Abstract
Chromosomal rearrangements of the human MLL/KMT2A gene are associated with infant, pediatric, adult and therapy-induced acute leukemias. Here we present the data obtained from 2345 acute leukemia patients. Genomic breakpoints within the MLL gene and the involved translocation partner genes (TPGs) were determined and 11 novel TPGs were identified. Thus, a total of 135 different MLL rearrangements have been identified so far, of which 94 TPGs are now characterized at the molecular level. In all, 35 out of these 94 TPGs occur recurrently, but only 9 specific gene fusions account for more than 90% of all illegitimate recombinations of the MLL gene. We observed an age-dependent breakpoint shift with breakpoints localizing within MLL intron 11 associated with acute lymphoblastic leukemia and younger patients, while breakpoints in MLL intron 9 predominate in AML or older patients. The molecular characterization of MLL breakpoints suggests different etiologies in the different age groups and allows the correlation of functional domains of the MLL gene with clinical outcome. This study provides a comprehensive analysis of the MLL recombinome in acute leukemia and demonstrates that the establishment of patient-specific chromosomal fusion sites allows the design of specific PCR primers for minimal residual disease analyses for all patients.
Introduction
Chromosomal rearrangements involving the human MLL gene are recurrently associated with the disease phenotype of acute leukemias.1, 2 The presence of distinct MLL rearrangements is an independent dismal prognostic factor, while very few MLL rearrangements display either a good or an intermediate outcome.3, 4 It became also clear from recent studies that the follow-up of patients during treatment and therapy adjustment based on minimal residual disease (MRD) monitoring has a very strong impact on outcome.5, 6, 7 For this purpose, we established a diagnostic network that allowed different study groups and clinical centers to obtain genomic MLL breakpoint sequences that can be directly used for quantifying MRD levels in patients. The current work flow to identify MLL rearrangements includes a prescreening step (cytogenetic analyses,8, 9 split-signal fluorescence in situ hybidization10, 11, 12 or reverse-transcription PCR) in combination with long-distance inverse PCR that was performed on small amounts (~1 μg) of isolated genomic DNA.13 This allowed us to readily identify reciprocal translocations, complex chromosomal rearrangements, gene internal duplications, deletions or inversions on chromosome 11q, and MLL gene insertions into other chromosomes, or vice versa, the insertion of partner chromosome material into the MLL gene located at 11q23.
To gain insight into the frequency of distinct MLL rearrangements, all prescreened samples of infant, pediatric and adult leukemia patients were sent for analysis to the Frankfurt Diagnostic Center of Acute Leukemia (DCAL) after cytogenetic prescreening. All prescreened MLL rearrangements were successfully analyzed at the Frankfurt DCAL, and patient-specific MLL fusion sequences for MRD monitoring were obtained.
Materials and methods
Patient material
Genomic DNA was isolated from bone marrow and/or peripheral blood samples of leukemia patients and sent to the DCAL (Frankfurt/Main, Germany). Patient samples were obtained from different study groups (the AMLCG-study group, Munich; the GMALL study group, Berlin; Polish Pediatric Leukemia and Lymphoma Study Group; Zabrze; I-BFM network) and diagnostic centers in Europe (Aarhus, Berlin, Barcelona, Bordeaux, Bratislava, Brest, Bristol, Catania, Copenhagen, Ekaterinburg, Frankfurt, Giessen, Granada, Graz, Grenoble, Haifa, Hamburg, Hanover, Heidelberg, Jena, Jerusalem, Kiel, Lille, Lisbon, Madrid, Minsk, Montpellier, Monza, Moscow, Munster, Munich, Nancy, Nantes, Newcastle upon Tyne, Olomouc, Padua, Paris, Porto, Prague, Reims, Rotterdam, Strasbourg, Tampere, Tel Hashomer, Toulouse, Turku, Ulm, Valenciennes, Vienna, Zabrze and Zurich) or centers located outside of Europe (Adelaide, Boston, Brisbane, Buenos Aires, Hong Kong, Houston, Rio de Janeiro, Seoul, Sydney and Tohoku), where acute leukemia patients are enrolled in local study groups. Informed consent was obtained from all patients or patients’ parents/legal guardians, and control individuals.
Long distance inverse PCR experiments
All DNA samples were treated and analyzed as described.13, 14, 15, 16 Briefly, 1 μg genomic patient DNA was digested with restriction enzymes and re-ligated to form DNA circles before long-distance inverse PCR analyses. Restriction polymorphic PCR amplimers were isolated from the gel and subjected to DNA sequence analyses to obtain the patient-specific fusion sequences. This genomic DNA fusion sequence is idiosyncratic for each leukemia patient and was made available to the sender of the DNA sample. The average processing time was around 5 working days.
Data evaluation and statistical analyses
All clinical and experimental patient data were implemented into a database program (FileMaker Pro, FileMaker Inc., Santa Clara, CA, USA) for further analysis. Information about all individual patients was used to compare all defined subgroups and to perform statistical analyses to retrieve important information or significant correlations. χ2-tests were performed to identify significant deviations from mean values.
Nomenclature
We are well aware about all the changes in the HUGO gene nomenclature over the past years. However, for the readability of the text, we use the following gene nomenclature throughout the text: MLL (KMT2A); AF4 (AFF1); LAF4 (AFF3); AF5 (AFF4); ENL (MLLT1); AF9 (MLLT3); AF6 (MLLT4); AF17 (MLLT6); AF10 (MLLT10); and AF1Q (MLLT11).
Results
The study cohort
To analyze the recombinome of the human MLL gene, 2381 prescreened acute leukemia samples were obtained from the above-mentioned centers from 2003 to 2016. In all cases, we first used PCR experiments combined with sequence analysis to diagnose the direct MLL fusion allele, and in case of failure or having a complex rearrangement, the reciprocal MLL fusion allele was analyzed. Successful analysis could be performed for all patient samples. In 31 cases we were only able to characterize the reciprocal MLL fusion allele to guarantee subsequent MRD experiments. Complete data were available on 2345/2381 cases (gender, age at diagnosis, disease type and subtype, or information about de novo or secondary leukemia). Genetic and clinical information of these 2345 patients are summarized in Table 1. The 36 excluded cases had the following MLL rearrangements: 9 × MLL–AF9; 8 × MLL–AF4; 4 × MLL–ENL; 4 × MLL–AF10; 3 × MLL–AF6; 2 × MLL–AF17; 2 × MLL–EPS15; 1 × MLL–GAS7; 1 × MLL–LOC100128568; 1 × MLL–CREBBP; and 1 × MLL–PTD. The exclusion of these 36 patients did not affected with the general conclusions made in this study.
Table 1. Overview about all investigated TPGs.
| # | Direct TPG |
Infant |
Pediatric |
Adult |
Total | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | AML | Other | ALL | AML | Other | ALL | AML | Other | |||
| 1 | AFF1/AF4 | 338 | 4 | 10 | 139 | 3 | 10 | 332 | 3 | — | 839 |
| 2 | MLLT3/AF9 | 113 | 40 | 5 | 56 | 132 | 3 | 9 | 90 | 1 | 449 |
| 3 | MLLT1/ENL | 154 | 2 | 4 | 56 | 21 | 1 | 50 | 14 | — | 302 |
| 4 | MLLT10/AF10 | 39 | 43 | 2 | 12 | 66 | 1 | 1 | 33 | — | 197 |
| 5 | PTD | — | — | — | — | 6 | — | 1 | 98 | 2 | 107 |
