Abstract
Mitochondrial dysregulation plays a central role in cancers and drives reactive oxygen species (ROS)-dependent tumor progression. We investigated the pro-tumoral roles of mitochondrial dynamics and altered intracellular ROS levels in pancreatic ductal adenocarcinoma (PDAC). We identified ‘family with sequence similarity 49 member B’ (FAM49B) as a mitochondria-localized protein that regulates mitochondrial fission and cancer progression. Silencing FAM49B in PDAC cells resulted in increased fission and mitochondrial ROS generation, which enhanced PDAC cell proliferation and invasion. Notably, FAM49B expression levels in PDAC cells were downregulated by the tumor microenvironment. Overall, the results of this study show that FAM49B acts as a suppressor of cancer cell proliferation and invasion in PDAC by regulating tumor mitochondrial redox reactions and metabolism.
Introduction
Pancreatic ductal adenocarcinoma (PDAC), whose 5-year survival rate is as low as 6%,1, 2 is one of the most aggressive malignancies, because the disease is often diagnosed at a late stage, and its treatment options are limited. PDAC has a very poor prognosis.3, 4, 5 Therefore, a better understanding of the mechanisms driving the progression of this cancer is needed. Approximately 90% of all PDACs acquire KRAS mutations,6 and the progression of these tumors is also accompanied by an increase in cellular oxidative stress levels.7, 8, 9
Mitochondria are the main source of reactive oxygen species (ROS), and their functional state is modified during tumor progression.10, 11, 12, 13 Mitochondrial ROS play an essential role in cell proliferation and tumorigenesis in PDAC.14, 15 In particular, mitochondrial fragmentation, a phenomenon known as ‘fission’, is associated with increased energy demands and increased ROS production.16, 17 Mitochondrial fission is also associated with the generation of new organelles. Fission is mainly regulated by dynamin-related protein 1 (DRP1). DRP1 recruitment around mitochondria results in the formation of spirals, which draw together both the inner and the outer mitochondrial membranes to allow mitochondrial division.18 Conversely, ‘fusion’, which is required to reduce stress, is regulated by mitofusins 1 and 2 (MFN1/2), which fuse the outer membrane, and optic atrophy 1, (OPA1), which fuses the inner membrane, creates elongated mitochondria.19, 20, 21 Metabolic changes in cells lead to the regulation of fission and fusion.22, 23, 24
Family with sequence similarity 49 member B (FAM49B) is encoded by a highly conserved gene in mammals. In humans, the FAM49B gene is localized on chromosome 8q24, encodes for a 37-kDa protein composed of 324 amino-acid residues,25 and contains a characteristic DUF1394 domain. Another FAM49B isoform of ~20 kDa lacks the first 123 amino acids due to alternative splicing of its transcript. None of the isoforms contain any other known functional motifs. To date, no functional data regarding this protein have been published, and its role in cancer is unknown.
In this study, we investigated the expression and role of FAM49B in PDAC. We demonstrated that FAM49B is highly expressed in PDAC cell lines and that this expression is downregulated in vivo by the surrounding tumor environment. In PDAC cells, FAM49B is predominantly localized in the mitochondria, and FAM49B gene knockdown leads to oxidative stress that enhances tumor proliferation and invasiveness. Thus, we have identified a novel ‘tumor suppressor’ gene that links the inflammatory environment to mitochondrial dynamics.
Results
FAM49B expression in PDAC
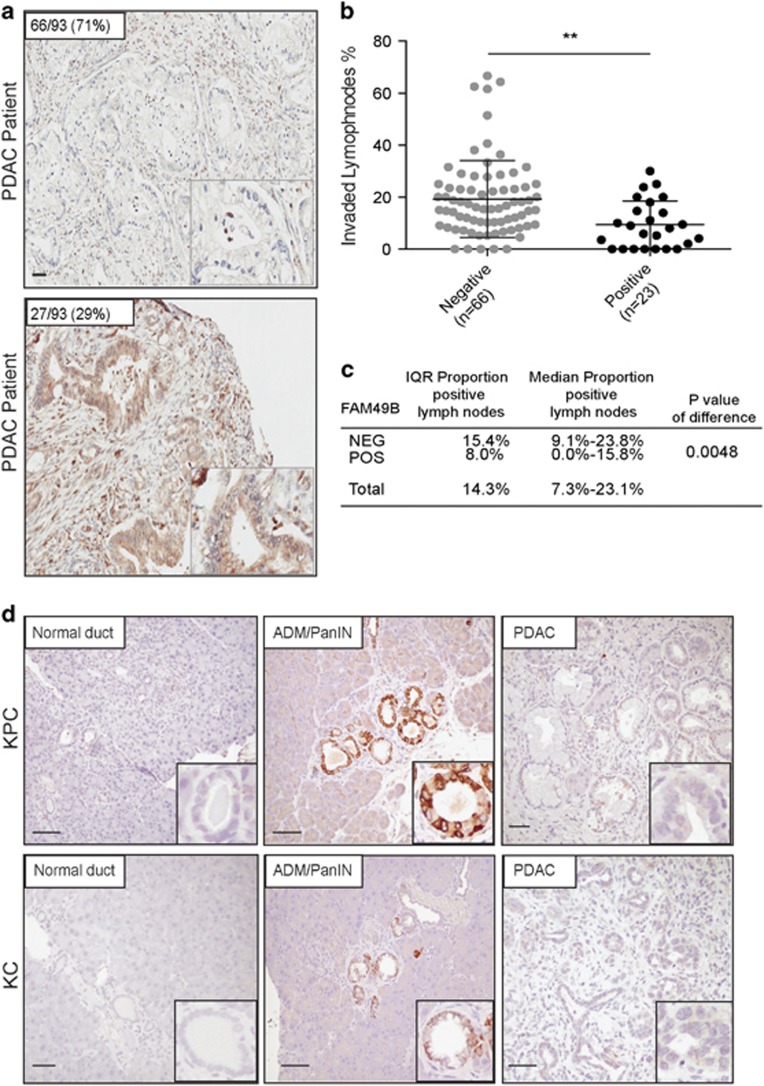
FAM49B expression levels in PDAC biopsy tissue samples (n=93) were analyzed by immunohistochemistry (Figure 1a). Most of the PDAC biopsies (71%) displayed negative staining for FAM49B (Figure 1a). However, the remaining biopsies (29%) displayed positive staining for FAM49B (Figure 1a). We subsequently evaluated the association between FAM49B expression and PDAC patient clinical and pathological characteristics (Supplementary Table 1). There were no significant differences in FAM49B expression levels among patients of different ages or survival times (Supplementary Figure 1A). However, there was a significant difference in the rate of tumor lymph node (LN) invasion between patients with positive and negative FAM49B expression. Specifically, the rate of LN invasion was much higher in patients who displayed negative FAM49B expression (P=0.004) than in patients who displayed positive FAM49B expression (Figures 1b and c). A similar pattern of low FAM49B expression was observed in the genetically engineered mouse (GEM) strains that spontaneously develop PDAC (LSL-KrasG12D/+, Pdx-1-Cre (KC), LSL-KrasG12D/+, LSL-Tp53R172H/+ and Pdx-1-Cre (KPC))26 (Figure 1d). To determine whether FAM49B expression is downregulated during disease progression we examined KC and KPC mouse tissues collected at different time points, namely 3, 6, 9 and 12 months of age (n=8/age). FAM49B expression was low in normal pancreatic ducts (Figure 1d, left panels) and strongly expressed in most of the ADM/ early pancreatic intraepithelial neoplasia (PanIN) lesions (Figure 1d, mid panels), which represents early stages of the disease. By contrast FAM49B staining was very faint in well-established PDAC (Figure 1d, right panels). These results suggest FAM49B expression is lost during the progression from early PanIN to adenocarcinoma and the loss of correlates with its invasiveness.
Figure 1.
FAM49B expression in PDAC and its correlation with LN invasion (a) Representative sections of tumors showing positive and negative immunostaining for FAM49B. The scale bars represent a 100-μm inset, × 600 magnification. (b) LN metastasis percentage in PDAC patients with positive and negative FAM49B expression. (c) Comparison of the proportions of a total of 89 patients with positive (n=23) and negative (n=65) FAM49B expression and positive lymph nodes. The results were analyzed with the Mann–Whitney U-test. (d) Representative sections of different grading derived from the PDAC GEM mouse models (LSL-KrasG12D/+, Pdx-1-Cre (KC); LSL- KrasG12D/+, LSL-Tp53R172H/+ and Pdx-1-Cre (KPC)) at different ages 3, 6, 9 and 12 months (n=8/age). The scale bar represents 100-μm.
