Abstract
The identification of specific genetic alterations as key oncogenic drivers and the development of targeted therapies are together transforming clinical oncology and creating a pressing need for increased breadth and throughput of clinical genotyping. Next-generation sequencing assays allow the efficient and unbiased detection of clinically actionable mutations. To enable precision oncology in patients with solid tumors, we developed Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT), a hybridization capture-based next-generation sequencing assay for targeted deep sequencing of all exons and selected introns of 341 key cancer genes in formalin-fixed, paraffin-embedded tumors. Barcoded libraries from patient-matched tumor and normal samples were captured, sequenced, and subjected to a custom analysis pipeline to identify somatic mutations. Sensitivity, specificity, reproducibility of MSK-IMPACT were assessed through extensive analytical validation. We tested 284 tumor samples with previously known point mutations and insertions/deletions in 47 exons of 19 cancer genes. All known variants were accurately detected, and there was high reproducibility of inter- and intrarun replicates. The detection limit for low-frequency variants was approximately 2% for hotspot mutations and 5% for nonhotspot mutations. Copy number alterations and structural rearrangements were also reliably detected. MSK-IMPACT profiles oncogenic DNA alterations in clinical solid tumor samples with high accuracy and sensitivity. Paired analysis of tumors and patient-matched normal samples enables unambiguous detection of somatic mutations to guide treatment decisions.
The identification of driver genetic alterations in key oncogenes and tumor suppressor genes plays an essential role in the diagnosis and treatment of many cancers. These mutations cause dysregulation of signaling pathways and cellular processes controlling proliferation, migration, metabolism, and apoptosis. In recent years, several targeted therapies have been approved that specifically inhibit and abrogate the tumorigenic effects of the aberrant proteins generated by these oncogenic mutations.1, 2, 3, 4, 5, 6, 7 It has thus become crucial to develop accurate, sensitive, and high-throughput genomic assays to accommodate the increasingly genotype-based therapeutic approaches.8 Massively parallel next-generation sequencing (NGS) technology fulfills this need because it enables the unbiased identification of mutations across the genome or across more targeted regions with high sensitivity and specificity. NGS assays can be developed to target the genome at various scales (whole genome, whole exome, targeted panels) and are a key component toward realizing effective stratified oncology. Panel-based targeted sequencing of selected cancer genes or mutational hotspots is a popular approach, and a number of groups have performed studies examining the sensitivity, specificity, precision, and reproducibility of their panel-based tests before routine use in a clinical setting.9, 10, 11, 12, 13, 14, 15, 16, 17, 18 These targeted NGS assays can generally be classified on the basis of the target enrichment method used (eg, amplicon PCR versus hybridization capture), sequencing chemistry, and scale of the sequencing platform.
There is considerable variability across targeted NGS panels implemented in different clinical laboratories in terms of the number and identities of genes tested, disease indication (solid tumors versus hematological malignancies), and sample throughput. However, the development of a custom NGS test requires significant operational and bioinformatics infrastructure investment that may not be feasible in many clinical labs. As a result, some groups have chosen to validate ready-made vendor solutions [Ion Torrent AmpliSeq Cancer Hotspot Panel9, 14, 19 (Life Technologies, Carlsbad, CA), Illumina TruSeq Amplicon Cancer Panel10 (Illumina, San Diego, CA)] as an expedient path toward implementing NGS profiling in the laboratory. These vendor solutions are generally amplicon PCR-based and target selected mutation hotspots in 1 to 50 cancer genes (not all exons of these genes are fully covered in most instances). The Ion Torrent AmpliSeq and Illumina TruSeq Amplicon solutions are designed for the PGM and MiSeq benchtop sequencers, and come with a generic analysis workflow installed (TorrentSuite or MiSeqReporter), which provides support for the detection of single nucleotide variants (SNVs) and short insertions/deletions (indels), and limited downstream variant annotation.
Some labs have developed tests using the vendor solutions as a starting point for customization, eg, Nikiforova et al20 validated a custom-designed AmpliSeq panel (ThyroSeq), targeting 284 hotspots across 12 thyroid cancer relevant genes. The composition of the gene panel was customized in this instance, but the assay leverages the same benchtop sequencing and analysis infrastructure as the AmpliSeq Cancer Hotspot Panel. Similarly, Luthra et al10 modified the TruSeq Amplicon Cancer Panel content for mutation screening in leukemias by spiking in primers for other genes relevant to acute myeloid leukemia and chronic lymphocytic leukemia. Taking the degree of customization a step further, in an earlier study, our group validated a 28-gene amplicon panel for the detection of mutations in myeloid malignancies, where both the gene list and analysis pipeline were custom designed and developed.21
Amplicon-based methods described above may be convenient for smaller gene panels but are susceptible to imbalanced sequence coverage across targets and to artifacts such as random sequence mismatches introduced by polymerase errors. Furthermore, because the coordinates of the amplicons are fixed and invariant, current methods do not provide accurate estimates of the number of unique input DNA molecules being sequenced as a consequence of PCR inflated coverage. Targeted enrichment using hybridization capture overcomes these shortcomings by providing a better estimate of unique coverage. The efficiency of fragment pull down is determined by the degree of hybridization to designed probes, and fragments are not constrained by start and stop positions. PCR is also used in hybridization capture workflows, but the primers used are specific to the ligated adapter sequences as opposed to the targeted regions themselves, allowing for disambiguation and removal of PCR duplicates. More genes can be incorporated into a hybridization panel design without risk of problematic primer–primer interactions, a problem frequently encountered in amplicon PCR approaches. Hybridization capture may also be more suitable for highly degraded formalin-fixed, paraffin-embedded (FFPE) DNA, because affinity-based pull down only requires partial overlap of capture probes with target regions, whereas amplicon PCR will fail to amplify DNA fragments where one or both of the primer binding sites are degraded. As a consequence, hybridization capture-based panels can provide more accurate estimates of copy number variation and can be used for detection of selected structural rearrangements. Several groups have developed clinical hybridization-capture assays using custom-designed biotinylated oligonucleotides, targeting a number of genes at varying scales (WuCAMP: 25 genes,12 UW-Oncoplex: 194 genes,13 Foundation Medicine: 287 genes17).
Here, we describe the analytical validation of a custom hybridization-capture based assay, Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) as a clinical test for interrogating somatic alterations in 341 oncogenes and tumor suppressors in FFPE tumor specimens.22
Custom biotinylated DNA probes (baits) were designed for targeted sequencing of all exons and selected introns of these 341 genes using the NimbleGen SeqCap system (Roche NimbleGen, Madison, WI). Baits were empirically redesigned through many iterations to achieve extremely high uniformity of coverage across targets. Captured libraries are sequenced on an Illumina HiSeq 2500 in rapid-run mode, and the sequencing output is processed using a custom analysis pipeline to detect SNVs, short indels (<30 bp), copy number aberrations and structural rearrangements. MSK-IMPACT is designed to analyze tumor-matched normal pairs for the detection of somatic alterations.
As part of the validation, we assessed the sensitivity of MSK-IMPACT on a validation set of 284 unique tumor samples with known SNVs and indels previously confirmed by independent methods. Intra- and interrun replicates were used to assess the precision and reproducibility of variant calling. Further, we demonstrated the ability of MSK-IMPACT to successfully detect known copy number aberrations and structural rearrangements in an independent set of selected samples. On the basis of the results of the validation, we have deployed MSK-IMPACT as a clinical test and are running it in real time to prospectively identify mutations in patient tumors. The breadth and sensitivity of MSK-IMPACT allow us to comprehensively profile actionable and other driver mutations in patients with advanced cancer to guide treatment decisions and match patients to the most appropriate clinical trials.
Materials and Methods
Panel Design and Capture Protocol
Custom DNA probes were designed for targeted sequencing of all exons and selected introns of 341 oncogenes, tumor suppressor genes, and members of pathways deemed actionable by targeted therapies. This included 4976 coding exons of canonical transcript isoforms, 104 exons of noncanonical transcripts, as well as probes targeting 33 introns of 14 recurrently rearranged genes. In addition, the panel contains probes that capture a 100-bp region in the TERT promoter where two recurrently mutated positions create de novo ETS transcription factor binding sites in melanoma,23 thyroid,24 gliomas,25 and other cancers.26 The panel contains probes that tile the positions of 1042 common single nucleotide polymorphisms (SNPs), which serve three purposes: i) the unique combination of genotypes at these polymorphic sites serve as a patient-specific fingerprint that can be cross-checked against another patient-matched sample (ie, blood normal) for concordance in cases where sample mix-up is suspected; ii) trace amounts of contaminating DNA can be detected through the presence of alternate alleles at homozygous sites; and iii) these probes mimic a low-density SNP tiling array with locations evenly distributed across the genome-coverage values at these positions are used to supplement the copy number analysis in genomic regions where few targeted genes are located (Supplemental Table S1).
Genomic DNA from tumor- and patient-matched normal samples were extracted using the Qiagen DNeasy Tissue kit and the EZ1 Advanced XL system (Qiagen, Valencia, CA), respectively. Genomic DNA was sheared using the Covaris E200 instrument (Covaris, Woburn, MA). Sequencing libraries were prepared using the KAPA HTP protocol (Kapa Biosystems, Wilmington, MA) and the Biomek FX system (Beckman Coulter, Brea, CA) through several enzymatic steps, including end repair, A-base addition, ligation of Illumina sequence adaptors followed by PCR amplification, and clean-up. Forty-eight adaptors containing 6-mer unique barcodes corresponding to the Illumina TruSeq barcoding system were used in the library preparation. Tumor and normal libraries were pooled at a 3:1 ratio. A mix of blocker oligos synthesized at IDT (Integrated DNA Technologies, Coralville, IA) and compatible with the TruSeq barcodes was also added to the pool to help prevent nonspecific binding and improve target capture specificity. Custom DNA probes targeting exons and selected introns of 341 genes were synthesized using the NimbleGen SeqCap EZ library custom oligo system and were biotinylated to allow for sequence enrichment by capture using streptavidin-conjugated beads. Pooled libraries containing captured DNA fragments were subsequently sequenced on the Illumina HiSeq 2500 system (rapid-run mode) as 2 × 100-bp paired-end reads.