| 6 | ELL | — | 24 | 1 | — | 24 | — | 1 | 45 | 2 | 97 |
| 7 | MLLT4/AF6 | 1 | 2 | — | 16 | 28 | — | 9 | 38 | 1 | 95 |
| 8 | EPS15 | 16 | 1 | 1 | 6 | 5 | — | 4 | 5 | — | 38 |
| 9 | MLLT11/AF1Q | 1 | 13 | — | — | 7 | — | — | 2 | — | 23 |
| 10 | no der(11) | 14 | 6 | 1 | 3 | 5 | 2 | 31 | |||
| 11 | SEPT6 | — | 5 | — | — | 10 | — | — | 2 | — | 17 |
| 12 | MLLT6/AF17 | — | — | — | 1 | 2 | — | — | 11 | — | 14 |
| 13 | SEPT9 | — | 2 | — | — | 5 | — | — | 6 | — | 13 |
| 14 | AFF3/LAF4 | 3 | — | — | 5 | — | — | — | — | — | 8 |
| 15 | TET1 | — | — | — | — | 1 | — | 2 | 3 | — | 6 |
| 16 | 11q23.3 | 1 | — | — | — | 1 | — | 2 | 1 | — | 5 |
| 17 | SEPT5 | — | 1 | — | 1 | 2 | — | — | 1 | — | 5 |
| 18 | ABI1 | — | 2 | — | — | 2 | — | — | — | — | 4 |
| 19 | KNL1 | — | — | — | — | 2 | — | — | — | 2 | 4 |
| 20 | MAML2 | — | — | — | 2 | — | — | 1 | — | 1 | 4 |
| 21 | MYO1F | — | 3 | — | — | 1 | — | — | — | — | 4 |
| 22 | PICALM | 1 | 1 | — | 1 | — | — | — | 1 | — | 4 |
| 23 | TNRC18 | 1 | — | — | 2 | — | — | 1 | — | — | 4 |
| 24 | FLNA | — | 2 | — | — | 1 | — | — | — | — | 3 |
| 25 | NEBL | 1 | 1 | — | — | 1 | — | — | — | — | 3 |
| 26 | ACTN4 | — | — | — | — | 1 | — | 1 | — | — | 2 |
| 27 | AFF4/AF5 | 2 | — | — | — | — | — | — | — | — | 2 |
| 28 | BTBD18 | 2 | — | — | — | — | — | — | — | — | 2 |
| 29 | CBL | — | — | — | 1 | — | — | — | 1 | — | 2 |
| 30 | CEP170B | — | — | — | — | — | — | — | 2 | — | 2 |
| 31 | CREBBP | — | — | — | — | — | 2 | — | — | — | 2 |
| 32 | DCP1A | 1 | — | — | — | 1 | — | — | — | — | 2 |
| 33 | FOXO3A | — | — | — | 2 | — | — | — | — | — | 2 |
| 34 | KIAS1524 | — | 2 | — | — | — | — | — | — | — | 2 |
| 35 | SEPT11 | — | — | — | 1 | 1 | — | — | — | — | 2 |
| 36 | ABI2 | — | 2 | — | — | — | — | — | — | — | 2 |
| 37 | ACACA | — | — | — | — | 1 | — | — | — | — | 1 |
| 38 | ACER1 | 1 | — | — | — | — | — | — | — | — | 1 |
| 39 | AKAP13 | — | — | — | — | — | — | — | 1 | — | 1 |
| 40 | AP2S2 | — | — | — | — | — | — | — | 1 | — | 1 |
| 41 | ARHGAP26 | — | — | — | — | 1 | — | — | — | — | 1 |
| 42 | ARHGEF12 | — | — | — | — | — | — | — | 1 | — | 1 |
| 43 | ARHGEF17 | — | — | — | — | 1 | — | — | — | — | 1 |
| 44 | BCL9L | — | — | — | 1 | — | — | — | — | — | 1 |
| 45 | BUD13 | — | — | — | — | 1 | — | — | — | — | 1 |
| 46 | C2CD3 | — | — | — | — | — | — | — | 1 | — | 1 |
| 47 | CASP8AP2 | — | — | — | — | — | — | — | 1 | — | 1 |
| 48 | CEP164 | — | — | — | — | — | — | 1 | — | — | 1 |
| 49 | CLTA | — | — | — | — | — | — | 1 | — | — | 1 |
| 50 | CLTC | — | — | — | — | 1 | — | — | — | — | 1 |
| 51 | CT45S2 | — | — | — | — | — | 1 | — | — | — | 1 |
| 52 | DCPS | — | — | — | — | — | — | — | 1 | — | 1 |
| 53 | EEFSEC | 1 | — | — | — | — | — | — | — | — | 1 |
| 54 | FNBP1 | — | 1 | — | — | — | — | — | — | — | 1 |
| 55 | GAS7 | — | 1 | — | — | — | — | — | — | — | 1 |
| 56 | GIGYF2 | — | — | — | — | — | — | — | — | — | 1 |
| 57 | GMPS | — | — | — | — | — | — | — | 1 | — | 1 |
| 58 | KIF2A | — | — | — | — | — | — | 1 | — | — | 1 |
| 59 | LAMC3 | — | — | — | — | 1 | — | — | — | — | 1 |
| 60 | LOC100131626 | — | — | — | — | — | — | — | — | 1 | 1 |
| 61 | ME2 | — | — | — | — | — | — | — | 1 | — | 1 |
| 62 | MKL1 | — | — | — | 1 | — | — | — | — | — | 1 |
| 63 | MYH11 | — | — | — | — | — | — | — | 1 | — | 1 |
| 64 | NOX4 | — | — | — | — | — | — | — | 1 | — | 1 |
| 65 | NRIP3 | — | 1 | — | — | — | — | — | — | — | 1 |
| 66 | NUP153 | — | — | — | 1 | — | — | — | — | — | 1 |
| 67 | PDS5A | — | — | — | — | — | — | — | 1 | — | 1 |
| 68 | PFDN4 | — | — | — | 1 | — | — | — | — | — | 1 |
| 69 | PRPF19 | 1 | — | — | — | — | — | — | — | — | 1 |
| 70 | PRRC1 | — | — | — | — | — | — | — | 1 | — | 1 |
| 71 | RABGAP1 | — | — | — | — | — | 1 | — | — | — | 1 |
| 72 | RUNDC3B | — | — | — | 1 | — | — | — | — | — | 1 |
| 73 | SEPT2 | — | — | — | — | 1 | — | — | — | — | 1 |
| 74 | SMAP1 | — | — | — | — | — | — | — | 1 | — | 1 |
| 75 | TCF12 | — | — | — | — | — | — | — | 1 | — | 1 |
| 76 | TOP3A | — | — | — | — | — | — | — | 1 | — | 1 |
| 77 | VAV1 | — | — | — | — | 1 | — | — | — | — | 1 |
| 78 | 1p13.1 | — | — | — | — | — | — | 1 | — | — | 1 |
| 79 | 6q27 | — | — | — | — | 1 | — | — | — | — | 1 |
| 80 | 9p13.3 | 1 | — | — | — | — | — | — | — | — | 1 |
| 81 | 11q24.3 | — | 1 | — | — | — | — | — | — | — | 1 |
| 82 | 21q22 | — | — | — | 1 | — | — | — | — | — | 1 |
| 83 | MLL internal inv | — | — | — | — | 1 | — | — | — | — | 1 |
| SUM | 692 | 160 | 24 | 313 | 339 | 19 | 415 | 373 | 10 | 2345 | |
Abbreviations: AML, acute lymphoblastic leukemia; AML, acute myeloid leukemia; DCAL, Diagnostic Center of Acute Leukemia; TPG, translocation partner gene.
All fusion genes that have been analyzed at the DCAL and their distribution between infant, pediatric and adult leukemia patients are shown. Total numbers are given for each patient group separated in ALL, AML and other diseases. The most frequent fusion partner genes were separated from the other genes that have been isolated less frequently by a line. Genes marked in bold represent out-of-frame MLL–X fusions.
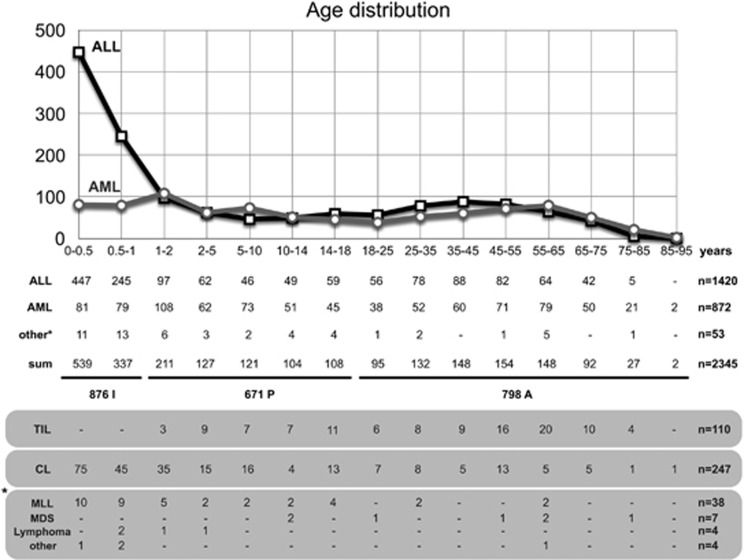
Age distribution according to clinical subtypes
We first analyzed our cohort according to the age at diagnosis. As displayed in Figure 1, the age distribution is quite similar to the expected distributions known from other cancer registries. MLL-r acute lymphoblastic leukemia (ALL) incidence has a peak in the first 2 years, then declines during the pediatric and young adult phase and then steadily increases again with age. A similar picture was observed with MLL-r acute myeloid leukemia (AML) patients, however, missing the postnatal peak seen for infant ALL. For the purpose of our study we separated our cohort into an ‘infant acute leukemia cohort’ (0.03–12 months; n=876: 692 ALL, 160 AML and 24NA) (not annotated), a ‘pediatric acute leukemia group’ (>12 months–18 years; n=671: 313 ALL, 339 AML and 19NA) and an ‘adult acute leukemia patient’ group (>18 years; n=798: 415 ALL, 373 AML and 10NA). As shown in Figure 1, we also added information about therapy-induced leukemia (n=110). Fifty-three patients could not be categorized into ‘ALL’ or ‘AML’ because they received other diagnoses (mixed lineage leukemia (MLL)=38, myelodysplastic syndrome=7 and lymphoma=4), or because we had no information from the corresponding center (unknown disease type=4).
Figure 1.
Age distribution of investigated patients. The age distribution of all analyzed patients (n=2345) is summarized. Upper part: diagram displaying ALL and AML patients. Age at diagnosis was divided into infants (0–1 year), pediatric (1–18 years) and adult patients (>18 years). The number of ALL, AML and other patients is listed below. We also added the information about therapy-induced leukemia (TIL) patients, the number of complex MLL rearrangements (CL) and specified the ‘non-ALL’ and ‘non-AML’ patients (MLL, MDS, lymphoma and other) in more detail for each age group. The precise number of all patient cases is summarized on the right.
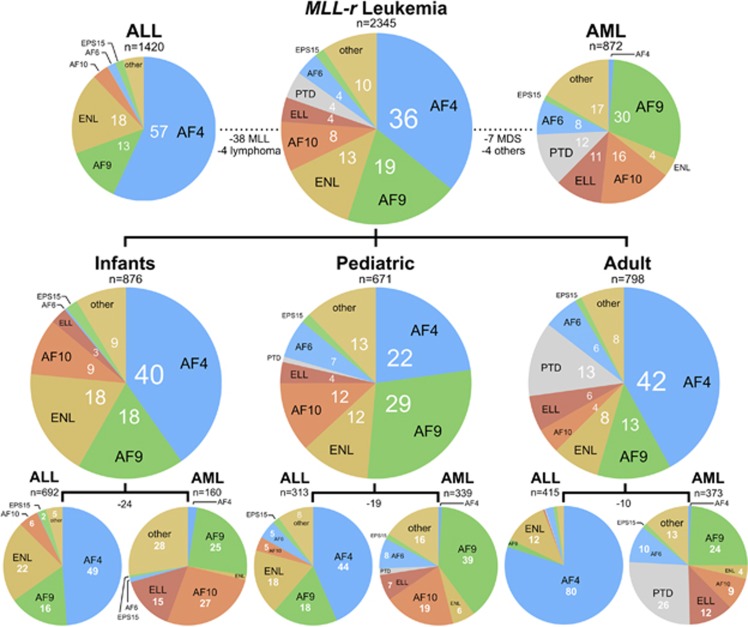
Identification of MLL rearrangements and their distribution in clinical subgroups
The most frequent MLL rearrangements in these six subgroups were summarized in Figure 2. Infant ALL (n=692) displayed 338 AF4, 113 AF9, 154 ENL, 39 AF10, 1 AF6 and 16 EPS15 gene fusions. Additional 31 MLL rearrangements were identified (9p13.3, 11q23.3, ACER, AF1Q, LAF4, AF5, BTBD18, CLTA, DCP1A, EEFSEC, 14 cases (NA) with no der(11) and only a reciprocal fusion allele, PICALM, PRPF19 and TNRC18).
