Abstract
The multistep process of TP53 mutation expansion during myeloproliferative neoplasm (MPN) transformation into acute myeloid leukemia (AML) has been documented retrospectively. It is currently unknown how common TP53 mutations with low variant allele frequency (VAF) are, whether they are linked to hydroxyurea (HU) cytoreduction, and what disease progression risk they carry. Using ultra-deep next-generation sequencing, we examined 254 MPN patients treated with HU, interferon alpha-2a or anagrelide and 85 untreated patients. We found TP53 mutations in 50 cases (0.2–16.3% VAF), regardless of disease subtype, driver gene status and cytoreduction. Both therapy and TP53 mutations were strongly associated with older age. Over-time analysis showed that the mutations may be undetectable at diagnosis and slowly increase during disease course. Although three patients with TP53 mutations progressed to TP53-mutated or TP53-wild-type AML, we did not observe a significant age-independent impact on overall survival during the follow-up. Further, we showed that complete p53 inactivation alone led to neither blast transformation nor HU resistance. Altogether, we revealed patient's age as the strongest factor affecting low-burden TP53 mutation incidence in MPN and found no significant age-independent association between TP53 mutations and hydroxyurea. Mutations may persist at low levels for years without an immediate risk of progression.
Introduction
Leukemic transformation of Ph-negative myeloproliferative neoplasms (MPN; polycythemia vera, PV; essential thrombocythemia, ET; primary myelofibrosis, PMF) is a relatively rare but fatal event. Several intrinsic risk factors have been suggested involving MPN phenotype (PMF>PV>ET), abnormal karyotype and higher age.1 The effect of MPN therapy has been widely discussed and alkylating agents, pipobroman and 32P were shown to be leukemogenic.1, 2, 3 Possible negative impacts of hydroxyurea (HU) remain controversial,1, 2, 4, 5, 6, 7, 8, 9 as summarized in.10 Great effort has been invested into the search for genetic changes predicting and/or triggering MPN transformation to AML and MDS.11, 12, 13, 14, 15, 16
Eliminating tumor suppressor TP53 during myelopoiesis helps escape from control mechanisms preventing differentiation loss, aberrant self-renewal, and large genome rearrangements.17, 18 In chronic MPN phase, TP53 gene defects have been extremely rarely detected using Sanger sequencing or cytogenetic analysis; on the contrary, they were shown to be common in post-MPN acute myeloid leukemia (AML).19, 20, 21 This pronounced difference is indicative of TP53 role in the transformation process. Retrospective analysis of individual cases of TP53-mutated post-MPN AML showed that TP53 mutations can be traced months or even years before leukemic transformation.11, 19, 22, 23 The level of mutation burden was shown to remain low until complete p53 inactivation by losing the second allele (17p defects or second mutation), followed by rapid clonal expansion.11, 22
TP53 mutations occurring at a level above detection limit of Sanger sequencing (10–20% variant allelic frequency; VAF) show negative prognostic and/or predictive impact in some types of cancer, especially in hematological malignancies.24, 25 Small TP53-mutated subclones below this sensitivity threshold were described to drive relapse or disease progression in many cases of chronic hematological malignancies,26, 27, 28, 29 but their impact is less clear in prospective setting.30 Cytotoxic agents support a minor TP53-mutated subclone overgrowth.26, 31, 32, 33 Previous therapy with hydroxyurea (HU), a ribonucleotide reductase inhibitor activating p53 response via replication stress,34, 35 has been associated with TP53/17p defects in post-MPN AML;4, 12, 22, 36 however, this observation has not been confirmed in a large unbiased study.
While minor TP53 mutations in MPN have been tracked retrospectively in individual cases and have been suggested as carrying an increased risk of leukemic transformation,11 the occurrence of low-burden TP53 mutations (<5%) has not been analyzed so far. Whether therapy or other factors affect their origin and outgrowth is unknown. To map TP53-mutated subclones' presence in MPN patients treated with cytoreductive drugs and study their evolution over time, we used an ultra-deep next-generation sequencing (NGS) approach.
Patients and methods
Patients and samples
Peripheral blood (PB) samples and clinical and routine laboratory data from MPN patients were collected from Czech hospitals (University Hospital Brno and local hospitals) and Vienna General Hospital, Austria. The study was approved by the Ethical Committee of University Hospital Brno. For all samples, written informed consent approved by the Ethics Committees of the respective institutions were available in accordance with the Declaration of Helsinki. Patients were diagnosed according to the revised World Health Organization criteria.37
In total, 339 MPN patients were included (Supplementary Table S1). Treated patients (N=254) were having or had discontinued cytoreductive therapy–HU, interferon alpha-2a (IFN) or anagrelide (ANG)—and had been diagnosed ⩾4 years (y) before sampling. To assess the effect of therapy, the treated patients were categorized as follows: (1) by administration of HU, IFN or ANG at any time during disease course (referred to as HU-yes/HU-no, IFN-yes/IFN-no, ANG-yes/ANG-no); (2) more strictly, in the HU-yes group, only patients fulfilling a criterion of HU treatment for ⩾4y were kept. This group was compared to HU-no patients. Besides these, 85 samples from patients with no cytoreductive therapy before sampling were analyzed. Retrospective and prospective samples from 31/50 patients with detected TP53 mutations were analyzed to describe mutation development.
Ultra-deep next-generation sequencing of TP53 amplicons
NGS analysis was performed as described previously26 with minor modifications. Briefly, 30 ng of leukocyte or granulocyte DNA was amplified with high-fidelity Q5 Polymerase (New England Biolabs, Ipswich, MA, USA) using primers specific for the TP53 exons 4–10. The indexed library was prepared with Nextera XT DNA Sample Preparation Kit (Illumina, San Diego, CA, USA) and sequenced using MiSeq Reagent Kit v2 (300 cycles; Illumina, San Diego, CA, USA) on a MiSeq instrument according to manufacturer recommendations. The coverage per base exceeded 5000 (⩾10 000 in 82% of exons); mean coverage reached 39535. For variant detection we used bioinformatics pipeline (Supplementary Figure S1) combining CLC Genomic Workbench version 7.5 (Qiagen, Hilden, Germany) and the deepSNV R-package.38, 39, 40, 41, 42 Samples containing variants above 0.2% VAF by either approach were subjected to validation from independent sampling and/or PCR amplification (Supplementary Table S2). For over-time monitoring and validation of previously identified mutation, cutoff 0.1% was applied (minimal coverage per base ⩾10 000).
Statistical analysis
Statistical analyses were performed within the R environment.42 The distribution normality was tested using the Kolmogorov-Smirnov normality test. Non-parametric tests were applied because of normality violation in most clinical variables (for example, age distribution). To analyze the relationship between the variables, the Spearman correlation test, Mann–Whitney test, Kruskal–Wallis test and Fisher’s exact test with simulated P-value (Monte-Carlo simulation) were used. Data sets were described with median and s.d. and/or range as indicated in the legends. The comparison of patients' survival was performed by log-rank test and visualized using Kaplan–Meier curves; Cox proportional hazards regression was used to model the effect of multiple predictor variables. Logistic regression models were applied to assess the significance of age and therapy in TP53 mutational status. Age adjustment was performed by adding the age covariate into the logistic model. Finally, the age-adjusted models were compared with a model with age parameter only by anova chi-square tests. The level of statistical significance was set P⩽0.05. All statistical tests were performed as two-sided. Plots were created with the GraphPad Prism version 5.00 for Windows (GraphPad Software, La Jolla, CA, USA).
Single-nucleotide polymorphism arrays
Genome-wide analysis was performed on CytoScan HD arrays (Affymetrix, Santa Clara, CA, USA) according to the manufacturer's instructions. CEL files were analyzed using the Chromosome Analysis Suite software, v3.1.0.15 (Affymetrix, Santa Clara, CA, USA) and annotated using NetAffx 33.1 annotation data set.
For details on methods see Supplementary Material.