FAM49B expression is downregulated by the tumor microenvironment
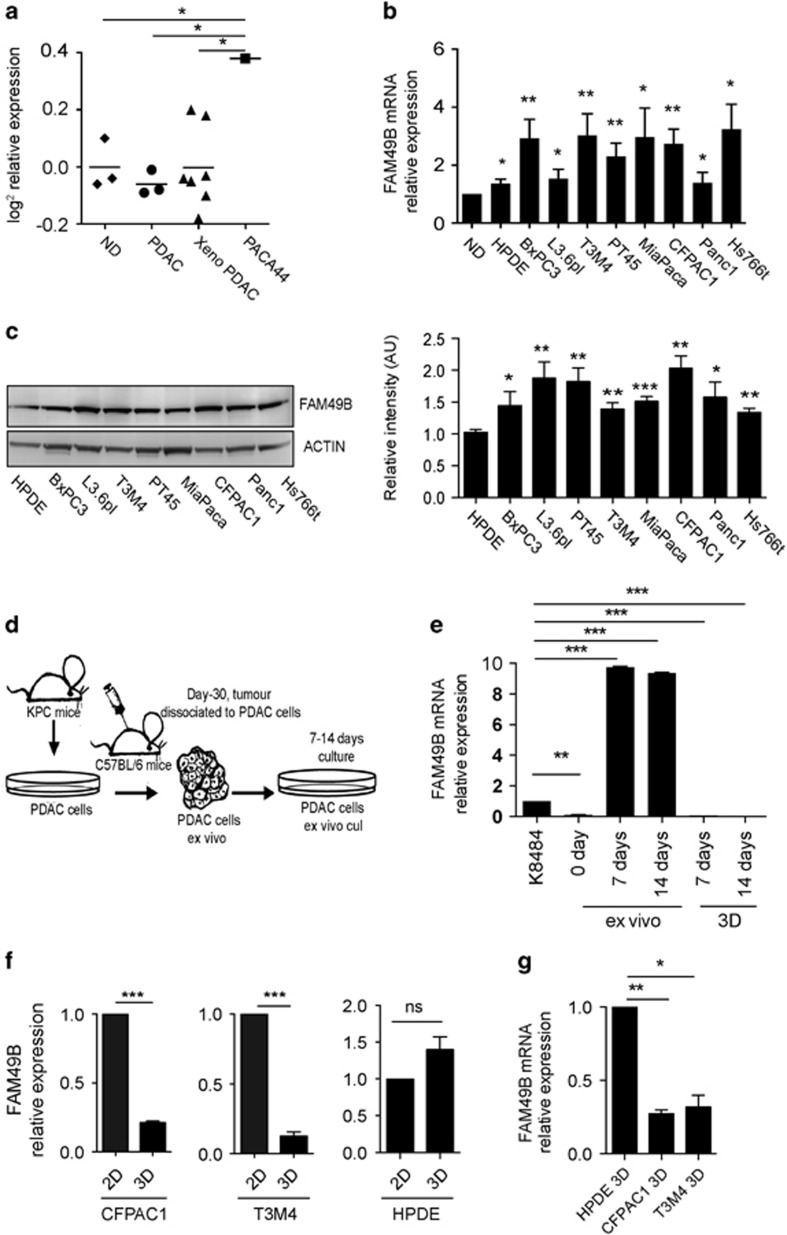
Microarray analysis of FAM49B mRNA expression showed that FAM49B was expressed at similarly low levels in normal pancreatic duct samples and PDAC biopsy samples from both patients and xeno-transplanted immunosuppressed (xeno-PDAC) mice. In contrast, FAM49B was highly expressed in the Paca44 PDAC cell line (Figure 2a). Additional mRNA analysis showed that FAM49B expression levels were two- to threefold higher in six human PDAC cell lines (BXPC3, T3M4, PT45, Mia-PA-CA2, CFPAC1 and Hs766T) and elevated to a lesser extent in another two cell lines (L3.6pl and PANC1), as well as in the primary human pancreatic duct epithelial (HPDE) non-tumor cell line (Figure 2b), compared to normal human pancreatic duct tissues. Western blot analysis confirmed that FAM49B expression levels were higher in all the PDAC cell lines than in the HPDE cell line (Figure 2c) and that FAM49B expression was not detectable in the normal pancreatic duct (Supplementary Figure 1B).
Figure 2.
FAM49B expression in PDAC is downregulated by the surrounding microenvironment (a) Differential expression of FAM49B in normal ducts (NDs), PDAC biopsies, PDAC xenotransplants and the PDAC cell line Paca44, according to the microarray data. The data are shown as base-two logarithm ratios of the intensity values to the median intensity values of normal pancreas tissue. (b) The relative expression levels of FAM49B mRNA in HPDE cells and different PDAC cell lines were evaluated by qPCR. β-actin was used as a reference gene. (c) FAM49B protein expression in different PDAC cell line and HPDE cell line was analyzed by western blotting. Actin was used as a loading control. (d) Experimental scheme showing the syngeneic orthotopic cell line-derived xenograft model. (e) FAM49B expression in the K8484 murine PDAC cell line was analyzed ex vivo (day 0) and after 7 and 14 days of in vitro culture and 3D culture by qPCR. Actin was used as a reference gene. (f) FAM49B expression in CFPAC1 and T3M4 PDAC cells and normal HPDE cells cultured in 3D Matrigel embedded for 14 days or in in vitro 2D monolayer cultures, expression levels was analyzed by qPCR. Actin was used as a reference gene. (g) FAM49B expression in CFPAC1, T3M4 PDAC cells cultured in 3D Matrigel for 14 days in comparison with the and Normal HPDE cell. All experiments were performed at least three times, and the data are represented as the mean±s.e.m. (*P<0.05, **P<0.001, and ***P<0.0001 are values significantly different from control or untreated samples).
Our finding that FAM49B expression levels were higher in PDAC cell lines than in tumor tissues suggests that FAM49B expression is downregulated by the tumor environment. To confirm this hypothesis, we kinetically evaluated FAM49B ex vivo expression in orthotopically injected PDAC cells. KPC-derived K8484 murine PDAC cells expressing FAM49B (Supplementary Figure 1C) were orthotopically injected into syngeneic mice. After 30 days, the tumors were excised and dissociated, and the cells were analyzed for FAM49B expression (Figure 2d). On day 0, ex vivo mRNA analysis showed that FAM49B transcription was almost completely absent. However, when the K8484 cells were cultured again over 7–14 days, FAM49B expression increased significantly (Figure 2e).
The extracellular matrix (ECM) can interact with tumor cells to influence their cellular behavior, such as migration, adhesion and proliferation. To evaluate the regulation of FAM49B expression by the ECM, we cultured CFPAC1 and T3M4 PDAC cell lines in a three-dimensional (3D) culture, by embedding cells in Matrigel or seeding cells on Matrigel coated plates, as the cell-cell and cell-ECM interactions that characterize this environment more closely mimic those of the natural environment found in vivo.27 After spheroid formation (Supplementary Figure 1D), RNA and protein were isolated from the cells and analyzed for FAM49B expression. PDAC cells cultured in 3D displayed an almost complete downregulation of FAM49B compared to PDAC cells cultured on a 2D monolayer (Figure 2f) or normal HPDE cells (Figure 2g). Normal HPDE cells did not display the same downregulation of FAM49B when cultured in 2D and 3D conditions (Figure 2f). PDAC cells cultured on Matrigel coated plates showed a similar trend in the downregulation of FAM49B expression, while no changes were observed for HPDE cells (Supplementary Figure 1E). Even the murine PDAC cell line K8484 cultured in 3D conditions showed no detectable levels of FAM49B transcript at both 7 and 14 days (Figure 2e). To check possibility that downregulation of FAM49B expression in tumor microenvironment was due to accelerated protein degradation, we treated the PDAC cell lines CFPAC1, T3M4, K8484 and normal HPDE cells with the proteasome inhibitor MG132. There was no increase in the levels of FAM49B upon proteasome inhibition both in 2D and 3D cultures (Supplementary Figures 2A and B). These data strongly suggest that FAM49B downregulation in tumor cells occur at transcript levels rather than due to its protein stability.