Figure 2.
Classification of patients according to age classes and disease type. Top: frequency of most frequent TPGs in the investigated patient cohort of MLL-r acute leukemia patients (n=2345). This patient cohort was divided into ALL (left) and AML patients (right). Gene names are written in black, percentages are indicated as white numbers. Fifty-three patients could not be classified into the ALL or the AML disease types, respectively. Middle: TPG frequencies for the infant, pediatric and adult patient group. Bottom: subdivision of all three age groups into ALL and AML patients. Negative numbers refer again to the number of patients that were neither classified to the ‘ALL’ nor to the ‘AML’ subgroup.
Infant AML (n=160) was represented by 4 AF4, 40 AF9, 2 ENL, 43 AF10, 24 ELL, 2 AF6 and 1 EPS15 gene fusion. Another 44 MLL rearrangements have been characterized (11q24, ABI1, ABI2, AF1Q, FLNA, FNBP1, GAS7, KIAS1524, MYO1F, 14 cases (NA) with no der(11) and only a reciprocal fusion allele, NEBL, NRIP3, PICALM, SEPT5, SEPT6 and SEPT9).
Pediatric ALL (n=313) displayed 139 AF4, 56 AF9, 56 ENL, 12 AF10, 16 AF6 and 6 EPS15 gene fusions. Another 31 MLL rearrangements were characterized (21q22, AF17, LAF4, BCL9L, CBL, FOXO3A, MAML2, MKL1, 5 cases (NA) with no der(11) and only a reciprocal fusion allele, NUP153, PFDN4, PICALM, RUNDC3B, SEPT5, SEPT11, TET1 and TNRC18).
Pediatric AML (n=339) displayed 3 AF4, 132 AF9, 21 ENL, 66 AF10, 24 ELL, 6 MLL PTDs, 28 AF6 and 5 EPS15 gene fusions. Another 54 MLL rearrangements have been diagnosed (6q27, 11q23.3, ABI1, ACACA, ACTN4, AF1Q, AF17, ARHGAP26, ARHGEF17, BUD13, CLTC, DCP1A, FLNA, KLN1, LAMC3, an MLL gene-internal deletion, MYO1F, 5 cases (NA) with no der(11) and only a reciprocal fusion allele, NEBL, SEPT2, SEPT5, SEPT6, SEPT9, SEPT11, TET1 and VAV1).
Adult ALL (n=415) displayed 332 AF4, 9 AF9, 50 ENL, 1 AF10, 1 ELL, 1 MLL PTD, 9 AF6 and 4 EPS15 gene fusions. Additional 8 MLL rearrangements were identified (11q23, ACTN4, CEP164, KIF2A, MAML2, PRRC1, PTD and TET1).
Adult AML (n=373) displayed 3 AF4, 90 AF9, 14 ENL, 33 AF10, 45 ELL, 98 MLL PTDs, 38 AF6 and 5 EPS15 gene fusions. Another 47 MLL rearrangements were detected (1p13.1, 11q23, AF1Q, AKAP13, AP2S2, ARHGEF12, C2CD3, CASP8AP2, CBL, CEP170B, DCPS, GMPS, ME2, AF17, MYH11, 2 cases (NA) with no der(11) and only a reciprocal fusion allele, NOX4, PDS5A, PICALM, SEPT5, SEPT6, SEPT9, SMAP1, TCF12, TET1 and TOP3A).
On the basis of the above distribution, about 96% of all ALL patients (n=1420) were characterized by the fusion genes MLL–AF4 (~57%), MLL–ENL (~18%), MLL–AF9 (~13%), MLL–AF10 (~4%), MLL–EPS15 (~2%) and MLL–AF6 (~2%). About 83% of all AML patients (n=872) were characterized by the fusion genes MLL–AF9 (~30%), MLL–AF10 (~16%), MLL–ELL (~11%), MLL PTDs (~12%), MLL–AF6 (~8%), MLL–ENL (~4%) and MLL–SEPT6 (~2%). These results are in line with recently published data about the frequency and distribution of different MLL fusion partner genes.16, 17, 18 This updated information is quite important for diagnosis and has already been used to establish a fast reverse-transcription PCR-based multiplex screening method.19 Additional information about the distribution of translocation partner genes (TPGs) in major disease subgroups (different B-cell developmental stages, T-ALL or French–American–British (FAB) M0–M7 for AML patients) have been summarized in Supplementary Figures S1 and S2. Here the different FAB classes in AML show a quite distinct pattern of fusion genes with some of the major fusion partners missing in distinct FAB groups M0–M7. For example, in FAB M0 AF4 and ELL are missing, in FAB M1 ENL is missing, in FAB M2 AF4, AF10 and AF6 are missing and so on. In FAB class M6 and M7, only certain fusion genes could be identified. In B-ALL stages, no such exclusion patterns were observed, rather a shift for specific fusion genes, while T-ALL is mainly composed of ENL and AF6 gene fusions (see also below).
Breakpoint distribution according to clinical subtypes
We also investigated the distribution of chromosomal breakpoints within the MLL breakpoint cluster region in all investigated clinical subgroups. Briefly, the major breakpoint cluster region is localizing between MLL exon 9 and MLL intron 11, where the majority of patients (93.5%) had their individual breakpoints (n=2192). Only 153 patients (6.5%) had their breakpoint outside of the major breakpoint cluster region (Supplementary Figures S3–S5 and Supplementary Table S6).
As the localization of breakpoints may have an impact on cancer biology and clinical behavior, we started to analyze the breakpoint distribution for all clinical subgroups and compared them with the ‘mean distribution’ (MD) observed for all 2345 patients. We decided not to use a ‘random distribution model’ of chromosomal breakpoints, because this is only based on the length of each DNA region. However specific features in MLL intron 9 (four Alu repetitive elements of which three are transcriptionally active) and MLL intron 11 (sensitivity against cytotoxic drugs, a DNase1 hypersensitive site,20 an apoptotic cleavage site,21 an RNA polymerase II-binding site22 and topoisomerase II-binding sites23) may account for a specific increase of DNA double-stranded breaks due to specific molecular features of the chromatin, or, breakpoints differ because of a selection process for resulting MLL fusion proteins.
For our analyses, we subdivided the MLL breakpoint cluster region into three subregions: (A) exon 9–intron 9=1761 bp; (B) exon 10–intron 10=679 bp; (C) exon 11–intron 11–exon 12–intron 12 and exon 13=5026 bp. The observed ‘MD’ for these three MLL breakpoint regions was A=37.0%, B=19.8% and C=40.1% for all 2345 patients listed in Supplementary Table S7. In these analyses, all patients were investigated for their fusion partner gene in correlation with age at diagnosis, gender, patient group, therapy-induced leukemia, complex genetic rearrangements, origin of patient and breakpoint distribution. Here a significant deviation from the ‘MD’ was observed for AF1Q, AF6, AF10, ENL, EPS15, SEPT6, SEPT9, AF17 and MLL PTDs. The fusion partner genes ENL and SEPT6 had preferentially MLL intron 11 breaks, while all others tend to bear MLL intron 9 recombination events. Of interest, also therapy-induced acute leukemias differ significantly in their ‘MD’, with a tendency for MLL intron 11 breaks.
A detailed analysis for the most frequent MLL fusion partner genes is depicted in Supplementary Table S8. Here we separated according to fusion partners and patient subgroup (infant I, pediatric P and adult A) with regard to several other parameters such as age, gender, therapy-induced, complex translocation, origin and disease type. The overall breakpoint distribution of all seven most frequent genetic aberrations with more than 2000 patients was not significantly deviating from the MD of all patients. However, significant changes were observed for patient subgroups bearing chromosomal translocations to AF4 (I and A), AF9 (A), ENL (I and P), AF10 (P), ELL (I, P and A), AF6 (I, P and A) and MLL PTD patients (P and A). This clearly demonstrates that certain fusion genes have a selective preference for distinct breakpoints, most likely because of the resulting function of a given fusion protein. As an example, AF6 fusions in ALL and AML patients are mostly occurring in MLL intron 9 (or even upstream), while infant AF4 and infant/pediatric ENL fusions tend to have breakpoints within MLL intron 11. Similar observations were made for the more rare fusion partner AF1Q (significantly toward MLL intron 9), AF17 (significantly toward MLL introns 8 and 9), EPS15 (significantly toward MLL intron 11 in adult patients), SEPT6 (significantly toward MLL intron 11 in pediatric and adult patients) and SEPT9 (significantly toward MLL introns 7–9).
To evaluate these data further, we correlated the breakpoint distribution with the age of patients. We have done so for ALL and AML patients (Supplementary Figures S9 and S10). These analyses revealed that the disease subtypes (ALL and AML) divide patients into two groups (ALL more in MLL intron 11 breakpoints; AML more in MLL intron 9 breakpoints). However, these breakpoint tendencies seem to change with age. Thus, young patients usually display MLL intron 11 breakage, while older patients have more breaks in MLL intron 9. This is true for all investigated subgroups (AF4, ENL and AF9) where we had enough patients to perform this type of analysis and to obtain a significant result. Vice versa, young AML patients usually prefer MLL intron 9 breakage, while older patients have more breaks in MLL intron 11. This has been done also for the AF9, AF10 and ELL subgroups. The cross-over points were 10–14 years in ALL patients and 75 years in AML patients. MLL–ELL patients within the AML group are somehow different from all other subgroups because they start very early on with a preference for MLL intron 11 (all patients above 1 year of age) and display no cross-over point. These breakpoint preferences and their change with age are indicating that two different molecular mechanisms are driving MLL rearrangements: one is a transcription-coupled hot spot that is quite sensible for external cytotoxic triggers (MLL intron 11), while the other is presumably based on transcriptionally active ALU repeats where POL III and POL II transcription is causing torsional DNA stress.