Results
Ultra-deep NGS analysis of TP53 gene in treated MPN patients
To screen for TP53 mutations and assess the effect of therapy, we analyzed 254 chronic-phase MPN patients using ultra-deep NGS. All patients were treated with one or more cytoreductive drugs (hydroxyurea, HU; interferon alpha-2a, IFN; anagrelide, ANG) and diagnosed ⩾4y before sampling (4.2–29.5; median 9.2y; Supplementary Table S1). TP53 mutations were identified in 41 patients (41/254; 16.1% Table 1) with VAF for the most abundant variant ranging between 0.2 and 11.6%. In a pronounced proportion of patients, more than one mutation was present (11/41; 26.8%). Colony-forming assay43, 44 performed in 3 patients confirmed the presence of TP53-mutated subclones within JAK2 or CALR-mutated populations (Supplementary Figure S2). To verify the mutations’ presence in myeloid lineage in patients examined from leukocyte DNA, the granulocyte sample was analyzed where available. No evident discrepancy was found (Supplementary Table S3).
Table 1. Clinical and laboratory data of patients treated with cytoreductive drugs and carrying TP53 mutations.
|
Disease type and course |
Therapy |
Mutations |
Follow-up |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Therapy group | Sample | Driver mutation a | Sex | Disease phenotype | Age at sampling | Time from dg to study enrollment (mo) | Total therapy length (mo) | Therapy at sampling | HU therapy (mo) | IFN therapy (mo) | ANG therapy (mo) | Busulfan or chemo/radio therapy | Most abundant mutation (VAF %) | TP53 mutations (VAF %) | Follow-up from study enrollment (mo) | Outcome |
| no HU | MP315 | JAK2 | M | PV | 62 | 109 | 89 | I | 0 | 0 | 0 | no | 0.8 | p.Y126D 0.79 | 39 | Alive |
| 048C | JAK2 | M | PV | 73 | 145 | 96 | I | 0 | 96 | 0 | no | 0.8 | p.E285K 0.82 | 41 | Alive | |
| JAK1716 | JAK2 | F | PV | 63 | 111 | 110 | I | 0 | 110 | 0 | no | 0.2 | p.C238S 0.20 | 83 | Alive | |
| MP189 | CALR+JAK2 | F | ET | 67 | 138 | 112 | A | 0 | 0 | 112 | no | 2.9 | p.P58A 2.90 | 51 | Alive | |
| MP247 | CALR | F | PMF | 56 | 122 | 119 | A | 0 | 5 | 115 | no | 0.3 | p.Y220C 0.26 | 48 | Alive | |
| MP155 | CALR | F | PMF | 61 | 122 | 121 | A | 0 | 0 | 121 | no | 0.2 | p.V143E 0.21 | 52 | Alive | |
| 227A | CALR | M | PMF | 72 | 154 | 123 | I | 0 | 123 | 0 | CH | 0.3 | p.A138V 0.26 | 10 | Dead | |
| 65A | CALR | F | ET | 70 | 158 | 152 | IA | 0 | 152 | 11 | no | 1.7 | c.376-2A>G 1.71; p.R273H 0.2 | 121 | Alive | |
| 186A | JAK2 | F | ET | 80 | 180 | 175 | I | 0 | 175 | 0 | no | 8.3 | c.454dupC 8.27 | 37 | Dead-AMLb | |
| 221A | CALR | M | ET | 69 | 180 | 179 | I | 0 | 179 | 0 | no | 2.2 | p.I162S 2.24 | 118 | Alive | |
| HU <4y | MP15 | JAK2 | M | PV | 66 | 84 | 84 | I | 17 | 77 | 0 | no | 4.4 | p.Y220C 4.4 | 62 | Alive |
| MP160 | JAK2 | M | ET | 69 | 61 | 18 | H | 18 | 0 | 0 | no | 0.2 | p.P142R 0.23 | 52 | Alive | |
| MP68 | JAK2 | F | MPN | 70 | 58 | 56 | H | 47 | 0 | 9 | no | 6.9 | p.R248Q 6.90; p.A159V 6.40; p.P151R 1.73; p.I195S 0.24; p.R273H 0.23 | 44 | Dead-AMLc | |
| HU ⩾4y | MP326 | JAK2 | F | PV | 58 | 80 | 48 | H | 48 | 0 | 0 | no | 0.6 | p.Y220C 0.55 | 39 | Alive |
| MP345 | JAK2 | M | PV | 68 | 50 | 48 | H | 48 | 0 | 0 | no | 1.1 | p.S215R 1.10 | 38 | Alive | |
| MP302 | JAK2 | F | PV | 71 | 231 | 65 | H | 56 | 0 | 0 | B | 0.4 | p.G199E 0.44 | 29 | Alive | |
| MP319 | JAK2 | M | PV | 61 | 62 | 57 | HA | 57 | 0 | 32 | no | 0.4 | p.R273H 0.37 | 28 | Alive | |
| MP168 | JAK2 | F | PV | 68 | 86 | 57 | H | 57 | 0 | 0 | no | 1.0 | p.H179R 0.95 | 49 | Alive | |
| MP153 | JAK2 | M | PV | 63 | 62 | 61 | HA | 60 | 0 | 61 | no | 0.2 | p.R213G 0.23 | 51 | Alive | |
| MP96 | JAK2 | F | PV | 55 | 64 | 62 | H | 62 | 0 | 0 | no | 0.5 | p.I195T 0.51; p.R282W 0.20 | 54 | Alive | |
| MP327 | JAK2 | F | ET | 73 | 125 | 64 | H | 64 | 0 | 0 | no | 0.5 | p.R273H 0.47 | 39 | Alive | |
| MP317 | CALR | M | PMF | 81 | 65 | 65 | H | 65 | 0 | 0 | no | 0.8 | p.V216M 0.83; p.H179R 0.25 | 17 | Dead | |
| MP63 | JAK2 | F | ET | 68 | 79 | 69 | H | 69 | 0 | 0 | no | 0.2 | p.M246V 0.20 | 53 | Alive | |
| MP5 | JAK2 | M | PV | 68 | 70 | 70 | H | 70 | 0 | 0 | no | 0.3 | p.I195T 0.26 | 64 | Alive | |
| MP10 | JAK2 | M | PMF | 79 | 77 | 76 | H | 76 | 0 | 0 | no | 10.5 | p.G245S 10.50; p.R158H 0.54; p.H168R 0.29; p.F134L 0.23; p.Y234H 0.21 | 46 | Dead-expansiond | |
| MP307 | MPL | F | PV | 82 | 103 | 76 | H | 76 | 0 | 0 | no | 0.2 | c.572_574del 0.21 | 40 | Alive | |
| MP369 | JAK2 | F | PV | 73 | 84 | 81 | H | 81 | 0 | 0 | no | 1.4 | p.T170M 1.39 | 35 | Alive | |
| MP246 | JAK2 | F | ET | 71 | 108 | 103 | A | 82 | 0 | 30 | no | 0.2 | p.D259H 0.2; p.G245S 0.2 | 48 | Alive | |
| MP314 | JAK2 | F | PV | 69 | 233 | 92 | H | 86 | 0 | 0 | B | 1.3 | p.C242Y 1.26; p.G245D 0.35 | 39 | Alive | |
| MP8 | JAK2 | M | PV | 82 | 92 | 90 | H | 90 | 0 | 0 | no | 0.2 | p.R175H 0.24 | 49 | Dead | |
| MP356 | JAK2 | F | ET | 69 | 109 | 91 | H | 91 | 0 | 0 | no | 1.6 | p.R248Q 1.64 | 37 | Alive | |
| MP230 | JAK2 | M | MPN | 70 | 96 | 96 | H | 96 | 0 | 0 | no | 0.5 | p.R248Q 0.50; c.512_514dup 0.22; p.V274G 0.20 | 22 | Dead | |
| MP289 | JAK2 | F | PV | 61 | 265 | 140 | no | 98 | 42 | 0 | no | 3.3 | p.R213* 3.29; p.I251T 0.93; p.Y234H 0.67 | 39 | Alive | |
| MP273 | JAK2 | F | ET | 75 | 111 | 111 | H | 111 | 0 | 0 | no | 0.3 | p.C176S 0.31; p.S240G 0.22 | 47 | Alive | |
| MP229 | JAK2 | F | PMF | 85 | 112 | 112 | H | 112 | 0 | 0 | no | 1.0 | c.376-1G>A 0.95 | 49 | Alive | |
| MP363 | JAK2 | M | ET | 87 | 117 | 112 | H | 112 | 0 | 0 | no | 0.3 | p.R248Q 0.29 | 37 | Alive | |
| MP7 | JAK2 | F | PV | 69 | 133 | 130 | H | 121 | 9 | 0 | no | 0.5 | p.I255T 0.46 | 64 | Alive | |
| MP329 | JAK2 | F | ET | 73 | 162 | 142 | H | 142 | 0 | 0 | no | 2.6 | p.R248W 2.61 | 27 | Alive | |
| MP250 | JAK2 | F | PV | 74 | 152 | 152 | H | 152 | 0 | 0 | no | 0.4 | p.W91* 0.36 | 11 | Dead | |
| MP2 | CALR | F | ET | 68 | 195 | 186 | HA | 169 | 11 | 110 | no | 11.6 | p.E286K 11.60; p.R248Q 0.45 | 67 | Alive | |
| MP324 | JAK2 | M | PV | 72 | 255 | 255 | H | 255 | 0 | 0 | no | 0.3 | p.T253A 0.26 | 15 | Dead | |
Abbreviations: A/ANG, anagrelide; B, busulphan; CH, chemotherapy; ET, essencial thrombocythemia; F, female; H/HU, hydroxyurea; I/IFN, interferon alpha; M, male; mo, months; PMF, primary myelofibrosis; PV, polycythemia vera; R, radiotherapy; y, years.