FAM49B knockdown enhances PDAC proliferation and invasion
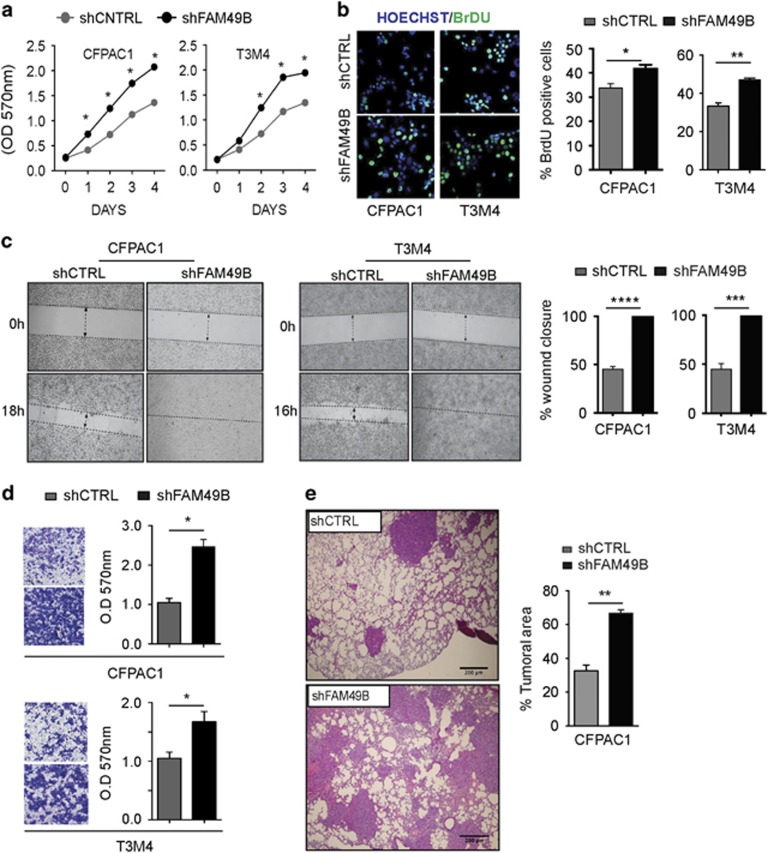
The low in vivo FAM49B expression levels, mentioned above, correlate with higher PDAC LN invasion (Figure 1c). In addition, culturing ex vivo-cultured PDAC cells in vitro strongly enhances FAM49B expression. Those data prompted us to investigate whether FAM49B expression limits PDAC progression and invasiveness. We stably knocked down FAM49B in CFPAC1, T3M4 and HPDE cells using a short hairpin RNA (shRNA) for FAM49B (shFAM49B) or a scrambled control (shCTRL) (Supplementary Figures 2C and D) and evaluated their proliferation and invasiveness Using MTT and BrdU assays, we observed that both FAM49B-silenced PDAC cell lines proliferated significantly more than shCTRL cells (Figures 3a and b). Wound healing and Matrigel invasion assays revealed that FAM49B-silenced cells displayed enhanced migration compared to control cells (Figures 3c and d). Notably, FAM49B-silenced HPDE cells did not display increased proliferation or migration (Supplementary Figures 3A and B). Finally, to investigate the effects of FAM49B silencing in vivo, we injected shFAM49B or shCTRL CFAPC1 cells into the tail veins of immune compromised mice. The FAM49B-silenced cells spread to the lungs of these mice, where they grew to a much greater extent than the shCTRL cells (Figure 3e).
Figure 3.
FAM49B functions as a metastasis suppressor. (a) shCTRL (gray circles) and shFAM49B (black circles) CFPAC1 and T3M4 cell proliferation was evaluated by MTT assay. The data are shown as the mean±s.e.m. OD at 570 nm of three independent experiments. (b) BrdU staining (green) in shCTRL and shFAM49B CFPAC1 and T3M4 cells. Cell nuclei are stained with Hoechst (blue). The graphs in the right panel represent the results of the quantification of BrDU-positive cells, as determined by immunofluorescence analysis of shCTRL (gray bars) and shFAM49B (black bars) CFPAC1 and T3M4 cells. Data are shown as the mean±s.e.m. of three independent experiments. (c) Wound-healing assays of shCTRL and shFAM49B CFPAC1 and T3M4 cells. The dotted lines indicate the wound edge at 0 h. Migration of individual cells over 16–24 h was tracked using ImageJ. The results are represented as wound closure percentage bars representing the mean values±s.d. of three experiments. (d) Statistical analysis of the results of the Matrigel invasion assays of shCTRL and shFAM49B cells. The results are expressed as the mean±s.e.m. of three independent experiments. (e) Lung metastasis assay of CFPAC1 cells transfected with shCTRL or shFAM49B. Representative hematoxylin–eosin (H&E) images of the lung metastases are shown scale bar represents 200 μm. Tumor metastases were quantified in the histological sections, n=4 per group, by ImageJ. Student’s t-test was used to determine the significance of the differences between the groups (*P<0.05, **P<0.001, ***P<0.0001, Student’s t-test).
FAM49B silencing alters mitochondrial dynamics
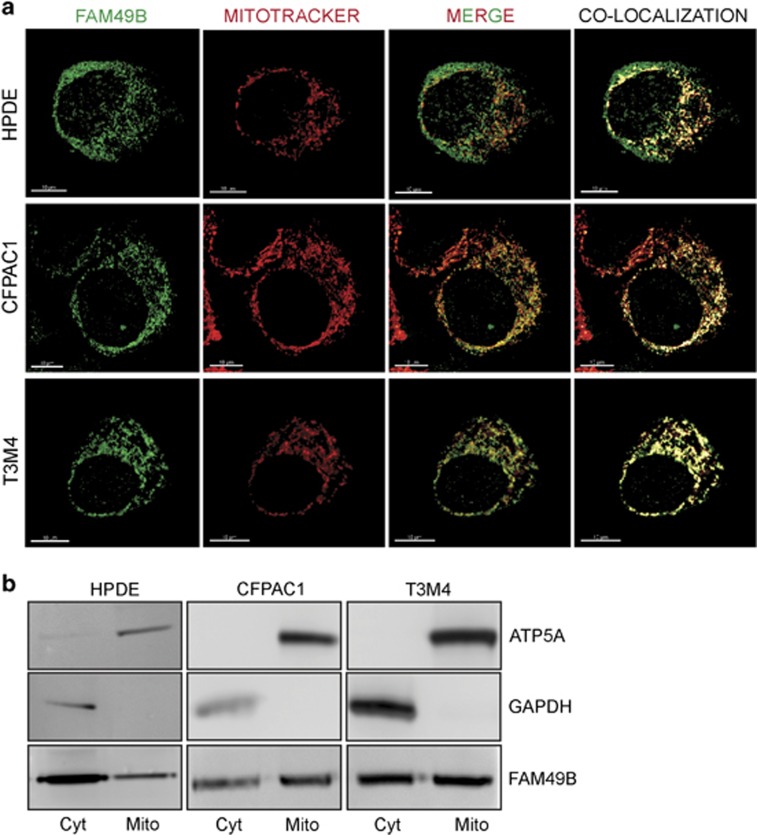
The intracellular distribution of FAM49B was also investigated. FAM49B-GFP fusion protein were overexpressed in HEK293 cells and subjected to fluorescence protease protection assay, which utilizes digitonin and trypsin digestion. Both treatments eliminated the majority of the cytoplasmic proteins in the above-mentioned cell line, but the cells overexpressing FAM49B-GFP partially retained the fluorescence signal, suggesting that in addition to localizing in the cytoplasm, FAM49B also localizes in organelles (Supplementary Figure 4A), particularly in mitochondria, as demonstrated by electron microscopy (Supplementary Figure 4B). Furthermore, confocal analysis showed that in CFPAC1 and T3M4 cells, as well as in HPDE cells, FAM49B (green) co-localized with the MitoTracker dye (red), a finding that confirmed its mitochondrial localization (Figure 4a). Of note, HPDE cells showed greater cytoplasmic FAM49B expression than PDAC cells. Western blot analysis of FAM49B expression confirmed its predominant mitochondrial localization (Figure 4b).
Figure 4.
Localization of FAM49B in the mitochondria. (a) Cells co-stained with Mitotracker (red) and anti-FAM49B antibody (green) was detected by immunofluorescence. Tissue sections were analyzed with a laser scanning confocal microscope (Zeiss LSM5 Pascal, Jena, Germany) using its multichannel acquisition mode to avoid fluorescence cross talk. Images were acquired with a × 100 oil-immersion objective, scale bar 10 μm. The pixels indicating co-localization are shown in yellow. (b) FAM49B expression in the cytosolic (Cyto) and mitochondria-enriched (Mito) fractions of HPDE, CFPAC1 and T3M4 cells was measured by western blot analysis. The mitochondrial marker ATP5A was used as a control of the quality of the mitochondrial fractions. GAPDH served as a cytosolic marker.
