification was performed with Q5 Hot Start High-Fidelity DNA Polymerase (New England Biolabs Inc., Ipswich, MA), C1000 Thermal cycler (Bio-Rad Laboratories Inc., Hercules, CA) and the following PCR program: first cycle at 95°C for 15 minutes, followed by an additional 40 cycles of a denaturation step at 95°C for 30 seconds, then annealing at 60°C for 30 seconds, and extension step at 72°C for 1 minute. PCR products were sequenced with primer sets described earlier.30 DNA sequence alignment was performed with National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/nuccore; GenBank accession numbers JN806226.1 and JN806227.1) and bioedit software version 7.0.9.0 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html; last accessed November 13, 2015).
RT-qPCR for Quantification of HCV RNA
The HCV RNA level in infected human hepatocytes was determined with RT-qPCR as described previously.31 Briefly, 1 μg of cellular RNA was used to amplify the 5′-untranslated region of the HCV genome, using sense (5′-TCTTCACGCAGAAAGCGTCTA-3′) and antisense (5′-CGGTTCCGCAGACCACTATG-3′) primers. The probe (5′-/56-FAM/TGAGTGTCG/ZEN/TGCAGCCTCCAGGA/3IBκFQ/-3′) labeled at the 5′ ends with a 6-carboxyfluorescein (FAM) fluorophore reporter molecule and ZEN-Iowa Black FQ double quenchers was used to reduce the background and to increase the signal (Integrated DNA Technologies Inc., Coralville, IA). The RT-qPCR assay was performed in 20 μL that contained 10 μL of iQ supermix (Bio-Rad Laboratories Inc.), 0.25 μmol/L of each primers, probe, and 4 μL of cDNA product obtained from the RT reaction. The amplification was performed with a standard program (48°C for 30 minutes, 95°C for 10 minutes, followed by 44 additional cycles); each cycle included a denaturation step at 95°C for 15 seconds, then an annealing and extension step at 60°C for 1 minute. Amplification, data acquisition, and analysis were performed with a CFX96 Real-Time instrument with CFX manager software version 3.0.1224.1015 (Bio-Rad Laboratories Inc.).
ISG Quantitation by RT-qPCR
Hepatoma cell lines were seeded at a density of 1 × 106 cells in 100-cm plates and incubated for 48 hours. The next day, cells were treated with 10 to 1000 UI/mL IFN-α or 1 to 100 ng/mL IFN-λ1 for 24 hours. Cells were harvested and lyzed, and RNA was isolated with the Pure Link RNA mini kit (Life Technology, Grand Island, NY). Total RNA (1 μg) was mixed with antisense primer, and cDNA was synthesized for 1 hour at 42°C. mRNA was reverse transcribed with a standard method established in our laboratory. The RT-qPCR assay was performed in 20 μL that contained 10 μL of iQ supermix (Bio-Rad Laboratories Inc.), 0.25 mmol/L of each primer, and 4 μL of cDNA product obtained from the RT reaction. Each reaction was run in triplicate. The oligonucleotide sequences for sense and antisense primer used for quantification of PKR forward, 5′-TGGAAAGCGAACAAGGAGTAAG-3′, reverse, 5′-CCATCCCGTAGGTCTGTGAA-3′; MxA forward, 5′-GCGGGCTGTGGATATGCTA-3′, reverse, 5′-TTTATCGAAACATCTGTGAAAGCAA-3′; OAS1 forward, 5′-AGAAGGCAGCTCACGAAACC-3′, reverse, 5′-ACAGCGAGGGTAAATCCTTGA-3′; and GAPDH forward, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′, reverse, 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′. The qPCR was performed with the CFX96 C1000 Real-Time instrument thermal cycler (Bio-Rad Laboratories Inc.). The amplification was performed with the following PCR program: first cycle at 50°C for 2 minutes, 95°C for 8 minutes, followed by additional 40 cycles. Each PCR cycle included a denaturation step at 95°C for 30 seconds, then annealing and extension step at melting temperature for 1 minute. Data acquisition and analysis were performed with CFX manager software (Bio-Rad Laboratories Inc.).
HCV-IRES Transfection and IFN-α, IFN-λ Treatment
In a previous study, we calibrated the biological activity of commercially available IFN-α and IFN-λ1 (IL29) with HCV-infected cell culture by comparing their 90% inhibitory concentration.23 HCV IRES-mediated translation was measured with a two-step transfection procedure described previously.7 IFN-α (10 to 1000 IU/mL) and IFN-λ1 (1 to 100 ng/mL) treatment was performed immediately after the transfection. After 24 hours, cells were washed with phosphate-buffered saline and lyzed, and Renilla luciferase activity was measured (Luman LB9507; EG & G, Berthold, Berlin, Germany). For Western blot analysis, cells were treated again for 30 minutes before harvesting to observe Stat proteins activation.
Western Blot Analysis
Cells were lyzed in ice-cold RIPA lysis buffer for 10 minutes. Protein concentration was determined by DCA protein assay. Samples were boiled for 10 minutes at 80°C in the presence of 1× SDS-PAGE loading buffer (250 mmol/L Tris-HCL pH 6.8, 40% glycerol, 8% SDS, 0.57 mol/L β-mercaptoethanol, 0.12% bromophenol blue). Protein (30 μg) was run on 12% SDS-PAGE and transferred into a nitrocellulose membrane (Hybond; Amersham Biosciences, Pittsburgh, PA). Membrane was blocked with blotting-grade blocker 5% (Bio-Rad Laboratories Inc.) for 1 hour then incubated with primary antibody. After overnight incubation, the primary antibody reactions were visualized with horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse IgG and the electrochemiluminescence detection system (Invitrogen, Pierce, Amersham).
siRNA Transfection
Huh-7.5.1 cells were cultured in 24-well plates up to 60% confluency in Dulbecco's Modified Eagle's Medium supplemented with 10% fetal bovine serum media without antibiotics. The next day, culture media was replaced with fresh Dulbecco's Modified Eagle's Medium with 2% fetal bovine serum, and cells were then transfected with 20 pmole of siRNA to IL29 (5′-UAAAUUAAGGAAGGUGUAGGG-3′) or siRNA control (Qiagen) with oligofectamine (Life Technology). After 24 hours cells were transfected with pHCV-IRES-Luc plasmid and then treated with 100 IU/mL IFN-α for another 24 hours. Knockdown efficiency was analyzed by Western blot analysis. IFN-α inhibition of HCV IRES was determined by the measurement of Renilla luciferase activity described in HCV-IRES Transfection and IFN-α, IFN-λ Treatment.
IFN-λ Overexpression
Huh7.5 cell line with IL28B rs12979860 C/T was seeded in 10-cm plates and infected the next day by overnight incubation with cell culture grown HCV particles at 0.1 multiplicity of infection. The next day, the infective virus was removed, and cells were cultured with growth media. After 7 days, infected cells were isolated by trypsin-EDTA and seeded in 24-well plates at a density of 2 × 104 cells per well. After 24 hours, they were transfected with different concentrations (50 ng/mL, 250 ng/mL, 500 ng/mL) of expression plasmid (pcDNA3.1, pEF2-IFNλ3-FLAG, pEF2-IFNλ1 FLAGL1, pEF2-IFNλ4-FLAG) with the Turbofect transfection reagent (ThermoFisher, Waltham, MA). After 24 hours, cells were treated with 100 IU/mL IFN-α. After 24 hours, cells were treated again for 30 minutes before harvesting to observe Stat proteins activation, then washed with phosphate-buffered saline and lyzed for Western blot analysis.
IL28, IL29, ISRE-Luciferase Promoter Activity Analysis
Huh7-CC (C/C) and Huh-7.5.1 (T/T) cells were seeded 2 × 104 cells per well in a 24-well plate. After 24 hours, cells were transfected with 1 μg of pISRE or pEGFP-Renilla plasmid (as control) and treated with either 10 to 1000 IU/mL IFN-α or 1 to 100 ng/mL IFN-λ1. After 24 hours, the cells were washed with phosphate-buffered saline, then lyzed for the measurement of Firefly luciferase (Luman LB9507; EG & G, Berthold, Berlin, Germany). First, the firefly luciferase values were normalized against Renilla luciferase and then they were normalized with 1 μg of protein. The effects of different IFN-λ cDNA expression on the promoter activity was studied in a co-transfection experiment.
Statistical Analysis
All experiments were performed three times, in triplicate. All results were expressed as means ± SEM and n = 3. Comparison between two groups was performed with a t-test. We assume that all measurements have normal probability distributions, which is expected for these types of data. P < 0.05 was considered significant using the t-test.
Results
IFN-α– and IFN-λ–Induced Antiviral Activity Is Impaired in HCV-Infected PHHs with IL28B T/T Genotype
We developed a HCV replication model in PHHs obtained from multiple donors to understand the impact of IL28B genotype distribution on HCV replication and IFN antiviral activity. PHHs obtained from seven donors were genotyped for IL28B rs12979860 and IFN-λ4 (ss469415590-ΔG/ΔG). Genotyping of PHHs indicated that the genetic variant (IFN-λ4 ΔG) that produces IFN-λ4 protein has 100% correlation with IL28B rs12979860-T/T variant (also called IFN-λ4 rs12979860) (Table 1).
Table 1.
IFN-λ4 Genotype and Characteristics of Primary Hepatocytes
| Batch of PHH | SNP rs12979860 | SNP ss469415590 | Sex | Race | Age, years |
|---|---|---|---|---|---|
| PHH-1 | C/C | TT/TT | F | W | 79 |
| PHH-2 | C/C | TT/TT | F | W | 66 |
| PHH-3 | C/C | TT/TT | M | W | 27 |
| PHH-4 | C/C | TT/TT | F | W | 62 |
| PHH-5 | T/T | −G/−G | F | AA | 45 |
| PHH-6 | T/T | −G/−G | M | AA | 59 |
| PHH-7 | T/T | −G/−G | F | AA | 54 |
F, female; M, male; AA, African American; IFN, interferon; PHH, primary human hepatocyte; SNP, single nucleotide polymorphism; W, white.