All three driver genes (JAK2 V617F+exon 12, CALR exon 9 and MPL exon 10) were sequenced in all patients with TP53 mutation.
Sample from leukemic transformation not available.
sAML JAK2-wt/TP53-wt.
Clonal expansion without leukemic transformation.
TP53 mutations in MPN are strongly associated with higher age
TP53 mutations were found in all disease subtypes, regardless of driver gene status and even if no HU had been administered (Figure 1; Table 2). Further, we performed thorough analysis of relationships between TP53 mutational status and disease parameters (Supplementary Table S4). The comparison of patients carrying TP53 mutation (TP53-mut) to patients without TP53 mutation (TP53-wt) revealed a highly significant association between TP53 mutations and higher age (P=5.54 × 10−5; median age at sampling 69.3 for TP53-mut and 62.4 for TP53-wt; Figure 2a). TP53 mutations were less frequent in patients who obtained ANG during disease course (9/104, 8.7% in ANG-no vs 32/150, 21.3% in ANG-yes; P=0.0087). Patients receiving HU at sampling carried the TP53 mutation more frequently (HU at sampling, 28/125, 22.4% vs without HU at sampling, 13/128, 10.2% P=0.0205) but associating the TP53 mutation with HU administration anytime during disease course did not reach significance (31/164; 18.9% in HU-yes vs 10/90; 11.1% in HU-no). As expected, the age at sampling was significantly associated to multiple therapy parameters, partially due to the frequent use of HU in older patients (Figure 2b, Supplementary Figure S3).
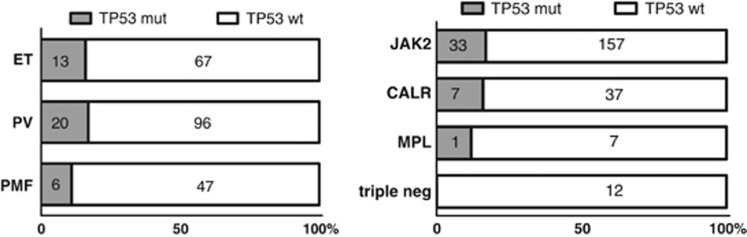
Figure 1.
Disease type and driver gene mutation status stratified according to TP53 mutation presence in patients treated with cytoreductive drugs (NS; Fisher exact test). Driver gene mutations examined in order of JAK2>CALR>MPL.
Table 2. Clinical characteristics of patients according to TP53 mutational status.
|
Treated (N=254) |
Untreated (N=85) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TP53 wt | % | TP53 mut | % | P | TP53 wt | % | TP53 mut | % | P | |
| N | 213 | 100 | 41 | 100 | 76 | 100 | 9 | 100 | ||
| Sex (male) | 91 | 43 | 16 | 39 | NS | 36 | 47 | 5 | 56 | NS |
| Age at study enrollment (median, range, s.d.) | 62 (25–90, 13.2) | 69 (55–87, 7.4) | 0.0002 | 70 (19–89, 13.8) | 70 (66–82, 5.4) | NS | ||||
| Time from diagnosis (mo; median, range, s.d.) | 109 (50–354, 54.9) | 111 (50–265, 54.5) | NS | 0 (0–255, 52.9) | 4 (0–53, 17.2) | NS | ||||
| Disease subtype | ||||||||||
| ET | 67 | 31 | 13 | 32 | NS | 21 | 28 | 1 | 11 | NS |
| PV | 96 | 45 | 20 | 49 | 20 | 26 | 2 | 22 | ||
| PMF | 47 | 22 | 6 | 15 | 30 | 39 | 6 | 67 | ||
| unclassified MPN | 3 | 1 | 2 | 5 | 5 | 7 | 0 | 0 | ||
| JAK2-mutated | 157 | 74 | 33 | 80 | NS | 54 | 72 | 8 | 89 | NS |
| JAK2-wt | 56 | 26 | 8 | 20 | 22 | 29 | 1 | 11 | ||
| CALR-mut | 37 | 17 | 7 | 17 | 13 | 17 | 1 | 11 | ||
| MPL-mut | 7 | 3 | 1 | 2 | 4 | 5 | 0 | 0 | ||
| Triple negative | 12 | 6 | 0 | 0 | 3 | 4 | 0 | 0 | ||
| Therapetutic history | ||||||||||
| Total therapy length (mo; median, range, s.d.) | 87 (24–265, 44.7) | 92 (18–255, 45.4) | NS | 0 | 0 | |||||
| HU yes | 133 | 62 | 31 | 76 | NS | 0 | 0 | 0 | 0 | |
| Length of HU in HU yes (mo; median, range, s.d.) | 65 (2–265, 53.9) | 76 (17–255, 46.1) | 0.0343 | 0 | 0 | |||||
| HU at study enrollment | 97 | 46 | 28 | 68 | 0.0120 | 0 | 0 | 0 | 0 | |
| HU⩾48 months | 94 | 44 | 28 | 68 | 0.0060 | 0 | 0 | 0 | 0 | |
| ANG yes | 95 | 45 | 9 | 22 | 0.0392 | 0 | 0 | 0 | 0 | |
| IFN yes | 72 | 34 | 12 | 29 | NS | 0 | 0 | 0 | 0 | |
| Busulfan/chemo-/radiotherapy | 9 | 4 | 3 | 7 | NS | 2 | 3 | 1 | 11 | NS |
Abbreviations: %, percentage of patients with given parameter in TP53-wt or TP53-mut group; mo, months; driver gene status considered in order JAK2>CALR>MPL; PMF, primary myelofibrosis; PV, polycythemia vera; ET, essential thrombocythemia; post-PV MF was grouped to PV, no post-PV patient carried TP53 mutation. MPL status was unknown in one untreated JAK2-wt/CALR-wt/TP53-wt patient
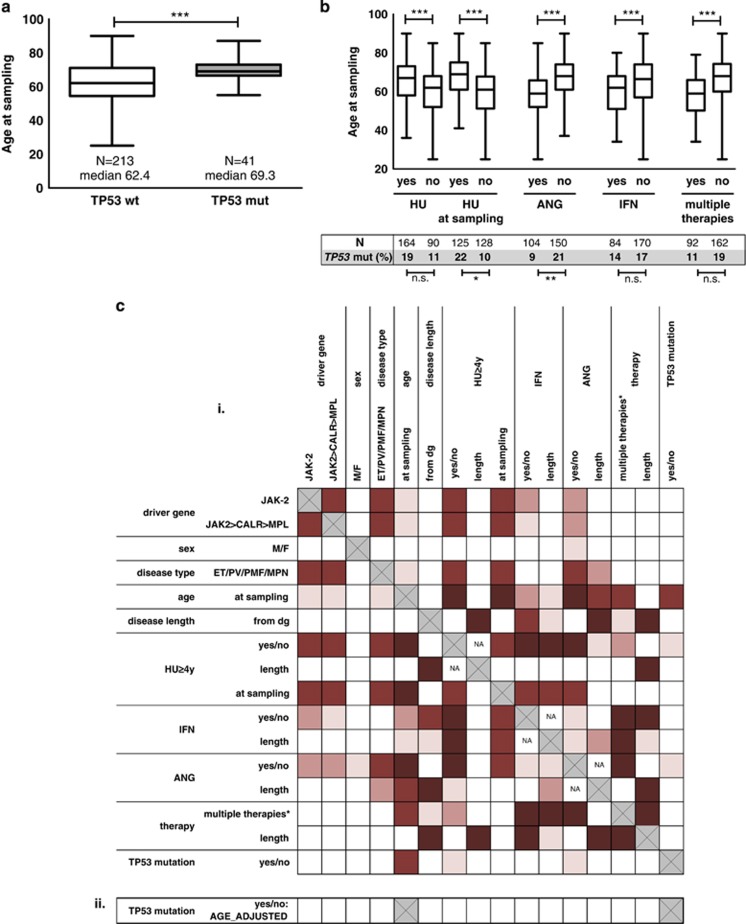
Figure 2.