Another important point is the outcome of patients that is linked to the distribution of chromosomal breakpoints within the MLL breakpoint cluster region.24 Basically, the outcome of leukemia patients with breakpoints in MLL intron 11 was worse compared to those patients with upstream breakpoints. A rational explanation for this observation came from functional studies of the plant homeodomain (PHD) domain of the MLL protein, encoded by MLL exons 11–16 (Supplementary Figure S11). This domain is built up by PHD1, PHD2 and an enhanced PHD3. The first three PHD domains are separated by the adjacent bromodomain and another enhanced PHD4. PHD3 has an important dual function, because it either binds to the CYP33/PPIE protein25, 26 or to methylated lysine-4 residues of histone H3.27 Binding of PHD3 to H3K4me2/3 peptides is greatly enhanced by the adjacent bromodomain,28 but binding of the prolyl-peptidyl isomerase CYP33/PPIE confers a cis–trans isomerization of proline-1665. This enables binding to BMI1 and associated repressor proteins (HDAC/CBX4/KDM5B). This molecular switch determines the human MLL protein of being a transcriptional activator/maintenance factor or a transcriptional repressor. Noteworthy, PHD2 and PHD3 also bind to E3-ligases (CDC34 and ASB2, respectively), which control the steady-state stability of the MLL protein.29, 30 As shown recently by our group, breakpoints within MLL intron 11 destroy the dimerization capacity of the PHD1–3 domain.31 This also disables binding to the BMI1 repressor complex.32 Thus, a breakpoint within MLL intron 11 has functional consequences for the resulting fusion proteins, which may explain the clinical finding of the altered outcome.24
The MLL recombinome
On the basis of the results obtained in the present and previous studies,13, 14, 15, 16 a total of 84 direct TPGs and their specific breakpoint regions have now been identified, all of which generate an in-frame MLL fusion protein (Table 2A). Additional 10 MLL fusions were characterized that were fused out of frame to translocation partner genes (Table 2B). In the latter cases, alternative splicing may still allow to generate an MLL-fusion protein, however, this was not investigated here. Another 6 loci have been cloned where the 5′-portion of MLL was not fused to another gene (Table 2C). 3′-Race and reverse-transcription PCR experiments with several exon combinations were performed to identify potential fusion transcripts. But no in-frame fusion RNAs could be identified. Therefore, these 16 unusual MLL rearrangements—where neither any dimerization nor a transcriptional activation domain is present—probably represent a subclass of MLL abnormalities, which have per se no or only a weak ability to transform hematopoietic cells and are only identified in the context of other genetic abnormalities in hematopoietic stem cells.33, 34
Table 2. Overview about the MLL recombinome 2017.
| # | Cytogenetic abnormality | Breakpoint | TPG | References | Leukemia type |
|---|---|---|---|---|---|
| A. MLL fusion in-frame | |||||
| 1 | t(1;11)(p32;q23) | 1p32 | EPS15/AF1P | Bernard et al. (1994) | ALL, BAL, AML, CML |
| 2 | t(1;11)(q21;q23) | 1q21 | MLLT11/AF1Q | Tse et al. (1995) | AML, t-AML, ALL, t-ALL, BAL |
| 3 | t(2;11)(p23.3;q23) | 2q23.3 | ASXL2 | Haferlach et al. (2016) | t-AML |
| 4 | ins(2;11)(q11.2;q23) | 2q11.2 | AFF3/LAF4 | von Bergh et al. (2003) | ALL |
| 5 | t(2;11)(q33;q23) | 2q33 | ABI2 | Coenen et al. (2012) | AML |
| 6 | t(2;11)(q37;q23) | 2q37 | SEPT2 | Cerveira et al. (2006) | t-AML, AML, t-MDS |
| 7 | t(2;11)(q37.1;q23) | 2q37.1 | GIGYF2 | Not published yet | ALL |
| 8 | t(3;11)(p21;q23) | 3p21 | NCKIPSD/AF3P21 | Sano et al. (2000) | t-AML |
| 9 | t(3;11)(p21.1;q23) | 3p21.1 | DCP1A | Meyer et al. (2008) | ALL, AML |
| 10 | t(3;11)(q13.13;q23) | 3q13.13 | KIAS1524 | Coenen et al. (2011) | AML |
| 11 | t(3;11)(q24;q23) | 3q24 | GMPS | Pegram et al. (2000) | t-AML, t-MDS |
| 12 | t(3;11)(q28;q23) | 3q28 | LPP | Daheron et al. (2001) | t-AML |
| 13 | t(4;11)(p14;q23) | 4p14 | PDS5A | Meyer et al. (2011) | t-AML, AML |
| 14 | t(4;11)(p11;q23) | 4p11 | FRYL | Hayette et al. (2006) | t-ALL, t-AML, t-MDS |
| 15 | t(4;11)(q21.1;q23) | 4q21.1 | SEPT11/FLJ10849 | Kojima et al. (2004) | T-ALL, CML, t-ALL, t-AML |
| 16 | t(4;11)(q21;q23) | 4q21 | AF4/AFF1 | Gu et al. (1992) | ALL, t-ALL, BAL, AML |
| 17 | t(4;11)(q35.1;q23) | 4q35.1 | SORBS2/ARGBP2 | Pession et al. (2006) | AML |
| 18 | t(5;11)(q12.1;q23) | 5q12.1 | KIF2A | Not published yet | ALL |
| 19 | complex abnormalities | 5q12.3 | CENPK/FKSG14 | Taki et al. (1996) | AML |
| 20 | t(5;11)(q23.2;q23) | 5q23.2 | PRRC1 | Douet-Guilbert et al. (2014) | t-ALL |
| 21 | ins(5;11)(q31;q13q23) | 5q31 | AFF4/AF5Q31 | Taki et al. (1999) | ALL |
| 22 | t(5;11)(q31;q23) | 5q31 | ARHGAP26/GRAF | Borkhardt et al. (2000) | JMML |
| 23 | t(6;11)(q13;q23) | 6q13 | SMAP1 | 13 | AML |
| 24 | t(6;11)(q15;q23) | 6q15 | CASP8AP2 | Park et al. (2009) | AML |
| 25 | t(6;11)(q21;q23) | 6q21 | FOXO3/AF6Q21 | Hillion et al. (1997) | t-AML, t-ALL |
| 26 | t(6;11)(q27;q23) | 6q27 | MLLT4/AF6 | Prasad et al. (1993) | T-ALL, AML, t-AML, ALL |
| 27 | t(7;11)(p22.1;q23) | 7p22.1 | TNRC18/KIAS1856 | Meyer et al. (2008) | T-ALL, ALL |
| 28 | t(7;11)(q11.23;q23) | 7q11.23 | CLIP2 | Not published yet | B-ALL |
| 29 | t(7;11)(q21.12;q23) | 7q21.12 | RUNDC3B | 16 | T-ALL |
| 30 | t(7;11)(q32.1;q23) | 7q32.1 | FLNC | Haferlach et al. (2016) | t-AML |
| 31 | t(9;11)(p13;q23) | 9p13 | CLTA | Not published yet | ALL |
| 32 | t(9;11)(p22;q23) | 9p22 | MLLT3/AF9 | Nakamura et al. (1993) | AML, t-AML, ALL, T-ALL, BAL |
| 33 | t(9;11)(q33.2;q23) | 9q33.2 | DAB2IP/AF9Q34 | von Bergh et al. (2004) | AML |
| 34 | t(9;11)(q34.11;q23) | 9q34.11 | RABGAP1 | Not published yet | ALL |
| 35 | ins(11;9)(q23;q34)inv(11)(q13)(q23) | 9q34 | FNBP1/FBP17 | Fuchs et al. (2001) | AML |
| 36 | t(9;11)(q34.12;q23) | 9q34.12 | LAMC3 | 15 | t-AML |
| 37 | ins(10;11)(p12;q23) | 10p12 | NEBL | Cóser et al. (2010) | AML |
| 38 | ins(10;11)(p12;q23q13) | 10p12 | MLLT10/AF10 | Chaplin et al. (1995) | AML, t-AML, ALL, T-ALL, BAL |
| 39 | t(10;11)(p11.2;q23) | 10p11.2 | ABI1 | Taki et al. (1998) | AML |
| 40 | t(10;11)(q21;q23) | 10q21 | TET1/LCX | Ono et al. (2002) | AML, ALL |
| 41 | inv(11)(p15.3q23) | 11p15.3 | NRIP3 | 3 | AML |
| 42 | inv(11)(q12.1q23) | 11q12.1 | BTBD18 | Alonso et al. (2012) | ALL |
| 43 | inv(11)(q12.2q23) | 11q12.2 | PRPF19 | 16 | ALL |
| 44 | t(11;11)(q13.4;q23) | 11q13.4 | ARHGEF17 | Teuffel et al. (2005) | AML |
| 45 | inv(11)(q13.4q23) | 11q13.4 | C2CD3 | 15 | AML |
| 46 | inv(11)(q14q23) | 11q14 | PICALM/CALM | Wechsler et al. (2003) | AML, ALL |
| 47 | inv(11)(q21q23) | 11q21 | MAML2 | 14 | T-ALL,t-T-ALL, t-AML, t-MDS |
| 48 | del(11)(q23q23.3) | 11q23.3 | CBL | Fu et al. (2003) | AML, t-AML, ALL |
| 49 | del(11)(q23q23.3) | 11q23.3 | ARHGEF12/LARG | Kourlas et al. (2000) | AML, t-AML |
| 50 | del(11))(q23q24.2) | 11q24.2 | DCPS | 13 | AML |
| 51 | t(11;12)(q23;p11.23) | 12p11.23 | ITPR2 | Haferlach et al. (2016) | t-MDS |
| 52 | t(11;12)(q23;q13.2) | 12q13.2 | SARNP/CIP29 | Hashii et al. (2004) | AML |
| 53 | t(11;14)(q23;q23.3) | 14q23.3 | GPHN | Kuwada et al. (2001) | AML, t-AML |
| 54 | t(11;14)(q23;q32.33) | 14q32.33 | CEP170B/KIAA0284 | Burmeister et al. (2008) | t-AML |
| 55 | t(11;15)(q23;q14) | 15q14 | KNL1/CASC5 | Hayette et al. (2000) | AML, t-MDS, ALL, |
| 56 | t(11;15)(q23;q15.1) | 15q15.1 | ZFYVE19/MPFYVE | Chinwalla et al. (2003) | AML |
| 57 | t(11;15)(q23;q21) | 15q21 | TCF12 | Not published yet | t-AML |