Demultiplexing and Read Alignment
BCL2FASTQ version 1.8.3 (Illumina) was used to demultiplex the base calls into individual FASTQ files using the following options: --force --no-eamss --fastq-cluster-count 0 --mismatches 1. Reads for which a matching index could not be identified were stored in a set of FASTQ files labeled Undetermined Indices. To monitor possible barcode contamination, a check was also performed to determine whether any known barcode indices were over-represented within the undetermined indices.
Vestigial adapter sequences were removed from the 3′ end of sequence reads before alignment using adaptor-trimming tools (TrimGalore and CutAdapt software version 0.2.5, http://www.bioinformatics.babraham.ac.uk/projects/trim_galore, last accessed October 1, 2013). Adaptor trimming was performed using the following options: 10% error and minimum of 3-bp match. Read pairs with insert size length <25 bp were discarded. Reads were aligned in paired-end mode to the hg19 b37 version of the human genome using BWA-MEM (Burrows-Wheeler Aligner software version 0.7.5a, http://arxiv.org/abs/1303.3997, last accessed October 1, 2013). Aligned reads were written to a Sequence Alignment/Map file, which was converted into Binary Alignment/Map format using tools available in Picard (http://broadinstitute.github.io/picard, last accessed November 20, 2014). PCR duplicates were marked for exclusion in subsequent analysis using the MarkDuplicates tool in Picard. IndelRealigner (GATK software version 2.7.227) was used to perform a local multiple sequence realignment of reads in regions where indels were present; samples from the same patient (eg, tumor and normal) were jointly realigned. BaseRecalibrator (GATK27) was used to adjust the reported quality scores based on the following covariates: read group, reported quality score, cycle, and local sequence context. Recalibrated quality scores were subjected to a base quality threshold of 20, corresponding to a 1/100 chance of error. Both IndelRealigner and BaseRecalibrator steps were performed on intervals corresponding to the targeted regions only.
Sample Quality Control Checks
The baits used for hybridization capture included custom intergenic and intronic probes targeting 1042 regions throughout the genome centered on common SNPs. The unique combination of SNPs specific to a given sample serves as a fingerprint for the identity of the corresponding patient. Genotype analysis of these fingerprint SNPs was used to identify potential sample and/or barcode mix-ups. The presence of reads bearing the alternate allele at sites where the patient is homozygous was used as an indicator of contamination involving DNA from a different individual or contamination among different barcoded adapters. Samples were flagged if the average minor allele frequency at homozygous sites was observed to be >1%. (Patient-matched normal samples, when available, were used to define which sites were homozygous.) Further, samples for which >55% of fingerprint sites were heterozygous were also eliminated, suggesting large-scale contamination of DNA from another individual.
Although these analyses enabled detection of mix-ups or contamination involving DNA from different patients, they did not address the issue of admixture of tumor and normal DNA from the same patient. Somatic mutations in a tumor sample can be missed if tumor and normal samples are mislabeled, or if a normal sample contains appreciable amounts of tumor DNA. To detect the potential presence of tumor DNA in normal samples, we subjected normal samples to genotyping for 299 known hotspot mutations from COSMIC version 6428 (variants with >5 mentions in COSMIC version 64) (Supplemental Table S2) to detect presence of mutant alleles. The sensitivity of this genotyping check was assessed in silico by deliberately spiking increasing numbers of tumor sample reads into their respective matched normal samples (Supplemental Table S3). The results of the in silico analysis showed that tumor contamination in its matched normal sample could be detected down to approximately 250,000 reads for a tumor with a moderate- or high-frequency hotspot mutation, or a million reads for a tumor with a low-frequency hotspot mutation. As an additional check for tumor contamination in normal samples, we compared sequenced normal samples against a set of diploid control normal samples to identify samples with abnormal copy number profiles. This included a previously run pool of standard normal FFPE controls, as well as a mixed normal control sample included in the same run (equal parts mixture of DNA from 10 normal FFPE samples). Normal samples with copy number gains and/or losses relative to the control normal samples were flagged as possibly contaminated with tumor DNA, and may be excluded from further analysis pending review.
SNVs and Indels in Variant Calling
We performed paired-sample variant calling on tumor samples and their respective matched normal samples to identify point mutations/SNVs and small indels (<30 bp in length). In instances where a matched normal sample was unavailable, or where the matched normal sample was sequenced with low coverage (<50×), tumor samples were considered as unmatched samples, and variant calling was performed using a within-batch mixed normal control sample instead. MuTect29 (version 1.1.4) was used for SNV calling, and SomaticIndelDetector,27 a tool in GATK version 2.3.9, was used for detecting indel events. The following standard filters were applied to the raw MuTect and SomaticIndelDetector output as a first pass (with more rigorous filters being applied at a subsequent stage): variant frequency in tumor/variant frequency in normal >5×, Number of mutant allele reads in tumor sample >5, variant frequency in tumor sample >1%. Variants were annotated using Annovar30 (version 527), and the output was reformatted using a custom script to ensure annotations of the cDNA and protein primary sequence changes are compliant with HGVS31 standards. Dinucleotide and trinucleotide substitutions identified by the pipeline were annotated manually because this functionality was not supported by the version of Annovar used. Only variant annotations relative to the canonical transcript for each gene (derived from a list of known canonical transcripts obtained from the UCSC Genome Browser32) were reported. In cases where variant calling was performed using an unmatched normal sample, variants with minor allele frequency >1% in the 1000 Genomes cohort33 were also removed because they were more likely to be common population polymorphisms than somatic mutations.
Filtering for High-Confidence SNVs and Indels
Annotated SNV and indel calls were subjected to a series of filtering steps to ensure that only high-confidence calls were admitted to the final step of manual review. Variant calls were filtered on the basis of the following listed parameters: i) evidence in literature for being an oncogenic or recurrent hotspot mutation; ii) occurrence of variant in previously run pool of normal controls (ie, reproducible assay artifacts); iii) technical characteristics of the variant call: coverage depth, number of mutant reads supporting the variant, and variant frequency; and iv) annotation-based filters: location (eg, exonic versus non-exonic) and effect (eg, nonsynonymous versus silent).
Evidence in Literature for Hotspot Mutations
Prior knowledge from the literature was incorporated in the analysis through a two-tiered variant filtering scheme: variants corresponding to known hotspot mutations with extensive supporting evidence in the literature were considered first-tier events. These variants were subjected to lower requirements on coverage, number of mutant reads, and variant frequency to be considered as high-confidence calls. Conversely, we required higher levels of evidence to consider novel variants with no supporting evidence in the literature as high-confidence calls. First-tier mutations included the following: i) hotspot SNVs reported in COSMIC version 6428; ii) mutation hotspots reported in The Cancer Genome Atlas (TCGA) project34; and iii) indels in selected exons of established oncogenes (ie, KIT exons 9 and 11, ERBB2 exon 20, EGFR exons 19 and 20). Variants listed in COSMIC version 64 were considered hotspot point mutations if they presented with ≥5 mentions and occurred in exons of the 341 genes targeted by MSK-IMPACT (Supplemental Table S4).
Occurrence of Variant in Pool of Standard Normal Controls
Some variants in the raw pipeline output may correspond to sequencing, hybridization-capture, or sequence alignment–related artifacts. These artifacts usually present with low variant frequencies, and can be distinguished from true positive mutations by their high levels of recurrence in unrelated samples, regardless of sample status (tumor or normal). To flag variant calls as possible artifacts, we profiled 10 normal FFPE control samples in duplicate with MSK-IMPACT to establish a standard normal database. Binary Alignment/Map files from these 20 standard normal samples were genotyped for variants reported by the pipeline on an incoming tumor sample: if a variant was detected (ie, ≥3 mutant reads and >1% variant frequency) in >20% of the standard normal samples, it was considered a likely artifact and was removed.
Filtering Based on Technical Characteristics of the Variant Call
Based on an empirical analysis of false-positive calls generated by comparing replicates of normal FFPE samples against each other (see Results), we determined thresholds on coverage depth, number of mutant reads, and variant frequency for rejecting almost all false-positive calls. First-tier variants (ie, well-characterized hotspot mutations) were considered in a separate class from novel second-tier variants. First-tier variants were filtered using the following criteria: coverage depth ≥20×, mutant reads ≥8, and variant frequency ≥2%, compared to second-tier variants: coverage depth ≥20×, mutant reads ≥10, and variant frequency ≥5%.
Filtering Based on Location and Effect of Variants
Because the objective of MSK-IMPACT is to identify clinically actionable mutations, variants with clear effects on protein function or transcription were prioritized for manual review. As such, non-exonic variants passing all previous criteria were redirected to a separate output file (intronic, untranslated region, intergenic, upstream) with the exception of TERT promoter variants that created new binding sites for ETS transcription factors.23 Synonymous (ie, silent) exonic variants were similarly redirected. Only calls that resulted in changes to the protein primary sequence (ie, nonsynonymous: missense and nonsense, splice site, frameshift indel, in-frame indel) were retained and sent to the final output file for manual review using the Integrated Genomics Viewer software version 2.3.36.35 Because functional mutations may exist among the unreported synonymous and non-exonic variants, these variants are not discarded but are stored in a database for retrospective research.
Copy Number Variant and Structural Variant Calling
Copy number aberrations were identified by comparing sequence coverage of targeted regions in a tumor sample relative to a standard diploid normal sample. Specifically, coverage of targeted regions (exonic and fingerprint regions) was computed using the GATK DepthOfCoverage27 tool, square-root transformed, and subsequently adjusted for GC content using a Loess normalization procedure. A set of normal FFPE and blood control samples is used for reference diploid genome comparison. Target regions in the lowest fifth percentile of coverage in ≥20% of all normal control samples were removed from analysis. Normalized coverage values from tumor samples were divided by corresponding values in normal samples, and log-transformed to yield log-ratios. A single tumor sample was compared against multiple normal sampless from the set of control normal samples to obtain different sets of log-ratio values.