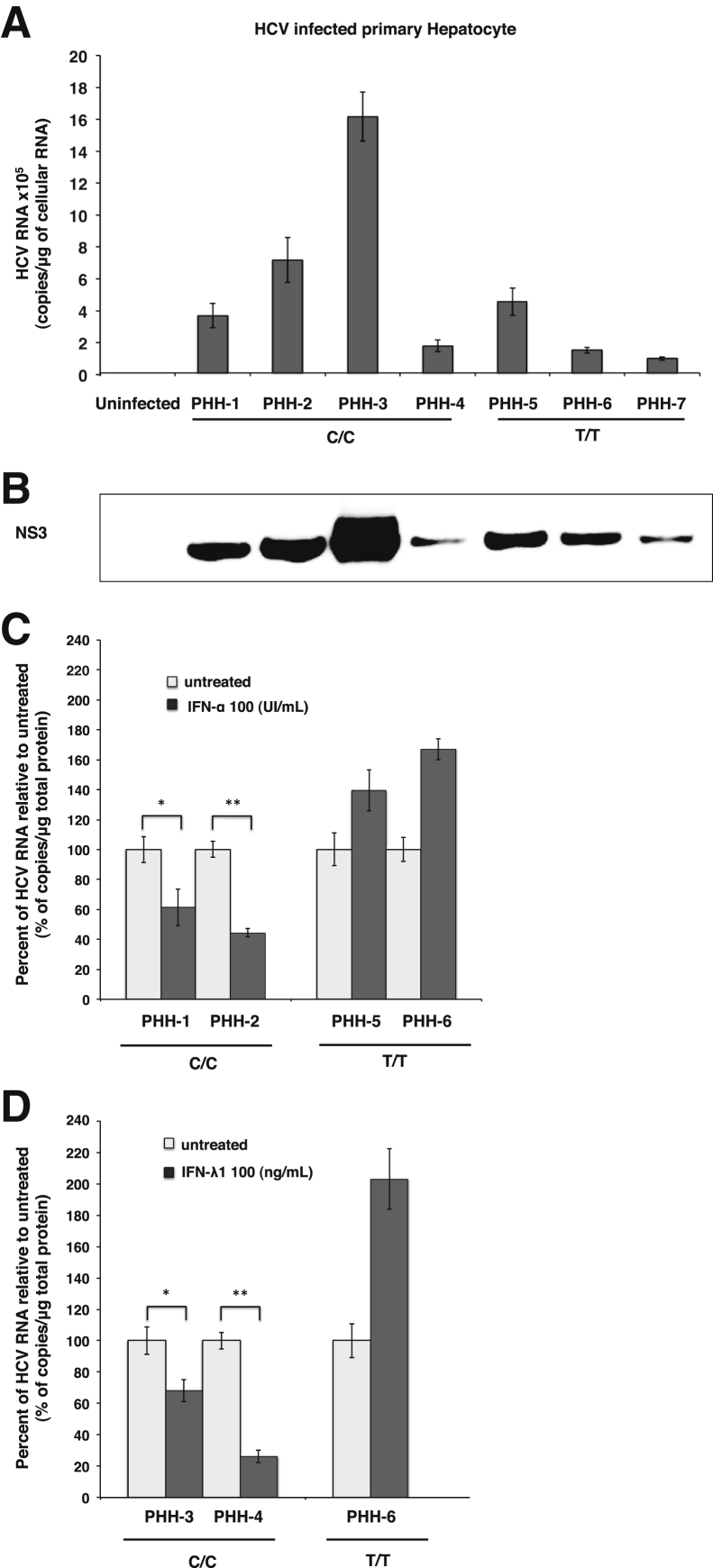
To determine whether the levels of HCV replication will differ among PHHs with IL28B C/C versus T/T, cells seeded in 6-well plates were infected with cell culture-derived HCV (genotype 2a), and HCV RNA levels between C/C and T/T genotype cells were compared on day 6 by RT-qPCR (Figure 1A). The levels of HCV replication in PHHs (108 copies/mg of cellular RNA) are comparable with chronically infected patients and chimpanzee (equivalent to 108–11 HCV genome per gram of liver tissue).32, 33 Western blot analysis was performed as an alternative method to real-time RT-PCR to confirm that HCV infection was successful in the PHHs (Figure 1B). We were unable to make a conclusion as to whether levels of HCV replication are different between C/C and T/T hepatocytes because they were not statistically significant. To determine whether there is any difference in the IFN-induced antiviral activity between C/C- and T/T-infected hepatocytes, the infected cultures at day 6 were treated with either IFN-α or IFN-λ for an additional 48 hours. The IFN-antiviral activity of infected culture was compared by the measurement of HCV RNA levels by real-time RT-PCR. The antiviral effect of IFN-α and IFN-λ1 was compared between two batches of C/C and T/T hepatocytes. Antiviral effect of IFN-α and IFN-λ1 were found to be stronger in HCV-infected PHHs of C/C genotype than T/T cells (Figure 1, C and D). Antiviral effect of IFN with an in vitro replication model of T/T PHH culture showed results consistent with clinical studies with chronic HCV patients treated with IFN-α.10
Figure 1.
IFN-α and IFN-λ induce HCV clearance in infected PHHs of IL28B rs12979860 C/C and T/T genotype. PHHs from donors were infected with culture-derived HCV supernatant fluid at a multiplicity of infection of 0.1. A: HCV RNA levels in the uninfected and infected untreated human hepatocytes at day 6, measured by RT-qPCR. B: Western blot analysis of HCV-infected PHHs from seven batches with NS3 antibody. C: Two batches of C/C and two batches of T/T culture at day 6 were treated with IFN-α for 48 hours. The HCV RNA levels between treated and untreated culture were measured by RT-qPCR. D: Two batches of C/C and only one batch of T/T culture at day 6 were treated with IFN-λ for the same time. HCV RNA levels between treated and untreated culture were compared. In all analysis, HCV RNA level was measured with 1 μg of cellular RNA, and values are normalized against untreated control. Data are expressed as means + SE. n = 7 donors. ∗P < 0.05, ∗∗P < 0.01. HCV, hepatitis C virus; IFN, interferon; PHH, primary human hepatocyte; RT-qPCR, quantitative real-time RT-PCR.
IL28BT/T Is Associated with High Basal Level of IFN-λs Protein Expression
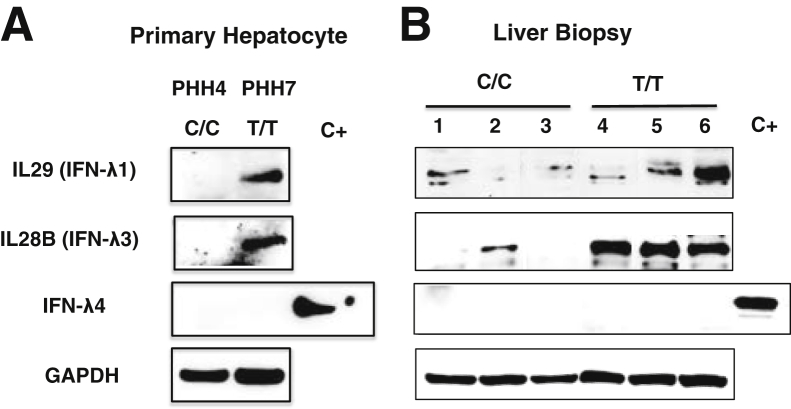
The association of IL28B genetic polymorphisms with HCV clearance provides evidence, indicating that expression of IFN-λ could be playing a major role in antiviral clearance. Thus, we investigated whether the IL28B SNPs could be related to the difference in the expression of IFN-λ (1 to 4) in primary hepatocytes and liver biopsies. The genotype distribution of liver biopsies used in this analysis is shown in Table 2. Interestingly, Western blot analysis found that the level of IFN-λ1 and IFN-λ3 proteins was higher in IL28B T/T SNP than in the C/C genotype among primary hepatocytes (representative of three batches of T/T compared with C/C) (Figure 2A) and liver biopsy (Figure 2B). IFN-λ4 protein expression was not detectable in PHHs or liver biopsies, as suggested previously.13 Western blot analyses demonstrated that the basal expression level of ISGs and IFNAR1, IFN-λs receptor (IL10Rβ) was higher in T/T hepatocytes than in C/C hepatocytes (Supplemental Figure S1). Western blot quantification of liver biopsies showed that the expression of IL29 and IL28B is high in IL28B T/T patients (Supplemental Figure S2). Taken together these results show that the IL28B T/T genotype is associated with high basal level of IFN-λs proteins, IFN receptor, and ISG expression.
Table 2.
IFN-λ4 Genotype and Characteristics of Liver Biopsies
| Patient | SNP rs12979860 | SNP ss469415590 | Sex | Race |
|---|---|---|---|---|
| 1 | C/C | TT/TT | M | W |
| 2 | C/C | TT/TT | F | AA |
| 3 | C/C | TT/TT | F | W |
| 4 | T/T | −G/−G | M | AA |
| 5 | T/T | −G/−G | M | AA |
| 6 | T/T | −G/−G | M | AA |
F, female; M, male; AA, African American; IFN, interferon; SNP, single nucleotide polymorphism; W, white.
Figure 2.
IFN-λ expression levels in PHHs and liver biopsies of chronic HCV patients with IL28B C/C and T/T genotype. A: Western blot analysis of IL-29, IL-28B, and IFN-λ4 between C/C and T/T uninfected human hepatocytes. B: Western blot analysis of IL-29, IL-28B, and IFN-λ4 in pretreatment liver biopsies obtained from three C/C and three T/T chronic HCV patients. GAPDH expression level was monitored as a loading control. HCV, hepatitis C virus; IFN, interferon; PHH, primary human hepatocyte.