| 58 | t(11;15)(q23;q25.3) | 15q25.3 | AKAP13 | 16 | t-AML |
| 59 | t(11;16)(q23;p13.3) | 16p13.3 | CREBBP/CBP | Taki et al. (1997) | t-MDS, t-AML, AML, t-ALL, t-CML |
| 60 | t(11;16)(q23;p13.11) | 16p13.11 | MYH11 | 16 | AML |
| 61 | t(11;17)(q23;p13.1) | 17p13.1 | GAS7 | Megonigal et al. (2000) | t-AML |
| 62 | t(11;17)(q23;p11.2) | 17p11.2 | TOP3A | Herbaux et al. (2012) | AML |
| 63 | t(11;17)(q23;q12) | 17q12 | LASP1 | Strehl et al. (2003) | AML |
| 64 | ins(11;17)(q23;q21) | 17q21 | ACACA | 13 | AML |
| 65 | t(11;17)(q23;q21) | 17q21 | MLLT6/AF17 | Prasad et al. (1994) | AML, ALL |
| 66 | t(11;17)(q23;q23.1) | 17q23.1 | CLTC | Not published yet | AML |
| 67 | t(11;17)(q23;q25) | 17q25 | SEPT9/AF17Q25 | Osaka et al. (1999) | t-AML, AML, MDS, ALL |
| 68 | t(11;18)(q23;q21) | 18q21 | ME2 | Szotkowski et al. (2015) | t-AML |
| 69 | t(11;19)(q23;p13.3) | 19p13.3 | MLLT1/ENL | Tkachuk et al. (1992) | ALL, T-ALL, AML, BAL, t-AL |
| 70 | t(11;19)(q23;p13.3) | 19p13.3 | ACER1/ASAH3 | Lo Nigro et al. (2002) | ALL |
| 71 | t(11;19)(q23;p13.3) | 19p13.3 | SH3GL1/EEN | So et al. (1997) | AML |
| 72 | ins(11;19)(q23;p13.3) | 19p13.3 | VAV1 | 15 | AML |
| 73 | t(11;19)(q23;p13.2) | 19p13.2 | MYO1F | Lo Nigro et al. (2002) | AML |
| 74 | t(11;19)(q23;p13.1) | 19p13.1 | ELL | Thirman et al. (1994) | ALL, BAL, AML, t-AML |
| 75 | t(11;19)(q23;q13) | 19q13 | ACTN4 | Burmeister et al. (2009) | t-ALL, t-AML |
| 76 | t(11;20)(q23;q11.21) | 20q11.21 | MAPRE1 | Fu et al. (2005) | ALL |
| 77 | t(11;20)(q23;q13.2) | 20q13.2 | PFDN4 | Not published yet | T-ALL |
| 78 | t(11;22)(q23;q11.21) | 22q11.21 | SEPT5/CDCREL | Megonigal et al. (1998) | AML, T-ALL |
| 79 | t(11;22)(q23;q13) | 22q13 | MKL1 | Not published yet | ALL |
| 80 | t(11;22)(q23;q13.2) | 22q13.2 | EP300/P300 | Ida et al. (1997) | t-AML |
| 81 | t(X;11)(q13.1;q23) | Xq13.1 | FOXO4/AFX | Parry et al. (1994) | T-ALL, ALL, t-ALL,CLL, AML |
| 82 | ins(X;11)(q24;q23) | Xq24 | SEPT6 | Borkhardt et al. (2001) | AML |
| 83 | ins(X;11)(q26.3;q23) | Xq26.3 | CT45S2 | Cerveira et al. (2010) | BAL |
| 84 | ins(11;X)(q23q28q13.1) | Xq28 | FLNA | De Braekeleer et al. (2009) | AML |
| B. MLL fusion not in-frame | |||||
| 1 | t(3;11)(p21.3;q23) | 3p21.3 | SACM1L | 34 | N/A |
| 2 | t(3;11)(q21.3;q23) | 3q21.3 | EEFSEC/SELB | 13 | ALL |
| 3 | t(6;11)(p22.3;q23) | 6p22.3 | NUP153 | Not published yet | ALL |
| 4 | inv(11)(p15.5q23) | 11p15.5 | AP2S2 | 16 | AML |
| 5 | complex | 11q14 | NOX4 | Not published yet | ALL |
| 6 | t(11;15)(q23.3;q21) | 11q23.3 | LOC100131626 | 33 | MDS |
| 7 | inv(11)(q23.3q23) | 11q23.3 | BUD13 | 16 | AML |
| 8 | del(11)(q23q23.3) | 11q23.3 | CEP164 | 16 | t-ALL |
| 9 | del(11)(q23q23.3) | 11q23.3 | BCL9L | 14 | ALL |
| 10 | t(2;11;19)(p23.3;q23;p13.3) | 19p13.3 | LOC100128568 | 15 | AML |
| C. No partner gene fused to 5′-MLL gene | |||||
| 1 | t(1;11)(p13.1;q23) | 1p13.1 | 16 | PMF | |
| 2 | t(6;11)(q27;q23) | 6q27 | Not published yet | AML | |
| 3 | t(9;11)(p13.3;q23) | 9p13.3 | 16 | t-ALL | |
| 4 | t(11;11)(q23;q23.3) | 11q23.3 | 16 | ALL, AML | |
| 5 | t(11;11)(q23;q24.3) | 11q24.3 | 16 | AML | |
| 6 | t(11;21)(q23;q22) | 21q22 | 16 | t-ALL | |
| D. Not characterized at the molecular level (published by others) | |||||
| 1 | t(1;11)(p36;q23) | ||||
| 2 | t(1;11)(q31;q23) | ||||
| 3 | t(1;11)(q32;q23) | ||||
| 4 | t(2;11)(p21;q23) | ||||
| 5 | t(2;11)(q37;q23) | ||||
| 6 | t(3;11)(p13;q23) | ||||
| 7 | t(4;11)(p11;q23) | ||||
| 8 | t(6;11)(q13;q23) | ||||
| 9 | t(7;11)(p15;q23) | ||||
| 10 | t(7;11)(q22;q23) | ||||
| 11 | t(7;11)(q32;q23) | ||||
| 12 | t(8;11)(q11;q23) | ||||
| 13 | t(8;11)(q21;q23) | ||||
| 14 | t(8;11)(q24;q23) | ||||
| 15 | t(9;11)(p11;q23) | ||||
| 16 | t(9;11)(q33;q23) | ||||
| 17 | t(10;11)(q25;q23) | ||||
| 18 | t(11;11)(q11;q23) | ||||
| 19 | t(11;11)(q13;q23) | ||||
| 20 | t(11;11)(q21;q23) | ||||
| 21 | t(11;12)(q23;p13) | ||||
| 22 | t(11;12)(q23;q13) | ||||
| 23 | t(11;12)(q23;q24) | ||||
| 24 | t(4;13;11)(q21;q34;q23) | ||||
| 25 | t(11;14)(q23;q11) | ||||
| 26 | t(11;14)(q23;q32) | ||||
| 27 | t(11;15)q23;q15) | ||||
| 28 | t(11;17)(q23;q11) | ||||
| 29 | t(11;17)(q23;q23) | ||||
| 30 | t(11;18)(q23;q12) | ||||
| 31 | t(11;18)(q23;q23) | ||||
| 32 | t(11;20)(q23;q13) | ||||
| 33 | t(11;21)(q23;q11) | ||||
| 34 | t(Y;11)(p11;q23) | ||||
| 35 | t(X;11)(q22;q23) | ||||
Abbreviations: AML, acute lymphoblastic leukemia; AML, acute myeloid leukemia; BAL, bilineal acute leukemia; CML, chronic myelogenous leukemia; DCAL, Diagnostic Center of Acute Leukemia; JMML, juvenile myelomonocytic leukemia; MLL, mixed lineage leukemia; t-ALL, therapy-related ALL; t-AML, therapy-related AML; t-MDS, therapy-related MDS; TPG, translocation partner gene.
List of the cytogenetic localization of all yet-characterized direct TPGs (n=94), the gene name, the appropriate reference and observed disease type. Genes marked as ‘not published yet’ are completely new. All references in italics have been identified at the DCAL during the last decade. In addition, 6 cloned gene loci and 35 cytogenetic chromosome loci have been identified.
In 31 additional cases we were not able to identify a der(11) fusion gene at all. This could be either attributed to a technical problem (for example, a too long genomic fragment) or to the fact that no der(11) exists in these few patients. However, in 22/31 cases we successfully identified a reciprocal MLL fusion allele, while 9 cases carried no detectable direct or reciprocal fusion gene. This subgroup (n=31) was included in the group of ‘complex MLL rearrangements’ (n=247) because of the extending class of ‘reciprocal MLL fusion genes’ (Supplementary Table S12). Within this group of patients with complex MLL rearrangements, a total of 32 reciprocal MLL fusions represent in-frame fusions (marked in red in Supplementary Table S12), while 215 fusions were out-of-frame fusions at the genomic DNA level (88 gene loci/127 partner genes; shown in black in Supplementary Table S12). Finally, there were still 35 chromosomal translocations of the human MLL gene that were characterized in the past by cytogenetic methods, but that were never analyzed at the molecular level (Table 2D). Thus, the MLL recombinome to date comprises 94 different ‘direct TPGs’ (decoding the MLL N terminus) and 6 different 5′-MLL genes fused only to genomic DNA. On the other hand we have now 247 ‘reciprocal TPGs’ (bearing the MLL C terminus) that are deriving from complex rearrangements with already-known ‘direct TPGs’. It is noteworthy that nearly all of these reciprocal MLL fusion (243 out of 247) are per se able to express only the 3′-MLL portion, named MLL*, due to a gene internal promoter located upstream of MLL exon 12.22
Novel translocation partner genes
Apart from the many new MLL fusion genes that have already been discovered at the DCAL and published in the last decade (Tables 2a–c; n=40), we present additional 11 novel TPGs (marked as ‘not published yet’): GIGYF2 (GRB10-interacting GYF; 2q37.1; 1299 aa); KIF2A (kinesin heavy chain member 2A; 5q12.1; 706 aa); CLIP2 (CAP-GLY domain-containing linker protein 2; 7q11.23; 1046 amino acids (aa)); CLTA (clathrin, light chain A; 9p13.3; 248 aa); RABGAP1 (RAB GTPase-activating protein 1; 9q33.2; 1069 aa); TCF12 (transcription factor 12; 15q21.3; 682 aa); CLTC (clathrin, heavy chain; 17q23.1; 1,675 aa); PFDN4 (prefoldin subunit 4; 20q13.2; 134 aa); MKL1 (megakaryoblastic leukemia (translocation) 1; 22q13.1; 931 aa); NUP153 (nucleoporin 153 kDa; 6p22.3; 1475 aa); and NOX4 (NADPH oxidase 4; 11q14.3; 578 aa).