The sum-squared log-ratio was computed for each normal sample compared against, and was used as a measure of signal to noise. The normal sample yielding the lowest sum-squared log-ratio was selected as the best comparator normal for the tumor sample analyzed. Log-ratio coverage values were subsequently segmented by circular binary segmentation,36 and segmented values were grouped into clusters using a separation threshold of 0.1 and a minimum cluster membership threshold of three segments. Target regions belonging to the segment cluster with mean segmented log-ratio closest to 0 were used to parameterize a null distribution for estimating significance of whole gene copy number events.
The following criteria were used to determine significance of whole-gene gain or loss events: fold change >2.0 (gain) or <−2.0 (loss), P < 0.05 (false discovery rate–corrected for multiple testing). Of note, the comparator normal yielding the lowest sum-squared log-ratio may not always be the matched normal for a given tumor sample. Matched normal samples are subjected to the same copy number variant calling algorithm, using the same set of normal FFPE and blood control samples, with modified thresholds for detecting germline events: fold change >1.3× (single copy gain, ideally 1.5×) or <−1.8× (single copy loss, ideally −2.0×), P < 0.05 (false discovery rate–corrected). The resulting germline calls are subtracted from the total set of copy number calls made on the tumor sample, to ensure the final set of copy number variants from the tumor sample are confirmed to be somatic.
DELLY37 version 0.3.3 was used to detect somatic structural variants from tumor and matched normal read-pair data. DELLY requires paired-read and split-read support to nominate rearrangement breakpoints; using a paired-sample calling procedure; each structural aberration detected in the tumor sample is evaluated in the comparator normal sample as well. This increases specificity by eliminating germline structural aberrations as well as false-positive events, such as systematic sequencing/mapping artifacts. All candidate somatic structural aberrations were filtered, annotated using in-house tools, and manually reviewed using the Integrative Genomics Viewer.35 Similar to SNVs and indels, known rearrangements with strong literature support are subjected to less stringent filtering criteria (ie, three paired or split reads, mapping quality ≥5, length >500 bp) compared to novel rearrangements (ie, five paired or split reads, mapping quality ≥20, length >500 bp).
Statistical Determination of Coverage Requirements
We assumed that alleles reported by reads at a given genomic position can be modeled by a Bernoulli random process, where mutant alleles are detected with P, corresponding to the true variant frequency of the variant. Assuming each read is an independent sample, the total number of reads supporting the mutant allele at a given chromosomal position should follow a Binomial distribution parameterized by P. The requirement to detect mutations with a quality score Q20 or greater implies a sequencing error rate of 1% or less. We thus assumed there is a 1% background probability that mismatches to the reference genome can occur due to random sequencing error. Using a cloglog parameterization of a binomial distribution, we performed a power analysis to compute the expected sample size (ie, coverage or total number of reads) needed to detect a mutation with true underlying variant frequency 2% or greater, for varying levels of power (0.8 to 0.99), assuming a fixed α (Type I error rate) of 0.05 (Supplemental Figure S1A and Supplemental Table S5). We also computed a 95% confidence interval representing the range of observable variant frequency values for a mutation with true underlying variant frequency of 2% or greater. Expectedly, the variability of observed variant frequencies decreased with increasing coverage. Coverage appeared to have the greatest effect on the variability of observed variant frequencies at lower coverage values (ie, <500×), but had minimal impact when coverage exceeded 500× (Supplemental Figure S1B).
The power analysis above enabled us to set a theoretical lower limit for coverage. From Supplemental Table S5, with 100× coverage, we were able to detect a mutation with true underlying variant frequency of 10% or greater, with 98% confidence (power) at an α of 0.05. This implies that if an exon is sequenced with coverage exceeding 100×, but no mutation is called, we can be confident that this exon does not contain any mutations with true underlying variant frequency of 10% or greater. In other words, we require exons to be covered to at least 100× to be sufficiently powered to call a negative result.
However, a mutation with true underlying variant frequency of 10% will not be detected at exactly 10% every time. Using a 95% confidence interval, we estimated the range of observed frequencies values for a 10% variant to be between 5.0% and 17.6% when the overall coverage is 100× (Supplemental Table S6). That is, by using a threshold of 5.0%, corresponding to the lower limit of possible observed variant frequencies, our assay will retain most variants of true underlying variant frequency of 10% or greater, which is powered to detect at 100× coverage.
Empirical Determination of Coverage Requirements
To confirm whether the results of our theoretical power calculations match what is observed in practice, we performed an experiment in which DNA from 10 normal (diploid) FFPE samples from unrelated individuals was mixed in equimolar parts and subjected to targeted resequencing using our assay. We expected each of the 10 individuals assayed to be either heterozygous (50% variant frequency) or homozygous (100% positive for reference or alternate allele) for a set of common SNPs, and by mixing them in equal parts, we created a pool in which the expected variant frequencies for these SNPs were known, and ranged from 5% to 100%. The objective of the experiment was to empirically measure the range of observed variant frequencies at these SNP locations, and compare them to their respective expected values.
Eight hundred and sixty-two common SNPs with minor allele frequency >1% in the 1000 Genomes cohort were considered for this experiment, and Supplemental Table S7 lists the breakdown of SNPs by expected variant frequency, as well as the range of observed variant frequencies obtained from genotyping. Supplemental Figure S2 shows the range of observed variant frequencies as a boxplot. The mean coverage of this sample was 480×. Of note, the observed variant frequencies range from 5.0% to 13.9% for a SNP with true underlying variant frequency of 10%. This range in values was roughly consistent with what we would expect from our power calculations for a coverage depth of 500× (7.5% to 13.0%), suggesting that the assumptions that our power calculations were based on are largely valid. Additionally, these values provide empirical support for using a 5% lower limit for detecting variant frequencies with true underlying frequency of 10%.
Results
Panel Design and Target Region Coverage
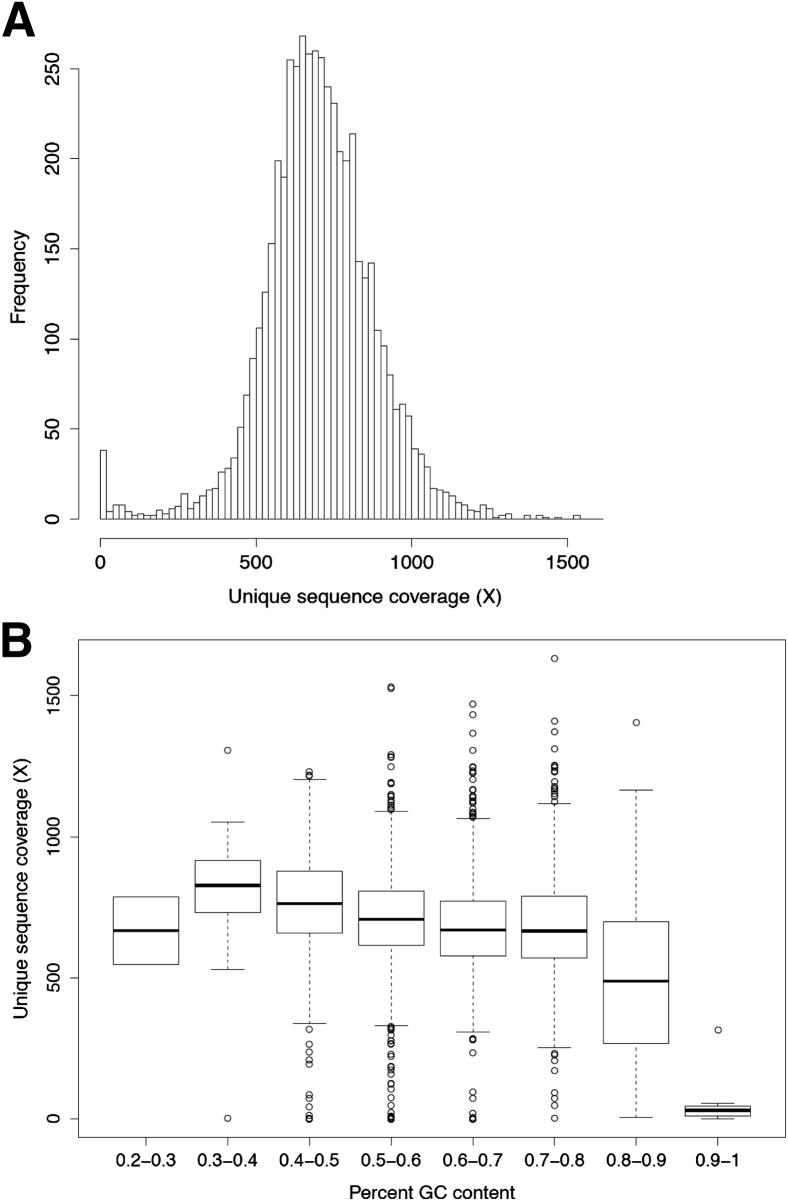
We designed a custom hybridization capture panel (MSK-IMPACT) for the targeted sequencing of all exons of 341 cancer relevant genes. The assay targets 4976 exons corresponding to canonical transcripts of the 341 genes, 104 exons of noncanonical transcripts, as well as 33 introns corresponding to sites of common oncogenic rearrangement events (eg, EML4-ALK, CD74-ROS1). A survey of coding mutations with at least five mentions in COSMIC version 64 showed that our assay was capable of interrogating 1245 known variants in the targeted exons (Supplemental Table S8). Ten normal (diploid) FFPE samples were profiled in duplicate using this assay (total = 20 replicates) and were multiplexed in a single Illumina HiSeq 2500 rapid run to generate summary statistics for coverage across all targeted exons (Supplemental Table S9). (The normal samples in this set were also used as reference diploid samples for copy number analysis, and for confirming recurrently called variants as systematic assay artifacts.) The mean unique coverage across all targeted exons for the normal samples was 700× (SD = ±182×) (Figure 1A). Ninety-seven percent of canonical exons were sequenced to at least half of the average sample coverage (350×). Forty-two exons presented with consistently low coverage (<5% of sample coverage), because of two reasons: 31 exons had poor mapping quality as a result of sequence similarity with other loci in the genome, and 11 exons had high GC content (average = 80%) and thus were challenging for library amplification and sequencing (Supplemental Table S9). These poorly covered regions did not contain any COSMIC variants with more than five mentions, so their lack of coverage should have minimal impact on the ability of the assay to interrogate known mutations. The mean unique coverage, excluding the 42 exons with persistent low coverage, was 706× (SD = ±171×). Although lower coverage was observed for regions with high GC content (ie, above 70%), we observed consistent, uniform coverage across regions with GC content between 20% and 70% GC (Figure 1B).