TP53 mutations: age and treatment in patients treated with cytoreductive drugs. (a) Age at sampling in TP53-mut and TP53-wt patients (P=5.54 × 10−5; Kruskal–Wallis test). (b) Age at sampling and TP53 mutation frequency according to therapy parameters (Kruskal–Wallis and Fisher exact test, respectively; age: HU-yes/no P=0.0007; HU at sampling yes/no P=3.96 × 10−7; ANG-yes/no P=1.32 × 10−8; IFN-yes/no P=0.0006; multiple therapies during disease course yes/no P=8.7 × 10−8). Lines within boxes indicate median, box limits—25th and 75th percentiles, whiskers—minimum and maximum. (c) Comparison of patients treated with HU for ⩾4y and patients treated with IFN or/and ANG only. (i) Relationship between clinical and laboratory parameters. Red boxes: significant, scaled from P⩽0.0001 (dark red) to 0.05⩾P>0.01 (light red). Statistical tests used for combinations of variables: continuous—Spearman correlation test, continuous vs categorical—Kruskal–Wallis test; categorical–Fisher’s exact test. (ii) Logistic regression model with the age adjustment did not reveal any age-independent significant difference. *Multiple therapies=more types of cytoreductive therapy during disease course. Length of the therapy=restricted to patients positive for given therapy type.
As the patient cohort was compiled of several hospitals' contributions, which may have introduced bias, we limited the analysis to University Hospital Brno patients (N=169; NTP53-mut=22), which lead to similar results (data not shown).
Some of the patients received HU for a short time period and were switched to other therapy or vice versa. Thus, to further disclose the relationship between TP53 mutations, HU, and age, we eliminated these patients from the analysis, using more stringent criteria to categorize patient therapy (Figure 2c; Supplementary Table S5): (1) patients who had obtained HU for at least 48 months (N=122) and (2) patients treated with IFN and/or ANG only (HU-no; N=90). Also in this subset, patient age was the most significant predictor of TP53 mutation (Supplementary Figure S4A; TP53-mut 69.5y vs TP53-wt 63.4y; P=0.0009) and TP53 mutations were more frequent in patients in the HU subgroup (HU⩾4y, 28/122, 22.9% vs HU-no, 10/90, 11.1% P=0.03). In parallel, the therapy category was strongly associated to age (Supplementary Figure S4B). To eliminate the influence of age, we applied a logistic regression model with the age adjustment (Supplementary Table S6); using this approach, the TP53 mutation frequency was not found to be significantly different in patients treated with various cytoreductive drugs. This is in agreement with the observation that in patients over 65y, the difference in proportion of TP53 mutation between HU⩾4y and HU-no therapy groups was much lower (23/75, 30.6% vs 6/30, 20.0% n.s.) even though the HU-no group was significantly younger (P=0.026; Supplementary Figure S5).
To further explore whether minor TP53 mutations occur independently of the therapy, we examined a set of 85 patients with no cytoreductive treatment (Supplementary Table S1) and found TP53 mutations in 9 of them (10.6% Supplementary Table S7, Supplementary Figure S6).
TP53 mutations in HU-treated and other patients
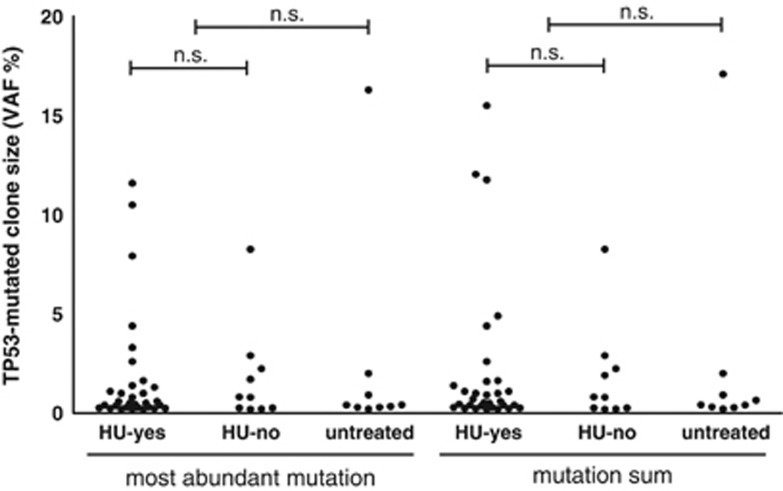
We did not see any difference in mutation spectra between patients treated with HU and the others (Supplementary Figure S7). Neither the VAF of the most abundant variant nor the cumulative size of the mutated population significantly differed between the therapy groups (Figure 3). Nevertheless, we observed a trend towards the presence of more than one mutation (⩾0.2% VAF) in HU-treated patients (10/31) compared to patients treated with non-HU drugs (1/10) and untreated patients (2/9) (n.s.). The mutations showed typical distribution, the vast majority of them were located within the DNA-binding domain and they clustered within characteristic hot-spot sites (Supplementary Figure S8, Supplementary Table S8). All but one patient carried mutations which have been described as non-functional or, rarely, partially functional. The exception was the mutation p.P58A (MP189; VAF 2.9%) which displays no significant loss of transactivation activity.45, 46 The mutation remained stable in all three samplings (7.7y). We have not excluded the mutation from the analysis as we could not rule out other effects on p53 function.
Figure 3.
Mutated clone size. VAFs of most abundant mutation and VAF sums do not significantly differ between therapy groups (Mann–Whitney test).
TP53 mutations may escape detection if examined at diagnosis
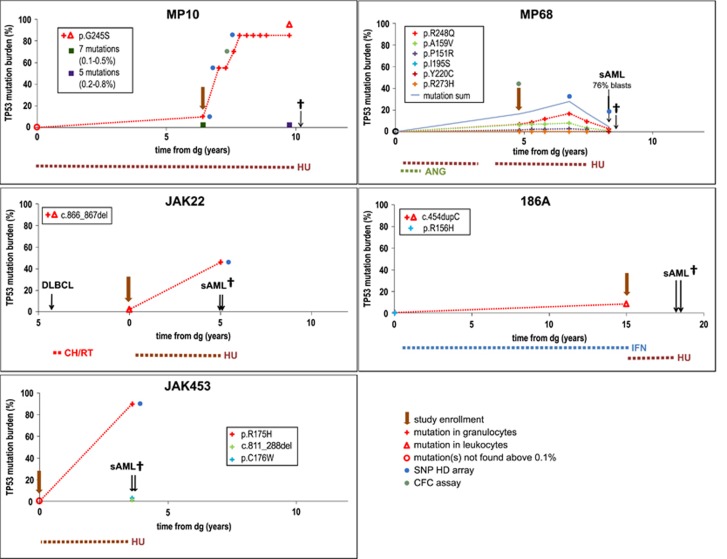
Since the majority of mutations we describe were identified in samples taken later during the disease course, we raised the question whether the mutations can be detected earlier, or even at diagnosis. Out of 50 patients with a mutated clone, at least one retrospective sample was available in 20 (Table 2). A later identified mutation was traceable in at least one sample in 14 patients. The sample from the time of diagnosis was available in 10 patients; no TP53 mutation was detected in 5 (50% VAF⩾0.1%) of them. Correspondingly, the mutation was detectable at diagnosis in only one of two patients who later developed TP53mut sAML (JAK22 and JAK453; Figure 4). In conclusion, despite some TP53 mutations being found at diagnosis with VAF⩾10% (JAK646: 16.3%, Supplementary Table S7; 221A: 10.2% Table 3), mutations frequently appear later in the disease course and may be undetectable (⩾0.1%) at diagnosis.
Figure 4.