Inhibition of HCV IRES-Mediated Translation of Renilla Luciferase by IFN-α and IFN-λ1 (IL29) Is Impaired in T/T Cell Lines
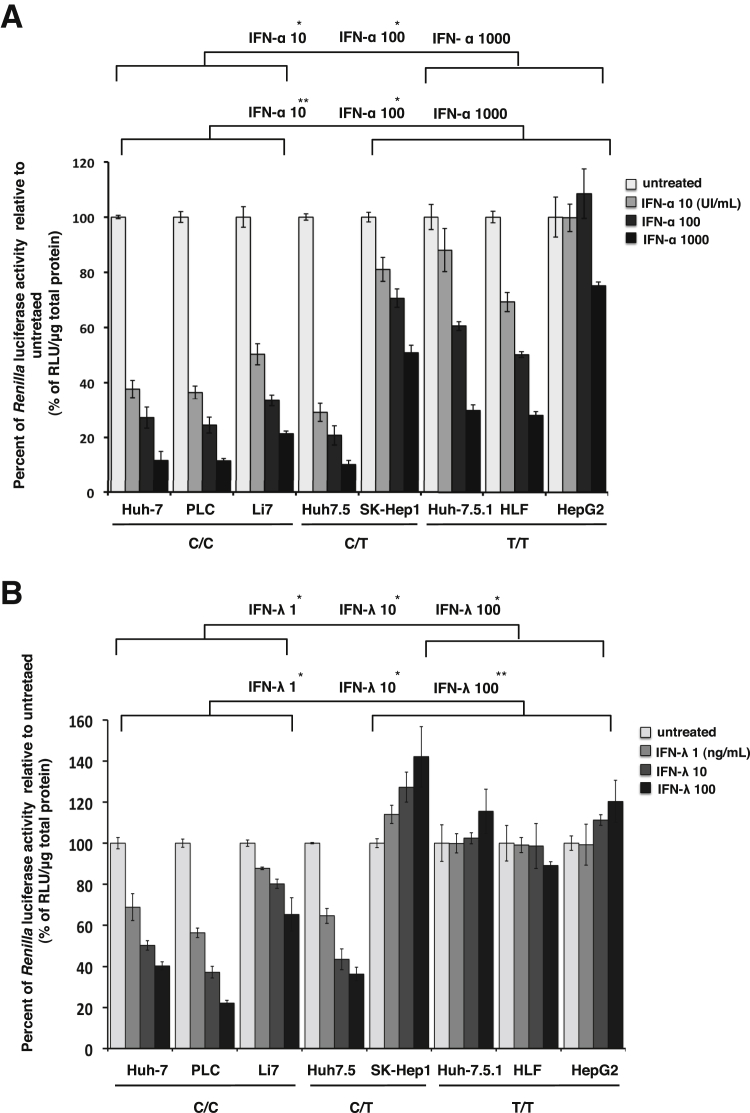
Previously, we showed that IFN-α, IFN-λ1, and RBV all inhibit HCV replication at the level of inhibiting IRES-mediated translation in hepatoma cell culture.7 In this study, we also collected panels of hepatoma cell lines and genotyped for IL28B and IFNL4 (Table 3). Because all hepatoma cell lines do not support HCV replication, we examined the effect of IFN treatment at the level of HCV mRNA translation inhibition in a transient transfection assay. Hepatoma cell lines were transfected with pHCV IRES-RLuc plasmid and then treated with equivalent concentrations of IFN-α, IFN-λ1, or RBV for 24 hours. The effect of IFN treatment on the HCV IRES mRNA translation was measured by luciferase assay. We found that IFN-α inhibits translation of HCV IRES-Renilla luciferase significantly more in a dose-dependent manner in hepatoma cell lines with the IL28B C/C than those with the T/T genotype (Figure 3A). Interestingly, IFN-λ–induced block at the IRES-mediated translation was impaired more than IFN-α in the T/T genotype cells (Figure 3B). No significant difference in the HCV IRES translation inhibition by RBV treatment was observed among hepatoma cell lines with IL28B genotype variation (data not shown). These results indicate that IL28B genotype studies that used HCV IRES clones in hepatoma cell lines also showed impaired antiviral activities of IFN-α and IFN-λ in T/T cell lines compared with C/C or C/T cell lines at the level of translation.
Table 3.
IFN-λ4 Genotype of Hepatocarcinoma Cell Lines
| Hepatocarcinoma cell lines | SNP rs12979860 | SNP ss469415590 |
|---|---|---|
| Huh-7 | C/C | TT/TT |
| PLC | C/C | TT/TT |
| Li-7 | C/C | TT/TT |
| Huh-7.5 | C/T | −G/TT |
| SK-Hep1 | C/T | −G/−T |
| Huh-7.5.1 | T/T | −G/−G |
| HLF | T/T | −G/−G |
| HepG2 | T/T | −G/−G |
IFN, interferon; SNP, single nucleotide polymorphism.
Figure 3.
IFN-α and IFN-λ mediated inhibition of HCV IRES-Renilla luciferase translation in eight different hepatoma cell lines (three C/C, two C/T, and three T/T genotype). Equal numbers of cell were seeded in 24-well plates. Next day, they were transfected with HCV IRES-luciferase plasmid and then treated with 10 to 1000 IU/mL IFN-α or 1 to 100 ng/mL IFN-λ for 24 hours. Renilla luciferase activity of HCV IRES translation between C/C and T/T was measured, and the values were normalized against untreated control and with 1 μg of cellular protein. A: Inhibition of HCV translation by IFN-α in hepatoma cell lines with different IL28B genotype. B: Inhibition of HCV translation by IFN-λ in hepatoma cell lines with different IL28B genotypes. Data are expressed as means + SE. ∗P < 0.05, ∗∗P < 0.01. HCV, hepatitis C virus; IFN, interferon; IRES, internal ribosome entry site; PHH, primary human hepatocyte; RLU, relative light unit.
IFN-α Antiviral Activity Is Blocked at the Level of PKR Activation in the T/T Cell Line
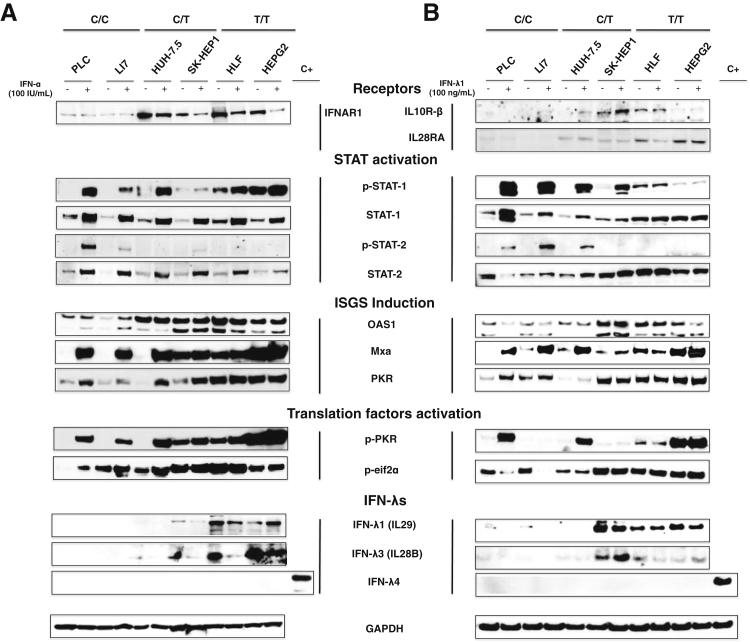
To investigate the mechanism of impaired antiviral activity of IFN-α in T/T cells, we tested the Jak-Stat pathway activation, using that panel of six cell lines. Hepatoma cell lines with different IL28B genotype were treated with increasingly equivalent concentrations of IFN-α (10 to 1000 IU/mL) for 24 hours, and the protein extracts were analyzed by Western blot analysis. It was found that IL28B T/T hepatoma cell lines show higher basal expression of IFNAR1, impaired Stat1 and Stat2 phosphorylation, ISG induction (OAS, MxA, PKR), activation of translation initiation factors (PKR and eiF2α), and increased expression of IL28B and IL29 (Figure 4A). IFN treatment induced Stat1 and Stat2 phosphorylation in all C/C cell lines, but the induction was attenuated in all T/T cell lines (Figure 4A), also observed in a dose-dependent manner (Supplemental Figure S3). It has been shown that IFN antiviral activity is associated with ISG expression induction and the translation inhibition in mammalian cells by phosphorylation of PKR and eIf2α.34, 35 We first showed that IFN-α treatment induced the activity of ISRE promotor and ISG mRNA, in a dose-dependent manner, in both the C/C cell line and the T/T cell line (Supplemental Figure S4). Then, we examined the regulation of ISG protein expression at the level of translation. Western blot analysis showed that the basal expression levels of ISGs (PKR, MxA) and the phosphorylation of PKR level were significantly higher in all hepatoma cell lines with IL28B T/T, but the IFN-α activity to induce ISG expression was attenuated in a concentration-dependent manner compared with C/C cell lines (Figure 4A, Supplemental Figure S3). Results of these analyses showed that IFN-α–induced STAT activation and ISG induction were impaired specifically in IL28B T/T cells (Supplemental Figure S5). Then we also investigated whether the IL28B SNPs were related to the difference in the expression of IFN-λ (1 to 4) proteins in the hepatoma cell lines. Interestingly, Western blot analysis found that hepatoma cell lines with IL28B T/T SNP expressed higher basal levels of IFN-λ1 and IFN-λ3 proteins than the C/C genotype. These results obtained from tumor cell lines are consistent with the results of PHHs (Figure 4A). Furthermore, IFN-α induced expression of IFN-λ1 and IFN-λ3 in the C/C cell line, whereas the expression was inhibited in the T/T cell line (Figure 4A, Supplemental Figures S3 and S5). IFN-λ4 protein was not detected by Western blot analysis in C/C or T/T genotype hepatoma cell lines. This observation was consistent with another recent study showing that IL28B induction by IFN-α was suppressed in the presence of IFN-λ4.36 Taken together, we have now confirmed that T/T genotype hepatoma cells express IFN-λ proteins and higher basal level of ISG protein. IFN-α–induced ISG induction was impaired specifically in T/T genotype cells compared with C/C cells.
Figure 4.
IFN-α and IFN-λ induce activation of Janus kinase-Stat pathway and ISG expression among two C/C, two C/T, and two T/T hepatoma cell lines. Hepatoma cells were treated with either 100 IU/mL IFN-α or 100 ng/mL IFN-λ for 24 hours. The induction of IFN-receptor, Stat phosphorylation, ISG induction, PKR activation, and IL-29, IL-28 expression were measured by Western blot analysis. A: IFN-α–treated and untreated culture. B: IFN-λ–treated and untreated culture. IFN, interferon; ISG, interferon-stimulated gene.
IFN-λ–Induced Stat1/Stat2 Phosphorylation Is Impaired in T/T Cell Lines
A similar experiment was performed to examine IFN-λ1–induced activation of Jak-Stat signaling between C/C and T/T hepatoma cell lines. Our results showed that Stat1 and Stat2 phosphorylation in T/T cell lines was impaired in T/T cell lines treated with 100 ng/mL IFN-λ1 (Figure 4B) whereby these cells showed higher level of IL10-Rβ receptors. These observations were confirmed in IFN-λ1 dose-dependent treatment (1 to 100 ng/mL) (Supplemental Figure S3). Likewise, IFN-λ1–induced ISG promoters mediated firefly luciferase expression in C/C cell lines, whereas the induction of ISRE promoter by IFN-λ1 treatment was impaired in T/T cell lines (Supplemental Figure S4). These observations were confirmed in measuring the mRNA level of PKR, MxA, and OAS1 by real-time RT-PCR assay (Supplemental Figure S4). IFN-λ1 was also unable to induce ISG translation and PKR, eiF2α phosphorylation in the T/T cell line, possibly as a consequence of Stat phosphorylation impairment (Figure 4B, Supplemental Figures S3 and S5). In conclusion, our results showed that IFN-λ1 was unable to induce Stat phosphorylation, in particularly Stat2, with the consequences of a lack of ISG mRNA induction in T/T cell line compared with C/C cells. We have shown that the IFN-λ antiviral action against HCV was predominately mediated by Stat2.37 These results may explain why the IFN-λ1 antiviral effect against HCV is impaired in T/T cell lines.