Figure 1.
Uniformity of sequence coverage. A: Distribution of mean coverage values across all canonical exons in 341 genes targeted by MSK-IMPACT. Coverage values were computed using a panel of 10 diploid, normal formalin-fixed, paraffin-embedded samples, each run in duplicate. B: Coverage for canonical exons binned by percent GC content.
Optimization of Mutation Calling Filters
The assay uses a paired-sample analysis pipeline (tumor versus matched normal) to identify somatic variants in the targeted exons and is designed to detect SNVs as well as small indels <30 bp in length. In cases where a patient-matched normal sample is unavailable, or is sequenced with low coverage (<50×), variant calling is performed against an unmatched, pooled normal control instead. Based on a statistical power analysis and empirical confirmation (see Materials and Methods), we estimate that 100× coverage is needed to detect mutations with true variant frequencies ≥10% with 98% power (α = 0.05). As such, we require samples to be sequenced to at least 200× mean unique coverage, which is the minimum sample coverage threshold we determined to be necessary for ≥98% of targeted exons to be covered at ≥100×. (In practice, we typically sequence to a depth of 500× to 1000× mean unique coverage.) The results of the power analysis imply that regions with <100× coverage are at increased risk of false negatives; some fraction of variants with underlying frequency at or below 10% may be missed because coverage is insufficient to adequately power their detection. Conversely, for variants that are observed at low allele frequencies or in regions of low coverage, precautions are taken to discriminate between real mutations and false-positive calls.
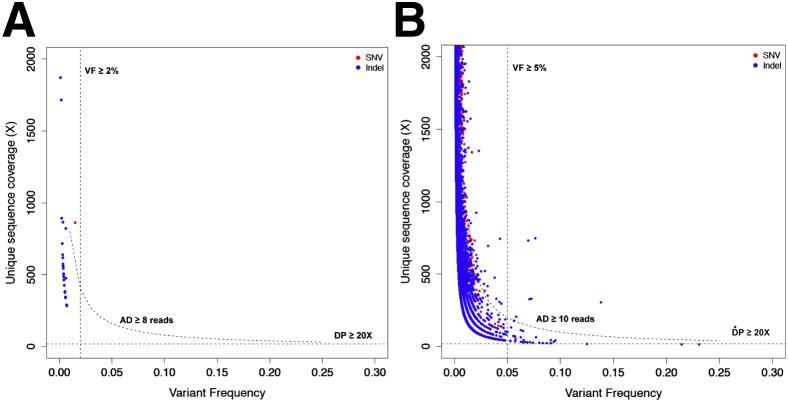
To determine thresholds on coverage, alternate allele depth, and variant frequency for rejecting potential false-positive calls, we performed variant calling on the set of 10 normal FFPE samples assayed in duplicate, where for each sample, variants calls were made by comparing normal replicate 1 (tumor) versus normal replicate 2 (normal), or vice versa. All variants identified through this process would necessarily be false positives, and by reviewing their distributions of coverage, mutant allele depth, and variant frequency values, we would be able to identify suitable thresholds to reject these false-positive calls. Permissive filters for MuTect and SomaticIndelDetector were deliberately used to select the most appropriate thresholds at this step: variant frequency in the tumor sample ≥1%, ratio of variant frequency in tumor versus normal ≥5. We restricted our false-positive analysis to exonic variants with clear effects on protein function (ie, nonsynonymous SNVs, frameshift, and in-frame indels). In addition, indel variants present in >20% of samples from a set of normal FFPE controls were regarded as artifacts and removed from analysis. The remaining false-positive variants were further classified into two groups: those occurring in locations corresponding to first-tier hotspot mutations (n = 28) and those occurring outside those locations as second-tier (n = 41,220). The following filtering thresholds were able to remove all but nine false-positive second-tier events (eight indels, one SNV remaining) for a 99.9% false-positive rejection rate: Coverage depth ≥20×, number of mutant reads ≥10, and variant frequency ≥5% (Figure 2B).
Figure 2.
Selection of variant calling filters. Coverage and variant frequency values are plotted for false-positive variant calls generated by comparing experimental replicates of normal formalin-fixed, paraffin-embedded samples against each other. A: First-tier false-positive calls occurring in hotspot regions. B: Second-tier false-positive calls occurring outside hotspot regions. Dotted lines indicate decision boundaries for rejecting false positives based on thresholds on coverage (DP ≥20×), number of mutant reads (AD ≥8 reads for first-tier events, AD ≥10 reads for second-tier events), and variant frequency (VF ≥2% for first-tier events, VF ≥5% for second-tier events). AD, number of mutant reads; DP, coverage depth; Indel, insertion/deletion; SNV, single nucleotide variation; VF, variant frequency.
First-tier events were subjected to more lenient criteria for filtering (coverage depth ≥20×, mutant reads ≥8, and variant frequency ≥2%), which did not compromise specificity despite being less restrictive: all first-tier false-positive events were rejected using this criteria (Supplemental Table S10 and Figure 2A). In summary, the power analysis established that 100× coverage is needed for a target region to avoid false negatives (ie, missing true variants with at least 10% variant frequency). It should be noted that a mutation might still be detected in a region with coverage <100×. Further, the normal replicate versus normal replicate variant calling analysis is a controlled experiment meant to generate false positives, and the thresholds on coverage, number of mutant reads, and variant frequency identified through this analysis serve as guidelines subject to reinterpretation in clinical practice, depending on aspects of input specimen quality, eg, in cases with low percentage tumor content or in a subclonal specimen with substantial heterogeneity.
Clinical Validation in Samples with Known SNVs and Indels
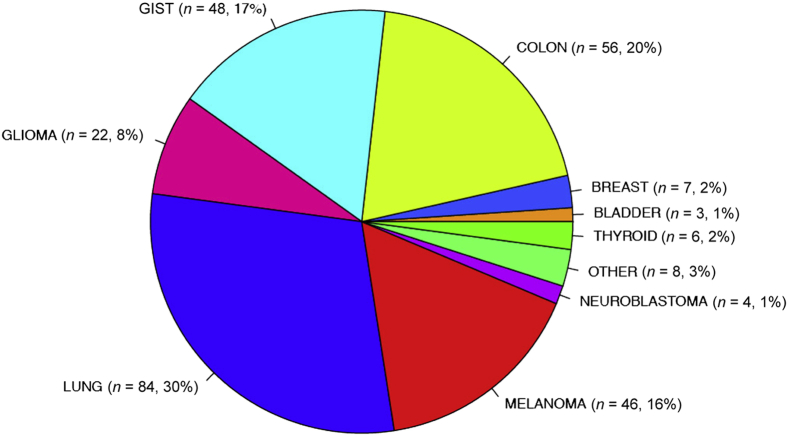
Two hundred and eighty-four unique tumor DNA samples and 75 matched control samples from FFPE sections were used for our clinical validation study. These samples had been previously genotyped or sequenced in our clinical laboratory by alternative methods and were confirmed to be positive for SNVs and indel mutations in 47 exons of 19 genes (Table 1). Tested samples were confirmed to be >10% tumor content by pathologist review. MSK-IMPACT was validated for more than 10 samples for mutations in each of the following exons: BRAF exons 11 and 15 (G469, D594, V600, and K601 hotspots), EGFR exons 18 to 21 (G719, T790M, L858R hotspots, exon 19 deletions, and exon 20 insertions), ERBB2 exon 20 insertions, IDH1 exon 4 (R132 hotspot), KIT exons 9, 11, and 13 (V654 hotspot, exon 9 insertions, and exon 11 deletions), KRAS exons 2 and 3 (G12, G13, and Q61 hotspots), NRAS exon 3 (Q61 hotspot), PDGFRA exon 18 (D842 hotspot), and PIK3CA exons 10 and 21 (E542, E545, H1047 hotspots). Figure 3 illustrates the distribution of tumor types for these samples used in the validation study. A full list of samples and their detected mutations can be found in Supplemental Table S11. For genes/exons that have <10 positive samples tested, further validation has been ongoing (Supplemental Table S12).
Table 1.
Overview of Samples with Known Mutations Profiled for the Validation Study
| Index | Gene | Exon/targets validated |
Target# (N = 47) | |
|---|---|---|---|---|
| ≥10 Samples tested | <10 Samples tested | |||
| 1 | AKT1 | 3 | 1 | |
| 2 | ALK | 23, 25 | 2 | |
| 3 | BRAF | 11, 15 | 2 | |
| 4 | EGFR | 18, 19,∗ 20,∗ 21 | 4 | |
| 5 | ERBB2 | 20∗ | 8, 19 | 3 |
| 6 | FGFR2 | 7, 9, 12 | 3 | |
| 7 | FGFR3 | 7, 9, 18 | 3 | |
| 8 | GNA11 | 5 | 1 | |
| 9 | GNAQ | 5 | 1 | |
| 10 | GNAS | 8 | 1 | |
| 11 | HRAS | 2, 3 | 2 | |
| 12 | IDH1 | 4 | 1 | |
| 13 | IDH2 | 4 | 1 | |
| 14 | KIT | 9, 11,∗ 13 | 17 | 4 |
| 15 | KRAS | 2, 3 | 4 | 3 |
| 16 | NRAS | 3 | 2 | 2 |
| 17 | PDGFRA | 18 | 12 | 2 |
| 18 | PIK3CA | 10, 21 | 2, 5, 8 | 5 |
| 19 | TP53 | 4, 5, 6,∗ 7, 8,∗ 10 | 6 | |
| Total | 17 | 30 | 47 | |
Samples validated for both single nucleotide variations and insertion/deletions.