Selected cases of leukemic transformation or clonal expansion in patients with TP53 mutations. Details on cytogenetic analysis using Single-nucleotide polymorphism (SNP) HD Array are shown in Supplementary Table S9. MP10: TP53G245S clonal expansion without leukemic transformation in PMF. TP53G245S development was monitored by Sanger sequencing. Diagnostic sample and two other samples (green and purple mark) were analyzed by NGS, for detail see Supplementary Table 1. Chromosome 17 analysis: cn-LOH(17)(p13.3p11.2) in 2nd and 3rd SNP array. MP68: JAK2wtTP53wt-AML outside multiple JAK2V617FTP53mut-clones. After transient increase of JAK2V617F/wtTP53R248Q subclone from 8 to 16%, all TP53mut clones decreased accompanied by JAK2V617F burden drop and transformation to sAML 3.5y from study enrollment. SNP array showed no aberrations on chromosome 17. JAK22: JAK2V617FTP53L289fs/L289fs sAML with complex karyotype changes including cn-LOH(17)(p13.3p11.2) developed from PV secondary to diffuse large B-cell lymphoma (DLBCL). Single mutation TP53L289fs was present at PV diagnosis (2%) and expanded in blast transformation. 186A: sAML with unknown TP53 status developed in JAK2mut ET 2.9y after study enrollment when TP53 mutation was present with VAF 8.3%. JAK453: JAK2V617FTP53R175H/- sAML with complex karyotype changes including del(17p) developed 3.6y after study enrollment at PMF diagnosis when no TP53 mutation and karyotype changes were found.
Table 3. Monitoring TP53 mutations over time.
| Sample ID | Retrospective (R) /prospective (P) analysis | First available sample: time from dg (y) | Study enrollment: time from dg (years) | Last available sample: time from dg (years) | Total follow-up (years) | MPN therapy before study enrollment | Therapy after study enrollment | Mutation ⩾0.1% in dg sample | The development of most abundant mutation (%) a | Mutation development during follow-up |
|---|---|---|---|---|---|---|---|---|---|---|
| 186A | R | 0.0 | 15.0 | 15.0 | 15.0 | IFN | HUb | yes | 0.4-8.3-NA | Increase |
| MP15 | P/R | 0.2 | 7.0 | 11.9 | 11.5 | HU, IFN | IFN | no | 0-0.8-2.1-2.6-3.8-4.4-10.7-11.3-11.2(W)13.8(G) | |
| MP10 | P/R | 0.0 | 6.4 | 9.8 | 9.8 | HU | HU | no | 0-10.1-85.5 | |
| JAK1716 | P/R | 6.0 | 9.3 | 16.2 | 10.2 | IFN | IFN→HU | NA | 0-0.1-0.1-0.2-0.7-2.2-2-2.2 | |
| MP326 | P/R | 0.0 | 6.7 | 9.1 | 9.1 | HU | HU | no | 0-0.6-0.5-0.5 | |
| MP96 | P/R | 0.0 | 5.4 | 8.8 | 8.8 | HU | HU | no | 0-0.5-0.6 | |
| MP168 | P/R | 2.4 | 7.1 | 10.5 | 8.1 | HU | HU | NA | 0.1-1-0.5-0.6 | |
| MP319 | P/R | 0.0 | 5.2 | 7.5 | 7.5 | HU, ANA | HU, ANA | yes | 0.1-0.4-0.4-0.8 | |
| MP369 | P | 0.0 | 0.0 | 7.0 | 7.0 | nonec | HU | NA | NA-0.13-1.4 | |
| 65A | P | 13.2 | 13.2 | 19.6 | 6.5 | IFN, ANA | IFN, ANA | NA | NA-1.7-5.4 | |
| MP327 | P/R | 4.9 | 10.4 | 11.2 | 6.3 | HU | HU | NA | 0-0.1-0.5-0.5 | |
| JAK22 | P | 0.0 | 0.0 | 5.0 | 5.0 | nonec | HU | yes | NA-2.0-46.1 | |
| MP273 | P | 9.3 | 9.3 | 12.0 | 2.8 | HU | HU | NA | NA-0.3/0-0.3/1.7 | |
| MP317 | P | 5.5 | 5.5 | 6.0 | 0.5 | HU | HU | NA | NA-0.8-1.9 | |
| MP289 | P/R | 15.2 | 22.1 | 24.6 | 9.3 | HU, IFN | none | NA | 0.2/1.4-3.3/0.7-4.4/0.4-6.3/0.6-3.5/0.4d | Increase/decrease |
| MP68 | P/R | 0.0 | 4.8 | 8.3 | 8.3 | HU, ANA | HU→none | no | 0-6.9-8.8-10.7-11.7-16.9-9.6-2.7 | |
| MP315 | P/R | 4.6 | 9.1 | 11.5 | 6.9 | IFN | IFN | NA | 0.2-0.8-0.4-0.2 | |
| MP230 | P | 8.0 | 8.0 | 9.2 | 1.2 | HU | HU | NA | NA-0.5-2.3-1.3 | |
| MP189 | P/R | 6.9 | 11.5 | 14.6 | 7.7 | ANA | HU, ANA | NA | 3.9-2.8-2.5 | Stable |
| MP155 | P/R | 5.7 | 10.2 | 13.0 | 7.3 | ANA | ANA | NA | 0.4-0.2-0.2 | |
| MP229 | P | 9.4 | 9.4 | 12.4 | 3.0 | HU | HU, ANA | NA | NA-1-0.5-0.6 | |
| MP302 | P | 19.3 | 19.3 | 21.7 | 2.4 | busulfan→HU | HU | NA | NA-0.4-0.4-0.7 | |
| MP329 | P | 13.5 | 13.5 | 15.7 | 2.2 | HU | HU, ANA | NA | NA-2.6-1.9-3.1/3.0 | |
| MP324 | P | 21.2 | 21.2 | 22.0 | 0.7 | HU | HU | NA | NA-0.3-0.4 | |
| 221A | P/R | 0.0 | 15.0 | 24.6 | 24.6 | IFN | IFN | yes | 10.2/ND-2.2/0.6-0.7/2.0d | Decrease |
| MP2 | P/R | 12.1 | 16.3 | 21.7 | 9.6 | HU, IFN, ANA | HU, ANA | NA | 11.5-10.6-11.6-2.9-2.2-1.9-2.1 | |
| MP63 | P/R | 1.5 | 6.6 | 10.0 | 8.5 | HU | HU | NA | 0-0.2/0.1-0.1/0.2-0/0d | |
| MP160 | P/R | 1.2 | 5.1 | 7.1 | 5.8 | HU | HU | NA | 0.5-0.2-0 | |
| MP307 | P | 8.6 | 8.6 | 10.2 | 1.6 | HU | HU | NA | 0.2-0 | |
| MP314 | P/R | 14.8 | 19.4 | 21.9 | 7.1 | busulfan→HU | HU | NA | 3.3/0.2-1.3/0.4-0.6/0.9-0.2/0.2d | |
| MP345 | P/R | 0.0 | 4.2 | 6.3 | 6.3 | HU | HU | yes | 0.1-1.1-0.9-0.5-0.6 |
Abbreviations: G, granulocytes; NA, not available; W, leukocytes.
Mutations with highest VAF are shown. "Study enrollment" describes the sampling from which the mutation was identified. For over-time monitoring and validation of previously identified mutation, cut-off 0.1% was applied (minimal coverage per base ⩾10000). VAF bellow 0.1% considered as a background (0%). Development was cathegorised as follows: increase—VAF twofold increase between first and last sample or no mutation at diagnosis; decrease—VAF decrease in VAF to half between first and last sample or no mutation at last sampling; increase/decrease—increase followed by decrease; stable—other. y, years; NA, not available; G, granulocytes; W, leukocytes.
Result from study enrollment is highlighted.
Samples not available.
Mutation identified at diagnosis.
Two mutations with the highest VAFs in distinct samplings.
Monitoring patients with TP53 mutations – dynamic behavior of mutated clones
To explore TP53 mutation evolution in MPN prospectively, disease course was monitored and serial samples were collected. Prospective samples were available in 30 of 50 MPN patients with TP53mut-subclones (median between study enrollment and the last serial sample 2.8y; 0.5–9.6y; Table 3). TP53 mutations remained present in all but three serial samples (MP63, MP160 and MP307) in which mutations originally identified and confirmed in 0.2% were not detected 3.4, 2 and 1.6 years later, respectively.