Silencing IL29 in T/T Hepatoma Cell Lines Improves IFN-α Antiviral Activity
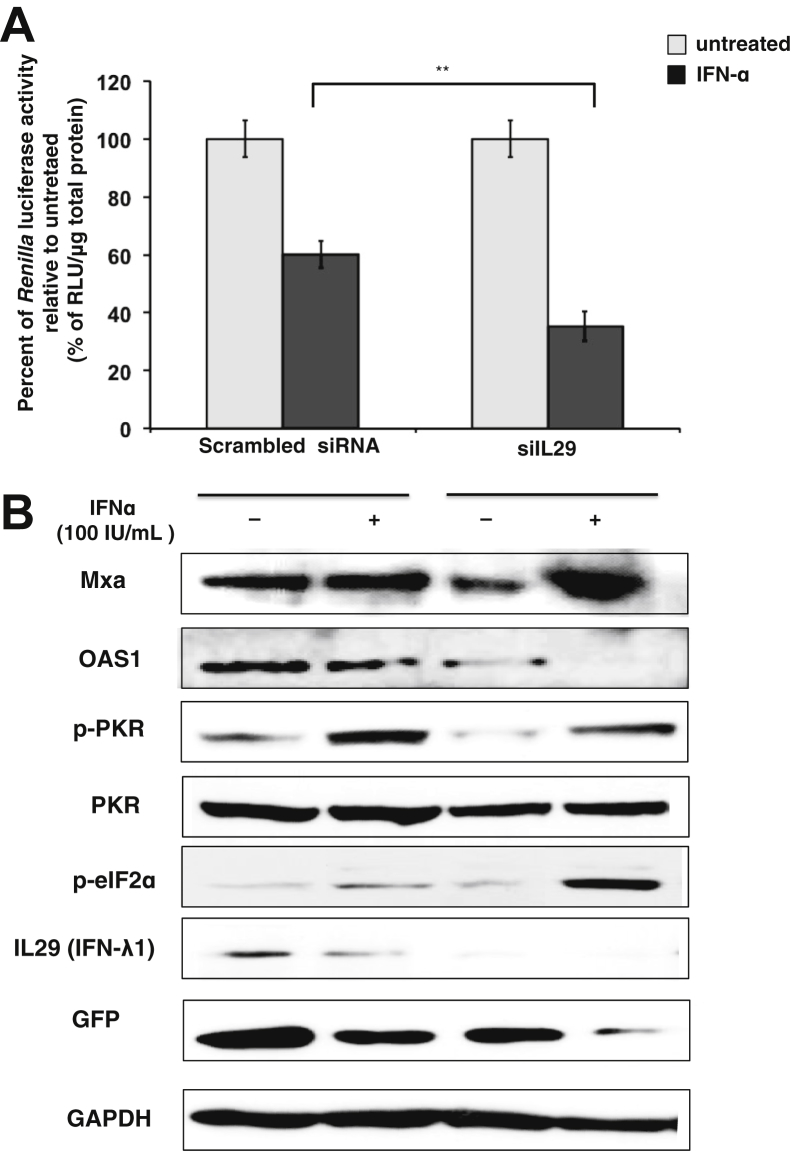
To verify the hypothesis that intracellular overexpression of IFN-λ in T/T hepatoma cells impairs IFN-α antiviral response, we measured IFN-α–mediated HCV IRES-luciferase inhibition with and without silencing IL29 (IFNL4 not being detected in Western blot analysis). Huh-7.5.1 (T/T) cell lines were transfected with siRNA to IL29 or scrambled siRNA (Qiagen) for 24 hours. Cells were then transfected with IRES-luciferase plasmid and treated with IFN-α for 24 hours. Renilla luciferase activity of cell lysate between IFN-α–treated and untreated cultures was compared. Silencing IL29 showed a significantly higher inhibition of HCV IRES-luciferase translation by IFN-α (Figure 5A). The effect of IL29 silencing on the ISG induction was compared between IFN-α–treated and untreated cell lysates after 24 hours. We observed that IL29 silencing reduced the basal expression of ISGs (PKR, MxA, and OAS1) and rescued the capacity to IFN-α to induce ISG expression (Figure 5B). Taken together, these results allowed us to establish the hypothesis that the IL28B T/T genotype was associated with high basal expression of IFN-λ, which preactivated the Jak-Stat pathway and impaired additional exogenous IFN treatment.
Figure 5.
Silencing IL-29 in a T/T cell line (Huh-7.5.1) improves the IFN-α response. A: The effect of IFN-α treatment on the expression of HCV IRES-Renilla luciferase in T/T cell after IL29 silencing. Huh-7.5.1 (T/T) cell line was transfected with siRNA to IL29 and a scrambled siRNA with lipofectamine, after 48 hours the same culture was transfected with HCV IRES-Renilla luciferase reporter plasmid and then treated with 100 IU/mL IFN-α for 24 hours. B: Western blot analysis of the cellular extract of IL29-silenced and IFN-treated culture showed the expression of ISGs and phosphorylation of translation initiation factors. GAPDH levels were monitored as a loading control. Data are expressed as means + SE. ∗∗P < 0.01. HCV, hepatitis C virus; IFN, interferon; IRES, internal ribosome entry site; ISG, IFN-stimulated gene; RLU, relative light unit.
Intracellular Expression of IFN-λ Preactivates the Jak-Stat Signaling and Inhibits HCV Protein Translation
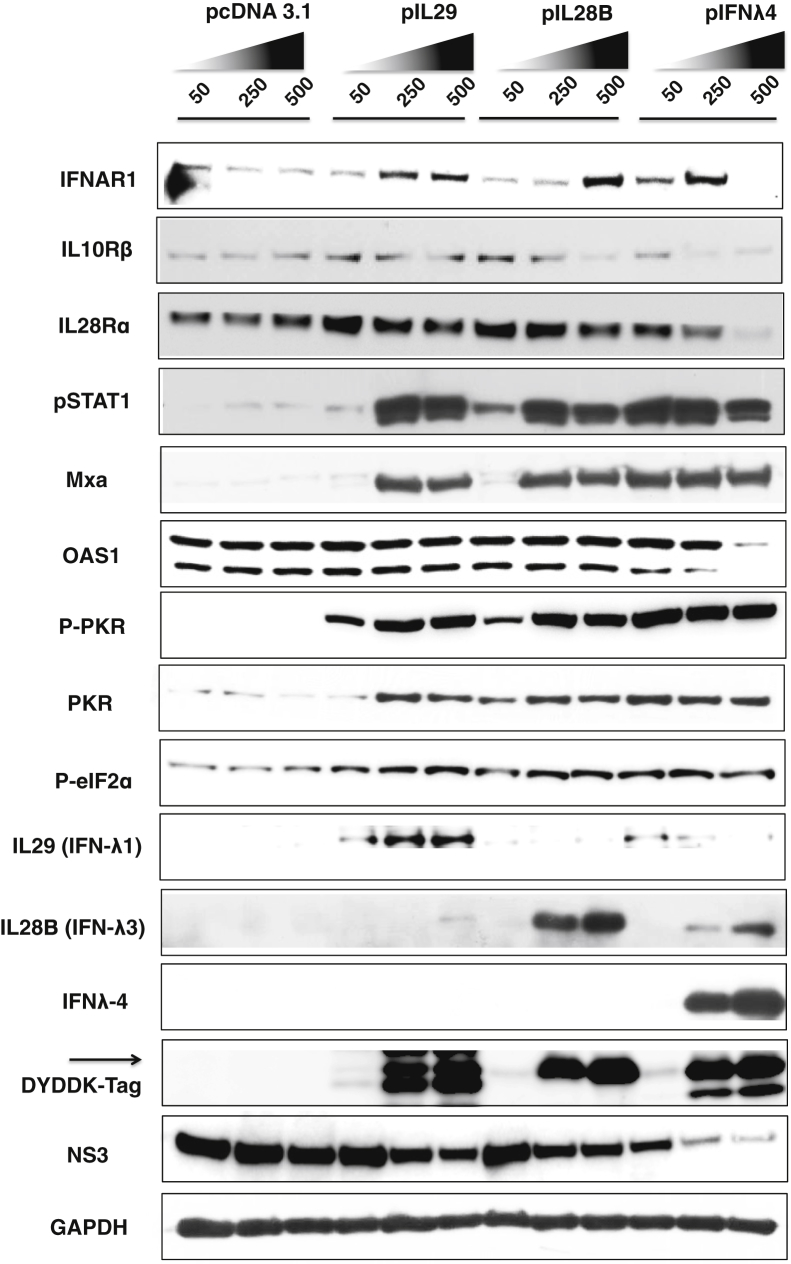
It has been reported that IL28B rs12979860 SNP T/T present a strong linkage disequilibrium with ss469415590-ΔG/ΔG variant, and the mechanism of impaired IFN-α response in this variant is related to the production of IFN-λ4 protein.13, 36 To test the hypothesis that the high protein level of IFN-λ is responsible for ISG overproduction, HCV-infected Huh-7.5 cells (Supplemental Figure S6) were transfected with increasing concentrations of individual IFN-λ expression plasmid (IL28B, IL29, or IFN-λ4) for 24 hours. We found that intracellular expression of IL29, IL28B, and IFN-λ4 inhibited HCV replication in a dose-dependent manner (Figure 6). These results are consistent with findings that demonstrated that intracellular IFN-λ expression inhibits influenza virus replication.38 We observed that the inhibition of HCV replication was stronger with IFN-λ4 expression. We also measured the effect of IFN-λ expression on the Jak-Stat signaling and ISG expression by Western blot analysis in the same lysate. Any intracellular IFN-λ–induced expression of pStat1, MxA, OAS1, and PKR levels was observed (Figure 6). These results indicated that intracellular expression of recombinant IFN-λ was able to activate the Jak-Stat signaling and to stimulate ISG expression that reduced viral replication. This could be the reason why IL28B T/T patients that show high ISG induction also support low-level viral replication compared with IL28B C/C genotype. Interestingly, we found that undetected levels of IFN-λ4 protein are able to induce a strong induction of ISGs and phosphorylation of PKR and eIF2α, associated with a stronger antiviral effect than IFN-λ3 and IFN-λ1 alone. Results from Western blot analysis showed that overexpression of IFN-λ4 induced the expression of IL28b and IL29 protein (Figure 6). These results provided a potential explanation for why IFN-λ expression was high in T/T cell lines. Finally, we tested the hypothesis of whether IFN-λ4 could induce the expression of IL29 and IL28B. These results indicated that low intracellular IFN-λ4 production was able to activate the IL28A/B, IL29 and ISRE promoter, whereas IL28B and IL29 were not (Supplemental Figure S7). These results indicated that IFN-λ4 could induce expression of other λ proteins and could activate the cellular Jak-Stat pathway with a high potency, consistent with a previous report.13
Figure 6.