Figure 3.
Distribution of tumor types among the 284 unique samples profiled for the clinical validation study.
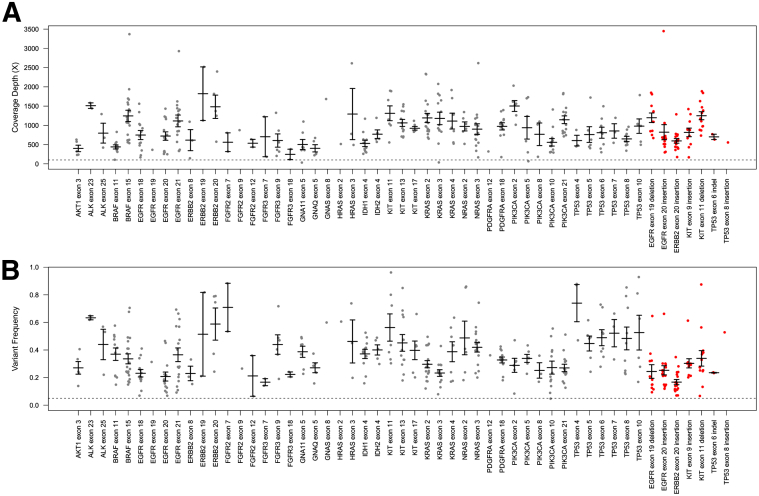
We were able to sequence 275 of 284 tested tumor samples to coverage exceeding 200×, for an overall probability for technical success of 96.8%. The median sample coverage was 753×. MSK-IMPACT was able to detect the known variant(s) associated with each sample in all 284 cases, including the nine tumors where coverage did not reach 200×. A total of 393 known variants were detected: 13 of the known variants were detected with frequencies <10%, demonstrating the sensitivity of the assay for low-frequency mutations. Known SNVs were detected with mean coverage of 912× (SD = ±529×) and variant frequency of 36% (SD = ±18%). Known indels were detected at similar coverage levels (897×, SD = ±514×), but presented with lower variant frequencies on average (26%, SD = ±15%) (Figure 4).
Figure 4.
Range of coverage depth values (A) and variant frequencies (B) for known variants detected in 47 exons of 19 genes tested. Gray indicates SNVs, red, indels. Bars indicate mean values, whiskers indicate SEM. indel, insertion/deletion; SNV, single nucleotide variation.
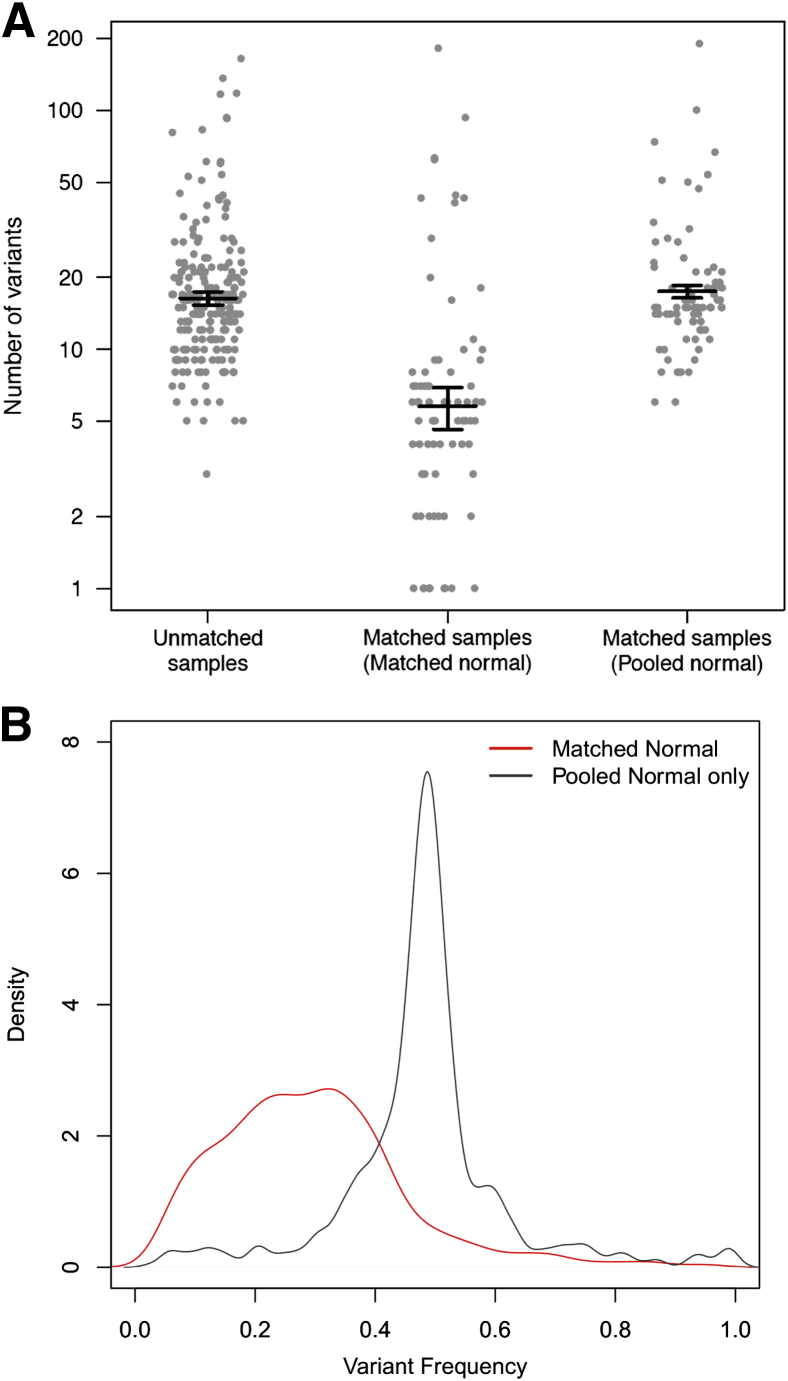
As expected, fewer total mutations were called when a patient-matched normal was used in variant calling because private germline SNVs were appropriately filtered out. Variant calling in tumors with patient-matched normal generated 6 somatic calls on average, compared to 15 calls on average for tumors where no matched normal was available (Figure 5A). To more precisely evaluate the effect of a matched normal on variant calling, we deliberately performed variant calling against a pool of unmatched normal DNAs for tumors where a matched normal was available. Doing so resulted in a gain of approximately 24 exonic, nonsynonymous germline calls per sample, of which 15 calls on average were highly polymorphic in the general population (minor allele frequency in 1000 Genomes data >1%). However, not all additional germline calls can be filtered out by comparing against the 1000 Genomes cohort data,33 and despite filtering on population minor allele frequency, unmatched variant calling still results in an additional nine private germline calls per sample on average. The variant frequency for these additional private germline mutations is approximately 48.3% (43.6% to 51.8%), compared to 28.3% (18.7% to 37.6%) for somatic mutations detected using a matched normal (Figure 5B). These private mutations also contain fewer indel calls (48 of 698, or 6.5%, compared to 171 of 958, or 17.8%, in somatic calls), which is consistent with the view that there is considerable negative selective pressure against indels in coding regions, because they are more likely to disrupt gene function than point mutations. Overall, these data highlight the importance of matched normal analysis to improve variant interpretation.
Figure 5.
Variant calling for unmatched tumors. A: Number of variants called on: i) 209 samples for which a matched normal was unavailable, 75 samples where matched normal samples were available; and ii) variant calling was performed against the matched normal; or iii) variant calling was deliberately performed using a generic pooled normal. B: Distribution of variant frequencies for somatic mutations (red line) and additional private germline mutations (black line) identified when a generic pooled normal is used. Bars indicate mean values, whiskers indicate SEM.
Assessment of Reproducibility and Analytic Sensitivity
Three samples with known point mutations (BRAF V600E, KRAS G12C, and EGFR L858R) and three samples with known indels (EGFR exon 19 deletion, ERBB2 exon 20 insertion, KIT exon 11 deletion) were assayed in triplicate as intra- and interrun replicates, to assess reproducibility of our variant calling pipeline. Different barcodes were used across replicates to minimize possible bias from barcode selection. The results are summarized in Table 2. The known variant was successfully detected in all cases, with similar variant frequencies and at similar levels of coverage (after adjusting for the overall sample coverage). The total number of variants called across both inter- and intrarun replicates was generally consistent, which is noteworthy given differences in sample coverage across replicates. Only one discrepancy was observed across all cases, which corresponded to a low-frequency mutation, detected with a frequency value close to the 5% frequency cutoff for nonhotspot mutations.
Table 2.