TP53 mutation expanded and became predominant tumor cell population in 2/30 patients. The expansion was accompanied by the second allele inactivation in both cases. While the JAK22 patient progressed to AML carrying biallelic TP53 inactivation (VAF 46%), the clonal biallelic expansion in patient MP10 (VAF 86%) did not result in leukemic transformation; this case is described further in detail (Figure 4; Supplementary Table S9).
When we considered all samples tested during the disease course, that is, retrospective as well as prospective samples, median interval between first and last sample was 7.1y (0.5–24.6; 31 patients). During this time, a slow mutation burden increase was the most frequent scenario (14 patients). We saw no clear association between the mutation burden changes and the VAF at study enrollment, therapy, other clinical data or mutation localization (Table 3, Supplementary Figure S8B and S10).
Impact of TP53 mutations on overall survival or leukemic transformation
TP53 mutation with VAF<5% did not impact overall survival (OS) during the follow-up when tested either from diagnosis or study enrollment (Supplementary Figure S9). The patients carrying TP53 mutations above 5% VAF at study enrollment had significantly shorter OS (P=0.0064 OS from sampling, P=0.0185 OS from diagnosis). However, the impact on OS was lost when adjusted for age (Cox proportional hazard regression model for both age and mutation: P=1.01 × 10−10 for age; P=0.121 for TP53 mutation>5%). Besides, their shortened survival was not attributed to TP53-mut AML (Supplementary Figure S9G).
In total, AML developed in three patients with TP53 mutation (3/50; 6% Figure 4). Patient JAK22 (p.289fs 2%) was diagnosed with PV 4y after chemo/radiotherapy for B-cell lymphoma. The patient was treated with HU and progressed to TP53mut-AML 5y later. On the contrary, patient MP68 (p.R248Q 6.9%) treated with HU developed JAK2wtTP53wt-AML outside multiple JAK2mutTP53mut-clones 3.5y from study enrollment (8.3y from diagnosis). Patient 186A (p.P153fs 8.27%) treated with IFN progressed to AML 2.9y after mutation detection (17.9y from diagnosis). The patient was switched to HU soon after study enrollment and an AML sample was not available, thus we cannot confirm the clonal expansion of TP53 mutation or the effect of the therapy.
Rapid TP53-mutated clone expansion accompanied by cn-LOH but no other karyotype changes resulted neither in AML transformation nor HU resistance
In the JAK2mut-PMF patient MP10, multiple TP53 low-burden mutations were present at study enrollment. Among them, a hot-spot TP53G245S mutation grew rapidly during prospective monitoring up to 95% in granulocytes (Figure 4), reflecting the loss of heterozygosity (LOH). Since complex karyotype changes have been described as very common in AML with mutated TP53,18 we analyzed the karyotype changes using CytoScan HD arrays. Only copy-neutral LOH (cn-LOH) in 17p13, including TP53 gene, and chromosome Y loss were detected in the expanded clone. To further examine the time course of allelic changes, we analyzed myeloid progenitors. CFC assay indicated that second allele inactivation occurred intra-clonally in the clone carrying monoallelic p.G245S mutation (Supplementary Figure S2). Interestingly, despite complete p53 inactivation and clonal expansion, the patient remained clinically stable without signs of blast transformation for next 23 months, showed no signs of HU resistance and died 10.2y from diagnosis.
Discussion
Previously published retrospectively analyzed cases showed that the development of TP53-mutated post-MPN AML is a multistep process. It likely involves mutation origin in the HSPC pool, mutated subclone propagation to level exceeding detection limit and persistence at low levels for an extended time period. Second allele inactivation was described as resulting in rapid clonal expansion and leukemic transformation.
We focused on the early phase of this process, that is, occurrence of low-burden TP53 mutations which, in theory, may carry increased risk of leukemic transformation. Using highly sensitive and previously verified methods enabled us to detect mutations as low as 0.2%.26 In total, we found mutations in 50 patients (14.7%). This is the first study using ultra-deep NGS to search for TP53 mutations in MPN at a level ⩽1%. Lundberg et al.11 found mutations in 5/197 (2.5%) patients using NGS with sensitivity of 5%, which roughly corresponds to our data (5 patients with mutations >5% out of all 339 examined, 1.5%). As our study aimed to compare patients on HU and non-HU therapies, the frequency in the general MPN cohort was out of the scope. However, some information may be gained from our analysis of 48 consecutive newly diagnosed MPNs examined partially within the untreated cohort and partially as retrospective samples (data not shown): only one mutation ⩾0.2% was found (2.1%). This observation, together with the slow increase in mutation load during disease course and strong age bias, points to the fact that TP53 mutations are probably rare in general cohorts at diagnosis.
We detected no mutation ⩾0.2% in 70 patients below 55y. In contrast, 41/179 (22.9%) patients above 65y carried TP53 mutation. This agrees with the hypothesis that TP53 mutations arose spontaneously and accumulate with age, as described in the elderly population without hematological abnormalities.47 Higher age brings inherent risks of MPN transformation1 and less vital progenitor pools in the elderly may favor cells carrying oncogenic mutations both under DNA-damaging and normal conditions.48 On the other hand, younger patients are a subgroup with the perspective of decades living with clonally shifted hematopoiesis and using cytoreductive drugs and should be examined in detail.
It has been described that not only single accidentally arising TP53-mutated clones but several coexisting in parallel may be present in myeloid precursors before expansion.49 We detected more than one mutation in one third of cases. This phenomenon, described as ‘convergence’,50 occurs for example, in chronic lymphocytic leukemia (CLL)51 and points to a selective pressure favoring the mutations in some but not all patients. In MPN, one may consider either the pressure of cytoreductive therapy, since multiple mutations tended to be more frequent in HU-treated patients, or, possibly, the effort of non-vital HPSC to survive and proliferate. The clonal competition among individual TP53-mutated subclones is difficult to foresee and likely depends on accompanying defects; hot-spot mutations with documented oncogenic properties may be overgrown by subclones carrying variants with lower oncogenic potential (for example, loss-of function mutations).25, 26
The murine model and clinical observations pointed to the leukemogenic potential of JAK2V617F overexpression in TP53-null background;23, 52 JAK2wtTP53mut AML following JAK2mut-MPN is however not exceptional.22 We observed no clear evidence supporting the leukemogenic potential of TP53mut and JAK2V617F combination. Admittedly, the JAK2 mutation was homozygous neither in patient MP68 developing JAK2wt-AML alongside multiple JAK2mutTP53mut subclones nor patient MP10 with complete TP53 loss within the JAK2V617F population.
TP53/17p-aberrant post-MPN AML has been repeatedly suggested - but never independently proven - to be associated with HU therapy.4, 12, 22, 36 Several findings support possible TP53mut-subclone selection by HU: (1) HU blocks cell division via ribonucleotide reductase inhibition, resulting in dNTP depletion.34 Replication stress then activates p53 and cell cycle arrest although these processes' p53 dependence is controversial and dose-dependent.35, 53, 54 (2) The expansion of low-burden TP53mut-subclones under TP53-triggering therapy was shown in CLL,26, 27, 28 myelodysplastic syndrome29 and secondary AML.32 Further, HSPC competition triggered by low-level DNA damage in the murine model led to TP53+/+ being outcompeted by TP53+/- cells via senescence-like changes.33 Similarly, clinically relevant low- (but not high) level replication stress induced p53-dependent senescence-like arrest in fibroblasts and led to TP53-aberant subpopulation selection.53 We suppose that if there was a proliferative and/or survival advantage favoring TP53mut-subclones during HU treatment in MPN, we would have observed a pronounced difference in the abundance and incidence of low-burden TP53 mutations after several years of therapy. Contrary to this assumption and observations from in vitro and in vivo models, we saw no significant age-independent difference between patients treated with HU and non-HU therapies; moreover, the mutations were present even if no cytoreductive therapy was given. Although long-term prospective monitoring of patients with TP53 mutations on various therapies is necessary to fully exclude any HU impact on the second allele loss and clonal expansion, the case of patient MP10 further weakens the advantage of MPN cells with aberrant p53 during HU therapy. While the TP53G245S/G245S clone replaced TP53wt myelopoiesis, the patient did not show any signs of HU resistance, blood counts remained unchanged and no disease progression was observed for next 2y, despite the patient being treated with a constant HU dose. The clonal competition in MPN both under and without HU treatment may differ from experimental data for several reasons: (1) chronic low-level replication stress may affect HPSC compartment differently to single dose DNA damage; (2) in highly sensitive myeloid cells, threshold for p53-dependent selection induced by low-level replication stress55 may be shifted; (3) competition may be affected by presence of oncogenic mutations; (4) p53 activity in MPN cells may not fully correspond to that of artificially manipulated p53 in murine and cell line models. In contrast, there seems to be a difference between subclones with monoallelic and biallelic p53 defects, first increasing slowly with the latter expanding rapidly.11
We showed that relatively high proportion of older-age MPN patients carry low-burden TP53 mutations. In contrast to retrospective reports, we did not observe correlation with disease progression accompanied with TP53 mutation clonal expansion. However, our study was not designed with the primary goal to assess the prognostic impact of low-burden TP53 mutations as we were aware that larger cohorts and long follow-up is definitely required to address this issue completely.