IFN-λ expression by transfection activates the induced pStat1 and ISG expression in HCV-infected Huh-7.5 cells. HCV-infected Huh-7.5 cells (C/T) at day 6 were transfected with mammalian expression plasmid that contained full-length cDNA clone for IFN-λ1 (IL29), IFN-λ3 (IL28B), or IFN-λ4 or pcDNA3.1 vector alone at various concentrations. After 24 hours, protein extracts were prepared and examined for the expression of IFN receptor, Stat phosphorylation, and ISG expression by Western blot analysis. The induction of IL28B, IL29 in IFN-λ4–transfected culture was examined by Western blot analysis. GAPDH levels were monitored as a loading control. HCV, hepatitis C virus; IFN, interferon; ISG, IFN-stimulated gene.
Overexpression of IFN-λ (1 to 4) in a Persistently HCV-Infected C/T Cell Line Induces ISG Expression and Impairs Antiviral Activity of Exogenous IFN-α
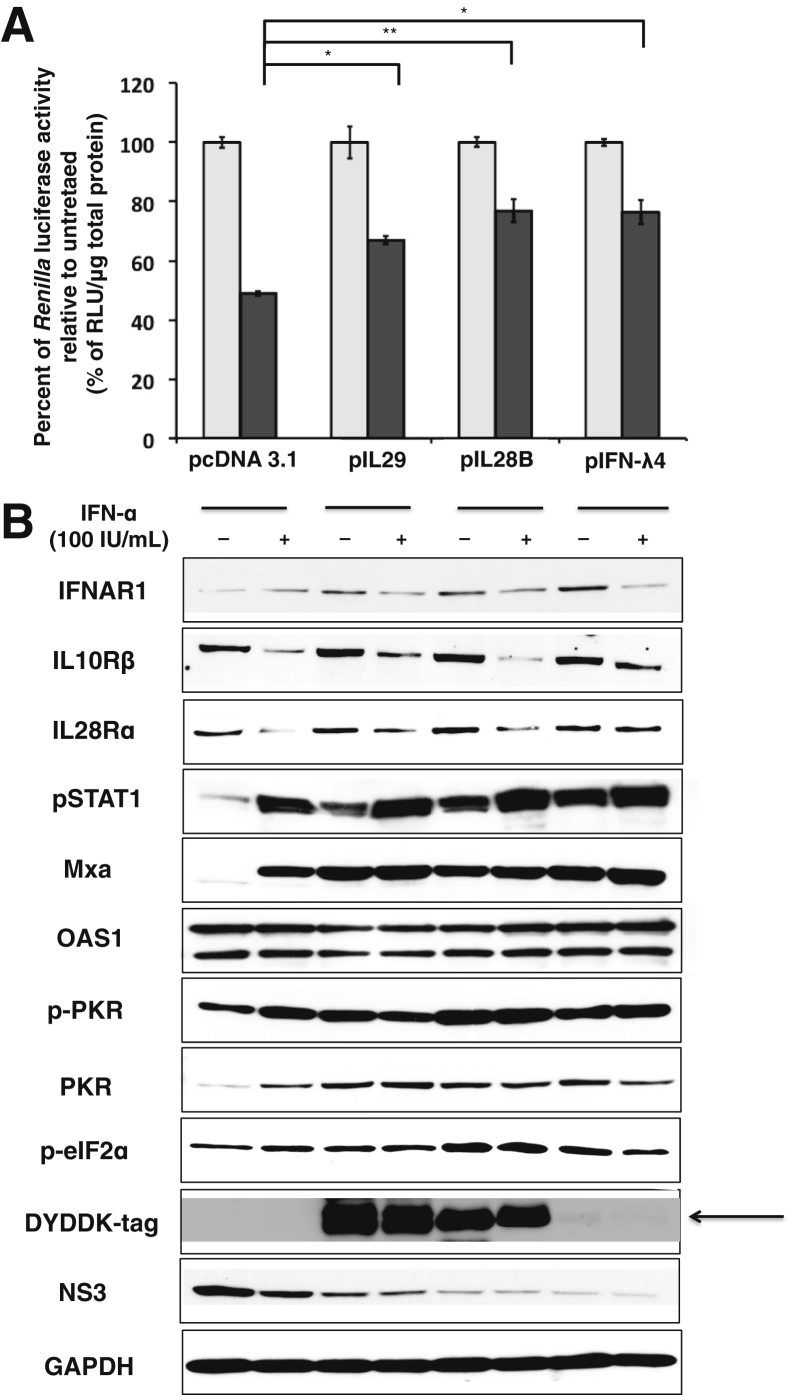
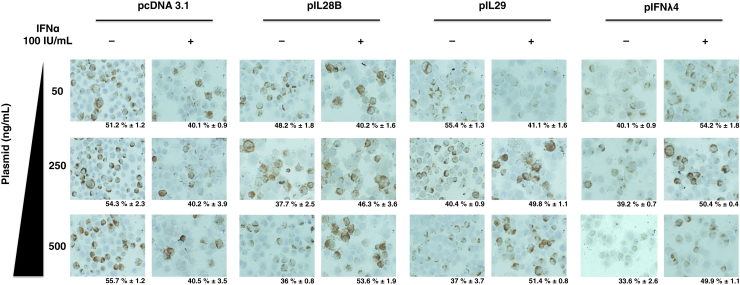
We then examined the effect of intracellular IFN-λ expression on the antiviral response of exogenously added IFN-α. HCV-infected culture was transfected with minimum concentration of IFN-λ plasmid inducing ISGs and then was treated with 100 IU/mL IFN-α for 24 hours. IFN-α and IFN-λ antiviral response against HCV was determined by the measurement of Renilla luciferase. We found that cells overexpressing recombinant IL29, IL28B, or IFN-λ4 protein prevented antiviral activity of exogenous IFN-α (Figure 7A). These results suggested that intracellular overexpression of IFN-λ inhibited antiviral response of exogenous IFN-α in HCV-infected culture. We observed that intracellular expression of IFN-λ in the infected cell showed increased expression of ISGs and high phosphorylation of PKR and eIF2-α, which prevented further activation by exogenous IFN-α treatment (Figure 7B). Immunostaining for HCV core antigen was performed as an alternative method to verify the impact of IFN-λ cDNA transfection on IFN-α antiviral activity. Intracellular expression of IFN-λs showed a dose-dependent decrease of HCV core positive cells. However, IFN-λ expression prevented antiviral activity of exogenous IFN-α, because the number of core positive cells was increased, compared with vector control (Figure 8). These results indicated that intracellular expression of IFN-λ impaired IFN-α antiviral activity. Taken together, these findings support the mechanism that low-level IFN-λ4 expression induces the production of IL28 and IL29, leading to the Jak-Stat signaling activation. The preactivation of Jak-Stat signaling results in the impaired antiviral activity of IFN-α and IFN-λ against HCV.
Figure 7.
Effect of IFN-λ expression on antiviral activity of exogenous IFN-α. HCV-infected Huh-7.5 cells (C/T) at day 6 were transfected with mammalian expression plasmid that contained full-length cDNA clone for IFN-λ1 (250 ng/mL), IFN-λ3 (250 ng/mL), IFN-λ4 (50 ng/mL), or pcDNA3.1 (250 ng/mL) vector alone for 24 hours and then treated with 100 IU/mL IFN-α for another 24 hours. A: IFN-α antiviral activity in the presence of intracellular λ expression. HCV Renilla luciferase was measured with equal amounts of protein, and values were normalized for untreated control. B: Expression of ISGs and phosphorylation of translation initiation factors with or without IFN-α treatment were tested by Western blot analysis. The arrow indicates the place of the thin band of DYKDDK-tag observed in cells transfected with only 50 ng/mL of IFN-λ4. Data are expressed as means + SE. ∗P < 0.05, ∗∗P < 0.01. HCV, hepatitis C virus; IFN, interferon; ISG, IFN-stimulated gene; RLU, relative light unit.
Figure 8.
Effect of IFN-λ expression on antiviral activity of exogenous IFN-α by immunostaining for HCV core. HCV-infected Huh-7.5 cells (C/T) after 6 days were transfected with mammalian expression plasmid that contained full-length cDNA clone for IFN-λ1 (250 ng/mL), IFN-λ3 (250 ng/mL), IFN-λ4 (50 ng/mL), or pcDNA3.1 (250 ng/mL) vector alone for 24 hours and then treated with 100 IU/mL IFN-α for another 24 hours. HCV core protein expression (brown) was detected by immunohistochemistry. Percentage of HCV core positive cells were counted in five different high-power fields (40×). Data are expressed as means + SE. HCV, hepatitis C virus; IFN, interferon.