Reproducibility of Variant Calls Comparing Intra- and Interrun Replicates
| Known variant | Barcode | Run | Sample statistics |
Known variant statistics |
|||
|---|---|---|---|---|---|---|---|
| Coverage (X) | Variants called | Coverage (X) | Variant frequency | Normalized coverage | |||
| BRAF exon 15 p.V600E | bc04 | 1 | 928 | 10 | 1409 | 0.45 | 1.52 |
| bc05 | 1 | 904 | 10 | 1392 | 0.43 | 1.54 | |
| bc06 | 1 | 864 | 10 | 1329 | 0.41 | 1.54 | |
| bc27 | 2 | 1397 | 10 | 1874 | 0.43 | 1.34 | |
| bc04 | 3 | 793 | 10 | 1153 | 0.41 | 1.45 | |
| KRAS exon 2 p.G12C | bc16 | 1 | 342 | 6 | 423 | 0.29 | 1.24 |
| bc17 | 1 | 310 | 6 | 386 | 0.35 | 1.25 | |
| bc18 | 1 | 310 | 6 | 337 | 0.31 | 1.09 | |
| bc14 | 2 | 741 | 6 | 919 | 0.3 | 1.24 | |
| bc14 | 3 | 1280 | 6 | 1639 | 0.3 | 1.28 | |
| EGFR exon 21 p.L858R | bc15 | 1 | 934 | 5 | 1468 | 0.22 | 1.57 |
| bc16 | 1 | 1025 | 5 | 1565 | 0.19 | 1.53 | |
| bc18 | 1 | 826 | 5 | 1269 | 0.17 | 1.54 | |
| bc05 | 2 | 834 | 5 | 1296 | 0.2 | 1.55 | |
| bc08 | 3 | 1033 | 4† | 1622 | 0.2 | 1.57 | |
| EGFR exon 19 p.746_750del | bc31 | 1 | 618 | 4 | 605 | 0.33 | 0.98 |
| bc32 | 1 | 433 | 4 | 406 | 0.32 | 0.94 | |
| bc33 | 1 | 579 | 4 | 618 | 0.31 | 1.07 | |
| bc05 | 2 | 758 | 4 | 670 | 0.32 | 0.88 | |
| bc08 | 3 | 718 | 4 | 661 | 0.34 | 0.92 | |
| ERBB2 exon 20∗ p.E770_A771insAYVM | bc28 | 1 | 376 | 17 | 520 | 0.14 | 1.38 |
| bc29 | 1 | 402 | 17 | 572 | 0.17 | 1.42 | |
| bc30 | 1 | 256 | 17 | 326 | 0.16 | 1.27 | |
| bc23 | 2 | 1002 | 17 | 1094 | 0.2 | 1.09 | |
| bc04 | 3 | 350 | 17 | 403 | 0.19 | 1.15 | |
| KIT exon 11 p.556_558del | bc31 | 1 | 821 | 4 | 1459 | 0.54 | 1.78 |
| bc32 | 1 | 822 | 4 | 1541 | 0.56 | 1.87 | |
| bc33 | 1 | 822 | 4 | 1466 | 0.55 | 1.78 | |
| bc19 | 2 | 725 | 4 | 1320 | 0.59 | 1.82 | |
| bc24 | 3 | 803 | 4 | 1386 | 0.56 | 1.73 | |
Matched normal not available for sample.
EGFR exon 21 HNF1A L312F missed in bc08 replicate was detected at 3%, below the 5% variant frequency threshold.
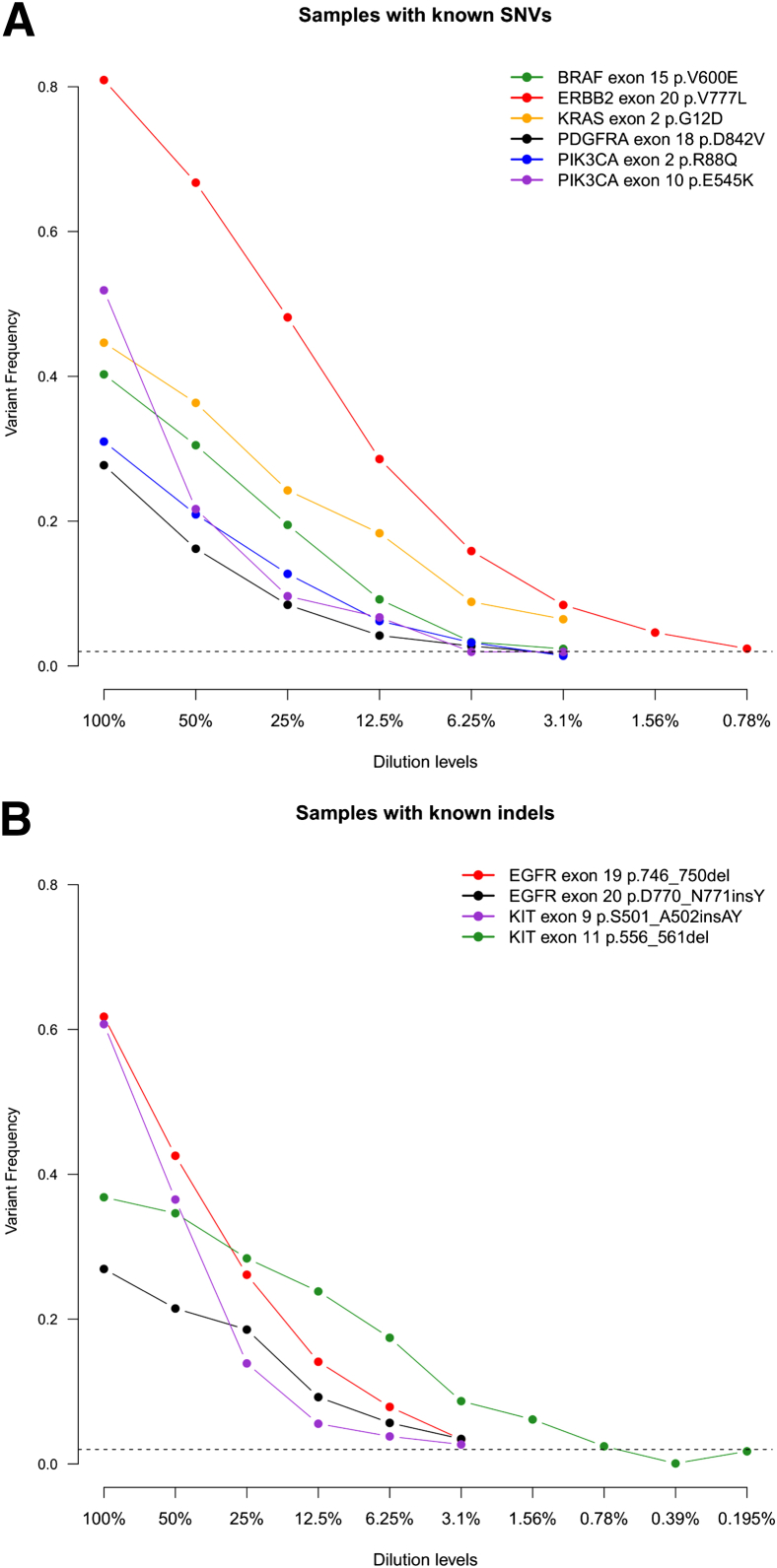
To determine the limit of detection of the assay for low-frequency mutations, ie, the analytic sensitivity, tumor DNA from samples positive for SNVs and indels was repeatedly diluted with normal DNA from the same individual if available, or normal DNA from an unrelated individual. The samples at various dilution levels were then profiled, and variant calling was performed to determine the dilution level at which the known variant failed to be detected. To examine the effect of coverage depth on analytic sensitivity, we performed the serial dilutions using six samples with known SNVs and four samples with known indels (Figure 6). For most cases, the assay and analysis pipeline were able to call the known variant down to the lowest dilution level tested (0.78% for ERBB2 V777L, 3.1% for other cases), except for the KIT exon 11 deletion where the known variant failed to be called at the 0.78% dilution level. In each case, when the particular known variant failed to be called, it was not because coverage had become limiting (mean coverage = 920× for all cases across dilution levels), but because the variant frequency had fallen below the 2% threshold used to reject potential false positives. That is, because of the deep coverage across targeted regions, MSK-IMPACT is limited by its specificity rather than its sensitivity, ie, the need to distinguish between true low-frequency mutations and false-positive artifacts.
Figure 6.
Variant frequencies of known SNV (A) and indel (B) calls tracked across successive serial dilutions. indel, insertion/deletion; SNV, single nucleotide variation.
Identification of Copy Number and Structural Variants
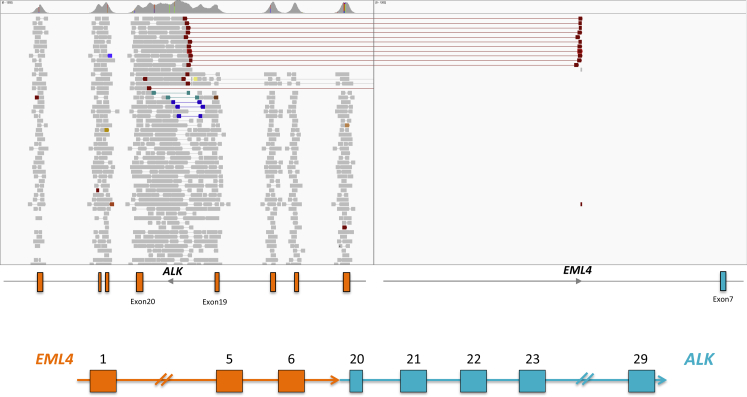
As a proof of concept, we asked whether MSK-IMPACT was capable of identifying copy number and structural variants in known positive samples. We tested 19 ERBB2-amplified samples (8 breast cancer samples, 11 gastric cancer samples) and 4 lung adenocarcinoma samples containing EML4-ALK translocations. These samples had previously tested positive for their respective mutations by a combination of fluorescent in situ hybridization and/or real-time quantitative PCR. Examples of ERBB2 amplification in breast and gastric cancer samples are shown in Figure 7, A and B, respectively. MSK-IMPACT was able to identify the ERBB2 amplification events in all known positive samples: the median value for ERBB2 amplification was 3.4-fold in breast cancer samples and 4.1-fold in gastric cancer samples, correspondingly. Similarly, MSK-IMPACT was able to retrieve the known EML4-ALK translocation in all four positive samples. There was strong paired-read and split-read support in all cases (approximately 10 paired reads and approximately 13 split reads on average, Figure 8). Taken together, these results demonstrate that MSK-IMPACT is capable of identifying copy number and structural variants from FFPE tumor DNA; additional work to validate and characterize the sensitivity, specificity, reproducibility, and limits of detection of MSK-IMPACT for copy number and structural variants is currently ongoing.