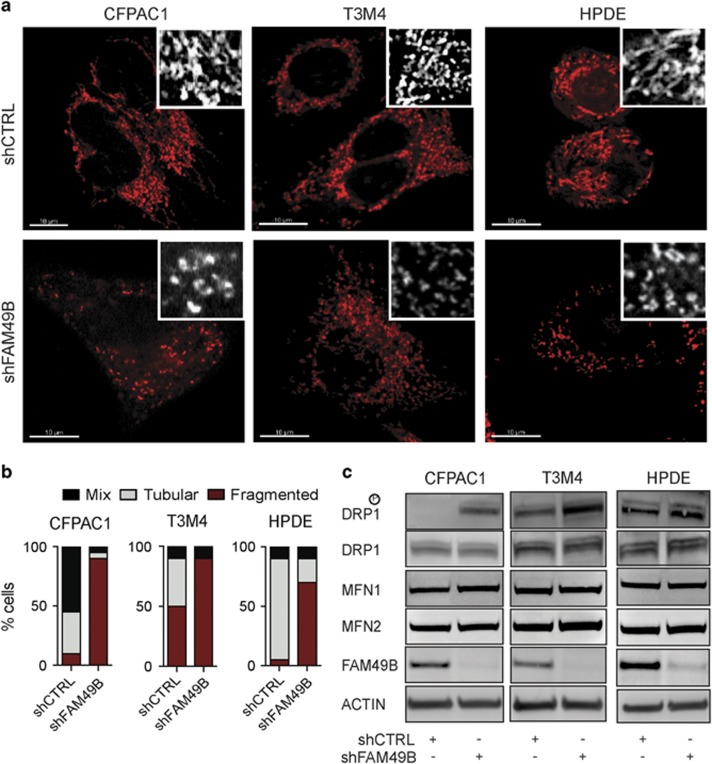
We employed MitoTracker RED staining to assess whether FAM49B silencing affected mitochondrial morphology. We found that shCTRL cells exhibited many more interconnected, elongated and tubular mitochondria than their counterparts, while shFAM49B cells, including HPDE cells, displayed more punctuated mitochondria than other cells (Figures 5a and b).
Figure 5.
FAM49B regulates mitochondrial dynamics. (a) MitoTracker staining was used to analyze mitochondrial morphology in CFPAC1, T3M4 and HPDE cells. The cells were analyzed with a laser-scanning microscope (Zeiss LSM5 Pascal). (b) Quantitation of the mitochondrial morphologies observed in the cells described in a, n>50 cells, was blindly performed by three people in three independent experiments. The bars represent the mean percentages of the mixed (black bars), tubular (gray bars) or fragmented (red bars) mitochondrial phenotypes from one representative experiment. (c) CFPAC1, T3M4 and HPDE cell lysates were analyzed for mitochondrial dynamics regulator expression by western blotting with anti-FAM49B, anti-DRP1, anti-MFN1, and anti-MFN2 Abs. Actin was used as a loading control.
Mitochondrial dynamics are driven by fission and fusion machinery. DRP1 phosphorylation, a hallmark of fission,28 was induced in shFAM49B CFPAC1 cells and was increased in shFAM49B T3M4 and HPDE cells (Figure 5c), whereas fusion-related protein, MFN1 and MFN2 expression,29 was not modified by FAM49B silencing (Figure 5c). These data suggest that FAM49B maintains mitochondrial health status, as its absence favors fission in normal and PDAC cells. These findings were also confirmed by our observation of a decrease in mitochondrial membrane potential (Δψm), which is considered a consequence of fission (Supplementary Figure 4C), but FAM49B silencing is not associated to the induction of apoptosis as no changes were observed in the cytochrome c release or caspase 9 and 3 activation (Supplementary Figures 4D and E). In addition, FAM49B re-expression in silenced cells restored the elongated and interconnected mitochondrial morphology (Supplementary Figure 4F) suggesting that FAM49B expression regulates the mitochondrial dynamics.
FAM49B regulates oxidative stress
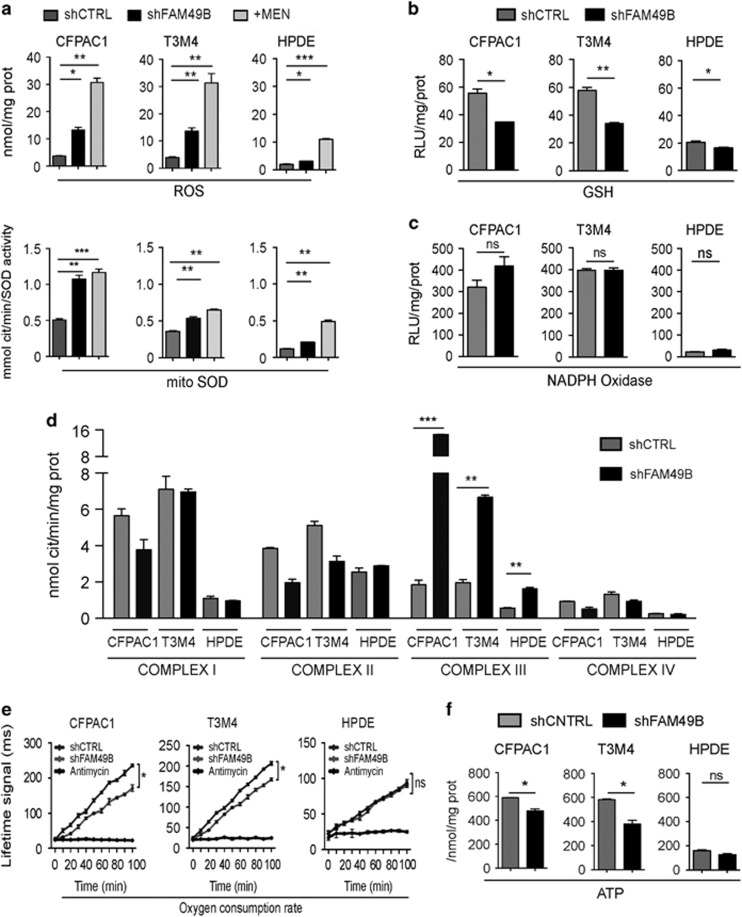
As FAM49B affects mitochondrial dynamics, and mitochondria are key regulators of redox balance,30 we investigated whether FAM49B silencing was accompanied by an increase in intracellular ROS levels and antioxidant pathway activation. Compared to shCTRL cells, FAM49B-silenced PDAC cells displayed much higher levels of intracellular ROS, as well as elevated superoxide dismutase (SOD) activity levels in their mitochondria (Figure 6a), but not in their cytosol (data not shown). A similar trend was also observed in HPDE cells, wherein ROS levels and SOD activity levels were lower than in PDAC cells (Figure 6a). Glutathione (GSH) expression levels decreased in shFAM49B cells concurrently with the above increases in ROS levels (Figure 6b). No significant changes were observed in NADPH oxidase activity (Figure 6c), which indicated this enzyme is not the source of the increased ROS levels that were detected in shFAM49B cells. As efficient oxidative phosphorylation is associated with increased mitochondrial ROS production,31 we investigated electron transport chain (ETC) complex activity. shFAM49B cells displayed aberrant complex III activity (Figure 6d). Consistent with the above findings, the extent of this phenomenon was greater in PDAC cells than in HPDE cells. Mitochondrial oxygen consumption and ATP generation were significantly decreased in FAM49B-silenced PDAC cells, but not in FAM49B-silenced HPDE cells (Figures 6e and f), suggesting that silencing FAM49B in PDAC cells uncouples electron transport and ATP synthesis. The pentose phosphate pathway (PPP), which is activated in response to intracellular ROS, fuels lipid and nucleotide biosynthesis and sustains antioxidant responses to support cell survival and proliferation.32 PPP activity levels were increased in FAM49B-silenced PDAC cells but not in FAM49B-silenced HPDE cells (Figure 7a).
Figure 6.
FAM49B expression alters mitochondrial function and cell redox states. (a) ROS levels (upper panels) and mitochondrial SOD activity levels (lower panels) were evaluated in shCTRL and shFAM49B CFPAC1, T3M4 and HPDE cells. (b, c) GSH and NADPH oxidase activity levels in shCTRL and shFAM49B CFPAC1, T3M4 and HPDE cells. (d) Analysis of mitochondrial respiratory chain complex I-IV activity levels in shCTRL and shFAM49B CFPAC1 and T3M4 cells, expressed as nmol NAD+/min/mg mitochondrial protein for complex I, nmol Cyt c reduced/min/mg mitochondrial protein for complexes II–III and nmol Cyt c oxidized/min/mg mitochondrial protein for complex IV. (e) shCTRL and shFAM49B CFPAC1, T3M4 and HPDE cell oxygen consumption rates (OCRs). Antimycin was used as a positive control for PCR inhibition. (f) Relative ATP levels in shCTRL and shFAM49B CFPAC1, T3M4 and HPDE cells. All the graphs illustrate the mean results of three independent experiments±s.e.m. *P<0.05, **P<0.01*** and P<0.001 values indicate significant differences from shCTRL.
Figure 7.