Discussion
This study was initiated to understand the cellular mechanism of IL28B T/T gene polymorphism that impairs HCV clearance by IFN. At present, three different hypotheses have been proposed to explain the mechanisms of impaired HCV clearance in the liver among IL28B T/T genotype patients by IFN-α–based antiviral therapy. The first hypothesis is supported because patients with no response to IFN treatment show up-regulated expression of ISGs. The expression of hepatic ISGs is strongly associated with treatment response and genetic variation of IL28B.39, 40, 41, 42, 43, 44 The second hypothesis is proposed from the report by Duong et al,45 indicating that IL28B genetic association with increased hepatic ISG expression is because of the high expression of IFN-λ receptor 1 (IFNLR1). The investigators showed significant induction of IFNLR1 mRNA and protein levels in hepatic cells after IFN-α treatment. The investigators showed that stable expression of IFNLR1 activated Jak-Stat signaling and ISG expression, which prevented IFN-α–induced Stat1 phosphorylation. The third hypothesis is related to the intracellular expression of IFN-λ4 protein. The National Cancer Institute group led by Prokunina-Olsson and O'Brien13 discovered that intracellular expression of IFN-λ4 is strongly associated with impaired HCV clearance by IFN-α antiviral therapy.46 IFN-λ4 is produced by individuals who carry the IFNL4-ΔG variant but not in individuals who are homozygous for the IFNL4-ΔG, due to a frame shift in the open reading frame. The SNP rs12979860 of the IL28B variant is actually located within the intron 1 of the IFNL4 gene. The association of IFNL4-ΔG with IFN-α treatment response and viral clearance was similar to rs12979860 in participants of the HALT-C and viral Hep-C trials. This IFNL4-ΔG genotype is associated more strongly with impaired antiviral clearance than rs12979860 among African Americans. These results indicate that the IFN-λ axis is associated with modulating the host's innate immunity to control HCV infection.46, 47, 48
Here, we investigated the association of IL28B genetic variation with IFN antiviral mechanisms at the level of HCV RNA replication and translation with a panel of PHHs, liver biopsies, and hepatoma cell lines. Our results showed that IL28B T/T variant cells show impaired inhibition of HCV replication and HCV IRES-translation by IFN-α and IFN-λ compared with the C/C cell types. The anti-HCV activity of IFN-λ was impaired significantly more in T/T variant cells compared with IFN-α, indicating IL28B genetic polymorphisms may differ at the level of IFN-λ signaling. Our results indicating that IFN-λ mediated phosphorylation of Stat2 was impaired in T/T cells compared with C/C cells. We found that IFN-α induced Stat1, Stat2 phosphorylation and ISRE promoter activation and ISG mRNA level (PKR, MXA, and OAS1) in T/T and C/C cells. IFN-α–mediated ISG induction was impaired in T/T cells compared with C/C cells, which is related to attenuation of ISG protein translation induction in T/T cells compared with C/C cells. We also found that basal expression of ISGs (PKR, MXA, and OAS1) was high in T/T cells compared with C/C cells. Moreover, the expression of IFN-λ proteins (IL29, IL28B) was found to be higher in IL28B T/T variant cells than in C/C cells, supporting the notion that impaired IFN response is related to IFN-λ expression. Our results show that intracellular expression of IFN-λ4 induces expression of IL29 and IL28B, suggesting that T/T genotype cells that produce IFN-λ4 may be involved in the production of IL28B and IL29 cells. These results are in agreement with prior studies of Honda et al,40 who have demonstrated high IFN-λ4 expression in liver biopsies of patients with IL28B minor allele associated with impaired response to IFN-α and RBV combination therapy.
We propose that intracellular IFN-λ overexpression activates the Jak-Stat signaling, including ISG expression, which impairs further exogenous activation by IFN, first at the level of PKR phosphorylation and second at the level of Stat phosphorylation. The role of PKR activation as resistance factor to IFN-α treatment in HCV infection has already been reported.49 We found that IFN-λ–induced Stat2 phosphorylation is impaired in T/T cells. Thus, we hypothesize that the high IFN-λ expression could saturate the IFNLR and/or lead to the down-regulation of IFN receptors, which will result in the impairment of Stat phosphorylation by exogenous IFN. Our results also support the second hypothesis proposed by Doung et al,45 IFN-λ receptor expression was found to be high in T/T cells, supporting the alternative mechanism proposed for the Jak-Stat activation.45, 50
Interestingly, the inhibition of HCV IRES translation by IFN-α in SK-Hep1 cell line with a C/T genotype was similar to hepatoma cell lines with IL28B T/T genotype. The SNP of IFN-λ4 for this cell line shows homozygosity for the deletion in the ss469415590 (IFNL4-ΔG) similar to rs12979860 T/T SNP (Table 3). These results support a prior report that indicated a more significant association of the responsiveness with the IFNL4 SNP ss469415590-ΔG/ΔG.13
Because the IFN-λ4 expression in T/T cells is expected to explain the mechanism of association between IL28B genotype with impaired HCV clearance,13, 36, 50 we examined the expression of IFN-λ4. The expression of endogenous IFN-λ4 was found to be low in T/T cells, which could not detected by Western blot analysis.13 However, we showed that low intracellular expression of recombinant IFN-λ4 by transfection induced the expression of other -λ proteins, their promoters, and ISG expression, indicating that IFN-λ4 is able to significantly induce the expression of IL28B and IL29 in T/T variant cells, associated with a strong antiviral effect. This provides an explanation for why T/T cells express high-level IL28B and IL29 compared with C/C cells without IFN-λ4 detection. Moreover, these results highlight a high potency of IFN-λ4, compared with the other IFN-λ proteins, which represents an innovative therapeutic interest. However, the molecular mechanism of this potency requires further investigation. To establish the cause-effect relation between intracellular IL29, ISG activation, and impaired HCV clearance by IFN, overexpression and knockdown experiments were performed in T/T and C/T cell lines. These results show that abolishing the expression of IL29 by siRNA enhanced IFN-α antiviral activity at the level of IRES-mediated translation inhibition. However, the intracellular expression of recombinant IL28B, IL29, and IFN-λ4 protein by plasmid transfection in the HCV-infected culture impaired exogenous IFN-α antiviral response. We propose that IFN-λ signaling also affects IFN-α signaling, because both type I and type III IFN share the same Jak-Stat signaling, through the engagement of Tyk2 to their receptors.
Conclusion
In summary, we have developed a cellular system to understand the mechanism by which IFN-λ genetics is related to IFN treatment resistance. We provide evidence that low intracellular expression of IFN-λ4 induced IFN-λ expression, leading to the pre-activation of the Jak-Stat signaling and ISG expression, which impaired further exogenous IFN-α activity.
Acknowledgments
We thank Loula Burton for critically reviewing this manuscript, Charles Rice for Huh-7.5 cells, and Shankar Japa and Swarmanala Ratnayaka for performing IL28B genotype analysis of hepatoma cell lines.
Footnotes
Supported by NIH grants CA089121, AI103106, and CA127481.
Disclosures: None declared.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2015.11.027.
Supplemental Data
IFN receptor and ISG expression in PHHs. Equal amounts of protein extract derived from uninfected human hepatocytes were separated on SDS-PAGE and transferred to nylon membrane and IFNAR1, IL10Rβ, IL28RA, ISGs, translation factor activation were detected by immunoblotting. GAPDH expression level was monitored as a loading control. This Western blot analysis is representative of three T/T PHHs compared with three C/C PHHs. IFN, interferon; ISG, interferon-stimulated gene; PHH, primary human hepatocyte.
Western blot quantification of IFN-λ protein level in human liver biopsy. Quantification of Western blot band intensity was performed with ImageJ software version 1.47v (NIH, Bethesda, MD; http://imagej.nih.gov/ij). First the band intensity was normalized with GAPDH. Second, the fold-induction was determined for untreated control.
Janus kinase-Stat activation, ISG expression, translation factor induction, and IFN-λ protein level in cell lines with SNP rs12979860 C/C or T/T of IL28B gene, after IFN-α and IFN-λ1 treatment in a dose-dependent manner. Huh-7/CC (C/C) and Huh-7.5.1 (T/T) cells were transfected with HCV IRES-luciferase plasmid and then treated with IFN-α or IFN-λ for 24 hours. To measure phosphorylated proteins, cultured cells were treated again for 30 minutes before harvesting. Expression of IFNAR1, IL-10R-β, pStat1, and pStat2, ISGs (MxA, PKR, OAS1), phosphorylation of translation factors, IFN-λ1 (IL-29), IFN-λ3 (IL-28B), and IFN-λ4 levels were measured by Western blot analysis. Cultures were treated for 24 hours with IFN-α (10 to 1000 IU/mL) (A) or IFN-λ1 (1 to 100 ng/mL) (B). GAPDH expression level was monitored as a loading control. HCV, hepatitis C virus; IFN, interferon; IRES, internal ribosome entry site; ISG, interferon-stimulated gene.
Activation of ISRE promoter activity by IFN. The activation of ISRE promoter activity was tested by transfection of ISRE-Renilla luciferase plasmid, and then cells were treated with IFN-α (10 to 1000 IU/mL) (A) or IFN-λ1 (1 to 100 ng/mL) (B) for 24 hours. Results were normalized with firefly luciferase (peFirefly-GFP) and with 1 μg of cellular protein. Quantification of ISG mRNA levels by real-time RT-PCR, after 24 hours of IFN dose-dependent treatment. Reverse transcription was performed with 1 μg total RNA with gene-specific primer, and the resulting cDNA was amplified by real-time PCR. PKR, Mxa, and OAS1 mRNA levels between C/C and T/T cell line after 10 to 1000 IU/mL IFN-α (C) or 1 to 100 ng/mL IFN-λ1 (D). Data are expressed as means ± SE. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. IFN, interferon; IRES, internal ribosome entry site; ISG, interferon-stimulated gene; ISRE, IFN-stimulated response element.
Western blot quantification of Janus kinase-Stat activation, IFN-ISGE, translation factor induction, and IFN-λ protein level among two C/C (PLC, Li7), two C/T (Huh-7.5, SK-Hep1), and two T/T (HLE, HepG2) hepatoma cell lines. Hepatoma cells were treated with 100 IU/mL IFN-α or 100 ng/mL IFN-λ1 for 24 hours. Quantification of Western blot band intensity was performed with ImageJ computer software. First, the band intensity was normalized with GAPDH. Second, the fold-induction was determined for untreated control. A: Basal level of each protein was normalized again GAPDH. B: Fold-change of IFN-α activity was normalized against untreated. C: Fold-change of IFN-λ1 C was normalized against untreated. IFN, interferon; ISGE, interferon-stimulated gene expression.
Kinetic of HCV infection in Huh-7.5. Huh-7.5 was infected with HCV-JFH-1 for 6 days. HCV RNA was detected by quantitative real-time RT-PCR at different time point (A) and confirmed by Western blot analysis (B). Data are expressed as means ± SE. HCV, hepatitis C virus; P.I., post infection.
IFN-λ promoter activity in HCV-infected Huh-7.5 overexpressing IFN-λ in cell culture. Effect of 50 ng/mL of IFN-λ1, IFN-λ3, IFN-λ4 protein expression on IL28B, IL29, and IFN-stimulated gene promoter activity relative to pcDNA control expression. Data are expressed as means ± SE. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. HCV, hepatitis C virus; IFN, interferon; ISRE, internal ribosome entry site; RLU, relative light unit.