The competition between an in-theory-adverse minor subclone and a major population may be more complex than one may assume from the retrospective studies. Observations on minor TP53 mutations in CLL, another non-acute hematological malignancy, show that the mutation does not have to expand despite several specific therapy lines in some patients.26 Further, a TP53-mutated subclone outgrowth occurs very rarely in patients that remain untreated, that is, strong selection pressure in the form of chemotherapy dramatically changes the TP53-wt vs TP53-mut clonal competition.26, 28 Apparently, cytoreduction regimens currently used in MPN do not create such strong pressure. More likely, other intrinsic factors (genomic instability, hematopoiesis exhaustion) lead to disease progression only in a proportion of patients carrying minor TP53-mutated clone.
Importantly, 3 out of 4 patients with clonal evolution (blast transformation or mut-TP53 clonal expansion) carried TP53 mutation(s) with VAF⩾5%. This indicates that TP53 mutations increased to a certain level may be the marker of clonal instability or even a poor prognosis, as demonstrated by shorter OS in patients with mutations ⩾5% VAF in our study which however cannot be attributed to TP53 mutation expansion followed by leukemic transformation. Of note, we recorded 6 patients whose prospective samples were available and VAF exceeded 5% at some point, but none developed TP53-mutated sAML during the follow-up. Moreover, one patient with no mutations ⩾0.1% VAF developed TP53-mutated sAML within 3.6y.
To conclude, we show that minor TP53 mutations are present in a significant proportion of MPN patients and their presence is strongly associated with age. We did not see any significant age-independent association with hydroxyurea therapy, disease type or MPN driver gene mutations. We also show that even a fully expanded biallelic hotspot mutation, leading to complete loss of TP53 transactivation activity,45, 46 does not a priori lead to leukemic transformation. Despite our findings do not support the assumption that there is unequivocal relation between TP53-mutated subclones, HU cytoreduction and leukemic transformation in MPN, larger sample sizes are warranted to definitively address this. TP53 minor mutations in MPN undoubtedly represent a pool for further clonal evolution; however their prognostic and predictive utilization requires further investigation to identify which patients are at risk and whether any risk factors are preventable.
Acknowledgments
We kindly thank the staff of local hospitals in Kromeriz, Znojmo, Breclav, Boskovice, Kyjov, Uherske Hradiste, Ostrava, Hodonin, Trebic, Nove Mesto na Morave for providing samples and clinical data, Hana Skabrahova for organizing blood collections, Jan Palecek for providing us methodical help and Matthew Smith for language editing. Supported by projects of Faculty of Medicine, Masaryk University MUNI/A/1106/2016 and ROZV/24/LF/2016 (SPav and JMal), by projects of MH CR 16-29447A, MEYS CR LM2015064 EATRIS, CEITEC 2020 (LQ1601) and TACR TE02000058. Also supported by Czech Leukemia Study Group for Life (CELL) and Genomics Core Facility CEITEC under MEYS CR project LM2011020.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
HG: AOP Orphan, Calgene, Novartis—Consultancy, Honoraria, Research Funding and Speakers Bureau; JC: Honoraria and Speakers Bureau; Baxalta-Consultancy and Honoraria. MJ: Research Funding from AOP Orphan and Novartis. KR: Research Funding from AOP Orphan, a member on an entity’s Board of Directors or advisory committees in Qiagen. PM: AOP Orphan and Novartis—Consultancy, Honoraria, Research Funding and Speakers Bureau. DM: AOP Orphan, Novartis—Consultancy and Honoraria. The remaining authors declare no conflict of interest.
Supplementary Material
References
- Finazzi G, Caruso V, Marchioli R, Capnist G, Chisesi T, Finelli C et al. Acute leukemia in polycythemia vera: an analysis of 1638 patients enrolled in a prospective observational study. Blood 2005; 105: 2664–2670. [DOI] [PubMed] [Google Scholar]
- Kiladjian JJ, Chevret S, Dosquet C, Chomienne C, Rain JD. Treatment of polycythemia vera with hydroxyurea and pipobroman: final results of a randomized trial initiated in 1980. J Clin Oncol 2011; 29: 3907–3913. [DOI] [PubMed] [Google Scholar]
- Berk PD, Goldberg JD, Silverstein MN, Weinfeld A, Donovan PB, Ellis JT et al. Increased incidence of acute leukemia in polycythemia vera associated with chlorambucil therapy. N Engl J Med 1981; 304: 441–447. [DOI] [PubMed] [Google Scholar]
- Sterkers Y, Preudhomme C, Laï JL, Demory JL, Caulier MT, Wattel E et al. Acute myeloid leukemia and myelodysplastic syndromes following essential thrombocythemia treated with hydroxyurea: high proportion of cases with 17p deletion. Blood 1998; 91: 616–622. [PubMed] [Google Scholar]
- Björkholm M, Derolf AR, Hultcrantz M, Kristinsson SY, Ekstrand C, Goldin LR et al. Treatment-related risk factors for transformation to acute myeloid leukemia and myelodysplastic syndromes in myeloproliferative neoplasms. J Clin Oncol 2011; 29: 2410–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A. Is hydroxyurea leukemogenic in essential thrombocythemia? Blood 1998; 92: 1459–1460, author reply 1460–1451. [PubMed] [Google Scholar]
- Nand S, Stock W, Godwin J, Fisher SG. Leukemogenic risk of hydroxyurea therapy in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Am J Hematol 1996; 52: 42–46. [DOI] [PubMed] [Google Scholar]
- Liozon E, Brigaudeau C, Trimoreau F, Desangles F, Fermeaux V, Praloran V et al. Is treatment with hydroxyurea leukemogenic in patients with essential thrombocythemia? An analysis of three new cases of leukaemic transformation and review of the literature. Hematol Cell Ther 1997; 39: 11–18. [DOI] [PubMed] [Google Scholar]
- Baz W, Najfeld V, Yotsuya M, Talwar J, Terjanian T, Forte F. Development of myelodysplastic syndrome and acute myeloid leukemia 15 years after hydroxyurea use in a patient with sickle cell anemia. Clin Med Insights Oncol 2012; 6: 149–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkholm M, Hultcrantz M, Derolf Å. Leukemic transformation in myeloproliferative neoplasms: therapy-related or unrelated? Best Pract Res Clin Haematol 2014; 27: 141–153. [DOI] [PubMed] [Google Scholar]
- Lundberg P, Karow A, Nienhold R, Looser R, Hao-Shen H, Nissen I et al. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood 2014; 123: 2220–2228. [DOI] [PubMed] [Google Scholar]
- Thoennissen NH, Krug UO, Lee DH, Kawamata N, Iwanski GB, Lasho T et al. Prevalence and prognostic impact of allelic imbalances associated with leukemic transformation of Philadelphia chromosome-negative myeloproliferative neoplasms. Blood 2010; 115: 2882–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Wahab O, Manshouri T, Patel J, Harris K, Yao J, Hedvat C et al. Genetic analysis of transforming events that convert chronic myeloproliferative neoplasms to leukemias. Cancer Res 2010; 70: 447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SJ, Rampal R, Manshouri T, Patel J, Mensah N, Kayserian A et al. Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood 2012; 119: 4480–4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Harada Y, Imagawa J, Kimura A, Harada H. AML1/RUNX1 point mutation possibly promotes leukemic transformation in myeloproliferative neoplasms. Blood 2009; 114: 5201–5205. [DOI] [PubMed] [Google Scholar]