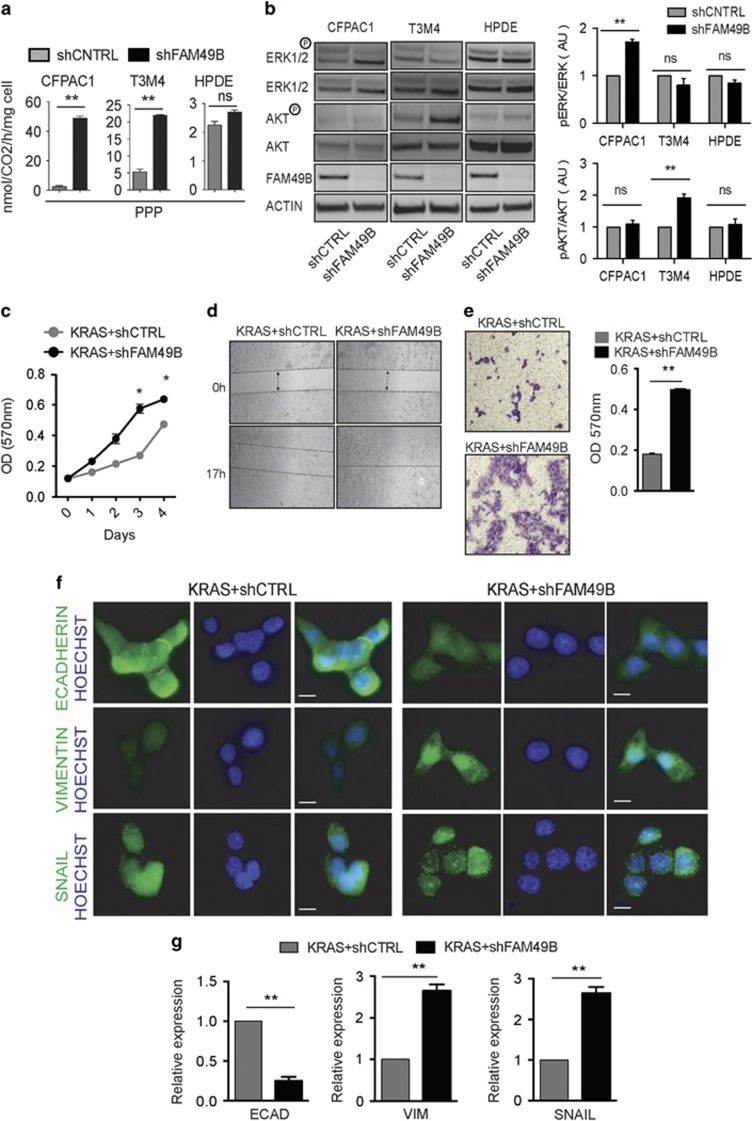
FAM49B downregulation enhances KRAS tumorigenesis. (a) Analysis of PPP flux in the shCTRL and shFAM49B CFPAC1, T3M4 cell lines. Data are shown as the mean±s.e.m. of three independent experiments. (b) ERK1/2 and AKT phosphorylation in shCTRL or shFAM49B cells was assessed by SDS–PAGE/ WB. Actin was used as a loading control. (c) shCTRL (gray circles) and shFAM49B (black circles) HPDE KRAS cell proliferation was evaluated by MTT assay. Data are shown as the mean±s.e.m. OD at 570 nm of three independent experiments. (d) Wound-healing assays of shCTRL and shFAM49B HPDE KRAS cells. The dotted lines indicate the wound edge at 0 h. Migration of individual cells over 18–24 h, which was represented as wound closure percentage bars representing the mean±s.d. of three experiments, was tracked using ImageJ. (e) Statistical analysis of the results of Matrigel invasion assays of shCTRL and shFAM49B cells. The results are expressed as the mean±s.e.m. of three independent experiments. (f) Cells co-stained with antibodies to various markers of EMT (green) and cell nuclei stained with Hoechst (blue), as demonstrated by immunofluorescence scale bar represent 10-μm. (g) Expression of various EMT markers, as demonstrated by real-time PCR in shCTRL and shFAM49B HPDE KRAS cells. Actin was used as a reference gene. All graphs illustrate the mean results of three independent experiments±s.e.m. (*P<0.05, **P<0.001, ***P<0.0001, Student’s t-test).
ROS act as secondary messengers that favor tumor proliferation and invasion through ERK and AKT signaling.14, 33, 34, 35, 36 Western blot analysis revealed the presence of increased ERK1/2 and AKT phosphorylation in shFAM49B PDAC cells (Figure 7b), but not in shFAM49B HPDE cells (Figure 7b), suggesting that the absence of FAM49B significantly upregulated the pro-tumoral functions of ROS in PDAC cells, but not in HPDE cells.
FAM49B ablation cooperates with KRAS to increase PDAC invasiveness
As activated KRAS is present in >90% of PDACs,37 to determine if FAM49B inactivation synergizes with KRAS oncogene activation, FAM49B was silenced the HPDE cell line expressing KRASG12V (KRAS+shFAM49B).
In constitutive KRAS activated HPDE, FAM49B inactivation was able to induce an increased cell proliferation, cell motility, invasiveness, and EMT marker expression (Figures 7c–g). These KRAS-mutated shFAM49B HPDE cells also displayed a decreased expression of key intracellular junction components, namely, the epithelial marker E-cadherin, and an increased expression of the mesenchymal marker Vimentin (Figure 7g). By contrast, no changes were observed in EMT marker expression with FAM49B-silenced or not parental HPDE (Supplementary Figure 3C).
In addition, the mesenchymal-associated transcriptional factor SNAIL was mostly localized into the nuclei of these cells compared to KRAS-mutated shCTRL HPDE cells, which in general displayed a more epithelial phenotype (Figures 7f and g). These findings indicate that FAM49B inactivation in the presence of abnormal KRAS activity promotes tumor progression.
Discussion
This study showed for the first time that the FAM49B protein is involved in mitochondrial fission/fusion regulation and acts as a metastasis suppressor in PDAC. Notably, FAM49B expression was downregulated by the PDAC tumor microenvironment, as demonstrated by the results of our ex vivo analysis of its transcription in tumor cells injected into mice. FAM49B expression was significantly decreased in cells analyzed immediately after excision and then sharply increased as early as 7 days after culture initiation. Tumor cells exhibit different morphological characteristics when cultured in 3D as opposed to a 2D monolayer, as 3D culture is known to preserve cell proliferation rates, gene expression profiles and drug sensitivities.38, 39 Downregulation of FAM49B expression in PDAC cells cultured in a 3D Matrigel environment provided evidence indicating the ECM interacts with PDAC cells, a critical interaction that subsequently regulates FAM49B expression. Therefore, the dynamic and reciprocal interactions between tumor and stromal cells, as well as between tumor cells and the ECM, are crucial for the downregulation of FAM49B expression, which is associated with PDAC metastasis. Most of the patients who had lower LN invasion percentages were positive for FAM49B, while the patients with higher LN invasion percentages were negative for FAM49B.
We show that the loss of FAM49B expression occurring in human PDAC correlates with the invasion of the PDAC. We used two mouse models of spontaneous and autochthonous PDAC, namely KC and KPC that fully recapitulate the human disease, Both mouse models shows similar progressive loss of FAM49B expression during pancreatic lesion progression: FAM49B expression was at lower levels in normal acinar cells consistent with results in human tissues, and increased in acinar-to-ductal metaplasia (ADM) and early PanIN. The cells undergoes metabolic changes, stress and plasticity during the ADM40 as later diminished FAM49B expression strongly correlated with increasing severity of later PanIN lesions and FAM49B was not detected in the tumor samples.
The tumor microenvironment is known to affect the regulation of genes, and the expression other tumor suppressor genes, such as BRAC1 and PTEN, is downregulated in the tumor environment.41, 42 Here, we showed that FAM49B plays a suppressive role in PDAC, as shFAM49B PDAC cells displayed higher proliferation and invasive ability than their counterparts, both in vitro and in vivo, confirming our initial hypothesis.
A unique feature of FAM49B is its prevalent mitochondrial localization. This observation was confirmed by confocal microscopy and western blot analysis. Interestingly, FAM49B silencing resulted in punctuated mitochondria, a finding reflective of an increase in fission, in conjunction with higher DRP1 phosphorylation, which is considered the hallmark of this process. However, MFN1/2 protein expression, which is associated with the fusion process, was not modified by FAM49B silencing, suggesting that the absence of FAM49B has a stronger impact on fission rather than on fusion. Mitochondrial dynamics have been shown to play an important role in maintaining mitochondrial morphology, stress responses, mitochondrial DNA stability, respiratory capacity and cellular apoptosis.21, 43, 44 It is worth noting that FAM49B re-expression after FAM49B silencing led to restoration of the elongated and tubular mitochondrial morphology.
Another phenomenon induced by FAM49B silencing in non-tumoral and neoplastic pancreatic cells is the modification of mitochondrial ETC complex activity, particularly complex III activity, which results in the uncoupling of this complex from subsequent ATP synthesis. This uncoupled oxidative phosphorylation may explain the increase in mitochondrial ROS, which has occurred in several cell models,30, 45 and the parallel attempts by cells to counter balance this ROS activity through increase in mitochondrial SOD and PPP activity. In addition to buffering ROS, the PPP also promotes tumor cell proliferation, prevents oxidative cell damage and paradoxically transforms the small amounts ROS still present from damage-inducing agents into pro-survival signals.46
Interestingly, we demonstrated that in the context of KRAS mutations, depleting FAM49B triggered the pro-tumoral functions of ROS, as demonstrated by the increases in ERK1/2 or AKT phosphorylation in PDAC cells. The consequences of triggering the pro-tumoral effects of ROS are higher cancer cell proliferation rates and invasiveness but not in non-tumoral HPDE cells, which has no mutation in the most commonly mutated genes as PDAC cell lines though p53 is nonfunctional and there is loss of the Rb gene.47 However, when KRAS is constitutively activated in non-tumoral HPDE cells, FAM49B depletion induced an increase in proliferation and invasion.