Figure 7.
MSK-IMPACT reveals copy number alterations. Log-ratios comparing tumor versus normal coverage values are calculated across all targeted regions for samples containing ERBB2 amplifications: breast cancer sample with ERBB2, GNAS, MAPK1 amplifications, and ATM loss (A), gastric cancer sample with EGFR, ERBB2, and PIM1 amplifications (B).
Figure 8.
MSK-IMPACT reveals structural rearrangements. Integrated Genomics Viewer screenshot of EML4-ALK translocation in a lung adenocarcinoma sample known to be positive for this translocation.
Discussion
In this study, we performed an extensive analytical validation of MSK-IMPACT, a hybridization capture-based assay targeting all coding regions of 341 oncogenes and tumor suppressors. We assessed the ability of MSK-IMPACT to detect SNVs and indels in 284 known positive tumor samples, 75 of which had a matched normal sample available for somatic variant calling. Not only was MSK-IMPACT able to successfully detect the known variant in each of the 284 cases, but the known variant was detected at coverage depths routinely exceeding 500×. Variant frequencies and coverage values for known variants detected were similar across intra- and interrun replicates of the same sample. Additionally, the number of variant calls was also consistent across replicates, confirming that assay results were indeed reproducible. Using samples positive for ERBB2 amplification and EML4-ALK fusion, we also demonstrated that MSK-IMPACT can identify somatic copy number aberrations and selected structural rearrangements, in addition to SNVs and indels.
Our analysis comparing variants called against a matched normal versus a generic pooled FFPE normal identified on average nine additional private exonic, nonsynonymous germline mutations per sample. These mutations cannot be easily distinguished from true somatic mutations by simply using polymorphisms identified in population genotyping studies as a filter [ie, 1000 Genomes cohort, NCBI Single Nucleotide Polymorphism database (dbSNPv137)]. The burden of such private exonic nonsynonymous mutations, in the event that a matched normal sample is unavailable, will only increase for larger targeted sequencing panels that cover more genes, or in whole-exome sequencing. More stringent filters for variants called in unmatched tumors (eg, prior evidence of recurrent somatic mutations at that site) can reduce the number of germline polymorphisms, at the expense of excluding novel somatic mutations. Since the clinical implementation of MSK-IMPACT at MSKCC, matched normal samples have been received for 955 of the first 1000 cases (95.5%), reaffirming that collection and analysis of patient-matched normal is feasible and worthwhile.
On the basis of statistical and empirical determinations, we established a threshold on coverage (100×) to detect variants with 10% frequency at 98% power (α = 0.05). In practice, however, by pooling 30 to 35 samples in a single Illumina HiSeq 2500 rapid run, we are able to achieve approximately 700× unique sequence coverage for tumor samples, and coverage depths in hotspot regions (47 exons of the 19 genes used in the validation study) can exceed 900×. Maintaining a requirement of 98% power, this suggests that MSK-IMPACT may even be sufficiently powered to detected variants with frequencies as low as 2% in hotspot exons of known oncogenes. The results of the serial dilution experiments are consistent with this estimate: in almost every case, we were able to detect the known variant in each sample down to the lowest dilution levels tested (0.78% to 3.1%). The assay failed to detect the known variant only after its frequency at the given dilution level fell below the 2% variant frequency threshold established to reject false-positive mutation calls. This suggests that owing to the high levels of coverage in the targeted regions, the ability of MSK-IMPACT to detect hotspot SNVs and indels is limited by requirements on specificity rather than sensitivity. It should be noted that the observed variant frequencies do not always decrease in a log-linear relationship, ie, exactly 0.5-fold, with each subsequent serial dilution (Supplemental Figure S3). These deviations could be a result of stoichiometric imperfections in mixing tumor and normal DNA, but could also be influenced by local copy number and ploidy differences between the tumor and its diluting normal.
Variant calls from MSK-IMPACT are reproducible comparing within and across-batch replicates. From Table 2, we observe tight SDs for variant frequencies, ranging from 0.01 to 0.025. The variability in variant frequencies observed across the intra- and interrun replicates reflects technical batch variation from operator and various wet-lab processes. There is greater variability between the observed and expected variant frequencies, for the experiment in which multiple FFPE normal samples are pooled, sequenced, and genotyped (Supplemental Table S7). Unlike the previous experiment, the pooling experiment does not measure technical batch variation, and the increased variability is likely due to imprecise pooling of DNA from various samples.
The implementation of MSK-IMPACT as a routine clinical test involved numerous workflow improvements. In terms of wet-lab operations, we optimized assay parameters (hybridization times) to minimize turn-around time, and implemented a laboratory information management system (LIMS; Sapio Sciences, Baltimore, MD) to efficiently track tumor and matched normal samples, as well as reagents used at various points in the workflow. In terms of data analysis, we implemented various systems and databases to automate pipeline kickoff, manage, track, and archive pipeline output [FASTQs, Binary Alignment/Maps (BAM files), and Variant Call Files (VCFs)], and present mutation calls in a format amenable to pathologist manual review and sign-out. Current efforts are focused on additional improvements to the analysis framework. Assembly-based methods can be used to improve local realignment of indels, which may allow detection of indels >30 bp in length. Other indel calling methods such as Pindel38 can be attempted to detect internal tandem duplications, medium-sized inversions from the data. We demonstrated that MSK-IMPACT is capable of detecting copy number alterations, but work to establish thresholds on fold change and P value to optimize for sensitivity and specificity is ongoing. Methods such as Absolute39 and PyClone40 may enable estimation of tumor purity and ploidy, and inference of subclonal populations.
In summary, our results confirm that as a clinical test, MSK-IMPACT can identify somatic mutations in FFPE samples with high levels of accuracy, sensitivity, and reproducibility. By including 30 to 36 samples (15 to 18 tumor matched normal pairs) in each pool, we can routinely obtain high levels of coverage (>500×) in tumor samples, which allow low-frequency mutations to be detected at adequate power. Our experience from implementing MSK-IMPACT in the clinical lab so far has indicated that comparative analysis of tumors and patient-matched normal samples is feasible in the clinical setting, and prospective targeted sequencing can comprehensively detect somatic mutations for use in guiding treatment decisions.
Acknowledgments
We thank Kety Huberman, Agnes Viale, Joanne Edington, Chris Pepper, Nicholas Socci, and other members of the Integrated Genomics Operation and Bioinformatics Core at MSKCC for their assistance in library prep, sequencing, and computing infrastructure development and Peter Ntiamoah, histology lab manager in pathology, for providing FFPE samples in an expedited manner for the validation effort.
Footnotes
Supported by the Department of Pathology, Memorial Sloan Kettering Cancer Center, the Farmer Family Foundation, the Geoffrey Beene Cancer Research Center (M.F.B.), and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology.
Disclosures: None declared.
Current address of T.M., The Jackson Laboratory for Genomic Medicine, Farmington, CT; of D.T.C., Illumina, Inc., Hayward, CA.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2014.12.006.
Supplemental Data
Power analysis using binomial model approximation. A: Unique sequencing coverage (number of reads) (y axis) required to detect a variant with true underlying variant frequency (x axis) for a given level of power and specified Type I error rate (α = 0.05). B: 95% confidence interval ranges of observed variant frequencies (y axis) for a variant of true underlying variant frequency (dotted line) 2%, 5%, 10%, and 20%, as a function of unique sequencing coverage (number of reads, x axis). DP, coverage depth; VF, variant frequency.
Boxplot showing the range of observed variant frequencies for 862 common single nucleotide polymorphisms genotyped in pooled normal sample, binned by their respective true underlying variant frequency (VF).
Variant frequencies of known SNV (A) and indel (B) calls tracked across successive serial dilutions, plotted in log2 units on the y axis. indel, insertion/deletion; SNV, single nucleotide variation.