- Green A, Beer P. Somatic mutations of IDH1 and IDH2 in the leukemic transformation of myeloproliferative neoplasms. N Engl J Med 2010; 362: 369–370. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Zuber J, Diaz-Flores E, Lintault L, Kogan SC, Shannon K et al. p53 loss promotes acute myeloid leukemia by enabling aberrant self-renewal. Genes Dev 2010; 24: 1389–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferlach C, Dicker F, Herholz H, Schnittger S, Kern W, Haferlach T. Mutations of the TP53 gene in acute myeloid leukemia are strongly associated with a complex aberrant karyotype. Leukemia 2008; 22: 1539–1541. [DOI] [PubMed] [Google Scholar]
- Harutyunyan A, Klampfl T, Cazzola M, Kralovics R. p53 lesions in leukemic transformation. N Engl J Med 2011; 364: 488–490. [DOI] [PubMed] [Google Scholar]
- Neri A, Fracchiolla NS, Radaelli F, Boletini A, Ribera S, Migliorini C et al. p53 tumour suppressor gene and RAS oncogenes: molecular analysis in the chronic and leukaemic phases of essential thrombocythaemia. Br J Haematol 1996; 93: 670–673. [DOI] [PubMed] [Google Scholar]
- Gaidano G, Guerrasio A, Serra A, Carozzi F, Cambrin GR, Petroni D et al. Mutations in the P53 and RAS family genes are associated with tumor progression of BCR/ABL negative chronic myeloproliferative disorders. Leukemia 1993; 7: 946–953. [PubMed] [Google Scholar]
- Beer PA, Delhommeau F, LeCouédic JP, Dawson MA, Chen E, Bareford D et al. Two routes to leukemic transformation after a JAK2 mutation-positive myeloproliferative neoplasm. Blood 2010; 115: 2891–2900. [DOI] [PubMed] [Google Scholar]
- Rampal R, Ahn J, Abdel-Wahab O, Nahas M, Wang K, Lipson D et al. Genomic and functional analysis of leukemic transformation of myeloproliferative neoplasms. Proc Natl Acad Sci USA 2014; 111: E5401–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles A, Harris C. Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harb Perspect Biol 2010; 2: a001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcikova J, Pavlova S, Kozubik KS, Pospisilova S. TP53 mutation analysis in clinical practice: lessons from chronic lymphocytic leukemia. Human Mutation 2014; 35: 663–671. [DOI] [PubMed] [Google Scholar]
- Malcikova J, Stano-Kozubik K, Tichy B, Kantorova B, Pavlova S, Tom N et al. Detailed analysis of therapy-driven clonal evolution of TP53 mutations in chronic lymphocytic leukemia. Leukemia 2015; 29: 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenz T, Krober A, Scherer K, Habe S, Buhler A, Benner A et al. Monoallelic TP53 inactivation is associated with poor prognosis in chronic lymphocytic leukemia: results from a detailed genetic characterization with long-term follow-up. Blood 2008; 112: 3322–3329. [DOI] [PubMed] [Google Scholar]
- Rossi D, Khiabanian H, Spina V, Ciardullo C, Bruscaggin A, Famà R et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood 2014; 123: 2139–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jädersten M, Saft L, Pellagatti A, Göhring G, Wainscoat JS, Boultwood J et al. Clonal heterogeneity in the 5q- syndrome: p53 expressing progenitors prevail during lenalidomide treatment and expand at disease progression. Haematologica 2009; 94: 1762–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belickova M, Vesela J, Jonasova A, Pejsova B, Votavova H, Merkerova MD et al. TP53 mutation variant allele frequency is a potential predictor for clinical outcome of patients with lower-risk myelodysplastic syndromes. Oncotarget 2016; 7: 36266–36279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusyk A, Porter CC, Zaberezhnyy V, DeGregori J. Irradiation selects for p53-deficient hematopoietic progenitors. PLoS Biol 2010; 8: e1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong TN, Ramsingh G, Young AL, Miller CA, Touma W, Welch JS et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature 2015; 518: 552–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondar T, Medzhitov R. p53-mediated hematopoietic stem and progenitor cell competition. Cell Stem Cell 2010; 6: 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakoff IH, Brown NC, Reichard P. Inhibition of ribonucleoside diphosphate reductase by hydroxyurea. Cancer Res 1968; 28: 1559–1565. [PubMed] [Google Scholar]
- Nayak BK, Das GM. Stabilization of p53 and transactivation of its target genes in response to replication blockade. Oncogene 2002; 21: 7226–7229. [DOI] [PubMed] [Google Scholar]
- Furgerson JL, Vukelja SJ, Baker WJ, O'Rourke TJ. Acute myeloid leukemia evolving from essential thrombocythemia in two patients treated with hydroxyurea. Am J Hematol 1996; 51: 137–140. [DOI] [PubMed] [Google Scholar]
- Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood 2009; 114: 937–951. [DOI] [PubMed] [Google Scholar]
- van Dongen JJ, Langerak AW, Brüggemann M, Evans PA, Hummel M, Lavender FL et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 2003; 17: 2257–2317. [DOI] [PubMed] [Google Scholar]
- Vandenbroucke I, Van Marck H, Verhasselt P, Thys K, Mostmans W, Dumont S et al. Minor variant detection in amplicons using 454 massive parallel pyrosequencing: experiences and considerations for successful applications. Biotechniques 2011; 51: 167–177. [DOI] [PubMed] [Google Scholar]
- Varley JM. Germline TP53 mutations and Li-Fraumeni syndrome. Hum Mutat 2003; 21: 313–320. [DOI] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res 2015; 43 (Database issue): D805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R_Core_Team. R: A Language and Environment for Statistical Computing, 2015. Available at https://www.R-project.org.
- Olcaydu D, Harutyunyan A, Jäger R, Berg T, Gisslinger B, Pabinger I et al. A common JAK2 haplotype confers susceptibility to myeloproliferative neoplasms. Nat Genet 2009; 41: 450–454. [DOI] [PubMed] [Google Scholar]
- Klampfl T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med 2013; 369: 2379–2390. [DOI] [PubMed] [Google Scholar]
- Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J et al. TP53 variations in human cancers: new lessons from the IARC TP53 Database and Genomics Data. Hum Mutat 2016; 37: 865–876. [DOI] [PubMed] [Google Scholar]
- Soussi T, Hamroun D, Hjortsberg L, Rubio-Nevado JM, Fournier JL, Béroud C. MUT-TP53 2.0: a novel versatile matrix for statistical analysis of TP53 mutations in human cancer. Hum Mutat 2010; 31: 1020–1025. [DOI] [PubMed] [Google Scholar]
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014; 371: 2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusyk A, DeGregori J. Declining cellular fitness with age promotes cancer initiation by selecting for adaptive oncogenic mutations. Biochim Biophys Acta 2008; 1785: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer PA, Ortmann CA, Stegelmann F, Guglielmelli P, Reilly JT, Larsen TS et al. Molecular mechanisms associated with leukemic transformation of MPL-mutant myeloproliferative neoplasms. Haematologica 2010; 95: 2153–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012; 366: 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jethwa A, Hüllein J, Stolz T, Blume C, Sellner L, Jauch A et al. Targeted resequencing for analysis of clonal composition of recurrent gene mutations in chronic lymphocytic leukaemia. Br J Haematol 2013; 163: 496–500. [DOI] [PubMed] [Google Scholar]
- Tsuruta-Kishino T, Koya J, Kataoka K, Narukawa K, Sumitomo Y, Kobayashi H et al. Loss of p53 induces leukemic transformation in a murine model of Jak2 V617F-driven polycythemia vera. Oncogene 2017; 36: 3300–3311. [DOI] [PubMed] [Google Scholar]
- Marusyk A, Wheeler LJ, Mathews CK, DeGregori J. p53 mediates senescence-like arrest induced by chronic replicational stress. Mol Cell Biol 2007; 27: 5336–5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottifredi V, Shieh S, Taya Y, Prives C. p53 accumulates but is functionally impaired when DNA synthesis is blocked. Proc Natl Acad Sci USA 2001; 98: 1036–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusyk A, DeGregori J. Replicational stress selects for p53 mutation. Cell Cycle 2007; 6: 2148–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.