Regulation of ‘fusion-fission’ mitochondrial dynamics is involved in several physiological and deregulated processes that are classical ‘hallmarks’ of cancer,48 such as tumor metastasis.10, 13 Other tumor suppressors that participate in mitochondrial functions have been described previously. The Fus1 gene, which modulates inflammatory responses and mitochondrial functions, such as ROS production and mitochondrial membrane potential, is downregulated, mutated or lost in most inflammatory thoracic malignancies.49 A mitochondrial outer membrane channel known as VDAC1 stabilizes HIF-1alpha by inducing ROS accumulation and favors the growth of Ras-transformed MEF, which indicates that the protein plays a tumor suppressive role.50 Another member of the ‘family with sequence similarity’ is FAM96A, which has been identified as a pro-apoptotic tumor suppressor in gastrointestinal stromal tumors and increases mitochondria-mediated apoptosis by binding apoptotic protease activating factor 1 (APAF1).51 In addition, the cyclin-dependent kinase inhibitor 1B (P27) maintains mitochondrial membrane integrity.52 These data are relevant to our findings regarding a novel tumor suppressor gene that is responsible for regulating mitochondrial dynamics and highlight the role of mitochondrial integrity and metabolism in the promotion of tumor growth and invasion.
Collectively, our findings indicate that FAM49B activity is important for preserving mitochondrial integrity and for protecting cells from oxidative stress. FAM49B is an emerging metastasis suppressor in PDAC cells, as its downregulation leads to mitochondria deregulation, increased cellular ROS levels and, consequently, genomic instability and thus benefits cancer cells. FAM49B is involved in regulating tumor metabolism and mitochondria morphology, which has not been previously described, and FAM49B expression is suppressed in PDAC tumors as a selective metabolic growth advantage.
An important finding of our study is that FAM49B gene is controlled by the tumor microenvironment, which influences tumor metastasis by downregulating FAM49B expression. ECM stiffness has been described as being critical for tumor progression, as it mechanically activates pro-tumorigenic signaling pathways and also alters the metabolism of the cancer cells.53, 54 ECM components negatively regulated FAM49B expression, which we have shown to reprogram the metabolic states of cancer cells by affecting mitochondrial dynamics, and the distribution of mitochondria as a bioenergetics source have been described to enhanced ECM-dependent invasion, migration and metastasis.13, 55 As mitochondrial dynamic changes have an impact on cancer metastatic potential and chemo-resistance,10, 11, 56 mitochondrial fission and other vulnerable metabolic functions that are controlled by FAM49B expression may be promising targets of new therapeutic approaches designed to treat PDAC.
Materials and methods
Cell culture
PDAC cell lines CFPAC1, MIAPaCa2, BxPC3, PANC1 (obtained from ECACC), T3M4, PT45, L3.6pl, HS766T, HPDE (kindly provided by Dr P Nisticò, Regina Elena National Cancer Institute, Rome, Italy), normal human ductal cells (kindly provided by Dr Lorenzo Piemonti, San Raffaele Hospital, Milan, Italy). K8484 murine PDAC cell line (Kindly provided by Dr K Olive, Columbia University, NY, USA). Cell lines were cultured in DMEM, RPMI and KSF (Lonza, Basel, Switzerland) accordingly and supplemented with 10% FBS (Lonza, Basel Switzerland) plus 50 μg/ml gentamicin (Sigma-Aldrich, St Louis, MO, USA).
Tissue microarray analysis
For tissue microarrays (TMAs) including a retrospective series of 93 formalin-fixed paraffin-embedded (FFPE) PDACs and 20 non-neoplastic controls were retrieved from the ARC-Net biobank at Verona University Hospital. All specimens were from naïve-treatment patients underwent surgery for curative intent. The materials have been collected under the Program 1885 approved by the Integrated University Hospital Trust (AOUI) Ethics Committee (Comitato Etico Azienda Ospedaliera Universitaria Integrata). Four 1-mm tissue-cores per case were analyzed. Immunohistochemical staining for FAM49B was performed as described.57 Two pathologists (AS, BR) scored each tissue core in a blinded manner. Cases were considered negative when having absent (score=0) or weak (score=1) staining, and positive when having intermediate (score=2) to strong (score=3) staining. Immunostaining was homogeneous among tissue-cores from the same case.
Murine IHC analysis
Immunohistochemical staining for FAM49B was done on 4-μm-thick FFPE Pancreatic tissues obtained from KC and KPC mouse at 3, 6, 9 and 12 months of age (n=8/age) as previously described.26 Slides were analyzed in double blinded fashion.
In vivo studies
Orthotropic injection into the pancreases of C57B16 mice (n=6) was performed with 1 × 105 K8484 cells. Thirty days later tumors were collected immediately processed or cultured them in vitro after subjected to standard enzymatic disaggregation.
For in vivo metastasis NOD-SCID IL2Rgammanull (NSG) were injected into the tail vein (i.v.) with shFAM49B or shCTRL cells 1 × 105 (in 0.1 ml PBS), After 28 days, mice were killed, necropsied, and examined for the presence of tumor masses. Mice (n=5) were used in each group. All animals were handled in accordance with European and institutional guidelines (Legislative Order No. 116/92, under protocol No 265/2015 PR) and maintained at the animal facility of the Molecular Biotechnology Center, University of Turin, Italy.
Three-dimensional culture
A total of 1.5 × 105 cells were seeded on transwell apparatuses in six-well plates containing a 1:1 ratio of cell preparation and Matrigel (Corning GF reduced) or were plated on plastic plates pre-coated with Matrigel. For the embedded 3D culture the cells were incubated at 37 °C for 20–30 min to allow the matrix to set before 1.8 ml of medium was added to the culture plates beneath the transwell apparatus. The 3D culture was then allowed to grow at 37 °C for ~7 days or 14 days, and fresh medium was added to the culture every 2 days. Imaging was performed via inverted microscopy. For RNA and protein extraction, the cells were isolated from the Matrigel by dispase (Sigma-Aldrich) digestion for ~2 h.
Transfection
Cells were transfected using calcium phosphate precipitation method and Lipofectamine 3000 (Life Technologies, Carlsbad, CA, USA), according to the manufacturer’s instructions.
Knockdown experiment
MISSION shRNAs targeting human FAM49B mRNA cloned into the pLKO.1-puro vector were purchased as glycerol stocks from Sigma-Aldrich (SHCLNG-NM_016623). The efficiency of five shRNA with respect to facilitating decreases in FAM49B mRNA levels was assessed by qRT-PCR in transiently transfected cells. The two most efficient shRNAs (TRCN0000168446 and TRCN0000168778) and control shRNA (SHC002, Sigma-Aldrich) were chosen to generate stable cell lines using lentiviral transduction as described.58
Protease protection assay
A GFP tag was added to the C terminus of the FAM49B delta stop codon to facilitate protein fusion using pEGFP plasmids. HEK293T cells were transfected with GFP-fusion FAM49B, FPP assay was performed as described.59
Quantitative RT–PCR
Total RNA was extracted using an RNeasy Mini Kit (Qiagen, Hilden, Germany), and reverse transcription was performed using 2 μg of total RNA using an iScript cDNA Synthesis Kit (BioRad, Hercules, CA, USA), according to the manufacturer’s instructions. Quantitative RT–PCR was performed using SYBR Green Dye (Life Technologies) and SsoAdvanced Universal SYBR green mix (BioRad) on a Thermal iCycler (BioRad). The PCR reactions were performed in triplicate, and the relative amounts of cDNA were calculated using the ΔΔCT method.
Protein lysate and western blot analysis
Protein lysate and western blot was performed as described.60
Migration assay
Wound-healing assay using Ibidi culture inserts (Thistle Scientific Ltd, UK) according to manufacture instructions. Images were acquired along the cell-free zone between time 0 and 24 h every 2 h. Cell migration was assessed using ImageJ software by measuring the area covered by the cells. Cell-free gap encroachment was determined by comparing the results at any time point to time 0.
Invasion assay
The cell lines, with or without FAM49B silencing were evaluated with respect to their invasion capacity using transwell Matrigel assay as described.58
MTT assay
Cells were seeded in 96-well plates at a density of 2 × 103 cells/well and synchronized using serum deprivation for 24 h cells were released using complete medium and were treated with 20 μl of MTT solution (Sigma-Aldrich; 5 mg/ml) according to manufacture instructions.