References
- 1.Lavanchy D. The global burden of hepatitis C. Liver Int. 2009;29(Suppl 1):74–81. doi: 10.1111/j.1478-3231.2008.01934.x. [DOI] [PubMed] [Google Scholar]
- 2.NIH Consensus Statement on Management of Hepatitis C: 2002. NIH Consens State Sci Statements. 2002;19:1–46. [PubMed] [Google Scholar]
- 3.Thomas D.L., Thio C.L., Martin M.P., Qi Y., Ge D., O'Huigin C., Kidd J., Kidd K., Khakoo S.I., Alexander G., Goedert J.J., Kirk G.D., Donfield S.M., Rosen H.R., Tobler L.H., Busch M.P., McHutchison J.G., Goldstein D.B., Carrington M. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang J.D., Roberts L.R. Hepatocellular carcinoma: a global view. Nat Rev Gastroenterol Hepatol. 2010;7:448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsuura K., Watanabe T., Tanaka Y. Role of IL28B for chronic hepatitis C treatment toward personalized medicine. J Gastroenterol Hepatol. 2014;29:241–249. doi: 10.1111/jgh.12475. [DOI] [PubMed] [Google Scholar]
- 6.Donnelly R.P., Dickensheets H., O'Brien T.R. Interferon-lambda and therapy for chronic hepatitis C virus infection. Trends Immunol. 2011;32:443–450. doi: 10.1016/j.it.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dash S., Prabhu R., Hazari S., Bastian F., Garry R., Zou W., Haque S., Joshi V., Regenstein F.G., Thung S.N. Interferons alpha, beta, gamma each inhibit hepatitis C virus replication at the level of internal ribosome entry site-mediated translation. Liver Int. 2005;25:580–594. doi: 10.1111/j.1478-3231.2005.01082.x. [DOI] [PubMed] [Google Scholar]
- 8.Liang T.J., Ghany M.G. Current and future therapies for hepatitis C virus infection. N Engl J Med. 2013;368:1907–1917. doi: 10.1056/NEJMra1213651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rauch A., Kutalik Z., Descombes P., Cai T., Di Iulio J., Mueller T., Bochud M., Battegay M., Bernasconi E., Borovicka J., Colombo S., Cerny A., Dufour J.F., Furrer H., Gunthard H.F., Heim M., Hirschel B., Malinverni R., Moradpour D., Mullhaupt B., Witteck A., Beckmann J.S., Berg T., Bergmann S., Negro F., Telenti A., Bochud P.Y. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–1345. doi: 10.1053/j.gastro.2009.12.056. 1345 e1–7. [DOI] [PubMed] [Google Scholar]
- 10.Ge D., Fellay J., Thompson A.J., Simon J.S., Shianna K.V., Urban T.J., Heinzen E.L., Qiu P., Bertelsen A.H., Muir A.J., Sulkowski M., McHutchison J.G., Goldstein D.B. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka Y., Nishida N., Sugiyama M., Kurosaki M., Matsuura K., Sakamoto N., Nakagawa M., Korenaga M., Hino K., Hige S., Ito Y., Mita E., Tanaka E., Mochida S., Murawaki Y., Honda M., Sakai A., Hiasa Y., Nishiguchi S., Koike A., Sakaida I., Imamura M., Ito K., Yano K., Masaki N., Sugauchi F., Izumi N., Tokunaga K., Mizokami M. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 12.Suppiah V., Moldovan M., Ahlenstiel G., Berg T., Weltman M., Abate M.L., Bassendine M., Spengler U., Dore G.J., Powell E., Riordan S., Sheridan D., Smedile A., Fragomeli V., Muller T., Bahlo M., Stewart G.J., Booth D.R., George J. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 13.Prokunina-Olsson L., Muchmore B., Tang W., Pfeiffer R.M., Park H., Dickensheets H., Hergott D., Porter-Gill P., Mumy A., Kohaar I., Chen S., Brand N., Tarway M., Liu L., Sheikh F., Astemborski J., Bonkovsky H.L., Edlin B.R., Howell C.D., Morgan T.R., Thomas D.L., Rehermann B., Donnelly R.P., O'Brien T.R. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet. 2013;45:164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu T.W., Kulkarni R., Gane E.J., Roberts S.K., Stedman C., Angus P.W., Ritchie B., Lu X.Y., Ipe D., Lopatin U., Germer S., Iglesias V.A., Elston R., Smith P.F., Shulman N.S. Effect of IL28B genotype on early viral kinetics during interferon-free treatment of patients with chronic hepatitis C. Gastroenterology. 2012;142:790–795. doi: 10.1053/j.gastro.2011.12.057. [DOI] [PubMed] [Google Scholar]
- 15.Meissner E.G., Bon D., Prokunina-Olsson L., Tang W., Masur H., O'Brien T.R., Herrmann E., Kottilil S., Osinusi A. IFNL4-DeltaG genotype is associated with slower viral clearance in hepatitis C, genotype-1 patients treated with sofosbuvir and ribavirin. J Infect Dis. 2014;209:1700–1704. doi: 10.1093/infdis/jit827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meissner E.G., Wu D., Osinusi A., Bon D., Virtaneva K., Sturdevant D., Porcella S., Wang H., Herrmann E., McHutchison J., Suffredini A.F., Polis M., Hewitt S., Prokunina-Olsson L., Masur H., Fauci A.S., Kottilil S. Endogenous intrahepatic IFNs and association with IFN-free HCV treatment outcome. J Clin Invest. 2014;124:3352–3363. doi: 10.1172/JCI75938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan T.R., O'Brien T.R. IL28B-genotype testing now and in the era of direct-acting antiviral agents. Clin Gastroenterol Hepatol. 2011;9:293–294. doi: 10.1016/j.cgh.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi X., Pan Y., Wang M., Wang D., Li W., Jiang T., Zhang P., Chi X., Jiang Y., Gao Y., Zhong J., Sun B., Xu D., Jiang J., Niu J. IL28B genetic variation is associated with spontaneous clearance of hepatitis C virus, treatment response, serum IL-28B levels in Chinese population. PLoS One. 2012;7:e37054. doi: 10.1371/journal.pone.0037054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langhans B., Kupfer B., Braunschweiger I., Arndt S., Schulte W., Nischalke H.D., Nattermann J., Oldenburg J., Sauerbruch T., Spengler U. Interferon-lambda serum levels in hepatitis C. J Hepatol. 2011;54:859–865. doi: 10.1016/j.jhep.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 20.Dolganiuc A., Kodys K., Marshall C., Saha B., Zhang S., Bala S., Szabo G. Type III interferons, IL-28 and IL-29, are increased in chronic HCV infection and induce myeloid dendritic cell-mediated FoxP3+ regulatory T cells. PLoS One. 2012;7:e44915. doi: 10.1371/journal.pone.0044915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang S., Kodys K., Li K., Szabo G. Human type 2 myeloid dendritic cells produce interferon-lambda and amplify interferon-alpha in response to hepatitis C virus infection. Gastroenterology. 2013;144:414–425.e7. doi: 10.1053/j.gastro.2012.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McFarland A.P., Horner S.M., Jarret A., Joslyn R.C., Bindewald E., Shapiro B.A., Delker D.A., Hagedorn C.H., Carrington M., Gale M., Jr., Savan R. The favorable IFNL3 genotype escapes mRNA decay mediated by AU-rich elements and hepatitis C virus-induced microRNAs. Nat Immunol. 2014;15:72–79. doi: 10.1038/ni.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandra P.K., Bao L., Song K., Aboulnasr F.M., Baker D.P., Shores N., Wimley W.C., Liu S., Hagedorn C.H., Fuchs S.Y., Wu T., Balart L.A., Dash S. HCV infection selectively impairs type I but not type III IFN signaling. Am J Pathol. 2014;184:214–229. doi: 10.1016/j.ajpath.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panigrahi R., Hazari S., Chandra S., Chandra P.K., Datta S., Kurt R., Cameron C.E., Huang Z., Zhang H., Garry R.F., Balart L.A., Dash S. Interferon and ribavirin combination treatment synergistically inhibit HCV internal ribosome entry site mediated translation at the level of polyribosome formation. PLoS One. 2013;8:e72791. doi: 10.1371/journal.pone.0072791. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Panigrahi R., Chandra P.K., Ferraris P., Kurt R., Song K., Garry R.F., Reiss K., Coe I.R., Furihata T., Balart L.A., Wu T., Dash S. Persistent hepatitis C virus infection impairs ribavirin antiviral activity through clathrin-mediated trafficking of equilibrative nucleoside transporter 1. J Virol. 2014;89:626–642. doi: 10.1128/JVI.02492-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandra P.K., Gunduz F., Hazari S., Kurt R., Panigrahi R., Poat B., Bruce D., Cohen A.J., Bohorquez H.E., Carmody I., Loss G., Balart L.A., Wu T., Dash S. Impaired expression of type I and type II interferon receptors in HCV-associated chronic liver disease and liver cirrhosis. PLoS One. 2014;9:e108616. doi: 10.1371/journal.pone.0108616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Israelow B., Narbus C.M., Sourisseau M., Evans M.J. HepG2 cells mount an effective antiviral interferon-lambda based innate immune response to hepatitis C virus infection. Hepatology. 2014;60:1170–1179. doi: 10.1002/hep.27227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osterlund P.I., Pietila T.E., Veckman V., Kotenko S.V., Julkunen I. IFN regulatory factor family members differentially regulate the expression of type III IFN (IFN-lambda) genes. J Immunol. 2007;179:3434–3442. doi: 10.4049/jimmunol.179.6.3434. [DOI] [PubMed] [Google Scholar]
- 29.Hamming O.J., Terczynska-Dyla E., Vieyres G., Dijkman R., Jorgensen S.E., Akhtar H., Siupka P., Pietschmann T., Thiel V., Hartmann R. Interferon lambda 4 signals via the IFNlambda receptor to regulate antiviral activity against HCV and coronaviruses. EMBO J. 2013;32:3055–3065. doi: 10.1038/emboj.2013.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bibert S., Roger T., Calandra T., Bochud M., Cerny A., Semmo N., Duong F.H., Gerlach T., Malinverni R., Moradpour D., Negro F., Mullhaupt B., Bochud P.Y., Swiss Hepatitis C Cohort Study IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J Exp Med. 2013;210:1109–1116. doi: 10.1084/jem.20130012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinzani P., Salvianti F., Cascella R., Massi D., De Giorgi V., Pazzagli M., Orlando C. Allele specific Taqman-based real-time PCR assay to quantify circulating BRAFV600E mutated DNA in plasma of melanoma patients. Clin Chim Acta. 2010;411:1319–1324. doi: 10.1016/j.cca.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 32.Sugano M., Hayashi Y., Yoon S., Kinoshita M., Ninomiya T., Ohta K., Itoh H., Kasuga M. Quantitation of hepatitis C viral RNA in liver and serum samples using competitive polymerase chain reaction. J Clin Pathol. 1995;48:820–825. doi: 10.1136/jcp.48.9.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bigger C.B., Guerra B., Brasky K.M., Hubbard G., Beard M.R., Luxon B.A., Lemon S.M., Lanford R.E. Intrahepatic gene expression during chronic hepatitis C virus infection in chimpanzees. J Virol. 2004;78:13779–13792. doi: 10.1128/JVI.78.24.13779-13792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sadler A.J., Williams B.R. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pai M., Prabhu R., Panebra A., Nangle S., Haque S., Bastian F., Garry R., Agrawal K., Goodbourn S., Dash S. Activation of interferon-stimulated response element in huh-7 cells replicating hepatitis C virus subgenomic RNA. Intervirology. 2005;48:301–311. doi: 10.1159/000085099. [DOI] [PubMed] [Google Scholar]