References
- 1.Garraway L.A. Genomics-driven oncology: framework for an emerging paradigm. J Clin Oncol. 2013;31:1806–1814. doi: 10.1200/JCO.2012.46.8934. [DOI] [PubMed] [Google Scholar]
- 2.MacConaill L.E., Van Hummelen P., Meyerson M., Hahn W.C. Clinical implementation of comprehensive strategies to characterize cancer genomes: opportunities and challenges. Cancer Discov. 2011;1:297–311. doi: 10.1158/2159-8290.CD-11-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor B.S., Ladanyi M. Clinical cancer genomics: how soon is now? J Pathol. 2011;223:318–326. doi: 10.1002/path.2794. [DOI] [PubMed] [Google Scholar]
- 4.Romano E., Schwartz G.K., Chapman P.B., Wolchock J.D., Carvajal R.D. Treatment implications of the emerging molecular classification system for melanoma. Lancet Oncol. 2011;12:913–922. doi: 10.1016/S1470-2045(10)70274-6. [DOI] [PubMed] [Google Scholar]
- 5.Chapman P.B., Hauschild A., Robert C., Haanen J.B., Ascierto P., Larkin J., Dummer R., Garbe C., Testori A., Maio M., Hogg D., Lorigan P., Lebbe C., Jouary T., Schadendorf D., Ribas A., O'Day S.J., Sosman J.A., Kirkwood J.M., Eggermont A.M., Dreno B., Nolop K., Li J., Nelson B., Hou J., Lee R.J., Flaherty K.T., McArthur G.A. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaw A.T., Kim D.W., Nakagawa K., Seto T., Crino L., Ahn M.J., De Pas T., Besse B., Solomon B.J., Blackhall F., Wu Y.L., Thomas M., O'Byrne K.J., Moro-Sibilot D., Camidge D.R., Mok T., Hirsh V., Riely G.J., Iyer S., Tassell V., Polli A., Wilner K.D., Janne P.A. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 7.Shaw A.T., Kim D.W., Mehra R., Tan D.S., Felip E., Chow L.Q., Camidge D.R., Vansteenkiste J., Sharma S., De Pas T., Riely G.J., Solomon B.J., Wolf J., Thomas M., Schuler M., Liu G., Santoro A., Lau Y.Y., Goldwasser M., Boral A.L., Engelman J.A. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med. 2014;370:1189–1197. doi: 10.1056/NEJMoa1311107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kris M.G., Johnson B.E., Berry L.D., Kwiatkowski D.J., Iafrate A.J., Wistuba I.I., Varella-Garcia M., Franklin W.A., Aronson S.L., Su P.F., Shyr Y., Camidge D.R., Sequist L.V., Glisson B.S., Khuri F.R., Garon E.B., Pao W., Rudin C., Schiller J., Haura E.B., Socinski M., Shirai K., Chen H., Giaccone G., Ladanyi M., Kugler K., Minna J.D., Bunn P.A. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsongalis G.J., Peterson J.D., de Abreu F.B., Tunkey C.D., Gallagher T.L., Strausbaugh L.D., Wells W.A., Amos C.I. Routine use of the Ion Torrent AmpliSeq Cancer Hotspot Panel for identification of clinically actionable somatic mutations. Clin Chem Lab Med. 2014;52:707–714. doi: 10.1515/cclm-2013-0883. [DOI] [PubMed] [Google Scholar]
- 10.Luthra R., Patel K.P., Reddy N.G., Haghshenas V., Routbort M.J., Harmon M.A., Barkoh B.A., Kanagal-Shamanna R., Ravandi F., Cortes J.E., Kantarjian H.M., Medeiros L.J., Singh R.R. Next-generation sequencing-based multigene mutational screening for acute myeloid leukemia using MiSeq: applicability for diagnostics and disease monitoring. Haematologica. 2014;99:465–473. doi: 10.3324/haematol.2013.093765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abel H.J., Al-Kateb H., Cottrell C.E., Bredemeyer A.J., Pritchard C.C., Grossmann A.H., Wallander M.L., Pfeifer J.D., Lockwood C.M., Duncavage E.J. Detection of gene rearrangements in targeted clinical next-generation sequencing. J Mol Diagn. 2014;16:405–417. doi: 10.1016/j.jmoldx.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cottrell C.E., Al-Kateb H., Bredemeyer A.J., Duncavage E.J., Spencer D.H., Abel H.J., Lockwood C.M., Hagemann I.S., O'Guin S.M., Burcea L.C., Sawyer C.S., Oschwald D.M., Stratman J.L., Sher D.A., Johnson M.R., Brown J.T., Cliften P.F., George B., McIntosh L.D., Shrivastava S., Nguyen T.T., Payton J.E., Watson M.A., Crosby S.D., Head R.D., Mitra R.D., Nagarajan R., Kulkarni S., Seibert K., Virgin H.W., 4th, Milbrandt J., Pfeifer J.D. Validation of a next-generation sequencing assay for clinical molecular oncology. J Mol Diagn. 2014;16:89–105. doi: 10.1016/j.jmoldx.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pritchard C.C., Salipante S.J., Koehler K., Smith C., Scroggins S., Wood B., Wu D., Lee M.K., Dintzis S., Adey A., Liu Y., Eaton K.D., Martins R., Stricker K., Margolin K.A., Hoffman N., Churpek J.E., Tait J.F., King M.C., Walsh T. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J Mol Diagn. 2014;16:56–67. doi: 10.1016/j.jmoldx.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh R.R., Patel K.P., Routbort M.J., Reddy N.G., Barkoh B.A., Handal B., Kanagal-Shamanna R., Greaves W.O., Medeiros L.J., Aldape K.D., Luthra R. Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J Mol Diagn. 2013;15:607–622. doi: 10.1016/j.jmoldx.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Duncavage E.J., Abel H.J., Szankasi P., Kelley T.W., Pfeifer J.D. Targeted next generation sequencing of clinically significant gene mutations and translocations in leukemia. Mod Pathol. 2012;25:795–804. doi: 10.1038/modpathol.2012.29. [DOI] [PubMed] [Google Scholar]
- 16.Kanagal-Shamanna R., Portier B.P., Singh R.R., Routbort M.J., Aldape K.D., Handal B.A., Rahimi H., Reddy N.G., Barkoh B.A., Mishra B.M., Paladugu A.V., Manekia J.H., Kalhor N., Chowdhuri S.R., Staerkel G.A., Medeiros L.J., Luthra R., Patel K.P. Next-generation sequencing-based multi-gene mutation profiling of solid tumors using fine needle aspiration samples: promises and challenges for routine clinical diagnostics. Mod Pathol. 2014;27:314–327. doi: 10.1038/modpathol.2013.122. [DOI] [PubMed] [Google Scholar]
- 17.Frampton G.M., Fichtenholtz A., Otto G.A., Wang K., Downing S.R., He J. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Allen E.M., Wagle N., Stojanov P., Perrin D.L., Cibulskis K., Marlow S. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med. 2014;20:682–688. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin M.T., Mosier S.L., Thiess M., Beierl K.F., Debeljak M., Tseng L.H., Chen G., Yegnasubramanian S., Ho H., Cope L., Wheelan S.J., Gocke C.D., Eshleman J.R. Clinical validation of KRAS, BRAF, and EGFR mutation detection using next-generation sequencing. Am J Clin Pathol. 2014;141:856–866. doi: 10.1309/AJCPMWGWGO34EGOD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikiforova M.N., Wald A.I., Roy S., Durso M.B., Nikiforov Y.E. Targeted next-generation sequencing panel (ThyroSeq) for detection of mutations in thyroid cancer. J Clin Endocrinol Metab. 2013;98:E1852–E1860. doi: 10.1210/jc.2013-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng D.T., Cheng J., Mitchell T.N., Syed A., Zehir A., Mensah N.Y., Oultache A., Nafa K., Levine R.L., Arcila M.E., Berger M.F., Hedvat C.V. Detection of mutations in myeloid malignancies through paired-sample analysis of microdroplet-PCR deep sequencing data. J Mol Diagn. 2014;16:504–518. doi: 10.1016/j.jmoldx.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Won H.H., Scott S.N., Brannon A.R., Shah R.H., Berger M.F. Detecting somatic genetic alterations in tumor specimens by exon capture and massively parallel sequencing. J Vis Exp. 2013:e50710. doi: 10.3791/50710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang F.W., Hodis E., Xu M.J., Kryukov G.V., Chin L., Garraway L.A. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landa I., Ganly I., Chan T.A., Mitsutake N., Matsuse M., Ibrahimpasic T., Ghossein R.A., Fagin J.A. Frequent somatic TERT promoter mutations in thyroid cancer: higher prevalence in advanced forms of the disease. J Clin Endocrinol Metab. 2013;98:E1562–E1566. doi: 10.1210/jc.2013-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Killela P.J., Reitman Z.J., Jiao Y., Bettegowda C., Agrawal N., Diaz L.A., Jr. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vinagre J., Almeida A., Populo H., Batista R., Lyra J., Pinto V., Coelho R., Celestino R., Prazeres H., Lima L., Melo M., da Rocha A.G., Preto A., Castro P., Castro L., Pardal F., Lopes J.M., Santos L.L., Reis R.M., Cameselle-Teijeiro J., Sobrinho-Simoes M., Lima J., Maximo V., Soares P. Frequency of TERT promoter mutations in human cancers. Nat Commun. 2013;4:2185. doi: 10.1038/ncomms3185. [DOI] [PubMed] [Google Scholar]
- 27.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forbes S.A., Bhamra G., Bamford S., Dawson E., Kok C., Clements J., Menzies A., Teague J.W., Futreal P.A., Stratton M.R. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr Protoc Hum Genet. 2008 doi: 10.1002/0471142905.hg1011s57. Chapter 10:Unit 10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horaitis O., Cotton R.G. The challenge of documenting mutation across the genome: the human genome variation society approach. Hum Mutat. 2004;23:447–452. doi: 10.1002/humu.20038. [DOI] [PubMed] [Google Scholar]
- 32.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., Antipin Y., Reva B., Goldberg A.P., Sander C., Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olshen A.B., Venkatraman E.S., Lucito R., Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 37.Rausch T., Zichner T., Schlattl A., Stutz A.M., Benes V., Korbel J.O. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye K., Schulz M.H., Long Q., Apweiler R., Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carter S.L., Cibulskis K., Helman E., McKenna A., Shen H., Zack T., Laird P.W., Onofrio R.C., Winckler W., Weir B.A., Beroukhim R., Pellman D., Levine D.A., Lander E.S., Meyerson M., Getz G. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roth A., Khattra J., Yap D., Wan A., Laks E., Biele J., Ha G., Aparicio S., Bouchard-Cote A., Shah S.P. PyClone: statistical inference of clonal population structure in cancer. Nat Methods. 2014;11:396–398. doi: 10.1038/nmeth.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Power analysis using binomial model approximation. A: Unique sequencing coverage (number of reads) (y axis) required to detect a variant with true underlying variant frequency (x axis) for a given level of power and specified Type I error rate (α = 0.05). B: 95% confidence interval ranges of observed variant frequencies (y axis) for a variant of true underlying variant frequency (dotted line) 2%, 5%, 10%, and 20%, as a function of unique sequencing coverage (number of reads, x axis). DP, coverage depth; VF, variant frequency.
Boxplot showing the range of observed variant frequencies for 862 common single nucleotide polymorphisms genotyped in pooled normal sample, binned by their respective true underlying variant frequency (VF).
Variant frequencies of known SNV (A) and indel (B) calls tracked across successive serial dilutions, plotted in log2 units on the y axis. indel, insertion/deletion; SNV, single nucleotide variation.