BrDu assay
Cells were seeded as MTT assay labeling for BrDu assay was performed by adding 10 μM BrDU (BD Biosciences, Franklin Lakes, NJ, USA) according to the manufacturer’s instructions. Hoechst was used to stain the nuclei. Image J was used to quantify the numbers of positive cells.
Mitochondrial membrane potential analysis
Cytofluorometric analysis of the mitochondrial membrane potential was performed on the shCNTRL and shFAM49B cell lines with 2.5 μm 5, 5′, 6, 6′-tetrachloro-1, 1′, 3, 3′-tetraethyl- imidacar-bocyanine iodide (JC-1) (Sigma Aldrich). Staining was performed in accordance with the manufacturer’s instructions, and 1 μm valinomycin (Sigma Aldrich) was used as a control.
Electron microscopy
Electron microscopy was performed on HEK293 cells expressing FAM49B GFP fusion protein. These cells were grown on Thermanox TMX coverslips (NUNC). Embedding and staining were performed as previously described.61
Proteasome Inhibitor analysis
MG132 (M7449 Sigma-Aldrich) used at 5 μm, at 6, 12, 24 and 48 h in 2D culture, For the treatment of 3D culture of PDAC cells and Normal HPDE spheroids were grown till 7 days in Matrigel and treated for 24 h with 5 μm of MG132.
Microarray analysis
RNA was obtained from three PDAC primary tumor samples and three adjacent normal pancreas (NP) tissue samples from the Surgery Department of the University of Verona, as well as from eight PDAC xenografts and one PDAC cell line (PaCa44). Microarray analysis was performed as described.62
Mitochondria staining and analysis
The mitochondrial structural network staining was done using MitoTracker Red (Invitrogen) according to manufacture’s instructions. Images were recorded with an LSM5 Pascal confocal microscope and super-resolution microscopy. The images were analyzed as previously described.63
Immunofluorescence
Cells were fixed permeabilized using Image It Kit (Life technologies) Followed with overnight incubation of primary antibodies, at dilution 1:500 and with an Alexa Fluor secondary antibody (Life Technologies) at a dilution of 1:200 for 1 h. Nuclei were stained with Hoechst (Life Technologies).
Mitochondrial fraction extraction
The mitochondrial fractions were extracted via sequential-detergent extraction with a cell fractionation kit (Mitosciences MS861, Eugene, OR, USA), according to the manufacturer’s instructions.
ROS, SOD, NADPH oxidase, ATP and PPP activity measurements
We evaluated ROS, SOD, NADPH, ATP and PPP activity levels with the shCTRL or shFAM49B transfected PDAC cell lines CFPAC1, T3M4, and HPDE, as previously described.64
Mitochondrial respiratory chain activity measurement
Complex I-III activity levels were measured using non-sonicated extracted mitochondria, as previously reported.65
Cytochrome c release
10 μg of proteins from cytosolic and mitochondrial extracts, prepared as reported above, were probed with an anti-cytochrome c antibody (mouse, clone 6H2.B4; BD Biosciences).
Caspase activity
The activity of caspase 9 and 3 was measuring by incubating 20 μg cell lysates with the respective fluorogenic substrates Ac-LEHD-7-amino-4-methylcumarine (LEHD-AMC) or DEVD-7-amino-4-methylcumarine (DEVD-AMC), as reported.66
Antibodies and primers
Antibodies used for this study are stated in (Supplementary Table 2). Primers stated in (Supplementary Table 3).
Statistical analysis
All experiments were repeated at least three times. Data are expressed as the mean±s.e.m. Statistical analysis were conducted using the two-tailed T-test or ANOVA with GraphPad Software. P values ⩽0.05 were considered statistically significant.
Acknowledgments
We thank Roberta Curto for technical support in the in vivo experiments, Elena Frola for technical support in the electron microscopy and Dr Radhika Srinivasan for critically reading the manuscript. The nature research language editing service edited this work. This work was supported by grants from the Associazione Italiana Ricerca sul Cancro (5 × mille no. 12182 to AS and FN, IG no. 15257 to FN, IG no. 15232 to AS); University of Turin-Progetti Ateneo 2014-Compagnia di San Paolo (PC-METAIMMUNOTHER to FN and PANTHER to PC), Italian Ministry of Health-Progetti Ricerca Finalizzata (RF-2013-02354892 to FN), Fondazione Ricerca Molinette Onlus, Fondazione Nadia Valsecchi to FN, European Community FP7 Grant Cam-Pac (agreement no: 602783) to AS; FIMP, Ministero Salute (CUP_J33G13000210001) to AS. MSC was supported by Fondazione Ursula e Giorgio Cytron, Torino.
Author contributions
MSC designed and performed the majority of the experiments, interpreted the results, generated the figures and tables and wrote the manuscript; CR designed and performed the metabolic functional assays and interpreted the results; MS performed the CFL and electron microscopy studies; MP, MMS, and CR performed the in vitro experiments; CC performed the immunohistochemistry on the mouse tissues; AE performed the biostatistics analysis of the PDAC patients; PA generated the HPDE overexpressing KRAS cell line; AS, RS and BR recruited PDAC patients, performed immunohistochemistry and analyzed results. PC supervised and designed the in vivo experiments, interpreted the results and wrote the manuscript; FN supervised the study, interpreted the results and wrote the manuscript.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: Cancer J Clin 2015; 65: 5–29. [DOI] [PubMed] [Google Scholar]
- Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends-an update. Cancer Epidemiol Biomarkers Prevention 2016; 25: 16–27. [DOI] [PubMed] [Google Scholar]
- Konstantinidis IT, Warshaw AL, Allen JN, Blaszkowsky LS, Castillo CF, Deshpande V et al. Pancreatic ductal adenocarcinoma: is there a survival difference for R1 resections versus locally advanced unresectable tumors? What is a ‘true’ R0 resection? Ann Surg 2013; 257: 731–736. [DOI] [PubMed] [Google Scholar]
- Krska Z, Svab J, Hoskovec D, Ulrych J. Pancreatic cancer diagnostics and treatment - current state. Prague Med Rep 2015; 116: 253–267. [DOI] [PubMed] [Google Scholar]
- Maitra A, Hruban RH. Pancreatic cancer. Ann Rev Pathol 2008; 3: 157–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishi A, Goggins M, Wood LD, Hruban RH. Pathological and molecular evaluation of pancreatic neoplasms. Semin Oncol 2015; 42: 28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Saati T, Clerc P, Hanoun N, Peuget S, Lulka H, Gigoux V et al. Oxidative stress induced by inactivation of TP53INP1 cooperates with KrasG12D to initiate and promote pancreatic carcinogenesis in the murine pancreas. Am J Pathol 2013; 182: 1996–2004. [DOI] [PubMed] [Google Scholar]
- Kodydkova J, Vavrova L, Stankova B, Macasek J, Krechler T, Zak A. Antioxidant status and oxidative stress markers in pancreatic cancer and chronic pancreatitis. Pancreas 2013; 42: 614–621. [DOI] [PubMed] [Google Scholar]
- Ogrunc M, Di Micco R, Liontos M, Bombardelli L, Mione M, Fumagalli M et al. Oncogene-induced reactive oxygen species fuel hyperproliferation and DNA damage response activation. Cell Death Differ 2014; 21: 998–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira-da-Silva A, Valacca C, Rios E, Populo H, Soares P, Sobrinho-Simoes M et al. Mitochondrial dynamics protein Drp1 is overexpressed in oncocytic thyroid tumors and regulates cancer cell migration. PLoS One 2015; 10: e0122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashatus JA, Nascimento A, Myers LJ, Sher A, Byrne FL, Hoehn KL et al. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol Cell 2015; 57: 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serasinghe MN, Wieder SY, Renault TT, Elkholi R, Asciolla JJ, Yao JL et al. Mitochondrial division is requisite to RAS-induced transformation and targeted by oncogenic MAPK pathway inhibitors. Mol Cell 2015; 57: 521–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhang J, Yu M, Xie Y, Huang Y, Wolff DW et al. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene 2013; 32: 4814–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg F, Chandel NS. Reactive oxygen species-dependent signaling regulates cancer. Cell Mol Life Sci 2009; 66: 3663–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci USA 2010; 107: 8788–8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Jhun BS, Yoon Y. High-glucose stimulation increases reactive oxygen species production through the calcium and mitogen-activated protein kinase-mediated activation of mitochondrial fission. Antiox Redox Signal 2011; 14: 425–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Robotham JL, Yoon Y. Increased production of reactive oxygen species in hyperglycemic conditions requires dynamic change of mitochondrial morphology. Proc Natl Acad Sci USA 2006; 103: 2653–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q et al. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol 1999; 1: 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eura Y, Ishihara N, Yokota S, Mihara K. Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J Biochem 2003; 134: 333–344. [DOI] [PubMed] [Google Scholar]