The Drosophila GIGYF2 protein ortholog was shown to be a modulator of autophagy that controls neuron and muscle homeostasis.35 GIGYF2 binds directly to AGO2 and is involved in siRNA-mediated post-transcriptional silencing.36 A quite specific feature of GIGYF2 is to build a complex together with eIF4E and ZNF598 to selectively block the process of translation of distinct capped mRNAs.37 Several papers have linked GIGYF2 also to Parkinson’s disease, however, these data are so far not sufficiently significant.
KIF2A is a member of the kinesin-13 family and involved in spindle assembly at the metaphase I–anaphase I transition of oocytes.38, 39 Moreover, genetic mutations in the motor domain of this protein is associated with cortical malformation syndromes such as microcephaly.40 Vice versa, overexpression of KIF2A has been diagnosed in different cancers, because KIF2A expression and phosphorylation influences microtubuli dynamics, which is important for tumor cell migration and metastasis.41
CLIP2 has been discovered as overexpressed biomarker after radiation in papillary thyroid carcinomas, usually accompanied by a gain of chromosome band 7q11.42 This disease has been frequently diagnosed as the main consequence of the Chernobyl accident.
CLTA, also named clathrin light chain A, is involved in vesicle trafficking and endocytosis. However, a recent paper demonstrated that CLTA has a role on the migration of tumor cells.43 This is in part due to the fact that CLTA interacts with Huntingtin-interacting protein, involved in the regulation of the actin cytoskeleton. Upon depletion of clathrin light chains a steady-state downregulation of β1-integrins was observed because of defects in vesicle recycling.
RABGAP1 is highly specific for RAB6A and has a role in microtubule nucleation at the centrosome. It also participates in a RAB6A-mediated pathway involved in the metaphase–anaphase transition (Mad2-spindle checkpoint).44
TCF12, also known as HeLa E-box binding protein, is quite interesting as it controls the osteogenic differentiation of mesenchymal stem cells in the bone marrow.45 This basic helix–loop–helix transcription factor was also found to be mutated in anaplastic oligodendroglioma.46 TCF12 is able to bind to TWIST1 and involved in the early differentiation pathways of thymic T cells (DN3->DN4 and ISP->DP).47
CLTC has been identified in complex chromosomal rearrangements causing the expression of the CLTC–ALK fusion in blastic plasmacytoid dendritic cell neoplasm.48 A more recent work has found CLTC mutations are associated with neuronal malformations and intellectual developmental delays.49 This is due to the fact that clathrin heavy and light chains (CLTA and CLTC) are involved in vesicle trafficking, vesicle recyling and neurotransmitter release.
PFDN4 has been linked to colorectal cancer, however, inversely correlated with outcome (low expression has poorer outcome). A knockdown of this gene was correlated with increased cell growth and invasiveness.50
MKL1 interacts with the transcription factor myocardin, a key regulator of smooth muscle cell differentiation. The encoded protein is predominantly nuclear and may help transduce signals from the cytoskeleton to the nucleus. This gene is involved in a specific translocation event that creates a fusion of this gene and the RNA-binding motif protein-15 gene. This specific t(1;22)(p13;q13) translocation has been associated with the development of acute megakaryocytic leukemia.51
NUP153 s a highly versatile protein, involved in nuclear pore functions, pore architecture, nuclear import and export, de novo pore formation after mitosis and destruction of NUP153 during apoptosis.52
NOX4 is NADPH oxidase 4 that is important in the regulation of glycolysis and glutamate metabolism. Disruption of NOX4 by CRISPR/Cas9 is inhibiting cell growth of HeLa cells, indicating that NOX4 is quite important as metabolic regulator in tumor cells.53 NOX4 has been identified in many tumors as a relevant gene.
T-ALL cases
A tiny fraction of investigated patients were diagnosed with a T-ALL (n=59) (Supplementary Figure S13). This group of patients is mainly characterized by MLL fusion with AF6 (n=23) and ENL (n=22). Other fusions were AF4, AF9, AF10, MAML2, PFDN4, RUNDC3B, SEPT5, SEPT11, TNRC18 and 1 reciprocal USP20–MLL fusion. Only in the cohort of MLL–AF6 patients, quite unusual MLL breakpoints were observed (n=4), where the chromosomal breakpoint in the MLL gene was diagnosed within intron 21 and 23. This is quite important because such a far away downstream breakpoint includes the complete PHD1–3, the bromodomain as well as the complete enhanced PHD4 domain of MLL into the fusion protein with AF6 (Supplementary Figure S11). These additional 581 amino acids could be an important hint for the importance of these MLL domains in T-ALL. The PHD1–3 and bromodomain exert important regulatory functions to the MLL N terminus, such as chromatin reading, protein stability or CYP33 binding. In the latter case, binding of the BMI1 repressor complex will reverse the function of the MLL–AF6 fusion by repressing gene transcription. This is quite interesting and provides a new research aspect for MLL–AF6.
Therapy-induced leukemia cases
We also investigated the therapy-induced patient cases (n=110; Supplementary Table S14). The dominant partner genes are AF9 (n=41), ELL (n=11), AF4 (n=11) and ENL (n=10). All other fusions (n=23) have been identified one to four times. To our surprise, the AF9 cases were shifting from MLL intron 9 breaks to MLL intron 11 breaks. Some MLL fusions can only be found in therapy-induced acute leukemia and not in patients with de novo diseases: ACTN4; AKAP13; ARHGEF12; FOXO3A; GMPS; LAMC3; ME2; PDS5A; PRRC1; and TCF12. As expected, therapy-induced acute leukemias were only diagnosed in pediatric and adult patients, not in infants.
Spliced fusions
Spliced fusions are rare events except for the ENL fusion gene (n=302). In the latter cases, about 50% of all breakpoints localize far upstream of ENL exon 1 (n=153; Supplementary Table S15). In these cases, no reciprocal fusion protein can be made, only an MLL–ENL fusion transcript.54 For the other cases, a similar scenario was found. In all these cases, a 3′-terminal truncated MLL was recombined upstream of PRPF19 (1 out of 1 case), ELL (8 out of 97 cases), MYO1F (1 out of 4 cases), EPS15 (9 out of 38 cases), AF4 (1 out of 839 cases), AF6 (3 out of 95 cases) and AF9 (2 out of 449 cases). A total of 180 cases were identified that show this unusual peculiarities.
Discussion
Here we present an update of the ‘MLL recombinome’ associated with different hematologic malignancies, and in particular with acute leukemia (ALL and AML). All our analyses were performed by using small amounts of genomic DNA that were isolated from bone marrow or peripheral blood samples (n=2345) of leukemia patients. In some cases, we analyzed cDNA from a given patient to validate the presence of MLL spliced fusions, or to investigate alternative splicing of RNA generated from the investigated MLL fusion genes. The results of this study allow to draw several conclusions.
The applied long-distance inverse PCR technique allowed to identify direct and reciprocal MLL fusions, MLL gene-internal duplications, chromosome 11 inversions, chromosomal 11 deletions and the insertion of chromosome 11 material into other chromosomes, or vice versa, the insertion of chromatin material of other chromosomes into the MLL gene. It is noteworthy to mention that no other technique (for example, next-generation sequencing) displays such a high identification of chromosomal fusion sites so far. Even paired-end mRNA analysis by next-generation sequencing has a discovery rate of 60–70% only, however, RNA-based technologies do not provide the patient-specific chromosomal fusion sequences that could be used for MRD analyses. Thus, this ‘old-fashioned’ method is still state of the art and will be used also in the future to gain additional information of the MLL recombinome.
Moreover, we extended our knowledge on complex MLL rearrangements (n=247) leading to a large collection of reciprocal MLL fusions (Supplementary Table S12). About 13% represent in-frame fusions that can be readily expressed into reciprocal fusion proteins. All other represented out-of-frame fusions with either a chromosomal locus or a reciprocal TPG. Out-of-frame fusions such as IKZF1–MLL, PBX1–MLL or JAK1–MLL most likely represent a situation where such TPGs were destroyed, creating a typical loss-of-function situation. However, even those out-of-frame MLL fusions still allow to transcribe and express a 5′-truncated MLL protein, termed MLL*.22 This shorter version of MLL has no ability to bind Menin1, LEDGF or MYB, but still carries all enzymatic functions necessary to carry out H4K16 acetylations by the associated MOF protein or H3K4 methylation by the SET domain complex.
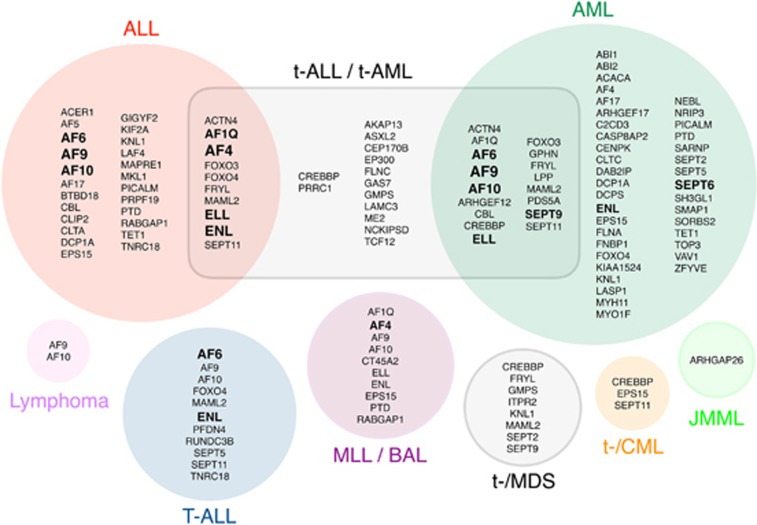
The analysis of 2345 MLL fusion alleles led to the discovery of 51 novel TPGs in the past 12 years, of which 40 have already been described (Tables 2a–c). Eleven TPGs are completely new and have not been published yet. Together with 49 MLL fusions that have been described by others, we can present today a total of 94 MLL fusions that have been characterized at the molecular level and 6 MLL translocations to different genetic loci (with no obvious gene fusion). All these MLL fusions provide a rich source for future analyses of oncogenic MLL protein variants. We have summarized all yet-known MLL fusion partner genes in Figure 3, according to their disease type/subtype in which they have been diagnosed.