- 36.Murakawa M., Asahina Y., Nakagawa M., Sakamoto N., Nitta S., Kusano-Kitazume A., Watanabe T., Kawai-Kitahata F., Otani S., Taniguchi M., Goto F., Nishimura-Sakurai Y., Itsui Y., Azuma S., Kakinuma S., Watanabe M. Impaired induction of interleukin 28B and expression of interferon lambda 4 associated with nonresponse to interferon-based therapy in chronic hepatitis C. J Gastroenterol Hepatol. 2015;30:1075–1084. doi: 10.1111/jgh.12902. [DOI] [PubMed] [Google Scholar]
- 37.Poat B., Hazari S., Chandra P.K., Gunduz F., Alvarez X., Balart L.A., Garry R.F., Dash S. Intracellular expression of IRF9 Stat fusion protein overcomes the defective Jak-Stat signaling and inhibits HCV RNA replication. Virol J. 2010;7:265. doi: 10.1186/1743-422X-7-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svetlikova D., Kabat P., Ohradanova A., Pastorek J., Betakova T. Influenza A virus replication is inhibited in IFN-lambda2 and IFN-lambda3 transfected or stimulated cells. Antiviral Res. 2010;88:329–333. doi: 10.1016/j.antiviral.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 39.Urban T.J., Thompson A.J., Bradrick S.S., Fellay J., Schuppan D., Cronin K.D., Hong L., McKenzie A., Patel K., Shianna K.V., McHutchison J.G., Goldstein D.B., Afdhal N. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology. 2010;52:1888–1896. doi: 10.1002/hep.23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Honda M., Shirasaki T., Shimakami T., Sakai A., Horii R., Arai K., Yamashita T., Sakai Y., Yamashita T., Okada H., Murai K., Nakamura M., Mizukoshi E., Kaneko S. Hepatic interferon-stimulated genes are differentially regulated in the liver of chronic hepatitis C patients with different interleukin-28B genotypes. Hepatology. 2014;59:828–838. doi: 10.1002/hep.26788. [DOI] [PubMed] [Google Scholar]
- 41.Dill M.T., Duong F.H., Vogt J.E., Bibert S., Bochud P.Y., Terracciano L., Papassotiropoulos A., Roth V., Heim M.H. Interferon-induced gene expression is a stronger predictor of treatment response than IL28B genotype in patients with hepatitis C. Gastroenterology. 2011;140:1021–1031. doi: 10.1053/j.gastro.2010.11.039. [DOI] [PubMed] [Google Scholar]
- 42.Asselah T., Bieche I., Narguet S., Sabbagh A., Laurendeau I., Ripault M.P., Boyer N., Martinot-Peignoux M., Valla D., Vidaud M., Marcellin P. Liver gene expression signature to predict response to pegylated interferon plus ribavirin combination therapy in patients with chronic hepatitis C. Gut. 2008;57:516–524. doi: 10.1136/gut.2007.128611. [DOI] [PubMed] [Google Scholar]
- 43.Chen L., Borozan I., Feld J., Sun J., Tannis L.L., Coltescu C., Heathcote J., Edwards A.M., McGilvray I.D. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology. 2005;128:1437–1444. doi: 10.1053/j.gastro.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 44.Honda M., Sakai A., Yamashita T., Nakamoto Y., Mizukoshi E., Sakai Y., Yamashita T., Nakamura M., Shirasaki T., Horimoto K., Tanaka Y., Tokunaga K., Mizokami M., Kaneko S., Hokuriku Liver Study Group Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology. 2010;139:499–509. doi: 10.1053/j.gastro.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 45.Duong F.H., Trincucci G., Boldanova T., Calabrese D., Campana B., Krol I., Durand S.C., Heydmann L., Zeisel M.B., Baumert T.F., Heim M.H. IFN-lambda receptor 1 expression is induced in chronic hepatitis C and correlates with the IFN-lambda3 genotype and with nonresponsiveness to IFN-alpha therapies. J Exp Med. 2014;211:857–868. doi: 10.1084/jem.20131557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Booth D., George J. Loss of function of the new interferon IFN-lambda4 may confer protection from hepatitis C. Nat Genet. 2013;45:119–120. doi: 10.1038/ng.2537. [DOI] [PubMed] [Google Scholar]
- 47.Asahina Y., Tsuchiya K., Muraoka M., Tanaka K., Suzuki Y., Tamaki N., Hoshioka Y., Yasui Y., Katoh T., Hosokawa T., Ueda K., Nakanishi H., Itakura J., Takahashi Y., Kurosaki M., Enomoto N., Nitta S., Sakamoto N., Izumi N. Association of gene expression involving innate immunity and genetic variation in interleukin 28B with antiviral response. Hepatology. 2012;55:20–29. doi: 10.1002/hep.24623. [DOI] [PubMed] [Google Scholar]
- 48.Makowska Z., Duong F.H., Trincucci G., Tough D.F., Heim M.H. Interferon-beta and interferon-lambda signaling is not affected by interferon-induced refractoriness to interferon-alpha in vivo. Hepatology. 2011;53:1154–1163. doi: 10.1002/hep.24189. [DOI] [PubMed] [Google Scholar]
- 49.Garaigorta U., Chisari F.V. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe. 2009;6:513–522. doi: 10.1016/j.chom.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terczynska-Dyla E., Bibert S., Duong F.H., Krol I., Jorgensen S., Collinet E., Kutalik Z., Aubert V., Cerny A., Kaiser L., Malinverni R., Mangia A., Moradpour D., Mullhaupt B., Negro F., Santoro R., Semela D., Semmo N., Swiss Hepatitis C Cohort Study Group. Heim M.H., Bochud P.Y., Hartmann R. Reduced IFNlambda4 activity is associated with improved HCV clearance and reduced expression of interferon-stimulated genes. Nat Commun. 2014;5:5699. doi: 10.1038/ncomms6699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
IFN receptor and ISG expression in PHHs. Equal amounts of protein extract derived from uninfected human hepatocytes were separated on SDS-PAGE and transferred to nylon membrane and IFNAR1, IL10Rβ, IL28RA, ISGs, translation factor activation were detected by immunoblotting. GAPDH expression level was monitored as a loading control. This Western blot analysis is representative of three T/T PHHs compared with three C/C PHHs. IFN, interferon; ISG, interferon-stimulated gene; PHH, primary human hepatocyte.
Western blot quantification of IFN-λ protein level in human liver biopsy. Quantification of Western blot band intensity was performed with ImageJ software version 1.47v (NIH, Bethesda, MD; http://imagej.nih.gov/ij). First the band intensity was normalized with GAPDH. Second, the fold-induction was determined for untreated control.
Janus kinase-Stat activation, ISG expression, translation factor induction, and IFN-λ protein level in cell lines with SNP rs12979860 C/C or T/T of IL28B gene, after IFN-α and IFN-λ1 treatment in a dose-dependent manner. Huh-7/CC (C/C) and Huh-7.5.1 (T/T) cells were transfected with HCV IRES-luciferase plasmid and then treated with IFN-α or IFN-λ for 24 hours. To measure phosphorylated proteins, cultured cells were treated again for 30 minutes before harvesting. Expression of IFNAR1, IL-10R-β, pStat1, and pStat2, ISGs (MxA, PKR, OAS1), phosphorylation of translation factors, IFN-λ1 (IL-29), IFN-λ3 (IL-28B), and IFN-λ4 levels were measured by Western blot analysis. Cultures were treated for 24 hours with IFN-α (10 to 1000 IU/mL) (A) or IFN-λ1 (1 to 100 ng/mL) (B). GAPDH expression level was monitored as a loading control. HCV, hepatitis C virus; IFN, interferon; IRES, internal ribosome entry site; ISG, interferon-stimulated gene.
Activation of ISRE promoter activity by IFN. The activation of ISRE promoter activity was tested by transfection of ISRE-Renilla luciferase plasmid, and then cells were treated with IFN-α (10 to 1000 IU/mL) (A) or IFN-λ1 (1 to 100 ng/mL) (B) for 24 hours. Results were normalized with firefly luciferase (peFirefly-GFP) and with 1 μg of cellular protein. Quantification of ISG mRNA levels by real-time RT-PCR, after 24 hours of IFN dose-dependent treatment. Reverse transcription was performed with 1 μg total RNA with gene-specific primer, and the resulting cDNA was amplified by real-time PCR. PKR, Mxa, and OAS1 mRNA levels between C/C and T/T cell line after 10 to 1000 IU/mL IFN-α (C) or 1 to 100 ng/mL IFN-λ1 (D). Data are expressed as means ± SE. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. IFN, interferon; IRES, internal ribosome entry site; ISG, interferon-stimulated gene; ISRE, IFN-stimulated response element.
Western blot quantification of Janus kinase-Stat activation, IFN-ISGE, translation factor induction, and IFN-λ protein level among two C/C (PLC, Li7), two C/T (Huh-7.5, SK-Hep1), and two T/T (HLE, HepG2) hepatoma cell lines. Hepatoma cells were treated with 100 IU/mL IFN-α or 100 ng/mL IFN-λ1 for 24 hours. Quantification of Western blot band intensity was performed with ImageJ computer software. First, the band intensity was normalized with GAPDH. Second, the fold-induction was determined for untreated control. A: Basal level of each protein was normalized again GAPDH. B: Fold-change of IFN-α activity was normalized against untreated. C: Fold-change of IFN-λ1 C was normalized against untreated. IFN, interferon; ISGE, interferon-stimulated gene expression.
Kinetic of HCV infection in Huh-7.5. Huh-7.5 was infected with HCV-JFH-1 for 6 days. HCV RNA was detected by quantitative real-time RT-PCR at different time point (A) and confirmed by Western blot analysis (B). Data are expressed as means ± SE. HCV, hepatitis C virus; P.I., post infection.
IFN-λ promoter activity in HCV-infected Huh-7.5 overexpressing IFN-λ in cell culture. Effect of 50 ng/mL of IFN-λ1, IFN-λ3, IFN-λ4 protein expression on IL28B, IL29, and IFN-stimulated gene promoter activity relative to pcDNA control expression. Data are expressed as means ± SE. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. HCV, hepatitis C virus; IFN, interferon; ISRE, internal ribosome entry site; RLU, relative light unit.