- Olichon A, Emorine LJ, Descoins E, Pelloquin L, Brichese L, Gas N et al. The human dynamin-related protein OPA1 is anchored to the mitochondrial inner membrane facing the inter-membrane space. FEBS Lett 2002; 523: 171–176. [DOI] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science 2012; 337: 1062–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol 2011; 13: 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold AS, Kostelecky B, Lippincott-Schwartz J. Fuse or die: Shaping mitochondrial fate during starvation. Communicat Integrat Biol 2011; 4: 752–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res 2004; 64: 985–993. [DOI] [PubMed] [Google Scholar]
- Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet 2004; 36: 40–45. [DOI] [PubMed] [Google Scholar]
- Cappello P, Rolla S, Chiarle R, Principe M, Cavallo F, Perconti G et al. Vaccination with ENO1 DNA prolongs survival of genetically engineered mice with pancreatic cancer. Gastroenterology 2013; 144: 1098–1106. [DOI] [PubMed] [Google Scholar]
- Vinci M, Gowan S, Boxall F, Patterson L, Zimmermann M, Court W et al. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol 2012; 10: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem 2007; 282: 11521–11529. [DOI] [PubMed] [Google Scholar]
- Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, Chan DC. Structural basis of mitochondrial tethering by mitofusin complexes. Science 2004; 305: 858–862. [DOI] [PubMed] [Google Scholar]
- Handy DE, Loscalzo J. Redox regulation of mitochondrial function. Antiox Redox Signal 2012; 16: 1323–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saborido A, Soblechero L, Megias A. Isolated respiring heart mitochondria release reactive oxygen species in states 4 and 3. Free Rad Res 2005; 39: 921–931. [DOI] [PubMed] [Google Scholar]
- Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 2012; 149: 656–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerkin JS, Naughton R, Quiney C, Cotter TG. Mechanisms of ROS modulated cell survival during carcinogenesis. Cancer Lett 2008; 266: 30–36. [DOI] [PubMed] [Google Scholar]
- Hancock JT. The role of redox mechanisms in cell signalling. Mol Biol 2009; 43: 162–166. [DOI] [PubMed] [Google Scholar]
- Hurd TR, DeGennaro M, Lehmann R. Redox regulation of cell migration and adhesion. Trends Cell Biol 2012; 22: 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li Y, Sarkar FH. Signaling mechanism(s) of reactive oxygen species in epithelial-mesenchymal transition reminiscent of cancer stem cells in tumor progression. Curr Stem Cell Res Ther 2010; 5: 74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016; 531: 47–52. [DOI] [PubMed] [Google Scholar]
- Birgersdotter A, Sandberg R, Ernberg I. Gene expression perturbation in vitro—a growing case for three-dimensional (3D) culture systems. Seminars in Cancer Biology 2005; 15: 405–412. [DOI] [PubMed] [Google Scholar]
- Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol 1997; 137: 231–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz P. Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat Rev Gastroenterol Rev 2017; 14: 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang S, Yao J, Lowery FJ, Zhang Q, Huang WC et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 2015; 527: 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Graves M, MacDonald G, Cipollone J, Mueller CR, Roskelley CD. Microenvironmental regulation of BRCA1 gene expression by c-Jun and Fra2 in premalignant human ovarian surface epithelial cells. Mol Cancer Res 2013; 11: 272–281. [DOI] [PubMed] [Google Scholar]
- Wai T, Langer T. Mitochondrial dynamics and metabolic regulation. Trends Endocrinol Metab 2016; 27: 105–117. [DOI] [PubMed] [Google Scholar]
- Willems PH, Rossignol R, Dieteren CE, Murphy MP, Koopman WJ. Redox homeostasis and mitochondrial dynamics. Cell Metab 2015; 22: 207–218. [DOI] [PubMed] [Google Scholar]
- Stowe DF, Camara AK. Mitochondrial reactive oxygen species production in excitable cells: modulators of mitochondrial and cell function. Antiox Redox Signal 2009; 11: 1373–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riganti C, Gazzano E, Polimeni M, Aldieri E, Ghigo D. The pentose phosphate pathway: an antioxidant defense and a crossroad in tumor cell fate. Free Rad Biol Med 2012; 53: 421–436. [DOI] [PubMed] [Google Scholar]
- Ouyang H, Mou L, Luk C, Liu N, Karaskova J, Squire J et al. Immortal human pancreatic duct epithelial cell lines with near normal genotype and phenotype. Am J Pathol 2000; 157: 1623–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Uzhachenko R, Issaeva N, Boyd K, Ivanov SV, Carbone DP, Ivanova AV. Tumour suppressor Fus1 provides a molecular link between inflammatory response and mitochondrial homeostasis. J Pathol 2012; 227: 456–469. [DOI] [PubMed] [Google Scholar]
- Brahimi-Horn MC, Giuliano S, Saland E, Lacas-Gervais S, Sheiko T, Pelletier J et al. Knockout of Vdac1 activates hypoxia-inducible factor through reactive oxygen species generation and induces tumor growth by promoting metabolic reprogramming and inflammation. Cancer Metab 2015; 3: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwamb B, Pick R, Fernandez SB, Volp K, Heering J, Dotsch V et al. FAM96A is a novel pro-apoptotic tumor suppressor in gastrointestinal stromal tumors. Int J Cancer 2015; 137: 1318–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Hou D, Luo Z, Chen P, Lv B, Wu L et al. The novel protective role of P27 in MLN4924-treated gastric cancer cells. Cell Death & Disease 2015; 6: e1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung JC, Barnes JM, Desai SR, Sistrunk C, Conklin MW, Schedin P et al. Tumor mechanics and metabolic dysfunction. Free Rad Biol Med 2015; 79: 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013; 19: 1423–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caino MC, Seo JH, Aguinaldo A, Wait E, Bryant KG, Kossenkov AV et al. A neuronal network of mitochondrial dynamics regulates metastasis. Nat Commun 2016; 7: 13730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Norris KL, Cleland MM, Jeong SY, Youle RJ. Role of Bax and Bak in mitochondrial morphogenesis. Nature 2006; 443: 658–662. [DOI] [PubMed] [Google Scholar]
- Kaistha BP, Honstein T, Muller V, Bielak S, Sauer M, Kreider R et al. Key role of dual specificity kinase TTK in proliferation and survival of pancreatic cancer cells. Br J Cancer 2014; 111: 1780–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Principe M, Ceruti P, Shih NY, Chattaragada MS, Rolla S, Conti L et al. Targeting of surface alpha-enolase inhibits the invasiveness of pancreatic cancer cells. Oncotarget 2015; 6: 11098–11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz H, Hailey DW, Lippincott-Schwartz J. Fluorescence protease protection of GFP chimeras to reveal protein topology and subcellular localization. Nat Methods 2006; 3: 205–210. [DOI] [PubMed] [Google Scholar]
- Tomaino B, Cappello P, Capello M, Fredolini C, Ponzetto A, Novarino A et al. Autoantibody signature in human ductal pancreatic adenocarcinoma. J Proteome Res 2007; 6: 4025–4031. [DOI] [PubMed] [Google Scholar]
- Sassoe-Pognetto M, Ottersen OP. Organization of ionotropic glutamate receptors at dendrodendritic synapses in the rat olfactory bulb. J Neurosci 2000; 20: 2192–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello P, Tomaino B, Chiarle R, Ceruti P, Novarino A, Castagnoli C et al. An integrated humoral and cellular response is elicited in pancreatic cancer by alpha-enolase, a novel pancreatic ductal adenocarcinoma-associated antigen. Int J Cancer 2009; 125: 639–648. [DOI] [PubMed] [Google Scholar]
- Viltono L, Patrizi A, Fritschy JM, Sassoe-Pognetto M. Synaptogenesis in the cerebellar cortex: differential regulation of gephyrin and GABAA receptors at somatic and dendritic synapses of Purkinje cells. J Comp Neurol 2008; 508: 579–591. [DOI] [PubMed] [Google Scholar]
- Riganti C, Costamagna C, Doublier S, Miraglia E, Polimeni M, Bosia A et al. The NADPH oxidase inhibitor apocynin induces nitric oxide synthesis via oxidative stress. Toxicol Appl Pharmacol 2008; 228: 277–285. [DOI] [PubMed] [Google Scholar]
- Campia I, Lussiana C, Pescarmona G, Ghigo D, Bosia A, Riganti C. Geranylgeraniol prevents the cytotoxic effects of mevastatin in THP-1 cells, without decreasing the beneficial effects on cholesterol synthesis. Br J Pharmacol 2009; 158: 1777–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzon-Daza M, Garzon R, Couraud P, Romero I, Weksler B, Ghigo D et al. The association of statins plus LDL receptor-targeted liposome-encapsulated doxorubicin increases in vitro drug delivery across blood-brain barrier cells. Br J Pharmacol 2012; 167: 1431–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.