Figure 3.
Classification of all yet known fusion partner genes by disease. All TPGs identified were grouped by their diagnosed disease type. Genes have been diagnosed in ALL, t-ALL, t-AML, AML, T-ALL, MLL, bilineal acute leukemia (BAL), MDS, t-MDS, chronic myelogenous leukemia (CML), t-CML, juvenile myelomonocytic leukemia (JMML) and lymphoma. Genes in the intersection belong to two different groups. Bold-marked TPGsb are the most frequent ones.
According to our data, the 7 most frequent rearrangements of the MLL gene differ significantly in the cohorts of infant, pediatric and adult leukemia patients. We also observed significant tendencies that correlate with fusion genes, gender or age at diagnosis. As an example MLL–AF10 (P=0.0024) occur more frequently in the male group of patients, while females were more affected by MLL–AF4 fusions (P=0.00576). The most striking finding was that breakpoint distributions differ significantly when concerning distinct TPGs and age groups. It is well-known that breakpoints in infants occur more frequently in MLL intron 11. We could validate this finding for infants with MLL–AF4, infant/pediatric patients with MLL–ENL fusions and pediatric/adult patients with MLL–ELL fusions. However, we observed a contrary situation in adult patients with MLL–AF9 or pediatric patients with MLL–AF10 fusions. Quite surprising was the breakpoint distribution for MLL–AF6 fusions that displayed a clear preference for MLL intron 9 recombinations. Again, these deviations from the observed mean breakpoint distribution (MD) are an argument for differences in the biology of the resulting fusion proteins with respect to oligomerization or factor-binding dependency. Alternatively, it may reflect differences in the biology of transformed cell types, or, reflect different situations during the onset of these translocations (in utero exposition with poisons vs postnatal acquirement).
An important translational aspect of this study is the establishment of patient-specific DNA sequences that can be used for monitoring MRD by quantitative PCR techniques. Because of the fact that a given MLL fusion allele is genetically stable and a mono-allelic marker for each tumor cell, a more reliable quantification and tracing of residual tumor cells becomes possible. For each of these 2384 acute leukemia patients at least one MLL fusion allele was identified and characterized by sequencing. Several prospective studies were already initiated and first published data verified the reliability of these genomic markers for MRD monitoring.4, 5, 6, 7 Therefore, the use of these MRD markers will contribute in the future to a better stratification of leukemia patients, which will help to further improve the outcome. In particular, for infant ALL patients, due to the relatively low numbers of potential IG/TR MRD-PCR targets, the availability of an MLL fusion DNA rearrangement has a high impact for the clinical application of MRD monitoring.
The analysis of the MLL recombinome allows to classify MLL fusion partner genes into functional categories. As discussed above, only very few TPGs are recurrently identified with a significant frequency. On the basis of the present study these TPGs are AF4, AF6, AF9, AF10, ELL and ENL. At least for the AF4, AF9, ENL and AF10 proteins exist a functional correlation, as all these proteins are organized within a protein complex (or different subcomplexes) that affect transcriptional elongation. AF4 is the docking platform for AF9 or ENL, which both interact (via AF10) to DOT1L.55, 56 DOT1L enable methylation of lysine-79 residues of histone H3 proteins, a prerequisite for the maintenance of RNA transcription57, 58 AF4 binds with its N-terminal portion to the P-TEFb kinase that phosphorylates the largest subunit of RNA polymerase II, DSIF, the NELF complex and UBE2A. This converts RNA POL A into POL E and allows gene transcription.59 As a result, increased and extended H3K79 methylation signatures seem to accompany the presence of several fusion proteins (MLL–AF4, AF4–MLL, MLL–AF9, MLL–ENL, MLL–AF10 and MLL–AF6),60 while an additional increase in H3K4 methylation was only demonstrated by the presence of the reciprocal AF4–MLL59 that causes proB ALL in C57Bl6 mice61 and was shown to cooperate with the RUNX1 protein.62 Thus, all the major MLL fusions share a common pathway, which is not only functionally related but offers new and interesting venues to develop new drugs against these leukemias, for example, by the development of DOT1L inhibitors.63 The fusion proteins MLL–ENL, MLL–AF9 and MLL–AF10 recruit thereby the AF4 complex, while the reciprocal AF4–MLL fusion protein is able to perform exactly the same actions on RNA polymerase II and DOT1L. Thus, future therapies addressing either the inhibition of DOT1L, P-TEFb, or blocking the interaction of the MLL N terminus with MENIN1/LEDGF/MYB are promising new ways to address these leukemias. In addition, the inhibition of Taspase1 would help to inactivate the AF4–MLL fusion protein, as the uncleaved fusion protein is rapidly degraded by SIAH1 and SIAH2.64
For all the other MLL fusion partners identified so far, a systematic classification about their function(s) has been described in great detail recently.65 However, further functional studies are necessary to study the mechanisms that are involved or causative for their leukemogenic activity. Such studies will provide the basis for developing new therapeutic strategies in the future.
Acknowledgments
We thank all local doctors and biologists who provided clinical information and material. This work was made possible by and conducted within the framework of the International BFM Study Group. TB and RM were supported by grant 13R/2016 and R14/02 from the Deutsche José Carreras Leukemia Stiftung.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
✠
The authors declare no conflict of interest.
Supplementary Material
References
- Pui CH, Gaynon PS, Boyett JM, Chessells JM, Baruchel A, Kamps W et al. Outcome of treatment in childhood acute lymphoblastic leukaemia with rearrangements of the 11q23 chromosomal region. Lancet 2002; 359: 1909–1915. [DOI] [PubMed] [Google Scholar]
- Pui CH, Chessells JM, Camitta B, Baruchel A, Biondi A, Boyett JM et al. Clinical heterogeneity in childhood acute lymphoblastic leukemia with 11q23 rearrangements. Leukemia 2003; 17: 700–706. [DOI] [PubMed] [Google Scholar]
- Balgobind BV, Raimondi SC, Harbott J, Zimmermann M, Alonzo TA, Auvrignon A et al. Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: results of an international retrospective study. Blood 2009; 114: 2489–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepański T, Harrison CJ, van Dongen JJ. Genetic aberrations in paediatric acute leukaemias and implications for management of patients. Lancet Oncol 2010; 11: 880–889. [DOI] [PubMed] [Google Scholar]
- Burmeister T, Marschalek R, Schneider B, Meyer C, Gökbuget N, Schwartz S et al. Monitoring minimal residual disease by quantification of genomic chromosomal breakpoint sequences in acute leukemias with MLL aberrations. Leukemia 2006; 20: 451–457. [DOI] [PubMed] [Google Scholar]
- van der Velden VH, Corral L, Valsecchi MG, Jansen MW, De Lorenzo P, Cazzaniga G et al. Prognostic significance of minimal residual disease in infants with acute lymphoblastic leukemia treated within the Interfant-99 protocol. Leukemia 2009; 23: 1073–1079. [DOI] [PubMed] [Google Scholar]
- Yeoh AE, Ariffin H, Chai EL, Kwok CS, Chan YH, Ponnudurai K et al. Minimal residual disease-guided treatment deintensification for children with acute lymphoblastic leukemia: results from the Malaysia-Singapore acute lymphoblastic leukemia 2003 study. J Clin Oncol 2012; 30: 2384–2392. [DOI] [PubMed] [Google Scholar]
- Johansson B, Moorman AV, Secker-Walker LM. Derivative chromosomes of 11q23-translocations in hematologic malignancies. European 11q23 Workshop participants. Leukemia 1998; 12: 828–833. [DOI] [PubMed] [Google Scholar]
- Heerema NA, Sather HN, Ge J, Arthur DC, Hilden JM, Trigg ME et al. Cytogenetic studies of infant acute lymphoblastic leukemia: poor prognosis of infants with t(4;11)—a report of the Children's Cancer Group. Leukemia 1999; 13: 679–686. [DOI] [PubMed] [Google Scholar]
- Van der Burg M, Beverloo HB, Langerak AW, Wijsman J, van Drunen E, Slater R et al. Rapid and sensitive detection of all types of MLL gene translocations with a single FISH probe set. Leukemia 1999; 13: 2107–2113. [DOI] [PubMed] [Google Scholar]
- van der Burg M, Poulsen TS, Hunger SP, Beverloo HB, Smit EM, Vang-Nielsen K et al. Split-signal FISH for detection of chromosome aberrations in acute lymphoblastic leukemia. Leukemia 2004; 18: 895–908. [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Moorman AV, Barber KE, Broadfield ZJ, Cheung KL, Harris RL et al. Interphase molecular cytogenetic screening for chromosomal abnormalities of prognostic significance in childhood acute lymphoblastic leukaemia: a UK Cancer Cytogenetics Group Study. Br J Haematol 2005; 129: 520–530. [DOI] [PubMed] [Google Scholar]
- Meyer C, Schneider B, Reichel M, Angermueller S, Strehl S, Schnittger S et al. Diagnostic tool for the identification of MLL rearrangements including unknown partner genes. Proc Natl Acad Sci USA 2005; 102: 449–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C, Schneider B, Jakob S, Strehl S, Schnittger S, Schoch C et al. The MLL recombinome of acute leukemias. Leukemia 2006; 20: 777–784. [DOI] [PubMed] [Google Scholar]
- Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J, Trka J et al. New insights into the MLL recombinome of acute leukemias. Leukemia 2009; 23: 1490–1499. [DOI] [PubMed] [Google Scholar]
- Meyer C, Hofmann J, Burmeister T, Gröger D, Park TS, Emerenciano M et al. The MLL recombinome of acute leukemia in 2013. Leukemia 2013; 27: 2165–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daser A, Rabbitts TH. The versatile mixed lineage leukaemia gene MLL and its many associations in leukaemogenesis. Semin Cancer Biol 2005; 15: 175–188. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer 2007; 7: 823–833. [DOI] [PubMed] [Google Scholar]
- Burmeister T, Meyer C, Gröger D, Hofmann J, Marschalek R. Evidence-based RT-PCR methods for the detection of the 8 most common MLL aberrations in acute leukemias. Leuk Res 2015; 39: 242–247. [DOI] [PubMed] [Google Scholar]
- Strissel PL, Strick R, Rowley JD, Zeleznik-Le NJ. An in vivo topoisomerase II cleavage site and a DNase I hypersensitive site colocalize near exon 9 in the MLL breakpoint cluster region. Blood 1998; 92: 3793–3803. [PubMed] [Google Scholar]
- Stanulla M, Wang J, Chervinsk DS, Thandla S, Aplan PD. DNA cleavage within the MLL breakpoint cluster region is a specific event which occurs as part of higher-order chromatin fragmentation during the initial stages of apoptosis. Mol Cell Biol 1997; 17: 4070–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf S, Zech J, Bursen A, Schraets D, Oliver PL, Kliem S et al. Transcription linked to recombination: a gene-internal promoter coincides with the recombination hot spot II of the human MLL gene. Oncogene 2007; 26: 1361–1371. [DOI] [PubMed] [Google Scholar]
- Felix CA. Leukemias related to treatment with DNA topoisomerase II inhibitors. Med Pediatr Oncol 2001; 36: 525–535. [DOI] [PubMed] [Google Scholar]
- Emerenciano M, Meyer C, Mansur MB, Marschalek R, Pombo-de-Oliveira MS, . The Brazilian Collaborative Study Group of Infant Acute Leukaemia. The distribution of MLL breakpoints correlates with outcome in infant acute leukaemia. Br J Haematol 2013; 161: 224–236. [DOI] [PubMed] [Google Scholar]
- Fair K, Anderson M, Bulanova E, Mi H, Tropschug M, Diaz MO. Protein interactions of the MLL PHD fingers modulate MLL target gene regulation in human cells. Mol Cell Biol 2001; 21: 3589–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia ZB, Anderson M, Diaz MO, Zeleznik-Le NJ. MLL repression domain interacts with histone deacetylases, the polycomb group proteins HPC2 and BMI-1, and the corepressor C-terminal-binding protein. Proc Natl Acad Sci USA 2003; 100: 8342–8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PY, Hom RA, Musselman CA, Zhu L, Kuo A, Gozani O et al. Binding of the MLL PHD3 finger to histone H3K4me3 is required for MLL-dependent gene transcription. J Mol Biol 2010; 400: 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Song J, Milne TA, Wang GG, Li H, Allis CD et al. Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression. Cell 2010; 141: 1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Muntean AG, Hess JL. ECSASB2 mediates MLL degradation during hematopoietic differentiation. Blood 2012; 119: 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Muntean AG, Wu L, Hess JL. A subset of mixed lineage leukemia proteins has plant homeodomain (PHD)-mediated E3 ligase activity. J Biol Chem 2012; 287: 43410–43416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rössler T, Marschalek R. An alternative splice process renders the MLL protein either into a transcriptional activator or repressor. Pharmazie 2013; 68: 601–607. [DOI] [PubMed] [Google Scholar]
- Grow EJ, Wysocka J. Flipping MLL1's switch one proline at a time. Cell 2010; 141: 1108–1110. [DOI] [PubMed] [Google Scholar]
- Meyer C, Kowarz E, Yip SF, Wan TS, Chan TK, Dingermann T et al. A complex MLL rearrangement identified five years after initial MDS diagnosis is causing out-of-frame fusions whithout progression to acute leukemia. Cancer Genet 2011; 204: 557–562. [DOI] [PubMed] [Google Scholar]
- Mori T, Nishimura N, Hasegawa D, Kawasaki K, Kosaka Y, Uchide K et al. Persistent detection of a novel MLL-SACM1L rearrangement in the absence of leukemia. Leuk Res 2010; 34: 1398–1401. [DOI] [PubMed] [Google Scholar]
- Kim M, Semple I, Kim B, Kiers A, Nam S, Park HW et al. Drosophila Gyf/GRB10 interacting GYF protein is an autophagy regulator that controls neuron and muscle homeostasis. Autophagy 2015; 11: 1358–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryszke MH, Adjeriou B, Liang F, Chen H, Dautry F. Post-transcriptional gene silencing activity of human GIGYF2. Biochem Biophys Res Commun 2016; 475: 289–294. [DOI] [PubMed] [Google Scholar]
- Morita M, Ler LW, Fabian MR, Siddiqui N, Mullin M, Henderson VC et al. A novel 4EHP-GIGYF2 translational repressor complex is essential for mammalian development. Mol Cell Biol 2012; 32: 3585–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Liu Y, Wang YL, Liu R, Xu BH, Zhang F et al. KIF2A regulates the spindle assembly and the metaphase I-anaphase I transition in mouse oocyte. Sci Rep 2016; 6: 39337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi ZY, Ma XS, Liang QX, Zhang T, Xu ZY, Meng TG et al. Kif2a regulates spindle organization and cell cycle progression in meiotic oocytes. Sci Rep 2016; 6: 38574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallin M, Bijlsma EK, El Morjani A, Moutton S, Peeters EA, Maillard C et al. Recurrent KIF2A mutations are responsible for classic lissencephaly. Neurogenetics 2017; 18: 73–79. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Kakeno M, Matsui T, Sugiyama I, Arimura N, Matsuzawa K et al. TTBK2 with EB1/3 regulates microtubule dynamics in migrating cells through KIF2A phosphorylation. J Cell Biol 2015; 210: 737–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmansberger M, Feuchtinger A, Zurnadzhy L, Michna A, Kaiser JC, Abend M et al. CLIP2 as radiation biomarker in papillary thyroid carcinoma. Oncogene 2015; 34: 3917–3925. [DOI] [PubMed] [Google Scholar]
- Majeed SR, Vasudevan L, Chen CY, Luo Y, Torres JA, Evans TM et al. Clathrin light chains are required for the gyrating-clathrin recycling pathway and thereby promote cell migration. Nat Commun 2014; 5: 3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miserey-Lenkei S, Couëdel-Courteille A, Del Nery E, Bardin S, Piel M, Racine V et al. A role for the Rab6A' GTPase in the inactivation of the Mad2-spindle checkpoint. EMBO J 2006; 25: 278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi S, Yu M, Yang S, Miron RJ, Zhang Y. Tcf12, a member of basic helix-loop-helix transcription factors, mediates bone marrow mesenchymal stem cell osteogenic differentiation in vitro and in vivo. Stem Cells 2017; 35: 386–397. [DOI] [PubMed] [Google Scholar]
- Labreche K, Simeonova I, Kamoun A, Gleize V, Chubb D, Letouzé E et al. TCF12 is mutated in anaplastic oligodendroglioma. Nat Commun 2015; 6: 7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein M, Anderson MK. HEB in the spotlight: Transcriptional regulation of T-cell specification, commitment, and developmental plasticity. Clin Dev Immunol 2012; 2012: 678705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda K, Eguchi-Ishimae M, Yagi C, Kawabe M, Moritani K, Niiya T et al. CLTC-ALK fusion as a primary event in congenital blastic plasmacytoid dendritic cell neoplasm. Genes Chromosomes Cancer 2014; 53: 78–89. [DOI] [PubMed] [Google Scholar]
- DeMari J, Mroske C, Tang S, Nimeh J, Miller R, Lebel RR. CLTC as a clinically novel gene associated with multiple malformations and developmental delay. Am J Med Genet A 2016; 170A: 958–966. [DOI] [PubMed] [Google Scholar]
- Miyoshi N, Ishii H, Mimori K, Nishida N, Tokuoka M, Akita H et al. Abnormal expression of PFDN4 in colorectal cancer: a novel marker for prognosis. Ann Surg Oncol 2010; 17: 3030–3036. [DOI] [PubMed] [Google Scholar]
- Ma Z, Morris SW, Valentine V, Li M, Herbrick JA, Cui X et al. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat Genet 2001; 28: 220–221. [DOI] [PubMed] [Google Scholar]
- Ball JR, Ullman KS. Versatility at the nuclear pore complex: lessons learned from the nucleoporin Nup153. Chromosoma 2005; 114: 319–330. [DOI] [PubMed] [Google Scholar]
- Jafari N, Kim H, Park R, Li L, Jang M, Morris AJ et al. CRISPR-Cas9 mediated NOX4 knockout inhibits cell proliferation and invasion in HeLa cells. PLoS One 2017; 12: e0170327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C, Burmeister T, Strehl S, Schneider B, Hubert D, Zach O et al. Spliced MLL fusions: a novel mechanism to generate functional chimeric MLL-MLLT1 transcripts in t(11;19)(q23;p13.3) leukemia. Leukemia 2007; 21: 588–590. [DOI] [PubMed] [Google Scholar]
- Zeisig DT, Bittner CB, Zeisig BB, García-Cuéllar MP, Hess JL, Slany RK. The eleven-nineteen-leukemia protein ENL connects nuclear MLL fusion partners with chromatin. Oncogene 2005; 24: 5525–5532. [DOI] [PubMed] [Google Scholar]
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet 2007; 16: 92–106. [DOI] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM et al. hDOT1L links histone methylation to leukemogenesis. Cell 2005; 121: 167–178. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 2001; 15: 2343–2360. [DOI] [PubMed] [Google Scholar]
- Benedikt A, Baltruschat S, Scholz B, Bursen A, Arrey TN, Meyer B et al. The leukemogenic AF4-MLL fusion protein causes P-TEFb kinase activation and altered epigenetic signatures. Leukemia 2011; 25: 135–144. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell 2008; 14: 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursen A, Schwabe K, Rüster B, Henschler R, Ruthardt M, Dingermann T et al. AF4-MLL is capable of inducing ALL in mice without requirement of MLL-AF4. Blood 2010; 115: 3570–3579. [DOI] [PubMed] [Google Scholar]
- Wilkinson AC, Ballabio E, Geng H, North P, Tapia M, Kerry J et al. RUNX1 Is a key target in t(4;11) leukemias that contributes to gene activation through an AF4-MLL complex interaction. Cell Rep 2013; 3: 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 2011; 20: 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursen A, Moritz S, Gaussmann A, Dingermann T, Marschalek R. Interaction of AF4 wildtype and AF4•MLL fusion protein with SIAH proteins: indication for t(4;11) pathobiology? Oncogene 2004; 23: 6237–6249. [DOI] [PubMed] [Google Scholar]
- Marschalek R. Systematic classification of mixed-lineage leukemia fusion partners predicts additional cancer pathways. Ann Lab Med 2016; 36: 85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.